 ,1,2
,1,2Applications of the CRISPR/Cas9 system in insects
Xiaoling Tong1,2, Chunyan Fang1,2, Tingting Gai1,2, Jin Shi1,2, Cheng Lu1,2, Fangyin Dai ,1,2
,1,2通讯作者:
编委: 张博
收稿日期:2017-10-16修回日期:2018-01-21网络出版日期:2018-04-20
| 基金资助: |
Editorial board:
Received:2017-10-16Revised:2018-01-21Online:2018-04-20
| Fund supported: |
作者简介 About authors
童晓玲,博士,副教授,硕士生导师,研究方向:昆虫发育遗传E-mail:
方春燕,硕士研究生,专业方向:昆虫发育遗传E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (529KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
童晓玲, 方春燕, 盖停停, 石津, 鲁成, 代方银. CRISPR/Cas9系统在昆虫中的应用. 遗传[J], 2018, 40(4): 266-278 doi:10.16288/j.yczz.17-263
Xiaoling Tong, Chunyan Fang, Tingting Gai, Jin Shi, Cheng Lu, Fangyin Dai.
昆虫种类繁多,形态各异,是地球上数量最多的动物群体,在生态系统中扮演着重要的角色,与人类生活密切相关。例如,绢丝类昆虫家蚕是丝绸业的生物基础;显花农作物的授粉主要由授粉性昆虫如蝴蝶、蜜蜂等完成;有的昆虫还是传播疾病的媒介,如按蚊传播的疟疾即是一种致命性传染疾病,《2017年世界疟疾报告》显示2016年全球共发生约2.2亿疟疾病例,其中死亡人数约44.5万人。
随着许多昆虫基因组测序的完成,为深入了解昆虫生长、发育、变态、进化和繁殖提供了全新的信息支持,相关基因功能解析取得了突破性的进展。在后基因组时代,通过基因组编辑(genome editing)阐释昆虫学的各种科学问题已成为昆虫学研究的重要手段。基因组编辑技术是一种在基因组水平对DNA序列进行改造的遗传操作技术,能够实现基因定点插入或缺失突变、基因敲除、多位点或多基因同时突变和片段删除等精确操作。目前,已报道的基因组编辑技术包括锌指核酸酶(zinc-finger nucleases, ZFN)[1]、类转录激活因子核酸酶(transcription activator like effector nucleases, TALEN)[2,3]和成簇规律间隔短回文重复序列系统(clustered regularly interspaced short palindromic repeat/CRISPR-associated nuclease 9, CRISPR/Cas9)[4]。ZFN和TALEN复合体由多个酶亚基组成,不同靶位点通过构建不同的工程核酸,利用蛋白质和DNA结合的方式执行靶位点的序列识别和切割活性,因其设计比较复杂,实验过程较为繁琐,从而限制了其更为广泛的应用。CRISPR/Cas系统作为第三代基因编辑技术,是基于简单的核苷酸互补配对方式结合在基因组靶位点实现定点突变,其具有实验过程简单、耗时短和工作量小等优势,已成功应用于多种生物的基因组编辑。本文主要阐述了CRISPR/Cas9的结构及作用原理,总结了CRISPR/Cas9导入昆虫的方法和应用及脱靶应对策略,以期对经济昆虫和有益昆虫的分子育种、害虫的生物技术防控等研究提供参考。
1 CRISPR/Cas9系统的结构及作用原理
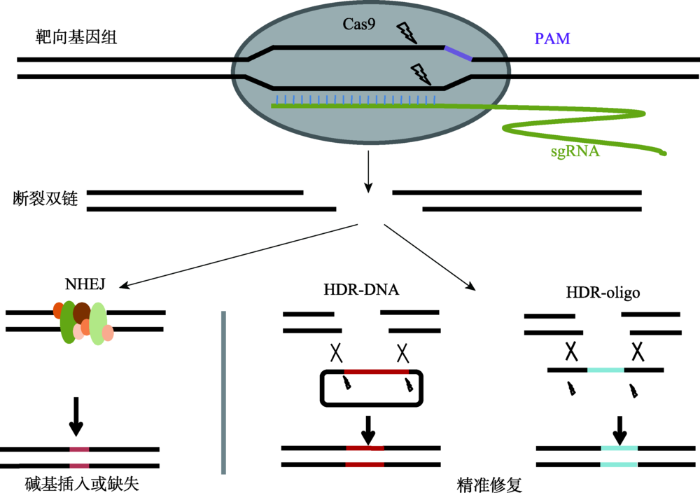
CRISPR/Cas系统是1987年由日本****在大肠杆菌中发现的一系列串联间隔短回文序列,这种短回文序列广泛存在于细菌和古细菌中,是一种后天性免疫系统[5]。CRISPR/Cas系统由CRISPR基因座,tracrRNA基因和一系列Cas蛋白基因组成,根据Cas序列及免疫机制的不同,CRISPR/Cas系统被分为Ⅰ型、Ⅱ型和Ⅲ型。目前,应用最为广泛的是肺炎链球菌S. pneumoniae Cas9(SpCas9)系统,即Ⅱ型系统。与Ⅰ型和Ⅲ型系统相比,Ⅱ型系统组成成分简单,由靶位点特异性CRISPR-derived RNA(crRNA)、反式激活crRNA(trans-activating crRNA, tracrRNA)和Cas9核酸内切酶3个组件构成。CRISPR/Cas9系统依赖Cas9蛋白靶向并切割基因组序列,其中crRNA和tracrRNA通过碱基互补配对形成二聚体,该二聚体与Cas9蛋白结合形成RNA-蛋白复合物,二聚体识别原型间隔序列毗邻基序(protospacer adjacent motif, PAM)前的靶位点序列[6]。Cas9蛋白含有HNH和RuvC两个内切酶结构域,其中HNH结构域切割与二聚体互补配对的靶序列,RuvC结构域切割含有PAM的序列,从而产生断裂双链(double-DNA strand breaks, DSBs)[7]。DSBs能够诱导细胞引发非同源末端连接修复(nonhomologous end joining, NHEJ)或同源重组修复机制(homologous-directed repair, HDR)对DSBs进行修复[8]。产生的DSBs在没有模板链的情况下会诱导NHEJ修复机制,这种修复机制通过碱基的随机插入、缺失或替换而导致基因功能丧失从而产生新的突变。HDR是通过细胞自身的姊妹染色单体或外源同源重组模板链介导的修复机制,可以实现靶基因的精准修复。正是通过HDR和NHEJ这两种修复机制实现了对靶序列的定点插入、敲除或修饰等编辑方式(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CRISPR/Cas9系统基因组编辑原理
CRISPR/Cas9系统在基因组特定位点形成DSB,DSB通过非同源末端连接修复(NHEJ)或者同源重组修复机制(HDR),NHEJ能在特定位点产生随机插入、缺失以导致移码突变,HDR可通过提供的模板链实现在基因组特定位点的精确编辑。
Fig. 1A schematic principle of CRISPR/Cas9-medited genome editing
2013年初,美国麻省理工学院张锋研究团队首次利用该系统针对EXM1和PACLB基因在哺乳动物细胞中实现了定点编辑,同时证明crRNA与tracrRNA的成熟与结合不需要体外的RNaseⅢ[4]。同时期,美国哈佛大学Church实验室在Science上发表的研究工作表明,经改造过的crRNA与tracrRNA形成的sgRNA同样能引导Cas9蛋白进行编辑[9]。作者针对AAVS1基因在HEK293T、人慢性粒细胞白血病K562细胞及多能干细胞中的编辑效率进行了比较,编辑效率从2%~25%不等,推测同一基因在不同细胞系中由于其表现状态不同而导致突变效率不同。随后美国加州大学伯克利分校Doudna实验室在eLife上发表论文,她们对gRNA序列的3端进行加长以稳定结构,表明较长的3端能提高编辑效率。正是这几篇论文的相继报道,将CRISPR/Cas9系统在真核生物中的应用推向高潮[10]。
2 CRISPR/Cas9系统导入昆虫的策略
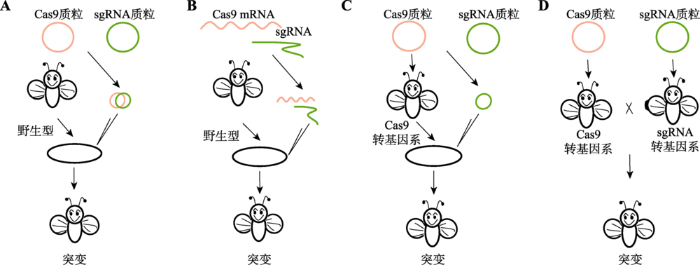
基因组编辑技术日趋完善,研究者们期望借助基因组编辑技术阐明基因在个体发育过程中的作用。在昆虫中,主要有以下4种策略可将CRISPR/ Cas9系统的两个组件导入昆虫体内从而实现目的基因的靶向突变:(1)通过共同注射Cas9蛋白质粒和sgRNA质粒到早期胚胎得到相应突变体(图2A)。Ren等[11]对CRISPR/Cas9系统进行优化,比较不同启动子启动Cas9蛋白及sgRNA序列长短对突变效率的影响,结果表明在nos启动子下表达Cas9蛋白的质粒和在U6b启动子下表达最短序列sgRNA的质粒共同注射后,突变效率最高约为3.2%,但突变遗传至下一代的效率较低。(2)将Cas9与sgRNA的表达盒在体外转录形成相应的mRNA,将其共同注射以达到编辑靶标基因的目的(图2B)。Bassett等[12]将Cas9蛋白和靶向yellow基因的sgRNA进行体外转录形成mRNA,注射进果蝇胚胎有88%的个体产生了至少一个可遗传突变。Yu等[13]利用相同的办法对yellow等7个基因进行定点修饰,其突变率甚至高达100%。且这种办法不受gRNA启动子序列限制,能在基因组上广泛应用,但就实验成本而言,体外转录需要更多的经费支持。(3)制作在生殖细胞中表达Cas9蛋白的转基因系,随后在该转基因系产生的早期胚胎中注射sgRNA,产生相应的突变体(图2C)。(4)通过转基因手段产生Cas9和sgRNA稳定的转基因品系,相互杂交产生突变后代(图2D)。为了提高遗传效率,可利用在生殖细胞中特异性表达的启动子如Vasa、nanos启动Cas9蛋白表达。利用该种办法突变效率较高,且可通过共表达两种sgRNA产生大片段缺失,Gratz等[14]利用Vasa驱动Cas9在果蝇生殖细胞中表达,实现特异性编辑靶标基因ry,其中有15%的个体能够将突变表型传递至下一代。利用不同的生殖细胞系特异性表达的启动子启动Cas9蛋白获得的突变效率不同,但均能获得较多的突变后代[15]。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CRISPR/Cas9系统导入昆虫的普遍策略
A:Cas9(橙色)与sgRNA(绿色)以质粒形式同时显微注射入野生型昆虫卵(黑色椭圆)中;B:Cas9(橙色)与sgRNA(绿色)体外转录形成RNA后同时显微注射入野生型昆虫卵中;C:首先获得Cas9转基因系,将sgRNA质粒注射入Cas9转基因系昆虫卵中;D:分别获得Cas9和sgRNA的转基因系,将两个转基因系杂交获得突变体。
Fig. 2Commonly used delivery strategies of the CRISPR system in insects
通过以上策略获得的突变体一般会存在马赛克现象。Cas9 mRNA翻译成蛋白需要一定的时间,此外Cas9蛋白和gRNA形成的复合体半衰期比较长,使其在受精卵卵裂后依然具有核酸内切酶的活性,导致在不同的卵裂细胞基因组中进行切割而产生不同的靶位点修饰,形成嵌合体。近期发表在Cell Research的研究表明,针对老鼠Tyr基因的第4个外显子设计5个毗邻的靶标位点,将其同时与Cas9转入老鼠胚胎细胞,G0代所有个体均表现为白化现象,而在注射单个靶位点的老鼠中,只存在少量的突变个体,且大部分是以嵌合或野生形态存在的,表明针对某个基因的关键外显子设计多个靶标位点能在G0代实现完全敲除的目的,消除马赛克现象[16]。在哺乳动物细胞中可通过电转的方式将Cas9蛋白和gRNA导入早期受精卵中,这种策略在一定程度上降低了嵌合体的产生,但是完全避免嵌合体的产生还需进一步探究[17]。
3 CRISPR/Cas9系统在昆虫中的应用及拓展
自2013年成功将CRISPR/Cas9系统应用于真核生物细胞以来,该技术已经被广泛应用于各种生物中。CRISPR/Cas9作为一种强大的基因编辑系统,已经成为科学研究的有力工具。同时研究者们利用天才般想象力不断完善CRISPR/Cas9系统并拓展其应用领域。迄今为止,利用CRISPR/Cas9系统已经在果蝇[18,19,20,21]、家蚕[5]、蚊子[26,27,28]和蝴蝶[29,30,31]等昆虫中开展了研究并取得了一定成果(表1)。Table 1
表1
表1 CRISPR/Cas9在昆虫中的应用
Table 1
| 昆虫 | 基因 | 导入策略 | 突变形式 | 参考文献 | |
|---|---|---|---|---|---|
| 黑腹果蝇 (Drosophila melanogaster) | Yellow, white | mRNA | Knock-out | [38] | |
| Yellow, rosy | Plasmid | Knock-out, Knock-in | [14] | ||
| Ast, capa, Ccap, Crz, Eh, Mip, npf, miR-219, miR-315, White | Transgene, plasmid | Knock-out | [15] | ||
| White | Transgene, plasmid | Knock-out | [11] | ||
| Phosphoglycerate kinase | Plasmid | Knock-out | [13] | ||
| Ebony, yellow, white | Cas9-transgene, plasmid | Knock-out, Knock-in | [39] | ||
| Sosy, DSH3PX1 | Cas9-transgene, plasmid | Knock-out, Knock-in | [40] | ||
| Nicotinic acetylcholine receptor | dCas9-VPR-transgene, plasmid | Transcriptional activation | [41] | ||
| Ebony, Singed | Cas9-protein, mRNA | Knock-out | [42] | ||
| Piwi | Cas9D10A-transgene, plasmid | Knock-out, Knock-in | [43] | ||
| Ebony, vermillion, yellow, white | Cas9-transgene, plasmid | Knock-out | [20] | ||
| EGFP, Mrfp | Cas9-transgene, plasmid | Knock-out | [21] | ||
| Ms(3)k81, white, yellow | Cas9-transgene, plasmid | Knock-out, Knock-in | [44] | ||
| Bam, nos, yellow, cid, notch | CRISPR/Cas9 conditional mutagenesis, transgene | Knock-out | [45] | ||
| Ebony, sepia, forked, curled, wnt | Cas9-transgene, plasmid | Knock-out | [19] | ||
| Dα6, G275E | Plasmid | Knock-out, Knock-in | [46] | ||
| AatII, M4790I | Cas9-transgene, plasmid | Knock-in | [47] | ||
| Wg, hnt, cut, scute | dCas9-VPR-transgene | Cas9 transcriptional activators | [48] | ||
| Ebony, lbk | nos-Cas9-transgene, plasmid | Co-CRISPR-knock-out | [49] | ||
| WingGFP-tan | Plasmid | scarless allele replacement | [50] | ||
| 斑翅果蝇 (D. suzukii) | White, Sex Lethal | Plasmid | Knock-out | [51] | |
| 埃及伊蚊 (Aedes aegypti) | miR-309 | mRNA | Knock-out | [52] | |
| Nix | Cas9-transgene, plasmid | Gene Drive | [53] | ||
| Nix | Cas9-protein, mRNA | Knock-out | [37] | ||
| ECFP | mRNA | Knock-out | [54] | ||
| Kmo, lig4, ku70, r2d2, lops | Cas9-protein, mRNA | Knock-out | [57] | ||
| 致乏库蚊 (Culex quinquefasciatus) | CYP9M10 | mRNA | Knock-out | [55] | |
| 冈比亚按蚊 (Anopheles gambiae) | AGAP005958, AGAP011377, AGAP007280) | Plasmid | Gene drive, Knock-out, Knock-in | [26] | |
| 史按蚊 (Anopheles stephensi) | Hydroxylasewhite | Plasmid | Gene drive, Knock-in | [56] | |
| 家蚕 (Bombyx mori) | BmBLOS2 | mRNA | Knock-out | [22] | |
| Kynu-2, yellow-e, ebony, BmBlos2, tyrosine hydroxylase, red egg, flugellos | Cas9-protein, mRNA, plasmid | Knock-out | [24] | ||
| Bmku70 | Plasmid | Knock-out | [25] | ||
| Bm-ok, BmKMO, BmTH, Bmtan | mRNA | Knock-out | [57] | ||
| BmBLOS2, EGFP | Plasmid | Knock-out | [32] | ||
| BmBlos2, BmP25, BmSericin1, BmSericin3 | SpCas9/SaCas9/AsCpf-protein, plasmid, mRNA | Knock-out | [33] | ||
| 昆虫 | 基因 | 导入策略 | 突变形式 | 参考文献 | |
| 柑橘凤蝶 (Papilio xuthus) | Abdominal-B, ebony, frizzled | mRNA | Knock-out | [29] | |
| 小红蛱蝶 (Vanessa cardui) | Spalt, distal-less | Cas9-protein, mRNA | Knock-out | [58] | |
| 鹿眼蛱蝶 (Junonia coenia) | Spalt, distal-less | Cas9-protein, mRNA | Knock-out | [31] | |
| 小菜蛾 (Plutella xylostella) | Abdominal-A | mRNA | Knock-out | [59] | |
| 斜纹夜蛾 (Spodoptera litura) | SlitPBP3 | mRNA | Knock-out | [60] | |
| Slabd-A | mRNA | Knock-out | [61] | ||
| 棉铃虫 (Helicoverpa armigera) | Cadherin | mRNA | Knock-out | [62] | |
| HarmPBP1 | mRNA | Knock-out | [63] | ||
| HaABCA2 | Cas9-protein, mRNA | Knock-out | [64] | ||
| HarmOR16 | mRNA | Knock-out | [65] | ||
| White, Brown, Scarlet, Ok | mRNA | Knock-out | [66] | ||
| 棉叶虫 (Spodoptera littoralis) | SlitOrco | Cas9-protein, mRNA | Knock-out | [67] | |
| 赤拟谷盗 (Tribolium castaneum) | EGFP | Plasmid, mRNA | Knock-out, Knock-in | [68] | |
| W, Ace2, TC010993 | Plasmid | Gene drive | [69] | ||
| Robo2/3 | nos-Cas9-transgene, plasmid | Knock-out, Knock-in | [70] | ||
| 飞蝗 (Locusta migratoria) | Orco | mRNA | Knock-out | [71] | |
| 双斑蟋 (Gryllus bimaculatus) | Dop1 | mRNA | Knock-out | [72] | |
新窗口打开|下载CSV
3.1 靶向敲除
黑腹果蝇(Drosophila melanogaster)作为奠定经典遗传学基础的重要模式生物之一,对其染色体的组成、基因编码和定位的认识,是其他生物无法比拟的。因此,基于清晰的遗传背景和便捷的遗传操纵,Gratz等[14]首次尝试将CRISPR/Cas系统应用于果蝇,分别在yellow基因5′端和3′端设计了2个gRNA位点,分子检测表明该基因发生了4.6 kb的大片段删除。同年,Yu等[3]将位于Y染色体上的基因kl-3成功敲除,且其敲除效率高达100%,由此证明CRISPR/Cas系统不仅对常染色体上的基因靶位点有效,而且对性染色体上的位点同样高效。家蚕(Bombyx mori)作为一种泌丝结茧类昆虫,具有悠久的驯化历史和很高的经济价值。家蚕作为研究鳞翅目昆虫的模式生物,许多****利用CRISPR/ Cas9系统对其进行基因编辑,从而实现探究基因功能和预防蚕病的目的。Wang等[22]首次在家蚕中成功应用CRISPR/Cas9系统,以BmBLOS2为靶基因,设计了针对BmBLOS2基因的两个靶位点,通过显微注射,将编码Cas9和sgRNA的mRNA混合注射进家蚕胚胎,获得了高突变率的可遗传的油蚕突变体,且在基因组上实现了大片段删除。Liu等[4]以BmBLOS2基因为靶标,将带有核定位信号的Cas9表达载体和sgRNA表达载体共转染BmN细胞,经检测发现CRISPR/Cas9系统能有效介导BmN细胞内的基因编辑,并且能实现多个基因的编辑。由于受RNA Pol Ⅲ启动子的限制,sgRNA的靶位点设计通常是以GN19NGG或GGN18NGG的形式进行的,也因此限制了该系统在基因组上找到最佳靶位点的范围。针对这一问题,Zeng等[32]对CRISPR/Cas9系统在家蚕中的应用进行了优化,以U6启动子为例,在家蚕中设计了以A、T、C、G为起始碱基的靶位点序列,分析发现N20NGG能实现在细胞及个体水平特异性敲除,并可稳定遗传。此外,研究人员以Bm-ok、BmKMO、BmTH、Bmtan和Bm-Wnt1为靶位点,将Cas9和SgRNA以mRNA的形式注射进胚胎,实现了靶基因的定点编辑。为了进一步提高CRISPR/ Cas9系统在家蚕中的敲除效率,Ma等[33]将在驱动Cas9表达的启动子前加上HR3 enhancer,敲除效率提高3.5倍,同时为了拓宽编辑位点在基因组上的范围,作者将SpCas9替换成SaCas9和AsCpf1,以BmBLOS2为靶位点,统计表明SpCas9的敲除效率从9.30%~14.13%不等, AsCpf1的敲除效率在8.7%和16.7%之间,SaCas9敲除效率为11.9%。病毒病作为蚕业生产上危害最为严重的疾病,培育抗性品种应对蚕病是目前有效地策略之一。Chen等[34]将BmNPV中early-1 (ie-1)和me53为靶标,用piggybac转座子在家蚕中导入Cas9和gRNA,形成稳定转基因系,分析表明能有效编辑BmNPV基因组,并抑制BmNPV在家蚕体内的复制和扩散。
蚊子作为传播各类疾病的媒介,与人类健康息息相关。Dong等[35]利用CRISPR/Cas9系统以ECFP为靶标,将ECFP转基因系蚊子中的ECFP基因破坏,获得相应突变体,突变率为5.5%;Kistler等通过改进CRISPR/Cas9系统以及调整Cas9浓度,将突变效率提升至24%[36];Hall等[30]应用CRISPR/Cas9系统对Nix进行研究,发现昆虫中第一个M因子[37]。在蝴蝶中,对Abd-B敲除后,高达92.5%个体出现突变表型;对生物钟基因cry2和clk敲除后,得到可遗传突变后代,同时说明clk能影响蝴蝶在迁移过程中的生理节律[31]。
3.2 时空特异性编辑
如果对一些机体发育所必须的基因进行敲除,会导致机体产生严重的生理缺陷而死亡。因此,这限制了对于探究某些基因在特定发育时间段的功能研究。为此,Xue等[45]为了达到在时间和空间特异性敲除特定基因的目的,在果蝇中构建了条件性诱变系统(CRISPR/Cas9-mediated conditional mutagenesis, CMCM)。该系统由在翅膀、卵巢、眼睛及精巢中特异性启动子驱动GAL4的表达的转基因系及含有Cas9和gRNA的表达盒的UAS转基因品系构成,G1代时将两品系杂交后获得相应的特异性敲除个体。在昆虫中,驱动gRNA表达最为广泛的启动子为U6、U3及T7等,这类启动子属于Ⅲ型启动子,其表达特征为全身性、全时间表达。Zhao等[73]在gRNA序列前后加上核酸酶序列,该结构可通过具有组织时期特异性Ⅱ型启动子驱动表达,转录表达的pre-gRNA利用加在两端的核酸酶将其剪切形成成熟的短的gRNA,gRNA引导Cas9酶切割目标序列。此外,在各个gRNA之间用tRNA串联之后还可实现时间空间的多位点编辑[74]。
3.3 靶向敲入
在基因组特定位点整合进外源DNA序列称为靶向敲入(targeted knock-in)。通常情况下,在插入基因两端需要添加与基因组靶标序列的同源臂,其长度在几十到几千个碱基之间,有研究表明其插入效率与同源臂长度之间存在正向线性关系[75]。在果蝇、家蚕以及蚊子等昆虫中,研究者通过ssODNs (single-stranded oligodeoxynucleotides)模板链、带有同源臂的PCR双链模板链或者质粒模板链对CRISPR/ Cas9系统产生的DSB进行外源基因的插入,从而实现外源基因的靶向敲入[39,56,76]。在细胞内,一旦DSB产生,会诱导NHEJ和HR两种修复机制,这两种机制会竞争性地与DSB处结合以完成修复,因此通过导入外源模板链实现对断裂双链的重组修复,其效率是极低的。研究表明,通过抑制与NHEJ修复途径相关蛋白(如Ku70、 Ku80和Ligase IV)的活性以提高HR修复机制,可实现外源基因的靶向插入[77]。Ma等[25]在家蚕Ku70敲除系中靶向插入3xp3-DsRed-sv40的表达盒,与在非Ku70敲除系中靶向插入3xp3-DsRed-sv40的个体中进行荧光强度检测,发现前者荧光强度更强,表明敲除Ku70后,提高了机体精准修复的效率。Liu等[24]在家蚕细胞系中敲除NHEJ途径相关因子,使HR途径修复提高7倍,此外也表明模板链的同源重组臂在100 bp时仍能实现精确的靶向插入。
靶向敲入标签蛋白基因使其与靶基因融合表达,可以实现追踪靶基因的时空表达,达到实时观测靶基因的表达部位、表达时期等目的,从而解析靶基因的作用机制。Mavor等[78]在Rab8基因的N末端插入Tag标签,通过标签对Rab8基因进行定位和功能研究,发现Rab8在早期果蝇胚胎的沟陷形成过程中起到重要的作用;Li等[79]利用同样的办法在Rn基因3′端加上蛋白标签,对其进行亚细胞定位发现其表达于前体嗅觉感受器,对前体受体细胞分化具有重要作用。此外还可在基因组特定位点插入整合酶位点如loxP、Ⅰ-Sce Ⅰ或phiC31以实现外源基因的单拷贝插入。
3.4 转录水平调控
Cas9蛋白有两个内切酶核酸结构域—RuvC1和HNH,分别切割DNA的两条链。对核酸酶进行点突变可获得缺失酶切活性的Cas9D10A和Cas9H840A两种突变体,这两种突变体只能对DNA的其中一条链进行切割。因此,如果将两个核酸内切酶结构域同时突变即可获得内切酶完全丧失的Cas9突变体即dCas9(dead Cas9)[80]。当dCas9结合于基因的编码序列或者调控序列区域即可有效地阻止基因转录,从而实现基因表达的敲低或沉默,这也就是所谓的CRISPR干扰(CRISPR interference, CRISPRi)技术。Ghosh等[81]利用CRISPRi技术干涉roX1和roX2基因的表达时发现,该技术可在在RNA水平显著下调两个基因的表达。将催化失活的dCas9和转录激活因子p65、和Rta和VP64融合表达形成dCas9-VPR复合体,在sgRNA的特异性引导下该复合体可激活特定基因的表达(CRISPR active, CRISPRa)。美国哈佛-麻省理工学院Broad研究所Konermann团队[82]对sgRNA骨架进行修饰,使其能招募RNA结合蛋白,该结合蛋白与转录激活因子融合表达,使sgRNA同样能与转录激活因子结合,作者将该系统命名为SAM(synergistic activation mediator system),同dCas9- VPR相比,SAM系统能大幅度上调基因的表达。有研究表明,当dCAS9上融合蓝光激活的CIB1和CRY2基因并连接上一个转录激活因子P65时,在蓝光(激发GFP)照射条件下,IL1RN基因就能够被过表达,从而使过表达成为一个可控的过程[83]。目前在酵母、人、鼠及果蝇细胞中已经证实了该技术的可行性。CRISPRi和CRISPRa技术已经成为研究基因及其顺式作用元件之间的调控关系的更为快速和有效的方法。3.5 基因驱动技术
基因驱动技术(gene drive)是指一个能将特定性状快速扩散到群体中的方法。基于基因驱动技术,可以将人为改造的基因比如性别决定因子、生长发育相关因子等扩散到野生生物群体中,以控制有害生物群体数量。目前,基于CRISPR/Cas9系统的基因驱动(CRISPR/Cas9 gene drive, CGD)技术在蚊子中的应用最具代表性[84]。研究表明,蚊子中Nix基因为位于Y染色体上的雄性决定因子,在雌性蚊子的胚胎中加入Nix基因可导致胚胎向雄性转换或者死亡,因此Hall等[37]尝试利用CRISPR/Cas9系统将Nix基因插入到雌性蚊子的胚胎基因组中,得到经过改造的雄性蚊子,将其释放到野外与雌性蚊子杂交后会得到更多的雄性蚊子或者不育的雄性蚊子,以减少雌性蚊子的数量,达到控制蚊子群体数量的目的。早在30多年前,科学家们就已经使用其他编辑工具如ZFN、TALEN等对昆虫基因组进行编辑,然而自CRISPR/Cas9这个突破性技术问世以来,因其简洁的编辑方式受到了极大的青睐,由其衍生的生物安全问题也受到全世界的关注。基因驱动技术也存在一系列的弊端,它既然能让蚊子灭绝,那么就不排除使包括人在内的其他物种遭受破坏;另一方面,如果真正实现有害物种的灭绝,那么可能会对生物链造成不可逆的影响。当然,研究者们也采用了一系列的措施来规避基因驱动技术带来的弊端,比如对基因驱动系统进行改造,将靶向靶基因的靶位点改为靶向Cas9,当机体自身Cas9蛋白表达时就能将插在基因组中的Cas9蛋白敲除,使其丧失驱动功能[85];将CRISPR与Cas9基因分开到不同的载体上,以降低驱动效率;在应用上实现严格可逆,比如拆分基因驱动系统(split gene drive systems)[86]。然而这种通过CRISPR/Cas9系统释放携带有驱动系统的昆虫必须要求严格的风险评估及可能产生的生态后果。
4 结语和展望
CRSIPR/Cas9系统以其设计简单、容易操作及价格低廉等优势,得到科研工作者的青睐和广泛的应用,但其在昆虫的研究和应用领域中仍然存在的一系列不容忽视的问题。首先,提高靶向编辑精准性和避免脱靶的问题。在人类基因组中每8~12个碱基即可出现NGG这种简单的三联碱基,而在鳞翅目昆虫家蚕中每6.5个碱基即可出现这种三联碱基,这种高频率为寻找靶位点提供了便利。然而相对于点突变、等位基因识别等编辑工作就需要对靶位点进行更精确的定位,对核苷酸需要更严格的分辨率,因此就需要选择性更广的PAM基序来引导Cas蛋白识别靶位点,比如脑膜炎奈瑟菌(Neisseriameningitides, NmeCas9)的CRISPR-Cas9需要较长的PAM识别基序NNNNGATT,虽然降低了可编辑位点,但是编辑精确度提高,脱靶率降低[87]。
其次,进一步提高基因组编辑的效率。在昆虫中,利用显微注射的策略导入外源片段时,其转化效率也与插入片段大小有关,片段越大其转化效率越低。张锋研究团队作为CRISPR/Cas9系统在真核生物中应用的先行者,一直在对该系统进行着精益求精的改进。2015年,他们通过搜寻整个细菌群体,在Staphylococcus aureus找到了更小的可替代的Cas9酶,根据菌属来源将其分别命名为Acidaminococcus sp. Cpf1 (AsCpf1)和Lachnospiraceae bacterium Cpf1 (LbCpf1)[88]。与传统的Cas9比较,这些核酸酶有如下几个特点:(1) Cpf1酶在识别靶位点过程中仅需要一条crRNA即可,而非像SpCas9酶需要crRNA/tracrRNA二聚体引导Cas9酶识别靶位点;(2) PAM识别基序TTTN位于3′端,而非5′端;(3) Cpf1酶切割双链时产生粘性末端(overhang),而非平齐末端(blunt-ended),可能有效提高精准插入外源DNA,实现HDR整合;(4) Cpf1切口远离靶位点,这表示即使靶位点已经发生基因编辑,仍可进行再次编辑。正是Cpf1酶的这些特性可能为更精确的基因编辑打开了一扇大门。
再者,社会对CRISPR/Cas系统编辑品种的认可问题。利用CRSIPR/Cas系统对基因组中的靶基因进行定点突变,具有与自然突变或者诱导突变相似的效果,可获得在基因组上无转基因痕迹的可遗传的突变品系,理论上可消除人们对传统转基因引入外源基因产生的恐惧。然而,其脱靶效应对物种产生的影响尚缺乏系统评价,因此各国对其推广持不同意见,目前还没有CRSIPR/Cas编辑新品种大面积推广的报道。
尽管CRISPR/Cas系统还存在一些问题,但通过CRISPR/Cas系统的不断改进以及其衍生出的各项技术,为昆虫基因功能、昆虫生物反应器、害虫防控、经济昆虫分子改良等领域的研究提供了新途径和新思路,必将在昆虫学研究领域带来新的应用和巨大发展。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
URLPMID:3152341 [本文引用: 1]

Abstract Recent studies indicate that the DNA recognition domain of transcription activator-like (TAL) effectors can be combined with the nuclease domain of FokI restriction enzyme to produce TAL effector nucleases (TALENs) that, in pairs, bind adjacent DNA target sites and produce double-strand breaks between the target sequences, stimulating non-homologous end-joining and homologous recombination. Here, we exploit the four prevalent TAL repeats and their DNA recognition cipher to develop a 'modular assembly' method for rapid production of designer TALENs (dTALENs) that recognize unique DNA sequence up to 23 bases in any gene. We have used this approach to engineer 10 dTALENs to target specific loci in native yeast chromosomal genes. All dTALENs produced high rates of site-specific gene disruptions and created strains with expected mutant phenotypes. Moreover, dTALENs stimulated high rates (up to 34%) of gene replacement by homologous recombination. Finally, dTALENs caused no detectable cytotoxicity and minimal levels of undesired genetic mutations in the treated yeast strains. These studies expand the realm of verified TALEN activity from cultured human cells to an intact eukaryotic organism and suggest that low-cost, highly dependable dTALENs can assume a significant role for gene modifications of value in human and animal health, agriculture and industry.
.
URL [本文引用: 2]
URL [本文引用: 3]
URLPMID:23360965 [本文引用: 2]

Abstract Here we use the clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated Cas9 endonuclease complexed with dual-RNAs to introduce precise mutations in the genomes of Streptococcus pneumoniae and Escherichia coli. The approach relies on dual-RNA:Cas9-directed cleavage at the targeted genomic site to kill unmutated cells and circumvents the need for selectable markers or counter-selection systems. We reprogram dual-RNA:Cas9 specificity by changing the sequence of short CRISPR RNA (crRNA) to make single- and multinucleotide changes carried on editing templates. Simultaneous use of two crRNAs enables multiplex mutagenesis. In S. pneumoniae, nearly 100% of cells that were recovered using our approach contained the desired mutation, and in E. coli, 65% that were recovered contained the mutation, when the approach was used in combination with recombineering. We exhaustively analyze dual-RNA:Cas9 target requirements to define the range of targetable sequences and show strategies for editing sites that do not meet these requirements, suggesting the versatility of this technique for bacterial genome engineering.
URLPMID:21048762 [本文引用: 1]

Bacteria and Archaea have developed several defence strategies against foreign nucleic acids such as viral genomes and plasmids. Among them, clustered regularly interspaced short palindromic repeats (CRISPR) loci together with cas (CRISPR-associated) genes form the CRISPR/Cas immune system, which involves partially palindromic repeats separated by short stretches of DNA called spacers, acquired from extrachromosomal elements. It was recently demonstrated that these variable loci can incorporate spacers from infecting bacteriophages and then provide immunity against subsequent bacteriophage infections in a sequence-specific manner. Here we show that the Streptococcus thermophilus CRISPR1/Cas system can also naturally acquire spacers from a self-replicating plasmid containing an antibiotic-resistance gene, leading to plasmid loss. Acquired spacers that match antibiotic-resistance genes provide a novel means to naturally select bacteria that cannot uptake and disseminate such genes. We also provide in vivo evidence that the CRISPR1/Cas system specifically cleaves plasmid and bacteriophage double-stranded DNA within the proto-spacer, at specific sites. Our data show that the CRISPR/Cas immune system is remarkably adapted to cleave invading DNA rapidly and has the potential for exploitation to generate safer microbial strains.
[本文引用: 1]
URLPMID:12016139 [本文引用: 1]

The DNA double-strand break (DSB) is the principle cytotoxic lesion for ionizing radiation and radio-mimetic chemicals but can also be caused by mechanical stress on chromosomes or when a replicative DNA polymerase encounters a DNA single-strand break or other type of DNA lesion. DSBs also occur as intermediates in various biological events, such as V(D)J recombination in developing lymphoid cells. Inaccurate repair or lack of repair of a DSB can lead to mutations or to larger-scale genomic instability through the generation of dicentric or acentric chromosomal fragments. Such genome changes may have tumourigenic potential. In other instances, DSBs can be sufficient to induce apoptosis. Because of the threats posed by DSBs, eukaryotic cells have evolved complex and highly conserved systems to rapidly and efficiently detect these lesions, signal their presence and bring about their repair. Here, I provide an overview of these systems, with particular emphasis on the two major pathways of DSB repair: non-homologous end-joining and homologous recombination. Inherited or acquired defects in these pathways may lead to cancer or to other human diseases, and may affect the sensitivity of patients or tumour cells to radiotherapy and certain chemotherapies. An increased knowledge of DSB repair and of other DNA DSB responses may therefore provide opportunities for developing more effective treatments for cancer.
URL [本文引用: 1]

A method of altering a eukaryotic cell is provided including transfecting the eukaryotic cell with a nucleic acid encoding RNA complementary to genomic DNA of the eukaryotic cell, transfecting the eukaryotic cell with a nucleic acid encoding an enzyme that interacts with the RNA and cleaves the genomic DNA in a site specific manner, wherein the cell expresses the RNA and the enzyme, the RNA binds to complementary genomic DNA and the enzyme cleaves the genomic DNA in a site specific manner.
URLPMID:23386978 [本文引用: 1]

Abstract Type II CRISPR immune systems in bacteria use a dual RNA-guided DNA endonuclease, Cas9, to cleave foreign DNA at specific sites. We show here that Cas9 assembles with hybrid guide RNAs in human cells and can induce the formation of double-strand DNA breaks (DSBs) at a site complementary to the guide RNA sequence in genomic DNA. This cleavage activity requires both Cas9 and the complementary binding of the guide RNA. Experiments using extracts from transfected cells show that RNA expression and/or assembly into Cas9 is the limiting factor for Cas9-mediated DNA cleavage. In addition, we find that extension of the RNA sequence at the 3' end enhances DNA targeting activity in vivo. These results show that RNA-programmed genome editing is a facile strategy for introducing site-specific genetic changes in human cells.DOI:http://dx.doi.org/10.7554/eLife.00471.001.
[本文引用: 1]
URLPMID:24576617 [本文引用: 1]

Abstract Genome engineering has revolutionised genetic analysis in many organisms. Here we describe a simple and efficient technique to generate and detect novel mutations in desired target genes in Drosophila melanogaster. We target double strand breaks to specific sites within the genome by injecting mRNA encoding the Cas9 endonuclease and in vitro transcribed synthetic guide RNA into Drosophila embryos. The small insertion and deletion mutations that result from inefficient non-homologous end joining at this site are detected by high resolution melt analysis of whole flies and individual wings, allowing stable lines to be made within 1 month. Copyright 脗漏 2014. Published by Elsevier Inc.
URL [本文引用: 1]
[本文引用: 2]
URLPMID:24002648 [本文引用: 1]

We report a simple yet extremely efficient platform for systematic gene targeting by the RNA-guided endonuclease Cas9 in Drosophila. The system comprises two transgenic strains: one expressing Cas9 protein from the germline-specific nanos promoter and the other ubiquitously expressing a custom guide RNA (gRNA) that targets a unique site in the genome. The two strains are crossed to form an active Cas9-gRNA complex specifically in germ cells, which cleaves and mutates the target site. We demonstrate rapid generation of mutants in seven neuropeptide and two microRNA genes in which no mutants have been described. Founder animals stably expressing Cas9-gRNA transmitted germline mutations to an average of 60% of their progeny, a dramatic improvement in efficiency over the previous methods based on transient Cas9 expression. Simultaneous cleavage of two sites by co-expression of two gRNAs efficiently induced internal deletion with frequencies of 4.3-23%. Our method is readily scalable to high-throughput gene targeting, thereby accelerating comprehensive functional annotation of the Drosophila genome.
URLPMID:28585534 [本文引用: 1]

The CRISPR/Cas9 system is an efficient gene-editing method, but the majority of gene-edited animals showed mosaicism, with editing occurring only in a portion of cells. Here we show that single gene or multiple genes can be completely knocked out in mouse and monkey embryos by zygotic injection of Cas9 mRNA and multiple adjacent single-guide RNAs (spaced 10-200 bp apart) that target only a single key exon of each gene. Phenotypic analysis of F0 mice following targeted deletion of eight genes on the Y chromosome individually demonstrated the robustness of this approach in generating knockout mice. Importantly, this approach delivers complete gene knockout at high efficiencies (100% onArntland 91% onPrrt2) in monkey embryos. Finally, we could generate a completePrrt2knockout monkey in a single step, demonstrating the usefulness of this approach in rapidly establishing gene-edited monkey models.
URLPMID:27474397 [本文引用: 1]

The CRISPR/Cas9 system is a powerful tool for elucidating the roles of genes in a wide variety of organisms including mice. To obtain genetically modified embryos or mice by this method, Cas9 mRNA and sgRNA are usually introduced into zygotes by microinjection or electroporation. However, most mutants generated with this method are genetically mosaic, composed of several types of cells carrying different mutations, which complicates phenotype analysis in founder embryos or mice. To simplify the analysis and to elucidate the roles of genes involved in developmental processes, a method for producing non-mosaic mutants is needed. Here, we established a method for generating non-mosaic mouse mutant embryos. We introduced Cas9 protein and sgRNA into in vitro fertilized (IVF) zygotes by electroporation, which enabled the genome editing to occur before the first replication of the mouse genome. As a result, all of the cells in the mutant carried the same set of mutations. This method solves the problem of mosaicism/allele complexity in founder mutant embryos or mice generated by the CRIPSR/Cas9 system.
[本文引用: 1]
In: Dahmann C, ed. Drosophila.
URL [本文引用: 1]
URLPMID:25437567 [本文引用: 1]

Abstract The CRISPR/Cas9 system has recently emerged as a0002powerful tool for functional genomic studies in Drosophila melanogaster. However, single-guide RNA (sgRNA) parameters affecting the specificity and efficiency of the system in flies are still not clear. Here, we found that off-target effects did not occur in regions of genomic DNA with three or more nucleotide mismatches to sgRNAs. Importantly, we document for a strong positive correlation between mutagenesis efficiency and sgRNA GC content of the six protospacer-adjacent motif-proximal nucleotides (PAMPNs). Furthermore, by injecting well-designed sgRNA plasmids at the optimal concentration we determined, we could efficiently generate mutations in four genes in one step. Finally, we generated null alleles of HP1a using optimized parameters through homology-directed repair and achieved an overall mutagenesis rate significantly higher than previously reported. Our work demonstrates a comprehensive optimization of sgRNA and promises to vastly simplify CRISPR/Cas9 experiments in Drosophila. Copyright 0008 2014 The Authors. Published by Elsevier Inc. All rights reserved.
URLPMID:3974895 [本文引用: 1]

The type II CRISPR/Cas9 system (clustered regularly interspaced short palindromic repeats/CRISPR-associated) has recently emerged as an efficient and simple tool for site-specific engineering of eukaryotic genomes. To improve its applications in Drosophila genome engineering, we simplified the standard two-component CRISPR/Cas9 system by generating a stable transgenic fly line expressing the Cas9 endonuclease in the germline (Vasa-Cas9 line). By injecting vectors expressing engineered target-specific guide RNAs into Vasa-Cas9 fly embryos, mutations were generated from site-specific DNA cleavages and efficiently transmitted into progenies. Because Cas9 endonuclease is the universal component of the type II CRISPR/Cas9 system, site-specific genomic engineering based on this improved platform can be achieved with lower complexity and toxicity, greater consistency, and excellent versatility.
URLPMID:24165890 [本文引用: 1]

Cell death and differentiation is a monthly research journal focused on the exciting field of programmed cell death and apoptosis. It provides a single accessible source of information for both scientists and clinicians, keeping them up-to-date with advances in the field. It encompasses programmed cell death, cell death induced by toxic agents, differentiation and the interrelation of these with cell proliferation.
URLPMID:24175911

Abstract Rapid advances in genome engineering tools, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced palindromic repeats/CRISPR-associated (CRISPR/Cas) system, have enabled efficient gene knockout experiments in a wide variety of organisms. Here, we review the recent progress in targeted gene disruption techniques in the silkworm, Bombyx mori. Although efficiency of targeted mutagenesis was very low in an early experiment using ZFNs, recent studies have shown that TALENs can induce highly efficient mutagenesis of desired target genes in Bombyx. Notably, mutation frequencies induced by TALENs can reach more than 50% of G0 gametes. Thus, TALENs can now be used as a standard tool for gene targeting studies, even when mutant phenotypes are unknown. We also propose guidelines for experimental design and strategy for knockout experiments in Bombyx. Genome editing technologies will greatly increase the usefulness of Bombyx as a model for lepidopteran insects, the major agricultural pests, and lead to sophisticated breeding of Bombyx for use in sericulture and biotechnology. 脗漏 2013 The Authors Development, Growth & Differentiation 脗漏 2013 Japanese Society of Developmental Biologists.
URLPMID:24698835 [本文引用: 1]

Abstract Bombyx mori is an economically important insect and a model organism for studying lepidopteran and arthropod biology. Using a highly efficient CRISPR/Cas9 system, we showed that this system could mediate highly efficient targeted genome editing of a single gene locus, large chromosomal deletion or inversion, and also multiplex genome editing of 6 genes simultaneously in BmNs cell line derived from B.02mori. The simplicity and high efficiency of our system provide unprecedented possibilities for researchers to implement precise and sophisticated manipulation of a chosen B.02mori gene in BmNs cells easily in a limited time course, and perhaps new opportunities for functional genomics of B.02mori and other lepidopteran insects. Copyright 08 2014 Elsevier Ltd. All rights reserved.
URLPMID:24671069 [本文引用: 1]

CRISPR/Cas9, a bacterial adaptive immune system derived genome-editing technique, has become to be one of the most compelling topics in biotechnology. Bombyx mori is an economically important insect and a model organism for studying lepidopteran and arthropod biology. Here we reported highly efficient and multiplex genome editing in B. mori cell line and heritable site-directed mutagenesis of Bmku70, which is required for NHEJ pathway and also related to antigen diversity, telomere length maintenance and subtelomeric gene silencing, using CRISPR/Cas9 system. We established a simple and practicable method and obtained several Bmku70 knockout B. mori lines, and showed that the frequency of HR was increased in embryos of the Bmku70 knockout B. mori. The mutant lines obtained in this study could be a candidate genetic resource for efficient knock-in and fundamental research of DNA repair in B. mori. We also provided a strategy and procedure to perform heritable genome editing of target genes with no significant phenotype effect.
URLPMID:4913862 [本文引用: 1]

Gene-drive systems that enable super-Mendelian inheritance of a transgene have the potential to modify insect populations over a timeframe of a few years [AU please provide a real estimate, this seems vague]. We describe CRISPR-Cas9 endonuclease constructs that function as gene-drive systems inAnopheles gambiae, the main vector for malaria [AU:OK?]. We identified three genes (AGAP005958,AGAP011377andAGAP007280) that confer a recessive female sterility phenotype upon disruption, and inserted into each locus CRISPR-Cas9 gene-drive constructs designed to target and edit each gene [AU:OK?]. For each locus targeted we observed strong gene drive at the molecular level, with transmission rates to progeny of 91 to 99.6%. Population modelling and cage experiments indicate that a CRISPR-Cas9 construct targeting one of these loci,AGAP007280,meets the minimum requirement for a gene drive targeting female reproduction in an insect population. These findings could expedite the development of gene drives to control suppress mosquito populations to levels that do not support malaria transmission.
URLPMID:25536399 [本文引用: 1]

61BAC transgenic reporter indicates noNodalexpression in primitive endoderm.61Visceral endoderm specific mutation ofNodalaffects DVE/AVE migration.61Visceral endoderm Nodal signals regulate epiblastNodalgene expression levels.
URLPMID:24992213 [本文引用: 1]

BackgroundIntrogressing anti-pathogen constructs into wild vector populations could reduce disease transmission. It is generally assumed that such introgression would require linking an anti-pathogen gene with a selfish genetic element or similar technologies. Yet none of the proposed transgenic anti-pathogen gene-drive mechanisms are likely to be implemented as public health measures in the near future. Thus, much attention now focuses instead on transgenic strategies aimed at mosquito population suppression, an approach generally perceived to be practical. By contrast, aiming to replace vector competent mosquito populations with vector incompetent populations by releasing mosquitoes carrying a single anti-pathogen gene without a gene-drive mechanism is widely considered impractical.Methodology/Principal FindingsHere we use Skeeter Buster, a previously published stochastic, spatially explicit model of Aedes aegypti to investigate whether a number of approaches for releasing mosquitoes with only an anti-pathogen construct would be efficient and effective in the tropical city of Iquitos, Peru. To assess the performance of such releases using realistic release numbers, we compare the transient and long-term effects of this strategy with two other genetic control strategies that have been developed in Ae. aegypti: release of a strain with female-specific lethality, and a strain with both female-specific lethality and an anti-pathogen gene. We find that releasing mosquitoes carrying only an anti-pathogen construct can substantially decrease vector competence of a natural population, even at release ratios well below that required for the two currently feasible alternatives that rely on population reduction. Finally, although current genetic control strategies based on population reduction are compromised by immigration of wild-type mosquitoes, releasing mosquitoes carrying only an anti-pathogen gene is considerably more robust to such immigration.Conclusions/SignificanceContrary to the widely held view that transgenic control programs aimed at population replacement require linking an anti-pathogen gene to selfish genetic elements, we find releasing mosquitoes in numbers much smaller than those considered necessary for transgenic population reduction can result in comparatively rapid and robust population replacement. In light of this non-intuitive result, directing efforts to improve rearing capacity and logistical support for implementing releases, and reducing the fitness costs of existing recombinant technologies, may provide a viable, alternative route to introgressing anti-pathogen transgenes under field conditions.
URLPMID:26354079 [本文引用: 1]

Abstract Butterflies are exceptionally diverse but their potential as an experimental system has been limited by the difficulty of deciphering heterozygous genomes and a lack of genetic manipulation technology. Here we use a hybrid assembly approach to construct high-quality reference genomes for Papilio xuthus (contig and scaffold N50: 492090009kb, 3.4090009Mb) and Papilio machaon (contig and scaffold N50: 81090009kb, 1.15090009Mb), highly heterozygous species that differ in host plant affiliations, and adult and larval colour patterns. Integrating comparative genomics and analyses of gene expression yields multiple insights into butterfly evolution, including potential roles of specific genes in recent diversification. To functionally test gene function, we develop an efficient (up to 92.5%) CRISPR/Cas9 gene editing method that yields obvious phenotypes with three genes, Abdominal-B, ebony and frizzled. Our results provide valuable genomic and technological resources for butterflies and unlock their potential as a genetic model system.
URLPMID:4825660 [本文引用: 2]

The eastern North American monarch butterfly,Danaus plexippus, is an emerging model system to study the neural, molecular, and genetic basis of animal long-distance migration and animal clockwork mechanisms. While genomic studies have provided new insight into migration-associated and circadian clock genes, the general lack of simple and versatile reverse-genetic methods has limitedin vivofunctional analysis of candidate genes in this species. Here, we report the establishment of highly efficient and heritable gene mutagenesis methods in the monarch butterfly using transcriptional activator-like effector nucleases (TALENs) and CRISPR-associated RNA-guided nuclease Cas9 (CRISPR/Cas9). Using two clock gene loci,cryptochrome 2andclock(clk), as candidates, we show that both TALENs and CRISPR/Cas9 generate high-frequency nonhomologous end-joining (NHEJ)-mediated mutations at targeted sites (up to 100%), and that injecting fewer than 100 eggs is sufficient to recover mutant progeny and generate monarch knockout lines in about 3 months. Our study also genetically defines monarch CLK as an essential component of the transcriptional activation complex of the circadian clock. The methods presented should not only greatly accelerate functional analyses of many aspects of monarch biology, but are also anticipated to facilitate the development of these tools in other nontraditional insect species as well as the development of homology-directed knock-ins.
URLPMID:805206 [本文引用: 2]

Butterfly eyespot colour patterns are a key example of how a novel trait can appear in association with the co-option of developmental patterning genes. Little is known, however, about how, or even whether, co-opted genes function in eyespot development. Here we use CRISPR/Cas9 genome editing to determine the roles of two co-opted transcription factors that are expressed during early eyespot determination. We found that deletions in a single gene, spalt, are sufficient to reduce or completely delete eyespot colour patterns, thus demonstrating a positive regulatory role for this gene in eyespot determination. Conversely, and contrary to previous predictions, deletions in Distal-less (Dll) result in an increase in the size and number of eyespots, illustrating a repressive role for this gene in eyespot development. Altogether our results show that the presence, absence and shape of butterfly eyespots can be controlled by the activity of two co-opted transcription factors.
[本文引用: 1]
URLPMID:28189747 [本文引用: 1]

Genome editing enabled unprecedented new opportunities for targeted genomic engineering of a wide variety of organisms ranging from microbes, plants, animals and even human embryos. The serial establishing and rapid applications of genome editing tools significantly accelerated Bombyx mori ( B.mori ) research during the past years. However, the only CRISPR system in B.mori was the commonly used SpCas9, which only recognize target sites containing NGG PAM sequence. In the present study, we first improve the efficiency of our previous established SpCas9 system by 3.5 folds. The improved high efficiency was also observed at several loci in both BmNs cells and B.mori embryos. Then to expand the target sites, we showed that two newly discovered CRISPR system, SaCas9 and AsCpf1, could also induce highly efficient site-specific genome editing in BmNs cells, and constructed an integrated CRISPR system. Genome-wide analysis of targetable sites was further conducted and showed that the integrated system cover 69,144,399 sites in B.mori genome, and one site could be found in every 6.5bp. The efficiency and resolution of this CRISPR platform will probably accelerate both fundamental researches and applicable studies in B.mori , and perhaps other insects.
URLPMID:5375672 [本文引用: 1]

We developed a novel antiviral strategy by combining transposon-based transgenesis and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system for the direct cleavage of Bombyx mori nucleopolyhedrovirus (BmNPV) genome DNA to promote virus clearance in silkworms. We demonstrate that transgenic silkworms constitutively expressing Cas9 and guide RNAs targeting the BmNPVimmediate early-1(ie-1) andme53genes effectively induce target-specific cleavage and subsequent mutagenesis, especially large (鈭7-kbp) segment deletions in BmNPV genomes, and thus exhibit robust suppression of BmNPV proliferation. Transgenic animals exhibited higher and inheritable resistance to BmNPV infection than wild-type animals. Our approach will not only contribute to modern sericulture but also shed light on future antiviral therapy. IMPORTANCEPathogen genome targeting has shown its potential in antiviral research. However, transgenic CRISPR/Cas9 system-mediated viral genome targeting has not been reported as an antiviral strategy in a natural animal host of a virus. Our data provide an effective approach against BmNPV infection in a real-world biological system and demonstrate the potential of transgenic CRISPR/Cas9 systems in antiviral research in other species.
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
URLPMID:23827738

Bassett, Liu, and colleagues present a simple and highly efficient method for generating and detecting mutations of any gene in Drosophila melanogaster through the use of the CRISPR/Cas9 system (clustered regularly interspaced palindromic repeats/CRISPR-associated). They show that injection of RNA into the Drosophila embryo can induce highly efficient mutagenesis of desired target genes in up to 88% of injected flies. These mutations can be transmitted through the germline, and this method offers the opportunity to generate a desired mutant fly line within one month.
URLPMID:4232553 [本文引用: 1]

The CRISPR-associated RNA-guided nuclease Cas9 has emerged as a powerful tool for genome engineering in a variety of organisms. To achieve efficient gene targeting rates in Drosophila, current approaches require either injection of in vitro transcribed RNAs or injection into transgenic Cas9-expressing embryos. We report a simple and versatile alternative method for CRISPR-mediated genome editing in Drosophila using bicistronic Cas9/sgRNA expression vectors. Gene targeting with this single-plasmid injection approach is as efficient as in transgenic nanos-Cas9 embryos and allows the isolation of targeted knock-out and knock-in alleles by molecular screening within 2 months. Our strategy is independent of genetic background and does not require prior establishment of transgenic flies.
URL
URLPMID:4025491

Bacterial Cas9 nuclease induces site-specific DNA breaks using small gRNA as guides. Cas9 has been successfully introduced into Drosophila for genome editing. Here, we improve the versatility of this method by developing a transgenic system that expresses Cas9 in the Drosophila germline. Using this system, we induced inheritable knock-out mutations by injecting only the gRNA into embryos, achieved highly efficient mutagenesis by expressing gRNA from the promoter of a novel non-coding RNA gene, and recovered homologous recombination-based knock-in of a fluorescent marker at a rate of 4.5% by co-injecting gRNA with a circular DNA donor.
URLPMID:4232542 [本文引用: 1]

Existing transgenic RNA interference (RNAi) methods greatly facilitate functional genome studies via controlled silencing of targeted mRNA in Drosophila. Although the RNAi approach is extremely powerful, concerns still linger about its low efficiency. Here, we developed a CRISPR/Cas9-mediated conditional mutagenesis system by combining tissue-specific expression of Cas9 driven by the Gal4/upstream activating site system with various ubiquitously expressed guide RNA transgenes to effectively inactivate gene expression in a temporally and spatially controlled manner. Furthermore, by including multiple guide RNAs in a transgenic vector to target a single gene, we achieved a high degree of gene mutagenesis in specific tissues. The CRISPR/Cas9-mediated conditional mutagenesis system provides a simple and effective tool for gene function analysis, and complements the existing RNAi approach.
URLPMID:27117524

61The nAChR alpha6 subunit ofDrosophila melanogasterhas been successfully mutatedin02vivoby CRISPR/Cas9 genome editing.61We demonstrate that the G275E mutation inDα6confers significant resistance to spinosad.61G275E inDα6resembles the phenotype reported for this particular mutation inFrankliniella occidentalis.
URLPMID:28669775

Diamide insecticides are used widely against lepidopteran pests, acting as potent activators of insect Ryanodine Receptors (RyRs) and thus inducing muscle contraction and eventually death. However, resistant phenotypes have recently evolved in the field, associated with the emergence of target site resistance mutations (G4946E/V and I4790M). We investigated the frequency of the mutations found in a resistant population of Tuta absoluta from Greece (G4946V鈥79% and I4790M鈥21%) and the associated diamide resistance profile: there are very high levels of resistance against chlorantraniliprole (9329-fold) and flubendiamide (4969-fold), but moderate levels against cyantraniliprole (191-fold). To further investigate functionally the contribution of each mutation in the resistant phenotype, we used CRISPR/Cas9 to generate genome modified Drosophila carrying alternative allele combinations, and performed toxicity bioassays against all three marketed diamides. Genome modified flies bearing the G4946V mutation exhibited high resistance to flubendiamide (91.3-fold) and chlorantraniliprole (194.7-fold), but low in cyantraniliprole (5.4-fold). Flies naturally bearing the I4790M mutation were moderately resistant to flubendiamide (15.3-fold) but less resistant to chlorantraniliprole (7.5-fold), and cyantraniliprole (2.3-fold). These findings provide in vivo functional genetic confirmation for the role and relative contribution of RyR mutations in diamide resistance and suggest that the three diamides employ different binding modes on the RyR protein.
URLPMID:5217126

Genome editing using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and associated nuclease (Cas9) enables specific genetic modifications, including deletions, insertions, and substitutions in numerous organisms, such as the fruit flyDrosophila melanogaster. One challenge of the CRISPR/Cas9 system can be the laborious and time-consuming screening required to find CRISPR-induced modifications due to a lack of an obvious phenotype and low frequency after editing. Here we apply the successful co-CRISPR technique inDrosophilato simultaneously target a gene of interest and a marker gene,ebony, which is a recessive gene that produces dark body color and has the further advantage of not being a commonly used transgenic marker. We found thatDrosophilabroods containing higher numbers of CRISPR-inducedebonymutations (“jackpot” lines) are significantly enriched for indel events in a separate gene of interest, while broods with few or noebonyoffspring showed few mutations in the gene of interest. Using two different PAM sites in our gene of interest, we report that 6561% (52–70%) of flies from theebony-enriched broods had an indel in DNA near either PAM site. Furthermore, this marker mutation system may be useful in detecting the less frequent homology-directed repair events, all of which occurred in theebony-enriched broods. By focusing on the broods with a significant number of ebony flies, successful identification of CRISPR-induced events is much faster and more efficient. The co-CRISPR technique we present significantly improves the screening efficiency in identification of genome-editing events inDrosophila.
URLPMID:27494619

Abstract Genome editing via the CRISPR/Cas9 RNA-guided nuclease system has opened up exciting possibilities for genetic analysis. However, technical challenges associated with homology-directed repair have proven to be roadblocks for producing changes in the absence of unwanted, secondary mutations commonly known as "scars". To address these issues, we developed a two-stage, marker-assisted strategy to facilitate precise, "scarless" edits in Drosophila with a minimal requirement for molecular screening. Using this method, we modified two base pairs in a gene of interest without altering the final sequence of the CRISPR cut sites. We executed this two-stage allele swap using a novel transformation marker that drives expression in the pupal wings, which can be screened for in the presence of common eye-expressing reporters. The tools we developed can be used to make a single change or a series of allelic substitutions in a region of interest in any D. melanogaster genetic background as well as in other Drosophila species.
URLPMID:26721433

61The CRISPR/Cas9 system generated mutations in thewhiteandSex-lethalgenes ofDrosophila suzukii.61Males hemizygous forwhitemutations developed white eyes.61Some of the females that developed from Cas9/Sxl gRNA-injected embryos showed abnormal genitalia.61TheSex-lethalCRISPR/Cas9 system could be modified to develop a gene drive system for suppression ofD.suzukiipopulations.
URLPMID:27489347

Obligatory blood-triggered reproductive strategy is an evolutionary adaptation of mosquitoes for rapid egg development. It contributes to the vectorial capacity of these insects. Therefore, understanding the molecular mechanisms underlying reproductive processes is of particular importance. Here, we report that microRNA-309 (miR-309) plays a critical role in mosquito reproduction. A spatiotemporal expression profile of miR-309 displayed its blood feeding-dependent onset and ovary-specific manifestation in female Aedes aegypti mosquitoes. Antagomir silencing of miR-309 impaired ovarian development and resulted in nonsynchronized follicle growth. Furthermore, the genetic disruption of miR-309 by CRISPR/Cas9 system led to the developmental failure of primary follicle formation. Examination of genomic responses to miR-309 depletion revealed that several pathways associated with ovarian development are down-regulated. Comparative analysis of genes obtained from the high-throughput RNA sequencing of ovarian tissue from the miR-309 antagomir-silenced mosquitoes with those from the in silico computation target prediction identified that the gene-encoding SIX homeobox 4 protein (SIX4) is a putative target of miR-309. Reporter assay and RNA immunoprecipitation confirmed that SIX4 is a direct target of miR-309. RNA interference of SIX4 was able to rescue phenotypic manifestations caused by miR-309 depletion. Thus, miR-309 plays a critical role in mosquito reproduction by targeting SIX4 in the ovary and serves as a regulatory switch permitting a stage-specific degradation of the ovarian SIX4 mRNA. In turn, this microRNA (miRNA)-targeted degradation is required for appropriate initiation of a blood feeding-triggered phase of ovarian development, highlighting involvement of this miRNA in mosquito reproduction.
URLPMID:25775608

Abstract Conventional control strategies for mosquito-borne pathogens such as malaria and dengue are now being complemented by the development of transgenic mosquito strains reprogrammed to generate beneficial phenotypes such as conditional sterility or pathogen resistance. The widespread success of site-specific nucleases such as transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 in model organisms also suggests that reprogrammable gene drive systems based on these nucleases may be capable of spreading such beneficial phenotypes in wild mosquito populations. Using the mosquito Aedes aegypti, we determined that mutations in the FokI domain used in TALENs to generate obligate heterodimeric complexes substantially and significantly reduce gene editing rates. We found that CRISPR/Cas9-based editing in the mosquito Ae. aegypti is also highly variable, with the majority of guide RNAs unable to generate detectable editing. By first evaluating candidate guide RNAs using a transient embryo assay, we were able to rapidly identify highly effective guide RNAs; focusing germ line-based experiments only on this cohort resulted in consistently high editing rates of 24-90%. Microinjection of double-stranded RNAs targeting ku70 or lig4, both essential components of the end-joining response, increased recombination-based repair in early embryos as determined by plasmid-based reporters. RNAi-based suppression of Ku70 concurrent with embryonic microinjection of site-specific nucleases yielded consistent gene insertion frequencies of 2-3%, similar to traditional transposon- or 脦娄C31-based integration methods but without the requirement for an initial docking step. These studies should greatly accelerate investigations into mosquito biology, streamline development of transgenic strains for field releases, and simplify the evaluation of novel Cas9-based gene drive systems.
URLPMID:25815482

In vivo targeted gene disruption is a powerful tool to study gene function. Thus far, two tools for genome editing in Aedes aegypti have been applied, zinc-finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN). As a promising alternative to ZFN and TALEN, which are difficult to produce and validate using standard molecular biological techniques, the clustered regularly interspaced short palindromic repeats/CRISPR-associated sequence 9 (CRISPR/Cas9) system has recently been discovered as a "do-it-yourself" genome editing tool. Here, we describe the use of CRISPR/Cas9 in the mosquito vector, Aedes aegypti. In a transgenic mosquito line expressing both Dsred and enhanced cyan fluorescent protein (ECFP) from the eye tissue-specific 3xP3 promoter in separated but tightly linked expression cassettes, we targeted the ECFP nucleotide sequence for disruption. When supplying the Cas9 enzyme and two sgRNAs targeting different regions of the ECFP gene as in vitro transcribed mRNAs for germline transformation, we recovered four different G1 pools (5.5% knockout efficiency) where individuals still expressed DsRed but no longer ECFP. PCR amplification, cloning, and sequencing of PCR amplicons revealed indels in the ECFP target gene ranging from 2-27 nucleotides. These results show for the first time that CRISPR/Cas9 mediated gene editing is achievable in Ae. aegypti, paving the way for further functional genomics related studies in this mosquito species.
URLPMID:27095599

Recently-emerging genome editing technologies have enabled targeted gene knockout experiments even in non-model insect species. For studies on insecticide resistance, genome editing technologies offer some advantages over the conventional reverse genetic technique, RNA interference, for testing causal relationships between genes of detoxifying enzymes and resistance phenotypes. There were relatively abundant evidences indicating that the overexpression of a cytochrome P450 geneCYP9M10confers strong pyrethroid resistance in larvae of the southern house mosquitoCulex quinquefasciatus. However, reverse genetic verification has not yet been obtained because of the technical difficulty of microinjection into larvae. Here, we tested two genome editing technologies, transcription activator-like effector nucleases (TALEN)s and clustered regularly interspaced short palindromic repeats (CRISPR/Cas9), to disruptCYP9M10in a resistant strain ofC. quinquefasciatus. Additionally, we developed a novel, effective approach to construct a TALE using the chemical cleavage of phosphorothioate inter-nucleotide linkages in the level 1 assembly. Both TALEN and CRISPR/Cas9 induced frame-shifting mutations in one or all copies ofCYP9M10in a pyrethroid-resistant strain. A line fixed with a completely disruptedCYP9M10haplotype showed more than 100-fold reduction in pyrethroid resistance in the larval stage.
URLPMID:26598698 [本文引用: 1]

Genetic engineering technologies can be used both to create transgenic mosquitoes carrying antipathogen effector genes targeting human malaria parasites and to generate gene-drive systems capable of introgressing the genes throughout wild vector populations. We developed a highly effective autonomous Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated protein 9 (Cas9)-mediated gene-drive system in the Asian malaria vector Anopheles stephensi, adapted from the mutagenic chain reaction (MCR). This specific system results in progeny of males and females derived from transgenic males exhibiting a high frequency of germ-line gene conversion consistent with homology-directed repair (HDR). This system copies an 鈭17-kb construct from its site of insertion to its homologous chromosome in a faithful, site-specific manner. Dual anti-Plasmodium falciparum effector genes, a marker gene, and the autonomous gene-drive components are introgressed into 鈭99.5% of the progeny following outcrosses of transgenic lines to wild-type mosquitoes. The effector genes remain transcriptionally inducible upon blood feeding. In contrast to the efficient conversion in individuals expressing Cas9 only in the germ line, males and females derived from transgenic females, which are expected to have drive component molecules in the egg, produce progeny with a high frequency of mutations in the targeted genome sequence, resulting in near-Mendelian inheritance ratios of the transgene. Such mutant alleles result presumably from nonhomologous end-joining (NHEJ) events before the segregation of somatic and germ-line lineages early in development. These data support the design of this system to be active strictly within the germ line. Strains based on this technology could sustain control and elimination as part of the malaria eradication agenda.
URLPMID:4094479

We report the establishment of an efficient and heritable gene mutagenesis method in the silkworm Bombyx mori using modified type II clustered regularly interspaced short palindromic repeats (CRISPR) with an associated protein (Cas9) system. Using four loci Bm-ok, BmKMO, BmTH, and Bmtan as candidates, we proved that genome alterations at specific sites could be induced by direct microinjection of specific guide RNA and Cas9-mRNA into silkworm embryos. Mutation frequencies of 16.7-35.0% were observed in the injected generation, and DNA fragments deletions were also noted. Bm-ok mosaic mutants were used to test for mutant heritability due to the easily determined translucent epidermal phenotype of Bm-ok-disrupted cells. Two crossing strategies were used. In the first, injected Bm-ok moths were crossed with wild-type moths, and a 28.6% frequency of germline mutation transmission was observed. In the second strategy, two Bm-ok mosaic mutant moths were crossed with each other, and 93.6% of the offsprings appeared mutations in both alleles of Bm-ok gene (compound heterozygous). In summary, the CRISPR/Cas9 system can act as a highly specific and heritable gene-editing tool in Bombyx mori.
URLPMID:27383790

Butterflies rely extensively on colour vision to adapt to the natural world. Most species express a broad range of colour-sensitive Rhodopsin proteins in three types of ommatidia (unit eyes), which are distributed stochastically across the retina. The retinas of Drosophila melanogaster use just two main types, in which fate is controlled by the binary stochastic decision to express the transcription factor Spineless in R7 photoreceptors. We investigated how butterflies instead generate three stochastically distributed ommatidial types, resulting in a more diverse retinal mosaic that provides the basis for additional colour comparisons and an expanded range of colour vision. We show that the Japanese yellow swallowtail (Papilio xuthus, Papilionidae) and the painted lady (Vanessa cardui, Nymphalidae) butterflies have a second R7-like photoreceptor in each ommatidium. Independent stochastic expression of Spineless in each R7-like cell results in expression of a blue-sensitive (Spineless(ON)) or an ultraviolet (UV)-sensitive (Spineless(OFF)) Rhodopsin. In P. xuthus these choices of blue/blue, blue/UV or UV/UV sensitivity in the two R7 cells are coordinated with expression of additional Rhodopsin proteins in the remaining photoreceptors, and together define the three types of ommatidia. Knocking out spineless using CRISPR/Cas9 (refs 5, 6) leads to the loss of the blue-sensitive fate in R7-like cells and transforms retinas into homogeneous fields of UV/UV-type ommatidia, with corresponding changes in other coordinated features of ommatidial type. Hence, the three possible outcomes of Spineless expression define the three ommatidial types in butterflies. This developmental strategy allowed the deployment of an additional red-sensitive Rhodopsin in P. xuthus, allowing for the evolution of expanded colour vision with a greater variety of receptors. This surprisingly simple mechanism that makes use of two binary stochastic decisions coupled with local coordination may prove to be a general means of generating an increased diversity of developmental outcomes.
URLPMID:27318252

Abstract The diamondback moth, Plutella xylostella (L.), is a worldwide agricultural pest that has developed resistance to multiple classes of insecticides. Genetics-based approaches show promise as alternative pest management approaches but require functional studies to identify suitable gene targets. Here we use the CRISPR/Cas9 system to target a gene, abdominal-A, which has an important role in determining the identity and functionality of abdominal segments. We report that P. xylostella abdominal-A (Pxabd-A) has two structurally-similar splice isoforms (A and B) that differ only in the length of exon II, with 15 additional nucleotides in isoform A. Pxabd-A transcripts were detected in all developmental stages, and particularly in pupae and adults. CRISPR/Cas9-based mutagenesis of Pxabd-A exon I produced 91% chimeric mutants following injection of 448 eggs. Phenotypes with abnormal prolegs and malformed segments were visible in hatched larvae and unhatched embryos, and various defects were inherited by the next generation (G1). Genotyping of mutants demonstrated several mutations at the Pxabd-A genomic locus. The results indicate that a series of insertions and deletions were induced in the Pxabd-A locus, not only in G0 survivors but also in G1 individuals, and this provides a foundation for genome editing. Our study demonstrates the utility of the CRISPR/Cas9 system for targeting genes in an agricultural pest and therefore provides a foundation the development of novel pest management tools. Copyright 脗漏 2016 Elsevier Ltd. All rights reserved.
URLPMID:27343383

Cadherins have been identified as receptors of Bacillus thuringiensis (Bt) Cry1A toxins in several lepidopteran insects including the cotton bollworm, Helicoverpa armigera . Disruption of the cadherin gene HaCad has been genetically linked to resistance to Bt toxin Cry1Ac in H . armigera . By using the CRISPR/Cas9 genome editing system (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9), HaCad from the Cry1Ac-susceptible SCD strain of H . armigera was successfully knocked out. A single positive CRISPR event with a frame shift deletion of 4 nucleotides was identified and made homozygous to create a knockout line named SCD-Cad. Western blotting confirmed that HaCad was no longer expressed in the SCD-Cad line while an intact HaCad of 210kDa was present in the parental SCD strain. Insecticide bioassays were used to show that SCD-Cad exhibited 549-fold resistance to Cry1Ac compared with SCD, but no significant change in susceptibility to Cry2Ab. Our results not only provide strong reverse genetics evidence for HaCad as a functional receptor of Cry1Ac, but also demonstrate that the CRISPR/Cas9 technique can act as a powerful and efficient genome editing tool to study gene function in a global agricultural pest, H . armigera .
URLPMID:5214861

Many insect pigments are localized in subcellular pigment granules, and transport of pigment precursors from the cytoplasm is accomplished by ABC proteins.Drosophila melanogasterhas three half-transporter genes (white, scarlet, andbrown, all affecting eye pigments) andBombyx morihas a fourth (ok). The White, Brown, Scarlet and Ok proteins each have one transmembrane and one cytoplasmic domain and they heterodimerize to form functional transporters with different substrate specificities. We used CRISPR/Cas9 to create somatic and germ-line knockout mutations of these four genes in the noctuid mothHelicoverpa armigera. Somatic knockouts ofwhiteblock pigmentation of the egg, first instar larva and adult eye, but germ-line knockouts ofwhiteare recessive lethal in the embryo. Knockouts ofscarletare viable and produce pigmentless first instar larvae and yellow adult eyes lacking xanthommatin. Knockouts ofbrownshow no phenotypic effects on viability or pigmentation. Knockouts ofokare viable and produce translucent larval cuticle and black eyes. CRISPR/Cas9-induced mutations are a useful tool for analyzing how essential and non-essential genes interact to produce the diversity of insect pigmentation patterns found in nature.
URLPMID:4940732

Lepidoptera suffer critical lack of genetic tools and heritable genome edition has been achieved only in a few model species. Here we demonstrate that the CRISPR/Cas9 system is highly efficient for genome editing in a non-model crop pest Lepidoptera, the noctuid mothSpodoptera littoralis. We knocked-out the olfactory receptor co-receptorOrcogene to investigate its function in Lepidoptera olfaction. We find that 89.6% of the injected individuals carriedOrcomutations, 70% of which transmitted them to the next generation. CRISPR/Cas9-mediatedOrcoknockout caused defects in plant odor and sex pheromone olfactory detection in homozygous individuals. Our work genetically defines Orco as an essential OR partner for both host and mate detection in Lepidoptera, and demonstrates that CRISPR/Cas9 is a simple and highly efficient genome editing technique in noctuid pests opening new routes for gene function analysis and the development of novel pest control strategies.
URLPMID:26160901

Gene-editing techniques are revolutionizing the way we conduct genetics in many organisms. The CRISPR/Cas nuclease has emerged as a highly versatile, efficient and affordable tool for targeting chosen sites in the genome. Beyond its applications in established model organisms, CRISPR technology provides a platform for genetic intervention in a wide range of species, limited only by our ability to deliver it to cells and to select mutations efficiently. Here, we test the CRISPR technology in an emerging insect model and pest, the beetle Tribolium castaneum. We use simple assays to test CRISPR/Cas activity, we demonstrate efficient expression of guide RNAs and Cas9 from Tribolium U6 and hsp68 promoters and we test the efficiency of knockout and knock-in approaches in Tribolium. We find that 55-80% of injected individuals carry mutations (indels) generated by non-homologous end joining, including mosaic bi-allelic knockouts; 71-100% carry such mutations in their germ line and transmit them to the next generation. We show that CRISPR-mediated gene knockout of the Tribolium E-cadherin gene causes defects in dorsal closure, which is consistent with RNAi-induced phenotypes. Homology-directed knock-in of marker transgenes was observed in 14% of injected individuals and transmitted to the next generation by 6% of injected individuals. Previous work in Tribolium mapped a large number of transgene insertions associated with developmental phenotypes and enhancer traps. We present an efficient method for re-purposing these insertions, via CRISPR-mediated replacement of these transgenes by new constructs.
URLPMID:5438214

Mating system and genetic variation impede the spread of gene drives, which target natural populations of disease-vectoring insects. Synthetic gene drives based on CRISPR/Cas9 have the potential to control, alter, or suppress populations of crop pests and disease vectors, but it is unclear how they will function in wild populations. Using genetic data from four populations of the flour beetleTribolium castaneum, we show that most populations harbor genetic variants in Cas9 target sites, some of which would render them immune to drive (ITD). We show that even a rare ITD allele can reduce or eliminate the efficacy of a CRISPR/Cas9-based synthetic gene drive. This effect is equivalent to and accentuated by mild inbreeding, which is a characteristic of many disease-vectoring arthropods. We conclude that designing such drives will require characterization of genetic variability and the mating system within and among targeted populations.
URLPMID:5455095

Axon guidance receptors of the Roundabout (Robo) family regulate a number of axon guidance outcomes in bilaterian animals in addition to their canonical role in Slit-dependent midline repulsion. In the fruit flyDrosophila melanogaster, three Robo paralogs (Robo1, Robo2, and Robo3) each have specialized roles in regulating midline crossing and the formation of longitudinal axon pathways in the embryonic ventral nerve cord. The number ofrobogenes differs in other insects, and it is unknown whether the roles and/or signaling mechanisms ofDrosophilaRobos are shared in other insect species. To directly compare the axon guidance activities of Robo receptors inDrosophilaand the flour beetleTribolium castaneum,I have used a CRISPR/Cas9-based approach to replaceDrosophila robo3withTribolium robo2/3. I show that when expressed from therobo3locus inDrosophilaembryos,TriboliumRobo2/3 (TcRobo2/3) protein is properly translated and localized to axons, where it reproduces the normal expression pattern ofDrosophilaRobo3. In embryos expressingTcRobo2/3in place ofrobo3, two distinct subsets of longitudinal axons are guided properly to their normal positions in the intermediate neuropile, indicating that TcRobo2/3 can promote Robo3-dependent axon guidance decisions in developingDrosophilaneurons. These observations suggest that the mechanism by whichDrosophilaRobo3 promotes longitudinal pathway formation is evolutionarily conserved inTribolium, where it is performed by TcRobo2/3. The CRISPR/Cas9-based gene replacement approach described here can be applied to comparative evolutionary developmental studies of otherDrosophilagenes and their orthologs in other species.
URLPMID:27744049

Locusts are important agricultural pests worldwide and regarded as study models for entomology. However, the absence of targeted gene manipulation systems for locusts has restricted their applications for research. Herein, we report the successful use of the CRISPR/Cas9 system to induce a targeted heritable mutagenesis of the migratory locust, Locusta migratoria . The target sequence of gRNA was designed to disrupt the gene encoding the odorant receptor co-receptor ( Orco ) and examine the roles of the odorant receptor pathway in the locust. Microinjection of the mixture of Cas9-mRNA and Orco -gRNA into the locust eggs resulted in efficient target-gene editing at a rate of 71.7% in G0 animals and achieved a germline efficiency of up to 88.1% in G1 animals. By a crossing strategy, we successfully established stable Orco mutant lines. EAGs and SSRs indicated that the fourth-instar nymphs of the Orco mutants showed severely impaired electrophysiological responses to multiple odors. The Orco mutant locusts lost an attraction response to aggregation pheromones under the crowding conditions. The locomotor activity and body coloration of the Orco mutant locusts did not significantly differ from those of the two other genotypes. This study provides an easy and effective approach by using the CRISPR/Cas9 system for generating loss-of-function mutants for functional genetic studies of locusts and for managing insect pests.
URLPMID:4629116

Elucidation of reinforcement mechanisms in associative learning is an important subject in neuroscience. In mammals, dopamine neurons are thought to play critical roles in mediating both appetitive and aversive reinforcement. Our pharmacological studies suggested that octopamine and dopamine neurons mediate reward and punishment, respectively, in crickets, but recent studies in fruit-flies concluded that dopamine neurons mediates both reward and punishment, via the type 1 dopamine receptor Dop1. To resolve the discrepancy between studies in different insect species, we producedDop1knockout crickets using the CRISPR/Cas9 system and found that they are defective in aversive learning with sodium chloride punishment but not appetitive learning with water or sucrose reward. The results suggest that dopamine and octopamine neurons mediate aversive and appetitive reinforcement, respectively, in crickets. We suggest unexpected diversity in neurotransmitters mediating appetitive reinforcement between crickets and fruit-flies, although the neurotransmitter mediating aversive reinforcement is conserved. This study demonstrates usefulness of the CRISPR/Cas9 system for producing knockout animals for the study of learning and memory.
URL [本文引用: 1]
URLPMID:27799472 [本文引用: 1]

Abstract Clustered regularly interspaced short palindromic repeat/Cas9 (CRISPR/Cas9) system has emerged in recent years as a highly efficient RNA-guided gene manipulation platform. Simultaneous editing or transcriptional activation/suppression of different genes becomes feasible with the co-delivery of multiple guide RNAs (gRNAs). Here, we report that multiple gRNAs linked with self-cleaving ribozymes and/or tRNA could be simultaneously expressed from a single U6 promoter to exert genome editing of dystrophin and myosin binding protein C3 in human and mouse cells. Moreover, this strategy allows the expression of multiple gRNAs for synergistic transcription activation of follistatin when used with catalytically inactive dCas9-VP64 or dCas9-p300core fusions. Finally, the gRNAs linked by the self-cleaving ribozymes and tRNA could be expressed from RNA polymerase type II (pol II) promoters such as generic CMV and muscle/heart-specific MHCK7. This is particularly useful for in vivo applications when the packaging capacity of recombinant adeno-associated virus is limited while tissue-specific delivery of gRNAs and Cas9 is desired. Taken together, this study provides a novel strategy to enable tissue-specific expression of more than one gRNAs for multiplex gene editing from a single pol II promoter. 脗漏 The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
URL [本文引用: 1]

传统转基因技术,如显微注射、转座子、慢病毒转染等将目的基因插入基因组内的整合方式是随机的,这些随机整合对后期转基因动物品系组建和育种带来诸多不利,因此有研究人员提出了定点整合转基因技术.目前该技术的定点整合效率非常低,主要取决于两个方面:一是靶位点产生DNA双链断裂(double-strand break,DSB)的效率;二是断裂后的靶位点与携带同源臂及外源基因的供体质粒发生同源重组的效率,其中同源重组修复(homologous recombination repair,HDR)是基因组定点整合最为依赖的修复机制.靶位点产生DSB后,机体的DNA修复既可能发生HDR,也可能发生非同源末端连接(nonhomologous end joining,NHEJ),并且两者之间存在竞争关系,因此激活HDR或抑制NEHJ都可提高定点整合转基因的效率.本文结合影响定点整合的因素,对提高定点整合效率最新探索方面进行了综述.
URL [本文引用: 1]

传统转基因技术,如显微注射、转座子、慢病毒转染等将目的基因插入基因组内的整合方式是随机的,这些随机整合对后期转基因动物品系组建和育种带来诸多不利,因此有研究人员提出了定点整合转基因技术.目前该技术的定点整合效率非常低,主要取决于两个方面:一是靶位点产生DNA双链断裂(double-strand break,DSB)的效率;二是断裂后的靶位点与携带同源臂及外源基因的供体质粒发生同源重组的效率,其中同源重组修复(homologous recombination repair,HDR)是基因组定点整合最为依赖的修复机制.靶位点产生DSB后,机体的DNA修复既可能发生HDR,也可能发生非同源末端连接(nonhomologous end joining,NHEJ),并且两者之间存在竞争关系,因此激活HDR或抑制NEHJ都可提高定点整合转基因的效率.本文结合影响定点整合的因素,对提高定点整合效率最新探索方面进行了综述.
[本文引用: 1]
URLPMID:25798939 [本文引用: 1]

Methods to introduce targeted double-strand breaks (DSBs) into DNA enable precise genome editing by increasing the rate at which externally supplied DNA fragments are incorporated into the genome through homologous recombination. The efficiency of these methods is limited by nonhomologous end joining (NHEJ), an alternative DNA repair pathway that competes with homology-directed repair (HDR). To promote HDR at the expense of NHEJ, we targeted DNA ligase IV, a key enzyme in the NHEJ pathway, using the inhibitor Scr7. Scr7 treatment increased the efficiency of HDR-mediated genome editing, using Cas9 in mammalian cell lines and in mice for all four genes examined, up to 19-fold. This approach should be applicable to other customizable endonucleases, such as zinc finger nucleases and transcription activator-like effector nucleases, and to nonmammalian cells with sufficiently conserved mechanisms of NHEJ and HDR.
URLPMID:26839362 [本文引用: 1]

Abstract One of the most fundamental changes in cell morphology is the ingression of a plasma membrane furrow. The Drosophila embryo undergoes several cycles of rapid furrow ingression during early development that culminate in the formation of an epithelial sheet. Previous studies have demonstrated the requirement for intracellular trafficking pathways in furrow ingression; however, the pathways that link compartmental behaviors with cortical furrow ingression events are unclear. Here, we show that Rab8 has striking dynamic behaviors in vivo. As furrows ingress, cytoplasmic Rab8 puncta are depleted and Rab8 accumulates at the plasma membrane in a location that coincides with known regions of directed membrane addition. We additionally use CRISPR/Cas9 technology to N-terminally tag Rab8, which is then used to address endogenous localization and function. Endogenous Rab8 displays partial coincidence with Rab11 and the Golgi, and this colocalization is enriched during the fast phase of cellularization. When Rab8 function is disrupted, furrow formation in the early embryo is completely abolished. We also demonstrate that Rab8 behaviors require the function of the exocyst complex subunit Sec5 as well as the recycling endosome protein Rab11. Active, GTP-locked Rab8 is primarily associated with dynamic membrane compartments and the plasma membrane, whereas GDP-locked Rab8 forms large cytoplasmic aggregates. These studies suggest a model in which active Rab8 populations direct furrow ingression by guiding the targeted delivery of cytoplasmic membrane stores to the cell surface through interactions with the exocyst tethering complex. 脗漏 2016. Published by The Company of Biologists Ltd.
URLPMID:4683652 [本文引用: 1]

Abstract The zinc-finger protein Rotund (Rn) plays a critical role in controlling the development of the fly olfactory system. However, little is known about its molecular function in vivo. Here, we added protein tags to the rn locus using CRISPR-Cas9 technology in Drosophila to investigate its subcellular localization and the genes that it regulates . We previously used a reporter construct to show that rn is expressed in a subset of olfactory receptor neuron (ORN) precursors and it is required for the diversification of ORN fates. Here, we show that tagged endogenous Rn protein is functional based on the analysis of ORN phenotypes. Using this method, we also mapped the expression pattern of the endogenous isoform-specific tags in vivo with increased precision. Comparison of the Rn expression pattern from this study with previously published results using GAL4 reporters showed that Rn is mainly present in early steps in antennal disc patterning, but not in pupal stages when ORNs are born. Finally, using chromatin immunoprecipitation, we showed a direct binding of Rotund to a previously identified regulatory element upstream of the bric-a-brac gene locus in the developing antennal disc. Copyright 脗漏 2015 Li et al.
URLPMID:3664290 [本文引用: 1]

Targeted gene regulation on a genome-wide scale is聽a powerful strategy for interrogating, perturbing, and engineering cellular systems. Here, we develop a method for controlling gene expression based on Cas9, an RNA-guided DNA endonuclease from a type II CRISPR system. We show that a catalytically dead Cas9 lacking endonuclease activity, when coexpressed with a guide RNA, generates a DNA recognition complex that can specifically interfere with transcriptional elongation, RNA polymerase binding, or transcription factor binding. This system, which we call CRISPR interference (CRISPRi), can efficiently repress expression of targeted genes in Escherichia coli, with no detectable off-target effects. CRISPRi can be used to repress multiple target genes simultaneously, and its effects are reversible. We also show evidence that the system can be adapted for gene repression in mammalian cells. This RNA-guided DNA recognition platform provides a simple approach for selectively perturbing gene expression on a genome-wide scale.
URLPMID:26850642 [本文引用: 1]

Long non-coding RNAs (lncRNAs) have emerged as regulators of gene expression across metazoa. Interestingly, some lncRNAs function independently of their transcripts 鈥 the transcription of the lncRNA locus itself affects target genes. However, current methods of loss-of-function analysis are insufficient to address the role of lncRNA transcription from the transcript which has impeded analysis of their function. Using the minimal CRISPR interference (CRISPRi) system, we show that coexpression of the catalytically inactive Cas9 (dCas9) and guide RNAs targeting the endogenousroXlocus in theDrosophilacells results in a robust and specific knockdown ofroX1androX2RNAs, thus eliminating the need for recruiting chromatin modifying proteins for effective gene silencing. Additionally, we find that the human andDrosophilacodon optimized dCas9 genes are functional and show similar transcription repressive activity. Finally, we demonstrate that the minimal CRISPRi system suppressesroXtranscription efficientlyin vivoresulting in loss-of-function phenotype, thus validating the method for the first time in a multicelluar organism. Our analysis expands the genetic toolkit available for interrogating lncRNA functionin situand is adaptable for targeting multiple genes across model organisms.
URLPMID:4420636 [本文引用: 1]

Systematic interrogation of gene function requires the ability to perturb gene expression in a robust and generalizable manner. Here we describe structure-guided engineering of a CRISPR-Cas9 complex to mediate efficient transcriptional activation at endogenous genomic loci. We used these engineered Cas9 activation complexes to investigate single-guide RNA (sgRNA) targeting rules for effective transcriptional activation, to demonstrate multiplexed activation of ten genes simultaneously, and to upregulate long intergenic non-coding RNA (lincRNA) transcripts. We also synthesized a library consisting of 70,290 guides targeting all human RefSeq coding isoforms to screen for genes that, upon activation, confer resistance to a BRAF inhibitor. The top hits included genes previously shown to be able to confer resistance, and novel candidates were validated using individual sgRNA and complementary DNA overexpression. A gene expression signature based on the top screening hits correlated with markers of BRAF inhibitor resistance in cell lines and patient-derived samples. These results collectively demonstrate the potential of Cas9-based activators as a powerful genetic perturbation technology.
URLPMID:4412021 [本文引用: 1]

Abstract Optogenetic systems enable precise spatial and temporal control of cell behavior. We engineered a light-activated CRISPR-Cas9 effector (LACE) system that induces transcription of endogenous genes in the presence of blue light. This was accomplished by fusing the light-inducible heterodimerizing proteins CRY2 and CIB1 to a transactivation domain and the catalytically inactive dCas9, respectively. The versatile LACE system can be easily directed to new DNA sequences for the dynamic regulation of endogenous genes.
URLPMID:26232409 [本文引用: 1]

ABSTRACT The use of recombinant genetic technologies for population manipulation has mostly...
URLPMID:26849513 [本文引用: 1]

With the advent of clustered, regularly interspaced, short palindromic repeats (CRISPR)鈥揅RISPR-associated protein 9 (Cas9) technology, researchers can construct gene drives that can bias the inheritance of edited alleles to alter entire populations. As demonstrated with the mutagenic chain reaction in Drosophila4, the CRISPR-Cas9 system can propagate genomic modification together with the genome-editing machinery itself. Although gene drives might have the potential to control insect-borne diseases and agricultural pests, substantial concerns have been raised over unanticipated ecological consequences as a result of drive use. Here we report the development of a potential Cas9-based gene drive 'brake' that remains inert in a wild-type genome but is activated by Cas9 to both cleave the genomic cas9 sequence and to convert an incoming cas9 allele into a brake. This means that the propagation of the brake is favored in a cas9-carrying population.
URL [本文引用: 1]
[本文引用: 1]
URLPMID:27096362 [本文引用: 1]

Abstract CRISPR-Cas systems that provide defence against mobile genetic elements in bacteria and archaea have evolved a variety of mechanisms to target and cleave RNA or DNA. The well-studied types I, II and III utilize a set of distinct CRISPR-associated (Cas) proteins for production of mature CRISPR RNAs (crRNAs) and interference with invading nucleic acids. In types I and III, Cas6 or Cas5d cleaves precursor crRNA (pre-crRNA) and the mature crRNAs then guide a complex of Cas proteins (Cascade-Cas3, type I; Csm or Cmr, type III) to target and cleave invading DNA or RNA. In type II systems, RNase III cleaves pre-crRNA base-paired with trans-activating crRNA (tracrRNA) in the presence of Cas9 (refs 13, 14). The mature tracrRNA-crRNA duplex then guides Cas9 to cleave target DNA. Here, we demonstrate a novel mechanism in CRISPR-Cas immunity. We show that type V-A Cpf1 from Francisella novicida is a dual-nuclease that is specific to crRNA biogenesis and target DNA interference. Cpf1 cleaves pre-crRNA upstream of a hairpin structure formed within the CRISPR repeats and thereby generates intermediate crRNAs that are processed further, leading to mature crRNAs. After recognition of a 5'-YTN-3' protospacer adjacent motif on the non-target DNA strand and subsequent probing for an eight-nucleotide seed sequence, Cpf1, guided by the single mature repeat-spacer crRNA, introduces double-stranded breaks in the target DNA to generate a 5' overhang. The RNase and DNase activities of Cpf1 require sequence- and structure-specific binding to the hairpin of crRNA repeats. Cpf1 uses distinct active domains for both nuclease reactions and cleaves nucleic acids in the presence of magnesium or calcium. This study uncovers a new family of enzymes with specific dual endoribonuclease and endonuclease activities, and demonstrates that type V-A constitutes the most minimalistic of the CRISPR-Cas systems so far described.
