 ,, 何玉兰, 郑学礼
,, 何玉兰, 郑学礼 ,南方医科大学公共卫生学院病原生物学系,广州 510515
,南方医科大学公共卫生学院病原生物学系,广州 510515Research progress in RNA interference against the infection of mosquito-borne viruses
Yong Wei ,, Yulan He, Xueli Zheng
,, Yulan He, Xueli Zheng ,Department of Pathogen Biology, School of Public Health, Southern Medical University, Guangzhou 510515, China
,Department of Pathogen Biology, School of Public Health, Southern Medical University, Guangzhou 510515, China通讯作者: 郑学礼,博士,教授,研究方向:传染病预防与控制。E-mail:zhengxueli2001@126.com
第一联系人:
编委: 岑山
收稿日期:2019-10-15修回日期:2019-12-14网络出版日期:2020-01-02
| 基金资助: |
Received:2019-10-15Revised:2019-12-14Online:2020-01-02
| Fund supported: |

摘要
蚊媒病因具有较高的发病率和传播率使其成为全球关注的重要公共卫生问题。蚊虫作为蚊媒病的传播媒介,研究其与蚊媒病毒两者之间的相互作用机制将有助于蚊媒病的防控。蚊虫抵御蚊媒病毒的先天免疫降低和病毒成功逃避蚊虫免疫屏障为病毒在蚊虫体内的持续感染和蚊媒病的暴发流行造成了潜在风险。RNA干扰(RNA interference, RNAi)途径作为蚊虫体内强大的抗病毒防御屏障,通过产生多种小RNA降解病毒RNA,从而达到抑制病毒复制和传播的目的。本文对小干扰RNA (small interfering RNA, siRNA)、微小RNA (microRNA, miRNA)、Piwi蛋白相作用RNA (Piwi-interacting RNA, piRNA)等3种小分子RNA在蚊虫体内发挥抗蚊媒病毒感染的先天免疫机制的相关研究进行了综述,以期为蚊媒病的防控提供理论参考。
关键词:
Abstract
Mosquito-borne diseases have become an important public health issue of global concern because of their high incidence and transmission rate. As a vector for mosquito-borne diseases, studying the interaction mechanism between mosquitoes and mosquito-borne viruses will help control mosquito-borne diseases. The impaired innate immunity and immune barriers evasion caused by mosquito-borne viruses in mosquitoes pose a potential risk for the persistent infection of the virus in mosquitoes and the outbreak of mosquito-borne diseases. The RNA interference (RNAi) pathway, as a powerful antiviral defense barrier in mosquitoes, can inhibit viral replication and transmission by producing a variety of small RNAs to degrade viral RNA. In this review, we summarize the related studies on the innate immune mechanism against mosquito- borne virus infection in mosquitoes about small interfering RNA (siRNA), microRNA (miRNA), and Piwi-interacting RNA (piRNA), aiming to provide a theoretical reference for the prevention and control of mosquito-borne diseases.
Keywords:
PDF (719KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
魏勇, 何玉兰, 郑学礼. RNAi在抗蚊媒病毒感染中的研究进展. 遗传[J], 2020, 42(2): 153-160 doi:10.16288/j.yczz.19-262
Yong Wei.
蚊媒病是一类由病媒蚊虫传播的自然疫源性疾病,常见的有流行性乙型脑炎、登革热、黄热病等。随着全球气候变暖、交通运输便捷和旅游业发展,蚊虫在全球范围内快速扩张,这为蚊媒病的暴发流行和传播扩散构成了潜在风险。绝大多数蚊媒病毒以及在全球发病率和死亡率占较大比重的均是RNA病毒,如黄病毒科黄病毒属(正链RNA病毒)、披膜病毒科甲病毒属(正链RNA病毒)、布尼亚病毒科白蛉病毒属(负链RNA病毒)[1,2]。黄病毒科黄病毒属包括黄热病病毒(yellow fever virus, YFV)、登革病毒(Dengue virus, DENV)、乙型脑炎病毒(Japanese encephalitis virus, JEV)、西尼罗河病毒(West Nile virus, WNV)和寨卡病毒(Zika virus, ZIKV)等[3],披膜病毒科甲病毒属包括基孔肯雅病毒(Chikungunya virus, CHIKV)、辛德毕斯病毒(Sindbis virus, SINV)、塞姆利基森林病毒(Semliki forest virus, SFV)、罗斯河病毒(Ross river virus, RRV)等[4],布尼亚病毒科白蛉病毒属包括托斯卡纳病毒(Toscana virus, TOSV)、裂谷热病毒(Rift Valley fever virus, RVFV)等[5]。在过去的几十年间,登革热至少在128个国家或地区暴发流行,每年平均有3.9亿人口感染登革热,登革热一直是重点关注的全球公共卫生问题[6]。寨卡病毒病是近年来新兴的蚊媒传播性疾病,2016年2月世界卫生组织(WHO)宣布将寨卡疫情列为全球紧急公共卫生事件。自从2015年巴西发生大规模寨卡病毒感染疫情后,目前已有64个国家和地区报告了寨卡疫情[7]。寨卡病毒感染引起的成人格林-巴利综合征(Guillain-Barre syndrome, GBS)和新生儿出生缺陷,如产前感染所致的小头畸形,在近年来引发了全球广泛关注[8]。因目前缺乏蚊媒病相应的有效疫苗,抑制病毒在蚊虫体内复制和阻断病毒传播将是控制蚊媒病暴发流行的有效途径[9]。因此,研究蚊虫体内抗蚊媒病毒感染的先天免疫机制将有助于制定相应的蚊媒病控制策略。本文将对目前蚊虫体内3大主要抗病毒的RNAi途径的相关研究进行综述,以期为蚊媒病的防控提供理论参考。
1 蚊虫体内三大主要的RNAi途径
蚊虫的先天免疫屏障和蚊媒病毒的免疫逃避机制是影响病毒在蚊虫体内成功复制和传播的关键因素。蚊虫体内抗病毒的效应分子(抗菌肽(antimicrobial peptide, AMP)、活性氧(reactive oxygen species, ROS)和酚氧化酶级联反应的组分等),以及效应分子所依赖的信号通路(JAK-STAT、Toll和Imd等信号通路)是蚊虫体内免疫防御的重要方面[10,11,12]。此外,RNA干扰(RNAi)途径是蚊虫体内最为强大的抗病毒防御体系[13]。RNAi是指由双链RNA(dsRNA)诱发的具有高度保守的小RNA片段可高效特异性降解同源mRNA的现象,是转录后水平的基因沉默(post- transcriptional gene silencing, PTGS)。RNAi途径主要包含3种类型的小RNA:小干扰RNA (siRNA)、微小RNA (miRNA)和Piwi蛋白相作用RNA (piRNA),其中siRNA为蚊虫体内抗病毒免疫的主要小RNA分子[13,14,15]。RNAi具有高度的序列特异性,严格按照碱基互补配对原则与同源基因的mRNA结合并进行降解,从而实现针对目的基因的精准沉默;RNAi具有高效抑制基因表达的特性,表型可达到缺失突变体表型的程度;RNAi还具有可遗传性和可传播 性,RNA干扰效应能稳定遗传给下一代,也可穿过细胞界限,传播至扩散处细胞乃至整个机体[16,17]。利用RNAi特性发展的RNAi技术可广泛地应用于基因功能的探索、传染性疾病和恶性肿瘤的治疗领域[18,19,20,21]。2 siRNA在蚊虫体内的抗病毒作用
siRNA途径可分为内源性通路和外源性通路。内源性siRNA一般由细胞内基因双向转录形成的部分互补配对或由RNA依赖性RNA聚合酶(RNA- dependent RNA polymerase, RDRP)进行的RNA链复制等形成的dsRNA经剪切修饰而产生,在生物体不同生长发育时期发挥调控作用。外源性siRNA一般是由转基因技术或病毒感染细胞后产生的复制中间体—dsRNA经过核糖核酸酶III(RNase III)家族中的Dicer-2核酸内切酶剪切成长度约为21~25 nt的双链小RNA分子[22,23]。细胞内siRNA通过与AGO、TRBP和PACT等蛋白分子结合形成RNA诱导的沉默复合物(RNA-induced silencing complex, RISC),siRNA的随从链被降解,引导链通过碱基互补配对的方式找到靶基因的mRNA或同源的病毒RNA,然后RISC中的Ago-2蛋白降解其mRNA或病毒RNA,阻止翻译表达或病毒复制(图1)[24,25]。
图1
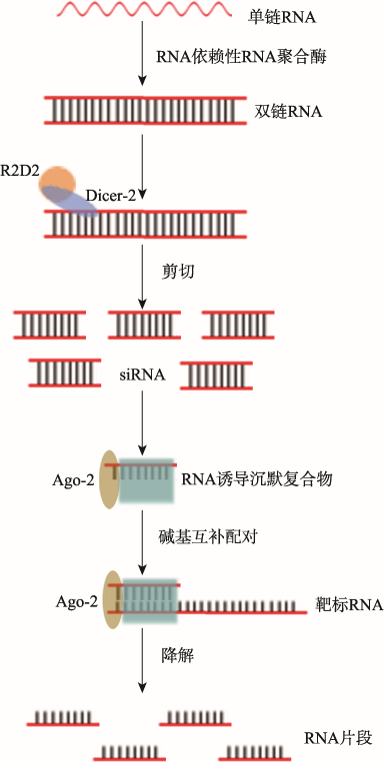 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1siRNA的生物合成和作用机制
Fig. 1The mechanism of siRNA biogenesis and functions
Li等[26]首次在冈比亚按蚊(Anopheles gambiae)单链RNA逆转录形成双链RNA,经Dicer-2核酸内切酶剪切成双链小RNA分子,其引导链与Ago-2等相应蛋白组成RNA诱导沉默复合物,通过碱基互补配对的方式结合靶标RNA,并将该靶标RNA降解。
细胞系中验证了siRNA的抗病毒作用,并表明其抑制效应依赖于Ago-2蛋白。Sánchez-Vargas等[27]研究发现沉默埃及伊蚊(Aedes aegypti) siRNA通路会增加蚊虫体内登革2型病毒的复制,这表明siRNA通路在蚊虫体内病毒复制过程中发挥抑制作用。Khoo等[28]通过转基因技术破坏埃及伊蚊中肠的siRNA通路,发现这样会增加中肠内辛德毕斯病毒(SINV)的复制和播散率。Basu等[29]通过基因敲除抑制埃及伊蚊Dcr-2酶的表达,发现蚊虫体内抗辛德毕斯病毒的免疫反应下降;另外,Dcr-2基因突变型的埃及伊蚊较野生型的蚊虫体内具有显著增加的黄热病病毒复制水平[30]。多项研究表明,在敲除siRNA通路的相关成分后,成蚊体内或培养的蚊虫细胞系中的病毒复制水平会显著升高[27,31]。成蚊或不同细胞系感染蚊媒病毒后会导致该病毒来源的siRNA (virus-derived small interfering RNA, vsiRNA)产生[32,33]。Myles等[34]研究表明vsiRNA对蚊虫体内抗甲病毒感染具有重要调控作用。能够表达dsRNA结合蛋白和病毒RNA沉默抑制因子(viral suppressors of RNA silencing, VSRs)的重组甲病毒感染埃及伊蚊和冈比亚按蚊后,蚊虫体内的vsiRNA显著下降,从而导致病毒复制量和蚊虫死亡率显著增加。
蚊媒病毒在受到蚊虫宿主先天免疫消除的同时,也在不断进化适应宿主的生理环境,并对抗媒介蚊虫的抗病毒免疫途径。通过鉴别蚊媒病毒基因组中编码的病毒RNA沉默抑制因子(VSRs),了解病毒蛋白对蚊虫免疫途径的拮抗作用,将有助于理解蚊媒病毒在蚊虫体内长期感染和传播的机制,并应用于蚊媒病的防控[35]。布尼亚维拉病毒(Bunyamwera virus, BUNV)S片段上的非结构蛋白(non-structure proteins, NSs)是VSRs,NSs缺陷性BUNV在Dcr-2缺陷性的蚊虫细胞系中的复制水平要高于在正常蚊虫细胞系中的复制水平,NSs缺陷性BUNV在埃及伊蚊体内的感染能力要低于野生型BUNV[36]。Kakumani等[37]通过体外实验证明登革热病毒(DENV) NS4B蛋白能够干扰Dicer对siRNA的加工处理过程。哺乳动物细胞中的基因沉默实验显示NS4B蛋白的跨膜结构域3 (transmembrane domain 3, TMD3)和TME5参与VSRs抑制病毒增殖的活性,其具体机制目前尚不明确。Samuel等[38]研究发现在Dcr-2缺陷性的蚊虫细胞系中表达黄病毒衣壳蛋白(yellow fever virus capsid, YFC)的SINV与不表达YFC的SINV的增殖能力和毒力相似,并且验证了YFC作为VSRs拮抗siRNA途径。YFC对siRNA途径拮抗作用可能是非特异性结合双链RNA(dsRNA),干扰Dicer产生vsiRNA。虽然VSRs有多种不同的蛋白,但它与dsRNA非特异性结合是探讨其拮抗作用的主要内容。
3 miRNA在蚊虫体内的抗病毒作用
miRNA是一类内源性的非编码小RNA (18~ 25 nt),通过降解mRNA或抑制mRNA翻译来调控靶基因转录后表达水平[39]。与siRNA途径相似,miRNA途径也始于dsRNA剪切成小的双链RNA,其中一条引导链加载到RISC中,然后RISC中Ago-2蛋白降解靶基因的mRNA或同源的病毒RNA[40]。miRNA与siRNA途径的主要区别在于所发生的亚细胞结构位置和所参与的效应蛋白分子[41]。siRNA的转录、剪切和加工过程主要发生在细胞质中,而编码miRNA的基因在宿主RNA聚合酶Ⅱ的作用下转录成miRNA初始转录物(primary miRNA, pri-miRNA),然后由Drosha剪切加工成miRNA前体物(precursor miRNA, pre-miRNA),这一过程均是在细胞核内完成。pre-miRNA输出到细胞质后,由Dicer-1进一步加工至成熟的miRNA,并加载到RISC中发挥RNA干扰作用(图2)[42]。图2
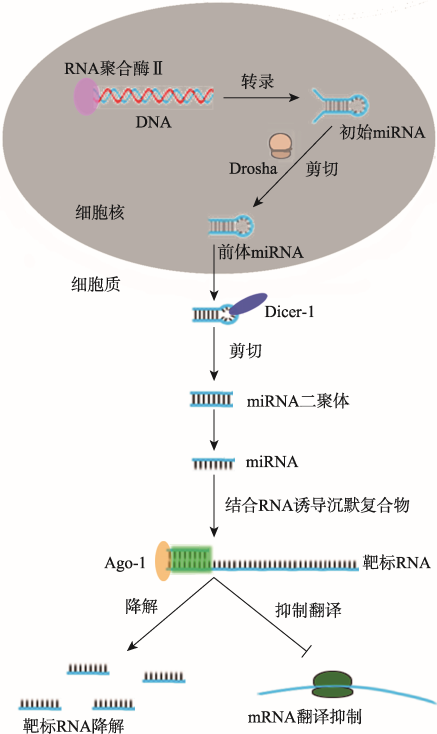 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2miRNA的生物合成和作用机制
含miRNA序列的DNA经过转录后形成初始miRNA,然后经Drosha和Dicer-1等蛋白剪切加工修饰后形成成熟的miRNA,成熟的miRNA与Ago-1等相应蛋白组成RNA诱导沉默复合物,通过碱基互补配对的方式结合靶标RNA,并将该靶标RNA降解或抑制mRNA翻译。
Fig. 2The mechanism of miRNA biogenesis and functions
多种蚊媒病毒均利用宿主miRNA通路来逃避宿主的抗病毒免疫反应,从而增加病毒复制和致病力[43]。Hussain等[44]在西尼罗河病毒(WNV)RNA序列的3°端非编码区发现了类似miRNA的小RNA片段,并通过Northern印记杂交的方法检测到感染WNV的埃及伊蚊和白纹伊蚊(Aedes albopictus)细胞系中存在一种病毒来源的成熟miRNA,命名为KUN-miR-1。KUN-miR-1通过上调转录因子GATA4的表达来促进病毒在蚊虫细胞内的增殖。Hussain 等[45]通过二代测序技术发现感染登革2型病毒(DENV-2)的埃及伊蚊体内含有6种病毒来源的miRNA样的小RNA,称为vsRNA 1~6;其中vsRNA5已证实与病毒增殖有关。蚊虫体内miRNA通路关键分子的功能丧失性突变将有助于鉴定其分子特性,并确定KUN-miR-1和DENV-2-vsRNA 1~6的生物发生机制和抗病毒免疫机制。
当蚊虫感染蚊媒病毒后体内会出现多种miRNA差异性表达[46,47]。Su等[48]利用含DENV-2的血餐以及不含DENV-2的血餐分别喂食白纹伊蚊,鉴定蚊虫中肠内差异性表达的miRNA,相比于喂食不含DENV-2血餐的白纹伊蚊,一共有43个miRNA上调,4个miRNA下调;并且上调的miRNA中aal-miR- 4728-5p瞬时转入C6/36蚊虫细胞后能够增强DENV-2在细胞内的复制。Su等[49]再次利用含DENV-2的血餐喂食白纹伊蚊,将感染上DENV-2与未感染上DENV-2的白纹伊蚊中肠内miRNA进行差异性分析,发现感染上DENV-2的白纹伊蚊中肠内有15个miRNA上调,2个miRNA下调。其中miR-1767和miR-276-3p能够增强DENV-2在C6/36细胞内的复制,而miR-4448抑制DENV-2在C6/36细胞内的复制。
4 piRNA在蚊虫体内的抗病毒作用
piRNA途径可在siRNA途径缺陷的条件下进行抗病毒免疫,是RNAi介导的抗病毒免疫应答的相互补充[50]。与siRNA/miRNA相比,piRNA的生成不依赖Dicer,而依赖PIWI亚家族蛋白;piRNA的长度约为26~32 nt,piRNA基因簇主要分别在转座子和重复序列等区域;piRNA的3'端会出现甲基化修饰,可能与其稳定性或功能有关[51,52,53]。piRNA的生成涉及3种PIWI蛋白,包括Piwi、Aub和Ago-3,共同形成piRNA诱导的沉默复合物(piRNA-induced silencing complex, piRISC)[54]。对于piRNA的生成,目前提出了“乒乓”循环扩增模型,piRNA从转录前体物中循环扩增[55]。反义链piRNA与Aub和Piwi结合形成具有核酸酶活性的piRNA复合物(piRNA complex, piRC),piRC能结合正义链前体piRNA并将其剪切加工成具有成熟5°端的前体piRNA,然后该正义链前体piRNA经Zuc等酶剪切3°末端形成成熟的正义链piRNA;反之,正义链piRNA与Ago-3结合形成piRC,然后以同样的方式形成反义链piRNA (图3)[56]。piRNA途径在生殖遗传、配子形成、胚胎发育、基因转座、基因沉默和病毒增殖等方面均有调控作用。图3
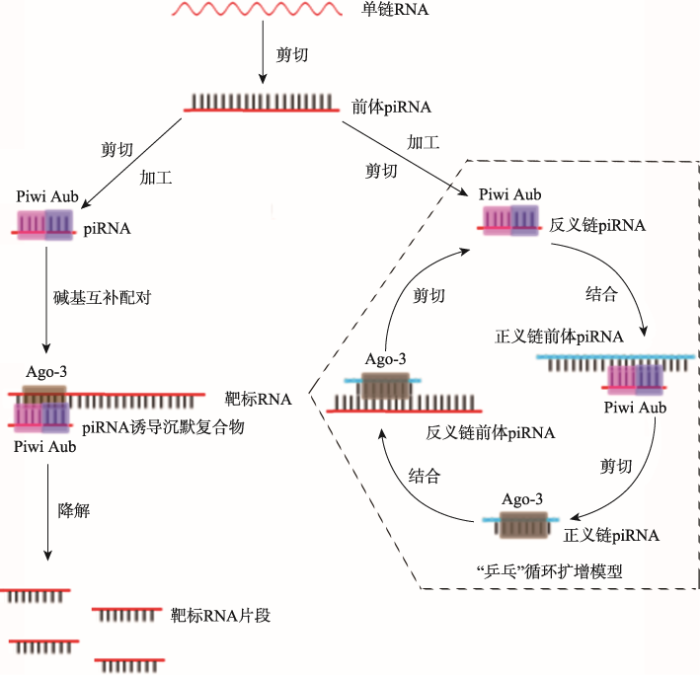 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3piRNA的生物合成和作用机制
单链RNA经剪切形成前体piRNA,然后经Zuc等酶剪切加工后形成成熟的piRNA,成熟的piRNA与Piwi、Aub和Ago-3等蛋白组成piRNA诱导沉默复合物,通过碱基互补配对的方式结合靶标RNA,并将该靶标RNA降解。虚线框内为piRNA的“乒乓”循环扩增模型。
Fig. 3The mechanism of piRNA biogenesis and functions
蚊虫或蚊虫细胞系感染蚊媒病毒后,能够产生一类依赖“乒乓”模型的病毒来源的piRNA (virus- derived piRNA, vpiRNA),这些vpiRNA不同于以往研究中来源于重复序列元件或piRNA簇的piRNA[57,58]。Morazzani等[57]在感染有基孔肯亚热病毒(CHIKV)的埃及伊蚊和白纹伊蚊体内检测到vpiRNA。Miesen等[59]用SINV感染埃及伊蚊细胞系后,通过免疫沉淀反应和小RNA的Northern免疫印迹检测到了Ago-3和Piwi-5蛋白特异性富集的vpiRNA。通过对这类vpiRNA测序分析,显示反义链vpiRNA倾向于与Piwi-5结合,而正义链vpiRNA倾向于与Ago-3结合,这也表明了这两种蛋白在“乒乓”模型中的作用机制。抑制vpiRNA在siRNA途径缺陷的蚊虫细胞系中表达,将会加重受病毒感染细胞的病变程度,这体现了piRNA途径在siRNA途径缺陷的蚊虫细胞系中的抗病毒作用[57]。Schnettler等[60]通过深度测序检测到塞姆利基森林病毒(SFV)来源的piRNA,敲除Piwi-4基因后会导致Aag2埃及伊蚊细胞内SFV复制增加,表明了piRNA途径在抗病毒免疫中的重要作用。
5 结语与展望
每年全球均有大量的蚊媒病暴发流行,并且目前大部分的蚊媒病缺乏有效的疫苗进行预防,所以阻止蚊媒病毒传播一直是蚊媒病预防控制的重要任务。目前有关化学杀虫剂和Wolbachia生物防治等方面的防控措施主要是通过控制蚊虫数量来控制蚊媒病的暴发流行。病毒感染和蚊虫防御机制以及两者动态平衡的演变过程一直是科研工作者关注和探索的问题,深入了解蚊虫先天免疫系统如何抵御病毒感染,病毒如何在蚊虫体内持续稳定增殖,以及不同蚊虫种类或病毒株如何影响疾病暴发流行等方面内容,将有助于提高现有策略的有效性,并提出新的蚊媒控制策略。RNAi途径作为蚊虫体内主要的抗病毒防御体系,目前我国科研工作者对蚊虫RNAi途径的研究主要集中在相关小RNA分子的作用机制,然而通过基因编辑加强蚊虫RNAi抗病毒免疫的相关研究较少,将转基因蚊广泛有效地应用于蚊媒病防控也是任重道远。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1111/j.1749-6632.2001.tb02681.xURLPMID:11797771 [本文引用: 1]

Viral diseases transmitted by blood-feeding arthropods (arboviral diseases) are among the most important of the emerging infectious disease public health problems facing the world at the beginning of the third millennium. There are over 534 viruses listed in the arbovirus catalogue, approximately 134 of which have been shown to cause disease in humans. These are transmitted principally by mosquitoes and ticks. In the last two decades of the twentieth century, a few new arboviral diseases have been recognized. More important, however, is the dramatic resurgence and geographic spread of a number of old diseases that were once effectively controlled. Global demographic and societal changes, and modern transportation have provided the mechanisms for the viruses to break out of their natural ecology and become established in new geographic locations where susceptible arthropod vectors and hosts provide permissive conditions for them to cause major epidemics. West Nile virus is just the the latest example of this type of invasion by exotic viruses. This paper will provide an overview of the medically important arboviruses and discuss several in more detail as case studies to illustrate our tenuous position as we begin the twenty-first century.
DOI:10.1212/CON.0000000000000240URLPMID:26633778 [本文引用: 1]

Arbovirus (arthropod-borne virus) infections are increasingly important causes of neurologic disease in the United States through both endemic transmission and travel-associated infections. This article reviews the major arbovirus infections that can cause neurologic disease likely to be encountered in the United States.
DOI:10.3389/fimmu.2018.02180URLPMID:30319635 [本文引用: 1]

Flaviviruses are emerging and re-emerging arthropod-borne pathogens responsible for significant mortality and morbidity worldwide. The genus comprises more than seventy small, positive-sense, single-stranded RNA viruses, which are responsible for a spectrum of human and animal diseases ranging in symptoms from mild, influenza-like infection to fatal encephalitis and haemorrhagic fever. Despite genomic and structural similarities across the genus, infections by different flaviviruses result in disparate clinical presentations. This review focusses on two haemorrhagic flaviviruses, dengue virus and yellow fever virus, and two neurotropic flaviviruses, Japanese encephalitis virus and Zika virus. We review current knowledge on host-pathogen interactions, virus entry strategies and tropism.
DOI:10.3390/v10020084URLPMID:29443908 [本文引用: 1]

Alphaviruses are arthropod-borne viruses and are predominantly transmitted via mosquito vectors. This vector preference by alphaviruses raises the important question of the determinants that contribute to vector competence. There are several tissue barriers of the mosquito that the virus must overcome in order to establish a productive infection. Of importance are the midgut, basal lamina and the salivary glands. Infection of the salivary glands is crucial for virus transmission during the mosquito's subsequent bloodfeed. Other factors that may contribute to vector competence include the microflora and parasites present in the mosquito, environmental conditions, the molecular determinants of the virus to adapt to the vector, as well as the effect of co-infection with other viruses. Though mosquito innate immunity is a contributing factor to vector competence, it will not be discussed in this review. Detailed understanding of these factors will be instrumental in minimising transmission of alphaviral diseases.
[本文引用: 1]
URLPMID:23995308 [本文引用: 1]

Arthropod-borne viruses (arboviruses) have become significant public health problems, with the emergence and re-emergence of arboviral diseases nearly worldwide. The most populated Southeast Asia region is particularly vulnerable. The arboviral diseases such as dengue (DEN), Japanese encephalitis (JE), West Nile virus (WNV), chikungunya fever (CHIK), hemorrhagic fevers such as Crimean-Congo hemorrhagic (CCHF) fever, Kyasanur forest disease virus (KFDV), etc. are on the rise and have spread unprecedentedly, causing considerable burden of disease. The emergence/re-emergence of these diseases is associated with complex factors, such as viral recombination and mutation, leading to more virulent and adaptive strains, urbanization and human activities creating more permissive environment for vector-host interaction, and increased air travel and commerce. Climate is a major factor in determining the geographic and temporal distribution of arthropods, the characteristics of arthropod life cycles, the consequent dispersal patterns of associated arboviruses, the evolution of arboviruses; and the efficiency with which they are transmitted from arthropods to vertebrate hosts. The present and future arboviral threats must be mitigated by priority actions such as improving surveillance and outbreak response, establishing collaboration and communication intersectorally, and strengthening the prevention and control programmes along with improving biosafety aspects with regards to highly infectious nature of these arboviral diseases. Evidence from research needs to be generated and priority areas for research defined.
DOI:10.3390/ijerph15010096URLPMID:29315224 [本文引用: 1]

The first confirmed case of Zika virus infection in the Americas was reported in Northeast Brazil in May 2015, although phylogenetic studies indicate virus introduction as early as 2013. Zika rapidly spread across Brazil and to more than 50 other countries and territories on the American continent. The Aedesaegypti mosquito is thought to be the principal vector responsible for the widespread transmission of the virus. However, sexual transmission has also been reported. The explosively emerging epidemic has had diverse impacts on population health, coinciding with cases of Guillain-Barré Syndrome and an unexpected epidemic of newborns with microcephaly and other neurological impairments. This led to Brazil declaring a national public health emergency in November 2015, followed by a similar decision by the World Health Organization three months later. While dengue virus serotypes took several decades to spread across Brazil, the Zika virus epidemic diffused within months, extending beyond the area of permanent dengue transmission, which is bound by a climatic barrier in the south and low population density areas in the north. This rapid spread was probably due to a combination of factors, including a massive susceptible population, climatic conditions conducive for the mosquito vector, alternative non-vector transmission, and a highly mobile population. The epidemic has since subsided, but many unanswered questions remain. In this article, we provide an overview of the discovery of Zika virus in Brazil, including its emergence and spread, epidemiological surveillance, vector and non-vector transmission routes, clinical complications, and socio-economic impacts. We discuss gaps in the knowledge and the challenges ahead to anticipate, prevent, and control emerging and re-emerging epidemics of arboviruses in Brazil and worldwide.
DOI:10.1016/j.antiviral.2016.03.010URLPMID:26996139 [本文引用: 1]

Zika virus (ZIKV), a previously obscure flavivirus closely related to dengue, West Nile, Japanese encephalitis and yellow fever viruses, has emerged explosively since 2007 to cause a series of epidemics in Micronesia, the South Pacific, and most recently the Americas. After its putative evolution in sub-Saharan Africa, ZIKV spread in the distant past to Asia and has probably emerged on multiple occasions into urban transmission cycles involving Aedes (Stegomyia) spp. mosquitoes and human amplification hosts, accompanied by a relatively mild dengue-like illness. The unprecedented numbers of people infected during recent outbreaks in the South Pacific and the Americas may have resulted in enough ZIKV infections to notice relatively rare congenital microcephaly and Guillain-Barré syndromes. Another hypothesis is that phenotypic changes in Asian lineage ZIKV strains led to these disease outcomes. Here, we review potential strategies to control the ongoing outbreak through vector-centric approaches as well as the prospects for the development of vaccines and therapeutics.
DOI:10.1016/S1473-3099(16)30471-6URLPMID:28185870 [本文引用: 1]

Advances in the development of new dengue control tools, including vaccines and vector control, herald a new era of desperately needed dengue prevention and control. The burden of dengue has expanded for decades, and now affects more than 120 countries. Complex, large-scale global forces have and will continue to contribute to the expansion of dengue, including population growth, unplanned urbanisation, and suboptimal mosquito control in urban centres. Although no so-called magic bullets are available, there is new optimism following the first licensure of a dengue vaccine and other promising vaccine candidates, and the development of novel vector control interventions to help control dengue and other expanding mosquito-borne diseases such as Zika virus. Implementation of effective and sustainable immunisation programmes to complement existing methods will add complexity to the health systems of affected countries, which have varying levels of robustness and maturity. Long-term high prioritisation and adequate resources are needed. The way forward is full commitment to addressing a complex disease with a set of solutions integrating vaccination and vector control methods. A whole systems approach is thus needed to integrate these various approaches and strategies for controlling dengue within the goal of universal health coverage. The ultimate objective of these interventions will be to reduce the disease burden in a sustainable and equitable manner.
DOI:10.1371/journal.ppat.1000098URLPMID:18604274 [本文引用: 1]

Aedes aegypti, the mosquito vector of dengue viruses, utilizes its innate immune system to ward off a variety of pathogens, some of which can cause disease in humans. To date, the features of insects' innate immune defenses against viruses have mainly been studied in the fruit fly Drosophila melanogaster, which appears to utilize different immune pathways against different types of viruses, in addition to an RNA interference-based defense system. We have used the recently released whole-genome sequence of the Ae. aegypti mosquito, in combination with high-throughput gene expression and RNA interference (RNAi)-based reverse genetic analyses, to characterize its response to dengue virus infection in different body compartments. We have further addressed the impact of the mosquito's endogenous microbial flora on virus infection. Our findings indicate a significant role for the Toll pathway in regulating resistance to dengue virus, as indicated by an infection-responsive regulation and functional assessment of several Toll pathway-associated genes. We have also shown that the mosquito's natural microbiota play a role in modulating the dengue virus infection, possibly through basal-level stimulation of the Toll immune pathway.
DOI:10.1073/pnas.0905006106URLPMID:19805194 [本文引用: 1]

Here, we show that the major mosquito vector for dengue virus uses the JAK-STAT pathway to control virus infection. Dengue virus infection in Aedes aegypti mosquitoes activates the JAK-STAT immune signaling pathway. The mosquito's susceptibility to dengue virus infection increases when the JAK-STAT pathway is suppressed through RNAi depletion of its receptor Domeless (Dome) and the Janus kinase (Hop), whereas mosquitoes become more resistant to the virus when the negative regulator of the JAK-STAT pathway, PIAS, is silenced. The JAK-STAT pathway exerts its anti-dengue activity presumably through one or several STAT-regulated effectors. We have identified, and partially characterized, two JAK-STAT pathway-regulated and infection-responsive dengue virus restriction factors (DVRFs) that contain putative STAT-binding sites in their promoter regions. Our data suggest that the JAK-STAT pathway is part of the A. aegypti mosquito's anti-dengue defense and may act independently of the Toll pathway and the RNAi-mediated antiviral defenses.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.cell.2007.07.039URLPMID:17693253 [本文引用: 2]

Plants and invertebrates can protect themselves from viral infection through RNA silencing. This antiviral immunity involves production of virus-derived small interfering RNAs (viRNAs) and results in specific silencing of viruses by viRNA-guided effector complexes. The proteins required for viRNA production as well as several key downstream components of the antiviral immunity pathway have been identified in plants, flies, and worms. Meanwhile, viral mechanisms to suppress this small RNA-directed immunity by viruses are being elucidated, thereby illuminating an ongoing molecular arms race that likely impacts the evolution of both viral and host genomes.
DOI:10.4161/21655979.2014.979701URLPMID:25424593 [本文引用: 1]

RNA interference (RNAi) in insects is a gene regulatory process that also plays a vital role in the maintenance and in the regulation of host defenses against invading viruses. Small RNAs determine the specificity of the RNAi through precise recognition of their targets. These small RNAs in insects comprise small interfering RNAs (siRNAs), micro RNAs (miRNAs) and Piwi interacting RNAs (piRNAs) of various lengths. In this review, we have explored different forms of the RNAi inducers that are presently in use, and their applications for an effective and efficient fundamental and practical RNAi research with insects. Further, we reviewed trends in next generation sequencing (NGS) technologies and their importance for insect RNAi, including the identification of novel insect targets as well as insect viruses. Here we also describe a rapidly emerging trend of using plant viruses to deliver the RNAi inducer molecules into insects for an efficient RNAi response.
DOI:10.1186/s13071-019-3433-8URLPMID:30975197 [本文引用: 1]

Mosquito-borne diseases are associated with major global health burdens. Aedes spp. and Culex spp. are primarily responsible for the transmission of the most medically important mosquito-borne viruses, including dengue virus, West Nile virus and Zika virus. Despite the burden of these pathogens on human populations, the interactions between viruses and their mosquito hosts remain enigmatic. Viruses enter the midgut of a mosquito following the mosquito's ingestion of a viremic blood meal. During infection, virus recognition by the mosquito host triggers their antiviral defense mechanism. Of these host defenses, activation of the RNAi pathway is the main antiviral mechanism, leading to the degradation of viral RNA, thereby inhibiting viral replication and promoting viral clearance. However, whilst antiviral host defense mechanisms limit viral replication, the mosquito immune system is unable to effectively clear the virus. As such, these viruses can establish persistent infection with little or no fitness cost to the mosquito vector, ensuring life-long transmission to humans. Understanding of the mosquito innate immune response enables the discovery of novel antivectorial strategies to block human transmission. This review provides an updated and concise summary of recent studies on mosquito antiviral immune responses, which is a key determinant for successful virus transmission. In addition, we will also discuss the factors that may contribute to persistent infection in mosquito hosts. Finally, we will discuss current mosquito transmission-blocking strategies that utilize genetically modified mosquitoes and Wolbachia-infected mosquitoes for resistance to pathogens.
DOI:10.1038/35888URLPMID:9486653 [本文引用: 1]

Experimental introduction of RNA into cells can be used in certain biological systems to interfere with the function of an endogenous gene. Such effects have been proposed to result from a simple antisense mechanism that depends on hybridization between the injected RNA and endogenous messenger RNA transcripts. RNA interference has been used in the nematode Caenorhabditis elegans to manipulate gene expression. Here we investigate the requirements for structure and delivery of the interfering RNA. To our surprise, we found that double-stranded RNA was substantially more effective at producing interference than was either strand individually. After injection into adult animals, purified single strands had at most a modest effect, whereas double-stranded mixtures caused potent and specific interference. The effects of this interference were evident in both the injected animals and their progeny. Only a few molecules of injected double-stranded RNA were required per affected cell, arguing against stochiometric interference with endogenous mRNA and suggesting that there could be a catalytic or amplification component in the interference process.
DOI:10.1038/nature07712URLPMID:19204732 [本文引用: 1]

Multicellular organisms evolved sophisticated defence systems to confer protection against pathogens. An important characteristic of these immune systems is their ability to act both locally at the site of infection and at distal uninfected locations. In insects, such as Drosophila melanogaster, RNA interference (RNAi) mediates antiviral immunity. However, the antiviral RNAi defence in flies seems to be a local, cell-autonomous process, as flies are thought to be unable to generate a systemic RNAi response. Here we show that a recently defined double-stranded RNA (dsRNA) uptake pathway is essential for effective antiviral RNAi immunity in adult flies. Mutant flies defective in this dsRNA uptake pathway were hypersensitive to infection with Drosophila C virus and Sindbis virus. Mortality in dsRNA-uptake-defective flies was accompanied by 100-to 10(5)-fold increases in viral titres and higher levels of viral RNA. Furthermore, inoculating naked dsRNA into flies elicited a sequence-specific antiviral immune response that required an intact dsRNA uptake pathway. These findings suggest that spread of dsRNA to uninfected sites is essential for effective antiviral immunity. Notably, infection with green fluorescent protein (GFP)-tagged Sindbis virus suppressed expression of host-encoded GFP at a distal site. Thus, similar to protein-based immunity in vertebrates, the antiviral RNAi response in flies also relies on the systemic spread of a virus-specific immunity signal.
DOI:10.1093/jme/tjv191URLPMID:26659858 [本文引用: 1]

Effective mosquito control is vital to curtail the devastating health effects of many vectored diseases. RNA interference (RNAi)-mediated control of mosquitoes is an attractive alternative to conventional chemical pesticides. Previous studies have suggested that transcripts for inhibitors of apoptosis (IAPs) may be good RNAi targets. To revisit and extend previous reports, we examined the expression of Aedes aegypti (L.) IAPs (AaeIAPs) 1, 2, 5, 6, 9, and a viral IAP-associated factor (vIAF) as well as Anopheles quadrimaculatus Say and Culex quinquefasciatus Say IAP1 homologs (AquIAP1 and CquIAP1) in adult females. Expression profiles of IAPs suggested that some older female mosquitoes had significantly higher IAP mRNA levels when compared to the youngest ones. Minor differences in expression of AaeIAPs were observed in mosquitoes that imbibed a bloodmeal, but the majority of the time points (up to 48?h) were not significantly different. Although in vitro experiments with the Ae. aegypti Aag-2 cell line demonstrated that the various AaeIAPs could be effectively knocked down within one day after dsRNA treatment, only Aag-2 cells treated with dsIAP1 displayed apoptotic morphology. Gene silencing and mortality were also evaluated after topical application and microinjection of the same dsRNAs into female Ae. aegypti. In contrast to previous reports, topical administration of dsRNA against AaeIAP1 did not yield a significant reduction in gene expression or increased mortality. Knockdown of IAP1 and other IAPs by microinjection did not result in significant mortality. In toto, our findings suggest that IAPs may not be suitable RNAi targets for controlling adult mosquito populations.
DOI:10.3347/kjp.2008.46.1.1URLPMID:18344671 [本文引用: 1]

Introduction of double-stranded RNA (dsRNA) into some cells or organisms results in degradation of its homologous mRNA, a process called RNA interference (RNAi). The dsRNAs are processed into short interfering RNAs (siRNAs) that subsequently bind to the RNA-induced silencing complex (RISC), causing degradation of target mRNAs. Because of this sequence-specific ability to silence target genes, RNAi has been extensively used to study gene functions and has the potential to control disease pathogens or vectors. With this promise of RNAi to control pathogens and vectors, this paper reviews the current status of RNAi in protozoans, animal parasitic helminths and disease-transmitting vectors, such as insects. Many pathogens and vectors cause severe parasitic diseases in tropical regions and it is difficult to control once the host has been invaded. Intracellularly, RNAi can be highly effective in impeding parasitic development and proliferation within the host. To fully realize its potential as a means to control tropical diseases, appropriate delivery methods for RNAi should be developed, and possible off-target effects should be minimized for specific gene suppression. RNAi can also be utilized to reduce vector competence to interfere with disease transmission, as genes critical for pathogenesis of tropical diseases are knockdowned via RNAi.
DOI:10.1016/j.jconrel.2014.09.001URLPMID:25204288 [本文引用: 1]

Chemotherapeutic agents have certain limitations when it comes to treating cancer, the most important being severe side effects along with multidrug resistance developed against them. Tumor cells exhibit drug resistance due to activation of various cellular level processes viz. activation of drug efflux pumps, anti-apoptotic defense mechanisms, etc. Currently, RNA interference (RNAi) based therapeutic approaches are under vibrant scrutinization to seek cancer cure. Especially small interfering RNA (siRNA) and micro RNA (miRNA), are able to knock down the carcinogenic genes by targeting the mRNA expression, which underlies the uniqueness of this therapeutic approach. Recent research focus in the regime of cancer therapy involves the engagement of targeted delivery of siRNA/miRNA in combinations with other therapeutic agents (such as gene, DNA or chemotherapeutic drug) for targeting permeability glycoprotein (P-gp), multidrug resistant protein 1 (MRP-1), B-cell lymphoma (BCL-2) and other targets that are mainly responsible for resistance in cancer therapy. RNAi-chemotherapeutic drug combinations have also been found to be effective against different molecular targets as well and can increase the sensitization of cancer cells to therapy several folds. However, due to stability issues associated with siRNA/miRNA suitable protective carrier is needed and nanotechnology based approaches have been widely explored to overcome these drawbacks. Furthermore, it has been univocally advocated that the co-delivery of siRNA/miRNA with other chemodrugs significantly enhances their capability to overcome cancer resistance compared to naked counterparts. The objective of this article is to review recent nanocarrier based approaches adopted for the delivery of siRNA/miRNA combinations with other anticancer agents (siRNA/miRNA/pDNA/chemodrugs) to treat cancer.
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/ni1335URLPMID:16554838 [本文引用: 1]

The fruit fly Drosophila melanogaster is a model system for studying innate immunity, including antiviral host defense. Infection with drosophila C virus triggers a transcriptional response that is dependent in part on the Jak kinase Hopscotch. Here we show that successful infection and killing of drosophila with the insect nodavirus flock house virus was strictly dependent on expression of the viral protein B2, a potent inhibitor of processing of double-stranded RNA mediated by the essential RNA interference factor Dicer. Conversely, flies with a loss-of-function mutation in the gene encoding Dicer-2 (Dcr-2) showed enhanced susceptibility to infection by flock house virus, drosophila C virus and Sindbis virus, members of three different families of RNA viruses. These data demonstrate the importance of RNA interference for controlling virus replication in vivo and establish Dcr-2 as a host susceptibility locus for virus infections.
DOI:10.1126/science.1125694URLPMID:16556799 [本文引用: 1]

Innate immunity against bacterial and fungal pathogens is mediated by Toll and immune deficiency (Imd) pathways, but little is known about the antiviral response in Drosophila. Here, we demonstrate that an RNA interference pathway protects adult flies from infection by two evolutionarily diverse viruses. Our work also describes a molecular framework for the viral immunity, in which viral double-stranded RNA produced during infection acts as the pathogen trigger whereas Drosophila Dicer-2 and Argonaute-2 act as host sensor and effector, respectively. These findings establish a Drosophila model for studying the innate immunity against viruses in animals.
DOI:10.1016/j.cell.2005.08.044URLPMID:16271386 [本文引用: 1]

In the Drosophila and mammalian RNA interference pathways, siRNAs direct the protein Argonaute2 (Ago2) to cleave corresponding mRNA targets, silencing their expression. Ago2 is the catalytic component of the RNAi enzyme complex, RISC. For each siRNA duplex, only one strand, the guide, is assembled into the active RISC; the other strand, the passenger, is destroyed. An ATP-dependent helicase has been proposed first to separate the two siRNA strands, then the resulting single-stranded guide is thought to bind Ago2. Here, we show that Ago2 instead directly receives the double-stranded siRNA from the RISC assembly machinery. Ago2 then cleaves the siRNA passenger strand, thereby liberating the single-stranded guide. For siRNAs, virtually all RISC is assembled through this cleavage-assisted mechanism. In contrast, passenger-strand cleavage is not important for the incorporation of miRNAs that derive from mismatched duplexes.
DOI:10.1101/gad.1370605URLPMID:16287716 [本文引用: 1]

Argonaute proteins play important yet distinct roles in RNA silencing. Human Argonaute2 (hAgo2) was shown to be responsible for target RNA cleavage ("Slicer") activity in RNA interference (RNAi), whereas other Argonaute subfamily members do not exhibit the Slicer activity in humans. In Drosophila, AGO2 was shown to possess the Slicer activity. Here we show that AGO1, another member of the Drosophila Argonaute subfamily, immunopurified from Schneider2 (S2) cells associates with microRNA (miRNA) and cleaves target RNA completely complementary to the miRNA. Slicer activity is reconstituted with recombinant full-length AGO1. Thus, in Drosophila, unlike in humans, both AGO1 and AGO2 have Slicer functions. Further, reconstitution of Slicer activity with recombinant PIWI domains of AGO1 and AGO2 demonstrates that other regions in the Argonautes are not strictly necessary for small interfering RNA (siRNA)-binding and cleavage activities. It has been shown that in circumstances with AGO2-lacking, the siRNA duplex is not unwound and consequently an RNA-induced silencing complex (RISC) is not formed. We show that upon addition of an siRNA duplex in S2 lysate, the passenger strand is cleaved in an AGO2-dependent manner, and nuclease-resistant modification of the passenger strand impairs RISC formation. These findings give rise to a new model in which AGO2 is directly involved in RISC formation as "Slicer" of the passenger strand of the siRNA duplex.
DOI:10.1073/pnas.0308308100URLPMID:14745017 [本文引用: 1]

Homology-dependent RNA silencing occurs in many eukaryotic cells. We reported recently that nodaviral infection triggers an RNA silencing-based antiviral response (RSAR) in Drosophila, which is capable of a rapid virus clearance in the absence of expression of a virus-encoded suppressor. Here, we present further evidence to show that the Drosophila RSAR is mediated by the RNA interference (RNAi) pathway, as the viral suppressor of RSAR inhibits experimental RNAi initiated by exogenous double-stranded RNA and RSAR requires the RNAi machinery. We demonstrate that RNAi also functions as a natural antiviral immunity in mosquito cells. We further show that vaccinia virus and human influenza A, B, and C viruses each encode an essential protein that suppresses RSAR in Drosophila. The vaccinia and influenza viral suppressors, E3L and NS1, are distinct double-stranded RNA-binding proteins and essential for pathogenesis by inhibiting the mammalian IFN-regulated innate antiviral response. We found that the double-stranded RNA-binding domain of NS1, implicated in innate immunity suppression, is both essential and sufficient for RSAR suppression. These findings provide evidence that mammalian virus proteins can inhibit RNA silencing, implicating this mechanism as a nucleic acid-based antiviral immunity in mammalian cells.
DOI:10.1371/journal.ppat.1000299URLPMID:19214215 [本文引用: 2]

A number of studies have shown that both innate and adaptive immune defense mechanisms greatly influence the course of human dengue virus (DENV) infections, but little is known about the innate immune response of the mosquito vector Aedes aegypti to arbovirus infection. We present evidence here that a major component of the mosquito innate immune response, RNA interference (RNAi), is an important modulator of mosquito infections. The RNAi response is triggered by double-stranded RNA (dsRNA), which occurs in the cytoplasm as a result of positive-sense RNA virus infection, leading to production of small interfering RNAs (siRNAs). These siRNAs are instrumental in degradation of viral mRNA with sequence homology to the dsRNA trigger and thereby inhibition of virus replication. We show that although dengue virus type 2 (DENV2) infection of Ae. aegypti cultured cells and oral infection of adult mosquitoes generated dsRNA and production of DENV2-specific siRNAs, virus replication and release of infectious virus persisted, suggesting viral circumvention of RNAi. We also show that DENV2 does not completely evade RNAi, since impairing the pathway by silencing expression of dcr2, r2d2, or ago2, genes encoding important sensor and effector proteins in the RNAi pathway, increased virus replication in the vector and decreased the extrinsic incubation period required for virus transmission. Our findings indicate a major role for RNAi as a determinant of DENV transmission by Ae. aegypti.
DOI:10.1186/1471-2180-10-130URLPMID:20426860 [本文引用: 1]

The RNA interference (RNAi) pathway acts as an innate antiviral immune response in Aedes aegypti, modulating arbovirus infection of mosquitoes. Sindbis virus (SINV; family: Togaviridae, genus: Alphavirus) is an arbovirus that infects Ae. aegypti in the laboratory. SINV strain TR339 encounters a midgut escape barrier (MEB) during infection of Ae. aegypti. The nature of this barrier is not well understood. To investigate the role of the midgut as the central organ determining vector competence for arboviruses, we generated transgenic mosquitoes in which the RNAi pathway was impaired in midgut tissue of bloodfed females. We used these mosquitoes to reveal effects of RNAi impairment in the midgut on SINV replication, midgut infection and dissemination efficiencies, and mosquito longevity.
DOI:10.1073/pnas.1502370112URLPMID:25775608 [本文引用: 1]

Conventional control strategies for mosquito-borne pathogens such as malaria and dengue are now being complemented by the development of transgenic mosquito strains reprogrammed to generate beneficial phenotypes such as conditional sterility or pathogen resistance. The widespread success of site-specific nucleases such as transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 in model organisms also suggests that reprogrammable gene drive systems based on these nucleases may be capable of spreading such beneficial phenotypes in wild mosquito populations. Using the mosquito Aedes aegypti, we determined that mutations in the FokI domain used in TALENs to generate obligate heterodimeric complexes substantially and significantly reduce gene editing rates. We found that CRISPR/Cas9-based editing in the mosquito Ae. aegypti is also highly variable, with the majority of guide RNAs unable to generate detectable editing. By first evaluating candidate guide RNAs using a transient embryo assay, we were able to rapidly identify highly effective guide RNAs; focusing germ line-based experiments only on this cohort resulted in consistently high editing rates of 24-90%. Microinjection of double-stranded RNAs targeting ku70 or lig4, both essential components of the end-joining response, increased recombination-based repair in early embryos as determined by plasmid-based reporters. RNAi-based suppression of Ku70 concurrent with embryonic microinjection of site-specific nucleases yielded consistent gene insertion frequencies of 2-3%, similar to traditional transposon- or ΦC31-based integration methods but without the requirement for an initial docking step. These studies should greatly accelerate investigations into mosquito biology, streamline development of transgenic strains for field releases, and simplify the evaluation of novel Cas9-based gene drive systems.
DOI:10.1073/pnas.1600544113URLPMID:27849599 [本文引用: 1]

Mosquito-borne flaviviruses, including yellow fever virus (YFV), Zika virus (ZIKV), and West Nile virus (WNV), profoundly affect human health. The successful transmission of these viruses to a human host depends on the pathogen's ability to overcome a potentially sterilizing immune response in the vector mosquito. Similar to other invertebrate animals and plants, the mosquito's RNA silencing pathway comprises its primary antiviral defense. Although a diverse range of plant and insect viruses has been found to encode suppressors of RNA silencing, the mechanisms by which flaviviruses antagonize antiviral small RNA pathways in disease vectors are unknown. Here we describe a viral suppressor of RNA silencing (VSR) encoded by the prototype flavivirus, YFV. We show that the YFV capsid (YFC) protein inhibits RNA silencing in the mosquito Aedes aegypti by interfering with Dicer. This VSR activity appears to be broadly conserved in the C proteins of other medically important flaviviruses, including that of ZIKV. These results suggest that a molecular "arms race" between vector and pathogen underlies the continued existence of flaviviruses in nature.
DOI:10.1073/pnas.0406983101URLPMID:15583140 [本文引用: 1]

RNA interference (RNAi) is triggered in eukaryotic organisms by double-stranded RNA (dsRNA), and it destroys any mRNA that has sequence identity with the dsRNA trigger. The RNAi pathway in Anopheles gambiae can be silenced by transfecting cells with dsRNA derived from exon sequence of the A. gambiae Argonaute2 (AgAgo2) gene. We hypothesized that RNAi may also act as an antagonist to alphavirus replication in A. gambiae because RNA viruses form dsRNA during replication. Silencing AgAgo2 expression would make A. gambiae mosquitoes more permissive to virus infection. To determine whether RNAi conditions the vector competence of A. gambiae for O'nyong-nyong virus (ONNV), we engineered a genetically modified ONNV that expresses enhanced GFP (eGFP) as a marker. After intrathoracic injection, ONNV-eGFP slowly spread to other A. gambiae tissues over a 9-day incubation period. Mosquitoes were then coinjected with virus and either control beta-galactosidase dsRNA (dsbetagal; note that "ds" is used as a prefix to indicate the dsRNA derived from a given gene throughout) or ONNV dsnsP3. Treatment with dsnsP3 inhibited virus spread significantly, as determined by eGFP expression patterns. ONNV-eGFP titers from mosquitoes coinjected with dsnsP3 were significantly lower at 3 and 6 days after injection than in mosquitoes coinjected with dsbetagal. Mosquitoes were then coinjected with ONNV-eGFP and dsAgAgo2. Mosquitoes coinjected with virus and AgAgo2 dsRNA displayed widespread eGFP expression and virus titers 16-fold higher than dsbetagal controls after 3 or 6 days after injection. These observations provide direct evidence that RNAi is an antagonist of ONNV replication in A. gambiae, and they suggest that the innate immune response conditions vector competence.
DOI:10.1371/journal.ppat.1000502URLPMID:19578437 [本文引用: 1]

West Nile virus (WNV) exists in nature as a genetically diverse population of competing genomes. This high genetic diversity and concomitant adaptive plasticity has facilitated the rapid adaptation of WNV to North American transmission cycles and contributed to its explosive spread throughout the New World. WNV is maintained in nature in a transmission cycle between mosquitoes and birds, with intrahost genetic diversity highest in mosquitoes. The mechanistic basis for this increase in genetic diversity in mosquitoes is poorly understood. To determine whether the high mutational diversity of WNV in mosquitoes is driven by RNA interference (RNAi), we characterized the RNAi response to WNV in the midguts of orally exposed Culex pipiens quinquefasciatus using high-throughput, massively parallel sequencing and estimated viral genetic diversity. Our data demonstrate that WNV infection in orally exposed vector mosquitoes induces the RNAi pathway and that regions of the WNV genome that are more intensely targeted by RNAi are more likely to contain point mutations compared to weakly targeted regions. These results suggest that, under natural conditions, positive selection of WNV within mosquitoes is stronger in regions highly targeted by the host RNAi response. Further, they provide a mechanistic basis for the relative importance of mosquitoes in driving WNV diversification.
DOI:10.4161/rna.6.4.8946URLPMID:19535909 [本文引用: 1]

The continual transmission in nature of many arthropod-borne viruses depends on the establishment of a persistent, nonpathogenic infection in a mosquito vector. The importance of antiviral immunity directed by small RNAs in the mechanism by which alphaviruses establish a persistent, nonpathogenic infection in the mosquito vector has recently been demonstrated. The origin of the small RNAs central to this RNA silencing response has recently been the subject of debate. Here we briefly summarize what is known about the mechanism of small RNA-directed immunity in invertebrates, and discuss current models for the viral triggers of this response. Finally, we summarize evidence indicating that alphavirus double-stranded replicative intermediates trigger an exogenous-siRNA pathway in mosquitoes resulting in the biogenesis of virus-derived siRNAs.
DOI:10.1073/pnas.0803408105URLPMID:19047642 [本文引用: 1]

Mosquito-borne viruses cause significant levels of morbidity and mortality in humans and domesticated animals. Maintenance of mosquito-borne viruses in nature requires a biological transmission cycle that involves alternating virus replication in a susceptible vertebrate and mosquito host. Although the vertebrate infection is acute and often associated with disease, continual transmission of these viruses in nature depends on the establishment of a persistent, nonpathogenic infection in the mosquito vector. An antiviral RNAi response has been shown to limit the replication of RNA viruses in flies. However, the importance of the RNAi pathway as an antiviral defense in mammals is unclear. Differences in the immune responses of mammals and mosquitoes may explain why these viruses are not generally associated with pathology in the invertebrate host. We identified virus-derived small interfering RNAs (viRNAs), 21 nt in length, in Aedes aegypti infected with the mosquito-borne virus, Sindbis (SINV). viRNAs had an asymmetric distribution that spanned the length of the SINV genome. To determine the role of viRNAs in controlling pathogenic potential, mosquitoes were infected with recombinant alphaviruses expressing suppressors of RNA silencing. Decreased survival was observed in mosquitoes in which the accumulation of viRNAs was suppressed. These results suggest that an exogenous siRNA pathway is essential to the survival of mosquitoes infected with alphaviruses and, thus, the maintenance of these viruses in nature.
DOI:10.1128/JVI.02144-14URL [本文引用: 1]

Insects are a reservoir for many known and novel viruses. We discovered an unknown virus, tentatively named mosinovirus (MoNV), in mosquitoes from a tropical rainforest region in Cote d'Ivoire. The MoNV genome consists of two segments of positive-sense RNA of 2,972 nucleotides (nt) (RNA 1) and 1,801 nt (RNA 2). Its putative RNA-dependent RNA polymerase shares 43% amino acid identity with its closest relative, that of the Pariacoto virus (family Nodaviridae). Unexpectedly, for the putative capsid protein, maximal pairwise identity of 16% to Lake Sinai virus 2, an unclassified virus with a nonsegmented RNA genome, was found. Moreover, MoNV virions are nonenveloped and about 50 nm in diameter, larger than any of the known nodaviruses. Mature MoNV virions contain capsid proteins of similar to 56 kDa, which do not seem to be cleaved from a longer precursor. Northern blot analyses revealed that MoNV expresses two subgenomic RNAs of 580 nt (RNA 3) and 292 nt (RNA 4). RNA 4 encodes a viral suppressor of RNA interference (RNAi) that shares its mechanism with the B2 RNAi suppressor protein of other nodaviruses despite lacking recognizable similarity to these proteins. MoNV B2 binds long double-stranded RNA (dsRNA) and, accordingly, inhibits Dicer-2-mediated processing of dsRNA into small interfering RNAs (siRNAs). Phylogenetic analyses indicate that MoNV is a novel member of the family Nodaviridae that acquired its capsid gene via reassortment from an unknown, distantly related virus beyond the family level.
IMPORTANCE;The identification of novel viruses provides important information about virus evolution and diversity. Here, we describe an unknown unique nodavirus in mosquitoes, named mosinovirus (MoNV). MoNV was classified as a nodavirus based on its genome organization and on phylogenetic analyses of the RNA-dependent RNA polymerase. Notably, its capsid gene was acquired from an unknown virus with a distant relationship to nodaviruses. Another remarkable feature of MoNV is that, unlike other nodaviruses, it expresses two subgenomic RNAs (sgRNAs). One of the sgRNAs expresses a protein that counteracts antiviral defense of its mosquito host, whereas the function of the other sgRNA remains unknown. Our results show that complete genome segments can be exchanged beyond the species level and suggest that insects harbor a large repertoire of exceptional viruses.
DOI:10.1371/journal.pntd.0001823URLPMID:23029584 [本文引用: 1]

Bunyamwera orthobunyavirus is both the prototype and study model of the Bunyaviridae family. The viral NSs protein seems to contribute to the different outcomes of infection in mammalian and mosquito cell lines. However, only limited information is available on the growth of Bunyamwera virus in cultured mosquito cells other than the Aedes albopictus C6/36 line.
DOI:10.1128/JVI.02774-12URLPMID:23741001 [本文引用: 1]

RNA interference (RNAi) is an important antiviral defense response in plants and invertebrates; however, evidences for its contribution to mammalian antiviral defense are few. In the present study, we demonstrate the anti-dengue virus role of RNAi in mammalian cells. Dengue virus infection of Huh 7 cells decreased the mRNA levels of host RNAi factors, namely, Dicer, Drosha, Ago1, and Ago2, and in corollary, silencing of these genes in virus-infected cells enhanced dengue virus replication. In addition, we observed downregulation of many known human microRNAs (miRNAs) in response to viral infection. Using reversion-of-silencing assays, we further showed that NS4B of all four dengue virus serotypes is a potent RNAi suppressor. We generated a series of deletion mutants and demonstrated that NS4B mediates RNAi suppression via its middle and C-terminal domains, namely, transmembrane domain 3 (TMD3) and TMD5. Importantly, the NS4B N-terminal region, including the signal sequence 2K, which has been implicated in interferon (IFN)-antagonistic properties, was not involved in mediating RNAi suppressor activity. Site-directed mutagenesis of conserved residues revealed that a Phe-to-Ala (F112A) mutation in the TMD3 region resulted in a significant reduction of the RNAi suppression activity. The green fluorescent protein (GFP)-small interfering RNA (siRNA) biogenesis of the GFP-silenced line was considerably reduced by wild-type NS4B, while the F112A mutant abrogated this reduction. These results were further confirmed by in vitro dicer assays. Together, our results suggest the involvement of miRNA/RNAi pathways in dengue virus establishment and that dengue virus NS4B protein plays an important role in the modulation of the host RNAi/miRNA pathway to favor dengue virus replication.
DOI:10.1073/pnas.1600544113URLPMID:27849599 [本文引用: 1]

Mosquito-borne flaviviruses, including yellow fever virus (YFV), Zika virus (ZIKV), and West Nile virus (WNV), profoundly affect human health. The successful transmission of these viruses to a human host depends on the pathogen's ability to overcome a potentially sterilizing immune response in the vector mosquito. Similar to other invertebrate animals and plants, the mosquito's RNA silencing pathway comprises its primary antiviral defense. Although a diverse range of plant and insect viruses has been found to encode suppressors of RNA silencing, the mechanisms by which flaviviruses antagonize antiviral small RNA pathways in disease vectors are unknown. Here we describe a viral suppressor of RNA silencing (VSR) encoded by the prototype flavivirus, YFV. We show that the YFV capsid (YFC) protein inhibits RNA silencing in the mosquito Aedes aegypti by interfering with Dicer. This VSR activity appears to be broadly conserved in the C proteins of other medically important flaviviruses, including that of ZIKV. These results suggest that a molecular "arms race" between vector and pathogen underlies the continued existence of flaviviruses in nature.
DOI:10.1038/nrg3965URLPMID:26077373 [本文引用: 1]

MicroRNAs (miRNAs) are a conserved class of small non-coding RNAs that assemble with Argonaute proteins into miRNA-induced silencing complexes (miRISCs) to direct post-transcriptional silencing of complementary mRNA targets. Silencing is accomplished through a combination of translational repression and mRNA destabilization, with the latter contributing to most of the steady-state repression in animal cell cultures. Degradation of the mRNA target is initiated by deadenylation, which is followed by decapping and 5'-to-3' exonucleolytic decay. Recent work has enhanced our understanding of the mechanisms of silencing, making it possible to describe in molecular terms a continuum of direct interactions from miRNA target recognition to mRNA deadenylation, decapping and 5'-to-3' degradation. Furthermore, an intricate interplay between translational repression and mRNA degradation is emerging.
DOI:10.1038/nature06205URLPMID:17943132 [本文引用: 1]

RNA interference through non-coding microRNAs (miRNAs) represents a vital component of the innate antiviral immune response in plants and invertebrate animals; however, a role for cellular miRNAs in the defence against viral infection in mammalian organisms has thus far remained elusive. Here we show that interferon beta (IFNbeta) rapidly modulates the expression of numerous cellular miRNAs, and that eight of these IFNbeta-induced miRNAs have sequence-predicted targets within the hepatitis C virus (HCV) genomic RNA. The introduction of synthetic miRNA-mimics corresponding to these IFNbeta-induced miRNAs reproduces the antiviral effects of IFNbeta on HCV replication and infection, whereas neutralization of these antiviral miRNAs with anti-miRNAs reduces the antiviral effects of IFNbeta against HCV. In addition, we demonstrate that IFNbeta treatment leads to a significant reduction in the expression of the liver-specific miR-122, an miRNA that has been previously shown to be essential for HCV replication. Therefore, our findings strongly support the notion that mammalian organisms too, through the interferon system, use cellular miRNAs to combat viral infections.
DOI:10.1016/s0092-8674(04)00261-2URLPMID:15066283 [本文引用: 1]

The RNase III enzyme Dicer processes RNA into siRNAs and miRNAs, which direct a RNA-induced silencing complex (RISC) to cleave mRNA or block its translation (RNAi). We have characterized mutations in the Drosophila dicer-1 and dicer-2 genes. Mutation in dicer-1 blocks processing of miRNA precursors, whereas dicer-2 mutants are defective for processing siRNA precursors. It has been recently found that Drosophila Dicer-1 and Dicer-2 are also components of siRNA-dependent RISC (siRISC). We find that Dicer-1 and Dicer-2 are required for siRNA-directed mRNA cleavage, though the RNase III activity of Dicer-2 is not required. Dicer-1 and Dicer-2 facilitate distinct steps in the assembly of siRISC. However, Dicer-1 but not Dicer-2 is essential for miRISC-directed translation repression. Thus, siRISCs and miRISCs are different with respect to Dicers in Drosophila.
URLPMID:369476 [本文引用: 1]
URLPMID:369476 [本文引用: 1]
DOI:10.1016/j.molmed.2016.11.003URLPMID:27989642 [本文引用: 1]

microRNAs (miRNAs) are non-coding RNAs that regulate many processes within a cell by manipulating protein levels through direct binding to mRNA and influencing translation efficiency, or mRNA abundance. Recent evidence demonstrates that miRNAs can also affect RNA virus replication and pathogenesis through direct binding to the RNA virus genome or through virus-mediated changes in the host transcriptome. Here, we review the current knowledge on the interaction between RNA viruses and cellular miRNAs. We also discuss how cell and tissue-specific expression of miRNAs can directly affect viral pathogenesis. Understanding the role of cellular miRNAs during viral infection may lead to the identification of novel mechanisms to block RNA virus replication or cell-specific regulation of viral vector targeting.
DOI:10.1093/nar/gkr848URLPMID:22080551 [本文引用: 1]

West Nile virus (WNV) belongs to a group of medically important single-stranded, positive-sense RNA viruses causing deadly disease outbreaks around the world. The 3' untranslated region (3'-UTR) of the flavivirus genome, in particular the terminal 3' stem-loop (3'SL) fulfils multiple functions in virus replication and virus-host interactions. Using the Kunjin strain of WNV (WNV(KUN)), we detected a virally encoded small RNA, named KUN-miR-1, derived from 3'SL. Transcription of WNV(KUN) pre-miRNA (3'SL) in mosquito cells either from plasmid or Semliki Forest virus (SFV) RNA replicon resulted in the production of mature KUN-miR-1. Silencing of Dicer-1 but not Dicer-2 led to a reduction in the miRNA levels. Further, when a synthetic inhibitor of KUN-miR-1 was transfected into mosquito cells, replication of viral RNA was significantly reduced. Using cloning and bioinformatics approaches, we identified the cellular GATA4 mRNA as a target for KUN-miR-1. KUN-miR-1 produced in mosquito cells during virus infection or from plasmid DNA, SFV RNA replicon or mature miRNA duplex increased accumulation of GATA4 mRNA. Depletion of GATA4 mRNA by RNA silencing led to a significant reduction in virus RNA replication while a KUN-miR-1 RNA mimic enhanced replication of a mutant WNV(KUN) virus producing reduced amounts of KUN-miR-1, suggesting that GATA4-induction via KUN-miR-1 plays an important role in virus replication.
DOI:10.1073/pnas.1320123111URLPMID:24550303 [本文引用: 1]

MicroRNAs (miRNAs) are small regulatory RNAs that play significant roles in most cellular processes. In the seemingly endless arms race between hosts and pathogens, viruses also encode miRNAs that facilitate successful infection. In search of functional miRNAs or viral small RNAs (vsRNAs) encoded by Dengue virus (DENV), deep sequencing data of virus-infected Aedes aegypti mosquitoes were used. From six vsRNAs, with candidate stem-loop structures in the 5' and 3' untranslated regions of the viral genomic RNA, inhibition of DENV-vsRNA-5 led to significant increases in viral replication. Silencing of RNA interference (RNAi)/miRNA pathways' associated proteins showed that Argonaute 2 is mainly involved in DENV-vsRNA-5 biogenesis. Cloning of the precursor stem loop, immunoprecipitations, ectopic expression and detection in RNAi-deficient C6/36, and the mammalian Vero cell lines further confirmed DENV-vsRNA-5 production. Furthermore, significant impact of synthetic mimic and inhibitor of DENV-vsRNA-5 on DENV RNA levels revealed DENV-vsRNA-5's role in virus autoregulation by targeting the virus nonstructural protein 1 gene. Notably, DENV-vsRNA-5 homologous mimics from DENV serotypes 1 and 4, but not 3, inhibited DENV-2 replication. The results revealed that DENV is able to encode functional vsRNAs, and one of those, which resembles miRNAs, specifically targets a viral gene, opening an avenue for possible utilization of the small RNA to limit DENV replication.
DOI:10.1111/imb.12070URL [本文引用: 1]

To define microRNA (miRNA) involvement during arbovirus infection of Aedes aegypti, we mined deep sequencing libraries of Dengue type 2 (DENV2)-exposed mosquitoes. Three biological replicates for each timepoint [2, 4 and 9 days post-exposure (dpe)] and treatment group allowed us to remove the outliers associated with sample-to-sample variability. Using edgeR (R Bioconductor), designed for use with replicate deep sequencing data, we determined the log fold-change (logFC) of miRNA levels [18-23 nucleotides (nt)]. The number of significantly modulated miRNAs increased from 5 at 2 and 4 dpe to 23 unique miRNAs by 9 dpe. Putative miRNA targets were predicted by aligning miRNAs to the transcriptome, and the list was reduced to include the intersection of hits found using the Miranda, PITA, and TargetScan algorithms. To further reduce false-positives, putative targets were validated by cross-checking them with mRNAs reported in recent DENV2 host response transcriptome reports; 4076 targets were identified. Of these, 464 gene targets have predicted miRNA-binding sites in 3 untranslated regions. Context-specific target functional groups include proteins involved in transport, transcriptional regulation, mitochondrial function, chromatin modification and signal transduction processes known to be required for viral replication and dissemination. The miRNA response is placed in context with other vector host response studies by comparing the predicted targets with those of transcriptome studies. Together, these data are consistent with the hypothesis that profound and persistent changes to gene expression occur in DENV2-exposed mosquitoes.
DOI:10.1128/JVI.00317-14URL [本文引用: 1]

West Nile virus (WNV) is an enveloped virus with a single-stranded positive-sense RNA genome from the Flaviviridae family. WNV is spread by mosquitoes and able to infect humans, causing encephalitis and meningitis that can be fatal; it therefore presents a significant risk for human health. In insects, innate response to RNA virus infection mostly relies on RNA interference and JAK/SAT pathways; however, some evidence indicates that it can also involve microRNAs (miRNAs). miRNAs are small noncoding RNAs that regulate gene expression at posttranscriptional level and play an important role in a number of processes, including immunity and antiviral response. In this study, we focus on the miRNA-mediated response to WNV in mosquito cells. We demonstrate that in response to WNV infection the expression of a mosquito-specific miRNA, aae-miR-2940, is selectively downregulated in Aedes albopictus cells. This miRNA is known to upregulate the metalloprotease m41 FtsH gene, which we have also shown to be required for efficient WNV replication. Correspondingly, downregulation of aae-miR-2940 reduced the metalloprotease level and restricted WNV replication. Thus, we have identified a novel miRNA-dependent mechanism of antiviral response to WNV in mosquitoes.
IMPORTANCE;A detailed understanding of vector-pathogen interactions is essential to address the problems posed by vector-borne diseases. Host and viral miRNAs play an important role in regulating expression of viral and host genes involved in endogenous processes, including antiviral response. There has been no evidence to date for the role of mosquito miRNAs in response to flaviviruses. In this study, we show that downregulation of aae-miR-2940 in mosquito cells acts as a potential antiviral mechanism in the mosquito host to inhibit WNV replication by repressing the expression of the metalloprotease m41 FtsH gene, which is required for efficient WNV replication. This is the first identification of an miRNA-dependent antiviral mechanism in mosquitoes, which inhibits replication of WNV. Our findings should facilitate identification of targets in the mosquito genome that can be utilized to suppress vector population and/or limit WNV replication.
DOI:10.1186/s13071-017-1966-2URLPMID:28159012 [本文引用: 1]

The midgut is the first barrier to dengue virus (DENV) infections of mosquitoes and therefore is a major bottleneck for the subsequent development of vector competence. However, the molecular mechanisms responsible for this barrier are unknown.
DOI:10.1186/s13071-018-3261-2URLPMID:30658692 [本文引用: 1]

The mosquito Aedes albopictus is an important vector for dengue virus (DENV) transmission. The midgut is the first barrier to mosquito infection by DENV, and this barrier is a critical factor affecting the vector competence of the mosquito. However, the molecular mechanism of the interaction between midgut and virus is unknown.
DOI:10.1128/mSphere.00144-17URLPMID:28497119 [本文引用: 1]

The small interfering RNA (siRNA) pathway is a major antiviral response in mosquitoes; however, another RNA interference pathway, the PIWI-interacting RNA (piRNA) pathway, has been suggested to be antiviral in mosquitoes. Piwi4 has been reported to be a key mediator of this response in mosquitoes, but it is not involved in the production of virus-specific piRNAs. Here, we show that Piwi4 associates with members of the antiviral exogenous siRNA pathway (Ago2 and Dcr2), as well as with proteins of the piRNA pathway (Ago3, Piwi5, and Piwi6) in an Aedes aegypti-derived cell line, Aag2. Analysis of small RNAs captured by Piwi4 revealed that it is predominantly associated with virus-specific siRNAs in Semliki Forest virus-infected cells and, to a lesser extent, with viral piRNAs. By using a Dcr2 knockout cell line, we showed directly that Ago2 lost its antiviral activity, as it was no longer bound to siRNAs, but Piwi4 retained its antiviral activity in the absence of the siRNA pathway. These results demonstrate a complex interaction between the siRNA and piRNA pathways in A.?aegypti and identify Piwi4 as a noncanonical PIWI protein that interacts with members of the siRNA and piRNA pathways, and its antiviral activities may be independent of either pathway. IMPORTANCE Mosquitoes transmit several pathogenic viruses, for example, the chikungunya and Zika viruses. In mosquito cells, virus replication intermediates in the form of double-stranded RNA are cleaved by Dcr2 into 21-nucleotide-long siRNAs, which in turn are used by Ago2 to target the virus genome. A different class of virus-derived small RNAs, PIWI-interacting RNAs (piRNAs), have also been found in infected insect cells. These piRNAs are longer and are produced in a Dcr2-independent manner. The only known antiviral protein in the PIWI family is Piwi4, which is not involved in piRNA production. It is associated with key proteins of the siRNA and piRNA pathways, although its antiviral function is independent of their actions.
DOI:10.1038/nature06263URLPMID:17952056 [本文引用: 1]

Heterochromatin, representing the silenced state of transcription, consists largely of transposon-enriched and highly repetitive sequences. Implicated in heterochromatin formation and transcriptional silencing in Drosophila are Piwi (P-element induced wimpy testis) and repeat-associated small interfering RNAs (rasiRNAs). Despite this, the role of Piwi in rasiRNA expression and heterochromatic silencing remains unknown. Here we report the identification and characterization of 12,903 Piwi-interacting RNAs (piRNAs) in Drosophila, showing that rasiRNAs represent a subset of piRNAs. We also show that Piwi promotes euchromatic histone modifications and piRNA transcription in subtelomeric heterochromatin (also known as telomere-associated sequence, or TAS), on the right arm of chromosome 3 (3R-TAS). Piwi binds to 3R-TAS and a piRNA uniquely mapped to 3R-TAS (3R-TAS1 piRNA). In piwi mutants, 3R-TAS loses euchromatic histone modifications yet accumulates heterochromatic histone modifications and Heterochromatin Protein 1a (HP1a). Furthermore, the expression of both the 3R-TAS1 piRNA and a white reporter gene in 3R-TAS becomes suppressed. A P element inserted 128 base pairs downstream of the 3R-TAS1 piRNA coding sequence restores the euchromatic histone modifications of 3R-TAS and the expression of 3R-TAS1 piRNA in piwi mutants, as well as partly rescuing their defects in germline stem-cell maintenance. These observations suggest that Piwi promotes the euchromatic character of 3R-TAS heterochromatin and its transcriptional activity, opposite to the known roles of Piwi and the RNA-mediated interference pathway in epigenetic silencing. This activating function is probably achieved through interaction with at least 3R-TAS1 piRNA and is essential for germline stem-cell maintenance.
DOI:10.1038/nsmb1218URLPMID:17384647 [本文引用: 1]

Piwi-interacting RNAs (piRNAs) are a novel class of small RNAs that are expressed specifically and abundantly in male germ cells. Here we report that the 3' termini of piRNAs are 2'-O-methylated; this modification may have important implications for the biogenesis and function of piRNAs.
DOI:10.16288/j.yczz.18-072URLPMID:29959117 [本文引用: 1]

Transposable elements (TEs) are DNA elements that can change their position within or between chromosomes. Most active TEs are retrotransposons, which transpose through a RNA intermediate. Since retrotransposons comprise high proportions in cell genomes, their frequent transposition may cause deleterious effects on the structure and function of the host cell genomes, thus causing serious genetic diseases such as cancer. Host cells therefore developed defense strategies to restrict these mobile elements. piRNA, which belongs to non-coding interfering RNAs, can efficiently decrease the level of retrotransposon RNA intermediates at transcriptional or post-transcriptional stages. In this review, we summarize the progress in mechanisms of piRNA-mediated retrotransposon inhibition. We hope that this review may shed light on the research of transposon and genome regulation.
DOI:10.16288/j.yczz.18-072URLPMID:29959117 [本文引用: 1]

Transposable elements (TEs) are DNA elements that can change their position within or between chromosomes. Most active TEs are retrotransposons, which transpose through a RNA intermediate. Since retrotransposons comprise high proportions in cell genomes, their frequent transposition may cause deleterious effects on the structure and function of the host cell genomes, thus causing serious genetic diseases such as cancer. Host cells therefore developed defense strategies to restrict these mobile elements. piRNA, which belongs to non-coding interfering RNAs, can efficiently decrease the level of retrotransposon RNA intermediates at transcriptional or post-transcriptional stages. In this review, we summarize the progress in mechanisms of piRNA-mediated retrotransposon inhibition. We hope that this review may shed light on the research of transposon and genome regulation.
DOI:10.1038/nrm3089URLPMID:21427766 [本文引用: 1]

PIWI-interacting RNAs (piRNAs) are a distinct class of small non-coding RNAs that form the piRNA-induced silencing complex (piRISC) in the germ line of many animal species. The piRISC protects the integrity of the genome from invasion by 'genomic parasites'--transposable elements--by silencing them. Owing to their limited expression in gonads and their sequence diversity, piRNAs have been the most mysterious class of small non-coding RNAs regulating RNA silencing. Now, much progress is being made into our understanding of their biogenesis and molecular functions, including the specific subcellular compartmentalization of the piRNA pathway in granular cytoplasmic bodies.
DOI:10.1016/j.cell.2007.01.043URLPMID:17346786 [本文引用: 1]

Drosophila Piwi-family proteins have been implicated in transposon control. Here, we examine piwi-interacting RNAs (piRNAs) associated with each Drosophila Piwi protein and find that Piwi and Aubergine bind RNAs that are predominantly antisense to transposons, whereas Ago3 complexes contain predominantly sense piRNAs. As in mammals, the majority of Drosophila piRNAs are derived from discrete genomic loci. These loci comprise mainly defective transposon sequences, and some have previously been identified as master regulators of transposon activity. Our data suggest that heterochromatic piRNA loci interact with potentially active, euchromatic transposons to form an adaptive system for transposon control. Complementary relationships between sense and antisense piRNA populations suggest an amplification loop wherein each piRNA-directed cleavage event generates the 5' end of a new piRNA. Thus, sense piRNAs, formed following cleavage of transposon mRNAs may enhance production of antisense piRNAs, complementary to active elements, by directing cleavage of transcripts from master control loci.
DOI:10.1038/nature20162URLPMID:27851737 [本文引用: 1]

Small regulatory RNAs guide Argonaute (Ago) proteins in a sequence-specific manner to their targets and therefore have important roles in eukaryotic gene silencing. Of the three small RNA classes, microRNAs and short interfering RNAs are processed from double-stranded precursors into defined 21- to 23-mers by Dicer, an endoribonuclease with intrinsic ruler function. PIWI-interacting RNAs (piRNAs)-the 22-30-nt-long guides for PIWI-clade Ago proteins that silence transposons in animal gonads-are generated independently of Dicer from single-stranded precursors. piRNA 5' ends are defined either by Zucchini, the Drosophila homologue of mitoPLD-a mitochondria-anchored endonuclease, or by piRNA-guided target cleavage. Formation of piRNA 3' ends is poorly understood. Here we report that two genetically and mechanistically distinct pathways generate piRNA 3' ends in Drosophila. The initiating nucleases are either Zucchini or the PIWI-clade proteins Aubergine (Aub) or Ago3. While Zucchini-mediated cleavages directly define mature piRNA 3' ends, Aub/Ago3-mediated cleavages liberate pre-piRNAs that require extensive resection by the 3'-to-5' exoribonuclease Nibbler (Drosophila homologue of Mut-7). The relative activity of these two pathways dictates the extent to which piRNAs are directed to cytoplasmic or nuclear PIWI-clade proteins and thereby sets the balance between post-transcriptional and transcriptional silencing. Notably, loss of both Zucchini and Nibbler reveals a minimal, Argonaute-driven small RNA biogenesis pathway in which piRNA 5' and 3' ends are directly produced by closely spaced Aub/Ago3-mediated cleavage events. Our data reveal a coherent model for piRNA biogenesis, and should aid the mechanistic dissection of the processes that govern piRNA 3'-end formation.
DOI:10.1371/journal.ppat.1002470URLPMID:22241995 [本文引用: 3]

The natural maintenance cycles of many mosquito-borne pathogens require establishment of persistent non-lethal infections in the invertebrate host. The mechanism by which this occurs is not well understood, but we have previously shown that an antiviral response directed by small interfering RNAs (siRNAs) is important in modulating the pathogenesis of alphavirus infections in the mosquito. However, we report here that infection of mosquitoes with an alphavirus also triggers the production of another class of virus-derived small RNAs that exhibit many similarities to ping-pong-dependent piwi-interacting RNAs (piRNAs). However, unlike ping-pong-dependent piRNAs that have been described previously from repetitive elements or piRNA clusters, our work suggests production in the soma. We also present evidence that suggests virus-derived piRNA-like small RNAs are capable of modulating the pathogenesis of alphavirus infections in dicer-2 null mutant mosquito cell lines defective in viral siRNA production. Overall, our results suggest that a non-canonical piRNA pathway is present in the soma of vector mosquitoes and may be acting redundantly to the siRNA pathway to target alphavirus replication.
DOI:10.1371/journal.ppat.1006017URLPMID:28033427 [本文引用: 1]

Vector mosquitoes are responsible for transmission of the majority of arthropod-borne (arbo-) viruses. Virus replication in these vectors needs to be sufficiently high to permit efficient virus transfer to vertebrate hosts. The mosquito immune response therefore is a key determinant for arbovirus transmission. Mosquito antiviral immunity is primarily mediated by the small interfering RNA pathway. Besides this well-established antiviral machinery, the PIWI-interacting RNA (piRNA) pathway processes viral RNA into piRNAs. In recent years, significant progress has been made in characterizing the biogenesis and function of these viral piRNAs. In this review, we discuss these developments, identify knowledge gaps, and suggest directions for future research.
DOI:10.1093/nar/gkv590URLPMID:26068474 [本文引用: 1]

The PIWI-interacting RNA (piRNA) pathway is essential for transposon silencing in many model organisms. Its remarkable efficiency relies on a sophisticated amplification mechanism known as the ping-pong loop. In Alphavirus-infected Aedes mosquitoes, piRNAs with sequence features that suggest ping-pong-dependent biogenesis are produced from viral RNA. The PIWI family in Aedes mosquitoes is expanded when compared to other model organisms, raising the possibility that individual PIWI proteins have functionally diversified in these insects. Here, we show that Piwi5 and Ago3, but none of the other PIWI family members, are essential for piRNA biogenesis from Sindbis virus RNA in infected Aedes aegypti cells. In contrast, the production of piRNAs from transposons relies on a more versatile set of PIWI proteins, some of which do not contribute to viral piRNA biogenesis. These results indicate that functional specialization allows distinct mosquito PIWI proteins to process RNA from different endogenous and exogenous sources.
DOI:10.1099/vir.0.053850-0URLPMID:23559478 [本文引用: 1]

The exogenous siRNA pathway is important in restricting arbovirus infection in mosquitoes. Less is known about the role of the PIWI-interacting RNA pathway, or piRNA pathway, in antiviral responses. Viral piRNA-like molecules have recently been described following infection of mosquitoes and derived cell lines with several arboviruses. The piRNA pathway has thus been suggested to function as an additional small RNA-mediated antiviral response to the known infection-induced siRNA response. Here we show that piRNA-like molecules are produced following infection with the naturally mosquito-borne Semliki Forest virus in mosquito cell lines. We show that knockdown of piRNA pathway proteins enhances the replication of this arbovirus and defines the contribution of piRNA pathway effectors, thus characterizing the antiviral properties of the piRNA pathway. In conclusion, arbovirus infection can trigger the piRNA pathway in mosquito cells, and knockdown of piRNA proteins enhances virus production.
