2.
3.
4.
Research advance and application in the gene therapy of gene editing technologies
Yunxiao Ren1,2,3, Rudan Xiao1,4, Xiaomin Lou1,2,3,4, Xiangdong Fang1,2,3,41. 2.
3.
4.
编委: 吴强
收稿日期:2018-08-16修回日期:2018-10-27网络出版日期:2018-12-06
| 基金资助: |
Received:2018-08-16Revised:2018-10-27Online:2018-12-06
| Fund supported: |
作者简介 About authors
任云晓,硕士研究生,专业方向:疾病组学与转化医学研究E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (477KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
任云晓, 肖茹丹, 娄晓敏, 方向东. 基因编辑技术及其在基因治疗中的应用[J]. 遗传, 2019, 41(1): 18-27 doi:10.16288/j.yczz.18-142
Yunxiao Ren, Rudan Xiao, Xiaomin Lou, Xiangdong Fang.
基因编辑技术是以改变目的基因序列为目的,实现定点突变、插入或敲除的技术。从20世纪末人们就开始对基因编辑技术进行探索,但直到2013年CRISPR/Cas9(clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9)技术成功用于哺乳动物细胞,才极大地推动了基因编辑技术的发展热潮[1]。
真核生物的基因组包含数十亿个碱基,对其基因组的操作一直面临挑战。同源重组技术(homologous recombination, HR)是最早的基因编辑技术,也是真核生物基因编辑的一个重大突破。其原理是将外源性目的基因导入受体细胞,通过同源序列交换,使外源性DNA片段取代原位点上的基因,从而达到使特定基因失活或修复缺陷基因的目的。但是对高等真核生物来说,外源DNA与目的DNA自然重组率非常低,只有10-7~10-6[2,3];若要得到稳定遗传的纯合体基因敲除模型,至少需要两代遗传,因此HR的大规模应用受到了一定的限制。
为应对这一挑战,一系列基于核酸酶的基因编辑技术相继出现,实现了在真核生物尤其是哺乳动物中精准有效的基因编辑。与传统的基因编辑技术相比,基于核酸酶的基因编辑技术减少了外源基因随机插入,提高了对基因组特定片段进行精确修饰的几率。目前基因编辑技术主要包括以下几种:人工介导的锌指核酸酶技术(zinc finger nucleases, ZFNs)、类转录激活因子效应核酸酶技术(transcription activator- like effectors nucleases, TALENs)、规律成簇的间隔短回文重复相关蛋白技术(CRISPR/Cas9)和单碱基编辑(base editor, BE)技术等[4]。
基因编辑技术掀起的研究热潮,一方面是因为基因编辑技术本身的发展,更为精准、高效、低成本的基因编辑技术不断地被开发出来;另一方面基因编辑技术作为一项重要的工具,在基因筛查、动物、细胞模型构建等基础研究中发挥着重要的作用,也为许多疾病的基因治疗提供了新的思路[2,3,4]。因此,本文从基因编辑技术的发展历程及其在基因治疗中的探索和应用进行概述,并对基因编辑技术面临的挑战和机遇进行讨论。
1 基因编辑技术研究进展
基于DNA核酸酶的基因编辑技术发展迅速,从第一代DNA核酸酶编辑系统ZFNs、第二代TALENs到第三代CRISPR/Cas9系统,基因编辑效率不断提高、成本逐渐降低,应用范围不断扩大。ZFNs、TALENs和CRISPR/Cas9等3种基因编辑技术都是在基因组靶标位点引起DNA双链断裂(double-strand breaks, DSBs),进而激活细胞内部修复机制的基础上建立的。细胞内DNA双链断裂的修复机制包括易引起随机插入、缺失的异源末端连接(non-homologous end joining, NHEJ)和需要同源模板存在才可以激活的同源重组修复(homology directed repair, HDR)[5,6]。2016年,BE技术的开发实现了在不引起DNA双链断裂和无需同源模板的情况下的单个碱基转换,有效地规避了基于双链DNA断裂后NHEJ和HDR修复的基因编辑技术的不足。1.1 ZFNs
20世纪90年代,Fok I酶的发现促进了ZFNs的出现[7,8]。1996年,美国约翰霍普金斯大学环境卫生科学系Chandrasegaran团队构建了基于Fok Ⅰ酶和锌指蛋白融合的ZFNs技术[9]。ZFNs包含两个结构域:DNA结合的锌指蛋白区域和限制性核酸内切酶FokⅠ的核酸酶切活性区域(图1)。锌指蛋白区域决定了ZFNs的序列特异性。锌指基序一般由30个氨基酸组成,其结合锌离子的保守区域通常为4个半胱氨酸或2个半胱氨酸和2个组氨酸,其空间结构由1个α螺旋和2个反向的β平行结构组成。α螺旋的1、3、6位的氨基酸分别特异性地识别并结合DNA序列中的3个连续的碱基。由于不同的锌指基序中α螺旋的1、3、6位氨基酸不同,因此由3~6个不同锌指基序组成的锌指蛋白区域与FokⅠ核酸酶区域连接就构成了可以特异识别DNA序列并进行切割的人工核酸酶。FokⅠ必须二聚化才具有活性。由于FokⅠ自身二聚化也能对DNA进行切割,但是切割效率低且易产生非特异切割,所以在设计ZFNs时可以对FokⅠ进行突变,使之不能形成同源二聚体。当两个结合不同靶序列的突变的FokⅠ被5~7 bp的spacers隔开就可以形成具有核酸酶的活性的异源二聚体[10,11]。这样设计的ZFNs可以增加其DNA序列识别的特异性。图1
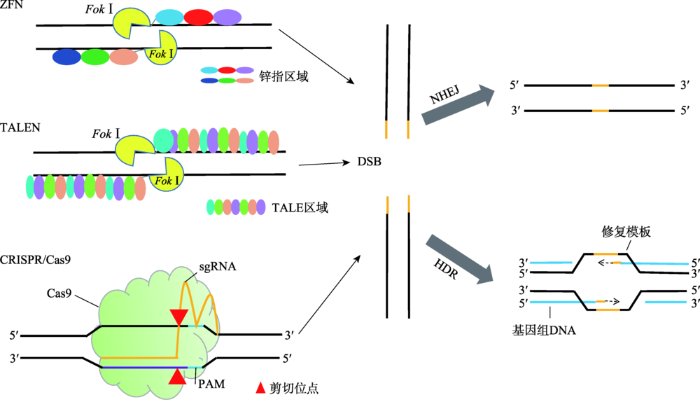 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1ZFN、TALEN和CRISPR/Cas9的结构和DNA断裂修复类型
Fig. 1The structure of ZFN, TALEN, CRISPR/Cas9 and the repair types of DNA DSBs
基于Chandrasegaran的工作,美国犹他大学医学院生物化学系的Dana Carrol团队使用ZFNs注入果蝇胚胎,第一次实现了在动物中的基因编辑[12,13]。随后,科学家用ZFNs技术在动物、植物和人类细胞中都实现了靶基因的编辑[10,14]。尽管ZFNs技术在多个物种中成功进行了基因编辑,但是锌指蛋白的设计费时费力,成本较高,限制了该方法的大规模应用[14,15]。
1.2 TALENs
2009年,美国爱荷华州立大学植物病理学与生物信息学系的Adam J. Bogdanove团队和德国马丁卢瑟大学生物研究所的Ulla Bonas团队分别发现了来自植物致病黄单胞菌属的转录激活效应蛋白(transcription-activator-like effector,TALE)和DNA的相互作用[16,17]。将TALE蛋白与FokⅠ酶区域结合构建了新一代的核酸酶编辑技术——TALENs[14,18]。TALENs的组成和ZFNs的相似之处是在其羧酸末端也含有FokⅠ核酸酶结构域,不同之处是TALENs的DNA结合域为TALE蛋白(图1)[19]。TALE蛋白中每个识别模块由34个氨基酸组成,除了第12和13位氨基酸外其余氨基酸序列都是保守的,第12和13位氨基酸称为可变的双氨基酸残基(repeat variable di-residue, RVD)。RVD决定了TALE识别并结合的DNA碱基。4种不同的碱基都有与之对应的TALE识别模块。因此构建TALEN人工核酸酶时,只需要按照目标序列的顺序将不同的TALE识别模块的序列连接起来,再与FokⅠ的编码序列融合即可。相对于ZFNs来说,TALENs的设计变得容易,对任意的DNA序列理论上都可以设计和构建一个特异的TALEN核酸酶。但是目标序列的每个碱基都需要一个TALE识别模块,因此TALENs的构建过程工作量较大。此外,TALENs在人类细胞中的细胞毒性较低[14,20]。2011年,Miller等[18]第一次使用TALENs在人类细胞中对NTF3 和CCR5 基因进行编辑,证明了TALEN核酸酶对内源靶向基因的调节和修饰作用。1.3 CRISPR/Cas9
2012年CRISPR/Cas9的体外重构和2013年在人类细胞中证明了其基因编辑功能,标志着新一代基因编辑时代的开始[21,22]。CRISPR/Cas9系统来源于细菌和古细菌的天然获得性免疫系统,通过CRISPR RNA (crRNA)和trans-activating crRNA (tracrRNA)以及Cas9蛋白组成的复合体抵御外源性DNA的入侵。CRISPR/Cas9系统发挥作用的基本过程可以分为3个阶段:第一个阶段为间隔序列获得期,质粒或噬菌体携带的DNA片段被宿主的核酸酶切割成短的DNA片段,符合条件的DNA片段整合进宿主CRISPR位点成为crRNA重复序列间的间隔序列;第二个阶段为CRISPR/Cas9表达期,Cas9蛋白表达,CRISPR序列由pre-crRNA 加工为成熟的crRNA,成熟的crRNA包含间隔序列,靶向结合于外来入侵的DNA;第三阶段为DNA干扰期,Cas9蛋白在向导crRNA的引导下识别靶向位点,并调节基因组的剪切[22]。根据Cas蛋白的不同,CRISPR/Cas系统可以分为5类:类型Ⅰ、Ⅲ和Ⅳ的CRISPR位点包含crRNA与多个Cas蛋白形成的复合物;类型Ⅱ (Cas9)和类型V(Cpf1)只需要RNA介导的核酸酶[23]。许多CRISPR系统都依赖于临近crRNA靶向位点的PAM (protospacer adjacent motif)序列,PAM序列的缺失将会导致Ⅰ型和Ⅱ型CRISPR系统的自我剪切[24]。广泛用于基因编辑的CRISPR/Cas9是Ⅱ型CRISPR系统,由Cas9蛋白和sgRNA(single guide RNA)组成。sgRNA是根据crRNA和tracrRNA形成的高级结构设计的,与Cas9核酸酶蛋白结合,指导其识别并剪辑靶向序列,靶向序列附近必需存在含有NGG或者NAG的PAM基序[25,26,27](图1)。
相比较ZFNs和TALENs,CRISPR/Cas9通过一段与目标DNA片段匹配的向导RNA引导核酸酶识别靶向位点,提高了Cas9核酸酶的特异性;同时,Cas9在sgRNA的引导下以单体蛋白的形式发挥功能,不像ZFNs和TALENs的FokⅠ酶只有二聚化才具有切割靶向DNA的活性,因此CRISPR/Cas9也避免了精细复杂的蛋白质设计或组装的需要。但是,由于CRISPR/Cas9来自于原核生物天然获得性免疫系统抵御外来遗传物质的防御系统,Cas9核酸酶可能继承了序列特异性低的特点,使其非特异性切割的几率增加,造成脱靶效应增多。不同的科研团队提出了不同的方法修饰或编辑Cas9和sgRNA以降低脱靶效应[28,29]。如将Cas9蛋白与FokⅠ核酸酶、锌指蛋白或者TALE蛋白结合提高Cas9的特异性[30,31,32];用失活的Cas9蛋白和FokⅠ区域融合形成新的核酸酶,使其只有在核酸酶二聚化时才具有活性;Fatih等[32]将锌指蛋白或者TALE蛋白与Cas9蛋白变异体融合增强核酸酶的特异性。另一种降低脱靶效应的方法是使用切口酶代替核酸酶,产生单链断裂而不是双链断裂,单链断裂不能诱导NHEJ的修复,仍然可以激活HR的精确修复[14]。单链断裂可以使脱靶效应降低,但修复效率也降低,因此有人提出了使用双切口酶的方法,既提高了基因编辑的特异性也提高了编辑的效率[33]。
1.4 BE
ZFNs、TALENs和CRISPR/Cas9技术都依赖于在靶位点诱导双链断裂进而激活DNA的NHEJ和HDR。NHEJ容易引起随机插入和缺失,造成移码突变,进而影响靶基因的功能;HDR尽管精确性高于NHEJ,但是其在细胞中的同源重组修复效率低,约为0.1%~5%。BE技术的出现有效地改善了以上问题[34,35]。2016年4月,美国哈佛大学David Liu实验室第一次发表了不需要DNA双链断裂也不需要同源模板即可进行单碱基转换的基因编辑技术——BE技术。该技术基于无核酸酶活性的dCas9 (Inactive, or dead Cas9)或有单链DNA切口酶活性的Cas9n (Cas9 nickase)、胞嘧啶脱氨酶、尿嘧啶糖基化酶抑制子(uracil DNA glycosylase inhibitor, UGI)以及sgRNA形成的复合体,在不引起双链DNA断裂的情况下,直接使靶向位点的胞嘧啶(Cytosine, C)脱氨基变成尿嘧啶(Uracil, U);由于尿嘧啶糖基化酶抑制子的存在,抑制了U的切除;随着DNA复制,U被胸腺嘧啶(Thymine, T)取代;同时,互补链上原来与C互补的鸟嘌呤(Guanine, G)将会替换为腺嘌呤(Adenine, A),最终实现了在一定的活性窗口内C到T和G到A的单碱基精准编辑。BE技术的出现促进了点突变基因编辑的有效性和使用范围[36]。
David Liu团队对4种胞嘧啶脱氨酶——hAID (human activation induced deaminase)、hAPOBEC3G (human apoliprotein B mRNA-editing enzyme-catalytic polypeptide-like-3G)、rAPOBEC1 (rat apolipoprotein B mRNA-editing enzyme 1)和七鳃鳗(Lethenteron camtschaticum)来源的AID类似物PmCDA1进行评估,发现rAPOBEC1具有最高的脱氨酶活性[36]。他们通过将rAPOBEC1与dCas9的N末端以及16个残基的XTEN连接体融合,组成了第一代碱基编辑器BE1 (rAPOBEC1-XTEN- dCas9),具有5 nt的活性窗口[36]。第二代碱基编辑器BE2 (APOBEC-XTEN- dCas9-UGI)融入了UGI抑制了U糖基化引起碱基切除修复,编辑效率在人类细胞比BE1高3倍[36]。第三代碱基编辑器BE3 (APOBEC-XTEN-dCas9(A840H)- UGI)恢复了Cas9 HNH区域840位置组氨酸的催化作用,可以剪切非编辑链上与编辑链上U互补的G碱基,对非编辑链的剪切使BE3的编辑效率比BE2高2~6倍[36]。
随着单碱基编辑技术的出现,日本科学家基于PmCDA1和dCas9或Cas9n以及鸟嘌呤糖苷化酶抑制子UGI的融合,也开发了PMCDATA-dCas9/Cas9n- UGI的单碱基编辑器[37]。中国科学院上海营养与健康研究院常兴研究组开发了基于hAID的dCas9- AIDx单碱基编辑系统,用于耐药突变的筛选[38]。
2017年,David Liu 实验室对BE技术做了多方面改进。首先针对常用的化脓性链球菌(Streptococcus pyogenes)的Cas9(SpCas9)蛋白靶向范围较窄,只识别含有NGG或NGA的PAM序列的靶位点的限制,他们使用金黄色葡萄球菌(Staphylococcus Aureus)的Cas9 (SaCas9)、SaCas9突变体(Sacas9-KKH)、SpCas9突变体(SpCas9-VQR、SpCas9-EQR、SpCas9-VRER)替代SpCas9,可以识别含有NGG、NGA、NGAN、NGAG、NGGG、NNGRRT和NNNRRT的PAM序列,从而显著提高了单碱基基因编辑的靶向范围。其次,他们通过突变胞嘧啶脱氨酶,降低酶的活性、改变底物的结合和构象,或者直接降低底物进入胞嘧啶脱氨酶活性区域的能力,缩小了单碱基编辑系统的活性窗口,从5个核苷酸窗口缩小至1~2个核苷酸活性窗口[39]。再次,David Liu实验室通过对BE3引入突变来减少脱靶效应,产生了高保真的碱基编辑器(high-fidelity base editor, HF-BE3)[40]。随后,他们又报道了基于E. coli TadA (ecTadA)的新型单碱基编辑器——腺嘌呤碱基编辑器(adenine base editors, ABEs),实现了A?T碱基对向G?C碱基对的转换。经过不断的改进,第7代的腺嘌呤碱基编辑器将靶向的A.T碱基转化为G?C碱基对可以达到约50%的效率(人类细胞),并且引入插入或缺失的频率低于0.1%[34]。
2 基因编辑技术在基因治疗中的应用
从1996年对ZFNs第一次进行体外验证到2012年CRISPR/Cas9技术的出现和之后的蓬勃发展(图2),基因编辑技术发展迅速,编辑效率和精确性不断提高,应用领域也不断拓宽。不仅可用于表达调控和基因功能的研究、细胞动物模型的构建、癌基因和药物靶点的筛选,在基因治疗中更是具有巨大的发展前景,为单基因遗传病、癌症等疾病提供了新的治疗方法[41,42]。图2
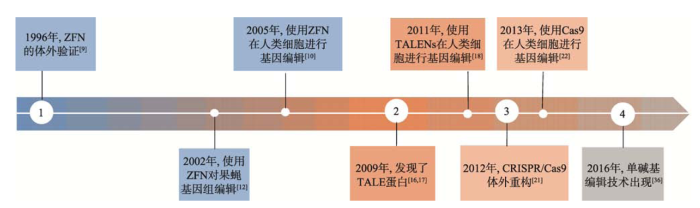 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基因编辑技术的发展和应用
Fig. 2Development and application of genome editing technologies
基因治疗通过导入正常基因或者编辑修复缺陷基因,实现治疗疾病的目的。目前,利用基因编辑技术在多种疾病,如单基因遗传病、眼科疾病、艾滋病及肿瘤等的基因治疗中得到了应用[43]。
镰状细胞病(sickle cell disease, SCD)是由于β-珠蛋白基因的第7个密码子的单基因点突变造成的。Hoban等[44]利用ZFNs特异性靶向于β-珠蛋白基因,并诱导CD34+造血干细胞和祖细胞中的DNA被切割。当ZFNs与整合酶缺陷型慢病毒载体或寡核苷酸供体一起传送进细胞时,有效地实现了β-珠蛋白基因座的基因矫正。Hoban的研究为镰状型细胞性贫血的基因治疗提供了重要的方法路径[44]。
基因编辑技术不仅应用于遗传性疾病的治疗,在非遗传性疾病中也有重大的突破。与年龄相关的黄斑衰退(age-related macular degeneration, AMD)是导致成人失明的主要原因,脉络膜新血管形成(choroidal neovascularization, CNV)是其主要病理学特征,血管生成因子如VEGFA (vascular endothelial growth factor A, VEGFA)基因的高表达是造成病变的主要原因[45,46]。Kim等[47]将预先设计好的Cas9核糖核蛋白(ribonucleoprotein, RNP)导入成年小鼠眼中,使视网膜色素上皮中VEGFA基因失活;并且在AMD的小鼠模型中发现 cas9 RNPs有效地减少了脉络膜新血管生成的面积。该研究表明CRISPR/Cas9技术有可能用于非遗传性退行性眼部疾病的局部治疗。
Table 1
表1
表1 ZFNs、TALENs、CRISPR/Cas9和BE技术的比较
Table 1
| 基因编辑技术 | 优点 | 缺点 |
|---|---|---|
| ZFNs | 靶向传递基因的效率高; 靶向结合效率高 | 核酸酶设计成功率低; 可能有较高的脱靶率; 不适合高通量靶向目的基因 |
| TALENs | 特异性高,容易设计; 靶向结合效率高; 核酸酶设计成功较高 | 靶向传递效率低; 重复序列可能造成非特异性剪切; 通量低 |
| CRISPR/Cas9 | 编辑效率更高; 操作简单,成本低; 通量上无限制 | 脱靶效率较高; 同源重组效率低 |
| BE | 脱靶效应低; 单碱基精准转换 | 不能进行敲除和敲入; 靶点临近的非靶向胞嘧啶的编辑 |
新窗口打开|下载CSV
基因编辑技术在艾滋病、肿瘤治疗等领域也取得了初步进展。第一次在人类中应用靶向核酸酶是利用ZFNs技术编辑CCR5基因抵抗HIV。研究者从HIV病人中提取T细胞,用ZFNs技术干扰T细胞中CCR5基因,由于CCR5基因是大部分HIV菌株尤其是早期感染菌株的辅助受体,干扰CCR5基因的表达可以抗HIV感染,表明基因编辑技术可能成为艾滋病治疗的新方向[48,49,50,51]。
基因编辑应用于肿瘤治疗,主要是与免疫治疗相结合,尤其是与CAR (chimeric antigen receptor) T 细胞,该方法在白血病、淋巴瘤和部分实体瘤中有巨大的发展前景[52,53]。CARs包括肿瘤细胞特异抗原的胞外单链可变片段和细胞内嵌合信号域,可以激活T细胞和杀伤肿瘤细胞[53]。Ren等[54]使用CRISPR/ Cas9系统同时破坏多个基因位点,产生的TCR (T cell receptor)和HLA-I (HLA class I)缺陷的CAR T细胞可作为通用的CAR T细胞,用于免疫治疗;除了产生通用的CAR T细胞,基因编辑技术也可通过敲除编码T细胞抑制受体或信号分子的基因如PD1 (programmed cell death protein 1)和CTLA4 (cytotoxic T lymphocyte-associated protein 4),用于产生增强型CAR T细胞[53,55,56]。
2016年,四川大学华西医院卢铀团队开展了CRISPR基因编辑技术的临床实验,从转移性非小细胞肺癌患者中分离出T细胞,并使用CRISPR/Cas9技术敲除细胞中的PD-1基因,在体外扩增到一定量后再重新输回患者体内,达到杀死肿瘤细胞的目的[57,58]。但是简单的敲除T细胞的抑制因子是一把双刃剑,真正投入临床使用还需要进一步研究敲除这些抑制因子是否会引起细胞的不可控制的增殖或者产生严重的自身免疫[53]。
由于BE技术是在不造成双链DNA断裂的情况下进行的精确碱基转换,这对于基因治疗而言无疑是一个非常有效的工具。β-地中海贫血是由于珠蛋白基因(hemoglobin-beta, HBB)突变所致,中国和东南亚地区的发病原因主要是HBB基因A到G的突变。 2017年,中山大学黄军就团队利用单碱基编辑技术在不能发育成熟的人类三元核胚胎中战对HBB的点突变进行编辑,即将碱基G改为A从而修正错误。该研究是第一个利用BE技术对遗传疾病突变位点进行精准修复的研究,为治疗新生儿β-型地中海贫血症,甚至为其他遗传性疾病的治疗打开了新窗口[59]。Chadwick团队也通过BE技术实现PCSK9基因的敲除,从而降低血浆胆固醇的水平[60]。
3 基因编辑技术面临的挑战
尽管基因编辑技术在基因治疗领域展现了广阔的应用前景,但是目前仍然面临着诸多挑战,如脱靶效应、传递系统的有效性和安全性、免疫排斥反应、伦理争论等。3.1 脱靶效应
脱靶效应会导致假表型,造成错误的理解和解读,是限制基因编辑技术应用的重要原因。脱靶效应面临两个问题:一是如何从技术本身上降低脱靶效应;二是如何提高检测方法的灵敏性。降低脱靶效应的可行性策略有减少NHEJ修复引入的插入和缺失,如使用单切口酶活性突变体和不需要双链断裂激活修复的BE技术等[33,58];改进sgRNA的设计,如在保证一定的sgRNA结合靶点效率的基础上,截短sgRNA的长度[59,61];提高核酸酶蛋白的特异性,如David Liu研究组通过在Cas9蛋白特定位点插入羟基他莫昔芬应激性内含肽(hydroxytamoxifen (4-HT)- responsive intein)产生小分子激活Cas9核酸酶,使Cas9靶向编辑的特异性提高了25倍[62]。目前脱靶效应的检测方法有基于全基因组范围内检测DSBs的方法,该方法可以无偏向性地评估Cas9剪切的特异性,如通过NHEJ将双链寡聚脱氧核苷酸整入DSBs,进而扩增和测序的GUIDE-Seq (genome-wide unbiased identification of DSBs enabled by sequencing)[63],以及用Cas9体外消化分离基因组DNA,然后进行全基因组测序的Digenome-Seq (digested genome sequencing)[29]。虽然两种方法都无偏向性,且敏感性较好,但是两者都需要参考基因组,GUIDE-Seq整合双链寡聚脱氧核苷酸进入DSBs的效率不高,Digenome-Seq检验gRNA费用昂贵,且检测多个gRNA时测序深度也受限制[64]。因此,脱靶效应仍然是基因编辑技术未来发展必需解决的问题之一。
3.2 核酸酶的传递效率和安全性
核酸酶的传送系统包括病毒、质粒、RNA和蛋白质的系统,其中病毒载体系统具有较高的编辑效率和插入突变,较低的脱靶效应和较高的免疫反应,如AAV (adeno-associated virus)载体和慢病毒载体广泛用于靶向基因向细胞的传送,但是其对传输物质大小有限制;质粒型载体在编辑效率、插入突变、脱靶效应和免疫反应均具有中等表现;RNA或蛋白质核酸复合物均不具有插入突变的功能,但普遍认为以蛋白核酸复合物进行传输具有起效快且无长期表达的特点,因而具有较低的脱靶效应[64]。就BE系统来说,需要Cas9与胞嘧啶脱氨酶、UGI、sgRNA的融合,序列组成较长。如果使用AAV载体,其对包装物质4.7 kb长度的限制,使BE系统无法被包装进去。可以考虑将BE系统拆分包装成两个病毒,但是系统拆分后的编辑效率是否会受到影响尚不得知。因此,提高核酸酶传送系统的有效性和安全性将会进一步促进基因编辑的应用,有研究表明脂质体可能在基因编辑传送系统中展现巨大的发展潜力[2]。3.3 基因编辑引起的副作用
美国斯坦福大学儿科血液学家Matthew Porteus和Kenneth Weinberg的研究小组发现人体中普遍存在Cas9抗体,如果使用CIRSPR/Cas9进行治疗,可能引发人体剧烈的免疫反应,最终导致治疗失败,但是目前有关基因编辑可能引起免疫反应的研究还相对较少[65]。近期,Nature Medicine发表了分别来自瑞典卡洛林斯卡研究所和美国剑桥诺华生物医学研究院的研究[66,67]。他们分别独立发现CRISPR/Cas9基因编辑过程造成的DNA双链断裂可以激活p53通路。这意味着基因编辑成功的细胞很可能成为潜在的癌细胞,利用CRISPR/Cas9进行临床治疗可能无意中增加患癌症的风险。
CRISPR/Cas9技术除了面临脱靶问题,可能还面临染色体结构异常的问题。Kosicki等[68]在Nature Biotechnology上报道了Cas9还可能在作用靶点附近导致大规模的DNA删除,在部分情况下,甚至引起复杂的DNA重排。研究人员通过长读长测序和大范围的PCR基因型鉴定发现在小鼠胚胎干细胞和人类造血祖系细胞内,可能出现数千DNA碱基的删除,导致临近基因或调控序列受到影响并改变细胞功能。因此引起的DNA重排问题也成为了CRISPR/Cas9技术面临的另一个安全隐患[68]。
除了上述挑战之外,基因编辑技术在基因治疗中的应用仍可能有很多意想不到的问题出现。但无论如何,技术的发展总会遇到瓶颈,瓶颈的攻克也将必然给技术的应用带来更广阔的空间。
3.4 基因编辑中的伦理问题
基因编辑技术为遗传性疾病、肿瘤等多种疾病带来了治疗的希望,但是人类胚胎细胞的基因编辑引起了社会上广泛的讨论和激烈的争议。一方面,对胚胎细胞的基因编辑有助于基础科学研究,促进我们了解人类胚胎发育的分子机制,有利于实现在早期胚胎发育时期治愈疾病的目标;另一方面基因编辑技术可能产生的脱靶或者其他基因编辑失误,从而导致慢性疾病或者严重的致残个体,这些对人类的繁衍和生活方式等都将会产生非常大的危害。然而,目前很多国家的法律对基因编辑技术应用于基因治疗的监管措施依然存在大量空白。4 结语与展望
基因编辑技术的快速发展极大地提高了我们对真核细胞基因组进行精准改变的能力。可编辑的核酸酶,尤其是编辑效率更高、操作简单、成本低的CRISPR/Cas系统,彻底改变了我们对基因组功能的研究;单碱基编辑技术的出现和不断改进,实现了单个碱基的精准转换,降低了脱靶效率。虽然目前基因编辑技术仍然面临着脱靶效率和潜在的免疫反应等副作用的问题,相信在未来多学科的交叉融合和科学家的共同合作下,新一代基因编辑技术将更加简便、高效、精确,目前存在的问题也会逐步得到解决[69]。随着人们对疾病的新的有效靶点的不断认识和新的基因编辑技术的不断发展,基因编辑治疗方案的大规模临床应用将成为可能,特别是对那些传统疗法难以治愈的疾病,可以通过纠正致病基因或引入有益突变来达到治愈疾病的目的。基因编辑技术的临床转化和应用研究值得拭目以待,这将成为下一代转化疗法和治疗范例的关键,也将促进个体化医疗的快速发展。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:23287722 [本文引用: 1]

Bacteria and archaea have evolved adaptive immune defenses termed clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems that use short RNA to direct degradation of foreign nucleic acids. Here, we engineer the type II bacterial CRISPR system to function with custom guide RNA (gRNA) in human cells. For the endogenous AAVS1 locus, we obtained targeting rates of 10 to 25% in 293T cells, 13 to 8% in K562 cells, and 2 to 4% in induced pluripotent stem cells. We show that this process relies on CRISPR components, is sequence-specific, and upon simultaneous introduction of multiple gRNAs, can effect multiplex editing of target loci. We also compute a genome-wide resource of ~190k unique gRNAs targeting ~40.5% of human exons. Our results establish an RNA-guided editing tool for facile, robust, and multiplexable human genome engineering.
URL [本文引用: 3]
URLPMID:24690881 [本文引用: 2]

Programmable nucleases - including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and RNA-guided engineered nucleases (RGENs) derived from the bacterial clustered regularly interspaced short palindromic repeat (CRISPR)-Cas (CRISPR-associated) system - enable targeted genetic modifications in cultured cells, as well as in whole animals and plants. The value of these enzymes in research, medicine and biotechnology arises from their ability to induce site-specific DNA cleavage in the genome, the repair (through endogenous mechanisms) of which allows high-precision genome editing. However, these nucleases differ in several respects, including their composition, targetable sites, specificities and mutation signatures, among other characteristics. Knowledge of nuclease-specific features, as well as of their pros and cons, is essential for researchers to choose the most appropriate tool for a range of applications.
URLPMID:3694601 [本文引用: 2]

Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) comprise a powerful class of tools that are redefining the boundaries of biological research. These chimeric nucleases are composed of programmable, sequence-specific DNA-binding modules linked to a nonspecific DNA cleavage domain. ZFNs and TALENs enable a broad range of genetic modifications by inducing DNA double-strand breaks that stimulate error-prone nonhomologous end joining or homology-directed repair at specific genomic locations. Here, we review achievements made possible by site-specific nuclease technologies and discuss applications of these reagents for genetic analysis and manipulation. In addition, we highlight the therapeutic potential of ZFNs and TALENs and discuss future prospects for the field, including the emergence of clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas-based RNA-guided DNA endonucleases.
URLPMID:29428910 [本文引用: 1]

Abstract Precise nucleic acid editing technologies have facilitated the research of cellular function and the development of novel therapeutics, especially the current programmable nucleases-based editing tools, such as the prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR)-associated nucleases (Cas). As CRISPR-based therapies are advancing toward human clinical trials, it is important to understand how natural genetic variation in the human population may affect the results of these trials and even patient safety. The development of "base-editing" technique allows the direct, stable transformation of target DNA base into an alternative in a programmable way, without DNA double strand cleavage or a donor template. Genome-editing techniques hold promises for the treatment of genetic disease at the DNA level by blocking the sequences associated with disease from producing disease-causing proteins. Currently, scientists can select the gene they want to modify, use the Cas9 as a "molecular cutter" to cut it out, and transform it into a more desirable version. In this review, we focus on the recent advances of CRISPR/Cas system by outlining the evolutionary and biotechnological implications of current strategies for improving the specificity and accuracy of these genome-editing technologies.
URLPMID:28614576 [本文引用: 1]

Abstract Importance Clustered regularly interspaced short palindromic repeats (CRISPR) CRISPR-associated protein 9 (CRISPR-Cas9) has garnered a great degree of attention since its first reported uses in mammalian cells in early 2013 due to its perceived impact with respect to potential research applications and, especially, therapeutic applications. Observations CRISPR-Cas9 is being widely used in the laboratory and has greatly improved the ability to generate genetically modified animal models of human diseases. First steps have been reported with respect to proof-of-concept cardiovascular therapeutic applications in mouse models—which might eventually lead to new treatments for patients—as well as modification of human embryos, which raises a host of social, ethical, and legal questions. Conclusions and Relevance CRISPR-Cas9 is already having a substantial effect on cardiovascular basic science research. Although the first steps toward potential therapies have been taken, there are substantial obstacles that will need to be overcome if CRISPR-Cas9 is to be used in the practice of cardiovascular medicine.
URLPMID:1584761 [本文引用: 1]

The PCR was used to alter transcriptional and translational signals surrounding the Flavobacterium okeanokoites restriction endonuclease (fokIR) gene, so as to achieve high expression in Escherichia coli. By changing the ribosome-binding site sequence preceding the fokIR gene to match the consensus E. coli signal and by placing a positive retroregulator stem-loop sequence downstream of the gene, Fok I yield was increased to 5-8% of total cellular protein. Fok I was purified to homogeneity with phosphocellulose, DEAE-Sephadex, and gel chromatography, yielding 50 mg of pure Fok I endonuclease per liter of culture medium. The recognition and cleavage domains of Fok I were analyzed by trypsin digestion. Fok I in the absence of a DNA substrate cleaves into a 58-kDa carboxyl-terminal and 8-kDa amino-terminal fragment. The 58-kDa fragment does not bind the DNA substrate. Fok I in the presence of a DNA substrate cleaves into a 41-kDa amino-terminal fragment and a 25-kDa carboxyl-terminal fragment. On further digestion, the 41-kDa fragment degrades into 30-kDa amino-terminal and 11-kDa carboxyl-terminal fragments. The cleaved fragments both bind DNA substrates, as does the 41-kDa fragment. Gel-mobility-shift assays indicate that all the protein contacts necessary for the sequence-specific recognition of DNA substrates are encoded within the 41-kDa fragment. Thus, the 41-kDa amino-terminal fragment constitutes the Fok I recognition domain. The 25-kDa fragment, purified by using a DEAE-Sephadex column, cleaves nonspecifically both methylated (pACYCfokIM) and nonmethylated (pTZ19R) DNA substrates in the presence of MgCl2. Thus, the 25-kDa carboxyl-terminal fragment constitutes the Fok I cleavage domain.
URL [本文引用: 1]
[本文引用: 1]
URLPMID:16251401 [本文引用: 2]

Custom-designed zinc finger nucleases (ZFNs), proteins designed to cut at specific DNA sequences, are becoming powerful tools in gene targeting--the process of replacing a gene within a genome by homologous recombination (HR). ZFNs that combine the non-specific cleavage domain (N) of FokI endonuclease with zinc finger proteins (ZFPs) offer a general way to deliver a site-specific double-strand break (DSB) to the genome. The development of ZFN-mediated gene targeting provides molecular biologists with the ability to site-specifically and permanently modify plant and mammalian genomes including the human genome via homology-directed repair of a targeted genomic DSB. The creation of designer ZFNs that cleave DNA at a pre-determined site depends on the reliable creation of ZFPs that can specifically recognize the chosen target site within a genome. The (Cys2His2) ZFPs offer the best framework for developing custom ZFN molecules with new sequence-specificities. Here, we explore the different approaches for generating the desired custom ZFNs with high sequence-specificity and affinity. We also discuss the potential of ZFN-mediated gene targeting for 'directed mutagenesis' and targeted 'gene editing' of the plant and mammalian genome as well as the potential of ZFN-based strategies as a form of gene therapy for human therapeutics in the future.
URLMagsci [本文引用: 1]

锌指核酸酶(ZFN)由锌指蛋白(ZFP)结构域和<em>Fok</em> I核酸内切酶的切割结构域人工融合而成, 是近年来发展起来的一种可用于基因组定点改造的分子工具。ZFN可识别并结合特定的DNA序列, 并通过切割这一序列的特定位点造成DNA的双链断裂(DSB)。在此基础上, 人们可以对基因组的特定位点进行各种遗传操作, 包括基因打靶、基因定点插入、基因修复等, 从而能够方便快捷地对基因组实现靶向遗传修饰。这种新的基因组定点修饰方法的突出优势是适用性好, 对物种没有选择性, 并且可以在细胞和个体水平进行遗传操作。文章综述了ZFN技术的研究进展及应用前景, 重点介绍ZFN的结构与作用机制、现有的靶点评估及锌指蛋白库的构建与筛选方法、基因组定点修饰的策略, 以及目前利用这一技术已成功实现突变的物种及内源基因, 为开展这一领域的研究工作提供参考。
URLMagsci [本文引用: 1]

锌指核酸酶(ZFN)由锌指蛋白(ZFP)结构域和<em>Fok</em> I核酸内切酶的切割结构域人工融合而成, 是近年来发展起来的一种可用于基因组定点改造的分子工具。ZFN可识别并结合特定的DNA序列, 并通过切割这一序列的特定位点造成DNA的双链断裂(DSB)。在此基础上, 人们可以对基因组的特定位点进行各种遗传操作, 包括基因打靶、基因定点插入、基因修复等, 从而能够方便快捷地对基因组实现靶向遗传修饰。这种新的基因组定点修饰方法的突出优势是适用性好, 对物种没有选择性, 并且可以在细胞和个体水平进行遗传操作。文章综述了ZFN技术的研究进展及应用前景, 重点介绍ZFN的结构与作用机制、现有的靶点评估及锌指蛋白库的构建与筛选方法、基因组定点修饰的策略, 以及目前利用这一技术已成功实现突变的物种及内源基因, 为开展这一领域的研究工作提供参考。
[本文引用: 1]
URLPMID:12730594 [本文引用: 1]

Author information: (1)Department of Biochemistry, University of Utah School of Medicine, 20 North 1900 East, Salt Lake City, UT 84132-3201, USA.
URLPMID:27490630 [本文引用: 5]

Abstract Genome editing harnesses programmable nucleases to cut and paste genetic information in a targeted manner in living cells and organisms. Here, I review the development of programmable nucleases, including zinc finger nucleases (ZFNs), TAL (transcription-activator-like) effector nucleases (TALENs) and CRISPR (cluster of regularly interspaced palindromic repeats)-Cas9 (CRISPR-associated protein 9) RNA-guided endonucleases (RGENs). I specifically highlight the key advances that set the foundation for the rapid and widespread implementation of CRISPR-Cas9 genome editing approaches that has revolutionized the field.
URL [本文引用: 1]
URLPMID:19933106 [本文引用: 1]

TAL effectors of plant pathogenic bacteria in the genus Xanthomonas bind host DNA and activate genes that contribute to disease or turn on defense. Target specificity depends on an effector-variable number of typically 34 amino acid repeats, but the mechanism of recognition is not understood. We show that a repeat-variable pair of residues specifies the nucleotides in the target site, one pair to one nucleotide, with no apparent context dependence. Our finding represents a previously unknown mechanism for protein-DNA recognition that explains TAL effector specificity, enables target site prediction, and opens prospects for use of TAL effectors in research and biotechnology.
URLPMID:19933107 [本文引用: 1]

TAL effectors are important virulence factors of bacterial plant pathogenic Xanthomonas, which infect a wide variety of plants including valuable crops like pepper, rice, and citrus. TAL proteins are translocated via the bacterial type III secretion system into host cells and induce transcription of plant genes by binding to target gene promoters. Members of the TAL effector family differ mainly in their central domain of tandemly arranged repeats of typically 34 amino acids each with hypervariable di-amino acids at positions 12 and 13. We recently showed that target DNA-recognition specificity of TAL effectors is encoded in a modular and clearly predictable mode. The repeats of TAL effectors feature a surprising one repeat-to-one-bp correlation with different repeat types exhibiting a different DNA base pair specificity. Accordingly, we predicted DNA specificities of TAL effectors and generated artificial TAL proteins with novel DNA recognition specificities. We describe here novel artificial TALs and discuss implications for the DNA recognition specificity. The unique TAL-DNA binding domain allows design of proteins with potentially any given DNA recognition specificity enabling many uses for biotechnology.
URL [本文引用: 2]
URLPMID:27741224 [本文引用: 1]

Abstract Molecular scissors engineered for site-specific modification of the genome hold great promise for effective functional analyses of genes, genomes and epigenomes and could improve our understanding of the molecular underpinnings of disease states and facilitate novel therapeutic applications. Several platforms for molecular scissors that enable targeted genome engineering have been developed, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and, most recently, clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated-9 (Cas9). The CRISPR/Cas9 system's simplicity, facile engineering and amenability to multiplexing make it the system of choice for many applications. CRISPR/Cas9 has been used to generate disease models to study genetic diseases. Improvements are urgently needed for various aspects of the CRISPR/Cas9 system, including the system's precision, delivery and control over the outcome of the repair process. Here, we discuss the current status of genome engineering and its implications for the future of biological research and gene therapy.
URLPMID:4792841 [本文引用: 1]

The manner by which eukaryotic genomes are packaged into nuclei while maintaining crucial nuclear functions remains one of the fundamental mysteries in biology. Over the last ten years, we have witnessed rapid advances in both microscopic and nucleic acid-based approaches to map genome architecture, and the application of these approaches to the dissection of higher-order chromosomal structures has yielded much new information. It is becoming increasingly clear, for example, that interphasechromosomesform stable, multilevel hierarchical structures. Among them, self-associating domains like so-called topologically associating domains (TADs) appear to be building blocks for large-scale genomic organization. This review describes features of these broadly-defined hierarchical structures, insights into the mechanisms underlying their formation, our current understanding of how interactions in the nuclear space are linked to gene regulation, and important future directions for the field.
URL [本文引用: 1]
URLPMID:23360966 [本文引用: 2]

We employ the CRISPR-Cas system of Streptococcus pyogenes as programmable RNA-guided endonucleases (RGENs) to cleave DNA in a targeted manner for genome editing in human cells. We show that complexes of the Cas9 protein and artificial chimeric RNAs efficiently cleave two genomic sites and induce indels with frequencies of up to 33%.
URLPMID:20863886 [本文引用: 1]

CRISPR/Cas and CRISPR/Cmr immune machineries of archaea and bacteria provide an adaptive and effective defence mechanism directed specifically against viruses and plasmids. Present data suggest that both CRISPR/Cas and Cmr modules can behave like integral genetic elements. They tend to be located in the more variable regions of chromosomes and are displaced by genome shuffling mechanisms including transposition. CRISPR loci may be broken up and dispersed in chromosomes by transposons with the potential for creating genetic novelty. Both CRISPR/Cas and Cmr modules appear to exchange readily between closely related organisms where they may be subjected to strong selective pressure. It is likely that this process occurs primarily via conjugative plasmids or chromosomal conjugation. It is inferred that interdomain transfer between archaea and bacteria has occurred, albeit very rarely, despite the significant barriers imposed by their differing conjugative, transcriptional and translational mechanisms. There are parallels between the CRISPR crRNAs and eukaryal siRNAs, most notably to germ cell piRNAs which are directed, with the help of effector proteins, to silence or destroy transposons. No homologous proteins are identifiable at a sequence level between eukaryal siRNA proteins and those of archaeal or bacterial CRISPR/Cas and Cmr modules.
URLPMID:28086097 [本文引用: 1]

Abstract Class 2 CRISPR-Cas systems are characterized by effector modules consisting of single, large, multidomain proteins that appear to have been derived from mobile genetic elements. Some Class 2 effector proteins, such as Cas9 and Cas12a (Cpf1), have been successfully repurposed for genome engineering. Published by Elsevier Inc.
URLPMID:28624197 [本文引用: 1]

With the expansion of the microbiology field of research, a new genome editing tool arises from the biology of bacteria that holds the promise of achieving precise modifications in the genome with a simplicity and versatility that surpasses previous genome editing methods. This new technique, commonly named CRISPR/Cas9, led to a rapid expansion of the biomedical field; more specifically, cancer characterization and modeling have benefitted greatly from the genome editing capabilities of CRISPR/Cas9. In this paper, we briefly summarize recent improvements in CRISPR/Cas9 design meant to overcome the limitations that have arisen from the nuclease activity of Cas9 and the influence of this technology in cancer research. In addition, we present challenges that might impede the clinical applicability of CRISPR/Cas9 for cancer therapy and highlight future directions for designing CRISPR/Cas9 delivery systems that might prove useful for cancer therapeutics.
URLPMID:28530842 [本文引用: 1]
URLPMID:19681174 [本文引用: 1]

The construction of mitotically stable yeast strains for heterologous gene or pathway expression often requires chromosomal integration. However, transcription levels vary between different chromosome regions. We therefore characterized 20 different integration sites of the Sacchromyces cerevisiae genome by inserting lacZ as a reporter gene under the control of two different promoters and determining expression levels through enzyme activity measurement. An up to 8.7-fold difference was detected between the sites conferring lowest and highest expression, respectively. This opens the opportunity for modulating gene expression levels while retaining promoter and culture conditions. Copyright 2009 John Wiley & Sons, Ltd.
[本文引用: 1]
URLPMID:25664545 [本文引用: 2]

Abstract Although RNA-guided genome editing via the CRISPR-Cas9 system is now widely used in biomedical research, genome-wide target specificities of Cas9 nucleases remain controversial. Here we present Digenome-seq, in vitro Cas9-digested whole-genome sequencing, to profile genome-wide Cas9 off-target effects in human cells. This in vitro digest yields sequence reads with the same 5' ends at cleavage sites that can be computationally identified. We validated off-target sites at which insertions or deletions were induced with frequencies below 0.1%, near the detection limit of targeted deep sequencing. We also showed that Cas9 nucleases can be highly specific, inducing off-target mutations at merely several, rather than thousands of, sites in the entire genome and that Cas9 off-target effects can be avoided by replacing 'promiscuous' single guide RNAs (sgRNAs) with modified sgRNAs. Digenome-seq is a robust, sensitive, unbiased and cost-effective method for profiling genome-wide off-target effects of programmable nucleases including Cas9.
URLPMID:4090141 [本文引用: 1]

Monomeric CRISPR-Cas9 nucleases are widely used for targeted genome editing but can induce unwanted off-target mutations with high frequencies. Here we describe dimeric RNA-guided FokI Nucleases (RFNs) that recognize extended sequences and can edit endogenous genes with high efficiencies in human cells. The cleavage activity of an RFN depends strictly on the binding of two guide RNAs (gRNAs) to DNA with a defined spacing and orientation and therefore show improved specificities relative to wild-type Cas9 monomers. Importantly, direct comparisons show that RFNs guided by a single gRNA generally induce lower levels of unwanted mutations than matched monomeric Cas9 nickases. In addition, we describe a simple method for expressing multiple gRNAs bearing any 5 end nucleotide, which gives dimeric RFNs a broad targeting range. RFNs combine the ease of RNA-based targeting with the specificity enhancement inherent to dimerization and are likely to be useful in applications that require highly precise genome editing.
[本文引用: 1]
URLPMID:4679368 [本文引用: 2]

The CRISPR-Cas9 system is commonly employed in biomedical research; however, the precision of Cas9 is sub-optimal for gene therapy applications that involve editing a large population of cells. Variations on the standard Cas9 system have yielded improvements in the precision of targeted DNA cleavage, but often restrict the range of targetable sequences. It remains unclear whether these variants can limit lesions to a single site within the human genome over a large cohort of treated cells. Here, we demonstrate that fusing a programmable DNA-binding domain (pDBD) to Cas9 combined with the attenuation of Cas9’s inherent DNA binding affinity produces a Cas9-pDBD chimera with dramatically improved precision and increased targeting range. Because the specificity and affinity of this framework is easily tuned, Cas9-pDBDs provide a flexible system that can be tailored to achieve extremely precise genome editing at nearly any genomic locus – characteristics that are ideal for gene therapy applications.
URLPMID:23907171 [本文引用: 2]

Prokaryotic type II CRISPR-Cas systems can be adapted to enable targeted genome modifications across a range of eukaryotes(1-7). Here we engineer this system to enable RNA-guided genome regulation in human cells by tethering transcriptional activation domains either directly to a nuclease-null Cas9 protein or to an aptamer-modified single guide RNA (sgRNA). Using this functionality we developed a transcriptional activation-based assay to determine the landscape of off-target binding of sgRNA: Cas9 complexes and compared it with the off-target activity of transcription activator-like (TALs) effectors(8,9). Our results reveal that specificity profiles are sgRNA dependent, and that sgRNA: Cas9 complexes and 18-mer TAL effectors can potentially tolerate 1-3 and 1-2 target mismatches, respectively. By engineering a requirement for cooperativity through offset nicking for genome editing or through multiple synergistic sgRNAs for robust transcriptional activation, we suggest methods to mitigate off-target phenomena. Our results expand the versatility of the sgRNA: Cas9 tool and highlight the critical need to engineer improved specificity.
URLPMID:29160308 [本文引用: 2]

Abstract The spontaneous deamination of cytosine is a major source of transitions from C090004G to T090004A base pairs, which account for half of known pathogenic point mutations in humans. The ability to efficiently convert targeted A090004T base pairs to G090004C could therefore advance the study and treatment of genetic diseases. The deamination of adenine yields inosine, which is treated as guanine by polymerases, but no enzymes are known to deaminate adenine in DNA. Here we describe adenine base editors (ABEs) that mediate the conversion of A090004T to G090004C in genomic DNA. We evolved a transfer RNA adenosine deaminase to operate on DNA when fused to a catalytically impaired CRISPR-Cas9 mutant. Extensive directed evolution and protein engineering resulted in seventh-generation ABEs that convert targeted A090004T base pairs efficiently to G090004C (approximately 50% efficiency in human cells) with high product purity (typically at least 99.9%) and low rates of indels (typically no more than 0.1%). ABEs introduce point mutations more efficiently and cleanly, and with less off-target genome modification, than a current Cas9 nuclease-based method, and can install disease-correcting or disease-suppressing mutations in human cells. Together with previous base editors, ABEs enable the direct, programmable introduction of all four transition mutations without double-stranded DNA cleavage.
URL [本文引用: 1]

近年发展起来的人工核酸酶可通过引起特定位点的DNA双链断裂实现对目的片段的有效编辑.为进一步提高碱基修改的效率和精确度,2016年研究者们利用CRISPR/Cas9识别特定DNA序列的功能,结合胞嘧啶脱氨酶的生化活性发明了将胞嘧啶高效转换为胸腺嘧啶(C>T)的嘧啶单碱基编辑系统(base editor).这一系统虽然能精准实现嘧啶直接转换,大大提高精确基因编辑效率,但美中不足的是无法对嘌呤进行修改.近期,Nature 报道了将细菌中的tRNA腺嘌呤脱氨酶定向进化形成具有催化DNA腺嘌呤底物的脱氨酶,将其与Cas9系统融合发明了具有高效催化腺嘌呤转换为鸟嘌呤的新工具—腺嘌呤单碱基编辑系统(ABEs,adenine base editors).本文总结了单碱基编辑工具的发展历程和最新研究进展,着重介绍ABEs的研发过程,并对单碱基编辑工具今后的应用方向和研发方向进行展望.
URL [本文引用: 1]

近年发展起来的人工核酸酶可通过引起特定位点的DNA双链断裂实现对目的片段的有效编辑.为进一步提高碱基修改的效率和精确度,2016年研究者们利用CRISPR/Cas9识别特定DNA序列的功能,结合胞嘧啶脱氨酶的生化活性发明了将胞嘧啶高效转换为胸腺嘧啶(C>T)的嘧啶单碱基编辑系统(base editor).这一系统虽然能精准实现嘧啶直接转换,大大提高精确基因编辑效率,但美中不足的是无法对嘌呤进行修改.近期,Nature 报道了将细菌中的tRNA腺嘌呤脱氨酶定向进化形成具有催化DNA腺嘌呤底物的脱氨酶,将其与Cas9系统融合发明了具有高效催化腺嘌呤转换为鸟嘌呤的新工具—腺嘌呤单碱基编辑系统(ABEs,adenine base editors).本文总结了单碱基编辑工具的发展历程和最新研究进展,着重介绍ABEs的研发过程,并对单碱基编辑工具今后的应用方向和研发方向进行展望.
URLPMID:27096365 [本文引用: 5]

Current genome-editing technologies introduce double-stranded (ds) DNA breaks at a target locus as the first step to gene correction.1,2Although most genetic diseases arise from point mutations, current approaches to point mutation correction are inefficient and typically induce an abundance of random insertions and deletions (indels) at the target locus from the cellular response to dsDNA breaks.1,2Here we report the development of base editing, a new approach to genome editing that enables the direct, irreversible conversion of one target DNA base into another in a programmable manner, without requiring dsDNA backbone cleavage or a donor template. We engineered fusions of CRISPR/Cas9 and a cytidine deaminase enzyme that retain the ability to be programmed with a guide RNA, do not induce dsDNA breaks, and mediate the direct conversion of cytidine to uridine, thereby effecting a C→T (or G→A) substitution. The resulting “base editors” convert cytidines within a window of approximately five nucleotides (nt), and can efficiently correct a variety of point mutations relevant to human disease. In four transformed human and murine cell lines, second- and third-generation base editors that fuse uracil glycosylase inhibitor (UGI), and that use a Cas9 nickase targeting the non-edited strand, manipulate the cellular DNA repair response to favor desired base-editing outcomes, resulting in permanent correction of 6515-75% of total cellular DNA with minimal (typically ≤ 1%) indel formation. Base editing expands the scope and efficiency of genome editing of point mutations.
URLPMID:27492474 [本文引用: 1]

The generation of genetic variation (somatic hypermutation) is an essential process for the adaptive immune system in vertebrates. We demonstrate the targeted single-nucleotide substitution of DNA using hybrid vertebrate and bacterial immune systems components. Nuclease-deficient type II CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated) and the activation-induced cytidine deaminase (AID) ortholog PmCDA1 were engineered to form a synthetic complex (Target-AID) that performs highly efficient target-specific mutagenesis. Specific point mutation was induced primarily at cytidines within the target range of five bases. The toxicity associated with the nuclease-based CRISPR/Cas9 system was greatly reduced. Although combination of nickase Cas9(D10A) and the deaminase was highly effective in yeasts, it also induced insertion and deletion (indel) in mammalian cells. Use of uracil DNA glycosylase inhibitor suppressed the indel formation and improved the efficiency.
URLPMID:27723754 [本文引用: 1]

Abstract A large number of genetic variants have been associated with human diseases. However, the lack of a genetic diversification approach has impeded our ability to interrogate functions of genetic variants in mammalian cells. Current screening methods can only be used to disrupt a gene or alter its expression. Here we report the fusion of activation-induced cytidine deaminase (AID) with nuclease-inactive clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (dCas9) for efficient genetic diversification, which enabled high-throughput screening of functional variants. Guided by single guide (sg)RNAs, dCas9-AID-P182X (AIDx) directly changed cytidines or guanines to the other three bases independent of AID hotspot motifs, generating a large repertoire of variants at desired loci. Coupled with a uracil-DNA glycosylase inhibitor, dCas9-AIDx converted targeted cytidines specifically to thymines, creating specific point mutations. By targeting BCR-ABL with dCas9-AIDx, we efficiently identified known and new mutations conferring imatinib resistance in chronic myeloid leukemia cells. Thus, targeted AID-mediated mutagenesis (TAM) provides a forward genetic tool to screen for gain-of-function variants at base resolution.
URLPMID:28191901 [本文引用: 1]

Base editing is a recently developed approach to genome editing that uses a fusion protein containing a catalytically defectiveStreptococcus pyogenesCas9, a cytidine deaminase, and an inhibitor of base excision repair to induce programmable, single-nucleotide changes in the DNA of living cells without generating double-strand DNA breaks, without requiring a donor DNA template, and without inducing an excess of stochastic insertions and deletions1. Here we report the development of five new C (or G ) base editors that use natural and engineered Cas9 variants with different protospacer-adjacent motif (PAM) specificities to expand the number of sites that can be targeted by base editing by 2.5-fold. Additionally, we engineered new base editors containing mutated cytidine deaminase domains that narrow the width of the apparent editing window from approximately 5 nucleotides to as little as 1 to 2 nucleotides, enabling the discrimination of neighboring C nucleotides that would previously be edited with comparable efficiency, thereby doubling the number of disease-associated target Cs that can be corrected preferentially over nearby non-target Cs. Collectively, these developments substantially increase the targeting scope of base editing and establish the modular nature of base editors.
URLPMID:28585549 [本文引用: 1]

We recently developed base editing, a genome-editing approach that enables the programmable conversion of one base pair into another without double-stranded DNA cleavage, excess stochastic insertions and deletions, or dependence on homology-directed repair. The application of base editing is limited by off-target activity and reliance on intracellular DNA delivery. Here we describe two advances that address these limitations. First, we greatly reduce off-target base editing by installing mutations into our third-generation base editor (BE3) to generate a high-fidelity base editor (HF-BE3). Next, we purify and deliver BE3 and HF-BE3 as ribonucleoprotein (RNP) complexes into mammalian cells, establishing DNA-free base editing. RNP delivery of BE3 confers higher specificity even than plasmid transfection of HF-BE3, while maintaining comparable on-target editing levels. Finally, we apply these advances to deliver BE3 RNPs into both zebrafish embryos and the inner ear of live mice to achieve specific, DNA-free base editingin vivo. Third-generation base editors consist of a catalytically disabled Cas9 fused to a cytidine deaminase and a base excision repair inhibitor, enabling efficient, precise editing of individual base pairs in DNA. Here the authors describe engineering and protein delivery of base editors to improve their DNA specificity and enable specific base editing in live animals.
URLPMID:5577055 [本文引用: 1]

Abstract PURPOSE OF REVIEW: Cell xenotransplantation has the potential to provide a safe, ethically acceptable, unlimited source for cell replacement therapies. This review focuses on genetic modification strategies aimed to overcome remaining hurdles standing in the way of clinical porcine islet transplantation and to develop neural cell xenotransplantation. RECENT FINDINGS: In addition to previously described genetic modifications aimed to mitigate hyperacute rejection, instant blood-mediated inflammatory reaction, and cell-mediated rejection, new data showing the possibility of increasing porcine islet insulin secretion by transgenesis is an interesting addition to the array of genetically modified pigs available for xenotransplantation. Moreover, combining multiple modifications is possible today thanks to new, improved genomic editing tools. SUMMARY: Genetic modification of large animals, pigs in particular, has come a long way during the last decade. These modifications can help minimize immunological and physiological incompatibilities between porcine and human cells, thus allowing for better tolerance and function of xenocells.
[本文引用: 1]
URLPMID:5473066 [本文引用: 1]

react-text: 477 The possibility to generate patient-specific induced pluripotent stem cells (iPSCs) offers an unprecedented potential of applications in clinical therapy and medical research. Human iPSCs and their differentiated derivatives are tools for diseases modelling, drug discovery, safety pharmacology, and toxicology. Moreover, they allow for the engineering of bioartificial tissue and are promising... /react-text react-text: 478 /react-text [Show full abstract]
URLPMID:25733580 [本文引用: 2]

Abstract Sickle cell disease (SCD) is characterized by a single point mutation in the seventh codon of the -globin gene. Site-specific correction of the sickle mutation in hematopoietic stem cells would allow for permanent production of normal red blood cells. Using zinc-finger nucleases (ZFNs) designed to flank the sickle mutation, we demonstrate efficient targeted cleavage at the -globin locus with minimal off-target modification. By co-delivering a homologous donor template (either an integrase-defective lentiviral vector or a DNA oligonucleotide), high levels of gene modification were achieved in CD34(+) hematopoietic stem and progenitor cells. Modified cells maintained their ability to engraft NOD/SCID/IL2r (null) mice and to produce cells from multiple lineages, although with a reduction in the modification levels relative to the in vitro samples. Importantly, ZFN-driven gene correction in CD34(+) cells from the bone marrow of patients with SCD resulted in the production of wild-type hemoglobin tetramers.
URLPMID:28740073 [本文引用: 1]

Angiogenesis, in which vascular endothelial growth factor receptor (VEGFR) 2 plays an essential role, is associated with a variety of human diseases including proliferative diabetic retinopathy and wet age-related macular degeneration. Here we report that a system of adeno-associated virus (AAV)-mediated clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease (Cas)9 from Streptococcus pyogenes (SpCas9) is used to deplete VEGFR2 in vascular endothelial cells (ECs), whereby the expression of SpCas9 is driven by an endothelial-specific promoter of intercellular adhesion molecule 2. We further show that recombinant AAV serotyp e 1 (rAAV1) transduces ECs of pathologic vessels, and that editing of genomic VEGFR2 locus using rAAV1-mediated CRISPR/Cas9 abrogates angiogenesis in the mouse models of oxygen-induced retinopathy and laser-induced choroid neovascularization. This work establishes a strong foundation for genome editing as a strategy to treat angiogenesis-associated diseases.
URL [本文引用: 1]
URLPMID:28209587 [本文引用: 1]

RNA-guided genome surgery using CRISPR-Cas9 nucleases has shown promise for the treatment of diverse genetic diseases. Yet, the potential of such nucleases for therapeutic applications in nongenetic diseases is largely unexplored. Here, we focus on age-related macular degeneration (AMD), a leading cause of blindness in adults, which is associated with retinal overexpression of, rather than mutations in, the VEGFA gene. Subretinal injection of preassembled, Vegfa gene pecific Cas9 ribonucleoproteins (RNPs) into the adult mouse eye gave rise to mutagenesis at the target site in the retinal pigment epithelium. Furthermore, Cas9 RNPs effectively reduced the area of laser-induced choroidal neovascularization (CNV) in a mouse model of AMD. Genome-wide profiling of Cas9 off-target effects via Digenome-seq showed that off-target mutations were rarely induced in the human genome. Because Cas9 RNPs can function immediately after in vivo delivery and are rapidly degraded by endogenous proteases, their activities are unlikely to be hampered by antibody- and cell-mediated adaptive immune systems. Our results demonstrate that in vivo genome editing with Cas9 RNPs has the potential for the local treatment for nongenetic degenerative diseases, expanding the scope of RNA-guided genome surgery to a new dimension.
URLPMID:27053530 [本文引用: 1]

Abstract HIV/AIDS has long been at the forefront of the development of gene- and cell-based therapies. While conventional gene therapy approaches typically involve the addition of anti-HIV genes to cells using semi-randomly integrating viral vectors, newer genome editing technologies based on engineered nucleases are now allowing more precise genetic manipulations. The possible outcomes of genome editing include gene disruption, which has been most notably applied to the CCR5 co-receptor gene, or the introduction of small mutations or larger whole gene cassette insertions at a targeted locus. Disruption of CCR5 using zinc-finger nucleases was the first-in-man application of genome editing and remains the most clinically advanced platform, with 7 completed or ongoing clinical trials in T cells and hematopoietic stem/progenitor cells (HSPCs). Here we review laboratory and clinical findings of CCR5 editing in T cells and HSPCs for HIV therapy, and summarize other promising genome editing approaches for future clinical development. In particular, recent advances in the delivery of genome editing reagents, and the demonstration of highly efficient homology-directed editing in both T cells and HSPCs, are expected to spur the development of even more sophisticated applications of this technology for HIV therapy. Copyright 2016 American Society of Hematology.
URLPMID:5267384 [本文引用: 1]

Natural killer T cells (NKT cells) play an important role in the immunity against viral infections. They produce cytokines or have direct cytolytic effects that can restrict virus replication. However, the exact function of NKT cells in retroviral immunity is not fully elucidated. Therefore, we analyzed the antiretroviral functions of NKT cells in mice infected with the Friend retrovirus (FV). After FV infection numbers of NKT cells remained unchanged but activation as well as improved effector functions of NKT cells were found. While the release of pro-inflammatory cytokines was not changed after infection, activated NKT cells revealed an elevated cytotoxic potential. Stimulation with -Galactosylceramide significantly increased not only total NKT cell numbers and activation but also the anti-retroviral capacity of NKT cells. We demonstrate a strong activation and a potent cytolytic function of NKT cells during acute retroviral infection. Therapeutic treatment with -Galactosylceramide could further improve the reduction of early retroviral replication by NKT cells, which could be utilized for future treatment against viral infections. The online version of this article (doi:10.1186/s12977-017-0327-8) contains supplementary material, which is available to authorized users.
URLPMID:27854119 [本文引用: 1]

Abstract Despite significant advances in HIV drug treatment regimens, which grant near-normal life expectancies to infected individuals who have good virological control, HIV infection itself remains incurable. In recent years, novel gene- and cell-based therapies have gained increasing attention due to their potential to provide a functional or even sterilizing cure for HIV infection with a one-shot treatment. A functional cure would keep the infection in check and prevent progression to AIDS, while a sterilizing cure would eradicate all HIV viruses from the patient. Genome editing is the most precise form of gene therapy, able to achieve permanent genetic disruption, modification, or insertion at a predesignated genetic locus. The most well-studied candidate for anti-HIV genome editing is CCR5, an essential coreceptor for the majority of HIV strains, and the lack of which confers HIV resistance in naturally occurring homozygous individuals. Genetic disruption of CCR5 to treat HIV has undergone clinical testing, with seven completed or ongoing trials in T cells and hematopoietic stem and progenitor cells, and has shown promising safety and potential efficacy profiles. Here we summarize clinical findings of CCR5 editing for HIV therapy, as well as other genome editing-based approaches under pre-clinical development. The anticipated development of more sophisticated genome editing technologies should continue to benefit HIV cure efforts.
URLPMID:28904745 [本文引用: 1]

Abstract Background The main approach to treat HIV-1 infection is combination antiretroviral therapy (cART). Although cART is effective in reducing HIV-1 viral load and controlling disease progression, it has many side effects, and is expensive for HIV-1 infected patients who must remain on lifetime treatment. HIV-1 gene therapy has drawn much attention as studies of genome editing tools have progressed. For example, zinc finger nucleases (ZFN), transcription activator like effector nucleases (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 have been utilized to successfully disrupt the HIV-1 co-receptors CCR5 or CXCR4, thereby restricting HIV-1 infection. However, the effects of simultaneous genome editing of CXCR4 and CCR5 by CRISPR-Cas9 in blocking HIV-1 infection in primary CD4+ T cells has been rarely reported. Furthermore, combination of different target sites of CXCR4 and CCR5 for disruption also need investigation. Results In this report, we designed two different gRNA combinations targeting both CXCR4 and CCR5, in a single vector. The CRISPR-sgRNAs-Cas9 could successfully induce editing of CXCR4 and CCR5 genes in various cell lines and primary CD4+ T cells. Using HIV-1 challenge assays, we demonstrated that CXCR4-tropic or CCR5-tropic HIV-1 infections were significantly reduced in CXCR4- and CCR5-modified cells, and the modified cells exhibited a selective advantage over unmodified cells during HIV-1 infection. The off-target analysis showed that no non-specific editing was identified in all predicted sites. In addition, apoptosis assays indicated that simultaneous disruption of CXCR4 and CCR5 in primary CD4+ T cells by CRISPR-Cas9 had no obvious cytotoxic effects on cell viability. Conclusions Our results suggest that simultaneous genome editing of CXCR4 and CCR5 by CRISPR-Cas9 can potentially provide an effective and safe strategy towards a functional cure for HIV-1 infection.
URL [本文引用: 1]
URLPMID:28434148 [本文引用: 4]

The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (CRISPR/Cas9) system, an RNA-guided DNA targeting technology, is triggering a revolution in the field of biology. CRISPR/Cas9 has demonstrated great potential for genetic manipulation. In this review, we discuss the current development of CRISPR/Cas9 technologies for therapeutic applications, especially chimeric antigen receptor (CAR) T cell-based adoptive immunotherapy. Different methods used to facilitate efficient CRISPR delivery and gene editing in T cells are compared. The potential of genetic manipulation using CRISPR/Cas9 system to generate universal CAR T cells and potent T cells that are resistant to exhaustion and inhibition is explored. We also address the safety concerns associated with the use of CRISPR/Cas9 gene editing and provide potential solutions and future directions of CRISPR application in the field of CAR T cell immunotherapy. As an integration-free gene insertion method, CRISPR/Cas9 holds great promise as an efficient gene knock-in platform. Given the tremendous progress that has been made in the past few years, we believe that the CRISPR/Cas9 technology holds immense promise for advancing immunotherapy.
URLPMID:27815355 [本文引用: 1]

Abstract PURPOSE: Using gene-disrupted allogeneic T cells as universal effector cells provides an alternative to and potentially improves current chimeric antigen receptor (CAR) T cell therapy against cancers and infectious diseases. EXPERIMENTAL DESIGN: The CRISPR/Cas9 system has recently emerged as a simple and efficient way for multiplex genome engineering. By combining the lentiviral delivery of CAR and CRISPR RNA electroporation to co-introduce RNA encoding the Cas9 and gRNAs targeting endogenous TCR, beta-2 microglobulin (B2M) and PD1 simultaneously, to generate gene-disrupted allogeneic CAR T cells deficient of TCR, HLA class I molecule and PD1. RESULTS: The CRISPR gene-edited CAR T cells showed potent anti-tumor activities, both in vitro and in animal models and were as potent as non-gene edited CAR T cells. In addition the TCR and HLA class I double deficient T cells had reduced alloreactivity and did not cause graft-versus-host disease. Finally, simultaneous triple genome editing by adding the disruption of PD1 led to enhanced in vivo anti-tumor activity of the gene-disrupted CART cells. CONCLUSIONS: Gene-disrupted allogeneic CAR and TCR T cells could provide an alternative as a universal donor to autologous T cells, which carry difficulties and high production costs. Gene-disrupted CAR and TCR T cells with disabled checkpoint molecules may be potent effector cells against cancers and infectious diseases. Copyright {copyright, serif}2016, American Association for Cancer Research.
URLPMID:23935598 [本文引用: 1]

Recent early stage clinical trials evaluating the adoptive transfer of patient CD8+ T-cells re-directed with antigen receptors recognizing tumors have shown very encouraging results. These reports provide strong support for further development of the therapeutic concept as a curative cancer treatment. In this respect combining the adoptive transfer of tumor-specific T-cells with therapies that increase their anti-tumor capacity is viewed as a promising strategy to improve treatment outcome. The ex vivo genetic engineering step that underlies T-cell re-direction offers a unique angle to combine antigen receptor delivery with the targeting of cell-intrinsic pathways that restrict T-cell effector functions. Recent progress in genome editing technologies such as protein- and RNA-guided endonucleases raise the possibility of disrupting gene expression in T-cells in order to enhance effector functions or to bypass tumor immune suppression. This approach would avoid the systemic administration of compounds that disrupt immune homeostasis, potentially avoiding autoimmune adverse effects, and could improve the efficacy of T-cell based adoptive therapies.
URLPMID:26965203 [本文引用: 1]

Abstract Since the regulatory approval of ipilimumab in 2011, the field of cancer immunotherapy has been experiencing a renaissance. This success is based on progress in both preclinical and clinical science, including the development of new methods of investigation. Immuno-oncology has become a sub-specialty within oncology owing to its unique science and its potential for substantial and long-term clinical benefit. Immunotherapy agents do not directly attack the tumour but instead mobilize the immune system - this can be achieved through various approaches that utilize adaptive or innate immunity. Therefore, immuno-oncology drug development encompasses a broad range of agents, including antibodies, peptides, proteins, small molecules, adjuvants, cytokines, oncolytic viruses, bi-specific molecules and cellular therapies. This Perspective summarizes the recent history of cancer immunotherapy, including the factors that led to its success, provides an overview of novel drug-development considerations, summarizes three generations of immunotherapies that have been developed since 2011 and, thus, illustrates the breadth of opportunities these new generations of immunotherapies represent.
URLPMID:27466105 [本文引用: 1]

Gene-editing technique to treat lung cancer is due to be tested in people in August.
URLPMID:27882996 [本文引用: 2]

CRISPR gene-editing tested in a person for the first time Nature 539, 7630 (2016). http://www.nature.com/doifinder/10.1038/nature.2016.20988 Author: David ...
URLPMID:28942539 [本文引用: 2]

β-Thalassemia is a global health issue, caused by mutations in theHBBgene. Among these mutations,HBB6128 (A>G) mutations is one of the three most common mutations in China and Southeast Asia patients with β-thalassemia. Correcting this mutation in human embryos may prevent the disease being passed onto future generations and cure anemia. Here we report the first study using base editor (BE) system to correct disease mutant in human embryos. Firstly, we produced a 293T cell line with an exogenousHBB6128 (A>G) mutant fragment for gRNAs and targeting efficiency evaluation. Then we collected primary skin fibroblast cells from a β-thalassemia patient with HBB 6128 (A>G) homozygous mutation. Data showed that base editor could precisely correctHBB6128 (A>G) mutation in the patient’s primary cells. To model homozygous mutation disease embryos, we constructed nuclear transfer embryos by fusing the lymphocyte or skin fibroblast cells with enucleatedin vitromatured (IVM) oocytes. Notably, the gene correction efficiency was over 23.0% in these embryos by base editor. Although these embryos were still mosaic, the percentage of repaired blastomeres was over 20.0%. In addition, we found that base editor variants, with narrowed deamination window, could promote G-to-A conversion atHBB6128 site precisely in human embryos. Collectively, this study demonstrated the feasibility of curing genetic disease in human somatic cells and embryos by base editor system. The online version of this article (doi:10.1007/s13238-017-0475-6) contains supplementary material, which is available to authorized users.
URLPMID:28751571 [本文引用: 1]

Abstract OBJECTIVE: High-efficiency genome editing to disrupt therapeutic target genes, such as PCSK9 (proprotein convertase subtilisin/kexin type 9), has been demonstrated in preclinical animal models, but there are safety concerns because of the unpredictable nature of cellular repair of double-strand breaks, as well as off-target mutagenesis. Moreover, precise knock-in of specific nucleotide changes-whether to introduce or to correct gene mutations-has proven to be inefficient in nonproliferating cells in vivo. Base editors comprising CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats [CRISPR]-CRISPR-associated 9) fused to a cytosine deaminase domain can effect the alteration of cytosine bases to thymine bases in genomic DNA in a sequence-specific fashion, without the need for double-strand DNA breaks. The efficacy of base editing has not been established in vivo. The goal of this study was to assess whether in vivo base editing could be used to modify the mouse Pcsk9 gene in a sequence-specific fashion in the liver in adult mice. APPROACH AND RESULTS: We screened base editors for activity in cultured cells, including human-induced pluripotent stem cells. We then delivered a base editor into the livers of adult mice to assess whether it could introduce site-specific nonsense mutations into the Pcsk9 gene. In adult mice, this resulted in substantially reduced plasma PCSK9 protein levels (>50%), as well as reduced plasma cholesterol levels ( 30%). There was no evidence of off-target mutagenesis, either cytosine-to-thymine edits or indels. CONCLUSIONS: These results demonstrate the ability to precisely introduce therapeutically relevant nucleotide variants into the genome in somatic tissues in adult mammals, as well as highlighting a potentially safer alternative to therapeutic genome editing. 2017 American Heart Association, Inc.
URLMagsci [本文引用: 1]

在细菌中发现的免疫系统CRISPR/Cas9, 已经成为最有效的基因工程编辑工具, 甚至大有希望可以治疗人类遗传性疾病。但是CRISPR/Cas9系统在使用时会产生严重的脱靶问题, 导致假表型和错误的解释。提高与靶点结合的高效率, 同时减少脱靶效应, 将是今后CRISPR/Cas9技术的挑战。综述关注与CRISPR/Cas9脱靶效应相关的内容, 总结了影响其靶点专一性的因素, 减少CRISPR/Cas9脱靶效应的可行性方法和设计工具等, 供大家学习讨论。
URLMagsci [本文引用: 1]

在细菌中发现的免疫系统CRISPR/Cas9, 已经成为最有效的基因工程编辑工具, 甚至大有希望可以治疗人类遗传性疾病。但是CRISPR/Cas9系统在使用时会产生严重的脱靶问题, 导致假表型和错误的解释。提高与靶点结合的高效率, 同时减少脱靶效应, 将是今后CRISPR/Cas9技术的挑战。综述关注与CRISPR/Cas9脱靶效应相关的内容, 总结了影响其靶点专一性的因素, 减少CRISPR/Cas9脱靶效应的可行性方法和设计工具等, 供大家学习讨论。
URLPMID:25848930 [本文引用: 1]

Directly modulating the activity of genome-editing has the potential to increase their specificity by reducing activity following target locus modification. We developed that are activated by the presence of a -permeable small molecule by inserting an evolved -responsive intein at specific positions in . In , conditionally active Cas9s modify target genomic sites with up to 25-fold higher specificity than wild-type .
URL [本文引用: 1]
URLPMID:28011075 [本文引用: 2]

The development of customizable sequence-specific nucleases such as TALENs, ZFNs and the powerful CRISPR/Cas9 system has revolutionized the field of genome editing. The CRISPR/Cas9 system is particularly versatile and has been applied in numerous species representing all branches of life. Regardless of the target organism, all researchers using sequence-specific nucleases face similar challenges: confirmation of the desired on-target mutation and the detection of off-target events. Here, we evaluate the most widely-used methods for the detection of on-target and off-target mutations in terms of workflow, sensitivity, strengths and weaknesses.
URLPMID:28263568 [本文引用: 1]

Abstract The successful use of clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-based gene editing for therapeutics requires efficient in vivo delivery of the CRISPR components. There are, however, major challenges on the delivery front. In this Topical Review, we will highlight recent developments in CRISPR delivery, and we will present hurdles that still need to be overcome to achieve effective in vivo editing.
URLPMID:29892062 [本文引用: 1]

Abstract CRISPR/Cas9 has revolutionized our ability to engineer genomes and conduct genome-wide screens in human cells 1-3 . Whereas some cell types are amenable to genome engineering, genomes of human pluripotent stem cells (hPSCs) have been difficult to engineer, with reduced efficiencies relative to tumour cell lines or mouse embryonic stem cells 3-13 . Here, using hPSC lines with stable integration of Cas9 or transient delivery of Cas9-ribonucleoproteins (RNPs), we achieved an average insertion or deletion (indel) efficiency greater than 80%. This high efficiency of indel generation revealed that double-strand breaks (DSBs) induced by Cas9 are toxic and kill most hPSCs. In previous studies, the toxicity of Cas9 in hPSCs was less apparent because of low transfection efficiency and subsequently low DSB induction 3 . The toxic response to DSBs was P53/TP53-dependent, such that the efficiency of precise genome engineering in hPSCs with a wild-type P53 gene was severely reduced. Our results indicate that Cas9 toxicity creates an obstacle to the high-throughput use of CRISPR/Cas9 for genome engineering and screening in hPSCs. Moreover, as hPSCs can acquire P53 mutations 14 , cell replacement therapies using CRISPR/Cas9-enginereed hPSCs should proceed with caution, and such engineered hPSCs should be monitored for P53 function.
URLPMID:29892067 [本文引用: 1]

Abstract Here, we report that genome editing by CRISPR-Cas9 induces a p53-mediated DNA damage response and cell cycle arrest in immortalized human retinal pigment epithelial cells, leading to a selection against cells with a functional p53 pathway. Inhibition of p53 prevents the damage response and increases the rate of homologous recombination from a donor template. These results suggest that p53 inhibition may improve the efficiency of genome editing of untransformed cells and that p53 function should be monitored when developing cell-based therapies utilizing CRISPR-Cas9.
[本文引用: 2]
URL [本文引用: 1]

基于CRISPR/Cas9系统介导的第三代基因组定点编辑技术,已被广泛应用于基因编辑和基因表达调控等研究领域.如何提高该技术对基因组编辑的效率与特异性、最大限度降低脱靶风险一直是该领域的难点.近年来,机器学习为解决CRISPR/Cas9系统所面临的问题提供了新思路,基于机器学习的CRISPR/Cas9系统已逐渐成为研究热点.本文阐述了CRISPR/Cas9的作用机理,总结了现阶段该技术面临的基因组编辑效率低、存在潜在的脱靶效应、前间区序列邻近基序(PAM)限制识别序列等问题,最后对机器学习应用于优化设计高效向导RNA(sgRNA)序列、预测sgRNA的活性、脱靶效应评估、基因敲除、高通量功能基因筛选等领域的研究现状与发展前景进行了展望,以期为基因组编辑领域的研究提供参考.
URL [本文引用: 1]

基于CRISPR/Cas9系统介导的第三代基因组定点编辑技术,已被广泛应用于基因编辑和基因表达调控等研究领域.如何提高该技术对基因组编辑的效率与特异性、最大限度降低脱靶风险一直是该领域的难点.近年来,机器学习为解决CRISPR/Cas9系统所面临的问题提供了新思路,基于机器学习的CRISPR/Cas9系统已逐渐成为研究热点.本文阐述了CRISPR/Cas9的作用机理,总结了现阶段该技术面临的基因组编辑效率低、存在潜在的脱靶效应、前间区序列邻近基序(PAM)限制识别序列等问题,最后对机器学习应用于优化设计高效向导RNA(sgRNA)序列、预测sgRNA的活性、脱靶效应评估、基因敲除、高通量功能基因筛选等领域的研究现状与发展前景进行了展望,以期为基因组编辑领域的研究提供参考.
