 ,2,*, 李烨
,2,*, 李烨 ,1,*, 姚明华2
,1,*, 姚明华2Genetic Diversity and Population Structure of Eggplant (Solanum melongena) Germplasm Resources Based on SRAP Method
Lifeng Xie1, Ning Li ,2,*, Ye Li
,2,*, Ye Li ,1,*, Minghua Yao2
,1,*, Minghua Yao2通讯作者:
收稿日期:2017-12-6接受日期:2018-08-21网络出版日期:2019-01-30
| 基金资助: |
Corresponding authors:
Received:2017-12-6Accepted:2018-08-21Online:2019-01-30

摘要
关键词:
Abstract
Keywords:
PDF (5770KB)摘要页面多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
引用本文
谢立峰, 李宁, 李烨, 姚明华. 茄子种质遗传多样性及群体结构的SRAP分析. 植物学报, 2019, 54(1): 58-63 doi:10.11983/CBB17237
Xie Lifeng, Li Ning, Li Ye, Yao Minghua.
茄子(Solanum melongena)是茄科(Solanaceae)茄属(Solanum)中重要的蔬菜作物之一。茄子起源于印度及东南亚热带地区, 古印度是其最早的驯化地, 中国是茄子的次生起源中心(李宁等, 2014)。我国是世界上茄子栽培面积最大的国家, 占世界茄子种植面积的50%以上。种质资源是种质创新、遗传改良和品种选育的基础, 资源评价是种质资源有效利用的前提。许多研究表明, 茄子栽培种遗传基础比较狭窄, 在一定程度上限制了茄子基础研究和育种工作的进程(李涛等, 2014)。因此, 开展茄子种质资源的收集及遗传多样性分析, 明确茄子种质的遗传结构, 对茄子遗传育种及其相关工作具有重要的指导意义。
1 材料与方法
1.1 实验材料
183份国内外茄子(Solanum melongena L.)种质由哈尔滨市农业科学院及湖北省农业科学院经济作物研究所提供。其中145份(编号为1-145)茄子种质为国内种质, 来源于东北、华北、华南和华中不同省市; 38份(编号为146-183)茄子种质是通过国际合作项目从日本、泰国及非洲引进。183份茄子种质来源见附录。于2016年3月在哈尔滨市农业科学院试验基地播种, 待茄子幼苗长至4-5片真叶时, 采集幼嫩叶片置于2.0 mL EP管中, 用液氮速冻后置于-20°C冰箱保存。1.2 方法
SRAP分析在东北农业大学番茄课题实验室进行。DNA提取采用CTAB法, 用1%琼脂糖凝胶电泳检测DNA的完整性。SRAP分析参照Li和Quiros (2001)的方法, 并略有改动。SRAP-PCR反应总体积为20 μL, 其中包含1 μL DNA模板(30 ng), E引物和M引物各1 μL, 0.2 μL dNTP, 2 μL Taq buffer, 0.2 μL Taq酶。SRAP-PCR扩增程序: 95°C预变性3分钟; 94°C变性1分钟, 35°C退火1分钟, 72°C延伸1分钟, 5个循环; 94°C变性1分钟, 50°C退火1分钟, 72°C延伸1分钟, 35个循环; 72°C延伸7分钟。SRAP-PCR产物的检测采用12%非变性聚丙烯酰胺凝胶电泳, 上样量为2.0 μL, 电泳上槽缓冲液为1×TBE, 恒功率为30 J?s-1, 电泳1小时。使用10%硝酸银染色。将每条SRAP引物组合扩增的每条带视为1个位点, 条带清晰可辨记为1, 缺失记为0; 统计位点总数和多态性位点数, 不具多态性的条带不统计。应用NTSYS 2.2软件(Rohlf, 1997)计算遗传相似系数。用DARwin 6.0软件(Perrier et al., 2003)进行主成分分析。利用Structure 2.3软件(Pritchard et al., 2007)分析供试材料的群体结构, 将MCMC (Markov Chain Monte Carlo)开始时的不作数迭代(length of burn-in period)设为10 000次, 不作数迭代后的MCMC迭代设为50 000次, 群体数目(K)设定为1-10, 重复运行3次。根据Flint-Garcia等(2003)的方法计算ΔK值, 并通过ΔK值的第1个峰值所对应的K值确定最佳组群数。
2 结果与讨论
2.1 引物多态性分析
利用筛选出的33对多态性高且重复性好的SRAP引物组合, 对供试的183份茄子种质进行扩增, 共扩增出215条清晰稳定的多态性条带, 多态性条带的比例为100%。平均每对引物组合产生7条多态性条带; 其中引物组合Me2Em5扩增出的多态性条带最多, 为13条; 引物组合Me1Em3、Me1Em4、Me1Em7、Me1Em9、Me5Em1和Me5Em8扩增出的多态性条带最少, 为4条。等位基因频率、基因多样性和多态性信息(PIC)指数变化范围分别为0.273 2-0.879 8、0.221 0-0.855 6和0.213 3-0.841 3, 平均值分别为 0.552 8、0.592和0.549 8 (表1)。引物组合Me1Em8对183份茄子种质扩增的结果见图1。Table 1
表1
表133对引物组合的多态性扩增结果
Table 1
| Primer combinations | Allele frequencies | Number of polymorphic bands | Genetic diversity | PIC |
|---|---|---|---|---|
| Me1Em3 | 0.6721 | 4 | 0.4962 | 0.4486 |
| Me1Em4 | 0.5519 | 4 | 0.5896 | 0.5215 |
| Me1Em7 | 0.8798 | 4 | 0.2210 | 0.2133 |
| Me1Em8 | 0.5792 | 5 | 0.5838 | 0.5283 |
| Me1Em9 | 0.7432 | 4 | 0.4176 | 0.3839 |
| Me2Em3 | 0.2732 | 11 | 0.8556 | 0.8413 |
| Me2Em4 | 0.5246 | 7 | 0.6770 | 0.6492 |
| Me2Em5 | 0.3525 | 13 | 0.7619 | 0.7295 |
| Me2Em6 | 0.6393 | 6 | 0.5606 | 0.5349 |
| Me3Em1 | 0.7268 | 6 | 0.4511 | 0.4289 |
| Me3Em2 | 0.3060 | 8 | 0.7718 | 0.7368 |
| Me3Em3 | 0.3060 | 8 | 0.7823 | 0.7509 |
| Me5Em1 | 0.4973 | 4 | 0.6209 | 0.5525 |
| Me5Em2 | 0.3333 | 12 | 0.7877 | 0.7598 |
| Me5Em7 | 0.3607 | 7 | 0.7106 | 0.6571 |
| Me5Em8 | 0.4208 | 4 | 0.6791 | 0.6205 |
| Me5Em9 | 0.4754 | 6 | 0.6854 | 0.6412 |
| Me5Em11 | 0.6120 | 5 | 0.5774 | 0.5400 |
| Me6Em7 | 0.4262 | 8 | 0.7518 | 0.7247 |
| Me6Em8 | 0.4590 | 6 | 0.6636 | 0.6035 |
| Me6Em9 | 0.5738 | 5 | 0.6127 | 0.5723 |
| Me6Em11 | 0.7268 | 6 | 0.4363 | 0.3987 |
| Me7Em3 | 0.6557 | 5 | 0.4901 | 0.4211 |
| Me7Em5 | 0.7486 | 7 | 0.4237 | 0.4058 |
| Me7Em8 | 0.6557 | 5 | 0.5184 | 0.4733 |
| Me7Em9 | 0.3224 | 7 | 0.7684 | 0.7338 |
| Me7Em11 | 0.5464 | 5 | 0.5994 | 0.5368 |
| Me7Em12 | 0.7486 | 7 | 0.4210 | 0.4001 |
| Me9Em1 | 0.6011 | 5 | 0.5344 | 0.4582 |
| Me9Em3 | 0.6284 | 6 | 0.5377 | 0.4839 |
| Me9Em13 | 0.6667 | 8 | 0.5197 | 0.4874 |
| Me8Em6 | 0.6885 | 12 | 0.5055 | 0.4860 |
| Me8Em7 | 0.5410 | 5 | 0.5252 | 0.4185 |
| Mean | 0.5528 | 7 | 0.5920 | 0.5498 |
新窗口打开|下载CSV
图1
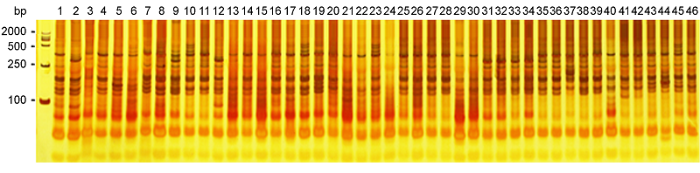 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1引物组合Me1Em8在部分茄子种质(编号1-46同附录)中的SRAP扩增结果
Figure 1SRAP amplification of primer Me1Em8 in some eggplant varieties (The number of 1-46 see Appendix)
2.2 遗传相似系数
利用NTSYS 2.2软件对茄子种质资源的遗传相似系数进行计算, 结果表明, 183份茄子种质的遗传相似系数介于0.276-0.813之间, 平均值为0.623。相似系数越大, 遗传距离越小。由此可见, 茄子种质资源间存在一定的遗传背景差异。2.3 基于SRAP标记的聚类分析
利用DARwin 6.0软件通过邻接法对183份茄子材料进行聚类和主成分分析(图2)。从聚类结果可以看出, 茄子材料间的亲缘关系比较复杂, 在遗传距离为0.345 6处可以划分为4组。第I组包含38份材料, 主要为长条形茄子材料, 占20.8%; 第II组包含51份材料, 主要为长棒形茄子材料, 占27.9%; 第III组包含27份材料, 主要为卵圆形茄子材料, 占14.8%; 第IV组包含67份材料, 均为卵圆形和圆球形茄子材料。不同组的分类总体上与茄子种质的果形性状相关性较高, 与茄子种质的地域来源相关性不大。图2
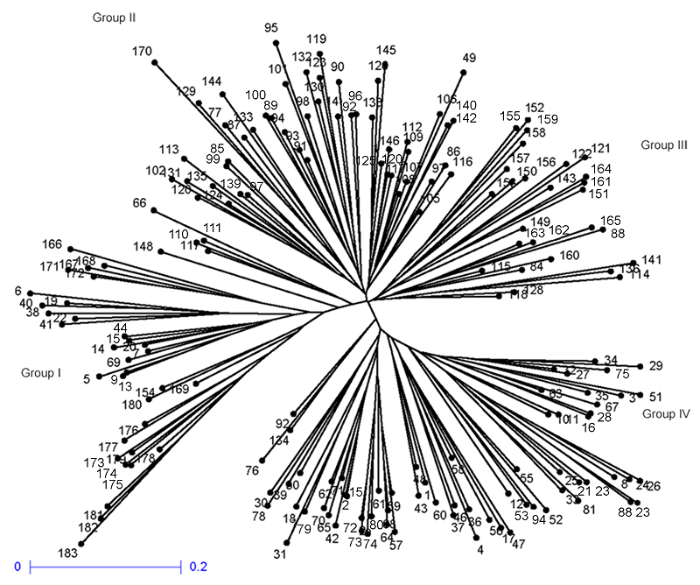 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2183份茄子种质(详细信息见附录)的聚类分析
Figure 2Cluster analysis for 183 eggplant varieties (See Appendix for the detailed information)
2.4 群体遗传结构分析
为揭示茄子种质的遗传背景, 我们利用SRAP引物组合对183份茄子种质进行了群体遗传结构分析。当样本的等位变异频率特征类型数K=4时, 其模型的后验概率最大, 根据K值将183份茄子种质划分为PopI、PopII、PopIII和PopIV 4个亚群(图3)。从群体结构图中可以看出, 收集的茄子种质可分为4个不同的遗传群体, 且存在中间类型材料, 不同群体间的界限十分明显。根据每个亚类群中主要颜色所占的比例(Q值), 将Q?0.6视为血缘单一, Q?0.6视为具有混合来源(刘丽华等, 2009)。从图3可以看出, 亚群间的基因渗透较高。PopI亚群中各份材料的Q值均大于0.6, 说明PopI亚群的种质遗传背景较单一, 与其它亚群间的基因交流不多; PopI亚群主要为卵圆形、紫色或紫黑色的茄子种质, 来源主要为河北和北京, 这也印证了Q值的分析结果, 说明该亚群的种质来源比较单一。PopII、PopIII和PopIV亚群部分种质的Q值小于0.6, 种质资源的基因渗透较高, 表明其具有混合来源, 遗传背景较复杂; 对亚群中茄子的果形、果色、果实萼片颜色及来源地进行综合分析, 同样也印证了Q值的分析结果, 即茄子的果形从长条形、长棒形和圆球形等均有分布, 果色从白色、紫红色、紫色到紫黑色均有分布, 来源包括我国东北、华中和西南以及日本和泰国。图3
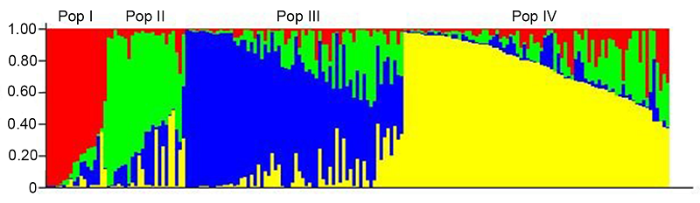 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3183份茄子种质资源的群体结构
Figure 3Population structure of 183 eggplant varieties
2.5 讨论
种质资源是开展育种工作的基础, 种质资源利用的前提是要对其进行多样性分析和遗传背景研究(An et al., 2008)。本研究采用33对SRAP引物组合对供试的183份茄子种质进行扩增, 平均每对引物组合产生7条多态性条带, 等位基因频率、基因多样性和多态性指数的平均值分别为0.552 8、0.592和0.549 8, 遗传相似系数平均值为0.623。与毛伟海等(2006)、冉进等(2007)、何倚剑(2013)及冯英娜等(2014)的研究结果相比, 本研究茄子种质资源间遗传背景相对丰富。Li等(2010)用55对SRAP引物组合对56份茄子种质进行了遗传多样性分析, 将供试的56份茄子材料分为3类, 即S. melongena、S. aethiopicum和S. surattense, 较好地区分开茄子栽培种(S. melongena)和野生种(S. aethiopicum和S. surattense), 同时进一步证明SRAP标记方法在茄子多样性分析中的可行性, 分析结果与供试材料的表型密切相关。本实验所用183份材料均为一年生茄子栽培种, 其中145份为国内资源, 其余为湖北省农科院通过国际合作引进的国外茄子种质资源, 地域分布更丰富。本研究表明, 在茄子的育种工作中, 要加大种质资源的交流力度, 提高种质资源多样性, 增加野生茄子资源的利用, 扩大我国茄子育种的种质资源基因库, 为培育优良新品种提供可选择亲本。聚类分析与群体结构分析是进行种质资源遗传多样性分析的有效手段。本研究采用邻接法将茄子种质划分为4类, 采用基于混合模型的群体结构分析方法同样将茄子种质划分为4个亚群, 划分结果基本一致。但与聚类分析方法相比, 群体结构分析能够反映出育种材料间的基因渗入和交流情况, 能够更好地揭示材料内在的遗传结构, 有助于育种工作者更准确掌握种质间的遗传关系。参照刘丽华等(2009)的描述, 本研究将Q?0.6视为具有混合来源以区分种质间存在基因交流。群体结构分析较直观明确地展示了种质间的遗传结构和基因渗入情况, 不同地域来源的茄子种质间存在基因渗入情况, 但基因渗入和群体结构的划分结果与地域相关性不大, 不同组的分类总体上与茄子种质的果形性状相关性较高。王秋锦等(2007)利用RAPD标记将34份茄子分为两大类型, 即圆茄类型和长茄类型, 与经典的形态学分类基本相符。孙源文等(2012)利用SSR标记对34份茄子材料进行聚类分析, 结果表明材料主要依果形归类。本研究利用SRAP标记方法进行聚类分析和群体结构分析, 获得类似的结果, 从分子水平上支持了以果形作为茄子品种分类指标的观点。
本研究采用SRAP标记方法对183份茄子种质资源的遗传关系和群体结构进行分析, 明确了茄子种质资源间遗传背景和群体间的基因渗透情况, 对今后育种材料的选用具有指导意义。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3969/j.issn.1000-4440.2014.04.024URL

为分析茄子的遗传多样性,并挖掘与茄子农艺性状和品质性状相关的分子标记位点,选用18对高 多态性SSR引物对70份国内外茄子材料进行多态性扫描、遗传多样性分析和群体结构分析。在此基础上采用Tassel 2.1 GLM方法进行标记位点与农艺性状和品质性状的关联分析。结果显示,18对SSR引物共检测到130个等位变异位点,平均每个引物7.2个等位变异。各种 质遗传相似系数为0.56~0.93,平均0.68。基因多样性指数和Shan-non指数变幅分别为0.0700~0.4044和 0.1036~0.5848。通过群体遗传结构分析将供试材料分为5个亚群。与农艺相关性状显著相关(P〈0.05)的位点有17个。供试材料之间存在一 定的遗传差异,但遗传基础比较狭窄。
DOI:10.7685/j.issn.1000-2030.2013.05.003URL

采用SRAP分子标记技术,对国内外70份茄子材料的遗传多样性和群体结构进行了分析。结果表明:21对引物共扩增出375条条带,平均每条引物扩增出17.8条条带,其中多态性条带共72个,多态性比例为19.2%,各种质问遗传相似系数变幅为0.3889—0.9583,平均值为0.7053,表明参试材料间存在着一定的遗传差异,但遗传基础比较狭窄。群体结构分析则得出84.29%的材料遗传结构相对比较单一,仅15.71%拥有复杂的遗传背景。从材料划分类群来看,遗传多样性和群体结构分析结果基本一致,只有少数遗传背景复杂的材料类群划分有些差异。
DOI:10.14088/j.cnki.issn0439-8114.2014.23.037URL [本文引用: 1]

以保存的201份茄子 (Solanum melongena L.)资源为材料,对首花节位、株高、株幅等19个形态性状进行遗传多样性、相关性及聚类分析。结果表明,茄子资源19个表型性状的遗传多样性指数平均值 为1.46,平均变异系数为32.9%,表明201份茄子种质资源具有丰富的遗传多样性。相关性分析表明,首花节位与早期产量显著负相关;株高、株幅和果 粗极显著相关;果长与果形指数、单果重、单株果数、早期产量和单产的相关系数达极显著正相关。聚类分析将201份茄子资源划分为两类,分别为野生茄子资源 和栽培茄子资源,在栽培茄子资源中亚洲和非洲来源的茄子种质能够被区分。
URL [本文引用: 1]

To make clear genetic diversity and relationship among eggplant germplasm, 18 accessions of eggplant including 10 from Southeast Asia, 3 from South Amercia and 5 from China were assessed with ISSR markers. The results showed that 430 locis were amplified by 17 ISSR primers, and 390 locis were polymorphic bands, the percentage of polymorphic loci was 92 %. The average observed number of alleles, average effective number of alleles, average Nei’s gene diversity index, and average Shannon’s information index were 1.8971, 1.4778, 0.2850, and 0.4336 respectively. The genetic similarity among all accessions was ranged from 0.55 to 0.81. The 18 accessions of eggplant were clustered into two groups by UPGMA when GSC was 0.67, and 10 from Southeast Asia were clustered in group I, 3 from South Amercia in group II. The result showed that they had rich genetic diversity in 18 accessions of eggplant and provided a theoretical basis for further research and breeding for eggplant.
URL [本文引用: 1]

To understand the genetic basis of the dual cross-line hybrid wheat restorers with thermo顎憄hotoperiod sensitive male sterile, a total of 100 restorers based on 49 genomic-SSR and 40 EST-SSR loci were used to evaluate the genetic diversity and the population structure. 530 allelic variations were detected with an average allele number of 5.96. The average PIC (polymorphism information content) and gene diversity values were determined to be 0.57 and 0.63, respectively, indicating a relatively high degree of genetic diversity among the restorers. By NJ cluster and population structure (Bayesian clustering) analyses, the 100 restorers were clustered into 6 major groups with a wide range of gene flows among the restorers from northern China and from the Huang Huai valley winter wheat production regions. The population structure analyses based on mixed models revealed the genetic composition of the 100 restorers, in which 58 restorers shared a likely single ancestor, and the rest were from mixed ancestors. These results have provided valuable information for the mapping of important agronomic traits, as well as for the breeding and exploit of new wheat restorers.
DOI:10.3321/j.issn:0513-353X.2006.05.040URL

从分子水平用ISSR标记法对南方长茄资源的遗传多样性进行分析,从100个ISSR引物中共筛选出12个多态性明显、条带清晰、反应稳定的引物,对57个样品DNA共扩增出116条谱带,平均每个引物扩增出9·67条带,其中多态性位点84个(71%)。品种间遗传相似系数在0·51~0·98之间,表明茄子栽培种内品种间的遗传基础相对较狭窄。利用UPGMA聚类分析,能将57个南方长茄品种划分为6个类群,类群的划分与地方来源没有很大的关系。
DOI:10.3969/j.issn.1001-4829.2007.04.029URL

利用RAPD技术对来源于国内外的53份茄子种质资源进行了遗传多样性分析,从80个引物中筛选出9个多态性引物,共扩增出81条带,其中有46条多态性条带,占56.79%。种质间遗传相似系数在0.72~0.99之间,表明参试种质遗传基础比较狭窄。UPGMA树状图显示,可将53份参试茄子种质分为五大类群。聚类结果与传统分类并不完全一致,与地理分布没有联系。
URL

用105对茄子SSR引物对34份茄子高代材料进行遗传多样性分 析.试验结果表明:47个标记在供试材料中有多态性,共检测出148个多态性位点.多态性信息含量(PIC)的变化范围是0.025 4~0.909 3,显示了较高的多态性.供试材料遗传相似系数在0.63 ~ 0.93之间,供试材料之间的遗传差异不大,遗传基础相对狭窄.利用UPGMA聚类分析,将供试材料归为两大类4个亚类,SSR分子标记与果实性状聚类分 析结果基本一致.
URL

用RAPD分子标记的方法对来自不同国家的34份茄子品种进行遗传多样性分析,从120条RAPD引物中筛选出有效的22条引物分别对34份茄子品种进行扩增,共检测出232个等位基因位点,每条引物平均检测出10.5个,其中192个为多态位点,多态位点比率为82.76%。POPGENE结果分析表明,Nei’s基因多样性H为0.2756,Shannon指数为0.4145,显示出丰富的遗传多样性。计算得出的Jaccard相似系数变化范围为0.331-0.805,根据Jaccard相似系数和组内连接法建立的系统聚类图,34份茄子大致可分为两大类型:圆茄类型和长茄类型,这与经典的形态学分类基本上相符,从而从分子水平上支持了以果形作为茄子品种分类指标的观点。
.
DOI:10.1007/s00122-008-0732-4URLPMID:18338155 [本文引用: 1]

R2R3-MYB transcription factors of plants are involved in the regulation of trichome length and density. Several of them are differentially expressed during initiation and elongation of cotton fibers. We report sequence phylogenomic characterization of the six MYB genes, their chromosomal localization, and linkage mapping via SNP marker in AD-genome cotton (2 n 52). Phylogenetic grouping and comparison to At- and Dt-genome putative ancestral diploid species of allotetraploid cotton facilitated differentiation between genome-specific polymorphisms (GSPs) and marker-suitable locus-specific polymorphisms (LSPs). The SNP frequency averaged one per 77 bases overall, and one per 106 and 30 bases in coding and non-coding regions, respectively. SNP-based multivariate relationships conformed to independent evolution of the six MYB homoeologous loci in the four tetraploid species. Nucleotide diversity analysis indicated that the six MYB loci evolved more quickly in the Dt- than At-genome. The greater variation in the Dt-D genome comparisons than that in At-A genome comparisons showed no significant bias among synonymous substitution, non-synonymous substitution, and nucleotide change in non-coding regions. SNPs were concordantly mapped by deletion analysis and linkage mapping, which confirmed their value as candidate gene markers and indicated the reliability of the SNP discovery strategy in tetraploid cotton species. We consider that these SNPs may be useful for genetic dissection of economically important fiber and yield traits because of the role of these genes in fiber development.
DOI:10.1111/j.1471-8286.2007.01758.xURLPMID:18784791

Dominant markers such as amplified fragment length polymorphisms (AFLPs) provide an economical way of surveying variation at many loci. However, the uncertainty about the underlying genotypes presents a problem for statistical analysis. Similarly, the presence of null alleles and the limitations of genotype calling in polyploids mean that many conventional analysis methods are invalid for many organisms. Here we present a simple approach for accounting for genotypic ambiguity in studies of population structure and apply it to AFLP data from whitefish. The approach is implemented in the program structure version 2.2, which is available from http://pritch.bsd.uchicago.edu/structure.html.
DOI:10.1146/annurev.arplant.54.031902.134907URLPMID:14502995

Abstract Future advances in plant genomics will make it possible to scan a genome for polymorphisms associated with qualitative and quantitative traits. Before this potential can be realized, we must understand the nature of linkage disequilibrium (LD) within a genome. LD, the nonrandom association of alleles at different loci, plays an integral role in association mapping, and determines the resolution of an association study. Recently, association mapping has been exploited to dissect quantitative trait loci (QTL). With the exception of maize and Arabidopsis, little research has been conducted on LD in plants. The mating system of the species (selfing versus outcrossing), and phenomena such as population structure and recombination hot spots, can strongly influence patterns of LD. The basic patterns of LD in plants will be better understood as more species are analyzed.
.
DOI:10.1007/s001220100570URL

We developed a simple marker technique called sequence-related amplified polymorphism (SRAP) aimed for the amplification of open reading frames (ORFs). It is based on two-primer amplification. The primers are 17 or 18 nucleotides long and consist of the following elements. Core sequences, which are 13 to 14 bases long, where the first 10 or 11 bases starting at the 5′ end, are sequences of no specific constitution (”filler” sequences), followed by the sequence CCGG in the forward primer and AATT in the reverse primer. The core is followed by three selective nucleotides at the 3′ end. The filler sequences of the forward and reverse primers must be different from each other and can be 10 or 11 bases long. For the first five cycles the annealing temperature is set at 35°C. The following 35 cycles are run at 50°C. The amplified DNA fragments are separated by denaturing acrylamide gels and detected by autoradiography. We tested the marker technique in a series of recombinant inbred and doubled-haploid lines of Brassica oleracea L. After sequencing, approximately 45% of the gel-isolated bands matched known genes in the Genbank database. Twenty percent of the SRAP markers were co-dominant, which was demonstrated by sequencing. Construction of a linkage map revealed an even distribution of the SRAP markers in nine major linkage groups, not differing in this regard to AFLP markers. We successfully tagged the glucosinolate desaturation gene BoGLS-ALK with these markers. SRAPs were also easily amplified in other crops such as potato, rice, lettuce, Chinese cabbage (Brassica rapa L.), rapeseed ( Brassica napus L.), garlic, apple, citrus, and celery. We also amplified cDNA isolated from different tissues of Chinese cabbage , allowing the fingerprinting of these sequences.
.
DOI:10.1016/j.scienta.2010.02.023URL

Collection and characterization of all sorts of germplasm resources are required for the development of new cultivars. Molecular characterization is more reliable than morphological characterization. Here, we employed sequence-related amplified polymorphism (SRAP) markers to evaluate genetic variation in a diverse collection of 56 Solanum accessions. Fifty-five SRAP primer combinations were used and a total of 635 polymorphic bands were observed. Cluster analysis by the unweighted pair-group method with arithmetic averages based on similarity matrices indicated that there were three clusters: (i) S. melongena; (ii) S. aethiopicum; (iii) S. surattense. The coefficients of genetic similarity among all the accessions ranged from 0.04 to 0.96 with an average of 0.73, and averaged 0.78 among S. melongena accessions originated from China, indicating extensive genetic variation. These results demonstrated that SRAP can be efficiently used to estimate genetic diversity and analyze phylogenetic relationship.
[本文引用: 2]
[本文引用: 1]
茄子SSR遗传多样性及其农艺性状的关联分析
0
2014
茄子种质资源遗传多样性和群体结构分析
0
2013
亚洲及非洲茄子种质资源主要农艺性状的遗传多样性分析
1
2014
... 茄子(Solanum melongena)是茄科(Solanaceae)茄属(Solanum)中重要的蔬菜作物之一.茄子起源于印度及东南亚热带地区, 古印度是其最早的驯化地, 中国是茄子的次生起源中心(
东南亚茄子种质资源ISSR遗传多样性分析
1
2014
... 茄子(Solanum melongena)是茄科(Solanaceae)茄属(Solanum)中重要的蔬菜作物之一.茄子起源于印度及东南亚热带地区, 古印度是其最早的驯化地, 中国是茄子的次生起源中心(
光温敏二系杂交小麦恢复系遗传多样性和群体结构分析
1
2009
... 为揭示茄子种质的遗传背景, 我们利用SRAP引物组合对183份茄子种质进行了群体遗传结构分析.当样本的等位变异频率特征类型数K=4时, 其模型的后验概率最大, 根据K值将183份茄子种质划分为PopI、PopII、PopIII和PopIV 4个亚群(
我国南方长茄种质资源的ISSR标记分析
0
2006
茄子(S. melongena L.)种质资源遗传多样性的RAPD分析
0
2007
基于SSR分子标记的栽培种茄子遗传多样性分析
0
2012
茄子品种遗传多样性的RAPD检测与聚类分析
0
2007
Cotton ( Gossypium spp.) R2R3-MYB transcription factors SNP identification, phylogenomic characterization, chromosome localization, and linkage mapping
1
2008
... 种质资源是开展育种工作的基础, 种质资源利用的前提是要对其进行多样性分析和遗传背景研究(
Inference of population structure using multilocus genotype data: dominant markers and null alleles
0
2007
Structure of linkage disequilibrium in plants
0
2003
Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica
0
2001
Analysis of genetic variation in eggplant and related Solanum species using sequence-related amplified polymorphism markers
0
2010
2
... 将每条SRAP引物组合扩增的每条带视为1个位点, 条带清晰可辨记为1, 缺失记为0; 统计位点总数和多态性位点数, 不具多态性的条带不统计.应用NTSYS 2.2软件(
... )进行主成分分析.利用Structure 2.3软件(
NTSYS-pc-Numerical Taxonomy and Multivariate Analysis System Version 2.02e
1
1997
... 将每条SRAP引物组合扩增的每条带视为1个位点, 条带清晰可辨记为1, 缺失记为0; 统计位点总数和多态性位点数, 不具多态性的条带不统计.应用NTSYS 2.2软件(
备案号: 京ICP备16067583号-21
版权所有 © 2021 《植物学报》编辑部
地址:北京香山南辛村20号 邮编:100093
电话:010-62836135 010-62836131 E-mail:cbb@ibcas.ac.cn
本系统由北京玛格泰克科技发展有限公司设计开发
