 ,, 冷奇颖, 郑嘉辉, AliHassanNawaz, 焦振海, 王府建, 张丽
,, 冷奇颖, 郑嘉辉, AliHassanNawaz, 焦振海, 王府建, 张丽 ,广东海洋大学滨海农业学院,湛江 524088
,广东海洋大学滨海农业学院,湛江 524088Cloning and expression of circular transcript of mouse growth hormone receptor gene
Weilu Zhang ,, Qiying Leng, Jiahui Zheng, Ali Hassan Nawaz, Zhenhai Jiao, Fujian Wang, Li Zhang
,, Qiying Leng, Jiahui Zheng, Ali Hassan Nawaz, Zhenhai Jiao, Fujian Wang, Li Zhang ,College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang 524088, China
,College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang 524088, China通讯作者: 张丽,博士,教授,研究方向:家禽遗传育种与繁殖。E-mail:zhangli761101@163.com
编委: 单革
收稿日期:2021-04-27修回日期:2021-07-21
| 基金资助: |
Received:2021-04-27Revised:2021-07-21
| Fund supported: |
作者简介 About authors
张为露,在读硕士研究生,专业方向:动物遗传育种与繁殖。E-mail:

摘要
根据文献报道并通过circBase数据库查找,发现小鼠生长激素受体基因(growth hormone receptor, GHR)存在8个环状转录本。为确定GHR基因环状转录本(circGHR)的真实存在,探究其表达规律,本研究以昆明小鼠(Mus musculus)为研究对象,通过PCR扩增和测序检测circGHR的真实存在,并筛选出一个circGHR作为目标分子,通过RNase R处理和反转录证实了circGHR的环形结构,利用qRT-PCR分析circGHR和GHR mRNA的时空表达规律。结果表明:小鼠circGHR全长为820 nt,由GHR基因外显子2~8环化形成。RNase R耐受性分析表明,小鼠circGHR具备环形分子的一般特征,不易被RNase R降解。与oligo-d(T)18引物相比,随机引物对circGHR具有较高的反转录效率,进一步说明circGHR是一个不含poly(A)的环状结构分子。组织表达谱结果表明,circGHR在1周龄和7周龄小鼠肝脏和肾脏高表达,在胸肌和腿肌中低表达;circGHR在肝脏和胸肌组织的时序表达谱结果表明circGHR表达无显著差异;circGHR在腿肌组织的时序表达谱结果表明,circGHR在小鼠5周龄之前为低表达,在7周龄以后表现为高表达。本研究结果证实了小鼠GHR基因存在一个环状转录本circGHR,并初步揭示了circGHR的表达规律,为后期深入开展小鼠circGHR的生物学功能及其在小鼠生长发育过程中的作用机制奠定基础。
关键词:
Abstract
Based on reports in the literature and search results on the circBase database, 8 circular transcripts of the mouse growth hormone receptor (GHR) gene were identified. In order to confirm the existence of the circular transcripts of the GHR gene (circGHRs) and to explore their expression patterns, the Kunming mouse (Mus musculus) was used as a research animal. This study detected the existence of circGHRs by RT-PCR amplification and sequencing, one of which was selected as circGHR for detailed analysis. The circular structure of circGHR was confirmed by RNase R treatment and reverse transcription. The spatiotemporal expression of circGHR and GHR mRNA was analyzed by qRT-PCR. The results showed that the full length of mouse circGHR was 820 nt, which was formed by circularization of exons 2-8 of the transcript of the GHR gene. RNase R tolerance analysis shows that mouse circGHR has the general characteristics of circular molecules and is not easily degraded by RNase R. Compared with oligo-d(T)18 primers, random primers have higher reverse transcription efficiency for circGHR, which further shows that circGHR is a poly(A)-free cyclic structure molecule. Tissue expression profile results show that circGHR is highly expressed in the liver and kidney of 1 week-old and 7-week old Kunming mice, but is low in pectoral muscles and leg muscles. The time-series expression profile of circGHR does not show any significant difference between the liver and pectoral muscle tissue. The circGHR expression in the leg muscle was low before 5 weeks of age but increased after 7 weeks of age. This study confirmed the existence of a circular transcript circGHR of the mouse GHR gene, and initially revealed the expression pattern of circGHR. The results of the study laid a foundation for in-depth developmental studies on the biological functions of the mouse circGHR and its mechanism of action regarding the growth and development of mice.
Keywords:
PDF (1314KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张为露, 冷奇颖, 郑嘉辉, AliHassanNawaz, 焦振海, 王府建, 张丽. 小鼠生长激素受体基因环状转录本的克隆及其表达规律. 遗传[J], 2021, 43(9): 890-900 doi:10.16288/j.yczz.21-156
Weilu Zhang.
生长激素(growth hormone, GH)是一种由脑垂体前叶嗜酸性细胞分泌的单一肽链蛋白质类激素。生长激素作为一种特殊的生物活性蛋白分子起着促进动物机体生长、肌肉发育、调节代谢等重要生理功能[1]。而GH发挥生理作用的第一步是与靶细胞膜表面的GH受体(growth hormone receptor, GHR)结合,由GHR介导将信号传入细胞内从而产生一系列生理效应[2]。GHR是一种单链跨膜糖蛋白,属于细胞分裂素/血细胞生成素受体超家族(cytokine/hematopoietic receptor superfamily)[3,4]。
环状RNA (circRNA)属于非编码RNA (ncRNA)的范畴,是一个既不带5ʹ帽结构也不带3ʹ聚腺苷酸尾巴的共价闭环[5]。1976年,Sanger等[6]最先通过分离植物类病毒发现了环状RNA,从此环状RNA被人所认知。circRNA具有一定的组织、时序特异性;由于其是封闭环状结构,因此具有高度保守性[7]。与线性RNA相比,circRNA更不易被RNase R降解,更加稳定[8,9],且多数 circRNA来源于外显子,也有包含外显子-内含子的circRNA (EI ciRNA)[10,11,12]。环状RNA发挥生物学功能的作用机制包括作为海绵吸附结合miRNA分子和蛋白质[13],调控母源基因或靶基因转录[14]和翻译蛋白质等。越来越多的证据表明环状RNA可以行使翻译的功能,Legnini等[15]首次在人类细胞中发现编码成肌细胞增殖相关蛋白的环状RNA—circ-ZNF609,该分子存在包含启动子和终止子的开放阅读框,并在体内和体外均能表达蛋白;另有研究表明在结肠癌中circPPP1R12A通过编码蛋白circPPP1R12A-aa激活Hippo-YAP信号通路进而促进结肠癌的发生和转移[16]。
由Ensembl数据库可知,小鼠GHR基因位于15号染色体负链上,由11个外显子和10个内含子组成,该基因有10个线性转录本。本课题组前期通过查阅文献以及查找circBase数据库,发现circGHR可以通过向后剪切反式形成8个环状转录本(circGHR1~circGHR8)。经过前期筛选后,本文选择circGHR3为研究对象,通过基因克隆技术验证了circGHR3的真实存在,分析了其环形结构特征及组织和时序表达规律,并对该基因可能编码蛋白质进行生物信息学分析。本研究结果为今后深入分析 circGHR在肌肉生长和动物机体发育过程中的作用奠定了基础。
1 材料与方法
1.1 实验动物和样品采集
在1、3、5和7周龄的昆明小鼠(为本实验饲养)中各抽取3只雄鼠,分别采集心脏、肝脏、胸肌、腿肌等组织,液氮速冻后置于-80℃冰箱保存备用。1.2 目标分子筛选与确定
根据Dong等[17]报道,在小鼠中共发现7个circGHR;同时在circBase数据库中对小鼠circGHR进行检索,发现1个circGHR序列,共8个序列。生物信息学分析这8个circGHR序列和结构组成,分别针对8个circGHR的接口(junction)位置设计特异性引物,用于筛选出目标分子进行后续实验,引物均通过primer-BLAST (Table 1
表1
表1小鼠8个circGHR的特异性验证引物
Table 1
| 引物名称 | 引物序列(5ʹ→3ʹ) | 复性温度(℃) | 扩增产物长度(bp) | 位置 | 用途 |
|---|---|---|---|---|---|
| circGHR1 | F:TAGTTTGACCGGGATTCGTGG | 59 | 171 | Exon7 | circGHR1 环状验证 |
| R: GGAACGACACTTGGTGAATCG | Exon4 | ||||
| circGHR2 | F:GGTGAGATCCAGACAACGGA | 59 | 134 | Exon8 | circGHR2 环状验证 |
| R: GACACTTGGTGAATCGAGGC | Exon4 | ||||
| circGHR3 | F:GGGATTCGTGGAGACATCCAA | 59 | 343 | Exon7 | circGHR3 环状验证 |
| R: GACTGCCAGTGCCAAGGTTA | Exon2 | ||||
| circGHR4 | F:GACCGGGATTCGTGGAGACATC | 61 | 422 | Exon7 | circGHR4 环状验证 |
| R: ACGACACTTGGTGAATCGAGG | Exon4 | ||||
| circGHR5 | F:CCATCCCATATGGTGGATCTGT | 59 | 142 | Intron1 | circGHR5 环状验证 |
| R: ATGGGAAAGGAGGTGATGGC | |||||
| circGHR6 | F:GCTGGACCAAAAATGTTTCACTGTT | 60 | 176 | Exon6 | circGHR6 环状验证 |
| R: CGTTGGCTTTCCCTTTTAGCA | Exon4,5 | ||||
| circGHR7 | F:CAGCGAAGTCCTCCGTGTAATA | 59 | 100 | Exon8 | circGHR7 环状验证 |
| R: CAGGGCATTCTTTCCATTCCTG | Exon6 | ||||
| circGHR8 | F: CAGTCACCAGCAGCACATTTT | 58 | 70 | Exon2 | circGHR8 环状验证 |
| R: AGGTTAAGAAGACCTGACAAAGAT |
新窗口打开|下载CSV
Table 2
表2
表2引物序列信息
Table 2
| 引物名称 | 引物序列(5'→3') | 复性温度(℃) | 扩增产物长度(bp) | 位置 | 用途 |
|---|---|---|---|---|---|
| circGHR-D | F:GGGATTCGTGGAGACATCCAA | 59 | 343 | Exon7 | 环状验证 |
| R: GACTGCCAGTGCCAAGGTTA | Exon2 | ||||
| circGHR-full | F:GTCTCAGGTATGGATCTTTGTCAGG | 57 | 820 | Exon2 | 全长克隆 |
| R: CTTCTTCACATGCTTCCAATATGTTC | Exon8 | ||||
| circGHR-DL | F:GGGATTCGTGGAGACATCCAA | 59 | 343 | Exon7 | circGHR定量 |
| R: GACTGCCAGTGCCAAGGTTA | Exon2 | ||||
| GHR-DL | F:GGGTGAGATCCAGACAACGG | 57 | 285 | Exon8 | GHR mRNA定量 |
| R: TCACCTCCTCCAACTTCCCT | Exon11 | ||||
| GAPDH | F:AGGTTGTCTCCTGCGACTTCA | 57 | 184 | Exon6 | 内参基因定量 |
| R: TGGTCCAGGGTTTCTTACTCC | Exon7 |
新窗口打开|下载CSV
1.3 小鼠circGHR和线性mRNA转录本定量和全长克隆引物设计
针对circGHR的环形接头位置,在接头位置两端设计3对引物,分别为:(1)分散引物circGHR-D,用于circGHR环状分子的验证;(2)全长扩增引物circGHR-full用于克隆circGHR分子全长序列;(3)定量引物circGHR-DL用于该分子的定量分析。其中分散引物与定量引物序列相同。此外,在GHR基因非circGHR序列来源处设计线性GHR mRNA (参考序列:MGI:95708)的定量引物(GHR-DL)。内参基因为GAPDH。引物均通过 primer-BLAST (图1
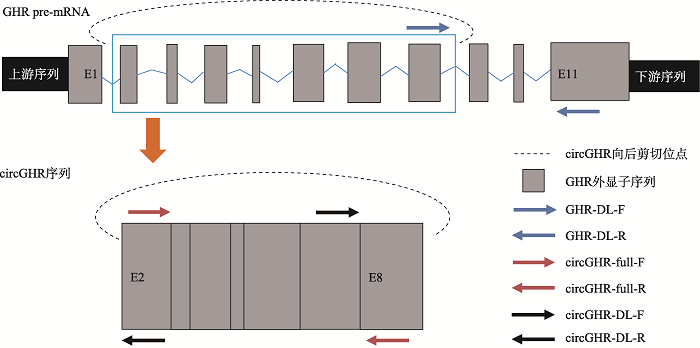 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1小鼠circGHR来源及引物位置示意图
E1~E11表示外显子Exon1~Exon11。
Fig. 1Diagram illustrating the origin of the mouse circGHR transcript and the locations of the primers used in this study
1.4 小鼠肝脏等组织总RNA提取及circGHR环状性质分析
按照 HiPure Universal RNA Mini Kit 试剂盒(广州美基生物科技有限公司)说明书分别提取1、3、5和7周龄小鼠肝脏、胸肌、腿肌、心脏等组织总RNA,并使用1%的变性琼脂糖凝胶电泳检测 RNA 的完整性,使用BioTek Epoch全波长酶标仪检测RNA浓度和纯度,依据PrimeScript RT reagent Kit With gDNA Eraser试剂盒(广州瑞真生物技术有限公司)说明书,使用随机引物(N9)将总RNA反转录成cDNA。将提取的7周龄小鼠肝脏组织总RNA分装为两份(每份5 ng),一份使用超纯水稀释至20 μL保存备用;另一份进行RNase R (广州吉赛生物股份有限公司)处理,反应体系包括RNA 5 ng、10× Reaction Buffer 2 μL、RNase R 20 U,最后使用去离子水将反应体系补至20 μL。反应程序为:37℃孵育30 min后85℃灭活RNase R 5 s,经RNase R处理后的RNA用于circGHR环状性质特征验证。依据PrimeScript RT reagent Kit With gDNA Eraser试剂盒说明书,分别使用随机引物(N9)和oligo-d(T)18将未经RNase R处理的7周龄小鼠肝脏组织总RNA反转录成cDNA。随机引物反转录合成的cDNA用于qRT-PCR定量分析circGHR的表达水平,定量使用TransStart Top Green qRT-PCR SuperMix试剂盒(北京全式金生物技术有限公司);随机引物和oligo- d(T)18引物反转录得到的cDNA用于qRT-PCR进行不同反转录引物的效率分析,进一步分析circGHR的环状结构特征。1.5 小鼠circGHR全长克隆
经RNase R处理的7周龄小鼠肝脏组织RNA,通过随机引物反转录合成cDNA,分别使用circGHR环状验证引物(circGHR-D)和全长引物进行PCR扩增,使用1.5%的琼脂糖凝胶电泳检测并对目标产物进行回收纯化,然后将获得的目标产物连接载体pMD18-T (广州瑞真生物技术有限公司)后转入E.coli DH5α感受态大肠杆菌(北京全式金生物技术有限公司)中,对菌株进行阳性鉴定后由生工生物工程(上海)股份有限公司进行测序,分析circGHR的序列信息和成环特征。1.6 小鼠circGHR的表达特征分析
以上述1.4中得到的cDNA为模板,采用 qRT-PCR分析circGHR的时空表达规律及环状特征。反应体系为20 μL:2×TransStart® Top Green qPCR supermix 10 μL,上、下游引物各0.5 μL,去离子水8 μL,cDNA 1 μL。反应程序:94℃预变性30 s;94℃变性5 s,59℃退火和延伸30 s,共40个循环。每个样本进行3次重复。1.7 数据统计与分析
使用2-ΔΔCT方法计算qRT-PCR结果,用IBMSPSS数据统计软件19进行显著性分析,结果以平均数(mean)±标准误(SEM)表示。2 结果与分析
2.1 小鼠8个环分子筛选和目的分子确定
通过分析Dong等[17]结果和circBase中注释的小鼠circGHR序列信息,本研究绘制出小鼠8个circGHR的结构示意图(图2)。针对每个环分子设计特异引物,用同一只7周龄雄鼠肝脏组织RNA作为模板,反转录得到cDNA后用表1中8对引物进行PCR扩增,PCR产物由生工生物工程(上海)股份有限公司进行测序。对测序结果进行分析后,发现引物2、3、4、6的测序结果序列一致,均对circGHR3的接口位置进行验证,因此后续分析选择circGHR3作为实验目标分子,并命名为circGHR。其余7个分子均未测序验证成功。图2
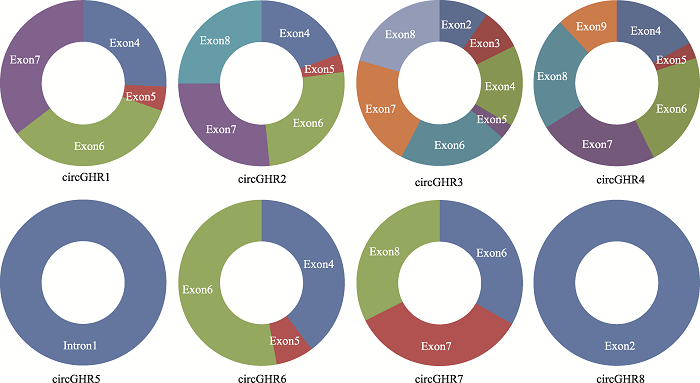 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2小鼠8个circGHR序列组成和结构示意图
Fig. 2Schematic diagram of the sequence composition and structure of eight mouse circGHR
2.2 小鼠circGHR连接成环验证及其全长序列分析
本研究针对circGHR连接成环的接头位置设计特异性分散引物,以经过RNase R处理后的7周龄小鼠肝脏组织总RNA反转录得到的cDNA为模板进行PCR扩增。结果表明,在343 bp位置产生了符合circGHR接头位置片段长度的预期目标片段,初步表明GHR基因存在向后剪切产物。测序结果显示,circGHR连接成环的接头片段序列来源于GHR基因外显子2和8 (图3,A和B),该结果与生物信息学预测分析circGHR3分子结果完全吻合。克隆结果表明,小鼠circGHR全长为820 bp,由GHR基因外显子2~8组成,向后剪切位点为外显子2和外显子8 (图3,C和D)。图3
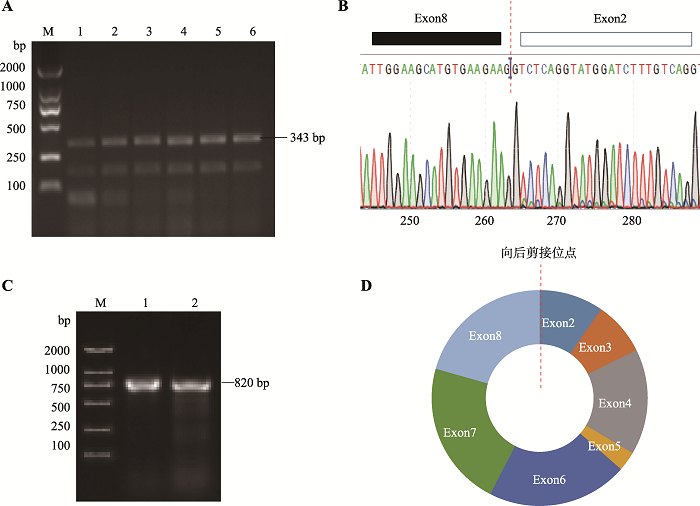 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3小鼠circGHR全长克隆及结构验证分析
A:circGHR 接头位置扩增结果。M:DL2000 maker;泳道1~6:以7周龄昆明小鼠肝脏组织cDNA为模板,circGHR-D引物扩增结果。B:circGHR-DL引物扩增产物测序结果。C:circGHR全长扩增结果。M:DL2000 maker;泳道1~2:以7周龄昆明小鼠肝脏组织cDNA为模板,circGHR-full引物扩增产物。D:小鼠circGHR结构示意图。
Fig. 3Full-length cloning and structure verification analysis of mouse circGHR
2.3 小鼠circGHR分子RNase R耐受性及不同反转录引物的反转录效率分析
环形RNA分子具有RNase R耐受性,不易被RNase R降解;而线性RNA分子不具备RNase R耐受性。为了确认小鼠circGHR具备环形分子的一般特征,本研究分析了小鼠circGHR的RNase R耐受性。以7周龄小鼠肝脏组织总RNA为材料,经RNase R处理后,使用随机引物(N9)反转录为cDNA,qRT-PCR结果显示,使用RNase R处理后,circGHR的表达丰度无明显下降(P>0.05),反而呈现增加的趋势,但是线性分子GHR mRNA经RNase R处理后下降极显著(P<0.01),说明circGHR具有较强的RNase R耐受性。此外,以7周龄小鼠肝脏组织总RNA为模板,分别使用oligo-d(T)18和随机引物(N9)反转录合成cDNA,qRT-PCR比较circGHR的反转录效率。结果表明,用oligo-d(T)18引物反转录后circGHR的表达丰度差异极显著(P<0.01)。因此,与oligo-d(T)18引物的反转录效果相比,随机引物对circGHR具有较好的反转录效果,同时也说明circGHR是一个不含poly(A)结构的环状分子。图4
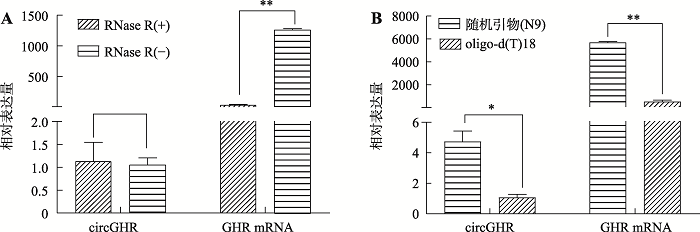 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4小鼠circGHR和GHR mRNA对RNase R耐受性及反转录特征分析
A:RNase R对cricGHR和GHR mRNA表达丰度的影响;B:不同反转录引物对circGHR和GHR mRNA表达丰度的影响。RNase R(-):未经RNase R处理过的总RNA;RNase R(+):经RNase R处理过的总RNA;n=3,用2-ΔΔCT 方法计算 qRT-PCR 结果;*表示P<0.05,差异显著;**表示P<0.01,差异极显著。
Fig. 4Resistance of the mouse circGHR and GHR mRNA to RNase R and analysis of reverse transcription characteristics
2.4 小鼠circGHR和线性GHR mRNA时空表达规律
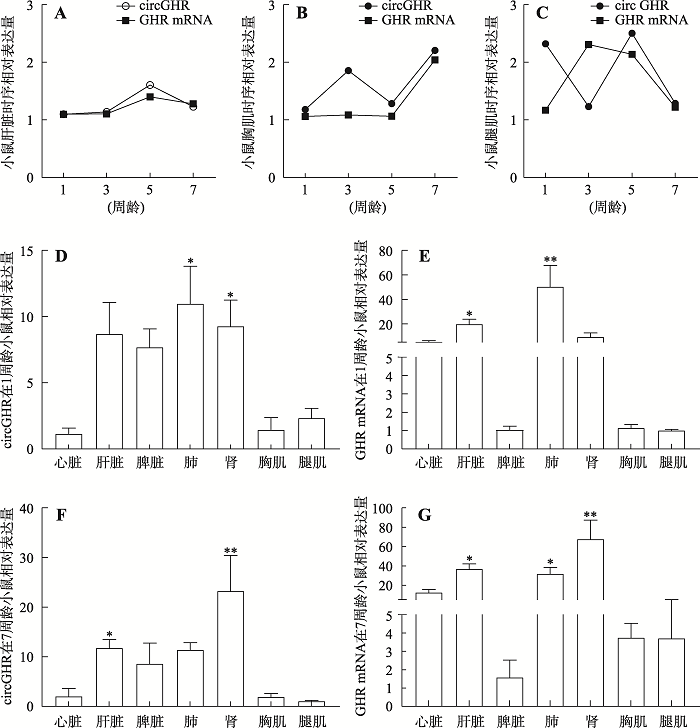
分别提取1、3、5和7周龄雄鼠肝脏、胸肌、腿肌、心脏等组织RNA,经反转录合成cDNA,利用qRT-PCR对circGHR和GHR mRNA的表达量进行分析。通过分析发现1、3、5和7周龄小鼠肝脏、胸肌中circGHR和GHR mRNA的时序表达量规律一致,且表达量都呈上升趋势,有可能是GHR基因表达量上升导致该结果,也暗示两种分子可能存在协同作用(图5,A和B);而在腿肌中3周龄以前两者呈负相关,3周龄以后呈正相关(图5C)。circGHR和GHR mRNA的组织表达谱显示,两者在肝脏、肾脏以及肺组织高表达,而在胸肌和腿肌组织低表达。结果表明,circGHR 在 1 周龄小鼠的肺和肾组织中高表达,而在心脏、腿肌和胸肌组织中低表达(图5D),circGHR 在 7周龄小鼠的肾脏中高表达,而在胸肌和腿肌中低表达(图5F);在1周龄小鼠中发现GHR mRNA在肝脏和肺组织中高表达,而在脾脏、胸肌和腿肌组织低表达(图5E);在7周龄小鼠中发现GHR mRNA在肝脏、肺和肾组织高表达,在脾脏、胸肌和腿肌组织低表达(图5G)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5小鼠circGHR和GHR mRNA的时空表达规律
A:1、3、5和7周龄小鼠肝脏组织中circGHR和GHR mRNA的表达规律;B:1、3、5和7周龄小鼠胸肌组织中circGHR和GHR mRNA的表达规律;C:1、3、5和7周龄小鼠腿肌组织中circGHR和GHR mRNA的表达规律;D:1周龄小鼠不同组织中circGHR的表达规律;E:1周龄小鼠不同组织中GHR mRNA的表达规律;F:7周龄小鼠不同组织中circGHR的表达规律;G:7周龄小鼠不同组织中GHR mRNA的表达规律。图A、B、C中n=12,图D、E中n=3,图F、G中n=3;使用2-ΔΔCT方法计算qRT-PCR结果;*表示P<0.05,差异显著;**表示P<0.01,差异极显著。
Fig. 5The spatiotemporal expression pattern of mouse circGHR and GHR mRNA
2.5 小鼠circGHR编码蛋白质的潜能分析
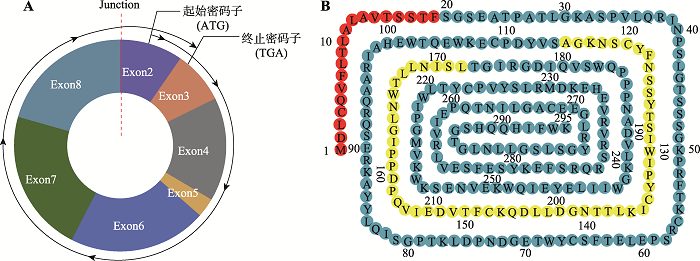
2.5.1 小鼠circGHR开放阅读框(ORF)分析小鼠circGHR全长为820 nt,由GHR基因外显子2~8环化形成,使用SnapGene软件对该段序列进行ORF预测,发现这段序列具有一个完整的ORF结构,长度为888 nt,可能具有蛋白质编码潜能。ORF结构从外显子2的第10个碱基开始编码(ATG),围绕环状RNA编码一周后,跨过junction位置,再次在外显子2的第77个碱基处终止(TGA),如图6A所示。开放阅读框编码一个长度为295 aa的多肽,如图6B所示。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6小鼠circGHR预测的ORF结构示意图及其编码的多肽功能区域分析
A:开放阅读框结构示意图。带箭头的黑线表示小鼠circGHR的ORF,从起始密码子开始编码,跨过Junction位置到终止密码子结束;B:氨基酸序列和功能区域示意图。红色氨基酸表示信号肽段,黄色表示内部核糖体进入位点序列(IRES)。
Fig. 6Schematic diagram of the ORF structure predicted by mouse circGHR and analysis of the functional regions of the encoded polypeptide
2.5.2 利用Singal P分析氨基酸信号肽
蛋白质合成后通过“信号肽”来实现分选或转运功能,每一个需要运输的多肽都含有一段氨基酸序列,称为信号肽序列(signal peptide, SP),引导多肽至不同的转运系统。对GHR基因的线性mRNA和circGHR编码氨基酸序列的信号肽进行分析,发现两者存在同一个信号肽(Sec/SPI),即“标准”分泌信号肽(图6B),由Sec转运蛋白转运并被信号肽酶I (Lep)切割。根据预测结果,分析circGHR基因可能存在编码功能,且合成的蛋白质与GHR基因编码蛋白质在核糖体合成后,通过信号肽转运发挥作用。
2.5.3 小鼠circGHR内部核糖体进入位点序列(IRES)预测分析
真核mRNA的翻译都需要5ʹ帽子来介导核糖体结合,但环状RNA没有帽子结构,因此具有一段较
短的RNA序列(约150~250 bp),这类RNA序列能折叠成类似于起始tRNA的结构,从而介导核糖体与RNA结合,起始蛋白质翻译[18],这段非翻译RNA被称为内部核糖体进入位点序列(internal ribosome entry site, IRES)。用 IRESfinder 软件对该序列进行预测,发现小鼠circGHR存在多个IRES,其中预测分值最高的IRES片段如图6B所示。
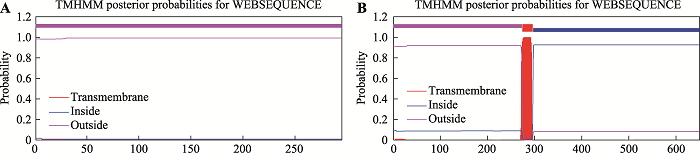
2.5.4 对比分析小鼠GHR环RNA和线性mRNA编码蛋白跨膜区
利用在线软件TMHMM Server v.2.0 (
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7小鼠circGHR和GHR mRNA编码蛋白跨膜区对比分析
A:circGHR编码的蛋白跨膜区预测结果;B:GHR mRNA编码的蛋白跨膜区预测结果。
Fig. 7Comparative analysis of transmembrane regions of mouse circGHR and GHR mRNA encoding proteins
3 讨论
在本课题组前期实验的基础上,本研究筛选出8个circGHR,并设计8对引物对其进行扩增,结果发现其中4对引物都能对circGHR3进行扩增,而其他7个分子均未验证成功。因此circGHR3可能是小鼠肝脏组织中存在最多,最为广泛的GHR基因环状转录本。通过对其进行克隆,发现circGHR由该基因外显子2到外显子8向后剪切形成。环状RNA具有不含 poly(A)序列的闭环结构,因此oligo-d(T)18引物反转录获得的circGHR丰度低于随机引物,提示在环形 RNA 反转录时宜选用随机引物进行反转录,同时该结果也间接表明circGHR是一个头尾相连的环状结构。RNase R是一种靶向线性RNA分子并对其具有切割作用的核酸酶,但对环形分子不敏感[19]。本研究表明,小鼠circGHR对RNase R表现出较强的耐受性,总RNA经RNase R处理后,circGHR表达量变化不显著,进一步说明小鼠circGHR具备环形分子的一般特征。
研究表明,基因的环形和线性转录本的表达存在相关性[20]。从本研究结果可以发现在肝脏和胸肌组织中,circGHR与GHR mRNA的时序表达基本一致,转录水平呈现持续上升,表明两者可能存在相互作用,也有可能是随着小鼠的发育整个基因组转录水平上升,导致circGHR与GHR mRNA同时上升;在腿肌组织中1~3周两者表达趋势相反,3周以后表达趋势相同,推测可能是由于不同个体样本间差异较大造成,也可能是由于基因定量的样本数少所致。此外,小鼠GHR mRNA与circGHR广泛存在于各种组织中,但表达具有一定的差异。有研究发现,GHR基因敲除后小鼠肺部发育相关蛋白表达明显降低[21];同样对20日龄鸡胚肺部组织进行检测,同样也发现少量GHR[22]。本研究中,1周龄小鼠中GHR mRNA与circGHR都是高表达,说明GHR可能参与早期肺部生长发育,并与肺部的氧化保护、脂质和能量代谢以及蛋白酶体活性等生理活动有关。在患糖尿病的动物肾脏中,GHR在肾脏皮质区和髓质区高表达[23];在对小鼠肾脏的研究中发现,肾脏的皮质和髓质区域存在GHR[24]。本研究中,7周龄小鼠GHR mRNA与circGHR在肾脏中高表达,分析这可能与肾小管的重吸收和上皮细胞的分泌功能有关。
circRNA具有miRNA海绵、调控转录、结合蛋白质和翻译等作用,本研究通过SnapGene软件预测发现circGHR存在一个ORF,并且对其翻译的蛋白进行了生物信息学分析。首先对与两种蛋白进行信号肽分析,发现两者存在同一个“标准”分泌信号肽,这提示两种蛋白都属于分泌性蛋白,即穿过合成所在的细胞到其他组织细胞去的蛋白质[25]。由于环状RNA缺乏帽子结构,所以环状RNA的翻译应通过IRES进行,IRES可促进核糖体与被翻译的环状RNA结合[26]。对该ORF进行IRES分析,发现有多段IRES,且有两段序列可能性超过80%,表明该circGHR可能通过核糖体介入位点介导翻译,因此可以通过检测这些IRES能否启动编码,间接验证circGHR能否被编码,为验证circGHR的编码能力提供方法。最后通过对两种蛋白跨膜区进行分析,发现circGHR编码蛋白不具有跨膜结构,氨基酸位于膜的外侧发挥作用;而GHR基因则具有一个跨膜结构,印证了GHR属于单链跨膜糖蛋白。通过该结果推测circGHR编码的蛋白质在细胞膜外发挥作用,与GHR mRNA编码的蛋白质作用位置不同。关于小鼠circGHR的作用机制尚需进一步研究证实,以期为将来阐述GHR涉及的调控网络及其调控动物生长发育理论奠定基础。
(责任编委: 单革)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
PMID:6356363 [本文引用: 1]

The promoter or regulatory region of the mouse gene for metallothionein-I was fused to the structural gene coding for human growth hormone. These fusion genes were introduced into mice by microinjection of fertilized eggs. Twenty-three (70 percent) of the mice that stably incorporated the fusion genes showed high concentrations of human growth hormone in their serum and grew significantly larger than control mice. Synthesis of human growth hormone was induced further by cadmium or zinc, which normally induce metallothionein gene expression. Transgenic mice that expressed human growth hormone also showed increased concentrations of insulin-like growth factor I in their serum. Histology of their pituitaries suggests dysfunction of the cells that normally synthesize growth hormone. The fusion genes were expressed in all tissues examined, but the ratio of human growth hormone messenger RNA to endogenous metallothionein-I messenger RNA varied among different tissues and different animals, suggesting that expression of the foreign genes is influenced by site of integration and tissue environment.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
PMID:8874495 [本文引用: 1]

Growth hormone (GH) has long been known to stimulate linear growth and regulate metabolism. The cellular mechanism by which GH elicits these effects has only recently begun to be understood. This review provides an overview of a current model of GH signaling. Briefly, binding of GH to GH receptor induces receptor dimerization and activation of the tyrosine kinase JAK2. Tyrosyl phosphorylation of GH receptor and JAK2 recruits and activates signaling molecules such as Stat transcription factors, SHC, and insulin receptor substrates 1 and 2 that lead to the release of second messengers such as diacylglycerol, calcium, and nitric oxide and the activation of enzymes such as mitogen-activated protein kinase, protein kinase C, phospholipase A2, and phosphatidylinositol 3'-kinase. These pathways regulate cellular function including gene transcription, metabolite transport, and enzymatic activity that result in the ability of GH to control body growth and metabolism.
[本文引用: 1]
[本文引用: 1]
DOI:10.1073/pnas.73.11.3852URL [本文引用: 1]
DOI:10.1261/rna.035667.112URL [本文引用: 1]
DOI:10.1093/nar/gkl151URL [本文引用: 1]
DOI:10.3390/ijms15069331URL [本文引用: 1]
DOI:10.1007/s11427-015-4855-yURL [本文引用: 1]
DOI:10.2174/1389202916666150707161554URL [本文引用: 1]
DOI:10.1080/21541264.2015.1071301URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/nsmb.2959URL [本文引用: 1]
DOI:S1097-2765(17)30132-6PMID:28344082 [本文引用: 1]

Circular RNAs (circRNAs) constitute a family of transcripts with unique structures and still largely unknown functions. Their biogenesis, which proceeds via a back-splicing reaction, is fairly well characterized, whereas their role in the modulation of physiologically relevant processes is still unclear. Here we performed expression profiling of circRNAs during in vitro differentiation of murine and human myoblasts, and we identified conserved species regulated in myogenesis and altered in Duchenne muscular dystrophy. A high-content functional genomic screen allowed the study of their functional role in muscle differentiation. One of them, circ-ZNF609, resulted in specifically controlling myoblast proliferation. Circ-ZNF609 contains an open reading frame spanning from the start codon, in common with the linear transcript, and terminating at an in-frame STOP codon, created upon circularization. Circ-ZNF609 is associated with heavy polysomes, and it is translated into a protein in a splicing-dependent and cap-independent manner, providing an example of a protein-coding circRNA in eukaryotes.Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
DOI:10.1186/s12943-019-1010-6PMID:30925892 [本文引用: 1]

It has been well established that circular RNAs (circRNAs) play an important regulatory role during tumor progression. Recent studies have indicated that even though circRNAs generally regulate gene expression through miRNA sponges, they may encode small peptides in tumor pathogenesis. However, it remains largely unexplored whether circRNAs are involved in the tumorigenesis of colon cancer (CC).The expression profiles of circRNAs in CC tissues were assessed by circRNA microarray. Quantitative real-time PCR, RNase R digestion assay and tissue microarray were used to confirm the existence and expression pattern of circPPP1R12A. The subcellular distribution of circPPP1R12A was analyzed by nuclear mass separation assay and fluorescence in situ hybridization (FISH). SDS-PAGE and LC/MS were employed to evaluate the protein-coding ability of circPPP1R12A. CC cells were stably transfected with lentivirus approach, and cell proliferation, migration and invasion, as well as tumorigenesis and metastasis in nude mice were assessed to clarify the functional roles of circPPP1R12A and its encoded protein circPPP1R12A-73aa. RNA-sequencing and Western blotting analysis were furthered employed to identify the critical signaling pathway regulated by circPPP1R12A-73aa.We firstly screened the expression profiles of human circRNAs in CC tissues and found that the expression of hsa_circ_0000423 (termed as circPPP1R12A) was significantly increased in CC tissues. We also found that circPPP1R12A was mostly localized in the cytoplasm of CC cells. Kaplan-Meier analysis showed that patients with higher levels of circPPP1R12A had a significantly shorter overall survival. By gain- and loss-of-function approaches, the results suggested that circPPP1R12A played a critical role in proliferation, migration and invasion of CC cells. Furthermore, we showed that circPPP1R12A carried an open reading frame (ORF), which encoded a functional protein (termed as circPPP1R12A-73aa). Next, we found that PPP1R12A-C, not circPPP1R12A, promoted the proliferation, migration and invasion abilities of CC in vitro and in vivo. Finally, we identified that circPPP1R12A-73aa promoted the growth and metastasis of CC via activating Hippo-YAP signaling pathway. In addition, the YAP specific inhibitor Peptide 17 dramatically alleviated the promotive effect of circPPP1R12A-73aa on CC cells.In the present study, we illustrated the coding-potential of circRNA circPPP1R12A in the progression of CC. Moreover, we identified that circPPP1R12A-73aa promoted the tumor pathogenesis and metastasis of CC via activating Hippo-YAP signaling pathway. Our findings might provide valuable insights into the development of novel potential therapeutic targets for CC.
DOI:10.1080/15476286.2016.1269999PMID:27982734 [本文引用: 2]

Circular RNAs (circRNAs) are broadly identified from precursor mRNA (pre-mRNA) back-splicing across various species. Recent studies have suggested a cell-/tissue- specific manner of circRNA expression. However, the distinct expression pattern of circRNAs among species and its underlying mechanism still remain to be explored. Here, we systematically compared circRNA expression from human and mouse, and found that only a small portion of human circRNAs could be determined in parallel mouse samples. The conserved circRNA expression between human and mouse is correlated with the existence of orientation-opposite complementary sequences in introns that flank back-spliced exons in both species, but not the circRNA sequences themselves. Quantification of RNA pairing capacity of orientation-opposite complementary sequences across circRNA-flanking introns by Complementary Sequence Index (CSI) identifies that among all types of complementary sequences, SINEs, especially Alu elements in human, contribute the most for circRNA formation and that their diverse distribution across species leads to the increased complexity of circRNA expression during species evolution. Together, our integrated and comparative reference catalog of circRNAs in different species reveals a species-specific pattern of circRNA expression and suggests a previously under-appreciated impact of fast-evolved SINEs on the regulation of (circRNA) gene expression.
[本文引用: 1]
[本文引用: 1]
PMID:16893880 [本文引用: 1]

RNase R is a processive, 3' to 5' hydrolytic exoribonuclease that together with polynucleotide phosphorylase plays an important role in the degradation of structured RNAs. However, RNase R differs from other exoribonucleases in that it can by itself degrade RNAs with extensive secondary structure provided that a single-stranded 3' overhang is present. Using a variety of specifically designed substrates, we show here that a 3' overhang of at least 7 nucleotides is required for tight binding and activity, whereas optimum binding and activity are achieved when the overhang is 10 or more nucleotides in length. In contrast, duplex RNAs with no overhang or with a 4-nucleotide overhang bind extremely poorly to RNase R and are inactive as substrates. A duplex RNA with a 10-nucleotide 5' overhang also is not a substrate. Interestingly, this molecule is bound only weakly, indicating that RNase R does not simply recognize single-stranded RNA, but the RNA must thread into the enzyme with 3' to 5' polarity. We also show that ribose moieties are required for recognition of the substrate as a whole since RNase R is unable to bind or degrade single-stranded DNA. However, RNA molecules with deoxyribose or dideoxyribose residues at their 3' termini can be bound and degraded. Based on these data and a homology model of RNase R, derived from the structure of the closely related enzyme, RNase II, we present a model for how RNase R interacts with its substrates and degrades RNA.
DOI:10.1016/j.molcel.2015.03.027PMID:25921068 [本文引用: 1]

Circular RNAs (circRNAs) are an endogenous class of animal RNAs. Despite their abundance, their function and expression in the nervous system are unknown. Therefore, we sequenced RNA from different brain regions, primary neurons, isolated synapses, as well as during neuronal differentiation. Using these and other available data, we discovered and analyzed thousands of neuronal human and mouse circRNAs. circRNAs were extraordinarily enriched in the mammalian brain, well conserved in sequence, often expressed as circRNAs in both human and mouse, and sometimes even detected in Drosophila brains. circRNAs were overall upregulated during neuronal differentiation, highly enriched in synapses, and often differentially expressed compared to their mRNA isoforms. circRNA expression correlated negatively with expression of the RNA-editing enzyme ADAR1. Knockdown of ADAR1 induced elevated circRNA expression. Together, we provide a circRNA brain expression atlas and evidence for important circRNA functions and values as biomarkers. Copyright © 2015 Elsevier Inc. All rights reserved.
DOI:10.1002/(ISSN)1615-9861URL [本文引用: 1]
DOI:10.1007/s00441-005-0040-0URL [本文引用: 1]
PMID:10990443 [本文引用: 1]

Growth hormone (GH) may have a role in the development of diabetic nephropathy. The effect of experimental diabetes on renal expression of the growth hormone receptor gene products, including the receptor itself (GHR) and its binding protein (GHBP) was examined. Adult female rats received i.v. streptozotocin and were killed at 7, 30, 90 and 180 days after the induction of diabetes. Diabetic animals had a pronounced increase in kidney weight and progressive albuminuria. In renal cortex, no change was seen in GHR mRNA levels throughout the observation period of 6 months, while a significant increase in cortical GHBP mRNA levels was observed after 1 month of diabetes and sustained for the rest of the study period. Immunohistochemical analysis of kidney sections revealed a stronger staining for GHBP at the cortical and inner medullary areas in the diabetic animals. These data indicate that although the GHR and GHBP mRNAs originate from the same gene, their renal levels are differentially regulated during the development of experimental diabetic kidney disease, suggesting a functional role for GHBP.
DOI:10.1016/j.ghir.2012.08.003URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.canlet.2020.10.002PMID:33038492 [本文引用: 1]

The human genome contains thousands of noncoding RNAs (ncRNAs), which are thought to lack open reading frames (ORFs) and cannot be translated. Some ncRNAs reportedly have important functions, including epigenetic regulation, chromatin remolding, protein modification, and RNA degradation, but the functions of most ncRNAs remain elusive. Through the application and development of ribosome profiling and sequencing technologies, an increasing number of studies have discovered the translation of ncRNAs. Although ncRNAs were initially defined as noncoding RNAs, a number of ncRNAs actually contain ORFs that are translated into peptides. Here, we summarize the available methods, tools, and databases for identifying and validating ncRNA-encoded peptides/proteins, and the recent findings regarding ncRNA-encoded small peptides/proteins in cancer are compiled and synthesized. Importantly, the role of ncRNA-encoding peptides/proteins has application prospects in cancer research, but some potential challenges remain unresolved. The aim of this review is to provide a theoretical basis that might promote the discovery of more peptides/proteins encoded by ncRNAs and aid the further development of novel diagnostic and prognostic cancer markers and therapeutic targets.Copyright © 2020 Elsevier B.V. All rights reserved.
