 ,1, 任沛东2, 叶文新2, 陆光涛
,1, 任沛东2, 叶文新2, 陆光涛 ,2
,2Analysis of transcriptional regulators HpaR1 and Clp regulating the expression of glycoside hydrolase-encoding gene in the Xanthomonas campestris pv. campestris
Guofang Liu ,1, Peidong Ren2, Wenxin Ye2, Guangtao Lu
,1, Peidong Ren2, Wenxin Ye2, Guangtao Lu ,2
,2通讯作者: 陆光涛,博士,研究员,研究方向:微生物学。E-mail:lugt@gxu.edu.cn
编委: 刘钢
收稿日期:2021-05-11修回日期:2021-07-5
| 基金资助: |
Received:2021-05-11Revised:2021-07-5
| Fund supported: |
作者简介 About authors
刘国芳,博士,副教授,研究方向:植物保护。E-mail:

摘要
十字花科黑腐病菌(Xanthomonas campestris pv. campestris, Xcc)是一种维管束致病菌,能够引起寄主的黑腐病,是研究植物病原细菌与植物互作的一种重要模式菌株。在Xcc中,GntR家族的全局性转录调控因子HpaR1参与调控Xcc的运动、胞外多糖和胞外酶的合成等许多细胞过程,并与Xcc的过敏反应(hypersensitive response, HR)和致病相关;全局性转录调控因子Clp则参与调控胞外酶和胞外多糖的分泌与合成,并与黄单胞菌的致病相关。前期研究发现,Xcc中的转录调控因子HpaR1和Clp均能与糖苷水解酶(glycoside hydrolase)编码基因(命名为ghy基因)的启动子区结合。为探究转录调控因子HpaR1和Clp共同调控ghy基因表达的分子机理,本研究首先通过凝胶阻滞分析(electrophoresis mobility shift assay, EMSA)发现HpaR1和Clp在体外能够结合在ghy基因的启动子区;利用染色质免疫共沉淀(chromatin immunoprecipitation, ChIP)方法,进一步证实HpaR1和Clp在细胞内能够结合在ghy基因启动子区。通过5ʹ-cDNA末端快速扩增(rapid amplication of 5ʹ-cDNA ends, 5ʹ- RACE)和DNase I保护实验(DNase I footprinting)确定HpaR1和Clp均结合在ghy基因启动子的-35区上游,并且HpaR1的结合位点位于Clp结合位点的上游。通过实时荧光定量PCR(real time fluorescence quantitative PCR, RT-qPCR)和体外转录的方法,发现HpaR1抑制ghy基因的转录,而Clp激活ghy基因的转录。当二者共同存在时,HpaR1能够抑制Clp对ghy基因的转录激活作用。HpaR1和Clp单独存在时,分别负调控和正调控ghy基因的转录,推测HpaR1尽管位于ghy基因启动子-35区上游,但可能通过抑制RNA聚合酶的活性来调控ghy基因的表达。
关键词:
Abstract
Xanthomonas campestris pv. campestris (Xcc) is a vascular pathogen that causes black rot in host. It is an important model strain for studying the interaction between the phytopathogen and plants. In Xcc, global transcription regulator HpaR1 that belongs to the GntR family regulates many cellular processes such as the movement and synthesis of extracellular polysaccharides and extracellular enzymes, and is associated with hypersensitive response (HR) and pathogenicity. On the other hand, the global transcriptional regulator Clp regulates the secretion and synthesis of extracellular enzymes and extracellular polysaccharides, and is associated with the pathogenicity of Xanthomonas. Previous studies have shown that both HpaR1 and Clp bind to the promoter region of the glycoside hydrolase encoding gene (named ghy gene). This study investigates the molecular mechanism of the co-regulation of HpaR1 and Clp on the expression of ghy gene. Through electrophoresis mobility shift assay (EMSA), we found that both HpaR1 and Clp bind to the promoter regions of gene ghy in vitro. Both HpaR1 and Clp also bind to the promoter regions of gene ghy in vivo by chromatin immunoprecipitation (ChIP) assays. DNase I footprinting and 5ʹ-RACE assays showed that both HpaR1 and Clp bind to the -35 region upstream of the ghy promoter. The HpaR1 binding site was located upstream of the Clp binding site. RT-qPCR and in vitro transcription assays showed that HpaR1 negatively while Clp positively regulates the transcription of gene ghy. Furthermore, HpaR1 inhibits the activation of Clp on the transcription of gene ghy in vitro. Our findings indicate that HpaR1 and Clp exhibit opposite effect on the transcription of gene ghy. It is speculated that HpaR1 may regulate the expression of gene ghy by inhibiting the activity of RNA polymerase.
Keywords:
PDF (1194KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘国芳, 任沛东, 叶文新, 陆光涛. 十字花科黑腐病菌中转录因子HpaR1与Clp调控一个糖苷水解酶基因表达的分析. 遗传[J], 2021, 43(9): 910-920 doi:10.16288/j.yczz.21-171
Guofang Liu.
细菌生活的环境是复杂多变的,经常会面对一些极端环境,如pH、渗透压骤变,营养物质匮乏等恶劣环境。为了维持自身的生长与生存,细菌必须开启不同的调控系统以适应环境条件变化。其中最常见的调控方式是通过转录调控因子与启动子的结合来调控基因转录的起始,进而调控基因的转录水平。在细菌中,已鉴定出多种转录调控因子,其中最为常见的是螺旋-转角-螺旋(helix-turn-helix, HTH)类群,包含一个保守的α螺旋-转角-α螺旋DNA结合结构域[1]。HTH转录调控因子中分布较为广泛的一类是GntR家族转录调控因子,此家族的许多转录调控因子已被鉴定,可以调控多种不同的生物学过程[2,3]。GntR家族转录调控因子包含两个功能域,分别是保守的N端DNA结合结构域和非保守的C端效应物结合结构域/寡聚化作用结构域。依据C端氨基酸序列将GntR家族转录调控因子分为7个亚家族,分别是AraR、DevA、FadR、HutC、MocR、PlmA和YtrA[4,5]。
黄单胞菌属中的十字花科黑腐病菌(Xanthomonas campestris pv. campestris,Xcc)是研究植物病原细菌与植物互作的一种重要模式菌株,能侵染包括萝卜、花椰菜、白菜、芥菜、油菜等多种十字花科植物[6,7]。该病原菌是一种维管束致病菌,主要是通过植物组织的水孔和伤口侵入寄主植物体内[8~11]并在其寄主体内繁殖,引起寄主的黑腐病。
在Xcc中,已知GntR家族YtrA亚家族的全局性转录调控因子HpaR1,除了与Xcc的过敏反应HR和致病相关外[12],还参与调控多种细胞过程[12,13],如运动性、胞外多糖和胞外酶的合成等。全局性转录调控因子Clp(cyclic AMP receptor protein-like protein)调控胞外酶和胞外多糖的分泌与合成,并与黄单胞菌的致病相关[14,15]。本课题组前期的工作发现,在Xcc 8004菌株的全基因组中,一个编号为XC_0026基因(命名为ghy基因)可能编码一个糖苷水解酶,该酶可能参与纤维素或木质素的降解,该基因的表达可能受全局性转录调控因子HpaR1和Clp的共同调控。
两个或两个以上转录因子共同调控靶基因在原核生物中普遍存在[16],但HpaR1和Clp调控ghy基因的机理并不清楚。本研究对HpaR1、Clp共同调控ghy基因的模式进行了分析,发现HpaR1、Clp均结合在ghy基因启动子-35区上游,并且HpaR1抑制ghy基因的转录,而Clp激活ghy基因的转录。进一步研究提示HpaR1可能通过抑制RNA聚合酶的活性,来调控ghy基因的转录。
1 材料与方法
1.1 菌株、质粒、培养条件和引物
本研究所用的菌株和质粒见表1;本研究所用引物见表2。Xcc在NY培养基中培养(NYGB:Peptone 5 g/L、Yeast extract 3 g/L、Glycerol 20 g/L,pH 7.0;NYGA:NYGB培养基中加入0.8% (W/V) 琼脂粉配置而成,用于Xcc固体培养),培养温度为28℃。大肠杆菌(E. coli)在L培养基中培养(LB:Trytone 10 g/L、Yeast extract 5 g/L、NaCl 5 g/L,pH 7.0;LA:LB培养基中加入0.8% (W/V)琼脂粉配制而成,用于E. coli固体培养),培养温度为37℃。培养E. coli时抗生素的用量为:利福平(Rif) 50 μg/mL、卡那霉素(Km)25 μg/mL、壮观霉素(Spc) 50 μg/mL、四环素(Tc)15 μg/mL、氨苄青霉素(Amp)100 μg/mL;培养Xcc时抗生素的用量为:Rif 50 μg/mL、Km 25 μg/mL、Spc 50 μg/mL、Tc 5 μg/mL。Table 1
表1
表1本研究所用的菌株与质粒
Table 1
| 菌株和质粒 | 相关特征 |
|---|---|
| 大肠杆菌 | |
| DH5α | F-,φ80d/lacZ△M15,Δ(lacZYA-argF)U169,deoR,recA1,endA1,hsdR17(rk-, mk+),phoA,supE44, λ-,thi-1,gyrA96,relA1 |
| M15 | Nals,trs,Rifs,Thi-,Lac-,Ar+a,Gal+,Mtl-,F-,RecA+,Uvr+,Lon+harboring pREP4 plasmid,Kmr |
| ED8767 | RecA,met,含有帮助质粒 pRK2073,Spcr |
| pQE-HpaR1/M15 | M15中含有质粒pQE-HpaR1,Kmr,Ampr |
| pQE-Clp/M15 | M15中含有质粒 pQE-Clp,Kmr,Ampr |
| pL3-HpaR1::3×Flg/ DH5α | DH5α中含有质粒pL3-HpaR1:: 3×Flg,Tcr |
| pL3-Clp:: 3×Flg/ DH5α | DH5α中含有质粒pL3-Clp:: 3×Flg,Tcr |
| 十字花科黑腐病菌 | |
| 8004 | 野生型,Rifr |
| ΔhpaR1 | hpaR1基因的缺失突变体,Rifr |
| Δclp | Clp基因的缺失突变体,Rifr |
| 8004/pLAFR3 | 8004中含有质粒pLAFR3,Rifr,Tcr |
| ΔhpaR1/pL3-HpaR1::3×Flg | ΔhpaR1中含有质粒 pL3-HpaR1::3×Flg,Rifr,Tcr |
| Δclp/pL3-Clp:: 3×Flg | Δclp中含有质粒 pL3-Clp:: 3×Flg,Rifr,Tcr |
| 质粒 | |
| pK18mob | 黄单胞菌属中的自杀质粒,Kmr |
| pRK2073 | 帮助质粒,Tra+,Mob+,ColE1,Spcr |
| pQE30 | 表达载体,Ampr |
| pQE30-HpaR1 | pQE30上含有hpaR1基因的360 bp ORF,Ampr |
| pQE30-Clp | pQE30上含有 clp基因的690 bp ORF,Ampr |
| pLAFR3 | PK2 replicon;Mob+,Tra-;cos+,Tcr |
| pL3-HpaR1::3×Flg | pLAFR3上含有 hpaR1::3×Flg,Tcr |
| pL3-Clp::3×Flg | pLAFR3上含有 clp::3×Flg,Tcr |
新窗口打开|下载CSV
1.2 蛋白质的表达纯化
根据十字花科黑腐病菌中的clp基因(XC_0486)序列,设计引物扩增clp完整的ORF,通过酶切、连接的方法,将PCR产物分别连接到含强启动子的高表达载体pQE30,将得到的重组质粒pQE30-Clp分别转化至E. coli M15菌株中,获得Clp表达菌株pQE-Clp/M15。将本研究构建的Clp表达菌株和从本课题组所保存的HpaR1表达菌株pQE-HpaR1/M15进行液体培养,经IPTG诱导后,分离纯化蛋白质产物。本研究纯化蛋白质所用的是Promega公司的MagneHisTM Protein Purification System试剂盒。1.3 实时荧光定量PCR (real time fluorescence quantitative PCR, RT-qPCR)
按1%的接菌量将Xcc野生型菌株8004、hpaR1突变体ΔhpaR1、clp突变体Δclp分别接种至NYGB培养基中,28℃培养24 h,提取RNA,通过RT-qPCR的方法检测ghy基因在Xcc 菌株中的转录水平。RNA的提取采用OMEGA公司的Bacterial RNA Kit。RT-qPCR采用的是Vazyme公司的ChamQTM SYBR Color qPCR Master Mix完成。1.4 凝胶阻滞分析(electrophoresis mobility shift assay, EMSA)
为了确定和研究HpaR1/Clp是否能够结合在ghy基因的启动子,需要通过EMSA实验来分析蛋白质和DNA的相互作用。通过PCR扩增ghy基因上游412 bp的DNA片段(DNA片段带有荧光标记),将其与梯度浓度的表达蛋白HpaR1或者Clp混合,然后采用非变性聚丙烯酰胺凝胶电泳分离DNA,观察DNA与不同浓度蛋白混合后,泳动率是否发生了变化,从而判断蛋白与基因间是否存在直接的转录调控关系。1.5 DNase I保护实验(DNase I footprinting)
本实验采用DNase I保护实验来鉴定HpaR1/ Clp在ghy基因启动子上所结合的DNA序列。在一系列的EP管中,每管加入10 µL binding buffer,并编号。每管加入不同浓度的HpaR1蛋白或者Clp蛋白,室温放置20 min;再加入适量的带有荧光标记的DNA (300 ng),室温放置20 min;然后加入10 µL DNase I buffer,适量的DNase I,37℃反应适当的时间;氯仿终止反应;抽提DNA,干燥,送测序公司(上海迈普生物),得到的结果进行软件分析。1.6 5ʹ-cDNA末端快速扩增(rapid amplication of 5ʹ-cDNA ends, 5ʹ-RACE )
提取Xcc 8004总RNA,合成第一链cDNA并纯化cDNA,cDNA加尾;然后进行常规PCR,取5 μL加尾产物作为模板,以AAP和GSP2为引物(表2),进行常规PCR。如果PCR产物量少,可再取0.5 μL PCR反应产物作为模板,以AUAP和GSP3为引物(表2),再进行常规PCR。将所得的PCR产物进行纯化并与pGEM-T载体连接,将所得的重组质粒转化到E. coli DH5α感受态细胞中,并涂在含有相应抗生素的LA筛选平板上。挑选出转化所得的阳性克隆并进行PCR验证,将PCR验证正确的克隆送北京华大测序,分析测序结果。Table 2
表2
表2本研究所用的引物
Table 2
| 用途 | 引物名称 | 引物序列(5′→3′) |
|---|---|---|
| 蛋白表达 | Clp-OF | ACAGTTGGATCC ATGAGCCTAGGGAACAGCAC |
| Clp-OR | ACAGTTAAGCTT TTAGCGCGTGCCGTACAACA | |
| 凝胶阻滞分析 | 0026 F | ACGGCAATCGATCAGTTCGC |
| 0026 R | CATGGAAACCACTCCTTCGACG | |
| DNase I保护实验 | 0026(HpaR1)F | GACAAGCCGCAGATGAAAACCC |
| 0026(HpaR1)R | GTTACGCTGCATCTGCCCGT | |
| 0026(Clp)F | GCAGCCGGCGCACGCAGTAGACCTGTACT | |
| 0026(Clp)R | CGACGGAAAAGATGCATCGCAGCCC | |
| 5ʹ-RACE | AAP | GGCCACGCGTCGACTAGTACGGGGGGGGGG |
| AUAP | GGCCACGCGTCGACTAGTAC | |
| 0026GSP1 | GTTCCCACAGGATCGGCAGG | |
| 0026GSP2 | TCCAGTCCCAGCGAAATAGC | |
| 0026GSP3 | TGTTGAGGACGCCAGGTTTT | |
| 0026GSPF | ATGTCGTGTTCTCTGTGGTTGC | |
| 体外转录 | 0026ivtF | GCGACAAGCCGCAGATGAAAACCCT |
| 0026ivtR | GATGACGCAAATTCCGCACCCGCC | |
| 3077ivtF | TCTCACTCTGTCTTGCAAACTGCGA | |
| 3077ivtR | AATGGGCATCGAAAACCAGAAGC | |
| 染色质免疫共沉淀 | Clp(ChIP)F | CGGGATCCGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGACTACAA-AGATGACGACGATAAAATGAGCCTAGGGAACACGACG |
| Clp(ChIP)R | CCAAGCTTTTAGCGCGTGCCGTACAACA | |
| HpaR1(ChIP)F | CGGGATCCGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGACTACAA-AGATGACGACGATAAAATGACTGACATCCAGTGGAGC | |
| HpaR1(ChIP)R | CCAAGCTTTCATGGTGTTTTCCCCTGTG | |
| 0026(ChIP)F | CGTCACGCTTTCGTCGCCTA | |
| 0026(ChIP)R | AACTGCGTTGTGGCCTGCAC | |
| 实时荧光定量PCR | 0026qRT-F | ATGTCGTGTTCTCTGTGGTTGC |
| 0026qRT-R | TGTTGAGGACGCCAGGTTTT |
新窗口打开|下载CSV
1.7 体外转录(in vitro transcription)
根据Xcc 8004全基因组序列,用Vector NTI软件设计引物,扩增包括ghy基因启动子区及起始密码子ATG下游约150 bp的DNA片段作为体外转录的模板;为除去HpaR1蛋白和Clp蛋白溶液中的咪唑对体外转录的影响,用磷酸缓冲液对蛋白液进行超滤;将DNA片段、HpaR1蛋白或者Clp蛋白、四种NTP (含生物素标记的UTP)和E. coli RNA聚合酶在转录缓冲液(40 mmol/L Tris-HCl pH 7.9,6 mmol/L MgCl2,2 mmol/L spermidine,10 mmol/L NaCl,5 mmol/L DTT,5% glycerol,50 mmol/L KCl,1 U RNase inhibitor)中温育30 min,终止反应后,用7 mol/L尿素,5%聚丙烯酰胺凝胶电泳分离反应混合物。1.8 染色质免疫共沉淀(chromatin immunoprecipitation, ChIP)
通过设计引物、酶切、转化、三亲本等实验方法,分别构建能够表达融合3×Flg标签的HpaR1蛋白的菌株ΔhpaR1/pL3-HpaR1::3×Flg、融合3×Flg标签的Clp蛋白的菌株Δclp/pL3-Clp::3×Flg以及对照菌株8004/pLAFR3。将所获得的菌株在NYGB,28℃,200 r/min条件下,培养24 h,各取300 mL培养液转移到另一洁净的培养瓶中,经过交联、解交联等一系列反应,富集回收DNA用于下一步PCR验证。2 结果与分析
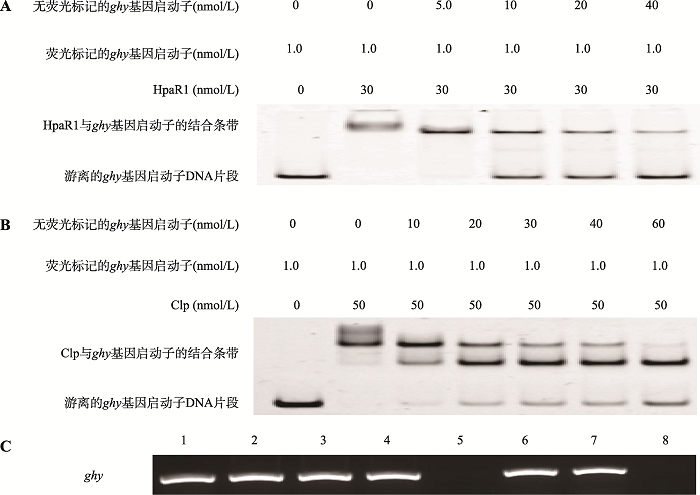
2.1 HpaR1和Clp均能结合在ghy基因启动子区
为了解HpaR1、Clp是否通过结合在ghy基因启动子直接调控ghy基因的表达,分别过量表达并纯化HpaR1蛋白和Clp蛋白,并以0026F/0026R为引物PCR扩增412 bp的ghy基因启动子区,将所获得的蛋白和DNA片段进行EMSA实验。结果显示,HpaR1和Clp均可以与ghy基因的启动子区在体外相结合;在EMSA反应体系中同时加入没有荧光标记和荧光标记的ghy基因启动子DNA片段时,随着没有荧光标记的ghy基因启动子DNA片段浓度的增加,体系中游离的荧光标记的ghy基因启动子DNA片段逐渐增多,说明HpaR1、Clp能够特异性的与ghy基因的启动子区在体外相结合(图1,A和B)。采用ChIP进一步分析HpaR1、Clp在细胞内是否能结合ghy基因的启动子区。为此,首先构建能够表达融合有3×Flag标签的HpaR1蛋白的重组质粒pL3-HpaR1::3×Flag,以及表达融合有3×Flag标签的Clp蛋白的重组质粒pL3-Clp::3×Flag,并分别导入到HpaR1缺失突变体ΔhpaR1和Clp缺失突变体Δclp,获得菌株ΔhpaR1/pL3-HpaR1::3×Flag和Δclp/pL3-Clp::3×Flag。同时,作为对照,将空载体pLAFR3导入Xcc野生型菌株8004,获得菌株8004/pLAFR3。
将所得到的菌株进行液体培养、甲醛固定、裂解以及DNA的免疫亲和层析分离。将所获得的DNA样品作为模板,PCR扩增154 bp ghy基因启动子区。结果发现:分别以ΔhpaR1/pL3-HpaR1::3×Flag和Δclp/pL3-Clp::3×Flag菌株中洗脱得到的DNA为模板时,能够扩增得到PCR产物;而以8004/pLAFR3菌株中洗脱得到的DNA为模板时,则不能得到PCR产物(图1C)。这表明,HpaR1和Clp在细胞内能与ghy基因启动子结合。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1HpaR1、Clp与ghy基因启动子区结合
A:HpaR1与ghy基因启动子区结合的体外EMSA实验。B:Clp与ghy基因启动子区结合的体外EMSA实验。C:HpaR1、Clp与ghy基因启动子区结合的体内ChIP实验。PCR检测洗脱DNA。1:8004/pLAFR3总DNA;2:8004/pLAFR3 input对照;3:ΔhpaR1/pL3-HpaR1::3×Flag input对照;4:Δclp/pL3-Clp::3×Flag input对照;5:8004/pLAFR3洗脱DNA;6:ΔhpaR1/pL3-HpaR1::3×Flag洗脱DNA;7:Δclp/pL3-Clp::3×Flag洗脱DNA;8:ddH2O。
Fig. 1Binding of HpaR1, Clp to ghy promoter region
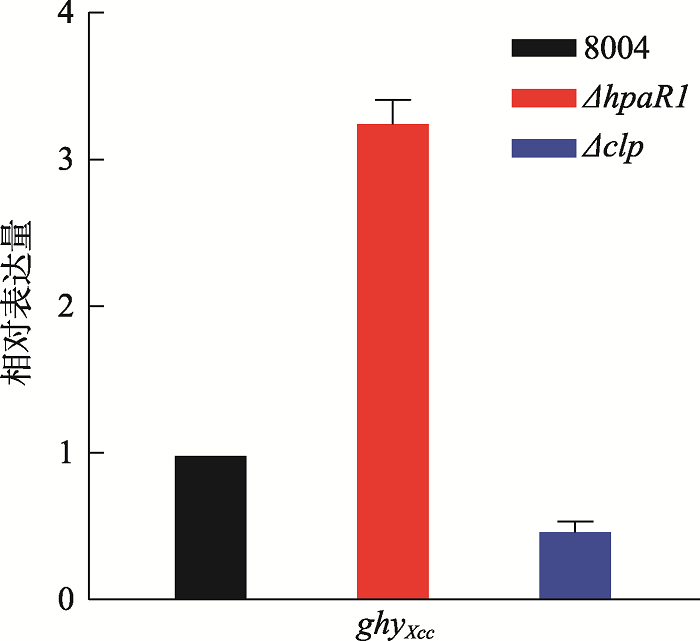
2.2 HpaR1、Clp调控ghy基因的表达
为明确HpaR1、Clp调控ghy基因表达的方式,通过RT-qPCR分析HpaR1、Clp对ghy基因转录水平的影响。按1%的接菌量将野生型菌株8004、hpaR1突变体ΔhpaR1、clp突变体Δclp分别接种至NYGB培养基中,28℃培养24 h,提取RNA,通过RT-qPCR检测ghy基因在Xcc 菌株中的转录水平。结果发现,ghy基因在突变体ΔhpaR1中的转录水平明显高于野生型菌株,而在突变体Δclp中的转录水平明显低于野生型菌株(P=0.01,t-test) (图2)。此结果证明,HpaR1负调控ghy基因的转录,Clp正调控ghy基因的转录。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2RT-qPCR检测ghy基因在野生型菌株8004、hpaR1和clp突变体中的转录水平
Fig. 2RT-qPCR analysis of transcription level of ghy in wild-type strain 8004, ΔhpaR1, and Δclp
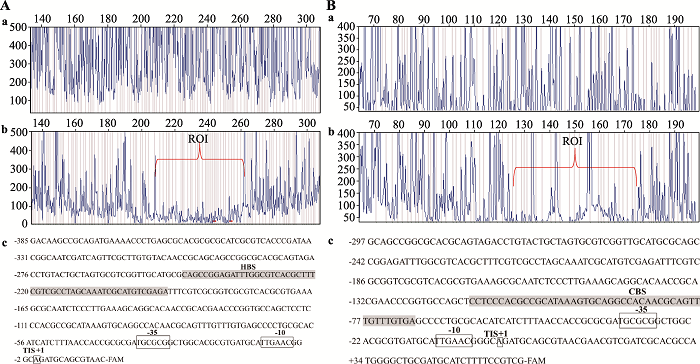
2.3 HpaR1、Clp在ghy基因启动子区结合位点的精确定位
通过分段EMSA的结果,初步确定了HpaR1、Clp结合在ghy基因启动子区的区域。根据结合位点附近的DNA序列,分别设计合适的引物,扩增ghy基因上游约300 bp的序列(包含靶标序列),进一步采用DNase I保护实验来精确定位HpaR1、Clp在ghy基因启动子区上的结合序列。结果显示:HpaR1结合在ghy基因启动子区一段约53 bp的序列“CAGCCGGAGATTTGGCGTCACGCTTTCGTCGCCTAGCA AATCGCATGTCGAGA” (图3A);Clp结合在ghy基因启动子区一段约47 bp的序列“CCTCCCACGCCGCATAAAGTGCAGGCCACAACGCAG-TTTGTTTGTGA” (图3B)。HpaR1与ghy基因启动子区的结合序列没有YtrA亚家族保守的结合序列“..GT…TA….TA…AC..”,说明HpaR1能识别另一种序列。上述DNase I保护实验确定了HpaR1、Clp的结合序列,但结合位点与启动子-10区和-35区的关系还需要进一步研究。本研究采用5ʹ-RACE的方法确定了ghy基因的转录起始位点,进而确定了其启动子的-10区与-35区。结果显示:ghy基因的转录起始位点位于翻译起始位点ATG上游77 bp的“A”处,从而确定了-10区“TTGAAC”和-35区“TGCGCG”(图4)。结合DNase I保护实验的结果可知, HpaR1和Clp均结合在ghy基因启动子的-35区上游,Clp对ghy基因的转录起促进作用,与已报道的转录调控因子调控机制一致;而HpaR1对ghy基因的转录起抑制作用,一般情况下转录调控因子结合在基因启动子-35区上游,会激活基因的转录。因此,我们推测HpaR1以一种新的模式抑制ghy基因的表达。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3HpaR1/Clp与ghy基因启动子区的结合序列
A:DNase I保护实验分析HpaR1与ghy基因启动子区结合序列。a:FAM标记的DNA经DNase I处理后的测序结果;b:FAM标记的DNA+HpaR1经DNase I处理后的测序结果;ROI:region of interest;c:FAM标记的DNA序列;HBS (HpaR1 binding site):HpaR1与ghy基因启动子区的结合序列。B:DNase I保护实验分析Clp与ghy基因启动子区结合序列。a:FAM标记的DNA经DNase I处理后的测序结果;b:FAM标记的DNA+Clp经DNase I处理后的测序结果;c:FAM标记的DNA序列;CBS (Clp binding site):Clp与ghy基因启动子区的结合序列。
Fig. 3DNase I footprinting analysis of HpaR1 and Clp binding to ghy promoter region
图4
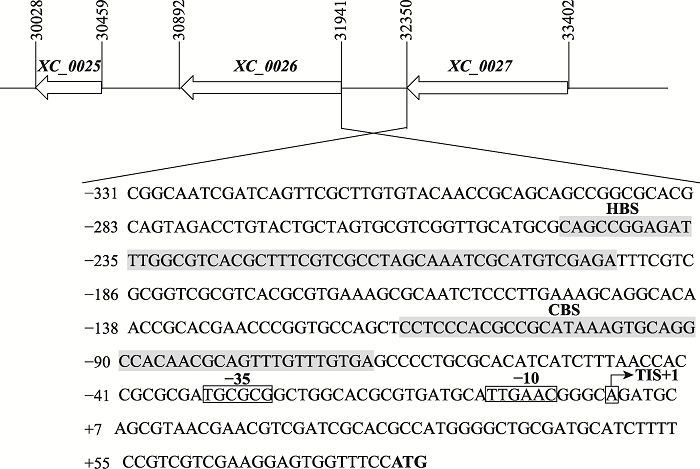 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图45ʹ-RACE确定ghy基因转录起始位点以及启动子区-35区、-10区
TIS:ghy基因启动子的转录起始位点(+1);矩形框标出的序列分别是ghy基因启动子区的-10区(TTGAAC)、-35区(TGCGCG);CBS(Clp binding site):Clp与ghy基因启动子区的结合序列;HBS (HpaR1 binding site):HpaR1与ghy基因启动子区的结合序列。
Fig. 4Determination of the transcription initiation site and -35, -10 promoter regions of ghy by 5ʹ-RACE
2.4 HpaR1、Clp在体外分别抑制和激活ghy基因的转录
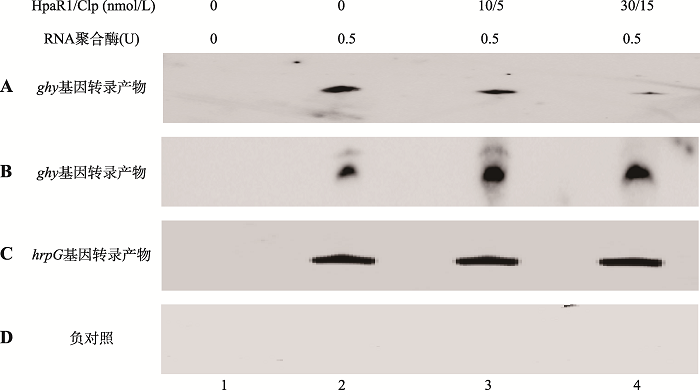
上述结果表明HpaR1在胞内负调控而Clp正调控ghy基因的表达。为了进一步确定HpaR1和Clp对ghy基因的直接调控作用,本研究又进行了体外转录实验。根据Xcc 8004全基因组序列,用Vector NTI软件设计引物( 0026ivtF/0026ivtR,引物序列见表2),扩增ghy基因启动子区和起始密码子ATG后约150 bp的片段。将所获得的DNA片段作为体外转录的模板。结果显示:没有HpaR1、Clp时,RNA聚合酶能够启动ghy基因的转录;当反应体系中加入HpaR1时,随着HpaR1的增加,ghy基因的转录产物逐渐减少(图5A);但对照片段hrpG基因的转录产物水平不受HpaR1的影响(图5C);当反应体系中加入Clp时,随着Clp的增加,ghy基因的转录产物逐渐增加(图5B);而启动子区缺失-35区,-10区和转录起始位点的ghy基因没有转录产物合成(图5D)。这表明,HpaR1能够通过结合在ghy基因启动子-35区的上游,来抑制ghy基因的转录水平;而Clp则能增强ghy基因的转录水平。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5HpaR1/Clp调控ghy基因体外转录结果
将体外转录模板与不同浓度HpaR1或Clp室温静置30 min,然后加入0.5 U的RNA聚合酶进行体外转录,1×TBE、7 mol/L尿素、5%聚丙烯酰胺凝胶电泳,转膜,扫描成像。1:无RNA聚合酶;2:体外转录模板加RNA聚合酶后转录产物;3和4:体外转录模板与HpaR1 (A、C、D)或者Clp(B)室温温育30 min后,加入RNA聚合酶后的转录产物。A:ghy基因体外转录产物随着HpaR1浓度的增加而减少;B:ghy基因体外转录产物随着Clp浓度的增加而增加;C:hrpG体外转录产物随着HpaR1浓度的增加没有明显变化;D:无-35区、-10区和TIS的 ghy基因启动子区无转录产物。
Fig. 5HpaR1/Clp regulate the transcription of gene ghy in vitro
2.5 HpaR1和Clp在ghy基因启动子上的相互作用分析
研究中发现,HpaR1和Clp分别结合在ghy基因启动子的-35区上游,HpaR1抑制了ghy基因的转录,Clp激活了ghy基因的转录,那么HapR1和Clp在调控ghy基因转录的过程中是否存在相互作用?本研究进行了初步探究。体外转录的结果发现:在体外转录体系中单独加入Clp后,ghy基因的转录被激活,Clp的浓度不变,随着HpaR1浓度的增加,ghy基因的转录逐渐减弱(图6)。此结果说明,HpaR1作为ghy基因的一个转录抑制子,能够降低转录激活子Clp对ghy基因的转录激活作用。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6HpaR1、Clp共同调控ghy基因的表达
Fig. 6HpaR1 and Clp co-regulate ghy expression
3 讨论
3.1 HpaR1、Clp结合靶点的鉴定
本研究对HpaR1、Clp调控ghy基因机理做了初步的研究。HpaR1和Clp作为十字花科黑腐病菌中的全局性转录调控因子,可以调控细菌的很多基础代谢反应,在细菌的代谢调控中扮演着重要的角色。HpaR1属于GntR转录调控因子家族中的YtrA亚家族,YtrA亚家族转录调控因子结合序列的保守结构为“..GT…TA….TA…AC..”[3]。HpaR1结合自身启动子序列为一段24 bp的反向重复序列“GGTGTTATAGTTGAATACAAAACC”(下划线碱基为HpaR1结合自身启动子序列的保守结构),该序列具有YtrA亚家族结合序列的保守结构[12]。大肠杆菌中的Clp同源蛋白CRP的结合序列是一段22 bp保守的回文结构“AAATGTGA-TCTAGA-TCACATTT” (黑色加粗的碱基为回文结构)[17],其中结合位点的5、6、7位置的碱基对转录调控因子CRP的结合是非常重要的,称为“GTG motif”。十字花科黑腐病菌中的Clp与CRP有相同的保守结合位点“GTG motif”,如Clp直接上调的启动子engA[15]、pehA、pelA1和xpsE也含有“GTG motif”[18~20]。本研究发现,Clp结合ghy基因启动子序列具有保守结合位点“GTG motif”。本研究发现,HpaR1结合ghy基因启动子序列与之前发现的HpaR1结合在自身启动子区的HpaR1-box序列没有相似之处,没有明显的反向重复序列。之前的研究发现,野生型8004菌株中HpaR1结合gumB启动子的序列也没有反向重复序列[13]。这说明在8004菌株中,HpaR1能识别多种不同类型的靶序列,从这些靶序列中找到HpaR1其他保守的结合序列具有重要意义。HpaR1与ghy基因启动子的结合位点位于-35区上游,但HpaR1对ghy基因转录起抑制作用,这与之前报道的转录调控因子位于启动子区-35区上游起转录激活作用不同。这说明HpaR1识别不同的启动子序列,调控模式可能也会有所差异。
3.2 HpaR1、Clp调控模式的探讨
基因转录抑制主要包括三种模式[21]:(1)转录因子结合在启动子的-10区附近,使得RNAP无法与启动子结合,进而抑制基因的转录。(2)转录因子结合在启动子的特定位置,使启动子区的构象发生改变,形成一个环,封闭了启动子的-35区和-10区,使得RNAP无法与启动子结合,阻止了转录的起始。(3)某些基因的转录需要转录激活因子,而转录抑制因子通过与转录激活因子相互作用,使得激活因子无法起到激活转录的作用,进而阻止转录的起始。本研究发现,HpaR1与ghy基因启动子的结合位点位于-35区上游,而HpaR1对ghy基因转录却起抑制作用,这与已报道的基因转录抑制的第一种模式是不同的,转录因子结合在基因启动子的-35区上游一般起转录激活作用。由此推测,HpaR1可能是通过抑制RNAP与ghy基因启动子的结合,进而抑制了ghy基因的转录。HpaR1结合位点离ghy基因启动子-35区较远,其是怎样和RNAP接触,进而抑制RNAP与ghy基因启动子的结合?我们推测,HpaR1与ghy基因启动子结合后,造成了DNA弯曲,与RNAP相互作用,抑制了RNAP与ghy基因启动子结合,进而抑制了ghy基因的转录。
传统的转录抑制模式是转录抑制子通过与其下游的转录激活子相互作用,使得激活因子无法起到激活转录的作用,间接地抑制了基因的转录[22]。而本研究发现转录抑制子HpaR1能够降低Clp对ghy基因的转录激活作用。这种降低转录激活因子的作用,可能除了通过与Clp的相互作用来实现外,也可能是通过HpaR1自身对靶基因的强烈抑制作用来实现。我们因此推测,HpaR1、Clp共同调控ghy基因表达的模式可能是一种新的未被发现的模式,需要人们更深入的研究。
(责任编委: 刘钢)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
PMID:2060763 [本文引用: 1]

A new family of bacterial regulatory proteins has been identified by sequence similarity. The family contains the repressor of the Bacillus subtilis gluconate operon (GntR), the regulators for histidine utilization in Pseudomonas putida (HutCPp) and Klebsiella aerogenes (HutCKa), the repressor (FadR) of fatty acid degradation in Escherichia coli, a regulator involved in the conjugal transfer of the broad host range plasmid pIJ101 (KorA), and three proteins of unidentified function in E. coli (GenA, P30 and PhnF). The proteins share amino acid sequence similarities in a 69-residue N-terminal region. A helix-turn-helix motif is predicted in the most highly-conserved segment of each protein suggesting that they are members of a new family of helix-turn-helix DNA-binding proteins.
DOI:10.1016/S0065-2164(09)69001-8PMID:19729089 [本文引用: 1]

One of the most abundant and widely distributed groups of Helix-turn-helix (HTH) transcription factors is the metabolite-responsive GntR family of regulators (>8500 members in the Pfam database; Jan 2009). These proteins contain a DNA-binding HTH domain at the N terminus of the protein and an effector-binding and/or oligomerisation domain at the C terminus, where upon on binding an effector molecule, a conformational change occurs in the protein which influences the DNA-binding properties of the regulator resulting in repression or activation of transcription. This review summarises what we know about the distribution, structure, function and classification of these regulators and suggests that they may have a future role in biotechnology.
DOI:10.1074/jbc.M110968200URL [本文引用: 2]
DOI:10.1186/1471-2164-8-289URL [本文引用: 1]
DOI:10.1371/journal.pone.0132618URL [本文引用: 1]
DOI:10.1094/MPMI-3-233URL [本文引用: 1]
DOI:10.1111/mpp.2011.12.issue-8URL [本文引用: 1]
campestris into hydathodes of Arabidopsis thaliana leaves: a system for studying early infections events in bacterial pathogenesis
DOI:10.1094/MPMI.1998.11.6.537URL
DOI:10.1371/journal.pone.0003828URL [本文引用: 1]
DOI:10.1094/MPMI-08-10-0180URL [本文引用: 3]
DOI:10.1038/srep19862URL [本文引用: 2]
DOI:10.1128/JB.01253-09URL [本文引用: 1]
DOI:10.1016/j.febslet.2005.05.023URL [本文引用: 2]
DOI:10.1111/mpp.2019.20.issue-1URL [本文引用: 1]
PMID:3045325 [本文引用: 1]

The statistics of base-pair usage within known recognition sites for a particular DNA-binding protein can be used to estimate the relative protein binding affinities to these sites, as well as to sites containing any other combinations of base-pairs. As has been described elsewhere, the connection between base-pair statistics and binding free energy is made by an equal probability selection assumption; i.e. that all base-pair sequences that provide appropriate binding strength are equally likely to have been chosen as recognition sites in the course of evolution. This is analogous to a statistical-mechanical system where all configurations with the same energy are equally likely to occur. In this communication, we apply the statistical-mechanical selection theory to analyze the base-pair statistics of the known recognition sequences for the cyclic AMP receptor protein (CRP). The theoretical predictions are found to be in reasonable agreement with binding data for those sequences for which experimental binding information is available, thus lending support to the basic assumptions of the selection theory. On the basis of this agreement, we can predict the affinity for CRP binding to any base-pair sequence, albeit with a large statistical uncertainty. When the known recognition sites for CRP are ranked according to predicted binding affinities, we find that the ranking is consistent with the hypothesis that the level of function of these sites parallels their fractional saturation with CRP-cAMP under in-vivo conditions. When applied to the entire genome, the theory predicts the existence of a large number of randomly occurring "pseudosites" with strong binding affinity for CRP. It appears that most CRP molecules are engaged in non-productive binding at non-specific or pseudospecific sites under in-vivo conditions. In this sense, the specificity of the CRP binding site is very low. Relative specificity requirements for polymerases, repressors and activators are compared in light of the results of this and the first paper in this series.
DOI:10.1007/s00284-007-9081-9URL
DOI:10.1099/mic.0.2007/012930-0URL
campestris
DOI:10.1021/jf900701nURL [本文引用: 1]
PMID:15035009 [本文引用: 1]

Bacteria use their genetic material with great effectiveness to make the right products in the correct amounts at the appropriate time. Studying bacterial transcription initiation in Escherichia coli has served as a model for understanding transcriptional control throughout all kingdoms of life. Every step in the pathway between gene and function is exploited to exercise this control, but for reasons of economy, it is plain that the key step to regulate is the initiation of RNA-transcript formation.
DOI:10.1146/annurev-micro-092611-150012URL [本文引用: 1]
