Cloning of the soybean E2 ubiquitin-conjugating enzyme GmUBC1 and its expression in Arabidopsis thaliana
Zhuozhuo Mao, Yu Gong, Guixia Shi, Yali Li, Deyue Yu, Fang HuangState Key Laboratory of Crop Genetics & Germplasm Enhancement, National Center for Soybean Improvement, Key Laboratory of Biology and Genetic Improvement of Soybean, Ministry of Agriculture, Nanjing Agricultural University, Nanjing 210095, China通讯作者: 黄方,博士,教授,博士生导师,研究方向:大豆遗传育种。E-mail:fhuang@njau.edu.cn
编委: 储成才
收稿日期:2020-05-21修回日期:2020-07-7网络出版日期:2020-08-20
| 基金资助: |
Received:2020-05-21Revised:2020-07-7Online:2020-08-20
| Fund supported: |
作者简介 About authors
毛卓卓,在读硕士研究生,专业方向:作物遗传育种。。

摘要
E2泛素结合酶(ubiquitin-conjugating enzyme)参与植物的抗逆、生长发育等多种生物途径的调控,其功能研究在拟南芥(Arabidopsis thaliana)中的报道较多,但在重要经济作物大豆(Glycine max)中鲜有报道。本研究从大豆“南农94-16”中克隆了1个与大豆子叶折叠突变体可能相关的基因Glyma.12G161200,序列分析结果表明该基因编码一个E2泛素结合酶,因此将其命名为GmUBC1。该基因编码区全长为462 bp,编码一个含153个氨基酸的蛋白,预测其分子量为17.25 kDa,等电点为6.74。利用qRT-PCR技术对GmUBC1在大豆不同组织中的表达模式及其对不同非生物胁迫和激素处理的响应模式进行分析,发现该基因在开花后40 d种子中表达量最高,在PEG、低温、JA和ABA处理下表达下调。亚细胞定位分析发现GmUBC1蛋白在整个细胞内都有表达。进一步在拟南芥中异源表达GmUBC1,发现转基因株系的千粒重和总氨基酸含量显著提高,表明异源过表达GmUBC1可以调控种子重量和氨基酸含量,为大豆品质改良提供了基因资源。
关键词:
Abstract
Plant E2 Ubiquitin-conjugating enzymes regulate various biological pathways such as stress resistance, growth and development. Reports on its functions are more frequent in Arabidopsis thaliana, but relatively rare in soybean (Glycine max), which is one of the most important economic crops. In this study, a gene Glyma.12G161200, which may be related to the soybean cotyledon folding mutant, was cloned from soybean “Nanong 94-16”. Analysis of its sequence suggested that it encodes an E2 ubiquitin binding enzyme, so it was named as GmUBC1. Its coding region is 462 bp in length, which encodes a protein of 153 amino acids with a predicted molecular mass of 17.25 kDa and an isoelectric point of 6.74. The expression pattern of GmUBC1 in different tissues of soybean and its response patterns to different stresses and hormone treatments were analyzed by real-time PCR. The results showed that the gene was expressed at the highest level in mutant seeds at 40 days after flowering. Moreover, the expression of the GmUBC1 gene was down-regulated by the treatments of PEG, cold, JA and ABA, respectively. Subcellular localization analysis of GmUBC1 revealed that the protein was expressed in the whole cell. When GmUBC1 was ectopically expressed in Arabidopsis, the 1000-grain weight and total amino acid content of some transgenic lines were found to be significantly increased. Collectively, heterologous overexpression of GmUBC1 can regulate seed weights and amino acid contents, which may provide genetic resources for soybean quality improvement.
Keywords:
PDF (1046KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
毛卓卓, 宫宇, 史贵霞, 李亚丽, 喻德跃, 黄方. 大豆E2泛素结合酶基因GmUBC1的克隆及在拟南芥中的异源表达. 遗传[J], 2020, 42(8): 788-798 doi:10.16288/j.yczz.20-141
Zhuozhuo Mao.
泛素/26S蛋白酶体途径是目前研究较多的植物蛋白降解系统机制之一。泛素是一种高度保守且由76个氨基酸组成的小分子蛋白,分子量约为8.5 kDa,几乎存在于所有真核生物体内[1]。该途径由一系列泛素酶及类泛素酶参与,如E1泛素激活酶(ubiquitin-activating enzyme)、E2泛素结合酶(ubiquitin-conjugating enzyme)、E3泛素连接酶(ubiquitin ligase E3)以及一些蛋白酶体等。该途径广泛参与植物激素信号转导[2,3,4],在维持细胞功能以及细胞周期运转、抵御环境胁迫、激素响应、胚胎发育和衰老等方面发挥着重要的作用[5,6,7,8,9,10,11,12,13,14]。植物泛素蛋白酶体途径在拟南芥中研究的最为深入,拟南芥中约有1400个基因编码的蛋白(约占总蛋白质数量的5%)参与蛋白泛素化系统[13]。其中编码E2泛素结合酶的基因有37个,编码E2泛素结合酶类似蛋白的基因为8个[8]。E2泛素结合酶由150个氨基酸组成的保守催化结构域组成,内含一个高度保守的半胱氨酸保守位点。研究发现,在真核生物中许多编码泛素结合酶的基因参与抗逆、生长发育等多种生物途径的调控,如在多种非生物胁迫处理下,泛素结合酶基因GmUBC13的表达量变化显著,同时该基因在烟草(Nicotiana tabacum)中过表达可以显著提高烟草的抗旱性[15]。陈超等[16]研究发现,大豆GmUBC46基因在酵母中异源表达降低了酵母对干旱和盐胁迫的耐受性。此外,Wan等[17]研究发现E2泛素结合酶在调节植物生长发育等多方面也发挥着重要的作用。Vierstra等[18]研究表明,Atubc1-1 Atubc2-1双突变体拟南芥能抑制FLC、MAF4和MAF5基因的表达,从而表现出早花的表型,同时该双突变体还表现出莲座叶数量显著减少、叶片表面增大、茎直径减小以及花序茎中的维管束数量减少等表型。Wang等[19]通过VIGS (virus induced gene silencing)技术对番茄(Solanum lycopersicum) E2泛素结合酶基因SlUBC32和SlUBC41进行沉默后,发现这两个基因参与了果实成熟的调控,使番茄果实成熟期延迟。在葡萄(Vitis vinifera)中的研究显示,编码E2泛素结合酶基因VvUBC3/7/12/34在葡萄浆果发育期间以及在收获后的生理过程中发挥着重要的作用[20]。尽管E2泛素结合酶在多种植物中的功能研究越来越多,但是其在大豆中的报道相对较少。
本课题组前期为了探索大豆子叶折叠突变体cco (curled-cotyledon)形成的分子机制,基于比较基因组杂交芯片(comparative genomic hybridization, CGH)[21]和RNA-seq[22]结果筛选出一个在大豆子叶折叠突变体cco与栽培大豆品种“南农94-16”中表达差异的基因。本研究进一步从大豆中克隆了该基因,序列分析结果显示其编码一个泛素结合酶,因此将其命名为GmUBC1。进一步利用荧光定量PCR技术分析了GmUBC1基因在组织器官和在各种胁迫处理(盐碱、干旱、低温、JA、ABA)下的表达特性,构建了该基因的植物表达载体并转化拟南芥,发现过表达GmUBC1的转基因拟南芥种子千粒重以及氨基酸含量显著提高。本研究结果为进一步研究大豆E2泛素结合酶参与植物发育和逆境中的作用提供参考。
1 材料与方法
1.1 植物材料、菌株及试剂
大豆栽培品种“南农94-16”(WT)由南京农业大学国家大豆改良中心种质资源库提供,大豆子叶折叠突变体(cco)由本实验室通过NaN3-60COγ诱变大豆栽培品种“南农94-16”获得,材料均种植于南京农业大学江浦试验场。出苗后1个月左右,分别取WT和cco的根、茎、叶、花和开花后7 d、10 d、15 d、30 d、40 d的豆荚和种子。取样后立即液氮速冻,-80℃备用。用于胁迫处理的大豆品种Williams82种植于本实验室光照培养箱内。拟南芥Columbia-0生态型种子由本实验室保存。根癌农杆菌EHA105及植物表达载体pMDC83由国家大豆改良中心实验室保存。
1.2 GmUBC1基因CDS全长序列的克隆
选取“南农94-16”花为材料,利用RNA提取试剂盒(天根生化科技(北京)有限公司)提取RNA,之后反转录成cDNA。利用Phytozome数据库预测其完整的基因序列,以cDNA为模板,进行PCR扩增。扩增引物序列如下:GmUBC1F1:5?-TTTGCTGATTCTCTCGGCGT-3?;GmUBC1R1:5?-AGGTAACTGGAAACTGCGAGG-3?。50 μL PCR扩增体系包括:KOD-Plus-Neo 0.25 μL,10×PCR Buffer 5 μL,25 mmol/L MgSO4 4 μL,2 mmol/L dNTPs 4 μL,cDNA 2 μL,上下游引物各2 μL,补灭菌ddH2O至50 μL。扩增条件:95℃预变性5 min;94℃变性30 s,57℃退火30 s,72℃延伸30 s,31个循环;最后再72℃延伸10 min,4℃保存备用。扩增产物用1%琼脂糖凝胶进行电泳分离(电压110 V),目的片段用胶回收试剂盒回收后检测浓度。胶回收产物首先用Taq酶加polyA尾,再连接到pMD19-T载体上。转化大肠杆菌DH5α感受态细胞,涂布于含有筛选抗生素的培养基上。挑取单克隆进行菌液PCR检测,测定正确后,进行测序验证。
1.3 GmUBC1基因生物信息学分析
通过Phytozome网站(1.4 GmUBC1基因组织表达分析以及在不同胁迫处理下的表达模式分析
提取大豆突变体cco和“南农94-16”不同时期的组织(根、茎、叶、花以及开花后7 d、10 d、15 d、30 d、40 d豆荚和种子)的RNA,反转录成cDNA。以cDNA为模板,根据大豆基因组Phytozome数据库中该基因的CDS序列,利用Prime5.0设计荧光定量PCR引物,序列为:GmUBC1F2:5?-GGCGTTGTTCATTTCTGTCTCA-3?,GmUBC1R2:5?-TCGAGTTCGATTGCAAAACGT-3?。以大豆组成型表达基因Tubulin (GenBank登录号:AY907703)作为内参,扩增引物序列为:GmtubullinF:5?-GGAGTTCACAGAGGCAGAG-3?,GmtubullinR:5?-CACTTACGCATCACATAGCA-3?。荧光定量PCR扩增程序为:95℃预变性1 min;95℃变性15 s,60℃退火15 s,72℃延伸45 s,40个循环;72℃延伸10 min终止反应。PCR扩增结束后通过荧光值变化曲线以及溶解曲线分析基因的表达情况。分别采集用15% 聚乙二醇(polyethylene glycol, PEG)处理0 h、0.5 h、2 h,4℃处理0 h、2 h、4 h以及250 mmol/L 氯化钠(NaCl)、50 mol/L 茉莉酸(jasmonic acid, JA)、100 μmol/L脱落酸(abscisic acid, ABA)胁迫诱导0 h、3 h、6 h后的大豆Williams82叶片立即液氮冷冻,-80℃保存备用。提取经过不同非生物胁迫以及激素处理后的大豆Williams82叶片RNA,然后反转录成cDNA。以cDNA为模板,以大豆组成型表达基因Tubulin作为内参,进行qRT- PCR,验证GmUBC1基因是否受胁迫诱导。
1.5 GmUBC1基因植物表达载体的构建
本研究选择pMDC83作为表达载体,首先以测序正确的连接有全长GmUBC1 cDNA的pMD19-T载体为模板,扩增GmUBC1基因的开放阅读框(ORF),包含终止子。所用引物序列为:GmUBC1F3:5?- ggggacaagtttgtacaaaaaagcaggcttcATGGCCAACAGCAACCTCCC-3?,GmUBC1R3:5?-ggggaccactttgtacaagaaagctgggtcTCAGACGCCACTAGCATATA-3? (下划线的小写字母为接头引物序列)。用Gateway系统的BP反应将其完整的ORF通过同源重组构建到入门载体pDORNOR221上。测序正确后进行LR反应,将入门载体克隆产物与表达载体通过重组交换,将目的基因GmUBC1转化到pMDC83载体上,即完成pMDC83-GmUBC1表达载体的构建。经过菌液PCR及酶切鉴定后,转化农杆菌EHA105感受态细胞,挑取单克隆,培养后进行PCR检测。1.6 大豆GmUBC1亚细胞定位
取新鲜的洋葱,在基因枪轰击的前一天在超净台内剥取合适的内表皮,紧密贴合在MS固体培养基平板上,黑暗过夜培养,第二天待用。以测序正确的连接有全长GmUBC1载体为模板,扩增GmUBC1基因的ORF,将其与GFP报告基因融合,形成一个GmUBC1-GFP的嵌合基因。质粒金粉处理后之后利用基因枪法打入洋葱表皮细胞中,进行基因枪轰击后的洋葱表皮在室温暗环境下培养1 d后可进行激光共聚焦显微镜观察。1.7 蘸花法转化拟南芥
将含有重组载体pMDC83-GmUBC1质粒的阳性农杆菌菌液在含有卡那霉素(Kan)、潮霉素(HygB)、利福平(Rif)的 YEB固体培养基上划线培养24 h后,接种于含有相应抗生素的YEB液体培养基中,28℃、200 r/min震荡培养12~14 h,按1∶10左右比例转接培养。当OD值达到0.8~1.0后,离心收集菌液,加入含有0.3%表面活性剂Silwet的蔗糖溶液悬浮菌液,重新调至OD600=0.8左右。于拟南芥开花期,利用蘸花法将构建好的过表达载体pMDC83-GmUBC1农杆菌菌液侵染野生型拟南芥,农杆菌侵染当代的植株为T0代。1.8 转基因拟南芥抗性筛选及阳性植株的鉴定
将收获的T0代种子用70%酒精浸泡1 min,30%H2O2消毒10 min,于超净台内用灭菌水清洗干净之后,均匀种植于固体MS培养基上,培养基带有潮霉素抗性,置于人工智能培养箱培养15 d左右。根系生长正常的幼苗初步确定为阳性T1代株系。提取转基因拟南芥及对照野生型拟南芥植株叶片的DNA。取1 μL DNA为模板,以构建表达载体时的引物(GmUBC1F3和GmUBC1R3)进行PCR鉴定。设置含有GmUBC1基因编码框的质粒为阳性对照,野生型拟南芥的DNA为阴性对照。收获的T1代植株的种子为T2代,继续种于含有潮霉素抗性的平板上筛选,最终得到3个纯合的T3代株系,记为U20-3、U20-5、U20-7。1.9 转基因拟南芥种子千粒重以及氨基酸含量测定
将T3代成熟种子放置于2 mL离心管中,置于30℃烘箱一周以上,而后进行转基因种子千粒重的统计和氨基酸含量的测定。使用天平(AL104-LC)称取每个株系的千粒重,重复3~5次。称取至少0.12 g的拟南芥T3代种子于液氮中研磨后加入5 mL HCl浸提液过滤后定容至10 mL。之后加入4%磺基水杨酸,15,000 r/min离心15 min,过滤上清液,用氨基酸分析仪(L-8900)测氨基酸含量。相同提取条件下,每个样品重复3次。1.10 不同胁迫处理下转基因拟南芥种子的萌发率
选取U20-3、U20-5、U20-7各3个株系的转基因拟南芥和野生型拟南芥(WT)种子,用70%酒精浸泡1 min,30%H2O2消毒10 min,于超净台内用灭菌水清洗干净之后,分别种植于不同逆境(70 mmol/L的NaCl、6%的PEG和0.1 μmol/L ABA)处理的固体MS培养基上,将胚根突破种皮1 mm视为萌发,培养7 d后对种子萌发率进行统计并重复3次。数据分析采用Excel 2007,绘图采用ORIGIN 8.0软件。2 结果与分析
2.1 GmUBC1基因克隆
本研究以栽培大豆品种“南农94-16”花cDNA为模板进行PCR扩增,得到一个长度为462 bp的CDS片段。通过ExPASy proteomics 网站预测该基因编码的蛋白包含153个氨基酸,蛋白分子量为17.25 kDa,等电点为6.74。在NCBI网站将其氨基酸序列进行对比,发现其具有UBCc结构域,属于E2泛素结合酶家族,因此将该基因命名为GmUBC1。通过phytozome网站进一步获得该基因的基因组序列,全长为6785 bp,包含8个外显子和7个内含子。将GmUBC1氨基酸序列与拟南芥、水稻(Oryza sativa)、苜蓿(Medicago truncatula)、玉米(Zea mays)、菜豆(Phaseolus vulgaris)、番茄(Solanum lycopersicum)氨基酸序列进行比对,发现不同物种的E2泛素结合酶氨基酸序列高度相似,均具有一个典型的泛素结合酶UBCc结构域,属于E2泛素结合酶家族。其中,GmUBC1与拟南芥(AT1G16890)和番茄(Solyc07g062570)的氨基酸相似性最高(一致率为99.35%,相似率为100%),此外与苜蓿Medtr2g078010 (99.39%,98.69%)、玉米GRMZM5G862131 (97.39%,99.35%)、水稻LOC_Os01g48280 (97.39%,98.69%)和菜豆中Phvul.005G066900 (97.39%,99.35%)相似度也较高,推测各物种同源蛋白可能具有高度相似的功能(图1)。进一步将拟南芥、水稻、玉米、苜蓿、菜豆、番茄的UBCc结构域蛋白与GmUBC1蛋白构建系统进化树(图2),结果发现GmUBC1与拟南芥的AtUBC36、番茄的SIUBC聚为一枝,表明他们的亲缘关系较近。此外,系统进化树的结果显示双子叶植物,如大豆、拟南芥、菜豆等UBCc结构域蛋白聚为一枝,而单子叶植物,如水稻、玉米等单独聚为另外一枝,说明单子叶和双子叶植物的UBCc蛋白功能发生了分化。
图1
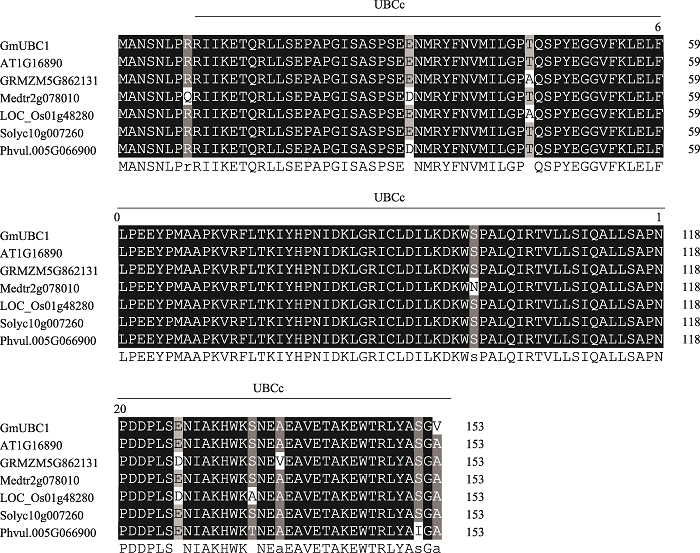 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1GmUBC1与其他植物的E2泛素结合酶的氨基酸序列比对
大豆:GmUBC1(Glyma.12G161200);苜蓿:Medtr2g078010;拟南芥:AT1G16890;菜豆:Phvul.005G066900;番茄:Solyc10g007260;玉米:GRMZM5G862131;水稻:LOC_Os01g48280。
Fig. 1Amino acid sequence alignment of GmUBC1 with other E2 ubiquitin binding enzyme homologs from different plants
图2
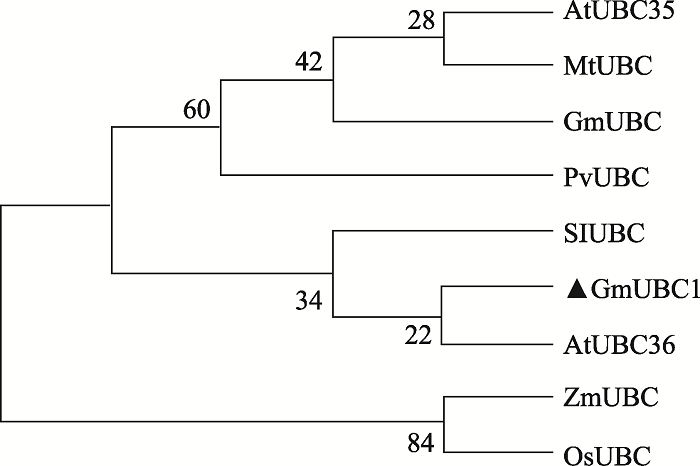 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2大豆GmUBC1与其他植物的系统进化树分析
大豆:GmUBC (Glyma.13G270800);苜蓿:MtUBC (Medtr2g078010);拟南芥:AtUBC35 (AT1G78870)、AtUBC36 (AT1G16890);菜豆:PvUBC (Phvul.005G066900);番茄:SIUBC2 (Solyc10g007260);玉米:ZmUBC1 (GRMZM5G862131);水稻:OsUBC (LOC_Os01g48280)。
Fig. 2Phylogenetic analysis of GmUBC1 of soybean and other plants
2.2 大豆GmUBC1基因在不同组织和胁迫诱导下的表达分析
为了研究GmUBC1基因在大豆栽培品种“南农94-16”(WT)和子叶折叠突变体(cco)不同组织中的表达模式,分别提取两种材料的根、茎、叶、花以及开花后10 d、15 d、30 d、40 d种子的RNA,反转成cDNA,利用qRT-PCR技术分析GmUBC1的表达情况。结果表明,GmUBC1基因在花和种子中的表达量相对较高,其中在开花后40 d的种子中表达量最高(图3)。本研究进一步分析了该基因在种子发育不同时期的表达量,结果表明与WT相比,GmUBC1在开花后30 d的子叶折叠突变体种子中表达量显著升高,而在开花后40 d的种子中表达量极显著降低(图4)。同时在种子发育过程中,GmUBC1在子叶折叠突变体种子发育早期10 d的种子中表达量最高,随后随着种子成熟该基因表达量下降,而在WT中GmUBC1在开花后40 d的种子中表达量达到最高。图3
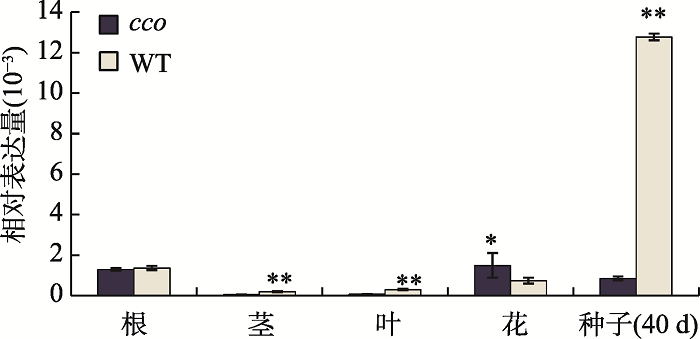 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3大豆GmUBC1在不同组织发育时期的表达分析
*表示P<0.05;**表示P<0.01。
Fig. 3Analysis of GmUBC1 expression in different tissue developmental stages
图4
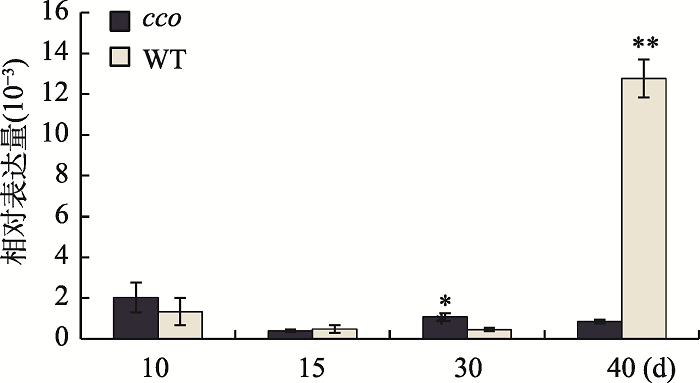 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4GmUBC1在大豆不同时期种子中的表达分析
*表示P<0.05;**表示P<0.01。
Fig. 4Expression analysis of GmUBC1 in soybean seeds at different periods
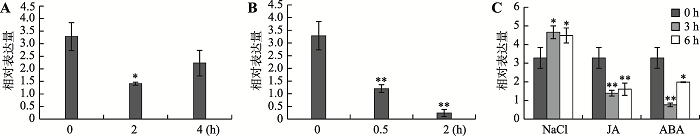
本研究进一步分析了GmUBC1在不同生物胁迫下的表达情况,结果显示在低温处理2 h时,GmUBC1基因表达量达到最低值,之后表达量逐渐升高(图5A)。经PEG处理后,GmUBC1基因的表达量在0.5 h时就极显著低于0 h,在2 h下降到更低的水平(图5B)。盐处理后3 h,基因表达量达到最大值,之后有所降低,但均显著高于对照未处理材料。在激素JA 处理3 h和6 h时,表达量均极显著低于对照。在ABA处理3 h时,表达量急剧下降,随着胁迫处理时间的延长,基因表达量有轻微上升,但仍显著低于对照(图5C)。以上结果说明,大豆GmUBC1基因的表达显著受到非生物胁迫低温、PEG以及激素JA、ABA的诱导下调,仅在盐处理条件下GmUBC1基因表达量上调。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5GmUBC1基因在不同胁迫条件下的表达模式
A:4℃低温处理;B:15% PEG处理;C:250 mmol/L NaCl、50 mol/L JA、100 μmol/L ABA处理。*表示P<0.05;**表示P<0.01。
Fig. 5Expression patterns of GmUBC1 under different stress treatments
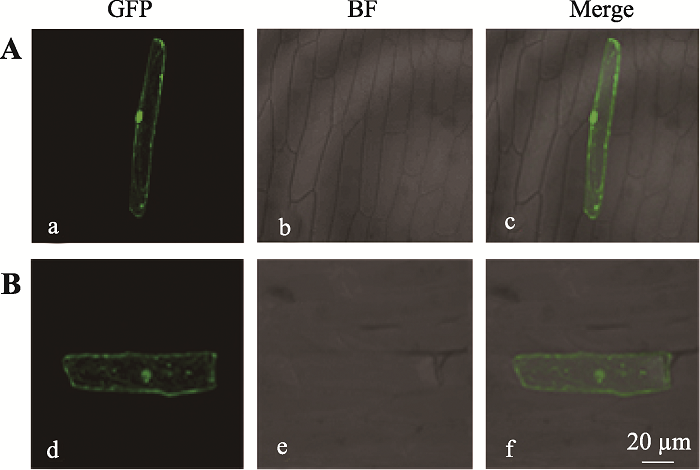
2.3 GmUBC1融合蛋白亚细胞定位
将测序正确的GmUBC1基因全长ORF(不含终止密码子)连接到pMDC83载体,通过基因枪法将pMDC83-GmUBC1-GFP转入到洋葱表皮细胞内。亚细胞定位结果表明,pMDC83-GFP融合蛋白绿色荧光分布在整个细胞(图6A),GmUBC1:GFP融合蛋白也分布在整个细胞(图6B),说明GmUBC1蛋白在细胞内均有表达。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6GmUBC1蛋白亚细胞定位
A:空载体pMDC83-GFP转化洋葱表皮细胞后GFP在细胞内的定位;B:植物过表达载体pMDC83-GmUBC1-GFP转化洋葱表皮细胞后GFP在细胞内的定位;a、d:绿色荧光;b、e:明视野;c、f:叠加图。
Fig. 6Subcellular location of GmUBC1
2.4 转GmUBC1基因拟南芥阳性植株的PCR检测
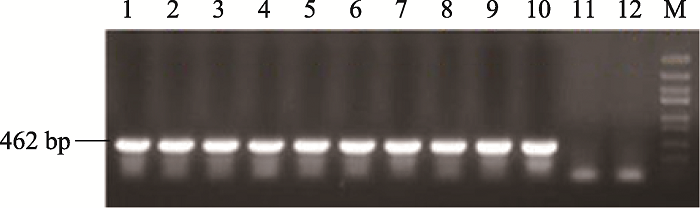
为了进一步探究GmUBC1基因的生物学功能,本研究通过蘸花法将GmUBC1转入拟南芥基因组中,获得了过表达GmUBC1转基因拟南芥。本研究提取9个具有潮霉素抗性的GmUBC1拟南芥转基因株系叶片DNA进行PCR检测,鉴定目的基因是否插入拟南芥基因组中。利用特异引物进行PCR扩增,以pMDC83-GmUBC1重组质粒为阳性对照,以野生型拟南芥DNA及灭菌水作为阴性对照。结果如图7所示,阴性对照(WT)中没有目的条带,阳性转基因株系扩增得到长度为462 bp的条带,说明pMDC83- GmUBC1已经成功插入到拟南芥基因组中。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7转基因拟南芥DNA的PCR检测
1~9:过表达GmUBC1拟南芥株系;10:阳性质粒;11:WT;12:水。
Fig. 7The PCR detection of transgenic Arabidopsis
2.5 GmUBC1转基因拟南芥千粒重和氨基酸含量
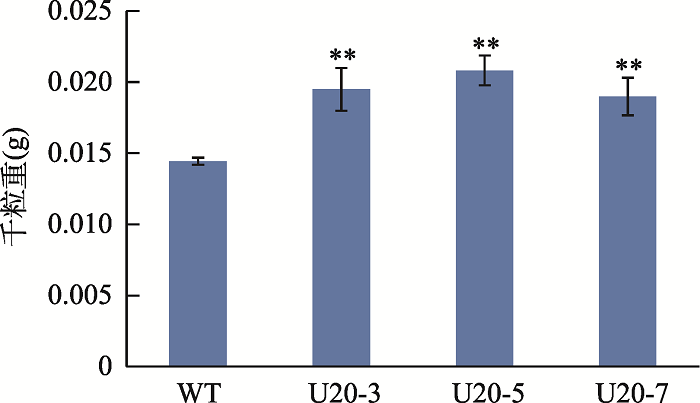
对T3代GmUBC1转基因拟南芥进行表型观察,发现与对照相比并没有明显的表型变化。为了探究GmUBC1基因与拟南芥果实发育的关系,本研究对转基因拟南芥种子的千粒重进行了统计分析。结果如图8所示,转基因株系U20-3、U20-5、U20-7的千粒重均比野生型植株有极显著提高。同时本研究还测定了转基因株系种子的总氨基酸含量,结果表明转基因株系U20-3、U20-5、U20-7的各组分氨基酸含量以及总氨基酸含量基本都比WT显著提高(图9),表明GmUBC1在种子的发育过程中可能发挥重要作用。图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8GmUBC1转基因拟南芥千粒重
Fig. 81000-seed weights of GmUBC1-transgenic Arabidopsis
图9
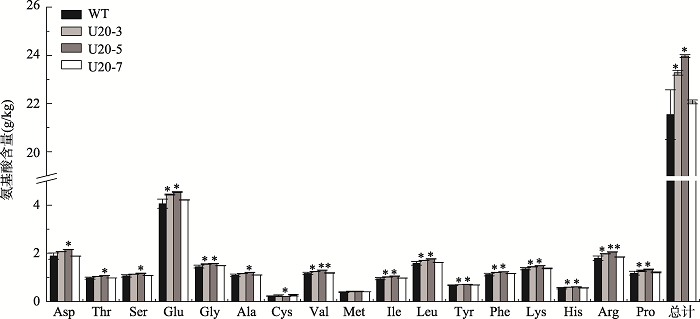 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9GmUBC1转基因拟南芥种子各组分氨基酸含量
*:表示P<0.05;**:表示P<0.01。Asp:天冬氨酸,Thr:苏氨酸,Ser:丝氨酸,Glu:谷氨酸,Ala:丙氨酸,Cys:半胱氨酸,Val:缬氨酸,Met:蛋氨酸,Ile:异亮氨酸,Leu:亮氨酸,Tyr:酪氨酸,Phe:苯丙氨酸,Lys:赖氨酸,His:组氨酸,Arg:精氨酸,Pro:脯氨酸。
Fig. 9Contents of each amino acid in GmUBC1- transgenic Arabidopsis seeds
2.6 GmUBC1转基因拟南芥萌发率
对转基因拟南芥种子和WT进行非生物胁迫处理一周,对萌发率进行统计。结果发现,经70 mmol/L NaCl处理后,转基因拟南芥种子的萌发率高于对照(图10A);经6% PEG和0.1 μmol/LABA处理,转基因拟南芥种子的萌发率与对照相比,均有不同程度的降低(图10,B和C)。该结果表明过表达GmUBC1可以使拟南芥对各种逆境有不同程度的响应。图10
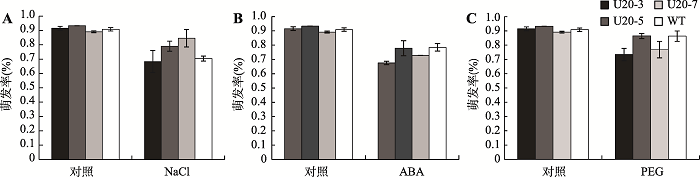 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10胁迫处理后GmUBC1转基因拟南芥种子萌发率
A:70 mmol/L NaCl处理后种子萌发率;B:0.1 μmol/L ABA处理后种子萌发率;C:6% PEG处理后种子萌发率。
Fig. 10Germination rate of GmUBC1-transgenic Arabidopsis thaliana seeds after stress treatment
3 讨论
E2泛素结合酶是一个多基因家族,在动物和人类中的数量涉及100多种。相关研究发现cco种子中IAA和ABA含量较高而发芽率和GA3含量较低。cco突变体的子叶具有更大的液泡和更多的膜状多层结构,以及更高的蛋氨酸和半胱氨酸含量,转录组测序结果显示cco中的多种植物激素生物合成和信号转导途径的相关基因存在显著差异[22],因此对差异表达基因GmUBC1的研究有助于了解大豆子叶折叠突变体的分子调控机制。本研究亚细胞定位结果与预测结果一致,GmUBC1蛋白在整个细胞内均有表达,且不存在信号肽。研究发现大多数泛素结合酶基因在植物的各个器官中都有表达,比如E2泛素结合酶基因TaE2在小麦(Triticum aestivum)的根、茎、叶和种子中均有表达[23]。本研究在提取大豆各个不同时期组织的RNA后,利用实时荧光定量PCR技术检测了WT及cco突变体不同组织中GmUBC1基因的表达量。结果表明GmUBC1基因在各个组织中均有表达,其中在开花后40 d的种子中表达量最高,且cco在开花后30 d的种子中表达量显著高于WT,但在开花后40 d的种子中表达量却极显著低于WT,且随着种子的成熟,基因表达量逐渐下降,表明GmUBC1可能主要在cco种子发育前期起作用。众所周知,基因序列的改变可能引起基因表达量的改变,从而引起生物表型的改变,包括SNPs、In/Del和结构上的变异等[24,25]。除此之外,表观遗传学也同样调控生物的表型。表观遗传学是在不改变基因序列的情况下,通过对基因进行可逆性修饰,产生可遗传的基因表达改变,导致表型变异的现象[26]。表观遗传学的调节机制包括DNA甲基化、组蛋白修饰和非编码RNA[27,28]。例如在番茄中,利用CRISPR/ Cas9技术获得异染色质转座子区CG、CHG位点严重低甲基化的Slddm1a和Slddm1b突变体,该突变体叶片呈杂色、花芽较小、花粉数量及活力显著降低,且无法产生可育后代[29]。拟南芥中Pol V可以合成非编码RNA,与一些抑制染色质修饰的沉默因子结合,从而导致基因沉默[30]。同时,在植物体内,启动子中顺式作用元件与上游转录因子的互作也可以控制基因的表达。如柽柳(Tamarix chinensis) ThWRKY2和ThWRKY8为ThERF1上游的调控基因,它们通过结合ThERF1启动子中的W-box元件来调控ThERF1基因的表达[31]。综上所述,基于GmUBC1基因在大豆栽培品种“南农94-16”和子叶折叠突变体(cco)中的表达量差异可能是由多种因素造成的,还有待进一步的研究进行验证。植物在生长发育过程中会遭受到各种非生物胁迫。研究表明泛素蛋白酶体途径在植物非生物胁迫中发挥着重要作用[32,33],如过表达绿豆(Vigna radiata)泛素结合酶基因VrUBC1改变ABA相关基因的表达,从而使转基因拟南芥渗透胁迫抗性增强[34]。玉米ZmUBC-76基因响应盐害和干旱的胁迫[35],小麦TaE2基因在干旱、高盐和ABA胁迫下均上调表达[23]。在拟南芥中异源过表达大豆GmUBC2增加转基因拟南芥逆境条件下脯氨酸含量,并且激活了逆境响应基因的表达,提高转基因拟南芥耐盐性[36]。为探究大豆GmUBC1基因是否同样受到非生物胁迫诱导,通过研究发现在低温、PEG以及激素JA、ABA胁迫处理下GmUBC1基因均处于下调状态,仅在盐处理条件下表达上调。低温处理2 h时,该基因的表达量达到最低值,之后逐渐升高。经PEG处理后,GmUBC1基因的表达量在0.5 h和2 h时均极显著低于对照。盐处理后3 h时,该基因表达量极显著上升,之后有所降低,但均显著高于对照。在激素JA 处理3 h和6 h时,表达量与对照相比呈下调趋势。在ABA处理3 h时,表达量急剧下降,但在6 h时基因表达量有轻微上升,但仍显著低于对照。这些结果说明大豆GmUBC1基因不同程度地响应低温、盐碱、干旱与JA和ABA胁迫处理。过表达GmUBC1基因拟南芥种子在PEG、ABA处理下萌发率降低,在NaCl处理下,转基因拟南芥种子萌发率较对照高,说明GmUBC1基因过表达使拟南芥对逆境有不同程度的响应。
以往研究表明E2泛素结合酶不仅在植物逆境中发挥一定的作用,同时也具有调节植物生长发育的作用[17]。在拟南芥中,AtUBC22参与雌配子体发育,UBC22的敲除突变体角果长度和种子数大大减少[37];UBC13基因能通过影响生长素信号传导和Aux/IAA蛋白的稳定性而在根发育中起作用[38]。同时,本实验室早期的研究表明,相比于WT子叶折叠突变体cco的百粒重显著降低,生育期明显延迟,氨基酸和蛋白含量提高[21,39]。本研究通过构建GmUBC1基因过表达载体,进行拟南芥遗传转化,进而对转基因拟南芥种子的千粒重和总氨基酸含量进行了测定,发现有些转基因株系的千粒重和总氨基酸含量均有显著提高,说明该基因主要对种子的生长发育以及氨基酸合成起到调控作用。通过探究过表达该基因对拟南芥产生的影响,为提高大豆品质提供参考。泛素结合酶蛋白在大豆生长发育中功能的研究尚少,因此克隆及研究大豆泛素结合酶功能意义重大。本研究初步研究了大豆GmUBC1基因的功能,有利于进一步了解泛素结合酶在植物生长发育和逆境中的作用。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1146/annurev.biochem.67.1.425URLPMID:9759494 [本文引用: 1]

The selective degradation of many short-lived proteins in eukaryotic cells is carried out by the ubiquitin system. In this pathway, proteins are targeted for degradation by covalent ligation to ubiquitin, a highly conserved small protein. Ubiquitin-mediated degradation of regulatory proteins plays important roles in the control of numerous processes, including cell-cycle progression, signal transduction, transcriptional regulation, receptor down-regulation, and endocytosis. The ubiquitin system has been implicated in the immune response, development, and programmed cell death. Abnormalities in ubiquitin-mediated processes have been shown to cause pathological conditions, including malignant transformation. In this review we discuss recent information on functions and mechanisms of the ubiquitin system. Since the selectivity of protein degradation is determined mainly at the stage of ligation to ubiquitin, special attention is focused on what we know, and would like to know, about the mode of action of ubiquitin-protein ligation systems and about signals in proteins recognized by these systems.
DOI:10.1038/nature03542URLPMID:15917798 [本文引用: 1]

Despite 100 years of evidence showing a pivotal role for indole-3-acetic acid (IAA or auxin) in plant development, the mechanism of auxin perception has remained elusive. Central to auxin response are changes in gene expression, brought about by auxin-induced interaction between the Aux/IAA transcriptional repressor proteins and the ubiquitin-ligase complex SCF(TIR1), thus targeting for them proteolysis. Regulated SCF-mediated protein degradation is a widely occurring signal transduction mechanism. Target specificity is conferred by the F-box protein subunit of the SCF (TIR1 in the case of Aux/IAAs) and there are multiple F-box protein genes in all eukaryotic genomes examined so far. Although SCF-target interaction is usually regulated by signal-induced modification of the target, we have previously shown that auxin signalling involves the modification of SCF(TIR1). Here we show that this modification involves the direct binding of auxin to TIR1 and thus that TIR1 is an auxin receptor mediating transcriptional responses to auxin.
DOI:10.1104/pp.110.166272URL [本文引用: 1]

The SLEEPY1 (SLY1) F-box gene is a positive regulator of gibberellin (GA) signaling in Arabidopsis (Arabidopsis thaliana). Loss of SLY1 results in GA-insensitive phenotypes including dwarfism, reduced fertility, delayed flowering, and increased seed dormancy. These sly1 phenotypes are partially rescued by overexpression of the SLY1 homolog SNEEZY (SNE)/SLY2, suggesting that SNE can functionally replace SLY1. GA responses are repressed by DELLA family proteins. GA relieves DELLA repression when the SCFSLY1 (for Skp1, Cullin, F-box) E3 ubiquitin ligase ubiquitinates DELLA protein, thereby targeting it for proteolysis. Coimmunoprecipitation experiments using constitutively expressed 35S:hemagglutinin (HA)-SLY1 and 35S:HA-SNE translational fusions in the sly1-10 background suggest that SNE can function similarly to SLY1 in GA signaling. Like HA-SLY1, HA-SNE interacted with the CULLIN1 subunit of the SCF complex, and this interaction required the F-box domain. Like HA-SLY1, HA-SNE coimmunoprecipitated with the DELLA REPRESSOR OF GA1-3 (RGA), and this interaction required the SLY1 or SNE carboxyl-terminal domain. Whereas HA-SLY1 overexpression resulted in a decrease in both DELLA RGA and RGA-LIKE2 (RGL2) protein levels, HA-SNE caused a decrease in DELLA RGA but not in RGL2 levels. This suggests that one reason HA-SLY1 is able to effect a stronger rescue of sly1-10 phenotypes than HA-SNE is because SLY1 regulates a broader spectrum of DELLA proteins. The FLAG-SLY1 fusion protein was found to coimmunoprecipitate with the GA receptor HA-GA-INSENSITIVE DWARF1b (GID1b), supporting the model that SLY1 regulates DELLA through interaction with the DELLA-GA-GID1 complex.
DOI:10.1111/j.1365-313X.2010.04112.xURLPMID:20409276 [本文引用: 1]

Plants utilize the ubiquitin-proteasome system (UPS) to modulate nearly every aspect of growth and development. Ubiquitin is covalently attached to target proteins through the action of three enzymes known as E1, E2, and E3. The ultimate outcome of this post-translational modification depends on the nature of the ubiquitin linkage and the extent of polyubiquitination. In most cases, ubiquitination results in degradation of the target protein in the 26S proteasome. During the last 10 years it has become clear that the UPS plays a prominent regulatory role in hormone biology. E3 ubiquitin ligases in particular actively participate in hormone perception, de-repression of hormone signaling pathways, degradation of hormone specific transcription factors, and regulation of hormone biosynthesis. It is certain that additional functions will be discovered as more of the nearly 1200 potential E3s in plants are elucidated.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1034/j.1399-3054.2001.1120401.xURLPMID:11473704 [本文引用: 1]

In recent years it has become obvious that protein degradation is an important catabolic process during development in plants and animals. One very conserved degradative system is the ubiquitin- and proteasome-dependent proteolytic pathway, which is found in all eukaryotes from yeast to mammals and plants. The pathway consists of two parts, one in which chains of ubiquitin are conjugated to substrate proteins, and one in which these substrate proteins are either degraded by the 26S proteasome or are de-ubiquitinated. The ubiquitin- and proteasome-dependent pathway degrades a wide range of proteins in the nucleus and cytoplasm. It is highly specific, but controls a large number of cellular events due to the diversity in the conjugating enzymes. This pathway is important for removal of abnormal/damaged proteins that have had their recognition sites exposed as well as for control of specific transcription factors and cell cycle regulators. In plants, ubiquitin- and proteasome-dependent proteolysis is known to be involved in regulation of the cell cycle and transcription factors as well as endoplasmic reticulum-associated protein degradation, stress response and developmental processes, such as xylogenesis and senescence.
DOI:10.1105/tpc.104.161220URLPMID:15579807 [本文引用: 2]
DOI:10.1146/annurev.arplant.55.031903.141801URL [本文引用: 1]
DOI:10.1093/aob/mcl255URLPMID:17220175 [本文引用: 1]

BACKGROUND: The covalent attachment of ubiquitin to a substrate protein changes its fate. Notably, proteins typically tagged with a lysine48-linked polyubiquitin chain become substrates for degradation by the 26S proteasome. In recent years many experiments have been performed to characterize the proteins involved in the ubiquitylation process and to identify their substrates, in order to understand better the mechanisms that link specific protein degradation events to regulation of plant growth and development. SCOPE: This review focuses on the role that ubiquitin plays in hormone synthesis, hormonal signalling cascades and plant defence mechanisms. Several examples are given of how targeted degradation of proteins affects downstream transcriptional regulation of hormone-responsive genes in the auxin, gibberellin, abscisic acid, ethylene and jasmonate signalling pathways. Additional experiments suggest that ubiquitin-mediated proteolysis may also act upstream of the hormonal signalling cascades by regulating hormone biosynthesis, transport and perception. Moreover, several experiments demonstrate that hormonal cross-talk can occur at the level of proteolysis. The more recently established role of the ubiquitin/proteasome system (UPS) in defence against biotic threats is also reviewed. CONCLUSIONS: The UPS has been implicated in the regulation of almost every developmental process in plants, from embryogenesis to floral organ production probably through its central role in many hormone pathways. More recent evidence provides molecular mechanisms for hormonal cross-talk and links the UPS system to biotic defence responses.
DOI:10.1105/tpc.106.048488URLPMID:17573536 [本文引用: 1]

Ubiquitination plays important roles in plant hormone signal transduction. We show that the RING finger E3 ligase, Arabidopsis thaliana SALT- AND DROUGHT-INDUCED RING FINGER1 (SDIR1), is involved in abscisic acid (ABA)-related stress signal transduction. SDIR1 is expressed in all tissues of Arabidopsis and is upregulated by drought and salt stress, but not by ABA. Plants expressing the ProSDIR1-beta-glucuronidase (GUS) reporter construct confirmed strong induction of GUS expression in stomatal guard cells and leaf mesophyll cells under drought stress. The green fluorescent protein-SDIR1 fusion protein is colocalized with intracellular membranes. We demonstrate that SDIR1 is an E3 ubiquitin ligase and that the RING finger conservation region is required for its activity. Overexpression of SDIR1 leads to ABA hypersensitivity and ABA-associated phenotypes, such as salt hypersensitivity in germination, enhanced ABA-induced stomatal closing, and enhanced drought tolerance. The expression levels of a number of key ABA and stress marker genes are altered both in SDIR1 overexpression and sdir1-1 mutant plants. Cross-complementation experiments showed that the ABA-INSENSITIVE5 (ABI5), ABRE BINDING FACTOR3 (ABF3), and ABF4 genes can rescue the ABA-insensitive phenotype of the sdir1-1 mutant, whereas SDIR1 could not rescue the abi5-1 mutant. This suggests that SDIR1 acts upstream of those basic leucine zipper family genes. Our results indicate that SDIR1 is a positive regulator of ABA signaling.
DOI:10.1111/j.1365-3040.2009.02065.xURLPMID:19895397 [本文引用: 1]

gamma-aminobutyric acid (GABA) is a four-carbon non-protein amino acid presented in a wide range of organisms. In this study, a suppression subtractive hybridization (SSH) library was constructed using roots of a legume shrub, Caragana intermedia, with the combined treatment of 300 mm NaCl and 300 mm NaCl + 10 mm GABA. We obtained 224 GABA-regulated unique expressed sequence tags (ESTs) including signal transduction, transcriptional regulation, hormone biosynthesis, reactive oxygen species (ROS) and polyamine metabolism, etc. The key H(2)O(2)-generated genes, NADPH oxidase (CaGR60), peroxidase (CaGR61) and amine oxidase (CaGR62), were regulated at the mRNA level by 10 mm GABA, which clearly inhibited H(2)O(2) accumulation brought about by NaCl stress in roots and leaves with the observation of 3,3'-diaminobenzidine (DAB) staining. Similarly, 10 mm GABA also regulated the expression of 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase (ACO) genes (CaGR30 and CaGR31) and ethylene production in NaCl-treated roots. Surprisingly, these H(2)O(2)-generated genes were enhanced at the mRNA level by a lower concentration of GABA, at 0.25 mm, but not other alternative nitrogen sources, and endogenous GABA accumulated largely just by the application of GABA at either concentration. Our results further proved that GABA, as a signal molecule, participates in regulating the expression of genes in plants under salt stress.
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2014.18.002URL [本文引用: 1]

【Objective】The objectives of this research are to identify the interacting proteins of stress related DREB like transcription factor, GmDREB5 in soybean (Glycine max), analyze the characteristics and biological function of its interaction protein, GmUBC13, and study the regulation mechanism that GmDREB5 enhanced the stress tolerance in plants. 【Method】 By yeast two hybrid system, AP2 domain of GmDREB5 was used as a bait to screen the interacting proteins of GmDREB5 from the drought-treated soybean cDNA library, and further interaction between GmDREB5 and its candidate interacting protein was confirmed through interacting testing in yeast and pull-down assay invitro, then, the properties of interacting protein, GmUBC13, such as phylogenetic tree, protein structure, subcellular localization were analyzed; expression profiles of GmUBC13 were tested under drought, salt, cold and ABA treatment by RT-PCR, and biological functions of GmUBC13 were also identified through transformation of tobacco in this research. 【Result】A GmDREB5-interacting protein, GmUBC13, belonging to the ubiquitin conjugating enzyme protein family, was obtained via screening drought-treated soybean cDNA library, and GmUBC13 contained UBCc (Ubiquitin-conjugating enzyme catalytic domain) domain, highly conserved E3 interaction residues and cysteine catalytic sites. Phylogenetic analyses showed that GmUBC13 shared the high homology with two members of the XV subgroup (containing total sixteen members) of ubiquitin conjugating enzyme like protein family, AtUBC13A (99%) and AtUBC13B (97%) in Arabidopsis and a ubiquitin conjugating enzyme like protein, Os01g0673600 (97%) in rice (Oryza sativa). Interaction between GmDREB5 and GmUBC13 was confirmed by interaction test in yeast and pull-down assay invitro. The expression of GmUBC13 gene in soybean was analyzed by drought, high salt, low temperature and ABA treatment. After ABA application, the transcript of GmUBC13 was induced after 1 h and reached the peak at 10 h, and slight down-regulated at 24 h. Under drought- and salt-treatment, the transcript of GmUBC13 was induced after 1 h and gradually increased along with the treatment time, and reached the peak at 24 h; under the low temperature, the expression of GmUBC13 induced rapidly and reached the peak at 5 h, and there was no expression at 10 h and 24 h. Further subcellular localization analysis showed that GmDREB5 was located on the nuclear and cytoplasm, and GmUBC13 was located on the nuclear. The analysis result of gene function showed that there was no difference among transgenic tobacco lines GmUBC13-1, GmUBC13-2 and wild type (W38) under normal MS medium, whereas exposed to different concentrations of PEG, the chlorophyll contents decreased, and the length of root and segment above ground were inhibited in all plants. However, the chlorophyll contents were higher, and the length of root and segment above ground was longer in GmDREB13 transgenic plants compared with W38, and there was a significant difference between transgenic lines GmUBC13-2 and W38 under treatment of 8% PEG. For the drought tolerance assay, two-week old transgenic plants and W38 were transferred into soil and withheld water for 21 days, and then re-watered for 6 days. Final results showed that transgenic lines grew healthier than W38, and the survival rates of transgenic lines were significantly higher than W38, suggesting that overexpression of GmUBC13 enhanced drought tolerance in transgenic plant.【Conclusion】 Soybean ubiquitin conjugating enzyme, GmUBC13 interacted with stress related transcription factor GmDREB5, and GmUBC13 was induced by various stress treatments, and overexpression of GmUBC13 enhanced drought tolerance in transgenic tobacco plants.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.jbiosc.2010.11.021URL [本文引用: 2]

Ubiquitin (Ub)-conjugating enzymes (UBCs) are key enzymes involved in ubiquitination. Although UBCs have been shown to play important roles in regulating various aspects of plant growth and development, the role of plant UBCs in abiotic stress response needs to be examined further. Here we report the characterization of a ubiquitin-conjugating enzyme gene AhUBC2 from dehydrated peanut plants. The expression of AhUBC2 gene in peanut plants is responsive to physiological water-stress induced by polyethylene glycol (PEG6000), high salinity, abscisic acid (ABA) or low temperature. The constitutive expression of AhUBC2 gene in wild-type Arabidopsis confers improved tolerance to water-stress induced by sorbitol or soil drought in 35S::AhUBC2 transgenic plants. Constitutive expression of AhUBC2 results in significantly increased expressions of three stress-responsive genes P5CS1, RD29A and KIN1 in 35S::AhUBC2 Arabidopsis grown under normal conditions, whereas the expressions of other four stress-responsive genes NCED3, ABF3, RD29B and RD22 are not affected. The proline level in 35S::AhUBC2 Arabidopsis is significantly higher than that in wild-type Arabidopsis under both soil-drought stressed and control conditions. In contrast, there is no significant difference in the levels of NCED3 transcript and endogenous ABA between wild-type and 35S::AhUBC2 Arabidopsis. These results suggest that constitutive expression of AhUBC2 in Arabidopsis confers improved water-stress tolerance likely through activating an ABA-independent signaling pathway, including regulating the expression of ABA-independent stress-responsive genes and promoting the synthesis of osmolyte proline to protect plants from water deficit. Copyright (C) 2010 The Society for Biotechnology, Japan. Published by Elsevier B.V.
DOI:10.1111/j.1365-313X.2008.03684.xURLPMID:18798874 [本文引用: 1]

Post-translational modifications of proteins by addition of ubiquitin can regulate protein degradation and localization, protein-protein interactions and transcriptional activation. In the ubiquitylation system, substrate specificity is primarily determined by the E2 ubiquitin-conjugating enzyme (UBC) and the E3 ubiquitin ligase. The Arabidopsis thaliana genome contains 37 genes encoding UBC homologs. However, the biological functions of these genes remain largely uncharacterized. Here, we report reverse genetic characterization of AtUBC1 and AtUBC2. While the loss-of-function single mutants Atubc1-1 and Atubc2-1 only show weak phenotypes, the double mutant Atubc1-1 Atubc2-1 shows a dramatically reduced number of rosette leaves and an early-flowering phenotype. Consistent with these results, the transcript levels of the floral repressor genes FLOWERING LOCUS C (FLC), MADS ASSOCIATED FLOWERING 4 (MAF4) and MAF5 are reduced in the double mutant. Loss-of-function mutants of HISTONE MONOUBIQUITINATION 1 (HUB1) and HUB2, which were previously reported to encode an E3 involved in histone H2B ubiquitylation, also show an early-flowering phenotype and reduced levels of FLC, MAF4 and MAF5 transcripts. In both Atubc1-1 Atubc2-1 and hub2-2 mutants, H2B mono-ubiquitylation is drastically reduced. Taken together, our results indicate that E2s AtUBC1/AtUBC2 and E3s HUB1/HUB2 together mediate H2B ubiquitylation, which is involved in the activation of floral repressor genes as well as in other processes as indicated by the pleiotropic phenotypes of the mutants.
DOI:10.1186/s13059-014-0548-2URLPMID:25464976 [本文引用: 1]

BACKGROUND: Fruits are unique to flowering plants and play a central role in seed maturation and dispersal. Molecular dissection of fruit ripening has received considerable interest because of the biological and dietary significance of fruit. To better understand the regulatory mechanisms underlying fruit ripening, we report here the first comprehensive analysis of the nuclear proteome in tomato fruits. RESULTS: Nuclear proteins were isolated from tomatoes in different stages of ripening, and subjected to iTRAQ (isobaric tags for relative and absolute quantification) analysis. We show that the proteins whose abundances change during ripening stages are involved in various cellular processes. We additionally evaluate changes in the nuclear proteome in the ripening-deficient mutant, ripening-inhibitor (rin), carrying a mutation in the transcription factor RIN. A set of proteins were identified and particular attention was paid to SlUBC32 and PSMD2, the components of ubiquitin-proteasome pathway. Through chromatin immunoprecipitation and gel mobility shift assays, we provide evidence that RIN directly binds to the promoters of SlUBC32 and PSMD2. Moreover, loss of RIN function affects protein ubiquitination in nuclei. SlUBC32 encodes an E2 ubiquitin-conjugating enzyme and a genome-wide survey of the E2 gene family in tomatoes identified five more E2s as direct targets of RIN. Virus-induced gene silencing assays show that two E2s are involved in the regulation of fruit ripening. CONCLUSIONS: Our results uncover a novel function of protein ubiquitination, identifying specific E2s as regulators of fruit ripening. These findings contribute to the unraveling of the gene regulatory networks that control fruit ripening.
DOI:10.1038/s41598-017-13513-xURLPMID:29038452 [本文引用: 1]

Ubiquitin-conjugating (UBC) E2 enzyme plays crucial roles in plant growth and development. Limited information can describe the function of UBC enzyme E2 in grapes. A total of 43 UBC enzyme E2 genes with conserved UBC domain were identified in grapes. These genes were divided into five groups based on phylogenetic tree with tomatoes. Sequence analyses indicated that VvUBCs in the same group possessed similar gene structures and conserved motifs. Gene distribution in chromosomes was uneven, and gene duplication existed in 36 VvUBCs. Transcriptome and qRT-PCR analysis indicated that most VvUBCs are involved in ripening and post-harvest stage, and feature functional roles in grape organs. According to the transcriptome and qRT-PCR results, seven and six VvUBCs in grape responded to cold and heat stress, respectively, whereas no remarkable VvUBCs change was noted under salt or water-deficit stress. This study provides new insights to physiological and developmental roles of these enzymes and regulation mechanism of E2 genes in grapes.
[本文引用: 2]
[本文引用: 2]
DOI:10.1186/1471-2164-15-510URL [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1086/514893URLPMID:9382090 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.18.00167URLPMID:29875274 [本文引用: 1]

In plants, cytosine methylation, an epigenetic mark critical for transposon silencing, is maintained over generations by key enzymes that directly methylate DNA and is facilitated by chromatin remodelers, like DECREASE IN DNA METHYLATION1 (DDM1). Short-interfering RNAs (siRNAs) also mediate transposon DNA methylation through a process called RNA-directed DNA methylation (RdDM). In tomato (Solanum lycopersicum), siRNAs are primarily mapped to gene-rich chromosome arms, and not to pericentromeric regions as in Arabidopsis thaliana Tomato encodes two DDM1 genes. To better understand their functions and interaction with the RdDM pathway, we targeted the corresponding genes via the CRISPR/Cas9 technology, resulting in the isolation of Slddm1a and Slddm1b knockout mutants. Unlike the single mutants, Slddm1a Slddm1b double mutant plants display pleiotropic vegetative and reproductive phenotypes, associated with severe hypomethylation of the heterochromatic transposons in both the CG and CHG methylation contexts. The methylation in the CHH context increased for some heterochromatic transposons and conversely decreased for others localized in euchromatin. We found that the number of heterochromatin-associated siRNAs, including RdDM-specific small RNAs, increased significantly, likely limiting the transcriptional reactivation of transposons in Slddm1a Slddm1b Taken together, we propose that the global production of siRNAs and the CHH methylation mediated by the RdDM pathway are restricted to chromosome arms in tomato. Our data suggest that both pathways are greatly enhanced in heterochromatin when DDM1 functions are lost, at the expense of silencing mechanisms normally occurring in euchromatin.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1101/gad.866801URLPMID:11297514 [本文引用: 1]

Low temperature is one of the most important environmental stimuli that control gene transcription programs and development in plants. In Arabidopsis thaliana, the HOS1 locus is a key negative regulator of low temperature-responsive gene transcription. The recessive hos1 mutation causes enhanced induction of the CBF transcription factors by low temperature as well as of their downstream cold-responsive genes. The hos1 mutant plants flower early, and this correlates with a low level of Flowering Locus C gene expression. The HOS1 gene was isolated through positional cloning. HOS1 encodes a novel protein with a RING finger motif near the amino terminus. HOS1 is ubiquitously expressed in all plant tissues. HOS1--GFP translational fusion studies reveal that HOS1 protein resides in the cytoplasm at normal growth temperatures. However, in response to low temperature treatments, HOS1 accumulates in the nucleus. Ectopic expression of HOS1 in wild-type plants causes cosuppression of HOS1 expression and mimics the hos1 mutant phenotypes.
DOI:10.1074/jbc.M101968200URLPMID:11557750 [本文引用: 1]

Proper folding of proteins (either newly synthesized or damaged in response to a stressful event) occurs in a highly regulated fashion. Cytosolic chaperones such as Hsc/Hsp70 are assisted by cofactors that modulate the folding machinery in a positive or negative manner. CHIP (carboxyl terminus of Hsc70-interacting protein) is such a cofactor that interacts with Hsc70 and, in general, attenuates its most well characterized functions. In addition, CHIP accelerates ubiquitin-dependent degradation of chaperone substrates. Using an in vitro ubiquitylation assay with recombinant proteins, we demonstrate that CHIP possesses intrinsic E3 ubiquitin ligase activity and promotes ubiquitylation. This activity is dependent on the carboxyl-terminal U-box. CHIP interacts functionally and physically with the stress-responsive ubiquitin-conjugating enzyme family UBCH5. Surprisingly, a major target of the ubiquitin ligase activity of CHIP is Hsc70 itself. CHIP ubiquitylates Hsc70, primarily with short, noncanonical multiubiquitin chains but has no appreciable effect on steady-state levels or half-life of this protein. This effect may have heretofore unanticipated consequences with regard to the chaperoning activities of Hsc70 or its ability to deliver substrates to the proteasome. These studies demonstrate that CHIP is a bona fide ubiquitin ligase and indicate that U-box-containing proteins may comprise a new family of E3s.
DOI:10.1371/journal.pone.0066056URLPMID:23824688 [本文引用: 1]

The ubiquitin conjugating enzyme E2 (UBC E2) mediates selective ubiquitination, acting with E1 and E3 enzymes to designate specific proteins for subsequent degradation. In the present study, we characterized the function of the mung bean VrUBC1 gene (Vigna radiata UBC 1). RNA gel-blot analysis showed that VrUBC1 mRNA expression was induced by either dehydration, high salinity or by the exogenous abscisic acid (ABA), but not by low temperature or wounding. Biochemical studies of VrUBC1 recombinant protein and complementation of yeast ubc4/5 by VrUBC1 revealed that VrUBC1 encodes a functional UBC E2. To understand the function of this gene in development and plant responses to osmotic stresses, we overexpressed VrUBC1 in Arabidopsis (Arabidopsis thaliana). The VrUBC1-overexpressing plants displayed highly sensitive responses to ABA and osmotic stress during germination, enhanced ABA- or salt-induced stomatal closing, and increased drought stress tolerance. The expression levels of a number of key ABA signaling genes were increased in VrUBC1-overexpressing plants compared to the wild-type plants. Yeast two-hybrid and bimolecular fluorescence complementation demonstrated that VrUBC1 interacts with AtVBP1 (A. thalianaVrUBC1 Binding Partner 1), a C3HC4-type RING E3 ligase. Overall, these results demonstrate that VrUBC1 plays a positive role in osmotic stress tolerance through transcriptional regulation of ABA-related genes and possibly through interaction with a novel RING E3 ligase.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11103-009-9575-xURL [本文引用: 1]

Previous studies have shown that ubiquitination plays important roles in plant abiotic stress responses. In the present study, the ubiquitin-conjugating enzyme gene GmUBC2, a homologue of yeast RAD6, was cloned from soybean and functionally characterized. GmUBC2 was expressed in all tissues in soybean and was up-regulated by drought and salt stress. Arabidopsis plants overexpressing GmUBC2 were more tolerant to salinity and drought stresses compared with the control plants. Through expression analyses of putative downstream genes in the transgenic plants, we found that the expression levels of two ion antiporter genes AtNHX1 and AtCLCa, a key gene involved in the biosynthesis of proline, AtP5CS, and the copper chaperone for superoxide dismutase gene AtCCS, were all increased significantly in the transgenic plants. These results suggest that GmUBC2 is involved in the regulation of ion homeostasis, osmolyte synthesis, and oxidative stress responses. Our results also suggest that modulation of the ubiquitination pathway could be an effective means of improving salt and drought tolerance in plants through genetic engineering.
DOI:10.1093/jxb/erw142URLPMID:27069118 [本文引用: 1]

Protein ubiquitination is critical for numerous processes in eukaryotes. The ubiquitin-conjugating enzyme (E2) is required for ubiquitination. The Arabidopsis genome has approximately 37 E2 genes, but in vivo functions for most of them remain unknown. In this study we observed that knockout mutants of Arabidopsis UBC22 had much-reduced silique length and seed number, with nearly 90% of ovules aborted. Analyses revealed that the majority of mutant embryo sacs displayed severe defects and often contained no gamete nuclei. There was no difference between mutant and wild-type Arabidopsis at the megaspore mother cell stage; however, the functional megaspore was either not present or appeared abnormal in a large portion of mutant ovules, suggesting that the defect started with functional megaspore degeneration in the mutants. Degeneration continued during megagametogenesis, such that the percentage of mature embryo sacs without any gamete nuclei was much greater than the percentage of developing ovules without a functional megaspore and, in addition, various abnormalities in megagametogenesis were observed. Additionally, heterozygous plants had only 13.1% of ovules aborted, indicating that the heterozygous sporophytic tissues could affect the development of the mutant female gametophyte. UBC22 is the sole member of an Arabidopsis E2 subfamily, and is more closely related to one type of E2s in animals that catalyzes Lys11-specific ubiquitination. Indeed, our results showed that Arabidopsis UBC22 could catalyze ubiquitin dimer formation in vitro in a Lys11-dependent manner, suggesting that it likely catalyzes Lys11-linked ubiquitination in plants. This study has thus identified one biochemical property of UBC22 and revealed a novel function in female gametophyte development.
DOI:10.1111/j.1365-313X.2010.04150.xURLPMID:20113438 [本文引用: 1]

Iron-deficiency responses comprise molecular, physiological and developmental adjustments, ultimately leading to an improved cellular Fe homeostasis. By using a proteomic approach, we identified the ubiquitin-conjugating enzyme UBC13 as being highly responsive to the Fe regime at the post-transcriptional level in the tips of cucumber (Cucumis sativus) roots. UBC13 has been shown to catalyze non-canonical Lys63-linked ubiquitin chains, playing important roles in signal transduction among eukaryotes. Ectopic expression of the cucumber UBC13 gene in Arabidopsis thaliana led to a more pronounced and Fe-responsive formation of branched root hairs, a key response of Arabidopsis roots to Fe deficiency. Plants carrying a mutation in the Arabidopsis ortholog UBC13A were unable to form branched root hairs upon Fe deficiency and showed a perturbed expression of Fe-regulated genes. Mutants defective in both Arabidopsis UBC13 genes, UBC13A and UBC13B, showed a marked reduction in root hair density. Mutations in the cognate E3 ligases RGLG2 and RGLG1 caused the constitutive formation of branched root hairs independent of the Fe supply, indicating the involvement of polyubiquitination in the altered differentiation of rhizodermal cells. It is concluded that UBC13, probably via the formation of Lys63-linked ubiquitin chains, has a critical function in epidermal cell differentiation and is crucial for the regulation of Fe-responsive genes and developmental responses to Fe deficiency.
[本文引用: 1]
[本文引用: 1]
