 ,先正达北京创新中心,北京 102206
,先正达北京创新中心,北京 102206Detection methods of genome editing in plants
Chunxia Liu, Lizhao Geng, Jianping Xu ,Syngenta Beijing Innovation Center, Beijing 102206, China
,Syngenta Beijing Innovation Center, Beijing 102206, China通讯作者:
编委: 高彩霞
收稿日期:2018-03-29修回日期:2018-08-18网络出版日期:2018-12-20
Received:2018-03-29Revised:2018-08-18Online:2018-12-20
作者简介 About authors
刘春霞,博士,专业方向:作物遗传育种,植物基因组编辑E-mail:chunxia.liu@syngenta.com。

摘要
关键词:
Abstract
Keywords:
PDF (569KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘春霞, 耿立召, 许建平. 植物基因组编辑检测方法[J]. 遗传, 2018, 40(12): 1075-1091 doi:10.16288/j.yczz.18-079
Chunxia Liu, Lizhao Geng, Jianping Xu.
随着分子生物学技术的发展以及大量物种基因组测序的完成,以序列特异性核酸酶(sequence- specific nucleases, SSNs)为基础的基因组编辑(genome editing)在植物中得到迅猛发展。序列特异性核酸酶可以在基因组的特定位置产生DNA双链断裂(double-strand breaks, DSBs),DSBs可以激发生物体的修复机制,从而在基因组的特定位置造成DNA序列的变化,进而改变基因的功能。近年来,成簇的规律间隔的短回文重复序列及其相关系统(clustered regularly interspaced short palindromic repeats/CRISPR- associated 9, CRISPR/Cas9 system)作为最新一代的基因组编辑工具,为植物基因功能研究和作物遗传育种带来了一场全新的技术革命。目前,CRISPR/ Cas9技术已经被广泛地应用于各种植物的研究,包括模式植物拟南芥、各种农作物和树木[1,2,3,4,5,6,7,8,9]。与转基因改良作物相比,基因组编辑技术在最终产品中并不会引入任何外源基因,更类似于传统诱变育种产生的品种,因此成为一种非常受欢迎的作物遗传改良手段。
植物细胞中有两种途径可以修复DSBs:非同源末端连接(non-homologous end joining, NHEJ)和同源重组(homologous recombination, HR)。NHEJ在植物中是一条主要的修复途径,也是一种易错的修复途径,在DNA修复过程中,连接处容易产生少数碱基的插入或缺失,导致基因失活。HR是一条次要的修复途径,需要以同源片段作为模板进行修复,是一条精确的修复途径[10]。由于HR在植物中的修复效率非常低,因此目前对基因功能的研究和作物改良主要借助NHEJ途径进行基因敲除。对于二倍体植物来说,NHEJ修复之后会产生5种可能的结果:(1) 没有产生任何突变;(2) 单等位基因突变,只有一个等位基因发生突变,也称杂合突变;(3) 双等位基因突变,两个等位基因都发生突变,但是突变的类型不一样;(4) 纯合突变,两个等位基因发生相同的突变;(5) 嵌合体(chimeric),同一个样品上有3种或3种以上的突变类型。这种修复的多样性和不确定性对基因编辑检测提出更高的要求。对于多倍体植物,由于存在两个以上的等位基因,基因组的情况更为复杂,这对基因编辑的检测带来更高的挑战。本文对目前植物基因组编辑在目标位点的检测方法进行了综述,对各种检测方法进行了详细的介绍和对比,并进一步对各种检测方法的应用趋势进行了分析,以期为植物基因组编辑的应用提供参考。
1 不同类型植物基因组编辑的检测方法
1.1 PCR/RE (restriction endonuclease)方法
1.1.1 PCR/RE方法PCR/RE是应用特异引物PCR和限制性酶切相结合而产生的一种检测方法,主要是通过对PCR扩增的DNA片段进行限制性酶切分析来区分目标位点是否被编辑。PCR/RE也被称为限制性片段长度多态性(restriction fragment length polymorphism, RFLP)分析[2,11,12]、酶切扩增多态性序列(cleaved amplified polymorphic sequence, CAPS)方法[13]。具体操作步骤总结如下:首先设计300~1700 bp的扩增子,扩增子包含所要突变的目标片段;之后用限制性内切酶消化该扩增子;最终通过琼脂糖凝胶电泳检测,基于电泳条带的带型进行区分。条带完全被切开,表示该植株没有产生突变;条带部分被切开,表示该植株为杂合突变或嵌合体;条带完全没有被切开,表示该植株为纯合突变或双等位基因突变(图1)。PCR/RE方法是目前植物基因组编辑检测中应用最为广泛的方法之一[2,5~7,11~71],尤其是应用在多倍体植物的检测中。目前,小麦基因组编辑的研究大都是应用这种方法检测[5,27,43,53,67]。与其他检测方法相比,PCR/RE方法优点如下:可以检测所有类型的突变,包括SNP和各种大小的插入缺失;具有很高的灵敏性;整个过程需要的时间也比较短,只需要几个小时;检测成本也较低;切割产物可以简单地通过琼脂糖凝胶来分辨,非常方便。PCR/RE方法最大的局限性是在基因组编辑核酸酶切割的位点处需要有限制性酶切位点。
图1
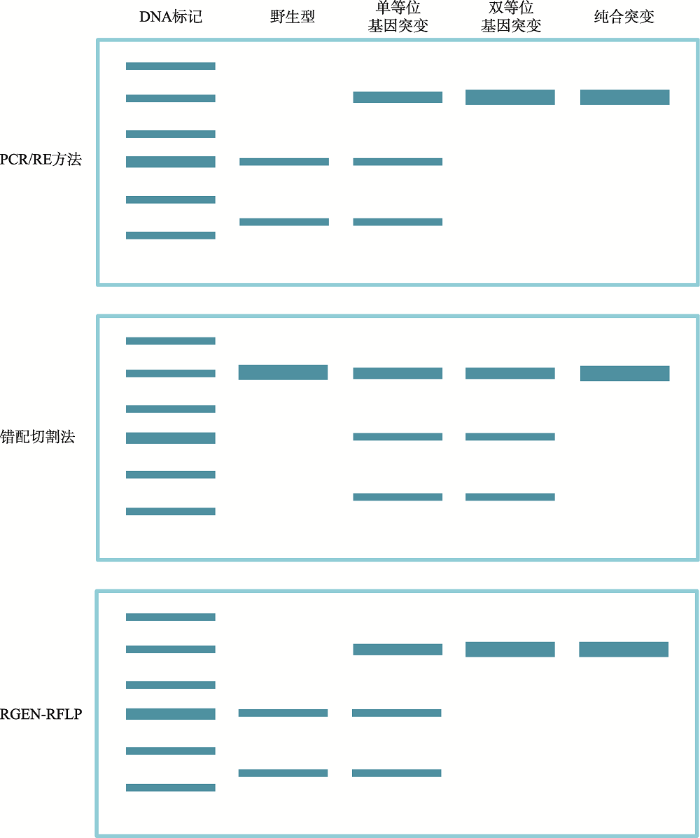 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1不同植物基因组编辑检测方法的比较
Fig. 1Comparison of different assays to detect mutations induced by genome editing
1.1.2 CAPS衍生方法
CAPS衍生方法(derived cleaved amplified polymorphic sequence, dCAPS),顾名思义,这种方法是在CAPS基础上发展起来的[72]。不同的是,需要在靠近突变位点的地方通过碱基错配引入一个酶切位点。酶切位点的引入可以通过dCAPS Finder 2.0软件(http://helix.wustl.edu/dcaps/dcaps.html)[73]来实现。设计dCAPS标记引物需要考虑以下几方面:首先引物中碱基的错配数目和位置都会影响扩增效率,错配越少,同时离引物3°端越远,扩增效率越高;其次,错配的空间特性也会影响扩增效率,因为嘌呤与嘌呤的错配没有嘧啶与嘧啶的错配稳定;值得注意的是,所产生的限制性内切酶的切割性能和价格也是一个需要考虑的因素。由于需要错配碱基的引入,所以dCAPS的扩增子一般不会太大,通常是200~300 bp。dCAPS方法的优点与PCR/RE方法类似,但是突破了PCR/RE的限制,对PCR/RE方法有一定的补充作用。不过在设计上比PCR/RE要复杂,所以应用并不是很多[55,74]。
1.1.3 限制性酶切PCR法
限制性酶切PCR法(restriction enzyme-PCR, RE-PCR)是与PCR/RE非常相似的一种方法,也是应用特异引物扩增和限制性酶切相结合产生的一种方法。不同之处在于,RE-PCR是对基因组先进行酶切,然后再对酶切产物进行PCR扩增。如果基因组发生突变,相应的片段就不会被切开,因此可以扩增出条带,而野生型因为被切断,所以无法扩增出条带。这种方法的灵敏度与PCR/RE一样,目前在植物中应用较多[4,33,75~80]。但也有研究将两种方法结合[22,35],先对植物基因组进行酶切,然后进行PCR扩增,扩增后的产物再次进行酶切(RE-PCR-RE),这样也增加了结果的准确性。RE-PCR方法的优点和PCR/RE类似,同样也受限制性酶切位点的限制。
1.2 错配切割法
错配切割法(mismatch cleavage assay)是通过错配切割酶识别并切割异源双链核酸分子进行检测的一种方法。具体操作步骤如下:首先是用PCR方法扩增基因组的目标区域,然后将扩增子变性、退火产生异源双链核酸分子。这种异源双链核酸分子可以被错配切割酶切开,但是同源双链核酸分子不能被切开,可以通过切开条带的亮度来估算插入缺失的频率(图1)。错配切割酶的优势在于不受限制性酶切位点的限制。但是该方法经常会低估突变频率,因为它只能检测出单等位基因突变和双等位基因突变类型,不能检测出纯合的基因突变类型。目前,可以应用的错配切割酶有很多种,包括T7EI、Surveyor (Cel I)和Cruiser。T7EI最为常见[23,24,27,36,57,67,75,81~91];其次是Surveyor[37,42,59,92~98];报道最少的是Cruiser[36]。这3种酶也各有优缺点:T7EI存在假阳性,因为它能够切割Holiday结构和十字(Cruciform)结构;Surveyor和Cruiser特异性稍高一些,但价格相对昂贵。每种方法的灵敏度也不同,T7EI是0.5%~5%[99]的突变率可以被检出,Surveyor大约是10%[100]的突变率才可以被检出。1.3 Sanger测序法
Sanger测序(Sanger sequencing)是基于一代测序技术来检测基因组编辑情况的方法。目前关于植物基因组编辑的研究中,该方法所占的比例很高[5,52,55,57,60,61,68,74,90,101~162](图2)。用来检测基因组编辑的方法分为两种:一种是PCR产物直接测序,通过测序图谱的峰值来判断是否发生编辑,该方法被称为Sanger sequencing chromatograms。多数情况下,这种方法只能知道是否发生了突变,不能判断突变的确切类型。Brinkman等[163]和Liu等[164]分别开发了相关的软件可以将测序的峰图进行解码,进而分辨出具体的突变类型。但是,这种方法只能分辨同时含有两种基因型的突变,超过两种以上的突变无法用该软件解码。由于这些软件的推广,目前越来越多的研究采取Sanger测序的方法来检测基因组编辑[129,137,138,144]。另外一种方法是将PCR产物进行克隆测序[9,162],这种方法得到的结果更为精确,可以确切判断具体的突变类型,同时也可以测定两种以上的突变类型。实际上,大部分的研究通过其他的方法进行初筛之后,最终还是选择Sanger测序来确定最终的突变类型[11~27,81~91]。图2
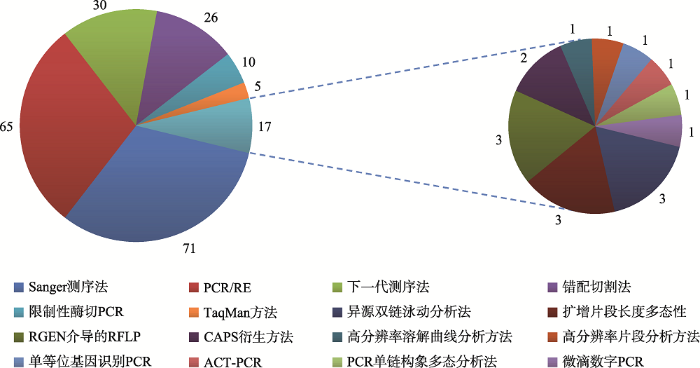 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2植物基因组编辑检测方法应用次数双饼分布图
Fig. 2Double pie distribution graphs showing application frequencies of genome editing detection methods
1.4 下一代测序法
随着测序技术的发展,费用更低、通量更高的下一代测序技术(next-generation sequencing, NGS)应运而生。下一代测序的核心思想是边合成边测序,通过捕捉新合成的末端的标记或者合成过程释放的特殊标记来确定DNA序列。目前仍在使用的平台有4个:Illumina平台、Roche 454平台(已经退市)、 Ion torrent-Proton/PGM平台和PacBio平台。植物基因组编辑主要使用Roche 454和Illumina两种精度更高的测序平台,大部分研究使用了Illumina平台[46,53~54,57,85~86,96,142,165~176],少数研究使用了Roche 454平台[118,177~180],极少数研究使用了PacBio平台[181]。这与测序技术的更新换代有着密切的联系,随着Roche 454平台的淡出,更多的基因编辑检测会使用Illumina平台。与Sanger测序相比,下一代测序可以同时产生成千上万的数据,通量大大提高。由于涉及到大量并行数据读取,下一代测序需要的时间会更长(Ion torrent除外),后续的数据分析也更复杂。并且,下一代测序技术读长都比较短,并不适合大片段缺失的检测。1.5 TaqMan方法
TaqMan方法是在PCR体系中加入一个与目标区域互补的荧光探针,探针的5′端标记一个发光基团,3′端标记一个淬灭基团。当进行PCR扩增时,探针与目标区域特异地结合在一起。随着PCR的延伸,聚合酶的5′外切活性会将探针切下,释放出发光基团,继而发出的荧光信号可以被检测到,从而通过荧光信号的强弱推测植物体内目标基因的拷贝数[182]。如果目标基因发生突变,拷贝数就会相应改变,从而可以推算出基因编辑的情况[183,184,185,186,187]。TaqMan的通量非常高,通常以96/384孔板的形式操作,灵敏度也很高,是一种可以高通量检测植物目标区域突变的方法。当然,这种方法也有其局限性,不能够区分双等位基因突变和纯合突变。同时,TaqMan需要昂贵的仪器和试剂,例如合成不同荧光染料标记的探针,并不适合少量样品操作。所以目前报道的研究中,只有几家大规模的农业生物公司如杜邦先锋[182,183,184,185]和先正达[186,187]应用了TaqMan方法。1.6 扩增片段长度多态性
扩增片段长度多态性(amplified fragment length polymorphism, AFLP)的基本原理:通过对基因组DNA酶切片段的选择性扩增来检测DNA酶切片段长度的多态性。现在用于基因组编辑检测的AFLP标记已经不完全等同于之前的AFLP,而是指特异的引物扩增出来的不同长度的DNA片段,通过片段的长度来揭示基因组目标位点是否发生突变。这种标记只适用于大片段的缺失,不适用于小片段的插入或缺失,也不能检测出碱基的替换,适用范围比较有限。目前只有少数几篇植物基因组编辑的文章用到了这种方法[94,188,189]。1.7 PCR单链构象多态分析法
单链构象多态分析法(single-strand conformational polymorphism, SSCP)是利用DNA单链构象具有多态性的特点,结合PCR技术进行多态分析的方法。在低温下,DNA单链呈现一种由内部分子相互作用形成的空间折叠,当其中一个碱基发生变化时,整条单链的空间构象也会发生变化,从而影响DNA分子在非变性胶中的迁移率,这样就可以区分不同的核酸分子。该方法可以有效地检测小片段的插入缺失和错配突变。Zheng等[190]于2016年将该方法成功地应用于水稻基因组编辑的检测,灵敏度可以达到10%。Zhou等[191]将该方法与RFLP相比,发现SSCP灵敏度更高。与其他DNA检测方法不同,SSCP检测的是单链DNA,几乎可以检测所有类型的突变,与PCR/RE相比,该方法不受酶切位点的限制,但是并不能确定最终的突变类型,只适用于初筛。1.8 RGEN介导的RFLP
RGEN (RNA-guided endonucleases)介导的RFLP是通过CRISPR/Cas9或者CRISPR/Cpf1对目标片段的切割来判断基因组编辑情况的方法[192,193]。RGEN是包含了Cas9蛋白或者Cpf1蛋白和gRNA的复合物,可以特异性切割基因组编辑的目的位点。具体操作步骤:扩增基因组编辑的目标区域,然后再加入RGEN进行切割,如果之前已经被编辑就不能切开,没有编辑的可以被切割,通过琼脂糖凝胶可以清楚地区分基因组被编辑的情况(图1)。与错配切割的方法相比较,该方法可以检测出纯合的突变类型,也能够区分单等位基因突变和双等位基因突变类型,但是不能够区分双等位基因突变和纯合突变的个体。当然该方法多用于检测CRISPR系统所产生的基因突变,少数情况下适用于TALEN产生的突变[193]。而且相比于限制性内切酶,CRISPR/Cas9酶或者CRISPR/Cpf1酶价格更高,所以该方法在植物基因组编辑的应用也很有限[85,193]。1.9 高分辨率片段分析方法
高分辨率片段分析(high-resolution fragment analysis, HRFA)是结合PCR和毛细管电泳的一种方法。具体操作步骤是采用96孔板进行DNA提取、PCR扩增,然后毛细管电泳,就可以分辨出不同类型的突变。或者在扩增引物上添加荧光标记,引物覆盖目标位点,加上荧光检测可以区分不同大小的产物。该方法的灵敏度比较高,可以区分小到1 bp的插入或缺失。另外,也可以区分多种不同大小的突变。这种方法的局限是不能检测出含有单碱基替换的突变,也不能区分具有相同大小插入缺失的突变。通过设计不同长度的扩增片段,同时结合不同的荧光标记,HRFA方法可以很容易地分析多个基因位点发生突变的情况。该方法更适用于多倍体植物的检测,Andersson等[194]利用该方法一次检测了4个基因位点的突变。1.10 高分辨率溶解曲线分析方法
高分辨率溶解曲线分析(high-resolution melting analysis, HRMA)方法是一种基于单核苷酸溶解温度不同而形成不同溶解曲线的检测方法。具体步骤:设计PCR引物进行扩增,扩增子大小一般是90~ 200 bp,应用实时定量PCR加上相应的荧光染料,然后通过分析溶解曲线的方法来分析突变类型。该方法具有极高的灵敏度,并且在PCR之后不需要酶切和电泳分析,可以在几分钟之内得到结果,特别适用于高通量操作。Thomas等[195]最早将这种方法应用在斑马鱼(Barchydanio rerio var)基因组编辑的检测,灵敏度可以达到5%。Hilioti等 [196]将该方法应用于植物基因组编辑的检测。1.11 异源双链泳动分析法
异源双链泳动分析法(heteroduplex mobility assay, HMA)是根据野生型和突变型DNA分子经过变性和退火后会产生异源双链DNA分子,按照同源双链分子和异源双链分子在非变性聚丙烯酰胺凝胶中电泳的泳动速度不同来区分基因组有没有发生突变。另外,也可以利用微芯片电泳系统来进行区分。与错配切割方法相比,省去了酶切步骤,避免了切割不完全导致的假阳性,但是容易错过大片段的缺失突变。Ota等[197]最早将该方法应用在斑马鱼基因组编辑的检测,之后该方法在植物基因组编辑检测中得到推广[41,42,49]。1.12 单等位基因识别PCR
单等位基因识别PCR(simple allele-discriminating PCR, SAP)的原理是基于ARMS (amplification refractory mutation system),即PCR的延伸是基于3′末端与模板的完全匹配,当3′端不完全匹配时候,会有不稳定的效应。在有些情况下,3′末位的单碱基错配并不能够很好地区分野生型和突变型,需要在倒数第二位引入错配来增加PCR的特异性。Bui等[198]最初用SAP方法区分拟南芥的突变体和野生型,之后Morineau等[120]将该方法用于基因组编辑检测。这种方法不受限制性内切酶的限制,但对PCR引物的设计有较高的要求。而且CRISPR/Cpf1系统的突变多为多碱基的缺失,所以不适合用这种方法进行检测。1.13 ACT-PCR
Hua等[199]通过控制PCR成功的两个关键因素,即特异的引物以及适宜的退火温度实现了基因组编辑的检测,这种方法被称为ACT-PCR (at critical temperature PCR)。在临界退火温度下,特异引物不能和突变体严格匹配,也就不能有效地进行扩增,因此可以用来区分野生型和基因编辑的突变体。该方法只需要进行一次常规的PCR反应,便可以很快地检测出成功编辑的个体,是一种简单准确、快速经济的方法,不过这也对PCR引物的设计和扩增条件提出较高要求。利用ACT-PCR,研究人员不仅在水稻中鉴定出基因编辑的个体,同时也在斑马鱼中成功鉴定出基因组编辑的个体,表明ACT-PCR的应用不受物种的限制[199]。1.14 微滴数字PCR
微滴数字PCR (droplet digital PCR, ddPCR)被称为第三代PCR,可以利用有限稀释、终点PCR和泊松分布来实现核酸浓度的绝对定量。该方法最早应用在人类细胞系基因组编辑检测方面[200,201],随后Gao等[202]将该方法应用在植物基因组编辑检测。与错配切割法相比,该方法需要很少的模板量,具有很高的灵敏度,适合高通量操作。2 植物基因组编辑检测方法的现状及发展趋势
本文对近几年发表在国内外具有较高影响力期刊中近200篇文章所涉及的检测方法进行了总结及分类归纳,发现不同方法在植物基因组编辑研究中的应用次数差别很大(图2)。PCR/RE和Sanger测序占据非常大的比例,下一代测序技术和错配切割法应用也比较广泛。其他方法的应用相对较少,有的方法仅被报道过一次[120,194,196,199],说明这些方法需要进一步优化。当然,大部分研究应用了不止一种方法,而是结合多种方法来分析不同的目标基因,说明每种方法各有优缺点,有的研究甚至使用了3种及以上的方法来检测基因组编辑的情况[42,55,57]。选择何种方法进行植物基因组编辑的检测,与诸多因素有关:
(1) 基因组编辑效率。如果突变频率很低,花费较高的Sanger测序方法不适用;如果突变频率高,高通量检测方法不具有优势,科研人员可以直接利用Sanger测序,从少量的样品中就可以得到需要的突变类型。
(2) 植物的倍性。有些检测方法不适用多倍体植物检测,例如TaqMan方法在多倍体小麦的检测中很难设计出合适的引物和探针,而PCR/RE和NGS却是不错的选择。目前,关于小麦基因组编辑的研究也大多利用这两种方法[5,27,43,53,67]。
(3) 基因组编辑工具。大部分方法都适用于CRISPR/Cas9系统,因为这些方法也大都是针对该酶而开发的,尤其是RGEN介导的RFLP。但是,如Base editing这样的工具,由于其造成的变化限定在特定的范围,科研人员更倾向于利用下一代测序的方法来检测。由于CRISPR/Cpf1系统一般会造成多碱基的缺失,很难用SAP或者ACT-PCR方法设计出合适的引物进行检测。
本文统计了2013~2017年间植物基因组编辑检测方法的发展趋势(图3)。为了便于统计,本文将PCR/RE、dCAPS和RE-PCR统一归为PCR+RE,而把数量很少的检测方法归为其他。从表3中可以看出,Sanger测序所占的比例由2013年的9.5%上升到2017年的45.2%。PCR+RE方法所占的比例在逐渐下降,由2013年的66.7%下降到2017年的27.4%。推测原因如下:(1) 测序技术的快速发展,成本有所降低;(2) Brinkman和Liu等[163,164]开发的解码软件大大简化了Sanger测序的流程,加快了Sanger测序直接用作基因编辑检测手段的推广;(3) 测序的结果能更直接地反映突变的类型,所有的筛选方法初筛之后最终都需要测序来确定突变类型,如果成本和通量差不多,那人们将会首选Sanger测序。
图3
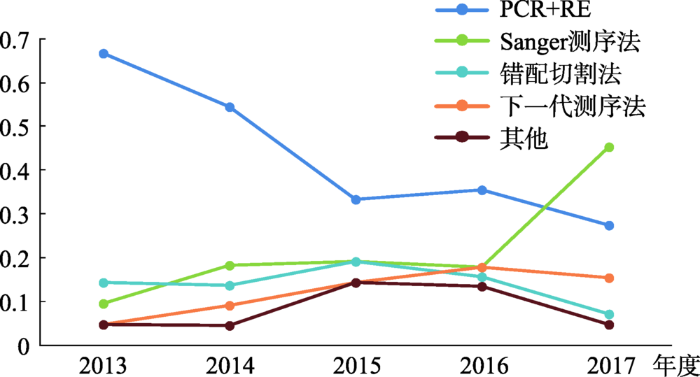 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3植物基因编辑检测方法发展趋势
每个点代表应用此种检测方法的研究占该年所有研究总数的比例。
Fig. 3The trend for detection methods in plant genome editing
下一代测序技术由于高准确性、高通量、高灵敏度以及低运行成本等突出优势,在基因组编辑检测中的应用也越来越广泛,呈现出一种上升的趋势。未来如果能将时间缩短,并且简化分析流程,该方法将是一个非常不错的选择。从图3也可以看出,2015年和2016年报道了很多新的方法用来检测植物基因组编辑,但是这些方法却没有得到推广,这也对科研人员开发新的检测方法提出了更高的要求。
随着越来越多植物基因组测序的完成,解读与改造基因的功能更为紧迫,从低等的苔藓到高等的树木,从二倍体的拟南芥到多倍体的小麦,以CRISPR/ Cas9为代表的基因组编辑技术无不显露出其强大的优势,这也对植物基因组编辑的检测方法提出更高的挑战。目前,还有一些检测方法在其他物种的基因组编辑中也有报道[203,204],但在植物中尚未尝试。将来可以根据植物基因组编辑的具体情况,考虑是否可以开发为植物基因组编辑的检测方法。
致谢
感谢先正达公司袁梦龙在文章图表制作上提供的帮助,感谢先正达公司陈钟颖和吕建在英文摘要修改上提供的帮助。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
URL [本文引用: 3]
URLPMID:24674878 [本文引用: 1]

The recent development of tools for precise editing of user-specified sequences is rapidly changing the landscape for plant genetics and biotechnology. It is now possible to target mutations and regulatory proteins to specific sites in a genome using zinc-finger nucleases (ZFNs), transcription activator-like endonucleases (TALENs), or the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system. Here we provide an update of recent developments in CRISPR/Cas9 technology and highlight online resources that will help biologists adopt new genome-editing tools.
URLPMID:3814374 [本文引用: 2]

The type II CRISPR/Cas system from Streptococcus pyogenes and its simplified derivative, the Cas9/single guide RNA (sgRNA) system, have emerged as potent new tools for targeted gene knockout in bacteria, yeast, fruit fly, zebrafish and human cells. Here, we describe adaptations of these systems leading to successful expression of the Cas9/sgRNA system in two dicot plant species, Arabidopsis and tobacco, and two monocot crop species, rice and sorghum. Agrobacterium tumefaciens was used for delivery of genes encoding Cas9, sgRNA and a non-fuctional, mutant green fluorescence protein (GFP) to Arabidopsis and tobacco. The mutant GFP gene contained target sites in its 5 coding regions that were successfully cleaved by a CAS9/sgRNA complex that, along with error-prone DNA repair, resulted in creation of functional GFP genes. DNA sequencing confirmed Cas9/sgRNA-mediated mutagenesis at the target site. Rice protoplast cells transformed with Cas9/sgRNA constructs targeting the promoter region of the bacterial blight susceptibility genes, OsSWEET14 and OsSWEET11, were confirmed by DNA sequencing to contain mutated DNA sequences at the target sites. Successful demonstration of the Cas9/sgRNA system in model plant and crop species bodes well for its near-term use as a facile and powerful means of plant genetic engineering for scientific and agricultural applications.
URLPMID:3852385 [本文引用: 5]

The clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) system has been used as an efficient tool for genome editing. We report the application of CRISPR-Cas-mediated genome editing to wheat (Triticum aestivum), the most important food crop plant with a very large and complex genome. The mutations were targeted in the inositol oxygenase (inox) and phytoene desaturase (pds) genes using cell suspension culture of wheat and in the pds gene in leaves of Nicotiana benthamiana. The expression of chimeric guide RNAs (cgRNA) targeting single and multiple sites resulted in indel mutations in all the tested samples. The expression of Cas9 or sgRNA alone did not cause any mutation. The expression of duplex cgRNA with Cas9 targeting two sites in the same gene resulted in deletion of DNA fragment between the targeted sequences. Multiplexing the cgRNA could target two genes at one time. Target specificity analysis of cgRNA showed that mismatches at the 3 end of the target site abolished the cleavage activity completely. The mismatches at the 5 end reduced cleavage, suggesting that the off target effects can be abolished in vivo by selecting target sites with unique sequences at 3 end. This approach provides a powerful method for genome engineering in plants.
URLPMID:24576457 [本文引用: 1]

Transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems have emerged as powerful tools for genome editing in a variety of species. Here, we report, for the first time, targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. We designed five TALENs targeting 4 genes, namely ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4, and obtained targeting efficiencies of up to 23.1% in protoplasts, and about 13.3% to 39.1% of the transgenic plants were somatic mutations. Also, we constructed two gRNAs targeting the ZmIPK gene in maize protoplasts, at frequencies of 16.4% and 19.1%, respectively. In addition, the CRISPR/Cas system induced targeted mutations in Z. mays protoplasts with efficiencies (13.1%) similar to those obtained with TALENs (9.1%). Our results show that both TALENs and the CRISPR/Cas system can be used for genome modification in maize.
URL [本文引用: 2]
URLPMID:5541054 [本文引用: 1]

Genome editing is an important tool for gene functional studies as well as crop improvement. The recent development of the CRISPR/Cas9 system using single guide RNA molecules (sgRNAs) to direct precise double strand breaks in the genome has the potential to revolutionize agriculture. Unfortunately, not all sgRNAs are equally efficient and it is difficult to predict their efficiency by bioinformatics. In crops such as cotton (Gossypium hirsutumL.), with labor-intensive and lengthy transformation procedures, it is essential to minimize the risk of using an ineffective sgRNA that could result in the production of transgenic plants without the desired CRISPR-induced mutations. In this study, we have developed a fast and efficient method to validate the functionality ofsgRNAsin cotton using a transient expression system. We have used this method to validate target sites for three different genesGhPDS, GhCLA1, andGhEF1and analyzed the nature of the CRISPR/Cas9-induced mutations. In our experiments, the most frequent type of mutations observed in cotton cotyledons were deletions ( 64%). We prove that the CRISPR/Cas9 system can effectively produce mutations in homeologous cotton genes, an important requisite in this allotetraploid crop. We also show that multiple gene targeting can be achieved in cotton with the simultaneous expression of several sgRNAs and have generated mutations inGhPDSandGhEF1at two target sites. Additionally, we have used the CRISPR/Cas9 system to produce targeted gene fragment deletions in theGhPDSlocus. Finally, we obtained transgenic cotton plants containing CRISPR/Cas9-induced gene editing mutations in theGhCLA1gene. The mutation efficiency was very high, with 80.6% of the transgenic lines containing mutations in theGhCLA1target site resulting in an intense albino phenotype due to interference with chloroplast biogenesis.
URLPMID:4507398 [本文引用: 2]

Abstract Recently, RNA-guided genome editing using the type II clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas) system has been applied to edit the plant genome in several herbaceous plant species. However, it remains unknown whether this system can be used for genome editing in woody plants. In this study, we describe the genome editing and targeted gene mutation in a woody species, Populus tomentosa Carr. via the CRISPR/Cas9 system. Four guide RNAs (gRNAs) were designed to target with distinct poplar genomic sites of the phytoene desaturase gene 8 (PtoPDS) which are followed by the protospacer-adjacent motif (PAM). After Agrobacterium-mediated transformation, obvious albino phenotype was observed in transgenic poplar plants. By analyzing the RNA-guided genome-editing events, 30 out of 59 PCR clones were homozygous mutants, 2 out of 59 were heterozygous mutants and the mutation efficiency at these target sites was estimated to be 51.7%. Our data demonstrate that the Cas9/sgRNA system can be exploited to precisely edit genomic sequence and effectively create knockout mutations in woody plants.
URL [本文引用: 1]

Cyanobacteria probably exhibit the widest range of diversity in growth habitats of all photosynthetic organisms. They are found in cold and hot, alkaline and acid, marine, freshwater, saline, terrestrial and symbiotic environments. In addition to this, they originated on earth at least 2.5 billion years ago and have evolved through periods of dramatic O2 increases, CO2 declines and temperature changes. One of the key problems they have faced through evolution and in their current environments is the capture of CO2 and its efficient use by Rubisco in photosynthesis. A central response to this challenge has been the development of a CO2 concentrating mechanism (CCM) that can be adapted to various environmental limitations. The functional elements of this CCM are: (i) the containment of Rubisco in carboxysome protein microbodies within the cell (the sites of CO2 elevation), and (ii) the presence of several inorganic carbon (Ci) transporters that deliver HCO3y intracellularly. Cyanobacteria show both species adaptation and acclimation of this mechanism. Between species, there are differ- ences in the suites of Ci transporters in each genome, the nature of the carboxysome structures and the functional roles of carbonic anhydrases. Within a species, different CCM activities can be induced depending on the Ci availability in the environment. This acclimation is largely based on the induction of multiple Ci transporters with different affinities and specificities for either CO2 or HCO3y as substrates. These features will be discussed in relation to our current knowledge of the genomes of diverse cyanobacteria and their ecological environments.
[本文引用: 3]
URLPMID:26524930 [本文引用: 1]
URLPMID:4523696 [本文引用: 1]

The CRISPR/Cas9 system is an efficient tool used for genome editing in a variety of organisms. Despite several recent reports of successful targeted mutagenesis using the CRISPR/Cas9 system in plants, in each case the target gene of interest, the Cas9 expression system and guide-RNA (gRNA) used, and the tissues used for transformation and subsequent mutagenesis differed, hence the reported frequencies of targeted mutagenesis cannot be compared directly. Here, we evaluated mutation frequency in rice using different Cas9 and/or gRNA expression cassettes under standardized experimental conditions. We introduced Cas9 and gRNA expression cassettes separately or sequentially into rice calli, and assessed the frequency of mutagenesis at the same endogenous targeted sequences. Mutation frequencies differed significantly depending on the Cas9 expression cassette used. In addition, a gRNA driven by the OsU6 promoter was superior to one driven by the OsU3 promoter. Using an all-in-one expression vector harboring the best combined Cas9/gRNA expression cassette resulted in a much improved frequency of targeted mutagenesis in rice calli, and bi-allelic mutant plants were produced in the T0 generation. The approach presented here could be adapted to optimize the construction of Cas9/gRNA cassettes for genome editing in a variety of plants. The online version of this article (doi:10.1007/s11103-015-0342-x) contains supplementary material, which is available to authorized users.
URLPMID:21464476

We performed targeted mutagenesis of a transgene and nine endogenous soybean (Glycine max) genes using zinc-finger nucleases (ZFNs). A suite of ZFNs were engineered by the recently described context-dependent assembly platform rapid, open-source method for generating zinc-finger arrays. Specific ZFNs targeting DICER-LIKE (DCL) genes and other genes involved in RNA silencing were cloned into a vector under an estrogen-inducible promoter. A hairy-root transformation system was employed to investigate the efficiency of ZFN mutagenesis at each target locus. Transgenic roots exhibited somatic mutations localized at the ZFN target sites for seven out of nine targeted genes. We next introduced a ZFN into soybean via whole-plant transformation and generated independent mutations in the paralogous genes DCL4a and DCL4b. The dcl4b mutation showed efficient heritable transmission of the ZFN-induced mutation in the subsequent generation. These findings indicate that ZFN-based mutagenesis provides an efficient method for making mutations in duplicate genes that are otherwise difficult to study due to redundancy. We also developed a publicly accessible Web-based tool to identify sites suitable for engineering context-dependent assembly ZFNs in the soybean genome.
URL

The ability to precisely engineer plant genomes offers much potential for advancing basic and applied plant biology. Here, we describe methods for the targeted modification of plant genomes using transcription activator-like effector nucleases (TALENs). Methods were optimized using tobacco (Nicotiana tabacum) protoplasts and TALENs targeting the acetolactate synthase (ALS) gene. Optimal TALEN scaffolds were identified using a protoplast-based single-strand annealing assay in which TALEN cleavage creates a functional yellow fluorescent protein gene, enabling quantification of TALEN activity by flow cytometry. Single-strand annealing activity data for TALENs with different scaffolds correlated highly with their activity at endogenous targets, as measured by high-throughput DNA sequencing of polymerase chain reaction products encompassing the TALEN recognition sites. TALENs introduced targeted mutations in ALS in 30% of transformed cells, and the frequencies of targeted gene insertion approximated 14%. These efficiencies made it possible to recover genome modifications without selection or enrichment regimes: 32% of tobacco calli generated from protoplasts transformed with TALEN-encoding constructs had TALEN-induced mutations in ALS, and of 16 calli characterized in detail, all had mutations in one allele each of the duplicate ALS genes (SurA and SurB). In calli derived from cells treated with a TALEN and a 322-bp donor molecule differing by 6 bp from the ALS coding sequence, 4% showed evidence of targeted gene replacement. The optimized reagents implemented in plant protoplasts should be useful for targeted modification of cells from diverse plant species and using a variety of means for reagent delivery.
URLPMID:23689819

Transcription activator-like effector nucleases (TALENs) enable targeted mutagenesis in a variety of organisms. The primary advantage of TALENs over other sequence-specific nucleases, namely zinc finger nucleases and meganucleases, lies in their ease of assembly, reliability of function, and their broad targeting range. Here we report the assembly of several TALENs for a specific genomic locus in barley. The cleavage activity of individual TALENs was first tested in vivo using a yeast-based, single-strand annealing assay. The most efficient TALEN was then selected for barley transformation. Analysis of the resulting transformants showed that TALEN-induced double strand breaks led to the introduction of short deletions at the target site. Additional analysis revealed that each barley transformant contained a range of different mutations, indicating that mutations occurred independently in different cells.
URLPMID:23288864
URLPMID:23929338

The article offers information on genome modification of crop plants using a CRISPR-Cas system. It states that genome editing technologies using zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can also generate genome modifications. Photographs related to genome editing in rice and wheat using an engineered type II CRISPR-Cas system are also presented.
URLPMID:23929340 [本文引用: 1]

The article offers information on mutagenesis in Nicotiana benthamiana using Cas9 RNA-guided endonuclease. It discusses the necessity of Sustainable intensification of crop production for food demand and supply. Photographs depicting assay scheme and DNA gel with PCR bands obtained upon amplification using primers flanking the target site within the PDS gene of N. benthamiana are also presented.
URLPMID:3789795 [本文引用: 1]

Tandemly arrayed genes (TAGs) or gene clusters are prevalent in higher eukaryotic genomes. For example, approximately 17% of genes are organized in tandem in the model plantArabidopsis thaliana. The genetic redundancy created by TAGs presents a challenge for reverse genetics. As molecular scissors, engineered zinc finger nucleases (ZFNs) make DNA double-strand breaks in a sequence-specific manner. ZFNs thus provide a means to delete TAGs by creating two double-strand breaks in the gene cluster. Using engineered ZFNs, we successfully targeted seven genes from three TAGs on two Arabidopsis chromosomes, including the well-knownRPP4gene cluster, which contains eight resistance (R) genes. The resulting gene cluster deletions ranged from a few kb to 55 kb with frequencies approximating 1% in somatic cells. We also obtained large chromosomal deletions of ~9 Mb at approximately one tenth the frequency, and gene cluster inversions and duplications also were achieved. This study demonstrates the ability to use sequence-specific nucleases in plants to make targeted chromosome rearrangements and create novel chimeric genes for reverse genetics and biotechnology.
[本文引用: 1]
URL
.
URLPMID:25232936 [本文引用: 4]

Targeted genome editing nucleases, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), are powerful tools for understanding gene function and for developing valuable new traits in plants. The clustered regularly interspersed short palindromic repeats (CRISPR)/Cas system has recently emerged as an alternative nuclease-based method for efficient and versatile genome engineering. In this system, only the 20-nt targeting sequence within the single-guide RNA (sgRNA) needs to be changed to target different genes. The simplicity of the cloning strategy and the few limitations on potential target sites make the CRISPR/Cas system very appealing. Here we describe a stepwise protocol for the selection of target sites, as well as the design, construction, verification and use of sgRNAs for sequence-specific CRISPR/Cas-mediated mutagenesis and gene targeting in rice and wheat. The CRISPR/Cas system provides a straightforward method for rapid gene targeting within 1-2 weeks in protoplasts, and mutated rice plants can be generated within 13-17 weeks.
URLPMID:25038773

Sequence-specific nucleases have been applied to engineer targeted modifications in polyploid genomes, but simultaneous modification of multiple homoeoalleles has not been reported. Here we use transcription activator-like effector nuclease (TALEN) and clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9 (refs. 4,5) technologies in hexaploid bread wheat to introduce targeted mutations in the three homoeoalleles that encode MILDEW-RESISTANCE LOCUS (MLO) proteins. Genetic redundancy has prevented evaluation of whether mutation of all three MLO alleles in bread wheat might confer resistance to powdery mildew, a trait not found in natural populations. We show that TALEN-induced mutation of all three TaMLO homoeologs in the same plant confers heritable broad-spectrum resistance to powdery mildew. We further use CRISPR-Cas9 technology to generate transgenic wheat plants that carry mutations in the TaMLO-A1 allele. We also demonstrate the feasibility of engineering targeted DNA insertion in bread wheat through nonhomologous end joining of the double-strand breaks caused by TALENs. Our findings provide a methodological framework to improve polyploid crops.
URLPMID:4262988

Background To accelerate the application of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9) system to a variety of plant species, a toolkit with additional plant selectable markers, more gRNA modules, and easier methods for the assembly of one or more gRNA expression cassettes is required. Results We developed a CRISPR/Cas9 binary vector set based on the pGreen or pCAMBIA backbone, as well as a gRNA (guide RNA) module vector set, as a toolkit for multiplex genome editing in plants. This toolkit requires no restriction enzymes besides BsaI to generate final constructs harboring maize-codon optimized Cas9 and one or more gRNAs with high efficiency in as little as one cloning step. The toolkit was validated using maize protoplasts, transgenic maize lines, and transgenic Arabidopsis lines and was shown to exhibit high efficiency and specificity. More importantly, using this toolkit, targeted mutations of three Arabidopsis genes were detected in transgenic seedlings of the T1 generation. Moreover, the multiple-gene mutations could be inherited by the next generation. Conclusions We developed a toolkit that facilitates transient or stable expression of the CRISPR/Cas9 system in a variety of plant species, which will facilitate plant research, as it enables high efficiency generation of mutants bearing multiple gene mutations.
URLPMID:25217528

Plant Physiol. 2014 Nov;166(3):1288-91. doi: 10.1104/pp.114.247593. Epub 2014 Sep 12. Research Support, Non-U.S. Gov't; Research Support, U.S. Gov't, Non-P.H.S.
URLPMID:25344637

Genome editing is one of the most powerful tools for revealing gene function and improving crop plants. Recently, RNA-guided genome editing using the type II clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas) system has been used as a powerful and efficient tool for genome editing in various organisms. Here, we report genome editing in tobacco ( Nicotiana tabacum) mediated by the CRISPR/Cas9 system. Two genes, NtPDS and NtPDR6, were used for targeted mutagenesis. First, we examined the transient genome editing activity of this system in tobacco protoplasts, insertion and deletion (indel) mutations were observed with frequencies of 16.2-20.3 % after transfecting guide RNA (gRNA) and the nuclease Cas9 in tobacco protoplasts. The two genes were also mutated using multiplexing gRNA at a time. Additionally, targeted deletions and inversions of a 1.8-kb fragment between two target sites in the NtPDS locus were demonstrated, while indel mutations were also detected at both the sites. Second, we obtained transgenic tobacco plants with NtPDS and NtPDR6 mutations induced by Cas9/gRNA. The mutation percentage was 81.8 % for NtPDS gRNA4 and 87.5 % for NtPDR6 gRNA2. Obvious phenotypes were observed, etiolated leaves for the psd mutant and more branches for the pdr6 mutant, indicating that highly efficient biallelic mutations occurred in both transgenic lines. No significant off-target mutations were obtained. Our results show that the CRISPR/Cas9 system is a useful tool for targeted mutagenesis of the tobacco genome.
URL [本文引用: 1]
URLPMID:25599829

Summary Fragrant rice is favoured worldwide because of its agreeable scent. The presence of a defective badh2 allele encoding betaine aldehyde dehydrogenase (BADH2) results in the synthesis of 2-acetyl-1-pyrroline (2AP), which is a major fragrance compound. Here, transcription activator-like effector nucleases (TALENs) were engineered to target and disrupt the OsBADH2 gene. Six heterozygous mutants (30%) were recovered from 20 transgenic hygromycin-resistant lines. Sanger sequencing confirmed that these lines had various indel mutations at the TALEN target site. All six transmitted the BADH2 mutations to the T1 generation; and four T1 mutant lines tested also efficiently transmitted the mutations to the T2 generation. Mutant plants carrying only the desired DNA sequence change but not the TALEN transgene were obtained by segregation in the T1 and T2 generations. The 2AP content of rice grains of the T1 lines with homozygous mutations increased from 0 to 0.35 0.75 mg/kg, which was similar to the content of a positive control variety harbouring the badh2-E7 mutation. We also simultaneously introduced three different pairs of TALENs targeting three separate rice genes into rice cells by bombardment and obtained lines with mutations in one, two and all three genes. These results indicate that targeted mutagenesis using TALENs is a useful approach to creating important agronomic traits.
URLPMID:26842992 [本文引用: 1]

CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system, which is a newly developed technology for targeted genome modification, has been successfully used in a number of species. In this study, we applied this technology to carry out targeted genome modification in maize. A marker gene Zmzb7was chosen for targeting. The sgRNA-Cas9 construct was transformed into maize protoplasts, and indel mutations could be detected. A mutant seedling with an expected albino phenotype was obtained from screening 120 seedlings generated from 10 callus events. Mutation efficiency in maize heterochromatic regions was also investigated. Twelve sites with different expression levels in maize centromeres or pericentromere regions were selected. The sgRNA-Cas9 constructs were transformed into protoplasts followed by sequencing the transformed protoplast genomic DNA. The results show that the genes in heterochromatic regions could be targeted by the CRISPR/Cas9 system efficiently, no matter whether they are expressed or not. Meanwhile, off-target mutations were not found in the similar sites having no PAM motif or having more than two mismatches. Together, our results show that the CRISPR/Cas9 system is a robust and efficient tool for genome modification in both euchromatic and heterochromatic regions in maize.
URLPMID:26603121 [本文引用: 2]

Gene targeting (GT) is of great significance for advancing basic plant research and crop improvement. Both TALENs (transcription activator-like effectors nucleases) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated 9) systems have been developed for genome editing in eukaryotes, including crop plants. In this work, we present the comparative analysis of these two technologies for two soybean genome editing targets,GmPDS11andGmPDS18. We found GT in soybean hairy roots with a single targeting efficiency range of 17.5–21.1% by TALENs, 11.7–18.1% by CRISPR/Cas9 using theAtU6-26promoter, and 43.4–48.1% by CRISPR/Cas9 using theGmU6-16g-1promoter, suggesting that the CRISPR/Cas9 using theGmU6-16g-1promoter is probably a much more efficient tool compared to the other technologies. Similarly, our double mutation GT efficiency experiment with these three technologies displayed a targeting efficiency of 6.25% by TALENs, 12.5% by CRISPR/Cas9 using theAtU6-26promoter, and 43.4–48.1% by CRISPR/Cas9 using theGmU6-16g-1promoter, suggesting that CRISPR/Cas9 is still a better choice for simultaneous editing of multiple homoeoalleles. Furthermore, we observed albino and dwarf buds (PDSknock-out) by soybean transformation in cotyledon nodes. Our results demonstrated that both TALENs and CRISPR/Cas9 systems are powerful tools for soybean genome editing.
URLPMID:26668334 [本文引用: 1]

Abstract Sequence-specific nucleases (SSNs) have been used successfully in homology-directed repair (HDR)-mediated gene targeting (GT) in many organisms. However, break-induced GT in plants remains challenging due to inefficient delivery of HDR templates and SSNs into plant nuclei. In many plants, including rice, Agrobacterium-mediated transformation is the most practical means of transformation because this biotic transformation system can deliver longer and more intact DNA payloads with less incorporation of fragmented DNA compared with physical transformation systems such as polyethylene glycol, electroporation, or biolistics. Following infection with Agrobacterium, transfer of transfer DNA (T-DNA) to the nucleus and its integration into the plant genome occur consecutively during cocultivation, thus timing the induction of DNA double-strand breaks (DSBs) on the target gene to coincide with the delivery of the HDR template is crucial. To synchronize DSB induction and delivery of the HDR template, we transformed a Cas9 expression construct and GT vector harboring the HDR template with guide RNAs (gRNAs) targeting the rice acetolactate synthase (ALS) gene either separately or sequentially into rice calli. When gRNAs targeting ALS were transcribed transiently from double-stranded T-DNA containing the HDR template, DSBs were induced in the ALS locus by the assembled Cas9/gRNA complex and homologous recombination was stimulated. Contrary to our expectations, there was no great difference in GT efficiency between Cas9-expressing and nonexpressing cells. However, when gRNA targeting DNA ligase 4 was transformed with Cas9 prior to the GT experiment, GT efficiency increased dramatically and more than one line exhibiting biallelic GT at the ALS locus was obtained. 2016 American Society of Plant Biologists. All Rights Reserved.
URLPMID:26768120
URLPMID:27212389

Dear Editor,\nThe Streptococcus pyogenes CRISPR-Cas9 system effectively mediates RNA-guided DNA double-strand breaks and is used for genome editing in many different organisms,including plants (Puchta,2016).CRISPR-Cas9 is a two-component system in which the Cas9 protein is expressed from a Pol Ⅱ promoter (Lowder et al.,2015).In contrast,the sgRNAs are typically expressed from Pol Ⅲ promoters,such as U6 and U3.Although the CRISPR-Cas9 system has been proven very powerful for genome editing,it has some limitations:(1) it is hard to achieve coordinated and/or inducible expression of Cas9 and the sgRNAs;(2) manipulating multiple sgRNAs for multiplexed gene editing can be tedious,requiring multiple Pol Ⅲ promoters;(3) in many non-model organisms,Pol Ⅲ promoters have not been well characterized,and heterologous Pol Ⅲ promoters often perform poorly (Sun et al.,2015).To overcome these limitations,we sought to drive the expression of both Cas9 and sgRNAs from a single Pol Ⅱ promoter (either inducible or constitutive) to achieve effective genome editing.This would allow a better spatiotemporal control of these gene targeting reagents,and would be applicable tothe organisms where Pol Ⅲ promoters are not well characterized.
URLPMID:27208253

Mutations generated by CRISPR/Cas9 in Arabidopsis (Arabidopsis thaliana) are often somatic and are rarely heritable. Isolation of mutations in Cas9-free Arabidopsis plants can ensure the stable transmission of the identified mutations to next generations, but the process is laborious and inefficient. Here, we present a simple visual screen for Cas9-free T2 seeds, allowing us to quickly obtain Cas9-free Arabidopsis mutants in the T2 generation. To demonstrate this in principle, we targeted two sites in the AUXIN-BINDING PROTEIN1 (ABP1) gene, whose function as a membrane-associated auxin receptor has been challenged recently. We obtained many T1 plants with detectable mutations near the target sites, but only a small fraction of T1 plants yielded Cas9-free abp1 mutations in the T2 generation. Moreover, the mutations did not segregate in Mendelian fashion in the T2 generation. However, mutations identified in the Cas9-free T2 plants were stably transmitted to the T3 generation following Mendelian genetics. To further simplify the screening procedure, we simultaneously targeted two sites in ABP1 to generate large deletions, which can be easily identified by PCR. We successfully generated two abp1 alleles that contained 1,141- and 711-bp deletions in the ABP1 gene. All of the Cas9-free abp1 alleles we generated were stable and heritable. The method described here allows for effectively isolating Cas9-free heritable CRISPR mutants in Arabidopsis.
URLPMID:27226350 [本文引用: 1]

The CRISPR/Cas9 system is an efficient and convenient tool for genome editing in plants. Cas9 nuclease derived fromStreptococcus pyogenes (Sp) is commonly used in this system. Recently,Staphylococcus aureusCas9 (SaCas9)-mediated genome editing was reported in human cells andArabidopsis. Because SaCas9 (1053 a.a.) is smaller than SpCas9 (1368 a.a.), SaCas9 could have substantial advantages for delivering and expressing Cas9 protein, especially using virus vectors. Since the protospacer adjacent motif (PAM) sequence of SaCas9 (5′-NNGRRT-3′) differs from that of SpCas9 (5′-NGG-3′), the use of this alternative Cas9 nuclease could expand the selectivity at potential cleavage target sites of the CRISPR/Cas9 system. Here we show that SaCas9 can mutagenize target sequences in tobacco and rice with efficiencies similar to those of SpCas9. We also analyzed the base preference for ‘T’ at the 6th position of the SaCas9 PAM. Targeted mutagenesis efficiencies in target sequences with non-canonical PAMs (5′-NNGRRV-3′) were much lower than those with a canonical PAM (5′-NNGRRT-3′). The length of target sequence recognized by SaCas9 is one or two nucleotides longer than that recognized by SpCas9. Taken together, our results demonstrate that SaCas9 has higher sequence recognition capacity than SpCas9 and is useful for reducing off-target mutations in crop.
URLPMID:4880914 [本文引用: 3]

Genome editing using the CRISPR/Cas9 system can be used to modify plant genomes, however, improvements in specificity and applicability are still needed in order for the editing technique to be useful in various plant species. Here, using genome editing mediated by a truncated gRNA (tru-gRNA)/Cas9 combination, we generated new alleles for OST2, a proton pump in Arabidopsis, with no off-target effects. By following expression of Cas9 and the tru-gRNAs, newly generated mutations in CRIPSR/Cas9 transgenic plants were detected with high average mutation rates of up to 32.8% and no off-target effects using constitutive promoter. Reducing nuclear localization signals in Cas9 decreased the mutation rate. In contrast, tru-gRNA Cas9 cassettes driven by meristematic- and reproductive-tissue-specific promoters increased the heritable mutation rate in Arabidopsis, showing that high expression in the germ line can produce bi-allelic mutations. Finally, the new mutant alleles obtained for OST2 exhibited altered stomatal closing in response to environmental conditions. These results suggest further applications in molecular breeding to improve plant function using optimized plant CRISPR/Cas9 systems.
URLPMID:27558837 [本文引用: 2]

Editing plant genomes is technically challenging in hard-to-transform plants and usually involves transgenic intermediates, which causes regulatory concerns. Here we report two simple and efficient genome-editing methods in which plants are regenerated from callus cells transiently expressing CRISPR/Cas9 introduced as DNA or RNA. This transient expression-based genome-editing system is highly efficient and specific for producing transgene-free and homozygous wheat mutants in the T0 generation. We demonstrate our protocol to edit genes in hexaploid bread wheat and tetraploid durum wheat, and show that we are able to generate mutants with no detectable transgenes. Our methods may be applicable to other plant species, thus offering the potential to accelerate basic and applied plant genome-engineering research. Plant genome editing typically relies upon transgenic intermediates, which is a concern given the current regulatory requirements concerning GMOs. Here, Zhanget al. describe a method to edit wheat genomes by transiently expressing CRISPR/Cas9 DNA or RNA, and are able to generate mutant plants with no detectable transgenes.
URLPMID:26808139

Summary Genome editing in plants has been boosted tremendously by the development of CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) technology. This powerful tool allows substantial improvement in plant traits in addition to those provided by classical breeding. Here, we demonstrate the development of virus resistance in cucumber ( Cucumis sativus L.) using Cas9/subgenomic RNA (sgRNA) technology to disrupt the function of the recessive eIF4E ( eukaryotic translation initiation factor 4E ) gene. Cas9/sgRNA constructs were targeted to the N and C termini of the eIF4E gene. Small deletions and single nucleotide polymorphisms (SNPs) were observed in the eIF4E gene targeted sites of transformed T1 generation cucumber plants, but not in putative off-target sites. Non-transgenic heterozygous eif4e mutant plants were selected for the production of non-transgenic homozygous T3 generation plants. Homozygous T3 progeny following Cas9/sgRNA that had been targeted to both eif4e sites exhibited immunity to Cucumber vein yellowing virus ( Ipomovirus ) infection and resistance to the potyviruses Zucchini yellow mosaic virus and Papaya ring spot mosaic virus-W . In contrast, heterozygous mutant and non-mutant plants were highly susceptible to these viruses. For the first time, virus resistance has been developed in cucumber, non-transgenically, not visibly affecting plant development and without long-term backcrossing, via a new technology that can be expected to be applicable to a wide range of crop plants.
URLPMID:27618611

Abstract Sequence-specific nucleases have been exploited to create targeted gene knockouts in various plants(1), but replacing a fragment and even obtaining gene insertions at specific loci in plant genomes remain a serious challenge. Here, we report efficient intron-mediated site-specific gene replacement and insertion approaches that generate mutations using the non-homologous end joining (NHEJ) pathway using the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9) system. Using a pair of single guide RNAs (sgRNAs) targeting adjacent introns and a donor DNA template including the same pair of sgRNA sites, we achieved gene replacements in the rice endogenous gene 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) at a frequency of 2.0%. We also obtained targeted gene insertions at a frequency of 2.2% using a sgRNA targeting one intron and a donor DNA template including the same sgRNA site. Rice plants harbouring the OsEPSPS gene with the intended substitutions were glyphosate-resistant. Furthermore, the site-specific gene replacements and insertions were faithfully transmitted to the next generation. These newly developed approaches can be generally used to replace targeted gene fragments and to insert exogenous DNA sequences into specific genomic sites in rice and other plants.
URLPMID:27614049 [本文引用: 1]

Targeted mutagenesis using programmableDNAendonucleases has broad applications for studying gene functionin plantaand developing approaches to improve crop yields. Recently, a genetic method that eliminates the need to emasculate the female inbred during hybrid seed production, referred to as Seed Production Technology, has been described. The foundation of this genetic system relied on classical methods to identify genes critical to anther and pollen development. One of these genes is a P450 gene which is expressed in the tapetum of anthers. Homozygous recessive mutants in this gene render maize and rice plants male sterile. While this P450 in maize corresponds to the male fertility geneMs26, male fertility mutants have not been isolated in other monocots such as sorghum and wheat. In this report, a custom designed homing endonuclease, Ems26+, was used to generatein plantamutations in the rice, sorghum and wheat orthologs of maizeMs26. Similar to maize, homozygous mutations in this P450 gene in rice and sorghum prevent pollen formation resulting in male sterile plants and fertility was restored in sorghum using a transformed copy of maizeMs26. In contrast, allohexaploid wheat plants that carry similar homozygous nuclear mutations in only one, but not all three, of their single genomes were male fertile. Targeted mutagenesis and subsequent characterization of male fertility genes in sorghum and wheat is an important step for capturing heterosis and improving crop yields through hybrid seed.
URLPMID:5399004

Summary The CRISPR/Cas9 nuclease system is a powerful and flexible tool for genome editing, and novel applications of this system are being developed rapidly. Here, we used CRISPR/Cas9 to target the FAD2 gene in Arabidopsis thaliana and in the closely related emerging oil seed plant, Camelina sativa, with the goal of improving seed oil composition. We successfully obtained Camelina seeds in which oleic acid content was increased from 16% to over 50% of the fatty acid composition. These increases were associated with significant decreases in the less desirable polyunsaturated fatty acids, linoleic acid (i.e. a decrease from ~16% to <4%) and linolenic acid (a decrease from ~35% to <10%). These changes result in oils that are superior on multiple levels: they are healthier, more oxidatively stable and better suited for production of certain commercial chemicals, including biofuels. As expected, A.??thaliana T2 and T3 generation seeds exhibiting these types of altered fatty acid profiles were homozygous for disrupted FAD2 alleles. In the allohexaploid, Camelina, guide RNAs were designed that simultaneously targeted all three homoeologous FAD2 genes. This strategy that significantly enhanced oil composition in T3 and T4 generation Camelina seeds was associated with a combination of germ???line mutations and somatic cell mutations in FAD2 genes in each of the three Camelina subgenomes.
URLPMID:27995308

CRISPR/Cas9 system can precisely edit genomic sequence and effectively create knockout mutations in T0 generation watermelon plants.Genome editing offers great advantage to reveal gene function and g
[本文引用: 1]
URLPMID:27940306
URLPMID:27943461

Abstract The ability to edit plant genomes through gene targeting (GT) requires efficient methods to deliver both sequence-specific nucleases (SSNs) and repair templates to plant cells. This is typically achieved using Agrobacterium T-DNA, biolistics or by stably integrating nuclease-encoding cassettes and repair templates into the plant genome. In dicotyledonous plants, such as Nicotinana tabacum (tobacco) and Solanum lycopersicum (tomato), greater than 10-fold enhancements in GT frequencies have been achieved using DNA virus-based replicons. These replicons transiently amplify to high copy numbers in plant cells to deliver abundant SSNs and repair templates to achieve targeted gene modification. In the present work, we developed a replicon-based system for genome engineering of cereal crops using a deconstructed version of the wheat dwarf virus (WDV). In wheat cells, the replicons achieve a 110-fold increase in expression of a reporter gene relative to non-replicating controls. Furthermore, replicons carrying CRISPR/Cas9 nucleases and repair templates achieved GT at an endogenous ubiquitin locus at frequencies 12-fold greater than non-viral delivery methods. The use of a strong promoter to express Cas9 was critical to attain these high GT frequencies. We also demonstrate gene-targeted integration by homologous recombination (HR) in all three of the homoeoalleles (A, B and D) of the hexaploid wheat genome, and we show that with the WDV replicons, multiplexed GT within the same wheat cell can be achieved at frequencies of ~1%. In conclusion, high frequencies of GT using WDV-based DNA replicons will make it possible to edit complex cereal genomes without the need to integrate GT reagents into the genome. 2016 The Authors The Plant Journal 2016 John Wiley & Sons Ltd.
URLPMID:27932049 [本文引用: 1]
URLPMID:28098143 [本文引用: 3]

Substantial efforts are being made to optimize the CRISPR/Cas9 system for precision crop breeding. The avoidance of transgene integration and reduction of off-target mutations are the most important targets for optimization. Here, we describe an efficient genome editing method for bread wheat using CRISPR/Cas9 ribonucleoproteins (RNPs). Starting from RNP preparation, the whole protocol takes only seven to nine weeks, with four to five independent mutants produced from 100 immature wheat embryos. Deep sequencing reveals that the chance of off-target mutations in wheat cells is much lower in RNP mediated genome editing than in editing with CRISPR/Cas9 DNA. Consistent with this finding, no off-target mutations are detected in the mutant plants. Because no foreign DNA is used in CRISPR/Cas9 RNP mediated genome editing, the mutants obtained are completely transgene free. This method may be widely applicable for producing genome edited crop plants and has a good prospect of being commercialized. Protocols for crop genome editing would ideally be quick, efficient and specific while avoiding integration of transgenes into the genome of edited plants. Here, Lianget al. show that CRISPR/Cas9 ribonucleoproteins can be used to generate genome edited wheat plants in as little as nine weeks.
URLPMID:28211909 [本文引用: 1]

Abstract Clustered regularly interspaced short palindromic repeats (CRISPR)-Cpf1 has emerged as an effective genome editing tool in animals. Here we compare the activity of Cpf1 from Acidaminococcus sp. BV3L6 (As) and Lachnospiraceae bacterium ND2006 (Lb) in plants, using a dual RNA polymerase II promoter expression system. LbCpf1 generated biallelic mutations at nearly 100% efficiency at four independent sites in rice T0 transgenic plants. Moreover, we repurposed AsCpf1 and LbCpf1 for efficient transcriptional repression in Arabidopsis, and demonstrated a more than tenfold reduction in miR159b transcription. Our data suggest promising applications of CRISPR-Cpf1 for editing plant genomes and modulating the plant transcriptome.
URLPMID:27918538 [本文引用: 3]

Plants evolved so that their flowering is triggered by seasonal changes in day lengths. However, day-length sensitivity in crops limits their geographical range of cultivation, and thus modification of the photoperiod response was critical for their domestication(2-11). Here we show that loss of day-length-sensitive flowering in tomato was driven by the florigen paralog and flowering repressor SELF-PRUNING 5G (SP5G). SP5G expression is induced to high levels during long days in wild species, but not in cultivated tomato because of cis-regulatory variation. CRISPR/Cas9-engineered mutations in SP5G cause rapid flowering and enhance the compact determinate growth habit of field tomatoes, resulting in a quick burst of flower production that translates to an early yield. Our findings suggest that pre-existing variation in SP5G facilitated the expansion of cultivated tomato beyond its origin near the equator in South America, and they provide a compelling demonstration of the power of gene editing to rapidly improve yield traits in crop breeding.
URLPMID:27747971

The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR‐associated protein 9 nuclease (Cas9) system depends on a guide RNA (gRNA) to specify its target. By efficiently co‐expressing multiple gRNAs that target different genomic sites, the polycistronic tRNA‐gRNA gene (PTG) strategy enables multiplex gene editing in the family of closely related rice mitogen‐activated protein kinase (MPK) genes. In this study, we identified MPK1 and MPK6 (Arabidopsis AtMPK6 and AtMPK4 orthologues, respectively) as essential genes for rice development by finding the preservation of MPK functional alleles and normal phenotypes in CRISPR‐edited mutants. The true knock‐out mutants of MPK1 were severely dwarfed and sterile, and homozygous mpk1 seeds from heterozygous parents were defective in embryo development. By contrast, heterozygous mpk6 mutant plants completely failed to produce homozygous mpk6 seeds. In addition, functional importance of specific MPK features could be evaluated by characterizing CRISPR‐induced allelic variation in the conserved kinase domain of MPK6. By simultaneously targeting two to eight genomic sites in the closely related MPK genes, we demonstrated 45 to 86% frequency of biallelic mutations and successful creation of single, double and quadruple gene mutants. Indels and fragment deletion were both stably inherited to the next generations and transgene‐free mutants of rice MPK genes were readily obtained via genetic segregation, thereby eliminating any positional effects of transgene insertions. Taken together, our study reveals essentiality of MPK1 and MPK6 in rice development and enables functional discovery of the previously inaccessible genes or domains because their phenotypes are masked by lethality or redundancy.
URLPMID:28244994 [本文引用: 4]

Single DNA base pairs are edited in wheat, rice and maize using a Cas9 nickase fusion protein.
URL

Genome editing in plants becomes popular since the advent of sequence-specific nucleases (SSNs) that are simple to set up and efficient in various plant species. Although transcription activator-like effector nucleases (TALENs) are one of the most prevalent SSNs and have a potential to provide higher target specificity by their dimeric property, TALENs are sensitive to methylated cytosines that are present not only in transposons but also in active genes in plants. In mammalian cells, the methylation sensitivity of TALENs could be overcome by using a base-recognition module (N65) that has a higher affinity to methylated cytosine. In contrast to mammals, plants carry DNA methylation at all cytosine contexts (CG, CHG, and CHH, where H represents A, C, or T) with various degrees and effectiveness of N65module in genome editing in plants has not been explored. In this study, we designed sets of TALENs with or without N65modules and examined their efficiency in genome editing of methylated regions in rice. Although improvement in genome editing efficiency was observed with N65-TALENs designed to a stably methylated target, another target carrying cytosines with various levels of methylation showed resistance to both normal and N65-TALENs. The results suggest that variability of cytosine methylation in target regions is an additional factor affecting the genome editing efficiency of TALENs.
URLPMID:5428692 [本文引用: 1]

Parthenocarpy in horticultural crop plants is an important trait with agricultural value for various industrial purposes as well as direct eating quality. Here, we demonstrate a breeding strategy to generate parthenocarpic tomato plants using the CRISPR/Cas9 system. We optimized the CRISPR/Cas9 system to introduce somatic mutations effectively intoSlIAA9—a key gene controlling parthenocarpy—with mutation rates of up to 100% in the T0 generation. Furthermore, analysis of off-target mutations using deep sequencing indicated that our customized gRNAs induced no additional mutations in the host genome. Regenerated mutants exhibited morphological changes in leaf shape and seedless fruit—a characteristic of parthenocarpic tomato. And the segregated next generation (T1) also showed a severe phenotype associated with the homozygous mutated genome. The system developed here could be applied to produce parthenocarpic tomato in a wide variety of cultivars, as well as other major horticultural crops, using this precise and rapid breeding technique.
URLPMID:28349304 [本文引用: 1]

The clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease 9 (CRISPR/Cas9) system has emerged as a promising technology for specific genome editing in many species. Here we constructed one vector targeting eight agronomic genes in rice using the CRISPR/Cas9 multiplex genome editing system. By subsequent genetic transformation and DNA sequencing, we found that the eight target genes have high mutation efficiencies in the T 0 generation. Both heterozygous and homozygous mutations of all editing genes were obtained in T 0 plants. In addition, homozygous sextuple, septuple, and octuple mutants were identified. As the abundant genotypes in T 0 transgenic plants, various phenotypes related to the editing genes were observed. The findings demonstrate the potential of the CRISPR/Cas9 system for rapid introduction of genetic diversity during crop breeding.
URLPMID:28346401 [本文引用: 1]

Targeted editing of single base pairs is achieved in monocot rice and dicot tomato using Target-AID (Cas9 activation-induced cytidine deaminase fusion).
URLPMID:28371831

Split-protein methods here a protein is split into two inactive fragments that must re-assemble to form an active protein an be used to regulate the activity of a given protein and reduce the size of gene transcription units. Here, we show that aStaphylococcus aureus Cas9(SaCas9) can be split, and thatsplit-SaCas9expressed fromAgrobacteriumcan induce targeted mutagenesis inNicotiana benthamiana. SinceSaCas9is smaller than the more commonly usedCas9derived fromStreptococcus pyogenes, the split-SaCas9 provides the smallest tool yet for clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) plant genome editing. Both sets of split-SaCas9 (_430N/431C and _739N/740C) exhibited genome-editing activity, and the activity of split-SaCas9_739N/740C was almost the same as that of full-length SaCas9. This result indicates that split-SaCas9_739N/740C is suitable for use in targeted mutagenesis. We also show that thesplit-SaCas9fragment expressed fromTomato mosaic viruscould induce targeted mutagenesis together with another fragment expressed fromAgrobacterium, suggesting that a split-SaCas9 system using a plant virus vector is a promising tool for integration-free plant genome editing. Split-SaCas9 has the potential to regulate CRISPR/Cas9-mediated genome editing activity in plant cells both temporally and spatially.
URLPMID:5436839

RNA-guided genome editing using the CRISPR/Cas9 CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) system has been applied successfully in several plant species. However, to date, there are few reports on the use of any of the current genome editing approaches in grape n important fruit crop with a large market not only for table grapes but also for wine. Here, we report successful targeted mutagenesis in grape (Vitis viniferaL., cv. Neo Muscat) using the CRISPR/Cas9 system. When a Cas9 expression construct was transformed to embryonic calli along with a synthetic sgRNA expression construct targeting theVitis viniferaphytoene desaturase (VvPDS) gene, regenerated plants with albino leaves were obtained. DNA sequencing confirmed that the VvPDS gene was mutated at the target site in regenerated grape plants. Interestingly, the ratio of mutated cells was higher in lower, older, leaves compared to that in newly appearing upper leaves. This result might suggest either that the proportion of targeted mutagenized cells is higher in older leaves due to the repeated induction of DNA double strand breaks (DSBs), or that the efficiency of precise DSBs repair in cells of old grape leaves is decreased.
URLPMID:28379609

Abstract The RNA-guided Cas9 system is a versatile tool for genome editing. Here, we established a RNA-guided endonuclease (RGEN) system as an in0002vivo desired-target mutator (DTM) in maize to reduce the linkage drag during breeding procedure, using the LIGULELESS1 (LG1) locus as a proof-of-concept. Our system showed 51.5%-91.2% mutation frequency in T0 transgenic plants. We then crossed the T1 plants stably expressing DTM with six diverse recipient maize lines and found that 11.79%-28.71% of the plants tested were mutants induced by the DTM effect. Analysis of successive F2 plants indicated that the mutations induced by the DTM effect were largely heritable. Moreover, DTM-generated hybrids had significantly smaller leaf angles that were reduced more than 50% when compared with that of the wild type. Planting experiments showed that DTM-generated maize plants can be grown with significantly higher density and hence greater yield potential. Our work demonstrate that stably expressed RGEN could be implemented as an in0002vivoDTM to rapidly generate and spread desired mutations in maize through hybridization and subsequent backcrossing, and hence bypassing the linkage drag effect in convention introgression methodology. This proof-of-concept experiment can be a potentially much more efficient breeding strategy in crops employing the RNA-guided Cas9 genome editing. 0008 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
URLPMID:28755320

Abstract In the present study, we utilized TALEN- and CRISPR/Cas9-induced mutations to analyze the promoter of the barley phytase gene HvPAPhy_a. The purpose of the study was dual, validation of the PAPhy_a enzyme as the main contributor of the mature grain phytase activity (MGPA), as well as validating the importance of a specific promoter region of the PAPhy_a gene which contains three overlapping cis-acting regulatory elements (GCN4, Skn1 and the RY-element) known to be involved in gene expression during grain filling. The results confirm that the barley PAPhy_a enzyme is the main contributor to the MGPA as grains of knock-out lines show very low MGPA. Additionally, the analysis of the HvPAPhy_a promoter region containing the GCN4/Skn1/RY motif highlights its importance for HvPAPhy_a expression as the MGPA in grains of plant lines with mutations within this motif is significantly reduced. Interestingly, lines with deletions located downstream of the motif show even lower MGPA levels, indicating that the GCN4/SKn1/RY motif is not the only element responsible for the level of PAPhy_a expression during grain maturation. Mutant grains with very low MPGA showed delayed germination as compared to grains of wild type barley. As grains with high levels of preformed phytases would provide more readily available phosphorous needed for a fast germination, this indicates that faster germination may be implicated in the positive selection of the ancient PAPhy gene duplication that lead to the creation of the PAPhy_a gene.
URLPMID:29226588

Summary We report that a single-sgRNA seed is capable of guiding CRISPR/Cas9 to simultaneously edit multiple genes AtRPL10A, AtRPL10B and AtRPL10C in Arabidopsis . Our results also demonstrate that it is possible to use CRISPR/Cas9 technology to create AtRPL10 triple mutants which otherwise cannot be generated by conventional genetic crossing. Compared to other conventional multiplex CRISPR/Cas systems, a single sgRNA seed has the advantage of reducing off-target gene-editing. Such a single sgRNA seed-induced gene editing system might be also applicable to modify other homologous genes or even less-homologous sequences for multiple gene-editing in plants and other organisms.
URLPMID:28918562 [本文引用: 3]

Abstract Genome editing has been a long-term challenge for molecular biology research, particularly for plants possess complex genome. The recently discovered Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is a versatile tool for genome editing which enables editing of multiple genes based on the guidance of small RNAs. Even though the efficiency of CRISPR/Cas9 system has been shown with several studies from diploid plants, its application remains a challenge for plants with polyploid and complex genome. Here, we applied CRISPR/Cas9 genome editing system in wheat protoplast to conduct the targeted editing of stress-responsive transcription factor genes, wheat dehydration responsive element binding protein 2 (TaDREB2) and wheat ethylene responsive factor 3 (TaERF3). Targeted genome editing of TaDREB2 and TaERF3 was achieved with transient expression of small guide RNA and Cas9 protein in wheat protoplast. The effectiveness of mutagenesis in wheat protoplast was confirmed with restriction enzyme digestion assay, T7 endonuclease assay, and sequencing. Furthermore, several off-target regions for designed sgRNAs were analyzed, and the specificity of genome editing was confirmed with amplicon sequencing. Overall results suggested that CRISPR/Cas9 genome editing system can easily be established on wheat protoplast and it has a huge potentiality for targeted manipulation of wheat genome for crop improvement purposes.
URL [本文引用: 1]

正Dear Editor,The CRISPR-Cas9(clustered regularly interspaced short palindromic repeats/Cas9)system has been widely used for a variety of applications,including targeted gene knockout,gene insertion,gene replacement and base editing.Despite its wide use,the genome editing using CRISPR-Cas9 is performed almost exclusively at sites containing canonical NGG protospacer adjacent motifs(PAMs).To overcome the PAM constraint of the CRISPR-Cas9 system,many attempts have been made to develop various Cas9 orthologs and
URLPMID:29332168

Abstract The first report presenting successful and efficient carrot genome editing using CRISPR/Cas9 system. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas9) is a powerful genome editing tool that has been widely adopted in model organisms recently, but has not been used in carrot-a model species for in vitro culture studies and an important health-promoting crop grown worldwide. In this study, for the first time, we report application of the CRISPR/Cas9 system for efficient targeted mutagenesis of the carrot genome. Multiplexing CRISPR/Cas9 vectors expressing two single-guide RNA (gRNAs) targeting the carrot flavanone-3-hydroxylase (F3H) gene were tested for blockage of the anthocyanin biosynthesis in a model purple-colored callus using Agrobacterium-mediated genetic transformation. This approach allowed fast and visual comparison of three codon-optimized Cas9 genes and revealed that the most efficient one in generating F3H mutants was the Arabidopsis codon-optimized AteCas9 gene with up to 90% efficiency. Knockout of F3H gene resulted in the discoloration of calli, validating the functional role of this gene in the anthocyanin biosynthesis in carrot as well as providing a visual marker for screening successfully edited events. Most resulting mutations were small Indels, but long chromosome fragment deletions of 116-119 nt were also generated with simultaneous cleavage mediated by two gRNAs. The results demonstrate successful site-directed mutagenesis in carrot with CRISPR/Cas9 and the usefulness of a model callus culture to validate genome editing systems. Given that the carrot genome has been sequenced recently, our timely study sheds light on the promising application of genome editing tools for boosting basic and translational research in this important vegetable crop.
URLPMID:29333573

Abstract Using a gRNA and Agrobacterium-mediated transformation, we performed simultaneous site-directed mutagenesis of two GmPPD loci in soybean. Mutations in GmPPD loci were confirmed in at least 33% of T 2 seeds. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated endonuclease 9 (Cas9) system is a powerful tool for site-directed mutagenesis in crops. Using a single guide RNA (gRNA) and Agrobacterium-mediated transformation, we performed simultaneous site-directed mutagenesis of two homoeologous loci in soybean (Glycine max), GmPPD1 and GmPPD2, which encode the orthologs of Arabidopsis thaliana PEAPOD (PPD). Most of the T 1 plants had heterozygous and/or chimeric mutations for the targeted loci. The sequencing analysis of T 1 and T 2 generations indicates that putative mutation induced in the T 0 plant is transmitted to the T 1 generation. The inheritable mutation induced in the T 1 plant was also detected. This result indicates that continuous induction of mutations during T 1 plant development increases the occurrence of mutations in germ cells, which ensures the transmission of mutations to the next generation. Simultaneous site-directed mutagenesis in both GmPPD loci was confirmed in at least 33% of T 2 seeds examined. Approximately 19% of double mutants did not contain the Cas9/gRNA expression construct. Double mutants with frameshift mutations in both GmPPD1 and GmPPD2 had dome-shaped trifoliate leaves, extremely twisted pods, and produced few seeds. Taken together, our data indicate that continuous induction of mutations in the whole plant and advancing generations of transgenic plants enable efficient simultaneous site-directed mutagenesis in duplicated loci in soybean.
URL [本文引用: 1]

【目的】光作为一种环境信号,可影响植物的基因表达、酶活性和形态建成。光敏色素互作因子在光信号传导过程中起着重要作用。本研究旨在构建水稻光敏色素互作因子OsPIL15的CRISPR/Cas9表达载体,创制OsPIL15突变体,挖掘水稻功能基因,丰富和完善水稻光信号调控分子机制。【方法】依据CRISPR/Cas9技术原理,设计OsPIL15突变靶点。将所设计靶序列在水稻基因组中进行比对,排除非特异性靶位点,同时使该靶序列含有常用酶切位点,方便后期突变体鉴定。化学合成靶位点寡核苷酸序列并与载体pBUN411连接构建CRISPR/Cas9表达载体,利用农杆菌介导法导入粳稻品种日本晴,以除草剂抗性标记筛选获得阳性转基因植株。利用酶切法判断T0代转基因植株是否发生突变,结合测序结果分析突变单株的突变基因型。将靶点序列在水稻全基因组中进行比对分析,选择5个与靶序列同源性较高且错配在4 bp以内的位点作为潜在脱靶位点进行脱靶效应评估,分析所设计靶序列特异性。【结果】所构建表达载体成功实现了对OsPIL15的定向编辑,酶切显示在选取的25株T0代转基因植株中获得15株突变体,其中包括5株纯合突变体、6株双等位突变体和4株杂合突变体,共10种不同突变基因型和11个突变株系。突变类型以单碱基插入或缺失为主,同时也得到2种56和66 bp较大片段缺失株系。对部分纯合突变、双等位突变和杂合突变体的T1代植株进行分析,结果表明,T0代产生的突变基因型绝大部分能稳定遗传给下一代。T0代纯合突变体后代为纯合突变单株,仅在株系14纯合突变体后代中检测到1株未突变单株;T0代双等位突变体后代可得到2种纯合突变型和1种双等位突变型;T0代杂合突变体后代则可得到纯合、杂合及未突变3种类型。对T0代未突变植株的后继世代酶切分析显示,62株T1代转基因植株均未发生突变,表17
URL [本文引用: 1]

【目的】光作为一种环境信号,可影响植物的基因表达、酶活性和形态建成。光敏色素互作因子在光信号传导过程中起着重要作用。本研究旨在构建水稻光敏色素互作因子OsPIL15的CRISPR/Cas9表达载体,创制OsPIL15突变体,挖掘水稻功能基因,丰富和完善水稻光信号调控分子机制。【方法】依据CRISPR/Cas9技术原理,设计OsPIL15突变靶点。将所设计靶序列在水稻基因组中进行比对,排除非特异性靶位点,同时使该靶序列含有常用酶切位点,方便后期突变体鉴定。化学合成靶位点寡核苷酸序列并与载体pBUN411连接构建CRISPR/Cas9表达载体,利用农杆菌介导法导入粳稻品种日本晴,以除草剂抗性标记筛选获得阳性转基因植株。利用酶切法判断T0代转基因植株是否发生突变,结合测序结果分析突变单株的突变基因型。将靶点序列在水稻全基因组中进行比对分析,选择5个与靶序列同源性较高且错配在4 bp以内的位点作为潜在脱靶位点进行脱靶效应评估,分析所设计靶序列特异性。【结果】所构建表达载体成功实现了对OsPIL15的定向编辑,酶切显示在选取的25株T0代转基因植株中获得15株突变体,其中包括5株纯合突变体、6株双等位突变体和4株杂合突变体,共10种不同突变基因型和11个突变株系。突变类型以单碱基插入或缺失为主,同时也得到2种56和66 bp较大片段缺失株系。对部分纯合突变、双等位突变和杂合突变体的T1代植株进行分析,结果表明,T0代产生的突变基因型绝大部分能稳定遗传给下一代。T0代纯合突变体后代为纯合突变单株,仅在株系14纯合突变体后代中检测到1株未突变单株;T0代双等位突变体后代可得到2种纯合突变型和1种双等位突变型;T0代杂合突变体后代则可得到纯合、杂合及未突变3种类型。对T0代未突变植株的后继世代酶切分析显示,62株T1代转基因植株均未发生突变,表17
URLPMID:29141013 [本文引用: 1]

Abstract Genetic manipulation of organisms using CRISPR/Cas9 technology generally produces small insertions/deletions (indels) that can be difficult to detect. Here, we describe a technique to easily and rapidly identify such indels. Sequence-identified mutations that alter a restriction enzyme recognition site can be readily distinguished from wild-type alleles using a cleaved amplified polymorphic sequence (CAPS) technique. If a restriction site is created or altered by the mutation such that only one allele contains the restriction site, a polymerase chain reaction (PCR) followed by a restriction digest can be used to distinguish the two alleles. However, in the case of most CRISPR-induced alleles, no such restriction sites are present in the target sequences. In this case, a derived CAPS (dCAPS) approach can be used in which mismatches are purposefully introduced in the oligonucleotide primers to create a restriction site in one, but not both, of the amplified templates. Web-based tools exist to aid dCAPS primer design, but when supplied sequences that include indels, the current tools often fail to suggest appropriate primers. Here, we report the development of a Python-based, species-agnostic web tool, called indCAPS, suitable for the design of PCR primers used in dCAPS assays that is compatible with indels. This tool should have wide utility for screening editing events following CRISPR/Cas9 mutagenesis as well as for identifying specific editing events in a pool of CRISPR-mediated mutagenesis events. This tool was field-tested in a CRISPR mutagenesis experiment targeting a cytokinin receptor (AHK3) in Arabidopsis thaliana. The tool suggested primers that successfully distinguished between wild-type and edited alleles of a target locus and facilitated the isolation of two novel ahk3 null alleles. Users can access indCAPS and design PCR primers to employ dCAPS to identify CRISPR/Cas9 alleles at http://indcaps.kieber.cloudapps.unc.edu/.
URLPMID:1244614021510300123000630008300 [本文引用: 1]

The detection of single nucleotide polymorphisms by PCR is necessary for many types of genetic analysis, from mapping genomes to tracking specific mutations. A web-based program, dCAPS Finder 2.0, facilitates the design of mismatched PCR primers to aid such analysis.
[本文引用: 2]
URLPMID:23956122 [本文引用: 2]

Targeted gene mutation was successfully achieved in rice using the CRISPRCas9 system. Experimental analyses of mutation efficiency and off-target effect as well as genome-wide prediction of specific guide RNA seeds suggest that the CRISPRCas9 system is a simple and effective tool for plant functional genomics and crop improvement.Precise and straightforward methods to edit the plant genome are much needed for functional genomics and crop improvement. Recently, RNA-guided genome editing using bacterial Type II cluster regularly interspaced short palindromic repeats (CRISPR)-associated nuclease (Cas) is emerging as an efficient tool for genome editing in microbial and animal systems. Here, we report the genome editing and targeted gene mutation in plants via the CRISPRCas9 system. Three guide RNAs (gRNAs) with a 2022-nt seed region were designed to pair with distinct rice genomic sites which are followed by the protospacer-adjacent motif (PAM). The engineered gRNAs were shown to direct the Cas9 nuclease for precise cleavage at the desired sites and introduce mutation (insertion or deletion) by error-prone non-homologous end joining DNA repairing. By analyzing the RNA-guided genome-editing events, the mutation efficiency at these target sites was estimated to be 38%. In addition, the off-target effect of an engineered gRNACas9 was found on an imperfectly paired genomic site, but it had lower genome-editing efficiency than the perfectly matched site. Further analysis suggests that mismatch position between gRNA seed and target DNA is an important determinant of the gRNACas9 targeting specificity, and specific gRNAs could be designed to target more than 90% of rice genes. Our results demonstrate that the CRISPRCas system can be exploited as a powerful tool for gene targeting and precise genome editing in plants.
URLPMID:25146436

Key message: Xanthomonas citri subsp. citri pretreatment before agroinfiltration could significantly promote transient expression in citrus leaves which were previously recalcitrant to agroinfiltration. Abstract: Transient expression via agroinfiltration is widely used in biotechnology but remains problematic in many economically important plants. Xanthomonas citri subsp. citri (Xcc)-facilitated agroinfiltration was employed to promote transient protein expression in Valencia sweet orange leaves, which are recalcitrant to agroinfiltration. However, it is unclear whether Xcc-facilitated agroinfiltration has broad application, i.e., whether Xcc-facilitated agroinfiltration could be used on other citrus varieties. In addition, we intended to investigate whether Xcc-facilitated agroinfiltration could be used to hasten transgene function assays, e.g., Cre/lox system and Cas9/sgRNA system. In this report, Xcc-facilitated agroinfiltration was further exploited to enhance -glucuronidase (GUS) expression in five citrus varieties. Xcc-facilitated agroinfiltration also significantly increased GFP expression in six citrus varieties tested. Both GUS and GFP assays indicated that Xcc-facilitated agroinfiltration had the best performance in grapefruit. After Xcc-facilitated agroinfiltration was carried out in grapefruit, protoplast analysis of the transformed cells indicated that there were more than 20 % leaf cells expressing GFP. In grapefruit, usefulness of Xcc-facilitated agroinfiltration was assayed in three case studies: (1) fast functional analysis of Cre/lox system, (2) the heat shock regulation of HSP70B promoter derived from Arabidopsis, and (3) Cas9/sgRNA-mediated genome modification.
URLPMID:26082432

The article discusses a study which described the production of gene knockouts in potato via the clustered regularly interspaced short palindromic repeats/associated protein 9 nuclease (CRISPR/CAS9) system. Particular focus is given to the growing role of the CRISPR/CAS9 system in plant genome editing, as well as the importance of potato in global food security. Also discussed are cloning of a native promoter for potato RNA, DNA extraction and polymerase chain reaction on transgenic plants.
URL
[本文引用: 1]
[本文引用: 1]
URLPMID:25655437 [本文引用: 2]

Restriction fragment length polymorphism (RFLP) analysis is one of the oldest, most convenient and least expensive methods of genotyping, but is limited by the availability of restriction endonuclease sites. Here we present a novel method of employing CRISPR/Cas-derived RNA-guided engineered nucleases (RGENs) in RFLP analysis. We prepare RGENs by complexing recombinant Cas9 protein derived from Streptococcus pyogenes with in vitro transcribed guide RNAs that are complementary to the DNA sequences of interest. Then, we genotype recurrent mutations found in cancer and small insertions or deletions (indels) induced in cultured cells and animals by RGENs and other engineered nucleases such as transcription activator-like effector nucleases (TALENs). Unlike T7 endonuclease I or Surveyor assays that are widely used for genotyping engineered nuclease-induced mutations, RGEN-mediated RFLP analysis can detect homozygous mutant clones that contain identical biallelic indel sequences and is not limited by sequence polymorphisms near the nuclease target sites.
URLPMID:25200087

The Cas9/sgRNA of the CRISPR/Cas system has emerged as a robust technology for targeted gene editing in various organisms, including plants, where Cas9/sgRNA-mediated small deletions/insertions at single cleavage sites have been reported in transient and stable transformations, although genetic transmission of edits has been reported only in Arabidopsis and rice. Large chromosomal excision between two remote nuclease-targeted loci has been reported only in a few non-plant species. Here we report in rice Cas9/sgRNA-induced large chromosomal segment deletions, the inheritance of genome edits in multiple generations and construction of a set of facile vectors for high-efficiency, multiplex gene targeting. Four sugar efflux transporter genes were modified in rice at high efficiency; the most efficient system yielding 87-100% editing in T0 transgenic plants, all with di-allelic edits. Furthermore, genetic crosses segregating Cas9/sgRNA transgenes away from edited genes yielded several genome-edited but transgene-free rice plants. We also demonstrated proof-of-efficiency of Cas9/sgRNAs in producing large chromosomal deletions (115-245 kb) involving three different clusters of genes in rice protoplasts and verification of deletions of two clusters in regenerated T0 generation plants. Together, these data demonstrate the power of our Cas9/sgRNA platform for targeted gene/genome editing in rice and other crops, enabling both basic research and agricultural applications.
URLPMID:27251395

CRISPR-Cas (clustered, regularly interspaced short palindromic repeats-CRISPR-associated proteins) is an adaptive immune system in many archaea and bacteria that cleaves foreign DNA on the basis of sequence complementarity. Here, using the geminivirus, beet severe curly top virus (BSCTV), transient assays performed in Nicotiana benthamiana demonstrate that the sgRNA-Cas9 constructs inhibit virus accumulation and introduce mutations at the target sequences. Further, transgenic Arabidopsis and N. benthamiana plants overexpressing sgRNA-Cas9 are highly resistant to virus infection.
URLPMID:26479191 [本文引用: 2]

Abstract Editing plant genomes without introducing foreign DNA into cells may alleviate regulatory concerns related to genetically modified plants. We transfected preassembled complexes of purified Cas9 protein and guide RNA into plant protoplasts of Arabidopsis thaliana, tobacco, lettuce and rice and achieved targeted mutagenesis in regenerated plants at frequencies of up to 46%. The targeted sites contained germline-transmissible small insertions or deletions that are indistinguishable from naturally occurring genetic variation.
URL [本文引用: 1]
URLPMID:27236699

Abstract KEY MESSAGE: The use of a meiosis I-specific promoter increased the efficiency of targeted mutagenesis and will facilitate the manipulation of homologous recombination. The CRISPR/Cas9 system has been harnessed for targeted engineering of eukaryotic genomes, including plants; however, CRISPR/Cas9 efficiency varies considerably in different plant tissues and species. In Arabidopsis, the generation of homozygous or bi-allelic mutants in the first (T1) generation is inefficient. Here, we used specific promoters to drive the expression of Cas9 during meiosis to maximize the efficiency of recovering heritable mutants in T1 plants. Our data reveal that the use of a promoter active in meiosis I resulted in high-efficiency (28 %) recovery of targeted mutants in the T1 generation. Moreover, this method enabled efficient simultaneous targeting of three genes for mutagenesis. Taken together, our results show that the use of meiosis-specific promoters will improve methods for functional genomic analysis and studying the molecular underpinnings of homologous recombination.
URLPMID:27875019

CRISPR‐Cpf1 is a newly identifiedCRISPR‐Cas system, and Cpf1 was recently engineered as a molecular tool for targeted genome editing in mammalian cells. To test whether the engineeredCRISPR‐Cpf1 system could induce the production of rice mutants, we selected two genome targets in theOsPDSandOsBELgenes. Our results show that both targets could be efficiently mutated in transgenic rice plants usingCRISPR‐Cpf1. We found that pre‐crRNAs with a full‐length direct repeat sequence exhibited considerably increased efficiencies compared with mature crRNAs. In addition, the specificity and transmission of the mutation were investigated, and the behaviours of crRNA‐Cpf1‐induced plant targeted genome mutagenesis were assessed. Taken together, our results indicate thatCRISPR‐Cpf1 expression via stable transformation can efficiently generate specific and heritable targeted mutations in rice and thereby constitutes a novel and important approach to specific and precise plant genome editing.
URLPMID:5599503 [本文引用: 1]

Precise genome editing of plants has the potential to reshape global agriculture through the targeted engineering of endogenous pathways or the introduction of new traits. To develop a CRISPR nuclease-based platform that would enable higher efficiencies of precise gene insertion or replacement, we screened the Cpf1 nucleases from Francisella novicida and Lachnospiraceae bacterium ND2006 for their capability to induce targeted gene insertion via homology directed repair. Both nucleases, in the presence of a guide RNA and repairing DNA template flanked by homology DNA fragments to the target site, were demonstrated to generate precise gene insertions as well as indel mutations at the target site in the rice genome. The frequency of targeted insertion for these Cpf1 nucleases, up to 8%, is higher than most other genome editing nucleases, indicative of its effective enzymatic chemistry. Further refinements and broad adoption of the Cpf1 genome editing technology have the potential to make a dramatic impact on plant biotechnology.
URL [本文引用: 2]

IAA2(Indole Acetic Acid 2)是拟南芥Aux/IAA生长素响应基因大家族中的一员,目前还没有它的突变体的报道,阻碍了对其功能和作用机制的深入研究。在CRISPR/Cas9基因组编辑技术中,1个sgRNA只能靶向基因的1个位点,有时基因敲除的效率并不高。为了提高敲除效率,本文在Golden-Gate克隆技术的基础上,通过两轮PCR扩增,将每3个sgRNA串联到同1个入门载体中,再将入门载体与含Cas9表达框的目标载体LR反应,获得最终的表达载体。结果表明,设计的6个sgRNA有4个发挥了作用,产生了碱基插入突变和大片段缺失突变等多种可遗传的突变。与单个sgRNA相比,多重sgRNA的基因敲除效率高、种系突变多;与其他构建多重sgRNA载体的方法相比,本方法具有快速、高效等优点。本文所得到的5个突变体为后续的IAA2功能研究提供了良好的材料。
URL [本文引用: 2]

IAA2(Indole Acetic Acid 2)是拟南芥Aux/IAA生长素响应基因大家族中的一员,目前还没有它的突变体的报道,阻碍了对其功能和作用机制的深入研究。在CRISPR/Cas9基因组编辑技术中,1个sgRNA只能靶向基因的1个位点,有时基因敲除的效率并不高。为了提高敲除效率,本文在Golden-Gate克隆技术的基础上,通过两轮PCR扩增,将每3个sgRNA串联到同1个入门载体中,再将入门载体与含Cas9表达框的目标载体LR反应,获得最终的表达载体。结果表明,设计的6个sgRNA有4个发挥了作用,产生了碱基插入突变和大片段缺失突变等多种可遗传的突变。与单个sgRNA相比,多重sgRNA的基因敲除效率高、种系突变多;与其他构建多重sgRNA载体的方法相比,本方法具有快速、高效等优点。本文所得到的5个突变体为后续的IAA2功能研究提供了良好的材料。
URLPMID:20508151 [本文引用: 1]

Site-directed mutagenesis in higher plants remains a significant technical challenge for basic research and molecular breeding. Here, we demonstrate targeted-gene inactivation for an endogenous gene in Arabidopsis using zinc finger nucleases (ZFNs). Engineered ZFNs for a stress-response regulator, the ABA-INSENSITIVE4 (ABI4) gene, cleaved their recognition sequences specifically in vitro, and ZFN genes driven by a heat-shock promoter were introduced into the Arabidopsis genome. After heat-shock induction, gene mutations with deletion and substitution in the ABI4 gene generated via ZFN-mediated cleavage were observed in somatic cells at frequencies as high as 3%. The homozygote mutant line zfn_abi4-1 1 for ABI4 exhibited the expected mutant phenotypes, i.e., ABA and glucose insensitivity. In addition, ZFN-mediated mutagenesis was applied to the DNA repair-deficient mutant plant, atku80. We found that lack of AtKu80, which plays a role in end-protection of dsDNA breaks, increased error-prone rejoining frequency by 2.6-fold, with increased end-degradation. These data demonstrate that an approach using ZFNs can be used for the efficient production of mutant plants for precision reverse genetics.
URLPMID:22565958

The article focuses on a study which examined the use of transcriptor activator-like (TAL) effector nucleases (TALEN) in editing a specific S gene in rice to prevent the virulence strategy of Xanthomonas oryzae. For TALEN-based disruption, the rice bacterial blight susceptibility gene was targeted. Results of the study showed that bacterial infection assays were resistant to infection by pathogenic Xoo.
URLPMID:3916745 [本文引用: 1]
URLPMID:26661595

Abstract The recently developed CRISPR/Cas9 system is a promising technology for targeted genome editing in a variety of species including plants. However, the first generation systems were designed to target one or two gene loci at a time. We designed a new multiplex CRISPR/Cas9 system that allows the co-expression of six sgRNA modules in one binary vector using a simple (three steps) cloning strategy in Arabidopsis. The transcription of the sgRNA modules is under the control of three different RNA Polymerase III-dependent promoters. We tested the efficiency of the new multiplex system by targeting six of the fourteen PYL families of ABA receptor genes in a single transformation experiment. One line with mutations in all six targeted PYLs was identified from 15 T1 plants. The mutagenesis frequency for the six individual PYL targets in the T1 lines ranged from 13 to 930002%. In the presence of ABA, the transgenic line identified as containing mutations in all six PYL genes produced the highest germination rate in the T2 progeny (370002%). Among these germinated seedlings, half of the analyzed plants (15/30) were homozygous mutants for at least four targeted genes and two plants (6.70002%) contained homozygous mutations in five of the targeted PYLs and the other targeted PYL had biallelic mutations. Homozygous sextuple mutants were identified in the T3 progeny and characterized together with previously described triple and sextuple PYL mutants. We anticipate that the application of this multiplex CRISPR/Cas9 system will strongly facilitate functional analysis of genes pathways and families.
URLPMID:26842991 [本文引用: 1]

CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins) is an adaptive immune system in bacteria and archaea to defend against foreign DNA fragments repeated invasions. Recently, it has been developed as a powerful targeted genome editing tool for a wide variety of species. However, its application on maize has only been tested in transiently expressed somatic cells or on limited number of stable transgenic T0plants. The exact efficiency and specificity of the CRISPR/Cas system in the highly complex maize genome has not been documented yet. Here we report an extensive study of the well-studied type II CRISPR-Cas9 system for targeted genome editing in maize, with the codon-optimized Cas9 protein and the short non-coding guide RNA generated through a functional maize U6 snRNA promoter. Targeted gene mutagenesis was detected for 90 loci by maize protoplast assay, with an average cleavage efficiency of 10.67%. Stable knockout transformants for maize phytoene synthase gene (PSY1) were obtained. Mutations occurred in germ cells can be stably inherited to the next generation. Moreover, no off-target effect was detected at the computationally predicted putative off-target loci. No significant difference between the transcriptomes of the Cas9 expressed and non-expressed lines was detected. Our results confirmed that the CRISPR-Cas9 could be successfully applied as a robust targeted genome editing system in maize.
URLPMID:27576893

The type II clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 system (CRISPR/Cas9) has been successfully applied to edit target genes in multiple plant species. However, it remains unknown whether this system can be used for genome editing in grape. In this study, we described genome editing and targeted gene mutation in ‘Chardonnay’ suspension cells and plants via the CRISPR/Cas9 system. Two single guide RNAs (sgRNAs) were designed to target distinct sites of the L-idonate dehydrogenase gene (IdnDH). CEL I endonuclease assay and sequencing results revealed the expected indel mutations at the target site, and a mutation frequency of 100% was observed in the transgenic cell mass (CM) as well as corresponding regenerated plants with expression of sgRNA1/Cas9. The majority of the detected mutations in transgenic CM were 1-bp insertions, followed by 1- to 3-nucleotide deletions. Off-target activities were also evaluated by sequencing the potential off-target sites, and no obvious off-target events were detected. Our results demonstrated that the CRISPR/Cas9 system is an efficient and specific tool for precise genome editing in grape.
URLPMID:28349358 [本文引用: 1]

Abstract KEY MESSAGE: CRISPR-Cas9/Cpf1 system with its unique gene targeting efficiency, could be an important tool for functional study of early developmental genes through the generation of successful knockout plants. The introduction and utilization of systems biology approaches have identified several genes that are involved in early development of a plant and with such knowledge a robust tool is required for the functional validation of putative candidate genes thus obtained. The development of the CRISPR-Cas9/Cpf1 genome editing system has provided a convenient tool for creating loss of function mutants for genes of interest. The present study utilized CRISPR/Cas9 and CRISPR-Cpf1 technology to knock out an early developmental gene EPFL9 (Epidermal Patterning Factor like-9, a positive regulator of stomatal development in Arabidopsis) orthologue in rice. Germ-line mutants that were generated showed edits that were carried forward into the T2 generation when Cas9-free homozygous mutants were obtained. The homozygous mutant plants showed more than an eightfold reduction in stomatal density on the abaxial leaf surface of the edited rice plants. Potential off-target analysis showed no significant off-target effects. This study also utilized the CRISPR-LbCpf1 (Lachnospiracae bacterium Cpf1) to target the same OsEPFL9 gene to test the activity of this class-2 CRISPR system in rice and found that Cpf1 is also capable of genome editing and edits get transmitted through generations with similar phenotypic changes seen with CRISPR-Cas9. This study demonstrates the application of CRISPR-Cas9/Cpf1 to precisely target genomic locations and develop transgene-free homozygous heritable gene edits and confirms that the loss of function analysis of the candidate genes emerging from different systems biology based approaches, could be performed, and therefore, this system adds value in the validation of gene function studies.
URLPMID:23417094 [本文引用: 1]

Transcription activator-like (TAL) effector nucleases (TALENs) can be readily engineered to bind specific genomic loci, enabling the introduction of precise genetic modifications such as gene knockouts and additions. Here we present a genome-scale collection of TALENs for efficient and scalable gene targeting in human cells. We chose target sites that did not have highly similar sequences elsewhere in the genome to avoid off-target mutations and assembled TALEN plasmids for 18,740 protein-coding genes using a high-throughput Golden-Gate cloning system. A pilot test involving 124 genes showed that all TALENs were active and disrupted their target genes at high frequencies, although two of these TALENs became active only after their target sites were partially demethylated using an inhibitor of DNA methyltransferase. We used our TALEN library to generate single- and double-gene-knockout cells in which NF-kappa B signaling pathways were disrupted. Compared with cells treated with short interfering RNAs, these cells showed unambiguous suppression of signal transduction.
URLPMID:4349094 [本文引用: 1]

Genome editing using engineered nucleases is used for targeted mutagenesis. But because genome editing does not target all loci with similar efficiencies, the mutation hit-rate at a given locus needs to be evaluated. The analysis of mutants obtained using engineered nucleases requires specific methods for mutation detection, and the enzyme mismatch cleavage method is used commonly for this purpose. This method uses enzymes that cleave heteroduplex DNA at mismatches and extrahelical loops formed by single or multiple nucleotides. Bacteriophage resolvases and single-stranded nucleases are used commonly in the assay but have not been compared side-by-side on mutations obtained by engineered nucleases. We present the first comparison of the sensitivity of T7E1 and Surveyor EMC assays on deletions and point mutations obtained by zinc finger nuclease targeting in frog embryos. We report the mutation detection limits and efficiencies of T7E1 and Surveyor. In addition, we find that T7E1 outperforms the Surveyor nuclease in terms of sensitivity with deletion substrates, whereas Surveyor is better for detecting single nucleotide changes. We conclude that T7E1 is the preferred enzyme to scan mutations triggered by engineered nucleases.
URLPMID:5335549 [本文引用: 1]

The complex allotetraploid genome is one of major challenges in cotton for repressing gene expression. Developing site-specific DNA mutation is the long-term dream for cotton breeding scientists. The clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system is emerging as a robust biotechnology for targeted-DNA mutation. In this study, two sgRNAs, GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, were designed in the identical genomic regions ofGhMYB25-like AandGhMYB25-like D, which were encoded by cotton A subgenome and the D subgenome, respectively, was assembled to direct Cas9-mediated allotetraploid cotton genome editing. High proportion (14.2 21.4%) CRISPR/Cas9-induced specific truncation events, either fromGhMYB25-like ADNA site or fromGhMYB25-like DDNA site, were detected in 50% examined transgenic cotton through PCR amplification assay and sequencing analyses. Sequencing results also demonstrated that 100% and 98.8% mutation frequency were occurred on GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 target site respectively. The off-target effect was evaluated by sequencing two putative off-target sites, which have 3 and 1 mismatched nucleotides with GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, respectively; all the examined samples were not detected any off-target-caused mutation events. Thus, these results demonstrated that CRISPR/Cas9 is qualified for generating DNA level mutations on allotetraploid cotton genome with high-efficiency and high-specificity.
URLPMID:23999856

Cell death and differentiation is a monthly research journal focused on the exciting field of programmed cell death and apoptosis. It provides a single accessible source of information for both scientists and clinicians, keeping them up-to-date with advances in the field. It encompasses programmed cell death, cell death induced by toxic agents, differentiation and the interrelation of these with cell proliferation.
URLPMID:24550464

The CRISPR (clustered regularly interspaced short palindromic repeat)/(CRISPR-associated) system has emerged as a powerful tool for targeted gene editing in many organisms, including . However, all of the reported studies in focused on either transient systems or the first generation after the CRISPR/system was stably transformed into . In this study we examined several plant generations with seven genes at 12 different target sites to determine the patterns, efficiency, specificity, and heritability of CRISPR/-induced gene mutations or corrections in . The proportion of bearing any mutations (chimeric, heterozygous, biallelic, or homozygous) was 71.2% at T1, 58.3% at T2, and 79.4% at T3 generations. CRISPR/-induced mutations were predominantly 1 bp insertion and short deletions. Gene modifications detected in T1 occurred mostly in somatic cells, and consequently there were no T1 that were homozygous for a gene modification event. In contrast, 22% of T2 were found to be homozygous for a modified gene. All homozygotes were stable to the next generation, without any new modifications at the target sites. There was no indication of any off-target mutations by examining the target sites and sequences highly homologous to the target sites and by in-depth whole-genome sequencing. Together our results show that the CRISPR/system is a useful tool for generating versatile and heritable modifications specifically at target genes in .
URLPMID:24854982

SummaryThe CRISPR/Cas9 system has been demonstrated to efficiently induce targeted gene editing in a variety of organisms including plants. Recent work showed that CRISPR/Cas9-induced gene mutations in Arabidopsis were mostly somatic mutations in the early generation, although some mutations could be stably inherited in later generations. However, it remains unclear whether this system will work similarly in crops such as rice. In this study, we tested in two rice subspecies 11 target genes for their amenability to CRISPR/Cas9-induced editing and determined the patterns, specificity and heritability of the gene modifications. Analysis of the genotypes and frequency of edited genes in the first generation of transformed plants (T0) showed that the CRISPR/Cas9 system was highly efficient in rice, with target genes edited in nearly half of the transformed embryogenic cells before their first cell division. Homozygotes of edited target genes were readily found in T0 plants. The gene mutations were passed to the next generation (T1) following classic Mendelian law, without any detectable new mutation or reversion. Even with extensive searches including whole genome resequencing, we could not find any evidence of large-scale off-targeting in rice for any of the many targets tested in this study. By specifically sequencing the putative off-target sites of a large number of T0 plants, low-frequency mutations were found in only one off-target site where the sequence had 1-bp difference from the intended target. Overall, the data in this study point to the CRISPR/Cas9 system being a powerful tool in crop genome engineering.
URL
URL
URLPMID:26089199

The CRISPR/Cas9 system is becoming an important genome editing tool for crop breeding. Although it has been demonstrated that target mutations can be transmitted to the next generation, their inheritance pattern has not yet been fully elucidated. Here, we describe the CRISPR/Cas9-mediated genome editing of four different rice genes with the help of online target-design tools. High-frequency mutagenesis and a large percentage of putative biallelic mutations were observed in T0 generations. Nonetheless, our results also indicate that the progeny genotypes of biallelic T0 lines are frequently difficult to predict and that the transmission of mutations largely does not conform to classical genetic laws, which suggests that the mutations in T0 transgenic rice are mainly somatic mutations. Next, we followed the inheritance pattern of T1 plants. Regardless of the presence of the CRISPR/Cas9 transgene, the mutations in T1 lines were stably transmitted to later generations, indicating a standard germline transmission pattern. Off-target effects were also evaluated, and our results indicate that with careful target selection, off-target mutations are rare in CRISPR/Cas9-mediated rice gene editing. Taken together, our results indicate the promising production of inheritable and 'transgene clean' targeted genome-modified rice in the T1 generation using the CRISPR/Cas9 system.
URLPMID:4507398

Abstract Recently, RNA-guided genome editing using the type II clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas) system has been applied to edit the plant genome in several herbaceous plant species. However, it remains unknown whether this system can be used for genome editing in woody plants. In this study, we describe the genome editing and targeted gene mutation in a woody species, Populus tomentosa Carr. via the CRISPR/Cas9 system. Four guide RNAs (gRNAs) were designed to target with distinct poplar genomic sites of the phytoene desaturase gene 8 (PtoPDS) which are followed by the protospacer-adjacent motif (PAM). After Agrobacterium-mediated transformation, obvious albino phenotype was observed in transgenic poplar plants. By analyzing the RNA-guided genome-editing events, 30 out of 59 PCR clones were homozygous mutants, 2 out of 59 were heterozygous mutants and the mutation efficiency at these target sites was estimated to be 51.7%. Our data demonstrate that the Cas9/sgRNA system can be exploited to precisely edit genomic sequence and effectively create knockout mutations in woody plants.
URLPMID:25917172

A robust CRISPR/Cas9 vector system for multiplex genome editing in monocot and dicot plants was developed. Multiple sgRNA expression cassettes can be assembled into the binary CRISPR/Cas9 vectors in one round of cloning. This system can uniformly, efficiently, and simultaneously produce multiple heritable mutations in rice and Arabidopsis by targeting multiple genes or genomic sites via single transformation events.
URLPMID:25747846

Dear Editor,The recent development of sequence-specific nuclease systems,i.e.,TALENs and CRISPR/Cas9,has made genomic targeting easier in many organisms including plants (Li et al.,2012;Cong et al.,2013;Joung and Sander,2013;Li,et al.,2013;Shan et al.,2013;Liang et al.,2014;Zhang et al.,2014).Mutations induced by CRISPR/Cas9 usually occur around the cleavage sites at three bases upstream of the protospacer-adjacent motif (PAM),producing insertion and deletion of nucleotides.For diploid organisms,such targeted mutations may happen in one or both homologous chromosomes.
URLPMID:26837606

Recently, CRISPR/Cas9 technology has emerged as a powerful approach for targeted genome modification in eukaryotic organisms from yeast to human cell lines. Its successful application in several plant species promises enormous potential for basic and applied plant research. However, extensive studies are still needed to assess this system in other important plant species, to broaden its fields of application and to improve methods. Here we showed that the CRISPR/Cas9 system is efficient in petunia (Petunia hybrid), an important ornamental plant and a model for comparative research. When PDS was used as target gene, transgenic shoot lines with albino phenotype accounted for 55.6%–87.5% of the total regenerated T0 Basta-resistant lines. A homozygous deletion close to 165kb in length can be readily generated and identified in the first generation. A sequential transformation strategy—introducing Cas9 and sgRNA expression cassettes sequentially into petunia—can be used to make targeted mutations with short indels or chromosomal fragment deletions. Our results present a new plant species amenable to CRIPR/Cas9 technology and provide an alternative procedure for its exploitation.
.
URLPMID:4955380

Genome editing using sequence-specific nucleases (SSNs) is rapidly being developed for genetic engineering in crop species. The utilization of zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated systems (CRISPR/Cas) for inducing double-strand breaks facilitates targeting of virtually any sequence for modification. Targeted mutagenesis via non-homologous end-joining (NHEJ) has been demonstrated extensively as being the preferred DNA repair pathway in plants. However, gene targeting via homologous recombination (HR) remains more elusive but could be a powerful tool for directed DNA repair. To overcome barriers associated with gene targeting, a geminivirus replicon (GVR) was used to deliver SSNs targeting the potatoACETOLACTATE SYNTHASE1(ALS1) gene and repair templates designed to incorporate herbicide-inhibiting point mutations within theALS1locus. Transformed events modified with GVRs held point mutations that were capable of supporting a reduced herbicide susceptibility phenotype, while events transformed with conventional T-DNAs held no detectable mutations and were similar to wild-type. Regeneration of transformed events improved detection of point mutations that supported a stronger reduced herbicide susceptibility phenotype. These results demonstrate the use of geminiviruses for delivering genome editing reagents in plant species, and a novel approach to gene targeting in a vegetatively propagated species.
URLPMID:4987624

Genome editing is a powerful technique for genome modification in molecular research and crop breeding, and has the great advantage of imparting novel desired traits to genetic resources. However, the genome editing of fruit tree plantlets remains to be established. In this study, we describe induction of a targeted gene mutation in the endogenous apple phytoene desaturase (PDS) gene using the CRISPR/Cas9 system. Four guide RNAs (gRNAs) were designed and stably transformed with Cas9 separately in apple. Clear and partial albino phenotypes were observed in 31.8% of regenerated plantlets for one gRNA, and bi-allelic mutations in applePDSwere confirmed by DNA sequencing. In addition, an 18-bp gRNA also induced a targeted mutation. These CRIPSR/Cas9 induced-mutations in the apple genome suggest activation of the NHEJ pathway, but with some involvement also of the HR pathway. Our results demonstrate that genome editing can be practically applied to modify the apple genome.
URLPMID:4982333

CRISPR/Cas9 genome editing strategy has been applied to a variety of species and the tRNA-processing system has been used to compact multiple gRNAs into one synthetic gene for manipulating multiple genes in rice. We optimized and introduced the multiplex gene editing strategy based on the tRNA-processing system into maize. Maize glycine-tRNA was selected to design multiple tRNA-gRNA units for the simultaneous production of numerous gRNAs under the control of one maize U6 promoter. We designed three gRNAs for simplex editing and three multiple tRNA-gRNA units for multiplex editing. The results indicate that this system not only increased the number of targeted sites but also enhanced mutagenesis efficiency in maize. Additionally, we propose an advanced sequence selection of gRNA spacers for relatively more efficient and accurate chromosomal fragment deletion, which is important for complete abolishment of gene function especially long non-coding RNAs (lncRNAs). Our results also indicated that up to four tRNA-gRNA units in one expression cassette design can still work in maize. The examples reported here demonstrate the utility of the tRNA-processing system-based strategy as an efficient multiplex genome editing tool to enhance maize genetic research and breeding. The online version of this article (doi:10.1186/s12896-016-0289-2) contains supplementary material, which is available to authorized users.
URLPMID:27543262

正Most of the important agronomic traits in crop plants,such as yield,quality and stress response,are quantitative and jointly controlled by many genomic loci or major genes.Improving these complex traits depends on the combination of beneficial alleles at the quantitative trait loci(QTLs).However,the conventional cross breeding method is extremely time-consuming and laborious for pyramiding multiple QTLs.In certain cases,this approach might
URLPMID:27628577

Grain yield is one of the most important and complex trait for genetic improvement in crops; it is known to be controlled by a number of genes known as quantitative trait loci(QTLs). In the past decade, many yield-contributing QTLs have been identified in crops.However, it remains unclear whether those QTLs confer the same yield performance in different genetic backgrounds. Here, we performed CRISPR/Cas_9-mediated QTL editing in five widely-cultivated rice varieties and revealed that the same QTL can have diverse, even opposing, effects on grain yield in different genetic backgrounds.
URLPMID:5062912 [本文引用: 1]

The ability to modulate levels of individual fatty acids within soybean oil has potential to increase shelf-life and frying stability and to improve nutritional characteristics. Commodity soybean oil contains high levels of polyunsaturated linoleic and linolenic acid, which contribute to oxidative instability – a problem that has been addressed through partial hydrogenation. However, partial hydrogenation increases levels oftrans-fatty acids, which have been associated with cardiovascular disease. Previously, we generated soybean lines with knockout mutations within fatty acid desaturase 2-1A (FAD2-1A) andFAD2-1Bgenes, resulting in oil with increased levels of monounsaturated oleic acid (18:1) and decreased levels of linoleic (18:2) and linolenic acid (18:3). Here, we stack mutations withinFAD2-1AandFAD2-1Bwith mutations in fatty acid desaturase 3A (FAD3A) to further decrease levels of linolenic acid. Mutations were introduced intoFAD3Aby directly delivering TALENs intofad2-1a fad2-1bsoybean plants. Oil fromfad2-1a fad2-1b fad3aplants had significantly lower levels of linolenic acid (2.502%), as compared tofad2-1a fad2-1bplants (4.702%). Furthermore, oil had significantly lower levels of linoleic acid (2.702% compared to 5.102%) and significantly higher levels of oleic acid (82.202% compared to 77.502%). Transgene-freefad2-1a fad2-1b fad3asoybean lines were identified. The methods presented here provide an efficient means for using sequence-specific nucleases to stack quality traits in soybean. The resulting product comprised oleic acid levels above 8002% and linoleic and linolenic acid levels below 302%. The online version of this article (doi:10.1186/s12870-016-0906-1) contains supplementary material, which is available to authorized users.
URLPMID:27874087

Abstract Hybrid rice breeding offers an important strategy to improve rice production, in which the cultivation of a male sterile line is the key to the success of cross-breeding. CRISPR/Cas9 systems have been widely used in target-site genome editing, whereas their application for crop genetic improvement has been rarely reported. Here, using the CRISPR/Cas9 system, we induced specific mutations in TMS5, which is the most widely applied thermo-sensitive genic male sterility (TGMS) gene in China, and developed new "transgene clean" TGMS lines. We designed 10 target sites in the coding region of TMS5 for targeted mutagenesis using the CRISPR/Cas9 system and assessed the potential rates of on- and off-target effects. Finally, we established the most efficient construct, the TMS5ab construct, for breeding potentially applicable "transgene clean" TGMS lines. We also discussed factors that affect the editing efficiency according to the characteristics of different target sequences. Notably, using the TMS5ab construct, we developed 11 new "transgene clean" TGMS lines with potential applications in hybrid breeding within only one year in both rice subspecies. The application of our system not only significantly accelerates the breeding of sterile lines but also facilitates the exploitation of heterosis.
[本文引用: 2]
URLPMID:28043782

Cpfl is a class 2/type V CRISPR effector that has been recently harnessed for genome editing(Zetsche et al.,2015;Hur et al.,2016;Kim et al.,2016).Cpfl recognizes thymidine-rich sequence as the protospacer-adjacent motif(PAM)at the 5'end of target sequences.In addition,Cpfl requires only a single shorter crRNA and
URLPMID:28049122

Abstract The most widely used gene editing technology-the CRISPR/Cas9 system-employs a bacterial monomeric DNA endonuclease known as clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) and single-guide RNA (sgRNA) that directs Cas9 to a complementary target DNA. However, introducing mutations into higher polyploid plant species, especially for species without genome information, has been difficult. Chrysanthemum morifolium (chrysanthemum) is one of the most important ornamental plants, but it is a hexaploid with a large genome; moreover, it lacks whole genome information. These characteristics hinder genome editing in chrysanthemum. In the present study, we attempted to perform gene editing using the CRISPR/Cas9 system to introduce mutations into chrysanthemum. We constructed transgenic chrysanthemum plants expressing the yellowish-green fluorescent protein gene from Chiridius poppei (CpYGFP) and targeted CpYGFP for gene editing. We compared the activity of a cauliflower mosaic virus (CaMV) 35S promoter and parsley ubiquitin promoter in chrysanthemum calli and chose the parsley ubiquitin promoter to drive Cas9 We selected two sgRNAs to target different positions in the CpYGFP gene and obtained transgenic calli containing mutated CpYGFP genes (CRISPR-CpYGFP-chrysanthemum). A DNA sequencing analysis and fluorescence observations indicated that cells containing the mutated CpYGFP gene grew independently of cells containing the original CpYGFP gene in one callus. We finally obtained the CRISPR-CpYGFP-chrysanthemum shoot containing a mutation in the CpYGFP sequence. This is the first report of gene editing using the CRISPR/Cas9 system in chrysanthemum and sheds light on chrysanthemum genome editing. The Author 2017. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
URL
URLPMID:28295376

Abstract Genetic imprinting refers to the unequal expression of paternal and maternal alleles of a gene in sexually reproducing organisms, including mammals and flowering plants. Although many imprinted genes have been identified in plants, the functions of these imprinted genes have remained largely uninvestigated. We report genome-wide analysis of gene expression, DNA methylation and small RNAs in the rice endosperm and functional tests of five imprinted genes during seed development using Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated gene9 (CRISPR/Cas9) gene editing technology. In the rice endosperm, we identified 162 maternally expressed genes (MEGs) and 95 paternally expressed genes (PEGs), which were associated with miniature inverted-repeat transposable elements, imprinted differentially methylated loci and some 21-22 small interfering RNAs (siRNAs) and long noncoding RNAs (lncRNAs). Remarkably, one-third of MEGs and nearly one-half of PEGs were associated with grain yield quantitative trait loci. Most MEGs and some PEGs were expressed specifically in the endosperm. Disruption of two MEGs increased the amount of small starch granules and reduced grain and embryo size, whereas mutation of three PEGs reduced starch content and seed fertility. Our data indicate that both MEGs and PEGs in rice regulate nutrient metabolism and endosperm development, which optimize seed development and offspring fitness to facilitate parental-offspring coadaptation. These imprinted genes and mechanisms could be used to improve the grain yield of rice and other cereal crops.
URLPMID:28303459

正Dear Editor,The CRISPR/Cas9(clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9)system is revolutionizing genome editing due to its high efficiency,low cost,design simplicity and versatility.However,introduction of a point mutation at a desired position remains a great challenge in plant genome engineering.Currently,point mutation in plants was achieved by incorporating a
URLPMID:27747971

The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR‐associated protein 9 nuclease (Cas9) system depends on a guide RNA (gRNA) to specify its target. By efficiently co‐expressing multiple gRNAs that target different genomic sites, the polycistronic tRNA‐gRNA gene (PTG) strategy enables multiplex gene editing in the family of closely related rice mitogen‐activated protein kinase (MPK) genes. In this study, we identified MPK1 and MPK6 (Arabidopsis AtMPK6 and AtMPK4 orthologues, respectively) as essential genes for rice development by finding the preservation of MPK functional alleles and normal phenotypes in CRISPR‐edited mutants. The true knock‐out mutants of MPK1 were severely dwarfed and sterile, and homozygous mpk1 seeds from heterozygous parents were defective in embryo development. By contrast, heterozygous mpk6 mutant plants completely failed to produce homozygous mpk6 seeds. In addition, functional importance of specific MPK features could be evaluated by characterizing CRISPR‐induced allelic variation in the conserved kinase domain of MPK6. By simultaneously targeting two to eight genomic sites in the closely related MPK genes, we demonstrated 45 to 86% frequency of biallelic mutations and successful creation of single, double and quadruple gene mutants. Indels and fragment deletion were both stably inherited to the next generations and transgene‐free mutants of rice MPK genes were readily obtained via genetic segregation, thereby eliminating any positional effects of transgene insertions. Taken together, our study reveals essentiality of MPK1 and MPK6 in rice development and enables functional discovery of the previously inaccessible genes or domains because their phenotypes are masked by lethality or redundancy.
URLPMID:28315751
URLPMID:28291639 [本文引用: 1]

@@
URLPMID:28315752
URLPMID:5428673

Genome editing has emerged as a technology with a potential to revolutionize plant breeding. In this study, we report on generating, in less than ten months, Tomelo, a non-transgenic tomato variety resistant to the powdery mildew fungal pathogen using the CRISPR/Cas9 technology. We used whole-genome sequencing to show that Tomelo does not carry any foreign DNA sequences but only carries a deletion that is indistinguishable from naturally occurring mutations. We also present evidence for CRISPR/Cas9 being a highly precise tool, as we did not detect off-target mutations in Tomelo. Using our pipeline, mutations can be readily introduced into elite or locally adapted tomato varieties in less than a year with relatively minimal effort and investment.
URLPMID:5404177

Targeted genome editing with the CRISPR/Cas9 system has been used extensively for the selective mutation of plant genes. Here we used CRISPR/Cas9 to disrupt the putative barley (Hordeum vulgarecv. olden Promise ) endo-N-acetyl- -D-glucosaminidase (ENGase) gene. Five single guide RNAs (sgRNAs) were designed for different target sites in the upstream part of the ENGase coding region. Targeted fragment deletions were induced by co-bombarding selected combinations of sgRNA with wild-type cas9 using separate plasmids, or by co-infection with separateAgrobacterium tumefacienscultures. Genotype screening was carried out in the primary transformants (T0) and their T1 progeny to confirm the presence of site-specific small insertions and deletions (indels) and genomic fragment deletions between pairs of targets. Cas9-induced mutations were observed in 78% of the plants, a higher efficiency than previously reported in barley. Notably, there were differences in performance among the five sgRNAs. The induced indels and fragment deletions were transmitted to the T1 generation, and transgene free (sgRNA:cas9 negative) genome-edited homozygous ENGase knock outs were identified among the T1 progeny. We have therefore demonstrated that mutant barley lines with a disrupted endogenous ENGase and defined fragment deletions can be produced efficiently using the CRISPR/Cas9 system even when this requires co-transformation with multiple plasmids by bombardment orAgrobacterium-mediated transformation. We confirm the specificity and heritability of the mutations and the ability to efficiently generate homozygous mutant T1 plants.
URLPMID:28509421

Abstract Flowering is an indication of the transition from vegetative growth to reproductive growth and has considerable effects on the life cycle of soybean (Glycine max). In this study, we employed the CRISPR/Cas9 system to specifically induce targeted mutagenesis of GmFT2a, an integrator in the photoperiod flowering pathway in soybean. The soybean cultivar Jack was transformed with three sgRNA/Cas9 vectors targeting different sites of endogenous GmFT2a via Agrobacterium tumefaciens-mediated transformation. Site-directed mutations were observed at all targeted sites by DNA sequencing analysis. T1 generation soybean plants homozygous for null alleles of GmFT2a frameshift mutated by a 1-bp insertion or short deletion exhibited late flowering under natural conditions (summer) in Beijing, China (N39 58', E116 20'). We also found that the targeted mutagenesis was stably heritable in the following T2 generation, and the homozygous GmFT2a mutants exhibited late flowering under both long-day and short-day conditions. We identified some "transgene-clean" soybean plants that were homozygous for null alleles of endogenous GmFT2a and without any transgenic element from the T1 and T2 generations. These "transgene-clean" mutants of GmFT2a may provide materials for more in-depth research of GmFT2a functions and the molecular mechanism of photoperiod responses in soybean. They will also contribute to soybean breeding and regional introduction. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
URLPMID:28688132

Abstract The CRISPR/Cas9-based genome editing tool has been used in diverse applications related to plant research, including for crop improvement (Sun et02al., 2016; Liu et02al., 2017). Mutant plants may be generated via transient transformations or DNA-free editing (Liang et02al., 2017). However, plant genomes are often edited during the production of transgenic plants, in a process that involves the identification of targeted edits in regenerated T 0 plants and the subsequent elimination of transgenes in T 1 plants (Sun et02al., 2016). This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
URLPMID:28502081

Wheat (Triticum aestivum L.) incurs significant yield losses from powdery mildew, a major fungal disease caused by Blumeria graminis f. sp. tritici, (Bgt). ENHANCED DISEASE RESISTANCE1 (EDR1) plays a negative role in the defense response against powdery mildew in Arabidopsis thaliana; however, the edr1 mutant does not show constitutively activated defense responses. This makes EDR1 an ideal target for approaches using new genome‐editing tools to improve powdery mildew resistance. We cloned TaEDR1 from hexaploid wheat and found high similarity among the three homoeologs of EDR1. Knock‐down of TaEDR1 by virus‐induced gene silencing (VIGS) or RNA interference (RNAi) enhanced resistance to powdery mildew, indicating that TaEDR1 negatively regulates powdery mildew resistance in wheat. We used CRISPR/Cas9 technology to generate Taedr1 wheat plants by simultaneous modification of the three homoeologs of wheat EDR1. No off‐target mutations were detected in the Taedr1 mutant plants. The Taedr1 plants were resistant to powdery mildew and did not show mildew‐induced cell death. Our study represents the successful generation of a potentially valuable trait using genome‐editing technology in wheat and provides germplasm for disease resistance breeding.
URLPMID:28584922

Key message Using CRISPR/Cas9, we successfully deleted large fragments of the yield-related gene DENSE AND ERECT PANICLE1 in Indica rice at relatively high frequency and generated gain-of-function...
URLPMID:28605576 [本文引用: 1]

Abstract Clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) is a revolutionary technology that enables efficient genomic modification in many organisms. Currently, the wide use of Streptococcus pyogenes Cas9 (SpCas9) primarily recognises sites harbouring a canonical NGG protospacer adjacent motif (PAM). The newly developed VQR (D1135V/R1335Q/T1337R) variant of Cas9 has been shown to cleave sites containing NGA PAM in rice, which greatly expanded the range of genome editing. However, the low editing efficiency of the VQR variant remains, which limits its wide application in genome editing. In this study, by modifying the single guide RNA (sgRNA) structure and using strong endogenous promoters, we significantly increased the editing efficiency of the VQR variant. The modified CRISPR-Cas9-VQR system provides a robust toolbox for multiplex genome editing at sites containing non-canonical NGA PAM. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
URL [本文引用: 1]
URLPMID:28645639
URLPMID:28928381

Abstract Quickly and precisely gain genetically enhanced breeding elites with value-adding performance traits is desired by the crop breeders all the time. The present of gene editing technologies, especially the CRISPR/Cas9 system with the capacities of efficiency, versatility and multiplexing provides a reasonable expectation towards breeding goals. For exploiting possible application to accelerate the speed of process at breeding by CRISPR/Cas9 technology, in this study, the Agrobacterium tumefaciens-mediated CRISPR/Cas9 system transformation method was used for obtaining tomato ALC gene mutagenesis and replacement, in absence and presence of the homologous repair template. The average mutation frequency (72.73%) and low replacement efficiency (7.69%) were achieved in T 0 transgenic plants respectively. None of homozygous mutation was detected in T 0 transgenic plants, but one plant carry the heterozygous genes (Cas9/*-ALC/alc) was stably transmitted to T 1 generations for segregation and genotyping. Finally, the desired alc homozygous mutants without T-DNA insertion (*/*-alc/alc) in T 1 generations were acquired and further confirmed by genotype and phenotype characterization, with highlight of excellent storage performance, thus the recessive homozygous breeding elites with the character of long-shelf life were generated. Our results support that CRISPR/Cas9-induced gene replacement via HDR provides a valuable method for breeding elite innovation in tomato.
URLPMID:28645638
URLPMID:28921815 [本文引用: 1]

Coeliac disease is an autoimmune disorder triggered in genetically predisposed individuals by the ingestion of gluten proteins from wheat, barley and rye. The -gliadin gene family of wheat contains four highly stimulatory peptides, of which the 33-mer is the main immunodominant peptide in patients with coeliac. We designed two sgRNAs to target a conserved region adjacent to the coding sequence for the 33-mer in the -gliadin genes. Twenty-one mutant lines were generated, all showing strong reduction in -gliadins. Up to 35 different genes were mutated in one of the lines of the 45 different genes identified in the wild type, while immunoreactivity was reduced by 85%. Transgene-free lines were identified, and no off-target mutations have been detected in any of the potential targets. The low-gluten, transgene-free wheat lines described here could be used to produce low-gluten foodstuff and serve as source material to introgress this trait into elite wheat varieties.
URLPMID:28864834

No Abstract available for this article.
URLPMID:28873302 [本文引用: 1]

Abstract Drought stress is one of the most destructive environmental factors that affect tomato plants adversely. Mitogen-activated protein kinases (MAPKs) are important signaling molecules that respond to drought stress. In this study, SlMAPK3 was induced by drought stress, and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (CRISPR/Cas9) system was utilized to generate slmapk3 mutants. Two independent T1 transgenic lines and wild-type (WT) tomato plants were used for analysis of drought tolerance. Compared with WT plants, slmapk3 mutants exhibited more severe wilting symptom, higher hydrogen peroxide content, lower antioxidant enzymes activities, and suffered more membrane damage under drought stress. Furthermore, knockout of SlMAPK3 led to up- or down-regulated expressions of drought stress-responsive genes including SlLOX, SlGST, and SlDREB. The results suggest that SlMAPK3 is involved in drought response in tomato plants by protecting cell membranes from oxidative damage and modulating transcription of stress-related genes.
URLPMID:5651273

CRISPR/Cas9 has become a powerful genome-editing tool for introducing genetic changes into crop species. In order to develop capacity for CRISPR/Cas9 technology in the tropical staple cassava (Manihot esculenta), thePhytoene desaturase(MePDS) gene was targeted in two cultivars using constructs carrying gRNAs targeting two sequences withinMePDSexon 13. AfterAgrobacterium-mediated delivery of CRISPR/Cas9 reagents into cassava cells, both constructs induced visible albino phenotypes within cotyledon-stage somatic embryos regenerating on selection medium and the plants regenerated therefrom. A total of 58 (cv. 60444) and 25 (cv. TME 204) plant lines were recovered, of which 38 plant lines (19 from each cultivar) were analyzed for mutagenesis. The frequency of plant lines showing albino phenotype was high, ranging from 90 to 100% in cv. TME 204. Observed albino phenotypes were comprised of full albinos devoid of green tissue and chimeras containing a mixture of white and green tissues. Sequence analysis revealed that 38/38 (100%) of the plant lines examined carried mutations at the targetedMePDSsite, with insertions, deletions, and substitutions recorded. One putatively mono-allelic homozygous line (1/19) was found from cv. 60444, while 1 (1/19) and 4 (4/19) putatively bi-allelic homozygous lines were found in 60444 and TME204, respectively. The remaining plant lines, comprised mostly of the chimeras, were found to be putatively heterozygous. We observed minor (1 bp) nucleotide substitutions and or deletions upstream of the 5 and or downstream of the 3 targetedMePDSregion. The data reported demonstrates that CRISPR/Cas9-mediated genome editing of cassava is highly efficient and relatively simple, generating multi-allelic mutations in both cultivars studied. Modification ofMePDSdescribed here generates visually detectable mutated events in a relatively short time frame of 6 8 weeks, and does not require sequencing to confirm editing at the target. It therefore provides a valuable platform to facilitate rapid assessment and optimization of CRISPR/Cas9 and other genome-editing technologies in cassava.
URLPMID:28919077

Major advances in crop yields are needed in the coming decades. However, plant breeding is currently limited by incremental improvements in quantitative traits that often rely on laborious selection of rare naturally occurring mutations in gene-regulatory regions. Here, we demonstrate that CRISPR/Cas9 genome editing of promoters generates diverse cis-regulatory alleles that provide beneficial quantitative variation for breeding. We devised a simple genetic scheme, which exploits trans-generational heritability of Cas9 activity in heterozygous loss-of-function mutant backgrounds, to rapidly evaluate the phenotypic impact of numerous promoter variants for genes regulating three major productivity traits in tomato: fruit size, inflorescence branching, and plant architecture. Our approach allows immediate selection and fixation of novel alleles in transgene-free plants and fine manipulation of yield components. Beyond a platform to enhance variation for diverse agricultural traits, our findings provide a foundation for dissecting complex relationships between gene-regulatory changes and control of quantitative traits.
URLPMID:29157799

正Tomato(Solanum lycopersicum)is the leading vegetable crop worldwide and an essential component of a healthy diet(Lin et al.,2014;Du et al.,2017).Fruit color is regarded as one of the most important commercial traits in tomato(The Tomato Genome Consortium,2012).Consumers in different regions have different color preferences.For example,European and American
URLPMID:29161464

Abstract The CRISPR/Cas9 system has greatly improved our ability to engineer targeted mutations in eukaryotic genomes. While CRISPR/Cas9 appears to work universally, the efficiency of targeted mutagenesis and the adverse generation of off-target mutations vary greatly between different organisms. In this study, we report that Arabidopsis plants subjected to heat stress at 3700°C show much higher frequencies of CRISPR-induced mutations compared to plants grown continuously at the standard temperature (2200°C). Using quantitative assays relying on GFP reporter genes, we found that targeted mutagenesis by CRISPR/Cas9 in Arabidopsis is increased by ~5-fold in somatic tissues and up to 100-fold in the germline upon heat treatment. This effect of temperature on the mutation rate is not limited to Arabidopsis, as we observed a similar increase in targeted mutations by CRISPR/Cas9 in Citrus plants exposed to heat stress at 3700°C. In vitro assays demonstrate that Cas9 from Streptococcus pyogenes (SpCas9) is more active in creating double-stranded DNA breaks at 3700°C than at 2200°C, thus indicating a potential contributing mechanism for the in vivo effect of temperature on CRISPR/Cas9. This study reveals the importance of temperature in modulating SpCas9 activity in eukaryotes, and provides a simple method to increase on-target mutagenesis in plants using CRISPR/Cas9. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
URLPMID:29223136

Effective weed control can protect yields of cassava (Manihot esculenta) storage roots. Farmers could benefit from using herbicide with a tolerant cultivar. We applied traditional transgenesis and gene editing to generate robust glyphosate tolerance in cassava. By comparing promoters regulating expression of transformed 5‐enolpyruvylshikimate‐3‐phosphate synthase (EPSPS) genes with various paired amino acid substitutions, we found that strong constitutive expression is required to achieve glyphosate tolerance duringin02vitroselection and in whole cassava plants. Using strategies that exploit homologous recombination (HR) and nonhomologous end‐joining (NHEJ)DNArepair pathways, we precisely introduced the best‐performing allele into the cassava genome, simultaneously creating a promoter swap and dual amino acid substitutions at the endogenousEPSPSlocus. PrimaryEPSPS‐edited plants were phenotypically normal, tolerant to high doses of glyphosate, with some free of detectable02T‐DNAintegrations. Our methods demonstrate an editing strategy for creating glyphosate tolerance in crop plants and demonstrate the potential of gene editing for further improvement of cassava.
URL
URLPMID:29210506

Abstract In rice, amylose content (AC) is controlled by a single dominant Waxy gene. We used CRISPR/Cas9 to introduce a loss-of-function mutation into the Waxy gene in two widely-cultivated elite japonica varieties. Our results show that mutations in the Waxy gene reduce AC and convert the rice into glutinous ones without affecting other desirable agronomic traits, offering an effective and easy strategy to improve glutinosity in elite varieties. Importantly, we successfully removed the transgenes from the progeny. Our study provides an example of generating improved crops with potential for commercialization, by editing a gene of interest directly in elite crop varieties.
URLPMID:29382569

ACCEPTED MANUSCRIPT codon-optimized for expression in rice (Figure S1) and attached to 5 terminal end of Cas9n-NLS using XTEN linker (Figure 1A and 1B). Therefore, it is likely that the editing window of hAID* would be restricted to the sgRNA-targeting site similar to the case of APOBEC1. To be noted, UGI was excluded at this time since more types of nucleotide conversions, produced in uracil N-glycosylase (UDG)-initiated base excision repair (BER), would be anticipated.The hAID* -XTEN-Cas9n-NLS chimeric gene (termed rBE5) were first tested in rice leaf sheath protoplast, with the sgRNAs targeting a GCAC-containing ApaLI restriction site in OsRLCK185 and a TCC-containing BamHI restriction site in OsCERK1, respectively. Sequencing data showed that distinct mutations with high frequency of C>T conversions occurred in the editing window (Figure S2A-S2H), suggesting rBE5 functions on GC, AC, TC as well as CC in rice cells.As a follow-up to our previous study of the rBE3 system (Ren et al., 2017), rBE3 gene in the binary vector pUbi:rBE3 was replaced by rBE5 gene, namely pUbi:rBE5, and then further tested for efficiency in stable transgenic rice plants through Agrobacterium-mediated transformation. Pi-d2, an agriculturally important rice blast R gene in which a GC-containing region was resistant to rBE3-mediated editing in our earlier experiment, was first chosen for targeting (Figure 1C). As reported previously, a single amino acid substitution at position 441 in the recessive allele of Pi-d2 gene results in loss of resistance to Magnaporthe oryzae (Chen et al., 2006). Therefore, we assumed that introducing a G>A mutation (M441I) in endogenous pi-d2kit using rBE5 could recover its biological function. The same protospacer for rBE3 was used in this experiment, totally 26 independent transgenic lines of Kitaake were obtained after transformation and genotyped subsequently by Sanger sequencing. 8 heterozygous lines (30.8% efficiency) carrying a desired single G to A conversion at position -17 upstream of PAM were identified (Figure 1D-1E, Figure S3A), suggesting that rBE5 is much more efficient on target C that immediately follow a G than rBE3 does. We assumed that the successful gene correction of pi-d2 would be attributable to both the specificity and the improved activity of hAID* . Remarkably, pi-d2 alleles with Indel mutations were also detected in 5 lines (Figure S3B-S3C).
URLPMID:29331077

Kiwifruit is an important fruit crop; however, technologies for its functional genomic and molecular improvement are limited. The clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR‐associated protein (Cas) system has been successfully applied to genetic improvement in many crops, but its editing capability is variable depending on the different combinations of the synthetic guide RNA (sgRNA) and Cas9 protein expression devices. Optimizing conditions for its use within a particular species is therefore needed to achieve highly efficient genome editing. In the present study, we developed a new cloning strategy for generating paired‐sgRNA/Cas9 vectors containing four sgRNAs targeting the kiwifruit phytoene desaturase gene (AcPDS). Comparing to the previous method of paired‐sgRNA cloning, our strategy only requires the synthesis of two gRNA‐containing primers which largely reduces the cost. We further compared efficiencies of paired‐sgRNA/Cas9 vectors containing different sgRNA‐expression devices, including both the polycistronic tRNA‐sgRNA cassette (PTG) and the traditional CRISPR expression cassette. We found the mutagenesis frequency of the PTG/Cas9 system were 10‐fold higher than that of the CRISPR/Cas9 system, coinciding with the relative expressions of sgRNAs in two different expression cassettes. In particular, we identified large chromosomal fragment deletions induced by the paired sgRNAs of the PTG/Cas9 system. Finally, as expected, we found both systems can successfully induce the albino phenotype of kiwifruit plantlets regenerated from the G418‐resistance callus lines. We conclude that the PTG/Cas9 system is a more powerful system than the traditional CRISPR/Cas9 system for kiwifruit genome editing, which provides valuable clues for optimizing CRISPR/Cas9 editing system in other plants.
URLPMID:29392632

Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (CRISPR/Cas9) technology provides an efficient tool for editing the genomes of plants, animals and...
URLPMID:29476916

Soil salinity is a major abiotic stress in plant agriculture worldwide. This has led to research into salt tolerance with the aim of improving crop plants. However, salt tolerance might have much wider implications because transgenic salt-tolerant plants often also tolerate other stresses including chilling, freezing, heat and drought. Unfortunately, suitable genetic model systems have been... [Show full abstract]
URL
URL

蔗糖非发酵相关激酶(sucrose non-fermenting related protein kinases,Sn RKs)是广泛存在于植物中的一类Ser/Thr蛋白激酶,在植物的生长、发育、代谢和抗逆等方面具有重要调节作用。大豆(Glycine max L.)基因组中含有4个Sn RK1同源基因,其中GmSnRK1.1和GmSnRK1.2为两个主要表达基因,可能参与大豆多种抗逆途径。为解析大豆GmSnRK1.1和GmSnRK1.2对ABA及碱胁迫的响应,本研究构建了双靶点CRISPR载体定向敲除GmSnRK1.1和GmSnRK1.2基因,利用发根农杆菌(Agrobacterium rhizogenes)介导大豆遗传转化,获得双基因敲除突变体毛状根,经测序鉴定双基因突变率为48.6%;同时,利用实验室前期构建的植物超量表达载体获得超量表达GmSnRK1基因大豆毛状根。经25μmol/L ABA处理15 d,对照组和超量表达毛状根的生长受到明显抑制,其根长与根鲜重均显著低于双基因敲除突变体毛状根;经50 mmol/L Na HCO3处理15 d,对照组和双基因敲除突变体毛状根的生长受到明显抑制,其根长与根鲜重均显著低于超量表达毛状根。本研究建立的CRISPR/Cas9系统能够有效地对大豆进行GmSnRK1.1和GmSnRK1.2双基因敲除,基因敲除突变降低了植物对ABA的敏感性及对碱胁迫的耐性,研究结果初步说明Sn RK1激酶在植物响应非生物胁迫中具有重要作用。
URL

蔗糖非发酵相关激酶(sucrose non-fermenting related protein kinases,Sn RKs)是广泛存在于植物中的一类Ser/Thr蛋白激酶,在植物的生长、发育、代谢和抗逆等方面具有重要调节作用。大豆(Glycine max L.)基因组中含有4个Sn RK1同源基因,其中GmSnRK1.1和GmSnRK1.2为两个主要表达基因,可能参与大豆多种抗逆途径。为解析大豆GmSnRK1.1和GmSnRK1.2对ABA及碱胁迫的响应,本研究构建了双靶点CRISPR载体定向敲除GmSnRK1.1和GmSnRK1.2基因,利用发根农杆菌(Agrobacterium rhizogenes)介导大豆遗传转化,获得双基因敲除突变体毛状根,经测序鉴定双基因突变率为48.6%;同时,利用实验室前期构建的植物超量表达载体获得超量表达GmSnRK1基因大豆毛状根。经25μmol/L ABA处理15 d,对照组和超量表达毛状根的生长受到明显抑制,其根长与根鲜重均显著低于双基因敲除突变体毛状根;经50 mmol/L Na HCO3处理15 d,对照组和双基因敲除突变体毛状根的生长受到明显抑制,其根长与根鲜重均显著低于超量表达毛状根。本研究建立的CRISPR/Cas9系统能够有效地对大豆进行GmSnRK1.1和GmSnRK1.2双基因敲除,基因敲除突变降低了植物对ABA的敏感性及对碱胁迫的耐性,研究结果初步说明Sn RK1激酶在植物响应非生物胁迫中具有重要作用。
URL

【目的】直链淀粉含量与稻米品质密切相关。Wx基因是控制水稻直链淀粉合成的主效基因,通过对Wx基因定点编辑以获得稳定遗传、直链淀粉含量适宜的突变体。【方法】构建CRISPR/Cas9表达载体p GK03-Wx-g RNA(靶点1和2分别在Wx基因第1和第2外显子),利用工程菌EHA105遗传转化超级稻楚粳27,潮霉素筛选获得转化株系,对转化株系及其后代进行分子检测、测序、基因表达和遗传稳定性分析以及直链淀粉含量测定。【结果】获得9个独立的T_0代转化株系,靶点1(L1~L5)5个株系,突变频率100%,靶点2(L6~L9)4个株系,突变频率75%。由T_0代突变体衍生出T_1和T_2代株系,测序发现T_0、T_1和T_2代株系出现缺失(单、双、多碱基缺失)和单碱基插入两种突变类型;T_0至T_1代部分株系(L1、L2、L3和L6)发生再编辑,T_1至T_2代遗传稳定。与野生型相比,突变株系RNA水平Wx基因表达量显著下降(P0.01),稻米直链淀粉含量显著降低(P0.01),从17.5%降到1.93%。【结论】利用CRISPR/Cas9系统成功编辑水稻Wx基因,获得了稳定遗传、低直链淀粉含量的突变体,为稻米品质改良提供了材料。
URL

【目的】直链淀粉含量与稻米品质密切相关。Wx基因是控制水稻直链淀粉合成的主效基因,通过对Wx基因定点编辑以获得稳定遗传、直链淀粉含量适宜的突变体。【方法】构建CRISPR/Cas9表达载体p GK03-Wx-g RNA(靶点1和2分别在Wx基因第1和第2外显子),利用工程菌EHA105遗传转化超级稻楚粳27,潮霉素筛选获得转化株系,对转化株系及其后代进行分子检测、测序、基因表达和遗传稳定性分析以及直链淀粉含量测定。【结果】获得9个独立的T_0代转化株系,靶点1(L1~L5)5个株系,突变频率100%,靶点2(L6~L9)4个株系,突变频率75%。由T_0代突变体衍生出T_1和T_2代株系,测序发现T_0、T_1和T_2代株系出现缺失(单、双、多碱基缺失)和单碱基插入两种突变类型;T_0至T_1代部分株系(L1、L2、L3和L6)发生再编辑,T_1至T_2代遗传稳定。与野生型相比,突变株系RNA水平Wx基因表达量显著下降(P0.01),稻米直链淀粉含量显著降低(P0.01),从17.5%降到1.93%。【结论】利用CRISPR/Cas9系统成功编辑水稻Wx基因,获得了稳定遗传、低直链淀粉含量的突变体,为稻米品质改良提供了材料。
URL [本文引用: 2]

TALEN(Transcription activator-like effector nucleases)系统和CRISPR/Cas9(Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9)系统是当前广泛应用的两大基因组编辑技术。本文比较分析了水稻(Oryza sativa L.)中这两大系统诱导突变的突变率、突变类型、突变位置、突变时间和遗传模式,发现TALEN系统和CRISPR/Cas9系统均可在T_0代水稻中有效诱发位点特异性突变,且CRISPR/Cas9的突变率更高。两大系统都以诱发10 bp以内的In Del突变为主,TALEN系统易诱导10 bp以内的缺失突变,而CRISPR/Cas9系统易诱导1 bp的插入突变,且CRISPR/Cas9系统诱发的DNA双链断裂(Double-strand breaks,DSBs)位置更加精确。此外,DSBs在修复过程中能以低频同源重组(Homologous recombination,HR)途径修复,产生DNA片段重复突变。对于相邻双靶点CRISPR/Cas9系统而言,双靶点间的DNA片段可发生缺失或倒位,这种双靶点间DNA片段突变的发生频率与双靶点各自的突变率无正相关性。两大系统诱导的突变最早发生在已转化的愈伤组织中,少量发生在水稻的体细胞中,导致了纯合突变、杂合突变、双等位突变和嵌合突变4种遗传模式,其中双等位突变比例最高,嵌合突变比例最低。除嵌合突变外,纯合突变、杂合突变、双等位突变的突变序列均可稳定遗传给下一代。
URL [本文引用: 2]

TALEN(Transcription activator-like effector nucleases)系统和CRISPR/Cas9(Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9)系统是当前广泛应用的两大基因组编辑技术。本文比较分析了水稻(Oryza sativa L.)中这两大系统诱导突变的突变率、突变类型、突变位置、突变时间和遗传模式,发现TALEN系统和CRISPR/Cas9系统均可在T_0代水稻中有效诱发位点特异性突变,且CRISPR/Cas9的突变率更高。两大系统都以诱发10 bp以内的In Del突变为主,TALEN系统易诱导10 bp以内的缺失突变,而CRISPR/Cas9系统易诱导1 bp的插入突变,且CRISPR/Cas9系统诱发的DNA双链断裂(Double-strand breaks,DSBs)位置更加精确。此外,DSBs在修复过程中能以低频同源重组(Homologous recombination,HR)途径修复,产生DNA片段重复突变。对于相邻双靶点CRISPR/Cas9系统而言,双靶点间的DNA片段可发生缺失或倒位,这种双靶点间DNA片段突变的发生频率与双靶点各自的突变率无正相关性。两大系统诱导的突变最早发生在已转化的愈伤组织中,少量发生在水稻的体细胞中,导致了纯合突变、杂合突变、双等位突变和嵌合突变4种遗传模式,其中双等位突变比例最高,嵌合突变比例最低。除嵌合突变外,纯合突变、杂合突变、双等位突变的突变序列均可稳定遗传给下一代。
URLPMID:4267669 [本文引用: 2]

Abstract The efficacy and the mutation spectrum of genome editing methods can vary substantially depending on the targeted sequence. A simple, quick assay to accurately characterize and quantify the induced mutations is therefore needed. Here we present TIDE, a method for this purpose that requires only a pair of PCR reactions and two standard capillary sequencing runs. The sequence traces are then analyzed by a specially developed decomposition algorithm that identifies the major induced mutations in the projected editing site and accurately determines their frequency in a cell population. This method is cost-effective and quick, and it provides much more detailed information than current enzyme-based assays. An interactive web tool for automated decomposition of the sequence traces is available. TIDE greatly facilitates the testing and rational design of genome editing strategies. The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
URL [本文引用: 2]
URLPMID:4365529 [本文引用: 1]

Background The ability to selectively alter genomic DNA sequences in vivo is a powerful tool for basic and applied research. The CRISPR/Cas9 system precisely mutates DNA sequences in a number of organisms. Here, the CRISPR/Cas9 system is shown to be effective in soybean by knocking-out a green fluorescent protein (GFP) transgene and modifying nine endogenous loci. Results Targeted DNA mutations were detected in 95% of 88 hairy-root transgenic events analyzed. Bi-allelic mutations were detected in events transformed with eight of the nine targeting vectors. Small deletions were the most common type of mutation produced, although SNPs and short insertions were also observed. Homoeologous genes were successfully targeted singly and together, demonstrating that CRISPR/Cas9 can both selectively, and generally, target members of gene families. Somatic embryo cultures were also modified to enable the production of plants with heritable mutations, with the frequency of DNA modifications increasing with culture time. A novel cloning strategy and vector system based on In-Fusion?? cloning was developed to simplify the production of CRISPR/Cas9 targeting vectors, which should be applicable for targeting any gene in any organism. Conclusions The CRISPR/Cas9 is a simple, efficient, and highly specific genome editing tool in soybean. Although some vectors are more efficient than others, it is possible to edit duplicated genes relatively easily. The vectors and methods developed here will be useful for the application of CRISPR/Cas9 to soybean and other plant species.
URL
URLPMID:26946469

Abstract CRISPR-Cas9 system is now widely used to edit a target genome in animals and plants. Cas9 protein derived from Streptococcus pyogenes (SpCas9) cleaves double-stranded DNA targeted by a chimeric single-guide RNA (sgRNA). For plant genome editing, Agrobacterium -mediated T-DNA transformation has been broadly used to express Cas9 proteins and sgRNAs under the control of CaMV 35S and U6/U3 promoter, respectively. We here developed a simple and high-throughput binary vector system to clone a 19 20 bp of sgRNA, which binds to the reverse complement of a target locus, in a large T-DNA binary vector containing an SpCas9 expressing cassette. Two-step cloning procedures: (1) annealing two target-specific oligonucleotides with overhangs specific to the Aar I restriction enzyme site of the binary vector; and (2) ligating the annealed oligonucleotides into the two Aar I sites of the vector, facilitate the high-throughput production of the positive clones. In addition, Cas9-coding sequence and U6/U3 promoter can be easily exchanged via the GatewayTM system and unique Eco RI/ Xho I sites on the vector, respectively. We examined the mutation ratio and patterns when we transformed these constructs into Arabidopsis thaliana and a wild tobacco, Nicotiana attenuata . Our vector system will be useful to generate targeted large-scale knock-out lines of model as well as non-model plant.
URL

Simultaneous multiplex mutation of large gene families using Cas9 has the potential to revolutionize agriculture and plant sciences. The targeting of multiple genomic sites at once raises concerns about the efficiency and specificity in targeting. The modelArabidopsis thalianais widely used in basic plant research. Previous work has suggested that the Cas9 off-target rate in Arabidopsis is undetectable. Here we use deep sequencing on pooled plants simultaneously targeting 14 distinct genomic loci to demonstrate that multiplex targeting in Arabidopsis is highly specific to on-target sites with no detectable off-target events. In addition, chromosomal translocations are extremely rare. The high specificity of Cas9 in Arabidopsis makes this a reliable method for clean mutant generation with no need to enhance specificity or adopt alternate Cas9 variants.
URLPMID:27848933

Targeted DNA double-strand breaks have been shown to significantly increase the frequency and precision of genome editing. In the past two decades, several double-strand break technologies have been developed. CRISPR–Cas9 has quickly become the technology of choice for genome editing due to its simplicity, efficiency and versatility. Currently, genome editing in plants primarily relies on delivering double-strand break reagents in the form of DNA vectors. Here we report biolistic delivery of pre-assembled Cas9–gRNA ribonucleoproteins into maize embryo cells and regeneration of plants with both mutated and edited alleles. Using this method of delivery, we also demonstrate DNA- and selectable marker-free gene mutagenesis in maize and recovery of plants with mutated alleles at high frequencies. These results open new opportunities to accelerate breeding practices in a wide variety of crop species. Genome editing in plants typically requires the expression of Cas9 and guide RNA from stably transformed plasmid DNA. Here, the authors show that successful editing can be achieved after delivery of the Cas9-guide RNA complex as a ribonucleoprotein to maize embryos via biolistics.
URLPMID:27936512

Abstract Citrus is a highly valued tree crop worldwide, while, at the same time, citrus production faces many biotic challenges, including bacterial canker and Huanglongbing (HLB). Breeding for disease???resistant varieties is the most efficient and sustainable approach to control plant diseases. Traditional breeding of citrus varieties is challenging due to multiple limitations, including polyploidy, polyembryony, extended juvenility and long crossing cycles. Targeted genome editing technology has the potential to shorten varietal development for some traits, including disease resistance. Here, we used CRISPR/Cas9/sgRNA technology to modify the canker susceptibility gene CsLOB1 in Duncan grapefruit. Six independent lines, DLOB2, DLOB3, DLOB9, DLOB10, DLOB11 and DLOB12, were generated. Targeted next???generation sequencing of the six lines showed the mutation rate was 31.58%, 23.80%, 89.36%, 88.79%, 46.91% and 51.12% for DLOB2, DLOB3, DLOB9, DLOB10, DLOB11 and DLOB12, respectively, of the cells in each line. DLOB2 and DLOB3 showed canker symptoms similar to wild???type grapefruit, when inoculated with the pathogen Xanthomonas citri subsp. citri (Xcc). No canker symptoms were observed on DLOB9, DLOB10, DLOB11 and DLOB12 at 4??days postinoculation (DPI) with Xcc. Pustules caused by Xcc were observed on DLOB9, DLOB10, DLOB11 and DLOB12 in later stages, which were much reduced compared to that on wild???type grapefruit. The pustules on DLOB9 and DLOB10 did not develop into typical canker symptoms. No side effects and off???target mutations were detected in the mutated plants. This study indicates that genome editing using CRISPR technology will provide a promising pathway to generate disease???resistant citrus varieties.
URLPMID:5170842

The combined availability of whole genome sequences and genome editing tools is set to revolutionize the field of fruit biotechnology by enabling the introduction of targeted genetic changes with unprecedented control and accuracy, both to explore emergent phenotypes and to introduce new functionalities. Although plasmid-mediated delivery of genome editing components to plant cells is very efficient, it also presents some drawbacks, such as possible random integration of plasmid sequences in the host genome. Additionally, it may well be intercepted by current process-based GMO regulations, complicating the path to commercialization of improved varieties. Here, we explore direct delivery of purified CRISPR/Cas9 ribonucleoproteins (RNPs) to the protoplast of grape cultivarChardonnayand apple cultivar such asGolden deliciousfruit crop plants for efficient targeted mutagenesis. We targetedMLO-7, a susceptible gene in order to increase resistance to powdery mildew in grape cultivar andDIPM-1, DIPM-2, andDIPM-4in the apple to increase resistance to fire blight disease. Furthermore, efficient protoplast transformation, the molar ratio of Cas9 and sgRNAs were optimized for each grape and apple cultivar. The targeted mutagenesis insertion and deletion rate was analyzed using targeted deep sequencing. Our results demonstrate that direct delivery of CRISPR/Cas9 RNPs to the protoplast system enables targeted gene editing and paves the way to the generation of DNA-free genome edited grapevine and apple plants.
URLPMID:28205546

Abstract Cpf1, a type V CRISPR effector, recognizes a thymidine-rich protospacer-adjacent motif and induces cohesive double-stranded breaks at the target site guided by a single CRISPR RNA (crRNA). Here we show that Cpf1 can be used as a tool for DNA-free editing of plant genomes. We describe the delivery of recombinant Cpf1 proteins with in vitro transcribed or chemically synthesized target-specific crRNAs into protoplasts isolated from soybean and wild tobacco. Designed crRNAs are unique and do not have similar sequences ( 3 mismatches) in the entire soybean reference genome. Targeted deep sequencing analyses show that mutations are successfully induced in FAD2 paralogues in soybean and AOC in wild tobacco. Unlike SpCas9, Cpf1 mainly induces various nucleotide deletions at target sites. No significant mutations are detected at potential off-target sites in the soybean genome. These results demonstrate that Cpf1-crRNA complex is an effective DNA-free genome-editing tool for plant genome editing.
URLPMID:28548094

Homologous recombination (HR) between parental chromosomes occurs stochastically. Here, we report on targeted recombination between homologous chromosomes upon somatic induction of DNA double-strand breaks (DSBs) via CRISPR-Cas9. We demonstrate this via a visual and molecular assay whereby DSB induction between two alleles carrying different mutations in the PHYTOENE SYNTHASE (PSY1) gene results in yellow fruits with wild type red sectors forming via HR-mediated DSB repair. We also show that in heterozygote plants containing one psy1 allele immune and one sensitive to CRISPR, repair of the broken allele using the unbroken allele sequence template is a common outcome. In another assay, we show evidence of a somatically induced DSB in a cross between a psy1 edible tomato mutant and wild type Solanum pimpinellifolium, targeting only the S. pimpinellifolium allele. This enables characterization of germinally transmitted targeted somatic HR events, demonstrating that somatically induced DSBs can be exploited for precise breeding of crops.
URLPMID:28522548

We report a comprehensive toolkit that enables targeted, specific modification of monocot and dicot genomes using a variety of genome engineering approaches. Our reagents, based on transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, are systematized for fast, modular cloning and accommodate diverse regulatory sequences to drive reagent expression. Vectors are optimized to create either single or multiple gene knockouts and large chromosomal deletions. Moreover, integration of geminivirus-based vectors enables precise gene editing through homologous recombination. Regulation of transcription is also possible. A Web-based tool streamlines vector selection and construction. One advantage of our platform is the use of the Csy-type (CRISPR system yersinia) ribonuclease 4 (Csy4) and tRNA processing enzymes to simultaneously express multiple guide RNAs (gRNAs). For example, we demonstrate targeted deletions in up to six genes by expressing 12 gRNAs from a single transcript. Csy4 and tRNA expression systems are almost twice as effective in inducing mutations as gRNAs expressed from individual RNA polymerase III promoters. Mutagenesis can be further enhanced 2.5-fold by incorporating the Trex2 exonuclease. Finally, we demonstrate that Cas9 nickases induce gene targeting at frequencies comparable to native Cas9 when they are delivered on geminivirus replicons. The reagents have been successfully validated in tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), Medicago truncatula, wheat (Triticum aestivum), and barley (Hordeum vulgare).
URLPMID:5573723

The CRISPR/Cas9 system has been applied in diverse eukaryotic organisms for targeted mutagenesis. However, targeted gene editing is inefficient and requires the simultaneous delivery of a DNA template for homology-directed repair (HDR). Here, we used CRISPR/Cas9 to generate targeted double-strand breaks and to deliver an RNA repair template for HDR in rice (Oryza sativa). We used chimeric single-guide RNA (cgRNA) molecules carrying both sequences for target site specificity (to generate the double-strand breaks) and repair template sequences (to direct HDR), flanked by regions of homology to the target. Gene editing was more efficient in rice protoplasts using repair templates complementary to the non-target DNA strand, rather than the target strand. We applied this cgRNA repair method to generate herbicide resistance in rice, which showed that this cgRNA repair method can be used for targeted gene editing in plants. Our findings will facilitate applications in functional genomics and targeted improvement of crop traits.
URLPMID:28499063 [本文引用: 1]

Gossypium hirsutum is an allotetraploid with a complex genome. Most genes have multiple copies that belong to At and Dt subgenomes. Sequence similarity is also very high between gene homologs. To efficiently achieve site/gene‐specific mutation is quite needed. Due to its high efficiency and robustness, the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 system has exerted broad site‐specific genome editing from prokaryotes to eukaryotes. In this study, we utilized a CRISPR/Cas9 system to generate two sgRNAs in a single vector to conduct multiple sites genome editing in allotetraploid cotton. An exogenously transformed gene Discosoma red fluorescent protein2(DsRed2)and an endogenous gene GhCLA1 were chosen as targets. The DsRed2 edited plants in T0 generation reverted its traits to wild type, with vanished red fluorescence the whole plants. Besides, the mutated phenotype and genotype were inherited to their T1 progenies. For the endogenous gene GhCLA1, 75% of regenerated plants exhibited albino phenotype with obvious nucleotides and DNA fragments deletion. The efficiency of gene editing at each target site is 66.7% to 100% .The mutation genotype were checked for both genes with Sanger sequencing. Barcode‐based high‐throughput sequencing, which could be highly efficient for genotyping to a population of mutants, was conducted in GhCLA1 edited T0 plants and it matched well with Sanger sequencing results. No off‐target editing was detected at the potential off‐target sites. These results proves that the CRISPR/Cas9 system is highly efficient and reliable for allotetraploid cotton genome editing.
URLPMID:19404259 [本文引用: 1]

Abstract Agricultural biotechnology is limited by the inefficiencies of conventional random mutagenesis and transgenesis. Because targeted genome modification in plants has been intractable, plant trait engineering remains a laborious, time-consuming and unpredictable undertaking. Here we report a broadly applicable, versatile solution to this problem: the use of designed zinc-finger nucleases (ZFNs) that induce a double-stranded break at their target locus. We describe the use of ZFNs to modify endogenous loci in plants of the crop species Zea mays. We show that simultaneous expression of ZFNs and delivery of a simple heterologous donor molecule leads to precise targeted addition of an herbicide-tolerance gene at the intended locus in a significant number of isolated events. ZFN-modified maize plants faithfully transmit these genetic changes to the next generation. Insertional disruption of one target locus, IPK1, results in both herbicide tolerance and the expected alteration of the inositol phosphate profile in developing seeds. ZFNs can be used in any plant species amenable to DNA delivery; our results therefore establish a new strategy for plant genetic manipulation in basic science and agricultural applications.
URLPMID:24836556

SummaryEngineered nucleases can be used to induce site-specific double-strand breaks (DSBs) in plant genomes. Thus, homologous recombination (HR) can be enhanced and targeted mutagenesis can be achieved by error-prone non-homologous end-joining (NHEJ). Recently, the bacterial CRISPR/Cas9 system was used for DSB induction in plants to promote HR and NHEJ. Cas9 can also be engineered to work as a nickase inducing single-strand breaks (SSBs). Here we show that only the nuclease but not the nickase is an efficient tool for NHEJ-mediated mutagenesis in plants. We demonstrate the stable inheritance of nuclease-induced targeted mutagenesis events in the ADH1 and TT4 genes of Arabidopsis thaliana at frequencies from 2.5 up to 70.0%. Deep sequencing analysis revealed NHEJ-mediated DSB repair in about a third of all reads in T1 plants. In contrast, applying the nickase resulted in the reduction of mutation frequency by at least 740-fold. Nevertheless, the nickase is able to induce HR at similar efficiencies as the nuclease or the homing endonuclease I ceI. Two different types of somatic HR mechanisms, recombination between tandemly arranged direct repeats as well as gene conversion using the information on an inverted repeat could be enhanced by the nickase to a similar extent as by DSB-inducing enzymes. Thus, the Cas9 nickase has the potential to become an important tool for genome engineering in plants. It should not only be applicable for HR-mediated gene targeting systems but also by the combined action of two nickases as DSB-inducing agents excluding off-target effects in homologous genomic regions.
URLPMID:25327456

Summary The CRISPR/Cas nuclease is becoming a major tool for targeted mutagenesis in eukaryotes by inducing double-strand breaks (DSBs) at pre-selected genomic sites that are repaired by non-homologous end joining (NHEJ) in an error-prone way. In plants, it could be demonstrated that the Cas9 nuclease is able to induce heritable mutations in Arabidopsis thaliana and rice. Gene targeting (GT) by homologous recombination (HR) can also be induced by DSBs. Using a natural nuclease and marker genes, we previously developed an in planta GT strategy in which both a targeting vector and targeting locus are activated simultaneously via DSB induction during plant development. Here, we demonstrate that this strategy can be used for natural genes by CRISPR/Cas-mediated DSB induction. We were able to integrate a resistance cassette into the ADH1 locus of A. thaliana via HR. Heritable events were identified using a PCR-based genotyping approach, characterised by Southern blotting and confirmed on the sequence level. A major concern is the specificity of the CRISPR/Cas nucleases. Off-target effects might be avoided using two adjacent sgRNA target sequences to guide the Cas9 nickase to each of the two DNA strands, resulting in the formation of a DSB. By amplicon deep sequencing, we demonstrate that this Cas9 paired nickase strategy has a mutagenic potential comparable with that of the nuclease, while the resulting mutations are mostly deletions. We also demonstrate the stable inheritance of such mutations in A. thaliana .
URLPMID:4999463 [本文引用: 1]

Sugarcane (Saccharumspp. hybrids) is a prime crop for commercial biofuel production. Advanced conversion technology utilizes both, sucrose accumulating in sugarcane stems as well as cell wall bound sugars for commercial ethanol production. Reduction of lignin content significantly improves the conversion of lignocellulosic biomass into ethanol. Conventional mutagenesis is not expected to confer reduction in lignin content in sugarcane due to its high polyploidy (x=6510–13) and functional redundancy among homo(eo)logs. Here we deploy transcription activator-like effector nuclease (TALEN) to induce mutations in a highly conserved region of thecaffeic acid O-methyltransferase(COMT) of sugarcane. Capillary electrophoresis (CE) was validated by pyrosequencing as reliable and inexpensive high throughput method for identification and quantitative characterization of TALEN mediated mutations. TargetedCOMTmutations were identified by CE in up to 7465% of the lines. In different events 8–9965% of the wild02typeCOMTwere converted to mutantCOMTas revealed by pyrosequencing. Mutation frequencies among mutant lines were positively correlated to lignin reduction. Events with a mutation frequency of 9965% displayed a 29–3265% reduction of the lignin content compared to non-transgenic controls along with significantly reduced S subunit content and elevated hemicellulose content. CE analysis displayed similar peak patterns between primaryCOMTmutants and their vegetative progenies suggesting that TALEN mediated mutations were faithfully transmitted to vegetative progenies. This is the first report on genome editing in sugarcane. The findings demonstrate that targeted mutagenesis can improve cell wall characteristics for production of lignocellulosic ethanol in crops with highly complex genomes. The online version of this article (doi:10.1007/s11103-016-0499-y) contains supplementary material, which is available to authorized users.
[本文引用: 1]
URLPMID:11464506 [本文引用: 2]

Abstract The development of transgenic events can be limited by many factors. These include expression levels, insert stability and inheritance, and the identification of simple insertion events. All of the factors can be related to the copy number of the transgene. Traditionally, copy number has been determined by laborious blotting techniques. We have developed an alternative approach that utilizes the fluorogenic 5' nuclease (TaqMan) assay to quantitatively determine transgene copy level in plants. Using this assay, hundreds of samples can be analyzed per day in contrast to the low throughput encountered with traditional methods. To develop the TaqMan copy number assay, we chose to utilize our highly efficient Agrobacterium-mediated transformation system of maize. This transformation procedure generates predominantly low copy number insertion events, which simplified assay development. We have also successful applied this assay to other crops and transformation systems.
URLPMID:26294043 [本文引用: 2]

Abstract Recently discovered bacteria and archaea adaptive immune system consisting of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) endonuclease has been explored in targeted genome editing in different species. Streptococcus pyogenes Cas9-guide RNA (gRNA) was successfully applied to generate targeted mutagenesis, gene integration, and gene editing in soybean (Glycine max). Two genomic sites, DD20 and DD43 on chromosome 4, were mutagenized with frequencies of 59% and 76%, respectively. Sequencing randomly selected transgenic events confirmed that the genome modifications were specific to the Cas9-gRNA cleavage sites and consisted of small deletions or insertions. Targeted gene integrations through homology-directed recombination were detected by border-specific polymerase chain reaction analysis for both sites at callus stage, and one DD43 homology-directed recombination event was transmitted to T1 generation. T1 progenies of the integration event segregated according to Mendelian laws and clean homozygous T1 plants with the donor gene precisely inserted at the DD43 target site were obtained. The Cas9-gRNA system was also successfully applied to make a directed P178S mutation of acetolactate synthase1 gene through in planta gene editing. 2015 American Society of Plant Biologists. All Rights Reserved.
URLPMID:26269544 [本文引用: 2]

Abstract Targeted mutagenesis, editing of endogenous maize (Zea mays) genes, and site-specific insertion of a trait gene using clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas)-guide RNA technology are reported in maize. DNA vectors expressing maize codon-optimized Streptococcus pyogenes Cas9 endonuclease and single guide RNAs were cointroduced with or without DNA repair templates into maize immature embryos by biolistic transformation targeting five different genomic regions: upstream of the liguleless1 (LIG1) gene, male fertility genes (Ms26 and Ms45), and acetolactate synthase (ALS) genes (ALS1 and ALS2). Mutations were subsequently identified at all sites targeted, and plants containing biallelic multiplex mutations at LIG1, Ms26, and Ms45 were recovered. Biolistic delivery of guide RNAs (as RNA molecules) directly into immature embryo cells containing preintegrated Cas9 also resulted in targeted mutations. Editing the ALS2 gene using either single-stranded oligonucleotides or double-stranded DNA vectors as repair templates yielded chlorsulfuron-resistant plants. Double-strand breaks generated by RNA-guided Cas9 endonuclease also stimulated insertion of a trait gene at a site near LIG1 by homology-directed repair. Progeny showed expected Mendelian segregation of mutations, edits, and targeted gene insertions. The examples reported in this study demonstrate the utility of Cas9-guide RNA technology as a plant genome editing tool to enhance plant breeding and crop research needed to meet growing agriculture demands of the future. 2015 American Society of Plant Biologists. All Rights Reserved.
URLPMID:27442592 [本文引用: 2]

MaizeARGOS8is a negative regulator of ethylene responses. A previous study has shown that transgenic plants constitutively overexpressingARGOS8have reduced ethylene sensitivity and improved grain yield under drought stress conditions. To explore the targeted use ofARGOS8native expression variation in drought‐tolerant breeding, a diverse set of over 400 maize inbreds was examined forARGOS8mRNAexpression, but the expression levels in all lines were less than that created in the originalARGOS8transgenic events. We then employed aCRISPR‐Cas‐enabled advanced breeding technology to generate novel variants ofARGOS8. The native maizeGOS2 promoter, which confers a moderate level of constitutive expression, was inserted into the 5′‐untranslated region of the nativeARGOS8gene or was used to replace the native promoter ofARGOS8. Precise genomicDNAmodification at theARGOS8locus was verified byPCRand sequencing. TheARGOS8variants had elevated levels ofARGOS8transcripts relative to the native allele and these transcripts were detectable in all the tissues tested, which was the expected results using theGOS2 promoter. A field study showed that compared to theWT, theARGOS8variants increased grain yield by five bushels per acre under flowering stress conditions and had no yield loss under well‐watered conditions. These results demonstrate the utility of theCRISPR‐Cas9 system in generating novel allelic variation for breeding drought‐tolerant crops.
URLPMID:28114299 [本文引用: 2]

Abstract Sexual reproduction in flowering plants involves double fertilization, the union of two sperm from pollen with two sex cells in the female embryo sac. Modern plant breeders increasingly seek to circumvent this process to produce doubled haploid individuals, which derive from the chromosome-doubled cells of the haploid gametophyte. Doubled haploid production fixes recombinant haploid genomes in inbred lines, shaving years off the breeding process. Costly, genotype-dependent tissue culture methods are used in many crops, while seed-based in vivo doubled haploid systems are rare in nature and difficult to manage in breeding programmes. The multi-billion-dollar maize hybrid seed business, however, is supported by industrial doubled haploid pipelines using intraspecific crosses to in vivo haploid inducer males derived from Stock 6, first reported in 1959 (ref. 5), followed by colchicine treatment. Despite decades of use, the mode of action remains controversial. Here we establish, through fine mapping, genome sequencing, genetic complementation, and gene editing, that haploid induction in maize (Zea mays) is triggered by a frame-shift mutation in MATRILINEAL (MTL), a pollen-specific phospholipase, and that novel edits in MTL lead to a 6.7% haploid induction rate (the percentage of haploid progeny versus total progeny). Wild-type MTL protein localizes exclusively to sperm cytoplasm, and pollen RNA-sequence profiling identifies a suite of pollen-specific genes overexpressed during haploid induction, some of which may mediate the formation of haploid seed. These findings highlight the importance of male gamete cytoplasmic components to reproductive success and male genome transmittance. Given the conservation of MTL in the cereals, this discovery may enable development of in vivo haploid induction systems to accelerate breeding in crop plants.
URLPMID:29988153 [本文引用: 2]
URLPMID:27579989 [本文引用: 1]

Abstract Genome editing facilitated by Cas9-based RNA-guided nucleases (RGNs) is becoming an increasingly important and popular technique for reverse genetics in both model and non-model species. So far, RGNs were mainly applied for the induction of point mutations, and one major challenge consists in the detection of genome-edited individuals from a mutagenized population. Also, point mutations are not appropriate for functional dissection of non-coding DNA. Here, the multiplexing capacity of a newly developed genome editing toolkit was exploited for the induction of inheritable chromosomal deletions at six different loci in Nicotiana benthamiana and Arabidopsis. In both species, the preferential formation of small deletions was observed, suggesting reduced efficiency with increasing deletion size. Importantly, small deletions (< 100 bp) were detected at high frequencies in N. benthamiana T0 and Arabidopsis T2 populations. Thus, targeting of small deletions by paired nucleases represents a simple approach for the generation of mutant alleles segregating as size polymorphisms in subsequent generations. Phenotypically selected deletions of up to 120 kb occurred at low frequencies in Arabidopsis, suggesting larger population sizes for the discovery of valuable alleles from addressing gene clusters or non-coding DNA for deletion by programmable nucleases. This article is protected by copyright. All rights reserved.
URLPMID:29157799 [本文引用: 1]

正Tomato(Solanum lycopersicum)is the leading vegetable crop worldwide and an essential component of a healthy diet(Lin et al.,2014;Du et al.,2017).Fruit color is regarded as one of the most important commercial traits in tomato(The Tomato Genome Consortium,2012).Consumers in different regions have different color preferences.For example,European and American
URLPMID:27007717 [本文引用: 1]

Key message A method based on DNA single-strand conformation polymorphism is demonstrated for effective genotyping of CRISPR/Cas9-induced mutants in rice.
URLPMID:28955376 [本文引用: 1]

Abstract MicroRNAs (miRNAs) are small non-coding RNAs that play important roles in plant development and stress responses. Loss-of-function analysis of miRNA genes has been traditionally challenging due to lack of appropriate knockout tools. In this study, single miRNA genes (OsMIR408 and OsMIR528) and miRNA gene families (miR815a/b/c and miR820a/b/c) in rice were targeted by CRISPR-Cas9. We showed single strand conformation polymorphism (SSCP) is a more reliable method than restriction fragment length polymorphism (RFLP) for identifying CRISPR-Cas9 generated mutants. Frequencies of targeted mutagenesis among regenerated T0 lines ranged from 48 to 89% at all tested miRNA target sites. In the case of miRNA528, three independent guide RNAs (gRNAs) all generated biallelic mutations among confirmed mutant lines. When targeted by two gRNAs, miRNA genes were readily to be deleted at a frequency up to 60% in T0 rice lines. Thus, we demonstrate CRISPR-Cas9 is an effective tool for knocking out plant miRNAs. Single-base pair (bp) insertion/deletion mutations (indels) in mature miRNA regions can lead to the generation of functionally redundant miRNAs. Large deletions at either the mature miRNA or the complementary miRNA * were found to readily abolish miRNA function. Utilizing mutants of OsMIR408 and OsMIR528 , we find that knocking out a single miRNA can result in expression profile changes of many other seemingly unrelated miRNAs. In a case study on OsMIR528 , we reveal it is a positive regulator in salt stress. Our work not only provides empirical guidelines on targeting miRNAs with CRISPR-Cas9, but also brings new insights into miRNA function and complex cross-regulation in rice.
URLPMID:25655437 [本文引用: 1]

Restriction fragment length polymorphism (RFLP) analysis is one of the oldest, most convenient and least expensive methods of genotyping, but is limited by the availability of restriction endonuclease sites. Here we present a novel method of employing CRISPR/Cas-derived RNA-guided engineered nucleases (RGENs) in RFLP analysis. We prepare RGENs by complexing recombinant Cas9 protein derived from Streptococcus pyogenes with in vitro transcribed guide RNAs that are complementary to the DNA sequences of interest. Then, we genotype recurrent mutations found in cancer and small insertions or deletions (indels) induced in cultured cells and animals by RGENs and other engineered nucleases such as transcription activator-like effector nucleases (TALENs). Unlike T7 endonuclease I or Surveyor assays that are widely used for genotyping engineered nuclease-induced mutations, RGEN-mediated RFLP analysis can detect homozygous mutant clones that contain identical biallelic indel sequences and is not limited by sequence polymorphisms near the nuclease target sites.
URL [本文引用: 3]

Abstract Despite the great achievements in genome editing, accurately detecting mutations induced by sequence﹕pecific nucleases is still a challenge in plants, especially in polyploidy plants. An efficient detection method is particularly vital when the mutation frequency is low or when a large population needs to be screened. Here, we applied purified CRISPR ribonucleoprotein complexes to cleave PCR products for genome〆dited mutation detection in hexaploid wheat and diploid rice. We show that this mutation detection method is more sensitive than Sanger sequencing and more applicable than PCR/RE method without the requirement for restriction enzyme site. We also demonstrate that this detection method is especially useful for genome editing in wheat, because target sites are often surrounded by single nucleotide polymorphisms. By using this screening method, we were also able to detect foreign DNA‐free tagw2 mutations induced by purified TALEN protein. Finally, we show that partial base editing mutations can also be detected using high fidelity SpCas9 variants or FnCpf1. The PCR/RNP method is lowヽost and widely applicable for rapid detection of genome〆dited mutation in plants. This article is protected by copyright. All rights reserved.
URLPMID:5206254 [本文引用: 2]

Altered starch quality with full knockout ofGBSSgene function in potato was achieved using CRISPR-Cas9 technology, through transient transfection and regeneration from isolated protoplasts. Site-directed mutagenesis (SDM) has shown great progress in introducing precisely targeted mutations. Engineered CRISPR-Cas9 has received increased focus compared to other SDM techniques, since the method is easily adapted to different targets. Here, we demonstrate that transient application of CRISPR-Cas9-mediated genome editing in protoplasts of tetraploid potato (Solanum tuberosum) yielded mutations in all four alleles in a single transfection, in up to 202% of regenerated lines. Three different regions of the gene encoding granule-bound starch synthase (GBSS) were targeted under different experimental setups, resulting in mutations in at least one allele in 2–1202% of regenerated shoots, with multiple alleles mutated in up to 6702% of confirmed mutated lines. Most mutations resulted in small indels of 1–1002bp, but also vector DNA inserts of 34–23602bp were found in 1002% of analysed lines. No mutations were found in an allele diverging one bp from a used guide sequence, verifying similar results found in other plants that high homology between guide sequence and target region near the protospacer adjacent motif (PAM) site is essential. To meet the challenge of screening large numbers of lines, a PCR-based high-resolution fragment analysis method (HRFA) was used, enabling identification of multiple mutated alleles with a resolution limit of 102bp. Full knockout of GBSS enzyme activity was confirmed in four-allele mutated lines by phenotypic studies of starch. One remaining wild-type (WT) allele was shown sufficient to maintain enough GBSS enzyme activity to produce significant amounts of amylose. The online version of this article (doi:10.1007/s00299-016-2062-3) contains supplementary material, which is available to authorized users.
URLPMID:4263700 [本文引用: 1]

Abstract With the goal to generate and characterize the phenotypes of null alleles in all genes within an organism and the recent advances in custom nucleases, genome editing limitations have moved from mutation generation to mutation detection. We previously demonstrated that High Resolution Melting (HRM) analysis is a rapid and efficient means of genotyping known zebrafish mutants. Here we establish optimized conditions for HRM based detection of novel mutant alleles. Using these conditions, we demonstrate that HRM is highly efficient at mutation detection across multiple genome editing platforms (ZFNs, TALENs, and CRISPRs); we observed nuclease generated HRM positive targeting in 1 of 6 (16%) open pool derived ZFNs, 14 of 23 (60%) TALENs, and 58 of 77 (75%) CRISPR nucleases. Successful targeting, based on HRM of G0 embryos correlates well with successful germline transmission (46 of 47 nucleases); yet, surprisingly mutations in the somatic tail DNA weakly correlate with mutations in the germline F1 progeny DNA. This suggests that analysis of G0 tail DNA is a good indicator of the efficiency of the nuclease, but not necessarily a good indicator of germline alleles that will be present in the F1s. However, we demonstrate that small amplicon HRM curve profiles of F1 progeny DNA can be used to differentiate between specific mutant alleles, facilitating rare allele identification and isolation; and that HRM is a powerful technique for screening possible off-target mutations that may be generated by the nucleases. Our data suggest that micro-homology based alternative NHEJ repair is primarily utilized in the generation of CRISPR mutant alleles and allows us to predict likelihood of generating a null allele. Lastly, we demonstrate that HRM can be used to quickly distinguish genotype-phenotype correlations within F1 embryos derived from G0 intercrosses. Together these data indicate that custom nucleases, in conjunction with the ease and speed of HRM, will facilitate future high-throughput mutation generation and analysis needed to establish mutants in all genes of an organism.
URLPMID:27473525 [本文引用: 2]

Key message A selection-free, highly efficient targeted mutagenesis approach based on a novel ZFN monomer arrangement for genome engineering in tomato reveals plant trait modifications.
URLPMID:23573916 [本文引用: 1]

The heteroduplex mobility assay (HMA) is widely used to characterize strain variants of human viruses. To determine whether it can detect small sequence differences in homologous templates, we constructed a series of deletion constructs (1 10 bp deletions) in the multiple cloning site (MCS) of pBluescript II. After PCR amplification of the MCS using a mixture of wild-type and one of the deletion constructs, the resulting PCR amplicons were electrophoresed using 15% polyacrylamide gels. Two types of heteroduplexes exhibited retarded electrophoretic migration compared with individual homoduplexes. Therefore, we applied this HMA to detect transcription activator-like effector nucleases (TALEN)-induced insertion and/or deletion (indel) mutations at an endogenous locus. We found that TALEN in vivo activity was easily estimated by the degree of multiple HMA profiles derived from TALEN-injected F0 embryos. Furthermore, TALEN-injected F0 founder fish produced several unique HMA profiles in F1 embryos. Sequence analysis confirmed that the different HMA profiles contained distinct indel mutations. Thus, HMA is a rapid and sensitive analytical method for the detection of the TALEN-mediated genome modifications.
URL [本文引用: 1]

Single nucleotide polymorphisms (SNPs) are widely observed between individuals, ecotypes, and species, serving as an invaluable molecular marker for genetic, genomic, ecological and evolutionary studies. Although, a large number of SNP-discriminating methods are currently available, few are suited for low-throughput and low-cost applications. Here, we describe a genotyping method namedSimpleAllele-discriminatingPCR (SAP), which is ideally suited for the small-scale genotyping and gene mapping routinely performed in small to medium research or teaching laboratories. We demonstrate the feasibility and application of SAP to discriminate wild type alleles from their respective mutant alleles inArabidopsis thaliana. Although the design principle was previously described, it is unclear if the method is technically robust, reliable, and applicable. Three primers were designed for each individual SNP or allele with two allele-discriminating forward primers (one for wild type and one for the mutant allele) and a common reverse primer. The two allele-discriminating forward primers are designed so that each incorporates one additional mismatch at the adjacent (penultimate) site from the SNP, resulting in two mismatches between the primer and its non-target template and one mismatch between the primer and its target template. The presence or absence of the wild type or the mutant allele correlates with the presence or absence of respective PCR product. The presence of both wild type-specific and mutant-specific PCR products would indicate heterozygosity. SAP is shown here to discriminate three mutant alleles (lug-3,lug-16, andluh-1) from their respective wild type alleles. In addition, the SAP principle is shown to work in conjunction with fluorophore-labeled primers, demonstrating the feasibility of applying SAP to high throughput SNP analyses. SAP offers an excellent alternative to existing SNP-discrimination methods such as Cleaved Amplified Polymorphic Sequence (CAPS) or derived CAPS (dCAPS). It can also be adapted for high throughput SNP analyses by incorporating fluorophore-labeled primers. SAP is reliable, cost-effective, fast, and simple, and can be applied to all organisms not limited toArabidopsis thaliana.
URLPMID:28416245 [本文引用: 3]

The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system provides a technological breakthrough in mutant generation. Several methods such as the polymerase chain reaction (PCR)/restriction enzyme (RE) assay, T7 endonuclease I (T7EI) assay, Surveyor nuclease assay, PAGE-based genotyping assay, and high-resolution melting (HRM) analysis-based assay have been developed for screening CRISPR/Cas9-induced mutants. However, these methods are time- and labour-intensive and may also be sequence-limited or require very expensive equipment. Here, we describe a cost-effective and sensitive screening technique based on conventional PCR, annealing at critical temperature PCR (ACT-PCR) for identifying mutants. ACT-PCR requires only a single PCR step followed by agarose gel electrophoresis. We demonstrated that ACT-PCR accurately distinguished CRISPR/Cas9-induced mutants from wild type in both rice and zebrafish. Moreover, the method can be adapted for accurately determining mutation frequency in cultured cells. The simplicity of ACT-PCR makes it particularly suitable for rapid, large-scale screening of CRISPR/Cas9-induced mutants in both plants and animals.
URLPMID:4063274 [本文引用: 1]

Author(s): Miyaoka, Y; Chan, AH; Judge, LM; Yoo, J; Huang, M; Nguyen, TD; Lizarraga, PP; So, PL; Conklin, BR | Abstract: Precise editing of human genomes in pluripotent stem cells by homology-driven repair of targeted nuclease-induced cleavage has been hindered by the difficulty of isolating rare clones. We developed an efficient method to ca pture rare mutational events, enabling isolation of mutant lines with single-base substitutions without antibiotic selection. This method facilitates efficient induction or reversion of mutations associated with human disease in isogenic human induced pluripotent stem cells. 2014 Nature America, Inc.
URLPMID:27089539 [本文引用: 1]

The rapid adoption of gene editing tools such as CRISPRs and TALENs for research and eventually therapeutics necessitates assays that can rapidly detect and quantitate the desired alterations. Currently, the most commonly used assay employs “mismatch nucleases” T7E1 or “Surveyor” that recognize and cleave heteroduplexed DNA amplicons containing mismatched base-pairs. However, this assay is prone to false positives due to cancer-associated mutations and/or SNPs and requires large amounts of starting material. Here we describe a powerful alternative wherein droplet digital PCR (ddPCR) can be used to decipher homozygous from heterozygous mutations with superior levels of both precision and sensitivity. We use this assay to detect knockout inducing alterations to stem cell associated proteins, NODAL and SFRP1, generated using either TALENs or an “all-in-one” CRISPR/Cas plasmid that we have modified for one-step cloning and blue/white screening of transformants. Moreover, we highlight how ddPCR can be used to assess the efficiency of varying TALEN-based strategies. Collectively, this work highlights how ddPCR-based screening can be paired with CRISPR and TALEN technologies to enable sensitive, specific, and streamlined approaches to gene editing and validation.
[本文引用: 1]
URLPMID:4997339 [本文引用: 1]

CRISPR/Cas9 genome-editing has emerged as a powerful tool to create mutant alleles in model organisms. However, the precision with which these mutations are created has introduced a new set of complications for genotyping and colony management. Traditional gene-targeting approaches in many experimental organisms incorporated exogenous DNA and/or allele specific sequence that allow for genotyping strategies based on binary readout of PCR product amplification and size selection. In contrast, alleles created by non-homologous end-joining (NHEJ) repair of double-stranded DNA breaks generated by Cas9 are much less amenable to such strategies. Here we describe a novel genotyping strategy that is cost effective, sequence specific and allows for accurate and efficient multiplexing of small insertion-deletions and single-nucleotide variants characteristic of CRISPR/Cas9 edited alleles. We show that ligation detection reaction (LDR) can be used to generate products that are sequence specific and uniquely detected by product size and/or fluorescent tags. The method works independently of the model organism and will be useful for colony management as mutant alleles differing by a few nucleotides become more prevalent in experimental animal colonies.
URLPMID:28860183 [本文引用: 1]

Abstract CRISPR-Cas9 based technology is currently the most flexible means to create targeted mutations by recombination or indel mutations by non-homologous end joining. During mouse transgenesis, recombinant and indel alleles are often pursued simultaneously. Multiple alleles can be formed in each animal to create significant genetic complexity that complicates the CRISPR-Cas9 approach and analysis. Currently, there are no rapid methods to measure the extent of on-site editing with broad mutation sensitivity. In this study, we demonstrate the allelic diversity arising from targeted CRISPR-editing in founder mice. Using this DNA sample collection, we validated specific, quantitative, and digital PCR methods (qPCR and dPCR, respectively) for measuring the frequency of on-target editing in founder mice. We found that locked nucleic acid (LNA) probes combined with an internal reference probe (Drop-Off Assay) provide accurate measurements of editing rates. The Drop-Off LNA Assay also detected on-target CRISPR-Cas9 gene editing in blastocysts with a sensitivity comparable to PCR-clone sequencing. Lastly, we demonstrate that the allele-specific LNA probes used in qPCR competitor assays can accurately detect recombinant mutations in founder mice. In summary, we show that LNA-based qPCR and dPCR assays provide a rapid method for quantifying the extent of on-target genome editing in vivo, testing RNA guides, and detecting recombinant mutations.
