 ,四川农业大学资源学院,成都 611130
,四川农业大学资源学院,成都 611130Evolution and the flanking sequences of β-lactamase resistance gene blaOKP
Wenwen Deng, Mei Long, Shengzhi Yang, Likou Zou ,College of Resources, Sichuan Agricultural University, Chengdu 611130, China
,College of Resources, Sichuan Agricultural University, Chengdu 611130, China第一联系人:
编委: 谢建平
收稿日期:2018-03-21修回日期:2018-05-3网络出版日期:2018-07-20
| 基金资助: |
Received:2018-03-21Revised:2018-05-3Online:2018-07-20
| Fund supported: |
作者简介 About authors
邓雯文,硕士研究生,专业方向:微生物分子生物学E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (498KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
邓雯文, 龙梅, 杨盛智, 邹立扣. β-内酰胺酶耐药基因blaOKP进化及其侧翼序列特征研究. 遗传[J], 2018, 40(7): 585-592 doi:10.16288/j.yczz.18-013
Wenwen Deng, Mei Long, Shengzhi Yang, Likou Zou.
β-内酰胺类抗生素是目前人类和动物临床应用最为广泛的一类抗生素,但由于临床的广泛及不合理使用,细菌对其耐药性逐年增加,给人类和动物的健康带来了极大的危害[1, 2]。产生β-内酰胺酶是细菌对β-内酰胺类抗生素耐药的主要方式,也是造成抗菌治疗失败的重要因素[3, 4]。β-内酰胺酶基因型种类较多,根据底物种类、基因介导情况等不同,可分为超广谱β-内酰胺酶(ESBLs)基因型(blaSHV、blaTEM、blaCTX-M等)、染色体介导的基因型(blaSHV、blaOKP、blaLEN等),以及耐酶抑制剂酶(IRTs)基因型(blaSHV、blaTEM等)[5,6,7]。
blaOKP可随宿主菌染色体复制并持续表达,介导宿主菌对阿莫西林、氨苄西林、羧苄西林、替卡西林及第一代头孢类抗生素等耐药[8]。blaOKP属于窄谱 β-内酰胺酶基因[9],来源于肺炎克雷伯氏菌(Klebsiella pneumoniae)[5, 9, 10]。目前,blaOKP基因进化的危害性已突显,进化出多个基因亚型[11, 12]。而与blaOKP基因共同进化祖先的blaSHV基因早期家族成员亦是首先发现于肺炎克雷伯菌株中,之后才在大肠杆菌等其他肠杆科细菌中逐渐被发现,并已进化出若干基因亚型[6, 13]。
肺炎克雷伯氏菌是一种常见的、目前被公认的导致人类和动物致病的条件致病菌,广泛分布于人和动物的呼吸道、消化道以及水、土壤和谷物等[14, 15]。该菌可引起人[11, 16]和动物[17, 18]的感染,并已对β-内酰胺类药物表现出较高程度的耐药[17, 19]。前期研究发现blaOKP基因的另一宿主为肠杆科的粘质沙雷氏菌(Serratia marcescens)[8],该菌亦为条件致病菌,可导致呼吸系统、泌尿系统、伤口感染及脑膜炎等,对大部分抗生素耐药,产生的ESBLs菌可产生严重的医源感染[20, 21]。
目前,对blaOKP基因进化及其侧翼序列分析的相关报道较少。本研究通过对1株检测到的新亚型blaOKP-A-16基因的肺炎克雷伯氏菌进行全基因组测序,进一步开展对blaOKP基因侧翼序列的研究,同时获取不同亚型的blaSHV基因的侧翼序列,分析blaOKP与blaSHV基因侧翼序列的遗传进化差异,可有助于了解blaOKP 基因的传播机制。
1 材料和方法
1.1 菌株来源
肺炎克雷伯氏菌(K. pneumoniae strain ws-2g) 分离于猪肉样品。该菌株对氨苄西林、阿莫西林、苯唑西林,阿莫西林克拉维酸、氨苄青霉素、头孢氨苄、头孢羟氨苄、头孢噻呋、头孢喹肟等抗生素全部耐药,并检测出blaOKP新亚型基因blaOKP-A-16。质控菌株为大肠埃希菌ATCC 25922和ATCC 35218。菌株详细信息见文献[18]。1.2 菌株基因组DNA提取
用移液器吸取 5 μL 保存菌液接种到20 mL LB 培养基中,180 r/min、37℃过夜培养约11 h至对数期。将菌悬液参照美国Omega公司E.Z.N.A Bacterial DNA Kit试剂盒提取肺炎克雷伯氏菌的基因组DNA。1.3 全基因组测序
将基因组DNA送于上海美吉医药科技有限公司采用Illumina Miseq测序平台进行全基因组测序。测序流程主要包括基因组DNA收集,基因组DNA片段化,文库构建,桥式PCR及Illumina Miseq测序。1.4 全基因组测序数据的处理和分析
将测序得到的原始图像数据经过Base Calling 转化为序列数据,通过生物信息统计学的方法,对所有测序Reads的每个Circle进行碱基分布和质量波动统计,并去除一些质量比较低的数据,继而对样品的DNA进行paired-end (PE)测序,构建500 bp文库。用Error Correction对数据进行校正,去除低频率的测序错误。其次,对校正后的数据进行Merge。最后,对Merge后的数据使用利用专业组装软件GS De Novo Assembler v2.8进行拼接。利用Glimmer 3.02 (http://www.cbcb.umd.edu/software/glimmer/)软件进行细菌的基因预测,将预测基因的蛋白序列分别与Nr、genes、string和GO数据库进行Blastp比对(BLAST 2.2.28+),从而获得预测基因的注释信息。1.5 β-内酰胺酶基因序列特征比较分析
在获取的blaOKP基因亚型基础上,在NCBI网站Nucleotide入口检索,下载部分已有报道的blaOKP、blaSHV、blaTEM基因序列,用ClustalX分别对基因及氨基酸序列进行比对。利用Modeltest 3.7软件筛选最佳替代模型和参数,运用Mega 5分析序列的碱基组成,计算遗传距离。分别以blaSHV、blaTEM基因为外群,采用NJ法(Neighbor-Joining)构建系统发育树。同时,筛选含blaSHV基因及其侧翼环境的基因序列进行分析,比较分析序列异同。2 结果与分析
2.1 全基因组测序概况
肺炎克雷伯氏菌经Illumina Miseq测序平台进行全基因组测序后,得到的基因组序列大小为6 688 729 bp,鸟嘌呤-胞嘧啶(GC)含量为57.31%。基因组编码了5741个开放阅读框(ORFs),5个rRNA (包括5S, 23S),82个tRNA。经预测,共有4428个基因具有COG功能分类,其中信息储存和处理基因共787个,细胞过程和信号转导基因共766个,代谢类基因共2148个,另有功能预测基因306个,功能未知基因421个。菌株共有3630个基因能够在GO数据库三大分支数据库中被注释到。功能注释结果显示共有43种不同功能分支,其中细胞学组件类(cellular component)有10个分支,共3267个基因注释结果,生物学途径类(biological process)有19个分支,共6177基因注释结果,分子功能类(molecular function)有14个分支,共2795个基因注释结果。在全基因组染色体序列中发现了一段含blaOKP序列,总长43 897 bp,并在GenBank中注册(登录号:KX827418),依据此绘制blaOKP基因侧翼序列示意图(图1)。研究发现blaOKP基因两侧分别与DeoR家族转录调控因子基因和钾离子转运ATP酶相连(KdpC),其周围结构还包括DNA重组蛋白(RecF)、转录调节器(LacY-LacZ-LacI)、转录调节器(LuxR- AraC)等。此外,在blaOKP基因的周围序列中未发现可移动遗传元件。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1blaOKP基因侧翼序列示意图
Fig. 1Diagram of genetic environment of the flanking sequences of blaOKP gene
2.2 β-内酰胺酶基因系统发育分析
利用Mega 5绘制blaOKP、blaSHV和blaTEM基因序列的系统发育分析(图2)。结果显示,同一种 β-内酰胺酶基因均被聚在一起,有3个明显的分支Group 1 (blaSHV), Group 2(blaOKP)和Group 3(blaTEM),说明不同基因间有一定的差异性。另外,相比blaTEM基因,blaOKP和blaSHV之间的亲缘关系更近。图2
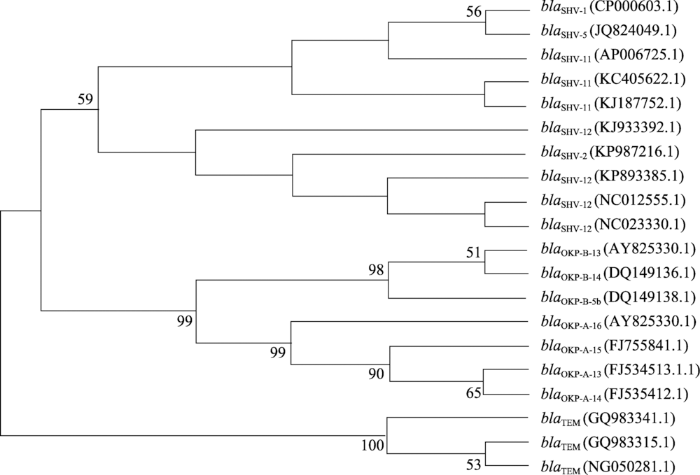 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2β-内酰胺酶基因系统发育树
Fig. 2Phylogenetic tree of β-lactamase genes
2.3 blaOKP与blaSHV基因环境比较
GenBank中部分不同亚型的blaSHV基因及其侧翼环境的菌株及序列相关信息见表1。由表1可知,blaSHV基因可由染色体或质粒介导,包括5种亚型(blaSHV-1、blaSHV-2、blaSHV-5、blaSHV-11和blaSHV-12),其检出国家主要分布在中国(3/5,包括中国大陆和中国台湾),美国(1/5)和法国(1/5)。序列提交时间分布在2007~2015年,检出种属为肠杆菌科的肺炎克雷伯菌(1/2)、鼠疫耶尔森氏菌(1/10)、弗氏柠檬酸杆菌(1/10)、斯氏普罗威登斯菌(1/10)、大肠杆菌(1/10)和阴沟肠杆菌(1/10)。Table 1
表1
表1 不同blaSHV、blaOKP和blaTEM基因基本信息
Table 1
| 基因型/亚型 | 种属 | 菌株/质粒名称 | GenBank号 | 定位 | 提交时间 | 国家/地区 |
|---|---|---|---|---|---|---|
| blaSHV-1 | 鼠疫耶尔森氏菌 (Yersinia pestis biovar Orientalis str.) | plasmid pIP1202 | CP000603.1 | 质粒 | 2007 | 美国 |
| blaSHV-5 | 斯氏普罗威登斯菌(Providencia stuartii) | plasmid pTC2 | JQ824049.1 | 质粒 | 2012 | 法国 |
| blaSHV-12 | 大肠杆菌(Escherichia coli) | plasmid pYD626E | KJ933392.1 | 质粒 | 2014 | 美国 |
| blaSHV-12 | 阴沟肠杆菌(Enterobacter cloacae) | plasmid pEC-IMP | NC_012555.1 | 质粒 | 2008 | 中国台湾 |
| blaSHV-12 | 肺炎克雷伯氏菌 (K. pneumoniae subsp. pneumoniae) | plasmid pKP1034 | KP893385.1 | 质粒 | 2015 | 中国大陆 |
| blaSHV-12 | 肺炎克雷伯氏菌(K. pneumoniae KPS77) | plasmid pKPS77 | NC_023330.1 | 质粒 | 2013 | 法国 |
| blaSHV-11 | 肺炎克雷伯氏菌(K. pneumoniae) | plasmid pKPC-LK30 | KC405622.1 | 质粒 | 2012 | 中国台湾 |
| blaSHV-11 | 肺炎克雷伯氏菌(K. pneumoniae 7433) | plasmid pTR2 | KJ187752.1 | 质粒 | 2014 | 中国台湾 |
| blaSHV-11 | 肺炎克雷伯氏菌 (K. pneumoniae subsp. pneumoniae) | NTUH-K2044 DNA | AP006725.1 | 基因组 | 2004 | 中国大陆 |
| blaOKP-B-13 | 肺炎克雷伯氏菌(K. pneumoniae) | plasmid kp622 | AY825330.1 | 质粒 | 2004 | 中国大陆 |
| blaOKP-B-14 | 肺炎克雷伯氏菌(K. pneumoniae) | 未命名 | DQ149136.1 | 基因组 | 2005 | 加拿大 |
| blaOKP-B-5b | 肺炎克雷伯氏菌(K. pneumoniae) | 未命名 | DQ149138.1 | 基因组 | 2005 | 加拿大 |
| blaOKP-A-13 | 肺炎克雷伯氏菌(K. pneumoniae) | clone S1L | FJ534513.1 | 基因组 | 2008 | 中国大陆 |
| blaOKP-A-14 | 肺炎克雷伯氏菌(K. pneumoniae) | clone S1L | FJ534512.1 | 基因组 | 2008 | 中国大陆 |
| blaOKP-A-15 | 肺炎克雷伯氏菌(K. pneumoniae) | clone S1L | FJ755841.1 | 基因组 | 2008 | 中国大陆 |
| blaTEM | 大肠杆菌(E. coli 22C) | 未命名 | GQ983315.1 | 质粒 | 2009 | 葡萄牙 |
| blaTEM | 阴沟肠杆菌(E. cloacae 68Q) | 未命名 | GQ983341.1 | 质粒 | 2009 | 葡萄牙 |
| blaTEM | 肺炎克雷伯氏菌 (K. pneumoniae subsp. pneumoniae) | CLSiS 3151/98 pbla-TEM-68 | NG_050281.1 | 基因组 | 2016 | 波兰 |
新窗口打开|下载CSV
图3和图4分别展示了染色体介导和质粒介导的blaSHV基因亚型及其两侧的序列结构。在染色体介导的基因blaSHV-11和blaOKP的两侧具有部分相同的序列结构,即基因一侧与DeoR家族转录调控因子基因相连,并具有LuxR-AraC结构,而另一侧含有RecF-LacY-LacZ-LacI结构,且都未发现可移动遗传元件。但仍存在差异,表现为blaOKP基因的一侧与KdpC相连,而不是blaSHV-11的RecF,blaSHV-11中还存在结构ygbN-ygbM-ygbK。此外,而在染色体或质粒介导的blaSHV基因周围序列中未发现KdpC。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3染色体介导的blaSHV基因侧翼序列示意图
Fig. 3Diagram of the flanking sequences of chromosome-encoded blaSHV gene
图4
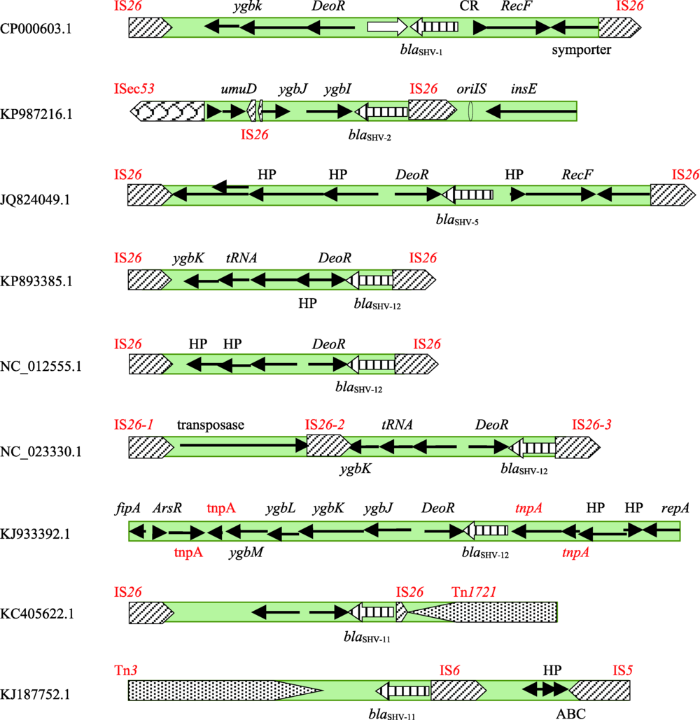 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4质粒介导的blaSHV基因侧翼序列示意图
Fig. 4Diagram of the flanking sequences of palsmid-encoded blaSHV genes
如图4所示,在质粒介导的blaSHV基因周围序列中均存在可移动遗传元件标记基因, 主要为插入序列遗传标记IS26,另外也检出了插入序列遗传标记ISec53,IS6和IS5 转座子遗传标记tnpA,转座子Tn1721和Tn3,其中blaSHV-11中含有多种不同的可移动遗传元件标记基因。然而,在染色体介导的blaSHV-11基因周围无可移动遗传元件。
在质粒介导的blaSHV基因中除blaSHV-1和blaSHV-5,其余blaSHV基因一侧均连接一个插入序列或转座子遗传标记基因。另外,blaSHV-5,blaSHV-12和染色体介导的blaSHV-11基因一侧与DeoR家族转录调控因子基因相连。说明不同亚型的blaSHV基因之间具有一定的相似性。
3 讨 论
国内外对blaOKP基因的研究起步较晚,最早报道于2004年。H?ggman等[9]首先命名了OKP酶(for other K. pneumoniae β-lactamase),发现4个blaOKP新亚型,并从等电点角度将OKP型β-内酰胺酶分为两组,分别为7.5~7.8和6.5~7.0。随后,blaOKP基因亚型不断被发现。目前,国内尽管已有关于肺炎克雷伯菌中blaOKP基因的检测报道[11, 22],但对于blaOKP基因侧翼序列的研究较少。因此,深入了解blaOKP基因的基因环境情况,有助于揭示 blaOKP基因亚型多样性及流行规律。本研究通过全基因测序技术,从1株肺炎克雷伯氏菌中获取了一个blaOKP基因及其侧翼序列。结果表明,在blaOKP基因的周围序列中无可移动遗传元件遗传标记基因,因此不具有水平传播性。H?ggman等[9]调查了肺炎克雷伯氏菌染色体型β-内酰胺酶基因的流行情况,分析结果显示blaSHV,blaOKP,blaLEN为主要基因型,它们来源于统一祖先,分属KpI,KpII及KpIII三个平行进化组,其中KpI和KpIII之间的进化分歧出现在6~28百万年前。其中,blaSHV基因早期家族成员亦是首先发现于肺炎克雷伯菌株中,随后才在大肠杆菌等其它肠杆科细菌中逐渐被发现,并已进化出若干基因亚型[6, 13]。已有研究证明TEM型β-内酰胺酶多样性、进化与临床抗生素的使用密切相关[23, 24]。通过将本研究中的blaOKP基因与已报道的染色体介导blaSHV-11基因比对,发现部分侧翼序列结构具有相似性且两个基因周围序列中都无可移动遗传元件遗传标记基因,提示其侧翼序列较为保守。
目前,由质粒介导的blaOKP基因较少,然而blaSHV基因已进化出由质粒介导的多种基因亚型。研究发现,在质粒介导的不同blaSHV基因的两侧都具可移动遗传元件标记基因, 主要为插入序列遗传标记IS26,可能参与blaSHV基因的广泛转移。Ford等[25]研究表明,blaSHV可以通过其侧翼序列IS-26 (含转座酶基因及重复序列)完成传播、定位。耐药 基因的传播需要依托转座子、插入序列或整合子等可移动遗传元件,这些元件往往位于耐药基因的两侧[26, 27]。插入序列是最简单的转座子,位于两个插入序列之间的基因序列也可随插入序列转移[28]。相对染色体介导耐药机制而言,由于质粒介导的耐药机制具有水平转移的特点,可在同种或不同菌株之间传播,容易引起耐药菌株的广泛流行。
综上所述,本研究通过全基因组测序技术对一株肺炎克雷伯氏菌进行检测,对blaOKP基因及其侧翼序列进行了探讨,并对blaOKP与blaSHV基因侧翼序列的遗传进化差异进行了研究对比。该研究结果可为blaOKP基因的流行趋势预测提供理论依据,并为耐药性的防控提供理论基础。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:28735050 [本文引用: 1]

Abstract OBJECTIVES: Klebsiella pneumoniae, which exists in the intestinal and respiratory tracts of humans and animals, is an important conditional pathogen in many animals. The aim of the current study was to investigate the antimicrobial resistance profiles and genotypes of extended-spectrum 0205-lactamase (ESBL)- and AmpC 0205-lactamase-producing K. pneumoniae isolated from dogs. METHODS: A total of 285 isolates, collected from faecal and urine samples of diseased dogs in a Veterinary Teaching Hospital in Beijing, were characterised by antimicrobial susceptibility testing and screened for ESBL and AmpC 0205-lactamase phenotypes. The relevant genes were identified by polymerase chain reaction and sequencing. RESULTS: All K. pneumoniae isolates were susceptible to meropenem, while the rates of resistance to the remaining 27 tested antimicrobials ranged from 24% to 97%. A total of 53% and 18% of K. pneumoniae isolates were positive for ESBL and AmpC 0205-lactamase production, respectively. ESBL/AmpC-producing strains were significantly resistant to more antimicrobial agents compared non-ESBL/AmpC-producing strains (P<0.05). CTX-M groups 1 and 9, and DHA-1 were the predominant genotypes of the ESBL/AmpC-producing K. pneumoniae isolates. CONCLUSIONS: In conclusion, the high percentage of drug resistance among K. pneumoniae isolates suggests that routine detection of ESBL production by reliable laboratory methods is required in small animal clinical practice. Copyright 0008 2017 International Society for Chemotherapy of Infection and Cancer. Published by Elsevier Ltd. All rights reserved.
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 2]
URL [本文引用: 3]
[本文引用: 1]
[本文引用: 2]
[本文引用: 4]
URL [本文引用: 1]
[本文引用: 3]
URL [本文引用: 1]
URLPMID:10511397 [本文引用: 2]

Abstract Resistance to beta-lactam antibiotics has been a problem for as long as these drugs have been used in clinical practice. In clinically significant bacteria the most important mechanism of resistance is the production of one or more beta-lactamases, enzymes that hydrolyse the beta-lactam bond characteristic of this family of antibiotics. Prominent among the beta-lactamases produced by the Enterobacteriaceae is the SHV family. The first reported SHV beta-lactamase had a narrow spectrum of activity. By the accumulation of point mutations at sites that affect the active site of the enzyme, a family of derivatives of SHV-1 has evolved. Derivatives of SHV-1 either have an extended spectrum of activity, capable of inactivating third-generation cephalosporins, or are resistant to beta-lactamase inhibitors. This review describes the evolution and spread of the SHV family of beta-lactamases, introducing the structure-function analysis made possible by DNA sequence analysis. It also reviews the methods used to characterize members of this family of beta-lactamases, indicating some of the difficulties involved.
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
URLPMID:18455821 [本文引用: 2]

Extended-spectrum β-lactamase (ESBL) and AmpC-producing Enterobacteriaceae are an increasing problem in human medicine and an emerging problem in the veterinary field. Our study, therefore, focused on assessing the prevalence of β-lactamases isolated from swine. Sixty-six Salmonella enterica serovar Typhimurium ( S. Typhimurium), 33 Salmonella enterica serovar Enteritidis ( S. Enteritidis), 26 Klebsiella pneumonia ( K. pneumoniae) and 130 Escherichia coli ( E. coli) pig isolates collected from 1999–2006 were screened for β-lactam resistance by the disk diffusion test (DDT) and micro-broth dilution. Among the isolates, five E. coli and five K. pneumoniae exhibited reduced susceptibility to the cephalosporins tested. PCR, plasmid profiling and Southern blot hybridization showed the presence of multiple β-lactamases in these isolates of animal origin. Hybridization patterns of the DHA-1 specific probe indicated that dissemination of DHA-1 related β-lactamases could be attributed to plasmids of one common size among the enteric microbes of animal origin. To the best of our knowledge, this study reports the first identification of SHV-28 and DHA-1 from microbes of animal origin.
[本文引用: 2]
[本文引用: 1]
URLPMID:28271652 [本文引用: 1]

We evaluated the impact of revised Clinical and Laboratory Standards Institute (CLSI) breakpoints for broad-spectrum cephalosporins (BSCs) on the susceptibilities of 1,742 isolates of Enterobacter species, Serratia marcescens, Citrobacter freundii, and Morganella morganii. The 2011 CLSI criteria for cefotaxime and ceftazidime reduced the rates of susceptibility by 2.9% and 5.9%, respectively. The 2014 CLSI criteria for cefepime reduced the rate of susceptibility by 13.9%, and categorized 11.8% isolates as susceptible-dose dependent (SDD) for cefepime. Among 183 isolates with extended-spectrum -lactamase (ESBL) phenotype, implementation of the new criteria reduced the rates of susceptibility to cefotaxime, ceftazidime, and cefepime by 2.8%, 14.8%, and 53.6%, respectively. The proportion of ESBL phenotype among BSC-susceptible isolates was low (0.9% for cefotaxime, 3.0% for ceftazidime, and 3.3% for cefepime). In summary, implementation of new CLSI criteria led to little change in susceptibility to cefotaxime and ceftazidime but a substantial change in susceptibility to cefepime. The recognition of revised CLSI criteria for BSC and SDD will help clinicians to select the optimal antibiotic and dosing regimen.
[本文引用: 1]
URLPMID:21035968 [本文引用: 1]

Little is known about the antimicrobial resistance mechanisms in Klebsiella pneumoniae from swine in China. Thus, this paper aims to demonstrate the β-lactam resistance phenotypes and genotypes of K. pneumoniae isolates from swine in southwestern China, detect possible new β-lactamase variants, and determine whether or not the variants differ in their antibiotic resistance. Isolates from 58 unrelated diseased swine were collected from 61 pig farms in southwestern China from 2007 to 2009. Among the 58 isolates, 75.8–100% were resistant to β-lactam, 62.0–68.97% to fluoroquinolone, 44.8–46.55% to aminoglycoside, and 8.62–17.24% to β-lactam inhibitors. PCR amplification and DNA sequencing showed that blaTEM-1 was detected in 100% (n=58) of the isolates, blaSHV in 82.76% (n=48), blaCTX-M in 39.66% (n=23), and blaOKP in 17.24% (n=10). The blaSHV types included blaSHV-1, blaSHV-11, blaSHV-12, and blaSHV-27. None of the isolates harbored blaKPC, blaLEN, or blaGES gene. Four novel variants (blaOKP-A-13, blaOKP-A-14, blaOKP-A-15, and blaOKP-A-16) were identified among the 10 OKP β-lactamase-producing K. pneumoniae isolates resistant to ampicillin, amoxicillin, oxacillin, cefalexin, and cefadroxil. Plasmid analysis and PCR amplification indicated that blaTEM-1 genes were detected in the total plasmid. Molecular typing by pulsed-field gel electrophoresis revealed the presence of 10 distinct pulsotypes of OKP producer isolates. Plasmid DNA digested with XbaI yielded two to six bands of ca. 0.15–30kb. Transformants of the 10 OKP producer isolates showed no differences in their antibiotic susceptibility, except for the pulsotype B transformant, which carried blaCTX-M. In China, β-lactam resistance appeared to be common among K. pneumoniae isolates from swine, suggesting that K. pneumoniae may be a reservoir for the dissemination of β-lactam resistance among Chinese pig farms.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]

目的:探讨耐药鲍氏不动杆菌的抗菌药物耐药基因及可移动遗传元件遗传标记的携带、blaOX A基因与插入序列连锁与菌株间的亲缘性。方法收集2012年1月-2013年8月IC U患者痰液标本中分离到的25株耐药鲍氏不动杆菌,经看家基因测序作BLAST 比对确认菌种,采用聚合酶链反应(PCR)及序列分析的方法分析A~D类共34种β‐内酰胺酶基因、15种氨基糖苷类耐药基因、2种喹诺酮类耐药基因、6种可移动遗传元件遗传标记,再做 ISaba1与blaOXA‐2、blaOXA‐23、blaOXA‐66连锁检测,扩增有阳性条带者再进行DNA测序验证,测序结果用Chromas作BLAST比对,最后对57种耐药基因测得结果作样本聚类分析。结果25株耐药鲍氏不动杆菌共检出7种β‐内酰胺类耐药基因、4种氨基糖苷类耐药基因、2种喹诺酮类耐药基因、4种可移动遗传元件遗传标记,且阳性率非常高;插入序列与 blaOXA 基因的连锁检测中,检测到 blaOXA‐23与 ISaba1连锁,但blaOXA‐2、blaOXA‐66与 ISaba1未连锁;样本聚类分析发现,1~18号株为同一克隆,20~25号株亦为同一克隆,19号株为孤立株。结论多药耐药鲍氏不动杆菌中携带的耐药基因导致本组菌对β‐内酰胺酶类、氨基糖苷类、喹诺酮类耐药,且耐药基因和耐药表型相对应,样本聚类分析提示1~18号株和20~25号株可能为医院感染菌株。
URL [本文引用: 1]

目的:探讨耐药鲍氏不动杆菌的抗菌药物耐药基因及可移动遗传元件遗传标记的携带、blaOX A基因与插入序列连锁与菌株间的亲缘性。方法收集2012年1月-2013年8月IC U患者痰液标本中分离到的25株耐药鲍氏不动杆菌,经看家基因测序作BLAST 比对确认菌种,采用聚合酶链反应(PCR)及序列分析的方法分析A~D类共34种β‐内酰胺酶基因、15种氨基糖苷类耐药基因、2种喹诺酮类耐药基因、6种可移动遗传元件遗传标记,再做 ISaba1与blaOXA‐2、blaOXA‐23、blaOXA‐66连锁检测,扩增有阳性条带者再进行DNA测序验证,测序结果用Chromas作BLAST比对,最后对57种耐药基因测得结果作样本聚类分析。结果25株耐药鲍氏不动杆菌共检出7种β‐内酰胺类耐药基因、4种氨基糖苷类耐药基因、2种喹诺酮类耐药基因、4种可移动遗传元件遗传标记,且阳性率非常高;插入序列与 blaOXA 基因的连锁检测中,检测到 blaOXA‐23与 ISaba1连锁,但blaOXA‐2、blaOXA‐66与 ISaba1未连锁;样本聚类分析发现,1~18号株为同一克隆,20~25号株亦为同一克隆,19号株为孤立株。结论多药耐药鲍氏不动杆菌中携带的耐药基因导致本组菌对β‐内酰胺酶类、氨基糖苷类、喹诺酮类耐药,且耐药基因和耐药表型相对应,样本聚类分析提示1~18号株和20~25号株可能为医院感染菌株。
