 ,, 李睿, 郭芳莹, 宋鸽鸽, 吴萌, 陈广文
,, 李睿, 郭芳莹, 宋鸽鸽, 吴萌, 陈广文 ,, 刘德增河南师范大学生命科学学院,新乡 453007
,, 刘德增河南师范大学生命科学学院,新乡 453007Functional analysis of autophagy-related gene Atg6 in planarian central nervous system regeneration
Kexue Ma ,, Rui Li, Fangying Guo, Gege Song, Meng Wu, Guangwen Chen
,, Rui Li, Fangying Guo, Gege Song, Meng Wu, Guangwen Chen ,, Dezeng LiuCollege of Life Sciences, Henan Normal University, Xinxiang 453007, China
,, Dezeng LiuCollege of Life Sciences, Henan Normal University, Xinxiang 453007, China通讯作者: 陈广文,博士,教授,博士生导师,研究方向:涡虫再生和资源保护。E-mail:Chengw0183@sina.com
编委: 林古法
收稿日期:2021-03-30修回日期:2021-06-21网络出版日期:2021-08-20
| 基金资助: |
Received:2021-03-30Revised:2021-06-21Online:2021-08-20
| Fund supported: |
作者简介 About authors
马克学,博士,副教授,硕士生导师,研究方向:涡虫再生和抗逆性的分子机制。E-mail:

摘要
细胞自噬基因Atg6在细胞自噬过程中发挥重要作用,其功能缺陷影响神经发生。涡虫是研究中枢神经系统(central nervous system, CNS)再生的良好模型,其头部切除后1周就能再生出一个新的头部。因此,研究Atg6基因在涡虫CNS再生中的作用对探究自噬调控神经发生具有重要意义。本研究首次报道了日本三角涡虫(Dugesia japonica) Atg6基因(DjAtg6)的分子特征,并利用RNAi技术研究了其在涡虫CNS再生中作用。结果显示:DjAtg6 cDNA全长1366 bp,编码423个氨基酸。DjATG6含有ATG6/Beclin 1蛋白家族的Coil-Coil结构域和β折叠α螺旋自噬功能结构域。涡虫沿咽前咽后切割后,DjAtg6表达量显著增加,其转录本主要在新再生的脑神经节表达。RNAi-DjAtg6引起涡虫头部再生迟缓、脑神经结构偏小,并下调神经相关基因的表达。此外,本研究还发现,RNAi-DjAtg6不影响涡虫干细胞的增殖,但下调细胞迁移相关基因mmp1和mmp2的表达,且干扰mmp1和mmp2的表达影响涡虫头再生。因此,本研究结果表明,DjAtg6在涡虫CNS再生的组织重构中发挥重要作用,干扰DjAtg6影响涡虫CNS再生可能与细胞迁移有关,其详细的分子机制尚需进行深入研究。
关键词:
Abstract
Autophagy-related gene 6 (Atg6) plays an essential role in autophagy, and loss of its function impairs neurogenesis. Planarian is a good model for the study of the central nervous system (CNS) regeneration. It can regenerate a new head de novo in 1 week following decapitation. Therefore, functional analysis of Atg6 in planarian CNS regeneration is very important for understanding of autophagy in the regulation of neurogenesis. In this work, we reported the molecular characteristics of Atg6 in Dugesia japonica (DjAtg6) for the first time and examined its function by RNAi. The full-length cDNA of DjAtg6 is 1366 bp encoding 423 amino acids. The deduced amino sequence of DjAtg6 contains the coil-coil domain and β-α-repeated autophagy-specific domain shared by ATG6/Beclin 1 family. Following amputation before and after the pharynx, DjAtg6 transcripts increased and were mainly distributed in the newly regenerated brain structure. RNAi-DjAtg6 delayed planarian head regeneration with a small size of brain, and decreased the expression levels of neural-related genes. In addition, our results revealed that RNAi-DjAtg6 did not affect the stem cell proliferation, but down-regulated the cell migration-related genes mmp1 and mmp2. Furthermore, RNAi-mmp1 and RNAi-mmp2 delayed planarian head regeneration. Therefore, our results suggest that DjAtg6 is important for planarian CNS regeneration. The abnormal CNS regeneration caused by RNAi-DjAtg6 may be related to cell migration, but the detailed mechanism needs to be further investigated.
Keywords:
PDF (1292KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
马克学, 李睿, 郭芳莹, 宋鸽鸽, 吴萌, 陈广文, 刘德增. 细胞自噬基因Atg6在涡虫中枢神经系统再生中的功能研究. 遗传[J], 2021, 43(8): 792-801 doi:10.16288/j.yczz.21-118
Kexue Ma.
细胞自噬是一种依赖溶酶体的蛋白质降解途径,广泛参与细胞增殖、分化、迁移和细胞命运决定等重要的生物学过程[1,2]。神经发生涉及细胞形态和功能的转变,细胞自噬参与其中[3,4]。研究表明,Beclin 1 (哺乳动物ATG6的同源蛋白)在小鼠(Mus musculus)侧脑室室下区表达,Beclin 1+/-小鼠神经球体面积缩小[5]。另有研究发现,Beclin 1下调表达与神经退行性疾病有关[6]。上述研究说明,Atg6/Beclin 1在神经发生和中枢神经(central nervous system, CNS)稳态维持过程中发挥重要作用。涡虫(Platyhelminthes, Turbellaria)是体内研究神经发生的理想实验动物,它能在一周内再生出一个全新的脑结构[7]。涡虫的神经再生吸引无数****关注,虽然研究人员已经发现了一些调控涡虫CNS再生的关键基因[8,9,10],但其神经发生的分子机制仍不十分清楚。涡虫CNS再生是细胞增殖、分化、迁移和细胞死亡等诸多细胞生物学过程整合的复杂机制。虽然早有研究人员提出细胞自噬参与其中[11],但并没有相关研究报道。为了研究细胞自噬调控神经发生的分子机制,本研究鉴定了日本三角涡虫(Dugesia japonica) Atg6同源基因,并用整体原位杂交和RNAi技术研究其在涡虫CNS再生过程中作用。
1 材料和方法
1.1 实验动物
日本三角涡虫来自本实验室无性繁殖,实验前至少饥饿7 d。再生实验的涡虫从咽前和咽后切成3段(头部片段、躯干片段和尾部片段),20℃避光培养。1.2 日本三角涡虫Atg6 cDNA克隆和生物信息学分析
随机挑选5~6条涡虫,在液氮中研磨成粉末,加入 1 mL Trizol试剂后按操作说明进行总RNA提取,然后取2 μg总RNA用于cDNA合成(PrimeScript 1st Strand cDNA Synthesis kit,日本TaKaRa公司)。根据从日本三角涡虫转录组数据库筛选出来的DjAtg6 EST序列设计3′-RACE和5′-RACE特异性引物(附表1),按照Clontech RACE cDNA试剂盒扩增cDNA片段。纯化的PCR片段连接pMD19-T后筛选阳性克隆进行测序分析。根据测序结果拼接出DjAtg6全长cDNA序列,并利用DNAstar 软件预测开放阅读框(ORF)并推导出编码的氨基酸序列。DjAtg6核苷酸序列和氨基酸序列同源性分析在NCBI (1.3 qRT-PCR分析
挑选躯干片段的再生涡虫按上述方法进行总RNA提取和cDNA合成。实时荧光定量PCR在ABI 7500系统中进行,反应体系为20 μL:10 μL 2 × TB Green Premix Ex Taq II (ROX plus,日本TaKaRa公司),上下游引物各 0.8 μL (10 μmol/L),cDNA模板 1 μL,灭菌超纯水 7.4 μL。PCR参数如下:95℃预变性5 min;95℃ 10 s,60℃ 30 s,40个循环。以Djef2为内参基因,以2-ΔΔCT法[12]计算mRNA 相对表达量,实验重复3次。本研究检测的干细胞相关基因(piwiA、mcm2、pcna、h2b)和神经相关基因(coe、soxB2、pax6、runt、hesl-3)的引物序列见附表1。采用SPASS 10.0软件进行单因子方差分析,P<0.05 表示显著性差异。1.4 整体原位杂交
原位杂交探针的合成采用 digoxigenin-labeling kit (SP6/T7, 美国Roche公司),扩增模板所用的引物序列见附表1。整体原位杂交(whole mount in situ hybridization, WISH)根据文献[13]操作并稍微修改。主要步骤如下:涡虫用2%HCl处死后立即用4%甲醛固定,然后经过6%H2O2漂白,蛋白酶K处理,然后加入400 ng/mL探针,56℃杂交>16 h。杂交信号用BCIP/NBT显色,用Leica DFC300FX显微镜拍照后用Adobe Photoshop处理。1.5 dsRNA干扰实验
双链RNA (double-stranded RNA,dsRNA) 合成使用MEGAscript RNAi Kit (Ambion 1626),扩增模板所用的引物序列见附表1。RNAi-DjAtg6采用注射法,纯化后的dsRNA按照文献[14, 15]中的操作方法注射涡虫,涡虫隔天注射一次,每次4×46 nL,共注射5次。注射结束后24 h切割涡虫,观察再生表型,用Leica DFC300FX显微镜拍照再生涡虫,保存图片并用系统附带的测量工具测量再生胚基长度。测量数据采用SPASS 10.0软件进行统计学处理。干扰组和对照组总RNA提取和qRT-PCR检测方法见1.2和1.3部分,检测相关基因的表达量所用引物见附表1。阴性对照为GFP dsRNA,阳性对照为β-catenin-1 dsRNA。RNAi实验至少重复3次。基质金属蛋白水解酶(matrix metalloproteinases, MMPs)调控细胞外基质动态变化,参与细胞迁移和细胞分化。已有研究表明,干扰mmp1和mmp2的表达影响涡虫干细胞迁移[16]。为进一步验证这个实验结果,本研究采用Rouhana等[17]提供的方法合成mmp1和mmp2 dsRNA (所用引物见附表1)并进行干扰实验。将合成的10 mg dsRNA与30 μL牛肝匀浆混合后喂食25条左右长度为3~4 mm涡虫,间隔3 d喂食一次,共喂食8次。喂食结束后次日切割涡虫观察再生表型,用Leica DFC300FX显微镜拍照再生虫子并测量再生胚基长度。实验结果采用SPASS 10.0软件进行统计学处理。
1.6 免疫荧光
整体免疫荧光按照文献[18]中的方法进行操作。磷酸化组蛋白H3(H3P)抗体(Millipore 06-870,1:200 diluted in PBST)用于检测有丝分裂细胞,anti- SYNORF1 (3C11)抗体(DSH, 1:20 diluted in PBST)用于显示涡虫CNS。二抗goat anti-rabbit Alexa Fluor 594和 goat anti-mouse Alexa Fluor 488 1:400倍稀释用于检测阳性信号,Stereo ?uorescence microscope (Axio Zoom. V16,德国蔡司公司)拍照并保存图片。统计H3P+阳性细胞的数量采用Image J 软件进行,按照细胞数量/面积进行分析,统计数据经SPASS 10.0软件进行分析处理。2 结果与分析
2.1 日本三角涡虫Atg6基因的鉴定和生物信息学分析
本研究采用RACE技术解析了日本三角涡虫细胞自噬基因Atg6 (命名为DjAtg6)的全长cDNA序列(GenBank登录号:MG209579)。DjAtg6 cDNA全长1366 bp,5′-末端有26 bp非翻译区,3′-末端有68 bp非翻译区并含有polyA尾巴,在polyA尾巴上游9 bp处可见AATAAA的加尾信号(图1)。经DNAstar 软件分析,该基因开放阅读框(ORF)长度1272 bp,编码423个氨基酸,分子量49 kDa。Pfam在线软件分析显示,DjATG6蛋白含有ATG6蛋白家族Coil-Coil结构域(图1,下划线显示位于104~229位氨基酸残基)和自噬功能结构域(图1,双下划线显示位于231~407位氨基酸残基)。亚细胞定位分析显示该蛋白定位于细胞质中的囊泡(图略)。Blast检索显示,DjATG6与已经报道的无脊椎动物和脊椎动物ATG6蛋白有很高的相似度(图略),说明DjATG6属于ATG6蛋白家族成员。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1日本三角涡虫Atg6基因核苷酸序列和编码区氨基酸序列
方框分别显示起始密码子、终止密码子和polyA加尾信号。下划线显示ATG6蛋白家族的标签序列Coil-Coil结构域,双下划线显示自噬功能结构域。
Fig. 1The nucleotide sequence of Atg6 gene and the amino acid sequence in the coding region from planarian Dugesia japonica
2.2 DjAtg6在涡虫再生过程中时空表达
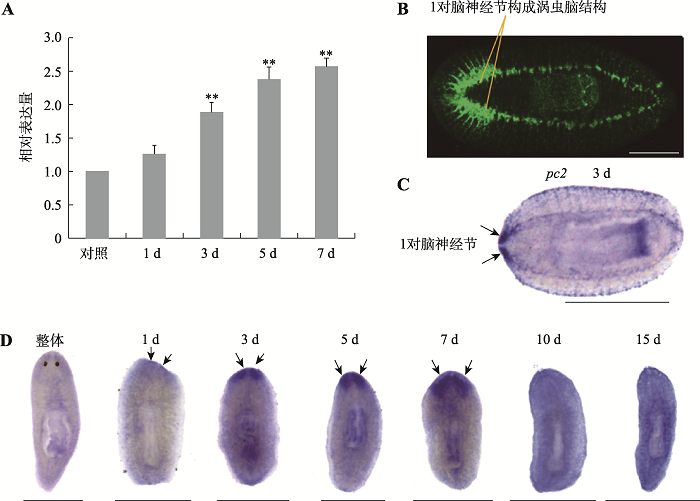
涡虫的脑神经结构简单,由1对脑神经节构成,呈倒“U”型(图2B)。为了研究DjAtg6在涡虫脑神经再生中的功能,本研究首先利用qRT-PCR方法检测了DjAtg6在涡虫再生过程中的表达变化,结果显示切割后再生3 d的DjAtg6表达量显著增加,再生5~7 d表达量维持在较高水平(图2A)。进一步用WISH技术检测了DjAtg6转录本的分布模式,结果显示涡虫再生1 d的DjAtg6阳性杂交信号开始在胚基前端出现(图2D)。涡虫再生3 d,杂交信号非常清晰的显示出1对脑神经节(图2D),与涡虫CNS marker基因pc2的表达模式非常相似(图2C)。涡虫再生5~7 d,特异性杂交信号清晰的显示出涡虫呈倒“U”型脑神经节(图2D)。涡虫再生10~15 d,特异性杂交信号减弱但脑神经节仍隐约可见(图2D)。结果表明,DjAtg6可能参与涡虫脑再生组织结构的重新构建。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2DjAtg6的时空表达模式
A:qRT-PCR 检测DjAtg6在涡虫再生过程中的表达变化。以再生0 h的涡虫为对照,收集躯干片段再生涡虫提取RNA进行检测,实验重复3 次,**P <0.01。B:anti-SYRNORF1 抗体显示涡虫脑结构:1对脑神经节组成。C:涡虫CNS marker基因pc2探针显示1对脑神经节(再生3 d)。D:整体和再生涡虫中DjAtg6表达模式。箭头显示1对脑神经节,标尺:1 mm。
Fig. 2Spatiotemporal expression pattern of DjAtg6
2.3 RNAi-DjAtg6对涡虫再生的影响
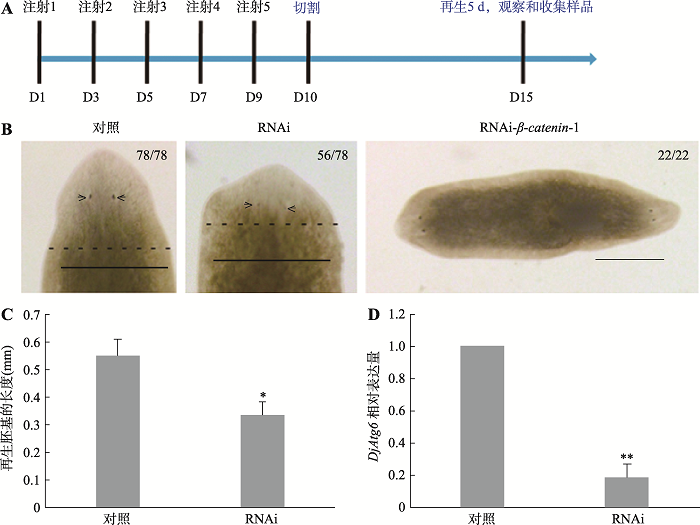
为研究DjAtg6在涡虫再生中的作用,采用RNAi技术抑制该基因的表达。结果显示,RNAi-DjAtg6导致大多数涡虫再生迟缓(图3B,56/78),形成一个较小的头部,且眼点不明显。本研究测量了对照组和RNAi组再生胚基的长度,结果表明对照组胚基长度是0.547±0.062 mm,而RNAi-DjAtg6实验组胚基长度是0.332±0.051 mm。与对照组相比,实验组新再生的胚基长度缩短了约40% (图3C)。本研究采用qRT-PCR检测了内源性DjAtg6的表达,结果显示RNAi组DjAtg6表达量降低了约80% (图3D),且干扰β-catenin-1的阳性对照组涡虫能够再生出双头涡虫表型(图 3B,22/22)。上述结果说明,本研究的干扰方法可靠,RNAi有效抑制了DjAtg6的表达。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3RNAi-DjAtg6对涡虫再生的影响
A:RNAi干扰方案和再生观察时间点示意图。B:涡虫再生(5 d)表型。56/78表示78条涡虫中有56条表现出再生迟缓表型。箭头指示眼点位置,黑色断续线显示新旧组织分界线。以RNAi-GFP为阴性对照,RNAi-β-catenin-1为阳性对照。标尺:0.5 mm。C:DjAtg6干扰组和阴性对照组胚基再生长度的比较。*P<0.05。D:qRT-PCR检测干扰后内源性DjAtg6相对表达量。GFP dsRNA 为对照,实验重复3次,**P<0.01。
Fig. 3Effects of RNAi-DjAtg6 on planarian regeneration
2.4 RNAi-DjAtg6对涡虫CNS再生和神经相关基因表达的影响
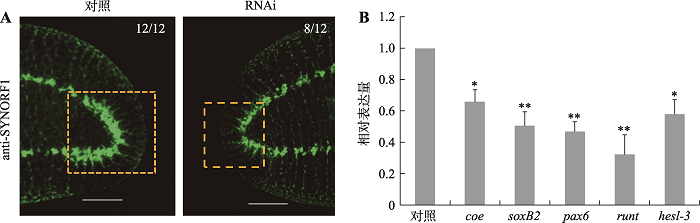
为进一步研究RNAi-DjAtg6对涡虫CNS再生的影响,本研究采用anti-SYNORF1(3C11)抗体显示涡虫脑神经结构[18]。结果如图4A所示,DjAtg6干扰组脑神经结构显著小于对照组,脑神经节较短且不明显。此外,本研究进一步采用qRT-PCR检测了几个与神经相关的转录因子的表达(图4B)。Coe基因编码的转录因子对动物CNS发育非常重要[19],干扰后其表达量下降了约30%,是对照水平的0.66倍。已有研究证明,soxB2、pax6和runt编码的转录因子对涡虫脑神经再生非常重要[10],干扰后它们的表达量分别是对照水平的0.51倍、0.47倍和0.33倍。Hesl-3编码的转录因子在涡虫脑神经节表达[10],干扰后其表达量是对照水平的0.58倍。可见,干扰DjAtg6降低了涡虫脑神经相关基因的表达。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4RNAi-DjAtg6对CNS再生和神经相关基因表达的影响
A:anti-SYNORF1免疫荧光显示涡虫脑神经结构。标尺: 200 μm。B:qRT-PCR检测神经相关基因的相对表达量。实验重复3次,*P<0.05,**P<0.01。
Fig. 4Effects of RNAi-DjAtg6 on CNS regeneration and expression of neural related genes
2.5 RNAi-DjAtg6对涡虫干细胞相关基因表达的影响
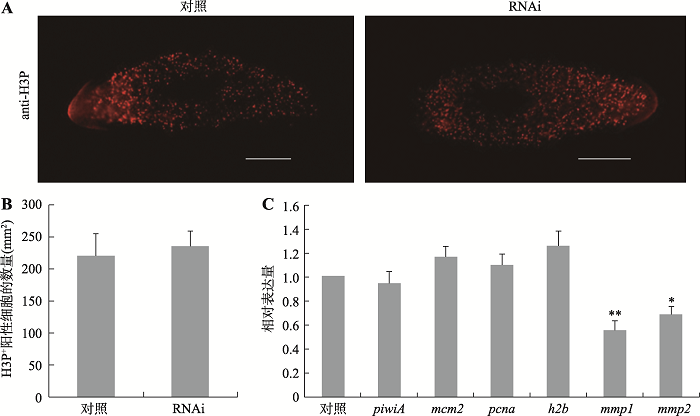
涡虫再生是以干细胞的增殖和分化为基础。为了验证干扰DjAtg6引起小头表型是否与干细胞的增殖活性降低有关,本研究首先检测了细胞有丝分裂指数的变化。H3P+阳性细胞是反映细胞有丝分裂数量的常用指标[20]。免疫荧光显示,H3P+阳性细胞的数量对照组是220±35/mm2,而实验组是235±24/mm2 (图5,A和B),两则相比没有显著性差异。本研究进一步采用qRT-PCR技术检测了涡虫干细胞相关基因的表达变化。结果显示,与对照组相比,干扰组涡虫干细胞markers基因(piwiA、mcm2、pcna、h2b)的表达量没有明显降低(图5C)。为了验证干扰DjAtg6是否影响了干细胞迁移,本研究检测了两个与细胞迁移相关的基因[16],结果显示mmp1和mmp2基因的表达量分别降低了约40%和30%,是对照水平的0.55倍和0.68倍。上述研究结果说明,干扰DjAtg6不影响干细胞的增殖活性,但可能影响干细胞的迁移和分化。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5RNAi-DjAtg6对有丝分裂指数和干细胞相关基因表达的影响
A:H3P+免疫荧光显示对照组和干扰组有丝分裂细胞。再生5 d涡虫,标尺:200 μm。B:定量统计对照组和干扰组H3P+阳性细胞数量,没有显著性差异。n=6。C:qRT-PCR检测干细胞相关基因的相对表达量。实验重复3次,*P<0.05,**P<0.01。
Fig. 5Effects of RNAi-DjAtg6 on mitotic index and expression of stem cell related genes
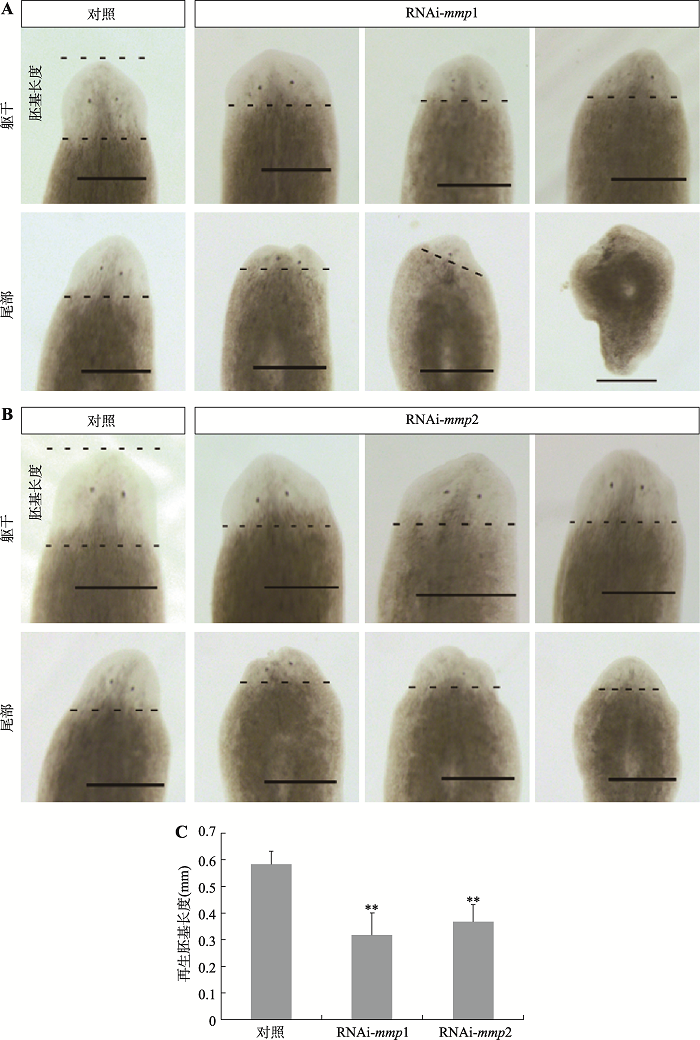
2.6 RNAi-mmp1和RNAi-mmp2对涡虫再生的影响
已有研究表明,RNAi-mmp1和RNAi-mmp2影响涡虫再生[16]。为了验证这一结果,本研究采用喂食dsRNA方法对涡虫mmp1和mmp2进行干扰实验。结果表明:RNAi-mmp1后约36%的躯干片段(7/19)头再生迟缓,约57%的尾部片段(11/19)头再生异常(图6A);RNAi-mmp2后约45%的躯干片段(10/22)头再生迟缓,约59%的尾部片段(13/22)头再生异常(图6B)。进一步测量了对照组和干扰组再生胚基的长度,结果表明对照组胚基长度是0.584±0.047 mm,RNAi-mmp1组胚基长度是0.318±0.082 mm,RNAi- mmp2组胚基长度是0.366±0.064 mm。与对照组相比,RNAi-mmp1组和RNAi-mmp2组新再生的胚基长度分别缩短了约50%和40%。本研究结果进一步证明,RNAi-mmp1和RNAi-mmp2影响涡虫再生。3 讨论
人和其他脊椎动物CNS再生能力很弱,受到创伤后很难修复。涡虫是少数CNS能够完全再生的动物。因此,涡虫CNS再生受到众多****关注并发现了大量影响涡虫CNS再生的基因[10]。涡虫CNS再生涉及到干细胞的增殖、分化、迁移等诸多细胞生物学过程。干细胞向神经前体细胞到神经细胞的转变涉及到细胞质动态的变化和不断重塑,而细胞自噬介导的蛋白质降解系统必定参与其中,但细胞自噬调控涡虫CNS再生相关的研究尚未见报道。本研究首先克隆了日本三角涡虫Atg6同源基因cDNA序列,该序列编码的蛋白质含有ATG6/Beclin 1蛋白家族的Coil-Coil结构域和自噬功能结构域[21]。其他研究表明,细胞自噬蛋白Beclin 1 (ATG6同源蛋白)参与调控神经发生[5]。因此,本文重点研究了自噬基因Atg6在涡虫CNS再生中的作用。研究发现,该基因在正常涡虫个体表达较弱,切割涡虫后其表达量随再生过程逐渐增加,且其转录本在新再生的CNS表达。由此推测,DjAtg6上调表达促进了涡虫CNS再生。干扰DjAtg6引起涡虫头部再生迟缓、脑神经节偏小,且干扰引起神经发育相关基因的表达下降。本研究结果揭示DjATG6是涡虫CNS再生的重要调控因子。但是,干扰DjAtg6并不影响涡虫干细胞的增殖和分裂,但影响与细胞迁移相关基因mmp1和mmp2的表达。本研究进一步证明,RNAi- mmp1和RNAi-mmp2影响涡虫头再生,且已有研究证明RNAi-mmp1和RNAi-mmp2影响涡虫干细胞迁移[16]。诸多研究表明,MMP通过调节细胞外基质的变化从而影响动物中枢神经发育[22,23]。值得关注的是,细胞自噬相关蛋白NBR1通过调节粘着斑的装配影响细胞迁移[24]。其他研究发现,过表达Beclin 1能够激活自噬,恢复敲降let-7引起的神经元细胞迁移障碍[25]。综合上述研究结果,本研究推测干扰DjAtg6引起涡虫CNS再生迟缓可能影响了细胞迁移,但具体的分子机制有待深入研究。总之,以涡虫为模型研究细胞自噬调控CNS再生的分子机制对理解高等动物神经发生和神经创伤修复具有重要的意义。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6RNAi-mmp1和RNAi-mmp2对涡虫头部再生的影响
A:RNAi-mmp1对涡虫躯干片段和尾部片段头部再生的影响。B:RNAi-mmp2对涡虫躯干片段和尾部片段头部再生的影响。C:干扰组和对照组胚基再生长度的比较。黑色断续线显示新旧组织分界线,标尺:500 μm,**P<0.01。
Fig. 6Effects of RNAi-mmp1 and RNAi-mmp2 on planarian head regeneration
附录:
附加材料详见文章电子版Supplementary Table 1
附表1
附表1本研究所用引物信息
Supplementary Table 1
| 引物名称 | 用途 | 序列 | 长度(bp) |
|---|---|---|---|
| 5GSP1 | 5ʹ-RACE | 5ʹ-GCTATTTCTCTATCTAACTGATC-3ʹ | 565 |
| 5GSP2 | 5ʹ-RACE | 5ʹ-CAGTAAACTCTGTTCTTCTTCTTCC-3ʹ | 512 |
| 3GSP1 | 3ʹ-RACE | 5ʹ-TACCTGAAACTCCAGTCGAATGGC-3ʹ | 562 |
| 3GSP2 | 3ʹ-RACE | 5ʹ-TTGCAGAATATTCTCAGGAAGATGG-3ʹ | 500 |
| qAtg6F | qPCR | 5ʹ-GGAACAATTATGGGAGATGC-3ʹ | 281 |
| qAtg6R | qPCR | 5ʹ-AATTCCGCCAGTAAACTCTG -3ʹ | |
| Atg6F1 | WISH | 5ʹ-GTTAATTGTAAAAAGTGTTCCTCACCGTT-3ʹ | 1212 |
| Atg6R1 | WISH | 5ʹ-ATACTGTTTGAATGATGCTTTGACC-3ʹ | |
| Atg6F2 | dsRNA | 5ʹ-ATCAACTTCAGATCATCCCATGTG-3ʹ | 946 |
| Atg6R2 | dsRNA | 5ʹ-ATACTGTTTGAATGATGCTTTGACC-3ʹ | |
| β-cateninF | dsRNA | 5ʹ-ACAACCATCGAATCTTATCCGCCAG-3ʹ | 1325 |
| β-cateninR | dsRNA | 5ʹ-CATTGTGTAACCGAATTATGTCTGT-3ʹ | |
| GFPF | dsRNA | 5ʹ-CGTGCAGTGCTTCAGCCGCTACCCC-3ʹ | 507 |
| GFPR | dsRNA | 5ʹ-AGCTCGTCCATGCCGTGAGTGATCC-3ʹ | |
| coeF | qPCR | 5ʹ-GCACCAGGAAGATTCGCATACAT-3ʹ | 284 |
| coeR | qPCR | 5ʹ-GTTAGGATTATTGGAGGCAGTAGAT-3ʹ | |
| soxB2F | qPCR | 5ʹ-AGTAAGTCCTCATTCAGCCAGT-3ʹ | 218 |
| soxB2R | qPCR | 5ʹ-CACCTGTTAGCATTCCACTCAT-3ʹ | |
| pax6F | qPCR | 5ʹ-ACGAGGTCATTCTGGAATCAATC-3ʹ | 246 |
| pax6R | qPCR | 5ʹ-ACAACTGAACTGGTAGCAACTC-3ʹ | |
| runtF | qPCR | 5ʹ-CCAATGCGAGGTGACTGACTTGAA-3ʹ | 291 |
| runtR | qPCR | 5ʹ-TGATTCTCCAATGTGAAGGTAACTG-3ʹ | |
| hesl-3F | qPCR | 5ʹ-CATCGTGAAGGAATTACCAGTC-3ʹ | 283 |
| hesl-3R | qPCR | 5ʹ-TACTCGTCTGTGCAGGATAATG-3ʹ | |
| pcnaF | qPCR | 5ʹ-AGCTACCGGAGATATTGGTAATGG-3ʹ | 168 |
| pcnaR | qPCR | 5ʹ-GAGACACGATAGGTGAAAGAGGC-3ʹ | |
| piwiAF | qPCR | 5ʹ-GGTTATTCCACAACTATTACAAGAG-3ʹ | 220 |
| piwiAR | qPCR | 5ʹ-AATCTACTTCGTCATTGATATCC-3ʹ | |
| mcm2F | qPCR | 5ʹ-GAGGAGGAGAAGAAGGATGT-3ʹ | 161 |
| mcm2R | qPCR | 5ʹ-GCTGTGCTCAAACTGGGACT-3ʹ | |
| mmp1F1 | qPCR | 5ʹ-ATGGCTGGAATAGAACAAGATGG-3ʹ | 202 |
| mmp1R1 | qPCR | 5ʹ-GACGAACTTCTCCTTCAGACATAG-3ʹ | |
| mmp2F1 | qPCR | 5ʹ-GAGCCTTAATAGTCGGTCTTCAAT-3ʹ | 259 |
| mmp2R2 | qPCR | 5ʹ-TCCTTCGGTCCATTCTTCAGCTG-3ʹ | |
| mmp1F2 | dsRNA | 5ʹ-TATGTCTGAAGGAGAAGTTCGTCG-3ʹ | 850 |
| mmp1R2 | dsRNA | 5ʹ-ATCGTGATACGAACTTTGTCTTGC-3ʹ | |
| mmp2F2 | dsRNA | 5ʹ-TGAGTTTTGCCGATGCTGAACACG-3ʹ | 746 |
| mmp2R2 | dsRNA | 5ʹ-GTCTTATCTCTCACGATTGCTGCG-3ʹ | |
| DjEF2F | qPCR | 5ʹ-TTAATGATGGGAAGATATGTTG-3ʹ | 250 |
| DjEF2R | qPCR | 5ʹ-GTACCATAGGATCTGACTTTGC-3ʹ |
新窗口打开|下载CSV
致谢:
感谢中国科学院上海营养与健康研究所荆清教授和清华大学吴畏教授在涡虫研究技术方面给予的帮助。
(责任编委: 林古法)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
PMID:27672021 [本文引用: 1]

Autophagy, a pathway for lysosomal-mediated cellular degradation, has recently been described as a regulator of cell migration. Although the molecular mechanisms underlying autophagy-dependent motility are only beginning to emerge, new work demonstrates that selective autophagy mediated by the autophagy cargo receptor, NBR1, specifically promotes the dynamic turnover of integrin-based focal adhesion sites during motility. Here, we discuss the detailed mechanisms through which NBR1-dependent selective autophagy supports focal adhesion remodeling, and we describe the interconnections between this pathway and other established regulators of focal adhesion turnover, such as microtubules. We also highlight studies that examine the contribution of autophagy to selective degradation of proteins that mediate cellular tension and to integrin trafficking; these findings hint at further roles for autophagy in supporting adhesion and migration. Given the recently appreciated importance of selective autophagy in diverse cellular processes, we propose that further investigation into autophagy-mediated focal adhesion turnover will not only shed light onto how focal adhesions are regulated but will also unveil new mechanisms regulating selective autophagy.© 2016. Published by The Company of Biologists Ltd.
DOI:10.1007/s00018-012-1032-3URL [本文引用: 1]
DOI:10.1016/j.tins.2020.07.003URL [本文引用: 1]
DOI:10.1073/pnas.1611282113URL [本文引用: 1]
DOI:10.1038/cddis.2014.358URL [本文引用: 2]
DOI:10.1172/JCI33585PMID:18497889 [本文引用: 1]

Autophagy is the principal cellular pathway for degradation of long-lived proteins and organelles and regulates cell fate in response to stress. Recently, autophagy has been implicated in neurodegeneration, but whether it is detrimental or protective remains unclear. Here we report that beclin 1, a protein with a key role in autophagy, was decreased in affected brain regions of patients with Alzheimer disease (AD) early in the disease process. Heterozygous deletion of beclin 1 (Becn1) in mice decreased neuronal autophagy and resulted in neurodegeneration and disruption of lysosomes. In transgenic mice that express human amyloid precursor protein (APP), a model for AD, genetic reduction of Becn1 expression increased intraneuronal amyloid beta (Abeta) accumulation, extracellular Abeta deposition, and neurodegeneration and caused microglial changes and profound neuronal ultrastructural abnormalities. Administration of a lentiviral vector expressing beclin 1 reduced both intracellular and extracellular amyloid pathology in APP transgenic mice. We conclude that beclin 1 deficiency disrupts neuronal autophagy, modulates APP metabolism, and promotes neurodegeneration in mice and that increasing beclin 1 levels may have therapeutic potential in AD.
DOI:10.1046/j.1440-169x.2002.00629.xURL [本文引用: 1]
DOI:10.1126/science.1202143URL [本文引用: 1]
DOI:10.1242/dev.098616URL [本文引用: 1]
DOI:10.7554/eLife.17002URL [本文引用: 4]
PMID:19164934 [本文引用: 1]

This review aims to demonstrate the importance of freshwater planarians as model organisms, particularly emphasizing those characteristics of the animal that make them a good model to study autophagy. The aim of this review is to provide a better understanding of autophagy in this model for the nonplanarian reader, and elucidate the relevance of autophagy research in this peculiar model organism. Furthermore, I will try to synthesize the evidence showing the importance of autophagy in planarian body remodeling, and I will discuss some ideas about the role of autophagy in stem cell biology. In light of these new developments, it is likely that the planarian field will make an important contribution to the study of the molecular mechanisms involved in autophagy in the future.
PMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.Copyright 2001 Elsevier Science (USA).
[本文引用: 1]
DOI:10.1073/pnas.96.9.5049URL
DOI:10.1371/journal.pgen.1004452URL
DOI:10.1371/journal.pone.0055649URL [本文引用: 4]
DOI:10.1002/dvdy.v242.6URL [本文引用: 1]
DOI:10.1242/dev.01941URL [本文引用: 2]
DOI:10.1371/journal.pgen.1004746URL [本文引用: 1]
DOI:10.1038/cr.2011.151URL [本文引用: 1]
DOI:10.1002/pro.v25.10URL [本文引用: 1]
DOI:10.1016/j.neulet.2020.134822URL [本文引用: 1]
DOI:10.1016/j.mcn.2015.03.015PMID:25827096 [本文引用: 1]

Matrix metalloproteinases (MMPs) are a family of highly conserved zinc-dependent proteases involved in both development and pathogenesis. The present study examines the role of MMP-2 (gelatinase A) and MMP-9 (gelatinase B) in adult neurogenesis, using the corpus cerebelli, a subdivision of the cerebellum, of knifefish (Apteronotus leptorhynchus) as a model system. Transcripts of five isoforms of these gelatinases were identified in the central nervous system of this species. Sequence similarity analysis and homology modeling indicated that functionally and structurally critical elements were highly conserved in knifefish gelatinases. Immunohistochemical staining revealed a differential distribution of MMP-2 and MMP-9 at both the cellular and subcellular level. MMP-2 expression was found mainly in Sox2-immunopositive stem/progenitor cells, both quiescent and mitotically active; and was localized in both the cytoplasmic compartment and the nucleus. By contrast, MMP-9 immunoreactivity was absent in neurogenic niches and displayed a more homogenous distribution, with low to moderate intensity levels, in the molecular and granular layers. MMP-9 expression appeared to be restricted to the extracellular space. In situ zymography indicated that gelatinase activity matched the cellular and subcellular distributions of the two MMPs. The observed patterns of gelatinase activity and expression support the hypothesis that MMP-2 is primarily involved in regulation of the activity of stem/progenitor cells that give rise to new granule neurons, whereas MMP-9 facilitates migration of the progeny of these cells by proteolysis of extracellular matrix proteins. Copyright © 2015 Elsevier Inc. All rights reserved.
PMID:27484104 [本文引用: 1]

Macroautophagy/autophagy has classically been recognized for its vital role in supporting cellular survival during various stresses. However, emerging work has demonstrated that selective autophagy has an impact on diverse cell biological processes by mediating the degradation of various cellular contents during normal cellular homeostasis. We recently established that selective autophagy supports cell migration by promoting the turnover of integrin-based cell-matrix adhesion sites, or focal adhesions (FAs). The autophagy cargo receptor NBR1 acts as a critical mediator of this pathway by promoting targeting of autophagosomes to FAs, leading to their disassembly via the sequestration of FA proteins. Our results demonstrate FAs as a new cellular target for selective autophagy.
DOI:10.15252/embj.201695235URL [本文引用: 1]
