 ,1,*
,1,*Recent Advances in the Regulation Mechanism of Transcription Factors and Metabolic Engineering of Anthocyanins
Xuewei Song1,2,3, Jiebing Wei1,2,3, Shaokang Di1, Yongzhen Pang ,1,*
,1,*通讯作者:
收稿日期:2018-01-15接受日期:2018-11-5网络出版日期:2019-01-30
| 基金资助: |
Corresponding authors:
Received:2018-01-15Accepted:2018-11-5Online:2019-01-30

摘要
关键词:
Abstract
Keywords:
PDF (1130KB)摘要页面多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
引用本文
宋雪薇, 魏解冰, 狄少康, 庞永珍. 花青素转录因子调控机制及代谢工程研究进展. 植物学报, 2019, 54(1): 133-156 doi:10.11983/CBB18016
Song Xuewei, Wei Jiebing, Di Shaokang, Pang Yongzhen.
花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物。花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(Zhao and Tao, 2015)。花青素在植物生长和生存过程中以多种方式发挥作用(Win- kel-Shirley, 2001; Dixon and Sumner, 2003; Zhu et al., 2017)。花青素具有保护植物免受生物侵害和吸引昆虫授粉的功能(Miller et al., 2011; Fan et al., 2016); 此外, 花青素可以响应生物与非生物胁迫(Shao et al., 2007)、清除氧自由基(Shih et al., 2007)以及保护植物免受高密度光照的伤害(Hughes et al., 2005)。近年来, 人们逐渐认识到花青素在抗癌、抗病和抗氧化方面的营养与保健功能(Zhu et al., 2011; Peiffer et al., 2016; Wei et al., 2018)。近期研究表明, 花青素可以降低血脂、减少胆固醇(Farrell et al., 2015)、提高青光眼视力(Shim et al., 2012)、治疗视网膜疾病(Tao et al., 2016)、修复记忆损伤(Jo et al., 2015)以及防治心血管疾病(Isaak et al., 2017)。花青素不仅在植物生长过程中发挥重要作用, 而且对人体健康有益, 因此受到越来越多的关注, 成为植物次生代谢领域的研究热点。
目前, 随着分子生物学的发展, 花青素生物合成途径已经逐渐被阐明, 转录因子单独或协同调控花青素生物合成的分子机制也正在被不断完善。前人已经对花青素的生物合成和转运、环境因子对花青素合成的影响进行了详细的总结(胡可等, 2010; 祝志欣和鲁迎青, 2016)。本文着重综述调控花青素生物合成的主要转录因子的研究进展, 重点阐述MYB、bHLH和WD40三类转录因子调控花青素生物合成的分子机制以及它们在花青素代谢工程中的应用。
1 花青素的生物合成途径
花青素来源于类黄酮化合物的合成途径: 衍生于香豆酰辅酶A (4-coumaroyl CoA)和丙二酰辅酶A (malon- yl CoA), 在查尔酮合酶(chalcone synthase, CHS)作用下合成查尔酮(chalcone)。查尔酮经查尔酮异构酶(chalcone isomerase, CHI)催化形成黄烷酮(flavo- nones)。黄烷酮再经过黄烷酮3-羟化酶(flavanone 3-hydroxylase, F3H)催化形成二氢黄酮醇(dihydro flavonols)。在花青素特异的分支途径中, 二氢黄酮醇还可以被黄酮羟化酶(flavonoid 3’-hydroxylase, F3’H和flavonoid 3’,5’-hydroxylase, F3’5’H)催化, 并在二氢黄酮醇还原酶(dihydroflavonol-4-reductase, DFR)作用下还原为无色花青素(leucoanthocyanidins), 无色花青素在花青素合成酶(anthocyanidin synthase, ANS)的催化作用下形成花青素苷元(anthocyani- dins), 不稳定的花青素苷元经糖基转移酶(glycosylt- ransferase, UGTs)修饰形成稳定的花青素苷(anthocyanins) (Holton and Cornish, 1995; Zhang et al., 2014b)。常见的花青素苷元包括天竺葵素(pelargonidin)、飞燕草素(delphinidin)和矢车菊素(cyanidin) 3种基本花青素苷元, 以及牵牛花素(petunidin)、芍药素(peonidin)和锦葵素(malvidin)等甲基化产物(Cabrita et al., 2000)。广义的花青素泛指各类花青素苷(Seeram and Nair, 2002)。Pelletier等(1997)将类黄酮途径的基因分为早期生物合成基因(early biosynthetic genes, EBGs)和晚期生物合成基因(late biosynthetic genes, LBGs)两类; 花青素生物合成途径中的EBGs即参与共同前体合成的基因, 包括CHS、CHI、F3’H和F3H; LBGs指花青素生物合成途径中的下游基因, 主要包括DFR、ANS和UGTs (图1)。
图1
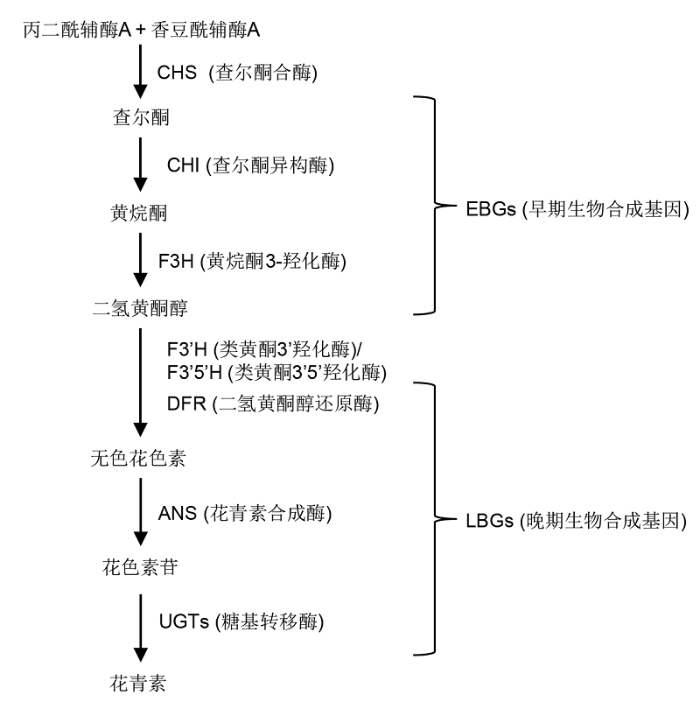 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1花青素的生物合成途径
Figure 1Simplified biosynthesis pathway of anthocyanins
2 花青素合成的转录调控因子
MYB、bHLH和WDR是目前研究最多的3类调控花青素合成的主要转录因子家族, 这3类转录因子的序列、表达特征和作用机制各不相同。它们广泛存在于模式植物、观赏植物和水果等经济作物中, 具有物种特异性和多样性。本文重点对这3类转录因子在不同类型植物中的调控机制进行归纳总结。2.1 MYB转录因子家族
MYB是植物中最大的转录因子家族, 在植物生长发育(Song et al., 2011)、次生代谢调控(Borevitz et al., 2000)、植物激素信号转导(Abe et al., 2003)和胁迫应答(Zhang et al., 2012)等诸多方面发挥作用。MYB转录因子的N端含有1个保守的MYB结构域, 根据MYB结构域的数量可将MYB转录因子分为4类: 1R-MYB、R2R3-MYB、3R-MYB和4R-MYB。其中, R2R3-MYB是数目最多的一类MYB转录因子(Dubos et al., 2010), 也是调控类黄酮途径的重要转录因子, 广泛参与调控花青素的生物合成。在拟南芥(Arabidopsis thaliana)、玉米(Zea mays)和矮牵牛(Petunia hybrida)等模式植物中均发现大量调控花青素生物合成的R2R3-MYB型转录因子。2.1.1 模式植物的MYB转录因子
第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(Cone et al., 1986; Paz-Ares et al., 1987; Pooma et al., 2002)。玉米中另一个不依赖光的基因PL (PI-Rhoades)同样也通过影响DFR基因的表达而调控花和叶中花青素的积累(Cone et al., 1993a, 1993b; Pooma et al., 2002)。在模式植物拟南芥中调控花青素生物合成的MYB转录因子研究得最为清楚, 其中PAP1 (Production of Anthocyanin Pigment 1)是调控花青素的关键转录因子。Borevitz等(2000)发现, 激活PAP1的过量表达诱导花青素的大量积累, 促使拟南芥叶片呈现深紫色(Borevitz et al., 2000)。过量表达PAP1影响CHS、CHI和ANS等基因的表达; 其中与花青素合成相关的2个糖基转移酶基因UGT78D2和UGT75C1均被PAP1诱导表达, 导致 花青素中的矢车菊素含量显著增加(Tohge et al., 2005)。
拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(Mitsunami et al., 2014; Nakabayashi et al., 2014; Onkokesung et al., 2014; Lee et al., 2016)。Maier等(2013)发现, 光应答元件COP1/SPA (CONSTITUTIVELY PHOTOMORPHO GENIC1/SUPPRESSOR OF PHYA-105)能与PAP1和PAP2互作, 在转录和转录后水平上影响花青素的合成。黑暗条件下, COP1/SPA降低PAP1和PAP2的表达, 抑制花青素的合成。另一个光信号途径的元件HY5 (LONG HYPOCOTYL 5)通过结合PAP1 启动子区域的G盒和ACE盒调控其表达(Shin et al., 2013)。同时, PAP1的表达也受到植物激素和蔗糖 水平的影响。例如, 生长素能够通过改变包括PAP1 在内的调控基因的表达水平, 进而影响pap1-D突变体红色细胞中矢车菊素的含量(Liu et al., 2014)。在拟南芥苗期进行蔗糖诱导时发现PAP1基因的表达量增加20倍, 花青素大量积累(Broeckling et al., 2016)。
拟南芥中参与调控花青素合成的主要MYB转录因子还包括MYB113和MYB114, 这2个基因在拟南芥中过量表达所产生的表型与过量表达PAP1所产生的表型相似; 然而在bHLH的突变体中过量表达MYB113和MYB114时, 花青素含量降低, 说明MYB113和MYB114是bHLH依赖型的转录因子(Gonzalez et al., 2008)。由于PAP1不能在豆科模式植物蒺藜苜蓿(Medicago truncatula)中诱导花青素的积累, Peel等(2009)鉴定了与PAP1同源的LAP1 (Legume Anthocyanin Production 1)基因, LAP1在紫花苜蓿(M. sativa)、蒺藜苜蓿和白三叶(Trifolium repens)中的过量表达均可大量积累矢车菊素类的花青素, 因此LAP1被认为是参与调控豆科植物花青素生物合成的关键转录因子。通过PAP1和LAP1的对比研究, 发现不同植物中调控花青素合成的关键MYB转录因子具有一定的物种特异性。
在另一模式植物矮牵牛中, MYB型转录因子AN2 (ANTHOCYANIN 2)和AN4都能够在转录水平影响DFR基因的表达。AN2在C端具有与玉米C1相似的结构域, 并且与C1功能相似, AN2调控矮牵牛花瓣中花青素的合成, 而AN4则调控花筒以及花药中花青素的合成(Quattrocchio et al., 1993, 1999)。除了AN2和AN4, 矮牵牛中调控花青素合成的MYB家族转录因子还包括DPL (DEEP PURPLE)和PHZ (PURPLE HAZE)。在矮牵牛中过量表达DPL和PHZ基因时, 植株整体呈现紫色, 飞燕草素、牵牛花素和锦葵素含量升高(Albert et al., 2011)。
在模式植物金鱼草(Antirrhinum majus)中, 调控花冠中花青素积累的Rosea1、Rosea2和Vensoa基因均编码MYB转录因子, 但是它们具有种间特异性, 分别调节不同的结构基因的表达水平: Rosea1能够提高F3H、DFR和ANS的表达水平, Rosea2仅调控F3’H的表达, Vensoa调控CHI、F3H、F3’5’H和ANS等多个结构基因的表达。这表明Rosea1、Rosea2和Vensoa等转录因子虽然存在一定的功能冗余, 但都各自调控相应的结构基因(Schwinn et al., 2006)。在烟草(Nicotiana tabacum)中特异表达NtAN2不仅能够激活CHS和DFR基因的表达, 还可以与bHLH型转录因子协同调控花青素的积累(Pattanaik et al., 2010)。上述模式植物中的研究表明, MYB家族转录因子在花青素合成途径中既可以独立调控DFR和(或) ANS的表达, 又可以作为MBW复合体的主要元件参与花青素的调控。
2.1.2 MYB转录因子调控果实中花青素
花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状。目前, 对果实中花青素合成机理的研究也日益增多。其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子。巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(Kobayashi et al., 2002)。VvMYB5a和VvMYB5b则通过激活ANS的表达调控葡萄浆果成熟过程中花青素的积累(Deluc et al., 2008)。VlmybA1-1、VlmybA1-2、VlmybA1-3和VlmybA2以及其它转录因子均与葡萄浆果花青素的合成相关, 但其详细的分子机制目前尚不明确(Kob- ayashi et al., 2004; Geekiyanage et al., 2007; Azuma et al., 2008; Cutanda-Perez et al., 2009; Costantini et al., 2015; Sun et al., 2016)。
与葡萄相似, 苹果果皮颜色与花青素的积累密切相关, 多个MYB转录因子参与调控苹果果皮花青素的合成。在苹果红色果皮中表达量较高的MdMYB1基因在光照条件下表达量升高, 可诱导果皮积累花青素; MdMYB1还可以激活DFR和UFGT基因的表达(Takos et al., 2006)。与苹果红色果皮颜色相关的MYB转录因子还包括MdMYBA和MdMYB3, Md- MYBA特异性地结合在ANS的启动子区调控花青素合成, 其表达受到UV-B辐射和低温的诱导; 应用基因枪技术将MdMYBA基因转入不积累花青素的苹果子叶时, 子叶因花青素积累而形成紫红色的斑点(Ban et al., 2007)。过量表达MdMYB3基因能够转录激活CHS和CHI基因的表达(Vimolmangkang et al., 2013)。苹果的另一个转录因子MdMYB10与PAP1等MYB转录因子在保守结构域上高度相似, Md- MYB10调控苹果果肉以及叶片中花青素的积累, 这一点不同于仅调控果皮花青素积累的转录因子(Espley et al., 2007)。过量表达MdMYB10的苹果愈伤组织中矢车菊素的含量升高, DFR基因的表达量也略有升高; 同时, MdMYB10还与MdbHLH3和Mdb- HLH33共表达促进花青素的生物合成(Espley et al., 2007)。在研究苹果红色果肉形成的过程中, Espley等(2009)发现, MdMYB10启动子区具有一段23 bp的串联重复序列, 这一特殊结构存在于各类红色果肉的苹果中, 而白色果肉苹果中则缺少这一序列; Md- MYB10能够结合到这一重复区域, 从而促进花青素的积累, 说明MdMYB10启动子区的这段重复序列在苹果红色果肉形成过程中具有至关重要的作用, 这一发现是花青素机制研究领域的一个重要突破。MdMYB10的同源基因MdMYB110也能够调控苹果果肉中花青素的生物合成, 并且很可能通过参与形成MBW复合体激活CHS基因的表达(Chagné et al., 2013)。苹果MdMYB9和MdMYB11可以结合到ANS的启动子区, 而MdbHLH3能通过结合到MdMYB9和MdMYB11的启动子区与其共同调控苹果花青素的合成。同时, An等(2014)发现茉莉酸酯信号途径中的MdJAZ (Jasmonate ZIM-domain)蛋白能与MdbHL- H3结合抑制其功能, 可见茉莉酸酯信号途径能够通过MdbHLH3转录因子影响花青素的合成调控。目前, 对苹果中调控花青素的转录因子的研究比较详尽, 研究结果对其它果实的相关研究具有一定的借鉴意 义。
除了葡萄和苹果这2种研究相对较多的果实外, 山竹(Garcinia mangostana)、草莓、甜樱桃(Prunus avium)和梨(Pyrus)等常见水果中以及其它研究相对较少的果实中调控花青素的转录因子也陆续被报道(表1), 但对其分子机制的研究并不完善。在对这些MYB转录因子的研究中发现, 大多数MYB均与苹果MdMYB10同源, 功能也相似; 这些MYB型转录因子更多地与bHLH型转录因子互作调控DFR的表达, 但也可独立调控ANS和UFGT的表达。以上各种研究表明, MYB10及其同源基因在常见水果果实中普遍存在, MYB10在果实中调控花青素合成的机制可能是保守的。
Table 1
表1
表1植物中调控花青素合成的MYB类转录因子
Table 1
| 物种名称 | 拉丁名 | 基因名称 | 类型 | 参考文献 |
|---|---|---|---|---|
| 玉米 | Zea mays | C1 | 激活 | Cone et al., 1986 |
| PL | 激活 | Cone et al., 1993a | ||
| PL-BH | 激活 | Cone et al., 1993b | ||
| 拟南芥 | Arabidopsis thaliana | AtCPC | 抑制 | Zhang et al., 2009 |
| AtMYB60 | 抑制 | Park et al., 2008 | ||
| AtMYBL2 | 抑制 | Matsui et al., 2008 | ||
| AtMYB75/PAP1 | 激活 | Borevitz et al., 2000 | ||
| AtMYB90/PAP2 | 激活 | Borevitz et al., 2000 | ||
| AtMYB113 | 激活 | Gonzalez et al., 2008 | ||
| AtMYB114 | 激活 | Gonzalez et al., 2008 | ||
| AtMYB56 | 激活 | Jeong et al., 2018 | ||
| 蒺藜苜蓿 | Medicago truncatula | LAP1 | 激活 | Peel et al., 2009 |
| 矮牵牛 | Petunia hybrida | AN2 | 激活 | Quattrocchio et al., 1999 |
| DPL | 激活 | Albert et al., 2011 | ||
| PHZ | 激活 | Albert et al., 2011 | ||
| MYBx | 抑制 | Albert et al., 2014 | ||
| MYB27 | 抑制 | Albert et al., 2014 | ||
| 金鱼草 | Antirrhinum majus | ROSEA1 | 激活 | Schwinn et al., 2006 |
| ROSEA2 | 激活 | Schwinn et al., 2006 | ||
| VENOSA | 激活 | Schwinn et al., 2016 | ||
| 烟草 | Nicotiana tabacum | NtAN2 | 激活 | Pattanaik et al., 2010 |
| 苹果 | Malus domestica | MdMYB1 | 激活 | Takos et al., 2006 |
| MdMYB3 | 激活 | Vimolmangkang et al., 2013 | ||
| MdMYB6 | 激活 | Gao et al., 2011 | ||
| MdMYB10 | 激活 | Espley et al., 2007 | ||
| MdMYB110a | 激活 | Chagné et al., 2013 | ||
| MdMYB9 | 激活 | An et al., 2014 | ||
| MdMYB11 | 激活 | An et al., 2014 | ||
| MdMYBA | 激活 | Ban et al., 2007 | ||
| MdMYB16 | 抑制 | Wang et al., 2010 | ||
| 葡萄 | Vitis labruscana | VlMYBA1-1 | 激活 | Kobayashi et al., 2002 |
| VlMYBA1-2 | 激活 | Cutanda-Perez et al., 2009 | ||
| VlMYBA1-3 | 激活 | Azuma et al., 2008 | ||
| VlMYBA2 | 激活 | Geekiyanage et al., 2007 | ||
| V. vinifera | VvMYB5a | 激活 | Deluc et al., 2006 | |
| VvMYB5b | 激活 | Deluc et al., 2008 | ||
| VvMYBA1 | 激活 | Kobayashi et al., 2004 | ||
| VvMYBA2 | 激活 | Kobayashi et al., 2004 | ||
| VvMYBPA1 | 激活 | Passeri et al., 2017 | ||
| VvMYBC2-L1 | 抑制 | Cavallini et al., 2015 | ||
| VvMYBC2-L3 | 抑制 | Cavallini et al., 2015 | ||
| 西洋梨 | Pyrus communis | PcMYB10 | 激活 | Pierantoni et al., 2010 |
| 沙梨 | P. pyrifolia | PyMYB10 | 激活 | Feng et al., 2010 |
| PyMYB114 | 激活 | Yao et al., 2017a | ||
| 物种名称 | 拉丁名 | 基因名称 | 类型 | 参考文献 |
| P. communis | PcMYB10 | 激活 | Wang et al., 2013 | |
| 山竹 | Garcinia mangostana | GmMYB10 | 激活 | Palapol et al., 2009 |
| 甜樱桃 | Prunus avium | PacMYBA | 激活 | Shen et al., 2014 |
| PaMYB10 | 激活 | Starkevi? et al., 2015 | ||
| 荔枝 | Litchi chinensis | LcMYB1 | 激活 | Lai et al., 2014 |
| 油桃 | Prunus persica | PpMYB10 | 激活 | Ravaglia et al., 2013 |
| 甜橙 | Citrus sinensis | CsRUBY | 激活 | Butelli et al., 2012 |
| 杨梅 | Myrica rubra | MrMYB1 | 激活 | Niu et al., 2010 |
| 草莓 | Fragaria ananassa | FaMYB10 | 激活 | Medina-Puche et al., 2014 |
| FaMYB1 | 抑制 | Aharoni et al., 2001 | ||
| 猕猴桃 | Actinidia chinensis | MYB110a | 激活 | Fraser et al., 2013 |
| 亚洲杂交百合 | Lilium spp. | LhMYB6 | 激活 | Yamagishi et al., 2010 |
| LhMYB12 | 激活 | Yamagishi et al., 2010 | ||
| LhMYB12-Lat | 激活 | Yamagishi et al., 2014 | ||
| 非洲菊 | Gerbera hybrida | GMYB10 | 激活 | Elomaa et al., 2003 |
| 菊花 | Chrysanthemum morifolium | CmMYB6 | 激活 | Liu et al., 2015 |
| 文心兰 | Oncidium gower | OgMYB1 | 激活 | Chiou et al., 2008 |
| 龙胆 | Gentian triflora | GtMYB3 | 激活 | Nakatsuka et al., 2008 |
| 紫苏 | Perilla frutescens | Myb-p1 | 激活 | Gong et al., 1999 |
| 番茄 | Lycopersicon esculentum | LeANT1 | 激活 | Mathews et al., 2003 |
| 红薯 | Ipomoea batatas | IbMYB1 | 激活 | Mano et al., 2007 |
| 马铃薯 | Solanum tuberosum | StAN1 | 激活 | Jung et al., 2009 |
| StAN2 | 激活 | Jung et al., 2009 | ||
| StMTF1 | 激活 | Rommens et al., 2008 | ||
| 结球甘蓝 | Brassica oleracea var. capitata | BoMYB2 | 激活 | Yuan et al., 2009 |
| 花椰菜 | B. oleracea var. botrytis | PURPLE | 激活 | Chiu and Yeh, 2010 |
| 紫背天葵 | Gynura bicolor | GbMYB1 | 激活 | Shimizu et al., 2011 |
| 羽衣甘蓝 | B. oeracea var. acephala | BoPAP1 | 激活 | Zhang et al., 2012 |
| 淫羊藿 | Epimedium sagittatum | EsMYBA1 | 激活 | Huang et al., 2013a |
| EsMYB9 | 激活 | Huang et al., 2017 | ||
| 胡萝卜 | Raphanus sativus | RsMYB1 | 激活 | Lim et al., 2016 |
| 洋葱 | Allium cepa | MYB1 | 激活 | Schwinn et al., 2016 |
| 小麦 | Triticum aestivum | TaPL1 | 激活 | Shin et al., 2016 |
| TaMYB3 | 激活 | Li et al., 2017 | ||
| 红掌 | Anthurium andraeanum | AaMYB2 | 激活 | Li et al., 2016a |
| 大豆 | Glycine max | GmMYB-G20-1 | 激活 | Takahashi et al., 2011 |
| 梅 | Prunus mume | PmMYBa1 | 激活 | Zhang et al., 2017 |
| 葡萄风信子 | Muscari armeniacum | MaAN2 | 激活 | Chen et al., 2017 |
| 智利草莓 | Fragaria chiloensis | FcMYB1 | 抑制 | Salvatierra et al., 2013 |
| 芜菁 | B. rapa | BrMYB4 | 抑制 | Zhang et al., 2014a |
| 银杏 | Ginkgo biloba | GbMYBF2 | 抑制 | Xu et al., 2014a |
| GbMYBFL | 激活 | Zhang et al., 2018 | ||
| 杨树 | Populus trichocarpa | PtrRML1 | 抑制 | Hu et al., 2016 |
新窗口打开|下载CSV
2.1.3 观赏植物中的MYB转录因子
植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素。花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少。非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(Elomaa et al., 2003)。GMYB10在不同组织中诱导的花青素种类具有特异性, GMYB10促进愈伤组织和营养组织中矢车菊素的积累, 而在雄蕊中则促进天竺葵素的积累(Laitinen et al., 2008)。文心兰(Oncidium gower)中的OgMYB1转录因子通过激活CHI和DFR促进红色花瓣中矢车菊素、飞燕草素、锦葵素和芍药素(Chiou and Yeh, 2008)的积累。龙胆(Gentian triflora)中的GtMYB3转录因子与GtbHLH1协同作用影响F3’5’H基因的表达, 进而促进花瓣中龙胆翠雀花素(gentiodelphin)的富集(Nakatsuka et al., 2008)。亚洲杂交百合(Lilium spp.)中的LhMYB6和LhMYB12与矮牵牛AN2同源, 能与LhbHLH2转录因子互作调控百合花中矢车菊素的生物合成, LhMYB6不仅影响百合花瓣上的花斑形成, 还受光诱导影响百合叶片花青素的积累; 而LhMYB- 12则调控百合花瓣和花药中花青素的合成(Yamagi- shi et al., 2010)。Yamagishi等(2014)还发现LhMYB12的等位基因LhMYB12-Lat也与百合花瓣上花青素局部积累形成的花斑相关。菊花(Chrysan- themum morifolium)中的CmMYB6主要调控紫色菊花发育时期花青素的合成, CmMYB6能够与bHLH型转录因子互作激活DFR的表达, 从而促进花青素的合成(Liu et al., 2015)。综上所述, 观赏植物中调控花青素生物合成的MYB型转录因子的调控具有组织特异性, 不同转录因子分别调控不同部位花青素的含量与种类。
2.1.4 其它植物中的MYB转录因子
除了水果和观赏植物之外, 蔬菜以及其它富含花青素的植物也是研究花青素调控机制的理想材料。目前, 已鉴定到影响花青素积累MYB转录因子的物种材料还包括各种蔬菜作物, 如马铃薯(Solanum tuberosum)、结球甘蓝(Brassica oleracea)、羽衣甘蓝(B. oleracea); 药用植物如淫羊藿(Epimedium sagittatum)和紫苏(Perilla frutescens); 作物如小麦(Triticum aestivum) (表1)。在这些转录因子基因中, 被证明通过影响DFR基因表达来调控花青素生物合成的MYB转录因子基因包括紫苏的Myb-p1和红掌(Anthurium andraeanum)的AaMYB2; 被证明通过与bHLH家族转录因子互作来促进花青素积累的包括结球甘蓝的BoMYB2、花椰菜的Pr、紫背天葵(Gynura bicolor)的GbMYB1和淫羊藿的EsMYBA1; 其它植物中的MYB转录因子基因仅被发现参与调控花青素的积累, 但都缺乏具体的分子机制研究。
2.2 bHLH转录因子家族
碱性螺旋-环-螺旋(basic helix-loop-helix, bHLH)蛋白在动植物中高度保守, 具有保守的氨基酸序列(Mor- genstern and Atchley, 1999)。bHLH转录因子被发现参与植物生长发育(Song et al., 2013)、信号转导(Zhang et al., 2009)和次生代谢(Zhang et al., 2003)等诸多生物学过程(Toledo-Ortiz et al., 2003)。2.2.1 模式植物中的bHLH转录因子
bHLH转录因子是调控花青素合成的另一大类转录因子家族(表2)。玉米中调控花青素合成的主要bHLH转录因子包括R1、B1、Lc (Leaf color )和Sn, 是最早发现的一批调控花青素合成的bHLH家族转录因子(Chandler et al., 1989; Ludwig et al., 1989; Consonni et al., 1992)。其中, Lc能够通过激活CHS和DFR的表达, 互补矮牵牛an2/an11双突变体中花青素缺失的表型(Quattrocchio et al., 1993)。金鱼草中的DELILA基因能够调控DFR的表达, 促进花青素的积累(Almeida et al., 1989)。
Table 2
Table 2 bHLH transcription factors (TFs) involved in the regulation of anthocyanin biosynthesis in plants
| 物种名称 | 拉丁名 | 基因名称 | 类型 | 参考文献 |
|---|---|---|---|---|
| 玉米 | Zea mays | R1 | 激活 | Chandler et al., 1989 |
| Lc | 激活 | Ludwig et al., 1989 | ||
| B1 | 激活 | Chandler et al., 1989 | ||
| Sn | 激活 | Tonelli et al., 1991 | ||
| 金鱼草 | Antirrhinum majus | DELILA | 激活 | Martin et al.,1991 |
| 矮牵牛 | Petunia hybrida | JAF13 | 激活 | Spelt et al., 2000 |
| AN1 | 激活 | Spelt et al., 2000 | ||
| 拟南芥 | Arabidopsis thaliana | GL3 | 激活 | Payne et al., 2000 |
| EGL3 | 激活 | Zhang et al., 2003 | ||
| MYC146 | 激活 | Ramsay et al., 2003 | ||
| TT8 | 激活 | Nesi et al., 2000 | ||
| 蒺藜苜蓿 | Medicago truncatula | MtTT8 | 激活 | Li et al., 2016b |
| 苹果 | Malus domestica | MdbHLH3 | 激活 | Xie et al., 2012 |
| MdbHLH33 | 激活 | Espley et al., 2007 | ||
| MdMYC2 | 激活 | An et al., 2016 | ||
| 烟草 | Nicotiana tabacum | NtAn1a | 激活 | Bai et al., 2011 |
| NtAn1b | 激活 | Bai et al., 2011 | ||
| 菊花 | Chrysanthemum morifolium | CmbHLH | 激活 | Xiang et al., 2015 |
| 非洲菊 | Gerbera hybrida | GMYC1 | 激活 | Elomaa et al., 2003 |
| 荔枝 | Litchi chinensis | LcbHLH1 | 激活 | Lai et al., 2016 |
| LcbHLH3 | 激活 | Lai et al., 2016 | ||
| 龙胆 | Gentian triflora | GtbHLH1 | 激活 | Nakatsuka et al., 2008 |
| 杨梅 | Myrica rubra | MrbHLH1 | 激活 | Liu et al., 2013 |
| 花椰菜 | Brassica oleracea var. botrytis | BobHLH1 | 激活 | Chiu and Li, 2012 |
| 结球甘蓝 | B. oleracea var. capitata | BoTT8 | 激活 | Yuan et al., 2009 |
| 紫背天葵 | Gynura bicolor | GbMYC1 | 激活 | Shimizu et al., 2011 |
| 杂交百合 | Lilium spp. | LhbHLH1 | 激活 | Nakatsuka et al., 2009 |
| 番茄 | Lycopersicon esculentum | AH | 激活 | Qiu et al., 2016 |
| 大丽花 | Dahlia variabilis | DvIVS | 抑制 | Ohno et al., 2011 |
| 圆叶牵牛 | Ipomoea purpurea | bHLH2 | 激活 | Park et al., 2007 |
| 甜橙 | Citrus sinensis | CsMYC2 | 激活 | Cultrone et al., 2010 |
| 紫苏 | Gynura bicolor | Myc-rp | 激活 | Gong et al., 1999 |
| 葡萄 | Vitis vinifera | VvbHLH1 | 激活 | Xu et al., 2014b |
| VvMYC1 | 激活 | Hichri et al., 2010 | ||
| VvMYCA1 | 激活 | Hichri et al., 2010 | ||
| Myc-F3G1 | 激活 | Yamazaki et al., 2003 | ||
| 甜樱桃 | Prunus avium | PabHLH3 | 激活 | Starkevi? et al., 2015 |
| 梨 | Pyrus pyrifolia | PybHLH3 | 激活 | Yao et al., 2017a |
| LhbHLH2 | 激活 | Nakatsuka et al., 2009 | ||
| 马铃薯 | Solanum tuberosum | StbHLH1 | 激活 | Payyavula et al., 2013 |
| StbHLH2 | 激活 | Payyavula et al., 2013 | ||
| AH | 激活 | Qiu et al., 2016 |
新窗口打开|下载CSV
拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源。TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(Baudry et al., 2004)。Zhang等(2003)发现gl3/egl3双突变体缺少花青素, 而且DFR的表达量远低于野生型。分别过量表达GL3和EGL3都能够回复ttg1突变体中花青素缺失的表型, 说明GL3和EGL3参与花青素的生物合成(Payne et al., 2000; Zhang et al., 2003)。EGL3还被证明能够与PAP1共表达促进花青素的积累, 可见EGL3在调控花青素合成的功能方面强于GL3 (Zhang et al., 2003)。Gonzalez等(2008)发现, 过量表达GL3还能够上调F3’H、DFR、ANS、TT8和PAP2的表达水平。GL3还参与氮胁迫环境条件下花青素的积累: 在缺氮条件下, GL3功能失活的植株中不合成花青素, 而且DFR的表达量远低于野生型(Feyissa et al., 2009)。模式植物矮牵牛中调控花青素合成的bHLH类转录因子主要包括JAF13和AN1, JAF13能够与AN2互作激活DFR的表达, 促进花青素合成(Quattrocchio et al., 1998)。AN1不仅能够与AN2结合激活DFR的表达, 还可以直接激活DFR的表达(Spelt et al., 2000)。AN1与AN2和AN4之间的协同互作具有组织特异性, AN2在矮牵牛叶片中的表达可以诱导AN1表达, 而在花药中AN1可以诱导AN4表达(Spelt et al., 2000)。
近年来, 其它模式植物中有关bHLH转录因子调控花青素合成的研究逐渐增多。MtTT8是蒺藜苜蓿中与拟南芥TT8同源的bHLH型转录因子。mttt8突变体表现为花青素缺失表型, 在突变体中过量表达MtTT8其毛状根中花青素的含量增加; 同时, 在拟南芥tt8突变体中表达MtTT8也能产生花青素, 恢复突变体花青素缺失的表型。MtTT8能与MtLAP1和MtWD40-1转录因子互作形成MBW复合体, 进而共同调控花青素的合成(Li et al., 2016b)。另一个模式植物烟草中的NtAn1a和NtAn1b转录因子能够在烟草花中促进花青素的积累, NtAn1a与烟草NtAn2互作激活DFR和CHS的表达, 从而调控花青素的合成(Bai et al., 2011)。模式植物中的这些研究表明, 绝大部分bHLH型转录因子需要与MYB互作, 独立行使功能的较少。虽然花青素生物合成途径包含CHS、CHI、F3H、DFR和ANS等多个基因, 但MYB和bHLH转录因子主要调控DFR和ANS基因的表达(尤其是DFR), 表明DFR在花青素合成途径中具有关键作用。
2.2.2 非模式植物中的bHLH转录因子
非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面。苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(Espley et al., 2007)。受低温诱导的MdbHLH3转录因子还能够结合到DFR的启动子区, 与MdMYB1共同促进花青素的积累(Xie et al., 2012)。MdMYBC还可以通过参与茉莉酸途径正调控花青素的合成(An et al., 2016)。荔枝(Litchi chinensis) LcbHLH1和LcbHLH3转录因子同样能与Lc- MYB1共同作用, 促进花青素积累(Lai et al., 2016)。非洲菊GMYC1特异地在花冠和心皮部位调控DFR活性, 与AN2和GMYB10互作促进DFR的表达(Elomaa et al., 2003)。菊花CmbHLH转录因子能够激活DFR的表达, 在与CmMYB6转录因子共表达时促进花青素的合成(Xiang et al., 2015)。能够与MYB共同作用调控花青素积累的bHLH类转录因子还包括龙胆的GtbHLH1 (Nakatsuka et al., 2008)、杨梅(Myrica rubra)的MrbHLH1 (Liu et al., 2013)、花椰菜(Brassica oleracea var. botrytis)的BobHLH1 (Chiu et al., 2010)、结球甘蓝(B. oleracea var. capitata)的BoTT8 (Yuan et al., 2009)和紫背天葵中的GbMYC1 (Shimizu et al., 2011)。
bHLH类转录因子基因也陆续在其它植物中被鉴定(表2), 但目前还未在其它植物中发现与其互作的MYB转录因子。目前, 对这些植物中bHLH的功能研究还不够深入, 它们在不同植物花青素调控网络中的上下游关系还不十分明确。
2.3 WD40重复蛋白家族
WD40重复蛋白(WD40 repeat proteins, WDR)主要存在于真核生物中, 具有保守而特异的二肽重复基序, 每个重复大概有40多个氨基酸残基; 重复的WD40基序在蛋白质互作时作为支架起固定作用(Mishra et al., 2012)。WDR参与植物非生物胁迫、生长发育和类黄酮的合成(Walker et al., 1999; Huang et al., 2008; Miller et al., 2016)。矮牵牛的AN11 (ANTHOCYANIN11)是第1个被发现调控花青素生物合成的WDR蛋白, 其相应的编码基因是通过转座子标签法被鉴定的。AN11作用于AN2上游, 可能通过参与1个信号转导串联体系激活AN2的表达, 从而调控矮牵牛花青素的合成(de Vetten et al., 1997)。拟南芥的TTG1 (TRANSPARENT TESTA GLABRA 1)与矮牵牛AN11转录因子同源, 其能够调控一系列生长发育过程, 包括表皮毛的生长、种子黏液质的形成和种子内含物的积累等; TTG1通过影响DFR的表达调控花青素的合成(Wal- ker et al., 1999); 酵母双杂交实验证明, TTG1可以与bHLH型转录因子GL3互作(Payne et al., 2000)。Gonzalez等(2008)进一步证实bHLH依赖型(bHLH- dependent) MYB转录因子和TTG1依赖型(TTG1- dependent) bHLH转录因子能够调控DFR及ANS的表达, 即PAP1和GL3/EGL3互作, GL3和TTG1互作调控花青素的生物合成; 从而提出了MYB/bHLH/ TTG1转录复合体调控花青素合成的机制。Shi和Xie (2011)在研究pap1-D突变体时发现, 在富含花青素的红色细胞中, PAP1、TT8、GL3和TTG1基因的表达量高于野生型, 同时DFR和ANS的表达量也升高; 他们认为在拟南芥的红色细胞中存在1个TTG1- GL3/TT8-PAP1复合体调控花青素的合成。随后, Zhou等(2012)发现在pap1-D突变体的红色细胞中, PAP1-GL3/TT8-TTG1复合体的表达受氮的影响, 氮水平可以调控花青素的合成。MYB-bHLH-WD40三元复合体的调控机制在除花青素之外的类黄酮分支途径中也广泛存在, 因此Albert等(2014)将调控花青素生物合成的MYB-bHLH-WDR三元复合体命名为MBW复合体。
拟南芥中WDR类TTG1参与MBW复合体调控类黄酮化合物合成的机制研究得较为详细, 其它物种中WDR蛋白调控的功能研究相对较少。目前已有报道的调控花青素合成的WDR均与AN11和/或TTG1同源, 例如, 玉米中的PAC1 (PALE ALEURONE COL- OR 1)和MP1基因(Carey et al., 2004), 紫苏中的PFWD也被证明通过与MYC互作进而调控花青素的积累(Sompornpailin et al., 2002)。作者所在课题组通过正向遗传筛选的方法在蒺藜苜蓿中鉴定了1个WDR类的MtWD40-1基因。MtWD40-1不仅可以互补拟南芥ttg1突变体, 而且可以回复蒺藜苜蓿Tnt1突变体的表型, 过量表达MtWD40-1导致蒺藜苜蓿毛状根中花青素含量回复, 证明MtWD40-1参与调控苜蓿花青素的生物合成; 但与TTG1不同的是, MtWD40-1虽然也参与种子原花青素和黏液质的积累, 但它并不影响表皮毛和根毛的形成, 这是MtWD40-1与TTG1功能显著不同之处(Pang et al., 2009)。其它植物中WDR调控花青素生物合成的研究相对较少, 仅限于对该类转录因子的挖掘(表3), 它们与TTG1或其它功能明确的WDR类蛋白在结构与功能方面的异同还有待进一步研究。
Table 3
表3
表3植物中调控花青素合成的WD40转录因子
Table 3
| 物种名称 | 拉丁名 | 基因名称 | 类型 | 参考文献 |
|---|---|---|---|---|
| 矮牵牛 | Petunia hybrida | AN11 | 激活 | De Vetten et al., 1997 |
| 拟南芥 | Arabidopsis thaliana | TTG1 | 激活 | Walker et al., 1999 |
| 玉米 | Zea mays | PAC1 | 激活 | Carey et al., 2004 |
| MP1 | 激活 | Carey et al., 2004 | ||
| 蒺藜苜蓿 | Medicago truncatula | WD40-1 | 激活 | Pang et al., 2009 |
| 苹果 | Malus domestica | MdTTG1 | 激活 | Brueggemann et al., 2010 |
| 石榴 | Punica granatum | PgWD40 | 激活 | Ben-Simhon et al., 2011 |
| 红薯 | Ipomoea batatas | IbWD40 | 激活 | Dong et al., 2014 |
| 葡萄 | Vitis vinifera | VvWDR1/2 | 激活 | Matus et al., 2010 |
| 马铃薯 | Solanum tuberosum | StWD40 | 激活 | Payyavula et al., 2013 |
| StAN11 | 激活 | Li et al., 2014 | ||
| 杨梅 | Myrica rubra | MrWD40-1 | 激活 | Liu et al., 2013 |
| 紫苏 | Gynura bicolor | PFWD | 激活 | Sompornpailin et al., 2002 |
| 甜樱桃 | Prunus avium | PaWD40 | 激活 | Starkevi? et al., 2015 |
| 圆叶牵牛 | Ipomoea purpurea | WDR1 | 激活 | Morita et al., 2006 |
| 花椰菜 | Brassica oleracea var. botrytis | BoWD40 | 激活 | Chiu and Li, 2012 |
| 苦荞麦 | Fagopyrum tataricum | FtWD40 | 激活 | Yao et al., 2017b |
新窗口打开|下载CSV
2.4 其它类型转录因子
2.4.1 bZIP类转录因子目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少。近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成。bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(Banerjee and Roychoudhury, 2017), 如植物生长发育(Gibalová et al., 2017)、环境胁迫应答(Wang et al., 2017)和光信号转导(Ang and Deng, 1994)。拟南芥中影响植物光形态建成的bZIP转录因子HY5是第1个被报道参与花青素合成调控途径的bZIP型转录因子。Lee等(2007)在研究HY5转录因子结合位点时, 发现HY5能够调控F3H和CHS基因的表达; 随后的研究还证明, HY5和PIF3 (Phytochrome Interacting Factor 3)共同作用, 直接结合在C4H、F3’H和DFR启动子的特异区域(Shin et al., 2007)。此外, HY5还可以结合到PAP1的启动子上正调控花青素的合成(Shin et al., 2013)。HY5还可以结合到MYBL2的启动子上, 通过组氨酸修饰抑制MYBL2的表达, 从而促进花青素合成(Ngu- yen et al., 2015)。miR858是HY5潜在的靶基因, 它可以与HY5共同作用参与调控花青素的合成(Wang et al., 2016)。An等(2017)通过同源克隆的方法获得了苹果MdHY5基因, MdHY5基因能够结合到MdMYB10的启动子区, 诱导苹果花青素的积累; 且MdHY5不仅与拟南芥HY5具有相同的功能, 都受光和脱落酸诱导, 而且还参与氮信号途径。bZIP型HY5转录因子与MYB、bHLH和WDR三类转录因子的区别在于HY5主要通过结合到保守启动子区行使功能, 通过调控参与花青素生物合成的转录因子促进花青素的积累, 而其它3类转录因子主要通过调控DFR和ANS基因的表达参与花青素的合成。bZIP型转录因子是否参与调控光、氮素和脱落酸等非生物因子介导的花青素代谢调控网络还有待进一步研究。
2.4.2 负转录因子
花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点。随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明。模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE)。mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(Dubos et al., 2008; Jun et al., 2015)。MYBL2转录因子具有独特的TLLLFR抑制结构域, 通过与TT8结合抑制DFR和TT8的表达(Matsui et al., 2008)。在拟南芥中过量表达CPC时, 花青素含量降低(Zhang et al., 2009; Zhu et al., 2009)。研究表明, CPC在氮胁迫下的花青素合成过程中具有反馈抑制作用(Nemie-Feyissa et al., 2014)。而SPL9转录因子通过破坏MBW复合体的稳定性抑制DFR的表达, 从而抑制花青素积累(Gou et al., 2011)。矮牵牛中MYB27通过与AN1互作抑制AN1的表达, 同时MYB27的表达在高光下受到抑制; 相反, MYBx作为MYB27的竞争性抑制因子, 却在高光条件下表达(Spelt et al., 2000; Albert et al., 2011)。Albert等(2014)的研究表明, MYB27还与JAF13和AN11互作, 通过结合bHLH型转录因子破坏MBW复合体的形成, 在花青素代谢网络中行使负调控功能。
除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物。葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(Cavallini et al., 2015)。在葡萄皮中特异表达VvMYB4-like能够下调DFR和ANS的表达; 在烟草中异源表达VvMYB4-like时, 花青素的积累受到抑制(Pérez-Díaz et al., 2016)。苹果MdMYB6作为一个负调控转录因子, 过表达时能够抑制包括DFR和CHS在内的多个结构基因的表达, 从而减少花青素的合成(Gao et al., 2011)。智利草莓(Fragaria chiloensis)中的FcMYB1能够抑制苹果表面花青素的积累, 当其表达量降低时, ANS的表达量升高(Salvatierra et al., 2013)。芜菁(Brassica rapa) BrMYB4基因的表达受UV-B诱导, 它可以结合到C4H的启动子区使其抑制花青素的合成(Zhang et al., 2014a)。杨树(Populus trichocarpa) PtrRML1 (R3 MYB-LIKE1)在拟南芥中过量表达时能够抑制GL3的表达, 从而抑制花青素的积累(Hu et al., 2016)。
综上所述, 目前花青素生物合成的负调控因子主要是MYB转录因子, 这类转录因子的分子调控机制可分为3类: 一是通过抑制DFR等结构基因的表达行使功能; 二是与其它正调控转录因子结合抑制正调控因子的表达, 从而减少花青素积累; 三是通过结合参与MBW复合体形成的转录因子, 破坏MBW复合体的稳定性, 进而行使其抑制功能。
3 花青素代谢工程
3.1 通过关键结构基因进行代谢工程改良
花色改良一直是花青素代谢工程的研究热点之一, 由于传统以杂交和诱变为主的观赏植物育种技术很难培育出富含飞燕草素的蓝色花朵, 而通过代谢工程的方法修饰花青素结构能够使花朵呈现纯正的蓝色, 更加贴合消费者的需求, 因此代谢工程为花色改良育种提供了新思路。其中, 通过改变关键结构基因的表达水平, 进而改变单个花青素产物是花色改良的一个重要策略。矮牵牛是一种常见的观赏植物, 也是研究花青素代谢调控的经典模式植物。Meyer等(1987)将玉米来源的DFR基因导入矮牵牛RL01突变体, 使含有矢车菊素和飞燕草素的浅粉色突变体花朵变为含有天竺葵素的砖红色花朵。Fukusaki等(2004)通过RNA干扰技术(RNA interference)抑制蝴蝶草(Torenia hybrida) CHS基因的表达, 使富含锦葵色素和芍药色素的蓝色蝴蝶草变为花青素缺失的白色蝴蝶草。Boase等(2010)将飞燕草素合成途径的关键酶基因F3’5’H反向转入仙客来(Cyclamen persicum)中, 抑制了F3’5’H基因的表达, 使花青素总量降低80%, 飞燕草素含量降低, 矢车菊素比例相对升高, 使花朵颜色由深粉色变浅。在康乃馨(Dianthus caryophyllus)、玫瑰(Rosa rugosa)和菊花中过量表达F3’5’H基因可以使花朵合成飞燕草素, 从而呈现蓝紫色(Kat- sumoto et al., 2007; Brugliera et al., 2013)。Noda等(2017)将风铃草(Campanula medium)来源的Cam- F3’5’H基因和蝶豆(Clitoria ternatea)来源的CtA3’- 5’GT基因转入菊花, 使其因飞燕草素积累而显现出纯正的蓝色, 从而改良了菊花颜色。另外, 利用微生物生产花青素作为化妆品和食品添加剂也是代谢工程的一个发展方向。Yan等(2005)将苹果来源的F3H和ANS、红掌来源的DFR以及矮牵牛来源的3-GT基因转入大肠杆菌, 第1次在大肠杆菌中产生了含量较低的天竺葵素和矢车菊素, 在微生物中重建了花青素的合成途径。该实验证明, 在充分阐明花青素代谢途径的基础上, 在微生物中目标导向性地重建花青素的合成途径切实可行。
3.2 通过调控关键基因进行代谢工程改良
异源过量表达转录因子基因是花青素代谢工程的另一种重要策略。来源于紫苏的Myc-rp (Gong et al., 1999)、葡萄的VlMYB2 (Geekiyanage et al., 2007)、苹果的MdMYB1 (Takos et al., 2006)、MdMYB110a (Chagné et al., 2013)、MdMYB3 (Vimolmangkang et al., 2013)、MdbHLH3 (Xie et al., 2017)和拟南芥的PAP1 (Gatica-Arias et al., 2012; Qiu et al., 2014)在烟草中异源表达时均能促进花青素的大量积累。山竹GmMYB10在烟草中与AtbHLH共表达时可以激活DFR的表达, 促进花青素的合成(Palapol et al., 2009)。荔枝LcMYB1也能够与LcbHLH1和LcbHLH3在烟草中共同作用, 从而促进烟草叶片中花青素的积累(Lai et al., 2016)。在模式植物烟草和拟南芥中, 探索外源花青素合成调控基因功能的一系列实验不断取得成功, 为代谢工程在作物中的蓬勃发展提供了具有借鉴意义的理论和实践基础。Butelli等(2008)将金鱼草的DELILA和ROSEA1基因转入番茄(Solanum lycopersicum cv. ‘MicroTom’), 使转基因番茄的果皮和果肉因大量富集酰基化花青素而呈现出深紫色, 转基因番茄的花青素含量达2.83 mg?g-1, 远高于富含花青素的蓝莓等水果; 将这些高花青素含量的转基因番茄粉添加到患癌症小鼠的饮食中, 发现小鼠的寿命延长, 该结果证明了花青素具有抗癌的特性, 引起了广泛关注。在本课题组的一项研究中, 将PAP1和Lc共表达转入稀有的野生药用植物天山雪莲(Saussurea involucrata), 转基因株系的愈伤组织和嫩枝中均出现至少4类矢车菊素的衍生物; 同时, 花青素合成途径中多数结构基因的表达受到诱导, 其中CHS基因表达量变化最为显著(Qiu et al., 2013)。He等(2017)将PAP1基因转入烟草, 在转基因烟草中检测到1种具有药用价值的花青素Cyanidin 3-O-rutinoside, 这种花青素的含量占总花青素含量的98%, 这种转基因烟草可以作为工业上提取大量具有药用功能的花青素的原料。Zhu等(2017)将来源于彩叶草(Solenostemon scutellarioi- des)的CHS、CHI、F3H、F3’H、DFR、ANS以及来源于玉米的转录因子基因Lc和Pl构建入同一个载体中, 在水稻胚乳特异表达启动子的驱动下转化水稻, 培育出了富含矢车菊素和芍药素的“紫晶米”(Zhu et al., 2017; 朱丽和钱前, 2017)。这一研究使在主要粮食作物中产生花青素成为可能, 同时富含花青素的转基因水稻的成功培育极大地促进了花青素代谢工程研究的发展。
4 展望
花青素作为类黄酮化合物代谢途径的1种重要分支产物, 在生物和非生物胁迫应答、人体保健、抗氧化以及抗病等方面具有重要的研究和应用价值。随着花青素代谢机制的逐渐阐明特别是调控花青素途径转录因子的分离和功能鉴定, 对转录因子的研究已经为花青素开发利用提供了目标基因和理论基础。调控花青素的3类主要转录因子MYB、bHLH和WDR形成1个复杂的调控网络; 其中, WDR是形成MBW复合体的保守元件, 目前已被鉴定的WDR蛋白均参与MBW复合体的形成, 但水果和观赏植物等非模式植物中的WDR及其功能还有待进一步研究。bHLH型转录因子主要通过与MYB转录因子互作的方式调控DFR和ANS的表达, 它们在MBW复合体中主要作为与MYB和WDR互作的功能元件。在MBW复合体中, 研究最充分的MYB转录因子在各种植物中广泛存在, 有的MYB转录因子可以独立调控以DFR为主的结构基因, 有的MYB转录因子则能够参与形成MBW复合体。MYB型转录因子在叶片、果实和花朵等不同组织器官中的表达具有一定的组织特异性, 尤其花青素在果实果皮与果肉中, 以及不同颜色观赏植物花瓣中的积累均受到不同MYB转录因子的调控。果实中花青素的积累通常还会受到光照、温度和植物激素等非生物因子的影响, 通过激活MYB和bHLH转录因子参与非生物因子介导的花青素合成调控; 然而, 目前除了HY5调控光照影响花青素积累的分子机制研究取得了一定进展之外, 更多的互作因子还有待发现, 光诱导调控花青素积累的信号通路还有待进一步明确(图2)。图2
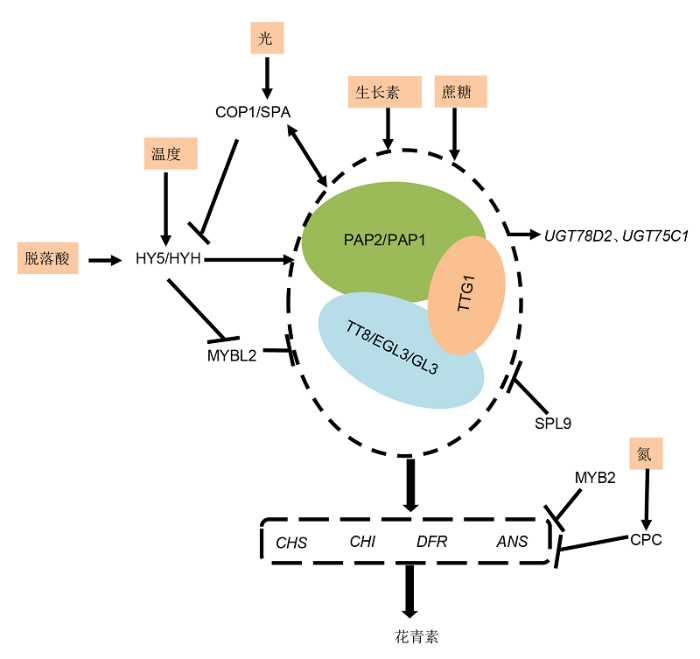 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2拟南芥MBW转录复合体参与花青素的代谢调控网络
拟南芥PAP1/PAP2、TT8/EGL3/GL3和TTG1组成的MBW复合体主要激活CHS、CHI、DFR和ANS的表达进而调控花青素的积累。MBW复合体的功能不仅受环境因子的影响, 还受负调控因子的调控。外源施加生长素和蔗糖直接影响花青素的积累。光信号直接作用于COP1/SPA受体, 温度和脱落酸通过诱导HY5/HYH基因的表达, 进而间接作用于MBW复合体中的MYB型转录因子。负调控因子MYBL2、SPL9、MYB2和CPC则通过负调控MBW复合体或者结构基因的表达, 进而抑制花青素的积累。
Figure 2Summary of MBW transcription complex in the regulation of anthocyanins in Arabidopsis
The MBW complex composes of PAP1/PAP2, TT8/EGL3/GL3 and TTG1, which mainly regulates anthocyanin accumulation in Arabidopsis thaliana by the regulation of the expression levels of key structural genes, including CHS, CHI, DFR and ANS. The function of the MBW complex is not only affected by environmental factors, but also by negative regulators. The application of auxin and sucrose can directly affect the accumulation of anthocyanins. The light signal directly acts on COP1/SPA receptor, and temperature and ABA induce the expression of HY5/HYH gene, which in turn affect the MYB transcription factor in the MBW complex. The negative regulatory factors such as MYBL2, SPL9, MYB2 and CPC, negatively regulate the expression levels of MBW complex or structural genes, and then reduce the accumulation of anthocyanins.
在花青素的转录调控网络中, 还有一些关键问题有待阐明。例如, 各种非生物因子是与单个转录因子作用影响花青素合成, 还是与多个转录因子协同作用影响花青素合成? 既影响结构基因又影响调控基因的负转录因子间是否存在一定的联系? 导入结构基因或编码MYB/bHLH型转录因子的基因可以明显提高花青素的含量, 也是花青素代谢工程中常见的策略, 过量表达WDR蛋白或者抑制负调控基因是否也能在花青素代谢工程中发挥作用, 导致花青素含量提高? 总而言之, 解决这些问题将有助于阐明花青素的合成及其调控模式, 有助于更好地采用代谢工程的方法调控植物花青素途径, 改良植物品质, 提高有益的花青素成分和含量, 创造有价值的产品, 进而造福于人类。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1093/mp/ssp118URLPMID:20118183

Previous results indicated that in grapevine (Vitis vinifera), regulation of the flavonoid pathway genes by MYB transcription factors depends on their interaction with basic helix-loop-helix proteins (bHLHs). The present study describes the first functional characterization of a bHLH factor from grapevine named VvMYC1. This transcription factor is phylogenetically related to Arabidopsis bHLH proteins, which participate in the control of flavonoid biosynthesis and epidermal cell fate. Transient promoter and yeast two-hybrid assays demonstrated that VvMYC1 physically interacts with MYB5a, MYB5b, MYBA1/A2, and MYBPA1 to induce promoters of flavonoid pathway genes involved in anthocyanin and/or proanthocyanidin (PA) synthesis. Additionally, transient promoter assays revealed that VvMYC1 is involved in feedback regulation of its own expression. Transcript levels of VvMYC1 during berry development correlate with the synthesis of anthocyanins and PAs in skins and seeds of berries, suggesting that VvMYC1 is involved in the regulation of anthocyanins and PA synthesis in these organs. Likewise, transient expression of VvMYC1 and VvMYBA1 induces anthocyanin synthesis in grapevine suspension cells. These results suggest that VvMYC1 is part of the transcriptional cascade controlling anthocyanin and PA biosynthesis in grapevine.
[本文引用: 1]
DOI:10.1007/s12374-015-0575-xURL [本文引用: 1]

Single-repeat R3 MYB transcription factors (R3 MYBs) regulate epidermal cell fate determination in Arabidopsis through a lateral inhibition mechanism. Previously we have shown that poplar R3 MYB genes regulate trichome formation when expressed in Arabidopsis. Here we report the identification and functional characterization of a poplar R3 MYB-like protein, Populus trichocarpa R3 MYB-LIKE1 (PtrRML1). So far all the MYB transcription factors identified have a highly conserved N-terminal DNA-binding domain composed of MYB repeats and a highly variable C-terminal domain. However, PtrRML1 has a single R3 MYB repeat at its C-terminal and an LxLxL repressor motif-containing N-terminal, and with amino acids about twice of that of the known R3 MYBs. PtrRML1 is localized in nuclear in transgenic Arabidopsis plants expressing PtrRML1-GFP . PtrRML1 repressed reporter gene expression in protoplasts, and it interacted with GL3 in plant cells. Expression of PtrRML1 in Arabidopsis resulted in glabrous phenotypes, increased number of root hairs, and decreased anthocyanin accumulation. Consist with these observation, the expression levels of some MBW component genes and some of their target genes involved in the regulation of epidermal cell fate determination and anthocyanin biosynthesis, including TRY , CPC , ETC1 , GL1 , GL2 , DFR , UF3GT and TT8 were reduced in the transgenic plants.
.
DOI:10.1016/j.gene.2008.07.027URLPMID:18755256 [本文引用: 1]

By analysis with microarray data, we found that a gene encoding a novel protein containing five WD40 repeats, was regulated by salt stress in rice and named as SRWD1 ( Salt responsive WD40 protein 1). By database searching, additional four SRWD1-like genes ( SRWD2鈥 SRWD5) were found in rice genome, and these five SRWD genes formed a novel WD40 subfamily. Phylogenetic analysis showed that plant SRWD proteins divided into four groups. The significant functional divergences during SRWD evolution were found. The tissue-specific and salt responsive expression profiling for SRWD genes was investigated based on microarray data. It was found that all five SRWD genes in rice were regulated by salt stress. Further, we found that SRWD1 was regulated with different patterns by salt stress in two rice cultivars responding differently to salt stress. Our study correlates WD40 proteins with salt stress in plants and provides fundamental information for the further investigation of plant SRWD proteins.
.
DOI:10.3389/fpls.2017.01274URLPMID:5515856

Epimediumspecies have been widely used both as traditional Chinese medicinal plants and ornamental perennials. Both flavonols, acting as the major bioactive components (BCs) and anthocyanins, predominantly contributing to the color diversity ofEpimediumflowers belong to different classes of flavonoids. It is well-acknowledged that flavonoid biosynthetic pathway is predominantly regulated by R2R3-MYB transcription factor (TF) as well as bHLH TF and WD40 protein at the transcriptional level.MYBTFs specifically regulating anthocyanin or flavonol biosynthetic pathway have been already isolated and functionally characterized fromEpimedium sagittatum, but aR2R3-MYBTF involved in regulating both these two pathways has not been functionally characterized to date inEpimediumplants. In this study, we report the functional characterization ofEsMYB9, aR2R3-MYBTF previously isolated fromE. sagittatum. The previous study indicated thatEsMYB9belongs to a small subfamily ofR2R3-MYBTFs containing grapeVvMYB5aandVvMYB5bTFs, which regulate flavonoid biosynthetic pathway. The present studies show that overexpression ofEsMYB9in tobacco leads to increased transcript levels of flavonoid pathway genes and increased contents of anthocyanins and flavonols. Yeast two-hybrid assay indicates that the C-terminal region ofEsMYB9contributes to the autoactivation activity, andEsMYB9interacts withEsTT8orAtTT8 bHLHregulator. Transient reporter assay shows thatEsMYB9slightly activates the expression ofEsCHS(chalcone synthase) promoter in transiently transformed leaves ofNicotiana benthamiana, but the addition ofAtTT8orEsTT8 bHLHregulator strongly enhances the transcriptional activation ofEsMYB9against five promoters of the flavonoid pathway genes exceptEsFLS(flavonol synthase). In addition, co-transformation ofEsMYB9andEsTT8in transiently transfected tobacco leaves strongly induces the expressions of flavonoid biosynthetic genes. The potential role ofEsMYB9in modulating the biosynthesis and accumulation of sucrose-induced anthocyanin and flavonol-derived BCs is also discussed. These findings suggest thatEsMYB9is a novelR2R3-MYBTF, which regulates the flavonoid biosynthetic pathway inEpimedium, but distinctly different with the anthocyanin or flavonol-specificMYBregulators identified previously inEpimediumplants.
.
DOI:10.1371/journal.pone.0070778URLPMID:23936468

Herba epimedii (Epimedium), a traditional Chinese medicine, has been widely used as a kidney tonic and antirheumatic medicine for thousands of years. The bioactive components in herba epimedii are mainly prenylated flavonol glycosides, end-products of the flavonoid pathway. Epimedium species are also used as garden plants due to the colorful flowers and leaves. Many R2R3-MYB transcription factors (TFs) have been identified to regulate the flavonoid and anthocyanin biosynthetic pathways. However, little is known about the R2R3-MYB TFs involved in regulation of the flavonoid pathway in Epimedium. Here, we reported the isolation and functional characterization of the first R2R3-MYB TF (EsMYBA1) from Epimedium sagittatum (Sieb. Et Zucc.) Maxim. Conserved domains and phylogenetic analysis showed that EsMYBA1 belonged to the subgroup 6 clade (anthocyanin-related MYB clade) of R2R3-MYB family, which includes Arabidopsis AtPAP1, apple MdMYB10 and legume MtLAP1. EsMYBA1 was preferentially expressed in leaves, especially in red leaves that contain higher content of anthocyanin. Alternative splicing of EsMYBA1 resulted in three transcripts and two of them encoded a MYB-related protein. Yeast two-hybrid and transient luciferase expression assay showed that EsMYBA1 can interact with several bHLH regulators of the flavonoid pathway and activate the promoters of dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS). In both transgenic tobacco and Arabidopsis, overexpression of EsMYBA1 induced strong anthocyanin accumulation in reproductive and/or vegetative tissues via up-regulation of the main flavonoid-related genes. Furthermore, transient expression of EsMYBA1 in E. sagittatum leaves by Agrobacterium infiltration also induced anthocyanin accumulation in the wounded area. This first functional characterization of R2R3-MYB TFs in Epimedium species will promote further studies of the flavonoid biosynthesis and regulation in medicinal plants.
DOI:10.3969/j.issn.1674-3466.2010.03.002URL [本文引用: 1]

花青素苷(anthocyanin)是决定被子植物花、果实和种皮等颜色的重要色素之一。花青素苷的合成与积累过程往往与植物发育过程密切相关,由内外因子共同控制。环境因子通过诱导植物体内花青素苷合成途径相关基因的表达来调控花青素苷的呈色反应。该文追踪了国内外相关研究,认为光是影响花青素苷呈色的主要环境因子之一,光质和光强均能在一定程度上影响花青素苷的合成,其中光质起着更为关键的作用;低温能诱导花青素苷的积累,高温则会加速花青素苷的降解;不同的糖类物质均能影响花青素苷的合成,大部分结构基因和调节基因的表达均受糖调控。关于花发育与花青素苷呈色的关系、观赏植物花色对环境因子的响应以及花青素苷抵御逆境的机理尚待深入研究。因此,综合考察花发育与植物花青素苷合成及其呈色之间的关系,特别是光周期对花发育的影响导致花青素苷合成及呈色的机理是花色研究的一个重要课题。利用环境因子调控花色将会极大地提高花卉的观赏价值。
DOI:10.11983/CBB17126URL [本文引用: 1]

随着转基因技术的日趋成熟,利用生物工程手段加快改良作物农艺性状,已经越来越显示出其巨大的应用潜力.在改良多基因调控的复杂农艺性状方面,单基因转化收效甚微,而长期以来多基因转化不仅受限于技术因素,而且在协调表达调控、代谢及修饰等一系列相关基因方面更是难于突破.近期,我国科学家首次利用自创的多基因垛叠表达系统,成功在水稻(Oryza sativa)胚乳中合成了具有抗氧化活性的花青素,在复杂性状多基因转化领域取得了突破性进展.
.
DOI:10.1111/j.1469-8137.2005.01546.xURLPMID:16313641 [本文引用: 1]

Summary Top of page Summary Introduction Materials and Methods Results Discussion Acknowledgements References 6168 High-light leaves of the evergreen herb Galax urceolata exhibit a striking color change from green to red during winter months due to anthocyanin synthesis in outermost mesophyll cells. Here we investigate three possible functions of this color change. 6168 To test the hypothesis that anthocyanins function as light attenuators, maximum photosystem II efficiency ( F v /F m ) of red and green leaves was measured during and after exposure to wavelengths either strongly or poorly absorbed by anthocyanin. To determine whether anthocyanins elevate radical-scavenging capacity, antioxidant activity of red and green leaves was assessed using the α,α-diphenyl-β-picrylhydrazyl assay. Nonstructural carbohydrate levels were analyzed to test the hypothesis that anthocyanins function as a carbon sink. 6168 Declines in F v /F m under white and green light were significantly greater for green than red leaves, but were comparable under red light. Anthocyanin content positively correlated with antioxidant activity. Although levels of anthocyanins did not appear to be related to nonstructural carbohydrate concentration, high levels of sugars may be necessary for their photoinduction. 6168 Results suggest that anthocyanins function as light attenuators and may also contribute to the antioxidant pool in winter leaves.
DOI:10.11983/CBB15059URL [本文引用: 1]

花青素是种子植物呈色的重要色素,由一系列结构基因编码的酶(CHS、CHI、F3H、F3'H、F3'5'H、DFR、ANS和3GT)催化而成,随后经过各种修饰被转运至液泡等部位储存.各类器官中差异表达的MYB、bHLH和WDR三种调控因子通过形成MBW复合体直接正调控以上结构基因的表达.这个过程涉及的基因变异常会导致植物的各种颜色变异.在生活中人们广泛利用这些变异品种,取其丰富色味.造成颜色变异的具体分子机制在很多情况下还不清楚,但日益积累的个例研究为其中的规律性提供了基础数据.该文概述了花青素的合成、转运过程及其转录调控机制,探讨了研究颜色变异品种的常用思路及方法.在总结近年工作的基础上,对生活中常见蔬菜、水果和花卉的颜色变异品种的分子机制进行了综述.
DOI:10.1105/tpc.006130URLPMID:12509522 [本文引用: 1]

In Arabidopsis, the induction of a dehydration-responsive gene, rd22, is mediated by abscisic acid (ABA). We reported previously that MYC and MYB recognition sites in the rd22 promoter region function as cis-acting elements in the drought- and ABA-induced gene expression of rd22. bHLH- and MYB-related transcription factors, rd22BP1 (renamed AtMYC2) and AtMYB2, interact specifically with the MYC and MYB recognition sites, respectively, in vitro and activate the transcription of the 尾-glucuronidase reporter gene driven by the MYC and MYB recognition sites in Arabidopsis leaf protoplasts. Here, we show that transgenic plants overexpressing AtMYC2 and/or AtMYB2 cDNAs have higher sensitivity to ABA. The ABA-induced gene expression of rd22 and AtADH1 was enhanced in these transgenic plants. Microarray analysis of the transgenic plants overexpressing both AtMYC2 and AtMYB2 cDNAs revealed that several ABA-inducible genes also are upregulated in the transgenic plants. By contrast, a Ds insertion mutant of the AtMYC2 gene was less sensitive to ABA and showed significantly decreased ABA-induced gene expression of rd22 and AtADH1. These results indicate that both AtMYC2 and AtMYB2 proteins function as transcriptional activators in ABA-inducible gene expression under drought stress in plants.
DOI:10.1139/cjpp-2016-0667URLPMID:28384410 [本文引用: 1]

Abstract Lingonberry grown in northern Manitoba, Canada, contains exceptionally high levels of anthocyanins and other polyphenols. Previous studies from our lab have shown that lingonberry anthocyanins can protect H9c2 cells from ischemia-reperfusion injury and anthocyanin-rich diets have been shown to be associated with decreased cardiovascular disease and mortality. Oxidative stress can impair function and trigger apoptosis in cardiomyocytes. This study investigated the protective effects of physiologically relevant doses of lingonberry extracts and pure anthocyanins against hydrogen-peroxide-induced cell death. Apoptosis and necrosis were detected in H9c2 cells after hydrogen peroxide treatment via flow cytometry using FLICA 660 caspase 3/7 combined with YO-PRO-1 and then confirmed with Hoechst staining and fluorescence microscopy. Each of the 3 major anthocyanins found in lingonberry (cyanidin-3-galactoside, cyanidin-3-glucoside, and cyanidin-3-arabinoside) was protective against hydrogen-peroxide-induced apoptosis in H9c2 cells at 10 ng mL -1 (20 nmol L -1 ) and restored the number of viable cells to match the control group. A combination of the 3 anthocyanins was also protective and a lingonberry extract tested at 3 concentrations produced a dose-dependent protective effect. Lingonberry anthocyanins protected cardiac cells from oxidative-stress-induced apoptosis and may have cardioprotective effects as a dietary modification.
.
DOI:10.14348/molcells.2018.2195URLPMID:29487277

Key message pap1 - D/fls1ko double mutant plants that produce substantial amounts of anthocyanin show tolerance to abiotic stress. Abstract Anthocyanins are flavonoids that are abundant in various plants and have beneficial effects on both plants and humans. Many genes in flavonoid biosynthetic pathways have been identified, including those in the MYB-bHLH-WD40 (MBW) complex. The MYB gene... [Show full abstract]
DOI:10.1046/j.1365-313X.2001.01154.xURLPMID:11722774

Summary Fruit ripening is characterized by dramatic changes in gene expression, enzymatic activities and metabolism. Although the process of ripening has been studied extensively, we still lack valuable information on how the numerous metabolic pathways are regulated and co-ordinated. In this paper we describe the characterization of FaMYB1 , a ripening regulated strawberry gene member of the MYB family of transcription factors. Flowers of transgenic tobacco lines overexpressing FaMYB1 showed a severe reduction in pigmentation. A reduction in the level of cyanidin 3-rutinoside (an anthocyanin) and of quercetin-glycosides (flavonols) was observed. Expression of late flavonoid biosynthesis genes and their enzyme activities were aversely affected by FaMYB1 overexpression. Two-hybrid assays in yeast showed that FaMYB1 could interact with other known anthocyanin regulators, but it does not act as a transcriptional activator. Interestingly, the C-terminus of FaMYB1 contains the motif pdLNL D / E Lxi G / S . This motif is contained in a region recently proposed to be involved in the repression of transcription by AtMYB4, an Arabidopsis MYB protein. Our results suggest that FaMYB1 may play a key role in regulating the biosynthesis of anthocyanins and flavonols in strawberry. It may act to repress transcription in order to balance the levels of anthocyanin pigments produced at the latter stages of strawberry fruit maturation, and/or to regulate metabolite levels in various branches of the flavonoid biosynthetic pathway.
DOI:10.4161/psb.29526URLPMID:24642943

The diversity of pigmentation patterns observed in plants occurs due to the spatial distribution and accumulation of colored compounds, which may also be associated with structural changes to the tissue. Anthocyanins are flavonoids that provide red/purple/blue coloration to plants, often forming complex patterns such as spots, stripes, and vein-associated pigmentation, particularly in flowers. These patterns are determined by the activity of MYB-bHLH-WDR (MBW) transcription factor complexes, which activate the anthocyanin biosynthesis genes, resulting in anthocyanin pigment accumulation. Recently, we established that the MBW complex controlling anthocyanin synthesis acts within a gene regulation network that is conserved within at least the Eudicots. This network involves hierarchy, reinforcement, and feedback mechanisms that allow for stringent and responsive regulation of the anthocyanin biosynthesis genes. The gene network and mobile nature of the WDR and R3-MYB proteins provide exciting new opportunities to explore the basis of pigmentation patterning, and to investigate the evolutionary history of the MBW components in land plants.
.
DOI:10.1111/j.1365-313X.2010.04465.xURLPMID:21235651 [本文引用: 2]

Abstract We present an investigation of anthocyanin regulation over the entire petunia plant, determining the mechanisms governing complex floral pigmentation patterning and environmentally induced vegetative anthocyanin synthesis. DEEP PURPLE (DPL) and PURPLE HAZE (PHZ) encode members of the R2R3-MYB transcription factor family that regulate anthocyanin synthesis in petunia, and control anthocyanin production in vegetative tissues and contribute to floral pigmentation. In addition to these two MYB factors, the basic helix-loop-helix (bHLH) factor ANTHOCYANIN1 (AN1) and WD-repeat protein AN11, are also essential for vegetative pigmentation. The induction of anthocyanins in vegetative tissues by high light was tightly correlated to the induction of transcripts for PHZ and AN1. Interestingly, transcripts for PhMYB27, a putative R2R3-MYB active repressor, were highly expressed during non-inductive shade conditions and repressed during high light. The competitive inhibitor PhMYBx (R3-MYB) was expressed under high light, which may provide feedback repression. In floral tissues DPL regulates vein-associated anthocyanin pigmentation in the flower tube, while PHZ determines light-induced anthocyanin accumulation on exposed petal surfaces (bud-blush). A model is presented suggesting how complex floral and vegetative pigmentation patterns are derived in petunia in terms of MYB, bHLH and WDR co-regulators. 2011 The Authors. The Plant Journal 2011 Blackwell Publishing Ltd.
.
[本文引用: 1]
.
[本文引用: 1]
.
DOI:10.1105/tpc.15.00476URLPMID:26410301 [本文引用: 1]

Abstract Accumulation of anthocyanins and proanthocyanidins (PAs) is limited to specific cell types and developmental stages, but little is known about how antagonistically acting transcriptional regulators work together to determine temporal and spatial patterning of pigmentation at the cellular level, especially for PAs. Here, we characterize MYB2, a transcriptional repressor regulating both anthocyanin and PA biosynthesis in the model legume Medicago truncatula. MYB2 was strongly upregulated by MYB5, a major regulator of PA biosynthesis in M. truncatula and a component of MYB-basic helix loop helix-WD40 (MBW) activator complexes. Overexpression of MYB2 abolished anthocyanin and PA accumulation in M. truncatula hairy roots and Arabidopsis thaliana seeds, respectively. Anthocyanin deposition was expanded in myb2 mutant seedlings and flowers accompanied by increased anthocyanin content. PA mainly accumulated in the epidermal layer derived from the outer integument in the M. truncatula seed coat, starting from the hilum area. The area of PA accumulation and ANTHOCYANIDIN REDUCTASE expression was expanded into the seed body at the early stage of seed development in the myb2 mutant. Genetic, biochemical, and cell biological evidence suggests that MYB2 functions as part of a multidimensional regulatory network to define the temporal and spatial pattern of anthocyanin and PA accumulation linked to developmental processes. 2015 American Society of Plant Biologists. All rights reserved.
DOI:10.1016/j.plaphy.2016.06.032URLPMID:27404131 [本文引用: 1]

61MdMYC2 is a homology ofArabidopsisMYC2 in apple, it has highly similarity in structure and function to theArabidopsisMYC2.61MdMYC2 protein is unstable and is rapidly degraded by the 26S-proteasomes pathway.61MdMYC2 is involved in anthocyanin biosynthesis and in a hypersensitization response to MeJA.
DOI:10.1038/hortres.2017.23URLPMID:5461414

Fruit development: ‘HY5’ for high-quality apples The regulatory gene HY5 plays an important role in two biochemical pathways controlling fruit quality and coloration in apple.
.
DOI:10.1007/s00122-009-1158-3URLPMID:19779693

Abstract A dominant allele at the D locus (also known as I in diploid potato) is required for the synthesis of red and purple anthocyanin pigments in tuber skin. It has previously been reported that D maps to a region of chromosome 10 that harbors one or more homologs of Petunia an2, an R2R3 MYB transcription factor that coordinately regulates the expression of multiple anthocyanin biosynthetic genes in the floral limb. To test whether D acts similarly in tuber skin, RT-PCR was used to evaluate the expression of flavanone 3-hydroxylase (f3h), dihydroflavonol 4-reductase (dfr) and flavonoid 3',5'-hydroxylase (f3'5'h). All three genes were expressed in the periderm of red- and purple-skinned clones, while dfr and f3'5'h were not expressed, and f3h was only weakly expressed, in white-skinned clones. A potato cDNA clone with similarity to an2 was isolated from an expression library prepared from red tuber skin, and an assay developed to distinguish the two alleles of this gene in a diploid potato clone known to be heterozygous Dd. One allele was observed to cosegregate with pigmented skin in an F(1) population of 136 individuals. This allele was expressed in tuber skin of red- and purple-colored progeny, but not in white tubers, while other parental alleles were not expressed in white or colored tubers. The allele was placed under the control of a doubled 35S promoter and transformed into the light red-colored cultivar D sir e, the white-skinned cultivar Bintje, and two white diploid clones known to lack the functional allele of D. Transformants accumulated pigment in tuber skin, as well as in other tissues, including young foliage, flower petals, and tuber flesh.
DOI:10.1093/pcp/pcm131URLPMID:17925311

Flower color is mainly determined by anthocyanins. Rosa hybrida lacks violet to blue flower varieties due to the absence of delphinidin-based anthocyanins, usually the major constituents of violet and blue flowers, because roses do not possess flavonoid 3',5'-hydoxylase (F3'5'H), a key enzyme for delphinidin biosynthesis. Other factors such as the presence of co-pigments and the vacuolar pH also affect flower color. We analyzed the flavonoid composition of hundreds of rose cultivars and measured the pH of their petal juice in order to select hosts of genetic transformation that would be suitable for the exclusive accumulation of delphinidin and the resulting color change toward blue. Expression of the viola F3'5'H gene in some of the selected cultivars resulted in the accumulation of a high percentage of delphinidin (up to 95%) and a novel bluish flower color. For more exclusive and dominant accumulation of delphinidin irrespective of the hosts, we down-regulated the endogenous dihydroflavonol 4-reductase (DFR) gene and overexpressed the Irisxhollandica DFR gene in addition to the viola F3'5'H gene in a rose cultivar. The resultant roses exclusively accumulated delphinidin in the petals, and the flowers had blue hues not achieved by hybridization breeding. Moreover, the ability for exclusive accumulation of delphinidin was inherited by the next generations.
.
DOI:10.1093/pcp/pcu205URLPMID:25527830

Anthocyanin and proanthocyanidin (PA) are important secondary metabolites and beneficial to human health. Their biosynthesis is induced by jasmonate (JA) treatment and regulated by MYB transcription factors (TFs). However, which and how MYB TFs regulate this process is largely unknown in apple. In this study, MdMYB9 and MdMYB11 which were induced by methyl jasmonate (MeJA) were functionally characterized. Overexpression of MdMYB9 or MdMYB11 promoted not only anthocyanin but also PA accumulation in apple calluses, and the accumulation was further enhanced by MeJA. Subsequently, yeast two-hybrid, pull-down and bimolecular fluorescence complementation assays showed that both MYB proteins interact with MdbHLH3. Moreover, Jasmonate ZIM-domain (MdJAZ) proteins interact with MdbHLH3. Furthermore, chromatin immunoprecipitation-quantitative PCR and yeast one-hybrid assays demonstrated that both MdMYB9 and MdMYB11 bind to the promoters of ANS, ANR and LAR, whereas MdbHLH3 is recruited to the promoters of MdMYB9 and MdMYB11 and regulates their transcription. In addition, transient expression assays indicated that overexpression of MdJAZ2 inhibits the recruitment of MdbHLH3 to the promoters of MdMYB9 and MdMYB11. Our findings provide new insight into the mechanism of how MeJA regulates anthocyanin and PA accumulation in apple.
DOI:10.1016/j.jplph.2012.01.015URLPMID:22405592

The abundance of anthocyanins and proanthocyanins in apples is tightly regulated by three classes of regulatory factors, MYB, bHLH and WD40 proteins, only some of which have been previously identified. In this study, we identified an apple WD40 protein (MdTTG1) that promotes the accumulation of anthocyanins. The biosynthetic genes required downstream in the flavonoid pathway were up-regulated when MdTTG1 was over-expressed in Arabidopsis. Consistent with its role as a transcriptional regulator, an MdTTG1-GFP fusion protein was observed only in the nucleus. We assayed the expression patterns of this gene in different organs and found that they were positively correlated with anthocyanin accumulation in the apple. Yeast two-hybrid and bimolecular fluorescence complementation assays demonstrated that MdTTG1 interacted with bHLH transcription factors (TFs) but not MYB protein, whereas bHLH was known to interact with MYB in apples. However, based on a ChIP assay, MdTTG1 does not appear to bind to the promoter of the anthocyanin biosynthetic genes MdDFR and MdUFGT. Taken together, these results suggest that the apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation.
.
[本文引用: 1]
.
[本文引用: 1]
DOI:10.1126/science.1095011URLPMID:15143274 [本文引用: 1]

The color of grape skins is determined by the accumulation of red plant pigments called anthocyanins. White cultivars of grape are thought to have arisen from different red cultivars by independent mutations ([ 1 ][1]), but the molecular bases of these color mutations are unknown. Myb -related genes
.
[本文引用: 1]
DOI:10.1007/s00425-011-1407-yURLPMID:21484270 [本文引用: 1]

Abstract expression driven by the NtAn1bNtAn1b, both subsets of early and late flavonoid pathway genes were up-regulated. Yeast two-hybrid assays showed that NtAn1 proteins interact with the previously characterized R2R3-MYB TF, NtAn2. The NtAn1 tAn2 complex activated the promoters of two key anthocyanin pathway genes, dihydroflavonol reductase and chalcone synthase. The promoter activation is severely repressed by dominant repressive forms of either NtAn1a or NtAn2, created by fusing the SRDX repressor domain to the TFs. Our results show that NtAn1 and NtAn2 act in concert to regulate the anthocyanin pathway in tobacco flowers and NtAn2 up-regulates NtAn1 gene expression.
DOI:10.1093/pcp/pcm066URLPMID:17526919 [本文引用: 1]

Red coloration of apple (Malus x domestica) skin is an important determinant of consumer preference and marketability. Anthocyanins are responsible for this coloration, and their accumulation is positively correlated with the expression level of anthocyanin biosynthetic genes. Regulation of expression of these genes is believed to be controlled by MYB transcription factors, and the MYB transcription factors involved in the activation of anthocyanin biosynthetic genes have been isolated in various plants. In the present study, we isolated and characterized a MYB transcription factor gene (MdMYBA) from apple skin. Characterization of MdMYBA demonstrated that (i) MdMYBA expression was specifically regulated depending on the tissue and cultivar/species; (ii) its expression level was much higher in a deep-red cultivar ('Jonathan') than in a pale-red cultivar ('Tsugaru'); (iii) when cauliflower mosaic virus 35S::MdMYBA was introduced into the cotyledons of apple seedlings by means of a transient assay, reddish-purple spots were induced, and MdMYBA also induced anthocyanin accumulation in reproductive tissues of transgenic tobacco; (iv) the expression of MdMYBA was induced by UV-B irradiation and low-temperature treatment, both of which are known to be important in the promotion of anthocyanin accumulation in apple skin; (v) MdMYBA bound specifically to an anthocyanidin synthase (MdANS) promoter region in a gel-shift assay; and (vi) MdMYBA was mapped to the near region of the BC226-STS (a1) marker for the red skin color locus (R(f)). These results suggest that MdMYBA is a key regulatory gene in anthocyanin biosynthesis in apple skin.
DOI:10.1007/s00709-015-0920-4URLPMID:26669319 [本文引用: 1]

One of the major causes of significant crop loss throughout the world is the myriad of environmental stresses including drought, salinity, cold, heavy metal toxicity, and ultraviolet-B (UV-B) rays. Plants as sessile organisms have evolved various effective mechanism which enable them to withstand this plethora of stresses. Most of such regulatory mechanisms usually follow the abscisic-acid (ABA)-dependent pathway. In this review, we have primarily focussed on the basic leucine zipper (bZIP) transcription factors (TFs) activated by the ABA-mediated signalosome. Upon perception of ABA by specialized receptors, the signal is transduced via various groups of Ser/Thr kinases, which phosphorylate the bZIP TFs. Following such post-translational modification of TFs, they are activated so that they bind to specific cis-acting sequences called abscisic-acid-responsive elements (ABREs) or GC-rich coupling elements (CE), thereby influencing the expression of their target downstream genes. Several in silico techniques have been adopted so far to predict the structural features, recognize the regulatory modification sites, undergo phylogenetic analyses, and facilitate genome-wide survey of TF under multiple stresses. Current investigations on the epigenetic regulation that controls greater accessibility of the inducible regions of DNA of the target gene to the bZIP TFs exclusively under stress situations, along with the evolved stress memory responses via genomic imprinting mechanism, have been highlighted. The potentiality of overexpression of bZIP TFs, either in a homologous or in a heterologous background, in generating transgenic plants tolerant to various abiotic stressors have also been addressed by various groups. The present review will provide a coherent documentation on the functional characterization and regulation of bZIP TFs under multiple environmental stresses, with the major goal of generating multiple-stress-tolerant plant cultivars in near future.
.
[本文引用: 1]
.
[本文引用: 2]
.
DOI:10.1371/journal.pone.0086293URLPMID:24466010

The red coloration of litchi fruit depends on the accumulation of anthocyanins. The anthocyanins level in litchi fruit varies widely among cultivars, developmental stages and environmental stimuli. Previous studies on various plant species demonstrate that anthocyanin biosynthesis is controlled at the transcriptional level. Here, we describe a litchi R2R3-MYB transcription factor gene, LcMYB1, which demonstrates a similar sequence as other known anthocyanin regulators. The transcription levels of the LcMYB1 and anthocyanin biosynthetic genes were investigated in samples with different anthocyanin levels. The expression of LcMYB1 was strongly associated with tissue anthocyanin content. LcMYB1 transcripts were only detected in anthocyanin-accumulating tissues and were positively correlated with anthocyanin accumulation in the pericarps of 12 genotypes. ABA and sunlight exposure promoted, whereas CPPU and bagging inhibited the expression of LcMYB1 and anthocyanin accumulation in the pericarp. Cis-elements associated with light responsiveness and abscisic acid responsiveness were identified in the promoter region of LcMYB1. Among the 6 structural genes tested, only LcUFGT was highly correlated with LcMYB1. These results suggest that LcMYB1 controls anthocyanin biosynthesis in litchi and LcUFGT might be the structural gene that is targeted and regulated by LcMYB1. Furthermore, the overexpression of LcMYB1 induced anthocyanin accumulation in all tissues in tobacco, confirming the function of LcMYB1 in the regulation of anthocyanin biosynthesis. The upregulation of NtAn1b in response to LcMYB1 overexpression seems to be essential for anthocyanin accumulation in the leaf and pedicel. In the reproductive tissues of transgenic tobacco, however, increased anthocyanin accumulation is independent of tobacco's endogenous MYB and bHLH transcriptional factors, but associated with the upregulation of specific structural genes.
.
DOI:10.1007/s00425-011-1438-4URLPMID:21643990

Anthocyanins are the major pigments responsible for the pomegranate ( Punica granatum L.) fruit skin color. The high variability in fruit external color in pomegranate cultivars reflects variations in anthocyanin composition. To identify genes involved in the regulation of anthocyanin biosynthesis pathway in the pomegranate fruit skin we have isolated, expressed and characterized the pomegranate homologue of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 ( TTG1 ), encoding a WD40-repeat protein. The TTG1 protein is a regulator of anthocyanins and proanthocyanidins (PAs) biosynthesis in Arabidopsis, and acts by the formation of a transcriptional regulatory complex with two other regulatory proteins: bHLH and MYB. Our results reveal that the pomegranate gene, designated PgWD40 , recovered the anthocyanin, PAs, trichome and seed coat mucilage phenotype in Arabidopsis ttg1 mutant. PgWD40 expression and anthocyanin composition in the skin were analyzed during pomegranate fruit development, in two accessions that differ in skin color intensity and timing of appearance. The results indicate high positive correlation between the total cyanidin derivatives quantity (red pigments) and the expression level of PgWD40 . Furthermore, strong correlation was found between the steady state levels of PgWD40 transcripts and the transcripts of pomegranate homologues of the structural genes PgDFR and PgLDOX . PgWD40 , PgDFR and PgLDOX expression also correlated with the expression of pomegranate homologues of the regulatory genes PgAn1 (bHLH) and PgAn2 (MYB). On the basis of our results we propose that PgWD40 is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development and that expression of PgWD40 , PgAn1 and PgAn2 in the pomegranate fruit skin is required to regulate the expression of downstream structural genes involved in the anthocyanin biosynthesis.
DOI:10.1186/1471-2229-10-107URLPMID:20540805

pAbstract/p pBackground/p pCyclamen is a popular and economically significant pot plant crop in several countries. Molecular breeding technologies provide opportunities to metabolically engineer the well-characterized flavonoid biosynthetic pathway for altered anthocyanin profile and hence the colour of the flower. Previously we reported on a genetic transformation system for cyclamen. Our aim in this study was to change pigment profiles and flower colours in cyclamen through the suppression of flavonoid 3, 5-hydroxylase, an enzyme in the flavonoid pathway that plays a determining role in the colour of anthocyanin pigments./p pResults/p pA full-length cDNA putatively identified as a itF35H /it(itCpF35H/it) was isolated from cyclamen flower tissue. Amino acid and phylogeny analyses indicated the itCpF35H /itencodes a F35H enzyme. Two cultivars of minicyclamen were transformed via itAgrobacterium tumefaciens /itwith an antisense itCpF35H /itconstruct. Flowers of the transgenic lines showed modified colour and this correlated positively with the loss of endogenous itF35H /ittranscript. Changes in observed colour were confirmed by colorimeter measurements, with an overall loss in intensity of colour (C) in the transgenic lines and a shift in hue from purple to red/pink in one cultivar. HPLC analysis showed that delphinidin-derived pigment levels were reduced in transgenic lines relative to control lines while the percentage of cyanidin-derived pigments increased. Total anthocyanin concentration was reduced up to 80% in some transgenic lines and a smaller increase in flavonol concentration was recorded. Differences were also seen in the ratio of flavonol types that accumulated./p pConclusion/p pTo our knowledge this is the first report of genetic modification of the anthocyanin pathway in the commercially important species cyclamen. The effects of suppressing a key enzyme, F35H, were wide ranging, extending from anthocyanins to other branches of the flavonoid pathway. The results illustrate the complexity involved in modifying a biosynthetic pathway with multiple branch points to different end products and provides important information for future flower colour modification experiments in cyclamen./p
.
DOI:10.1093/jxb/ern216URLPMID:2561154 [本文引用: 1]

Genetic modification of the flavonoid pathway has been used to produce novel colours and colour patterns in ornamental plants as well as to modify the nutritional and pharmaceutical properties of food crops. It has been suggested that co-ordinate control of multiple steps of the pathway with the help of regulatory genes would lead to a more predictable control of metabolic flux. Regulation of anthocyanin biosynthesis has been studied in a common ornamental plant, Gerbera hybrida (Asteraceae). An R2R3-type MYB factor, GMYB10, shares high sequence similarity and is phylogenetically grouped together with previously characterized regulators of anthocyanin pigmentation. Ectopic expression of GMYB10 leads to strongly enhanced accumulation of anthocyanin pigments as well as to an altered pigmentation pattern in transgenic gerbera plants. Anthocyanin analysis indicates that GMYB10 specifically induces cyanidin biosynthesis in undifferentiated callus and in vegetative tissues. Furthermore, in floral tissues enhanced pelargonidin production is detected. Microarray analysis using the gerbera 9K cDNA array revealed a highly predicted set of putative target genes for GMYB10 including new gene family members of both early and late biosynthetic genes of the flavonoid pathway. However, completely new candidate targets, such as a serine carboxypeptidase-like gene as well, as two new MYB domain factors, GMYB11 and GMYB12, whose exact function in phenylpropanoid biosynthesis is not clear yet, were also identified.
DOI:10.1105/tpc.106.047688URLPMID:17337630

The transcription factor LONG HYPOCOTYL5 (HY5) acts downstream of multiple families of the photoreceptors and promotes photomorphogenesis. Although it is well accepted that HY5 acts to regulate target gene expression, in vivo binding of HY5 to any of its target gene promoters has yet to be demonstrated. Here, we used a chromatin immunopre-cipitation procedure to verify suspected in vivo HY5 binding sites. We demonstrated that in vivo association of HY5 with promoter targets is not altered under distinct light qualities or during light-to-dark transition. Coupled with DNA chip hybridization using a high-density 60-nucleotide oligomer microarray that contains one probe for every 500 nucleotides over the entire Arabidopsis thaliana genome, we mapped genome-wide in vivo HY5 binding sites. This analysis showed that HY5 binds preferentially to promoter regions in vivo and revealed >3000 chromosomal sites as putative HY5 binding targets. HY5 binding targets tend to be enriched in the early light-responsive genes and transcription factor genes. Our data thus support a model in which HY5 is a high hierarchical regulator of the transcriptional cascades for photomorphogenesis.
DOI:10.2307/3871236URLPMID:11148285 [本文引用: 2]

Plants produce a wide array of natural products, many of which are likely to be useful bioactive structures. Unfortunately, these complex natural products usually occur at very low abundance and with restricted tissue distribution, thereby hindering their evaluation. Here, we report a novel approach for enhancing the accumulation of natural products based on activation tagging by Agrobacterium-mediated transformation with a T-DNA that carries cauliflower mosaic virus 35S enhancer sequences at its right border. Among 5000 Arabidopsis activation-tagged lines, we found a plant that exhibited intense purple pigmentation in many vegetative organs throughout development. This upregulation of pigmentation reflected a dominant mutation that resulted in massive activation of phenylpropanoid biosynthetic genes and enhanced accumulation of lignin, hydroxycinnamic acid esters, and flavonoids, including various anthocyanins that were responsible for the purple color. These phenotypes, caused by insertion of the viral enhancer sequences adjacent to an MYB transcription factor gene, indicate that activation tagging can overcome the stringent genetic controls regulating the accumulation of specific natural products during plant development. Our findings suggest a functional genomics approach to the biotechnological evaluation of phytochemical biodiversity through the generation of massively enriched tissue sources for drug screening and for isolating underlying regulatory and biosynthetic genes.
.
[本文引用: 1]
.
[本文引用: 1]
.
DOI:10.1007/s00299-016-2040-9URLPMID:27562381 [本文引用: 1]

Key message: pap1-D/fls1kodouble mutant plants that produce substantial amounts of anthocyanin show tolerance to abiotic stress.Abstract: Anthocyanins are flavonoids that are abundant in various plants and have beneficial effects on both plants and humans. Many genes in flavonoid biosynthetic pathways have been identified, including those in the MYB-bHLH-WD40 (MBW) complex. The MYB gene Production of Anthocyanin Pigment 1 (PAP1) plays a particularly important role in anthocyanin accumulation. PAP1 expression in many plant systems strongly increases anthocyanin levels, resulting in a dark purple color in many plant organs. In this study, we generated double mutant plants that harbor fls1ko in the pap1-D background (i.e., pap1-D/fls1ko plants), to examine whether anthocyanins can be further enhanced by blocking flavonol biosynthesis under PAP1 overexpression. We also wanted to examine whether the increased anthocyanin levels contribute to defense against osmotic stresses. The pap1-D/fls1ko mutants accumulated higher anthocyanin levels than pap1-D plants in both control and sucrose-treated conditions. However, flavonoid biosynthesis genes were slightly down-regulated in the pap1-D/fls1ko seedlings as compared to their expression in pap1-D seedlings. We also report the performance of pap1-D/fls1ko seedlings in response to plant osmotic stresses.
DOI:10.1007/s00299-016-2025-8URLPMID:27424029

A R2R3-MYB geneAaMYB2was isolated fromAnthurium andraeanum(Hort.) and was functionally characterized to be a positive transcriptional regulator for anthocyanin biosynthesis in the spathes and leaves.
DOI:10.1105/tpc.111.095232URLPMID:22427337

Traditionally, Sicilian blood oranges (Citrus sinensis) have been associated with cardiovascular health, and consumption has been shown to prevent obesity in mice fed a high-fat diet. Despite increasing consumer interest in these health-promoting attributes, production of blood oranges remains unreliable due largely to a dependency on cold for full color formation. We show that Sicilian blood orange arose by insertion of a Copia-like retrotransposon adjacent to a gene encoding Ruby, a MYB transcriptional activator of anthocyanin production. The retrotransposon controls Ruby expression, and cold dependency reflects the induction of the retroelement by stress. A blood orange of Chinese origin results from an independent insertion of a similar retrotransposon, and color formation in its fruit is also cold dependent. Our results suggest that transposition and recombination of retroelements are likely important sources of variation in Citrus.
DOI:10.1038/nbt.1506URLPMID:18953354

Abstract Dietary consumption of anthocyanins, a class of pigments produced by higher plants, has been associated with protection against a broad range of human diseases. However, anthocyanin levels in the most commonly eaten fruits and vegetables may be inadequate to confer optimal benefits. When we expressed two transcription factors from snapdragon in tomato, the fruit of the plants accumulated anthocyanins at levels substantially higher than previously reported for efforts to engineer anthocyanin accumulation in tomato and at concentrations comparable to the anthocyanin levels found in blackberries and blueberries. Expression of the two transgenes enhanced the hydrophilic antioxidant capacity of tomato fruit threefold and resulted in fruit with intense purple coloration in both peel and flesh. In a pilot test, cancer-susceptible Trp53(-/-) mice fed a diet supplemented with the high-anthocyanin tomatoes showed a significant extension of life span.
.
.
DOI:10.1111/nph.13816URLPMID:26725247 [本文引用: 1]

Summary The MYB- basic helix–loop–helix (bHLH)-WD40 complexes regulating anthocyanin and proanthocyanidin (PA) biosynthesis in plants are not fully understood. Here Medicago truncatula bHLH MtTT8 was characterized as a central component of these ternary complexes that control anthocyanin and PA biosynthesis. Mttt8 mutant seeds have a transparent testa phenotype with reduced PAs and anthocyanins. MtTT8 restores PA and anthocyanin productions in Arabidopsis tt8 mutant. Ectopic expression of MtTT8 restores anthocyanins and PAs in mttt8 plant and hairy roots and further enhances both productions in wild-type hairy roots. Transcriptomic analyses and metabolite profiling of mttt8 mutant seeds and M.02truncatula hairy roots ( mttt8 mutant, mttt8 mutant complemented with MtTT8 , or MtTT8 overexpression lines) indicate that MtTT8 regulates a subset of genes involved in PA and anthocyanin biosynthesis. MtTT8 is genetically regulated by MtLAP1, MtPAR and MtWD40-1. Combinations of MtPAR, MtLAP1, MtTT8 and MtWD40-1 activate MtTT8 promoter in yeast assay. MtTT8 interacts with these transcription factors to form regulatory complexes. MtTT8, MtWD40-1 and an MYB factor, MtPAR or MtLAP1, interacted and activated promoters of anthocyanidin reductase and anthocyanidin synthase to regulate PA and anthocyanin biosynthesis, respectively. Our results provide new insights into the complex regulation of PA and anthocyanin biosynthesis in M.02truncatula .
DOI:10.1016/S0308-8146(99)00170-3URL [本文引用: 1]

This study on anthocyanin stability and colour variation ( λ max, ε) in the pH range 1–12 during a period of 60 days storage at 10 and 23°C, was conducted on the 3-glucosides of the six common anthocyanidins. It was mostly in the alkaline region that differences in colour and stability became significant. Although it has been generally accepted that anthocyanins are stable only at low pH values, this study revealed that, for some of the anthocyanin 3-glucosides (e.g. malvidin 3-glucoside), the bluish colours were rather intense and stability relatively high in the alkaline region. Thus, they can be regarded as potential colorants for some slightly alkaline food products.
.
[本文引用: 1]
DOI:10.1104/pp.114.256172URLPMID:25659381 [本文引用: 1]

Abstract Because of the vast range of functions that phenylpropanoids possess, their synthesis requires precise spatiotemporal coordination throughout plant development and in response to the environment. The accumulation of these secondary metabolites is transcriptionally controlled by positive and negative regulators from the MYB and basic helix-loop-helix protein families. We characterized four grapevine (Vitis vinifera) R2R3-MYB proteins from the C2 repressor motif clade, all of which harbor the ethylene response factor-associated amphiphilic repression domain but differ in the presence of an additional TLLLFR repression motif found in the strong flavonoid repressor Arabidopsis (Arabidopsis thaliana) AtMYBL2. Constitutive expression of VvMYB4a and VvMYB4b in petunia (Petunia hybrida) repressed general phenylpropanoid biosynthetic genes and selectively reduced the amount of small-weight phenolic compounds. Conversely, transgenic petunia lines expressing VvMYBC2-L1 and VvMYBC2-L3 showed a severe reduction in petal anthocyanins and seed proanthocyanidins together with a higher pH of crude petal extracts. The distinct function of these regulators was further confirmed by transient expression in tobacco (Nicotiana benthamiana) leaves and grapevine plantlets. Finally, VvMYBC2-L3 was ectopically expressed in grapevine hairy roots, showing a reduction in proanthocyanidin content together with the down-regulation of structural and regulatory genes of the flavonoid pathway as revealed by a transcriptomic analysis. The physiological role of these repressors was inferred by combining the results of the functional analyses and their expression patterns in grapevine during development and in response to ultraviolet B radiation. Our results indicate that VvMYB4a and VvMYB4b may play a key role in negatively regulating the synthesis of small-weight phenolic compounds, whereas VvMYBC2-L1 and VvMYBC2-L3 may additionally fine tune flavonoid levels, balancing the inductive effects of transcriptional activators. 2015 American Society of Plant Biologists. All Rights Reserved.
DOI:10.1104/pp.112.206771URLPMID:23096157 [本文引用: 2]

Anthocyanin accumulation is coordinated in plants by a number of conserved transcription factors. In apple (Malus 3 domestica), an R2R3 MYB transcription factor has been shown to control fruit flesh and foliage anthocyanin pigmentation (MYB10) and fruit skin color (MYB1). However, the pattern of expression and allelic variation at these loci does not explain all anthocyanin-related apple phenotypes. One such example is an open-pollinated seedling of cv Sangrado that has green foliage and develops red flesh in the fruit cortex late in maturity. We used methods that combine plant breeding, molecular biology, and genomics to identify duplicated MYB transcription factors that could control this phenotype. We then demonstrated that the red-flesh cortex phenotype is associated with enhanced expression of MYB110a, a paralog of MYB10. Functional characterization of MYB110a showed that it was able to up-regulate anthocyanin biosynthesis in tobacco (Nicotiana tabacum). The chromosomal location of MYB110a is consistent with a whole-genome duplication event that occurred during the evolution of apple within the Maloideae. Both MYB10 and MYB110a have conserved function in some cultivars, but they differ in their expression pattern and response to fruit maturity.
.
DOI:10.1111/jipb.12136URLPMID:24304603

Anthocyanins are a class of products of plant secondary metabolism and are responsible for tubers color in potato. The biosynthesis of anthocyanins is a complex biological process, in which multiple genes are involved including structural genes and regulatory genes. In this study, StAN11, a WD40-repeat gene, was cloned from potato cultivar Chieftain (Solanum tuberosum L.). StAN11 (HQ599506) contained no intron and its open reading frame (ORF) was 1,029090009bp long, encoding a putative protein of 342 amino acids. In order to verify its role in anthocyanin biosynthesis, StAN11 was inserted behind the CaMV-35S promoter of pCMBIA1304 and the recombination vector was introduced into the potato cultivar D0108sir0108e plants by Agrobacterium-mediated transformation. The color of transgenic tuber skin was significantly deepened, compared to the wild-type control, which was highly consistent with the accumulation of anthocyanin and expression of StAN11 in transgenic lines tuber skin. Further analysis on the expression of Flavonone-3-hydroxylase (F3H), Dihydroflavonol reductase (DFR), Anthocyanidin synthase (ANS), and Flavonoid 3-O-glucosyl transferase (3GT) in transgenic plants revealed that only DFR was upregulated. This result suggested that StAN11 regulated anthocyanin biosynthesis in potato by controlling DFR expression and accumulation of anthocyanin could be increased through overexpression of StAN11 in the tubers with the genetic background of anthocyanin biosynthesis.
.
DOI:10.1007/s00299-015-1909-3URLPMID:26703384

Key message RsMYB1, a MYB TF of red radish origin, was characterized as a positive regulator to transcriptionally activate the anthocyanin biosynthetic machinery by itself in Arabidopsisand tobacco...
.
DOI:10.1105/tpc.1.12.1175URL [本文引用: 1]

Genetic studies in maize have identified several regulatory genes that control the tissue-specific synthesis of the purple anthocyanin pigments during development. Two such genes, R and B, exhibit extensive allelic diversity with respect to the tissue specificity and developmental timing of anthocyanin synthesis. Previous genetic studies demonstrated that certain B alleles can substitute for R function, and in these cases only one functional allele at either locus is required for pigment synthesis in the aleurone. In addition, biochemical studies have shown that both genes act on the same biosynthetic pathway, suggesting that the genes are functionally duplicate. In this report we describe DNA hybridization experiments that demonstrate that the functionally duplicate nature of B and R is reflected in DNA sequence similarity between the two genes. We took advantage of this homology and used the R genomic sequences to clone B. Two different strategies were pursued and two genomic clones isolated, a 2.5-kilobase BgIII fragment linked to the b allele in W23 inbred stocks and a 1.0-kilobase HindIII fragment linked to the B allele in CM37 stocks. Examination of several independent transposable element insertion mutations in B and revertant derivatives demonstrated that our clones recognize the functional B gene. Genomic clones representing the entire B-Peru allele were isolated, and a detailed restriction map was prepared. Using these clones we have identified a 2.2-kilobase mRNA in husks from plants containing either B-I or B-Peru alleles, but no B mRNA was detected in plants containing a b allele. The transcript is at least 100 times more abundant in strongly pigmented B-I husks than in weakly pigmented B-Peru husk tissue. Expression of functional B alleles in husk tissue correlates with the coordinate increase in mRNA levels of two structural genes of the pathway, A1 and Bz1, consistent with the postulated role of B as a regulatory gene.
.
DOI:10.1016/j.scienta.2015.08.018URL [本文引用: 1]

Metabolism of anthocyanin in Chrysanthemum (Chrysanthemum morifolium Ramat.) is catalyzed by several biosynthetic enzymes, however, the underlying transcriptional regulatory mechanisms remain unknown. In the present research, four MYB transcription factors, CmMYB3-6, were isolated from ‘Amadea’ Chrysanthemum, by RNA-seq and RACE. Among the four CmMYBs, CmMYB3 and CmMYB6 were expressed concurrently with the expression of biosynthetic genes and accumulation of anthocyanin during flower development. In order to study the transcription regulatory role of CmMYB3 and CmMYB6 in anthocyanin biosynthesis, the promoter region of CmDFR was isolated. Dual luciferase assay showed that CmMYB6 significantly activated the CmDFR promoter more than 8-fold. Furthermore, the combination of CmMYB6 and MrbHLH1 (from Myrica rubra involved in anthocyanin biosynthesis) resulted in approximately 34-fold induction of the CmDFR promoter. Using a transient over-expression system in Nicotiana tabacum leaves, CmMYB6 and MrbHLH1 co-expression lead to transient anthocyanin accumulation. Thus, CmMYB6 is proposed as a novel transcription factor in Chrysanthemum anthocyanin biosynthesis.
DOI:10.1007/s11240-013-0361-8URL [本文引用: 1]

Anthocyanins, being important for both plant functions and human health, were transcriptionally regulated by the MYB–bHLH–WD40 transcription complex. The key MYB regulator for Chinese bayberry (...
.
[本文引用: 1]
.
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
.
DOI:10.1073/pnas.83.24.9631URLPMID:3025847 [本文引用: 1]

The C1 gene of maize plays a regulatory role in the production of anthocyanin pigments in the aleurone layer of the endosperm. As an initial step toward understanding the molecular details of how C1 controls pigment biosynthesis, we cloned the C1 gene. This was accomplished by first cloning a mutable allele of C1, c1-m5, which contains the transposable element Spm. A combination of molecular and genetic analysis was used to identify the Spm at the C1 locus. Individual genomic DNAs from a population in which the c1-mutable phenotype was segregating with the recessive c1 phenotype were digested with methyl-sensitive restriction enzymes and probed with a small DNA fragment derived from a defective Spm. One Sal I restriction fragment complementary to the Spm probe was shown to be present in the DNA of individuals with the c1-m5 phenotype but absent from DNA of individuals with a recessive c1 phenotype. Subsequent cloning and restriction analysis of this fragment revealed sequences flanking the Spm that proved to be C1-specific. A DNA fragment derived from the flanking sequences was then used as a probe to clone the wild-type C1 gene and several additional alleles of C1, including one stable recessive, two mutations caused by Ds insertions, one mutation induced by insertion of a defective Spm, and two dominant mutations, C1-S and C1-I. RNA blot hybridization analysis of three C1 alleles indicates that C1 regulation of the Bz1 and A1 structural genes in the anthocyanin biosynthetic pathway is at the transcriptional level.
.
DOI:10.2307/3869695URLPMID:8305872 [本文引用: 1]

Genetic studies in maize have identified several regulatory genes that control the tissue-specific synthesis of purple anthocyanin pigments in the plant. c1 regulates pigmentation in the aleurone layer of the kernel, whereas pigmentation in the vegetative and floral tissues of the plant body depends on pl. c1 encodes a protein with the structural features of eukaryotic transcription factors and functions to control the accumulation of transcripts for the anthocyanin biosynthetic genes. Previous genetic and molecular observations have prompted the hypothesis that c1 and pl are functionally duplicate, in that they control the same set of anthocyanin structural genes but in distinct parts of the plant. Here, we show that this proposed functional similarity is reflected by DNA sequence homology between c1 and pl. Using a c1 DNA fragment as a hybridization probe, genomic and cDNA clones for pl were isolated. Comparison of pl and c1 cDNA sequences revealed that the genes encode proteins with 90% or more amino acid identity in the amino- and carboxylterminal domains that are known to be important for the regulatory function of the C1 protein. Consistent with the idea that the pl gene product also acts as a transcriptional activator is our finding that a functional pl allele is required for the transcription of at least three structural genes in the anthocyanin biosynthetic pathway.
.
DOI:10.2307/3869696URLPMID:8305873 [本文引用: 1]

The pl gene encodes a regulatory protein that controls the transcription of a number of structural genes of the anthocyanin biosynthetic pathway in maize. pl alleles have been classified phenotypically into two categories: dominant (Pl) alleles lead to intense, light-independent pigmentation in vegetative and floral organs of the plant; recessive "sun-red" alleles (pl) lead to light-dependent red pigmentation in which only tissues exposed to light become pigmented. Based on these observations, two alternate pathways leading to anthocyanin synthesis in the plant have been proposed: one requiring light and the other bypassing the light requirement through the action of Pl. To evaluate this hypothesis, we have analyzed light-independent and light-dependent alleles of pl. Sequence analysis revealed that the two types of alleles have very distinct promoters but have the capacity to encode very similar proteins. The protein encoded by one recessive allele was shown to be functional in transient assays. Measurements of husk mRNA levels by quantitative polymerase chain reaction showed that sun-red pl alleles are expressed at much lower levels than a Pl allele, but their expression is increased approximately sixfold by exposure to light. These results lead to the conclusion that the sun-red pl alleles are not null; instead, they synthesize functional mRNA and protein. We propose that the light-dependent pigmentation observed in pl plants is the result of a threshold effect in which light exposure boosts pl mRNA expression past a crucial level necessary to generate sufficient PL protein molecules to activate transcription of the anthocyanin structural genes.
DOI:10.1111/tpj.12153URLPMID:23425305

Anthocyanins are natural pigments that accumulate only in light-grown and not in dark-grown Arabidopsis plants. Repression of anthocyanin accumulation in darkness requires the CONSTITUTIVELY PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 (COP1/SPA) ubiquitin ligase, as cop1 and spa mutants produce anthocyanins also in the dark. Here, we show that COP1 and SPA proteins interact with the myeloblastosis (MYB) transcription factors PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP)1 and PAP2, two members of a small protein family that is required for anthocyanin accumulation and for the expression of structural genes in the anthocyanin biosynthesis pathway. The increased anthocyanin levels in cop1 mutants requires the PAP1 gene family, indicating that COP1 functions upstream of the PAP1 gene family. PAP1 and PAP2 proteins are degraded in the dark and this degradation is dependent on the proteasome and on COP1. Hence, the light requirement for anthocyanin biosynthesis results, at least in part, from the light-mediated stabilization of PAP1 and PAP2. Consistent with this conclusion, moderate overexpression of PAP1 leads to an increase in anthocyanin levels only in the light and not in darkness. Here we show that SPA genes are also required for reducing PAP1 and PAP2 transcript levels in dark-grown seedlings. Taken together, these results indicate that the COP1/SPA complex affects PAP1 and PAP2 both transcriptionally and post-translationally. Thus, our findings have identified mechanisms via which the COP1/SPA complex controls anthocyanin levels in Arabidopsis that may be useful for applications in biotechnology directed towards increasing anthocyanin content in plants.
.
DOI:10.1111/j.1365-313X.1991.00037.xURLPMID:1844879

The intensity and pattern of anthocyanin biosynthesis in Antirrhinum flowers is controlled by several genes. We have isolated six cDNA clones encoding enzymes in the pathway committed to flavonoid biosynthesis and used these to assay how the regulatory genes that modify colour pattern affect the expression of bio-synthetic genes. The biosynthetic genes of the later part of the pathway appear to be co-ordinately regulated by two genes, Delila (Del) , and Eluta (El) , while the early steps (which also lead to flavone synthesis) are controlled differently. This division of control is not the same as control of anthocyanin biosynthesis by the regulatory genes R(S) and C1 in maize aleurone, and may result from the adaptive significance of different flavonoids in flowers and seeds, reflecting their attractiveness to insects and mammals respectively. EI and del are probably involved in transcriptional control and both genes appear to be able to repress expression of some biosynthetic genes and activate expression of others.
DOI:10.1111/j.1365-313X.2008.03565.xURLPMID:18532977 [本文引用: 1]

In Arabidopsis, MYB transcription factors regulate flavonoid biosynthesis via the formation of protein complexes with a basic helix-loop-helix (bHLH) transcription factor and a WD40 repeat protein. Several R3-type single-MYB proteins (R3-MYB), such as CPC and TRY, act as negative regulators of the development of epidermal cells. However, such regulators of flavonoid biosynthesis have not yet been reported, to our knowledge. We show here that an R3-MYB protein, AtMYBL2, acts as a transcriptional repressor and negatively regulates the biosynthesis of anthocyanin in Arabidopsis. In an AtMYBL2 knockout line ( mybl2 ), the expression of the DFR and TT8 genes was enhanced and resulted in the ectopic accumulation of anthocyanin, while ectopic expression of AtMYBL2 or of a chimeric repressor that is a dominant negative form of AtMYBL2 suppressed the expression of DFR and TT8, and the biosynthesis of anthocyanin. The expression of AtMYBL2 was detected in various tissues but not in those in which anthocyanin accumulated or TT8 was expressed. The minimal repression domain of AtMYBL2 was found to be the six amino acids (TLLLFR) at the carboxyl terminus, and TLLLFR appears to be a novel repression motif that is different from the ERF-associated amphiphilic repression (EAR) motif. The defective phenotype of mybl2 mutants was complemented by 35S:AtMYBL2 but enhanced by a truncated form of AtMYBL2 from which the repression domain had been deleted. AtMYBL2 bound directly to TT8 protein and this complex suppressed the expression of DFR and TT8 . The repression activity of AtMYBL2 appears to play a critical role in the regulation of anthocyanin biosynthesis.
.
DOI:10.1038/330677a0URLPMID:3683587

Petunia hybrida is one of the classical subjects of investigation in plants in which the pathway of anthocyanin biosynthesis has been analysed genetically and biochemically. In petunia cyanidin- and delphinidin-derivatives, but no pelargonidin-derivatives are produced as pigments. This is due to the substrate specificity of the dihydroflavonol 4-reductase of petunia, which cannot reduce dihy-drokaempferol. The petunia mutant RL01, which accumulates dihydrokaempferol, shows no flower pigmentation. RL01 served as a recipient for the transfer of the A 1 gene of Zea mays encoding dihydroquercetin 4-reductase, which can reduce dihydrokaempferol and thereby provided the intermediate for pelargonidin biosynthesis. Transformation of RL01 with a vector p35Al, containing the A 1 -complementary DNA behind the 35S promotor leads to red flowers of the pelargonidin-type. Thus a new flower pigmentation pathway has been established in these plants.
[本文引用: 1]
DOI:10.1016/j.optlastec.2008.12.018URL [本文引用: 1]

While there is a range of colours found in plants the predominant colour is green. Pigments in plants have several roles e.g. photosynthesis and signalling. If colour is to be used as a signal then it must stand out from green. However, one should be aware that there are also coloured compounds where we have not yet fully investigated the role of colour in their functions hey may have roles in, for example, defence or heat exchange. In this paper, we will describe the basic chemistry of the major pigments found in plants and especially floral pigments. We will then discuss their locations in parts of the flower (such as sepals, petals, pollen and nectar), the cells in which they are found and their sub-cellular locations. Floral pigments have a large role to play in pollination of flowers by animals. They can and are modified in many ways during the development of flowers in nature, for example, at emergence and post-pollination. There are a range of biochemical mechanisms of colour change both within flowers and in isolated pigments. Some of the factors influencing colour are temperature, co-pigments, pH, metals, sugars, anthocyanin stacking and cell shape. There is a renewed interest in analysing floral pigments and how they are modified partly because of advances in recombinant DNA technologies, but also because of pollinators and their significance to biodiversity and for evolutionary studies. There is continued strong interest from the horticultural industry for the introduction of new colours e.g. the blue rose and for the exploitation of natural dyes. Funding in this area may impact future research in a potentially beneficial way but it must not deflect us from science-based conservation.
DOI:10.1007/s13562-012-0134-1URL [本文引用: 1]

AbstractPlants have been gifted with intricate regulatory networks to carry on with their sessile life form. Often such networks involve delicate association between various proteins. The WD40 proteins, which are present abundantly in several eukaryotes, act as scaffolding molecules assisting proper activity of other proteins. They comprise several stretches of 44鈥60 amino acid residues and often terminate with a WD dipeptide. They function in several cellular, metabolic and molecular pathways, biologically playing important roles in plant development and also during stress signaling. Moreover, some WD40 (named DWD) proteins also function as substrate receptors in Cullin4 RING dependent E3 ubiquitin ligase mediated proteosomal degradation and DNA damage repair mechanism. In this review, we have discussed the various aspects of these proteins that affect their highly diversified functions in plants.
.
[本文引用: 1]
DOI:10.1093/oxfordjournals.molbev.a026079URLPMID:10605108 [本文引用: 1]

Abstract Multidomain proteins usually contain several conserved and apparently independently evolved domains. As a result, classifications based on only a single small domain may obscure the true evolutionary relationships of the proteins. The current classification of basic helix-loop-helix (bHLH) domain-containing proteins is based on the conserved bHLH domain alone. Herein, we explore whether sequence homology and, therefore, evolutionary relationships can be detected among the flanking or non-bHLH components of the amino acid sequences of 122 bHLH proteins. These 122 proteins were the same proteins previously used to construct the existing classification of the bHLH-domain-containing proteins. Several possible scenarios are examined in order to explain the observed patterns of sequence divergence, including (1) monophyly, (2) convergent evolution, (3) addition of functional components to the bHLH domain, and (4) modular evolution with domain shuffling. Drawing on several lines of evidence, we suggest that modular evolution by domain shuffling may have played an important role in the evolution of this large group of transcriptional regulators.
.
DOI:10.1093/pcp/pcj012URLPMID:16446312

The transcriptional regulators for anthocyanin biosynthesis include members of proteins containing an R2R3-MYB domain, a bHLH (basic helix-loop-helix) domain and conserved WD40 repeats (WDRs). Spacial and temporal expression of the structural genes encoding the enzymes for anthocyanin biosynthesis is thought to be determined by combinations of the R2R3-MYB, bHLH and WDR factors and their interactions. While the wild-type Japanese morning glory () exhibits blue flowers with colored stems and dark-brown seeds, the mutants display white flowers with red stems and colored seeds, and the mutants exhibit white flowers with green stems and ivory seeds. Here, we characterize the tissue-specific expression of three genes, three genes and two genes in . We also show that the recessive and alleles are frameshift mutations caused by a 2bp deletion and 7bp insertions in the genes for the R2R3-MYB and WDR transcriptional regulators designated as InMYB1 and InWDR1, respectively. In addition to defects in flower, stem and seed pigmentations, the mutants were found to show reduced trichome formation in seeds but to produce leaf and stem trichomes and root hairs normally. Except for the gene for chalcone synthase E in the mutant, all structural genes tested were coordinately reduced in both and mutant flower limbs. However, slight but significant expression of the genes for chalcone synthase D, chalcone isomerase and flavanone 3-hydroxylase in the pathway for flavonol biosynthesis was detectable in and mutants, whereas no such residual expression could be observed in other genes involved in the later anthocyanin biosynthesis pathway. The biological roles of the and genes in epidermal traits and their evolutionary implications are also discussed.
DOI:10.1111/tpj.12388URLPMID:4282528 [本文引用: 1]

The notion that plants use specialized metabolism to protect against environmental stresses needs to be experimentally proven by addressing the question of whether stress tolerance by specialized metabolism is directly due to metabolites such as flavonoids. We report that flavonoids with radical scavenging activity mitigate against oxidative and drought stress in Arabidopsis thaliana. Metabolome and transcriptome profiling and experiments with oxidative and drought stress in wild-type, single overexpressors of MYB12/PFG1 (PRODUCTION OF FLAVONOL GLYCOSIDES1) or MYB75/PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT1), double overexpressors of MYB12 and PAP1, transparent testa4 (tt4) as a flavonoid-deficient mutant, and flavonoid-deficient MYB12 or PAP1 overexpressing lines (obtained by crossing tt4 and the individual MYB overexpressor) demonstrated that flavonoid overaccumulation was key to enhanced tolerance to such stresses. Antioxidative activity assays using 2,2-diphenyl-1-picrylhydrazyl, methyl viologen, and 3,3-diaminobenzidine clearly showed that anthocyanin overaccumulation with strong in vitro antioxidative activity mitigated the accumulation of reactive oxygen species in vivo under oxidative and drought stress. These data confirm the usefulness of flavonoids for enhancing both biotic and abiotic stress tolerance in crops.
DOI:10.1016/j.scienta.2009.01.008URL

Transcription of anthocyanin biosynthetic genes is usually controlled by transcription factors such as basic-helix-loop-helix (bHLH) and R2R3-MYB. To know the regulatory mechanisms of anthocyanin accumulation in Asiatic hybrid lily ( Lilium spp.), cDNA clones encoding bHLH transcription factors were isolated from tepals and their expression was examined in this study. This is the first report about cDNAs for bHLH genes among the species in monocots other than Gramineae. Two cDNAs corresponding to LhbHLH1 and LhbHLH2 genes were isolated; the former encoded the protein having the similarity to AmDELILA and PhJAF13 and the latter one did to PhAN1. LhbHLH1 and LhbHLH2 were expressed in tepals, stems and leaves. In filaments and pistils, only LhbHLH2 was transcribed. During tepal development, transcriptions of LhbHLH genes were always detected, and the peak of LhbHLH2 expression preceded the peak of LhDFR expression. When lily buds were exposed to light, anthocyanin accumulation was induced in leaves and peaked at 4 days after light exposure. The transcription of LhbHLH2 peaked at 2 days after light exposure and decreased after that. LhbHLH1 transcription was affected by light to a lesser extent than LhbHLH2 at 2 days. These expressional changes preceded the change of anthocyanin amount. Similarly in tepals, both anthocyanin amount and the transcription of LhDFR and LhbHLH2 decreased in the dark after 6 days, but LhbHLH1 transcription was not affected. These results suggest that LhbHLH1 and LhbHLH2, especially LhbHLH2, are involved in anthocyanin biosynthesis, and that response to light exposure was different between LhbHLH1 and LhbHLH2.
DOI:10.1093/pcp/pcn163URLPMID:18974195 [本文引用: 2]

Gentian plants have vivid blue-colored flowers, caused by accumulation of a polyacylated anthocyanin 'gentiodelphin'. We previously performed expression analysis of gentiodelphin biosynthetic genes, and hypothesized that the white-flowered gentian cultivar 'Polarno White' might have resulted from the mutation of certain regulatory factors responsible for anthocyanin biosynthesis in flower petals. In this study, we isolated 26 R2R3-MYB gene fragments including four full-length cDNAs (GtMYB2a, GtMYB2b, GtMYB3 and GtMYB4) and one basic helix-loop-helix (bHLH) gene (GtbHLH1) from blue-flowered gentian by degenerate PCR and rapid amplification of cDNA ends (RACE). Phylogenetic tree analysis showed that GtMYB3 was categorized into a clade involved in anthocyanin biosynthesis including petunia AN2 and Arabidopsis PAP1. On the other hand, GtbHLH1 exhibited high identity with petunia AN1 based on both phylogenetic and genomic structural analyses. Temporal profiles of GtMYB3 and GtbHLH1 transcript levels corresponded well with those of gentiodelphin accumulation and their biosynthetic genes in petals. Yeast two-hybrid analysis showed that GtbHLH1 interacted with GtMYB3. Moreover, transient expression analysis indicated that the co-expression of GtMYB3 and GtbHLH1 could enhance the promoter activities of late anthocyanin biosynthetic genes in tobacco BY2 cells. We also revealed that in cv. 'Polarno White' the GtMYB3 genes were mutated by insertions of transposable elements or uncharacterized sequences, indicating that the white coloration was caused by GtMYB3 mutation. These results strongly suggested that GtMYB3 and GtbHLH1 are involved in the regulation of gentiodelphin biosynthesis in gentian flowers.
DOI:10.1016/j.phytochem.2013.12.006URLPMID:24388610 [本文引用: 1]

Ternary complexes consisting of a R2R3-MYB, a bHLH and a WD40 protein (MBW complexes) regulate trichome formation and anthocyanin synthesis in plants. Small R3-MYBs interact with the MBW complexes to exert a negative feedback, and thereby participate in regulation of epidermal cell fate, for example trichome numbers and clustering in leaves. In Arabidopsis thaliana, GL3, a bHLH transcription factor, is important in the MBW complex regulating trichome formation as well as in the MBW complex induced by nitrogen depletion and promoting anthocyanin formation. The small R3-MYBs: CPC, TRY, ETC1, ETC2, ETC3/CPL3, TCL1, MYBL2, are all known to interact with GL3. We here investigated these R3-MYBs in leaves of Arabidopsis rosette stage plants under nitrogen depletion to examine if the small MYBs would interfere with anthocyanin accumulation in plants under normal (autotrophic) growth conditions. CPC expression was enhanced two-fold in response to nitrogen depletion, and ETC3/CPL3 expression was enhanced by almost an order of magnitude (9). Knockout of ETC3/CPL3 did not influence anthocyanin accumulation, but the results establish ETC3/CPL3 as a nitrate regulated gene and a putative candidate for being involved in nitrate status signaling and root development. Other R3-MYBs tested were not significantly influenced by nitrogen depletion. In conclusion, only CPC expression increased and clearly exerted a negative feedback on anthocyanin accumulation during nitrogen starvation in rosette leaves.
.
DOI:10.1111/tpj.13077URLPMID:26576746 [本文引用: 1]

Summary Photomorphogenesis is an essential program in plant development. This process is effected by the balanced cooperation of many factors under light and dark conditions. In a previous study, we showed that MYB hypocotyl elongation-related (MYBH) is involved in cell elongation. To expand our understanding of MYBH function, we performed a yeast two-hybrid assay and identified an MYB-like Domain transcription factor (MYBD). In this study, we investigated the function of MYBD, which is an MYBH homolog involved in anthocyanin accumulation. MYBD expression increased in response to light or cytokinin, and MYBD enhanced anthocyanin biosynthesis via repression of MYBL2 , which encodes a transcription factor that has a negative effect on this process. In addition, MYBD binding in vivo to the MYBL2 promoter and the lower level of histone H3K9 acetylation at the upstream region of MYBL2 in MYBD over-expressing plants in comparison with wild-type plants imply that MYBD represses MYBL2 expression via an epigenetic mechanism. HY5 directly binds to the MYBD promoter, which indicates that MYBD acts on HY5-downstream in light- or cytokinin-triggered signaling pathways, leading to anthocyanin accumulation. Our results suggest that, although MYBD and MYBH are homologs, they act in opposite ways during plant photomorphogenesis, and these functions should be examined in further studies.
.
DOI:10.1007/s00425-009-1095-zURLPMID:20183921

Chinese bayberry (Myrica rubra) is a fruit crop with cultivars producing fruit ranging from white (Shuijing, SJ) to red (Dongkui, DK) and dark red-purple (Biqi, BQ), as a result of different levels of anthocyanin accumulation. Genes encoding the anthocyanin biosynthesis enzymes chalcone synthase, chalcone isomerase, flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS) and UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT), as well as MrMYB1, a R2R3 MYB transcription factor homologous to known activators of anthocyanin biosynthesis, were isolated from ripe fruit of BQ. Differences in mRNA abundance of MrF3H, MrF3′H, MrDFR1, MrANS and MrUFGT were highly correlated with differential accumulation of anthocyanins between cultivars, suggesting coordinated regulation by transcription factors. The transcript level of MrMYB1 was strongly associated with the anthocyanin content in ripe fruit of the three cultivars, as well as different anthocyanin containing tissues of BQ fruit. Fruit bagging strongly inhibited anthocyanin accumulation in fruit as well as the expression of all anthocyanin biosynthetic genes and MrMYB1. Overexpression of MrMYB1 stimulated both anthocyanin accumulation and activated an Arabidopsis-DFR promoter in tobacco (Nicotiana tabacum). MrMYB1d, an allele with a 1 bp deletion at nucleotide 30 of coding sequence, was observed in SJ and DK fruit, suggesting that a nonsense mutation of the MYB1 protein may be responsible for no or low expression of MYB1 in the white and red fruit. These results show that coordinated expression of multiple biosynthetic genes is involved in anthocyanin accumulation in Chinese bayberry fruit, and this is regulated by MrMYB1.
DOI:10.1126/sciadv.1602785URLPMID:28782017

Coexpression of two anthocyanin modification genes elicits blue flower coloration through interaction with colorless flavonoids. Various colored cultivars of ornamental flowers have been bred by hybridization and mutation breeding; however, the generation of blue flowers for major cut flower plants, such as roses, chrysanthemums, and carnations, has not been achieved by conventional breeding or genetic engineering. Most blue-hued flowers contain delphinidin-based anthocyanins; therefore, delphinidin-producing carnation, rose, and chrysanthemum flowers have been generated by overexpression of the gene encoding flavonoid 3′,5′-hydroxylase (F3′5′H), the key enzyme for delphinidin biosynthesis. Even so, the flowers are purple/violet rather than blue. To generate true blue flowers, blue pigments, such as polyacylated anthocyanins and metal complexes, must be introduced by metabolic engineering; however, introducing and controlling multiple transgenes in plants are complicated processes. We succeeded in generating blue chrysanthemum flowers by introduction of butterfly pea UDP (uridine diphosphate)–glucose:anthocyanin 3′,5′-O-glucosyltransferase gene, in addition to the expression of the Canterbury bellsF3′5′H. Newly synthesized 3′,5′-diglucosylated delphinidin-based anthocyanins exhibited a violet color under the weakly acidic pH conditions of flower petal juice and showed a blue color only through intermolecular association, termed “copigmentation,” with flavone glucosides in planta. Thus, we achieved the development of blue color by a two-step modification of the anthocyanin structure. This simple method is a promising approach to generate blue flowers in various ornamental plants by metabolic engineering.
DOI:10.1093/jxb/err216URLPMID:21765172

Dahlias (Dahlia variabilis) exhibit a wide range of flower colours because of accumulation of anthocyanin and other flavonoids in their ray florets. Two lateral mutants were used that spontaneously occurred in ‘Michael J’ (MJW) which has yellow ray florets with orange variegation. MJOr, a bud mutant producing completely orange ray florets, accumulates anthocyanins, flavones, and butein, and MJY, another mutant producing completely yellow ray florets, accumulates flavones and butein. Reverse transcription–PCR analysis showed that expression ofchalcone synthase 1(DvCHS1),flavanone 3-hydroxylase(DvF3H),dihydroflavonol 4-reductase(DvDFR),anthocyanidin synthase(DvANS), andDvIVSencoding a basic helix–loop–helix transcription factor were suppressed, whereas that ofchalcone isomerase(DvCHI) andDvCHS2, anotherCHSwith 69% nucleotide identity withDvCHS1, was not suppressed in the yellow ray florets of MJY. A 5.465kb CACTA superfamily transposable element,transposable element of Dahlia variabilis 1(Tdv1), was found in the fourth intron of theDvIVSgene of MJW and MJY, and footprints ofTdv1were detected in the variegated flowers of MJW. It is shown that only one type ofDvIVSgene was expressed in MJOr, whereas these plants are likely to have three types of theDvIVSgene. On the basis of these results, the mechanism regulating the formation of orange and yellow ray florets in dahlia is discussed.
.
[本文引用: 1]
.
DOI:10.1007/s00425-009-0917-3URLPMID:19306102 [本文引用: 1]

Mangosteen (Garcinia mangostana L.) fruit undergo rapid red colour development, both on the tree and after harvest, resulting in high anthocyanin production in the pericarp. Here, we report the isolation of three full-length mangosteen MYB transcription factors (GmMYB1, GmMYB7 and GmMYB10) and all the anthocyanin biosynthetic pathway genes (GmPal to GmUFGT). Phylogenetic analysis at the protein level of the R2R3-MYB transcription factor family showed GmMYB10 had a high degree of similarity with production of anthocyanin pigment1 in Arabidopsis and as well as sequences from other plant species related to the elevation of anthocyanin pigmentation. In transient transactivation assays, GmMYB10, co-expressed with AtbHLH2, strongly activated the GmDFR and AtDFR promoters. Transcripts of GmMYB10 and GmUFGT were highly abundant with onset of pigmentation and subsequently during red colouration. Our results suggest that GmMYB10 plays an important role in regulating anthocyanin biosynthesis both on the tree and after harvest, while GmUFGT may be a key biosynthetic gene in mangosteen pigmentation. The expression patterns of GmMYB10 and GmUFGT correlated with ethylene production that increased linearly until stage 5 (dark purple) and decreased thereafter. 1-Methycyclopropene (1-MCP) clearly delayed red colouration with resulting down-regulation of GmMYB10. These results suggest that the effect of ethylene on anthocyanin biosynthesis may be via the regulation of GmMYB10 expression.
.
[本文引用: 1]
.
DOI:10.1007/s00299-008-0521-1URLPMID:18317777

The MYB transcription factors play important roles in the regulation of many secondary metabolites at the transcriptional level. We evaluated the possible roles of theArabidopsisR2R3-MYB transcription factors in flavonoid biosynthesis because they are induced by UV-B irradiation but their associated phenotypes are largely unexplored. We isolated their genes by RACE-PCR, and performed transgenic approach and metabolite analyses in lettuce (Lactuca sativa). We found that one member of this protein family, AtMYB60, inhibits anthocyanin biosynthesis in the lettuce plant. Wild-type lettuce normally accumulates anthocyanin, predominantly cyanidin and traces of delphinidin, and develops a red pigmentation. However, the production and accumulation of anthocyanin pigments inAtMYB60-overexpressing lettuce was inhibited. Using RT-PCR analysis, we also identified the complete absence or reduction of dihydroflavonol 4-reductase (DFR) transcripts inAtMYB60-overexpressing lettuce (AtMYB60-117 and AtMYB60-112 lines). The correlation between the overexpression ofAtMYB60and the inhibition of anthocyanin accumulation suggests that the transcription factorAtMYB60 controls anthocyanin biosynthesis in the lettuce leaf. Clarification of the roles of theAtMYB60transcription factor will facilitate further studies and provide genetic tools to better understand the regulation in plants of the genes controlled by the MYB-type transcription factors. Furthermore, the characterization of AtMYB60 has implications for the development of new varieties of lettuce and other commercially important plants with metabolic engineering approaches.
.
DOI:10.1111/j.1365-313X.2006.02988.xURLPMID:17270013

The transcriptional regulators for anthocyanin pigmentation include proteins containing R2R3-MYB domains, bHLH domains and conserved WD40 repeats, and their interactions determine the set of genes to be expressed. Spontaneous ivory seed ( ivs ) mutants of Ipomoea purpurea displaying pale pigmented flowers and ivory seeds are caused by insertions of DNA transposons into the bHLH2 gene that encodes a bHLH transcriptional regulator. A partial reduction in the expression of all structural genes encoding enzymes for anthocyanin biosynthesis was observed in the young flower buds of these ivs mutants. The DFR-B and ANS transcripts were completely abolished in the ivs seed coats, whereas the early biosynthetic genes for flavonol biosynthesis remained active. The production and accumulation of both proanthocyanidin and phytomelanin pigments in the ivory seed coats were drastically reduced. Moreover, the unbranched trichomes in the ivory seeds were smaller in size and fewer in number than those in the wild-type dark-brown seeds, and the surface of the epidermis without trichomes in the dark-brown seeds looked rougher, due to the protruding tangential walls, than that in the ivory seeds. Although the I. purpurea bHLH2 gene is the most closely related to the petunia AN1 gene, whose mutation is known to confer white flowers and to be deficient in acidification of their vacuoles, the vacuolar alkalization in the epidermal flower limbs of I. purpurea ivs mutants appears to occur normally. These results are discussed with regard to the function of bHLH transcriptional regulators controlling flower and seed pigmentation as well as other epidermal traits.
.
DOI:10.1007/s00425-017-2667-yURLPMID:28299441

This work shows that, in tobacco, the ectopic expression ofVvMYBPA1, a grape regulator of proanthocyanidin biosynthesis, up- or down-regulates different branches of the phenylproanoid pathway, in a st
[本文引用: 1]
[本文引用: 2]
DOI:10.1093/jxb/ert303URLPMID:3830490

Much remains unknown about how transcription factors and sugars regulate phenylpropanoid metabolism in tuber crops like potato (Solanum tuberosum). Based on phylogeny and protein similarity to known regulators of phenylpropanoid metabolism, 15 transcription factors were selected and their expression was compared in white, yellow, red, and purple genotypes with contrasting phenolic and anthocyanin profiles. Red and purple genotypes had increased phenylalanine ammonia lyase (PAL) enzyme activity, markedly higher levels of phenylpropanoids, and elevated expression of most phenylpropanoid structural genes, including a novel anthocyanin O-methyltransferase. The transcription factors Anthocyanin1 (StAN1), basic Helix Loop Helix1 (StbHLH1), and StWD40 were more strongly expressed in red and purple potatoes. Expression of 12 other transcription factors was not associated with phenylpropanoid content, except for StMYB12B, which showed a negative relationship. Increased expression of AN1, bHLH1, and WD40 was also associated with environmentally mediated increases in tuber phenylpropanoids. Treatment of potato plantlets with sucrose induced hydroxycinnamic acids, flavonols, anthocyanins, structural genes, AN1, bHLH1, WD40, and genes encoding the sucrose-hydrolysing enzymes SUSY1, SUSY4, and INV2. Transient expression of StAN1 in tobacco leaves induced bHLH1, structural genes, SUSY1, SUSY4, and INV1, and increased phenylpropanoid amounts. StAN1 infiltration into tobacco leaves decreased sucrose and glucose concentrations. In silico promoter analysis revealed the presence of MYB and bHLH regulatory elements on sucrolytic gene promoters and sucrose-responsive elements on the AN1 promoter. These findings reveal an interesting dynamic between AN1, sucrose, and sucrose metabolic genes in modulating potato phenylpropanoids.
.
DOI:10.1002/j.1460-2075.1987.tb02684.xURLPMID:3428265 [本文引用: 1]

The structure of the wild-type c1 locus of Zea mays was determined by sequence analysis of one genomic and two cDNA clones. The coding region is composed of three exons (150 bp, 129 bp and one, at least 720 bp) and two small introns (88 bp and 145 bp). Transcription of the mRNAs corresponding to the two cDNA clones cLC6 (1.1 kb) and cLC28 (2.1 kb) starts from the same promoter. Both cDNAs are identical except that cLC28 extends further at its 3' end. A putative protein, 273 amino acids in length was deduced from the sequence of both transcripts. It contains two domains, one basic and the other acidic and might function as a transcriptional activator. The basic domain of this c1-encoded protein shows 40% sequence homology to the protein products of animal myb proto-oncogenes.
.
DOI:10.1111/j.1365-313X.2009.03885.xURLPMID:19368693

MYB transcription factors help to control anthocyanin biosynthesis in plants, and ectopic expression of the Arabidopsis Production of Anthocyanin Pigment 1 (PAP1) transcription factor activates the anthocyanin pathway in tobacco, suggesting the general utility of such factors for metabolic engineering of anthocyanins and anthocyanin-derived compounds such as proanthocyanidins (condensed tannins). However, PAP1 does not activate anthocyanin biosynthesis in the model legume Medicago truncatula or in alfalfa ( Medicago sativa ). A related Legume Anthocyanin Production 1 ( LAP1 ) gene was identified from the genome of M. truncatula . When constitutively expressed in transgenic alfalfa, M. truncatula or white clover, LAP1 induced massive accumulation of anthocyanin pigments comprising multiple glycosidic conjugates of cyanidin. Oligomeric/polymeric compounds with some diagnostic characteristics of proanthocyanidins also accumulated in LAP1 -expressing plants, but these compounds were not composed of (epi)catechin units. Over 260 and 70 genes were up-regulated in leaves of alfalfa or M. truncatula , respectively , in response to constitutive expression of LAP1 , many of which are involved in anthocyanin biosynthesis. In particular, the glucosyltransferase UGT78G1, previously identified as showing preference for isoflavonoid substrates in vitro , was strongly up-regulated by LAP1, and appears to function as an anthocyanin glycosyltransferase in vivo . Over-expression of UGT78G1 in transgenic alfalfa resulted in increased anthocyanin accumulation when plants were exposed to abiotic stress.
DOI:10.1158/2326-6066.CIR-15-0091URLPMID:26603620 [本文引用: 1]

Abstract Freeze-dried black raspberries (BRB), their component anthocyanins (AC), and a metabolite of BRB ACs, protocatechuic acid (PCA), inhibit the development of esophageal cancer in rats induced by the carcinogen, N-nitrosomethylbenzylamine (NMBA). All three components reduce inflammation in the esophagus and in plasma. The present study determined the relation of changes in inflammatory markers to infiltration of innate immune cells into NMBA-treated esophagus. Rats were injected with NMBA (0.35 mg/kg) for 5 weeks while on control diet. Following NMBA treatment, rats were fed diets containing 6.1% BRB powder, an AC-rich fraction of BRBs (3.8 0204mol/g), or 500 ppm PCA. At weeks 15, 25, and 35, inflammatory biomarker expression in the plasma and esophagus was quantified, and infiltration of immune cells in the esophagus was examined. At all three time points, BRB, AC, and PCA similarly affected cytokine production in the esophagus and plasma of NMBA-treated rats, relative to the NMBA-only control. These included decreased expression of the proinflammatory cytokine IL10205 and increased expression of the anti-inflammatory cytokine IL10. Moreover, all three diets also increased the expression of IL12, a cytokine that activates both cytolytic natural killer and CD8(+) T cells. In addition, the three diets also decreased infiltration of both macrophages and neutrophils into the esophagus. Overall, our results suggest that another mechanism by which BRBs, ACs, and PCA inhibit NMBA-induced esophageal tumorigenesis is by altering cytokine expression and innate immune cell trafficking into tumor tissues. 00082015 American Association for Cancer Research.
[本文引用: 1]
DOI:10.1007/s11103-015-0394-yURLPMID:26497001 [本文引用: 1]

In grapevine, anthocyanins and proanthocyanidins are the main flavonoids in berries, which are associated to organoleptic properties in red wine such as color and astringency. Flavonoid pathway is...
.
DOI:10.1016/j.plaphy.2010.09.002URLPMID:20951056

‘Max Red Bartlett’ is a red bud mutation of the yellow pear (-glucosyltransferase (UFGT). Expression of the genes encoding these five enzymes was examined in pear fruit skin in order to elucidate the molecular mechanism for red coloration. In addition, the gene Research highlights? The expression level of anthocyanin-related genes during fruit development of the European pear ‘Williams’ and its red-skinned sport ‘Max Red Bartlett’ was determined. ? The transcription factor PcMYB10 is most likely involved in the anthocyanin production in the early stages of ‘Max Red Bartlett’ fruit development. ? The red phenotype of ‘Max Red Bartlett’ however was not originated by a mutation of PcMYB10, as the gene does not co-map with the red colour trait.
.
DOI:10.1017/S001667230200561XURL [本文引用: 2]

The understanding of control of gene regulation in higher eukaryotes relies heavily on results derived from non-in vivo studies, but rarely can the significance of these approximations be established in vivo. Here, we investigated the effect of Mutator and Spm insertions on the expression of the flavonoid biosynthetic gene a1, independently regulated by the transcription factors C1 and P. The a1-mum2 and a1-m2 alleles carry Mu1 and Spm insertions, respectively, in a cis-element (ARE) of unknown function located between the P- and C1-binding sites. We show that the insertions of Mu1 and Spm similarly influence the expression of a1 controlled by C1 or P. The P-controlled a1 expression in a1-m2 is Spm dependent, and the mutant phenotype of a1-mum2 is suppressed in the pericarp in the absence of the autonomous MuDR element. Footprints within the ARE affect the regulation of a1 by C1 and P differently, providing evidence that these factors control a1 expression using distinct cis-acting regulatory elements. Together, our findings contribute significantly to one of the best-described plant regulatory systems, while stressing the need to complement with in vivo experiments current approaches used for the study of control of gene expression.
.
DOI:10.1371/journal.pone.0070665URLPMID:23976949 [本文引用: 1]

The rare wild species of snow lotus Saussurea involucrata is a commonly used medicinal herb with great pharmacological value for human health, resulting from its uniquely high level of phenylpropanoid compound production. To gain information on the phenylpropanid biosynthetic pathway genes in this critically important medicinal plant, global transcriptome sequencing was performed. It revealed that the phenylpropanoid pathway genes were well represented in S. involucrata. In addition, we introduced two key phenylpropanoid pathway inducing transcription factors (PAP1 and Lc) into this medicinal plant. Transgenic S. involucrata co-expressing PAP1 and Lc exhibited purple pigments due to a massive accumulation of anthocyanins. The over-expression of PAP1 and Lc largely activated most of the phenylpropanoid pathway genes, and increased accumulation of several phenylpropanoid compounds significantly, including chlorogenic acid, syringin, cyanrine and rutin. Both ABTS (2,2-azinobis-3-ethylbenzotiazo-line-6-ulfonicacid) and FRAP (ferric reducing anti-oxidant power) assays revealed that the antioxidant capacity of transgenic S. involucrata lines was greatly enhanced over controls. In addition to providing a deeper understanding of the molecular basis of phenylpropanoid metabolism, our results potentially enable an alternation of bioactive compound production in S. involucrata through metabolic engineering.
.
[本文引用: 1]
.
.
DOI:10.2134/agronj1997.00021962008900030003xURLPMID:10449578 [本文引用: 1]

The shape and color of flowers are important for plant reproduction because they attract pollinators such as insects and birds. Therefore, it is thought that alterations in these traits may result in the attraction of different pollinators, genetic isolation, and ultimately, (sympatric) speciation. Petunia integrifolia and P. axillaris bear flowers with different shapes and colors that appear to be visited by different insects. The anthocyanin2 (an2) locus, a regulator of the anthocyanin biosynthetic pathway, is the main determinant of color differences. Here, we report an analysis of molecular events at the an2 locus that occur during Petunia spp evolution. We isolated an2 by transposon tagging and found that it encodes a MYB domain protein, indicating that it is a transcription factor. Analysis of P. axillaris subspecies with white flowers showed that they contain $an2^{-}$ alleles with two alternative frameshifts at one site, apparently caused by the insertion and subsequent excision of a transposon. A third $an2^{-}$ allele has a nonsense mutation elsewhere, indicating that it arose independently. The distribution of polymorphisms in $an2^{-}$ alleles suggests that the loss of an2 function and the consequent changes in floral color were not the primary cause for genetic separation of P. integrifolia and P. axillaris. Rather, they were events that occurred late in the speciation process, possibly to reinforce genetic isolation and complete speciation.
DOI:10.2307/3869734URLPMID:12271045 [本文引用: 2]

In this study, we demonstrate that in petunia at least four regulatory genes (anthocyanin-1 [an1], an2, an4, and an11) control transcription of a subset of structural genes from the anthocyanin pathway by using a combination of RNA gel blot analysis, transcription run-on assays, and transient expression assays. $an2^{-}$ and $an11^{-}$ mutants could be transiently complemented by the maize regulatory genes Leaf color (Lc) or Colorless-1 (C1), respectively, whereas $an1^{-}$ mutants only by Lc and C1 together. In addition, the combination of Lc and C1 induces pigment accumulation in young leaves. This indicates that Lc and C1 are both necessary and sufficient to produce pigmentation in leaf cells. Regulatory pigmentation genes in maize and petunia control different sets of structural genes. The maize Lc and C1 genes expressed in petunia differentially activate the promoters of the chalcone synthase genes chsA and chsJ in the same way that the homologous petunia genes do. This suggests that the regulatory proteins in both species are functionally similar and that the choice of target genes is determined by their promoter sequences. We present an evolutionary model that explains the differences in regulation of pigmentation pathways of maize, petunia, and snapdragon.
[本文引用: 1]
.
DOI:10.1023/A:1024852021124URLPMID:12956536

Basic helix-loop-helix ( bHLH ) proteins, similar to mammalian Myc transcription factors, regulate the anthocyanin biosynthetic pathway in both monocots and dicots. Two Arabidopsis bHLH genes, GLABRA3 ( GL3 ) and MYC-146 , encode proteins that are similar throughout the predicted amino acid sequence to R and DELILA, which regulate anthocyanin production in maize and snapdragon, respectively. Northern blot analysis indicates that MYC-146 is most highly expressed in flower buds and flowers. Expression of a MYC-146 cDNA from the CaMV 35S promoter was unable to complement the anthocyanin deficiency in a ttg1 mutant of Arabidopsis and resulted in no obvious phenotypic change in Columbia plants. However, transient expression of GL3 and MYC-146 upon microprojectile bombardment of petals of a white-flowered mutant of Matthiola incana was able to complement anthocyanin deficiency. The lack of anthocyanin-deficient Arabidopsis mutants mapping to the locations of GL3 and MYC-146 suggests that the two bHLH proteins may be partially redundant and overlap in function.
.
DOI:10.1186/1471-2229-13-68URLPMID:3648406

Flavonoids such as anthocyanins, flavonols and proanthocyanidins, play a central role in fruit colour, flavour and health attributes. In peach and nectarine (Prunus persica) these compounds vary during fruit growth and ripening. Flavonoids are produced by a well studied pathway which is transcriptionally regulated by members of the MYB and bHLH transcription factor families. We have isolated nectarine flavonoid regulating genes and examined their expression patterns, which suggests a critical role in the regulation of flavonoid biosynthesis. In nectarine, expression of the genes encoding enzymes of the flavonoid pathway correlated with the concentration of proanthocyanidins, which strongly increases at mid-development. In contrast, the only gene which showed a similar pattern to anthocyanin concentration wasUDP-glucose-flavonoid-3-O-glucosyltransferase(UFGT), which was high at the beginning and end of fruit growth, remaining low during the other developmental stages. Expression offlavonol synthase(FLS1)correlated with flavonol levels, both temporally and in a tissue specific manner. The pattern ofUFGTgene expression may be explained by the involvement of different transcription factors, which up-regulate flavonoid biosynthesis (MYB10,MYB123, andbHLH3), or repress (MYB111andMYB16) the transcription of the biosynthetic genes. The expression of a potential proanthocyanidin-regulating transcription factor,MYBPA1, corresponded with proanthocyanidin levels. Functional assays of these transcription factors were used to test the specificity for flavonoid regulation. MYB10 positively regulates the promoters ofUFGTanddihydroflavonol 4-reductase(DFR) but notleucoanthocyanidin reductase(LAR). In contrast, MYBPA1 trans-activates the promoters ofDFRandLAR, but notUFGT. This suggests exclusive roles of anthocyanin regulation by MYB10 and proanthocyanidin regulation by MYBPA1. Further, these transcription factors appeared to be responsive to both developmental and environmental stimuli.
DOI:10.1111/j.1467-7652.2008.00362.xURLPMID:18662373

Flavonols and caffeoylquinates represent important groups of phenolic antioxidants with health-promoting activities. The genetic potential of potato ( Solanum tuberosum ) to produce high levels of these dietary compounds has not been realized in currently available commodity varieties. In this article, it is demonstrated that tuber-specific expression of the native and slightly modified MYB transcription factor gene StMtf1 M activates the phenylpropanoid biosynthetic pathway. Compared with untransformed controls, transgenic tubers contained fourfold increased levels of caffeoylquinates, including chlorogenic acid (CGA) (1.80 mg/g dry weight), whilst also accumulating various flavonols and anthocyanins. Subsequent impairment of anthocyanin biosynthesis through silencing of the flavonoid-3',5'-hydroxylase ( F3'5'h ) gene resulted in the accumulation of kaempferol-rut (KAR) to levels that were approximately 100-fold higher than in controls (0.12 mg/g dry weight). The biochemical changes were associated with increased expression of both the CGA biosynthetic hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase ( Hqt ) gene and the upstream chorismate mutase ( Cm ) and prephenate dehydratase ( Pdh ) genes. Field trials indicated that transgenic lines produced similar tuber yields to the original potato variety Bintje. Processed products of these lines retained most of their phenylpropanoids and were indistinguishable from untransformed controls in texture and taste.
.
[本文引用: 1]
.
[本文引用: 1]
.
DOI:10.1021/jf025671qURLPMID:12207466 [本文引用: 1]

Abstract The antioxidant activities of a series of commonly consumed and biogenetically related plant phenolics, namely, anthocyanidins, anthocyanins, and catechins, in a liposomal model system have been investigated. The antioxidant efficacies of the compounds were evaluated on their abilities to inhibit the fluorescence intensity decay of an extrinsic probe, 3-[p-(6-phenyl)-1,3,5-hexatrienyl]phenylpropionic acid, caused by free radicals generated during metal ion-induced peroxidation. Distinct structure-activity relationships were revealed for the antioxidant abilities of these structurally related compounds. Whereas antioxidant activity increased with an increasing number of hydroxyl substituents present on the B-ring for anthocyanidins, the converse was observed for catechins. However, substitution by methoxyl groups diminished the antioxidant activity of the anthocyanidins. Substitution at position 3 of ring C played a major role in determining the antioxidant activity of these classes of compounds. The anthocyanidins, which possess a hydroxyl group at position 3, demonstrated potent antioxidant activities. For the cyanidins, an increasing number of glycosyl units at position 3 resulted in decreased antioxidant activity. Similarly, the substitution of a galloyl group at position 3 of the flavonoid moiety resulted in significantly decreased antioxidant activity for the catechins. Among catechins, cis-trans isomerism, epimerization, and racemization did not play a role in overall antioxidant activity. The antioxidant activities of test compounds (at 40 microM concentrations) were compared to the commercial antioxidants tert-butylhydroquinone, butylated hydroxytoluene, butylated hydroxyanisole, and vitamin E (all at 10 microM concentrations).
DOI:10.1111/j.1744-7909.2007.00527.xURL [本文引用: 2]

Chlorophyll fluorescence and antioxidative capability in detached leaves of the wild type Arabidopsis thaliana L. ecotype Landsberg erecta ( Ler ) and three mutants deficient in anthocyanins biosynthesis ( tt3 , tt4 , and tt3tt4 ) were investigated during treatment with temperatures ranging 2509000945 00°C. In comparison with the wild type, chlorophyll fluorescence parameters Fv/Fm, 0207 PSII , electron transport rate (ETR), Fv/Fo and qP in three anthocyanin-deficient mutants showed a more rapidly decreasing rate when the temperature was over 35 00°C. Non-photochemical quenching (NPQ) in these mutants was almost completely lost at 44 00°C, whereas the content of heat stable protein dropped and the rate of the membrane leakage increased. Fo-temperature curves were obtained by monitoring Fo levels with gradually elevated temperatures from 22 00°C to 72 00°C at 0.5 00°C/min. The inflexion temperatures of Fo were 45.8 00°C in Ler , 45.1 00°C in tt3 , 44.1 00°C in tt4 and 42.3 00°C in tt3tt4 , respectively. The temperatures of maximal Fo in three mutants were 1.90900093.8 00°C lower than the wild type plants. Meanwhile, three mutants had lower activities of superoxide dismutase (SOD) and ascorbate peroxidase (APX) and an inferior scavenging capability to DPPH (1.1-diphenyl-2-picrylhy.drazyl) radical under heat stress, and in particular tt3tt4 had the lowest antioxidative potential. The results of the diaminobenzidine-H 2 O 2 histochemical staining showed that H 2 O 2 was accumulated in the leaf vein and mesophyll cells of mutants under treatment at 40 00°C, and it was significantly presented in leaf cells of tt3tt4 . The sensitivity of Arabidopsis anthocyanins-deficient mutants to high temperatures has revealed that anthocyanins in normal plants might provide protection from high temperature injury, by enhancing its antioxidative capability under high temperature stress.
.
DOI:10.1093/pcp/pcu013URLPMID:24443499

The MYB transcription factors and plant hormone ABA have been suggested to play a role in fruit anthocyanin biosynthesis, but supporting genetic evidence has been lacking in sweet cherry. The present study describes the first functional characterization of an R2R3-MYB transcription factor, PacMYBA, from red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Transient promoter assays demonstrated that PacMYBA physically interacted with several anthocyanin-related basic helix-loop-helix (bHLH) transcription factors to activate the promoters of PacDFR, PacANS and PacUFGT, which are thought to be involved in anthocyanin biosynthesis. Furthermore, the immature seeds of transgenic Arabidopsis plants overexpressing PacMYBA exhibited ectopic pigmentation. Silencing of PacMYBA, using a Tobacco rattle virus (TRV)-induced gene silencing technique, resulted in sweet cherry fruit that lacked red pigment. ABA treatment significantly induced anthocyanin accumulation, while treatment with the ABA biosynthesis inhibitor nordihydroguaiaretic acid (NDGA) blocked anthocyanin production. PacMYBA expression peaked after 2 h of pre-incubation in ABA and was 15.2-fold higher than that of sweet cherries treated with NDGA. The colorless phenotype was also observed in the fruits silenced in PacNCED1, which encodes a key enzyme in the ABA biosynthesis pathway. The endogenous ABA content as well as the transcript levels of six structural genes and PacMYBA in PacNCED1-RNAi (RNA interference) fruit were significantly lower than in the TRV vector control fruit. These results suggest that PacMYBA plays an important role in ABA-regulated anthocyanin biosynthesis and ABA is a signal molecule that promotes red-colored sweet cherry fruit accumulating anthocyanin.
.
DOI:10.1007/s00425-010-1335-2URLPMID:21210143

http://link.springer.com/article/10.1007%2Fs00425-010-1335-2
DOI:10.1021/jf071933iURLPMID:17935293 [本文引用: 1]

Abstract Reactive oxygen species (ROS)-induced cell damage is inevitable and severe and is involved in numerous diseases, including cancer. Reducing oxidative stress is one of the strategies of chemoprevention. Anthocyanins are naturally occurring flavonoids that show multiple benefits. We first pointed out the effects of anthocyanins in the contributions to activation of phase II antioxidant and detoxifying enzymes, chemopreventive potency, and involved transcriptional regulation. Our results obtained in rat liver Clone 9 cells showed that treatment of anthocyanins leads to positive effects on elevating the antioxidant capacity, including activated expression of glutathione-related enzymes (glutathione reductase, glutathione peroxidase, and glutathione S-transferase) and recruited GSH content. In addition, the activity of NAD(P)H: quinone oxidoreductase (NQO1) was also promoted under the treatment of anthocyanin. This influential functions as the defense system against programmed cell death induced by H2O2. The capacity for induction of luciferase expression by anthocyanins in cells transfected with rat nqo1-promoter constructed plasmid was further investigated; we found that the molecular mechanism is related to the activation of antioxidant response element (ARE) upstream of genes that are involved in antioxidation and detoxification. Our data suggest that natural anthocyanins are recommended as chemopreventive phytochemicals and could stimulate the antioxidant system to resist oxidant-induced injury. And, more important, the promoting effect of anthocyanins on ARE-regulated phase II enzyme expression seems to be a critical point in modulating the defense system against oxidative stress.
.
DOI:10.1093/nar/20.2.373URLPMID:1741268 [本文引用: 1]

We prove general optimal euclidean Sobolev and Gagliardo-Nirenberg inequalities by using mass transportation and convex analysis results. Explicit extremals and the computation of some optimal constants are also provided. In particular we extend the optimal Gagliardo-Nirenberg inequality proved by Del Pino and Dolbeault 2003 and the optimal inequalities proved by Cordero-Erausquin et al. 2004.
DOI:10.1093/jxb/erv159URLPMID:4507754 [本文引用: 1]

The integration of metabolic, molecular marker, and transcriptomic data from a segregating grapevine progeny provides novel insights into the genetic control of anthocyanin content and composition in ripe berries. In the last decade, great progress has been made in clarifying the main determinants of anthocyanin accumulation in grape berry skin. However, the molecular details of the fine variation among cultivars, which ultimately contributes to wine typicity, are still not completely understood. To shed light on this issue, the grapes of 170 F1 progeny from the cross ‘Syrah’×’Pinot Noir’ were characterized at the mature stage for the content of 15 anthocyanins during four growing seasons. This huge data set was used in combination with a dense genetic map to detect genomic regions controlling the anthocyanin pathway both at key enzymatic points and at particular branches. Genes putatively involved in fine tuning the global regulation of anthocyanin biosynthesis were identified by exploring the gene predictions in the QTL (quantitative trait locus) confidence intervals and their expression profile during berry development in offspring with contrasting anthocyanin accumulation. New information on some aspects which had scarcely been investigated so far, such as anthocyanin transport into the vacuole, or completely neglected, such as acylation, is provided. These genes represent a valuable resource in grapevine molecular-based breeding programmes to improve both fruit and wine quality and to tailor wine sensory properties according to consumer demand.
DOI:10.1089/jmf.2012.2241URLPMID:22870951 [本文引用: 1]

Ginkgo biloba extract (GBE) and anthocyanins are considered beneficial for various vascular diseases. This study was performed to evaluate the effect of GBE and anthocyanins on visual function in patients with normal tension glaucoma (NTG) based on the vascular theory of mechanisms of glaucomatous optic nerve damage. Retrospective analysis was carried out by a chart review of 332 subjects (209 men and 123 women) who were treated with anthocyanins (n=132), GBE (n=103), or no medication (control, n=97). Humphrey Visual Field (HVF) test, logarithm of the minimal angle of resolution best-corrected visual acuity (logMAR BCVA), intraocular pressure, blood pressure, and fasting blood glucose were determined before and after treatment. Complete ocular and systemic examinations were performed. The mean follow-up duration was 23.82±9.84 (range, 12-59) months; the mean anthocyanin treatment duration was 24.32±10.43 (range, 6-53) months, and the mean GBE treatment duration was 23.81±10.36 months (range, 6-59) months. After anthocyanin treatment, the mean BCVA for all eyes improved from 0.16 (±0.34) to 0.11 (±0.18) logMAR units (P=.008), and HVF mean deviation improved from -6.44 (±7.05) to -5.34 (±6.42) (P=.001). After GBE treatment, HVF mean deviation improved from -5.25 (±6.13) to -4.31 (±5.60) (P=.002). A generalized linear model demonstrated that the final BCVA was not affected by demographic differences among the groups. These results suggest that anthocyanins and GBE may be helpful in improving visual function in some individuals with NTG.
.
[本文引用: 1]
DOI:10.1016/j.febslet.2013.03.037URLPMID:23583450 [本文引用: 2]

Several positive transcription factors regulate Arabidopsis anthocyanin biosynthesis. HY5, a component of light-signaling pathways, and PAP1, an R2R3-MYB transcription factor, share common regulatory targets on anthocyanin biosynthesis genes. The epistatic interactions between the two transcription factors are currently unknown. To address this problem, we analyzed crosses between hy5 and pap1 mutants (hy5pap1) or pap1D overexpressors (hy5pap1D), performed chromatin immunoprecipitation-qPCR, and determined the PAP1 promoter region through deletion analysis. The results show that HY5 regulates PAP1 expression via direct binding to G- and ACE-boxes in the promoter region, which suggests bifurcate regulation of anthocyanin biosynthesis by HY5 via transcriptional activation of PAP1. (C) 2013 Federation of European Biochemical Societies. Published by Elsevier B. V. All rights reserved.
DOI:10.1016/j.bbrc.2015.12.001URLPMID:26692488

Transcriptional activation of anthocyanin biosynthesis genes in vegetative tissues of monocotyledonous plants is mediated by cooperative activity of one component from each of the following two transcription factor families: MYB encoded byPURPLE PLANT1/COLORED ALEURONE1(PL1/C1), and basic helix-loop-helix (bHLH) encoded byRED/BOOSTER(R1/B1). In the present study, putativePLcDNA was cloned from the wheat (Triticum aestivum) cultivar Iksan370, which preferentially expresses anthocyanins in coleoptiles. Phylogenetic tree analysis of deduced amino acid sequences showed that a putative TaPL1 is highly homologous to barley (Hordeum vulgare) HvPL1, but is distinct from wheat TaC1. TransgenicArabidopsis thalianastably expressing putativeTaPL1accumulated anthocyanin pigments in leaves and up-regulated structural genes involved in both early and late anthocyanin biosynthesis steps.TaPL1transcript levels in Iksan370 were more prominent in vegetative tissues such as young coleoptiles than in reproductive tissues such as spikelets.TaPL1expression was significantly up-regulated by environmental stresses including cold, salt, and light, which are known to induce anthocyanin accumulation. These combined results suggest that TaPL1 is an active positive regulator of anthocyanin biosynthesis in wheat coleoptiles.
DOI:10.1111/j.1365-313X.2006.03021.xURLPMID:17319847 [本文引用: 1]

Abstract Phytochromes are red/far-red light receptors that regulate various light responses by initiating the transcriptional cascades responsible for changing the expression patterns of 10-30% of the entire plant transcriptome. Several transcription factors that are thought to participate in this process have been identified, but the functional relationships among them have not yet been fully elucidated. Here we investigated the functional relationship between two such transcription factors, PIF3 and HY5, and their effects on anthocyanin biosynthesis. Our results revealed that PIF3 and HY5 do not regulate each other at either the transcriptional or the protein levels in continuous light conditions, suggesting that they are not directly linked within phytochrome-mediated signaling. We found that both PIF3 and HY5 positively regulate anthocyanin biosynthesis by activating the transcription of the same anthocyanin biosynthetic genes, but the positive effects of PIF3 required functional HY5. Chromatin immunoprecipitation analyses indicated that both PIF3 and HY5 regulate anthocyanin biosynthetic gene expression by directly binding to different regions of the gene promoters in vivo. Additional experiments revealed that PIF3 bound the promoters regardless of light and HY5. Collectively, these data show that PIF3 and HY5 regulate anthocyanin biosynthesis by simultaneously binding anthocyanin biosynthetic gene promoters at separate sequence elements.
.
DOI:10.1023/A:1019850921627URLPMID:12369624 [本文引用: 1]

The WD-repeat proteins are found in eukaryotes and play an important role in the regulation of a wide variety of cellular functions such as signal transduction, transcription, and proliferation. In this study, we have isolated a cDNA encoding a novel WD-repeat protein, PFWD , from the anthocyanin-pigmented leaves of Perilla frutescens using AN11 cDNA from Petunia hybrida as the probe. The C-terminal region of PFWD contains a WD repeat that is highly conserved in homologous proteins from a variety of organisms that do not produce anthocyanin such as yeast, nematodes and mammals. Transgenic Arabidopsis plants overexpressing PFWD exhibited phenotypic changes including enhancement of anthocyanin production and reduced viability. A study of the interaction between PFWD and anthocyanin regulatory proteins using a yeast two-hybrid system showed strong interaction between PFWD and MYC-RP, a MYC-like protein from P.frutescens . PFWD fusion proteins transiently expressed in onion epidermal cells were localized in the cytosol under both dark and light conditions. However, co-expression of PFWD and MYC-RP fusion proteins resulted in nuclear localization of PFWD. We propose a model of genetic regulation in which the PFWD protein acts in signal transduction process in a variety of pathways through protein interaction with MYC proteins.
[本文引用: 1]
DOI:10.1105/tpc.111.083089URLPMID:21447791 [本文引用: 1]

The Arabidopsis thaliana F-box protein CORONATINE INSENSITIVE1 (COl1) perceives jasmonate (JA) signals and subsequently targets the Jasmonate-ZIM domain proteins (JAZs) for degradation by the SCF COl1 -26S proteasome pathway to mediate various jasmonate-regulated processes, including fertility, root growth, anthocyanin accumulation, senescence, and defense. In this study, we screened JAZ-interacting proteins from an Arabidopsis cDNA library in the yeast two-hybrid system. MYB21 and MYB24, two R2R3-MYB transcription factors, were found to interact with JAZ1, JAZ8, and JAZ11 in yeast and in planta. Genetic and physiological experiments showed that the myb21 myb24 double mutant exhibited defects specifically in pollen maturation, anther dehiscence, and filament elongation leading to male sterility. Transgenic expression of MYB21 in the coi1-1 mutant was able to rescue male fertility partially but unable to recover JA-regulated root growth inhibition, anthocyanin accumulation, and plant defense. These results demonstrate that the R2R3-MYB transcription factors MYB21 and MYB24 function as direct targets of JAZs to regulate male fertility specifically. We speculate that JAZs interact with MYB21 and MYB24 to attenuate their transcriptional function; upon perception of JA signal, COI1 recruits JAZs to the SCF COM complex for ubiquitination and degradation through the 26S proteasome; MYB21 and MYB24 are then released to activate expression of various genes essential for JA-regulated anther development and filament elongation.
DOI:10.1105/tpc.12.9.1619URLPMID:11006336 [本文引用: 3]

Abstract The petunia loci anthocyanin1 (an1), an2, an4, and an11 are required for the transcription of anthocyanin biosynthetic genes in floral organs. The an2 and an11 loci were recently cloned and shown to encode a MYB-domain transcriptional activator and a cytosolic WD40 protein, respectively. Here, we report the isolation of an1 by transposon tagging. an1 encodes a new member of the basic helix-loop-helix family of transcription factors that is functionally and evolutionarily distinct from JAF13, the apparent petunia ortholog of maize RED1 and snapdragon DELILA. We provide genetic evidence that the transcription factors encoded by an1, an2, and an4 operate in an unexpectedly complex regulatory hierarchy. In leaves, ectopic expression of AN2 induces an1 expression, whereas in anthers, an1 expression depends on an4, encoding (or controlling) a MYB protein that is paralogous to AN2. Experiments with transgenic plants expressing a post-translationally controlled AN1-GLUCOCORTICOID RECEPTOR fusion protein indicated that independent of protein synthesis, AN1 directly activates the expression of the dfrA gene encoding the enzyme dihydroflavonol 4-reductase and of Pmyb27 encoding a MYB-domain protein of unknown function.
.
DOI:10.1371/journal.pone.0126991URLPMID:4433224

Anthocyanins are essential contributors to fruit coloration, an important quality feature and a breed determining trait of a sweet cherry fruit. It is well established that the biosynthesis of anthocyanins is regulated by an interplay of specific transcription factors belonging to MYB and bHLH families accompanied by a WD40 protein. In this study, we isolated and analyzed PaWD40, PabHLH3, PabHLH33, and several closely related MYB10 gene variants from different cultivars of sweet cherry, analyzed their expression in fruits with different anthocyanin levels at several developmental stages, and determined their capabilities to modulate anthocyanin synthesis in leaves of two Nicotiana species. Our results indicate that transcription level of variant PaMYB10.1-1 correlates with fruit coloration, but anthocyanin synthesis in Nicotiana was induced by another variant, PaMYB10.1-3, which is moderately expressed in fruits. The analysis of two fruit-expressed bHLH genes revealed that PabHLH3 enhances MYB-induced anthocyanin synthesis, whereas PabHLH33 has strong inhibitory properties.
.
DOI:10.1186/s41065-016-0021-1URLPMID:28096779 [本文引用: 1]

The color of berry skin is an important economic trait for grape and is essentially determined by the components and content of anthocyanins. The fruit color of Chinese wild grapes is generally black, and the profile of anthocyanins in Chinese wild grapes is significantly different from that ofVitis vinifera. However,V. davidiiis the only species that possesses white berry varieties among Chinese wild grape species. Thus, we performed a transcriptomic analysis to compare the difference of transcriptional level in black and whiteV. davidii, in order to find some key genes that are related to anthocyanins accumulation inV. davidii. The results of anthocyanins detection revealed that 3,5-O-diglucoside anthocyanins is the predominant anthocyanins inV. davidii. It showed obvious differences fromV. viniferain the profile of the composition of anthocyanins. The transcriptome sequencing by Illumina mRNA-Seq technology generated an average of 57 million 100-base pair clean reads from each sample. Differential gene expression analysis revealed thousands of differential expression genes (DEGs) in the pairwise comparison of different fruit developmental stages between and within black and whiteV. davidii. After the analysis of functional category enrichment and differential expression patterns of DEGs, 46 genes were selected as the candidate genes. Some genes have been reported as being related to anthocyanins accumulation, and some genes were newly found in our study as probably being related to anthocyanins accumulation. We inferred that3AT(VIT_03s0017g00870) played an important role in anthocyanin acylation,GST4(VIT_04s0079g00690) andAM2(VIT_16s0050g00910) played important roles in anthocyanins transport inV. davidii. The expression of some selected DEGs was further confirmed by quantitative real-time PCR (qRT-PCR). The present study investigated the transcriptomic profiles of berry skin from black and white spine grapes at three fruit developmental stages by Illumina mRNA-Seq technology. It revealed the variety specificity of anthocyanins accumulation inV. davidiat the transcriptional level. The data reported here will provide a valuable resource for understanding anthocyanins accumulation in grapes, especially inV. davidii. The online version of this article (doi:10.1186/s41065-016-0021-1) contains supplementary material, which is available to authorized users.
DOI:10.1093/jhered/esr028URLPMID:21566002

Abstract Previous studies revealed that the recessive allele of the W2 locus generated purple-blue color and high vacuolar pH of flower petals in soybean. The location of W2 gene was reportedly close to simple sequence repeat marker Satt318 in molecular linkage group B2. We used information from the soybean genome to clone a candidate gene for W2. An MYB transcription factor gene belonging to G20 group was found in the vicinity of Satt318. Full-length cDNAs were cloned from purple-flowered cultivar Harosoy (W2 allele) and purple-blue flowered cultivars, Nezumisaya and w2-20 (w2 allele), by reverse transcription-PCR and designated as GmMYB-G20-1. Its open reading frame was 1083 bp long that encoded 361 amino acids in Harosoy. GmMYB-G20-1 had 53.7% similarity in amino acid sequence with the PH4 gene of petunia controlling blueness and vacuolar pH of flower petals. GmMYB-G20-1 of Nezumisaya and w2-20 had 3 base substitutions compared with that of Harosoy. The first substitution generated a stop codon in the MYB domain, resulting in truncated polypeptides. Cleaved amplified polymorphic sequence (CAPS) marker was developed to detect the base substitution. The polymorphic CAPS marker co-segregated with alleles at the W2 locus in the F(2) population. These results suggest that GmMYB-G20-1 might correspond to the W2 gene.
.
DOI:10.1104/pp.106.088104URLPMID:17012405 [本文引用: 2]

Anthocyanins are secondary metabolites found in higher plants that contribute to the colors of flowers and fruits. In apples (Malus domestica Borkh.), several steps of the anthocyanin pathway are coordinately regulated, suggesting control by common transcription factors. A gene encoding an R2R3 MYB transcription factor was isolated from apple (cv Cripps' Pink) and designated MdMYB1. Analysis of the deduced amino acid sequence suggests that this gene encodes an ortholog of anthocyanin regulators in other plants. The expression of MdMYB1 in both Arabidopsis (Arabidopsis thaliana) plants and cultured grape cells induced the ectopic synthesis of anthocyanin. In the grape (Vitis vinifera) cells MdMYB1 stimulated transcription from the promoters of two apple genes encoding anthocyanin biosynthetic enzymes. In ripening apple fruit the transcription of MdMYB1 was correlated with anthocyanin synthesis in red skin sectors of fruit. When dark-grown fruit were exposed to sunlight, MdMYB1 transcript levels increased over several days, correlating with anthocyanin synthesis in the skin. MdMYB1 gene transcripts were more abundant in red skin apple cultivars compared to non-red skin cultivars. Several polymorphisms were identified in the promoter of MdMYB1. A derived cleaved amplified polymorphic sequence marker designed to one of these polymorphisms segregated with the inheritance of skin color in progeny from a cross of an unnamed red skin selection (a sibling of Cripps' Pink) and the non-red skin cultivar Golden Delicious. We conclude that MdMYB1 coordinately regulates genes in the anthocyanin pathway and the expression level of this regulator is the genetic basis for apple skin color.
DOI:10.18240/ijo.2016.01.25URLPMID:4768496 [本文引用: 1]

Retinitis pigmentosa(RP) is a group of heterogeneous inherited retinal diseases that is characterized by primary death rod photoreceptors and the secondary loss of cones. The degeneration of cones causes gradual constriction of visual fields, leaving the central islands that are eventually snuffed out. Studies indicate that the hyperoxia causes oxidative damage in the retina and contributes to the cone death of RP. Moreover, abundant reactive oxidative species(ROS) which are generated in cones may result in mitochondria membrane depolarization, which has been ascribed a central role in the apoptotic process and has been proposed to act as a forward feeding loop for the activation of downstream cascades. Anthocyanin is a potent antioxidant which has been evidenced to be able to counteract oxidative damages, scavenge surplus ROS, and rectify abnormities in the apoptotic cascade. Taken together with its ability to attenuate inflammation which also contributes to the etiology of RP, it is reasonable to hypothesize that the anthocyanin could act as a novel therapeutic strategy to retard or prevent cone degeneration in RP retinas,particularly if the treatment is timed appropriately and delivered efficiently. Future pharmacological investigations will identify the anthocyanin as an effective candidate for PR therapy and refinements of that knowledge would ignite the hope of restoring the visual function in RP patients.
DOI:10.1111/j.1365-313X.2005.02371.xURLPMID:15807784 [本文引用: 1]

The integration of metabolomics and transcriptomics can provide precise information on gene-to-metabolite networks for identifying the function of unknown genes unless there has been a post-transcriptional modification. Here, we report a comprehensive analysis of the metabolome and transcriptome of Arabidopsis thaliana over-expressing the PAP1 gene encoding an MYB transcription factor, for the identification of novel gene functions involved in flavonoid biosynthesis. For metabolome analysis, we performed flavonoid-targeted analysis by high-performance liquid chromatography-mass spectrometry and non-targeted analysis by Fourier-transform ion-cyclotron mass spectrometry with an ultrahigh-resolution capacity, This combined analysis revealed the specific accumulation of cyanidin and quercetin derivatives, and identified eight novel anthocyanins from an array of putative 1800 metabolites in PAP1 over-expressing plants. The transcriptome analysis of 22 810 genes on a DNA microarray revealed the induction of 38 genes by ectopic PAP1 overexpression. In addition to well-known genes involved in anthocyanin production, several genes with unidentified functions or annotated with putative functions, encoding putative glycosyltransferase, acyltransferase, glutathione S-transferase, sugar transporters and transcription factors, wore induced by PAP1. Two putative glycosyltransferase genes (At5g17050 and At4g14090) induced by PAP1 expression were confirmed to encode flavonoid 3-O-glucosyltransferase and anthocyanin 5-O-glucosyltransferase, respectively, from the enzymatic activity of their recombinant proteins in vitro and results of the analysis of anthocyanins in the respective T-DNA-inserted mutants. The functional genomics approach through the integration of metabolomics and transcriptomics presented here provides an innovative means of identifying novel gene functions involved in plant metabolism.
DOI:10.1105/tpc.013839URLPMID:12897250 [本文引用: 1]

The basic/helix-loop-helix (bHLH) proteins are a superfamily of transcription factors that bind as dimers to specific DNA target sites and that have been well characterized in nonplant eukaryotes as important regulatory components in diverse biological processes. Based on evidence that the bHLH protein PIF3 is a direct phytochrome reaction partner in the photoreceptor's signaling network, we have undertaken a comprehensive computational analysis of the Arabidopsis genome sequence databases to define the scope and features of the bHLH family. Using a set of criteria derived from a previously defined consensus motif, we identified 147 bHLH protein-encoding genes, making this one of the largest transcription factor families in Arabidopsis. Phylogenetic analysis of the bHLH domain sequences permits classification of these genes into 21 subfamilies. The evolutionary and potential functional relationships implied by this analysis are supported by other criteria, including the chromosomal distribution of these genes relative to duplicated genome segments, the conservation of variant exon/intron structural patterns, and the predicted DNA binding activities within subfamilies. Considerable diversity in DNA binding site specificity among family members is predicted, and marked divergence in protein sequence outside of the conserved bHLH domain is observed. Together with the established propensity of bHLH factors to engage in varying degrees of homodimerization and heterodimerization, these observations suggest that the Arabidopsis bHLH proteins have the potential to participate in an extensive set of combinatorial interactions, endowing them with the capacity to be involved in the regulation of a multiplicity of transcriptional programs. We provide evidence from yeast two-hybrid and in vitro binding assays that two related phytochrome-interacting members in the Arabidopsis family, PIF3 and PIF4, can form both homodimers and heterodimers and that all three dimeric configurations can bind specifically to the G-box DNA sequence motif CACGTG. These data are consistent, in principle, with the operation of this combinatorial mechanism in Arabidopsis.
.
DOI:10.1007/BF00261680URLPMID:1673220

The Sn locus of maize is functionally similar to the R and B loci, in that Sn differentially controls the tissue-specific deposition of anthocyanin pigments in certain seedling and plant cells. We show that Sn shows molecular similarity to the R gene and have used R DNA probes to characterize several Sn alleles. Northern analysis demonstrates that all Sn alleles encode a 2.5 kb transcript, which is expressed in a tissue-specific fashion consistent with the distribution of anthocyanins. Expression of the Sn gene is light-regulated. However, the Sn: bol3 allele allows Sn mRNA transcription to occur in the dark, leading to pigmentation in dark-grown seedlings and cob integuments. We report the isolation of genomic and cDNA clones of the light-independent Sn: bol3 allele. Using Sn cDNA as a probe, the spatial and temporal expression of Sn has been examined. The cell-specific localization of Sn mRNA has been confirmed by in situ hybridization using labelled antisense RNA probes. According to its proposed regulatory role, expression of Sn precedes and, in turn, causes a coordinate and tissue-specific accumulation of mRNA of structural genes for pigment synthesis and deposition, such as A1 and C2 . The functional and structural relationship between R, B, Lc and Sn is discussed in terms of an evolutionary derivation from a single ancestral gene which gave rise this diverse gene family by successive duplication events.
DOI:10.1007/BF00019112URLPMID:8980509

Abstract In plants, MYC-related proteins function as transcription factors involved in anthocyanin production and trichome development. We cloned a gene, Atmyc1, and its corresponding cDNA, that encodes for a MYC-related protein from Arabidopsis thaliana. The putative protein has a basic/helix-loop-helix motif at the C-terminus and a highly homologous region with that of the maize B/R family at the N-terminus. The promoter region of Atmyc1 contains a Sph box (CATGCATG) that is known as a cis-regulatory element conferring seed-specific expression. In fact, Atmyc1 transcripts were more abundant in developing seeds than in stems and leaves where trichomes are normally expressed. Restriction fragment length polymorphism mapping demonstrated that Atmyc1 is located on the upper region of chromosome 4, which clearly indicates that Atmyc1 is distinct from the ttg (transparent testa glabrous) locus that affects both trichome development and anthocyanin biosynthesis.
DOI:10.1186/1471-2229-13-176URLPMID:24199943 [本文引用: 2]

Abstract BACKGROUND: Red coloration of fruit is an important trait in apple, and it is mainly attributed to the accumulation of anthocyanins, a class of plant flavonoid metabolites. Anthocyanin biosynthesis is genetically determined by structural and regulatory genes. Plant tissue pigmentation patterns are mainly controlled by expression profiles of regulatory genes. Among these regulatory genes are MYB transcription factors (TFs), wherein the class of two-repeats (R2R3) is deemed the largest, and these are associated with the anthocyanin biosynthesis pathway. Although three MdMYB genes, almost identical in nucleotide sequences, have been identified in apple, it is likely that there are other R2R3 MYB TFs that are present in the apple genome that are also involved in the regulation of coloration of red color pigmentation of the skin of apple fruits. RESULTS: In this study, a novel R2R3 MYB gene has been isolated and characterized in apple. This MYB gene is closely related to the Arabidopsis thaliana AtMYB3, and has been designated as MdMYB3. This TF belongs to the subgroup 4 R2R3 family of plant MYB transcription factors. This apple MdMYB3 gene is mapped onto linkage group 15 of the integrated apple genetic map. Transcripts of MdMYB3 are detected in all analyzed tissues including leaves, flowers, and fruits. However, transcripts of MdMYB3 are higher in excocarp of red-skinned apple cultivars than that in yellowish-green skinned apple cultivars. When this gene is ectopically expressed in Nicotiana tabacum cv. Petite Havana SR1, flowers of transgenic tobacco lines carrying MdMYB3 have exhibited increased pigmentation and accumulate higher levels of anthocyanins and flavonols than wild-type flowers. Overexpression of MdMYB3 has resulted in transcriptional activation of several flavonoid pathway genes, including CHS, CHI, UFGT, and FLS. Moreover, peduncles of flowers and styles of pistils of transgenic plants overexpressing MdMYB3 are longer than those of wild-type plants, thus suggesting that this TF is involved in regulation of flower development. CONCLUSIONS: This study has identified a novel MYB transcription factor in the apple genome. This TF, designated as MdMYB3, is involved in transcriptional activation of several flavonoid pathway genes. Moreover, this TF not only regulates the accumulation of anthocyanin in the skin of apple fruits, but it is also involved in the regulation of flower development, particularly that of pistil development.
.
[本文引用: 2]
DOI:10.1007/s00425-017-2704-xURLPMID:28474114 [本文引用: 1]

ABP9, encoding a bZIP transcription factor from maize, enhances tolerance to multiple stresses and may participate in the ABA signaling pathway in transgenic cotton by altering physiological and bioch
DOI:10.1186/1471-2229-10-50URLPMID:2923524

pAbstract/p pBackground/p pThe control of plant anthocyanin accumulation is via transcriptional regulation of the genes encoding the biosynthetic enzymes. A key activator appears to be an R2R3 MYB transcription factor. In apple fruit, skin anthocyanin levels are controlled by a gene called itMYBA /itor itMYB1/it, while the gene determining fruit flesh and foliage anthocyanin has been termed itMYB10/it. In order to further understand tissue-specific anthocyanin regulation we have isolated orthologous itMYB /itgenes from all the commercially important rosaceous species./p pResults/p pWe use gene specific primers to show that the three MYB activators of apple anthocyanin (itMYB10/MYB1/MYBA) /itare likely alleles of each other. MYB transcription factors, with high sequence identity to the apple gene were isolated from across the rosaceous family (e.g. apples, pears, plums, cherries, peaches, raspberries, rose, strawberry). Key identifying amino acid residues were found in both the DNA-binding and C-terminal domains of these MYBs. The expression of these itMYB10 /itgenes correlates with fruit and flower anthocyanin levels. Their function was tested in tobacco and strawberry. In tobacco, these MYBs were shown to induce the anthocyanin pathway when co-expressed with bHLHs, while over-expression of strawberry and apple genes in the crop of origin elevates anthocyanins./p pConclusions/p pThis family-wide study of rosaceous R2R3 MYBs provides insight into the evolution of this plant trait. It has implications for the development of new coloured fruit and flowers, as well as aiding the understanding of temporal-spatial colour change./p
.
DOI:10.1016/j.molp.2016.07.003URLPMID:27450422 [本文引用: 1]

This work demonstrates that miR858a positively regulates anthocyanin accumulation in Arabidopsis seedlings.MIR858aandHY5co-repress the expression ofMYBL2, the negative regulator of anthocyanin biosynthesis, in response to changing environmental cues. These results reveal a molecular mechanism incorporating both transcriptional and post-transcriptional regulations for controlling anthocyanin biosynthesis in plants.
.
DOI:10.1104/pp.113.214700URLPMID:23629835

Varieties of the European pear (Pyrus communis) can produce trees with both red- and green-skinned fruits, such as the Max Red Bartlett (MRB) variety, although little is known about the mechanism behind this differential pigmentation. In this study, we investigated the pigmentation of MRB and its green-skinned sport (MRB-G). The results suggest that a reduction in anthocyanin concentration causes the MRB-G sport. Transcript levels of PcUFGT (for UDP-glucose:flavonoid 3-O-glucosyltransferase), the key structural gene in anthocyanin biosynthesis, paralleled the change of anthocyanin concentration in both MRB and MRB-G fruit. We cloned the PcMYB10 gene, a transcription factor associated with the promoter of PcUFGT. An investigation of the 2-kb region upstream of the ATG translation start site of PcMYB10 showed the regions 2604 to 2911 bp and 21,218 to 21,649 bp to be highly methylated. A comparison of the PcMYB10 promoter methylation level between the MRB and MRB-G forms indicated a correlation between hypermethylation and the green-skin phenotype. An Agrobacterium tumefaciens infiltration assay was conducted on young MRB fruits by using a plasmid constructed to silence endogenous PcMYB10 via DNA methylation. The infiltrated fruits showed blocked anthocyanin biosynthesis, higher methylation of the PcMYB10 promoter, and lower expression of PcMYB10 and PcUFGT. We suggest that the methylation level of PcMYB10 is associated with the formation of the green-skinned sport in the MRB pear. The potential mechanism behind the regulation of anthocyanin biosynthesis is discussed.
DOI:10.3892/ijmm.2018.3378URLPMID:29328429 [本文引用: 1]

Oxidative stress is an important contributory factor resulting the development of kidney injury in patients with diabetes. Numerousin vitroandin vivostudies have suggested that anthocyanins, natural phenols commonly existing in numerous fruits and vegetables, exhibit important anti-oxidative, anti-inflammatory and antihyperlipidemic effects; however, their effects and underlying mechanisms on diabetic nephropathy (DN) have not yet been fully determined. In the present study, the regulation of apoptosis metabolism and antioxidative effects exhibited by anthocyanins [grape seed procyanidin (GSPE) and cyanidin-3-O-glucoside chloride (C3G)] were investigated, and the molecular mechanism underlying this process was investigatedin vivoandin vitro. GSPE administration was revealed to suppress renal cell apoptosis, as well as suppress the expression of Bcl-2 in diabetic mouse kidneys. Furthermore, GSPE administration was demonstrated to suppress the expression of thioredoxin interacting protein (TXNIP), in addition to enhancing p38 mitogen-activation protein kinase (MAPK) and extracellular signal-regulated kinase 1/2 (ERK1/2) oxidase activity in diabetic mouse kidneys.In vitroexperiments using HK-2 cells revealed that C3G suppressed the generation of HG-mediated reactive oxygen species, cellular apoptosis, the expression of cleaved caspase-3 and the Bax/Bcl-2 ratio; and enhanced the expression of cytochromecreleased from mitochondria. Furthermore, treatment with C3G was revealed to suppress the expression of TXNIP, in addition to the phosphorylation of p38 MAPK and ERK1/2 oxidase activity in HK-2 cells under HG conditions. In addition, treatment with C3G was revealed to attenuate the HG-induced suppression of the biological activity of thioredoxin, and to enhance the expression of thioredoxin 2 in HK-2 cells under HG conditions. In conclusion, the present study demonstrated that anthocyanins may exhibit protective effects against HG-induced renal injury in DN via antioxidant activity.
[本文引用: 1]
.
DOI:10.1371/journal.pone.0143892URLPMID:4664390 [本文引用: 1]

Chrysanthemums (Chrysanthemum morifoliumRamat.) exhibit a variety of flower colors due to their differing abilities to accumulate anthocyanins. OneMYBmember,CmMYB6, has been verified as a transcription regulator of chrysanthemum genes involved in anthocyanin biosynthesis; however, the co-regulators forCmMYB6remain unclear in chrysanthemum. Here, the expression pattern ofCmbHLH2, which is clustered in the IIIf bHLH subgroup, was shown to be positively correlated with the anthocyanin content of cultivars with red, pink and yellow flower colors, respectively.CmbHLH2significantly upregulated theCmDFRpromoter and triggered anthocyanin accumulation when co-expressed withCmMYB6. Yeast one-hybrid analyses indicated thatCmbHLH2was able to bind directly to theCmDFRpromoter. Moreover, yeast two-hybrid assays indicated protein-protein interaction betweenCmbHLH2andCmMYB6. These results suggest thatCmbHLH2is the essential partner forCmMYB6in regulating anthocyanin biosynthesis in chrysanthemum.
DOI:10.1111/j.1365-3040.2012.02523.xURLPMID:22519753 [本文引用: 1]

Low environmental temperatures promote anthocyanin accumulation and fruit colouration by up-regulating the expression of genes involved in anthocyanin biosynthesis and regulation in many fruit trees. However, the molecular mechanism by which fruit trees regulate this process in response to low temperature (LT) remains largely unknown. In this study, the cold-induced bHLH transcription factor gene MdbHLH3 was isolated from an apple tree and was found to interact physically and specifically through two regions (amino acids 1–23 and 186–228) at the N terminus with the MYB partner MdMYB1 (allelic to MdMYB10). Subsequently, MdbHLH3 bound to the promoters of the anthocyanin biosynthesis genes MdDFR and MdUFGT and the regulatory gene MdMYB1 to activate their expression. Furthermore, the MdbHLH3 protein was post-translationally modified, possibly involving phosphorylation following exposure to LTs, which enhanced its promoter-binding capacity and transcription activity. Our results demonstrate the molecular mechanism by which MdbHLH3 regulates LT-induced anthocyanin accumulation and fruit colouration in apple.
.
DOI:10.1007/s11240-017-1275-7URL [本文引用: 1]

The apple ( Malus domestica ) MdMYB1 gene encodes an R2R3-MYB transcription factor (TF) that regulates anthocyanin biosynthesis. The transient and stable expression of the apple basic helix-loop-helix 3 gene, MdbHLH3 , produces anthocyanin pigmentation in apple fruit and calli. In order to understand the MdMYB1- and MdbHLH3 -related regulatory mechanisms of anthocyanin biosynthesis, we established and characterized the expression of those two genes in stably transformed tobacco ( Nicotiana tabacum NC89) plants, including the phenotypic differences in anthocyanin-containing flowers, ovaries, anthers and anthocyanin-free leaves. A real-time PCR analysis showed that the expression levels of the MdMYB1 and MdbHLH3 transgenes were up-regulated in all tobacco organs. However, the bHLH and MYB TFs, NtAN1 and NtAN2 , were not detected in tobacco leaves, respectively, indicating that neither can individually activate anthocyanin biosynthesis. However, MdMYB1 induced pigment accumulation in ovaries because NtAN1 is active in that organ. Thus, the endogenous bHLH TF is necessary for anthocyanin biosynthesis in tobacco. Furthermore, MdMYB1 binding to the promoters of the anthocyanin biosynthesis genes MdDFR and MdUFGT activated their expression in apple. However, because NtAN2 is not activated in leaves or ovaries, the pigmentation patterns were not significantly altered in these organs in MdbHLH3 -overexpressing tobacco. Thus, the data indicated that MdMYB1 regulated anthocyanin biosynthesis directly by binding the MdDFR and MdUFGT promoters, and interacted with the tobacco bHLH TF, triggering anthocyanin accumulation.
.
DOI:10.1007/s11033-014-3404-2URLPMID:24859977

Basic helix-loop-helix ( bHLH)-type transcription factors play diverse roles in plant physiological response and stress-adaptive regulation network. Here, we identified one grapevine bHLH transcription factor from a cold-tolerant accession 'Heilongjiang seedling' of Chinese wild Vitis amurensis ( VabHLH1) as a transcriptional activator involved in cold stress. We also compared with its counterpart from a cold-sensitive Vitis vinifera cv. Cabernet Sauvignon ( VvbHLH1). These two putative proteins are characterized by the presence of the identically conserved regions of 54 amino acid residues of bHLH signature domain, and shared 99.1 % amino acid identity, whereas several stress-related cis-regulatory elements located in both promoter regions differed in types and positions. Expressions of two bHLHs in grapevine leaves were induced by cold stress, but evidently differ between two grapevine genotypes upon cold exposure. Two grapevine bHLH proteins were exclusively localized to the nucleus and exhibited strong transcriptional activation activities in yeast cells. Overexpression of either VabHLH1 or VvbHLH1 transcription factor did not affect the growth and development of transgenic Arabidopsis plants, but enhanced tolerance to cold stress. The improved tolerance in VabHLH1- or VvbHLH1-overexpressing Arabidopsis plants is associated with multiple physiological and biochemical changes that occurred during the time-course cold stress. These most common changes include the evaluated levels of proline, decreased amounts of malondialdehyde and reduced membrane injury as reflected by electrolyte leakage. VabHLH1 and VvbHLH1 displayed overlapping, but not identical, roles in activating the corresponding CBF cold signaling pathway, especially in regulating the expression of CBF3 and RD29A. Our findings demonstrated that two grapevine bHLHs act as positive regulators of the cold stress response, modulating the level of COR gene expression, which in turn confer tolerance to cold stress.
.
DOI:10.1093/pcp/pcq011URLPMID:20118109 [本文引用: 1]

Anthocyanins are secondary metabolites that contribute to colors of flowers, fruits and leaves. Asiatic hybrid lily (Lilium spp.) accumulates cyanidin anthocyanins in flower tepals, tepal spots and leaves of juvenile shoots. To clarify their mechanisms of regulation of anthocyanin pigmentation, two full-length cDNAs of R2R3-MYB (LhMYB6 and LhMYB12) were isolated from the anthocyanin-accumulating tepals of cultivar 'Montreux'. Analysis of the deduced amino acid sequences indicated they have homology with petunia AN2, homologous sequences of which had not been isolated in species of monocots. Yeast two-hybrid analysis showed that LhMYB6 and LhMYB12 interacted with the Lilium hybrid basic helix-loop-helix 2 (LhbHLH2) protein. Transient expression analysis indicated that co-expression of LhMYB6 and LhbHLH2 or LhMYB12 and LhbHLH2, introduced by a microprojectile, activated the transcription of anthocyanin biosynthesis genes in lily bulbscales. Spatial and temporal transcription of LhMYB6 and LhMYB12 was analyzed. The expression of LhMYB12 corresponded well with anthocyanin pigmentation in tepals, filaments and styles, and that of LhMYB6 correlated with anthocyanin spots in tepals and light-induced pigmentation in leaves. These results indicate that LhMYB6 and LhMYB12 positively regulate anthocyanin biosynthesis and determine organ- and tissue-specific accumulation of anthocyanin.
DOI:10.1111/nph.12572URLPMID:24180488

Many angiosperm families develop spatially regulated anthocyanin spots on their flowers. The Asiatic hybrid lily (Lilium spp.) cv 090004Latvia090005 develops splatter-type spots on its tepals. The splatters arise simply from the deposition of anthocyanin pigments in the tepal epidermis.To determine how splatter development was regulated, we analysed the transcription of anthocyanin biosynthesis genes, and isolated and characterized an R2R3-MYB gene specific to splatter pigmentation.All anthocyanin biosynthesis genes were expressed in splatter-containing regions of tepals, but not in other regions, indicating that splatter pigmentation is caused by the transcriptional regulation of biosynthesis genes. Previously characterized LhMYB12 regulators were not involved in splatter pigmentation, but, instead, a new allele of the LhMYB12 gene, LhMYB12-Lat, isolated in this study, contributed to splatter development. In 090004Latvia090005 and other lily plants expressing splatters, LhMYB12-Lat was preferentially transcribed in the splatter-containing region of tepals. Progeny segregation analysis showed that LhMYB12-Lat genotype and splatter phenotype were co-segregated among the F1 population, indicating that LhMYB12-Lat determines the presence or absence of splatters.LhMYB12-Lat contributes to splatter development, but not to full-tepal pigmentation and raised spot pigmentation. As a result of its unique sequences and different transcription profiles, this new allele of LhMYB12 should be a novel R2R3-MYB specifically associating with splatter spot development.
.
.
.
DOI:10.1007/s00425-009-1013-4URLPMID:19756724 [本文引用: 1]

Abstract The color of red cabbage (Brassica oleracea var. capitata) is due to anthocyanin accumulation. To investigate the regulatory control of anthocyanin production in red cabbage, the expression of anthocyanin biosynthetic and regulatory genes from eight commercial cultivars was examined. While the four green varieties had negligible amount of anthocyanins under normal growth condition, the four red cultivars contained up to 1.60 mg g(-1) fresh weight. HPLC analysis of the four red cultivars revealed that they produced similar composition of various forms of cyanidin glucosides but at different concentrations. Molecular analysis indicated that all the red cabbage shared common mechanism of regulatory control for anthocyanin biosynthesis. Except CHI which showed similar expression levels between green and red cultivars, the other structural genes, CHS, F3H, F3'H, DFR, LDOX, and GST, were constitutively up-regulated during all stages of vegetative growth in red varieties. The expression of these structural genes was also dramatically increased in green and red cabbage under nutrient stresses. The increased expression of the structural genes coincided with a coordinated increase in transcript levels of a bHLH gene, BoTT8, and a MYB transcription factor, BoMYB2. These results indicate that activation of these two regulatory factors by unknown mechanisms constitutively up-regulates nearly the entire pathway genes for the onset of anthocyanin biosynthesis in red cabbage. Moreover, the amount of total anthocyanins in red cabbage was found to be positively correlated with total antioxidant power, implicating the potential health benefit of red cabbage to human health.
.
[本文引用: 1]
DOI:10.1242/dev.00681URLPMID:12917293 [本文引用: 3]

GLABRA3 (GL3) encodes a bHLH protein that interacts with the WD repeat protein, TTG1. GL3 overexpression suppresses the trichome defect of the pleiotropic ttg1 mutations. However, single gl3 mutations only affect the trichome pathway with a modest trichome number reduction. A novel unlinked bHLH-encoding locus is described here, ENHANCER OF GLABRA3 (EGL3). When mutated, egl3 gives totally glabrous plants only in the gl3 mutant background. The double bHLH mutant, gl3 egl3, has a pleiotropic phenotype like ttg1 having defective anthocyanin production, seed coat mucilage production, and position-dependent root hair spacing. Furthermore, the triple bHLH mutant, gl3 egl3 tt8, phenocopies the ttg1 mutation. Yeast two-hybrid and plant overexpression studies show that EGL3, like GL3, interacts with TTG1, the myb proteins GL1, PAP1 and 2, CPC and TRY, and it will form heterodimers with GL3. These results suggest a combinatorial model for TTG1-dependent pathway regulation by this trio of partially functionally redundant bHLH proteins.
[本文引用: 1]
DOI:10.1105/tpc.109.070441URLPMID:20009022 [本文引用: 2]

In rice (Oryza sativa), brassinosteroids (BRs) induce cell elongation at the adaxial side of the lamina joint to promote leaf bending. We identified a rice mutant (ili1-D) showing an increased lamina inclination phenotype similar to that caused by BR treatment. The ili1-D mutant overexpresses an HLH protein homologous to Arabidopsis thaliana Paclobutrazol Resistance1 (PRE1) and the human Inhibitor of DNA binding proteins. Overexpression and RNA interference suppression of ILI1 increase and reduce, respectively, rice laminar inclination, confirming a positive role of ILI1 in leaf bending. ILI1 and PRE1 interact with basic helix-loop-helix (bHLH) protein IBH1 (ILI1 binding bHLH), whose overexpression causes erect leaf in rice and dwarfism in Arabidopsis. Overexpression of ILI1 or PRE1 increases cell elongation and suppresses dwarf phenotypes caused by overexpression of IBH1 in Arabidopsis. Thus, ILI1 and PRE1 may inactivate inhibitory bHLH transcription factors through heterodimerization. BR increases the RNA levels of ILI1 and PRE1 but represses IBH1 through the transcription factor BZR1. The spatial and temporal expression patterns support roles of ILI1 in laminar joint bending and PRE1/At IBH1 in the transition from growth of young organs to growth arrest. These results demonstrate a conserved mechanism of BR regulation of plant development through a pair of antagonizing HLH/bHLH transcription factors that act downstream of BZR1 in Arabidopsis and rice.
.
DOI:10.1007/s11240-017-1294-4URL

Flower color is an important economic trait of Prunus mume , a famous ornamental plant widely cultivated in East Asia. Anthocyanins, acting as major pigments of flower coloration, are biosynthesized via transcriptional regulation of transcription factors (TFs). Many R2R3-MYB TFs have been identified to regulate anthocyanin biosynthetic pathways. However, very little is known about the role of R2R3-MYB TFs regulating anthocyanin biosynthesis in P. mume . In our study, the first R2R3-MYB TF ( PmMYBa1 ) from P. mume has been isolated and characterized. Sequence analysis revealed that PmMYBa1 contains conserved R2R3 MYB domain and belongs to anthocyanin-related subgroup 6 of R2R3-MYB family. Overexpression of PmMYBa1 in tobacco contributed to anthocyanin accumulation by activating endogenous anthocyanin-relating genes in the flowers and fruits of transgenic lines. Gene expression analysis showed that almost all of the endogenous structural genes of anthocyanin biosynthesis were obviously up-regulated, as well as bHLH TFs An1a and An1b in the flowers and fruits of PmMYBa1 -overexpressing tobacco. In contrast, only three structural genes NtCHS, NtF3H , and NtANS were up-regulated in the leaves. In addition, the expression level of PmMYBa1 was higher in the anthocyanin-rich red flowers than in white ones and strongly correlated with anthocyanin content in the developing petals of P. mume . These findings strongly suggest that PmMYBa1 is involved in regulating anthocyanin biosynthesis in P. mume . Moreover, PmDFR and PmANS might be potential target genes regulated by PmMYBa1 in P. mume according to the correlation analysis. Isolation and functional characterization of PmMYBa1 laid a foundation for further exploring the mechanisms of anthocyanin synthesis and provide a potential candidate gene for breeding to manipulate flower colors in P. mume .
.
[本文引用: 1]
[本文引用: 1]
.
DOI:10.1093/mp/ssp030URLPMID:1982565621 [本文引用: 1]

Single-repeat R3 MYB transcription factors like CPC (CAPRICE) are known to play roles in developmental processes such as root hair differentiation and trichome initiation. However, none of the six Arabidopsis single-repeat R3 MYB members has been reported to regulate flavonoid biosynthesis. We show here that CPC is a negative regulator of anthocyanin biosynthesis. In the process of using CPC to test GAL4-dependent driver lines, we observed a repression of anthocyanin synthesis upon GAL4-mediated CPC overexpression. We demonstrated that this is not due to an increase in nutrient uptake because of more root hairs. Rather, CPC expression level tightly controls anthocyanin accumulation. Microarray analysis on the whole genome showed that, of 37 000 features tested, 85 genes are repressed greater than three-fold by CPC overexpression. Of these 85, seven are late anthocyanin biosynthesis genes. Also, anthocyanin synthesis genes were shown to be down-regulated in 35S::CPC overexpression plants. Transient expression results suggest that CPC competes with the R2R3 YB transcription factor PAP1/2, which is an activator of anthocyanin biosynthesis genes. This report adds anthocyanin biosynthesis to the set of programs that are under CPC control, indicating that this regulator is not only for developmental programs (e.g. root hairs, trichomes), but can influence anthocyanin pigment synthesis.
DOI:10.1016/j.molp.2017.05.008URLPMID:28666688 [本文引用: 2]
DOI:10.1373/clinchem.2011.167361URLPMID:21926181 [本文引用: 1]

Abstract BACKGROUND: Anthocyanins have been shown to improve endothelial function in animal models. However, whether these compounds have similar beneficial effects in humans is largely unknown. METHODS: In a short-term crossover study, 12 hypercholesterolemic individuals were given oral anthocyanins (320 mg) isolated from berries or placebo. Brachial artery flow-mediated dilation (FMD) was assessed before and after the intervention. In a long-term intervention trial (12 weeks), 150 hypercholesterolemic individuals were given anthocyanins (320 mg/day, n = 75) or placebo (n = 75), after which we measured FMD, plasma cGMP, and other serum biomarkers. Another short-term intervention was conducted in the presence of NO-cGMP inhibitors in 6 people and in a rat aortic ring model (n = 8). RESULTS: Significant increases of FMD from 8.3% (0.6%) at baseline to 11.0% (0.8%) at 1 h and 10.1% (0.9%) at 2 h were observed after short-term anthocyanin consumption, concomitantly with increases of plasma anthocyanin concentrations (P < 0.05). In the study participants who received long-term anthocyanin intervention, compared with the control group, we observed significant increases in the FMD (28.4% vs 2.2%), cGMP (12.6% vs -1.2%), and HDL-cholesterol concentrations, but decreases in the serum soluble vascular adhesion molecule-1 and LDL cholesterol concentrations (P < 0.05). The changes in the cGMP and HDL cholesterol concentrations positively correlated with FMD in the anthocyanin group (P < 0.05). In the presence of NO-cGMP inhibitors, the effects of anthocyanin on endothelial function were abolished in human participants and in a rat aortic ring model. CONCLUSIONS: Anthocyanin supplementation improves endothelium-dependent vasodilation in hypercholesterolemic individuals. This effect involves activation of the NO-cGMP signaling pathway, improvements in the serum lipid profile, and decreased inflammation.
.
DOI:10.1007/s11295-009-0232-yURL

Members of the MYB and MYC family regulate the biosynthesis of phenylpropanoids in several plant species. Two sequences, called CsMYB8 and CsMYC2 , were identified from Citrus sinensis , and both the cDNA and the genomic clones were isolated and characterized from the flesh of common and blood oranges. Analysis by real-time polymerase chain reaction showed that the expression pattern of CsMYC2 is generally higher in rind than in flesh and in blood oranges than in common ones. In contrast, no significant difference in expression was observed for CsMYB8 . The expression pattern of the structural genes chalcone synthase , anthocyanidin synthase , and UDP-glucose lavonoid 3-O-glucosyltransferase , which code for three enzymes involved in the anthocyanin biosynthetic pathway, was also analyzed and correlated with CsMYC2 , in flesh, rind, and leaf of the common and blood oranges, and in leaf of Citrus limon cultivars (characterized by anthocyanin absence or variable content). Surprisingly, CsMYC2 is highly expressed in the leaf and expression is correlated with UFGT expression in this organ. These results suggest that CsMYC2 is involved in the regulation of the flavonoid biosynthetic pathway in Citrus .
.
DOI:10.1007/s11103-008-9446-xURLPMID:19096760 [本文引用: 1]

The colour of the red wine is essentially due to the release of anthocyanins from the red skin of grape berries during the process of wine making. Anthocyanins are synthesized during ripening of the berries under the control of VvMYBA1 transcription factor that controls the expression of UFGT. In order to identify the whole set of downstream regulated genes, we targeted constitutive ectopic expression of VlmybA1-2 into grapevine hairy roots and plants. The ectopic expression of VlmybA1-2 triggered de novo production and storage of anthocyanins in all transgenic vegetative organs, leading to a very intense red coloration, and did not interfere with proanthocyanidin (PA) biosynthesis. The ectopic red pigmentation was due to the accumulation of anthocyanins in vacuoles and anthocyanin vacuolar inclusion (AVIs) in all organs but only in specific tissues. A transcriptomic analysis using a 14K oligoarray revealed that the ectopic expression of VlmybA1-2 activated only few genes, most of which are involved in both PA and anthocyanin biosynthesis, while the expression of BAN and LAR (two specific genes of the PA biosynthesis pathway) was unaffected. Among these, 4 genes emerged given the amplitude of their up-regulation, quantitatively similar to VlmybA1-2 itself. In addition to the previously described UFGT, this set comprised an isogen of GST, an O-methyltransferase, both of which are supposed to play a role in the anthocyanin biosynthesis pathway, as well as a candidate gene putatively involved in the vacuolar anthocyanin transport in grapevine (anthoMATE). Together, these results suggest that MybA1 activates the last steps of anthocyanin synthesis and transport through the regulation of a narrow, specific spectrum of genes regulated as a cluster.
.
DOI:10.1101/gad.11.11.1422URLPMID:9192870 [本文引用: 1]

In petunia flowers, the loci an1, an2, and an11 control the pigmentation of the flower by stimulating the transcription of anthocyanin biosynthetic genes. The an1 and an2 locus were recently cloned and encode a basic helix-loop-helix (bHLH) and MYB-domain transcriptional activator, respectively. Here, we report the isolation of the an11 locus by transposon tagging. RNA gel blot experiments show that an11 is expressed independently from an1 and an2 throughout plant development, as well as in tissues that do not express the anthocyanin pathway. It encodes a novel WD-repeat protein that is highly conserved even in species that do not produce anthocyanins such as yeast, nematodes, and mammals. The observation that the human an11 homolog partially complements the an11 petunia mutant in transient assays shows that sequence similarity reflects functional conservation. Overexpression of an2 in an11- petals restored the activity of a structural anthocyanin gene in transient assays, indicating that AN11 acts upstream of AN2. Cell fractionation experiments show that the bulk of the AN11 protein is localized in the cytoplasm. Taken together, this indicates that AN11 is a cytoplasmic component of a conserved signal transduction cascade that modulates AN2 function in petunia, thereby linking cellular signals with transcriptional activation.
DOI:10.1104/pp.105.067231URLPMID:16384897

The ripening of grape (Vitis vinifera) berry is characterized by dramatic changes in gene expression, enzymatic activities, and metabolism that lead to the production of compounds essential for berry quality. The phenylpropanoid metabolic pathway is one of the components involved in these changes. In this study, we describe the cloning and functional characterization of VvMYB5a, a cDNA isolated from a grape L. cv Cabernet Sauvignon berry library. VvMYB5a encodes a protein belonging to a small subfamily of R2R3-MYB transcription factors. Expression studies in grapevine indicate that the VvMYB5a gene is mainly expressed during the early steps of berry development in skin, flesh, and seeds. Overexpression of VvMYB5a in tobacco (Nicotiana tabacum) affects the expression of structural genes controlling the synthesis of phenylpropanoid and impacts on the metabolism of anthocyanins, flavonols, tannins, and lignins. Overexpressing VvMYB5a induces a strong accumulation of several phenolic compounds, including keracyanin (cyanidin-3-rhamnoglucoside) and quercetin-3-rhamnoglucoside, which are the main anthocyanin and flavonol compounds in tobacco. In addition, VvMYB5a overexpression increases the biosynthesis of condensed tannins and alters lignin metabolism. These findings suggest that VvMYB5a may be involved in the control of different branches of the phenylpropanoid pathway in grapevine.
DOI:10.1104/pp.108.118919URL [本文引用: 1]

Among the dramatic changes occurring during grape berry (Vitis vinifera) development, those affecting the flavonoid pathway have provoked a number of investigations in the last 10 years. In addition to producing several compounds involved in the protection of the berry and the dissemination of the seeds, final products of this pathway also play a critical role in berry and wine quality. In this article, we describe the cloning and functional characterization of VvMYB5b, a cDNA isolated from a grape berry (V. vinifera 'Cabernet Sauvignon') library. VvMYB5b encodes a protein belonging to the R2R3-MYB family of transcription factors and displays significant similarity with VvMYB5a, another MYB factor recently shown to regulate flavonoid synthesis in grapevine. The ability of VvMYB5a and VvMYB5b to activate the grapevine promoters of several structural genes of the flavonoid pathway was confirmed by transient expression of the corresponding cDNAs in grape cells. Overexpression of VvMYB5b in tobacco (Nicotiana tabacum) leads to an up-regulation of genes encoding enzymes of the flavonoid pathway and results in the accumulation of anthocyanin- and proanthocyanidin-derived compounds. The ability of VvMYB5b to regulate particularly the anthocyanin and the proanthocyanidin pathways is discussed in relation to other recently characterized MYB transcription factors in grapevine. Taken together, data presented in this article give insight into the transcriptional mechanisms associated with the regulation of the flavonoid pathway throughout grape berry development.
DOI:10.1104/pp.102.017319URLPMID:12644640 [本文引用: 1]

Article on legume natural products and understanding and manipulating complex pathways for human and animal health.
DOI:10.1007/s11738-014-1487-yURL

Many transcriptional factors including the R2R3-MYB domain, basic helix-loop-helix (bHLH) domain and WD40 repeat proteins, which regulate flavonoid biosynthesis, have been identified in various plant species. However, there is little information on WD40 proteins in underground organs. In this study, a WD40-repeat protein gene was isolated from purple-fleshed sweet potato ( Ipomoea batatas (L.) Lam. cv. Yamakawamurasaki) ( IbWD40 ). The expression patterns of this gene were positively correlated with anthocyanin accumulation in different sweet potato cultivars. An IbWD40-GFP fusion protein was observed only in the nucleus of onion epidermal cells, which was consistent with its role as a transcriptional regulator. Stable transformation analysis revealed that IbWD40 was up-regulated in Arabidopsis thaliana seedlings, which accumulated anthocyanins, with possible additional effects on the formation of other flavonoid compounds in other tissues. These results suggest that in storage roots of purple-fleshed sweet potato the activity of IbWD40 plays a critical role in the regulation of anthocyanin biosynthesis.
.
[本文引用: 1]
[本文引用: 1]
.
DOI:10.2307/4281500URLPMID:14605235 [本文引用: 2]

We have identified an R2R3-type MYB factor, GMYB10, from Gerbera hybrida (Asteraceae) that shares high sequence homology to and is phylogenetically grouped together with the previously characterized regulators of anthocyanin pigmentation in petunia (Petunia hybrida) and Arabidopsis. GMYB10 is able to induce anthocyanin pigmentation in transgenic tobacco (Nicotiana tabacum), especially in vegetative parts and anthers. In G. hybrida, GMYB10 is involved in activation of anthocyanin biosynthesis in leaves, floral stems, and flowers. In flowers, its expression is restricted to petal epidermal cell layers in correlation with the anthocyanin accumulation pattern. We have shown, using yeast (Saccharomyces cerevisiae) two-hybrid assay, that GMYB10 interacts with the previously isolated bHLH factor GMYC1. Particle bombardment analysis was used to show that GMYB10 is required for activation of a late anthocyanin biosynthetic gene promoter, PGDFR2. cis-Analysis of the target PGDFR2 revealed a sequence element with a key role in activation by GMYB10/GMYC1. This element shares high homology with the anthocyanin regulatory elements characterized in maize (Zea mays) anthocyanin promoters, suggesting that the regulatory mechanisms involved in activation of anthocyanin biosynthesis have been conserved for over 125 million years not only at the level of transcriptional regulators but also at the level of the biosynthetic gene promoters.
DOI:10.1105/tpc.108.059329URL

Mutations in the genes encoding for either the biosynthetic or transcriptional regulation of the anthocyanin pathway have been linked to color phenotypes. Generally, this is a loss of function resulting in a reduction or a change in the distribution of anthocyanin. Here, we describe a rearrangement in the upstream regulatory region of the gene encoding an apple (Malus 脳 domestica) anthocyanin-regulating transcription factor, MYB10. We show that this modification is responsible for increasing the level of anthocyanin throughout the plant to produce a striking phenotype that includes red foliage and red fruit flesh. This rearrangement is a series of multiple repeats, forming a minisatellite-like structure that comprises five direct tandem repeats of a 23-bp sequence. This MYB10 rearrangement is present in all the red foliage apple varieties and species tested but in none of the white fleshed varieties. Transient assays demonstrated that the 23-bp sequence motif is a target of the MYB10 protein itself, and the number of repeat units correlates with an increase in transactivation by MYB10 protein. We show that the repeat motif is capable of binding MYB10 protein in electrophoretic mobility shift assays. Taken together, these results indicate that an allelic rearrangement in the promoter of MYB10 has generated an autoregulatory locus, and this autoregulation is sufficient to account for the increase in MYB10 transcript levels and subsequent ectopic accumulation of anthocyanins throughout the plant.
DOI:10.1111/j.1365-313X.2006.02964.xURLPMID:1865000 [本文引用: 3]

Anthocyanin concentration is an important determinant of the colour of many fruits. In apple ( Malus domestica ), centuries of breeding have produced numerous varieties in which levels of anthocyanin pigment vary widely and change in response to environmental and developmental stimuli. The apple fruit cortex is usually colourless, although germplasm does exist where the cortex is highly pigmented due to the accumulation of either anthocyanins or carotenoids. From studies in a diverse array of plant species, it is apparent that anthocyanin biosynthesis is controlled at the level of transcription. Here we report the transcript levels of the anthocyanin biosynthetic genes in a red-fleshed apple compared with a white-fleshed cultivar. We also describe an apple MYB transcription factor, MdMYB10 , that is similar in sequence to known anthocyanin regulators in other species. We further show that this transcription factor can induce anthocyanin accumulation in both heterologous and homologous systems, generating pigmented patches in transient assays in tobacco leaves and highly pigmented apple plants following stable transformation with constitutively expressed MdMYB10. Efficient induction of anthocyanin biosynthesis in transient assays by MdMYB10 was dependent on the co-expression of two distinct bHLH proteins from apple, MdbHLH3 and MdbHLH33. The strong correlation between the expression of MdMYB10 and apple anthocyanin levels during fruit development suggests that this transcription factor is responsible for controlling anthocyanin biosynthesis in apple fruit; in the red-fleshed cultivar and in the skin of other varieties, there is an induction of MdMYB10 expression concurrent with colour formation during development. Characterization of MdMYB10 has implications for the development of new varieties through classical breeding or a biotechnological approach.
.
[本文引用: 1]
DOI:10.1039/c4fo01036aURLPMID:25758596 [本文引用: 1]

Abstract Serum high-density lipoprotein-cholesterol (HDL-C) is a risk factor considered to be protective of atherosclerosis. However, atherosclerosis is an inflammatory disease and contributes to impairment in high-density lipoprotein (HDL) function, including reductions in HDL-C, HDL antioxidant and anti-inflammatory activities. Anthocyanins are polyphenols that have demonstrated antioxidant and anti-inflammatory properties. The objective of this study was to determine whether an anthocyanin-rich black elderberry extract (Sambucus nigra) (BEE) (13% anthocyanins) would protect against inflammation-related impairments in HDL function and atherosclerosis in apoE(-/-) mice, a mouse model of hyperlipidemia and HDL dysfunction. We fed an AIN-93M diet supplemented with 1.25% (w/w) BEE or control diet to 10 week old male apoE(-/-) mice for 6 weeks. The BEE fed to mice was rich in cyanidin 3-sambubioside (9.8% w/w) and cyanidin 3-glucoside (3.8% w/w). After 6 weeks, serum lipids did not differ significantly between groups, while aspartate transaminase (AST) and fasting glucose were reduced in BEE-fed mice. Hepatic and intestinal mRNA changes with BEE-feeding were consistent with an improvement in HDL function (Apoa1, Pon1, Saa1, Lcat, Clu) and a reduction in hepatic cholesterol levels (increased Ldlr and Hmgcr, reduced Cyp7a1). In BEE-fed mice, serum paraoxonase-1 (PON1) arylesterase activity was significantly higher. In addition, mice fed BEE had significantly lower serum chemokine (C-C motif) ligand 2 (CCL2) compared to control-fed mice. Notably, we observed significant reductions in total cholesterol content of the aorta of BEE-fed mice, indicating less atherosclerosis progression. This study suggests that black elderberry may have the potential to influence HDL dysfunction associated with chronic inflammation by impacting hepatic gene expression.
.
DOI:10.1016/j.epsl.2008.07.045URLPMID:19621239 [本文引用: 1]

The bHLH transcription factors EGL3 (ENHANCER OF GLABRA3) and its close homologue GL3 (GLABRA3) are important regulators of the anthocyanin pathway in Arabidopsis thaliana, and together with TTG1 (a WD40 repeat protein) and MYB transcription factors regulate specific genes in the pathway. In response to nitrogen depletion, the MYB genes PAP1/PAP2 (production of anthocyanin pigment 1/2) and GL3 are strongly induced, and anthocyanin synthesis is activated in seedlings and rosette stage plants. In this study we show that anthocyanins accumulate in both wild type and egl3, but not in gl3 loss-of-function mutants when depleted of nitrogen. Several structural genes of flavonoid metabolism including CHS (chalcone synthase), FLS1 (flavonol synthase 1) and ANS (anthocyanidin synthase) were induced in response to nitrogen depletion in wild type as well as in the egl3 and gl3 mutants. Strikingly, in the gl3 mutant DFR (dihydroflavonol-4-reductase) transcript level was only 2% of the levels in wild type or egl3 mutant. Hence, low expression of DFR appears to be the bottleneck preventing anthocyanin synthesis in the gl3 mutant. The specific effect on DFR, but not ANS is compatible with involvement of the MYBL2 inhibitor.
DOI:10.1186/1471-2164-14-28URLPMID:3618344

Red colour in kiwifruit results from the presence of anthocyanin pigments. Their expression, however, is complex, and varies among genotypes, species, tissues and environments. An understanding of the biosynthesis, physiology and genetics of the anthocyanins involved, and the control of their expression in different tissues, is required. A complex, the MBW complex, consisting of R2R3-MYB and bHLH transcription factors together with a WD-repeat protein, activates anthocyanin 3-O-galactosyltransferase (F3GT1) to produce anthocyanins. We examined the expression and genetic control of anthocyanins in flowers of Actinidia hybrid families segregating for red and white petal colour. Four inter-related backcross families between Actinidia chinensis Planch. var. chinensis and Actinidia eriantha Benth. were identified that segregated 1:1 for red or white petal colour. Flower pigments consisted of five known anthocyanins (two delphinidin-based and three cyanidin-based) and three unknowns. Intensity and hue differed in red petals from pale pink to deep magenta, and while intensity of colour increased with total concentration of anthocyanin, no association was found between any particular anthocyanin data and hue. Real time qPCR demonstrated that an R2R3 MYB, MYB110a, was expressed at significant levels in red-petalled progeny, but not in individuals with white petals. A microsatellite marker was developed that identified alleles that segregated with red petal colour, but not with ovary, stamen filament, or fruit flesh colour in these families. The marker mapped to chromosome 10 in Actinidia. The white petal phenotype was complemented by syringing Agrobacterium tumefaciens carrying Actinidia 35S::MYB110a into the petal tissue. Red pigments developed in white petals both with, and without, co-transformation with Actinidia bHLH partners. MYB110a was shown to directly activate Actinidia F3GT1 in transient assays. The transcription factor, MYB110a, regulates anthocyanin production in petals in this hybrid population, but not in other flower tissues or mature fruit. The identification of delphinidin-based anthocyanins in these flowers provides candidates for colour enhancement in novel fruits.
.
DOI:10.1016/j.jbiotec.2004.02.019URLPMID:15246659

Suppression of biosynthetic genes involved in flower color formation is an important approach for obtaining target flower colors. Here we report that flower color of the garden plant Torenia hybrida was successfully modulated by RNA interference (RNAi) against a gene of chalcone synthase (CHS), a key enzyme for anthocyanin and flavonoid biosynthesis. By using each of the coding region and the 3-untranslated region of the CHS mRNA as an RNAi target, exhaustive and gene-specific gene silencing were successfully induced, and the original blue flower color was modulated to white and pale colors, respectively. Our results indicate that RNAi is quite useful for modulations of flower colors of commercially important garden plants.
[本文引用: 1]
.
[本文引用: 1]
.
DOI:10.1007/s11816-006-0001-4URL [本文引用: 2]

An myb-related transcription factor gene of the anthocyanin biosynthetic pathway, VlmybA2, from the Kyoho grape (Vitis labruscana) was introduced into tobacco and Arabidopsis under the control of the cauliflower mosaic virus 35S promoter. The 35S: VlmybA2-induced anthocyanin production was prominent in transformed tobacco calli, and the regenerated tobacco plants were completely purple. Except for the color, the transgenic plants were apparently not different from the control plants. During plant growth in pots, the purple color was not uniformly distributed but appeared as patches in the leaves, whereas the flowers showed intense pigmentation. In Arabidopsis, T1 transformants showing two prominent phenotypes: completely purple seedlings and seedlings with green leaves and purple roots. The partially purple seedlings grown in pots produced fertile and viable seeds of two distinguishable colors, purple and brown. VlmybA2 alone, without the aid of other myc-related genes, could induce complete pigmentation in tobacco and Arabidopsis, indicating its potential over other previously used myb- and myc-related genes.
DOI:10.1007/s00497-016-0295-5URLPMID:27896439 [本文引用: 1]

Abstract Key messagebZIP TF network in pollen. AbstractTranscriptional control of gene expression represents an important mechanism guiding organisms through developmental processes and providing plasticity towards environmental stimuli. Because of their sessile nature, plants require effective gene regulation for rapid response to variation in environmental and developmental conditions. Transcription factors (TFs) provide such control ensuring correct gene expression in spatial and temporal manner. Our work reports the interaction network of six bZIP TFs expressed in Arabidopsis thaliana pollen and highlights the potential functional role for AtbZIP18 in pollen. AtbZIP18 was shown to interact with three other pollen-expressed bZIP TFs tbZIP34, AtbZIP52, and AtbZIP61 in yeast two-hybrid assays. AtbZIP18 transcripts are highly expressed in pollen, and at the subcellular level, an AtbZIP18-GFP fusion protein was located in the nucleus and cytoplasm/ER. To address the role of AtbZIP18 in the male gametophyte, we performed phenotypic analysis of a T-DNA knockout allele, which showed slightly reduced transmission through the male gametophyte. Some of the phenotype defects in atbzip18 pollen, although observed at low penetrance, were similar to those seen at higher frequency in the T-DNA knockout of the interacting partner, AtbZIP34. To gain deeper insight into the regulatory role of AtbZIP18, we analysed atbzip18/ pollen microarray data. Our results point towards a potential repressive role for AtbZIP18 and its functional redundancy with AtbZIP34 in pollen.
.
[本文引用: 1]
DOI:10.1111/j.1365-313X.2007.03373.xURLPMID:18036197 [本文引用: 1]

In all higher plants studied to date, the anthocyanin pigment pathway is regulated by a suite of transcription factors that include Myb, bHLH and WD-repeat proteins. However, in Arabidopsis thaliana , the Myb regulators remain to be conclusively identified, and little is known about anthocyanin pathway regulation by TTG1-dependent transcriptional complexes. Previous overexpression of the PAP1 Myb suggested that genes from the entire phenylpropanoid pathway are targets of regulation by Myb/bHLH/WD-repeat complexes in Arabidopsis, in contrast to other plants. Here we demonstrate that overexpression of Myb113 or Myb114 results in substantial increases in pigment production similar to those previously seen as a result of over-expression of PAP1 , and pigment production in these overexpressors remains TTG1- and bHLH-dependent. Also, plants harboring an RNAi construct targeting PAP1 and three Myb candidates ( PAP2 , Myb113 and Myb114 ) showed downregulated Myb gene expression and obvious anthocyanin deficiencies. Correlated with these anthocyanin deficiencies is downregulation of the same late anthocyanin structural genes that are downregulated in ttg1 and bHLH anthocyanin mutants. Expression studies using GL3:GR and TTG1:GR fusions revealed direct regulation of the late biosynthetic genes only. Functional diversification between GL3 and EGL3 with regard to activation of gene targets was revealed by GL3:GR studies in single and double bHLH mutant seedlings. Expression profiles for Myb and bHLH regulators are also presented in the context of pigment production in young seedlings.
[本文引用: 1]
DOI:10.1111/ppl.12475URLPMID:27229540

Abstract In this investigation, we report metabolic engineering of anthocyanins in two dark tobacco crops (Narrow Leaf Madole and KY171) and evaluate effects of anthocyanin engineering on physiological features of plant photosynthesis. Arabidopsis PAP1 (production of anthocyanin pigment 1) gene (AtPAP1) encodes a R2R3-type MYB transcript factor that is a master component of regulatory complexes controlling anthocyanin biosynthesis. AtPAP1 was introduced to Narrow Leaf Madole and KY171 plants. Multiple transgenic plants developed red/purple pigmentation in different tissues. qRT-PCR analysis showed that the expression levels of six pathway genes were increased 2-8 fold in AtPAP1 transgenic plants compared to vector control plants. Dihydroflavonol reductase and anthocyanidin synthase genes that were not expressed in wild-type plants were activated. Spectrophotometric measurement showed that anthocyanins were 400 to 800 g g(-1) FW in AtPAP1 transgenic plants. High performance liquid chromatograph (HPLC) analysis showed that one main anthocyanin molecule accounted for approximately 98% of the total anthocyanins. Tandem MS/MS analysis using HPLC coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry identified the main anthocyanin as cyanidin 3-O-rutinoside, an important medicinal anthocyanin. Analysis of photosynthesis rate, chlorophylls and carotenoids contents showed no differences between red/purple transgenic and control plants, indicating that this metabolic engineering did not alter photosynthetic physiological traits. This study shows that AtPAP1 is of significance for metabolic engineering of anthocyanins in crop plants for value-added traits.
The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine
0
2010
Genetics and biochemistry of anthocyanin biosynthesis
1
1995
... 花青素来源于类黄酮化合物的合成途径: 衍生于香豆酰辅酶A (4-coumaroyl CoA)和丙二酰辅酶A (malon- yl CoA), 在查尔酮合酶(chalcone synthase, CHS)作用下合成查尔酮(chalcone).查尔酮经查尔酮异构酶(chalcone isomerase, CHI)催化形成黄烷酮(flavo- nones).黄烷酮再经过黄烷酮3-羟化酶(flavanone 3-hydroxylase, F3H)催化形成二氢黄酮醇(dihydro flavonols).在花青素特异的分支途径中, 二氢黄酮醇还可以被黄酮羟化酶(flavonoid 3’-hydroxylase, F3’H和flavonoid 3’,5’-hydroxylase, F3’5’H)催化, 并在二氢黄酮醇还原酶(dihydroflavonol-4-reductase, DFR)作用下还原为无色花青素(leucoanthocyanidins), 无色花青素在花青素合成酶(anthocyanidin synthase, ANS)的催化作用下形成花青素苷元(anthocyani- dins), 不稳定的花青素苷元经糖基转移酶(glycosylt- ransferase, UGTs)修饰形成稳定的花青素苷(anthocyanins) (
A repressor motif-containing poplar R3 MYB-like transcription factor regulates epidermal cell fate determination and antho- cyanin biosynthesis in Arabidopsis
1
2016
... 除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物.葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(
SRWD: a novel WD40 protein subfamily regulated by salt stress in rice ( Oryza sativa L.)
1
2008
... WD40重复蛋白(WD40 repeat proteins, WDR)主要存在于真核生物中, 具有保守而特异的二肽重复基序, 每个重复大概有40多个氨基酸残基; 重复的WD40基序在蛋白质互作时作为支架起固定作用(
Functional characteri- zation of a novel R2R3-MYB transcription factor modula- ting the flavonoid biosynthetic pathway from Epimedium sagittatum
0
2017
A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway
0
2013
环境因子调控植物花青素苷合成及呈色的机理
1
2010
... 目前, 随着分子生物学的发展, 花青素生物合成途径已经逐渐被阐明, 转录因子单独或协同调控花青素生物合成的分子机制也正在被不断完善.前人已经对花青素的生物合成和转运、环境因子对花青素合成的影响进行了详细的总结(
突破复杂性状多基因转化技术壁垒, 首创胚乳花青素高积累的水稻新种质
1
2017
... Butelli等(2008)将金鱼草的DELILA和ROSEA1基因转入番茄(Solanum lycopersicum cv. ‘MicroTom’), 使转基因番茄的果皮和果肉因大量富集酰基化花青素而呈现出深紫色, 转基因番茄的花青素含量达2.83 mg?g-1, 远高于富含花青素的蓝莓等水果; 将这些高花青素含量的转基因番茄粉添加到患癌症小鼠的饮食中, 发现小鼠的寿命延长, 该结果证明了花青素具有抗癌的特性, 引起了广泛关注.在本课题组的一项研究中, 将PAP1和Lc共表达转入稀有的野生药用植物天山雪莲(Saussurea involucrata), 转基因株系的愈伤组织和嫩枝中均出现至少4类矢车菊素的衍生物; 同时, 花青素合成途径中多数结构基因的表达受到诱导, 其中CHS基因表达量变化最为显著(
b). Differential activation of anthocyanin biosyn- thesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry
0
2013
Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata
1
2005
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
花青素代谢途径与植物颜色变异
1
2016
... 目前, 随着分子生物学的发展, 花青素生物合成途径已经逐渐被阐明, 转录因子单独或协同调控花青素生物合成的分子机制也正在被不断完善.前人已经对花青素的生物合成和转运、环境因子对花青素合成的影响进行了详细的总结(
Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling
1
2003
... MYB是植物中最大的转录因子家族, 在植物生长发育(
Lingonberry anthocyanins protect cardiac cells from oxidative-stress-induced apoptosis
1
2017
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
AtMyb56 regulates anthocyanin levels via the modulation of AtGPT2 expression in response to sucrose in Arabidopsis
0
2018
The strawberry FaMYB1 transcription factor suppresses anthocyaninand flavonol accumulation in transgenic tobacco
0
2001
A conserved network of trans- criptional activators and repressors regulates anthocyanin pigmentation in eudicots
0
2014
Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning
2
2011
... 在另一模式植物矮牵牛中, MYB型转录因子AN2 (ANTHOCYANIN 2)和AN4都能够在转录水平影响DFR基因的表达.AN2在C端具有与玉米C1相似的结构域, 并且与C1功能相似, AN2调控矮牵牛花瓣中花青素的合成, 而AN4则调控花筒以及花药中花青素的合成(
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
Genetic interactions underlying flower color pat- terns in Antirrhinum majus
1
1989
... bHLH转录因子是调控花青素合成的另一大类转录因子家族(
Effect of anthocyanins from rabbit-eye blueberry ( Vacci- nium virgatum) on cognitive function in mice under trime- thyltin-induced neurotoxicity
1
2015
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
The transcrip- tional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula
1
2015
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple
1
2016
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in ap- ple
0
2017
The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin
0
2009
Engineering of the rose flavonoid biosynthetic pathway successfully gene- rated blue-hued flowers accumulating delphinidin
0
2007
MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosyn- thesis of anthocyanin and proanthocyanidin in apples
0
2014
The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation
0
2012
Regulatory hierarchy of photo- morphogenic loci: allele-specific and light-dependent inte- raction between the HY5 and COP1 loci
1
1994
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
Geno- mic and genetic analysis of Myb-related genes that regu- late anthocyanin biosynthesis in grape berry skin
1
2008
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
Retrotransposon-induced mutations in grape skin color
1
2004
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
Myb-related genes of the Kyoho grape ( Vitis labruscana) regulate anthocyanin biosynthesis
1
2002
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
Flavonoid-related basic helix-loop-helix regu- lators, NtAn1a and NtAn1b of tobacco have originated from two ancestors and are functionally active
1
2011
... 近年来, 其它模式植物中有关bHLH转录因子调控花青素合成的研究逐渐增多.MtTT8是蒺藜苜蓿中与拟南芥TT8同源的bHLH型转录因子.mttt8突变体表现为花青素缺失表型, 在突变体中过量表达MtTT8其毛状根中花青素的含量增加; 同时, 在拟南芥tt8突变体中表达MtTT8也能产生花青素, 恢复突变体花青素缺失的表型.MtTT8能与MtLAP1和MtWD40-1转录因子互作形成MBW复合体, 进而共同调控花青素的合成(
Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin
1
2007
... 与葡萄相似, 苹果果皮颜色与花青素的积累密切相关, 多个MYB转录因子参与调控苹果果皮花青素的合成.在苹果红色果皮中表达量较高的MdMYB1基因在光照条件下表达量升高, 可诱导果皮积累花青素; MdMYB1还可以激活DFR和UFGT基因的表达(
Abscisic-acid-depen- dent basic leucine zipper (bZIP) transcription factors in plant abiotic stress
1
2017
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana
1
2004
... 拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源.TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(
Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinen- sis during anthocyanin accumulation
2
2016
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis
0
2014
A pomegranate ( Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pome- granate fruit development
0
2011
Isolation and antisense suppression of flavonoid 3',5'-hydroxylase modifies flower pigments and colour in cyclamen
0
2010
Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida
1
2008
... 植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素.花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少.非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(
Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development
0
2007
Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis
2
2000
... MYB是植物中最大的转录因子家族, 在植物生长发育(
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
Intronic sequence regulates sugar-dependent ex- pression of Arabidopsis thaliana Production of Antho- cyanin Pigment-1/MYB75
1
2016
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
A WD40-repeat gene from Malus × domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TES- TA GLABRA 1
0
2010
Violet/blue chrysanthem ums—metabolic engineering of the anthocyanin biosyn- thetic pathway results in novel petal colors
1
2013
... 花色改良一直是花青素代谢工程的研究热点之一, 由于传统以杂交和诱变为主的观赏植物育种技术很难培育出富含飞燕草素的蓝色花朵, 而通过代谢工程的方法修饰花青素结构能够使花朵呈现纯正的蓝色, 更加贴合消费者的需求, 因此代谢工程为花色改良育种提供了新思路.其中, 通过改变关键结构基因的表达水平, 进而改变单个花青素产物是花色改良的一个重要策略.矮牵牛是一种常见的观赏植物, 也是研究花青素代谢调控的经典模式植物.Meyer等(1987)将玉米来源的DFR基因导入矮牵牛RL01突变体, 使含有矢车菊素和飞燕草素的浅粉色突变体花朵变为含有天竺葵素的砖红色花朵.Fukusaki等(2004)通过RNA干扰技术(RNA interference)抑制蝴蝶草(Torenia hybrida) CHS基因的表达, 使富含锦葵色素和芍药色素的蓝色蝴蝶草变为花青素缺失的白色蝴蝶草.Boase等(2010)将飞燕草素合成途径的关键酶基因F3’5’H反向转入仙客来(Cyclamen persicum)中, 抑制了F3’5’H基因的表达, 使花青素总量降低80%, 飞燕草素含量降低, 矢车菊素比例相对升高, 使花朵颜色由深粉色变浅.在康乃馨(Dianthus caryophyllus)、玫瑰(Rosa rugosa)和菊花中过量表达F3’5’H基因可以使花朵合成飞燕草素, 从而呈现蓝紫色(
Drastic anthocyanin increase in response to PAP1 overexpression in fls1 knockout mutant confers en- hanced osmotic stress tolerance in Arabidopsis thaliana
1
2016
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
a). Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum(Hort.).
0
2016
Retrotransposons control fruit-specific, cold-dependent accu- mulation of anthocyanins in blood oranges
0
2012
Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors
0
2008
TaMYB3, encoding a functional MYB transcriptor, isolated from the purple pericarp of Triticum aestivum
0
2017
Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8
1
2016
... 近年来, 其它模式植物中有关bHLH转录因子调控花青素合成的研究逐渐增多.MtTT8是蒺藜苜蓿中与拟南芥TT8同源的bHLH型转录因子.mttt8突变体表现为花青素缺失表型, 在突变体中过量表达MtTT8其毛状根中花青素的含量增加; 同时, 在拟南芥tt8突变体中表达MtTT8也能产生花青素, 恢复突变体花青素缺失的表型.MtTT8能与MtLAP1和MtWD40-1转录因子互作形成MBW复合体, 进而共同调控花青素的合成(
Colour and stability of the six common anthocyanidin 3-glucosides in aqueous solutions
1
2000
... 花青素来源于类黄酮化合物的合成途径: 衍生于香豆酰辅酶A (4-coumaroyl CoA)和丙二酰辅酶A (malon- yl CoA), 在查尔酮合酶(chalcone synthase, CHS)作用下合成查尔酮(chalcone).查尔酮经查尔酮异构酶(chalcone isomerase, CHI)催化形成黄烷酮(flavo- nones).黄烷酮再经过黄烷酮3-羟化酶(flavanone 3-hydroxylase, F3H)催化形成二氢黄酮醇(dihydro flavonols).在花青素特异的分支途径中, 二氢黄酮醇还可以被黄酮羟化酶(flavonoid 3’-hydroxylase, F3’H和flavonoid 3’,5’-hydroxylase, F3’5’H)催化, 并在二氢黄酮醇还原酶(dihydroflavonol-4-reductase, DFR)作用下还原为无色花青素(leucoanthocyanidins), 无色花青素在花青素合成酶(anthocyanidin synthase, ANS)的催化作用下形成花青素苷元(anthocyani- dins), 不稳定的花青素苷元经糖基转移酶(glycosylt- ransferase, UGTs)修饰形成稳定的花青素苷(anthocyanins) (
Mutations in the pale aleurone color 1 regulatory gene of the Zea mays anthocyanin pathway have distinct pheno- types relative to the functionally similar TRANSPARENT TESTA GLABRA 1 gene in Arabidopsis thaliana
1
2004
... 拟南芥中WDR类TTG1参与MBW复合体调控类黄酮化合物合成的机制研究得较为详细, 其它物种中WDR蛋白调控的功能研究相对较少.目前已有报道的调控花青素合成的WDR均与AN11和/或TTG1同源, 例如, 玉米中的PAC1 (PALE ALEURONE COL- OR 1)和MP1基因(
The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine
1
2015
... 除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物.葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(
An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes
2
2013
... 与葡萄相似, 苹果果皮颜色与花青素的积累密切相关, 多个MYB转录因子参与调控苹果果皮花青素的合成.在苹果红色果皮中表达量较高的MdMYB1基因在光照条件下表达量升高, 可诱导果皮积累花青素; MdMYB1还可以激活DFR和UFGT基因的表达(
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation
0
2014
Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1
0
2016
Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R geno- mic sequences
1
1989
... bHLH转录因子是调控花青素合成的另一大类转录因子家族(
Corrigendum: ectopic expression of the grape hyacinth (Muscari armeniacum) R2R3-MYB transcription factor gene, MaAN2, induces anthocyanin accumulation in tobacco
0
2017
The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chry- santhemum flowers
1
2015
... 植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素.花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少.非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(
The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosyn- thetic genes in tobacco and Chinese bayberry(Myrica rubra) during anthocyanin biosynthesis
1
2013
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
Differential expression of MYB gene ( OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey
1
2008
... 植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素.花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少.非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(
Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosyn- thesis in purple cauliflower
0
2012
Regulation of anthocyanin biosynthesis in Arabidopsis thaliana red pap1-D cells metabolically programmed by auxins
1
2014
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc- homology region
1
1989
... bHLH转录因子是调控花青素合成的另一大类转录因子家族(
The purple cauliflower arises from activation of a MYB transcription factor
1
2010
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
Molecular analysis of the maize anthocyanin regulatory locus C1
1
1986
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
a). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant
1
1993
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
Role of the regulatory gene pl in the photocontrol of maize anthocyanin pigmentation
1
1993
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis
0
2013
Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato
0
2007
Control of anthocyanin biosynthesis in flowers of Antirrhinum majus
0
1991
Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport
0
2003
AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis
1
2008
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine(Vitis vinifera L.).
0
2010
MYB10 plays a major role in the regulation of flavonoid/ phenylpropanoid metabolism during ripening of Fragaria× ananassa fruits
0
2014
A new petunia flower colour generated by transformation of a mutant with a maize gene
0
1987
Ternary WD40 repeat-containing protein complexes: evolution, compo- sition and roles in plant immunity
1
2016
... WD40重复蛋白(WD40 repeat proteins, WDR)主要存在于真核生物中, 具有保守而特异的二肽重复基序, 每个重复大概有40多个氨基酸残基; 重复的WD40基序在蛋白质互作时作为支架起固定作用(
Plants and colour: flowers and pollination
1
2011
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Structure and regulatory networks of WD40 protein in plants
1
2012
... WD40重复蛋白(WD40 repeat proteins, WDR)主要存在于真核生物中, 具有保守而特异的二肽重复基序, 每个重复大概有40多个氨基酸残基; 重复的WD40基序在蛋白质互作时作为支架起固定作用(
Overexpression of the PAP1 transcription factor reveals a complex regulation of flavonoid and phenyl- propanoid metabolism in Nicotiana tabacum plants at- tacked by Spodoptera litura
1
2014
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
Evolution of bHLH transcription factors: modular evolution by domain shuf- fling?
1
1999
... 碱性螺旋-环-螺旋(basic helix-loop-helix, bHLH)蛋白在动植物中高度保守, 具有保守的氨基酸序列(
Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory
0
2006
Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids
1
2014
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosyn- thesis in flowers and leaves of asiatic hybrid lily
0
2009
Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers
2
2008
... 植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素.花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少.非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves
1
2014
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
The TT8 gene encodes a basic helix- loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques.
0
2000
MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis
1
2015
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
Coordinated regulation of anthocyanin biosynthesis in Chinese bay- berry ( Myrica rubra) fruit by a R2R3 MYB transcription factor
0
2010
Generation of blue chrysan- themums by anthocyanin B-ring hydroxylation and glu- cosylation and its coloration mechanism
0
2017
A bHLH transcription factor,DvIVS, is involved in regulation of anthocyanin synthesis indahlia(Dahlia variabilis)
0
Modulation of flavonoid metabolites in Arabidopsis thaliana through overex- pression of the MYB75 transcription factor: role of kaem- pferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae
1
2014
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen ( Garcinia mangostana L.) fruit during ripening
1
2009
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proan- thocyanidin biosynthesis but not for trichome development
1
2009
... 拟南芥中WDR类TTG1参与MBW复合体调控类黄酮化合物合成的机制研究得较为详细, 其它物种中WDR蛋白调控的功能研究相对较少.目前已有报道的调控花青素合成的WDR均与AN11和/或TTG1同源, 例如, 玉米中的PAC1 (PALE ALEURONE COL- OR 1)和MP1基因(
Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce ( Lactuca sativa)
0
2008
A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosyn- thesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation
0
2007
The R2R3MYB VvMYBPA1 from grape reprograms the phenylpropanoid pathway in tobacco flowers
0
2017
Isolation and functional char- acterization of a floral tissue-specific R2R3 MYB regulator from tobacco
1
2010
... 在模式植物金鱼草(Antirrhinum majus)中, 调控花冠中花青素积累的Rosea1、Rosea2和Vensoa基因均编码MYB转录因子, 但是它们具有种间特异性, 分别调节不同的结构基因的表达水平: Rosea1能够提高F3H、DFR和ANS的表达水平, Rosea2仅调控F3’H的表达, Vensoa调控CHI、F3H、F3’5’H和ANS等多个结构基因的表达.这表明Rosea1、Rosea2和Vensoa等转录因子虽然存在一定的功能冗余, 但都各自调控相应的结构基因(
GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1
2
2000
... 拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源.TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(
... 矮牵牛的AN11 (ANTHOCYANIN11)是第1个被发现调控花青素生物合成的WDR蛋白, 其相应的编码基因是通过转座子标签法被鉴定的.AN11作用于AN2上游, 可能通过参与1个信号转导串联体系激活AN2的表达, 从而调控矮牵牛花青素的合成(
Trans- cription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism
0
2013
The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators
1
1987
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
The LAP1 MYB transcription factor orchestrates anthocyanidin bio- synthesis and glycosylation in Medicago
0
2009
Dietary consumption of black raspberries or their anthocyanin constituents alters innate immune cell trafficking in esophageal cancer
1
2016
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Charac- terization of flavonol synthase and leucoanthocyanidin dioxygenase genes in arabidopsis: further evidence for differential regulation of "Early" and "Late" genes
1
1997
...
New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco
1
2016
... 除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物.葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(
Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear, Pyrus communis
0
2010
Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1
2
2002
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
... ;
Metabolic engineering of the phenylpropanoid pathway enhances the antioxidant capacity of Saussurea involucrata
1
2013
... Butelli等(2008)将金鱼草的DELILA和ROSEA1基因转入番茄(Solanum lycopersicum cv. ‘MicroTom’), 使转基因番茄的果皮和果肉因大量富集酰基化花青素而呈现出深紫色, 转基因番茄的花青素含量达2.83 mg?g-1, 远高于富含花青素的蓝莓等水果; 将这些高花青素含量的转基因番茄粉添加到患癌症小鼠的饮食中, 发现小鼠的寿命延长, 该结果证明了花青素具有抗癌的特性, 引起了广泛关注.在本课题组的一项研究中, 将PAP1和Lc共表达转入稀有的野生药用植物天山雪莲(Saussurea involucrata), 转基因株系的愈伤组织和嫩枝中均出现至少4类矢车菊素的衍生物; 同时, 花青素合成途径中多数结构基因的表达受到诱导, 其中CHS基因表达量变化最为显著(
Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum
1
2014
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
The tomato Hoffman’s Anthocyaninless gene encodes a bHLH transcription factor involved in ant- hocyanin biosynthesis that is developmentally regulated and induced by low temperatures
0
2016
Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color
1
1999
... 在另一模式植物矮牵牛中, MYB型转录因子AN2 (ANTHOCYANIN 2)和AN4都能够在转录水平影响DFR基因的表达.AN2在C端具有与玉米C1相似的结构域, 并且与C1功能相似, AN2调控矮牵牛花瓣中花青素的合成, 而AN4则调控花筒以及花药中花青素的合成(
Regulatory genes controlling anthocyanin pigmen- tation are functionally conserved among plant species and have distinct sets of target genes
2
1993
... 在另一模式植物矮牵牛中, MYB型转录因子AN2 (ANTHOCYANIN 2)和AN4都能够在转录水平影响DFR基因的表达.AN2在C端具有与玉米C1相似的结构域, 并且与C1功能相似, AN2调控矮牵牛花瓣中花青素的合成, 而AN4则调控花筒以及花药中花青素的合成(
... bHLH转录因子是调控花青素合成的另一大类转录因子家族(
Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are cau- sed by divergent evolution of target anthocyanin genes
1
1998
... 拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源.TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(
Two basic-helix-loop-helix genes ( MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant
0
2003
Transcriptional regulation of flavonoid biosynthesis in nectarine ( Prunus persica) by a set of R2R3 MYB transcription factors
0
2013
Engineered native path- ways for high kaempferol and caffeoylquinate production in potato
0
2008
Increased accumulation of anthocyanins in Fra- garia chiloensis fruits by transient suppression of FcMYB1 gene
1
2013
... 除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物.葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(
A small family of MYB-regulatory genes controls floral pig- mentation intensity and patterning in the genus Antir- rhinum
1
2006
... 在模式植物金鱼草(Antirrhinum majus)中, 调控花冠中花青素积累的Rosea1、Rosea2和Vensoa基因均编码MYB转录因子, 但是它们具有种间特异性, 分别调节不同的结构基因的表达水平: Rosea1能够提高F3H、DFR和ANS的表达水平, Rosea2仅调控F3’H的表达, Vensoa调控CHI、F3H、F3’5’H和ANS等多个结构基因的表达.这表明Rosea1、Rosea2和Vensoa等转录因子虽然存在一定的功能冗余, 但都各自调控相应的结构基因(
The onion ( Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis
0
2016
Inhibition of lipid peroxidation and structure—activity-related studies of the dietary con- stituents anthocyanins, anthocyanidins, and catechins
1
2002
... 花青素来源于类黄酮化合物的合成途径: 衍生于香豆酰辅酶A (4-coumaroyl CoA)和丙二酰辅酶A (malon- yl CoA), 在查尔酮合酶(chalcone synthase, CHS)作用下合成查尔酮(chalcone).查尔酮经查尔酮异构酶(chalcone isomerase, CHI)催化形成黄烷酮(flavo- nones).黄烷酮再经过黄烷酮3-羟化酶(flavanone 3-hydroxylase, F3H)催化形成二氢黄酮醇(dihydro flavonols).在花青素特异的分支途径中, 二氢黄酮醇还可以被黄酮羟化酶(flavonoid 3’-hydroxylase, F3’H和flavonoid 3’,5’-hydroxylase, F3’5’H)催化, 并在二氢黄酮醇还原酶(dihydroflavonol-4-reductase, DFR)作用下还原为无色花青素(leucoanthocyanidins), 无色花青素在花青素合成酶(anthocyanidin synthase, ANS)的催化作用下形成花青素苷元(anthocyani- dins), 不稳定的花青素苷元经糖基转移酶(glycosylt- ransferase, UGTs)修饰形成稳定的花青素苷(anthocyanins) (
Antioxidation of anthocyanins in photosynthesis under high temperature stress
2
2007
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
... 花色改良一直是花青素代谢工程的研究热点之一, 由于传统以杂交和诱变为主的观赏植物育种技术很难培育出富含飞燕草素的蓝色花朵, 而通过代谢工程的方法修饰花青素结构能够使花朵呈现纯正的蓝色, 更加贴合消费者的需求, 因此代谢工程为花色改良育种提供了新思路.其中, 通过改变关键结构基因的表达水平, 进而改变单个花青素产物是花色改良的一个重要策略.矮牵牛是一种常见的观赏植物, 也是研究花青素代谢调控的经典模式植物.Meyer等(1987)将玉米来源的DFR基因导入矮牵牛RL01突变体, 使含有矢车菊素和飞燕草素的浅粉色突变体花朵变为含有天竺葵素的砖红色花朵.Fukusaki等(2004)通过RNA干扰技术(RNA interference)抑制蝴蝶草(Torenia hybrida) CHS基因的表达, 使富含锦葵色素和芍药色素的蓝色蝴蝶草变为花青素缺失的白色蝴蝶草.Boase等(2010)将飞燕草素合成途径的关键酶基因F3’5’H反向转入仙客来(Cyclamen persicum)中, 抑制了F3’5’H基因的表达, 使花青素总量降低80%, 飞燕草素含量降低, 矢车菊素比例相对升高, 使花朵颜色由深粉色变浅.在康乃馨(Dianthus caryophyllus)、玫瑰(Rosa rugosa)和菊花中过量表达F3’5’H基因可以使花朵合成飞燕草素, 从而呈现蓝紫色(
A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng ( Prunus avium L.)
0
2014
Engineering of red cells of Arabidop- sis thaliana and comparative genome-wide gene expres- sion analysis of red cells versus wild-type cells
0
2011
Anthocyanins induce the activation of phase II enzymes through the antioxidant response element pathway against oxidative stress-indu- ced apoptosis
1
2007
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
cDNA nucleotide sequence of Sn, a regulatory gene in maize
1
1992
... bHLH转录因子是调控花青素合成的另一大类转录因子家族(
New candidate genes for the fine regulation of the colour of grapes
1
2015
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
Ginkgo biloba extract and bilberry anthocyanins improve visual function in patients with normal tension glaucoma
1
2012
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Co-expression of GbMYB1 and GbMYC1 induces antho- cyanin accumulation in roots of cultured Gynura bicolor DC
1
2011
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
HY5 regulates anthocyanin biosyn- thesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis
2
2013
... 拟南芥中花青素的积累还与PAP1是否受到生物或非生物因子诱导有关(
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
A wheat R2R3-MYB protein PURPLE PLANT 1 (TaPL1) functions as a positive regulator of anthocyanin biosynthesis
0
2016
PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis
1
2007
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
A WD-repeat-containing putative regulatory protein in antho- cyanin biosynthesis in Perilla frutescens
1
2002
... 拟南芥中WDR类TTG1参与MBW复合体调控类黄酮化合物合成的机制研究得较为详细, 其它物种中WDR蛋白调控的功能研究相对较少.目前已有报道的调控花青素合成的WDR均与AN11和/或TTG1同源, 例如, 玉米中的PAC1 (PALE ALEURONE COL- OR 1)和MP1基因(
The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant de- fense and development
1
2013
... 碱性螺旋-环-螺旋(basic helix-loop-helix, bHLH)蛋白在动植物中高度保守, 具有保守的氨基酸序列(
The jasmonate-ZIM domain proteins interact with the R2R3- MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis
1
2011
... MYB是植物中最大的转录因子家族, 在植物生长发育(
antho- cyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural antho- cyanin genes
3
2000
... 拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源.TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(
... ).AN1与AN2和AN4之间的协同互作具有组织特异性, AN2在矮牵牛叶片中的表达可以诱导AN1表达, 而在花药中AN1可以诱导AN4表达(
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
Expression and anthocyanin biosynthesis-modulating potential of sweet cherry ( Prunus avium L.) MYB10 and bHLH genes
0
2015
Transcriptome analysis of genes involved in anthocyanins biosynthesis and transport in berries of black and white spine grapes ( Vitis davidii)
1
2016
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
Nonsense mutation of an MYB transcription factor is associated with purple-blue flower color in soybean
0
2011
Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples
2
2006
... 与葡萄相似, 苹果果皮颜色与花青素的积累密切相关, 多个MYB转录因子参与调控苹果果皮花青素的合成.在苹果红色果皮中表达量较高的MdMYB1基因在光照条件下表达量升高, 可诱导果皮积累花青素; MdMYB1还可以激活DFR和UFGT基因的表达(
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
Anthocyanin can arrest the cone photoreceptor degeneration and act as a novel treatment for retinitis pigmentosa
1
2016
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor
1
2005
... 第1个被发现调控花青素生物合成的R2R3-MYB转 录因子是玉米中的C1 (Colorless-1), C1依赖光特异性地调控玉米糊粉层中CHS和DFR基因的表达(
The Arabidopsis basic/helix-loop-helix transcription factor family
1
2003
... 碱性螺旋-环-螺旋(basic helix-loop-helix, bHLH)蛋白在动植物中高度保守, 具有保守的氨基酸序列(
Genetic and molecular analysis of Sn, a light-inducible, tissue specific regulatory gene in maize
0
1991
Molecular cloning and characteri- zation of a gene that encodes a MYC-related protein in Arabidopsis
0
1996
An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower deve- lopment
2
2013
... 与葡萄相似, 苹果果皮颜色与花青素的积累密切相关, 多个MYB转录因子参与调控苹果果皮花青素的合成.在苹果红色果皮中表达量较高的MdMYB1基因在光照条件下表达量升高, 可诱导果皮积累花青素; MdMYB1还可以激活DFR和UFGT基因的表达(
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
The TRANSPARENT TESTA GLABRA 1 locus, which regulates trichome differentiation and antho- cyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein
2
1999
... WD40重复蛋白(WD40 repeat proteins, WDR)主要存在于真核生物中, 具有保守而特异的二肽重复基序, 每个重复大概有40多个氨基酸残基; 重复的WD40基序在蛋白质互作时作为支架起固定作用(
... 矮牵牛的AN11 (ANTHOCYANIN11)是第1个被发现调控花青素生物合成的WDR蛋白, 其相应的编码基因是通过转座子标签法被鉴定的.AN11作用于AN2上游, 可能通过参与1个信号转导串联体系激活AN2的表达, 从而调控矮牵牛花青素的合成(
ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in trans- genic cotton
1
2017
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae
0
2010
Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis
1
2016
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear
0
2013
Anthocyanins inhibit high glucose-induced renal tubular cell apoptosis caused by oxidative stress in db/db mice
1
2018
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and bio- technology
1
2001
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums ( Chrysanthemum morifolium Ramat.)
1
2015
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples
1
2012
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
The ectopic expression of apple MYB1 and bHLH3 differentially activates anthocyanin biosynthesis in tobacco
1
2017
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
a). An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba
0
2014
b). The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis
0
2014
Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily
1
2010
... 植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素.花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少.非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(
The novel allele of the LhMYB12 gene is involved in splatter-type spot for- mation on the flower tepals of Asiatic hybrid lilies(Lilium spp.).
0
2014
Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var
0
2003
Metabolic engineering of anthocyanin biosynthe- sis in Escherichia coli
0
2005
a). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis
0
2017
b)
0
2017
Transcriptional regulation of anthocyanin biosynthesis in red cabbage
1
2009
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
A putative functional MYB transcription factor induced by low temperature regulates anthocyanin bio- synthesis in purple kale ( Brassica Oleracea var
1
2012
... MYB是植物中最大的转录因子家族, 在植物生长发育(
A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis
3
2003
... 碱性螺旋-环-螺旋(basic helix-loop-helix, bHLH)蛋白在动植物中高度保守, 具有保守的氨基酸序列(
... 拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源.TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(
... ).EGL3还被证明能够与PAP1共表达促进花青素的积累, 可见EGL3在调控花青素合成的功能方面强于GL3 (
a). BrMYB4, a suppressor of genes for phenyl- propanoid and anthocyanin biosynthesis, is down-regul- ated by UV-B but not by pigment-inducing sunlight in turnip cv. Tsuda
1
2014
... 除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物.葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(
Antagonistic HLH/bHLH transcription factors me- diate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis
2
2009
... 碱性螺旋-环-螺旋(basic helix-loop-helix, bHLH)蛋白在动植物中高度保守, 具有保守的氨基酸序列(
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
Isolation and functional characterization of a R2R3-MYB regulator of Prunus mu- me anthocyanin biosynthetic pathway
0
2017
Charac- terization and functional analysis of a MYB gene ( Gb- MYBFL) related to ?avonoid accumulation in Ginkgo biloba
0
2018
b). Engineering antho- cyanin biosynthesis in plants
1
2014
... 花青素来源于类黄酮化合物的合成途径: 衍生于香豆酰辅酶A (4-coumaroyl CoA)和丙二酰辅酶A (malon- yl CoA), 在查尔酮合酶(chalcone synthase, CHS)作用下合成查尔酮(chalcone).查尔酮经查尔酮异构酶(chalcone isomerase, CHI)催化形成黄烷酮(flavo- nones).黄烷酮再经过黄烷酮3-羟化酶(flavanone 3-hydroxylase, F3H)催化形成二氢黄酮醇(dihydro flavonols).在花青素特异的分支途径中, 二氢黄酮醇还可以被黄酮羟化酶(flavonoid 3’-hydroxylase, F3’H和flavonoid 3’,5’-hydroxylase, F3’5’H)催化, 并在二氢黄酮醇还原酶(dihydroflavonol-4-reductase, DFR)作用下还原为无色花青素(leucoanthocyanidins), 无色花青素在花青素合成酶(anthocyanidin synthase, ANS)的催化作用下形成花青素苷元(anthocyani- dins), 不稳定的花青素苷元经糖基转移酶(glycosylt- ransferase, UGTs)修饰形成稳定的花青素苷(anthocyanins) (
Recent advances on the deve- lopment and regulation of flower color in ornamental plants
1
2015
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-pro- grammed red cells of Arabidopsis thaliana
0
2012
CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis
1
2009
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency trans- gene stacking system
2
2017
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
... Butelli等(2008)将金鱼草的DELILA和ROSEA1基因转入番茄(Solanum lycopersicum cv. ‘MicroTom’), 使转基因番茄的果皮和果肉因大量富集酰基化花青素而呈现出深紫色, 转基因番茄的花青素含量达2.83 mg?g-1, 远高于富含花青素的蓝莓等水果; 将这些高花青素含量的转基因番茄粉添加到患癌症小鼠的饮食中, 发现小鼠的寿命延长, 该结果证明了花青素具有抗癌的特性, 引起了广泛关注.在本课题组的一项研究中, 将PAP1和Lc共表达转入稀有的野生药用植物天山雪莲(Saussurea involucrata), 转基因株系的愈伤组织和嫩枝中均出现至少4类矢车菊素的衍生物; 同时, 花青素合成途径中多数结构基因的表达受到诱导, 其中CHS基因表达量变化最为显著(
Purified anthocyanin supplemen- tation improves endothelial function via NO-cGMP acti- vation in hypercholesterolemic individuals
1
2011
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Cloning and molecular characterization of R2R3-MYB and bHLH- MYC transcription factors from Citrus sinensis
0
2010
Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport
1
2009
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals
1
1997
... 矮牵牛的AN11 (ANTHOCYANIN11)是第1个被发现调控花青素生物合成的WDR蛋白, 其相应的编码基因是通过转座子标签法被鉴定的.AN11作用于AN2上游, 可能通过参与1个信号转导串联体系激活AN2的表达, 从而调控矮牵牛花青素的合成(
Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway
0
2006
The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries
1
2008
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
Legume natural products: understanding and manipulating complex pathways for human and animal health
1
2003
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Isolation of a WD40-repeat gene regulating anthocyanin biosynthesis in storage roots of purple-fleshed sweet potato
0
2014
MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana
1
2008
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
MYB transcription factors in Arabi- dopsis
1
2010
... MYB是植物中最大的转录因子家族, 在植物生长发育(
Activation of anthocyanin biosynthesis in Gerbera hybrida(Asteraceae) suggests conserved protein- protein and protein-promoter interactions between the anciently diverged monocots and eudicots
2
2003
... 植物缤纷的颜色大多源于花青素的积累, 花青素的种类和含量决定了花瓣颜色, 而花瓣颜色是衡量鲜花品质的一个重要性状, 也是决定鲜花经济价值的重要因素.花色形成的分子机制一直是花青素研究领域的重点, 然而观赏植物中被报道的调控花青素合成的转录因子相对较少.非洲菊(Gerbera hybrida)中GMYB10的表达与其叶片和花中的花青素积累相关; GMYB10能与bHLH型转录因子GMYC1互作, 通过激活DFR调控花瓣中花青素的合成(
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
Multiple repeats of a promoter segment causes transcription factor autoregu- lation in red apples
0
2009
Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10
3
2007
... 与葡萄相似, 苹果果皮颜色与花青素的积累密切相关, 多个MYB转录因子参与调控苹果果皮花青素的合成.在苹果红色果皮中表达量较高的MdMYB1基因在光照条件下表达量升高, 可诱导果皮积累花青素; MdMYB1还可以激活DFR和UFGT基因的表达(
... 基因的表达量也略有升高; 同时, MdMYB10还与MdbHLH3和Mdb- HLH33共表达促进花青素的生物合成(
... 非模式植物中bHLH型转录因子调控花青素的机制与模式植物相同, 主要包括调控DFR基因的表达以及与MYB转录因子互作两个方面.苹果MdbHLH3和MdbHLH33共同与MdMYB10转录因子互作, 促进果实颜色变红(
Anthocyanin accumulation enhanced in Lc-transgenic cotton under light and increased resistance to bollworm
1
2016
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Anthocyanin-rich black elderberry extract improves mar- kers of HDL function and reduces aortic cholesterol in hyperlipidemic mice
1
2015
... 花青素(anthocyanins)与黄酮(flavones)、异黄酮(isoflavones)、黄酮醇(flavonols)和原花色素(proanthocyanidins)都属于类黄酮(flavonoids)化合物, 它们是广泛存在于植物中的一大类重要的次生代谢产物.花青素主要在植物的花、叶片和果实等器官中积累, 赋予植物丰富多彩的颜色, 使其具有重要的观赏价值和经济价值(
Anthocyanin biosynthesis in pears is regulated by a R2R3- MYB transcription factor PyMYB10
0
2010
The endogenous GL3, but not EGL3 gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves
1
2009
... 拟南芥中调控花青素合成的bHLH家族转录因子TT8 (TRANSPARENT TESTA 8)、GL3 (GLABRA3)和EGL3 (ENHANCER OF GLABRA 3)均与玉米的R转录因子同源.TT8作为调控花青素的重要bHLH型转录因子, 主要调控幼苗和荚果中DFR的表达, 参与形成MBW复合体(
An R2R3 MYB transcription factor determines red petal colour in an Actinidia(kiwifruit) hybrid population.
0
2013
Flower color modulations of Torenia hybrida by downregulation of chalcone syn- thase genes with RNA interference
0
2004
The MYB transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis
1
2011
... 除了模式植物, 非模式植物中调控花青素途径的负转录因子也逐渐被报道, 特别是富含花青素的经济作物.葡萄中的VvMYBC2-L1和VvMYBC2-L3在矮牵牛中表达时能够抑制花瓣中花青素的积累, 两者均可以与AN1结合, 影响MBW复合体的稳定性; MYBA1在VvMYBC2-L1抑制花青素合成过程中起到平衡作用(
Flavonoid production in transgenic hop ( Humulus lupulus L.) altered by PAP1/ MYB75 from Arabidopsis thaliana L
1
2012
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
Anthocyanin production by over-expression of grape transcription factor gene VlmybA2 in transgenic tobacco and Arabidopsis
2
2007
... 花青素赋予果实鲜艳的色彩, 其含量是许多果实如苹果(Malus domestica)、葡萄(Vitis vinifera)和草莓(Fragaria ananassa)的重要品质性状.目前, 对果实中花青素合成机理的研究也日益增多.其中, 葡萄作为一种富含花青素的果实, 其基因组测序已经完成, 对葡萄基因组转录因子的挖掘也鉴定了不少调控花青素的MYB型转录因子.巨峰葡萄(V. labruscana) 的MybA基因能够促进与葡萄浆果花青素积累相关 的糖基转移酶UFGT基因的表达, 在葡萄体细胞胚胎上诱导出现紫红色斑点(
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte
1
2017
... 目前, 对花青素途径转录因子的研究主要集中在MYB、bHLH和WDR三大类转录因子家族, 对其它类型的转录因子家族研究相对较少.近年来, bZIP (basic leucine zipper)家族的转录因子也被证明参与调控花青素的生物合成.bZIP是植物中一大类转录因子家族, 由1个亮氨酸拉链二聚体和1个DNA结合结构域构成, 参与调控诸多生物学过程(
A light-inducible Myb-like gene that is specifically expressed in red Perilla frutescens and presumably acts as a determining factor of the anthocyanin forma
1
1999
... 异源过量表达转录因子基因是花青素代谢工程的另一种重要策略.来源于紫苏的Myc-rp (
Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings
1
2008
... 拟南芥中参与调控花青素合成的主要MYB转录因子还包括MYB113和MYB114, 这2个基因在拟南芥中过量表达所产生的表型与过量表达PAP1所产生的表型相似; 然而在bHLH的突变体中过量表达MYB113和MYB114时, 花青素含量降低, 说明MYB113和MYB114是bHLH依赖型的转录因子(
Negative regulation of anthocyanin biosynthesis in Ara- bidopsis by a miR156-targeted SPL transcription factor
1
2011
... 花青素生物合成正调控因子的分子调控机制一直是花青素领域研究的热点.随着研究的深入, 更多的负调控因子也被逐渐鉴定, 它们的作用机制也正在被阐明.模式植物拟南芥中调控花青素合成的负转录因子主要包括MYBL2、MYB2、SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9)和CPC (CAPRICE).mybl2和myb2突变体中的花青素含量增加, F3H、DFR和ANS的表达量升高(
Metabolic engineer- ing of anthocyanins in dark tobacco varieties
0
2017
备案号: 京ICP备16067583号-21
版权所有 © 2021 《植物学报》编辑部
地址:北京香山南辛村20号 邮编:100093
电话:010-62836135 010-62836131 E-mail:cbb@ibcas.ac.cn
本系统由北京玛格泰克科技发展有限公司设计开发
