 ,, 丁晓东
,, 丁晓东 ,东北农业大学农业生物功能基因重点实验室,哈尔滨 150030
,东北农业大学农业生物功能基因重点实验室,哈尔滨 150030Preliminary analysis of the role of GmSnRK1.1 and GmSnRK1.2 in the ABA and alkaline stress response of the soybean using the CRISPR/Cas9-based gene double-knockout system
Huiqing Li, Chao Chen, Ranran Chen, Xuewei Song, Jina Li, Yanming Zhu ,, Xiaodong Ding
,, Xiaodong Ding ,Key Laboratory of Agricultural Biological Functional Genes, Northeast Agricultural University, Harbin 150030, China
,Key Laboratory of Agricultural Biological Functional Genes, Northeast Agricultural University, Harbin 150030, China第一联系人:
编委: 高彩霞
收稿日期:2017-12-27修回日期:2018-05-16网络出版日期:2018-06-20
| 基金资助: |
Received:2017-12-27Revised:2018-05-16Online:2018-06-20
| Fund supported: |
作者简介 About authors
李慧卿,硕士研究生,专业方向:植物基因工程与分子生物学E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1528KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李慧卿, 陈超, 陈冉冉, 宋雪薇, 李佶娜, 朱延明, 丁晓东. 利用CRISPR/Cas9双基因敲除系统初步解析大豆GmSnRK1.1和GmSnRK1.2对ABA及碱胁迫的响应. 遗传[J], 2018, 40(6): 496-507 doi:10.16288/j.yczz.17-424
Huiqing Li, Chao Chen, Ranran Chen, Xuewei Song, Jina Li, Yanming Zhu, Xiaodong Ding.
蔗糖非发酵相关蛋白激酶(sucrose non-fermenting related protein kinases,SnRKs)是广泛存在于植物中的一类Ser/Thr蛋白激酶,能直接参与植物生长、发育、代谢和抗逆等途径[1]。根据序列和功能,SnRKs分为SnRK1、SnRK2和SnRK3家族,其中SnRK2和SnRK3亚家族成员已被广泛研究。据报道,SnRK2具有参与ABA调控的生理功能,包括气孔的开关、种子和根系的发育、抗盐、抗寒、抗旱等[2]。Zhu等[3]证明野生大豆(Glycine soja)GsAPK属于SnRK2家族激酶,能够提高植物对ABA和高盐胁迫的耐受性。SnRK3在植物抗盐的信号转导途径SOS3-SOS2-SOS1中扮演重要角色,被激活的SOS2 (SnRK3激酶)磷酸化并开启位于细胞膜上的Na+/H+泵,将盐逆向泵出细胞从而保护细胞不受盐份伤害[4]。此外,SnRK3在调控气孔开关、植物激素以及病源免疫等方面的功能也有报道[1]。相比于SnRK2和SnRK3,SnRK1可能具有更多的功能。在水稻(Oryza sativa L.)和野生大豆(Glycine soja)中,SnRK1比SnRK2和SnRK3拥有更多种类的互作蛋白,可能广泛参与基因转录调控,并且在多种植物激素信号转导途径中扮演关键角色[5,6]。植物中SnRK1激酶在进化和功能上与酵母(Saccharomyces)SNF1激酶最为相似,并能互补酵母的snf1突变功能[7]。Liu等[8]在苹果(Malus domestica)中沉默MtSnRK1.1基因,导致转基因苹果的愈伤组织对ABA敏感性降低。相比于SnRK2和SnRK3,SnRK1的同源基因数量较少,如在栽培大豆中只含有4个同源基因,遗传相对简单。目前,大豆GmSnRK1在响应ABA及碱胁迫应答中的功能却鲜有报道,阻碍了其作用机制的研究。
由于大豆基因组高度重复且十分复杂[9,10],使用传统技术沉默靶基因所产生的非特异性随机突变制约着植物基因功能验证及其分子机理的研究。近年来,CRISPR/Cas9基因组定点编辑技术能够精准、高效、快速地编辑特定单个或多个基因,已成为具有广阔应用前景的基因组编辑操作系统。CRISPR/ Cas9系统属于Ⅱ型CRISPR系统,核酸内切酶Cas9通过合成的向导RNA(gRNA)引导,识别靶序列并对靶位点进行切割,造成DNA双链断裂,从而激活细胞的同源定向修复或非同源末端连接自我修复机 制[11]。当碱基插入或缺失(InDels)引发移码突变或蛋白结构被改变时,靶基因的功能就会被破坏[11,12]。目前,CRISPR/Cas9系统已经应用到许多重要农作物中,如水稻[13,14,15,16]、番茄(Solanum lycopersicum L.)[17]、小麦(Triticum aestivum L.)[18]和玉米(Zea mays L.)[19]等。2015年,Jacobs等[20]首次利用CRISPR/Cas9技术在大豆中实现定点编辑;同年,Cai等[21]在大豆中快速检测CRISPR/Cas9系统介导定点编辑效率的方法,证明CRISPR/Cas9技术可对外源导入基因和内源基因进行定点敲除;2016年Du等[22]以GmPDS11和GmPDS18为靶基因,比较TALENs和CRISPR/Cas9技术在大豆中的编辑效率,结果表明CRISPR/Cas9系统更适用于同时编辑多个大豆同源基因。2017年,Cai等[23]利用CRISPR/Cas9技术定点敲除大豆开花调控关键基因GmFT2a,成功创制出无外源基因并稳定遗传的大豆晚花突变体材料。通过该系统可获得靶基因功能完全丧失的突变体,为开展大豆基因功能研究奠定了理论基础。
为解析大豆SnRK1激酶在响应ABA和碱胁迫中的功能,本研究构建了CRISPR/Cas9载体用于靶向敲除GmSnRK1.1和GmSnRK1.2双基因,并利用实验室前期构建的超量表达GmSnRK1基因载体,通过发根农杆菌K599介导大豆子叶节遗传转化,从基因敲除突变和超量表达两个方向,分析转基因毛状根在非生物胁迫下的表型,最终阐明植物SnRK1在非生物胁迫信号传导通路中的作用和功能。
1 材料和方法
1.1 材料
大豆栽培品种Williams 82由中国农业科学院韩天富研究员提供。发根农杆菌K599、35S强启动子诱导的GmSnRK1超量表达载体pCambia2300- SnRK1由东北农业大学植物生物工程实验室制备并保存。pTC217载体由美国明尼苏达大学惠赠[17]。1.2 GmSnRK1基因表达模式分析
选取Williams 82幼苗期根、茎、叶组织,分别提取各组织总RNA,使用GoldScript cDNA试剂盒(Invitrogen, 美国)反转录成cDNA。利用Primer Premier5.0设计基因特异性引物(表1),通过qRT- PCR分析GmSnRK1的4个同源基因在大豆不同组织中的表达量。将生长21 d长势一致的大豆幼苗分别置于25 μmol/L ABA和50 mmol/L NaHCO3条件下处理0 h、1 h、3 h、6 h、12 h、24 h后迅速切取幼嫩根尖部位,分别提取总RNA,并反转录成cDNA,利用qRT-PCR分析GmSnRK1.1和GmSnRK1.2在ABA处理和碱胁迫下的表达特性。PCR扩增体系为:5 μL 2×SYBR Mix (ABI Applied Biosystems,美国),1.5 μL cDNA模板,上下游引物(10 μmol/L)各0.5 μL,用灭菌水补足至10 μL。PCR扩增条件:94℃ 2 min;94℃ 15 s,60℃ 30 s,72℃ 1 min,40个循环;72℃ 1 min;4℃保存。以GmGAPDH为内参基因,数据处理采用比较CT法(2-ΔΔCT)定量分析基因相对表达量。实验设置3次生物学重复和3次技术重复进行数据标准化处理[24]。Table 1
表1
表1 qRT-PCR引物序列
Table 1
| 引物名称 | 引物序列(5°→3°) |
|---|---|
| Q-GmSnRK1.1 | F:CCAGTTCAATTGCCGTAGAAA |
| R:GCTTCGAGGAATTAAGCTAAGC | |
| Q-GmSnRK1.2 | F:CAAGACACAGAACTCCCATTTG |
| R:AAAGGTGAAAAGCAGCCTTAAG | |
| Q-GmSnRK1.3 | F:CAGTCATCTATCTCCCAATGCT |
| R:CTCAGGTATGGTCATTCTCCTC | |
| Q-GmSnRK1.4 | F:TCACTCTTCCTTGCCATCTATC |
| R:CTCAGGTATGGTCATCCTCTTC | |
| Q-GmGAPDH | F:GTCACAAGCCGTAGGAATCA |
| R:GCGGAAGTTGGTGAGAGATAG |
新窗口打开|下载CSV
1.3 基于大豆GmSnRK1.1和GmSnRK1.2基因靶点设计
为提高CRISPR/Cas9系统对豆科植物双靶基因的敲除效率,分别从GmSnRK1.1和GmSnRK1.2基因第1个外显子处、起始密码子下游300 bp以内,利用CRISPR-P (http://cbi.hzau.edu.cn/crispr/)[25]在线设计软件,获得正向链和反向链中紧邻5′-NGG (PAM)的所有潜在gRNA靶标序列(20 bp),通过对比大 豆基因组数据库选择特异性高的序列作为gRNA (表2)。Table 2
表2
表2 合成gRNA的引物序列
Table 2
| 引物名称 | 引物序列(5′→3′) |
|---|---|
| GmSnRK1.1-gRNA1 | F:gattgCAAGATTAAGAACATGGAAA |
| R:aaaacTTTCCATGTTCTTAATCTTG | |
| GmSnRK1.2-gRNA1 | F:gattgACATGTGTTGACTGGCCATA |
| R:aaaacTATGGCCAGTCAACACATGT |
新窗口打开|下载CSV
1.4 CRISPR/Cas9植物表达载体的构建
以pTC217载体为模板,利用高保真DNA聚合酶PrimeSTAR? HS (TaKaRa,北京)特异扩增出两段分别含有BsmBⅠ和BsaⅠ酶切位点的gRNA表达框,用T4连接酶(TaKaRa,北京)将经PNK磷酸化(NEB,美国)的gRNA表达框依次与线性化的骨架载体pTC217连接,获得含两个gRNA表达框的载体pPBELp-CRISPR/Cas9。将酶切位点BsmBⅠ突变的Bar基因经PNK磷酸化后,与经限制性内切酶PmlⅠ(NEB,美国)线性化并去磷酸化(CIP) (NEB,美国)的载体pPBELp-CRISPR/cas9酶连整合,获得pPBELp-CRISPR/Cas9-Bar载体。将5 μL gRNA寡核苷酸链的正反向引物(10 μmol/L)与5 μL 10×T4 ligase Buffer (TaKaRa,北京)混合,用灭菌水补足至50 μL,沸水浴处理5 min,缓慢冷却至室温完成退火获得双链gRNA (使用前稀释20倍);将载体pPBELp- CRISPR/Cas9-Bar经BsmBⅠ限制性内切酶(NEB,美国)线性化后与GmSnRK1.1-gRNA1双链接头进行酶连整合,获得重组载体后经BsaⅠ限制性内切酶(NEB,美国)线性化后与GmSnRK1.2-gRNA1双链接头进行酶连整合,最终获得可表达Cas9蛋白、双靶点gRNA和Bar筛选标记基因的pPBELp-CRISPR/ Cas9-Ⅰ植物表达载体(图1)。图1
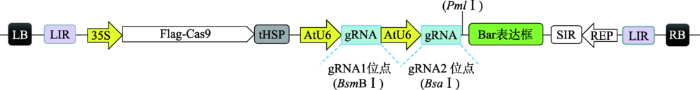 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CRISPR/Cas9介导的双基因敲除载体示意图
Fig. 1Schematic diagram of the CRISPR/Cas9 vector mediating gene double-knockout
连接产物均通过转化大肠杆菌Trans1-T1感受态(全式金,北京)细胞、37℃倒置培养过夜、挑取单菌落、菌落PCR鉴定阳性质粒后摇菌,使用质粒提取试剂盒(全式金,北京)抽提质粒,测序(金维智,苏州)鉴定正确的载体进行下一步操作。将植物表达载体利用冻融法转入发根农杆菌K599中,经过PCR鉴定正确的单菌落用于大豆毛状根诱导。
1.5 大豆毛状根诱导
选择饱满的Williams 82大豆种子,用氯气进行表面灭菌(16~20 h),置于萌发培养基(B5盐,20 g/L蔗糖,8 g/L琼脂,pH 5.8)中,在25℃ (16 h光照/ 8 h黑暗)萌发5 d。将含有载体的发根农杆菌进行二次活化,直至OD600为0.6~0.9。使用保留约3 mm下胚轴的子叶作为外植体,剥掉种皮去除幼芽,用解剖刀在子叶与下胚轴交接处轻划数痕,放到二次活化后的菌液中浸泡30 min,期间轻轻摇晃几次。侵染后倒掉菌液,将外植体切面朝下置于铺有灭 菌滤纸的共培养培养基(1/10 MS盐,30 g/L蔗糖,3.9 g/L MES,7 g/L琼脂,150 mg/L DTT,200 mmol/L乙酰丁香酮,pH 5.4)上,23℃暗培养4 d,促进T-DNA从质粒转移到植物细胞中。将共培养后的子叶利用液体除菌培养基(1/2 MS盐,30 g/L蔗糖,0.6 g/L MES,100 mg/L 阿莫西林克拉维酸钾,0.5 mg/L草甘膦,pH 5.8)冲洗4~5次,用灭菌滤纸吸净残留液体,并将子叶倾斜式插入到毛状根诱导培养基(1/2 MS盐,30 g/L蔗糖,0.6 g/L MES,100 mg/L 阿莫西林克拉维酸钾,0.5 mg/L草甘膦,8 g/L琼脂,pH 5.8)上,25℃ (16 h光照/ 8 h黑暗)培养7 d左右即可见毛状根。1.6 转基因毛状根PCR检测与突变体目的基因靶位点鉴定
利用植物基因组DNA提取试剂盒(全式金,北京)提取大豆新鲜毛状根的基因组DNA。利用Bar基因特异引物(表3)进行转基因毛状根PCR检测,初步确定转基因根系;以转基因阳性根的基因组DNA为模板,利用靶位点特异引物(表3)扩增目的基因靶位点区域序列,进行靶位点基因型鉴定,PCR产物长度约为500 bp。PCR扩增体系:10 μL 5× PrimeSTAR Buffer (Mg2+ Plus),4 μL dNTP Mixture (2.5 mmol/L),上下游引物(10 μmol/L)各1 μL,1 μL模板DNA,0.5 μL PrimeSTAR HS DNA Polymerase (2.5 U/μL)灭菌水补足至50 μL。PCR扩增条件:98℃ 8 min;98℃ 10 s,60℃ 10 s,72℃ 1 min,30个循环;72℃延伸10 min。扩增产物回收后连入pEASY- Blunt3 (全式金,北京)平末端载体,转化大肠杆菌Trans1-T1后随机挑取单克隆测序。利用序列比对分析软件Sequencher 4.7对Sanger测序结果进行分析。Table 3
表3
表3 PCR引物序列
Table 3
| 引物名称 | 引物序列(5′→3′) |
|---|---|
| GmSnRK1.1 | F:AACACTCGGCATTGGGTCCT |
| R:GGATGACCGCAATTAAGGCA | |
| GmSnRK1.2 | F:TATTCGCTGAACCCCTCTCT |
| R:AGACTGAGCAGGAGGTAACAT | |
| GmSnRK1.3 | F:GAGGTGGTGCTGGACTGGACA |
| R:ATGAACAAGACTGAGCACGAG | |
| GmSnRK1.4 | F:AAACTCTTGGCATTGGATCC |
| R:TGTTGTCACTCCATCTCTTTC | |
| Bar | F:CCTTATCTGGGAACTACTCACAC |
| R:CACCATCGTCAACCACTACAT |
新窗口打开|下载CSV
1.7 表型观察
将诱导生长10 d左右、长势一致的毛状根继代培养在分别含有25 μmol/L ABA和50 mmol/L NaHCO3培养基上,在25℃、16 h光照/8 h黑暗光周期下培养15 d后观察毛状根生长情况并记录根长与根鲜重。所有表型分析均设置3次重复,利用SPSS20.0检验显著性。2 结果与分析
2.1 靶基因的确定
2.1.1 GmSnRK1基因组织表达模式分析通过qRT-PCR对大豆GmSnRK1的4个同源基因(GmSnRK1.1、GmSnRK1.2、GmSnRK1.3、GmSnRK1.4)进行组织表达模式分析(图2)。结果表明,GmSnRK1的4个同源基因在根、茎、叶中均有表达,但不同组织中的表达量存在明显差异。其中GmSnRK1.1和GmSnRK1.2在各组织中的表达量均明显高于其他同源基因,并且在根中GmSnRK1.1和GmSnRK1.2的相对表达量占总相对表达量的45.15%和36.95%为主导表达基因,而GmSnRK1.3和GmSnRK1.4的相对表达量很低。因此,本研究选择GmSnRK1.1和GmSnRK1.2作为靶基因。
图2
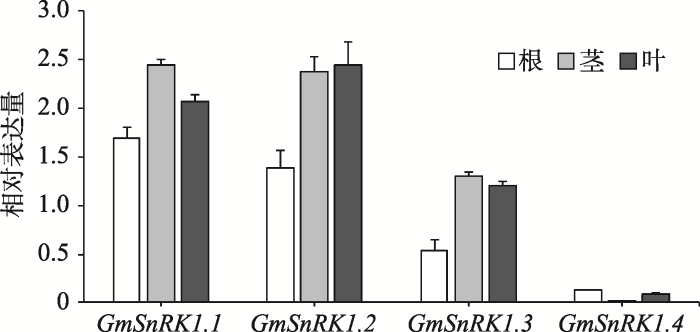 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2大豆GmSnRK1亚家族基因成员在根、茎、叶中的表达2.1.2 ABA及碱胁迫处理下GmSnRK1.1和GmSnRK1.2的表达模式分析
Fig. 2Spatial expression profiles of GmSnRK1 members in roots, stems and leaves of the soybean
为选择响应ABA及碱胁迫的靶基因,利用qRT-PCR分析GmSnRK1.1和GmSnRK1.2基因在ABA及碱胁迫处理下不同时间点转录水平上动态表达量的变化。结果表明,在25 μmol/L ABA处理后,两个基因均受ABA诱导表达(图3A),其中GmSnRK1.1和GmSnRK1.2均在ABA处理后3 h时表达量最高;在50 mmol/L NaHCO3胁迫处理后,两个基因均受碱胁迫上调表达(图3B),其中GmSnRK1.1在碱胁迫诱导后6 h时表达量最高,而GmSnRK1.2 在1 h时表达量最高。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3ABA及碱胁迫处理下GmSnRK1.1和GmSnRK1.2基因表达特性
A:ABA处理下基因表达特性;B:NaHCO3处理下基因表达特性。
Fig. 3Temporal expression profiles of GmSnRK1.1 and GmSnRK1.2 under ABA and alkaline treatments
以上结果说明,GmSnRK1.1和GmSnRK1.2在根中为两个主导表达基因,其表达受ABA和碱胁迫诱导,因此可作为基因敲除靶基因,并推测GmSnRK1.1和GmSnRK1.2可能参与大豆响应ABA和碱胁迫信号转导通路。
2.2 pPBELp-CRISPR/Cas9-Ⅰ载体获得及抗性毛状根分子生物学鉴定
为敲除双靶基因GmSnRK1.1和GmSnRK1.2,在BsmBⅠ和BsaⅠ位点上分别引入2个靶点gRNA,测序结果表明与设计序列一致(图4),表明包含双靶点gRNA的pPBELp- CRISPR/Cas9-Ⅰ载体构建成功。图4
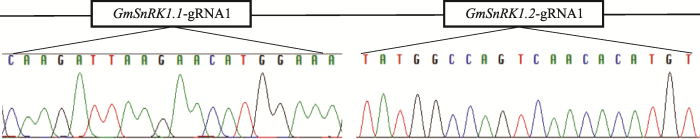 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4pPBELp-CRISPR/Cas9-Ⅰ载体靶点测序结果
Fig. 4pPBELp-CRISPR/Cas9-Ⅰvector sequencing results at vector targets
采用发根农杆菌介导的子叶节侵染法对大豆进行遗传转化。将不含gRNA的空载体pPBELp- CRISPR/Cas9-Bar (对照组)、超量表达载体pCambia2300-SnRK1以及双基因敲除载体pPBELp- CRISPR/Cas9-Ⅰ利用冻融法分别转入发根农杆菌K599中,鉴定出的阳性菌落用于侵染大豆子叶节。携带植物表达载体的发根农杆菌平均从每个大豆子叶节点处诱导4条抗性毛状根,通过PCR对抗性毛状根进行Bar基因检测。以含Bar基因的重组载体作为阳性对照,非转基因毛状根DNA作为阴性对照,部分凝胶电泳结果如图5所示。阳性对照和大部分抗性毛状根均扩增出预计大小条带,而阴性对照无扩增条带,初步说明T-DNA已整合到大豆基因组中。经PCR检测,对照组、超量表达型以及基因敲除型抗性毛状根阳性率分别为87.5% (70/80)、92.5% (74/80)、90% (72/80)。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5转基因抗性毛状根PCR检测
M:DL2000;+:阳性对照;-:阴性对照;1~14:转基因毛状根PCR扩增产物。
Fig. 5The PCR identification of resistant hairy roots
2.3 CRISPR/Cas9系统介导的大豆毛状根在ABA及碱胁迫下的表型分析
非生物胁迫会影响植株的生长发育,导致植物产生一系列的生理应答。为初步解析GmSnRK1.1和GmSnRK1.2对ABA及碱胁迫的响应,本研究将诱导生长10 d且长势一致的转基因毛状根转入分别含有25 μmol/L ABA和50 mmol/L NaHCO3的培养基中继续培养,15 d后观察毛状根生长情况。结果表明,在正常培养基上,基因敲除毛状根与其他毛状根根系相比生长状态正常,无明显差异。在含25 μmol/L ABA培养基中,基因敲除毛状根长势良好受影响较轻、仍能维持正常生长,而对照组和超量表达GmSnRK1毛状根生长发育受到明显抑制(图6A),基因敲除毛状根根长和根鲜重均显著高于其他毛状根根系(P<0.01) (图6:B和C)。图6
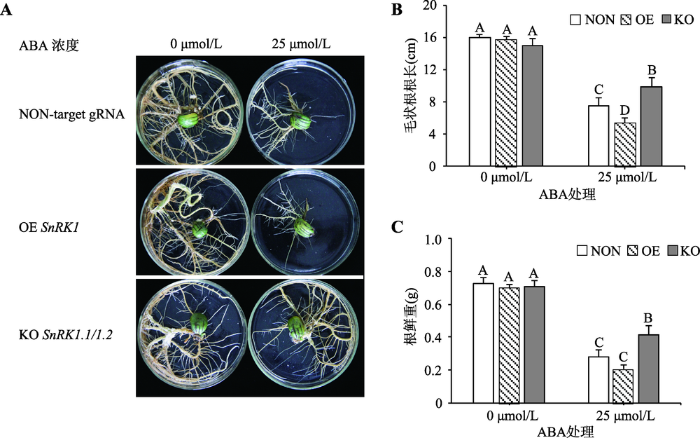 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不同基因型毛状根在ABA处理下的表型分析
A:大豆毛状根在ABA 处理下的表型;B:ABA处理对大豆毛状根根长的影响;C:ABA处理对大豆毛状根根鲜重的影响。柱型图上方的大写字母表示在P<0.01水平上差异显著;NON表示无靶点对照组;OE表示超量表达;KO表示基因敲除。
Fig. 6Phenotypic analysis of different genotypic hairy roots under ABA treatment
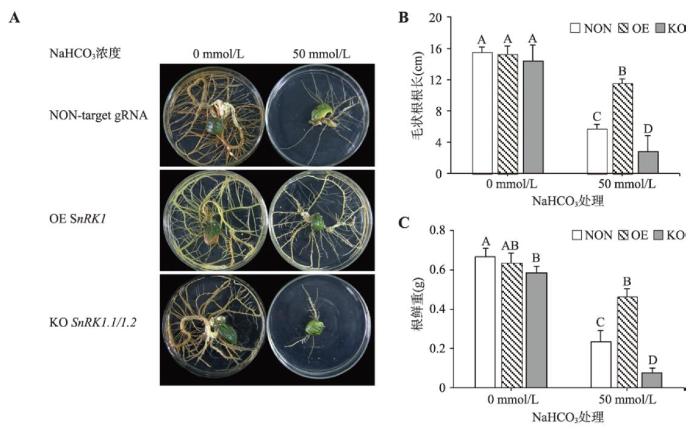
在含50 mmol/L NaHCO3培养基中,超量表达GmSnRK1毛状根长势受影响最轻,而对照组和基因敲除毛状根生长受到明显抑制(图7A),其根长和根鲜重显著低于超量表达毛状根(P<0.01) (图7:B和C),基因敲除毛状根表现出对碱胁迫高度敏感。以上结果说明,基因敲除毛状根在非生物胁迫下的表型可能与GmSnRK1.1/1.2基因突变,引发GmSnRK1功能丧失有关,而GmSnRK1基因的超量表达可以增强大豆耐碱能力;同时,利用本研究建立的CRISPR/ Cas9系统可以有效地将GmSnRK1.1和GmSnRK1.2进行基因敲除,所得的突变体材料初步解析了SnRK1激酶在ABA和碱胁迫中的功能。
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7不同基因型毛状根在在NaHCO3胁迫下的表型分析
A:大豆毛状根在NaHCO3胁迫下的表型;B:NaHCO3胁迫对大豆毛状根根长的影响;C:NaHCO3胁迫对大豆毛状根根鲜重的影响。柱型图上方的大写字母表示在P<0.01水平上差异显著;NON表示无靶点对照组;OE表示超量表达;KO表示基因敲除。
Fig. 7Phenotypic analysis of different genotypic hairy roots under NaHCO3 treatment
2.4 GmSnRK1.1及GmSnRK1.2靶位点基因型鉴定
本研究随机选取35条经PCR鉴定Bar基因阳性的抗性毛状根,分别提取每条毛状根的基因组DNA,对靶位点区域进行PCR克隆测序鉴定其基因型。测序结果表明,GmSnRK1.1在27条毛状根中存在基因突变(图8A),其中25条为有效突变(缺失或增加碱基数不是3的倍数),在GmSnRK1.1靶位点处发生了由碱基插入和碱基缺失引起的移码突变(图9:A和B);GmSnRK1.2在24条毛状根中存在基因突变(图8B),其中21条为有效突变,在GmSnRK1.2靶位点处均发生由碱基缺失引起的移码突变(图9C);真正有效的双基因突变有17条,均是由碱基缺失引起的移码突变(图9:D和E)。统计结果见表4。以上结果说明,测序结果与靶基因对ABA及碱胁迫的响应结果相一致,本研究成功获得了GmSnRK1.1及GmSnRK1.2双基因突变毛状根。图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8CRISPR/Cas9介导的大豆GmSnRK1.1和GmSnRK1.2双突变基因型多态性
A:GmSnRK1.1突变基因型多态性;B:GmSnRK1.2突变基因型多态性。红色虚线表示缺失碱基;红色字体表示增加碱基;下划线表示gRNA靶位点;加粗表示PAM序列。
Fig. 8Polymorphic mutant genotypes of soybean GmSnRK1.1 and GmSnRK1.2 genes mediated by the CRISPR/Cas9 system
图9
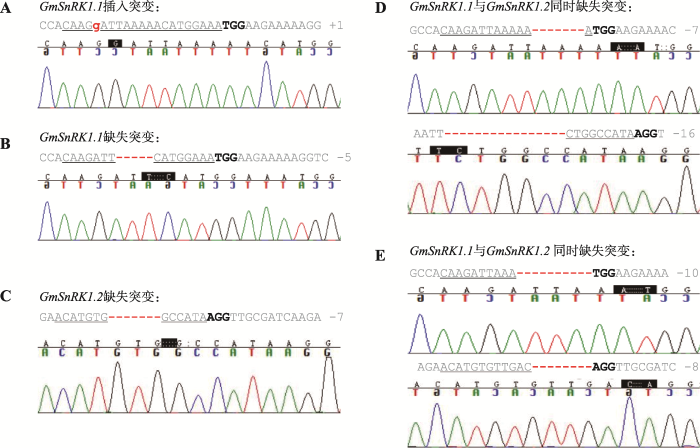 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9靶位点突变类型及其测序色谱分析
A,B:GmSnRK1.1单基因突变;C:GmSnRK1.2单基因突变;D,E:单条毛状根中双基因突变。红色虚线表示缺失碱基;红色字体表示增加碱基;下划线表示gRNA靶位点;加粗表示PAM序列;黑色框内表示突变位点。
Fig. 9Analyses of sequencing chromatogram data of the target mutant sites
Table 4
表4
表4 转基因毛状根突变率
Table 4
| 靶基因 | 转基因毛状根数 | 基因突变毛状根数 | 突变率(%) | 双基因突变毛状根数 | 双基因突变率(%) |
|---|---|---|---|---|---|
| GmSnRK1.1 | 35 | 27 | 77.1 | 17 | 48.6 |
| GmSnRK1.2 | 35 | 24 | 68.6 |
新窗口打开|下载CSV
3 讨 论
3.1 CRISPR/Cas9技术对双靶基因的编辑效率
在研究植物基因功能时,由于存在多个同源基因功能,或者多个不同源基因参与平行的信号传导[26],往往需要得到多个基因同时突变的材料,多靶点基因编辑系统则为基因功能研究提供更方便准确的选择。有研究表明,利用两个以上的靶点进行基因组定点编辑时,能够提高获得纯和突变体的概率[27]。本研究构建了CRISPR/Cas9系统介导的双靶点基因敲除体系,对大豆内源基因GmSnRK1.1和GmSnRK1.2进行定点敲除,获得了双基因突变的大豆毛状根根系。通过Sanger测序对靶位点编辑效率进行分析,结果表明不同靶点的突变率存在一定差异,GmSnRK1.1靶位点突变率为77.1%,GmSnRK1.2靶位点突变率为68.6%,双靶点同时发生突变的效率为48.6%。这可能是由于不同gRNA对靶序列的亲合能力不同,导致Cas9核酸内切酶与特定位置DNA序列的结合效率不同,从而影响突变率[28,29]。另外,在选择靶位点时,靶序列的碱基分布、GC含量等都对gRNA和靶标序列间的亲和力有影响[30]。
3.2 CRISPR/Cas9技术中的脱靶现象
随着基因编辑技术的快速发展,已有研究发现CRISPR/Cas9系统存在一定的脱靶现象[31]。张锋等[32]通过改变化脓性链球菌Cas9蛋白氨基酸序列中的3个氨基酸,显著降低了脱靶效应。Lee等[33]将Sth Cas9 替换成Spy Cas9蛋白用来识别较短的PAM序列,从而提高打靶效率。目前,已有多款软件针对CRISPR/Cas9技术的脱靶效应来辅助设计gRNA[34]。本研究通过CRISPR-GE (http://skl.scau.edu.cn/)[35]在线预测潜在脱靶位点,结果表明可能发生脱靶事 件的位点位于靶基因的同源基因GmSnRK1.3和GmSnRK1.4上,利用特异性引物(表3)对预测位点进行PCR扩增,经Sanger测序分析,结果显示没有发生基因突变,说明本研究使用的gRNA具有一定特异性。CRISPR/Cas9技术中导致脱靶事件发生的因素有很多,如何降低脱靶风险还需要进行更加深入的研究探讨。3.3 大豆GmSnRK1基因功能
SnRK1是一类广泛参与胁迫抗性等途径的激酶。Ding等[5]利用水稻SnRK1a和SnRK1b作为诱饵对水稻cDNA文库进行酵母二元杂交筛选,获得SnRK1复合体中的β、βγ亚单元、WRKY、AP2/ EREBP、B-box锌指蛋白等转录因子、植物衰老相关蛋白等,说明SnRK1广泛参与基因的转录调控。 水稻中SnRK1直接参与植物的水化合物代谢并能提高植物对非生物胁迫的抗性[36]。本研究利用CRISPR/Cas9双基因敲除系统获得了较为精准的功能基因敲除突变体,从突变体和超量表达角度初步探究GmSnRK1.1和GmSnRK1.2对ABA及碱胁迫的响应。研究发现,在含25 μmol/L ABA的培养基中基因敲除型毛状根对ABA敏感性很低,可维持正常生长,而超表达型毛状根生长受到明显抑制,说明SnRK1可能直接参与了ABA信号传导途径;在含50 mmol/L NaHCO3的培养基中基因敲除型毛状根长势受到明显抑制,而超表达型毛状根能够提高植物对碱胁迫的抗性,这与酵母SNF1能够提高酵母耐碱性相似[7],说明该类蛋白激酶在功能上的保守性。本研究初步确定GmSnRK1.1和GmSnRK1.2在ABA信号转导和大豆耐碱机制中发挥着重要作用,而具体分子机制还需进一步研究。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:20974737 [本文引用: 2]

Sucrose non-fermenting-1 (SNF1)-related protein kinases (SnRKs) take their name from their fungal homologue, SNF1, a global regulator of carbon metabolism. The plant family has burgeoned to comprise 38 members which can be subdivided into three sub-families: SnRK1, SnRK2, and SnRK3. There is now good evidence that this has occurred to allow plants to link metabolic and stress signalling in a way that does not occur in other organisms. The role of SnRKs, focusing in particular on abscisic acid-induced signalling pathways, salinity tolerance, responses to nutritional stress and disease, and the regulation of carbon metabolism and, therefore, yield, is reviewed here. The key role that SnRKs play at the interface between metabolic and stress signalling make them potential candidates for manipulation to improve crop performance in extreme environments.
URLPMID:19309312 [本文引用: 1]

The phosphorylation and dephosphorylation of proteins, catalysed by protein kinases and phosphatases, is the major mechanism for the transduction of intracellular signals in eukaryotic organisms. Signalling pathways often comprise multiple phosphorylation/dephosphorylation steps and a long-standing hypothesis to explain this phenomenon is that of the protein kinase cascade, in which a signal is amplified as it is passed from one step in a pathway to the next. This review represents a re-evaluation of this hypothesis, using the signalling network in which the SnRKs [Snf1 (sucrose non-fermenting-1)-related protein kinases] function as an example, but drawing also on the related signalling systems involving Snf1 itself in fungi and AMPK (AMP-activated protein kinase) in animals. In plants, the SnRK family comprises not only SnRK1, but also two other subfamilies, SnRK2 and SnRK3, with a total of 38 members in the model plant Arabidopsis. This may have occurred to enable linking of metabolic and stress signalling. It is concluded that signalling pathways comprise multiple levels not to allow for signal amplification, but to enable linking between pathways to form networks in which key protein kinases, phosphatases and target transcription factors represent hubs on/from which multiple pathways converge and emerge.
URLPMID:3306294 [本文引用: 1]

Plant Snf1 (sucrose non-fermenting-1) related protein kinase (SnRK), a subfamily of serine/threonine kinases, has been implicated as a crucial upstream regulator of ABA and osmotic signaling as in many other signaling cascades. In this paper, we have isolated a novel plant specific ABA activated calcium independent protein kinase (GsAPK) from a highly salt tolerant plant, Glycine soja (50109), which is a member of the SnRK2 family. Subcellular localization studies using GFP fusion protein indicated that GsAPK is localized in the plasma membrane. We found that autophosphorylation and Myelin Basis Protein phosphorylation activity of GsAPK is only activated by ABA and the kinase activity also was observed when calcium was replaced by EGTA, suggesting its independence of calcium in enzyme activity. We also found that cold, salinity, drought, and ABA stress alter GsAPK gene transcripts and heterogonous overexpression of GsAPK in Arabidopsis alters plant tolerance to high salinity and ABA stress. In summary, we demonstrated that GsAPK is a Glycine soja ABA activated calcium independent SnRK-type kinase presumably involved in ABA mediated stress signal transduction.
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
URLPMID:17438333 [本文引用: 2]

The Saccharomyces cerevisiae Snf1 protein kinase, a member of the Snf1/AMPK (AMP-activated protein kinase) family, has important roles in metabolic control, particularly in response to nutrient stress. Here we have addressed the role of Snf1 in responses to other environmental stresses. Exposure of cells to sodium ion stress, alkaline pH, or oxidative stress caused an increase in Snf1 catalytic activity and phosphorylation of Thr-210 in the activation loop, whereas treatment with sorbitol or heat shock did not. Inhibition of respiratory metabolism by addition of antimycin A to cells also increased Snf1 activity. Analysis of mutants indicated that the kinases Sak1, Tos3, and Elm1, which activate Snf1 in response to glucose limitation, are also required under other stress conditions. Each kinase sufficed for activation in response to stress, but Sak1 had the major role. In sak1Delta tos3Delta elm1Delta cells expressing mammalian Ca(2+)/calmodulin-dependent protein kinase kinase alpha, Snf1 was activated by both sodium ion and alkaline stress, suggesting that stress signals regulate Snf1 activity by a mechanism that is independent of the upstream kinase. Finally, we showed that Snf1 protein kinase is regulated differently during adaptation of cells to NaCl and alkaline pH with respect to both temporal regulation of activation and subcellular localization. Snf1 protein kinase becomes enriched in the nucleus in response to alkaline pH but not salt stress. Such differences could contribute to specificity of the stress responses.
URLPMID:29016962 [本文引用: 1]

Abstract ABA is a crucial phytohormone for development and stress responses in plants. Snf1-related protein kinase 1.1 (SnRK1.1) is involved in the ABA response. However, the molecular mechanism underlying the SnRK1.1 response to ABA is largely unknown. Here, it was found that overexpression of the apple MdSnRK1.1 gene enhanced ABA sensitivity in both transgenic apple calli and Arabidopsis seedlings. Subsequently, a yeast two-hybrid screen demonstrated that MdCAIP1 (C2-domain ABA Insensitive Protein1) interacted with MdSnRK1.1. Their interaction was further confirmed by pull-down and co-immunoprecipitation assays. Expression of the MdCAIP1 gene was positively induced by ABA. Its overexpression enhanced ABA sensitivity in transgenic apple calli. Furthermore, it was found that MdSnRK1.1 phosphorylated the MdCAIP1 protein in vivo and promoted its degradation in vitro and in vivo. As a result, MdSnRK1.1 inhibited MdCAIP1-mediated ABA sensitivity, and MdCAIP1 partially reduced MdSnRK1.1-mediated ABA sensitivity. Our findings indicate that MdSnRK1.1 plays an important role in the ABA response, partially by controlling the stability of the MdCAIP1 protein. The Author 2017. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists. All rights reserved. For permissions, please email: journals.permissions@oup.com.
URLPMID:2077340 [本文引用: 1]

Background Soybean, Glycine max(L.) Merr., is a well documented paleopolyploid. What remains relatively under characterized is the level of sequence identity in retained homeologous regions of the...
URLPMID:25108243 [本文引用: 1]

The recent whole-genome sequencing of soybean (Glycine max) revealed that soybean experienced whole-genome duplications 59 million and 13 million years ago, and it has an octoploid-like genome in spite of its diploid nature. We analyzed a natural green-cotyledon mutant line, Tenshin-daiseitou. The physiological analysis revealed that Tenshin-daiseitou shows a non-functional stay-green phenotype in senescent leaves, which is similar to that of the mutant of Mendel's green-cotyledon gene I, the ortholog of SGR in pea. The identification of gene mutations and genetic segregation analysis suggested that defects in GmSGR1 and GmSGR2 were responsible for the green-cotyledon/stay-green phenotype of Tenshin-daiseitou, which was confirmed by RNA interference (RNAi) transgenic soybean experiments using GmSGR genes. The characterized green-cotyledon double mutant d1d2 was found to have the same mutations, suggesting that GmSGR1 and GmSGR2 are D1 and D2. Among the examined d1d2 strains, the d1d2 strain K144a showed a lower Chl a/b ratio in mature seeds than other strains but not in senescent leaves, suggesting a seed-specific genetic factor of the Chl composition in K144a. Analysis of the soybean genome sequence revealed four genomic regions with microsynteny to the Arabidopsis SGR1 region, which included the GmSGR1 and GmSGR2 regions. The other two regions contained GmSGR3a/GmSGR3b and GmSGR4, respectively, which might be pseudogenes or genes with a function that is unrelated to Chl degradation during seed maturation and leaf senescence. These GmSGR genes were thought to be produced by the two whole-genome duplications, and they provide a good example of such whole-genome duplication events in the evolution of the soybean genome.
URL [本文引用: 2]
URLPMID:25200087 [本文引用: 1]

Abstract The Cas9/sgRNA of the CRISPR/Cas system has emerged as a robust technology for targeted gene editing in various organisms, including plants, where Cas9/sgRNA-mediated small deletions/insertions at single cleavage sites have been reported in transient and stable transformations, although genetic transmission of edits has been reported only in Arabidopsis and rice. Large chromosomal excision between two remote nuclease-targeted loci has been reported only in a few non-plant species. Here we report in rice Cas9/sgRNA-induced large chromosomal segment deletions, the inheritance of genome edits in multiple generations and construction of a set of facile vectors for high-efficiency, multiplex gene targeting. Four sugar efflux transporter genes were modified in rice at high efficiency; the most efficient system yielding 87-100% editing in T0 transgenic plants, all with di-allelic edits. Furthermore, genetic crosses segregating Cas9/sgRNA transgenes away from edited genes yielded several genome-edited but transgene-free rice plants. We also demonstrated proof-of-efficiency of Cas9/sgRNAs in producing large chromosomal deletions (115-245 kb) involving three different clusters of genes in rice protoplasts and verification of deletions of two clusters in regenerated T0 generation plants. Together, these data demonstrate the power of our Cas9/sgRNA platform for targeted gene/genome editing in rice and other crops, enabling both basic research and agricultural applications. The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
URLPMID:23929338 [本文引用: 1]

The article offers information on genome modification of crop plants using a CRISPR-Cas system. It states that genome editing technologies using zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can also generate genome modifications. Photographs related to genome editing in rice and wheat using an engineered type II CRISPR-Cas system are also presented.
URLPMID:4052633 [本文引用: 1]

Background The type II clustered, regularly interspaced, short palindromic repeat (CRISPR)/ CRISPR-associated protein 9 (Cas9) system is a novel molecular tool for site-specific genome modification....
URLPMID:3814374 [本文引用: 1]

The type II CRISPR/Cas system from Streptococcus pyogenes and its simplified derivative, the Cas9/single guide RNA (sgRNA) system, have emerged as potent new tools for targeted gene knockout in bacteria, yeast, fruit fly, zebrafish and human cells. Here, we describe adaptations of these systems leading to successful expression of the Cas9/sgRNA system in two dicot plant species, Arabidopsis and tobacco, and two monocot crop species, rice and sorghum. Agrobacterium tumefaciens was used for delivery of genes encoding Cas9, sgRNA and a non-fuctional, mutant green fluorescence protein (GFP) to Arabidopsis and tobacco. The mutant GFP gene contained target sites in its 5 coding regions that were successfully cleaved by a CAS9/sgRNA complex that, along with error-prone DNA repair, resulted in creation of functional GFP genes. DNA sequencing confirmed Cas9/sgRNA-mediated mutagenesis at the target site. Rice protoplast cells transformed with Cas9/sgRNA constructs targeting the promoter region of the bacterial blight susceptibility genes, OsSWEET14 and OsSWEET11, were confirmed by DNA sequencing to contain mutated DNA sequences at the target sites. Successful demonstration of the Cas9/sgRNA system in model plant and crop species bodes well for its near-term use as a facile and powerful means of plant genetic engineering for scientific and agricultural applications.
URLMagsci [本文引用: 1]

<p>利用CRISPR/Cas9技术对调控水稻产量千粒重基因<em>TGW6</em>定点编辑,获得了一套有重要育种价值的<em>tgw6</em>突变体。设计了分别由U3、U6a和U6b启动子驱动、长20 bp的guide RNA (gRNA)靶点以靶向编辑<em>TGW6</em>基因的外显子,首先将这3个靶点一起组装到pYLCRISPR/Cas9-MT(I)载体上,然后利用农杆菌介导侵染水稻材料H447 (R819/玉针香//R819的BC<sub>3</sub>F<sub>6</sub>);提取T<sub>0</sub>代转基因植株的基因组DNA并对编辑位点附近的DNA片段进行PCR检测及测序分析。结果表明,T<sub>0</sub>代材料中<em>tgw6</em>的突变频率高达90%,其中纯合缺失突变率约占51%。对T<sub>1</sub>代纯合缺失突变体的千粒重性状的调查分析结果表明,部分<em>tgw6</em>的缺失突变能显著提高千粒重(大于5%)。不同类型<em>tgw6</em>突变体的成功创建不仅丰富了<em>tgw6</em>的变异类型,为水稻的高产稳产奠定了重要的材料基础,还证实了CRISPR/Cas9技术在水稻基因工程育种中高效、易操作的特点,具有重要的理论与实践意义。</p>
URLMagsci [本文引用: 1]

<p>利用CRISPR/Cas9技术对调控水稻产量千粒重基因<em>TGW6</em>定点编辑,获得了一套有重要育种价值的<em>tgw6</em>突变体。设计了分别由U3、U6a和U6b启动子驱动、长20 bp的guide RNA (gRNA)靶点以靶向编辑<em>TGW6</em>基因的外显子,首先将这3个靶点一起组装到pYLCRISPR/Cas9-MT(I)载体上,然后利用农杆菌介导侵染水稻材料H447 (R819/玉针香//R819的BC<sub>3</sub>F<sub>6</sub>);提取T<sub>0</sub>代转基因植株的基因组DNA并对编辑位点附近的DNA片段进行PCR检测及测序分析。结果表明,T<sub>0</sub>代材料中<em>tgw6</em>的突变频率高达90%,其中纯合缺失突变率约占51%。对T<sub>1</sub>代纯合缺失突变体的千粒重性状的调查分析结果表明,部分<em>tgw6</em>的缺失突变能显著提高千粒重(大于5%)。不同类型<em>tgw6</em>突变体的成功创建不仅丰富了<em>tgw6</em>的变异类型,为水稻的高产稳产奠定了重要的材料基础,还证实了CRISPR/Cas9技术在水稻基因工程育种中高效、易操作的特点,具有重要的理论与实践意义。</p>
URLPMID:26541286 [本文引用: 2]

Abstract BACKGROUND: The use of homologous recombination to precisely modify plant genomes has been challenging, due to the lack of efficient methods for delivering DNA repair templates to plant cells. Even with the advent of sequence-specific nucleases, which stimulate homologous recombination at predefined genomic sites by creating targeted DNA double-strand breaks, there are only a handful of studies that report precise editing of endogenous genes in crop plants. More efficient methods are needed to modify plant genomes through homologous recombination, ideally without randomly integrating foreign DNA. RESULTS: Here, we use geminivirus replicons to create heritable modifications to the tomato genome at frequencies tenfold higher than traditional methods of DNA delivery (i.e., Agrobacterium). A strong promoter was inserted upstream of a gene controlling anthocyanin biosynthesis, resulting in overexpression and ectopic accumulation of pigments in tomato tissues. More than two-thirds of the insertions were precise, and had no unanticipated sequence modifications. Both TALENs and CRISPR/Cas9 achieved gene targeting at similar efficiencies. Further, the targeted modification was transmitted to progeny in a Mendelian fashion. Even though donor molecules were replicated in the vectors, no evidence was found of persistent extra-chromosomal replicons or off-target integration of T-DNA or replicon sequences. CONCLUSIONS: High-frequency, precise modification of the tomato genome was achieved using geminivirus replicons, suggesting that these vectors can overcome the efficiency barrier that has made gene targeting in plants challenging. This work provides a foundation for efficient genome editing of crop genomes without the random integration of foreign DNA.
URLPMID:24122057 [本文引用: 1]

Abstract The clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) system has been used as an efficient tool for genome editing. We report the application of CRISPR-Cas-mediated genome editing to wheat (Triticum aestivum), the most important food crop plant with a very large and complex genome. The mutations were targeted in the inositol oxygenase (inox) and phytoene desaturase (pds) genes using cell suspension culture of wheat and in the pds gene in leaves of Nicotiana benthamiana. The expression of chimeric guide RNAs (cgRNA) targeting single and multiple sites resulted in indel mutations in all the tested samples. The expression of Cas9 or sgRNA alone did not cause any mutation. The expression of duplex cgRNA with Cas9 targeting two sites in the same gene resulted in deletion of DNA fragment between the targeted sequences. Multiplexing the cgRNA could target two genes at one time. Target specificity analysis of cgRNA showed that mismatches at the 3' end of the target site abolished the cleavage activity completely. The mismatches at the 5' end reduced cleavage, suggesting that the off target effects can be abolished in vivo by selecting target sites with unique sequences at 3' end. This approach provides a powerful method for genome engineering in plants.
URLPMID:24576457 [本文引用: 1]

Transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems have emerged as powerful tools for genome editing in a variety of species. Here, we report, for the first time, targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. We designed five TALENs targeting 4 genes, namely ZmPDS, ZmIPK1A, ZmIPK, ZmMRP4, and obtained targeting efficiencies of up to 23.1% in protoplasts, and about 13.3% to 39.1% of the transgenic plants were somatic mutations. Also, we constructed two gRNAs targeting the ZmIPK gene in maize protoplasts, at frequencies of 16.4% and 19.1%, respectively. In addition, the CRISPR/Cas system induced targeted mutations in Z. mays protoplasts with efficiencies (13.1%) similar to those obtained with TALENs (9.1%). Our results show that both TALENs and the CRISPR/Cas system can be used for genome modification in maize.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:26603121 [本文引用: 1]

Abstract Gene targeting (GT) is of great significance for advancing basic plant research and crop improvement. Both TALENs (transcription activator-like effectors nucleases) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated 9) systems have been developed for genome editing in eukaryotes, including crop plants. In this work, we present the comparative analysis of these two technologies for two soybean genome editing targets, GmPDS11 and GmPDS18. We found GT in soybean hairy roots with a single targeting efficiency range of 17.5-21.1% by TALENs, 11.7-18.1% by CRISPR/Cas9 using the AtU6-26 promoter, and 43.4-48.1% by CRISPR/Cas9 using the GmU6-16g-1 promoter, suggesting that the CRISPR/Cas9 using the GmU6-16g-1 promoter is probably a much more efficient tool compared to the other technologies. Similarly, our double mutation GT efficiency experiment with these three technologies displayed a targeting efficiency of 6.25% by TALENs, 12.5% by CRISPR/Cas9 using the AtU6-26 promoter, and 43.4-48.1% by CRISPR/Cas9 using the GmU6-16g-1 promoter, suggesting that CRISPR/Cas9 is still a better choice for simultaneous editing of multiple homoeoalleles. Furthermore, we observed albino and dwarf buds (PDS knock-out) by soybean transformation in cotyledon nodes. Our results demonstrated that both TALENs and CRISPR/Cas9 systems are powerful tools for soybean genome editing. Copyright 2015 Elsevier B.V. All rights reserved.
[本文引用: 1]
URLPMID:17000637 [本文引用: 1]

The largely unused uracil-excision molecular cloning technique has excellent features in most aspects compared to other modern cloning techniques. Its application has, however, been hampered by incompatibility with proof-reading DNA polymerases. We have advanced the technique by identifying PfuCx as a compatible proof-reading DNA polymerase and by developing an improved vector design strategy. The original features of the technique, namely simplicity, speed, high efficiency and low cost are thus combined with high fidelity as well as a transparent, simple and flexible vector design. A comprehensive set of vectors has been constructed covering a wide range of different applications and their functionality has been confirmed.
URLPMID:24719468 [本文引用: 1]

Dear Editor, Precise and efficient genome editing is very important for gene functional characterization.In recent years,sequence-specific DNA nucleases have been developed to increase the efficiency of gene targeting or genome editing in animals and plants.Among them,Zinc-Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs) are two most commonly used sequencespecific chimeric proteins (Gaj et al.,2013).Recently,a breakthrough gene-targeting tool based on RNA-guided Cas9 nuclease from type 鈪 prokaryotic Cluster Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated system has been developed (Jinek et al.,2012).CRISPR/Cas system is an adaptive defense system in prokaryotes to fight against alien nucleic acids (Horvath and Barrangou,2010).The CRISPR loci are variable short spacers separated by short repeats,which are transcribed into synthetic single-guide RNA (sgRNA).The sgRNA forms a functional complex with CRISPR-associated nuclease (Cas9) and guide the nuclease to genomic loci matching a 20-bp complementary invading DNA,cleaving it immediately upstream of a required 5'-NGG Protospacer Adjacent Motif (PAM).A chimeric sgRNA that mimics the natural synthetic sgRNA can be used to target Cas9 for genome editing in eukaryotic cells.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:26089199 [本文引用: 1]

The CRISPR/Cas9 system is becoming an important genome editing tool for crop breeding. Although it has been demonstrated that target mutations can be transmitted to the next generation, their inheritance pattern has not yet been fully elucidated. Here, we describe the CRISPR/Cas9-mediated genome editing of four different rice genes with the help of online target-design tools. High-frequency mutagenesis and a large percentage of putative biallelic mutations were observed in T0 generations. Nonetheless, our results also indicate that the progeny genotypes of biallelic T0 lines are frequently difficult to predict and that the transmission of mutations largely does not conform to classical genetic laws, which suggests that the mutations in T0 transgenic rice are mainly somatic mutations. Next, we followed the inheritance pattern of T1 plants. Regardless of the presence of the CRISPR/Cas9 transgene, the mutations in T1 lines were stably transmitted to later generations, indicating a standard germline transmission pattern. Off-target effects were also evaluated, and our results indicate that with careful target selection, off-target mutations are rare in CRISPR/Cas9-mediated rice gene editing. Taken together, our results indicate the promising production of inheritable and 'transgene clean' targeted genome-modified rice in the T1 generation using the CRISPR/Cas9 system.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:26063738 [本文引用: 1]

Abstract The CRISPR/Cas9 system has revolutionized mammalian somatic cell genetics. Genome-wide functional screens using CRISPR/Cas9-mediated knockout or dCas9 fusion-mediated inhibition/activation (CRISPRi/a) are powerful techniques for discovering phenotype-associated gene function. We systematically assessed the DNA sequence features that contribute to single guide RNA (sgRNA) efficiency in CRISPR-based screens. Leveraging the information from multiple designs, we derived a new sequence model for predicting sgRNA efficiency in CRISPR/Cas9 knockout experiments. Our model confirmed known features and suggested new features including a preference for cytosine at the cleavage site. The model was experimentally validated for sgRNA-mediated mutation rate and protein knockout efficiency. Tested on independent data sets, the model achieved significant results in both positive and negative selection conditions and outperformed existing models. We also found that the sequence preference for CRISPRi/a is substantially different from that for CRISPR/Cas9 knockout and propose a new model for predicting sgRNA efficiency in CRISPRi/a experiments. These results facilitate the genome-wide design of improved sgRNA for both knockout and CRISPRi/a studies. 2015 Xu et al.; Published by Cold Spring Harbor Laboratory Press.
URL [本文引用: 1]

基因组编辑技术已经在多个模式植物、动物以及其他生物中得到成功应用。基因组编辑是利用序列特异核酸酶(Sequence-specific nucleases,SSNs)在基因组特定位点产生DNA双链断裂(Double-strand breaks,DSBs),从而激活细胞自身修复机制——非同源末端连接(Non-homologous end joining,NHEJ)或同源重组(Homologous recombination,HR),实现基因敲除、染色体重组以及基因定点插入或替换等。锌指核酸酶(Zinc finger nuclease,ZFN)、TALEN(Transcription activator-like effector nuclease)和CRISPR/Cas9(Clustered regularly interspaced short palindromic repeats/CRISPR-associated 9)系统是最主要的3类SSNs。ZFN和TALEN是利用蛋白与DNA结合方式靶向特定的基因组位点,而最新的CIRISPR/Cas9系统则是利用更简单的核苷酸互补配对方式结合在基因组靶位点,其构建简单、效率更高效,因而促进了基因组编辑在植物中的广泛应用。利用基因组编辑技术除了实现植物基因定点突变外,还可以将SSNs的DNA结合域与其他功能蛋白融合,实现基因的靶向激活、抑制和表观调控等衍生技术。本文从基因组编辑技术的原理与优势、SSNs组成及构建方法、基因组编辑及衍生技术在植物中应用、优化SSNs突变效率和减少脱靶突变方法等方面进行了系统介绍,并对未来需要迫切解决的一些问题进行了分析和展望。
URL [本文引用: 1]

基因组编辑技术已经在多个模式植物、动物以及其他生物中得到成功应用。基因组编辑是利用序列特异核酸酶(Sequence-specific nucleases,SSNs)在基因组特定位点产生DNA双链断裂(Double-strand breaks,DSBs),从而激活细胞自身修复机制——非同源末端连接(Non-homologous end joining,NHEJ)或同源重组(Homologous recombination,HR),实现基因敲除、染色体重组以及基因定点插入或替换等。锌指核酸酶(Zinc finger nuclease,ZFN)、TALEN(Transcription activator-like effector nuclease)和CRISPR/Cas9(Clustered regularly interspaced short palindromic repeats/CRISPR-associated 9)系统是最主要的3类SSNs。ZFN和TALEN是利用蛋白与DNA结合方式靶向特定的基因组位点,而最新的CIRISPR/Cas9系统则是利用更简单的核苷酸互补配对方式结合在基因组靶位点,其构建简单、效率更高效,因而促进了基因组编辑在植物中的广泛应用。利用基因组编辑技术除了实现植物基因定点突变外,还可以将SSNs的DNA结合域与其他功能蛋白融合,实现基因的靶向激活、抑制和表观调控等衍生技术。本文从基因组编辑技术的原理与优势、SSNs组成及构建方法、基因组编辑及衍生技术在植物中应用、优化SSNs突变效率和减少脱靶突变方法等方面进行了系统介绍,并对未来需要迫切解决的一些问题进行了分析和展望。
URL [本文引用: 1]
URLPMID:4786937 [本文引用: 1]

The clustered regularly-interspaced short palindromic repeats (CRISPR) RISPR-associated (Cas) system from Streptococcus pyogenes (Spy) has been successfully adapted for RNA-guided genome editing in a wide range of organisms. However, numerous reports have indicated that Spy CRISPR-Cas9 systems may have significant off-target cleavage of genomic DNA sequences differing from the intended on-target site. Here, we report the performance of the Neisseria meningitidis ( Nme ) CRISPR-Cas9 system that requires a longer protospacer-adjacent motif for site-specific cleavage, and present a comparison between the Spy and Nme CRISPR-Cas9 systems targeting the same protospacer sequence. The results with the native crRNA and tracrRNA as well as a chimeric single guide RNA for the Nme CRISPR-Cas9 system were also compared. Our results suggest that, compared with the Spy system, the Nme CRISPR-Cas9 system has similar or lower on-target cleavage activity but a reduced overall off-target effect on a genomic level when sites containing three or fewer mismatches are considered. Thus, the Nme CRISPR-Cas9 system may represent a safer alternative for precision genome engineering applications.
URL [本文引用: 1]

基于CRISPR/Cas9系统介导的第三代基因组编辑技术,已成功应用于动物、植物和微生物等诸多物种的基因组改造。如何提高CRISPR/Cas9技术的基因组编辑效率和最大限度降低脱靶风险一直是本领域的研究热点,而使用高效且特异的sg RNA(Small guide RNA)是基因组改造成功的关键性因素之一。目前,已有多款针对CRISPR/Cas9技术的sg RNA设计和/或脱靶效应评估软件,但不同的软件各有优缺点。本文重点对16款sg RNA设计和脱靶效应评估在线和单机版软件的特点进行了阐述,通过制定38项评估指标对不同软件进行了比较分析,最后对11种用于检测基因组编辑效率和脱靶的实验方法,以及如何筛选高效且特异的sg RNA进行了归纳总结。
URL [本文引用: 1]

基于CRISPR/Cas9系统介导的第三代基因组编辑技术,已成功应用于动物、植物和微生物等诸多物种的基因组改造。如何提高CRISPR/Cas9技术的基因组编辑效率和最大限度降低脱靶风险一直是本领域的研究热点,而使用高效且特异的sg RNA(Small guide RNA)是基因组改造成功的关键性因素之一。目前,已有多款针对CRISPR/Cas9技术的sg RNA设计和/或脱靶效应评估软件,但不同的软件各有优缺点。本文重点对16款sg RNA设计和脱靶效应评估在线和单机版软件的特点进行了阐述,通过制定38项评估指标对不同软件进行了比较分析,最后对11种用于检测基因组编辑效率和脱靶的实验方法,以及如何筛选高效且特异的sg RNA进行了归纳总结。
URL [本文引用: 1]
URLPMID:24569770 [本文引用: 1]

In plants, source-sink communication plays a pivotal role in crop productivity, yet the underlying regulatory mechanisms are largely unknown. The SnRK1A protein kinase and transcription factor MYBS1 regulate the sugar starvation signaling pathway during seedling growth in cereals. Here, we identified plant-specific SnRK1A-interacting negative regulators (SKINs). SKINs antagonize the function of SnRK1A, and the highly conserved GKSKSF domain is essential for SKINs to function as repressors. Overexpression of SKINs inhibits the expression of MYBS1 and hydrolases essential for mobilization of nutrient reserves in the endosperm, leading to inhibition of seedling growth. The expression of SKINs is highly inducible by drought and moderately by various stresses, which is likely related to the abscisic acid (ABA)-mediated repression of SnRK1A under stress. Overexpression of SKINs enhances ABA sensitivity for inhibition of seedling growth. ABA promotes the interaction between SnRK1A and SKINs and shifts the localization of SKINs from the nucleus to the cytoplasm, where it binds SnRK1A and prevents SnRK1A and MYBS1 from entering the nucleus. Our findings demonstrate that SnRK1A plays a key role regulating source-sink communication during seedling growth. Under abiotic stress, SKINs antagonize the function of SnRK1A, which is likely a key factor restricting seedling vigor.
