 ,, 李金环, 黄海燕
,, 李金环, 黄海燕 ,, 吴强
,, 吴强 ,上海交通大学系统生物医学研究院比较生物医学研究中心,系统生物医学教育部重点实验室,上海 200240
,上海交通大学系统生物医学研究院比较生物医学研究中心,系统生物医学教育部重点实验室,上海 200240Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers
Ling Wang ,, Jinhuan Li, Haiyan Huang
,, Jinhuan Li, Haiyan Huang ,, Qiang Wu
,, Qiang Wu ,Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China
,Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China通讯作者: 黄海燕,博士,副研究员,研究方向:药物分子遗传学。E-mail:hy_huang@sjtu.edu.cn吴强,博士,教授,研究方向:基因表达调控及神经发育。E-mail:qwu123@gmail.com
编委: 李大力
收稿日期:2021-04-11修回日期:2021-05-12网络出版日期:2021-08-20
| 基金资助: |
Received:2021-04-11Revised:2021-05-12Online:2021-08-20
| Fund supported: |
作者简介 About authors
王玲,硕士研究生,专业方向:生物学。E-mail:

摘要
三维基因组染色质架构蛋白CTCF (CCCTC-binding factor)能够介导增强子与基因启动子的远距离染色质相互作用,也可以结合调控区域的绝缘子发挥增强子绝缘功能,对发育中的基因表达调控具有重要作用。同源框基因家族(Homeobox gene family, Hox)编码一类控制动物发育的关键转录因子,在发育中主要沿胚胎首尾轴(head-to-tail axis)呈时空线性表达。在哺乳动物中,Hox基因分为HoxA、HoxB、HoxC和HoxD四个基因簇,在中枢神经系统、骨骼和四肢发育中发挥重要功能。HoxD基因簇主要调控四肢发育,受位于其两侧调控域内的增强子调节,沿肢体近远轴(proximal-distal axis)呈时空线性表达。在人类基因组中,HOXD基因簇及其两侧的调控区域分布有串联排列的CTCF结合位点(简称CTCF位点),参与9个HOXD基因的表达调控。本研究以HOXD基因簇为模式基因,探究CTCF对发育基因(developmental genes)转录调控的影响。利用CRISPR DNA片段编辑技术在人HEK293T细胞中获得一系列的串联反向CTCF位点删除的单细胞克隆株。RNA-seq实验揭示CTCF位点删除后HOXD基因表达下降。定量高分辨率染色体构象捕获实验显示,HOXD与上游增强子簇的远距离染色质相互作用增强,与下游增强子簇的远距离染色质相互作用减弱。综上所述,串联反向的CTCF位点通过其绝缘子功能维持上下游增强子簇对HOXD基因簇表达调控的平衡,为探究动物发育过程中Hox基因表达的精准调控机制提供参考。
关键词:
Abstract
The genome architectural protein CTCF (CCCTC-binding factor) not only mediates long-distance chromatin interactions between distal enhancers and target promoters, but also functions as an important insulator-binding factor to block improper enhancer activation of non-target promoters, and is thus of great significance to transcriptional regulation of developmental genes. The Hox (Homeobox) gene family plays an important role in the development of the brain, bones, and limbs. The spatiotemporal colinear expression of the HOXD cluster along the proximal-distal axis of limbs is regulated by two clusters of enhancers known as super-enhancers located in the flanking regulatory regions. We focused on the HOXD cluster to explore the architectural role of CTCF in transcriptional regulation of developmental genes. The HOXD cluster contains 9 paralogous genes intermixed with a series of CBS (CTCF-binding site) elements. Using the CRISPR DNA-fragment editing system, we generated a series of single-cell HEK293T clones with deletion of increasing numbers of reverse CBS elements. RNA-seq experiments revealed decreased levels of HOXD gene expression. In addition, chromosome conformation capture experiments revealed increased long-distance chromatin interactions between HOXD and the upstream enhancer cluster and corresponding decreased interactions between HOXD and the downstream enhancer cluster. Thus, tandem reverse CTCF sites function as insulators to maintain HOXD regulatory balance between the upstream and downstream enhancer clusters. This study has interesting implications on the precise gene expression control of the Hox family during animal development.
Keywords:
PDF (1959KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡. 遗传[J], 2021, 43(8): 775-791 doi:10.16288/j.yczz.21-132
Ling Wang.
染色质架构蛋白CTCF(CCCTC-binding factor)是一类结合DNA的锌指蛋白,在哺乳动物中可与基因组上调控区域内大量的绝缘子结合,对发育基因的精准调控起到关键作用[1,2,3,4,5,6]。CTCF最初在鸡中被发现是一种转录抑制因子或共抑制因子[7],CTCF结合位点(CTCF-binding site,CBS)序列在脊椎动物中高度保守,超过30多万的CBS广泛分布在整个基因组中[8]。CTCF与cohesin复合物通过环挤出模型(loop extrusion model)共同介导远距离染色质相互作用,参与基因的调控[3,9,10]。CBS序列的方向性直接影响染色质远距离互作,分别被正向和反向CBS元件锚定的增强子和启动子在CTCF/cohesin介导下形成远距离染色质环[2,3,5,11~14]。成对的反向-正向CBS (背靠背)通常分布在拓扑相关结构域(topological- associated domain, TAD)边界,发挥绝缘子的作用,阻止增强子跨区域异位激活基因表达[3]。CTCF除了能与cohesin一起介导染色质环,还能与其他的非编码调控元件及架构蛋白(如YY1)共同搭建三维基因组结构[1,15,16]。CTCF参与调控的染色质三维空间结构是生物体发挥正常生理功能的基础,包括神经系统中原钙粘蛋白(protocadherin, Pcdh)基因簇启动子的选择[4,13,14,17]、尿苷二磷酸葡萄糖醛酸转移酶1 (UDP-glucuronosyltransferase 1, UGT1)的表达[12]、免疫球蛋白(Immunoglobulin, Ig)和T细胞受体(T cell receptor, Tcr)的V(D)J重组[18,19,20]、四肢发育过程中的同源框基因家族(homeobox gene family, Hox)控制[21,22,23]以及生物学的其他方面[1,24]。
脊椎动物和果蝇Hox基因簇内的CTCF结合位点经过数亿年进化仍然存在,这暗示了在两侧对称动物进化历程中CTCF参与Hox基因表达调控的古老起源[25]。Hox基因簇编码的转录因子调控着细胞分化的多个方面,影响生物体四肢发育、器官形成等[26]。在哺乳动物中,Hox基因分布在四个簇中,分别命名为HoxA、HoxB、HoxC和HoxD,根据序列同源性及在簇中的位置,Hox基因分为13组(Hox1~Hox13) (图1A),躯干和四肢的不同区域表达Hox基因的不同组合[27]。在发育的最初阶段,Hox基因簇不表达,在发育过程中主要沿胚胎首尾轴(head-to-tail axis)呈时空线性表达[28]。
HoxD基因簇包括HoxD1、HoxD3、HoxD4、HoxD8~HoxD13,整体位于两个拓扑结构域边界,受上游增强子簇(upstream enhancer cluster)和下游增强子簇(downstream enhancer cluster)调控(图1,A和B)。在小鼠胚胎肢芽(limb bud)发育早期,位于基因簇3′端粒侧TAD(telomeric domain, T-DOM)处于激活状态,控制HoxD1~HoxD11在四肢的近端转录[29];随后,在胚胎肢芽(limb bud)远端细胞中,位于5′中心粒侧TAD (centromeric domain, C-DOM)处于激活状态,负责HoxD9~HoxD13在手指细胞中的表达[30,31]。在小鼠胚胎肢芽中,上游增强子簇中的5个增强子(Island1~5)包含许多保守的非编码调控元件,同时富集有H3K4me1、H3K27ac(活性增强子标记),与HoxD10~HoxD13有很强的互作[31]。删除部分增强子只会影响它们与HoxD10~HoxD13基因的互作,但对HoxD10~HoxD13基因的表达影响不大;当完全删除上游增强子簇,HoxD10~HoxD13基因表达才会完全消失。这些研究表明这些Island之间可能存在冗余性和互补性[31]。在小鼠生殖器结节(genital tubercle,GT)中,上游增强子簇中的GT1和GT2与HoxD13启动子也有远距离DNA相互作用,但GT2仅在生殖器中表现出较强的活性,在肢端细胞中不能显著激活HoxD基因转录[32]。在小鼠肢芽近端细胞中,下游增强子簇中的CS38-41 (conserved sequence element 38-41)参与调控HoxD1~HoxD11的表达[21]。这些上下游增强子簇通过转录因子或架构蛋白介导的染色质环与HoxD基因启动子实现远程互作,从而完成特定的HoxD基因调控[33,34,35]。然而,目前对于HoxD基因簇调控机制的研究主要借助于在小鼠中对其调控区域或者基因进行大片段删除或反转[21,22,31,32,36],对于人细胞中HOXD基因簇调控机制的研究较少。
本研究以人HOXD基因簇为模式基因,探索CTCF对发育基因(developmental genes)转录调控的影响。利用CRISPR DNA片段编辑系统[37,38,39,40]在人HEK293T细胞中获得一系列的串联反向CBS删除的单细胞克隆株。进行RNA-seq实验检测CBS删除后HOXD基因表达水平的变化,最后通过定量高分辨率染色体构象捕获(quantitative high resolution chromosome conformation capture copy, QHR-4C)[13]进一步观测CBS删除对HOXD基因簇三维基因组结构的影响,发现成串反向排列的CTCF位点具有平衡上下游增强子簇对HOXD基因簇转录调控的作用,为揭示哺乳动物HoxD基因转录的复杂调控提供参考。
1 材料与方法
1.1 实验材料
人胚胎肾细胞系HEK293T从中国科学院细胞库购买;胰酶、青霉素/链霉素双抗购自美国Gibco公司;DMEM培养基购自美国HyClone公司;胎牛血清购自南美ExCell公司;Lipofectamine 3000购自美国Invitrogen公司;pcDNA3.1-Cas9-WT质粒来源于北京大学席建忠教授;BsaⅠ限制性内切酶、T4 DNA连接酶、DpnⅡ限制性核酸内切酶、RNA文库制备试剂盒、Q5 Hot Start HiFi PCR Master Mix均从美国NEB公司采购;pGL3-U6-sgRNA-Puromycin- BsaⅠ质粒由上海科技大学黄行许教授赠予;质粒小量提取试剂盒、DNA凝胶回收试剂盒均从美国AXYGEN公司采购;MinElute Gel Extraction kit从德国QIAGEN公司采购;Green Taq Mix、dNTPs、高保真Phanta DNA聚合酶购自南京诺唯赞生物科技有限公司;pClone007 Versatile Simple Vector购自北京擎科新业生物技术有限公司;sgRNA、测序引物由上海生物工程(上海)股份有限公司合成;Triton X-100、10%SDS、DNA提取液、NP40等从美国SIGMA公司采购;三氯甲烷、硼酸和乙醇购自上海沪试实验室器材股份有限公司;异丙醇和氯化钠购自上海凌峰化学试剂有限公司;链霉亲和素磁珠从美国Invitrogen公司采购;PCR文库纯化试剂盒从瑞士Roche公司采购;甲醛溶液、RNA酶A、糖原等均从美国Thermo公司采购;TRIzol Reagent从美国Ambion公司采购;AMPure XP Beads从美国Beckman Coulter公司采购;CTCF抗体从英国Abcam公司采购。图1
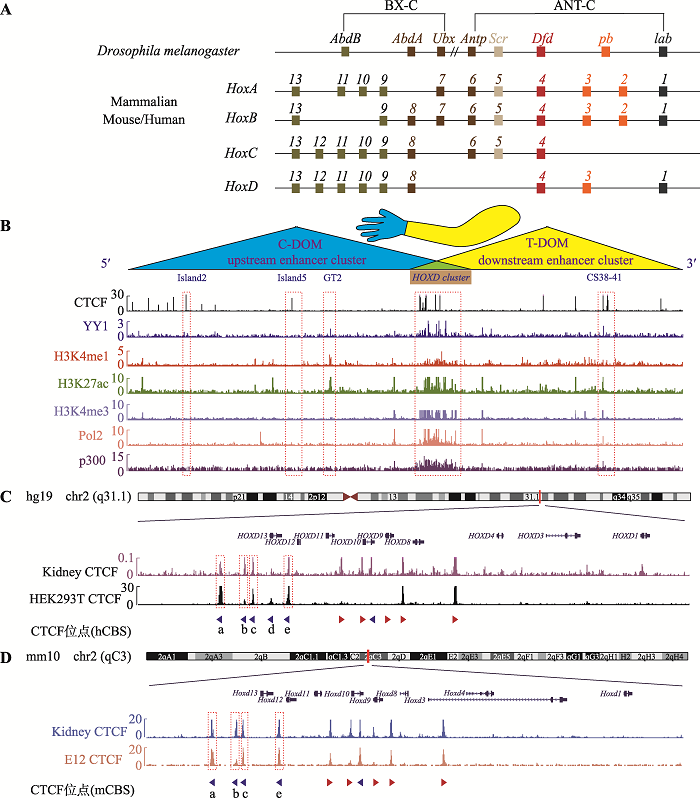 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CTCF位点在Hox基因簇及其调控区域内的分布
A:果蝇及哺乳动物Hox基因家族的基因组结构。果蝇Hox基因簇分为触足复合群(ANT-C)和双胸复合群(BX-C),其中ANT-C包含Antp、Scr、Dfd、pb和lab基因,BX-C包含AbdB、AbdA和Ubx基因。哺乳动物具有4个Hox基因簇:HoxA、HoxB、HoxC和HoxD,共包含39个基因,其中HoxD基因簇包括HoxD1、HoxD3、HoxD4、HoxD8~HoxD13。果蝇及哺乳动物相同颜色的Hox基因为直系同源基因(orthologues),它们起源于同一祖先基因。B:人胚胎肾细胞系HEK293T中的CTCF蛋白、架构蛋白YY1、增强子标记H3K4me1和H3K27ac、启动子标记H3K4me3、转录活性标记PolⅡ和p300在人HOXD基因簇及其调控区域内的分布。红色虚线框指示增强子Island2、Island5、GT2、CS38-41和HOXD基因簇所在区域。哺乳动物的HoxD基因簇位于3′端粒侧TAD (T-DOM)和5′中心粒侧TAD (C-DOM)交界处,并受到上下游增强子簇(upstream enhancer cluster和downstream enhancer cluster)的调控。C:人类HOXD基因簇区域CTCF位点的分布。人肾脏和胚胎肾细胞系HEK293T的CTCF ChIP-seq结合峰分布图:HOXD基因簇C-DOM和T-DOM交界区域均具有串联排列的CTCF位点。红色虚线框依次指示CBS a、b、c和e所在区域。D:小鼠HoxD基因簇区域CTCF位点的分布。小鼠肾脏和第12.5天胚胎肢芽CTCF ChIP-seq数据显示小鼠HoxD基因簇中心粒侧对应的位置具有串联反向排列的4个CTCF位点。图C和D中箭头代表CTCF位点,其中红色箭头代表正向CTCF位点,蓝色箭头代表反向CTCF位点。
Fig. 1Distribution of CTCF sites in the Hox cluster and its regulatory landscapes
1.2 细胞培养
HEK293T细胞的培养基为89% DMEM完全培养液、10%胎牛血清和1%青霉素/链霉素双抗的混合液,在37℃、5% CO2的恒温培养箱中培养。1.3 HEK293T细胞CTCF的ChIP-seq
收集2×107细胞后,首先用1%甲醛交联细胞,再加入终浓度为0.125 mol/L的甘氨酸溶液终止反应,然后用预冷的ChIP缓冲液裂解细胞两次(裂解条件均为4℃,缓慢旋转10 min),离心后再用预冷的ChIP缓冲液重悬细胞,冰上孵育10 min。用非接触式超声仪进行超声,凝胶电泳鉴定超声后DNA片段长度为100~10,000 bp。将细胞悬液离心后的上清转移至新的离心管中并加入50 µL的protein A/G磁珠孵育2 h。用磁力架弃去磁珠,将上清转移至新的离心管中加入4 µg的CTCF抗体,4℃缓慢旋转孵育过夜。第二天加入50 µL的protein A/G磁珠,4℃缓慢旋转孵育3 h。然后用ChIP缓冲液、高盐缓冲液、无盐缓冲液、LiCl缓冲液清洗磁珠(每次4℃缓慢旋转孵育10 min),最后用洗脱缓冲液洗脱DNA。含有DNA的溶液经过RNA酶A (37℃震荡孵育2 h)、蛋白酶K (55℃静置孵育2 h)及酚氯仿试剂纯化后,DNA沉淀用无核酸酶的水溶解,最后用诺唯赞公司的ChIP-seq试剂盒ND607-02构建DNA文库。文库通过苏州金唯智生物科技有限公司的Illumina HiSeq平台进行测序,根据index序列(P7-index序列见表1)对数据进行拆分,通过Bowtie2比对到GRCh37/hg19基因组,用BamCoverage进行归一化处理,标准化到RPKM (reads per kilobase per million mapped reads),共做两个重复,每个样品文库测序约2000万条序列。Table 1
表1
表1引物序列
Table 1
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| PCR | a1F | TTCCAGCACCTCGGCTTTGTC |
| a1R | CCCACTTTCCACCTCTGTCCTG | |
| b1F | GTCCGCCCGTGAGCTTCTGAA | |
| b1R1 | CTCACAGCAGCCGAAACCG | |
| c1F | TGATGCAGCCTCTGTGACCG | |
| c1R | AGTTTTCCCGTGGCGTCTGA | |
| e1F | TTCCCTGTCCCAGCTTGATTTC | |
| e1R | TCAACAGTGAAGGGCGGTGC | |
| e2F | CAAGCCACTCTCCCGCCACTA | |
| e2R | TCGCTCTCGTCCTCTCTTGGG | |
| e2R1 | GGCTCCTGCACTGAGACCACA | |
| sgRNA | sgRNA a1F | ACCGCGAAGAGTGCGGGAGAACGG |
| sgRNA a1R | AAACCCGTTCTCCCGCACTCTTCG | |
| sgRNA b1F | ACCGGGCGCATCAGGAATGTAAG | |
| sgRNA b1R | AAACCTTACATTCCTGATGCGCC | |
| sgRNA c1F | ACCGCAGGCGAAGTGCGGTTTCCA | |
| sgRNA c1R | AAACTGGAAACCGCACTTCGCCTG | |
| sgRNA e1F | ACCGAACTGTGCTCAAACGCTCTC | |
| sgRNA e1R | AAACGAGAGCGTTTGAGCACAGTT | |
| sgRNA e2F | ACCGGAGGCGCAAACAGCTGTTGT | |
| sgRNA e2R | AAACACAACAGCTGTTTGCGCCTC | |
| Index | Island5-bioprimer | 5′biotin-AAACACAAATGCATCAACCTG |
| GT2-bioprimer | 5′biotin-GAGCCAAACTGTACCCCTAGC | |
| HOXD9-bioprimer | 5′biotin-ACCGACTAGTTCGCAGGCT | |
| Island5-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCATCTCATGAAGCTGGCATCT | |
| GT2-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTAACTTTAGCTAAACCAAGGCCT | |
| HOXD9-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTGCAGCCTCCACCATTG | |
| P7-index-1 | CAAGCAGAAGACGGCATACGAGATCGAGTAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-2 | CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-3 | CAAGCAGAAGACGGCATACGAGATAATGAGCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-4 | CAAGCAGAAGACGGCATACGAGATGGAATCTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-5 | CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-6 | CAAGCAGAAGACGGCATACGAGATACGAATTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-7 | CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-8 | CAAGCAGAAGACGGCATACGAGATGCGCATTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-9 | CAAGCAGAAGACGGCATACGAGATCATAGCCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-10 | CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-11 | CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-12 | CAAGCAGAAGACGGCATACGAGATCTATCGCTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-13 | CAAGCAGAAGACGGCATACGAGATAGAGTACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-14 | CAAGCAGAAGACGGCATACGAGATGCTCCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-15 | CAAGCAGAAGACGGCATACGAGATCATGAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-16 | CAAGCAGAAGACGGCATACGAGATTGAATCGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-17 | CAAGCAGAAGACGGCATACGAGATGTCTGACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-18 | CAAGCAGAAGACGGCATACGAGATCTGAATGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-19 | CAAGCAGAAGACGGCATACGAGATCGCTTCTGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-20 | CAAGCAGAAGACGGCATACGAGATTCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-21 | CAAGCAGAAGACGGCATACGAGATAATAGCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-22 | CAAGCAGAAGACGGCATACGAGATGTCGCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-23 | CAAGCAGAAGACGGCATACGAGATACGCGATAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-24 | CAAGCAGAAGACGGCATACGAGATTGATCGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-25 | CAAGCAGAAGACGGCATACGAGATCCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-26 | CAAGCAGAAGACGGCATACGAGATCCACAATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-27 | CAAGCAGAAGACGGCATACGAGATGATGTTCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
新窗口打开|下载CSV
1.4 HEK293T细胞中组蛋白修饰、转录活性标志及CTCF的ChIP-seq数据分析
人胚胎肾细胞系HEK293T的H3K4me1和H3K27ac ChIP-seq数据来源于ENCODE数据库,数据编号分别为ENCSR000FCG和ENCSR000FCH;YY1、H3K4me3、PolⅡ、p300来源于GEO,数据编号分别为GSM3636215、GSM945288、GSM935534、GSM1239071;人肾脏CTCF ChIP-seq数据的GEO编号为GSM1006886;小鼠肾脏及第12.5天胚胎肢芽CTCF ChIP-seq数据的GEO编号分别为GSE91529、GSM4665698;将数据分别比对到GRCh37/hg19及GRCm38/mm10基因组进行分析。1.5 构建pGL3-U6-sgRNA质粒
按照表1合成单链sgRNA (共10条,分别合成双链sgRNA a1、sgRNA b1、sgRNA c1、sgRNA e1和sgRNA e2),按照如下体系(总体积为20 µL,)进行退火步骤:2 µL sgRNA-F,2 µL sgRNA-R,2 µL 10 × NEB Buffer 2,14 µL ddH2O;退火条件:95℃预变性5 min,缓慢降温(从第二个循环开始,每个循环20 s、降温0.2℃,共351个循环)。环状pGL3- U6-sgRNA-Puromycin-BsaⅠ质粒经过BsaⅠ限制性核酸内切酶处理后,得到线性的pGL3-U6-sgRNA- Puromycin载体。将退火后的双链sgRNA引物与线性化的pGL3-U6-sgRNA-Puromycin载体在室温进行连接,经感受态细胞转化培养后,挑取单菌落进行培养(每种sgRNA至少挑取3个单菌落),用质粒小量提取试剂盒纯化质粒并测序。1.6 细胞转染与单克隆化
待12孔板内的HEK293T细胞汇合度达到70%~ 90%左右时进行转染,一个离心管中加入Lipofectamine 3000与MEM混合均匀室温静置5 min,另一个管中加入Cas9质粒、两种sgRNA质粒、p3000试剂和MEM培养基混合均匀后室温静置5 min (此步骤中sgRNA组合:sgRNA a1/sgRNA e2或sgRNA b1/sgRNA e2或sgRNA c1/sgRNA e2或sgRNA e1/sgRNA e2),然后将Lipofectamine 3000加入其中,混合均匀后室温孵育约15 min,滴入培养细胞中。待转染时间达到48 h后,换用含有2 μg/mL嘌呤霉素的培养基对转染细胞进行药物筛选。药物筛选大约4天后,更换为DMEM完全培养基继续培养2天,然后用胰酶消化收集适量细胞至PCR管中,加入10 µL碱裂解液,混匀置于PCR仪中,98℃ 30 min使细胞裂解,再加入10 µL中和液,获得细胞基因组。在sgRNA上下游位置设计PCR引物,鉴定转染后的细胞中是否含有目的片段删除的细胞,若存在,则对12孔板中剩余的转染细胞进行计数,用培养基稀释至浓度约为1个细胞每100 µL培养液,转移至96孔板中培养。待细胞生长一周左右,显微镜下查看单一细胞团并做好标记。sgRNA和PCR引物序列见表1。
1.7 单细胞克隆株鉴定
将标记的单细胞克隆株(共1056个)用胰酶消化,转移适量细胞到含有10 µL碱裂解液的PCR小管中进行裂解反应获取细胞基因组。两个sgRNA对大片段DNA进行删除时,会造成两个切口,需要通过特异性引物鉴定排除片段反转的情况[37,41]。通过PCR反应(PCR引物序列见表1)及DNA琼脂糖凝胶电泳实验挑选出目的片段删除的纯合子单细胞克隆株,将细胞目的片段删除型的扩增产物通过凝胶电泳分离,切胶回收并送测。若测序结果为双峰,则需要进行TA克隆测序鉴定单细胞克隆株的基因型。按照TA克隆产品推荐用量将PCR产物与pClone007 Versatile Simple Vector混合均匀,室温孵育约10 min;在离心管中将连接产物与50 µL感受态细胞混合均匀,冰上静置30 min后,进行42℃热激45 s,然后迅速转移至冰上,2 min后加入500 µL的LB无抗培养基,然后将离心管放在37℃摇床上培养1 h。离心后去除450 µL上清,重悬细胞后均匀涂布在含有氨苄抗性的LB固体培养基上,37℃恒温箱中培养约16 h,次日挑取8~10个单菌落测序,然后将桑格测序结果与目的片段序列比对,确定单细胞克隆株基因型。最终获得CBS e删除、CBS c-e删除、CBS b-e删除和CBS a-e删除的单细胞克隆株各两株。
1.8 RNA-seq实验
待12孔板中处于对数生长期的HEK293T细胞、单细胞克隆株汇合度达到90%时,先用1 × PBS清洗2遍,然后加入500 µL/孔TRIzol溶液处理15 min,然后转移至无核酸酶的离心管中,用三氯甲烷、异丙醇、乙醇提取纯化总RNA。根据试剂说明书进行如下操作:使用Poly(A) mRNA Magnetic Isolation Module试剂盒提取mRNA,利用磁珠对mRNA片段进行分离与片段化;通过PCR反应先进行cDNA第一链合成,再进行cDNA第二链合成反应;双链cDNA连接Adaptor之后,用AMPure XP Beads和新鲜配制的80%的乙醇对DNA进行纯化,进行文库扩增反应;扩增后产物用AMPure XP Beads和新鲜配制的80%的乙醇进行纯化,最后用无核酸酶的水洗脱DNA文库。用Qubit3 Fluorometer测量终产物浓度后送至苏州金唯智生物科技有限公司的Illumina HiSeq平台进行测序,根据特异性index序列(表1)拆分数据,用STAR、cufflinks等软件分析数据[42]。本研究共有30个RNA-seq样品,每个样品文库测序约1000万条序列。1.9 定量高分辨率染色体构象捕获实验QHR-4C
用10 cm培养皿扩增培养HEK293T细胞、单细胞克隆株,待细胞汇合度达到80%~90%且生长良好时,用胰酶消化离心收集细胞;用1%甲醛交联细胞,再加入终浓度为0.125 mol/L的甘氨酸溶液终止反应;用预冷的1 × PBS重复清洗细胞,然后配制4C裂解液(1 × inhibitor、pH 7.5 50 mmol/L Tris-HCl、150 mmol/L NaCl、5 mmol/L EDTA、0.5% NP-40和1% Triton X-100)重复裂解细胞;通过Triton X-100、DpnⅡ限制性内切酶处理使DNA片段化,用T4 DNA连接酶捕捉相互邻近的DNA片段;纯化连接后的DNA,然后进行超声处理(不同长度的DNA片段经琼脂糖凝胶电泳鉴定呈弥散状分布且主要片段长度小于1000 bp)、PCR线性扩增反应及接头连接反应,最后进行DNA文库扩增。文库送至苏州金唯智生物科技有限公司的Illumina HiSeq平台进行测序,根据index(序列见表1)对测序结果进行拆分,通过Bowtie2、Samtools、r3C-seq等方法处理数据[13]。本研究每个实验三个重复,共有78个4C样品,每个样品文库测序约800万条序列。1.10 高通量测序数据信息
本研究的相关结果数据已收录在国家基因库生命大数据平台(CNGBdb)[43]的国家基因库序列归档系统(CNSA)[44],项目编号:CNP0001773。2 结果与分析
2.1 CTCF位点在HOXD基因簇的分布高度保守
Hox基因簇最初因引起黑腹果蝇翅膀数目改变而被发现,在两侧对称动物中高度保守[45]。果蝇基因组中含有一个Hox基因簇,由8个基因组成,分为触足复合群(ANT-C)和双胸复合群(BX-C),其中ANT-C包含Antp、Scr、Dfd、pb和lab基因,BX-C包含AbdB、AbdA和Ubx基因(图1A)。而哺乳动物基因组具有四个Hox基因簇:HoxA、HoxB、HoxC和HoxD,共包括39个基因[27] (图1A),这可能是由进化过程中的两次DNA片段复制过程产生[46,47]。本研究以人HEK293T细胞为模型,通过分析ChIP-seq数据发现位于C-DOM和T-DOM边界的HOXD基因簇串联排列着多个CTCF位点,提示CTCF可能对HOXD的转录调控发挥重要作用(图1B)。利用ChIP-seq数据进一步分析了与转录调控密切相关的转录因子和各组蛋白修饰在HOXD基因簇及其两侧调控区域内富集的情况,包括CTCF蛋白、架构蛋白YY1、增强子标记H3K4me1、H3K27ac,启动子标记H3K4me3,转录活性标记PolⅡ、p300(图1B):HOXD基因簇所在的区域有大量的CTCF蛋白、YY1蛋白富集以及H3K27ac、H3K4me3组蛋白修饰,是一个转录活跃区域;Island2、Island5处有CTCF蛋白结合峰,但是没有相应的组蛋白修饰,也没有转录活性标记(PolⅡ和p300),提示Island2、Island5在HEK293T细胞中可能没有增强子活性。GT2处没有CTCF蛋白结合,有YY1的结合峰、H3K4me1和H3K27ac等组蛋白修饰(图1B),说明在HEK293T中增强子GT2可能不完全依赖CTCF激活HOXD基因转录;CS38-41处有CTCF、YY1结合以及H3K27ac组蛋白修饰,同时富集PolⅡ和p300,因此在HEK293T细胞中具有较强的增强子活性。
为了分析HOXD基因簇处CTCF分布的保守性,本研究比较了人肾脏和人胚胎肾细胞系HEK293T中CTCF的ChIP-seq结合峰分布,发现HOXD基因簇C-DOM和T-DOM交界区域均分布着成簇排列的CBS,靠近C-DOM处具有5个串联排列的反向CBS,分别记为CBS a、b、c、d和e(图1C)。此外,分析小鼠肾脏及第12.5天胚胎肢芽CTCF的ChIP-seq数据发现,小鼠Hox基因簇中心粒侧对应的位置串联排列着类似的CBS簇(图1D)。综上所述,C-DOM和T-DOM边界处串联排列的反向CBS簇在哺乳动物中高度保守,CTCF可能通过结合该CBS簇参与指导两侧调控区域内的增强子对HOXD基因簇的转录表达调控。
2.2 边界处单个CTCF位点删除影响HOXD基因簇表达
为了研究反向串联排列的CBS a、b、c、d和e对人HOXD基因簇调控的作用,首先用CRISPR/ Cas9编辑技术对CBS e进行删除。我们针对CBS e设计一对sgRNA (sgRNA e1和sgRNA e2),Cas9核酸酶在sgRNA介导下特异性识别CBS e两侧靶向序列后进行切割,经过修复后能够获得CBS e删除的编辑细胞(图2A)。利用PCR筛选单细胞克隆株(图2B),对获得的单细胞克隆株进行TA克隆并测序确定其基因型(图2C)。获得的e#113存在两种不同的染色体基因型,Allele 1是853 bp片段删除的基因型,其切口连接处有小片段DNA删除,Allele 2是Cas9蛋白在PAM位点上游第3位切割后精确DNA修复产生的831 bp片段删除的基因型[41,48,49];e#126也存在两种不同的染色体基因型,其中一种是Cas9蛋白精准切割在PAM位点上游第3位后修复,另一种可能是Cas9蛋白切割在PAM位点上游第3位和第4位[48]:基因型鉴定及测序结果均证明e#113和e#126细胞中CBS e已被删除(图2C)。图2
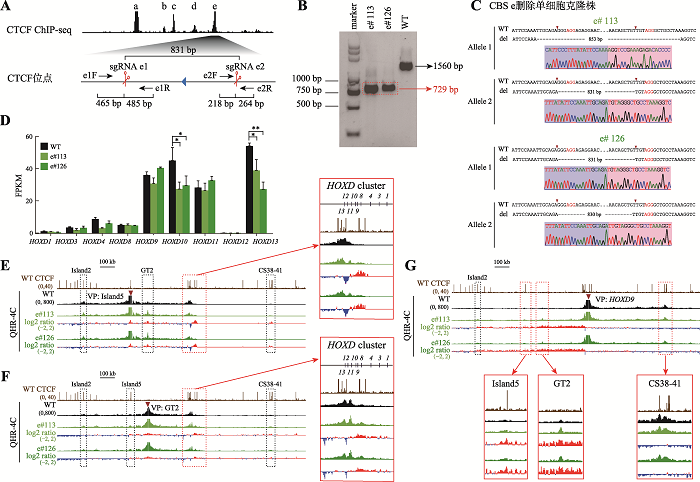 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CBS e删除改变增强子与启动子间的远程互作从而影响HOXD基因簇的基因表达
A:利用CRISPR DNA片段编辑技术获得CBS e删除的单细胞克隆株(e#113和e#126)的示意图。针对CBS e设计一对sgRNA(sgRNA e1和sgRNA e2),Cas9核酸酶在sgRNA e1和sgRNA e2的介导下特异性识别CBS e两侧靶向序列后进行切割,形成的两个切口被修复后连接在一起获得CBS e删除的编辑细胞。B:采用特异性引物PCR后进行凝胶电泳实验鉴定单细胞克隆株。引物对e1F/e2R在野生型(WT)细胞中扩增出1560 bp的片段,在e#113和e#126细胞株中扩增出CBS e删除后的729 bp片段(红色虚线框指示目的条带)。C:TA克隆并进行桑格测序确定单细胞克隆株的基因型。引物对e1F/e2R在e#113和e#126细胞株中扩增的产物经TA克隆后的测序结果图。D:RNA-seq数据分析比较WT细胞、e#113和e#126细胞株中HOXD基因簇表达水平。*:P<0.05;**:P<0.01;FPKM:fragments per kilobase of transcript per million mapped reads。E:在染色质构象捕获(QHR-4C)实验中,以增强子Island5为观测点(viewpoint,VP),分析e#113和e#126细胞株中Island5与HOXD基因簇启动子及调控元件Island2、GT2和CS38-41的远程互作。将e#113和e#126细胞株的数据分别与WT进行log2处理,红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以增强子GT2为VP,分析CBS e删除后GT2与HOXD基因簇启动子的远程互作。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,分析e#113和e#126细胞株中调控元件Island2、Island5、GT2、CS38-41与HOXD9启动子之间的染色质相互作用。黑色虚线框指示Island2,红色实线框内分别为Island5、GT2和CS38-41与HOXD9启动子之间的染色质相互作用放大图。
Fig. 2Alterations of long-distance chromatin interactions between enhancers and promoters upon CBS e deletion affect gene expression of the HOXD cluster
为了研究单个CBS e删除是否会影响HOXD基因转录,对获得的单细胞克隆株进行RNA-seq实验。在HEK293T细胞中,HOXD9~HOXD11以及HOXD13表达量较高;CBS e删除后HOXD10及HOXD13的表达水平降低最显著(图2D)。为了探究CBS e删除对HOXD基因表达影响的机制,对获得的e#113和e#126单细胞克隆株进行QHR-4C实验,分别以Island5、GT2及HOXD9为观测点,分析CBS e删除后细胞内HOXD基因簇染色质高级结构变化。在WT细胞中,增强子Island5、GT2与HOXD基因簇之间存在显著的DNA相互作用(图2,E和F),HOXD9启动子主要与下游增强子簇中的CS38-41有远距离DNA相互作用(图2G)。删除CBS e后,HOXD8~HOXD11启动子与Island5之间相互作用有显著增加(图2E)。除了HOXD12与HOXD13,其余HOXD基因启动子与GT2的相互作用略微增加(图2F)。HOXD9启动子与Island5、GT2的远程互作有明显增加,与前述结果一致,其与CS38-41的相互作用略微减少(图2G)。综上所述,单个CBS e删除引起HOXD基因簇启动子与增强子之间的远距离DNA相互作用改变,导致HOXD10及HOXD13表达水平降低。
2.3 边界处多个CTCF位点删除对HOXD基因簇远距离DNA相互作用的影响具有叠加效应
为了探究多个CBS删除对HOXD基因转录水平的影响,我们对CBS c-e (图3)和CBS b-e (图4)进行删除。分别设计一对sgRNA (sgRNA c1/sgRNA e2和sgRNA b1/sgRNA e2)对HEK293T细胞进行编辑(图3A和图4A),再用特异性引物PCR进行凝胶电泳鉴定(图3B和图4B)。将目的条带切胶纯化、TA克隆并进行桑格测序鉴定细胞的基因型,最终获得了CBS c-e删除的c-e#49和c-e#54单细胞克隆株(图3C)以及CBS b-e删除的b-e#59和b-e#80单细胞克隆株(图4C)。通过RNA-seq实验发现,在CBS c-e和CBS b-e删除的细胞(这些细胞的HOXD13已删除)中,HOXD9、HOXD10及HOXD11的转录水平下降,其中HOXD9下降最显著;HOXD9的转录水平分别降低了约40%和56%(图3D和图4D)。QHR-4C实验显示,无论在CBS c-e还是CBS b-e删除的细胞中,HOXD8~HOXD11与Island5之间的远程互作均增加(图3E和图4E),与GT2的互作也增加,且幅度比CBS e单独删除型细胞大(图3F和图4F)。在CBS b-e删除的细胞中,HOXD9启动子与增强子Island5及GT2的远程互作相对CBS c-e删除型细胞进一步增强(图3G和图4G),并且在Island5介导的远程互作中新增了下游增强子簇(CS38-41)(图3E和图4E);HOXD9启动子与CS38-41之间的相互作用没有进一步减弱(图3G和图4G)。综上所述,边界处CBS删除的数量对上游增强子簇与HOXD基因簇远程互作的影响有叠加效应,但不会递进式地影响整个HOXD基因(除了HOXD9)的转录表达水平。
图3
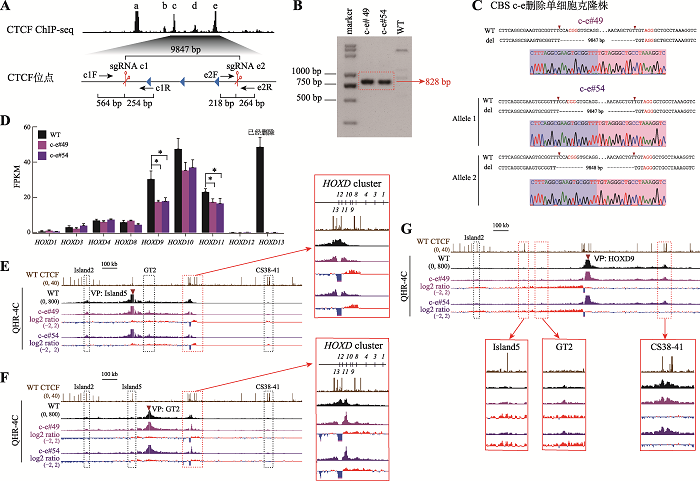 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3CBS c-e删除引起上游增强子与近端HOXD基因相互作用的增强
A:设计一对sgRNA(sgRNA c1和sgRNA e2)对HEK293T野生型细胞进行编辑获得CBS c-e删除的单细胞克隆株(c-e#49和c-e#54)的示意图。B:用引物c1F和e2R PCR鉴定c-e#49和c-e#54单细胞克隆株的凝胶电泳图。C:TA克隆鉴定c-e#49和c-e#54单细胞克隆株基因型的测序结果图。D:RNA-seq数据分析比较WT细胞、c-e#49和c-e#54细胞中HOXD基因簇表达水平。E:QHR-4C实验中,以Island5为VP,分析c-e#49和c-e#54细胞株中Island5与HOXD基因簇启动子的远程互作。红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以GT2为VP,分析CBS c- e删除后GT2与HOXD基因簇启动子的远程互作。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,c-e#49和c-e#54细胞株中Island2、Island5、GT2、CS38-41与HOXD9启动子之间的染色质相互作用。黑色虚线框指示Island2,红色实线框内分别为Island5、GT2和CS38-41与HOXD9启动子之间的染色质相互作用放大图。
Fig. 3Increases of long-distance chromatin interactions between enhancers and promoters upon CBS c-e deletion
图4
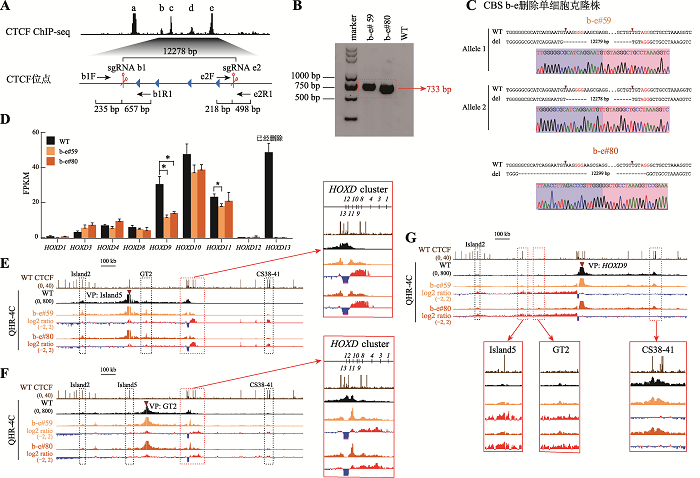 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4串联排列CTCF位点对增强子远程互作和HOXD基因表达具有叠加效应
A:利用CRISPR/Cas9编辑系统获得CBS b-e删除的单细胞克隆株(b-e#59和b-e#80)的示意图。B:用引物b1F和e2R1进行PCR鉴定b-e#59和b-e#80单细胞克隆株的凝胶电泳图。C:TA克隆鉴定b-e#59和b-e#80单细胞克隆株基因型的测序结果图。D:RNA-seq数据分析比较WT细胞、b-e#59和b-e#80细胞株细胞中HOXD基因簇表达水平。E:QHR-4C实验中,以Island5为VP,分析CBS b-e删除对Island5与HOXD基因簇启动子远程互作的影响。红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以GT2为VP,分析b-e#59和b-e#80细胞株中GT2与HOXD基因簇启动子的远程互作。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,分析b-e#59和b-e#80细胞株中Island5、GT2、CS38-41与HOXD9启动子间的染色质相互作用。黑色虚线框指示Island2,红色实线框内分别为Island5、GT2和CS38-41与HOXD9启动子之间的染色质相互作用放大图。
Fig. 4Additive effects on chromatin interactions and HOXD expression revealed by tandem CTCF sites deletion
2.4 边界处CTCF位点的全部删除破坏了增强子调控HOXD基因簇表达的平衡
CTCF常富集于TAD区域边界,对维持相关拓扑结构域的稳定具有重要作用。为了探究整个CBS a-e对HOXD基因簇表达调控的影响,设计一对sgRNA(sgRNA a1和sgRNA e2)将CBS a-e删除(图5A),进行PCR鉴定(图5B)、TA克隆并进行桑格测序(图5C)获得a-e#93和a-e#108单细胞克隆株。对CBS a-e删除的细胞进行RNA-seq实验发现,HOXD9、HOXD10与HOXD11的基因表达显著降低,其中HOXD9最显著,降低了约70% (图5D)。QHR-4C实验发现,Island5与下游增强子簇(CS38-41)的互作进一步增加,原本与CBS a-e整体的互作进一步转移至下游的一个反向CBS附近,从而使其与HOXD基因簇内启动子的互作从HOXD8~HOXD13扩展至HOXD4~HOXD11 (图5E);GT2与HOXD1~HOXD11启动子的互作增强,同时与T-DOM的染色质远距离互作增强(图5F)。以Island5、GT2为观测点发现一个有意思的现象:有CTCF结合的Island5与HOXD基因簇的染色质相互作用区域具有明显的边界(图2E、3E、4E和5E),而没有CTCF结合的增强子GT2与HOXD基因簇区域的染色质相互作用呈现广泛弥散式分布(图2F、3F、4F和5F),没有明显的边界,这提示具有CTCF结合的调控元件以及没有CTCF结合的调控元件与基因启动子之间的染色质相互作用模式是不同的。以HOXD9启动子为VP发现,在HEK293T野生型细胞中,HOXD9启动子主要与下游增强子簇有DNA相互作用;而在CBS a-e删除的细胞中,HOXD9启动子与上游增强子簇(Island2、Island5、GT2、GCR和Prox)的远程互作普遍显著增强,与下游增强子簇(CS38-41)的远程互作普遍减弱,CBS a-e删除改变了HOXD9的转录调控模式,由最初主要受下游增强子簇调控转变为上下游两个增强子簇调控(图5G),原有的调控平衡被破坏,这可能是导致HOXD9~HOXD11转录水平急剧降低的原因。图5
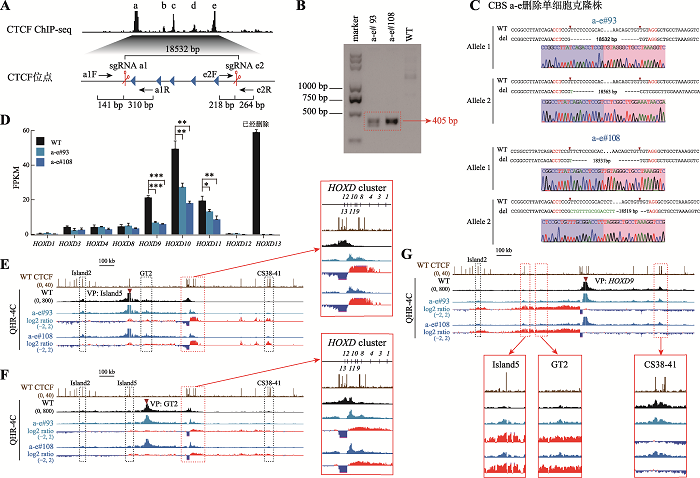 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5删除全部反向CTCT位点破坏增强子调控HOXD基因表达的平衡
A:获得CBS a-e删除的单细胞克隆株(a-e#93和a-e#108)的示意图。B:用引物a1F和e2R PCR鉴定a-e#93和a-e#108细胞株的凝胶电泳图。C:TA克隆鉴定a-e#93和a-e#108单细胞克隆株基因型的测序结果图。D:RNA-seq数据分析比较WT细胞、a-e#93和a-e#108细胞株细胞中HOXD基因簇转录水平。E:QHR-4C实验中,以Island5为VP,分析CBS a-e删除对Island5与HOXD基因簇启动子及两侧调控区域染色质相互作用的影响。红色实线框内为Island5与HOXD基因簇启动子之间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、GT2和CS38-41。F:以GT2为VP,分析a-e#93和a-e#108细胞株中GT2与HOXD基因簇启动子的染色质相互作用。红色实线框内为GT2与HOXD基因簇启动子间的染色质相互作用放大图,黑色虚线框依次指示调控元件Island2、Island5和CS38-41。G:以HOXD9启动子为VP,分析CBS a-e删除后HOXD9的表达调控模式。黑色虚线框指示Island2,红色实线框内为Island5、GT2、CS38-41与HOXD9基因簇启动子间的染色质相互作用放大图。
Fig. 5Regulatory balance of HOXD expression revealed by tandem reverse CTCF sites completely deletion
以上结果说明,C-DOM和T-DOM边界处的反向串联排列的CBS主要调控HOXD9~HOXD11及HOXD13的基因表达。在HEK293T细胞中,Island2、Island5没有增强子活性,GT2不完全依赖于CTCF调控HOXD9~HOXD11的表达,上游增强子簇与HOXD基因簇启动子的远程互作增加,削弱了下游增强子簇中的CS38-41对HOXD基因簇的激活强度。边界处单个CBS删除就会影响HOXD基因簇原有的三维基因组结构,改变HOXD10和HOXD13的基因表达。多个CBS删除对HOXD基因簇三维基因组结构以及HOXD9转录的影响具有叠加效应,但是没有大幅度改变其他HOXD基因的表达。当边界处整个CBS a-e全部删除后,HOXD基因簇与CS38-41的染色质相互作用进一步减弱,HOXD9~HOXD11转录水平急剧降低,并且上下游增强子簇之间出现了远距离相互作用。综上所述,HOXD基因簇靠近中心粒侧区域的反向串联排列CTCF位点具有绝缘子功能,通过阻断上游增强子簇与HOXD9~HOXD11的染色质远程互作,维持HOXD基因簇表达调控的平衡,使HOXD基因能够精准有序地表达。
图6
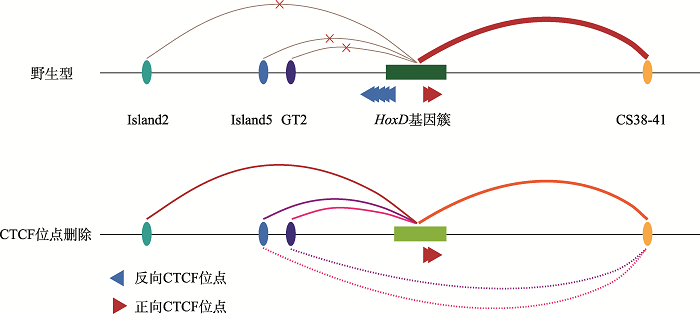 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6HOXD基因簇的表达调控模式图
A:野生型(WT)细胞中HOXD基因簇的表达调控示意图。在野生型细胞中,CTCF位点具有绝缘子作用,阻碍了上游增强子簇中的Island2、Island5、GT2与HOXD基因簇启动子的远程相互作用,HOXD基因簇主要受下游增强子簇中的CS38-41调控。B:CTCF位点删除的细胞中HOXD基因簇的表达调控示意图。在CBS删除的细胞中,CTCF位点绝缘子作用消失,上游增强子簇与HOXD基因簇之间产生远距离DNA相互作用,导致HOXD基因簇双边调控的平衡被破坏、HOXD基因表达降低。
Fig. 6A model for HOXD gene regulation
3 讨论
在哺乳动物中,增强子的数量远远超过基因的数量,复杂基因的表达往往受多个增强子组合调控,防止在其适当范围之外的异位表达[50]。增强子活性的经典模式是相对独立的、自主的并且可叠加的,但是顺式调控区域内存在功能有冗余性(redundancy)的增强子[51,52]。增强子激活需要结合多个转录因子[53],转录因子结合增强子后,对基因表达起抑制作用或激活作用取决于结合的转录因子[54]。例如,转录因子RFX5与增强子的结合对原钙粘蛋白基因簇的表达起抑制作用[55]。增强子活性的强弱取决于招募的共激活蛋白对染色质的修饰和增强子的重塑,包括组蛋白乙酰基转移酶(例如p300/CBP)、组蛋白甲基转移酶(例如MLL3/4和CARM1)、染色质重塑因子(例如Brg1和CHD7),以及在启动子处促进转录的相关因子(例如中介体复合物)[56]。本研究中,CBS a-e删除后,HOXD基因簇与上游增强子簇中的Island5、GT2的远程互作显著增加,而HOXD9~ HOXD11的表达下降,这表明Island5、GT2与启动子之间有互作,但是它们没有增强子活性,这可能是因为在这两个增强子上结合了抑制性的共激活蛋白或转录因子[35,57]。小鼠胚胎发育过程中Hox基因的表达就需要结合不同的转录因子[58]。CTCF与cohesin的结合是一个动态过程[59],成串的CBS可以阻碍渗透的cohesin滑动,形成类似于“葫芦”结构的染色质环[1,17],由此介导的远距离增强子与启动子的相互作用可有效持久地调控基因的表达。我们先前利用小鼠和多种细胞模型对原钙粘蛋白和珠蛋白模式基因进行系统的研究[13],发现单个CTCF位点可确保适当的增强子绝缘和启动子激活,而串联排列CTCF位点的绝缘作用可维持基因组三维空间的动态平衡及准确的启动子选择。在本研究中,我们进一步利用HEK293T细胞模型对HOXD基因簇中的CTCF位点进行了系统的组合性删除实验,发现串联排列CTCF位点通过其绝缘功能维持上下游增强子簇调控HOXD基因表达的平衡。另外我们观察到一个有趣的现象,包含CTCF位点的增强子Island5与HOXD基因的染色质相互作用具有明显的边界,而不包含CTCF位点的增强子GT2与HOXD基因的染色质相互作用区域则分布较广泛,这和原钙粘蛋白基因簇中的增强子HS5-1 (包含CTCF位点)、HS7 (不包含CTCF位点)与Pcdh基因染色质互作[13]的特点一致。
在本研究中,我们将HOXD基因簇C-DOM与T-DOM边界处CTCF位点进行逐步删除,随着剩余的CTCF位点数量减少,HOXD基因簇启动子与上游增强子簇DNA远程互作逐渐增强,与下游增强子簇的DNA远程互作逐渐减弱。当边界串联反向的CTCF位点全部删除后,HOXD9基因启动子由最初与下游增强子簇有相互作用转变为与上下游增强子簇均有相互作用,并且上下游增强子簇之间出现了DNA远程互作(图6)。
综上所述,HOXD基因簇中串联反向CTCF位点扮演着绝缘子的角色,阻碍上游增强子簇跨区域调控近端HOXD基因的表达,对HOXD基因簇在肢体发育中时空线性表达的精细调控具有重要意义,为深入研究Hox基因簇复杂调控网络提供了参考。
(责任编委: 李大力)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.jgg.2020.06.008URL [本文引用: 4]
DOI:10.1073/pnas.1219280110URL [本文引用: 2]
DOI:10.1016/j.cell.2015.07.038PMID:26276636 [本文引用: 4]

CTCF and the associated cohesin complex play a central role in insulator function and higher-order chromatin organization of mammalian genomes. Recent studies identified a correlation between the orientation of CTCF-binding sites (CBSs) and chromatin loops. To test the functional significance of this observation, we combined CRISPR/Cas9-based genomic-DNA-fragment editing with chromosome-conformation-capture experiments to show that the location and relative orientations of CBSs determine the specificity of long-range chromatin looping in mammalian genomes, using protocadherin (Pcdh) and β-globin as model genes. Inversion of CBS elements within the Pcdh enhancer reconfigures the topology of chromatin loops between the distal enhancer and target promoters and alters gene-expression patterns. Thus, although enhancers can function in an orientation-independent manner in reporter assays, in the native chromosome context, the orientation of at least some enhancers carrying CBSs can determine both the architecture of topological chromatin domains and enhancer/promoter specificity. These findings reveal how 3D chromosome architecture can be encoded by linear genome sequences. Copyright © 2015 Elsevier Inc. All rights reserved.
[本文引用: 2]
[本文引用: 2]
DOI:10.1038/cr.2017.131URL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
PMID:8649389 [本文引用: 1]

We have isolated and analyzed human CTCF cDNA clones and show here that the ubiquitously expressed 11-zinc-finger factor CTCF is an exceptionally highly conserved protein displaying 93% identity between avian and human amino acid sequences. It binds specifically to regulatory sequences in the promoter-proximal regions of chicken, mouse, and human c-myc oncogenes. CTCF contains two transcription repressor domains transferable to a heterologous DNA binding domain. One CTCF binding site, conserved in mouse and human c-myc genes, is found immediately downstream of the major P2 promoter at a sequence which maps precisely within the region of RNA polymerase II pausing and release. Gel shift assays of nuclear extracts from mouse and human cells show that CTCF is the predominant factor binding to this sequence. Mutational analysis of the P2-proximal CTCF binding site and transient-cotransfection experiments demonstrate that CTCF is a transcriptional repressor of the human c-myc gene. Although there is 100% sequence identity in the DNA binding domains of the avian and human CTCF proteins, the regulatory sequences recognized by CTCF in chicken and human c-myc promoters are clearly diverged. Mutating the contact nucleotides confirms that CTCF binding to the human c-myc P2 promoter requires a number of unique contact DNA bases that are absent in the chicken c-myc CTCF binding site. Moreover, proteolytic-protection assays indicate that several more CTCF Zn fingers are involved in contacting the human CTCF binding site than the chicken site. Gel shift assays utilizing successively deleted Zn finger domains indicate that CTCF Zn fingers 2 to 7 are involved in binding to the chicken c-myc promoter, while fingers 3 to 11 mediate CTCF binding to the human promoter. This flexibility in Zn finger usage reveals CTCF to be a unique "multivalent" transcriptional factor and provides the first feasible explanation of how certain homologous genes (i.e., c-myc) of different vertebrate species are regulated by the same factor and maintain similar expression patterns despite significant promoter sequence divergence.
DOI:10.1371/journal.pone.0041374URL [本文引用: 1]
PMID:11700297 [本文引用: 1]

The separation of sister chromatids at the metaphase to anaphase transition is one of the most dramatic of all cellular events and is a crucial aspect of all sexual and asexual reproduction. The molecular basis for this process has until recently remained obscure. New research has identified proteins that hold sisters together while they are aligned on the metaphase plate. It has also shed insight into the mechanisms that dissolve sister chromatid cohesion during both mitosis and meiosis. These findings promise to provide insights into defects in chromosome segregation that occur in cancer cells and into the pathological pathways by which aneuploidy arises during meiosis.
DOI:10.1126/science.aaz4475URL [本文引用: 1]
DOI:10.1007/s13238-019-00656-yURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s13059-020-01984-7URL [本文引用: 5]
[本文引用: 2]
DOI:S0092-8674(16)30073-3PMID:26967279 [本文引用: 1]

Proper expression of genes requires communication with their regulatory elements that can be located elsewhere along the chromosome. The physics of chromatin fibers imposes a range of constraints on such communication. The molecular and biophysical mechanisms by which chromosomal communication is established, or prevented, have become a topic of intense study, and important roles for the spatial organization of chromosomes are being discovered. Here we present a view of the interphase 3D genome characterized by extensive physical compartmentalization and insulation on the one hand and facilitated long-range interactions on the other. We propose the existence of topological machines dedicated to set up and to exploit a 3D genome organization to both promote and censor communication along and between chromosomes. Copyright © 2016 Elsevier Inc. All rights reserved.
DOI:10.1038/s41594-018-0135-4URL [本文引用: 1]
DOI:10.1007/s12264-020-00578-4URL [本文引用: 2]
[本文引用: 1]
DOI:10.1038/ni.3232 [本文引用: 1]

Chen, Liang; Carico, Zachary; Shih, Han-Yu; Krangel, Michael S. Duke Univ, Med Ctr, Dept Immunol, Durham, NC 27708 USA.
[本文引用: 1]
[本文引用: 3]
[本文引用: 2]
[本文引用: 1]
DOI:10.3389/fnins.2020.587819URL [本文引用: 1]
DOI:10.1073/pnas.1111941109URL [本文引用: 1]
DOI:10.1038/276565a0URL [本文引用: 1]
[本文引用: 2]
DOI:10.1126/science.1085753URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 4]
[本文引用: 2]
DOI:10.1016/j.cell.2016.09.018URL [本文引用: 1]
DOI:10.1038/s41576-019-0128-0PMID:31086298 [本文引用: 1]

Spatiotemporal gene expression programmes are orchestrated by transcriptional enhancers, which are key regulatory DNA elements that engage in physical contacts with their target-gene promoters, often bridging considerable genomic distances. Recent progress in genomics, genome editing and microscopy methodologies have enabled the genome-wide mapping of enhancer-promoter contacts and their functional dissection. In this Review, we discuss novel concepts on how enhancer-promoter interactions are established and maintained, how the 3D architecture of mammalian genomes both facilitates and constrains enhancer-promoter contacts, and the role they play in gene expression control during normal development and disease.
DOI:10.1016/j.molcel.2019.08.010URL [本文引用: 2]
[本文引用: 1]
DOI:10.1093/jmcb/mjv016URL [本文引用: 2]
DOI:10.1038/cr.2013.45URL [本文引用: 1]
DOI:10.1126/science.1258096URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1038/nprot.2012.016PMID:22383036 [本文引用: 1]

Recent advances in high-throughput cDNA sequencing (RNA-seq) can reveal new genes and splice variants and quantify expression genome-wide in a single assay. The volume and complexity of data from RNA-seq experiments necessitate scalable, fast and mathematically principled analysis software. TopHat and Cufflinks are free, open-source software tools for gene discovery and comprehensive expression analysis of high-throughput mRNA sequencing (RNA-seq) data. Together, they allow biologists to identify new genes and new splice variants of known ones, as well as compare gene and transcript expression under two or more conditions. This protocol describes in detail how to use TopHat and Cufflinks to perform such analyses. It also covers several accessory tools and utilities that aid in managing data, including CummeRbund, a tool for visualizing RNA-seq analysis results. Although the procedure assumes basic informatics skills, these tools assume little to no background with RNA-seq analysis and are meant for novices and experts alike. The protocol begins with raw sequencing reads and produces a transcriptome assembly, lists of differentially expressed and regulated genes and transcripts, and publication-quality visualizations of analysis results. The protocol's execution time depends on the volume of transcriptome sequencing data and available computing resources but takes less than 1 d of computer time for typical experiments and ~1 h of hands-on time.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
PMID:16341070 [本文引用: 1]

With their power to shape animal morphology, few genes have captured the imagination of biologists as the evolutionarily conserved members of the Hox clusters have done. Recent research has provided new insight into how Hox proteins cause morphological diversity at the organismal and evolutionary levels. Furthermore, an expanding collection of sequences that are directly regulated by Hox proteins provides information on the specificity of target-gene activation, which might allow the successful prediction of novel Hox-response genes. Finally, the recent discovery of microRNA genes within the Hox gene clusters indicates yet another level of control by Hox genes in development and evolution.
DOI:10.1016/j.febslet.2015.04.024URL [本文引用: 1]
[本文引用: 1]
DOI:S1097-2765(18)30466-0PMID:30033371 [本文引用: 2]

Chromosomal rearrangements including large DNA-fragment inversions, deletions, and duplications by Cas9 with paired sgRNAs are important to investigate genome structural variations and developmental gene regulation, but little is known about the underlying mechanisms. Here, we report that disrupting CtIP or FANCD2, which have roles in alternative non-homologous end joining, enhances precise DNA-fragment deletion. By analyzing the inserted nucleotides at the junctions of DNA-fragment editing of deletions, inversions, and duplications and characterizing the cleaved products, we find that Cas9 endonucleolytically cleaves the noncomplementary strand with a flexible scissile profile upstream of the -3 position of the PAM site in vivo and in vitro, generating double-strand break ends with 5' overhangs of 1-3 nucleotides. Moreover, we find that engineered Cas9 nucleases have distinct cleavage profiles. Finally, Cas9-mediated nucleotide insertions are nonrandom and are equal to the combined sequences upstream of both PAM sites with predicted frequencies. Thus, precise and predictable DNA-fragment editing could be achieved by perturbing DNA repair genes and using appropriate PAM configurations.Copyright © 2018 Elsevier Inc. All rights reserved.
DOI:10.1038/nbt.2623URL [本文引用: 1]
DOI:10.1073/pnas.1109873108URL [本文引用: 1]
DOI:10.1038/nature09158URL [本文引用: 1]
DOI:10.1002/bies.201100121PMID:22083793 [本文引用: 1]

This paper, in the form of a frequently asked questions page (FAQ), addresses outstanding questions about "shadow enhancers", quasi-redundant cis-regulatory elements, and their proposed roles in transcriptional control. Questions include: What exactly are shadow enhancers? How many genes have shadow/redundant/distributed enhancers? How redundant are these elements? What is the function of distributed enhancers? How modular are enhancers? Is it useful to study a single enhancer in isolation? In addition, a revised definition of "shadow enhancers" is proposed, and possible mechanisms of shadow enhancer function and evolution are discussed.Copyright © 2012 WILEY Periodicals, Inc.
DOI:10.1016/j.tig.2012.02.008URL [本文引用: 1]
DOI:10.1038/nature15518URL [本文引用: 1]
.
DOI:10.16288/j.yczz.20-184PMID:32952112 [本文引用: 1]

Gene expression and three-dimensional (3D) genome organization are thought to be closely related. The protocadherin (Pcdh) clusters are central for neuronal self-avoidance in brain development and have been used as model genes to explore the role of 3D genome structures in gene regulation. Transcription factor RFX5 (regulatory factor x 5) is a member of the winged-helix subfamily of the helix-turn-helix superfamily proteins. The RFX5 protein contains four domains: oligomerization, DNA-binding, helical, and transactivation domains. RFX5 plays an important role in regulating expression of the major histocompatibility complex class II (MHC II) gene complex. Here we report a role of RFX5 in the regulation of clustered Pcdh genes. We first knocked out the RFX5 gene by using CRISPR/Cas9 DNA-fragment editing in HEC-1-B cell line. By RNA-seq experiments, we found that RFX5 deletion results in a significant increase of expression levels of the Pcdhα6, Pcdhα12 and Pcdhαc2 genes. By ChIP-nexus experiments, we found that RFX5 deletion leads to the increased binding of CTCF and RAD21 in the Pcdhα cluster. Finally, through quantitative high-resolution chromosome conformation capture copy (QHR-4C) experiments, we found that RFX5 deletion results in a significant increase of long-distance chromatin interactions between the HS5-1 enhancer and its target promoters in the Pcdhα cluster. In conclusion, RFX5 regulates gene expression of the Pcdhα cluster through higher-order chromatin structure.
DOI:10.16288/j.yczz.20-184PMID:32952112 [本文引用: 1]

Gene expression and three-dimensional (3D) genome organization are thought to be closely related. The protocadherin (Pcdh) clusters are central for neuronal self-avoidance in brain development and have been used as model genes to explore the role of 3D genome structures in gene regulation. Transcription factor RFX5 (regulatory factor x 5) is a member of the winged-helix subfamily of the helix-turn-helix superfamily proteins. The RFX5 protein contains four domains: oligomerization, DNA-binding, helical, and transactivation domains. RFX5 plays an important role in regulating expression of the major histocompatibility complex class II (MHC II) gene complex. Here we report a role of RFX5 in the regulation of clustered Pcdh genes. We first knocked out the RFX5 gene by using CRISPR/Cas9 DNA-fragment editing in HEC-1-B cell line. By RNA-seq experiments, we found that RFX5 deletion results in a significant increase of expression levels of the Pcdhα6, Pcdhα12 and Pcdhαc2 genes. By ChIP-nexus experiments, we found that RFX5 deletion leads to the increased binding of CTCF and RAD21 in the Pcdhα cluster. Finally, through quantitative high-resolution chromosome conformation capture copy (QHR-4C) experiments, we found that RFX5 deletion results in a significant increase of long-distance chromatin interactions between the HS5-1 enhancer and its target promoters in the Pcdhα cluster. In conclusion, RFX5 regulates gene expression of the Pcdhα cluster through higher-order chromatin structure.
[本文引用: 1]
DOI:10.1242/dev.171736URL [本文引用: 1]
DOI:S0012-1606(16)30864-8PMID:28728680 [本文引用: 1]

Hox genes are crucial players in the generation and pattering of the vertebrate trunk and posterior body during embryogenesis. Their initial expression takes place shortly after the establishment of the primitive streak, in the posterior-most part of the mouse embryo and is a determinant step for setting up the definitive Hox expression boundaries along the antero-posterior body axis. The developmental signals and epigenetic mechanisms underlying this early activation remained unsolved until recently. The development of novel embryo-derived model systems, combined with methods that examine chromatin status and chromosome conformation, led to deeper understanding of the process of Hox activation in the early embryo. Here we summarize how the early Hox cis-regulatory landscape becomes active upon receiving the appropriate developmental signal, and we discuss the importance of the local topological segmentation of the HoxA cluster during early Hox activation.Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
DOI:10.1021/acs.jpclett.8b01440URL [本文引用: 1]
