 ,广东海洋大学农学院,湛江 524088
,广东海洋大学农学院,湛江 524088Cloning and expression analysis of chicken circular transcript of insulin degrading enzyme gene
Qiying Leng, Jiahui Zheng, Haidong Xu, Patricia Adu-Asiamah, Ying Zhang, Bingwang Du, Li Zhang ,College of Agriculture, Guangdong Ocean University, Zhanjiang 524088, China
,College of Agriculture, Guangdong Ocean University, Zhanjiang 524088, China通讯作者: 张丽,博士,教授,研究方向:家禽遗传育种与繁殖。E-mail:zhangli761101@163.com
编委: 李辉
收稿日期:2019-05-30修回日期:2019-11-17网络出版日期:2019-12-20
| 基金资助: |
Editorial board:
Received:2019-05-30Revised:2019-11-17Online:2019-12-20
| Fund supported: |
作者简介 About authors
冷奇颖,硕士研究生,专业方向:动物遗传育种与繁殖。E-mail:lengqiying1995@163.com。

摘要
本课题组前期通过高通量测序发现,鸡胰岛素降解酶(insulin degrading enzyme, IDE)基因可能存在一个环状转录本。为确定IDE基因环状转录本(circIDE)的真实存在,探究其表达规律,本研究以吉林芦花鸡为研究对象,通过PCR扩增和测序验证了circIDE的真实存在,通过RNase R处理和反转录证实了circIDE的环形结构,通过qRT-PCR分析circIDE和IDE mRNA的时空表达规律,并对比分析了circIDE和线性IDE mRNA在正常体型芦花鸡和GHR突变的矮小体型芦花鸡中的表达差异。结果表明:鸡circIDE全长为1332 nt,由IDE基因外显子2~11环化形成。RNase R耐受性分析表明,鸡circIDE具备环形分子的一般特征,不易被RNase R降解。与oligo-d(T)18引物相比,随机引物对circIDE具有较高的反转录效率,进一步说明circIDE是一个不含poly(A)的环状结构分子。组织表达谱结果表明,circIDE在1周龄和12周龄正常体型芦花鸡肝脏和心脏高表达,在胸肌和腿肌中低表达;circIDE在肝脏组织的时序表达谱结果表明,circIDE在鸡6周龄之前为低表达,在8周龄以后表现为高表达;正常与矮小体型芦花鸡品系间circIDE表达量对比分析表明,正常体型芦花鸡circIDE表达水平高于矮小体型芦花鸡,其中在肝脏组织中差异显著(P<0.05)。本研究证实了鸡IDE基因存在一个环状转录本circIDE,并初步揭示了circIDE的表达规律,circIDE在正常体型和矮小体型芦花鸡肝脏组织中表达量存在差异,本研究结果为深入开展鸡circIDE的生物学功能及其在鸡生长发育过程中的作用机制奠定了基础。
关键词:
Abstract
Insulin-degrading enzyme (IDE) is a highly conserved metallopeptidase that functions in the catabolism of bioactive peptides. In our previous study, we identified a putative circular transcript in that chicken insulin-degrading enzyme (IDE) gene through analyzing a high throughput sequencing result. Here we set to confirm the circular transcript of IDE (circIDE) and explore its expression regularity in normal barred Plymouth chicken. The circIDE was confirmed by PCR amplification and sequencing. The circular structure of circIDE was determined by RNase R processing and reverse transcription experiments. Then we analyzed the spatiotemporal expression pattern of circIDE and IDE mRNA and compared the differential expression of circIDE and IDE mRNA in the normal barred Plymouth chicken and the dwarf ones. The results showed that the full length of chicken circIDE was 1332 nt, divided form exon 2-11 of the IDE gene. RNase R tolerance analysis showed that chicken circIDE had the general characteristics of circular molecule, and was highly resistant to RNase R. The random primers had higher transcription efficiency than the oligo-d(T)18 primers, confirming that circIDE is a circular structured molecule without poly(A). circIDE was highly expressed in the liver and heart tissues but less in the muscle tissues of leg and breast in normal chickens at the age of 1 and 12 weeks. The expression profile of circIDE in liver tissue showed that circIDE level was lower in1 to 6 weeks and then became higher after 8 weeks of age. The expression of circIDE in liver tissue was significantly higher in normal chicken than that in dwarf barred Plymouth chicken (P<0.05). This study confirmed a circIDE strucutre in chicken IDE gene and uncovered its expression regularity. We demonstrated that the expression level of circIDE in the liver tissue was higher in normal barred Plymouth chicken compared to dwarf species. This study paves the way for further understanding the biological function of chicken circIDE, including its roles in regulating chicken growth and development.
Keywords:
PDF (544KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
冷奇颖, 郑嘉辉, 徐海冬, PatriciaAdu-Asiamah, 张颖, 杜炳旺, 张丽. 鸡胰岛素降解酶基因环状转录本克隆及其表达规律. 遗传[J], 2019, 41(12): 1129-1137 doi:10.16288/j.yczz.19-157
Qiying Leng.
胰岛素降解酶(insulin degrading enzyme, IDE)是一种在进化上保守的中性巯基金属内肽酶,在多种细胞内的胰岛素降解过程中发挥重要作用[1,2,3]。IDE对胰岛素的分解作用是胰岛素在体内分解代谢的主要途径[4,5,6]。此外,研究显示IDE还具有降解β-淀粉样蛋白、胰岛淀粉样多肽和胰高血糖素等多肽的能力[7]。IDE通过对多肽的降解作用来维持血糖平衡。由于IDE具有降解β-淀粉样蛋白的作用,因此也被认为是一种对抗阿兹海默病的新分子[8,9]。
基因转录的前体mRNA(pre-mRNA)既可以通过剪接作用形成线性mRNA,也可以通过反向剪接作用引起pre-mRNA的3′端和5′端发生共价连接从而形成环状RNA (circular RNA, circRNA)[10,11]。因此,circRNA与线性RNA相比,circRNA具有不含5′帽子及3′尾巴结构特征[12],这一特征使circRNA具有较高的稳定性[13]。此外,circRNA还具有时空表达特异性[14]、序列保守性[15]、序列来源多为外显子[16]等特点。circRNA的生成受多种因素调控,最新研究已经证实,内含子反向互补序列、RNA结合蛋白、先天性免疫、细胞的全能性等一系列因素可以调控circRNA的生成[17]。同时,circRNA以多种方式调控机体的生理活动[18,19,20,21],其中对母源基因的调控作用是circRNA发挥作用的重要方式[17]。由于基因的环状转录本和线性转录本均来源于同一个母源基因,circRNA和mRNA可能存在竞争调控关系[20]。已有证据表明,circRNA也参与胰岛素代谢调控[22]。
鸡IDE基因定位于6号染色体,全长55 kb,包含25个外显子。本课题组前期对鸡肝脏细胞进行高通量测序(GenBank登录号:PRJNA554754),发现IDE基因可能存在环状转录本(circIDE)。因此,本研究通过基因克隆技术验证了circIDE的真实存在,分析了其环形结构特征及组织和时序表达规律,并对比分析了其在正常体型芦花鸡和GHR突变的矮小体型芦花鸡中的表达差异。本研究结果为今后深入分析circIDE在胰岛素代谢和动物机体发育过程中的作用奠定了基础。
1 材料与方法
1.1 材料
1日龄正常体型芦花鸡和GHR基因突变而矮化的矮小体型吉林芦花鸡均由吉林省农业科学院提供。HiPure Universal RNA Mini Kit(R4130)和DNase On Column Kit(R4911)购自广州美基生物科技有限公司;RNase R(R0301)购自广州吉赛生物股份有限公司;PrimeScript RT reagent Kit With gDNA Eraser (Perfect Real Time) (RR047A, TaKaRa)和pMD18-T载体(6011)购自广州瑞真生物技术有限公司;TransStart Top Green qRT-PCR SuperMix试剂盒(AQ101- 03)、TransStart KD Plus DNA Polymerase(AP301-02)和E.coli DH5α感受态大肠杆菌(CD201-01)购自北京全式金生物技术有限公司。1.2 动物饲养及组织采集
1日龄正常体型芦花鸡和矮小体型芦花鸡佩戴翅号后,混笼饲养于广东海洋大学农学院,采用自由采食和饮水的方式进行饲养(日粮标准见表1),并进行常规免疫。在1、2、3、4、5、6、8和12周龄的最后1 d随机抽取6只芦花鸡母鸡(正常/矮小鸡各3只)进行样品采集,分别采集胸肌、腿肌、肝脏、十二指肠和心脏等组织,液氮速冻后置于-80℃冰箱保存备用。1.3 鸡IDE基因环形和线性转录本引物设计
针对circIDE的环形接头位置,在接头位置两端设计3对引物,分别为(1)分散引物circiDE-D,用于circIDE环状分子的验证;(2)全长扩增引物circIDE- full,用于克隆circIDE分子全长序列;(3)定量引物circIDE-DL用于该分子的定量分析。此外,在IDE基因非circIDE序列来源处设计线性IDE mRNA (参考序列:XM_004942157.3)的定量引物(IDE-DL)。Table 1
表1
表1试验鸡只日粮营养标准
Table 1
| 年龄(周) | 能量(MJ/kg) | 粗蛋白(%) | 钙(%) | 有效磷(%) | 赖氨酸(%) | 蛋氨酸(%) |
|---|---|---|---|---|---|---|
| 0~6 | 11.66 | 19.90 | 1.00 | 0.37 | 0.90 | 0.44 |
| 7~12 | 11.98 | 18.00 | 0.90 | 0.33 | 0.90 | 0.34 |
| 12~ | 12.11 | 16.98 | 0.86 | 0.33 | 0.90 | 0.34 |
新窗口打开|下载CSV
内参基因为GAPDH。引物均通过primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/)设计,引物信息见表2,引物在IDE基因上的位置及扩增示意图见图1。
Table 2
表2
表2引物序列信息
Table 2
| 引物名称 | 引物序列(5°→3°) | 复性温度(℃) | 扩增产物长度(bp) | 位置 | 用途 |
|---|---|---|---|---|---|
| circIDE-D | F: AGATAAAGAACGACCACG | 50 | 265 | Exon10 | 环状验证 |
| R: CAATTCCAGTCCACGATA | Exon2 | ||||
| circIDE-full | F: TCTTCAGAAGGAGACAA | 46 | 1326 | Exon2 | 全长克隆 |
| R: CGAATGTTTTCTGGTC | Exon11 | ||||
| circIDE-DL | F: GAAGAAGTATTGGCTGCTG | 54 | 198 | Exon11 | circIDE定量 |
| R: TTCCAGTCCACGATATTCTC | Exon2 | ||||
| IDE-DL | F: TGGCGTATCTCAAGACCCTG | 57 | 175 | Exon22 | IDE mRNA定量 |
| R: GCTGGTGCCAAGTTCACATC | Exon24 | ||||
| GAPDH | F: AGGACCAGGTTGTCTCCTGT | 57 | 153 | Exon7, 8 | 内参基因定量 |
| R: CCATCAAGTCCACAACACGG | Exon9 |
新窗口打开|下载CSV
图1
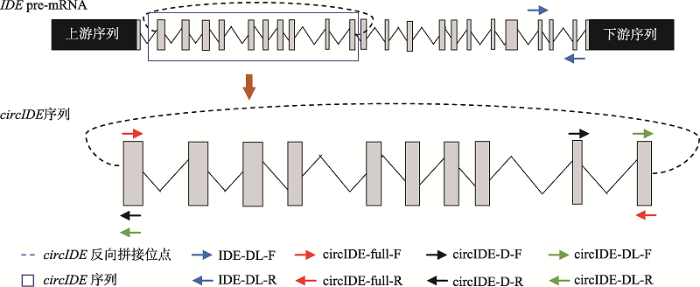 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1鸡circIDE来源及引物位置示意图
Fig. 1Diagram of chicken circIDE gene and the locations of the primers used in this study
1.4 芦花鸡肝脏等组织总RNA提取及环状性质分析
按照HiPure Universal RNA Mini Kit试剂盒说明书分别提取1、2、3、4、5、6、8、10和12周龄正常体型芦花鸡肝脏、胸肌、腿肌、心脏和十二指肠等组织总RNA,并使用1%的变性琼脂糖凝胶电泳检测RNA的完整性,使用BioTek Epoch全波长酶标仪检测RNA浓度和纯度,依据PrimeScript RT reagent Kit With gDNA Eraser试剂盒说明书,使用随机引物(N9)将总RNA反转录成cDNA。将提取的1周龄正常体型芦花鸡肝脏组织总RNA分装为两份(每份5 ng),一份使用超纯水稀释至20 μL保存备用;另一份进行RNase R处理,反应体系包括RNA 5 ng、10× Reaction Buffer 2 μL、RNase R 20 U,最后使用去离子水将反应体系补至20 μL。反应程序为:37℃孵育30 min后85℃灭活RNase R 5 s,经RNase R处理后的RNA用于circIDE环状性质特征验证。依据PrimeScript RT reagent Kit With gDNA Eraser试剂盒说明书,分别使用随机引物(N9)和oligo-d(T) 18 将未经RNase R处理的1周龄正常体型芦花鸡肝脏组织总RNA反转录成cDNA。随机引物反转录合成的cDNA用于qRT-PCR定量分析circIDE的表达水平;随机引物和oligo-d(T)18引物反转录得到的cDNA用于qRT-PCR 进行不同反转录引物的效率分析,进一步分析circIDE的环状结构特征。
1.5 芦花鸡circIDE环状特征验证及其克隆
经RNase R处理的1周龄正常体型芦花鸡肝脏组织RNA,通过随机引物反转录合成cDNA,分别使用circIDE环状验证引物(circIDE-D)和全长引物进行PCR扩增,使用1.5%的琼脂糖凝胶电泳检测并对目标产物进行回收纯化,然后将获得的目标产物连接载体pMD18-T后转入感受态大肠杆菌中,对菌株进行阳性鉴定后由生工生物工程(上海)股份有限公司进行测序,分析circIDE的序列信息和成环特征。1.6 芦花鸡circIDE的表达特征分析
以1.4中获得的cDNA为模板,采用qRT-PCR分析circIDE的时空表达规律及环状特征。反应体系为20 μL:2×TransStart? Top Green qPCR supermix 10 μL,上、下游引物各0.5 μL,去离子水 8 μL,cDNA 1 μL。反应程序:94℃预变性30 s;94℃变性5 s,59℃退火和延伸30 s,共40个循环。每个试验样本进行3次重复。1.7 数据统计与分析
使用2-ΔΔCT方法计算qRT-PCR结果,用IBM SPSS 数据统计软件19进行显著性分析,结果以平均数(mean)±标准误(SEM)表示。2 结果与分析
2.1 鸡circIDE连接成环验证及其全长序列分析
依据本课题组前期高通量测序结果,本研究针对circIDE连接成环的接头位置设计特异性分散引物,以经过RNase R处理后的1周龄正常体型芦花鸡肝脏组织总RNA反转录得到的cDNA为模板进行PCR扩增。结果表明,在265 bp位置生产了符合circIDE接头位置片段长度的预期目标的片段(图2A),初步表明IDE基因存在反向剪接产物。测序结果显示,circIDE连接成环的接头片段序列来源于IDE基因的外显子2和11 (图2B),说明circIDE的接头位置由IDE外显子2和外显子11共价连接形成。进一步使用circIDE全长扩增引物经PCR扩增和测序比对后发现,正常芦花鸡circIDE序列包含IDE基因外显子2~11 (图2C),全长为1332 nt,由IDE基因外显子2的5°端和外显子11的3′端反向剪接成环(GenBank登录号:MN183275)。图2
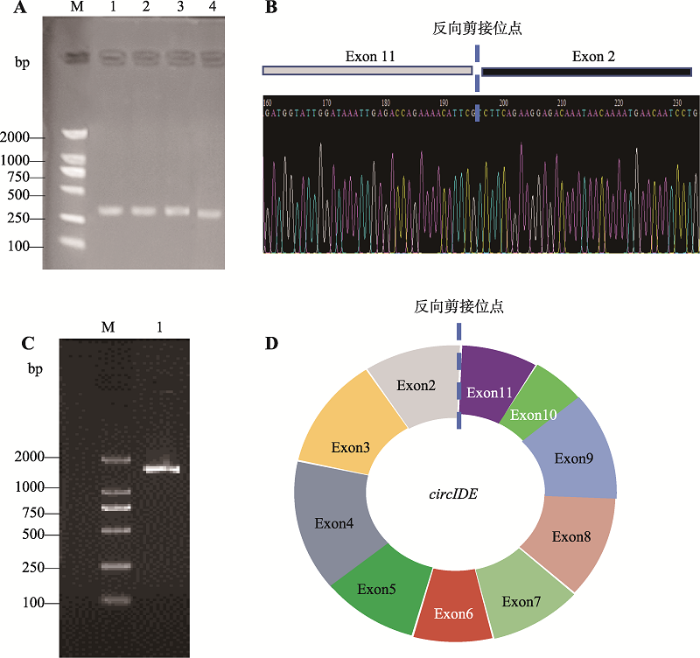 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2鸡circIDE全长克隆及结构验证
A:circIDE接头位置扩增结果。M:DL2000 maker;泳道1~4:以1周龄正常体型芦花鸡肝脏组织cDNA为模板,circIDE-D引物扩增结果。B:circIDE-D引物扩增产物测序结果。C:circIDE全长扩增结果。M:DL2000 maker;泳道1:以1周龄正常体型芦花鸡肝脏组织cDNA为模板,circIDE-full引物扩增产物。D:鸡circIDE结构示意图。
Fig. 2Cloning of the full length of chicken circIDE gene and validation of the circIDE structure
2.2 鸡circIDE分子RNase R耐受性及不同反转录引物的反转录效率分析
环形RNA分子具有RNase R耐受性,不易被RNase R降解;而线性RNA分子不具备RNase R耐受性。为了确认鸡circIDE具备环形分子的一般特征,本研究分析了鸡circIDE的RNase R耐受性。以1周龄正常体型芦花鸡组织总RNA为材料,经RNase R处理后,使用随机引物(N9)反转录为cDNA,qRT-PCR结果显示,使用RNase R处理后,circIDE的表达丰度无明显下降(P>0.05),反而呈现增加的趋势,但是线性分子IDE mRNA 经RNase R处理后下降极显著(P<0.01) (图3A),说明circIDE具有较强的RNase R耐受性。此外,以1周龄正常体型芦花鸡肝脏组织总RNA为模板,分别使用oligo-d(T)18和随机引物(N9)反转录合成cDNA,qRT-PCR比较circIDE的反转录效率。结果表明,用oligo-d (T)18引物反转录后circIDE的表达丰度仅为随机引物的2%,差异极显著(P<0.01)。而线性IDE mRNA的表达丰度不受反转录引物的影响(图3B,P>0.05)。因此,与oligo-d (T)18引物的反转录效果相比,随机引物对circIDE具有较好的反转录效果,同时也说明circIDE是一个不含poly(A)结构的环状分子。图3
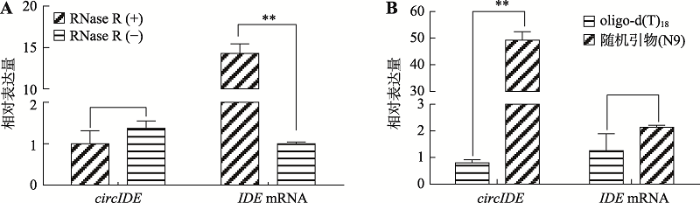 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3鸡circIDE和IDE mRNA对RNase R耐受性及反转录特征分析
A:RNase R对circIDE和IDE mRNA表达丰度的影响;B:不同反转录引物对circIDE 和 IDE mRNA 表达丰度的影响。RNase R(-):未经RNase R处理的总RNA;RNase R(+):经RNase R处理后的总RNA;**表示P<0.01,差异极显著。
Fig. 3RNase R tolerance and reverse transcription efficiency analysis of chicken circIDE and IDE mRNA
2.3 鸡circIDE和线性IDE mRNA的时空表达规律
分别提取1~12周龄正常体型芦花鸡肝脏、胸肌、腿肌、心脏和十二指肠等组织的RNA,经反转录合成cDNA,利用qRT-PCR对circIDE和IDE mRNA的表达量进行分析。结果表明,circIDE在1周龄正常体型芦花鸡的肝脏和心脏组织中高表达而在腿肌和胸肌组织中低表达(图4A),circIDE在12周龄正常体型芦花鸡的十二指肠、心脏和肝脏中高表达,而在胸肌和腿肌中低表达(图4B)。通过分析1~12周龄正常体型芦花鸡肝脏中circIDE的时序表达量发现,circIDE在鸡6周龄之前为低表达,在8周龄以后表现为高表达(图4C)。而鸡线性IDE mRNA 在1~12周龄期间随鸡的生长发育表达量呈逐步上升趋势(图4D)。图4
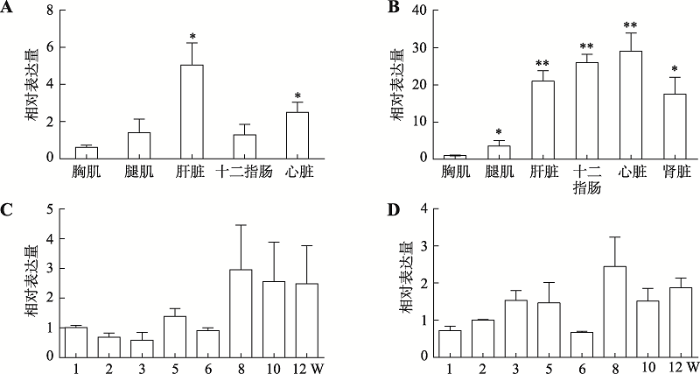 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4鸡circIDE和IDE mRNA的时空表达规律
A:1周龄正常体型芦花鸡不同组织中circIDE 的表达规律;B:12周龄正常体型芦花鸡不同组织中circIDE的表达规律;C:1~12周龄正常体型芦花鸡肝脏组织中circIDE的表达规律;D:1~12周龄正常体型芦花鸡肝脏中IDE mRNA的表达规律。*表示P<0.05,差异显著;**表示P<0.01,差异极显著。
Fig. 4Spatiotemporal expression of chicken circIDE and IDE mRNA
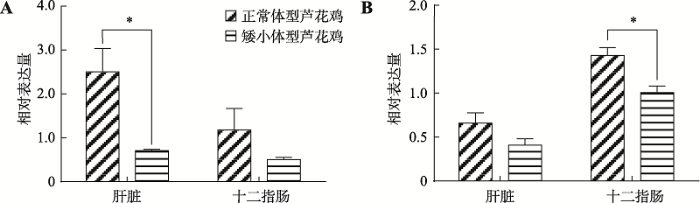
2.4 circIDE和线性IDE mRNA在正常体型和矮小体型芦花鸡中的表达分析
以1周龄正常体型芦花鸡中circIDE高表达的肝脏和十二指肠组织的cDNA为模板,qRT-qPCR对比分析1周龄正常体型芦花鸡和1周龄矮小体型芦花鸡circIDE和线性IDE mRNA的表达量。结果表明,circIDE和线性IDE mRNA在正常和矮小体型芦花鸡品系间存在表达差异。circIDE和线性IDE mRNA在正常体型芦花鸡中的表达水平均高于矮小体型芦花鸡,其中circIDE在正常体型芦花鸡肝脏组织中表达量显著高于矮小体型芦花鸡(P<0.05) (图5A),而线性IDE mRNA在正常体型芦花鸡十二指肠组织中的表达量显著高于矮小体型芦花鸡(P<0.05) (图5B)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5circIDE、IDE mRNA在正常和矮小体型芦花鸡中的表达对比
A:1周龄正常体型和矮小体型芦花鸡circIDE 的表达差异;B:1周龄正常体型和矮小体型芦花鸡IDE mRNA的表达差异。*表示P<0.05,差异显著。
Fig. 5Comparison the expression level of circIDE and IDE mRNA between the normal barred plymouth chickens and the dwarf ones
3 讨论
本研究克隆了鸡IDE基因的环状转录本circIDE,发现circIDE由该基因外显子2和外显子11反向剪接形成。查阅cirBase数据库(http://circrna.org/)发现人和小鼠的IDE基因也存在环状RNA[23,24]。人IDE基因产生的28个环状RNA中,有11个环状RNA是由外显子2与其他外显子反向剪接形成,其中hsa_circ_0094470与鸡circIDE一样,也是由外显子2和11环化形成。与人和鸡IDE基因序列相比,小鼠IDE基因缺少一个外显子。在cirBase数据库中注释的小鼠IDE基因有6个环状RNA,其中4个环状RNA是由外显子1与其他外显子反向剪接形成。因此,IDE基因成环的特征在鸡、人和小鼠之间具有物种保守性,外显子2可能是一个保守性的反向剪接位点,表明circIDE可能在人和动物发育过程中具有重要作用。环状RNA具有不含poly(A)序列的闭环结构,因此oligo-d(T)18引物反转录获得的circIDE丰度低于随机引物,提示在环形RNA反转录时宜选用随机引物进行反转录,同时该结果也间接表明circIDE是一个头尾相连的环状结构。RNase R是一种靶向线性RNA分子并对其具有切割作用的核酸酶,其对环形分子不敏感[25]。本研究表明,鸡circIDE对RNase R表现出较强的耐受性,经RNase R处理后总RNA,circIDE表达量变化不显著,进一步说明鸡circIDE具备环形分子的一般特征。
研究表明,基因的环形和线性转录本的表达存在相关性[24],本研究通过qRT-PCR分析表明,鸡circIDE的时序表达规律与其线性IDE mRNA相似。此外,鸡circIDE的表达具有时序规律性,circIDE在1周龄正常芦花鸡中的肝脏组织表现高表达,而在12周龄正常体型芦花鸡中的十二指肠组织中表现为高表达。IDE具有分解胰岛素样生长因子的作用[2],这可能与鸡消化系统的发育有关。鸡circIDE在多种组织均有表达,但存在组织差异性。circIDE在肝脏组织中高表达而在肌肉组织中低表达。
环状RNA可以通过多种模式对其母源基因或其靶基因发挥调控作用:(1)通过与U1 snRNP结合,正向调控母源的转录[26];(2)直接与线性mRNA竞争pre-mRNA,调控线性mRNA的表达[17];(3)与母源基因结合,形成DNA-RNA结合,抑制母源基因的转录[19];(4)环状RNA可以吸附miRNA,发挥分子海绵功能,参与基因调控[11]。circIDE来源于IDE基因的外显子2~11,属于外显子来源的环状RNA,由于circIDE属于EciRNA环状RNA,其序列来源不含有内含子序列,因此,circIDE无法与U1 snRNP结合促进母源基因表达的功能[26]。circIDE基因序列包含了IDE基因的10个外显子,含有大量的IDE基因结合位点,推测circIDE可能通过与IDE基因结合形成DNA-RNA复合物,调控IDE基因的转录发挥生物学功能。此外,已有两个circRNA分子被证实可以通过发挥海绵机制调控胰岛素分泌,Cdr1as可以发挥miR-7的分子海绵,调节胰岛素的代谢[22,27,28],circHIPK3也可以通过海绵机制吸附miR-124-3p和miR-338-3p调控胰岛素的代谢[29]。在TargetScan数据库(http://www.targetscan.org)对鸡circIDE进行miRNA靶向预测,发现circIDE存在小分子miRNA Let-7b的结合位点,推测 circIDE有可能充当Let-7b的分子海绵参与靶基因的调控。关于鸡circIDE的作用机制尚需进一步研究证实,以期为将来挖掘鸡分子育种新靶点,精确调控鸡生长发育奠定基础。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1371/journal.pone.0193101URLPMID:29447281 [本文引用: 1]

Insulin-degrading enzyme (IDE) is an atypical zinc-metalloendopeptidase that hydrolyzes insulin and other intermediate-sized peptide hormones, many of which are implicated in skin health and wound healing. Pharmacological inhibitors of IDE administered internally have been shown to slow the breakdown of insulin and thereby potentiate insulin action. Given the importance of insulin and other IDE substrates for a variety of dermatological processes, pharmacological inhibitors of IDE suitable for topical applications would be expected to hold significant therapeutic and cosmetic potential. Existing IDE inhibitors, however, are prohibitively expensive, difficult to synthesize and of undetermined toxicity. Here we used phage display to discover novel peptidic inhibitors of IDE, which were subsequently characterized in vitro and in cell culture assays. Among several peptide sequences tested, a cyclic dodecapeptide dubbed P12-3A was found to potently inhibit the degradation of insulin (Ki = 2.5 ± 0.31 μM) and other substrates by IDE, while also being resistant to degradation, stable in biological milieu, and highly selective for IDE. In cell culture, P12-3A was shown to potentiate several insulin-induced processes, including the transcription, translation and secretion of alpha-1 type I collagen in primary murine skin fibroblasts, and the migration of keratinocytes in a scratch wound migration assay. By virtue of its potency, stability, specificity for IDE, low cost of synthesis, and demonstrated ability to potentiate insulin-induced processes involved in wound healing and skin health, P12-3A holds significant therapeutic and cosmetic potential for topical applications.
URLPMID:8724818 [本文引用: 2]

The authors review recent research on an enzyme hypothesized to play a major role in the degradation of insulin. After binding to its receptor on the cell surface, insulin is internalized by receptor-mediated endocytosis and degraded within components of the endosomal apparatus. Degradation of insulin is important in the termination of signaling and clearance of the circulating hormone. It has been proposed that insulin-degrading enzyme (IDE), an evolutionarily conserved, neutral thiol-metalloendopeptidase, plays a crucial role in the degradation of internalized insulin in many types of cells. Despite the substantial evidence supporting the importance of IDE in cellular insulin degradation, there is controversy over its mode and site of action, mainly because of its cytosolic location. Its physiological location in cells has recently been elucidated through subcellular fractionation of liver parenchyma and through immunofluorescence microscopy of stably transfected Chinese hamster ovary cells that overexpress IDE. These experiments have excluded the presence of the enzyme in endosomes and have defined a peroxisomal location, consistent with the presence of a peroxisomal targeting sequence at the carboxyl terminus of the protein. Recently, researchers have demonstrated the functional significance of peroxisome-associated IDE (type I peroxisomal enzyme) in degrading cleaved leader peptides of peroxisomal proteins targeted by the type II motif. IDE is the first cloned and characterized proteinase to be localized to peroxisomes. Moreover, IDE appears to be a member of a newly identified superfamily of metalloendopeptidases that has an HXXEH active-site motif. Although fundamental questions concerning the biological role of IDE remain, its high degree of evolutionary conservation suggests that it must have important functions and multifaceted biological significance.
DOI:10.1016/j.neurobiolaging.2005.01.004URLPMID:16399206 [本文引用: 1]

Clinical and epidemiological studies have found that type 2 diabetes, and hyperinsulinaemia, increased the risk of developing Alzheimer's disease (AD) in the elderly. The link between hyperinsulinaemia and AD may be insulin-degrading enzyme (IDE). This enzyme degrades both insulin and amylin, peptides related to the pathology of type 2 diabetes, along with amyloid-beta peptide (Abeta), a short peptide found in excess in the AD brain. We review the current evidence, which suggests that hyperinsulinaemia may elevate Abeta through insulin's competition with Abeta for IDE. Genetic studies have also shown that IDE gene variations are associated with the clinical symptoms of AD as well as the risk of type 2 diabetes. The deficiency of IDE can be caused by genetic variation or by the diversion of IDE from the metabolism of Abeta to the metabolism of insulin. It is intriguing to notice that both hyperinsulinaemia and IDE gene variations are related to the risk of AD when the Apolipoprotein E4 (ApoE4) allele, the major risk factor of late-onset AD, is not present. Further studies of the role of IDE in the pathogenesis of AD, which may uncover potential treatment target, are much needed.
DOI:10.1080/10409238.2017.1337707URLPMID:28635330 [本文引用: 1]

Insulin-degrading enzyme (IDE) is a ubiquitous zinc peptidase of the inverzincin family, which has been initially discovered as the enzyme responsible for insulin catabolism; therefore, its involvement in the onset of diabetes has been largely investigated. However, further studies on IDE unraveled its ability to degrade several other polypeptides, such as β-amyloid, amylin, and glucagon, envisaging the possible implication of IDE dys-regulation in the &quot;aggregopathies&quot; and, in particular, in neurodegenerative diseases. Over the last decade, a novel scenario on IDE biology has emerged, pointing out a multi-functional role of this enzyme in several basic cellular processes. In particular, latest advances indicate that IDE behaves as a heat shock protein and modulates the ubiquitin-proteasome system, suggesting a major implication in proteins turnover and cell homeostasis. In addition, recent observations have highlighted that the regulation of glucose metabolism by IDE is not merely based on its largely proposed role in the degradation of insulin in vivo. There is increasing evidence that improper IDE function, regulation, or trafficking might contribute to the etiology of metabolic diseases. In addition, the enzymatic activity of IDE is affected by metals levels, thus suggesting a role also in the metal homeostasis (metallostasis), which is thought to be tightly linked to the malfunction of the &quot;quality control&quot; machinery of the cell. Focusing on the physiological role of IDE, we will address a comprehensive vision of the very complex scenario in which IDE takes part, outlining its crucial role in interconnecting several relevant cellular processes.
DOI:10.2337/diab.23.6.536URLPMID:4834293 [本文引用: 1]
DOI:10.1074/jbc.M900068200URLPMID:19321446 [本文引用: 1]

Insulin is a hormone vital for glucose homeostasis, and insulin-degrading enzyme (IDE) plays a key role in its clearance. IDE exhibits a remarkable specificity to degrade insulin without breaking the disulfide bonds that hold the insulin A and B chains together. Using Fourier transform ion cyclotron resonance (FTICR) mass spectrometry to obtain high mass accuracy, and electron capture dissociation (ECD) to selectively break the disulfide bonds in gas phase fragmentation, we determined the cleavage sites and composition of human insulin fragments generated by human IDE. Our time-dependent analysis of IDE-digested insulin fragments reveals that IDE is highly processive in its initial cleavage at the middle of both the insulin A and B chains. This ensures that IDE effectively splits insulin into inactive N- and C-terminal halves without breaking the disulfide bonds. To understand the molecular basis of the recognition and unfolding of insulin by IDE, we determined a 2.6-A resolution insulin-bound IDE structure. Our structure reveals that IDE forms an enclosed catalytic chamber that completely engulfs and intimately interacts with a partially unfolded insulin molecule. This structure also highlights how the unique size, shape, charge distribution, and exosite of the IDE catalytic chamber contribute to its high affinity ( approximately 100 nm) for insulin. In addition, this structure shows how IDE utilizes the interaction of its exosite with the N terminus of the insulin A chain as well as other properties of the catalytic chamber to guide the unfolding of insulin and allowing for the processive cleavages.
DOI:10.1080/07853890.2016.1197416URLPMID:27320287 [本文引用: 1]

Insulin-degrading enzyme (IDE) is a major enzyme responsible for insulin degradation. In addition to insulin, IDE degrades many targets including glucagon, atrial natriuretic peptide, and beta-amyloid peptide, regulates proteasomal degradation and other cell functions. IDE represents a pathophysiological link between type 2 diabetes (T2DM) and late onset Alzheimer's disease (AD). Potent and selective modulators of IDE activity are potential drugs for therapies of both diseases. Acute treatment with a novel IDE inhibitor was recently tested in a mouse study as a therapeutic approach for the treatment of T2DM. In contrast, effective IDE activators can be used for the AD treatment. However, because of the pleiotropic IDE action, the sustained treatment with systemic IDE modulators should be carefully tested in animal studies. Development of substrate-selective IDE modulators could overcome possible adverse effects of IDE modulators associated with multiplicity of IDE targets. KEY MESSAGES Insulin-degrading enzyme (IDE) represents a pathophysiological link between type 2 diabetes (T2DM) and Alzheimer's disease (AD). Selective modulators of IDE activity are potential drugs for both T2DM and AD treatment. Development of substrate-selective IDE modulators could overcome possible adverse effects of IDE modulators associated with multiplicity of IDE targets.
DOI:10.1016/j.tips.2017.10.008URLPMID:29132916 [本文引用: 1]

After decades of research and clinical trials there is still no cure for Alzheimer's disease (AD). While impaired clearance of amyloid beta (Aβ) peptides is considered as one of the major causes of AD, it was recently complemented by a potential role of other toxic amyloidogenic species. Insulin-degrading enzyme (IDE) is the proteolytic culprit of various β-forming peptides, both extracellular and intracellular. On the basis of demonstrated allosteric activation of IDE against Aβ, it is possible to propose a new strategy for the targeted IDE-based cleansing of different toxic aggregation-prone peptides. Consequently, specific allosteric activation of IDE coupled with state-of-the-art compound delivery and CRISP-Cas9 technique of transgene insertion can be instrumental in the fight against AD and related neurodegenerative maladies.
DOI:10.1038/srep12453URLPMID:26211738 [本文引用: 1]

Among the identified thousands of circular RNAs (circRNA) in humans and animals, Cdr1as (also known as CiRS-7) was recently demonstrated to act as a powerful miR-7 sponge/inhibitor in developing midbrain of zebrafish, suggesting a novel mechanism for regulating microRNA functions. MiR-7 is abundantly expressed in islet cells, but overexpressing miR-7 in transgenic mouse β cells causes diabetes. Therefore, we infer that Cdr1as expression may inhibit miR-7 function in islet cells, which in turn improves insulin secretion. Here, we show the first characterization of Cdr1as expression in islet cells, which was upregulated by long-term forskolin and PMA stimulation, but not high glucose, indicating the involvement of cAMP and PKC pathways. Remarkably, both insulin content and secretion were significantly increased by overexpression of Cdr1as in islet cells. We further identified a new target Myrip in the Cdr1as/miR-7 pathway that regulates insulin granule secretion, and also another target Pax6 that enhances insulin transcription. Taken together, our findings revealed the effects of the strongly interacting pair of Cdr1as/miR-7 on insulin secretion, which may become a new target for improving β cell function in diabetes.
DOI:10.1261/rna.047126.114URL [本文引用: 1]

It is now clear that there is a diversity of circular RNAs in biological systems. Circular RNAs can be produced by the direct ligation of 5' and 3' ends of linear RNAs, as intermediates in RNA processing reactions, or by "backsplicing," wherein a downstream 5' splice site (splice donor) is joined to an upstream 3' splice site (splice acceptor). Circular RNAs have unique properties including the potential for rolling circle amplification of RNA, the ability to rearrange the order of genomic information, protection from exonucleases, and constraints on RNA folding. Circular RNAs can function as templates for viroid and viral replication, as intermediates in RNA processing reactions, as regulators of transcription in cis, as snoRNAs, and as miRNA sponges. Herein, we review the breadth of circular RNAs, their biogenesis and metabolism, and their known and anticipated functions.
[本文引用: 2]
[本文引用: 2]
DOI:10.1021/jf60211a012URLPMID:67134 [本文引用: 1]
DOI:10.1021/jf60211a012URLPMID:67134 [本文引用: 1]
DOI:10.1261/rna.035667.112URL [本文引用: 1]

Circular RNAs composed of exonic sequence have been described in a small number of genes. Thought to result from splicing errors, circular RNA species possess no known function. To delineate the universe of endogenous circular RNAs, we performed high-throughput sequencing (RNA-seq) of libraries prepared from ribosome-depleted RNA with or without digestion with the RNA exonuclease, RNase R. We identified >25,000 distinct RNA species in human fibroblasts that contained non-colinear exons (a "backsplice") and were reproducibly enriched by exonuclease degradation of linear RNA. These RNAs were validated as circular RNA (ecircRNA), rather than linear RNA, and were more stable than associated linear mRNAs in vivo. In some cases, the abundance of circular molecules exceeded that of associated linear mRNA by >10-fold. By conservative estimate, we identified ecircRNAs from 14.4% of actively transcribed genes in human fibroblasts. Application of this method to murine testis RNA identified 69 ecircRNAs in precisely orthologous locations to human circular RNAs. Of note, paralogous kinases HIPK2 and HIPK3 produce abundant ecircRNA from their second exon in both humans and mice. Though HIPK3 circular RNAs contain an AUG translation start, it and other ecircRNAs were not bound to ribosomes. Circular RNAs could be degraded by siRNAs and, therefore, may act as competing endogenous RNAs. Bioinformatic analysis revealed shared features of circularized exons, including long bordering introns that contained complementary ALU repeats. These data show that ecircRNAs are abundant, stable, conserved and nonrandom products of RNA splicing that could be involved in control of gene expression.
DOI:10.3168/jds.2015-10381URLPMID:27040791 [本文引用: 1]

Recent studies have revealed that, in addition to hormones and other protein factors, noncoding RNA molecules play an important regulatory role in milk protein synthesis. Circular RNAs (circRNAs) are universally expressed noncoding RNA species that have been proposed recently to regulate the expression of their parental genes. In the present study, the deep RNA-sequencing technique known as RNA-seq was used to compare expression profiles of circRNAs from 2 pooled RNA samples from cow mammary gland on d 90 and 250 postpartum and to identify the key circRNAs involved in lactation. A total of 4,804 and 4,048 circRNAs were identified in the cow mammary gland on d 90 and 250 postpartum, respectively, of which only 2,231 circRNAs were co-expressed at both lactation stages, suggesting high stage specificity in the circRNAs. The enrichment of some Gene Ontology terms for the circRNA parental genes differed between lactation stages. Among the top 10 enriched Gene Ontology terms, vesicle, endoplasmic reticulum, and mitochondrial lumen were more common on lactation d 90. All 4 casein-coding genes (CSN1S1, CSN1S2, CSN2, and CSN3) produced circRNAs in the cattle mammary gland. In total, 80 circRNAs were identified from these 4 genes; circRNAs from CSN1S1 had very high abundance, and 3 of them accounted for 36% of all the circRNAs expressed in the mammary gland on lactation d 90. Three circRNAs from CSN1S1, 1 circRNA from CSN1S2, and 1 circRNA from CSN2 were all more highly expressed on lactation d 90 than on lactation d 250, as confirmed by quantitative PCR. These circRNAs had several target sites for the microRNA miR-2284 family and were predicted to target CSN1S1 and CSN2 mRNA, suggesting their potential involvement in regulating expression of the casein genes.
DOI:10.1371/journal.pone.0030733URLPMID:22319583 [本文引用: 1]

Most human pre-mRNAs are spliced into linear molecules that retain the exon order defined by the genomic sequence. By deep sequencing of RNA from a variety of normal and malignant human cells, we found RNA transcripts from many human genes in which the exons were arranged in a non-canonical order. Statistical estimates and biochemical assays provided strong evidence that a substantial fraction of the spliced transcripts from hundreds of genes are circular RNAs. Our results suggest that a non-canonical mode of RNA splicing, resulting in a circular RNA isoform, is a general feature of the gene expression program in human cells.
DOI:10.1101/gr.202895.115URLPMID:27365365 [本文引用: 1]

Circular RNAs (circRNAs) derived from back-spliced exons have been widely identified as being co-expressed with their linear counterparts. A single gene locus can produce multiple circRNAs through alternative back-splice site selection and/or alternative splice site selection; however, a detailed map of alternative back-splicing/splicing in circRNAs is lacking. Here, with the upgraded CIRCexplorer2 pipeline, we systematically annotated different types of alternative back-splicing and alternative splicing events in circRNAs from various cell lines. Compared with their linear cognate RNAs, circRNAs exhibited distinct patterns of alternative back-splicing and alternative splicing. Alternative back-splice site selection was correlated with the competition of putative RNA pairs across introns that bracket alternative back-splice sites. In addition, all four basic types of alternative splicing that have been identified in the (linear) mRNA process were found within circRNAs, and many exons were predominantly spliced in circRNAs. Unexpectedly, thousands of previously unannotated exons were detected in circRNAs from the examined cell lines. Although these novel exons had similar splice site strength, they were much less conserved than known exons in sequences. Finally, both alternative back-splicing and circRNA-predominant alternative splicing were highly diverse among the examined cell lines. All of the identified alternative back-splicing and alternative splicing in circRNAs are available in the CIRCpedia database (http://www.picb.ac.cn/rnomics/circpedia). Collectively, the annotation of alternative back-splicing and alternative splicing in circRNAs provides a valuable resource for depicting the complexity of circRNA biogenesis and for studying the potential functions of circRNAs in different cells.
DOI:10.1016/j.molcel.2018.06.034URLPMID:30057200 [本文引用: 3]

Covalently closed circular RNAs (circRNAs) are produced by precursor mRNA back-splicing of exons of thousands of genes in eukaryotes. circRNAs are generally expressed at low levels and often exhibit cell-type-specific and tissue-specific patterns. Recent studies have shown that their biogenesis requires spliceosomal machinery and can be modulated by both cis complementary sequences and protein factors. The functions of most circRNAs remain largely unexplored, but known functions include sequestration of microRNAs or proteins, modulation of transcription and interference with splicing, and even translation to produce polypeptides. However, challenges exist at multiple levels to understanding of the regulation of circRNAs because of their circular conformation and sequence overlap with linear mRNA counterparts. In?this review, we survey the recent progress on circRNA biogenesis and function and discuss technical obstacles in circRNA studies.
DOI:10.3892/ijo.2018.4265URLPMID:29431182 [本文引用: 1]

Circular RNAs (circRNAs) are key regulators in the development and progression of human cancers; however their roles in breast tumorigenesis are not yet well understood. Thus, the present study aimed to investigate the expression profiles and potential modulatory effects of circRNAs on breast carcinogenesis. A human circRNA microarray analysis was performed to screen for abnormally expressed circRNAs in breast cancer tissue and circRNA-000911 was identified as a circRNA which was significantly downregulated in breast cancer cells. Mechanistic investigations suggested that the enhanced expression of circRNA-000911 suppressed cell proliferation, migration and invasion, and promoted the apoptosis of breast cancer cells. By using a biotin-labeled circRNA-000911 probe to perform RNA precipitation in breast cancer cells, we identified miR?449a as the circRNA?000911-associated microRNA. Gain- and loss-of-function assays indicated that miR?449a antagonized circRNA-000911 to regulate breast cancer progression. Subsequently, Notch1 was identified as the functional target of miR?449a, and the overexpression of circRNA-000911 in breast cancer elevated Notch1 expression. Furthermore, Cignal Signal Transduction Reporter Array and western blot analysis identified nuclear factor-κB?(NF-κB) signaling as a functional target of the circRNA-000911/miR?449a pathway. On the whole, our findings indicate that circRNA-000911 plays an anti-oncogenic role in breast cancer and may thus serve as a promising therapeutic target for patients with breast cancer. Therefore, the overexpression of circRNA-000911 may provide a future direction which may aid in the development of a novel treatment strategy for breast cancer.
DOI:10.1038/nplants.2017.53URLPMID:28418376 [本文引用: 2]

Circular RNAs (circRNAs) are a diverse and abundant class of hyper-stable, non-canonical RNAs that arise through a form of alternative splicing (AS) called back-splicing. These single-stranded, covalently-closed circRNA molecules have been identified in all eukaryotic kingdoms of life1, yet their functions have remained elusive. Here, we report that circRNAs can be used as bona fide biomarkers of functional, exon-skipped AS variants in Arabidopsis, including in the homeotic MADS-box transcription factor family. Furthermore, we demonstrate that circRNAs derived from exon 6 of the SEPALLATA3 (SEP3) gene increase abundance of the cognate exon-skipped AS variant (SEP3.3 which lacks exon 6), in turn driving floral homeotic phenotypes. Toward demonstrating the underlying mechanism, we show that the SEP3 exon 6 circRNA can bind strongly to its cognate DNA locus, forming an RNA:DNA hybrid, or R-loop, whereas the linear RNA equivalent bound significantly more weakly to DNA. R-loop formation results in transcriptional pausing, which has been shown to coincide with splicing factor recruitment and AS2-4. This report presents a novel mechanistic insight for how at least a subset of circRNAs probably contribute to increased splicing efficiency of their cognate exon-skipped messenger RNA and provides the first evidence of an organismal-level phenotype mediated by circRNA manipulation.
DOI:10.1016/j.molcel.2014.08.019URLPMID:25242144 [本文引用: 2]

Circular RNAs (circRNAs) are widely expressed noncoding RNAs. However, their biogenesis and possible functions are poorly understood. Here, by studying circRNAs that we identified in neuronal tissues, we provide evidence that animal circRNAs are generated cotranscriptionally and that their production rate is mainly determined by intronic sequences. We demonstrate that circularization and splicing compete against each other. These mechanisms are tissue specific and conserved in animals. Interestingly, we observed that the second exon of the splicing factor muscleblind (MBL/MBNL1) is circularized in flies and humans. This circRNA (circMbl) and its flanking introns contain conserved muscleblind binding sites, which are strongly and specifically bound by MBL. Modulation of MBL levels strongly affects circMbl biosynthesis, and this effect is dependent on the MBL binding sites. Together, our data suggest that circRNAs can function in gene regulation by competing with linear splicing. Furthermore, we identified muscleblind as a factor involved in circRNA biogenesis.
DOI:10.1038/s41467-018-06862-2URLPMID:30367041 [本文引用: 1]

Circular RNAs (circRNAs) are a large class of transcripts in the mammalian genome. Although the translation of circRNAs was reported, additional coding circRNAs and the functions of their translated products remain elusive. Here, we demonstrate that an endogenous circRNA generated from a long noncoding RNA encodes regulatory peptides. Through ribosome nascent-chain complex-bound RNA sequencing (RNC-seq), we discover several peptides potentially encoded by circRNAs. We identify an 87-amino-acid peptide encoded by the circular form of the long intergenic non-protein-coding RNA p53-induced transcript (LINC-PINT) that suppresses glioblastoma cell proliferation in vitro and in vivo. This peptide directly interacts with polymerase associated factor complex (PAF1c) and inhibits the transcriptional elongation of multiple oncogenes. The expression of this peptide and its corresponding circRNA are decreased in glioblastoma compared with the levels in normal tissues. Our results establish the existence of peptides encoded by circRNAs and demonstrate their potential functions in glioblastoma tumorigenesis.
DOI:10.1038/srep12453URLPMID:26211738 [本文引用: 2]

Among the identified thousands of circular RNAs (circRNA) in humans and animals, Cdr1as (also known as CiRS-7) was recently demonstrated to act as a powerful miR-7 sponge/inhibitor in developing midbrain of zebrafish, suggesting a novel mechanism for regulating microRNA functions. MiR-7 is abundantly expressed in islet cells, but overexpressing miR-7 in transgenic mouse β cells causes diabetes. Therefore, we infer that Cdr1as expression may inhibit miR-7 function in islet cells, which in turn improves insulin secretion. Here, we show the first characterization of Cdr1as expression in islet cells, which was upregulated by long-term forskolin and PMA stimulation, but not high glucose, indicating the involvement of cAMP and PKC pathways. Remarkably, both insulin content and secretion were significantly increased by overexpression of Cdr1as in islet cells. We further identified a new target Myrip in the Cdr1as/miR-7 pathway that regulates insulin granule secretion, and also another target Pax6 that enhances insulin transcription. Taken together, our findings revealed the effects of the strongly interacting pair of Cdr1as/miR-7 on insulin secretion, which may become a new target for improving β cell function in diabetes.
DOI:10.1371/journal.pgen.1003777URLPMID:24039610 [本文引用: 1]

Thousands of loci in the human and mouse genomes give rise to circular RNA transcripts; at many of these loci, the predominant RNA isoform is a circle. Using an improved computational approach for circular RNA identification, we found widespread circular RNA expression in Drosophila melanogaster and estimate that in humans, circular RNA may account for 1% as many molecules as poly(A) RNA. Analysis of data from the ENCODE consortium revealed that the repertoire of genes expressing circular RNA, the ratio of circular to linear transcripts for each gene, and even the pattern of splice isoforms of circular RNAs from each gene were cell-type specific. These results suggest that biogenesis of circular RNA is an integral, conserved, and regulated feature of the gene expression program.
DOI:10.1016/j.molcel.2015.03.027URLPMID:25921068 [本文引用: 2]

Circular RNAs (circRNAs) are an endogenous class of?animal RNAs. Despite their abundance, their function and expression in the nervous system are unknown. Therefore, we sequenced RNA from different brain regions, primary neurons, isolated synapses, as well as during neuronal differentiation. Using these and other available data, we discovered and analyzed thousands of neuronal human and mouse circRNAs. circRNAs were extraordinarily enriched in the mammalian brain, well conserved in sequence, often expressed as circRNAs in both human and mouse, and sometimes even detected in Drosophila brains. circRNAs were overall upregulated during neuronal differentiation, highly enriched in synapses, and often differentially expressed compared to their?mRNA isoforms. circRNA expression correlated?negatively with expression of the RNA-editing enzyme ADAR1. Knockdown of ADAR1 induced elevated circRNA expression. Together, we provide a circRNA brain expression atlas and evidence for?important circRNA functions and values as biomarkers.
DOI:10.1074/jbc.M606744200URLPMID:16893880 [本文引用: 1]

RNase R is a processive, 3' to 5' hydrolytic exoribonuclease that together with polynucleotide phosphorylase plays an important role in the degradation of structured RNAs. However, RNase R differs from other exoribonucleases in that it can by itself degrade RNAs with extensive secondary structure provided that a single-stranded 3' overhang is present. Using a variety of specifically designed substrates, we show here that a 3' overhang of at least 7 nucleotides is required for tight binding and activity, whereas optimum binding and activity are achieved when the overhang is 10 or more nucleotides in length. In contrast, duplex RNAs with no overhang or with a 4-nucleotide overhang bind extremely poorly to RNase R and are inactive as substrates. A duplex RNA with a 10-nucleotide 5' overhang also is not a substrate. Interestingly, this molecule is bound only weakly, indicating that RNase R does not simply recognize single-stranded RNA, but the RNA must thread into the enzyme with 3' to 5' polarity. We also show that ribose moieties are required for recognition of the substrate as a whole since RNase R is unable to bind or degrade single-stranded DNA. However, RNA molecules with deoxyribose or dideoxyribose residues at their 3' termini can be bound and degraded. Based on these data and a homology model of RNase R, derived from the structure of the closely related enzyme, RNase II, we present a model for how RNase R interacts with its substrates and degrades RNA.
DOI:10.1038/nsmb0217-194aURLPMID:28170000 [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.13865/j.cnki.cjbmb.2016.09.03URL [本文引用: 1]

In eukaryotic cells, circular RNA (circRNA) is a non-linear RNA with characteristics such as high stability, specificity, and evolutionary conservation. It is found that circRNA regulates the expression of its target genes and mainly functions as microRNA sponges. Compared with the linear structure of coding RNA, the closed structure of circular RNA makes it more conserved. The cerebellar degeneration-related protein 1 antisense (CDR1as), a competing endogenous RNA, indirectly regulates miR-7 and is involved in the development of multiple diseases through a variety of pathways. In this paper, the discovery, origin, features, and functions of CDR1as, as well as its related diseases were summarized.
DOI:10.13865/j.cnki.cjbmb.2016.09.03URL [本文引用: 1]

In eukaryotic cells, circular RNA (circRNA) is a non-linear RNA with characteristics such as high stability, specificity, and evolutionary conservation. It is found that circRNA regulates the expression of its target genes and mainly functions as microRNA sponges. Compared with the linear structure of coding RNA, the closed structure of circular RNA makes it more conserved. The cerebellar degeneration-related protein 1 antisense (CDR1as), a competing endogenous RNA, indirectly regulates miR-7 and is involved in the development of multiple diseases through a variety of pathways. In this paper, the discovery, origin, features, and functions of CDR1as, as well as its related diseases were summarized.
DOI:10.1016/j.molmet.2018.01.010URLPMID:29396373 [本文引用: 1]

There is strong evidence for an involvement of different classes of non-coding RNAs, including microRNAs and long non-coding RNAs, in the regulation of β-cell activities and in diabetes development. Circular RNAs were recently discovered to constitute a substantial fraction of the mammalian transcriptome but the contribution of these non-coding RNAs in physiological and disease processes remains largely unknown. The goal of this study was to identify the circular RNAs expressed in pancreatic islets and to elucidate their possible role in the control of β-cells functions.
