 ,1
,1Research progress in the mechanism of translation initiation of eukaryotic mRNAs
Chaoxing Zheng1, Xiaofeng Ma1, Yonghua Zhang1, Hongjie Li2, Genfa Zhang ,1
,1通讯作者:
编委: 张飞雄
收稿日期:2018-04-9修回日期:2018-06-25网络出版日期:2018-08-16
| 基金资助: |
Received:2018-04-9Revised:2018-06-25Online:2018-08-16
| Fund supported: |
作者简介 About authors
郑超星,硕士研究生,专业方向:植物分子遗传学E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (695KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
郑超星, 马小凤, 张永华, 李洪杰, 张根发. 真核生物mRNA翻译起始机制研究进展[J]. 遗传, 2018, 40(8): 607-619 doi:10.16288/j.yczz.17-393
Chaoxing Zheng, Xiaofeng Ma, Yonghua Zhang, Hongjie Li, Genfa Zhang.
在真核生物中,mRNA翻译是一个不断循环的过程,包括起始、延伸和终止3个步骤。其中,起始阶段在基因表达调控中尤为重要。研究表明,此过程涉及到不同种类的翻译起始因子(eukaryotic initiation factors, eIFs)和不同特点的mRNA,它们在翻译调控中均发挥着重要的作用[1]。依赖于m7G帽结构的核糖体扫描机制(m7G cap-dependent scanning mechanism)是真核生物进行翻译起始的主要形式,也被公认为是最经典的分子作用机制,其过程主要包括43S前起始复合物的组装、43S前起始复合物与mRNA 5°末端m7G帽的结合、起始密码子的选择、60S核糖体亚基的加入和80S亚基的形成等[1]。
当经典翻译起始机制受到抑制时,一些mRNA则会通过其他机制进行翻译,以维持蛋白质的合成效率,保证细胞在逆境中的生存和恢复。在这些非典型的翻译起始案例当中,大多数却并不依赖于5°末端的m7G帽结构,因此被称为非帽依赖性翻译 起始机制(cap-independent mechanism)。例如,40S核糖体亚基可以通过特殊的内部核糖体进入位点(internal ribosome entry site, IRES)直接被招募到mRNA上游的某个位置,或者直接与起始密码子结合。此外,还有一种非典型机制既不依赖帽结构,也不依赖IRES,而是通过一种特殊的RNA结构完成核糖体亚基的招募,这种特殊结构被称为非帽依赖性翻译增强子(cap-independent translational enhancer, CITE)[2]。
真核生物的mRNA翻译起始是一个极其复杂的过程,尤其是当细胞处于胁迫状态时,各种替代式的非典型起始机制不断被发现。本文在总结经典的帽依赖性扫描机制的相关研究进展的基础上,对由IRES和CITE介导的两种非典型翻译起始机制进行了介绍,期望为更好地探索多种翻译起始方式及它们的调控方式提供参考。
1 经典的帽依赖性扫描机制
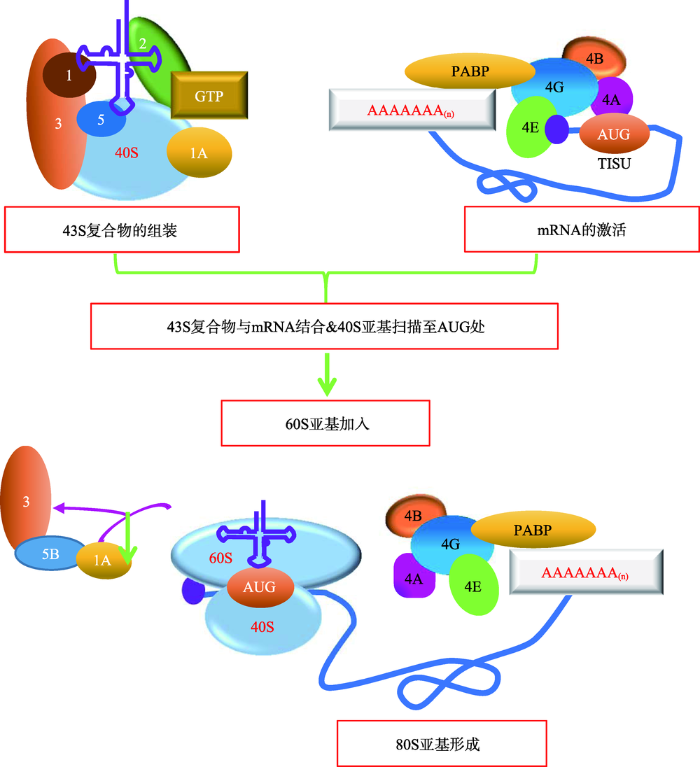
真核生物mRNA的典型结构是5°末端的m7G帽和3'末端的poly (A)尾,它们在mRNA的翻译和稳定性中发挥了重要作用[1],帽依赖性扫描机制是绝大多数拥有m7G帽和poly (A)尾的真核生物mRNA进行翻译起始的经典方式。该机制由Kozak[3]在1989年首次提出,经过不断完善和总结,主要可以分别两大步骤:其一,在翻译起始因子eIF1、eIF1A、eIF3和eIF5的作用下,三元复合物(eIF2-GTP-Met-tRNAiMet)与40S核糖体亚基结合形成43S前起始复合物(preinitiation complex, PIC);eIF4E、eIF4G、eIF4B、eIF4A和PABP等与mRNA结合并将其激活。其中,当eIF4E(帽结合蛋白)与mRNA 5°末端的m7G帽结构发生绑定时,激活过程正式开始。其二,组装好的43S复合物与激活的mRNA相互结合,核糖体40S亚基将沿着mRNA从5°至3°的方向扫描5°UTR,直到扫描至开放式阅读框(open reading frames, ORFs)的起始密码子AUG处,60S亚基加入后,80S延伸复合物形成,延伸过程开始。其间还需要真核起始因子(eIFs)及辅助因子的协作(图1)[4,5,6,7,8,9],它们可以调控mRNA、tRNA和核糖体亚基彼此间复杂的相互作用(表1)。扫描机制下的翻译起始效率极高,在一个多聚核糖体mRNA上,最快每6 s发生一次,在扫描的过程中也需要大量翻译起始因子的参与[9]。
图1
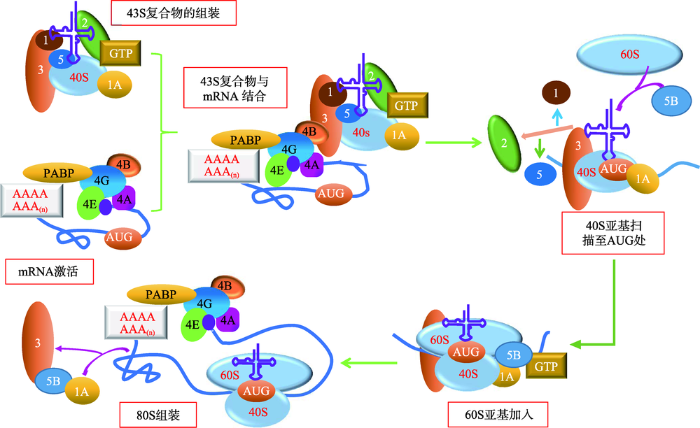 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1真核生物帽依赖性翻译起始机制的模型
Fig. 1A model of cap-dependent translation initiation in eukaryotes
真核生物帽依赖性翻译起始机制分为以下几个步骤:(1) 43S前起始复合物的组装:包括40S亚基、eIF1、eIF1A、eIF3、eIF2-GTP- Met-tRNAiMet以及可能存在的eIF5;(2) mRNA的激活:涉及eIF4E、eIF4G、eIF4B、eIF4A和PABP等与mRNA 5'帽的结合;(3) 43S复合物与激活的mRNA区域结合;(4) 43S复合物从5'到3'方向扫描5'UTR直至识别起始密码子AUG,AUG通过密码子—反密码子相互作用与Met-tRNAi进行碱基互补配对;(5) 60S亚基加入,eIF1、eIF2、eIF5从复合物上释放;(6) eIF5B、eIF1A和eIF3释放,延伸复合物80S亚基形成。参考文献[1]修改绘制。
Table 1
表 1
表 1 表1真核细胞翻译起始因子
Table 1
| 种类 | 名称 | 功能 | |
|---|---|---|---|
| 核心起始 因子 | eIF2 | 形成与40S亚基结合的eIF2-GTP-Met-tRNAi三元复合物,从而介导Met-tRNAi募集核糖体 | |
| eIF3 | 与40S亚基、eIF1、eIF4G和eIF5结合;促进eIF2-GTP-Met-tRNAi与40S亚基的结合;与eIF4G作用,促进43S复合物与mRNA的连接和扫描;具有核糖体解离和抗缔合活性,阻止40S和60S亚基的连接 | ||
| eIF1 | 确保起始密码子选择的忠实性;促进核糖体扫描;刺激eIF2-GTP-Met-tRNAi与40S亚基的结合;防止由eIF5介导的与eIF2结合的GTP提前水解释放Pi | ||
| eIF1A | 刺激eIF2-GTP-Met-tRNAi与40S亚基的结合;与eIF1共同促进核糖体扫描和起始密码子的选择 | ||
| eIF4E | 与mRNA m7G 5°帽结构结合,活性主要受mTOR激酶调节 | ||
| eIF4A* | DEAD-box ATP酶、ATP依赖性的RNA解旋酶 | ||
| eIF4G+ | eIF4E和eIF4A及其他蛋白eIF3、PABP、Mnk1/2激酶结合的支架;增强eIF4A的解旋酶活性 | ||
| eIF4F | 由eIF4E、eIF4A和eIF4G形成的复合物;识别m7GpppG帽结构,展开mRNA 5°近端区域并介导43S复合物的附着(植物eIF4F由eIF4E和eIF4G异二聚体组成,不包括eIF4A,另有eIF4F同系物eIF (iso) 4F,由eIF (iso) 4E和eIF (iso) 4G组成) | ||
| eIF4B | RNA结合蛋白,可增强eIF4A的解旋酶活性 | ||
| 种类 | 名称 | 功能 | |
| 核心起始 因子 | eIF4H | eIF4B的同源片段,可增强eIF4A的解旋酶活性 | |
| eIF5 | GTP酶激活蛋白,特异性诱导在识别起始密码子时与eIF2结合的GTP水解 | ||
| eIF5B | 核糖体依赖性GTP酶,介导核糖体亚基的介入 | ||
| eIF2B | 鸟苷核苷酸交换因子,促进GDP与GTP的互换 | ||
| 辅助因子 | DHX29 | DExH-box结合蛋白,直接与40S亚基结合,帮助扫描具有较长高度结构化5'UTRs区的哺乳动物mRNAs,促进核糖体与其结合 | |
| Ded1 | 与DEAD-box结合的NTP酶、RNA解旋酶 | ||
| eIF6 | 抗缔合因子,可与60S亚基结合,从而阻止其与40S亚基的连接 | ||
| p97 | 与eIF4G羧基末端的2/3处紧密相关;与eIF4A和eIF3结合;以一种潜在的mRNA特异性方式促进翻译起始 | ||
| PABP | 与mRNA的3° poly (A)尾、eIF4G、eRF3结合;增强eIF4F与帽端的结合;并可能协助招募终止后的40S亚基再循环回到mRNA 5'端 | ||
新窗口打开|下载CSV
Jang等[10]发现,40S亚基识别起始密码子的方式除了“核糖体扫描”外,可能还有“RNA looping”现象,他们认为位于43S复合物和起始密码子AUG之间的mRNA片段(即5°UTR)会发生环化,促使二者可以不经过核糖体扫描直接结合(图2)。“RNA looping”存在的一个有力证据是5°UTR(untranslated region)的长度会影响翻译的效率[10],当5°UTR的长度为70个碱基左右时,mRNA的翻译效率最高,当5°UTR的长度逐渐偏离最佳长度时,mRNA的翻译效率会极速下降,但是这一最佳长度却比核糖体与mRNA正常结合时所覆盖的长度长很多,因此推测43S亚基与起始密码子结合时二者之间的mRNA形成了环状结构。
图2
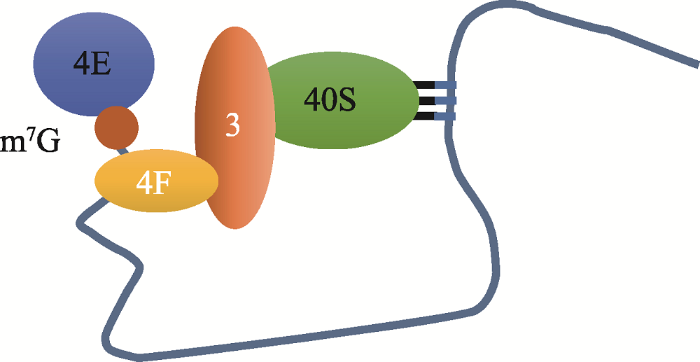 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图243S前起始复合物与起始密码子间的“RNA looping”示意图
Fig. 2“RNA looping” between the 43S precursor complex and the start codon
4E与mRNA m7G 5°帽结构结合,4F识别m7GpppG帽结构,展开5°近端区域并介导43S复合物的附着,43S复合物和起始密码子AUG之间的mRNA片段发生环化形成“RNA looping”。参考文献[10]修改绘制。
5°UTR是翻译起始的高度敏感区,其长短、茎环结构以及主要开放式阅读框(main open reading frames, mORFs)上游AUG(uAUG)的数量都会影响翻译起始的效率[11]。5°UTR的长度一般是100~200个核苷酸[12, 13],少于20个碱基时会发生遗漏扫描(Leaky scanning)[14, 15]。关于为什么最少需要20个碱基,最合理的解释是当40S核糖体亚基的P位点被AUG结合后,起始复合物需要与AUG上游的17个碱基和下游的11个碱基相互作用[16]。茎环结构对翻译起始的抑制作用取决于它在5°UTR的位置、稳定性及其与特定调控蛋白的结合能力[17, 18]。当一个稳定的茎环结构出现在接近m7G帽的位置时,会对翻译起始复合物的组装造成空间干扰,严重影响起始效率;当茎环结构出现在5°UTR下游,43S核糖体的扫描会被抑制,但这种抑制作用可以被与核糖体结合的解旋酶消除;当在AUG的下游出现弱茎环结构时,反而会增加翻译的忠实性,这可能是因为弱茎环结构会造成40S亚基的暂停,为P位点与AUG的结合提供充足的时间[19]。
2 非帽依赖性翻译机制
细胞在生理或病理胁迫环境中会迅速产生应激反应以保护自身,蛋白质合成的重新调整是促进这种反应的主要进程之一[20, 21]。与重新转录出新的mRNA经翻译得到所需蛋白质相比,改变现有mRNA的翻译方式进而产生新的蛋白质的这种途径显然更为快捷。因此,当通过帽依赖性翻译起始机制起始的蛋白质合成大幅减少时,非帽依赖性起始机制就会成为翻译起始的主导,启动保护细胞免受刺激的蛋白质的mRNA的正常翻译[20]。Braunstein等[22]发现,eIF4E结合蛋白(eIF4E-binding protein, 4E-BP)的激活可以响应缺氧和mTORC1抑制,调控帽依赖性机制向非帽依赖性机制的转换,从而有助于肿瘤细胞生长和血管生成,在晚期乳腺癌细胞中翻译调控蛋白4E-BP和起始因子eIF4G会发生过表达,继而导致mRNA进行非帽依赖性翻译起始。生物体mRNA中uAUG的存在非常普遍,约 50%的人类mRNA和果蝇mRNA的5°UTR上含有uAUGs[23]。由于帽依赖性扫描机制从5°UTR的近m7G帽开始扫描,所以一些uAUGs会抑制主要开放式阅读框(main open reading frames, mORFs)的翻译,而要确保核糖体到达目的AUG,就会利用遗漏扫描、重新起始(reinitiation)和核糖体绕道(ribosomal shunting)等方法[24, 25]。另外,近期发现的具有肽编码能力的小开放式阅读框除了存在于正常mRNA的5°UTR外,也多分布于CDS、3°UTR以及非编码RNA上。此时,sORFs及其周围的mORFs的翻译会受到不同类型的非典型翻译起始机制的调控,最终翻译出不同的蛋白质来适应外界环境的刺激[26]。除了上述的遗漏扫描、重新起始和核糖体绕道3种方式之外,当它所在的mRNAs没有5°帽端时,核糖体还可以依赖内部核糖体进入位点或非帽依赖性翻译增强子来介导翻译起始[27]。以下将对这两种非帽依赖性的翻译起始机制进行介绍。
2.1 内部核糖体进入位点介导的翻译起始
1988年,Jang等[28]通过构建双顺反子系统研究发现脑心肌炎病毒(encephalomyocarditis virus, EMCV) RNA序列的5°端UTR区域包含有一个潜在的内部核糖体进入位点(internal ribosome entry sites, IRES),且该序列的260~484位核苷酸对翻译起重要作用。同年,Pelletier等[29]也发现脊髓灰质炎病毒(poliovirus, PV)的5°UTR也具有核糖体结合作用。随后,Chen等[30]发现将微小核糖核酸病毒(picornavirus)的IRES连接到一个缺少5°末端的环状RNA上,该RNA能够被正常翻译,证实了IRES能够起始翻译。IRES是存在于一些mRNA 5'UTR的特异性结构元件,能在mRNA 5°末端缺失的情况下将起始前复合物招募到起始密码子上。IRES介导的翻译起始是一些正义单链RNA病毒翻译起始的主要方式,这些病毒在侵染宿主细胞后,会通过清除起始因子eIF4G、eIF4A、PABP、eIF3亚基或使eIF4E失活等方式降低细胞内的mRNA招募核糖体亚基的效率,导致宿主细胞中的蛋白质合成下调,而病毒mRNA凭借自身含有的IRES结构高效表达。
1991年,Macejak和Sarnow[31]首次在真核细胞中编码免疫球蛋白重链结合蛋白(Bip)的mRNA上发现了IRES。但关于真核细胞中是否也存在IRES介导的翻译起始机制的说法一直没有定论。目前来看,认为真核生物中存在该机制的观点处于上风。最初实验发现,当真核细胞处于内质网应激[32]、缺氧[33]、缺糖损伤[34]、有丝分裂[35]和细胞分化[36]等环境中时,蛋白质的合成会受到全面抑制,但在多项研究中均发现那些5°UTR高度结构化的mRNA仍然可以继续翻译。因此,有人据此推测是因为这些mRNA上的IRES发挥了作用,不过反对者则认为,上述研究中的检测手段受到了现有生物学技术的限制,而且均是在实验室内比较理想的条件下完成的,因此在自然条件下实验可能无法重复[37]。另外一项独立的实验显示,丙型肝炎病毒(hepatitis C virus, HCV)、蟋蟀麻痹病毒(cricket paralysis virus, CrPV)、艾滋病病毒(human immunodeficiency virus, HIV-1)和脑心肌炎病毒EMCV中的IRESs及一些细胞中的IRESs,都可与40S亚基结合,而且40S亚基的头部还需要与核糖体蛋白eS25结合以保证翻译活性,说明IRESs和40S亚基之间存在一种特别普遍的相互作用[38]。近期,Weingarten等[39]通过高通量系统分类方法发现,在随机选择的人类mRNA的5°UTR中,约10%都含有IRES元件。这些潜在的IRES元件可以分为两类:(1)具有整体RNA折叠结构,如二级结构(图3 A);(2)具有招募核糖体的短序列元件,比如能与18s rRNA互补配对的序列结构(图3 B)。此外,细胞中IRES元件的活性有望在Hox mRNAs的翻译调控中得到验证[39],Hox mRNAs是一种复合型RNA调节子,在其折叠的长5°UTR中有一段能阻碍帽依赖性蛋白合成的抑制序列,研究者普遍认为,在时空条件受到限制时,5°UTR中的IRES能够通过核糖体上的eL38招募特定的核糖体而不依赖于5°帽结构(图3 C)。
图3
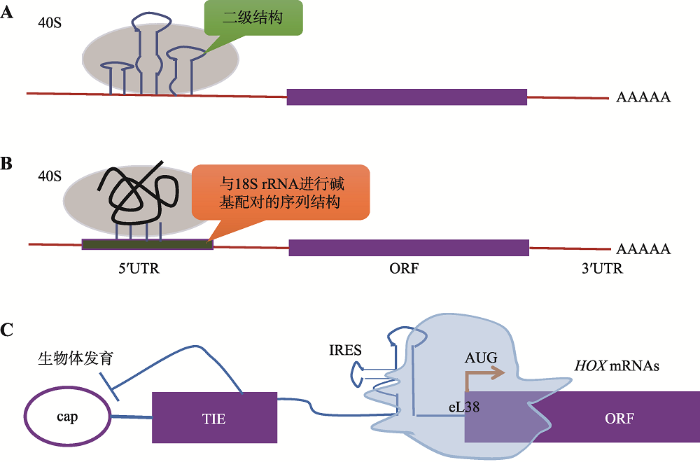 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3真核细胞mRNAs中IRES的预测分类与作用机制
Fig. 3Predictive classification and the mechanism of IRES in eukaryotic mRNAs
IRESs与40S亚基结合促进翻译的起始,IRES元件分为复杂的整体RNA折叠和短序列元件两类。A: IRESs具有整体RNA折叠结构,如二级结构;B: 具有招募核糖体的短序列元件,如能与18S rRNA互补配对的序列结构;C: 复合型RNA调节子Hox mRNAs的5°UTR中的IRES通过核糖体上的eL38招募特定的核糖体,使其以一种非帽依赖性的方式完成翻译。参考文献[39]修改绘制。大多数IRES位于mRNAs的5°UTR,但也有一些位于编码序列(coding sequence, CDS)[40, 41]。关于哪些5°UTR更容易包含IRES,目前尚无明确的划分依据。有实验证明5°UTR的长度、二级结构、上游AUGs的数量、GC含量以及一种Y形结构都不能作为分类依据[42, 43]。IRESs在与40S亚基相互作用时,对典型翻译起始因子的需求降低,但需要特定的IRESs反式作用因子(IRES trans-acting factors, ITAFs)的辅助,这一点在病毒和真核细胞中非常相似(图4)[44, 45]。许多ITAFs都是核不均一核糖核蛋白(hnRNP A1、C1/C2、I、E1/E2、K和L),它们可以在细胞核和细胞质之间穿梭。一些ITAFs,比如hnRNP A1、La、PTB、Unr和ITAF45会响应细胞代谢中的微小变化,并影响IRES的活性[46, 47]。由于几乎在所有编码蛋白质的mRNA中都有IRES出现,为了解哪类蛋白更可能由该机制翻译得到,Lacerda等[2]按照蛋白的功能进行了划分,发现含IRES的mRNA大多编码转录因子、转运蛋白、受体或通道以及生长因子;其次,与细胞凋亡相关的蛋白、热休克蛋白、肿瘤抑制因子、细胞骨架相关蛋白、癌症相关蛋白、RNA结合蛋白、细胞周期蛋白和翻译因子等其他几类蛋白质也都通过IRES依赖的翻译起始机制合成。这些蛋白质在合成时都需要进行微调节,因为它们在某种程度上与细胞至关重要的适应过程和生存过程存在联系。
图4
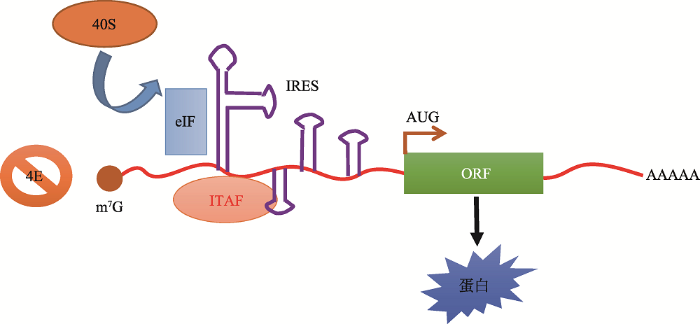 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4内部核糖体进入位点(IRES)依赖性翻译起始的模型
Fig. 4A model of internal ribosome entry site (IRES)-dependent translation initiation
IRESs在与40S亚基相互作用时,需要IRESs反式作用因子ITAFs的辅助,用茎环结构表示的mRNA二级结构直接将40S核糖体亚基招募到开放式阅读框(ORF)的起始密码子(AUG)上或其附近,该过程中,一些典型的起始因子(eIFs)和IRES反式作用因子(ITAF)起到直接或间接的帮助。考文献[2]修改绘制。
2.2 非帽依赖性翻译增强子介导的翻译起始
Terenin等[48]通过实验发现,当将一个病毒mRNA中IRES上的eIF4G结合序列插入到mRNAs的5°UTR后,发现该mRNA在哺乳动物细胞中可以不依赖帽结构通过典型的扫描机制大量翻译。在双顺反子实验中,第2个顺反子不能正常起始翻译,说明此类mRNA的翻译需要5°末端与43S前起始复合物的共同结合,这也证实了以上机制不同于IRES介导机制。但是这种机制虽然不依赖于帽端结构,却仍需要5°末端和非帽依赖性翻译增强子(cap-independent translation enhancer, CITE)的参与[48]。关于CITE能增强翻译起始,有两项证据:(1) CITE能与eIF4F复合物(植物中为eIFiso4F)相互作用,或直接与核糖体亚基结合[49];(2)病毒RNA的CITE与5°端序列频繁发生互补,可以帮助转移由CITE招募的翻译起始因子[50]。CITE分为5°CITE和3°CITE两种,分别位于5°UTR和3°UTR。5°CITE可能与扫描机制中的起始因子结合,但结合能力比较弱,目前研究的比较多的是3°CITE[51,52,53,54]。
3°CITE最早在卫星烟草坏死病毒(satellite tobacco necrosis virus, STNV)的翻译增强结构域(translation enhancer domain, TED)中被发现。STNV由一条1239 bp的亚病毒RNA和外壳蛋白组成,其mRNA 3°UTR区的627~737 bp序列在功能上可以代替5°帽在翻译起始中发挥作用[54]。绝大多数不含5°帽和3°poly (A)尾的植物病毒在3°UTR区至少有一个CITE,可以与翻译起始因子(eIF4E、eIF4G和eIF4F)及核糖体结合来增强翻译起始。根据碱基序列和二级结构的不同,目前已发现的3°CITE可被分为6大类,分别是BTE、PTE、TED、I-shaped、Y-shaped和T-shaped (表2)[55, 56]。Ogawa等[57]将6类不同病毒的最优3°CITE和对应的5°UTR分别添加到小麦胚芽提取物mRNA的3°UTR区,发现选自烟草坏死卫星病毒的120 nt 5°UTR和131 nt 3°CITE增强翻译起始的功能最佳。
Table 2
表2
表2 6类典型3°CITE的特征
Table 2
| 名称 | BTE | PTE | TED | I-shaped | Y-shaped | T-shaped |
|---|---|---|---|---|---|---|
| 代表病毒 | 大麦黄矮病毒 | 黍花叶坏死病毒 | 烟草坏死卫星病毒 | 甜瓜坏死斑病毒 | 番茄丛矮病毒 | 芜菁黄花叶病毒 |
| 二级结构 |  | 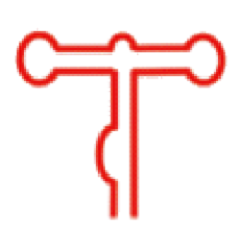 |  | 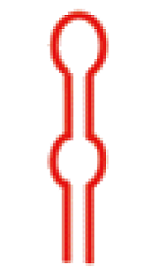 | 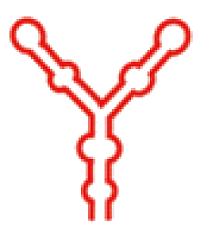 | 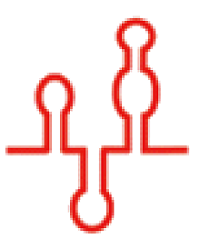 |
| 配体 | eIF4G | eIF4E | eIF4G/E | 核糖体60S亚基 | — | eIF4E/G |
新窗口打开|下载CSV
在翻译起始过程中,3°CITE可以与5°UTR通过RNA-RNA碱基互补配对进行结合,同时与相应的eIF结合,帮助招募43S核糖体亚基进入mRNA 5°端,进而40S亚基对5°UTR进行扫描直至开放式阅读框的起始密码子AUG处,待60S核糖体亚基加入形成80S延伸复合物后继续延伸[55](图5)。有证据显示,一些3°CITE可以发生互换,自然条件下的重组允许模块化的CITE基序进行互换,这可以帮助病毒克服遗传学抗性,拓宽宿主范围[50]。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图53°CITE介导的翻译起始模型
Fig. 5A model of translation initiation mediated by 3°CITE
非帽依赖性翻译增强子3°CITE与5°UTR通过RNA-RNA碱基互补配对进行结合,同时与eIF结合,招募43S核糖体亚基进入mRNA 5°端,进而40S亚基对5°UTR进行扫描直至ORF的起始密码子AUG处。“—”表示该病毒的相应配体尚未被发现。
真核生物的mRNA都拥有帽子结构,在正常情况下不需要通过IRESs来进行翻译起始,但是很有可能需要CITE的辅助[58]。人类APAF-1 (apoptotic protease activating factor 1) mRNA在帽结合因子eIF4E受到抑制时就通过这种机制起始翻译,也就是说APAF-1的5°UTR可以不依赖m7G帽端、但依赖5°末端进行扫描[59]。
3 翻译起始方式的区分
帽依赖性扫描、内部起始(IRES介导)和5°末端依赖性起始(CITE介导)3种翻译起始机制在很多方面存在不同点,特别是与5°帽相关的一些特征(表3)。此外,IRES和CITE的共同点是它们都会与翻译起始过程中的一些元件相结合,然后通过非帽依赖性方式来促进翻译起始;不同点是IRES介导的翻译起始完全不依赖m7G帽结构和5°末端,而CITE介导的翻译起始可以不依赖m7G帽结构,但是需要有独立的5°末端[60]。所以,区分IRES介导和CITE介导的关键在于确认是否依赖5°末端。因此可以通过阻止5°端依赖性翻译起始来区分IRES介导和CITE介导的翻译。因此,有3种方法可以通过阻止5°端依赖性翻译起始来区分IRES介导和CITE介导的翻译:(1)将mRNA环化,5°端将不复存在,这种方法最彻底;(2)利用双顺反子mRNA,当两个顺反子间缺少翻译耦合,不能同时翻译时(比如再起始),则必须由非5°端依赖性的翻译机制进行翻译;(3)在5°端加 入一个稳定的茎环结构以阻止核糖体招募,从而 消除5°端的作用[60]。其中采用较多的是构建双顺 反子[58, 61~63]。Table 3
表3
表3 3种翻译起始的特征
Table 3
| 类别 | 帽依赖性 | IRES | CITE |
|---|---|---|---|
| eIF4G解离 | 敏感 | 不敏感a | 不敏感a |
| mTORC1抑制剂 | 敏感 | 不敏感 | 不敏感a |
| 4E-BP或m7GTP | 敏感 | 不敏感a | 不敏感 |
| 双顺反子活性 | 无 | 有 | 无 |
| 帽依赖程度 | 强或一致 | 弱b | 弱 |
| 含帽mRNA活性 | 弱或一致 | 弱或一致或强 | 强或一致 |
新窗口打开|下载CSV
关于如何有效辨别mRNA是通过哪种机制进行翻译的,Terenin等[60]认为,首先将所要研究的mRNA的5°UTR克隆到双顺反子表达载体上;然后进行双顺反子表达、无启动子质粒表达以及RNAi (第一个顺反子沉默)表达试验,初步判断翻译起始类型;最后在mRNA水平上构建不同的顺反子,根据帽依赖性的强弱和第二个顺反子的表达情况确认具体的翻译起始方式(图6)。以DNA为基础的试验都不可避免的要进行以RNA为基础的试验,所以可以直接从RNA试验开始。
图6
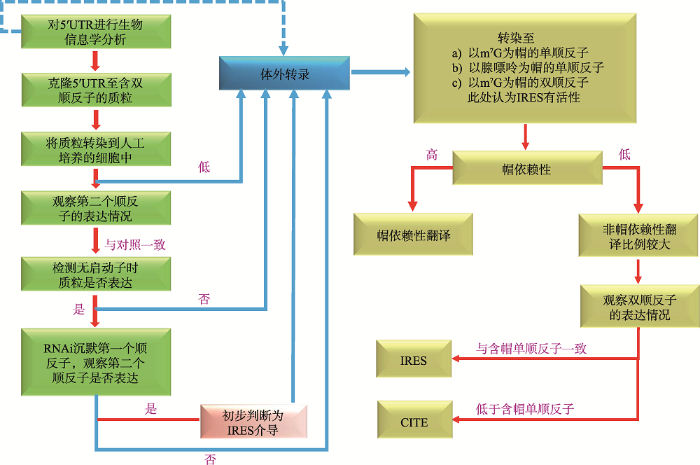 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6区分翻译起始方式的方法
Fig. 6A generalized workflow of how to address internal initiation
通过分子试验分析mRNA的翻译机制,分别构建相应的双顺反子表达载体;进行双顺反子表达、无启动子质粒表达以及RNAi表达,初步判断翻译起始类型;最后在mRNA水平上明确翻译起始方式。参考文献[60]修改绘制。
4 结语与展望
在一个哺乳动物细胞中,同一时间约有5000条不同的mRNAs在接近核糖体并进行翻译。起始过程是翻译的开端,也是翻译调控的关键。最早发现的、最经典的翻译起始方式是5°帽依赖性扫描机制,但是当细胞受到胁迫时,这种机制的作用将会受到大幅度的抑制。此时,非帽依赖性机制将成为主要的翻译起始方式,从而保证细胞在逆境中的生存和恢复。其中研究得最多的是IRES介导的翻译起始,但到目前为止,真核生物mRNA中的IRES是否能介导翻译起始仍没有确凿证据,Hox mRNA在人为控制的条件下可以由IRES介导翻译,但是在自然条件下是否确实如此,还需要有进一步的实验证据。当然,还有许多其他的非经典翻译起始机制逐渐被发现。2008年,Elfakess等[64]在用生物信息学工具搜索启动子邻近区域的调控元件时,发现了一种SAASATGGCGGC (S为C或G)序列,命名为短5°UTRs翻译起始因子(translation initiator of short 5′UTRs, TISU)。TISU存在于4.5%的哺乳动物蛋白质编码基因中,特别是缺少TATA-box启动子的基因,并严格定位于转录起始位点(transcription start site, TSS)下游+5至+30处。最初发现它是转录过程中的重要元件,活性受YY1转录因子的介导。此外,TISU序列中有一个恒定的ATG,实验证明它也能在翻译过程中发挥作用,在64%含有TISU序列的基因中,其中的AUG被识别为起始密码子。重要的是这些基因转录得到的mRNA的5′UTR极短,平均长度只有12个碱基。后来Elfakess等[65, 66]继续证实了TISU介导的翻译起始实际上是一种依赖m7G帽结构但无需扫描的机制,43S前起始复合物被招募后,可以不经过核糖体的扫描,也没有“RNA looping”的帮助,而是直接与起始密码子TISU序列中的AUG进行结合,继而起始翻译过程(图7)。
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7含TISU元件mRNA的帽依赖性不扫描翻译起始机制
Fig. 7A scheme describing cap-dependent and scannin-independent translation initiation of mRNAs bearing the TISU element
TISU序列中的AUG被识别为起始密码子,由于5°UTR极短,所以AUG的位置就在m7G帽端附近,43S前起始复合物不经过核糖体的扫描,直接与TISU序列中的AUG进行结合起始翻译。参考文献[1]修改绘制。
此外,许多真核细胞mRNAs都有被N6-甲基腺苷(N6-methyladenosine, m6A)残基修饰的现象,这是一种普遍的碱基修饰方式,在3°UTR、蛋白质编码区、5°UTR处均存在。近期有证据显示,5°UTR上的m6A可以代替5°帽起到促进翻译起始的作用,它们可以直接与eIF3和43S核糖体亚基结合以起始翻译,而且不需要帽结合蛋白eIF4E的协助,该位点也被称为是“m6A介导的核糖体结合位点(m6A-induced ribosome engagement sites, MIRES)”,当腺苷甲基化被抑制时,5°UTR上含有m6A的mRNA的翻译会被选择性抑制。而且,在热激反应中,Hsp70 mRNA会通过提高自身的m6A水平来调控其非帽依赖性翻译机制。但是由m6A介导的翻译起始仍然需要核 糖体亚基对5°UTR进行扫描,而且m6A是如何被 识别并促进非帽依赖性翻译起始的,仍然需要深入研究[67, 68]。
综上所述,针对于真核生物mRNA翻译起始机制的研究在未来将集中在两个方面:一是深入探究已发现的翻译起始机制的分子机理及其调控方式,二是挖掘更多的翻译起始机制。如果能够揭示生物体在不同环境条件下,特别是在响应环境胁迫或病理状态时,不同的翻译方式是如何交替发挥作用的,将对深入认识基因在翻译水平上的表达调控有重要意义。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:26381322 [本文引用: 5]

61Canonical translation involves mRNA cap recognition and ribosomal scanning.61Some mRNAs direct translation initiation that deviates from the canonical mechanism.61This review describes various mechanisms of cap-dependent translation initiation.61A particular focus is given to mRNAs with extremely short or highly complex 5′ UTRs.61These mRNAs drive cap-dependent but scanning free translation initiation.
URL [本文引用: 3]
URLPMID:2645293 [本文引用: 1]

The small (40S) subunit of eukaryotic ribosomes is believed to bind initially at the capped 5'-end of messenger RNA and then migrate, stopping at the first AUG codon in a favorable context for initiating translation. The first-AUG rule is not absolute, but there are rules for breaking the rule. Some anomalous observations that seemed to contradict the scanning mechanism now appear to be artifacts. A few genuine anomalies remain unexplained.
URLPMID:29414693 [本文引用: 1]

Eukaryotic cells evolved highly complex and accurate protein synthesis machinery that is finely tuned by various signaling pathways. Dysregulation of translation is a hallmark of many diseases, including cancer, and thus pharmacological approaches to modulate translation become very promising. While there has been much progress in our understanding of mammalian mRNA-specific translation control, surprisingly, relatively little is known about whether and how the protein components of the translation machinery shape translation of their own mRNAs. Here we analyze mammalian mRNAs encoding components of the translation initiation machinery for potential regulatory features such as 5 OP motifs, TISU motifs, poor start codon nucleotide context and upstream open reading frames.
URLPMID:20922269 [本文引用: 1]

Maintenance of cell homeostasis and regulation of cell proliferation depend importantly on regulating the process of protein synthesis. Many disease states arise when disregulation of protein synthesis occurs. This review focuses on mechanisms of translational control and how disregulation results in cell malignancy. Most translational controls occur during the initiation phase of protein synthesis, with the initiation factors being the major target of regulation through their phosphorylation. In particular, the recruitment of mRNAs through the m7G-cap structure and the binding of the initiator methionyl-tRNAi are frequent targets. However, translation, especially of specific mRNAs, may also be regulated by sequestration into processing bodies or stress granules, by trans-acting proteins or by microRNAs. When the process of protein synthesis is hyper-activated, weak mRNAs are translated relatively more efficiently, leading to an imbalance of cellular proteins that promotes cell proliferation and malignant transformation. This occurs, for example, when the cap-binding protein, eIF4E, is overexpressed, or when the methionyl-tRNAi-binding factor, eIF2, is too active. In addition, enhanced activity of eIF3 contributes to oncogenesis. The importance of the translation initiation factors as regulators of protein synthesis and cell proliferation makes them potential therapeutic targets for the treatment of cancer.
URLPMID:26901872 [本文引用: 1]

Eukaryotic cells restrict protein synthesis under various stress conditions, by inhibiting the eukaryotic translation initiation factor 2B (eIF2B). eIF2B is the guanine nucleotide exchange factor for eIF2, a heterotrimeric G protein consisting of α-, β- and γ-subunits. eIF2B exchanges GDP for GTP on the γ-subunit of eIF2 (eIF2γ), and is inhibited by stress-induced phosphorylation of eIF2α. eIF2B is a heterodecameric complex of two copies each of the α-, β-, γ-, δ- and ε-subunits; its α-, β- and δ-subunits constitute the regulatory subcomplex, while the γ- and ε-subunits form the catalytic subcomplex. The three-dimensional structure of the entire eIF2B complex has not been determined. Here we present the crystal structure of Schizosaccharomyces pombe eIF2B with an unprecedented subunit arrangement, in which the αβδhexameric regulatory subcomplex binds two γε dimeric catalytic subcomplexes on its opposite sides. A structure-based in vitro analysis by a surface-scanning site-directed photo-cross-linking method identified the eIF2α-binding and eIF2γ-binding interfaces, located far apart on the regulatory and catalytic subcomplexes, respectively. The eIF2γ-binding interface is located close to the conserved ‘NF motif’, which is important for nucleotide exchange. A structural model was constructed for the complex of eIF2B with phosphorylated eIF2α, which binds to eIF2B more strongly than the unphosphorylated form. These results indicate that the eIF2α phosphorylation generates the ‘nonproductive’ eIF2–eIF2B complex, which prevents nucleotide exchange on eIF2γ, and thus provide a structural framework for the eIF2B-mediated mechanism of stress-induced translational control.
URLPMID:16246168 [本文引用: 1]

Abstract In response to environmental stresses, a family of protein kinases phosphorylate eIF2 (eukaryotic initiation factor 2) to alleviate cellular injury or alternatively induce apoptosis. Phosphorylation of eIF2 reduces global translation, allowing cells to conserve resources and to initiate a reconfiguration of gene expression to effectively manage stress conditions. Accompanying this general protein synthesis control, eIF2 phosphorylation induces translation of specific mRNAs, such as that encoding the bZIP (basic leucine zipper) transcriptional regulator ATF4 (activating transcription factor 4). ATF4 also enhances the expression of additional transcription factors, ATF3 and CHOP (CCAAT/enhancer-binding protein homologous protein)/GADD153 (growth arrest and DNA-damage-inducible protein), that assist in the regulation of genes involved in metabolism, the redox status of the cells and apoptosis. Reduced translation by eIF2 phosphorylation can also lead to activation of stress-related transcription factors, such as NF-kappaB (nuclear factor kappaB), by lowering the steady-state levels of short-lived regulatory proteins such as IkappaB (inhibitor of NF-kappaB). While many of the genes induced by eIF2 phosphorylation are shared between different environmental stresses, eIF2 kinases function in conjunction with other stress-response pathways, such as those regulated by mitogen-activated protein kinases, to elicit gene expression programmes that are tailored for the specific stress condition. Loss of eIF2 kinase pathways can have important health consequences. Mice devoid of the eIF2 kinase GCN2 [general control non-derepressible-2 or EIF2AK4 (eIF2alpha kinase 4)] show sensitivity to nutritional deficiencies and aberrant eating behaviours, and deletion of PEK [pancreatic eIF2alpha kinase or PERK (RNA-dependent protein kinase-like endoplasmic reticulum kinase) or EIF2AK3] leads to neonatal insulin-dependent diabetes, epiphyseal dysplasia and hepatic and renal complications.
URLPMID:25110031 [本文引用: 1]

Abstract The cap-binding translation initiation factor eIF4E (eukaryotic initiation factor 4E) is central to protein synthesis in eukaryotes. As an integral component of eIF4F, a complex also containing the large bridging factor eIF4G and eIF4A RNA helicase, eIF4E enables the recruitment of the small ribosomal subunit to the 5' end of mRNAs. The interaction between eIF4E and eIF4G via a YXXXXL脧聲 motif is regulated by small eIF4E-binding proteins, 4E-BPs, which use the same sequence to competitively bind eIF4E thereby inhibiting cap-dependent translation. Additional eIF4E-binding proteins have been identified in the last 10-15 years, characterized by the YXXXXL脧聲 motif, and by interactions (many of which remain to be detailed) with RNA-binding proteins, or other factors in complexes that recognize the specific mRNAs. In the present article, we focus on the metazoan 4E-T (4E-transporter)/Cup family of eIF4E-binding proteins, and also discuss very recent examples in yeast, fruitflies and humans, some of which predictably inhibit translation, while others may result in mRNA decay or even enhance translation; altogether considerably expanding our understanding of the roles of eIF4E-binding proteins in gene expression regulation.
URLPMID:4461372 [本文引用: 3]

Protein synthesis is principally regulated at the initiation stage (rather than during elongation or termination), allowing rapid, reversible and spatial control of gene expression. Progress over recent years in determining the structures and activities of initiation factors, and in mapping their interactions in ribosomal initiation complexes, have advanced our understanding of the complex translation initiation process. These developments have provided a solid foundation for studying the regulation of translation initiation by mechanisms that include the modulation of initiation factor activity (which affects almost all scanning-dependent initiation) and through sequence-specific RNA-binding proteins and microRNAs (which affect individual mRNAs).
[本文引用: 3]
URLPMID:5382211 [本文引用: 1]

Viral protein synthesis is completely dependent upon the host cell's translational machinery. Canonical translation of host mRNAs depends on structural elements such as the 5 cap structure and/or the 3 poly(A) tail of the mRNAs. Although many viral mRNAs are devoid of one or both of these structures, they can still translate efficiently using non-canonical mechanisms. Here, we review the tools utilized by positive-sense single-stranded (+ss) RNA plant viruses to initiate non-canonical translation, focusing oncis-acting sequences present in viral mRNAs. We highlight how these elements may interact with host translation factors and speculate on their contribution for achieving translational control. We also describe other translation strategies used by plant viruses to optimize the usage of the coding capacity of their very compact genomes, including leaky scanning initiation, ribosomal frameshifting and stop-codon readthrough. Finally, future research perspectives on the unusual translational strategies of +ssRNA viruses are discussed, including parallelisms between viral and host mRNAs mechanisms of translation, particularly for host mRNAs which are translated under stress conditions.
URLPMID:18298820 [本文引用: 1]

pAbstract/p pBackground/p pDiversity in rates of gene expression is essential for basic cell functions and is controlled by a variety of intricate mechanisms. Revealing general mechanisms that control gene expression is important for understanding normal and pathological cell functions and for improving the design of expression systems. Here we analyzed the relationship between general features of genes and their contribution to expression levels./p pResults/p pGenes were divided into four groups according to their core promoter type and their characteristics analyzed statistically. Surprisingly we found that small variations in the TATA box are linked to large differences in gene length. Genes containing canonical TATA are generally short whereas long genes are associated with either non-canonical TATA or TATA-less promoters. These differences in gene length are primarily determined by the size and number of introns. Generally, gene expression was found to be tightly correlated with the strength of the TATA-box. However significant reduction in gene expression levels were linked with long TATA-containing genes (canonical and non-canonical) whereas intron length hardly affected the expression of TATA-less genes. Interestingly, features associated with high translation are prevalent in TATA-containing genes suggesting that their protein production is also more efficient./p pConclusion/p pOur results suggest that interplay between core promoter type and gene size can generate significant diversity in gene expression./p
URLPMID:11591473 [本文引用: 1]

The crucial role of the non-coding portion of genomes is now widely acknowledged. In particular, mRNA untranslated regions are involved in many post-transcriptional regulatory pathways that control mRNA localization, stability and translation efficiency. We review in this paper the major structural and compositional features of eukaryotic mRNA untranslated regions and provide some examples of bioinformatic analyses for their functional characterization.
URLPMID:1820208 [本文引用: 1]

The functional consequences of unusually short 5' noncoding sequences on eukaryotic mRNAs are explored here by using an in vitro transcription and translation system. As the distance of the first AUG codon from the m7G cap was decreased from 32 to 3 nucleotides, the yield of protein initiated from the first AUG codon progressively decreased, with a corresponding increase in initiation from the second AUG codon. The leakiness attributable to a too-short leader sequence was offset, however, by introducing secondary structure downstream from the first AUG codon.
URLPMID:1820209 [本文引用: 1]

Lengthening the 5' noncoding sequence on SP6-derived transcripts can increase their translational efficiency by an order of magnitude under some conditions of translation in reticulocyte lysates. This effect was observed upon reiterating three different synthetic oligonucleotides, the sequences of which were designed simply to preclude secondary structure. It seems unlikely that such arbitrarily designed sequences are recognized by sequence-specific translational enhancer proteins. Rather, long 5' leader sequences appear to accumulate extra 40S ribosomal subunits, which may account for their translational advantage. The buildup of 40S subunits on long, unstructured leader sequences is predicted by the scanning model for initiation. Leader sequences such as these may be ideal for in vitro expression vectors.
URL [本文引用: 1]
URLPMID:12459250 [本文引用: 1]

Selection of the translational initiation site in most eukaryotic mRNAs appears to occur via a scanning mechanism which predicts that proximity to the 5′ end plays a dominant role in identifying the start codon. This ‘position effect’ is seen in cases where a mutation creates an AUG codon upstream from the normal start site and translation shifts to the upstream site. The position effect is evident also in cases where a silent internal AUG codon is activated upon being relocated closer to the 5′ end. Two mechanisms for escaping the first-AUG rule – reinitiation and context-dependent leaky scanning – enable downstream AUG codons to be accessed in some mRNAs. Although these mechanisms are not new, many new examples of their use have emerged. Via these escape pathways, the scanning mechanism operates even in extreme cases, such as a plant virus mRNA in which translation initiates from three start sites over a distance of 900 nt. This depends on careful structural arrangements, however, which are rarely present in cellular mRNAs. Understanding the rules for initiation of translation enables understanding of human diseases in which the expression of a critical gene is reduced by mutations that add upstream AUG codons or change the context around the AUG START codon. The opposite problem occurs in the case of hereditary thrombocythemia: translational efficiency is increased by mutations that remove or restructure a small upstream open reading frame in thrombopoietin mRNA, and the resulting overproduction of the cytokine causes the disease. This and other examples support the idea that 5′ leader sequences are sometimes structured deliberately in a way that constrains scanning in order to prevent harmful overproduction of potent regulatory proteins. The accumulated evidence reveals how the scanning mechanism dictates the pattern of transcription – forcing production of monocistronic mRNAs – and the pattern of translation of eukaryotic cellular and viral genes.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:19917642 [本文引用: 2]

Abstract The IRESite (http://www.iresite.org) presents carefully curated experimental evidence of many eukaryotic viral and cellular internal ribosome entry site (IRES) regions. At the time of submission, IRESite stored >600 records. The IRESite gradually evolved into a robust tool providing (i) biologically meaningful information regarding the IRESs and their experimental background (including annotation of IRES secondary structures and IRES trans-acting factors) as well as (ii) thorough concluding remarks to stored database entries and regularly updated evaluation of the reported IRES function. A substantial portion of the IRESite data results purely from in-house bioinformatic analyses of currently available sequences, in silico attempts to repeat published cloning experiments, DNA sequencing and restriction endonuclease verification of received plasmid DNA. We also present a newly implemented tool for displaying RNA secondary structures and for searching through the structures currently stored in the database. The supplementary material contains an updated list of reported IRESs.
URLPMID:20965418 [本文引用: 1]

A number of stresses, including nutrient stress, temperature shock, DNA damage, and hypoxia, can lead to changes in gene expression patterns caused by a general shutdown and reprogramming of protein synthesis. Each of these stress conditions results in selective recruitment of ribosomes to mRNAs whose protein products are required for responding to stress. This recruitment is regulated by elements within the 5′ and 3′ untranslated regions of mRNAs, including internal ribosome entry segments, upstream open reading frames, and microRNA target sites. These elements can act singly or in combination and are themselves regulated by trans-acting factors. Translational reprogramming can result in increased life span, and conversely, deregulation of these translation pathways is associated with disease including cancer and diabetes.
URLPMID:17996713 [本文引用: 1]

Translational regulation is critical in cancer development and progression. Translation sustains tumor growth and development of a tumor vasculature, a process known as angiogenesis, which is activated by hypoxia. Here we first demonstrate that a majority of large advanced breast cancers overexpress translation regulatory protein 4E-BP1 and initiation factor eIF4G. Using model animal and cell studies, we then show that overexpressed 4E-BP1 and eIF4G orchestrate a hypoxia-activated switch from cap-dependent to cap-independent mRNA translation that promotes increased tumor angiogenesis and growth at the level of selective mRNA translation. Elevated levels of 4E-BP1 trigger hypoxia inhibition of cap-dependent mRNA translation at high-oxygen levels and, with eIF4G, increase selective translation of mRNAs containing internal ribosome entry sites (IRESs) that include key proangiogenic, hypoxia, and survival mRNAs. The switch from cap-dependent to cap-independent mRNA translation facilitates tumor angiogenesis and hypoxia responses in animal models.
URLPMID:21663794 [本文引用: 1]

78 Drosophila SXL protein cooperates with a uORF to regulate msl-2 mRNA translation 78 SXL-mediated regulation targets translation initiation, not elongation or termination 78 SXL promotes ribosome recognition of a uORF, thereby inhibiting msl-2 translation 78 Protein-regulated uORFs may define a general mechanism for the control of translation
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:26684391 [本文引用: 1]

Small open reading frames (sORFs) that encode peptides that are 100 amino acids or fewer in length are underrepresented in genome annotations. Ribosome profiling techniques, along with protein mass spectrometry and new bioinformatics approaches, are being used to support annotation of these features. sORFs have been found in coding and noncoding RNA. In plants and animals, several sORFs have been shown to initiate with non-AUG start codons.
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]

mRNA翻译的起动有两种机制:帽结构依赖性的翻译和IRES介导的非帽结构依赖的翻译。在RNA病毒和细胞内的部分mRNA中,蛋白合成的起始是由包含在5’端非翻译区(5’-TuR)结构中的IRES介导的,IRES能够在一些反式作用因子的参与下,募集核糖体40s亚单位,开始mRNA的翻译过程。不同病毒的IRES在结构和功能上,既存在保守性,又有各自的特点。病毒IRES序列在病毒基因的表达、复制和致病性上具有至关重要作用,对病毒IRES功能和调控的研究是当前病毒学研究热点之一。本文概述了近年来RNA病毒IRES的几种分型及各自的特点以及IRES的应用。
URLPMID:1652694 [本文引用: 1]

A Robosome-scanning model has been proposed to explain the initiation of eukaryotic messenger RNAs in which binding of the 43S ternary ribosomal subunit near or at the 5' end of the mRNA is facilitated by an interaction between the methylated cap-structure at the end of the mRNA and the cap-binding protein complex eIF-4F. But picornaviral mRNAs do not have a 5' terminal cap structure and are translated by internal ribosome binding. A cellular mRNA, encoding the immunoglobulin heavy-chain binding protein, can be translated in poliovirus-infected cells at a time when cap-dependent translation of host cell mRNAs is inhibited. We report here that the 5' leader of the binding protein mRNA can directly confer internal ribosome binding to an mRNA in mammalian cells, indicating that translation initiation by an internal ribosome-binding mechanism is used by eukaryotic mRNAs.
URL [本文引用: 1]
Sci Signal,
[本文引用: 1]
URLPMID:25721046 [本文引用: 1]

Tumor suppressor protein p53 is a master transcription regulator, indispensable for controlling several cellular pathways. Earlier work in our laboratory led to the identification of dual internal ribosome entry site (IRES) structure of p53 mRNA that regulates translation of full-length p53 and 40p53. IRES-mediated translation of both isoforms is enhanced under different stress conditions that induce DNA damage, ionizing radiation and endoplasmic reticulum stress, oncogene-induced senescence and cancer. In this study, we addressed nutrient-mediated translational regulation of p53 mRNA using glucose depletion. In cell lines, this nutrient-depletion stress relatively induced p53 IRES activities from bicistronic reporter constructs with concomitant increase in levels of p53 isoforms. Surprisingly, we found scaffold/matrix attachment region-binding protein 1 (SMAR1), a predominantly nuclear protein is abundant in the cytoplasm under glucose deprivation. Importantly under these conditions polypyrimidine-tract-binding protein, an established p53 ITAF did not show nuclear-cytoplasmic relocalization highlighting the novelty of SMAR1-mediated control in stress. In vivo studies in mice revealed starvation-induced increase in SMAR1, p53 and 40p53 levels that was reversible on dietary replenishment. SMAR1 associated with p53 IRES sequences ex vivo, with an increase in interaction on glucose starvation. RNAi-mediated-transient SMAR1 knockdown decreased p53 IRES activities in normal conditions and under glucose deprivation, this being reflected in changes in mRNAs in the p53 and 40p53 target genes involved in cell-cycle arrest, metabolism and apoptosis such as p21, TIGAR and Bax. This study provides a new physiological insight into the regulation of this critical tumor suppressor in nutrient starvation, also suggesting important functions of the p53 isoforms in these conditions as evident from the downstream transcriptional target activation.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:28684008 [本文引用: 1]

react-text: 466 The protein synthetic machinery is a highly complex apparatus that offers many potential sites for functional interference and represents a major target in the cell for antibiotics. The knowledge of ribosomal structure and function has progressed enormously in recent years, which has, in turn, accelerated our understanding of the mechanism of drug action. Conversely, drugs have been used as... /react-text react-text: 467 /react-text [Show full abstract]
URL [本文引用: 1]
[本文引用: 3]
URL [本文引用: 1]
URLPMID:23318444 [本文引用: 1]

Abstract Translational regulation of the p53 mRNA can determine the ratio between p53 and its N-terminal truncated isoforms and therefore has a significant role in determining p53-regulated signaling pathways. Although its importance in cell fate decisions has been demonstrated repeatedly, little is known about the regulatory mechanisms that determine this ratio. Two internal ribosome entry sites (IRESs) residing within the 5'UTR and the coding sequence of p53 mRNA drive the translation of full-length p53 and 020040p53 isoform, respectively. Here, we report that DAP5, a translation initiation factor shown to positively regulate the translation of various IRES containing mRNAs, promotes IRES-driven translation of p53 mRNA. Upon DAP5 depletion, p53 and 020040p53 protein levels were decreased, with a greater effect on the N-terminal truncated isoform. Functional analysis using bicistronic vectors driving the expression of a reporter gene from each of these two IRESs indicated that DAP5 preferentially promotes translation from the second IRES residing in the coding sequence. Furthermore, p53 mRNA expressed from a plasmid carrying this second IRES was selectively shifted to lighter polysomes upon DAP5 knockdown. Consequently, 020040p53 protein levels and the subsequent transcriptional activation of the 14-3-30303 gene, a known target of 020040p53, were strongly reduced. In addition, we show here that DAP5 interacts with p53 IRES elements in in vitro and in vivo binding studies, proving for the first time that DAP5 directly binds a target mRNA. Thus, through its ability to regulate IRES-dependent translation of the p53 mRNA, DAP5 may control the ratio between different p53 isoforms encoded by a single mRNA.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:15749702 [本文引用: 1]

Abstract Although studies on viral gene expression were essential for the discovery of internal ribosome entry sites (IRESs), it is becoming increasingly clear that IRES activities are present in a significant number of cellular mRNAs. Remarkably, many of these IRES elements initiate translation of mRNAs encoding proteins that protect cells from stress (when the translation of the vast majority of cellular mRNAs is significantly impaired). The purpose of this review is to summarize the progress on the discovery and function of cellular IRESs. Recent findings on the structures of these IRESs and specifically regulation of their activity during nutritional stress, differentiation, and mitosis will be discussed.
URLPMID:28115626 [本文引用: 1]

The type II arginine methyltransferase PRMT5 is responsible for the symmetric dimethylation of histone to generate the H3R8me2s and H4R3me2s marks, which correlate with the repression of transcription. However, the protein level of a number of genes (MEP50, CCND1, MYC, HIF1a, MTIFandCDKN1B) are reported to be downregulated by the loss of PRMT5, while their mRNA levels remain unchanged, which is counterintuitive for PRMT5's proposed role as a transcription repressor. We noticed that the majority of the genes regulated by PRMT5, at the posttranscriptional level, express mRNA containing an internal ribosome entry site (IRES). Using an IRES-dependent reporter system, we established that PRMT5 facilitates the translation of a subset of IRES-containing genes. The heterogeneous nuclear ribonucleoprotein, hnRNP A1, is an IRES transacting factor (ITAF) that regulates the IRES-dependent translation of Cyclin D1 and c-Myc. We showed that hnRNP A1 is methylated by PRMT5 on two residues, R218 and R225, and that this methylation facilitates the interaction of hnRNP A1 with IRES RNA to promote IRES-dependent translation. This study defines a new role for PRMT5 regulation of cellular protein levels, which goes beyond the known functions of PRMT5 as a transcription and splicing regulator.
URL [本文引用: 1]
URL [本文引用: 2]
URL [本文引用: 1]
URLPMID:23682606 [本文引用: 2]

In the absence of a 5 ' cap, plant positive-strand RNA viruses have evolved a number of different elements in their 3 ' untranslated region (UTR) to attract initiation factors and/or ribosomes to their templates. These 3 ' cap-independent translational enhancers (3 ' CITEs) take different forms, such as I-shaped, Y-shaped, T-shaped, or pseudoknotted structures, or radiate multiple helices from a central hub. Common features of most 3 ' CITEs include the ability to bind a component of the translation initiation factor eIF4F complex and to engage in an RNA-RNA kissing-loop interaction with a hairpin loop located at the 5 ' end of the RNA. The two T-shaped structures can bind to ribosomes and ribosomal subunits, with one structure also able to engage in a simultaneous long-distance RNA-RNA interaction. Several of these 3 ' CITEs are interchangeable and there is evidence that natural recombination allows exchange of modular CITE units, which may overcome genetic resistance or extend the virus's host range.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:25662021 [本文引用: 1]

Carmovirus is a genus of small, single-stranded, positive-strand RNA viruses in the Tombusviridae. One member of the carmoviruses, Turnip crinkle virus (TCV), has been used extensively as a model for examining the structure and function of RNA elements in 3′UTR as well as in other regions of the virus. Using a variety of genetic, biochemical and computational methods, a structure for the TCV 3′UTR has emerged where secondary structures and tertiary interactions combine to adopt higher order 3-D structures including an internal, ribosome-binding tRNA-shaped configuration that functions as a 3′ cap-independent translation enhancer (3′CITE). The TCV 3′CITE also serves as a scaffold for non-canonical interactions throughout the 3′UTR and extending into the upstream open reading frame, interactions that are significantly disrupted upon binding by the RNA-dependent RNA polymerase. Long-distance interactions that connect elements in the 3′UTR with both the 5′ end and the internal ribosome recoding site suggest that 3′UTR of carmoviruses are intimately involved in multiple functions in the virus life cycle. Although carmoviruses share very similar genome organizations, lengths of 5′ and 3′UTRs, and structural features at the 3′ end, the similarity rapidly breaks down the further removed from the 3′ terminus revealing different 3′CITEs and unique virus-specific structural features. This review summarizes 20 years of work dissecting the structure and function of the 3′UTR of TCV and other carmoviruses. The astonishing structural complexity of the 3′UTRs of these simple carmoviruses provides lessons that are likely applicable to many other plant and animal RNA viruses.
URL [本文引用: 2]
URLPMID:22440838 [本文引用: 2]

Highlights? Plant viruses utilize unique mechanisms to hijack the host translational machinery. ? 3′CITEs are a class of RNA element that performs this function in Tombusviridae. ? 3′CITEs operate by binding to translation initiation factors or ribosomal subunits. ? Major advances in the field as well as future prospects are discussed herein.
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]
URL [本文引用: 1]
URLPMID:27853833 [本文引用: 5]

The idea of internal initiation is frequently exploited to explain the peculiar translation properties or unusual features of some eukaryotic mRNAs. In this review, we summarize the methods and argume
URLPMID:3419317 [本文引用: 1]

Transcriptional regulation of gene expression has been widely studied. More recently, there has been increasing appreciation of the role that translational regulation plays in gene expression, resulting in a number of new fields engaging in translational studies. Regulation of protein synthesis is critical for cell growth, development, and survival, and is primarily controlled at the initiation step. Eukaryotic cells utilize multiple mechanisms to initiate translation, depending on cell stress, growth conditions, viral infection, or the sequences present in the mRNA. While the vast majority of mRNAs are translated in a cap-dependent manner, an important subset of mRNAs uses an alternative mechanism, whereby ribosomes are recruited internally to the message to initiate cap-independent translation. Some of these mRNAs contain an internal ribosome entry site (IRES) located in the 5芒聙虏 untranslated region (UTR). However, establishing that an RNA element is a functional IRES requires a number of carefully executed experiments with specific controls. This review will clearly explain the required experiments, and the pros and cons of various assays, used to determine whether (or not) an RNA element functions as an IRES to promote initiation of translation. We hope that demystifying the accepted methods for assaying IRES activity will open the study of this important mechanism to the broader community. WIREs RNA 2012 doi: 10.1002/wrna.1129For further resources related to this article, please visit the WIREs website.
URLPMID:2937932

Internal ribosome entry sites (IRESs) are specialized mRNA elements that allow recruitment of eukaryotic ribosomes to naturally uncapped mRNAs or to capped mRNAs under conditions in which cap-dependent translation is inhibited. Putative cellular IRESs have been proposed to play crucial roles in stress responses, development, apoptosis, cell cycle control, and neuronal function. However, most of the evidence for cellular IRES activity rests on bicistronic reporter assays, the reliability of which has been questioned. Here, the mechanisms underlying cap-independent translation of cellular mRNAs and the contributions of such translation to cellular protein synthesis are discussed. I suggest that the division of cellular mRNAs into mutually exclusive categories of "cap-dependent" and "IRES-dependent" should be reconsidered and that the implications of cellular IRES activity need to be incorporated into our models of cap-dependent initiation.
URLPMID:3552511 [本文引用: 1]

Internal ribosome entry sites/segments (IRESs) were first discovered over 20 years ago in picornaviruses, followed by the discovery of two other types of IRES in hepatitis C virus (HCV), and the dicistroviruses, which infect invertebrates. In the meantime, reports of IRESs in eukaryotic cellular mRNAs started to appear, and the list of such putative IRESs continues to grow to the point in which it now stands at ~100, 80% of them in vertebrate mRNAs. Despite initial skepticism from some quarters, there now seems universal agreement that there is genuine internal ribosome entry on the viral IRESs. However, the same cannot be said for cellular mRNA IRESs, which continue to be shrouded in controversy. The aim of this article is to explain why vertebrate mRNA IRESs remain controversial, and to discuss ways in which these controversies might be resolved.
URL [本文引用: 1]
URLPMID:3177215 [本文引用: 1]

Translation Initiator of Short 5' UTR (TISU) is a unique regulatory element of both transcription and translation initiation. It is present in a sizable number of genes with basic cellular functions and a very short untranslated region (5' UTR). Here, we investigated translation initiation from short 5' UTR mRNAs with AUG in various contexts. Reducing 5' UTR length to the minimal functional size increases leaky scanning from weak and strong initiators but hardly affects translation initiation and ribosomal binding directed by TISU. Ribosome interaction with TISU mRNA is cap dependent and involves AUG downstream nucleotides that compensate for the absent 5' UTR contacts. Interestingly, eIF1 inhibits cap-proximal AUG selection within weak or strong contexts but not within TISU. Furthermore, TISU-directed translation is unaffected by inhibition of the RNA helicase eIF4A. Thus, TISU directs efficient cap-dependent translation initiation without scanning, a mechanism that would be advantageous when intracellular levels of eIF1 and eIF4A fluctuate.
URLPMID:21983420 [本文引用: 1]

The major strategy for cap dependent translation involves ribosomal scanning. In the scanning mechanism the small ribosomal subunit is recruited to the mRNA through the m7G cap and then scans the 5′ UTR until it reaches an AUG codon. This short review focuses on a recently discovered alternative strategy of cap-dependent translation that operates without scanning, but nonetheless is highly efficient and accurate. This non-scanning translation is directed by the Translation Initiator of Short 5′ UTR (TISU) element. TISU is strictly located close to the 5′ end of the mRNA, resulting in a very short 5′ UTR. It is present in a sizable number of mammalian genes, many of them with fundamental cellular functions. In addition to its unique translational activity, TISU is also a transcription regulatory element that is specifically enriched in TATA-less promoters. Thus TISU represents a prototype regulatory element that links mammalian transcription to a specific mode of translation initiation.
URL [本文引用: 1]
URLPMID:26593424 [本文引用: 1]

N6-methyladenosine (m6A) residues within the 5′ UTR of mRNAs promote translation initiation through a mechanism that does not require the 5′ cap or cap-binding proteins. Diverse cellular stresses selectively increase the levels of m6A within 5′ UTRs, suggesting that 5′ UTR m6A is important for mediating stress-induced translational responses.
