 ,, 李晓宇
,, 李晓宇 ,中国医学科学院&北京协和医学院,医药生物技术研究所免疫生物学室,北京 100050
,中国医学科学院&北京协和医学院,医药生物技术研究所免疫生物学室,北京 100050Molecular mechanisms of genetic transposition inhibition by piRNA
Qipeng Liu, Ni An, Shan Cen ,, Xiaoyu Li
,, Xiaoyu Li ,Department of Immunology, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100050, China
,Department of Immunology, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100050, China第一联系人:
编委: 苗龙
收稿日期:2018-03-22修回日期:2018-05-16网络出版日期:2018-06-20
| 基金资助: |
Received:2018-03-22Revised:2018-05-16Online:2018-06-20
| Fund supported: |
作者简介 About authors
刘启鹏,硕士研究生,专业方向:宿主因子对转座子的调控机制的研究E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (441KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘启鹏, 安妮, 岑山, 李晓宇. piRNA抑制基因转座的分子机制. 遗传[J], 2018, 40(6): 445-450 doi:10.16288/j.yczz.18-072
Qipeng Liu, Ni An, Shan Cen, Xiaoyu Li.
转座子是一类可以在染色体上或不同染色体间自由移动的DNA,广泛存在于各类生物中[1]。根据其转座方式可分为DNA转座子和RNA转座子两类。DNA转座子能利用自身编码的转座酶将自身序列从原有位置上切除并插入到新的基因组位点,所以该转座过程并不增加拷贝数。DNA转座子在人类细胞基因组中所占的比例很少(3%),且处于失活状态。RNA转座子又称为逆转录转座子(retrotransposon),其转座过程需要先通过转录产生RNA中间体,再以该RNA为模板利用自身编码的逆转录酶生成cDNA后插入到新的基因组位点。该过程不仅保留了原有拷贝,并且在基因组中整合了新拷贝,从而使其在基因组中的拷贝数不断增加,因此人类基因组中逆转录转座子序列的比例可高达45%,且部分逆转录转座子仍具有转座活性。在逆转录转座子中,能够编码转座所必需的酶和蛋白并能独立完成转座过程的被称为自主型转座子,而需要借助于自主型转座子才能完成转座的被称为非自主型转座子。逆转录转座子长散布元件-1 (long interspersed elements-1, LINE-1)属于自主型逆转录转座子,是人类基因组中占比最高且唯一具有独立转座能力的转座子。LINE-1除自身可以转座外,还可以协助不具有自主转座能力的非自主型转座子短散布元件 (short interspersed elements, SINEs)进行转座[2]。
逆转录转座子的转座会造成插入位点突变,并影响周围基因表达;同时在其转座过程中造成的DNA双链断裂也会导致基因组不稳定,进而产生高频突变和重组[3,4,5]。通过这些方式,活跃的逆转录转座子会严重影响基因组的稳定性,并可能引发包括癌症在内的多种基因疾病[6,7,8]。宿主细胞在长期的进化中形成了多种抑制转座子活性的自我保护机制,其中piRNA(PIWI-interacting RNA)参与的转座子沉默途径是新近发现的一种重要的调控方式。本文将总结piRNA在抑制转座子转座方面的主要研究进展,为转座子相关研究提供思路。
1 piRNA:一种非编码小RNA
非编码小RNA是指细胞中一类长度小于50 nt且不编码蛋白质的RNA,其本身或与某些蛋白结合形成的复合体能发挥一定的生物学功能。piRNA是非编码小RNA的一种,在大多数动物中它与微小RNA (microRNA, miRNA)、小干扰RNA (small interfering RNA, siRNA)共称为小RNA干扰的3大机制[9]。piRNAs主要来源于含有大量转座子和重复序列的基因间区(intergenic region),又称之为piRNA簇[10]。其长度约为23~31 nt,5°端首位通常为单磷酸的尿嘧啶核糖核酸,3°端进行了甲基化修饰,通常与Ago (Argonaute)蛋白家族的PIWI亚家族蛋白相互结合共同行使功能[11]。与miRNA和siRNA相比,piRNA除长度明显长于两者外,它还起源于染色体着丝粒附近piRNA基因簇转录出的单链RNA,而非发夹结构的双链RNA,成熟过程也不依赖核酸酶Dicer,并且能在转录和转录后水平调控靶基因的表达[9]。在生殖细胞发育过程中对逆转录转座子进行调控是piRNA主要功能之一。在正常细胞中,甲基化作用是对逆转录转座子的最有效的控制手段之一;然而在生殖细胞和胚胎发育的早期阶段,为了使这些细胞具有发育的全能性,机体会进行表观遗传重编程(epigenetic reprogramming),致使基因组发生大规模的去甲基化现象,导致大量逆转录转座子RNA的转录被激活,转座活性增强。piRNA则是在这个阶段对逆转录转座子进行控制的有效手段,可以保证传递给子代的基因组保持应有的完整性及稳定 性[12]。近年来对线虫(C. elegans)模型的研究表明,piRNA的调控范围涉及生殖细胞中几乎所有的mRNA,它与目的基因的结合模式类似于miRNA,允许个别的不匹配现象。同时其结合效率取决于结合能,而非piRNA的丰度,这就使其在极低浓度下也能发挥作用。若通过转基因手段改变目的基因与piRNA的识别匹配能力,失去调控的目的基因则会持续、稳定、大量地表达,这表明piRNA对生殖细胞基因表达调控的广谱性和有效性[13, 14]。如果PIWI蛋白发生突变或piRNA通路受阻,则会使生殖细胞发育受到严重阻滞[15, 16]。多年的研究表明,piRNA对转座子转座的调控方式是多方面、多层次的。
2 piRNA抑制基因转座的分子机制
2.1 piRNA抑制转座子转录
piRNA诱导特殊位点的组蛋白修饰,是其抑制转座子转座的一种主要方式[17, 18]。在果蝇(Drosophila)卵泡体细胞中,piRNA首先与Piwi蛋白结合形成复合物,通过识别转座子的初生转录本而定位在其转录位点,进而招募组蛋白甲基转移酶Su (var)3-9和组蛋白H1。在Su (var)3-9的作用下,H3K9发生甲基化并与HP1a (heterochromatic protein 1a)结合。HP1a是在转座子沉默机制中扮演了重要角色的一种高度保守的染色质因子,当其减少时转座抑制作用会被削弱[19, 20]。而后,在Piwi蛋白下游效应器Mael (maelstrom)蛋白和聚集的H1组蛋白的共同作用下,HP1a会引起染色质发生紧缩而使转座子转录受到阻碍[18, 21, 22](图1A)。Piwi蛋白在此途径中有着关键的作用,敲除Piwi后,转座子基因区域的RNA聚合酶Ⅱ含量会增加,而H3K9三甲基化信号则会减弱,转座子活性也会明显增强[17]。此外,研究还表明,Piwi蛋白也能直接与组蛋白H1相互作用,引起一系列转座子基因位点处H1含量增加而使染色质发生紧缩[23]。图1
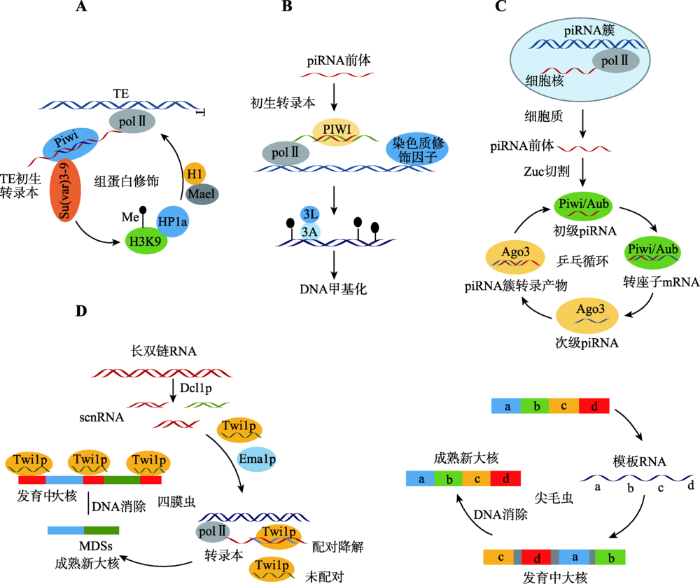 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1piRNA抑制基因转座的途径
A:组蛋白修饰途径。piRNA能引起组蛋白修饰,导致染色质紧缩,从而抑制转座子转座;B:DNA甲基化途径。piRNA能引起H3K9甲基化和H3K4去甲基化,随后招募DNA甲基化转移酶,引起LINE-1 DNA的甲基化而抑制LINE-1的转录;C:乒乓循环途径。次级piRNA扩增的同时,转座子mRNA作为底物被大量消耗,导致转座被抑制;D:DNA消除途径。四膜虫接合期间,在母体小核中产生的scnRNA与Twi1p复合体能介导发育中的大核IESs区域的整体删除;尖毛虫接合期间,母本大核中形成的模板RNA进入发育中的大核,作为模板指导IESs删除和乱序的MDSs重新排序。两种方式都能删除转座子基因序列,导致转座被抑制。
Fig. 1The pathway of genetic transposition inhibition by piRNA
piRNA还可以通过诱导LINE-1 DNA发生甲基化而抑制其转座[24, 25]。在哺乳动物小鼠(Mus musculus)精细胞中,由piRNA基因簇转录出的前体piRNA与PIWI蛋白形成piRNA-PIWI复合物后进入细胞核,识别LINE-1的初生转录本,进而招募染色质修饰物,引起转座子序列相关的组蛋白H3K9甲基化和H3K4去甲基化,随后,这种组蛋白信号变化会招募DNA甲基化转移酶Dnmt3A/Dnmt3L复合物,引起LINE-1 DNA的甲基化,从而抑制LINE-1的转录[26](图1B)。研究发现,将Miwi2 (PIWI蛋白的鼠源同源蛋白)进行突变之后,次级piRNA会发生缺失,进而降低了减数分裂过程中转座子甲基化水平,最终导致转座子表达上调[27];当piRNA基因发生突变之后,H3K9甲基化和H3K4去甲基化信号减弱;Dnmt3A/Dnmt3L的缺失也会降低转座子甲基化水平,导致转座子转座活性增强[25]。
2.2 piRNA在转录后水平抑制转座子转座
大量研究发现,在piRNA的生成过程中伴随着在转录后水平对转座子转座的抑制[28]。piRNA的生成途径可分为两种,即初级生成途径和次级生成途径。在初级生成途径中,piRNA簇转录生成的piRNA前体经Zuc酶等切割生成成熟的piRNA,称为初级piRNA。在随后的次级生成途径中,初级piRNA首先与Piwi或Aub (Aubergine)蛋白相互作用形成Piwi/Aub piRISCs复合物,该复合物能够识别并剪切转座子的mRNA,形成次级piRNA的5°端,并经3°端剪切和甲基化修饰等过程,形成与初级piRNA互补的次级piRNA。次级piRNA与AGO3蛋白形成的AGO3 piRISCs复合物可以识别并剪切piRNA簇转录产物,产生与初级piRNA完全相同的piRNA,然后进入下一轮循环。如此反复可形成一个正向放大的次级 piRNA生成循环通路,该过程被称为“乒乓循环(ping-pang cycle)”[29, 30]。通过“乒乓循环”,piRNA大量扩增,与此同时转座子转录本被大量消耗,进而导致转座活性被抑制[31, 32](图1C)。2.3 piRNA引起转座子基因消除
近年,在低等真核生物纤毛虫(Ciliate)中又发现了一种piRNA抑制转座子转座的新机制——转座子基因消除[33]。纤毛虫属于单细胞低等真核生物,具有明显的核二态性。大核(又称体细胞核)负责营养生长期的基因表达;小核(又称生殖核)负责接合(纤毛虫有性生殖方式)时遗传信息的交换,其基因组DNA序列可以分为两种——大核命运序列(macronuclear destined sequences, MDSs)和内部删除序列(internal eliminated sequences, IESs)。在接合和有丝分裂之后,子代小核会产生一个新的体细胞基因组代替亲本生殖细胞基因组,并通过基因组重排直接从体细胞大核基因组中删除转座子[33]。Mochizuki等[34] 在四膜虫(Tetrahymena)中发现了参与转座子基因消除的PIWI家族蛋白Twi1p及其所运载的piRNA。接合期间,在母体小核中产生的双链RNA被一种Dicer酶Dcl1p (Dicer-like protein 1)切割成短双链RNA,即scnRNA (scan RNA)。它能与Twi1p结合成复合体,在Emap1p蛋白的作用下进入母本大核中扫描转录本,能够与转录本匹配的scnRNA与转录本结合并被降解,而不能匹配的scnRNA则进入新的大核特异性地与IESs区域结合,在染色质删除小体的作用下将这些区域集体删 除[35](图1D)。敲除Twi1p或Emap1p基因会导致机体无法进行正常的DNA消除[36]。
与四膜虫相比较,尖毛虫(Oxytricha)中参与DNA消除的PIWI家族蛋白为Otiwi1[37, 38]。接合期间,在Otiwi1参与下母本大核基因转录形成长转录本(模板RNA),随后它进入发育中的大核,作为模板指导IESs删除和乱序的MDSs重新排序[39, 40](图1D)。Otiwi1对于尖毛虫生存极其重要,敲除Otiwi1基因会导致无法产生模板RNA,甚至机体无法存 活[37]。利用RNA干扰(RNAi)技术下调模板RNA会导致大核形成异常;将人工合成的大核染色体导入母本大核,在有性生殖的子代里会出现与人工序列相同的新型大核[41]。虽然四膜虫和尖毛虫基因组重排方式不同,但两种方式都会导致转座子基因序列被删除,从而在源头上沉默转座子。
3 结语和展望
除以上介绍的piRNA控制转座子转座的机制外,近期还有研究报导,piRNA能参与转座子的可变剪接[42],以及通过对转座子的调控来抑制DNA损伤,减缓衰老等[43]。目前具体的机制还在进一步完善中。逆转录转座子LINE-1在肿瘤组织中的过量表达已成为多种肿瘤中的普遍现象,如乳腺癌、直肠癌和肾癌,而近期临床研究显示,这些肿瘤组织样本中也普遍存在PIWI蛋白的异常表达[44, 45]。肿瘤组织中LINE-1活性的增强与PIWI蛋白的异常表达是否有直接的关系,以及其中的机制如何还需要进一步的研究。piRNA和PIWI蛋白能否成为癌症等疾病的预后生物标志物也正在积极探索中。随着核酸测序、基因编辑、晶体结构测定等生物技术的不断发展,未来对于piRNA的研究工作必定会取得引人瞩目的成果,也希望这些研究成果能够早日应用到医疗中,为患者带来福音。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:15430309 [本文引用: 1]

In the course of an experiment designed to reveal the genic composition of the short arm of chromosome 9, a phenomenon of rare occurrence (or recognition) in maize began to appear with remarkably high frequencies in the cultures. The terms mutable genes, unstable genes, variegation, mosaicism, mutable loci or osition-effect have been applied to this phenomenon. Its occurrence in a wide variety of organisms has been recognized. The most extensive investigations of this phenomenon have been undertaken in Drosophila melanogaster.1 In this organism, the conditions associated with the origin of genic instability have been well defined. The part played by the heterochromatic materials of the chromosomes, in inducing and controlling the type of variegation and its time and frequency of occurrence, has been established. It has not been generally recognized that the instability of genic expression in other organisms may be essentially the same as that occurring in Drosophila. As stated above, a large number of mutable loci have recently arisen in the maize cultures and are continuing to arise anew. The loci affect variegation for many different kinds of plant characters, each locus being concerned with a particular character or occasionally several characters. Some of these loci are c, yg2, wx, a2, y, pyd, which are well-investigated units in maize.2 Others involve previously unknown genetic units. The same types of genic instability appearing in the maize cultures have been described in many other organisms. The behavior of these new mutable loci in maize cannot be considered peculiar to this organism. The author believes that the mechanism underlying the phenomenon of variegation is basically the same in all organisms. The reasons for this conclusion will be made apparent in the discussion.
URLPMID:17024082 [本文引用: 1]

The article features the transposable elements (TEs) which are otherwise refered as jumping genes. They have been considered harmful as they could trigger mutations and thus lead to diseases, however, the recent findings prove otherwise. They act as gene regulatory elements, to the host and potentially carry the elements of genetic innovation of the organism. The article focuses on its classifications such as DNA and LTR retrotransposons, their gentic expressions and environment.
URL [本文引用: 1]
URLPMID:16490214 [本文引用: 1]

Long interspersed element-1 (L1) is an autonomous retroelement that is active in the human genome. The proposed mechanism of insertion for L1 suggests that cleavage of both strands of genomic DNA is required. We demonstrate that L1 expression leads to a high level of double-strand break (DSB) formation in DNA using immunolocalization of gamma-H2AX foci and the COMET assay. Similar to its role in mediating DSB repair in response to radiation, ATM is required for L1-induced gamma-H2AX foci and for L1 retrotransposition. This is the first characterization of a DNA repair response from expression of a non-long terminal repeat (non-LTR) retrotransposon in mammalian cells as well as the first demonstration that a host DNA repair gene is required for successful integration. Notably, the number of L1-induced DSBs is greater than the predicted numbers of successful insertions, suggesting a significant degree of inefficiency during the integration process. This result suggests that the endonuclease activity of endogenously expressed L1 elements could contribute to DSB formation in germ-line and somatic tissues.
URL [本文引用: 1]

Gametogenesis is a highly regulated process in all organisms. In Drosophila , a meiotic checkpoint which monitors double-stranded DNA breaks and involves Drosophila ATR and Chk2 coordinates the meiotic cell cycle with signaling events that establish the axis of the egg and embryo. Checkpoint activity regulates translation of the transforming growth-factor-alpha-like Gurken signaling molecule which induces dorsal cell fates in the follicle cells 1 , 2 and 3 . We found that mutations in the Drosophila gene cutoff (cuff) affect germline cyst development and result in ventralized eggs as a result of reduced Grk protein expression. Surprisingly, cuff mutations lead to a marked increase in the transcript levels of two retrotransposable elements, Het-A and Tart. We found that small interfering RNAs against the roo element are still produced in cuff mutant ovaries. These results indicate that Cuff is involved in the rasiRNA pathway and most likely acts downstream of siRNA biogenesis. The eggshell and egg-laying defects of c uff mutants are suppressed bya mutation in chk2 . We also found that mutations in aubergine (aub), another gene implicated in the rasiRNA pathway, are significantly suppressed by the chk2 mutation. Our results indicate that mutants in rasiRNA pathways lead to elevated transposition incidents in the germline, and that this elevation activates a checkpoint that causes a loss of germ cells and a reduction of Gurken protein in the remaining egg chambers.
.
URLPMID:29130960 [本文引用: 1]

Abstract P-Element induced wimpy testis (PIWI)-interacting RNAs (piRNAs) are a type of noncoding RNAs (ncRNAs) and interact with PIWI proteins. piRNAs were primarily described in the germline, but emerging evidence revealed that piRNAs are expressed in a tissue-specific manner among multiple human somatic tissue types as well and play important roles in transposon silencing, epigenetic regulation, gene and protein regulation, genome rearrangement, spermatogenesis and germ stem-cell maintenance. PIWI proteins were first discovered in Drosophila and they play roles in spermatogenesis, germline stem-cell maintenance, self-renewal, retrotransposons silencing and the male germline mobility control. A growing number of studies have demonstrated that several piRNA and PIWI proteins are aberrantly expressed in various kinds of cancers and may probably serve as a novel biomarker and therapeutic target for cancer treatment. Nevertheless, their specific mechanisms and functions need further investigation. In this review, we discuss about the biogenesis, functions and the emerging role of piRNAs and PIWI proteins in cancer, providing novel insights into the possible applications of piRNAs and PIWI proteins in cancer diagnosis and clinical treatment. 2017 The Author(s). Published by S. Karger AG, Basel.
[本文引用: 1]
[本文引用: 1]
.
URL [本文引用: 1]

LINE-1是现今人体内存在的唯一具有自主转座活性的转座子,约有500 000个拷贝,占人类基因组总量的17%。LINE-1是通过转录和逆转录在内的转座过程产生新的DNA拷贝,并使新产生的DNA拷贝插入基因组的不同位置。LINE-1转座会影响基因组中其他基因的表达或调控,因而会对基因组的稳定性产生影响,从而导致基因疾病或肿瘤的发生。本文总结了近年来国际上对LINE-1转座与肿瘤的发生和发展之间关系的研究进展,为肿瘤的治疗和机制研究提供一些线索。
URL [本文引用: 1]

LINE-1是现今人体内存在的唯一具有自主转座活性的转座子,约有500 000个拷贝,占人类基因组总量的17%。LINE-1是通过转录和逆转录在内的转座过程产生新的DNA拷贝,并使新产生的DNA拷贝插入基因组的不同位置。LINE-1转座会影响基因组中其他基因的表达或调控,因而会对基因组的稳定性产生影响,从而导致基因疾病或肿瘤的发生。本文总结了近年来国际上对LINE-1转座与肿瘤的发生和发展之间关系的研究进展,为肿瘤的治疗和机制研究提供一些线索。
URLPMID:2792755 [本文引用: 2]

Transposons populate the landscape of all eukaryotic genomes. Often considered purely genomic parasites, transposons can also benefit their hosts, playing roles in gene regulation and in genome organization and evolution. Peaceful coexistence with mobile elements depends upon adaptive control mechanisms, since unchecked transposon activity can impact long-term fitness and acutely reduce the fertility of progeny. Here, we review the conserved roles played by small RNAs in the adaptation of eukaryotes to coexist with their genomic colonists. An understanding of transposon-defense pathways has uncovered recurring themes in the mechanisms by which genomes distinguish "self" from "non-self" and selectively silence the latter.
URLPMID:25895683 [本文引用: 1]

PIWI-interacting RNAs (piRNAs) are a large family of small, single-stranded, non-coding RNAs present throughout the animal kingdom. They form complexes with several members of the PIWI clade of...
URL [本文引用: 1]
URLPMID:5681665 [本文引用: 1]

Transposable elements (TEs) contribute to the large amount of repetitive sequences in mammalian genomes and have been linked to species-specific genome innovations by rewiring regulatory circuitries. However, organisms need to restrict TE activity to ensure genome integrity, especially in germline cells to protect the transmission of genetic information to the next generation. This review features our current understandings of mammalian PIWI-interacting RNAs (piRNAs) and their role in TE regulation in spermatogenesis. Here we discuss functional implication and explore additional molecular mechanisms that inhibit transposon activity and altogether illustrate the paradoxical arms race between genome evolution and stability. Transposable elements can be activated during germ cell maturation, potentially leading to genome instability and rewiring of the genetic circuitry. In this review, the authors discuss how the piRNA machinery suppresses these elements to ensure accurate spermatogenesis.
URLPMID:29456082 [本文引用: 1]

piRNAs (Piwi-interacting small RNAs) engage Piwi Argonautes to silence transposons and promote fertility in animal germlines. Genetic and computational studies have suggested that C. elegans piRNAs tolerate mismatched pairing and in principle could target every transcript. Here we employ in vivo cross-linking to identify transcriptomewide interactions between piRNAs and target RNAs. We show that piRNAs engage all germline mRNAs and that piRNA binding follows microRNA-like pairing rules. Targeting correlates better with binding energy than with piRNA abundance, suggesting that piRNA concentration does not limit targeting. In mRNAs silenced by piRNAs, secondary small RNAs accumulate at the center and ends of piRNA binding sites. In germline-expressed mRNAs, however, targeting by the CSR-1 Argonaute correlates with reduced piRNA binding density and suppression of piRNA-associated secondary small RNAs. Our findings reveal physiologically important and nuanced regulation of individual piRNA targets and provide evidence for a comprehensive posttranscriptional regulatory step in germline gene expression.
URLPMID:29420292 [本文引用: 1]

By silencing transposons, Piwi-interacting RNAs (piRNAs) protect the stability of animal genomes in germ lines. However, many piRNAs do not map to transposons, and their functions have remained undefined. Zhang et al. described the piRNA targeting logic in Caenorhabditis elegans and identified an intrinsic sequence signal in endogenous germline genes that confer resistance to piRNA silencing. Thus, diverse piRNAs silence foreign nucleic acids but spare self genes to defend the C. elegans genome. In addition, multiple foreign transgenes can be engineered to escape piRNA targeting, allowing successful expression in the germline.
URL [本文引用: 1]
URLPMID:28552346 [本文引用: 1]

Abstract Genetic studies have elucidated critical roles of0002Piwi0002proteins in germline development in animals,0002but whether Piwi is an actual disease gene in human infertility remains unknown. We report germline mutations in human Piwi (Hiwi) in patients with azoospermia that prevent its ubiquitination and degradation. By modeling such mutations0002in Piwi (Miwi) knockin mice, we demonstrate0002that the genetic defects are directly responsible0002for male infertility. Mechanistically, we show that MIWI binds the histone ubiquitin ligase RNF8 in a Piwi-interacting RNA0002(piRNA)-independent0002manner, and MIWI stabilization sequesters RNF8 in the cytoplasm of late spermatids. The resulting aberrant sperm show histone retention, abnormal morphology, and severely compromised activity, which can be functionally0002rescued via blocking RNF8-MIWI interaction0002in0002spermatids with an RNF8-N peptide. Collectively, our findings identify Piwi as a factor0002in human infertility0002and reveal its role in regulating the histone-to-protamine exchange during spermiogenesis. Copyright 0008 2017 Elsevier Inc. All rights reserved.
URLPMID:23392610 [本文引用: 2]

In the metazoan germline, piwi proteins and associated piwi-interacting RNAs (piRNAs) provide a defense system against the expression of transposable elements. In the cytoplasm, piRNA sequences guide piwi complexes to destroy complementary transposon transcripts by endonucleolytic cleavage. However, some piwi family members are nuclear, raising the possibility of alternative pathways for piRNA-mediated regulation of gene expression. We found that Drosophila Piwi is recruited to chromatin, colocalizing with RNA polymerase II (Pol II) on polytene chromosomes. Knockdown of Piwi in the germline increases expression of transposable elements that are targeted by piRNAs, whereas protein-coding genes remain largely unaffected. Derepression of transposons upon Piwi depletion correlates with increased occupancy of Pol II on their promoters. Expression of piRNAs that target a reporter construct results in a decrease in Pol II occupancy and an increase in repressive H3K9me3 marks and heterochromatin protein 1 (HP1) on the reporter locus. Our results indicate that Piwi identifies targets complementary to the associated piRNA and induces transcriptional repression by establishing a repressive chromatin state when correct targets are found.
URLPMID:23159368 [本文引用: 2]

Eukaryotic genomes are colonized by transposons whose uncontrolled activity causes genomic instability. The piRNA pathway silences transposons in animal gonads, yet how this is achieved molecularly remains controversial. Here, we show that the HMG protein Maelstrom is essential for Piwi-mediated silencing in Drosophila. Genome-wide assays revealed highly correlated changes in RNA polymerase II recruitment, nascent RNA output, and steady-state RNA levels of transposons upon loss of Piwi or Maelstrom. Our data demonstrate piRNA-mediated trans-silencing of hundreds of transposon copies at the transcriptional level. We show that Piwi is required to establish heterochromatic H3K9me3 marks on transposons and their genomic surroundings. In contrast, loss of Maelstrom affects transposon H3K9me3 patterns only mildly yet leads to increased heterochromatin spreading, suggesting that Maelstrom acts downstream of or in parallel to H3K9me3. Our work illustrates the widespread influence of transposons and the piRNA pathway on chromatin patterns and gene expression.
URL [本文引用: 1]
URLPMID:23434410 [本文引用: 1]

Huang et02al. characterize genomic recruitment of epigenetic factors guided by piRNAs. They find that Piwi/piRNAs represent a major epigenetic mechanism exerting strong global effects. This work addresses a central enigma in epigenetics—how epigenetic factors, most of which do not bind DNA, are guided to specific genomic sites.
[本文引用: 1]
URL [本文引用: 1]
URLPMID:27425411 [本文引用: 1]

PIWI-interacting RNAs (piRNAs) mediate transcriptional and post-transcriptional silencing of transposable element (TE) in animal gonads. In Drosophila ovaries, Piwi-piRNA complexes (Piwi-piRISCs) repress TE transcription by modifying the chromatin state, such as by H3K9 trimethylation. Here, we demonstrate that Piwi physically interacts with linker histone H1. Depletion of Piwi decreases H1 density at a subset of TEs, leading to their derepression. Silencing at these loci separately requires H1 and H3K9me3 and heterochromatin protein 1a (HP1a). Loss of H1 increases target loci chromatin accessibility without affecting H3K9me3 density at these loci, while loss of HP1a does not impact H1 density. Thus, Piwi-piRISCs require both H1 and HP1a to repress TEs, and the silencing is correlated with the chromatin state rather than H3K9me3 marks. These findings suggest that Piwi-piRISCs regulate the interaction of chromatin components with target loci to maintain silencing of TEs through the modulation of chromatin accessibility.
URLPMID:28630288 [本文引用: 1]

The PIWI-interacting RNA (piRNA) pathway is essential for retrotransposon silencing. In piRNA-deficient mice, L1-overexpressing male germ cells exhibit excessive DNA damage and meiotic defects. It remains unknown whether L1 expression simply highlights piRNA deficiency or actually drives the germ-cell demise. Specifically, the sheer abundance of genomic L1 copies prevents reliable quantification of new insertions. Here, we developed a codon-optimized L1 transgene that is controlled by an endogenous mouse L1 promoter. Importantly, DNA methylation dynamics of a single-copy transgene were indistinguishable from those of endogenous L1s. Analysis of Mov10l161/61 testes established that de novo methylation of the L1 transgene required the intact piRNA pathway. Consistent with loss of DNA methylation and programmed reduction of H3K9me2 at meiotic onset, the transgene showed 1,400-fold increase in RNA expression and consequently 70-fold increase in retrotransposition in postnatal day 14 Mov10l161/61 germ cells compared with the wild-type. Analysis of adult Mov10l161/61 germ-cell fractions indicated a stage-specific increase of retrotransposition in the early meiotic prophase. However, extrapolation of the transgene data to endogenous L1s suggests that it is unlikely insertional mutagenesis alone accounts for the Mov10l161/61 phenotype. Indeed, pharmacological inhibition of reverse transcription did not rescue the meiotic defect. Cumulatively, these results establish the occurrence of productive L1 mobilization in the absence of an intact piRNA pathway but leave open the possibility of processes preceding L1 integration in triggering meiotic checkpoints and germ-cell death. Additionally, our data suggest that many heritable L1 insertions originate from individuals with partially compromised piRNA defense.
URLPMID:18413711 [本文引用: 2]

A biweekly scientific journal publishing high-quality research in molecular biology and genetics, cancer biology, biochemistry, and related fields
URLPMID:18922463 [本文引用: 1]

Abstract piRNAs and Piwi proteins have been implicated in transposon control and are linked to transposon methylation in mammals. Here we examined the construction of the piRNA system in the restricted developmental window in which methylation patterns are set during mammalian embryogenesis. We find robust expression of two Piwi family proteins, MIWI2 and MILI. Their associated piRNA profiles reveal differences from Drosophila wherein large piRNA clusters act as master regulators of silencing. Instead, in mammals, dispersed transposon copies initiate the pathway, producing primary piRNAs, which predominantly join MILI in the cytoplasm. MIWI2, whose nuclear localization and association with piRNAs depend upon MILI, is enriched for secondary piRNAs antisense to the elements that it controls. The Piwi pathway lies upstream of known mediators of DNA methylation, since piRNAs are still produced in dnmt3L mutants, which fail to methylate transposons. This implicates piRNAs as specificity determinants of DNA methylation in germ cells.
URL [本文引用: 1]
URLPMID:19812547 [本文引用: 1]

PIWI-interacting RNAs (piRNAs) silence retrotransposons in Drosophila germ lines by associating with the PIWI proteins Argonaute 3 (AGO3), Aubergine (Aub) and Piwi. piRNAs in Drosophila are produced from intergenic repetitive genes and piRNA clusters by two systems: the primary processing pathway and the amplification loop. The amplification loop occurs in a Dicer-independent, PIWI-Slicer-dependent manner. However, primary piRNA processing remains elusive. Here we analysed piRNA processing in a Drosophila ovarian somatic cell line where Piwi, but not Aub or AGO3, is expressed; thus, only the primary piRNAs exist. In addition to flamenco, a Piwi-specific piRNA cluster, traffic jam (tj), a large Maf gene, was determined as a new piRNA cluster. piRNAs arising from tj correspond to the untranslated regions of tj messenger RNA and are sense-oriented. piRNA loading on to Piwi may occur in the cytoplasm. zucchini, a gene encoding a putative cytoplasmic nuclease, is required for tj-derived piRNA production. In tj and piwi mutant ovaries, somatic cells fail to intermingle with germ cells and Fasciclin III is overexpressed. Loss of tj abolishes Piwi expression in gonadal somatic cells. Thus, in gonadal somatic cells, tj gives rise simultaneously to two different molecules: the TJ protein, which activates Piwi expression, and piRNAs, which define the Piwi targets for silencing.
URLPMID:17346786 [本文引用: 1]

Drosophila Piwi-family proteins have been implicated in transposon control. Here, we examine piwi-interacting RNAs (piRNAs) associated with each Drosophila Piwi protein and find that Piwi and Aubergine bind RNAs that are predominantly antisense to transposons, whereas Ago3 complexes contain predominantly sense piRNAs. As in mammals, the majority of Drosophila piRNAs are derived from discrete genomic loci. These loci comprise mainly defective transposon sequences, and some have previously been identified as master regulators of transposon activity. Our data suggest that heterochromatic piRNA loci interact with potentially active, euchromatic transposons to form an adaptive system for transposon control. Complementary relationships between sense and antisense piRNA populations suggest an amplification loop wherein each piRNA-directed cleavage event generates the 5' end of a new piRNA. Thus, sense piRNAs, formed following cleavage of transposon mRNAs may enhance production of antisense piRNAs, complementary to active elements, by directing cleavage of transcripts from master control loci.
URL [本文引用: 1]

Piwi-interacting RNAs (piRNAs) and Piwi proteins have the evolutionarily conserved function of silencing of repetitive genetic elements in germ lines. The founder of the Piwi subfamily, Drosophila nuclear Piwi protein, was also shown to be required for the maintenance of germ-line stem cells (GSCs). Hence, null mutant piwi females exhibit two types of abnormalities, overexpression of transposons and severely underdeveloped ovaries. It remained unknown whether the failure of GSC maintenance is related to transposon derepression or if GSC self-renewal and piRNA silencing are two distinct functions of the Piwi protein. We have revealed a mutation, piwiNt removing the nuclear localization signal of the Piwi protein. piwiNt females retain the ability of GSC self-renewal and a near-normal number of egg chambers in the ovarioles but display a drastic transposable element derepression and nuclear accumulation of their transcripts in the germ line. piwiNt mutants are sterile most likely because of the disturbance of piRNA-mediated transposon silencing. Analysis of chromatin modifications in the piwiNt ovaries indicated that Piwi causes chromatin silencing only of certain types of transposons, whereas others are repressed in the nuclei without their chromatin modification. Thus, Piwi nuclear localization that is required for its silencing function is not essential for the maintenance of GSCs. We suggest that the Piwi function in GSC self-renewal is independent of transposon repression and is normally realized in the cytoplasm of GSC niche cells.
[本文引用: 1]
[本文引用: 1]
URLPMID:3199450 [本文引用: 2]

Many transposon-related sequences are removed from the somatic macronucleus of ciliates during sexual reproduction. In the ciliate Tetrahymena, an RNAi-related mechanism produces small noncoding RNAs that induce heterochromatin formation, which is followed by DNA elimination. Because RNAi-related mechanisms repress transposon activities in a variety of eukaryotes, the DNA elimination mechanism of ciliates might have evolved from these types of transposon-silencing mechanisms. Nuclear dimorphism allows ciliates to identify any DNA that has invaded the germ-line micronucleus using small RNAs and a whole genome comparison of the micronucleus and the somatic macronucleus.
URLPMID:15598983 [本文引用: 1]

Abstract Previous studies indicated that genome rearrangement involving DNA sequence elimination that occurs at late stages of conjugation in Tetrahymena is epigenetically controlled by siRNA-like scan (scn) RNAs produced from nongenic, heterogeneous, bidirectional, micronuclear transcripts synthesized at early stages of conjugation. Here, we show that Dcl1p, one of three Tetrahymena Dicer-like enzymes, is required for processing the micronuclear transcripts to scnRNAs. DCL1 is also required for methylation of histone H3 at Lys 9, which, in wild-type cells, specifically occurs on the sequences (IESs) being eliminated. These results argue that Dcl1p processes nongenic micronuclear transcripts to scnRNAs and is required for IES elimination. This is the first evidence linking nongenic micronuclear transcripts, scnRNAs, and genome rearrangement. Dcl1p also is required for proper mitotic and meiotic segregation of micronuclear chromosomes and for normal chromosome alignment in meiotic prophase, suggesting that DCL1 has multiple functions in regulating chromosome dynamics.
URLPMID:12297043 [本文引用: 1]

During development of the somatic macronucleus from the germline micronucleus in ciliates, chromosome rearrangements occur in which specific regions of DNA are eliminated and flanking regions are healed, either by religation or construction of telomeres. We identified a gene, TWI1, in Tetrahymena thermophila that is homologous to piwi and is required for DNA elimination. We also found that small RNAs were specifically expressed prior to chromosome rearrangement during conjugation. These RNAs were not observed in TWI1 knockout cells and required PDD1, another gene required for rearrangement, for expression. We propose that these small RNAs function to specify sequences to be eliminated by a mechanism similar to RNA-mediated gene silencing.
URLPMID:23374338 [本文引用: 1]

Ciliates are an ancient and diverse group of microbial eukaryotes that have emerged as powerful models for RNA-mediated epigenetic inheritance. They possess extensive sets of both tiny and long noncoding RNAs that, together with a suite of proteins that includes transposases, orchestrate a broad cascade of genome rearrangements during somatic nuclear development. This Review emphasizes three important themes: the remarkable role of RNA in shaping genome structure, recent discoveries that unify many deeply diverged ciliate genetic systems, and a surprising evolutionary "sign change" in the role of small RNAs between major species groups.
URLPMID:3678556 [本文引用: 2]

Genome duality in ciliated protozoa offers a unique system to showcase their epigenome as a model of inheritance. In Oxytricha, the somatic genome is responsible for vegetative growth, whereas the germline contributes DNA to the next sexual generation. Somatic nuclear development removes all transposons and other so-called "junk" DNA, which comprise ~95% of the germline. We demonstrate that Piwi-interacting small RNAs (piRNAs) from the maternal nucleus can specify genomic regions for retention in this process. Oxytricha piRNAs map primarily to the somatic genome, representing the ~5% of the germline that is retained. Furthermore, injection of synthetic piRNAs corresponding to normally deleted regions leads to their retention in later generations. Our findings highlight small RNAs as powerful transgenerational carriers of epigenetic information for genome programming.
URLPMID:12389817 [本文引用: 1]

The patterns of expression of two metallothionein (MT) genes, MT-1 and MT-2 , previously identified as Cd-MT and Cu-MT , were analysed in Tetrahymena pigmentosa in response to metal inducers cadmium, copper and zinc and to a mixture of copper and cadmium at appropriate concentrations. Co-treatment induces synergistic accumulation of both metals and higher expression of MT -mRNAs in the first few hours. mRNA levels were observed not to completely correlate with MT-protein levels, suggesting that post-transcriptional regulation may be involved in MT induction. MT-1 is induced to higher levels than MT-2 . Zinc does not induce any MT expression. The lowest level of mRNA was observed for MT-2 , induced only by copper. Cadmium is a powerful inducer of the MT-1 gene, although a very low transcription rate by copper occurs in the first hour.
URLPMID:12732478 [本文引用: 1]

Abstract The micronuclear versions of genes in stichotrichous ciliates are interrupted by multiple, short, non-coding DNA segments called internal eliminated segments, or IESs. IESs divide a gene into macronuclear destined segments, or MDSs. In some micronuclear genes MDSs are in a scrambled disorder. During development of a micronucleus into a macronucleus after cell mating the IESs are excised from micronuclear genes and the MDSs are spliced in the sequentially correct order. Pairs of short repeat sequences in the ends of MDSs undergo homologous recombination to excise IESs and splice MDSs. However, the repeat sequences are too short to guide unambiguously their own alignment in preparation for recombination. Based on experiments by others on the distantly related ciliate, Paramecium, we propose a molecular model of template-guided recombination to explain the excision of the 100,000-150,000 IESs and splicing of MDSs, including unscrambling, in the genome of stichotrichous ciliates. The model solves the problem of correct pairing of pointers, precisely identifies MDS-IES junctions, and provides for irreversible recombination.
URL [本文引用: 1]
URLPMID:2647009 [本文引用: 1]

Nature is the international weekly journal of science: a magazine style journal that publishes full-length research papers in all disciplines of science, as well as News and Views, reviews, news, features, commentaries, web focuses and more, covering all branches of science and how science impacts upon all aspects of society and life.
URLPMID:29211718 [本文引用: 1]

Abstract Transposable elements can drive genome evolution, but their enhanced activity is detrimental to the host and therefore must be tightly regulated. The Piwi-interacting small RNA (piRNA) pathway is vital for the regulation of transposable elements, by inducing transcriptional silencing or post-transcriptional decay of mRNAs. Here we show that piRNAs and piRNA biogenesis components regulate precursor mRNA splicing of P-transposable element transcripts in vivo, leading to the production of the non-transposase-encoding mature mRNA isoform in Drosophila germ cells. Unexpectedly, we show that the piRNA pathway components do not act to reduce transcript levels of the P-element transposon during P-M hybrid dysgenesis, a syndrome that affects germline development in Drosophila. Instead, splicing regulation is mechanistically achieved together with piRNA-mediated changes to repressive chromatin states, and relies on the function of the Piwi-piRNA complex proteins Asterix (also known as Gtsf1) and Panoramix (Silencio), as well as Heterochromatin protein 1a (HP1a; encoded by Su(var)205). Furthermore, we show that this machinery, together with the piRNA Flamenco cluster, not only controls the accumulation of Gypsy retrotransposon transcripts but also regulates the splicing of Gypsy mRNAs in cultured ovarian somatic cells, a process required for the production of infectious particles that can lead to heritable transposition events. Our findings identify splicing regulation as a new role and essential function for the Piwi pathway in protecting the genome against transposon mobility, and provide a model system for studying the role of chromatin structure in modulating alternative splicing during development.
URL [本文引用: 1]

This article discusses the proposed role of the PIWI-piRNA pathway in setting the pace of aging as well as the possible mechanisms underlying this process.
URLPMID:16287078 [本文引用: 1]

Abstract Stem cell genetics research may be critical to our understanding of carcinogenesis, as both stem cells and cancer cells possess the ability to self-renew. Recent discoveries have indicated that the piwi family of genes plays an essential role in stem cell self-renewal in diverse organisms. The hiwi gene, the human homolog of the piwi family, participates in germ cell proliferation and its overexpression may cause the development of germ cell malignancy, but its expression and function in epithelial solid cancers have not been explored. In the present study, we investigated whether there was an association between hiwi expression and human gastric cancer and its potential mechanism. RT-PCR findings demonstrated that hiwi was expressed in different gastric cancer cell lines. To identify the HIWI protein in gastric cancer, we developed a specific monoclonal antibody against HIWI and immunohistochemistry was performed on various gastric tissues. We found that the expression ratio of hiwi in normal gastric tissues, atrophic gastritis, intestinal metaplasia and gastric cancers was 10% (5/50), 36% (18/50), 36% (18/50) and 76% (38/50), respectively, which was consistent with precancerous development. Notably, the expression pattern of hiwi in gastric cancer tissues was similar to that of Ki67, which was used as a marker of proliferation. Moreover, the suppression of hiwi by antisense or RNAi inhibited the growth of gastric cancer cells and induced cell cycle arrest in G 2 /M phase. These results suggest that hiwi may be involved in the development of gastric cancer and is a potential target for cancer therapy. 2005 Wiley-Liss, Inc.
URLPMID:21989785 [本文引用: 1]

Abstract Top of page Abstract INTRODUCTION MATERIALS AND METHODS RESULTS DISCUSSION FUNDING SUPPORT REFERENCES BACKGROUND: PIWI protein family was found to play an important role in stem cell self-renewal. Overexpression of HIWI, the human homolog of PIWI family proteins, was found in several solid tumors, although the role of HIWI in hepatocellular carcinoma (HCC) and its prognostic value remain unclear. METHODS: HIWI expression was measured in stepwise metastatic HCC cell lines (HCCLM3, MHCC97H, MHCC97L, SMMC7721, and HepG2), the normal liver cell line (L02), and HCC tissue samples (n = 20). Proliferation and invasion were investigated in HCC cell lines undergoing HIWI target small interfering RNA transfection. Also explored was HIWI expression in HCC tissue microarrays (n = 168) for survival analysis. RESULTS: Levels of HIWI protein and mRNA were up-regulated in highly metastatic HCC cell lines (HCCLM3, MHCC97H, and MHCC97L), whereas their proliferation and invasion significantly decreased after depletion of HIWI. Intratumoral HIWI expression was higher than that of peritumoral tissue ( P < .001) and positively associated with proliferating cell nuclear antigen expression ( P < .001). Positive expression of intratumoral HIWI was associated with larger tumor size ( P = .047) and intrahepatic metastasis ( P = .027) and was an independent risk factor for overall survival ( P = .007) and recurrence-free survival ( P = .036), particularly in patients with low serum -fetoprotein and low Edmondson-Steiner grade. CONCLUSIONS: HIWI may play a key role in HCC proliferation and metastasis and can be a potential prognostic factor for HCC after curative resection, particularly with well-differentiated HCC. Cancer 2011. 2011 American Cancer Society.
