 ,中国大熊猫保护研究中心,大熊猫国家公园珍稀动物保护生物学国家林业和草原局重点实验室,成都 611830
,中国大熊猫保护研究中心,大熊猫国家公园珍稀动物保护生物学国家林业和草原局重点实验室,成都 611830Progress on miRNA in giant panda (Ailuropoda melanoleuca)
Yan Zhu, Ming Wei, Xiao Zhou, Linhua Deng, Jian Qiu, Guo Li, Shiqiang Zhou, Hao Xie, Desheng Li, Chengdong Wang ,State Forestry and Grassland Administration Key Laboratory of Conservation Biology for Rare Animals of the Giant Panda State Park, China Conservation and Research Center for the Giant Panda, Chengdu 611830, China
,State Forestry and Grassland Administration Key Laboratory of Conservation Biology for Rare Animals of the Giant Panda State Park, China Conservation and Research Center for the Giant Panda, Chengdu 611830, China通讯作者: 王承东,博士,正高级工程师,研究方向:大熊猫疾病防控及繁育研究。E-mail:wolongpanda@qq.com
编委: 于黎
收稿日期:2021-06-11修回日期:2021-07-28
| 基金资助: |
Received:2021-06-11Revised:2021-07-28
| Fund supported: |
作者简介 About authors
朱艳,硕士,研究方向:大熊猫遗传与生态。E-mail:

摘要
MicroRNA (miRNA)是一类广泛存在于真核生物、长约22 nt的内源性非编码RNA。miRNA通过与靶基因mRNA特异性结合影响基因的表达,进而参与调控多种生物学过程。大熊猫(Ailuropoda melanoleuca)是我国特有的珍稀动物,备受全世界的关注。近年来,随着二代测序技术(next-generation sequencing, NGS)的普及,大熊猫miRNA陆续被发现和鉴定。本文综述了miRNA在大熊猫免疫反应、乳腺发育、精子冷冻耐受及其他生物学过程的研究进展,并探讨了大熊猫miRNA的研究前景,以期为深入研究大熊猫miRNA的调控机制和促进大熊猫繁育与保护工作提供科学参考和新思路。
关键词:
Abstract
MicroRNAs (miRNAs), a family of endogenous non-coding RNAs with a length of about 22 nucleotides, are widely found in eukaryotes. miRNAs can affect gene expression through specific bindings with mRNAs of target genes and participate in the regulation of a variety of biological processes. Giant panda is not only a unique rare animal in China, but also the focus of attention on wildlife preservation worldwide. In recent years, with the popularization of next-generation sequencing (NGS) technology, miRNAs in giant panda have been discovered and identified one after another. In this review, we focus on the research progress on miRNAs in giant panda, involved in immune response, mammary gland development, sperm freezing tolerance and other biological processes, and then discuss future research directions of miRNAs in giant panda, and thus providing the scientific references and new ideas for studying the regulatory mechanisms of miRNAs and promoting the breeding and protection of giant panda.
Keywords:
PDF (576KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
朱艳, 魏明, 周晓, 邓林华, 仇剑, 李果, 周世强, 谢浩, 李德生, 王承东. 大熊猫(Ailuropoda melanoleuca) miRNA研究进展. 遗传[J], 2021, 43(9): 849-857 doi:10.16288/j.yczz.21-209
Yan Zhu.
大熊猫是全球生物多样性保护的旗舰物种,我国特有的珍稀野生动物,有“国宝”之称。一直以来,关于大熊猫进化和保护方面的研究从未间断[1,2,3,4,5,6,7,8]。近年来,随着二代测序技术和生物信息学的发展,挖掘大熊猫基因组与转录组信息成为研究热点,为进一步揭示大熊猫潜在分子机制提供了有效手段。microRNA (miRNA)是一类内源性非编码小RNA,通过靶向抑制基因表达调控生物学过程。20世纪90年代,Lee等[9]首次发现lin-4参与调控秀丽隐杆线虫(Caenorhabditis elegans)的时序性发育,从此揭开了miRNA的研究序幕。目前,已有38,589条miRNA序列被发现并收录于miRBase.22数据库中(
1 miRNA概述
1.1 miRNA的发现
随着全基因组测序技术(whole genome sequencing, WGS) 的发展,人们发现全基因组中仅有1%的序列能够编码蛋白,而约99%的非编码序列是“无用的”[12]。但随着技术的进步和完善,研究表明这些所谓的“无用”序列实际发挥着重要作用,它们转录生成的非编码小RNA (如miRNA、lncRNA和circRNA)参与调控基因的表达进而影响生物学功能,其中miRNA是最早被发现也是目前的研究热点。1993年Lee等[9]在秀丽隐杆线虫中首次发现lin-4通过与lin-14 mRNA的3ʹ UTR特异性结合,进而抑制lin-14基因的表达,最终负调控LIN-14蛋白合成。Reinhart等[13]发现了另一个miRNA—let-7,调控秀丽隐杆线虫生长发育相关基因的表达且具有较高的物种保守性,从此揭开了miRNA的研究序幕。1.2 miRNA的调控机制
miRNA调控靶基因表达的作用机制十分复杂,miRNA的种子序列(5ʹ端第2~8个碱基) 与靶基因mRNA 3ʹ UTR互补配对,促使mRNA降解或抑制其翻译[14,15,16]。植物细胞中,miRNA与靶基因mRNA 3ʹ UTR完全互补配对从而直接降解靶基因[17],而在动物细胞中,识别位点大多不能完全互补配对,形成的凸泡结构(bulges)使miRNA只能在转录后调控翻译过程[18]。通常,一个miRNA可以调控多个靶基因表达,而一个靶基因也可以同时受到多个miRNA调控,这就形成了一个复杂的调控网络体系。研究预测miRNA能调控超过1/3的基因表达[19,20],但目前只有小部分miRNA进行了功能验证[21,22,23,24,25]。2 大熊猫miRNA相关研究
随着miRNA研究的不断深入,一些常见物种包括鱼类[26,27,28,29]、鸟类[30,31]和农业动物[32,33,34,35]均进行了转录组测序分析,并对miRNA的表达模式及调控功能进行了验证。近年来利用RNA-seq技术对大熊猫miRNA的研究也发展迅速[36~42] (图1),miRNA在大熊猫免疫反应[36,38,42]、乳腺发育[37,38]、精子冷冻耐受[40]及其他生物过程[39,41]等方面发挥着重要的调控作用(表1)。图1
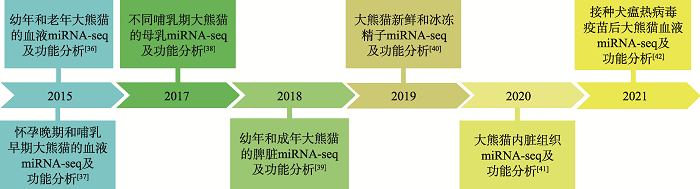 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1大熊猫miRNA研究进展
Fig. 1Research progress of miRNA in giant panda
Table 1
表1
表1大熊猫miRNA调控作用
Table 1
| 生物学功能 | 组织 | 研究策略 | 参考文献 |
|---|---|---|---|
| 调控免疫反应,包括血液免疫、母乳免疫 | 血液、乳汁 | 对不同年龄大熊猫的血液进行miRNA-seq;对接种犬瘟热病毒疫苗后的大熊猫血液进行miRNA-seq;对不同哺乳期的大熊猫母乳外泌体进行miRNA-seq | [36,38,42] |
| 调控乳腺发育及新生幼崽生长发育 | 血液、乳汁 | 对怀孕后期和哺乳早期大熊猫血液进行miRNA-seq;对不同哺乳期母乳外泌体进行miRNA-seq | [37,38] |
| 调控精子冷冻耐受 | 精子 | 对大熊猫新鲜和冷冻的精子外泌体进行miRNA-seq | [40] |
| 调控其他多种生物学过程 | 内脏(心、肝、脾、肺、肾) | 对不同内脏组织进行miRNA-seq;对新生和成年大熊猫的脾脏进行miRNA-seq | [39,41] |
新窗口打开|下载CSV
2.1 miRNA调控免疫反应
血液是免疫系统的主要组成部分,许多疾病都可以通过血液鉴定[43],因此对血液组织进行转录组测序分析具有重要意义。2015年,Yang等[36]利用Illumina二代测序技术分析了4只大熊猫血液中miRNA的表达模式,共检测到276个保守miRNA和51个新miRNA,差异表达分析发现7个miRNA的表达量在幼年大熊猫中明显高于成年大熊猫。此外,雌性大熊猫中有2个miRNA表达量上调,雄性个体中有1个miRNA表达量上调。靶基因预测表明大熊猫miRNA可能与4602个下游基因的表达有关,随后对预测的靶基因进行KEGG通路分析,发现这些基因主要参与宿主免疫应答,包括Ras信号通路、PI3K-Akt信号通路和MAPK信号通路。同年,Du等[44]利用Illumina HiSeq 2000测序技术对3只大熊猫的血液转录组进行鉴定,共注释38,522条转录本 (41.6%),其中有25,142条和8272条转录本分别被富集到GO (gene ontology)和COG (clusters of orthologous group )。KEGG(Kyoto Encyclopedia of genes and genomes)富集分析显示,9098 (9.83%)条转录本富集于324条KEGG通路,其中信号转导和免疫系统功能最具代表性。后来,Du等[45]利用RNA-seq技术研究了大熊猫血液组织中随着年龄的增长而出现的差异表达基因和通路。结果共获得210个差异表达基因,包括146个表达上调基因和64个表达下调基因,其中ISG15、STAT1、IRF7和DDX58是蛋白互作网络中的中枢基因,它们的表达量随大熊猫年龄增长而上调,并在应对病原菌入侵中发挥重要作用。功能富集分析表明这些表达量上调的基因主要参与先天免疫应答,而表达量下调的基因主要与B细胞激活有关。综上所述,大熊猫血液中的miRNA主要参与调控免疫反应,但具体的调控机制需进一步研究。
Sun等[42]为了研究miRNA在犬瘟热病毒(canine distemper virus, CDV)疫苗免疫应答中的作用,对5只已接种两次疫苗(间隔21天)的大熊猫幼仔血液进行miRNA-seq。结果共鉴定出187个保守miRNA和96个新miRNA,并发现29个差异表达miRNA,其中miR-16、miR-182、miR-30b和miR-101的表达量上调,表明先天免疫力可能增强,而miR-155和miR-181a的表达量下调,表明大熊猫产生抗体和B细胞的能力可能减弱。研究还发现20对miRNA- mRNA存在负调控关系,其中miR-204表达量的下调增加了TLR6基因的表达进而增强大熊猫幼崽先天免疫力,miR-330表达量的下调可能通过增加TMEM106A的表达激活巨噬细胞并调节免疫应答。综上所述,miRNA参与调控大熊猫的免疫应答反应,但具体的分子调控机制仍有待进一步研究。
2.2 miRNA调控乳腺发育及新生幼仔生长发育
母乳是哺乳动物婴儿理想的营养来源,它含有丰富且平衡的营养物质,满足婴儿大脑发育和生长需要,与配方奶相比,具有显著的健康益处[46]。研究发现,人(Homo sapiens)[47]、牛(Bos taurus)[48]和猪(Sus scrofa)[49]的乳汁中都存在miRNA表达且高丰度表达的miRNA大多参与免疫应答和神经系统相关的组织发育[50]。2015年,Wang等[37]通过RNA-seq技术从大熊猫外周血液中鉴定出202个成熟的miRNA以及27个miRNA家族和29个miRNA簇,在大熊猫怀孕后期和哺乳期早期阶段,通过PCR检测到12个与哺乳相关的miRNA表达。功能富集分析表明,这些miRNA在乳腺发育和代谢变化中发挥着重要调控作用。2017年,Ma等[38]对大熊猫乳汁外泌体中的miRNA进行了高通量测序,阐述了不同哺乳期大熊猫乳汁外泌体中miRNA的表达特征,并发现在初乳和成熟乳中富集与免疫和发育相关的内源性miRNA,这些miRNA在胞外囊泡的保护作用下稳定存在,以应对某些苛刻的环境条件。研究还发现母乳可能会促进大熊猫幼崽从饮食中摄入母亲的miRNA,以调节产后发育。此外,在大熊猫母乳外泌体中检测到来自大熊猫主要食物来源(竹子)的外源性植物miRNA,这些miRNA与基础代谢和神经元发育的调节作用相关,提示外源性植物miRNA被宿主细胞吸收,随后作为潜在的跨界调节因子被分泌到体液中。因此,大熊猫母乳外泌体中的miRNA是至关重要的母性调节因子,有助于促进新生幼崽的生长发育。2.3 miRNA调控精子冷冻耐受
精子生成是一个复杂的过程,原始生殖细胞增殖分化形成精原细胞,精原细胞有丝分裂形成初级精母细胞,接着减数分裂形成精子细胞,最后形成成熟精子[51,52]。研究发现许多miRNA参与调控小鼠 (Mus. musculus)[53,54,55]和人[56,57]的精子生成过程。但目前关于miRNA调控大熊猫精子生成的研究还未见报道。精子低温保存和人工授精是大熊猫繁育和保存现有遗传多样性的重要手段,但对精子低温损伤的分子机制及抗冻性等影响因素尚不清楚。Ran等[40]分析了miRNA对大熊猫精子冷冻耐受能力的调控作用,通过高通量测序技术比较了大熊猫新鲜精子和冷冻精子外泌体miRNA表达图谱,共鉴定出899个成熟miRNA,其中显著差异表达的miRNA有284个,包括195个表达量上调和89个表达量下调。GO富集分析发现这些差异表达miRNA的靶点主要分布在膜信号转导通路中,涉及能量代谢、嗅觉传导通路、炎症反应和细胞因子相互作用的差异miRNA可能参与调控精子的冷冻耐受机制,但具体的调控功能有待进一步验证。2.4 miRNA调控多种生物学过程
目前利用高通量测序技术对大熊猫内脏组织的研究报道还很少。Wang等[41]利用RNA-seq技术对4只大熊猫的5种内脏组织(心脏、肝脏、脾脏、肺和肾脏) miRNA进行测序分析,结果共鉴定出1256个已知miRNA和12个新的miRNA。在心脏、肝脏、脾脏、肺和肾脏中分别筛选出215、131、185、83和126个组织特异性差异miRNA,分别包括miR- 1b-5p、miR-122-5p、miR-143、miR-126-5p和miR- 10b-5p,预测的靶基因(包括MYL2、LRP5、MIF、CFD和PEBP1) 分别与组织特异性生物学功能密切相关。Peng等[39]通过转录组测序技术分别对新生和成年大熊猫脾脏miRNA进行测序分析,结果共鉴定出737个miRNA,且新生和成年脾脏miRNA的表达水平高度一致。此外,共筛选出503个差异表达miRNA,且大多数差异表达miRNA (81.1%)在新生脾脏中表达量上调,而成年脾脏中只有95个miRNA (18.9%)高表达。通过对预测的靶基因进行GO富集分析,结果发现靶基因主要富集在生物过程,其次是细胞成分和分子功能。KEGG富集分析显示差异表达miRNA的靶基因共涉及11条通路,包括细胞外基质受体相互作用(ECM-receptor interaction)、轴突导向(axon guidance)、T细胞受体信号通路(T cell receptor signaling pathway)、TGF-β信号通路(TGF-β signaling pathway)、血管内皮生长因子信号通路(VEGF signaling pathway)、弓形虫病(Toxoplasmosis)、基底细胞癌(basal cell carcinoma)、小细胞肺癌(small cell lung cancer)、细胞周期(cell cycle)、急性髓细胞白血病(acute myeloid leukemia)和破骨细胞分化(osteoclast differentiation),其中只有T细胞受体信号通路(T cell receptor signaling pathway)与免疫相关,说明miRNA不仅调控免疫应答,还可能参与调节其他生物过程,但具体的调控机制有待进一步研究。3 结语与展望
随着高通量测序技术的成熟,对非编码RNA进行转录组测序已成为当前研究热点。miRNA作为真核生物转录后基因调控因子,在哺乳动物各组织中广泛表达,并通过特异性结合靶基因调控生物学功能[58,59]。大熊猫作为珍稀保护物种,近年来关于其miRNA的研究发展迅速。miRNA在大熊猫血液、乳汁、精子和内脏组织中广泛表达且功能富集分析发现这些miRNA在大熊猫免疫反应、乳腺发育、精子冷冻耐受及其他生物学过程等方面发挥着重要调控作用。然而这些研究仅停留在鉴定和筛选层面,对预测的靶基因未做进一步生物学功能验证,因此今后应着重关注大熊猫功能性miRNA的验证和调控机制探究,以及如何应用于大熊猫繁育和保护工作。具体如下:
3.1 建立成熟的大熊猫原代细胞分离培养体系
由于大熊猫物种的珍稀性,目前关于分离培养大熊猫原代细胞的研究十分有限。2005年,Zhang等[60,61]利用酶消化法分离出大熊猫皮肤成纤维细胞,并利用表皮生长因子和胰岛素筛选出具有较好生物学特性的皮肤成纤维细胞。2015年,Yu等[62]利用组织块法和差速贴壁法成功分离出大熊猫骨骼肌来源的原代细胞。然而,这些研究涉及到的组织种类比较单一,且这些原代细胞在数次传代后,是否依然能保持较好的细胞活性?这些细胞活性如何变化?传代次数的极限是多少?如何保持传代后细胞的稳定性?这些问题都需要人们进一步探究。因此,针对不同组织来源的原代细胞(例如来源于脂肪的成脂细胞、来源于心脏的心肌细胞和来源于胚胎的胚胎干细胞等)分别建立起一套成熟的原代细胞分离培养体系,并且在多次传代后仍然保持较好的细胞活性。培养出稳定的大熊猫细胞系有利于后续的功能验证或克隆,也可为其他研究者提供珍稀和丰富的研究材料,同时建立起的大熊猫细胞库也将成为世界上珍贵的遗传资源保护系统。
3.2 验证免疫等生物学等功能,完善调控网络
为了进一步验证miRNA及其靶基因免疫等生物学功能,通过合成miRNA过表达和抑制剂、构建靶基因过表达和抑制载体分别在细胞和分子水平进行功能验证。将miRNA过表达和抑制剂分别转染至大熊猫原代细胞,验证miRNA对免疫等生物学功能的影响,同样的方法也用于验证靶基因功能。结合PCR技术分别定量miRNA、靶基因及其关联基因的表达情况,同时利用双荧光素酶报告系统(dual- luciferase reporter system) 再次验证miRNA与靶基因的靶标关系,进一步说明miRNA对大熊猫相关生物学功能的调控作用。此外,合成大熊猫相关抗体,利用免疫印迹(Western blot, WB)技术检验其蛋白表达水平。通过分子和蛋白水平的功能验证,明确miRNA对大熊猫免疫等生物学功能的调控机制,进一步完善调控互作网络,探讨如何利用miRNA抵抗病原菌入侵,提高大熊猫免疫能力,减少免疫相关的疾病发生,降低大熊猫在野外放归和重引入过程中因免疫疾病带来的死亡率。
3.3 构建大熊猫精子冷冻模型
Ran等[40]研究表明miRNA可能参与调控精子细胞的冷冻耐受机制,但未做进一步的功能验证。对此,建立一套大熊猫精子细胞冷冻模型,在不同冷冻温度或不同冷冻时间条件下,研究miRNA的表达情况及调控机制。同时,构建不同发育阶段的精子细胞冷冻模型,研究miRNA在大熊猫精子发育过程中对精子冷冻耐受的作用,深入挖掘miRNA对大熊猫精子冷冻耐受的调控机制,为进一步提高精液冷冻耐受和质量,完善精液保存技术,提高圈养大熊猫的繁殖率提供新思路。3.4 探究CRISPR/Cas9介导的基因编辑技术
基因编辑技术是对目标基因组进行定点修饰的一种基因工程技术,其中CRISPR/Cas9介导的基因编辑技术在设计和构建上相对简单,应用最广泛。目前,研究表明该技术已成功应用于人[63]、斑马鱼(Danio rerio)[64]、小鼠[65]、猪[66]、绵羊(Ovis arise)[67]等物种。研究发现,利用CRISPR/Cas9技术引入绵羊等大型家畜所需的SNP,可以促进肌肉生长,也能缩短育种年限[68,69]。Wu等[65]通过CRISPR/Cas9技术使引起小鼠白内障的一个单碱基发生突变,结果部分突变体受精卵发育成无白内障疾病的正常小鼠。因此,CRISPR/Cas9介导的基因编辑技术是否也能应用于大熊猫这一珍稀物种?该技术在大熊猫基因功能探究和基因疾病治疗方面是否能有新发现?这些都需要人们进一步探究。综上所述,建立大熊猫原代细胞分离培养体系,利用大熊猫精子冷冻模型及CRISPR/Cas9技术,验证大熊猫miRNA功能及作用机制,完善调控互作网络,探索其生物学功能对大熊猫繁育和保护工作具有重要意义。
(责任编委: 于黎)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.landusepol.2012.12.003URL [本文引用: 1]
DOI:10.1007/s11427-020-1750-7URL [本文引用: 1]
DOI:10.1371/journal.pone.0207433URL [本文引用: 1]
DOI:10.1016/j.biocon.2018.12.031 [本文引用: 1]

Small features in ecological systems are often underrepresented in conservation monitoring, management and policy. Tree cavities and other forms of refuge play disproportionately large ecological roles due to their importance for shelter and rearing vulnerable offspring. Giant panda (Ailuropoda melanoleuca) cubs are highly altricial, and dependent on dens. In Fengtongzai-a reserve with cavity-containing old growth forest-we measured 19 structural and microhabitat variables in potential tree dens. We also established data loggers in rock cavities in Foping Nature Reserve (which due to logging does not contain old growth) and tree cavities in Fengtongzai to monitor temperature and humidity inside and outside dens to evaluate microclimatic buffering. Fengtongzai pandas selected tree dens that were better concealed, with large interiors and entrances but smaller entrance to interior ratios. Microclimate inside dens differed dramatically from ambient conditions outside: in cold weather dens were warmer, in hot weather dens were cooler, dens were less humid and dens had more stable microclimates. Dens used by maternal pandas were warmer, drier and less variable than tree and rock cavities that were not used. Tree dens showed better capacity to buffer against extremes of temperature and humidity than did cave dens. Our findings have important conservation implications, including the value of den sites and the need for better monitoring and management. Specifically, management practices that preserve large old trees may increase carrying capacity and any experimentation with artificial dens as a conservation intervention should reference our findings on structural and microclimatic characteristics of preferred den sites.
DOI:10.1016/j.ecolind.2019.105886URL [本文引用: 1]
DOI:S0960-9822(19)30004-1PMID:30713107 [本文引用: 1]

The ancestral panda Ailurarctos lufengensis, excavated from the late Miocene, is thought to be carnivorous or omnivorous [1]. Today, giant pandas exclusively consume bamboo and have distinctive tooth, skull, and muscle characteristics adapted to a tough and fibrous bamboo diet during their long evolution [1, 2]. A special feature, the pseudo-thumb, has evolved to permit the precise and efficient grasping of bamboo [3, 4]. Unlike those of extant pandas, little is known about the diet and habitat preferences of extinct pandas. Prevailing studies suggest that the panda shifted to specialized bamboo feeding in the Pleistocene [5, 6]; however, this remains questionable. Pandas now survive in a fraction of their historical habitat [7], but no specific information has been reported. Stable isotope analyses can be used to understand diet- and habitat-related changes in animals [8]. Isotopic signals in bone collagen reflect dietary compositions of ancient human diets [9, 10] and dietary changes between historical and modern animal populations [11, 12]. Here, we conduct stable isotope analyses of bone and tooth samples from ancient and modern pandas and from sympatric fauna. We show that pandas have had a diet dominated by C resources over time and space and that trophic niches of ancient and modern pandas are distinctly different. The isotopic trophic and ecological niche widths of ancient pandas are approximately three times larger than those of modern pandas, suggesting that ancient pandas possibly had more complex diets and habitats than do their modern counterparts. Our findings provide insight into the dietary evolution and habitat contraction of pandas.Copyright © 2019 Elsevier Ltd. All rights reserved.
DOI:10.1007/s00114-010-0748-xPMID:21132275 [本文引用: 1]

In this study, landmark-based methods of geometric morphometrics are used for investigating the main aspects of cranial shape transformation in the evolution of the giant panda, Ailuropoda melanoleuca. Specifically, we explore if the highly derived cranial adaptations for bamboo feeding of the living panda were developed early in the panda's lineage. Results obtained show that the overall cranial morphologies of the oldest known panda, the "pygmy" Ailuropoda microta, and the late Pleistocene Ailuropoda baconi are both very similar to that of their closest living relative, A. melanoleuca, which agrees with a previous proposal based on qualitative criteria. However, we also describe several differences between the crania of A. microta, A. baconi, and A. melanoleuca, including the development of the postorbital process, the orientation of the occipital region, and the expansion of the braincase. As a result, the cranial morphology of A. microta shows a less specialized morphology toward a fibrous and durophagous diet compared to the giant panda. These results are confirmed by a comparative analysis of the dimensions of the upper teeth in bears, which has revealed differences in relative tooth size between A. microta and A. melanoleuca, most probably as a result of mosaic evolution. Therefore, we conclude that cranial shape did not remain essentially uniform in the Ailuropoda lineage, as previously thought, but underwent a number of changes during more than 2 Myr.
DOI:10.1016/j.gene.2016.07.029URL [本文引用: 1]
PMID:8252621 [本文引用: 2]

lin-4 is essential for the normal temporal control of diverse postembryonic developmental events in C. elegans. lin-4 acts by negatively regulating the level of LIN-14 protein, creating a temporal decrease in LIN-14 protein starting in the first larval stage (L1). We have cloned the C. elegans lin-4 locus by chromosomal walking and transformation rescue. We used the C. elegans clone to isolate the gene from three other Caenorhabditis species; all four Caenorhabditis clones functionally rescue the lin-4 null allele of C. elegans. Comparison of the lin-4 genomic sequence from these four species and site-directed mutagenesis of potential open reading frames indicated that lin-4 does not encode a protein. Two small lin-4 transcripts of approximately 22 and 61 nt were identified in C. elegans and found to contain sequences complementary to a repeated sequence element in the 3' untranslated region (UTR) of lin-14 mRNA, suggesting that lin-4 regulates lin-14 translation via an antisense RNA-RNA interaction.
DOI:10.1093/nar/gky1141 [本文引用: 1]

miRBase catalogs, names and distributes microRNA gene sequences. The latest release of miRBase (v22) contains microRNA sequences from 271 organisms: 38589 hairpin precursors and 48860 mature microRNAs. We describe improvements to the database and website to provide more information about the quality of microRNA gene annotations, and the cellular functions of their products. We have collected 1493 small RNA deep sequencing datasets and mapped a total of 5.5 billion reads to microRNA sequences. The read mapping patterns provide strong support for the validity of between 20% and 65% of microRNA annotations in different well-studied animal genomes, and evidence for the removal of >200 sequences from the database. To improve the availability of microRNA functional information, we are disseminating Gene Ontology terms annotated against miRBase sequences. We have also used a text-mining approach to search for microRNA gene names in the full-text of open access articles. Over 500 000 sentences from 18 542 papers contain microRNA names. We score these sentences for functional information and link them with 12 519 microRNA entries. The sentences themselves, and word clouds built from them, provide effective summaries of the functional information about specific microRNAs. miRBase is publicly and freely available at http://mirbase.org/.
DOI:10.1093/bioinformatics/bts496URL [本文引用: 1]
PMID:24846995 [本文引用: 1]

With the completion of Human Genome Project (HGP), it was revealed that among the 3 billion base pairs in human genome, only 1.5% of them encodes proteins. The remaining 98.5% of the sequence does not encode any protein, and was once regarded as accumulated junk sequences during evolution. However, in the subsequently initiated ENCODE project, it was unexpectedly found that about 75% of the human genome was transcribed into RNAs. Seventy-four percent of them are non-protein-coding RNAs (non-coding RNAs, ncRNAs). In this RNA category, most of the transcripts are longer than 200 nucleotides and thus named as long non-coding RNAs (lncRNAs). ncRNAs regulate gene expression at the transcriptional and post-transcriptional levels, function in fundamental biological processes including cell differentiation and organ development, and are closely associated with many human diseases. In this paper, we review the recent progress in the discovery, classification, expression, and function study of lncRNAs, as well as their roles in the pathogenesis of hu-man diseases.
PMID:24846995 [本文引用: 1]

With the completion of Human Genome Project (HGP), it was revealed that among the 3 billion base pairs in human genome, only 1.5% of them encodes proteins. The remaining 98.5% of the sequence does not encode any protein, and was once regarded as accumulated junk sequences during evolution. However, in the subsequently initiated ENCODE project, it was unexpectedly found that about 75% of the human genome was transcribed into RNAs. Seventy-four percent of them are non-protein-coding RNAs (non-coding RNAs, ncRNAs). In this RNA category, most of the transcripts are longer than 200 nucleotides and thus named as long non-coding RNAs (lncRNAs). ncRNAs regulate gene expression at the transcriptional and post-transcriptional levels, function in fundamental biological processes including cell differentiation and organ development, and are closely associated with many human diseases. In this paper, we review the recent progress in the discovery, classification, expression, and function study of lncRNAs, as well as their roles in the pathogenesis of hu-man diseases.
DOI:10.1038/35002607URL [本文引用: 1]
DOI:10.1146/annurev-biochem-060308-103103URL [本文引用: 1]
DOI:10.1261/rna.1565109PMID:19286629 [本文引用: 1]

MicroRNAs (miRNAs) are an abundant class of approximately 22 nucleotide (nt) long noncoding RNAs that negatively regulate gene expression post-transcriptionally through imperfect base-pairing interactions with sequences in the target messenger RNA (mRNA). We examined the interactions of the bantam miRNA with the 3' untranslated region (UTR) of the hid mRNA, and a synthetic derivative, in Drosophila S2 cells in order to define the relative contributions of proposed bantam binding sites. The contribution of the bantam miRNA to repression of reporter constructs carrying different 3' UTRs was evaluated by measuring derepression of reporter expression following the transfection of bantam complementary oligoribonucleotides (anti-bantam). Systematic excision of bantam miRNA target sequences in the hid 3' UTR identified by commonly used miRNA target prediction programs failed to relieve repression to the extent predicted by the anti-bantam experiment. However, removal of additional bantam complementary sequences (with a "seed" match to nucleotide 3-9) derepressed the reporter constructs to the full extent, arguing for a less narrow definition of the seed sequence. Further support for the potential contribution of the 3-9 seed register to microRNA-mediated gene regulation is provided by the experimental validation of several novel bantam targets identified with a more relaxed search algorithm.
DOI:10.1016/j.cell.2009.01.002PMID:19167326 [本文引用: 1]

MicroRNAs (miRNAs) are endogenous approximately 23 nt RNAs that play important gene-regulatory roles in animals and plants by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression. This review outlines the current understanding of miRNA target recognition in animals and discusses the widespread impact of miRNAs on both the expression and evolution of protein-coding genes.
PMID:15200956 [本文引用: 1]

MicroRNAs (miRNAs) are approximately 21-nucleotide RNAs, some of which have been shown to play important gene-regulatory roles during plant development. We developed comparative genomic approaches to systematically identify both miRNAs and their targets that are conserved in Arabidopsis thaliana and rice (Oryza sativa). Twenty-three miRNA candidates, representing seven newly identified gene families, were experimentally validated in Arabidopsis, bringing the total number of reported miRNA genes to 92, representing 22 families. Nineteen newly identified target candidates were confirmed by detecting mRNA fragments diagnostic of miRNA-directed cleavage in plants. Overall, plant miRNAs have a strong propensity to target genes controlling development, particularly those of transcription factors and F-box proteins. However, plant miRNAs have conserved regulatory functions extending beyond development, in that they also target superoxide dismutases, laccases, and ATP sulfurylases. The expression of miR395, the sulfurylase-targeting miRNA, increases upon sulfate starvation, showing that miRNAs can be induced by environmental stress.
DOI:10.1371/journal.pbio.0030085URL [本文引用: 1]
DOI:10.1038/ng1536URL [本文引用: 1]
DOI:10.1016/j.cell.2004.12.035URL [本文引用: 1]
DOI:10.1016/j.cell.2009.07.011PMID:19665978 [本文引用: 1]

Human breast tumors contain a breast cancer stem cell (BCSC) population with properties reminiscent of normal stem cells. We found 37 microRNAs that were differentially expressed between human BCSCs and nontumorigenic cancer cells. Three clusters, miR-200c-141, miR-200b-200a-429, and miR-183-96-182 were downregulated in human BCSCs, normal human and murine mammary stem/progenitor cells, and embryonal carcinoma cells. Expression of BMI1, a known regulator of stem cell self-renewal, was modulated by miR-200c. miR-200c inhibited the clonal expansion of breast cancer cells and suppressed the growth of embryonal carcinoma cells in vitro. Most importantly, miR-200c strongly suppressed the ability of normal mammary stem cells to form mammary ducts and tumor formation driven by human BCSCs in vivo. The coordinated downregulation of three microRNA clusters and the similar functional regulation of clonal expansion by miR-200c provide a molecular link that connects BCSCs with normal stem cells.
DOI:10.1016/j.cell.2007.03.030URL [本文引用: 1]
[本文引用: 1]
DOI:10.1080/15384101.2020.1864941URL [本文引用: 1]
DOI:10.24099/vet.arhivURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.3390/ijms19041209URL [本文引用: 1]
DOI:10.1007/s10142-019-00683-2URL [本文引用: 1]
DOI:10.1007/s10695-019-00654-1PMID:31115741 [本文引用: 1]

Flatfish pigmentation is a complex process, affected by environmental factors including light, nutrients, and hormones. Of those, the thyroid hormone has been reported to increase the albinism rate of Japanese flounder (Paralichthys olivaceus). However, the underlying mechanism remains unclear. In the present study, triiodothyronine (T3), thyroxine, and thiourea were introduced into P. olivaceus larvae from 16 to 57 days after hatching (DAH). By comparison of albinism rate, T3 treatment and control larvae of 42 DAH were chosen for mRNA and miRNA high-throughput sequencing analyses. A total of 337 miRNAs were identified via miRNA-seq, and 12 miRNAs exhibited significantly differential expression patterns in D42_T3 versus D42_Con (TPM >?10, fold change ≥?1.5 or ≤?0.67 and q?≤?0.05). These differentially expressed miRNAs targeted 3658 putative genes, which further enriched to 10 GO terms (q?<?0.05). RNA-seq identified 146 differentially expressed genes (DEGs) in D42_T3 versus D42_Con (|log fold change|?>?1 and q?<?0.005), including pigmentation-related genes such as the receptor tyrosine-protein kinase erbB-3, pro-opiomelanocortin A, and melanotransferrin, and the growth-related gene somatotropin. These DEGs were significantly enriched to 15 GO terms and 8 KEGG pathways (q?<?0.05), which included several sugar metabolic pathways (glycolysis/gluconeogenesis and the pentose phosphate pathway). Integrated analysis revealed that 26 overlapping genes between DEGs and mRNAs were targeted by miRNAs. Furthermore, seven mRNA-miRNA pairs exhibited reversed regulation patterns. This provides important clues to understand the role of thyroid hormones in flatfish pigmentation.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1371/journal.pone.0143242URL [本文引用: 4]
DOI:10.4238/2015.November.13.5PMID:26600479 [本文引用: 3]

The giant panda (Ailuropoda melanoleuca) is one of the world's most endangered mammals, and it has evolved several unusual biological and behavioral traits. During puberty, pregnancy, lactation, and involution, the mammary gland undergoes profound morphological and functional changes. A large number of microRNAs (miRNAs) have been identified to be involved in mammary gland development and lactation. In this study, we identified 202 conserved mature miRNAs, corresponding to 147 pre-miRNAs, in giant panda peripheral blood using a small RNA-sequencing approach. In addition, 27 miRNA families and 29 miRNA clusters were identified. We analyzed the arm selection preference of pre-miRNAs and found that: 1) most giant panda pre-miRNAs generated one-strand miRNAs, and the 5p-arm only miRNAs have a higher expression level than 3p-arm only miRNAs; 2) there were more 5p-arm dominant miRNAs than 3p-arm dominant miRNAs; and 3) 5p-arm dominant miRNAs have a larger fold change within miRNA pairs than 3p-arm dominant miRNAs. Expression of 12 lactation-related miRNAs was detected across late pregnancy and early lactation stages by qPCR, and seven miRNAs were identified as clustered in one significant model. Most of these clustered miRNAs exhibited inhibitory roles in proliferation and differentiation of mammary epithelial cells. Functional analysis highlighted important roles of the seven as signed miRNAs in mammary development and metabolic changes, including blood vessel morphogenesis, macromolecule biosynthesis, cell cycle regulation, and protein transport.
DOI:10.1038/s41598-017-03707-8URL [本文引用: 5]
DOI:10.1159/000488837PMID:29669315 [本文引用: 3]

Giant pandas, an endangered species, are a powerful symbol of species conservation. Giant pandas may suffer from a variety of diseases. Owing to their highly specialized diet of bamboo, giant pandas are thought to have a relatively weak ability to resist diseases. The spleen is the largest organ in the lymphatic system. However, there is little known about giant panda spleen at a molecular level. Thus, clarifying the regulatory mechanisms of spleen could help us further understand the immune system of the giant panda as well as its conservation.The two giant panda spleens were from two male individuals, one newborn and one an adult, in a non-pathological condition. The whole transcriptomes of mRNA, lncRNA, miRNA, and circRNA in the two spleens were sequenced using the Illumina HiSeq platform. EBseq and IDEG6 were used to observe the differentially expressed genes (DEGs) between these two spleens. Gene Ontology and KEGG analyses were used to annotate the function of DEGs. Furthermore, networks between non-coding RNAs and protein-coding genes were constructed to investigate the relationship between non-coding RNAs and immune-associated genes.By comparative analysis of the whole transcriptomes of these two spleens, we found that one of the major roles of lncRNAs could be involved in the regulation of immune responses of giant panda spleens. In addition, our results also revealed that microRNAs and circRNAs may have evolved to regulate a large set of biological processes of giant panda spleens, and circRNAs may function as miRNA sponges.To our knowledge, this is the first report of lncRNAs and circRNAs in giant panda, which could be a useful resource for further giant panda research. Our study reveals the potential functional roles of miRNAs, lncRNAs, and circRNAs in giant panda spleen.© 2018 The Author(s). Published by S. Karger AG, Basel.
DOI:10.3390/biom9090432URL [本文引用: 4]
DOI:10.1016/j.gene.2020.145206URL [本文引用: 3]
DOI:10.1089/dna.2020.5942URL [本文引用: 4]
DOI:10.1016/j.semcancer.2008.01.005PMID:18291670 [本文引用: 1]

The discovery of microRNAs (miRNAs) is one of the major scientific breakthroughs in recent years and has revolutionized the way we look at gene regulation. Although we are still at a very early stage in understanding their impact on immunity, miRNAs are changing the way we think about the development of the immune system and regulation of immune functions. MiRNAs are implicated in establishing and maintaining the cell fate of immune cells (e.g. miR-181a and miR-223), and they are involved in innate immunity by regulating Toll-like receptor signaling and ensuing cytokine response (e.g. miR-146). Moreover, miRNAs regulate central elements of the adaptive immune response such as antigen presentation (e.g. miR-155) and T cell receptor signaling (miR-181a). Recent evidence showing altered miRNA expression in chronic inflammatory diseases (e.g. miR-203 and miR-146) suggests their involvement in immune-mediated diseases. Furthermore, miRNAs have been implicated in viral immune escape and anti-viral defense (e.g. miR-196). In this review, we will summarize the latest findings about the role of miRNAs in the development of the immune system and regulation of immune functions and inflammation.
DOI:10.1111/1755-0998.12367URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/1758-907X-1-7URL [本文引用: 1]
DOI:S0022-0302(12)00497-3PMID:22916887 [本文引用: 1]

We previously reported that microRNA (miRNA) is present in human breast milk. Recently, other groups have reported that bovine milk also contains miRNA; however, these reports are few. We therefore investigated bovine milk miRNA using microarray and quantitative PCR analyses to identify the differences between colostrum and mature milk. The RNA concentration in a colostrum whey fraction was higher than that in a mature milk whey fraction. In total, 102 miRNA were detected in bovine milk by microarray analysis (100 in colostrum and 53 in mature milk; 51 were common to both). Among these miRNA, we selected several immune- and development-related miRNA, including miR-15b, miR-27b, miR-34a, miR-106b, miR-130a, miR-155, and miR-223. These miRNA were detected in bovine milk by quantitative PCR, and each of these miRNA was significantly more highly expressed in colostrum than in mature milk. We also confirmed the presence of some mRNA in bovine milk. Nevertheless, synthesized miRNA spiked in the raw milk whey were degraded, and naturally existing miRNA and mRNA in raw milk were resistant to acidic conditions and RNase treatment. The RNA molecules in milk were stable. We also detected miRNA and mRNA in infant formulas purchased from Japanese markets. It is still unknown whether milk-derived RNA molecules play biological roles in infants; however, if milk-derived RNA do show functions in infants, our data will help guide future studies.Copyright © 2012 American Dairy Science Association. Published by Elsevier Inc. All rights reserved.
DOI:10.4238/2012.June.18.3PMID:22782633 [本文引用: 1]

Determination of an optimal set/number of internal control microRNA (miRNA) genes is a critical, but often undervalued, detail of quantitative gene expression analysis. No validated internal genes for miRNA quantitative PCR (q-PCR) in pig milk were available. We compared the expression stability of six porcine internal control miRNA genes in pig milk from different lactation periods (1 h, 3 days, 7 days, 14 days, 21 days, and 28 days postpartum), using an EvaGreen q-PCR approach. We found that using the three most stable internal control genes to calculate the normalization factor is sufficient for producing reliable q-PCR expression data. We also found that miRNAs are superior to ribosomal RNA (rRNA) and snRNA, which are commonly used as internal controls for normalizing miRNA q-PCR data. In terms of economic and experimental feasibility, we recommend the use of the three most stable internal control miRNA genes (miR-17, -107 and -103) for calculating the normalization factors for pig milk samples from different lactation periods. These results can be applied to future studies aimed at measuring miRNA abundance in porcine milk.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.diff.2010.08.002URL [本文引用: 1]
DOI:10.1038/emboj.2010.319URL [本文引用: 1]
DOI:10.1093/molehr/gaq058URL [本文引用: 1]
DOI:10.1038/ng.2990URL [本文引用: 1]
DOI:10.1186/s13058-015-0593-0URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1089/bio.2014.0073PMID:26035009 [本文引用: 1]

The giant panda Ailuropoda melanoleuca is an endangered species and is a symbol for wildlife conservation. Although efforts have been made to protect this rare and endangered species through breeding and conservative biology, the long-term preservation of giant panda genome resources (gametes, tissues, organs, genomic libraries, etc.) is still a practical option. In this study, the giant panda skeletal muscle-derived cell line was successfully established via primary explants culture and cryopreservation techniques. The population doubling time of giant panda skeletal cells was approximately 33.8?h, and this population maintained a high cell viability before and after cryopreservation (95.6% and 90.7%, respectively). The two skeletal muscle-specific genes SMYD1 and MYF6 were expressed and detected by RT-PCR in the giant panda skeletal muscle-derived cell line. Karyotyping analysis revealed that the frequencies of giant panda skeletal muscle cells showing a chromosome number of 2n=42 ranged from 90.6~94.2%. Thus, the giant panda skeletal muscle-derived cell line provides a vital resource and material platform for further studies and is likely to be useful for the protection of this rare and endangered species.
DOI:10.1126/science.1232033URL [本文引用: 1]
DOI:10.1038/cr.2013.45URL [本文引用: 1]
DOI:10.1016/j.stem.2013.11.002URL [本文引用: 2]
DOI:10.1016/0014-4827(61)90435-9URL [本文引用: 1]
DOI:10.15302/J-FASE-2014007URL [本文引用: 1]
DOI:10.1038/ng1810URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
