 ,1,2
,1,2Mutant generation of the testis genes and phenotype analyses in Drosophila
Junbo Tang1, Haowei Cao1, Rui Xu1, Dandan Zhang1, Juan Huang ,1,2
,1,2第一联系人:
编委: 史庆华
收稿日期:2018-03-2修回日期:2018-04-27网络出版日期:2018-06-20
| 基金资助: |
Received:2018-03-2Revised:2018-04-27Online:2018-06-20
| Fund supported: |
作者简介 About authors
唐浚博,硕士研究生,专业方向:雄性生殖E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1183KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
唐浚博, 曹浩伟, 许蕊, 张丹丹, 黄娟. 果蝇睾丸基因敲除突变体的构建及表型分析. 遗传[J], 2018, 40(6): 478-487 doi:10.16288/j.yczz.18-055
Junbo Tang, Haowei Cao, Rui Xu, Dandan Zhang, Juan Huang.
雄性动物生殖系统的发育需要许多基因的参与并共同发挥作用。这些基因功能的异常会直接影响睾丸的发育及精子的成熟,最终导致育性的变化[1,2,3,4,5,6,7]。因此,对这些基因的功能研究有助于人们了解精子发生机制并为诊断和治疗不育症提供理论基础。
果蝇与人类的精子发生过程有诸多相似之处,可大致分为精原干细胞有丝分裂、精母细胞减数分裂和精子形成3个阶段[8]。同时,果蝇与人的基因在序列、结构和功能上具有很高的保守性,人类睾丸发育、精细胞分化、减数分裂和精子形成等过程相关的很多基因在果蝇中有同源基因[9]。例如,dBoule是保守的DAZ家族在果蝇中的成员,编码RNA结合蛋白,并在睾丸中优势表达;其功能缺失会引起精母细胞减数分裂阻滞,进而造成精子的缺乏;在果蝇突变体中过表达它的人类同源基因hBoule却可以挽救精子减数分裂的缺陷[10]。果蝇中U2A参与mRNA的剪接,其突变可导致有丝分裂后的精原细胞无法分化为精母细胞及成熟精子,从而造成不育;而在突变体中表达其人类同源基因SNRPA1可使果蝇恢复育性[11]。果蝇和人类基因在功能上的相似性使得果蝇成为研究人类基因功能的良好材料。此外,果蝇还具有生殖周期短、生殖系统简单、易于进行遗传学操作等特点,因此,近年来果蝇在生殖生物学的研究中发挥了重要的作用。
尽管已有的研究已经揭示了很多基因在雄性生殖系统中的功能,但仍有大量基因功能未知,有待进一步研究。目前,CRISPR/Cas9(clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9)技术的应用为实现高效基因打靶、研究基因功能奠定了基础[12,13,14,15]。本研究利用这一技术对果蝇中的8个功能未知基因(CG4161、CG11475、CG2921、CG10541、CG7276、CG3800、CG8117和CG16779)分别进行了敲除,获得了纯合的基因敲除果蝇品系,为基因功能研究提供了便捷的动物模型。同时,对突变体果蝇进行了雄性生殖功能的表型分析,为进一步研究这些基因的作用机制提供了参考。
1 材料和方法
1.1 材料
果蝇w1118、FM7a、Bc/CyO和TM3/TM6b均来自于Bloomington果蝇库,Nos-Cas9[w-]/CyO(2nd chr)和Nos-Cas9[w-](3rd chr)为本实验室果蝇库所有。同源臂及sgRNA载体质粒为本实验室构建。1.2 候选基因的选择
根据基因组数据库(NCBI)及果蝇数据库(FLYBASE)信息,参考文献报道,在果蝇中选择了8个候选基因(CG4161、CG11475、CG2921、CG10541、CG7276、CG3800、CG8117和CG16779)。这些基因均满足以下条件:在睾丸中有表达、雄性生殖方面功能未知、在小鼠和人中有高度同源基因。1.3 sgRNA质粒和供体DNA质粒构建
利用美国威斯康星大学果蝇CRISPR网站(http://flycrispr.molbio.wisc.edu)的TargetFinder功能,依据最少脱靶、切点距离靶基因最近和不破坏上下游相邻基因的原则在靶基因的起始和末尾分别寻找最优sgRNA。根据sgRNA序列设计引物,退火为双链DNA片段,并连接在sgRNA载体质粒上获得表达sgRNA的载体。携带有同源臂的供体质粒通过将靶基因前后两个约2 kb的同源臂分别插入同源臂载体质粒的两个多克隆位点(MCS)获得。所有引物合成及测序由南京金斯瑞生物科技有限公司负责。1.4 基因敲除果蝇纯合突变体的获得
分别提取sgRNA质粒和供体DNA质粒后用酚/氯仿抽提纯化混合,使两个sgRNA质粒终浓度分别为250 ng/μL,供体DNA质粒终浓度为500 ng/μL。收集Nos-Cas9[w-]/CyO(2nd chr)和Nos-Cas9[w-](3rd chr)果蝇胚胎分别用于3号染色体或2号染色体靶基因的显微注射,孵化出的雌蝇和雄蝇分别与w1118的雄蝇和处女蝇杂交,后代筛选红色荧光标记并与该染色体对应的平衡品系杂交。分别在基因组和插入的标记上设计引物用于跨同源臂PCR鉴定,扩增约3 kb大小的片段,经1%琼脂糖凝胶电泳检测确认正确后使带有平衡染色体品系自交,获得纯合基因敲除突变体果蝇;再在敲除区域内设计引物,扩增约400 bp大小的片段用于Negative PCR鉴定,经电泳检测正确后的果蝇用于后续实验研究。1.5 候选基因组织表达半定量PCR
取野生型果蝇头、胸、腹(去除生殖腺)、睾丸组织cDNA,及小鼠心、肺、脑、肠和睾丸组织cDNA,在基因外显子区域设计引物,PCR后经2%琼脂糖凝胶电泳。电泳图像利用Photoshop进行条带灰度分析,定量各基因在各组织中相对表达量。1.6 雄性果蝇育性及生育力测试
在果蝇生育力测试中,为保持各基因型雄性果蝇发育时期一致,所有品系果蝇在同样条件下培养,收集相同日期出生的雄蝇和w1118处女蝇25℃单独培养2天后取单只雄蝇与3只处女蝇杂交,杂交2天后取出亲本果蝇置于新瓶中,重复该操作5次。果蝇放入瓶中之日起第15天统计瓶中后代成蝇数量。依据5次统计结果绘制雄蝇生育力随时间变化曲线。在雄蝇育性实验中,不清空亲本,15天后观察是否有包括幼虫、蛹和成蝇在内的后代产生,统计没有后代的瓶子数量,计算不育率。1.7 果蝇睾丸形态学观察
根据需求选取羽化后4天未交配的雄蝇或羽化后交配2天再单独培养2天的雄蝇,解剖分离睾丸组织压片,立即在相差显微镜下观察并拍照。1.8 果蝇睾丸荧光染色及观察
解剖后的睾丸迅速放入4%多聚甲醛溶液(PFA)中固定20 min;0.1% PBST洗3次,每次10 min。之后放入1∶50稀释的鬼笔环肽(Phalloidin, Invitrogen, 美国),常温避光染色50 min;0.1% PBST洗3次,每次10 min。将睾丸转移至载玻片上,滴加5 μL按照1∶1000稀释的Hoechst 33342 (Invitrogen,美国),避光染色5 min。滴加80%甘油并缓慢覆盖盖玻片后指甲油封片,激光共聚焦显微镜观察并拍照。2 结果与分析
2.1 候选基因的同源性分析及表达水平验证
在基因组数据库(NCBI)及果蝇数据库(FLYBASE)中,搜集了8个果蝇候选基因及它们在人和小鼠中的同源基因的氨基酸序列,并对这些序列进行了分析,比较了它们之间的一致性和同源性(表1)。结果显示8个候选基因中除CG16779与其人和小鼠同源基因一致性(21%和21%)和相似性(32%和31%)较差以外,其他基因与同源基因相似性均可达到或接近50%,一致性均在30%以上。Table 1
表1
表1 候选果蝇基因与人和小鼠基因蛋白质同源性分析
Table 1
| 果蝇基因编号 | 人类同源基因 | 一致性(%) | 相似性(%) | 小鼠同源基因 | 一致性(%) | 相似性(%) |
|---|---|---|---|---|---|---|
| CG4161 | ARMT1 | 32 | 50 | Armt1 | 33 | 51 |
| CG11475 | ARMT1 | 31 | 51 | Armt1 | 34 | 52 |
| CG2921 | ARMT1 | 35 | 54 | Armt1 | 35 | 53 |
| CG10541 | TEKT1 | 32 | 54 | Tekt1 | 31 | 53 |
| CG7276 | TCTEX1D2 | 35 | 55 | Tctex1d2 | 33 | 49 |
| CG3800 | CNBP | 38 | 49 | Cnbp | 38 | 49 |
| CG8117 | TCEA1 | 49 | 72 | Tcea1 | 49 | 72 |
| CG16779 | ELMSAN1 | 21 | 32 | Elmsan1 | 21 | 31 |
新窗口打开|下载CSV
果蝇数据库信息显示其中5个基因(CG4161、CG11475、CG2921、CG10541和CG7276)在睾丸中优势表达,1个基因CG3800在全身各组织中表达量都较高,另外2个基因CG8117和CG16779在各组织中表达量均较低(图1A)。为验证这些数据,分别提取了果蝇头、胸、腹和睾丸组织中的RNA进行反转录,利用半定量PCR,以Rpl32为内参,对这些基因在果蝇不同组织的表达水平进行了检测。半定量分析电泳显示,结果与数据库信息一致(图1:B和C)。
图1
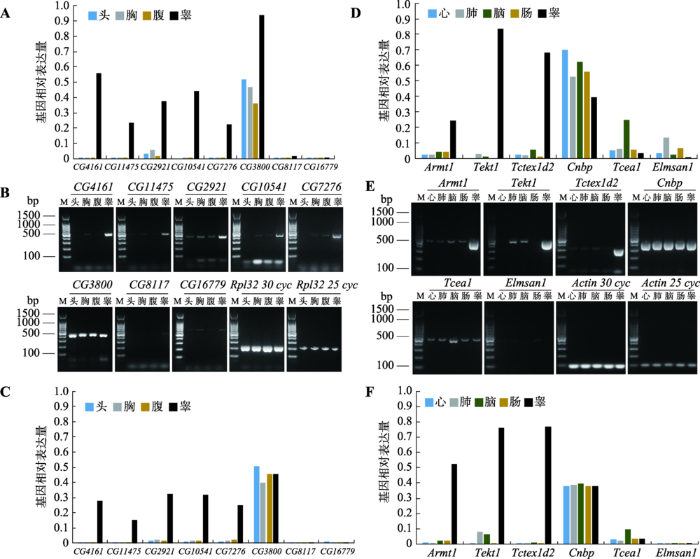 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1候选基因各组织表达水平分析
A:8个候选基因在果蝇头、胸、腹、雄性生殖组织中的相对表达水平。头:头部组织;胸:胸腹侧神经节和心脏组织;腹:肠组织;睾:睾丸组织或生殖腺。按照最高表达量取值(数据来源于果蝇数据库网站http://flybase.org)。B:8个候选基因在果蝇头、胸、腹、睾丸组织中的半定量PCR结果。头:全部头部组织;胸:全部胸部组织;腹:去除生殖器的腹部组织;睾:睾丸组织。M:100 bp marker;C:基于灰度计算的8个候选基因在果蝇头、胸、腹、睾丸组织中的相对表达水平。D:6个同源基因在小鼠心、肺、脑、大肠、睾丸中的相对表达水平(数据来源于NCBI)。E:6个同源基因在小鼠心、肺、脑、大肠、睾丸中的半定量PCR结果。F:基于灰度计算的6个同源基因在小鼠心、肺、脑、大肠、睾丸中的相对表达水平。
Fig. 1Expression analysis of candidate genes in different tissues
同时,利用NCBI查询了小鼠中相应同源基因在各组织中的表达情况。数据库信息显示,在果蝇睾丸中优势表达的基因,它们的同源基因Armt1、Tekt1和Tctex1d2也在小鼠睾丸中优势表达(CG4161、CG11475和CG2921对应同一个同源基因Armt1);在果蝇全身组织中都有高表达的CG3800,其同源基因Cnbp在小鼠各组织中表达量也比CG8117和CG16779的同源基因Tcea1和Elmsan1要高很多(图1D)。同样,利用半定量PCR,以Actin为内参,对这些基因在小鼠心、肺、脑、肠和睾丸组织中的表达水平进行了验证。结果也表明数据库信息真实可靠(图1:E和F)。
2.2 基因敲除突变体的获得及鉴定
为获得基因完全缺失的突变体,选择利用同源重组将目标基因编码区完全敲除,同源臂及sgRNA的选择见示意图2A。针对每个候选基因,设计了两个sgRNA位点,分别位于目的基因上下游,上游同源臂终止于5°sgRNA切割位点,而下游同源臂起始于3°sgRNA切割位点,2个sgRNA之间为敲除区域。同源重组后目标基因被替换为红色荧光标记基因(图2A)。获得候选突变体品系后,通过PCR对其进行分子鉴定。首先,为确认插入片段是否正确替换敲除区域,分别在同源臂外侧基因组和标记基因上设计PCR引物,保证只有同源重组的发生才能获得相应的条带,而非特异插入则无法获得(图2A)。电泳结果显示,8个基因敲除品系果蝇均扩增出两侧同源臂,负对照野生型均无条带(图2B),说明同源重组正确发生。其次,为确认目的区域已被完全敲除以及果蝇品系确为纯合,对目的基因中的外显子区域设计引物进行Negative PCR扩增,如仍可获得扩增产物,则说明基因并未敲除成功或突变体并未纯合(图2A)。结果显示,8个基因敲除品系果蝇均未扩增得到条带,野生型均有条带(图2C),说明目标基因敲除纯合突变体果蝇品系成功建立。图2
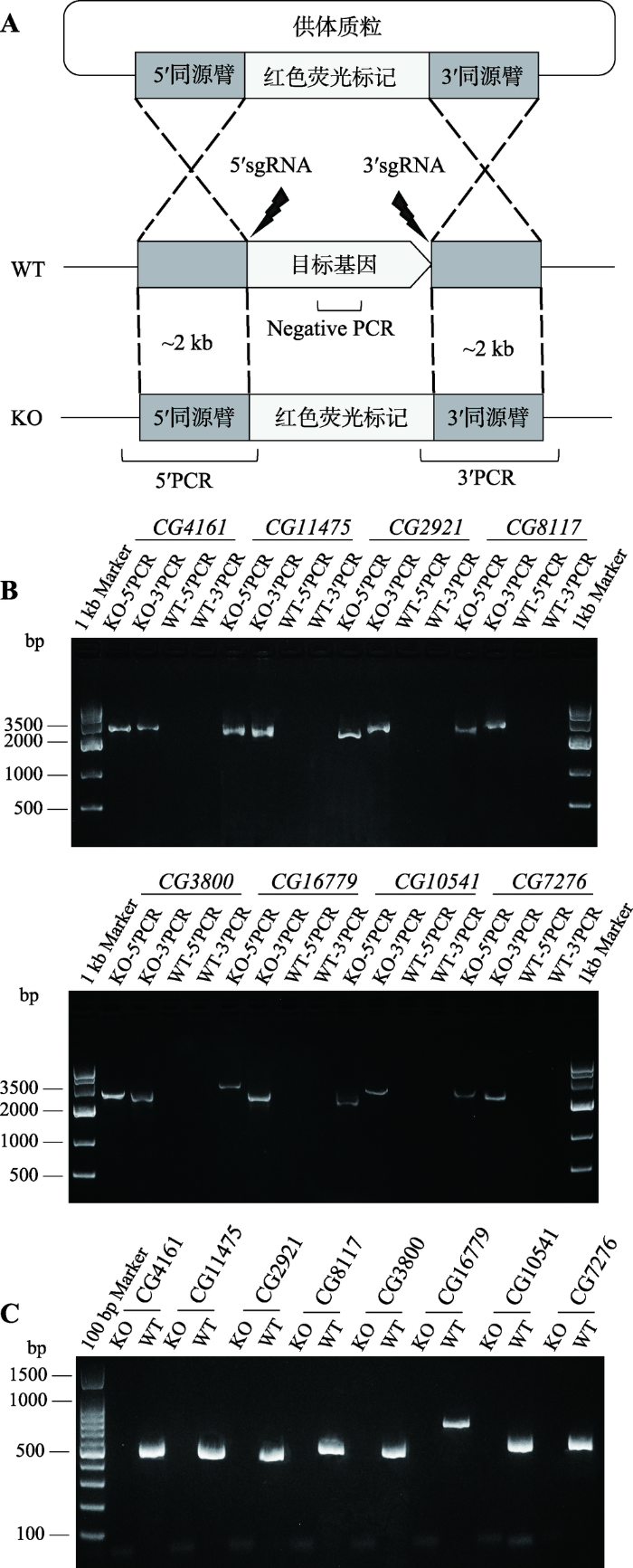 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基因敲除果蝇鉴定
A:CRISPR/Cas9介导同源指导性修复(HDR)敲除目的基因及PCR鉴定示意图。WT:野生型亲本果蝇;KO:基因敲除果蝇。 B:各基因敲除果蝇跨同源臂PCR鉴定结果。C:各基因敲除果蝇纯合品系Negative PCR鉴定结果。
Fig. 2Molecular verification of knockout flies
2.3 基因敲除雄蝇的育性测试
为探究这些基因对雄性生殖系统功能的影响,对8个纯合敲除品系雄蝇进行了生育力测试。测试结果显示,CG7276和CG3800的缺失会造成雄蝇部分不育,其中CG7276-/-雄蝇不育比例为48.9%,CG3800-/-雄蝇不育比例为38.4%,其他基因突变体育性正常(图3A)。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3基因敲除雄蝇生育力测试
A:各基因敲除纯合品系雄蝇育性测试结果。各品系果蝇n≥40。B:生育能力与时间关系实验示意图。每隔2天倒瓶1次,从放入果蝇当天起14天后统计后代数量。C:基因敲除果蝇纯合品系生育能力与时间关系。各组数据=平均值±标准差(n=20)。横坐标:倒瓶时间,与B图对应。D:各基因敲除纯合品系雄蝇后代总数统计。各组数据=平均值±标准差,方差分析t检验,** P<0.01,*** P<0.001,**** P<0.0001。WT:野生型w1118雄蝇。
Fig. 3Male fertility test of knockout flies
为考察基因敲除对雄蝇生育能力的影响,将突变体雄蝇与w1118处女蝇杂交后,每隔2天取出亲本果蝇置于新瓶中,重复该操作5次,分别统计每瓶中14天后的后代数量,不育雄蝇不计入统计数据(图3B)。结果显示,CG7276-/-雄蝇的后代数量较其他果蝇明显减少,而且这一减少随时间推移更加显著;其余基因突变体与野生型相比没有显著差异,且 生育力能维持相当长时间(图3C)。对可育雄蝇后 代总数进行统计则发现,CG3800-/-、CG7276-/-和CG11475-/-后代数量出现不同程度减少,其中CG3800-/-后代数量仅为野生型的58.3%,而CG7276-/-减少更为显著,后代数量仅为野生型的10.0% (图3D)。
以上结果表明,CG7276和CG3800的功能丧失不仅影响了雄蝇的育性,也严重影响了雄蝇的生育能力。
2.4 基因敲除雄蝇睾丸表型分析
为研究基因突变对雄蝇睾丸形态及发育的影响,将出生后雄蝇与野生型处女蝇共置2天以检测是否可育,再单独放置2天以积累精子后解剖睾丸,并在相差显微镜下观察精囊形态、精子发生各阶段细胞形态以及精子数量。结果显示,CG7276-/-和CG3800-/-可育雄蝇同野生型及其他基因敲除突变体雄蝇类似,精子发生过程正常,精原干细胞分化正常,可见成簇长形精细胞,精囊内精子饱满(图4:A~I)。在CG7276-/-不育雄蝇中,部分果蝇精囊与可育雄蝇相比明显缩小,精原干细胞数量相对减少(图4:J和J°);而CG3800-/-不育雄蝇精囊中虽然充满精子,但睾丸中各类型细胞杂乱分布,精原干细胞下移,长形精子上移(图4:K和K°)。这一结果从细胞学方面证实了CG7276和CG3800在雄性生殖系统发育过程中的作用,并提示了突变体不育的可能原因。图4
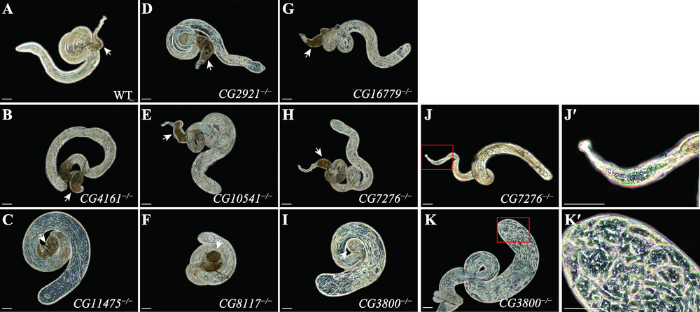 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4基因敲除雄蝇睾丸解剖分析
A~I:野生型(WT)及各基因敲除纯合可育雄蝇睾丸解剖观察。箭头所指为精囊,其中充满精子。J:CG7276-/-不育雄蝇睾丸,红色框区为精囊。J°:红框区域放大;K:CG3800-/-不育雄蝇睾丸,红色框区为睾丸头部;K°:红框区域放大。标尺:100 μm。
Fig. 4Testis anatomical analyses of knockout flies
2.5 不育雄蝇睾丸荧光染色分析
睾丸相差观察显示不育的CG7276-/-和CG3800-/-突变体表现出不同的细胞学表型。为详细观察这一表型,对突变体雄蝇的睾丸分别用Phalloidin和Hoechst33342进行了染色。结果显示,CG7276-/-和CG3800-/-可育雄蝇睾丸内各类型细胞分布正常,每64个精子DNA头部聚集成簇,64个环绕DNA的F-actin椎体组成个性化复合体(individualization complex, IC),从DNA头部沿着轴丝向尾部运动,使精子完成脱胞质及成熟过程(图5:A~A″,C~C″)。CG7276-/-不育雄蝇中,部分个性化复合体移动不同步,精囊显著缩小,提示精子未能完成成熟过程进入精囊(图5:B~B″);而CG3800-/-不育雄蝇则表现出精细胞的散乱分布,无法聚集成簇(图5:D~D″)。以上结果提示CG7276和CG3800在精子头部的聚集及同步化复合体的形成过程中发挥一定作用,它们的缺失影响了精子的成熟,因此造成了部分雄蝇不育,对可育雄蝇的生育能力也产生了一定影响。图5
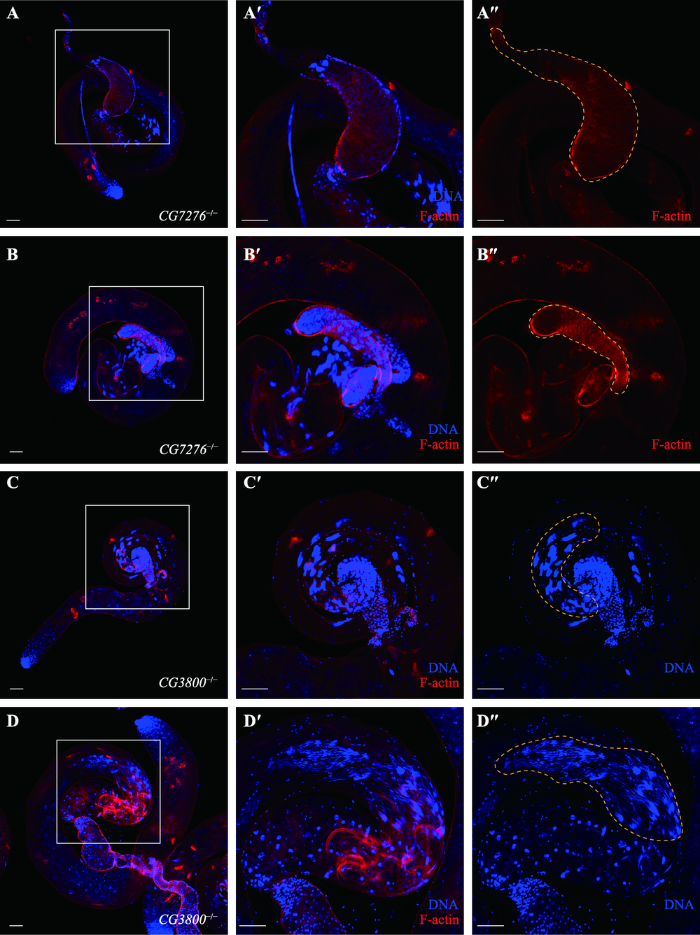 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5CG3800和CG7276基因敲除雄蝇荧光染色分析
A~A″:CG7276-/-可育雄蝇睾丸染色结果。B~B″:CG7276-/-不育雄蝇睾丸染色结果。黄色框内为果蝇精囊。C~C″:CG3800-/-可育雄蝇睾丸染色结果。D~D″:CG3800-/-不育雄蝇睾丸染色结果。黄色框内为睾丸第十区段。A°、B°、C°、D°分别为A、B、C、D白框区域放大。红色为F-actin(Phalloidin),蓝色为细胞核DNA(Hoechst33342)。标尺:50 μm。
Fig. 5Fluorescence staining of CG3800 and CG7276 knockout flies
3 讨 论
在过去的几年中,基于CRISPR/Cas9的基因编辑技术在细胞株和模式生物研究中都取得了长足的发展,使得大规模的基因编辑成为可能。这一系统在果蝇中也得到了广泛应用及不断改进。目前,将带有sgRNA的质粒显微注射进内源表达Cas9的转基因果蝇是最简单、最迅速、最廉价的方法,因为在高通量的情况下,体外RNA的合成步骤复杂且易降解,而质粒操作却简单易行[16, 17]。与其他多细胞模式生物相比,在果蝇中建立稳定的Cas9转基因品系相对比较容易。本研究中使用sgRNA和供体DNA质粒直接注射生殖细胞表达Cas9蛋白的转基因果蝇胚胎,这种果蝇借由nanos或vasa等生殖系统特异性启动子驱动Cas9表达[18],避免了体细胞表达可能导致的致死[19],制造可遗传变异更加可靠。Cas9是一种可以产生DNA双链断裂(DNA double- strand breaks, DSBs)的DNA核酶,在发生DSBs后, 可以通过非同源末端连接(non-homologous end joining, NHEJ)或者同源定向修复HDR产生突变。虽然NHEJ通路不需要在CRISPR系统中添加额外的成分,效率较高,但产生突变随机性较大[20]。而本研究中选用HDR策略,尽管需要额外提供同源重组模板,但其在基因组编辑中可以进行更有针对性更多样化的设计,且可用标记基因进行筛选。一般来说,基因倾向于在它发挥功能的部位表达,因此其表达部位可能与它的功能相关。本研究中选择的基因在雄性生殖系统睾丸中均有表达,因此预期这些基因应在生殖系统发育及功能中扮演一定角色。然而,在选择的8个基因中,仅CG7276和CG3800的缺失显示出较严重的生殖系统缺陷,其他基因功能缺失后雄蝇的生育能力并未受到显著影响,睾丸形态也未有变化。这提示生命体的功能调控是复杂多样的,不可一概而论。未有明显表型出现有多方面原因:可能是由于单一基因的缺失通过其他途径获得了功能上的补偿;可能是这些基因需要协同作用才能发挥功能;可能是这些基因在精子发生过程中只是起支撑性作用;也可能是它们在精子发生以外的过程中对果蝇的生殖行为进行着 调控。
综上所述,本研究构建了8个功能未知基因的敲除果蝇模型并对这些基因在生殖方面的功能进行了初步解析,它们的具体功能及作用机制还有待进一步研究。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:3431660 [本文引用: 1]

Bipolar mitotic spindle organization is fundamental to faithful chromosome segregation. Furry (Fry) is an evolutionarily conserved protein implicated in cell division and morphology. In human cells, Fry localizes to centrosomes and spindle microtubules in early mitosis, and depletion of Fry causes multipolar spindle formation. However, it remains unknown how Fry controls bipolar spindle organization. This study demonstrates that Fry binds to polo-like kinase 1 (Plk1) through the polo-box domain of Plk1 in a manner dependent on the cyclin-dependent kinase 1-mediated Fry phosphorylation at Thr-2516. Fry also binds to Aurora A and promotes Plk1 activity by binding to the polo-box domain of Plk1 and by facilitating Aurora A-mediated Plk1 phosphorylation at Thr-210. Depletion of Fry causes centrosome and centriole splitting in mitotic spindles and reduces the kinase activity of Plk1 in mitotic cells and the accumulation of Thr-210-phosphorylated Plk1 at the spindle poles. Our results suggest that Fry plays a crucial role in the structural integrity of mitotic centrosomes and in the maintenance of spindle bipolarity by promoting Plk1 activity at the spindle poles in early mitosis.
URLPMID:4620998 [本文引用: 1]

Translational control is crucial for proper timing of developmental eventsthat take place in the absence of transcription, as in meiotic activation inoocytes, early embryogenesis in many organisms, and spermatogenesis. Here weshow that a novel form of the translation initiation complex component eIF4Gin Drosophila, eIF4G2, is required specifically for male germ cellsto undergo meiotic division and proper spermatid differentiation. Flies mutantfor eIF4G2 are viable and female fertile but male sterile.Spermatocytes form, but the germ cells in mutant males skip the major eventsof the meiotic divisions and form aberrant spermatids with large nuclei.Consistent with the failure to undergo the meiotic divisions, function ofeIF4G2 is required post-transcriptionally for normal accumulation of the corecell cycle regulatory proteins Twine and CycB in mature spermatocytes. Loss ofeIF4G2 function also causes widespread defects in spermatid differentiation.Although differentiation markers Dj and Fzo are expressed in late-stageeIF4G2 mutant germ cells, several key steps of spermatiddifferentiation fail, including formation of a compact mitochondrialderivative and full elongation. Our results suggest that an alternate form ofthe translation initiation machinery may be required for regulation andexecution of key steps in male germ cell differentiation.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:5543815 [本文引用: 1]

Abstract Alternative splicing has essential roles in development. Remarkably, spermatogenic cells express more alternatively spliced RNAs compared to most whole tissues; however, regulation of these RNAs remains unclear. Here, we characterize the alternative splicing landscape during spermatogenesis and reveal an essential function for the RNA-binding protein Ptbp2 in this highly regulated developmental program. We found that Ptbp2 controls a network of genes involved in cell adhesion, migration, and polarity, suggesting that splicing regulation by Ptbp2 is critical for germ cell communication with Sertoli cells (multifunctional somatic cells necessary for spermatogenesis). Indeed, Ptbp2 ablation in germ cells resulted in disorganization of the filamentous actin (F-actin) cytoskeleton in Sertoli cells, indicating that alternative splicing regulation is necessary for cellular crosstalk during germ cell development. Collectively, the data delineate an alternative splicing regulatory network essential for spermatogenesis, the splicing factor that controls it, and its biological importance in germ-Sertoli communication. Copyright 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
URLPMID:26391954 [本文引用: 1]

RNA binding proteins (RBPs) are increasingly recognized as essential factors in tissue development and homeostasis. The polypyrimidine tract binding (PTB) protein family of RBPs are important posttranscriptional regulators of gene expression. In the nervous system, the function and importance of PTB protein 2 (Ptbp2) as a key alternative splicing regulator is well established. Ptbp2 is also abundantly expressed during spermatogenesis, but its role in this developmental program has not been explored. Additionally, the importance of alternative splicing regulation in spermatogenesis is unclear. Here, we demonstrate that Ptbp2 is essential for spermatogenesis. We also describe an improved dual fluorescence flow cytometry strategy to discriminate, quantify, and collect germ cells in different stages of development. Using this approach, in combination with traditional histological methods, we show that Ptbp2 ablation results in germ cell loss due to increased apoptosis of meiotic spermatocytes and postmeiotic arrest of spermatid differentiation. Furthermore, we show that Ptbp2 is required for alternative splicing regulation in the testis, as in brain. Strikingly, not all of the alternatively spliced RNAs examined were sensitive to Ptbp2 loss in both tissues. Collectively, the data provide evidence for an important role for alternative splicing regulation in germ cell development and a central role for Ptbp2 in this process.
URLPMID:29453757 [本文引用: 1]

Spermatogenesis starts within the seminiferous tubules of the testis by mitotic division of spermatogonia that produces spermatocytes. Meiotic division of these spermatocytes produces haploid spermatids that differentiate into spermatozoa. In this study, we examined the expression of ENaC and CFTR (a Cl 鈭 channel) in rat testicular sections using confocal microscopic immunofluorescence. The structural integrity of the seminiferous tubule sections was verified by precise phalloidin staining of the actin fibers located abundantly at both basal and adluminal tight junctions. The acrosome forming regions in the round spermatids were stained using an FITC coupled lectin (wheat germ agglutinin). In all phases of the germ cells (spermatogonia, spermatocytes, and spermatids) ENaC was localized in cytoplasmic pools. Prior to spermiation, ENaC immunofluorescence appeared along the tails of the spermatids. In spermatozoa isolated from the epididymis, ENaC was localized at the acrosome and a central region of the sperm flagellum. The mature sperm are transcriptionally silent. Hence, we suggest that ENaC subunits in cytoplasmic pools in germ cells serve as the source of ENaC subunits located along the tail of spermatozoa. The locations of ENaC is compatible with a possible role in the acrosomal reaction and sperm mobility. In contrast to ENaC, CFTR immunofluorescence was most strongly observed specifically within the Sertoli cell nuclei. Based on the nuclear localization of CFTR we suggest that, in addition to its role as an ion channel, CFTR may have an independent role in gene regulation within the nuclei.
URLPMID:11097781 [本文引用: 1]

Abstract Subfertility in men is a heterogeneous syndrome, its pathophysiology remaining unknown in the majority of affected men. A large number of genes and loci are associated with sterility in experimental animals, but the human homologues of most of these genes have not been characterized. A British study suggested that, in a large proportion of men with idiopathic infertility, the disorder is inherited as an autosomal recessive trait; this provocative hypothesis needs confirmation. Because normal germ cell development requires the temporally and spatially co-ordinated expression of a number of gene products at the hypothalamic, pituitary and testicular levels, it is safe to predict that a large number of autosomal, as well as X- and Y-linked, genes will probably be implicated in different subsets of male subfertility. Copyright 2000 Harcourt Publishers Ltd.
URLPMID:12499397 [本文引用: 1]

Abstract Defects in human germ cell development are common and yet little is known of genes required for germ cell development in men and women. The pathways that develop germ cells appear to be conserved broadly, at least in outline, in organisms as diverse as flies and humans beginning with allocation of cells to the germ cell lineage, migration of these cells to the fetal gonad, mitotic proliferation and meiosis of the germ cells, and maturation into sperm and eggs. In model organisms, a few thousand genes may be required for germ cell development including meiosis. To date, however, no genes that regulate critical steps of reproduction have been shown to be functionally conserved from flies to humans. This may be due in part to strong selective pressures that are thought to drive reproductive genes to high degrees of divergence. Here, we investigated the micro- and macro-evolution of the BOULE gene, a member of the human DAZ (deleted in azoospermia) gene family, within primates, within mammals and within metazoans. We report that sequence divergence of BOULE is unexpectedly low and that rapid evolution is not detectable. We extend the evolutionary analysis of BOULE to the level of phyla and show that a human BOULE transgene can advance meiosis in infertile boule mutant flies. This is the first demonstration that a human reproductive gene can rescue reproductive defects in a fly. These studies lend strong support to the idea that BOULE may encode a key conserved switch that regulates progression of germ cells through meiosis in men.
URLPMID:27035939 [本文引用: 1]

Processing of pre-mRNA into mRNA is an important regulatory mechanism in eukaryotes that is mediated by the spliceosome, a huge and dynamic ribonucleoprotein complex. Splicing defects are implicated in a spectrum of human disease, but the underlying mechanistic links remain largely unresolved. Using a genome-wide association approach, we have recently identified single nucleotide polymorphisms in humans that associate with nonobstructive azoospermia (NOA), a common cause of male infertility. Here, using genetic manipulation of corresponding candidate loci in Drosophila, we show that the spliceosome component SNRPA1/U2A is essential for male fertility. Loss of U2A in germ cells of the Drosophila testis does not affect germline stem cells, but does result in the accumulation of mitotic spermatogonia that fail to differentiate into spermatocytes and mature sperm. Lack of U2A causes insufficient splicing of mRNAs required for the transition of germ cells from proliferation to differentiation. We show that germ cell-specific disruption of other components of the major spliceosome manifests with the same phenotype, demonstrating that mRNA processing is required for the differentiation of spermatogonia. This requirement is conserved, and expression of human SNRPA1 fully restores spermatogenesis in U2A mutant flies. We further report that several missense mutations in human SNRPA1 that inhibit the assembly of the major spliceosome dominantly disrupt spermatogonial differentiation in Drosophila. Collectively, our findings uncover a conserved and specific requirement for the major spliceosome during the transition from spermatogonial proliferation to differentiation in the male testis, suggesting that spliceosome defects affecting the differentiation of human spermatogonia contribute to NOA.
URLPMID:28073824 [本文引用: 1]

Expression profiling and genomic sequencing methods enable the accumulation of vast quantities of data that relate to the expression of genes during the maturation of male germ cells from primordial germ cells to spermatozoa and potential mutations that underlie male infertility. However, the determination of gene function in specific aspects of spermatogenesis or linking abnormal gene function with infertility remain rate limiting, as even in an era of CRISPR analysis of gene function in mammalian models, this still requires considerable resources and time. Comparative developmental biology studies have shown the remarkable conservation of spermatogenic developmental processes from insects to vertebrates and provide an avenue of rapid assessment of gene function to inform the potential roles of specific genes in rodent and human spermatogenesis. The vinegar fly, , has been used as a model organism for developmental genetic studies for over one hundred years, and research with this organism produced seminal findings such as the association of genes with chromosomes, the chromosomal basis for sexual identity, the mutagenic properties of X-irradiation and the isolation of the first tumour suppressor mutations. researchers have developed an impressive array of sophisticated genetic techniques for analysis of gene function and genetic interactions. This review focuses on how these techniques can be utilised to study spermatogenesis in an organism with a generation time of 9 days and the capacity to introduce multiple mutant alleles into an individual organism in a relatively short time frame.
URLPMID:25953352 [本文引用: 1]
URLPMID:25002478 [本文引用: 1]

Abstract The type II clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system has emerged recently as a powerful method to manipulate the genomes of various organisms. Here, we report a toolbox for high-efficiency genome engineering of Drosophila melanogaster consisting of transgenic Cas9 lines and versatile guide RNA (gRNA) expression plasmids. Systematic evaluation reveals Cas9 lines with ubiquitous or germ-line-restricted patterns of activity. We also demonstrate differential activity of the same gRNA expressed from different U6 snRNA promoters, with the previously untested U6:3 promoter giving the most potent effect. An appropriate combination of Cas9 and gRNA allows targeting of essential and nonessential genes with transmission rates ranging from 25-100%. We also demonstrate that our optimized CRISPR/Cas tools can be used for offset nicking-based mutagenesis. Furthermore, in combination with oligonucleotide or long double-stranded donor templates, our reagents allow precise genome editing by homology-directed repair with rates that make selection markers unnecessary. Last, we demonstrate a novel application of CRISPR/Cas-mediated technology in revealing loss-of-function phenotypes in somatic cells following efficient biallelic targeting by Cas9 expressed in a ubiquitous or tissue-restricted manner. Our CRISPR/Cas tools will facilitate the rapid evaluation of mutant phenotypes of specific genes and the precise modification of the genome with single-nucleotide precision. Our results also pave the way for high-throughput genetic screening with CRISPR/Cas.
URLPMID:24478335 [本文引用: 1]

Abstract We and others recently demonstrated that the readily programmable CRISPR/Cas9 system can be used to edit the Drosophila genome. However, most applications to date have relied on aberrant DNA repair to stochastically generate frameshifting indels and adoption has been limited by a lack of tools for efficient identification of targeted events. Here we report optimized tools and techniques for expanded application of the CRISPR/Cas9 system in Drosophila through homology-directed repair (HDR) with double-stranded DNA (dsDNA) donor templates that facilitate complex genome engineering through the precise incorporation of large DNA sequences, including screenable markers. Using these donors, we demonstrate the replacement of a gene with exogenous sequences and the generation of a conditional allele. To optimize efficiency and specificity, we generated transgenic flies that express Cas9 in the germline and directly compared HDR and off-target cleavage rates of different approaches for delivering CRISPR components. We also investigated HDR efficiency in a mutant background previously demonstrated to bias DNA repair toward HDR. Finally, we developed a web-based tool that identifies CRISPR target sites and evaluates their potential for off-target cleavage using empirically rooted rules. Overall, we have found that injection of a dsDNA donor and guide RNA-encoding plasmids into vasa-Cas9 flies yields the highest efficiency HDR and that target sites can be selected to avoid off-target mutations. Efficient and specific CRISPR/Cas9-mediated HDR opens the door to a broad array of complex genome modifications and greatly expands the utility of CRISPR technology for Drosophila research.
URLPMID:25437567 [本文引用: 1]

Abstract The CRISPR/Cas9 system has recently emerged as a0002powerful tool for functional genomic studies in Drosophila melanogaster. However, single-guide RNA (sgRNA) parameters affecting the specificity and efficiency of the system in flies are still not clear. Here, we found that off-target effects did not occur in regions of genomic DNA with three or more nucleotide mismatches to sgRNAs. Importantly, we document for a strong positive correlation between mutagenesis efficiency and sgRNA GC content of the six protospacer-adjacent motif-proximal nucleotides (PAMPNs). Furthermore, by injecting well-designed sgRNA plasmids at the optimal concentration we determined, we could efficiently generate mutations in four genes in one step. Finally, we generated null alleles of HP1a using optimized parameters through homology-directed repair and achieved an overall mutagenesis rate significantly higher than previously reported. Our work demonstrates a comprehensive optimization of sgRNA and promises to vastly simplify CRISPR/Cas9 experiments in Drosophila. Copyright 0008 2014 The Authors. Published by Elsevier Inc. All rights reserved.
URLPMID:24191015 [本文引用: 1]

The ability to engineer genomes in a specific, systematic, and cost-effective way is critical for functional genomic studies. Recent advances using the CRISPR-associated single-guide RNA system (Cas9/sgRNA) illustrate the potential of this simple system for genome engineering in a number of organisms. Here we report an effective and inexpensive method for genome DNA editing in Drosophila melanogaster whereby plasmid DNAs encoding short sgRNAs under the control of the U6b promoter are injected into transgenic flies in which Cas9 is specifically expressed in the germ line via the nanos promoter. We evaluate the off-targets associated with the method and establish a Web-based resource, along with a searchable, genome-wide database of predicted sgRNAs appropriate for genome engineering in flies. Finally, we discuss the advantages of our method in comparison with other recently published approaches.
URLPMID:24002648 [本文引用: 1]

We report a simple yet extremely efficient platform for systematic gene targeting by the RNA-guided endonuclease Cas9 in Drosophila. The system comprises two transgenic strains: one expressing Cas9 protein from the germline-specific nanos promoter and the other ubiquitously expressing a custom guide RNA (gRNA) that targets a unique site in the genome. The two strains are crossed to form an active Cas9-gRNA complex specifically in germ cells, which cleaves and mutates the target site. We demonstrate rapid generation of mutants in seven neuropeptide and two microRNA genes in which no mutants have been described. Founder animals stably expressing Cas9-gRNA transmitted germline mutations to an average of 60% of their progeny, a dramatic improvement in efficiency over the previous methods based on transient Cas9 expression. Simultaneous cleavage of two sites by co-expression of two gRNAs efficiently induced internal deletion with frequencies of 4.3-23%. Our method is readily scalable to high-throughput gene targeting, thereby accelerating comprehensive functional annotation of the Drosophila genome.
URLPMID:3988796 [本文引用: 1]

Modifying the genomes of many organisms is becoming as easy as manipulating DNA in test tubes, which is made possible by two recently developed techniques based on either the customizable DNA binding protein, TALEN, or the CRISPR/Cas9 system. Here, we describe a series of efficient applications derived from these two technologies, in combination with various homologous donor DNA plasmids, to manipulate the Drosophila genome: (1) to precisely generate genomic deletions; (2) to make genomic replacement of a DNA fragment at single nucleotide resolution; and (3) to generate precise insertions to tag target proteins for tracing their endogenous expressions. For more convenient genomic manipulations, we established an easy-to-screen platform by knocking in a white marker through homologous recombination. Further, we provided a strategy to remove the unwanted duplications generated during the "ends-in" recombination process. Our results also indicate that TALEN and CRISPR/Cas9 had comparable efficiency in mediating genomic modifications through HDR (homology-directed repair); either TALEN or the CRISPR/Cas9 system could efficiently mediate in vivo replacement of DNA fragments of up to 5 kb in Drosophila, providing an ideal genetic tool for functional annotations of the Drosophila genome.
[本文引用: 1]
[本文引用: 1]
