 ,哈尔滨医科大学组织学与胚胎学教研室,哈尔滨 150081
,哈尔滨医科大学组织学与胚胎学教研室,哈尔滨 150081The epigenetic regulation of ribosomal DNA and tumorigenesis
Xiangrong Cheng, Xinglin Hu, Qi Jiang, Xingwei Huang, Nan Wang, Lei Lei ,Department of Histology and Embryology, Harbin Medical University, Harbin 150081, China
,Department of Histology and Embryology, Harbin Medical University, Harbin 150081, China通讯作者:
编委: 宋旭
收稿日期:2018-10-15修回日期:2019-01-15网络出版日期:2019-02-25
| 基金资助: |
Received:2018-10-15Revised:2019-01-15Online:2019-02-25
| Fund supported: |
作者简介 About authors
程香荣,在读硕士研究生,专业方向:基础医学E-mail:xiangrongcheng@foxmail.com。

摘要
关键词:
Abstract
Keywords:
PDF (345KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
程香荣, 胡兴琳, 姜琦, 黄星卫, 王楠, 雷蕾. 核糖体DNA转录的表观调控与肿瘤发生[J]. 遗传, 2019, 41(3): 185-192 doi:10.16288/j.yczz.18-244
Xiangrong Cheng, Xinglin Hu, Qi Jiang, Xingwei Huang, Nan Wang, Lei Lei.
早在19世纪晚期科学家们就发现癌细胞中核仁数量增多和比例增大,20世纪初已将这种大核仁特征列为恶性肿瘤的确诊依据。人非瘤病灶细胞中出现巨大核仁的表现与致瘤风险高度相关。核糖体蛋白S19 (ribosomal protein S19, RPS19)突变的遗传性“核糖体病人”,其早期易患先天性纯红细胞再生障碍性贫血(Diamond-Blackfan anemia, DBA),后期可发展为细胞高度增殖性疾病(癌症),提示核糖体缺陷可能与肿瘤发生有关[1]。因而人们提出核糖体生物合成过程中质和量的改变可能致瘤的假说[2]。目前,核糖体异常如何致瘤是生命科学研究领域的热点方向之一[3]。
核糖体DNA (ribosomal DNA, rDNA)转录为rRNA的过程是核糖体生物合成的限速步骤[4]。基因组中rDNA基因有数百个拷贝,具有重复序列的特征,其转录水平的调控主要有2种方式[5]:(1)通过影响细胞的生长增殖信号通路调节rDNA特异的RNA聚合酶Ⅰ (RNA polymeraseⅠ, PolⅠ)的转录效率; (2)通过表观分子作用机制调节活跃态rDNA拷贝数量的所占比例[6]。前者属于短期调节方式,细胞的营养、生长因子、致癌因素等会上调PolⅠ的转录效率,而基因毒性、代谢压力、饥饿、病毒感染、肿瘤抑制因素等会下调PolⅠ的转录效率;后者属于长期稳定的调节方式,通过建立新的表观遗传修饰状态来调控rDNA的转录潜能,在细胞的生长分化、转化过程中至关重要[6]。本文通过阐述核糖体生物合成异常与癌症发生发展的联系机制,讨论了rDNA的表观调控机制的缺陷可能对肿瘤发生的诱导或促进作用,以期为针对rDNA转录调控机制的药物研发提供新的理论支持。
1 核仁结构与核糖体生物合成过程
核糖体是由4种rRNA分子和约80种不同的核糖体蛋白(ribosomal proteins, RPs)组成的直径为25~30 nm的复合体微粒,负责“中心法则”的mRNA到蛋白质这一翻译过程,与细胞的生长增殖活动息息相关。核仁是细胞核糖体生物合成的重要场所,核仁结构的增大反映出核糖体合成速率的增加。H&E染色(hematoxylin-eosin staining)显示核仁通常表现为单一或多个匀质的球形小体,电子显微镜显示核仁包含3个主要结构:纤维中心、致密纤维组分和颗粒组分[7]。rDNA位于纤维中心,而从rDNA新合成出的rRNA分子主要集中在致密纤维组分,在颗粒组分部位继续加工成熟后,与RPs形成核糖体亚单位,最后被转运至胞质中。核仁的纤维组分包含所有rDNA转录过程所需的物质,包括PolⅠ、上游结合因子(upstream binding factor, UBF)、核仁素和核仁磷酸蛋白等,可以采用银染的方法使其选择性显色。银染核仁组织区域(Ag-stained nucleolar organizing region, AgNOR)的面积大小直接反映出rDNA的转录速率,继而反映出核糖体生物合成的速率[8]。大约400拷贝的rDNA基因根据表观修饰特点和转录功能被分为活跃态、沉默态[9]和中间准备态(poised state)[10],在不同分化状态的细胞中分配着不同的比例。活跃态rDNA在PolⅠ和至少3种基本因子[Rrn3 (TIF-IA)、SL1/TIF-IB (selectivity factor 1, SL1)、UBF]的辅助下转录合成47S pre-rRNA,随后加工形成成熟的18S、5.8S、28S rRNA。另一种rRNA分子5S rRNA在核质中由RNA聚合酶Ⅲ合成,随后被转运到核仁中。各种核糖体蛋白由RNA聚合酶Ⅱ转录,在胞质中翻译成成熟蛋白质后被转运到核仁中。随后28S、5.8S、5S rRNA和49种RPs组装形成核糖体大亚基60S,而18S和33种核糖体蛋白组装形成核糖体小亚基40S,最后,大小亚基都被转运至胞质中构成最终的80S核糖体微粒。在整个过程中,rDNA转录速率是核糖体生物合成过程中的限制性步骤[4]。2 核糖体生物合成与肿瘤发生的关系
自从发现癌症细胞存在核仁过度肥大且外形不规则的特点后,人们就一直在探索核仁改变与癌症发生之间的因果关系。一个快速增殖过程中的真核细胞每分钟就可产生多达2000个核糖体,而高度增殖的癌细胞更加依赖于核糖体的产生过程。肿瘤细胞为增加其核糖体生物合成的速率,经常突变缺失多个负调控rDNA转录过程的肿瘤抑制基因(CDKN2A、TP53、RB1和PTEN等)[3]。RPs除了作为分子组分和rRNA的分子伴侣参与核糖体生物合成外,还在凋亡、细胞周期停滞、细胞增殖、细胞迁移和侵袭、DNA损伤修复、维持基因组稳定等过程中发挥着重要作用[11]。对于这些“核糖体外功能”(extra-ribosomal function),即核糖体蛋白参与的与核糖体生物合成或总体蛋白质翻译过程无关的其它细胞生理过程[12]),主要是通过p53-MDM2通路介导和调节的,调节机制的异常时也会导致肿瘤发生[11,13,14]。此外,除了p53依赖的信号通路外,RPs还通过c-Myc、E2F-1、ATF4和NF-κB等途径影响到核糖体外功能,这些途径的异常也会诱导肿瘤的发生[11]。本文主要以p53依赖的途径为主,重点阐述核糖体生物合成与肿瘤发生的关系。2.1 核糖体生物合成与细胞周期
当细胞开始增殖时,蛋白合成的需求就会迅猛增加,以满足细胞分裂时所需的大量结构和功能组分。蛋白质合成速率的增加是通过上调核糖体合成速率来实现的[15],只有G1期达到足够多的核糖体储备时才会允许细胞经过G1-S期限制点[16]。事实上,有丝分裂原和生长因子主要通过以下3种途径刺激细胞增殖过程(图1):(1)通过MAPK/ERK信号通路,激活PolⅠ和RNA聚合酶Ⅲ的转录[6];(2)激活MYC基因,Myc蛋白正向调节核糖体生物合成过程[17];(3)激活mTOR信号通路,诱导促进PolⅠ和RNA聚合酶Ⅲ的转录过程[18]。细胞增殖信号可以激活核糖体的生物合成,同时核糖体生物合成的增加也能促进细胞增殖。在细胞静息状态下,较高水平的p53阻碍pRb的磷酸化过程,使pRb保持低磷酸化水平。低磷酸化的pRb与E2Fs结合,阻止其激活E2F靶基因。E2F的靶基因产物是进入S期所必须的,因此肿瘤抑制蛋白pRb与p53在细胞周期的调控中起了重要作用[19]。在细胞正常生理状态下,核糖体蛋白与rRNA分子结合形成核糖体,留下少量游离的RPs。其中,游离的大亚基核糖体蛋白RPL5、RPL11与5S rRNA形成复合物,通过结合MDM2 (mouse double minute 2, MDM2)而抑制其E3泛素连接酶的功能,剩余未被结合的MDM2降解p53而使其保持稳定水平[20,21]。当抑制或扰乱rDNA转录或成熟过程,如DNA损伤、RP基因突变、药物作用、饥饿状态或致癌基因激活造成核仁压力(核糖体压力)时,多余游离的RPs将更多地结合上MDM2,阻碍P53的蛋白酶降解过程,导致p53水平增高,细胞周期停滞[22,23]。图1
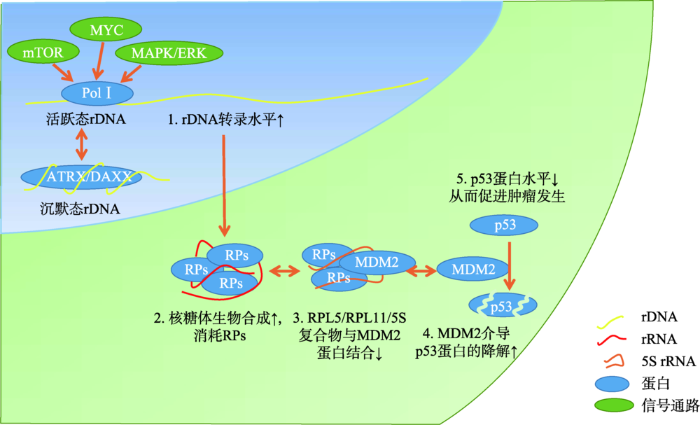 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1核糖体蛋白-MDM2-p53通路
Fig. 1Ribosomal proteins-MDM2-p53 pathway
2.2 核糖体生物合成与肿瘤发生
肿瘤细胞会利用各种方式来提高核糖体的生物合成水平。例如,p53基因的失活是肿瘤细胞中最常见的基因突变方式。p53蛋白负性调节rDNA转rDNA转录是细胞活动的中心。mTOR、MYC与MAPK/ERK通路对聚合酶Ⅰ(PolⅠ)的转录具有促进作用,促进核糖体生物合成。核糖体蛋白(RPs)参与核糖体生物合成过程,因而导致游离的核糖体蛋白RPL5、RPL11与5S RNA组成的复合物减少,从而释放出更多的MDM2。游离的MDM2导致p53的降解增多,p53蛋白水平的下降将促进细胞周期进程,从而促进肿瘤发生。本课题组发现ATRX/DAXX复合物与沉默rDNA相关,经常突变ATRX/DAXX基因的肿瘤细胞中,可能是通过开放更多活跃rDNA,从而促进肿瘤发生的重要途径之一。录,所以在p53突变缺失的肿瘤中rDNA的转录水平更高[7]。除了p53,肿瘤抑制基因(如ARF、pRb、GSK3β和PTEN)与致癌基因(如Myc、NPM)分别对 PolⅠ转录起着抑制和促进的调控作用[6]。如果抑癌基因和致癌基因发生突变失衡,或在癌症细胞中给予胰岛素或IL-6处理,增强rRNA的转录,就会消耗更多的RPs用于核糖体生物合成,从而减少了与MDM2结合抑制的RPs,导致p53蛋白降解增多,从而游离的p53蛋白水平下降。最终,肿瘤细胞将得益于核糖体合成水平的增高和细胞周期限制的解除,更加快速地增殖和侵袭[20,24]。核糖体生物合成的“量变”或者“质变”都有可能升高患癌风险。对人类疾病的研究数据表明,患有慢性炎症、二型糖尿病、肥胖的人群中细胞恶性转化的出现频率更高[25,26,27],这些人具有更高的IL-6或血浆胰岛素水平,IL-6或胰岛素将刺激rDNA的转录过程[20,24]。另外,大量实验研究数据证明了核糖体生物合成在诱导肿瘤发生过程中的重要性。敲除TIP5 (rDNA相关沉默复合物NoRC的大亚基)将导致rDNA的转录水平升高,诱导正常NIH3T3细胞出现转化表型[28]。解除MTG16a蛋白对核糖体DNA的抑制作用,将诱导人正常乳腺上皮细胞出现乳腺癌发生时的形态表型和分子特征[29]。IL-6通过刺激核糖体生物合成过程,诱导人正常结肠黏膜上皮细胞系NCM460出现上皮间质转化、侵袭的特征[24]。因此,核糖体生物合成速率增高的细胞将会有更高的肿瘤发生风险。此外,核糖体生物合成“质变”的“核糖体病人”(如DBA和5q-综合征患者)因其具有核糖体生物合成缺陷而也会具有更高的患癌风险[30,31]。RPs缺陷通常导致细胞低增殖性的表型:如贫血。低增殖特点给细胞带来选择压力,驱使细胞发生二次突变,从而获得高增殖的特性,导致细胞异常克隆并增殖成癌[1,3]。
3 调控rDNA转录的表观遗传异常可能参与肿瘤发生
已分化细胞中大约一半的rDNA基因处于异染色质化的沉默状态[32]。最新研究提示,影响rDNA转录的表观调控机制异常可能是癌症发生的驱动因素,rDNA的表观遗传调控基因突变也可触发肿瘤发生[28,33]。敲减H3K4me3/H3K36me2的去甲基化酶JHDM1B (JmjC domain-containing histone demethylase 1B, JHDM1B)能够诱导rDNA的表观遗传修饰发生重塑,导致转化与未转化的乳腺上皮细胞更具侵袭性[33]。NoRC复合物负责建立和维持rDNA基因的沉默态,敲除其亚基成分TIP5将导致沉默rDNA及大小卫星序列的不稳定,rDNA的转录水平增高,从而诱导正常NIH3T3细胞出现转化表型[28]。外显子测序结果显示,超过60%的胰腺神经内分泌瘤(pancreatic neuroendocrine tumor, PNETs)突变缺失了至少以下3种基因之一:MEN1 (multiple endocrine neoplasia type 1)、DAXX (death domain- associated protein)、ATRX (alpha- thalassemia/mental retardation X-linked syndrome protein)[34]。免疫荧光原位杂交实验证实,这些突变与肿瘤的ALT表型(alternative lengthening of telomeres,肿瘤细胞为达到永生而采取的端粒延长机制)相关[34,35]。与胰腺神经内分泌瘤具有相似突变特点的是胶质瘤。例如在儿童多形性胶质母细胞瘤(glioblastoma multiforme, GBM)中,ATRX、DAXX与H3F3A (编码组蛋白变体H3.3的基因之一)经常突变,突变频率可高达45%,且与GBM细胞的ALT表型有很强相关性[36]。又如在低级别胶质瘤(low-grade gliomas, LGGs)和继发性胶质母细胞瘤中,ATRX突变还经常与IDH1 (Isocitrate dehydrogenase 1/NADP+)、p53突变伴随发生[37]。MEN1的蛋白产物Menin是组蛋白甲基转移酶复合物的组成成分,参与组蛋白甲基转移酶MLL (mixed lineage leukemia)、PRMT5 (protein arginine methylransferase 5)、SUV39H1 (suppressor of variegation 3-9 homolog protein 1)的功能[38];ATRX是SWI/SNF家族成员的染色质重塑ATP酶,与H3.3特异分子伴侣DAXX组成复合物,将H3.3沉积于异染色质区,建立H3K9me3修饰,从而维持染色质结构稳定[39,40,41];IDH1突变将会促使全基因组范围的DNA和组蛋白的高甲基化状态[42]。考虑到MEN1、ATRX、DAXX和IDH1等基因都具有表观遗传调控的作用,因此该类基因的突变可能重塑了某些肿瘤相关基因的表观遗传修饰状态,有利于肿瘤的发生发展[43]。
虽然ATRX突变可以通过诱导重复序列的异常DNA重组而导致ALT表型的出现,使肿瘤细胞永生化[44,45],但ALT并不是肿瘤发生的机制。ATRX突变缺失促进肿瘤发生的机制可能是通过诱导基因转录水平改变、DNA修复机制异常来实现的[39]。ATRX综合征患者显示亚端粒区和rDNA基因的DNA甲基化水平降低,特别是rDNA的CpG岛区,这说明ATRX的染色质重塑活性与rDNA的甲基化修饰相关,ATRX突变可能对rDNA的表达造成影响[46]。本实验室利用NCBI-GEO数据库中小鼠胚胎干细胞(mouse embryonic stem cell, mESC)的H3.3与H3K9me3的reChIP-seq (连续染色质免疫共沉淀与测序技术)数据[GSE59189][47],采取最近生物信息学分析方法[48]重新分析单个rDNA序列单元上的富集信息。通过分析H3K9甲基转移酶SETDB1野生型和敲除时H3.3与H3K9me3共富集的情况,结果发现H3.3与H3K9me3主要共富集于rDNA的启动子和编码区,这种共富集状态明显受SETDB1敲除的影响。本实验室还证明ATRX及DAXX能够在rDNA的启动子和编码区特异性富集,且随细胞分化过程中沉默rDNA的比例增加而增多。本实验室证明了沉默态rDNA的建立和维持需要ATRX/DAXX复合物与H3.3介导的H3K9me3修饰,敲除DAXX将导致rDNA启动子区的甲基化水平降低,UBF结合的活跃rDNA拷贝数量增加等现象(待发表)。在其他报道中,ATRX缺失明显促进胶质母细胞瘤的生长、缩短小鼠的生存期[49]。在胰腺神经内分泌瘤的临床样本中,DAXX低表达与更高的Ki-67(增殖相关的基因)、更高的WHO分级明显相关[50]。以上结果共同提示,ATRX或DAXX缺失会诱导rDNA转录活性增加,可能是肿瘤发生发展的新机制(图1)。
4 结语与展望
核糖体的快速合成是癌细胞增殖侵袭的基础。核糖体蛋白除了被认为参与核糖体的生物合成外,还具有多种涉及肿瘤发生的核糖体外功能,因此核糖体生物合成过程直接或间接的与肿瘤发生相关。多种肿瘤细胞通过基因突变、代谢方式改变来促进核糖体的生物合成过程,满足细胞代谢需求。ATRX/ DAXX复合物与rDNA的沉默相关,然而其对rDNA转录的表观调控作用是否与ATRX或DAXX经常突变的肿瘤发生直接相关,还需更多的探索。目前,抑制核糖体生物合成已成为抗癌药物研发的重要路径。抑制核糖体生物合成将有利于大量游离的RPs结合MDM2,激活抑癌基因p53并抑制致癌基因Myc;另外核糖体生物合成不足将阻碍肿瘤细胞的蛋白质合成过程,通过影响细胞周期、细胞凋亡等其他途径,综合抑制肿瘤细胞的生长[51]。抑制核糖体生物合成的药物比起通过引发DNA损伤来激活p53、诱导细胞凋亡的传统方式,其基因毒性的副作用更小[52]。目前抑制核糖体生物合成的治疗药物主要靶向抑制PolⅠ、翻译起始因子EIF4A、EIF4e、EIF2S1和信号通路mTOR/PI3K,从而抑制核糖体的产生和蛋白翻译的起始阶段[53],未来还可根据rDNA转录的表观遗传调控环节的异常来研发新药。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:4342353 [本文引用: 2]

Ribosomopathies are largely congenital diseases linked to defects in ribosomal proteins or biogenesis factors. Some of these disorders are characterized by hypoproliferative phenotypes such as bone marrow failure and anemia early in life, followed by elevated cancer risks later in life. This transition from hypo- to hyperproliferation presents an intriguing paradox in the field of hematology known as "Dameshek's riddle." Recent cancer sequencing studies also revealed somatically acquired mutations and deletions in ribosomal proteins in T-cell acute lymphoblastic leukemia and solid tumors, further extending the list of ribosomopathies and strengthening the association between ribosomal defects and oncogenesis. In this perspective, we summarize and comment on recent findings in the field of ribosomopathies. We explain how ribosomopathies may provide clues to help explain Dameshek's paradox and highlight some of the open questions and challenges in the field.
URL [本文引用: 1]
URLPMID:28923911 [本文引用: 3]

Abstract A wealth of novel findings, including congenital ribosomal mutations in ribosomopathies and somatic ribosomal mutations in various cancers, have significantly increased our understanding of the relevance of ribosomes in oncogenesis. Here, we explore the growing list of mechanisms by which the ribosome is involved in carcinogenesis-from the hijacking of ribosomes by oncogenic factors and dysregulated translational control, to the effects of mutations in ribosomal components on cellular metabolism. Of clinical importance, the recent success of RNA polymerase inhibitors highlights the dependence on "onco-ribosomes" as an Achilles' heel of cancer cells and a promising target for further therapeutic intervention. Significance: The recent discovery of somatic mutations in ribosomal proteins in several cancers has strengthened the link between ribosome defects and cancer progression, while also raising the question of which cellular mechanisms such defects exploit. Here, we discuss the emerging molecular mechanisms by which ribosomes support oncogenesis, and how this understanding is driving the design of novel therapeutic strategies. Cancer Discov; 7(10); 1-19. 2017 AACR. 2017 American Association for Cancer Research.
URLPMID:17108330 [本文引用: 2]

Abstract Pre-rRNA synthesis and processing are key steps in ribosome biogenesis. Although recent evidence in yeast suggests that these two processes are coupled, the nature of their association is unclear. In this report, we analyze the coordination between rDNA transcription and pre-rRNA processing in mammalian cells. We found that pol I transcription factor UBF interacts with pre-rRNA processing factors as analyzed by immunoprecipitations, and the association depends on active rRNA synthesis. In addition, injections of plasmids containing the human rDNA promoter and varying lengths of 18S rDNA into HeLa nuclei show that pol I transcription machinery can be recruited to rDNA promoters regardless of the product that is transcribed, whereas subgroups of pre-rRNA processing factors are recruited to plasmids only when specific pre-rRNA fragments are produced. Our observations suggest a model for sequential recruitment of pol I transcription factors and pre-rRNA processing factors to elongating pre-rRNA on an as-needed basis rather than corecruitment to sites of active transcription.
URL [本文引用: 1]

: rDNA是控制细胞核糖体生物合成的串联重复基因,影响着整体蛋白质的翻译水平,与细胞生长代谢息息相关.由于rDNA序列具有多拷贝的重复特征,其转录除了受一般转录机制的调节外,还受多重表观调控机制的调节,精细调控着rDNA的转录状态.一般认为rDNA分为活跃和沉默两种状态,分别与活跃染色质标记和异染色质标记相关.近些年来,发现一种平衡态rDNA的存在,更加丰富了rDNA表观机制的研究.H3.3是一种H3组蛋白变体,是近些年来的研究热点,已有报道H3.3可能在分子伴侣HIRA的介导下整合进入活跃rDNA,然而沉默rDNA的维持是否也与H3.3的作用相关需要更多的探索.CTCF是rDNA重复单元间的绝缘子成分,与H3.3相关但并不清楚是否也调控着rDNA的转录.该综述讨论了几种调控rDNA表观状态的机制,并对可能参与该过程的新机制提出了设想.
URL [本文引用: 1]

: rDNA是控制细胞核糖体生物合成的串联重复基因,影响着整体蛋白质的翻译水平,与细胞生长代谢息息相关.由于rDNA序列具有多拷贝的重复特征,其转录除了受一般转录机制的调节外,还受多重表观调控机制的调节,精细调控着rDNA的转录状态.一般认为rDNA分为活跃和沉默两种状态,分别与活跃染色质标记和异染色质标记相关.近些年来,发现一种平衡态rDNA的存在,更加丰富了rDNA表观机制的研究.H3.3是一种H3组蛋白变体,是近些年来的研究热点,已有报道H3.3可能在分子伴侣HIRA的介导下整合进入活跃rDNA,然而沉默rDNA的维持是否也与H3.3的作用相关需要更多的探索.CTCF是rDNA重复单元间的绝缘子成分,与H3.3相关但并不清楚是否也调控着rDNA的转录.该综述讨论了几种调控rDNA表观状态的机制,并对可能参与该过程的新机制提出了设想.
URL [本文引用: 4]

All cells, from prokaryotes to vertebrates, synthesize enormous amounts of rRNA to produce 1 2 million ribosomes per cell cycle, which are required to maintain the protein synthesis capacity of the daughter cells. In recent years, considerable progress has been made in the elucidation of the basic principles of transcriptional regulation and the pathways that adapt cellular rRNA synthesis to metabolic activity, a process that is essential for understanding the link between nucleolar activity, cell growth, proliferation, and apoptosis. I will survey our present knowledge of the highly coordinated networks that regulate transcription by RNA polymerase I, coordinating rRNA gene transcription and ribosome production with environmental cues. Moreover, I will discuss the epigenetic mechanisms that control the chromatin structure and transcriptional activity of rRNA genes, in particular the role of noncoding RNA in DNA methylation and transcriptional silencing.
URL [本文引用: 2]
URL [本文引用: 1]
URLPMID:18616426 [本文引用: 1]

Annu Rev Cell Dev Biol. 2008;24:131-57. doi: 10.1146/annurev.cellbio.24.110707.175259. Research Support, Non-U.S. Gov't; Review
URLPMID:22570494 [本文引用: 1]

rRNA genes (rDNA) exist in two distinct epigenetic states, active promoters being unmethylated and marked by euchromatic histone modifications, whereas silent ones are methylated and exhibit heterochromatic features. Here we show that the nucleosome remodeling and deacetylation (NuRD) complex establishes a specific chromatin structure at rRNA genes that are poised for transcription activation. The promoter of poised rRNA genes is unmethylated, associated with components of the preinitiation complex, marked by bivalent histone modifications and covered by a nucleosome in the "off" position, which is refractory to transcription initiation. Repression of rDNA transcription in growth-arrested and differentiated cells correlates with elevated association of NuRD and increased levels of poised rRNA genes. Reactivation of transcription requires resetting the promoter-bound nucleosome into the "on" position by the DNA-dependent ATPase CSB (Cockayne syndrome protein B). The results uncover a unique mechanism by which ATP-dependent chromatin remodeling complexes with opposing activities establish a specific chromatin state and regulate transcription.
URLPMID:27294833 [本文引用: 3]

Ribosomal proteins (RPs), the essential components of the ribosome, are a family of RNA-binding proteins, which play prime roles in ribosome biogenesis and protein translation. Recent studies revealed that RPs have additional extra-ribosomal functions, independent of protein biosynthesis, in regulation of diverse cellular processes. Here, we review recent advances in our understanding of how RPs regulate apoptosis, cell cycle arrest, cell proliferation, neoplastic transformation, cell migration and invasion, and tumorigenesis through both MDM2/p53-dependent and p53-independent mechanisms. We also discuss the roles of RPs in the maintenance of genome integrity via modulating DNA damage response and repair. We further discuss mutations or deletions at the somatic or germline levels of some RPs in human cancers as well as in patients of Diamond-Blackfan anemia and 5q- syndrome with high susceptibility to cancer development. Moreover, we discuss the potential clinical application, based upon abnormal levels of RPs, in biomarker development for early diagnosis and/or prognosis of certain human cancers. Finally, we discuss the pressing issues in the field as future perspectives for better understanding the roles of RPs in human cancers to eventually benefit human health.
URLPMID:2679180 [本文引用: 1]

Ribosomal proteins are ubiquitous, abundant, and RNA binding: prime candidates for recruitment to extraribosomal functions. Indeed, they participate in balancing the synthesis of the RNA and protein components of the ribosome itself. An exciting new story is that ribosomal proteins are sentinels for the self-evaluation of cellular health. Perturbation of ribosome synthesis frees ribosomal proteins to interface with the p53 system, leading to cell-cycle arrest or to apoptosis. Yet in only a few cases can we clearly identify the recruitment of ribosomal proteins for other extraribosomal functions. Is this due to a lack of imaginative evolution by cells and viruses, or to a lack of imaginative experiments by molecular biologists?
URLPMID:3494370 [本文引用: 1]

Abstract The oncoprotein MDM2 is both the transcriptional target and the predominant antagonist of the tumor suppressor p53. MDM2 inhibits the functions of p53 via a negative feedback loop that can be circumvented by several ribosomal proteins in response to nucleolar or ribosomal stress. Stress conditions in the nucleolus can be triggered by a variety of extracellular and intracellular insults that impair ribosomal biogenesis and function, such as chemicals, nutrient deprivation, DNA damaging agents, or genetic alterations. The past decade has witnessed a tremendous progress in understanding this previously underinvestigated ribosomal stress-MDM2-p53 pathway. Here, we review the recent progress in understanding this unique signaling pathway, discuss its biological and pathological significance, and share with readers our insight into the research in this field.
URLPMID:24375388 [本文引用: 1]

Ribosome biogenesis is the most demanding energetic and metabolic expenditure of the cell. The nucleolus, a nuclear compartment, coordinates rRNA transcription, maturation, and assembly into ribosome subunits. The transcription process is highly coordinated with ribosome biogenesis. In this context, ribosomal proteins (RPs) play a crucial role. In the last decade, an increasing number of studies have associated RPs with extraribosomal functions related to proliferation. Importantly, the expression of RPs appears to be deregulated in several human disorders due, at least in part, to genetic mutations. Although the deregulation of RPs in human malignancies is commonly observed, a more complex mechanism is believed to be involved, favoring the tumorigenic process, its progression and metastasis. This review explores the roles of the most frequently mutated oncogenes and tumor suppressor genes in human cancer that modulate ribosome biogenesis, including their interaction with RPs. In this regard, we propose a new focus for novel therapies.
URLPMID:10806485 [本文引用: 1]

Abstract Although the importance of cell growth for cell-cycle progression has been recognized for thirty years, the molecular basis of this relationship is poorly understood. However, researchers have begun to tease apart these two processes in model systems. This commentary focuses on one potential mechanism by which ribosome biogenesis antagonizes cell-cycle progression until the cell has grown to an adequate size.
URLPMID:15389582 [本文引用: 1]

Cell growth is closely related to cell proliferation and an adequate ribosome biogenesis appears to be necessary for cell duplication. In the present study, we have investigated the relationship between rRNA synthesis and cell cycle progression. For this purpose, in a first set of experiments, we evaluated the effect of rRNA synthesis variation on cycle duration in asynchronously growing H4-II-E-C3 rat hepatoma cells. Cells were either treated with insulin or insulin plus actinomycin D (AMD). The hormone stimulated ribosome biogenesis, which was later followed by an increased synthesis of DNA and a shortening of cell doubling time (DT). Bivariate flow cytometry indicated that the reduced length of the cell cycle was mainly due to the shorter G1-phase. AMD, at the concentration of 0.04 microg/ml, hindered ribosome biogenesis without affecting heterogeneous RNA production. A 12-h reduction in ribosome biogenesis level by AMD caused a lowering of DNA synthesis and a lengthening of cell DT with a longer G1-phase. In a second set of experiments, we analyzed the cell content variations of 28S and 18S rRNA transcripts during G1 phase in H4-II-E-C3 cells, synchronized by serum deprivation, and then stimulated by serum, serum plus insulin, and serum plus insulin and AMD. In control cells, a progressive increase in rRNA content occurred until the highest value of rRNA content was reached 21 h after serum stimulation. In insulin-treated cells, the highest rRNA value was reached at 12 h whereas in AMD-treated cells, the rRNA quantity was constantly low until 18 h and then sharply increased at 21 h. In the three experimental conditions, the highest values of rRNA amount were reached at the end of G1 phase and were quite similar to one another. We also evaluated, by real-time RT-PCR, cyclin E mRNA expression, which appeared to sharply increase at those times in which the maximum increase in the rRNA content was observed. Our results indicated that the achievement of an appropriate amount of rRNA allows G1/S phase transition, probably by modulating the expression of cyclin E mRNA.
URLPMID:18451027 [本文引用: 1]

Mad1, a member of the Myc/Max/Mad family, suppresses Myc-mediated transcriptional activity by competing with Myc for heterodimerization with its obligatory partner, Max. The expression of Mad1 suppresses Myc-mediated cell proliferation and transformation. The levels of Mad1 protein are generally low in many human cancers, and Mad1 protein has a very short half-life. However, the mechanism that regulates the turnover of Mad1 protein is poorly understood. In this study, we showed that Mad1 is a substrate of p90 ribosomal kinase (RSK) and p70 S6 kinase (S6K). Both RSK and S6K phosphorylate serine 145 of Mad1 upon serum or insulin stimulation. Ser-145 phosphorylation of Mad1 accelerates the ubiquitination and degradation of Mad1 through the 26S proteasome pathway, which in turn promotes the transcriptional activity of Myc. Our study provides a direct link between the growth factor signaling pathways regulated by PI3 kinase/Akt and MAP kinases with Myc-mediated transcription.
URLPMID:17041624 [本文引用: 1]

The target of rapamycin (TOR) signal-transduction pathway is an important mechanism by which eucaryotic cells adjust their protein biosynthetic capacity to nutrient availability. Both in yeast and in mammals, the TOR pathway regulates the synthesis of ribosomal components, including transcription and processing of pre-rRNA, expression of ribosomal proteins and the synthesis of 5S rRNA. Expression of the genes encoding the numerous constituents of ribosomes requires transcription by all three classes of nuclear RNA polymerases. In this review, we summarize recent advances in understanding the interplay among nutrient availability, transcriptional control and ribosome biogenesis. We focus on transcription in response to nutrients, detailing the relevant downstream targets of TOR in yeast and mammals. The critical role of TOR in linking environmental queues to ribosome biogenesis provides an efficient means by which cells alter their overall protein biosynthetic capacity.
URLPMID:9039259 [本文引用: 1]

Cell. 1997 Feb 7;88(3):323-31. Review
URLPMID:21399665 [本文引用: 3]

Data on the relationship between ribosome biogenesis and p53 function indicate that the tumour suppressor can be activated by either nucleolar disruption or ribosomal protein defects. However, there is increasing evidence that the induction of p53 does not always require these severe cellular changes, and data are still lacking on a possible role of ribosome biogenesis in the downregulation of p53. Here, we studied the effect of the up- and downregulation of the rRNA transcription rate on p53 induction in mammalian cells. We found that a downregulation of rRNA synthesis, induced by silencing the POLR1A gene coding for the RNA polymerase I catalytic subunit, stabilised p53 without altering the nucleolar integrity in human cancer cells. p53 stabilisation was due to the inactivation of the MDM2-mediated p53 degradation by the binding of ribosomal proteins no longer used for ribosome building. p53 stabilisation did not occur when rRNA synthesis downregulation was associated with a contemporary reduction of protein synthesis. Furthermore, we demonstrated that in three different experimental models characterised by an upregulation of rRNA synthesis, cancer cells treated with insulin or exposed to the insulin-like growth factor 1, rat liver stimulated by cortisol and regenerating rat liver after partial hepatectomy, the p53 protein level was reduced due to a lowered ribosomal protein availability for MDM2 binding. It is worth noting that the upregulation of rRNA synthesis was responsible for a decreased p53-mediated response to cytotoxic stresses. These findings demonstrated that the balance between rRNA and ribosomal protein synthesis controls the function of p53 in mammalian cells, that p53 can be induced without the occurrence of severe changes of the cellular components controlling ribosome biogenesis, and that conditions characterised by an upregulated rRNA synthesis are associated with a reduced p53 response.
URLPMID:3928573 [本文引用: 1]

Noncoding 5S rRNA is now shown to be required, along with the 60S ribosomal proteins RPL5 and RPL11, for the inhibition of Hdm2 and stabilization of p53 following impaired ribosome biogenesis. Thomas and colleagues further demonstrate that all three ribosomal components are mutually dependent on one another for both binding and inhibiting Hdm2. Finally, they show that upon impaired ribosome biogenesis, it is the nascent RPL5/RPL11/5S rRNA preribosomal complex that is redirected from assembly into 60S ribosomes to the binding of Hdm2.
URL [本文引用: 1]
PMID:5297773 [本文引用: 1]

The nucleolus is the site of ribosome biogenesis, a complex process that requires the coordinate activity of all three RNA polymerases and hundreds of non-ribosomal factors that participate in the maturation of ribosomal RNA (rRNA) and assembly of small and large subunits. Nevertheless, emerging studies have highlighted the fundamental role of the nucleolus in sensing a variety of cellular stress stimuli that target ribosome biogenesis. This condition is known as nucleolar stress and triggers several response pathways to maintain cell homeostasis, either p53-dependent or p53-independent. The mouse double minute (MDM2)-p53 stress signaling pathways are activated by multiple signals and are among the most important regulators of cellular homeostasis. In this review, we will focus on the role of ribosomal proteins in p53-dependent and p53-independent response to nucleolar stress considering novel identified regulators of these pathways. We describe, in particular, the role of ribosomal protein uL3 (rpL3) in p53-independent nucleolar stress signaling pathways.
URLPMID:24531714 [本文引用: 3]

Chronic inflammation is an established risk factor for the onset of cancer, and the inflammatory cytokine IL-6 has a role in tumorigenesis by enhancing proliferation and hindering apoptosis. As factors stimulating proliferation also downregulate p53 expression by enhancing ribosome biogenesis, we hypothesized that IL-6 may cause similar changes in inflamed tissues, thus activating a mechanism that favors neoplastic transformation. Here, we showed that IL-6 downregulated the expression and activity of p53 in transformed and untransformed human cell lines. This was the consequence of IL-6-dependent stimulation of c-MYC mRNA translation, which was responsible for the upregulation of rRNA transcription. The enhanced rRNA transcription stimulated the MDM2-mediated proteasomal degradation of p53, by reducing the availability of ribosome proteins for MDM2 binding. The p53 downregulation induced the acquisition of cellular phenotypic changes characteristic of epithelial-mesenchymal transition, such as a reduced level of E-cadherin expression, increased cell invasiveness and a decreased response to cytotoxic stresses. We found that these changes also occurred in colon epithelial cells of patients with ulcerative colitis, a very representative example of chronic inflammation at high risk for tumor development. Histochemical and immunohistochemical analysis of colon biopsy samples showed an upregulation of ribosome biogenesis, a reduced expression of p53, together with a focal reduction or absence of E-cadherin expression in chronic colitis in comparison with normal mucosa samples. These changes disappeared after treatment with anti-inflammatory drugs. Taken together, the present results highlight a new mechanism that may link chronic inflammation to cancer, based on p53 downregulation, which is activated by the enhancement of rRNA transcription upon IL-6 exposure.
URLPMID:19824817 [本文引用: 1]

Abstract Body mass index, as an approximation of body adiposity, is associated with increased risk of several common and less common malignancies in a sex- and site-specific manner. These findings implicate sex- and cancer site-specific biological mechanisms underpinning these associations, and it is unlikely that there is a "one system fits all" mechanism. Three main candidate systems have been proposed-insulin and the insulin-like growth factor-I axis, sex steroids, and adipokines-but there are shortfalls to these hypotheses. In this review, three novel candidate mechanisms are proposed: obesity-induced hypoxia, shared genetic susceptibility, and migrating adipose stromal cells. While public health policies aimed at curbing the underlying causes of the obesity epidemic are being implemented, there is a parallel need to better understand the biological processes linking obesity and cancer as a prerequisite to the development of new approaches to prevention and treatment.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:20168299 [本文引用: 3]

Maintenance of specific heterochromatic domains is crucial for genome stability. In eukaryotic cells, a fraction of the tandem-repeated ribosomal RNA (rRNA) genes is organized in the heterochromatic structures. The principal determinant of rDNA silencing is the nucleolar remodelling complex, NoRC, that consists of TIP5 (TTF-1-interacting protein-5) and the ATPase SNF2h. Here we showed that TIP5 not only mediates the establishment of rDNA silencing but also the formation of perinucleolar heterochromatin that contains centric and pericentric repeats. Our data indicated that the TIP5-mediated heterochromatin is indispensable for stability of silent rRNA genes and of major and minor satellite repeats. Moreover, depletion of TIP5 impairs rDNA silencing, upregulates rDNA transcription levels and induces cell transformation. These findings point to a role of TIP5 in protecting genome stability and suggest that it can play a role in the cellular transformation process.
URLPMID:19961547 [本文引用: 1]

Human MTG16a (CBFA2T3), a chromatin repressor with nucleolar localization, was described to act as a suppressor of breast tumourigenesis. Here we show that MTG16a is a novel ribosomal gene repressor, which can counteract MYC-driven activation of ribosomal RNA (rRNA) transcription. We also show that either knocking down MTG16a by RNA interference, or sequestering MTG16a outside the nucleolus of human breast epithelial cells, hampers acinar morphogenesis concomitant with up-regulation of rRNA synthesis and increased ribogenesis. This is the first demonstration that loss of MTG16a function in the nucleolus of breast epithelial cells can induce morphological and molecular changes typical of breast cancer initiation.
URLPMID:20980806 [本文引用: 1]

Myelodysplastic Syndromes (MDS) are a heterogeneous group of acquired clonal bone marrow disorders, characterised by ineffective hematopoiesis. The mechanisms underlying many of these blood disorders have remained elusive due to the difficulty in pinpointing specific gene mutations or haploinsufficencies, which can occur within large deleted regions. However, there is an increasing interest in the classification of some of these diseases as ribosomopathies. Indeed, studies have implicated Ribosomal Protein (RP) S14 as a strong candidate for haploinsufficiency in 5q-syndrome, a particular form of MDS. Recently, two novel mouse models have provided evidence for the involvement of both RPS14 and the p53 pathway, and specific miRNAs in 5q-syndrome. In this review we will discuss: 5q-syndrome mouse models, the possible mechanisms underlying this blood disorder with respect to the candidate genes and comparisons with other ribosomopathies and the involvement of the p53 pathway in these diseases.
URLPMID:20116044 [本文引用: 1]

Diamond-Blackfan anemia (DBA), an inherited bone marrow failure syndrome characterized by anemia that usually presents before the first birthday or in early childhood, is associated with birth defects and an increased risk of cancer. Although anemia is the most prominent feature of DBA, the disease is also characterized by growth retardation and congenital malformations, in particular craniofacial, upper limb, heart, and urinary system defects that are present in 30% 50% of patients. DBA has been associated with mutations in seven ribosomal protein (RP) genes, RPS19, RPS24, RPS17, RPL35A, RPL5, RPL11, and RPS7, in about 43% of patients. To continue our large-scale screen of RP genes in a DBA population, we sequenced 35 ribosomal protein genes, RPL15, RPL24, RPL29, RPL32, RPL34, RPL9, RPL37, RPS14, RPS23, RPL10A, RPS10, RPS12, RPS18, RPL30, RPS20, RPL12, RPL7A, RPS6, RPL27A, RPLP2, RPS25, RPS3, RPL41, RPL6, RPLP0, RPS26, RPL21, RPL36AL, RPS29, RPL4, RPLP1, RPL13, RPS15A, RPS2, and RPL38, in our DBA patient cohort of 117 probands. We identified three distinct mutations of RPS10 in five probands and nine distinct mutations of RPS26 in 12 probands. Pre-rRNA analysis in lymphoblastoid cells from patients bearing mutations in RPS10 and RPS26 showed elevated levels of 18S-E pre-rRNA. This accumulation is consistent with the phenotype observed in HeLa cells after knockdown of RPS10 or RPS26 expression with siRNAs, which indicates that mutations in the RPS10 and RPS26 genes in DBA patients affect the function of the proteins in rRNA processing.
URLPMID:17613545 [本文引用: 1]

Eukaryotic cells contain several hundred ribosomal RNA (rRNA) genes (rDNA), a fraction of them being silenced by epigenetic mechanisms. The presence of two epigenetically distinct states of rRNA genes provides a unique opportunity to decipher the molecular mechanisms that establish the euchromatic, i.e. transcriptionally active, and the heterochromatic, i.e. transcriptionally silent, state of rDNA. This article summarizes our knowledge of the epigenetic mechanisms that control rDNA transcription and emphasizes how DNA methyltransferases and histone-modifying enzymes work in concert with chromatin-remodeling complexes and RNA-guided mechanisms to establish a specific chromatin structure that defines the transcriptional state of rRNA genes. These studies exemplify the mutual dependence and complex crosstalk among different epigenetic players in the alteration of the chromatin structure during the process of gene activation or silencing.
URLPMID:28415746 [本文引用: 2]

Abstract The alterations of ribosome biogenesis and protein synthesis play a direct role in the development of tumors. The accessibility and transcription of ribosomal genes is controlled at several levels, with their epigenetic regulation being one of the most important. Here we explored the JmjC domain-containing histone demethylase 1B (JHDM1B) function in the epigenetic control of rDNA transcription. Since JHDM1B is a negative regulator of gene transcription, we focused on the effects induced by JHDM1B knock-down (KD). We studied the consequences of stable inducible JHDM1B silencing in cell lines derived from transformed and untransformed mammary epithelial cells. In these cellular models, prolonged JHDM1B downregulation triggered a surge of 45S pre-rRNA transcription and processing, associated with a re-modulation of the H3K36me2 levels at rDNA loci and with changes in DNA methylation of specific CpG sites in rDNA genes. We also found that after JHDM1B KD, cells showed a higher ribosome content: which were engaged in mRNA translation. JHDM1B KD and the consequent stimulation of ribosomes biogenesis conferred more aggressive features to the tested cellular models, which acquired a greater clonogenic, staminal and invasive potential. Taken together, these data indicate that the reduction of JHDM1B leads to a more aggressive cellular phenotype in mammary gland cells, by virtue of its negative regulatory activity on ribosome biogenesis.
URL [本文引用: 2]
URL [本文引用: 1]
URLPMID:22286061 [本文引用: 1]

Glioblastoma multiforme (GBM) is a lethal brain tumour in adults and children. However, DNA copy number and gene expression signatures indicate differences between adult and paediatric cases1 4. To explore the genetic events underlying this distinction, we sequenced the exomes of 48 paediatricGBMsamples.Somaticmutations in the H3.3-ATRX-DAXX chromatin remodelling pathway were identified in 44% of tumours (21/48). Recurrent mutations in H3F3A, which encodes the replication-independent histone 3 variant H3.3, were observed in 31% of tumours, and led to amino acid substitutions at two critical positions within the histone tail (K27M, G34R/G34V) involved in key regulatory post-translational modifications.Mutations inATRX (a-thalassaemia/mental retardation syndrome X-linked)5 and DAXX (death-domain associated protein), encoding two subunits of a chromatin remodelling complex required for H3.3 incorporation at pericentric heterochromatin and telomeres6,7, were identified in 31% of samples overall, and in 100% of tumours harbouring a G34R or G34V H3.3 mutation. Somatic TP53 mutations were identified in 54% of all cases, and in 86% of samples with H3F3A and/or ATRX mutations. Screening of a large cohort of gliomas of various grades and histologies (n5784) showedH3F3A mutations to be specific toGBM and highly prevalent in children and young adults. Furthermore, the presence of H3F3A/ATRX-DAXX/TP53 mutations was strongly associated with alternative lengthening of telomeres and specific gene expression profiles. This is, to our knowledge, the first report to highlight recurrent mutations in a regulatory histone in humans, and our data suggest that defects of the chromatin architecture underlie paediatric and young adult GBM pathogenesis.
[本文引用: 1]
URLPMID:28811300 [本文引用: 1]

Abstract There is a trend of increasing prevalence of neuroendocrine tumors (NETs), and the inherited multiple endocrine neoplasia type 1 (MEN1) syndrome serves as a genetic model to investigate how NETs develop and the underlying mechanisms. Menin, encoded by the MEN1 gene, at least partly acts as a scaffold protein by interacting with multiple partners to regulate cellular homeostasis of various endocrine organs. Menin has multiple functions including regulating several important signaling pathways by controlling gene transcription. Here, we focus on reviewing the recent progress in elucidating the key biochemical role of menin in epigenetic regulation of gene transcription and cell signaling, as well as posttranslational regulation of menin itself. In particular, we will review the progress in studying structural and functional interactions of menin with various histone modifiers and transcription factors such as MLL, PRMT5, SUV39H1 and other transcription factors including c-Myb and JunD. Moreover, the role of menin in regulating cell signaling pathways such as TGF-beta, Wnt, and Hedgehog, as well as miRNA biogenesis and processing will be described. Further, the regulation of the MEN1 gene transcription, posttranslational modifications and stability of menin protein will be reviewed. These various modes of regulation by menin as well as regulation of menin by various biological factors broaden the view regarding how menin controls various biological processes in neuroendocrine organ homeostasis.
URLPMID:28062559 [本文引用: 2]

Recent genome sequencing efforts in a variety of cancers have revealed mutations and/or structural alterations in ATRX and DAXX, which together encode a complex that deposits histone variant H3.3 into repetitive heterochromatin. These regions include retrotransposons, pericentric heterochromatin, and telomeres, the latter of which show deregulation in ATRX/DAXX-mutant tumors. Interestingly, ATRX and DAXX mutations are often found in pediatric tumors, suggesting a particular developmental context in which these mutations drive disease. Here we review the functions of ATRX and DAXX in chromatin regulation as well as their potential contributions to tumorigenesis. We place emphasis on the chromatin remodeler ATRX, which is mutated in the developmental disorder for which it is named, -thalassemia, mental retardation, X-linked syndrome, and at high frequency in a number of adult and pediatric tumors.
URLPMID:20211137 [本文引用: 1]

78 H3.3 localization at specific genes and regulatory regions is cell-type specific 78 Hira controls H3.3 localization to genes and some regulatory regions 78 H3.3 localization to many regulatory elements and telomeres is Hira independent 78 Atrx is required for H3.3 telomeric localization, and for telomeric RNA repression
URL [本文引用: 1]

组蛋白是真核生物中一类进化上相对保守的蛋白质。由组蛋白八聚体及缠绕其上的DNA构成的核小体是真核生物染色质的基本组成单位。核小体使DNA保持固缩状态,既能维持基因组的稳定性,又能保证DNA序列可以正确地进行复制、转录、重组和修复。核小体调控细胞的生物过程除了通过组蛋白翻译后修饰,还可以通过组蛋白变体替换的方式进行。研究发现,组蛋白H3变体H3.3与常规组蛋白H3尽管仅有几个氨基酸的区别,但H3.3却能由特异的分子伴侣介导,整合进入染色质的特定区域,从而发挥不同的作用。同时,H3.3作为一种母源因子在正常受精和体细胞核移植等细胞重编程过程中也发挥着重要作用。本文总结了H3.3的结构特点和富集情况,探讨了特异的分子伴侣及其在细胞重编程中的作用,以期为提高体细胞重编程效率提供新思路,为体细胞重编程的应用奠定基础。
URL [本文引用: 1]

组蛋白是真核生物中一类进化上相对保守的蛋白质。由组蛋白八聚体及缠绕其上的DNA构成的核小体是真核生物染色质的基本组成单位。核小体使DNA保持固缩状态,既能维持基因组的稳定性,又能保证DNA序列可以正确地进行复制、转录、重组和修复。核小体调控细胞的生物过程除了通过组蛋白翻译后修饰,还可以通过组蛋白变体替换的方式进行。研究发现,组蛋白H3变体H3.3与常规组蛋白H3尽管仅有几个氨基酸的区别,但H3.3却能由特异的分子伴侣介导,整合进入染色质的特定区域,从而发挥不同的作用。同时,H3.3作为一种母源因子在正常受精和体细胞核移植等细胞重编程过程中也发挥着重要作用。本文总结了H3.3的结构特点和富集情况,探讨了特异的分子伴侣及其在细胞重编程中的作用,以期为提高体细胞重编程效率提供新思路,为体细胞重编程的应用奠定基础。
URLPMID:26147657 [本文引用: 1]

In recent years, frequent isocitrate dehydrogenase 1/2 (IDH1/IDH2) gene mutations were found in a variety of tumors, which specifically alter arginine residues of catalytic active site in IDH1/IDH2 and confer new enzymatic function of directly catalyzing alpha-ketoglutarate ( -KG) to R-2-hydroxyglutarate (2-HG). 2-HG could competitively inhibit -KG-dependent enzymes and might therefore contribute to tumorigenesis. In addition, mutation status of IDH1/IDH2 is closely related to the progress and prognosis of certain tumors. Thus IDH1/IDH2 is considered to be a promising biomarker for early diagnosis and prognosis and targeted therapy. In this study, the current research on IDH1/IDH2 mutation, especially the mechanisms and clinical characteristics related to tumor, are reviewed.
URL [本文引用: 1]
URLPMID:4501375 [本文引用: 1]

Fifteen per cent of cancers maintain telomere length independently of telomerase by the homologous recombination (HR)-associated alternative lengthening of telomeres (ALT) pathway. A unifying feature of these tumours are mutations in ATRX. Here we show that expression of ectopic ATRX triggers a suppression of the pathway and telomere shortening. Importantly ATRX-mediated ALT suppression is dependent on the histone chaperone DAXX. Re-expression of ATRX is associated with a reduction in replication fork stalling, a known trigger for HR and loss of MRN from telomeres. A G-quadruplex stabilizer partially reverses the effect of ATRX, inferring ATRX may normally facilitate replication through these sequences that, if they persist, promote ALT. We propose that defective telomere chromatinization through loss of ATRX promotes the persistence of aberrant DNA secondary structures, which in turn present a barrier to DNA replication, leading to replication fork stalling, collapse, HR and subsequent recombination-mediated telomere synthesis in ALT cancers.
URL [本文引用: 1]

Gliomas are the most common primary malignant brain tumor in humans. Lower grade gliomas are usually less aggressive but many cases eventually progress to a more aggressive secondary glioblastoma (GBM, WHO Grade IV), which has a universally fatal prognosis despite maximal surgical resection and concurrent chemo-radiation. With the identification of molecular markers, however, there is promise for improving diagnostic and therapeutic strategies. One of the key molecular alterations in gliomas is the alpha thalassemia/mental retardation syndrome X-linked (ATRX) gene, which is frequently mutated. One-third of pediatric GBM cases are also found to have the ATRX mutation and the genetic signatures are different from adult cases. The exact role of ATRX mutations in gliomagenesis, however, is unclear. In this review, we describe the normal cellular function of the ATRX gene product followed by consequences of its dysfunction. Furthermore, its possible association with the alternative lengthening of telomeres (ALT) phenotype is outlined. Lastly, therapeutic options potentiated through a better understanding of ATRX and the ALT phenotype are explored.
URLPMID:10742099 [本文引用: 1]

A goal of molecular genetics is to understand the relationship between basic nuclear processes, epigenetic changes and the numerous proteins that orchestrate these effects. One such protein, ATRX, contains a highly conserved plant homeodomain (PHD)-like domain, present in many chromatin-associated proteins, and a carboxy-terminal domain which identifies it as a member of the SNF2 family of helicase/ATPases. Mutations in ATRX give rise to characteristic developmental abnormalities including severe mental retardation, facial dysmorphism, urogenital abnormalities and alpha-thalassaemia. This circumstantial evidence suggests that ATRX may act as a transcriptional regulator through an effect on chromatin. We have recently shown that ATRX is localized to pericentromeric heterochromatin during interphase and mitosis, suggesting that ATRX might exert other chromatin-mediated effects in the nucleus. Moreover, at metaphase, some ATRX is localized at or close to the ribosomal DNA (rDNA) arrays on the short arms of human acrocentric chromosomes. Here we show that mutations in ATRX give rise to changes in the pattern of methylation of several highly repeated sequences including the rDNA arrays, a Y-specific satellite and subtelomeric repeats. Our findings provide a potential link between the processes of chromatin remodelling, DNA methylation and gene expression in mammalian development.
URLPMID:25938714 [本文引用: 1]

Transposable elements (TEs) comprise roughly forty per cent of mammalian genomes1. TEs have played an active role in genetic variation, adaptation, and evolution through the duplication or deletion of genes or their regulatory elements2-4and TEs themselves can act as alternative promoters for nearby genes resulting in non-canonical regulation of transcription5,6. However, TE activity can lead to detrimental genome instability7, and hosts have evolved mechanisms to appropriately silence TE mobility8,9. Recent studies have demonstrated that a subset of TEs, endogenous retroviral elements (ERVs) containing long terminal repeats (LTRs), are silenced through trimethylation of histone H3 on lysine 9 (H3K9me3) by ESET (also known as SETDB1, SET domain bifurcated 1, or KMT1E)10and a co-repressor complex containing KAP1 (KRAB-associated protein 1, also known as tripartite motif-containing protein 28, TRIM28)11in mouse embryonic stem cells (ESCs). Here we show that the replacement histone variant H3.3 is enriched at class I and class II ERVs, notably early transposon (ETn)/MusD and intracisternal A-type particles (IAPs). Deposition at a subset of these elements is dependent upon the H3.3 chaperone complex containing ATRX (alpha thalesemia/mental retardation syndrome X)12and DAXX (Death-associated protein 6)12-14. We demonstrate that recruitment of DAXX, H3.3, and KAP1 to ERVs are co-dependent and upstream of ESET, linking H3.3 to ERV-associated H3K9me3. Importantly, H3K9me3 is reduced at ERVs upon H3.3 deletion, resulting in derepression and dysregulation of adjacent, endogenous genes, along with increased retrotransposition of IAPs. Our study identifies a unique heterochromatin state marked by the presence of both H3.3 and H3K9me3 and establishes an important role for H3.3 in control of ERV retrotransposition in ESCs.
URL [本文引用: 1]
URLPMID:4639137 [本文引用: 1]

Recent work in human glioblastoma (GBM) has documented recurrent mutations in the histone chaperone protein ATRX. We developed an animal model of ATRX-deficient GBM and showed that loss of ATRX reduces median survival and increases genetic instability. Further, analysis of genome-wide data for human gliomas showed that ATRX mutation is associated with increased mutation rate at the single-nucleotide variant (SNV) level. In mouse tumors, ATRX deficiency impairs nonhomologous end joining and increases sensitivity to DNA-damaging agents that induce double-stranded DNA breaks. We propose that ATRX loss results in a genetically unstable tumor, which is more aggressive when left untreated but is more responsive to double-stranded DNA-damaging agents, resulting in improved overall survival.
URL [本文引用: 1]

Patients with pancreatic cancer typically develop tumor invasion and metastasis in the early stage. These malignant behaviors might be originated from cancer stem cells (CSCs), but the responsible target is less known about invisible CSCs especially for invasion and metastasis. We previously examined the proteasome activity of CSCs and constructed a real-time visualization system for human... [Show full abstract]
URLPMID:22391559 [本文引用: 1]

The MDM2-p53 feedback loop is crucially important for restricting p53 level and activity during normal cell growth and proliferation, and is thus subjected to dynamic regulation in order for cells to activate p53 upon various stress signals. Several ribosomal proteins, such as RPL11, RPL5, RPL23, RPL26 or RPS7, have been shown to have a role in regulation of this feedback loop in response to ribosomal stress. Here, we identify another ribosomal protein S14, which is highly associated with 5q-syndrome, as a novel activator of p53 by inhibiting MDM2 activity. We found that RPS14, but not RPS19, binds to the central acidic domain of MDM2, similar to RPL5 and RPL23, and inhibits its E3 ubiquitin ligase activity toward p53. This RPS14-MDM2 binding was induced upon ribosomal stress caused by actinomycin D or mycophenolic acid. Overexpression of RPS14, but not RPS19, elevated p53 level and activity, leading to G1 or G2 arrest. Conversely, knockdown of RPS14 alleviated p53 induction by these two reagents. Interestingly, knockdown of either RPS14 or RPS19 caused a ribosomal stress that led to p53 activation, which was impaired by further knocking down the level of RPL11 or RPL5. Together, our results demonstrate that RPS14 and RPS19 have distinct roles in regulating the MDM2-p53 feedback loop in response to ribosomal stress. Oncogene (2013) 32, 388-396; doi:10.1038/onc.2012.63; published online 5 March 2012
URLPMID:25464032 [本文引用: 1]

Recent studies have highlighted the fundamental role that key oncogenes such as MYC, RAS and PI3K occupy in driving RNA Polymerase I transcription in the nucleolus. In addition to maintaining essential levels of protein synthesis, hyperactivated ribosome biogenesis and nucleolar function plays a central role in suppressing p53 activation in response to oncogenic stress. Consequently, disruption of ribosome biogenesis by agents such as the small molecule inhibitor of RNA Polymerase I transcription, CX-5461, has shown unexpected, potent, and selective effects in killing tumour cells via disruption of nucleolar function leading to activation of p53, independent of DNA damage. This review will explore the mechanism of DNA damage-independent activation of p53 via the nucleolar surveillance pathway and how this can be utilised to design novel cancer therapies. Non-genotoxic targeting of nucleolar function may provide a new paradigm for treatment of a broad range of oncogene-driven malignancies with improved therapeutic windows. This article is part of a Special Issue entitled: Translation and Cancer.
URLPMID:29192214 [本文引用: 1]

The ribosome is a complex molecular machine composed of numerous distinct proteins and nucleic acids and is responsible for protein synthesis in every living cell. Ribosome biogenesis is one of the most multifaceted and energy- demanding processes in biology, involving a large number of assembly and maturation factors, the functions of which are orchestrated by multiple cellular inputs, including mitogenic signals and nutrient availability. Although causal associations between inherited mutations affecting ribosome biogenesis and elevated cancer risk have been established over the past decade, mechanistic data have emerged suggesting a broader role for dysregulated ribosome biogenesis in the development and progression of most spontaneous cancers. In this Opinion article, we highlight the most recent findings that provide new insights into the molecular basis of ribosome biogenesis in cancer and offer our perspective on how these observations present opportunities for the design of new targeted cancer treatments.
