0 引言
【研究意义】植物在生长发育过程中不可避免的会受到干旱、高温、土壤盐碱化等非生物胁迫因素的影响,其中盐胁迫是影响作物生长发育并最终导致减产的主要因素之一[1]。植物为了减少土壤中高盐离子浓度胁迫对自身造成的伤害,通过感知环境中离子浓度变化,精确调控相关基因表达,产生一系列的应答反应以应对盐胁迫。逆境胁迫诱导的相关基因表达主要通过转录因子进行调控,转录因子抑制或激活逆境胁迫相关基因的表达是植物响应逆境胁迫的重要过程[1]。谷子起源于中国,是耐贫瘠、抗逆性强的单子叶作物,已成为研究单子叶作物抗逆机制的重要材料[2],因此,研究谷子相关抗逆基因的功能对于筛选优良抗逆基因用于作物抗逆遗传改良具有重要意义。【前人研究进展】AP2/ERF转录因子家族的主要特征是蛋白序列中含有一个高度保守的58或59个氨基酸的AP2结构域[3]。SAKUMA等[4]根据AP2/ERF结构域将拟南芥基因组中的AP2/ERF类蛋白分为5个亚家族,分别为AP2、RAV、DREB、ERF亚家族及一个特异蛋白AL079349[5]。作为蛋白质,AP2亚家族调节生长发育系统中的糖含量,并且涉及运输、信号传递[6]。拟南芥AP2类蛋白在花发育的ABC模型中起着重要的作用,其中ANT(AINTEGUMENTA)类转录因子,是植物特有的一类DNA结合蛋白,由与乙烯应答元件结合的2个AP2结构域组成[7],在转录水平上调节基因的表达。ANT类基因是器官生长的关键调节剂,参与胚珠发育、花器官生长发育和器官大小的调节,在调节植物生长发育的过程中起着非常重要的作用[7,8,9,10,11,12]。拟南芥ANT突变体显示多效的作用,包括减少花器官和叶子的大小[7-8,11],过表达ANT导致细胞增殖的持续时间增加,并增大叶、花和长角果中的器官[11]。在拟南芥转录因子RAV1中,N端AP2结构域结合5′-CAACA-3′序列,而C端高度保守的B3结构域结合5′-CACCTG-3′序列[13],参与响应生物胁迫和非生物胁迫[14]。DREB亚家族成员可以识别干旱和冷诱导响应元件(DRE/ CRT,A/GCCGAC),在植物抵抗非生物胁迫过程中起着非常重要的作用[15,16],如水稻OsDREB2A显著影响转基因大豆耐盐性[17]。ERF亚家族成员能识别GCC盒(AGCCGCC)[18],在水稻中过表达JcERF011增加了其对盐胁迫的敏感性[19],水稻ERF转录因子OsERF922负调控稻瘟病菌的耐药性和耐盐性[20]。从谷子中克隆到ABA响应DREB(脱水响应元件结合)蛋白基因SiARDP,谷子过表达SiARDP增强了抗旱性,拟南芥组成型表达SiARDP增强了其种子萌发和幼苗发育期的抗旱性和耐盐性[21]。目前关于谷子耐旱性的报道,还有NAC类基因等,如谷子转录因子SiNAC18通过ABA信号途径正向调控干旱条件下的种子萌发[22]。在功能标记开发方面,谷子受激素诱导表达、参与器官大小控制的基因SiARGOS1,从其启动子区开发的SSR标记AP-2,可用于谷子穗重和穗粒重等产量性状相关优异等位变异的鉴定和筛选[23]。【本研究切入点】中国农业科学院作物科学研究所马有志课题组从谷子品种豫谷一号中克隆ANT类转录因子基因SiANT1(Si034722m.g,XP 004985124.1),位于第9染色体,全长4 196 bp,包含相间分布的6个外显子和5个内含子,cDNA序列长1 863 bp,编码620个氨基酸,蛋白质等电点为6.82,分子量为66.86 kD,该蛋白有2个AP2的DNA结合区,分别在280—346和382—440区域,长度为67个和59个氨基酸,即SiANT1有一个ANT结构域。目前,关于AP2/ERF转录因子参与植物生长发育和增强抗逆性的报道较多,而ANT类转录因子是否参与植物胁迫响应过程,尚不清楚。【拟解决的关键问题】本研究通过在水稻中过表达SiANT1,分析过表达SiANT1水稻与野生型水稻分别在萌芽期、室内苗期、田间成熟期经0.9%NaCl处理后的表型及部分农艺性状的差异,确定过表达SiANT1提高了水稻的耐盐性,进一步通过分子试验验证其可能的耐盐过程。1 材料与方法
1.1 谷子SiANT1的分析及进化树构建
使用生物信息学工具、在线网站Phytozome(https:// phytozome.jgi.doe.gov/pz/portal.html)分析SiANT1在search in搜索框中选中Setaria italica v2.2,并且keywords搜索框中输入SiANT1序列号Si034722m.g,查找到基因组序列、CDS序列、蛋白质序列,2个AP2结构域的位置。使用DNAMAN软件查询该蛋白分子量大并预测等电点。在NCBI(https://www.ncbi.nlm. nih.gov/)中的Protein blast比对该蛋白序列,下载比对结果中同源性较高并含有ERF、AP2、DREB、RAV结构域的蛋白序列,用MEGA6软件打开蛋白序列并使用Neighbor- joining方法构建进化树。1.2 不同胁迫条件下谷子幼苗中SiANT1的表达分析
将豫谷一号种子(由山西农业大学农学院孙黛珍课题组赠送)25℃清水浸泡4 d后(每天换2次水)移栽至配制的营养土(营养土﹕蛭石=2﹕1)中,置于地下温室(22℃,相对湿度65%、光照周期16 h/8 h)培养2周,然后取生长健康并且大小一致的幼苗分别用100 mmol·L-1 NaCl培养液、0.2 μmol·L-1低氮培养液、10% PEG培养液进行胁迫处理,并于处理后的0、1、6、12、24和48 h分别取样,液氮冷冻,-80℃保存备用。采用RNA提取试剂盒(TIANGEN公司)提取总RNA,并反转录成cDNA。通过NCBI的Primer Blast设计SiANT1和Actin引物(SiANT1-F:5′-CAAGTCT GGTGGCGTCAACT-3′,SiANT1-R:5′-AAAGGGAGG TGGGAAACGAC-3′;Actin-F:5′-GGCAAACAGGGA GAAGATGA-3′,Actin-R:5′-GAGGTTGTCGGTAAG GTCACG-3′)。反应体系为10 μL 2×Super Mix、0.5 μL 50×reference Dye Ⅱ、正反向引物各0.5 μL、7.5μL ddH2O和1μL cDNA。采用三步法反应程序:95℃ 15 min;95℃ 5 s,60℃ 20 s,72℃ 32 s后收集荧光信号,40个循环。Real-time PCR使用Real Master Mix(SYBR Green)试剂盒,ABI7500进行扩增。采用2-△△Ct法根据各样品在特定荧光阈值下的Ct值计算基因在不同样品中的相对表达量。
1.3 谷子SiANT1的表达载体构建及水稻的遗传转化
使用BamHⅠ酶切体系将适量LP047 1118-Bar- ubi-EDLL载体质粒30℃恒温酶切5 h以上,以豫谷一号的cDNA为模板,设计引物进行扩增含有BamHⅠ接头的SiANT1 CDS序列。用In-Fusion连接法将上述2个片段连接(50℃、20 min),转化TOP10大肠杆菌感受态,筛选阳性克隆测序。使用冻融法转化农杆菌,在农杆菌感受态中加入1.5 μL上述质粒,冰浴45 min,液氮中速冻1 min,并在37℃恒温水浴3 min。加800 μL的LB,28℃,200 r/min,培养3 h。涂于含利福平(50 mg·L-1)和卡那霉素(50 mg·L-1)的LB固体培养基上。筛选阳性菌落测序。按照蔗糖﹕siwet=5%﹕0.03%(v/v)配制农杆菌悬浮液。取5 μL的阳性菌液与1.5 mLLB液体培养基(含50 mg·L-1利福平和50 mg·L-1卡那霉素)混合,28℃ 250 r/min培养10—12 h。按1﹕100转移到三角瓶中。培养至OD600=0.6—0.8,6 000 r/min室温离心15 mim。再用悬浮液悬浮菌液调至OD600=0.8,用于水稻的遗传转化。利用农杆菌转化方法[22],将剥去壳的水稻成熟种子消毒、洗净后,利用组织培养的方法,经过诱导培养基-继代培养基-共培养培养基-选择培养基-生根培养基-移植到花盆,得到T0代植株,繁种后收获T1种子,本研究过表达SiANT1水稻是T3代种子。1.4 过表达SiANT1水稻的耐盐性鉴定
1.4.1 萌芽期盐处理试验 用浓度为0.9%的NaCl溶液分别浸泡过表达SiANT1水稻3个株系及野生型Kitaake种子,各50粒。同时设空白对照,自来水浸泡上述种子。期间每天换2次相应溶液,直至0.9%的NaCl溶液处理的水稻开始露白,统计发芽率,每隔12 h统计1次,取5个时间点,并拍照记录相应差异。1.4.2 培养室内水稻盐处理 将过表达SiANT1水稻各株系及受体发芽至5 cm左右移至多孔板上,每个株系40株,Hogland营养液培养10 d,之后用含有0.9%NaCl的Hogland营养液培养10 d,将各株系每5株烘干称重、重复3组并记录数据。试验于2016年在中国农业科学院作物科学研究所品种资源楼进行。
1.4.3 田间水稻盐处理 2016年在河北唐海自然盐碱地上进行耐盐性鉴定,土壤盐碱度相对均匀。每个株系20粒种子,旱播,待水稻苗长到30 cm左右开始灌水(60 d),在水稻孕穗前进行一次盐处理,正常水(545 μs·cm-1)﹕海水(59 ms·cm-1)=4﹕1,约为0.9% NaCl浓度,处理后,同一地块盐碱度提高。待水稻成熟后调查水稻籽粒产量,评价转基因水稻的耐盐能力,筛选耐盐株系。每个株系收取5株,统计每株的单株分蘖数、株高、单株籽粒重,筛除各株系极端值,即每个株系统计3株。
1.5 谷子SiANT1及可能的下游基因在转基因水稻中的表达分析
将过表达SiANT1水稻株系和野生型Kitaake种子在30℃条件下,用清水浸泡3 d后(每天换2次水),移至截去底部的96孔板上,置于22℃、相对湿度65%、光照周期16 h/8 h的温室水培至三叶一心期,取生长健康且大小一致的幼苗,液氮冷冻,-80℃保存备用。过表达SiANT1水稻和野生型Kitaake种子均来自于中国农业科学院作物科学研究所马有志课题组。在DNA水平上检测过表达SiANT1水稻各株系是否属于阳性。设计水稻Actin(F:5′-GAAGATCA CTGCCTTGCTCC-3′,R:5′-CGATAACAGCTCCTC TTGGC-3′[25])和SiANT1的RT-PCR引物。利用半定量RT-PCR的方法检测过表达SiANT1水稻的3个株系是否在RNA水平上表达SiANT1,进一步做Real time-PCR确定3个株系间相对表达量是否有差异。
通过在NCBI(https://www.ncbi.nlm.nih.gov/)查找相关的盐胁迫基因并下载相应CDS序列(salt overly sensitive 1 SOS1、protein kinase complex SOS2、calcium sensing protein SOS3、cation transporter HTK1、mitogen-activated protein kinases MAPK5a、zinc finger protein ZFP182等),设计引物进行Real-time PCR分析,明确过表达SiANT1水稻与受体Kitaake中的上述盐胁迫相关内源基因的相对表达情况。
2 结果
2.1 谷子SiANT1蛋白与AP2/ERF类转录因子各亚族间亲缘关系
谷子SiANT1全长4 196 bp,CDS长为1 863 bp,编码620个氨基酸,蛋白质等电点为6.82,分子量为66.86 kD,该蛋白有2个AP2的DNA结合区,分别在280—346和382—440区域,长度为67个和59个氨基酸,即SiANT1有一个ANT结构域。利用MEGA6软件分析谷子SiANT1蛋白在AP2/ERF类转录因子各亚族间的进化位置结果表明,水稻、小麦、玉米、高粱、谷子的AP2/ERF类转录因子分为DREB、ERF、AP2、RAV等4个亚族(图1),其中DREB亚族和ERF亚族之间亲缘关系较近。谷子SiANT1蛋白(XP004985124.1,SiANT1)属于AP2亚族,与高粱(XP021318293.1,SbANT1)和玉米(XP008658933.1,ZmAP2)亲缘关系较近。
2.2 逆境胁迫下谷子SiANT1的表达模式
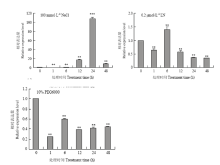
通过对100 mmol·L-1 NaCl、0.2 μmol·L-1低氮、10% PEG处理的豫谷一号幼苗进行实时荧光定量PCR分析,结果表明,在100 mmol·L-1 NaCl处理下(图2-A),与对照相比,谷子SiANT1在1和6 h时的表达量明显下降,在12 h时的表达量显著上升,在24 h时的表达量达到100倍以上,之后在48 h谷子SiANT1的表达量明显下降,说明谷子SiANT1受到高盐胁迫诱导上调表达。在0.2 μmol·L-1低氮(LN)处理下(图2-B),与对照相比,谷子SiANT1在1 h时表达量有所下降,在6 h时表达量达到1.5倍,之后12和48 h下降明显,并且24 h之后表达量几乎不变。在10%PEG处理下(图2-C),与对照相比,谷子SiANT1表达量在1 h时明显下降,6 h稍微上升,但仍远低于0 h,12 h之后相对表达量几乎不随处理时间变化,说明在10%PEG6000条件下SiANT1表达下调。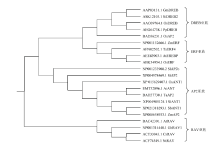 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图1利用部分植物AP2/ERF类转录因子构建的进化树
-->Fig. 1Phylogenetic tree constructed using part of the plant AP2/ERF like transcription factor
-->
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图2不同胁迫处理三叶一心期豫谷一号后SiANT1的相对表达量
-->Fig. 2The expression profile of SiANT1 gene in Yugu 1 under various stresses conditions including 100 mmol·L-1 NaCl, 0.2 μmol·L-1 LN, 10% PEG6000
-->
2.3 谷子SiANT1的表达载体构建与水稻遗传转化
经过BamHⅠ酶切的基因SiANT1和LP047 1118-Bar-ubi-EDLL载体连接,利用农杆菌转化法将目的基因SiANT1片段转移到水稻的基因组中,通过组织培养最终筛选过表达SiANT1水稻阳性T0植株,进而繁殖两代得到T3代种子。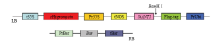 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图3LP047 1118-Bar-ubi-EDLL载体的T-DNA区段
-->Fig. 3T-DNA segment of LP047 1118-Bar-ubi-EDLL vector
-->
2.4 过表达SiANT1水稻株系的鉴定与表现
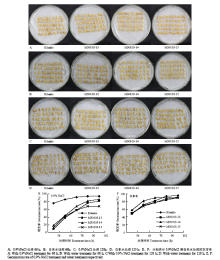
2.4.1 过表达SiANT1水稻各株系萌芽期表型及发芽率 通过对0.9%NaCl处理后的种子发芽率进行统计。0.9%NaCl处理66 h时,野生型Kitaake的发芽率(以露白为准)明显高于过表达SiANT1水稻株系(图4-A)。0.9%NaCl处理120 h时,野生型Kitaake的发芽率与过表达SiANT1水稻株系发芽率相差不大,但形态大小上差异明显(图4-C)。种子发芽120 h时,用自来水萌发的种子芽部大小整体大于相应的0.9%盐水处理(图4-D)。过表达SiANT1水稻株系和野生型Kitaake用自来水发芽时无太大差异,各株系发芽率随时间变化几乎一致(图4-B和图4-F)。然而,用0.9%NaCl盐水萌发种子54 h时,野生型Kitaake发芽率明显高于此时自来水浸泡下的萌发率,说明高盐胁迫加快了野生型Kitaake的萌发;但是,过表达SiANT1水稻株系的发芽率明显低于此时自来水浸泡下的萌发率,表明高盐胁迫抑制了过表达SiANT1水稻株系的萌发,即SiANT1对高盐胁迫是敏感的、有响应的。2.4.2 过表达SiANT1水稻各株系幼苗干重的变化
过表达SiANT1水稻各株系及受体苗期室内盐胁迫处理后,统计各株系地上部干重、根干重,并计算出单株干重(表1),重复3组,从表中可看出:过表达SiANT1水稻各株系地上部干重较受体野生型增加0.49%—21.07%,其中M30103-15株系与受体野生型之间无显著差异;过表达SiANT1水稻各株系根干重较受体野生型增加57.07%—100.51%(P<0.01);过表达SiANT1水稻各株系单株干重较受体野生型增加14.11%—37.42%(P<0.01)。这说明过表达SiANT1在一定程度上提高了水稻苗期的耐盐性,根部差异更为突出。
Table 1
表1
表1培养室内过表达SiANT1水稻各株系及受体Kitaake苗期盐处理后数据统计
Table 1Data of indoor post-salt treatment of SiANT1 overexpressing rice lines and recipient Kitaake during seedling period
| 材料 Material | 地上部干重 Shoot and leaves dry weight (g/5 strains) | 根干重 Root dry weight (g/5 strains) | 单株干重 Dry weight per plant (g) |
|---|---|---|---|
| Kitaake | 0.0617±0.0010B | 0.0198±0.0011C | 0.0163±0.0010C |
| M30103-13 | 0.0747±0.0007A | 0.0372±0.0011A | 0.0224±0.0009A |
| M30103-14 | 0.0708±0.0011A | 0.0397±0.0013A | 0.0221±0.0012A |
| M30103-15 | 0.0620±0.0009B | 0.0311±0.0010B | 0.0186±0.0009B |
新窗口打开
2.4.3 过表达SiANT1水稻各株系的田间表现 由表2可看出,在田间土样离子浓度差异不显著的前提下,过表达SiANT1水稻株系M30103-13、M30103-14的单株分蘖数增多,而M30103-15减少但与受体野生型差异不显著;3个株系株高较受体野生型增加3.33%—7.67%,与受体野生型株高差异不显著;过表达SiANT1水稻株系单株籽粒重较受体野生型增加56.12%—76.58%,且呈显著水平。表明过表达SiANT1水稻在田间相对于受体水稻耐盐性显著提高。
Table 2
表2
表2田间全生育期农艺性状调查
Table 2field survey of agronomic traits during the whole growth period
| 材料 Material | 土样离子浓度 Soil sample ion concentration (μs·cm-1) | 单株分蘖数 Number of tillers per plant | 株高 Plant height (cm) | 单株籽粒重 Grain weight per plant (g) |
|---|---|---|---|---|
| Kitaake | 882±28.58a | 20±1.63b | 60±1.41a | 10.46±1.13b |
| M30103-13 | 903±8.98a | 25±4.97ab | 64.6±4.71a | 18.47±2.56a |
| M30103-14 | 889±13.88a | 25±2.16a | 63±5.37a | 17.00±2.34a |
| M30103-15 | 885±29.39a | 19±2.05ab | 62±2.28a | 16.33±1.41a |
新窗口打开
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图4过表达SiANT1水稻各株系及野生型Kitaake的种子分别在0.9%NaCl和自来水处理下的表型及发芽率
-->Fig. 4Phenotypes and germination rates of over-expressing SiANT1 rice seeds and wild Kitaake seeds treated with 0.9% NaCl and tap water, respectively
-->
2.5 过表达SiANT1水稻的分子检测及相关基因表达模式分析
2.5.1 过表达SiANT1水稻株系阳性植株检测 通过对过表达SiANT1水稻株系和野生型水稻进行PCR检测(图5)。结果显示,过表达SiANT1水稻3个株系都是阳性,即谷子SiANT1已转入水稻中。 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图5过表达SiANT1水稻及受体Kitaake的阳性检测
-->Fig. 5Positive detection of overexpression of SiANT1 in rice and its receptor Kitaake
-->
2.5.2 过表达SiANT1水稻的半定量和定量PCR分析 通过对转基因株系中SiANT1进行半定量RT-PCR分析(图6-A),在Actin亮度一致的前提下,过表达SiANT1株系(M30103-13、M30103-14和M30103-15)均扩增出大小一致的条带,但受体野生型无带,说明野生型水稻中不存在谷子基因SiANT1,并且这三个过表达SiANT1水稻株系都在RNA水平上表达SiANT1。进一步定量PCR检测(图6-B),过表达SiANT1水稻中SiANT1的相对表达量为受体的178—10 615倍,其中M30103-13株系的相对表达量最高,M30103-15次之,M30103-14最低,该结果与半定量结果一致。
利用农杆菌转化法将SiANT1转入水稻中,但是,SiANT1是否插入水稻DNA中并形成重组DNA,插入位点及插入个数均不确定。已证明水稻株系M30103-13、M30103-14、M30103-15的DNA内含有SiANT1,并且都在RNA水平上表达,只是RNA相对表达量有所差异,因此,推测3个过表达SiANT1水稻株系之间相对表达量的差异可能是由于SiANT1拷贝数不同造成的。
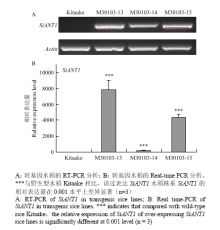 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图6过表达SiANT1水稻及受体Kitaake的半定量(A)及定量PCR分析(B)
-->Fig. 6Semi-quantitative(A) and quantitative PCR(B) of SiANT1 in transgenic rice lines and its receptor Kitaake
-->
2.5.3 可能的下游基因的表达 根据前面的推测,进一步检测细胞分裂素等植物激素形成和耐盐性相关基因在转基因株系中的表达水平。通过对水稻OsSOS1和OsZFP182进行实时荧光定量PCR检测(图7-A和图7-B),与受体Kitaake比较,水稻OsSOS1的相对表达量在M30103-13、M30103-14提高1.5倍以上,M30103-15提高1.1倍(图7-A);OsZFP182在M30103-13、M30103-14和M30103-15中的相对表达量均提高1.6倍以上(图7-B),呈显著增加水平。结果表明,在过表达SiANT1水稻株系OsSOS1和OsZFP182的相对表达量相对于受体野生型显著提高,结合水稻OsSOS1和OsZFP182[26,27,28]已被前人验证可以提高作物耐盐性,说明过表达SiANT1水稻株系可能增强了内源OsSOS1和OsZFP182的表达,进而提高了耐盐性。
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图7下游基因的相对表达量
-->Fig. 7The relative expression of downstream genes in transgenic rice
-->
3 讨论
盐胁迫增加了土壤中Na+和Cl-浓度,使植物受到渗透胁迫和离子毒害,造成植物生长发育迟缓甚至死亡。形态方面:叶面积减小、叶片卷曲干枯、植株矮小;生理生化方面:单株干重降低,光合作用受抑制、营养物质摄取受阻、活性氧增多、质膜解体、代谢毒物积累等[29]。植物为了降低盐胁迫对自身的伤害,增加糖类、脯氨酸、甜菜碱等渗透调节物质的合成,这起到保护细胞膜表面的作用,提高了细胞的耐逆性;大量的膜蛋白被激活将Na+最终转移到液泡中,重建胞内、胞外、液泡的离子平衡[29,30]。植物盐应答的分子包括两类:一类是直接参与代谢变化等生化应答过程,产生耐盐效应的效应分子;另一类是位于效应分子上游,介导信号传递,包括转录因子和信号级联系统中各种激酶的调控分子[29,31]。转录因子是能够与真核基因启动子区域中的顺式作用元件发生特异相互作用的结合蛋白,通过它们之间的相互作用,激活或抑制转录,进而激活或抑制下游基因表达[32]。转录因子在植物对逆境的应答过程中起着重要的调控作用,通过它可以精细地调控下游功能基因适时适量地表达,从而实现自身生长发育和繁殖的需要以及对各种生物或非生物逆境的应答[32,33]。MYB、MYC、bZIP、AP2/ERF and zinc finger protein等转录因子都参与逆境胁迫相关基因表达[32]。本文在水稻中过表达SiANT1,SiANT1蛋白属于AP2/ERF转录因子里的AP2亚家族。目前关于AP2亚族的报道,大多与植物生长发育相关[7]。本文主要研究SiANT1是否响应盐胁迫,是否可以提高作物耐盐性,如若可以,可能是通过怎样的方式调控。SiANT1是一个转录因子,将谷子SiANT1利用农杆菌转化法转到水稻中,SiANT1随着载体质粒的T-DNA区进入水稻,是否转入、插入位置、以及插入个数等都是不确定的,后续通过DNA检测筛选阳性植株,并筛选在RNA水平上表达的植株,本文未对SiANT1表达的蛋白进行研究。本文在高盐胁迫条件下,过表达SiANT1水稻种子萌芽期受抑制,然而,在自来水中过表达SiANT1水稻种子正常发芽,说明谷子SiANT1对高盐敏感。萌芽期的高盐胁迫试验结果,过表达SiANT1水稻种子萌芽受抑制,这说明可能SiANT1在萌芽期不具有耐盐性,但是,与野生型水稻种子相比,最终(0.9%NaCl处理120 h)两者发芽率相差不大,又说明可能过表达SiANT1水稻种子露白推迟是一种耐盐胁迫应答反应。过表达SiANT1水稻株系苗期干重和田间籽粒重相对于受体水稻显著增加,证明过表达SiANT1可以提高转基因水稻苗期及成熟期耐盐性。苗期干重和田间籽粒重都属于生物量,较野生型而言,在0.9%NaCl处理下,过表达SiANT1水稻株系生物量增加,可能是由于SiANT1的过表达导致细胞分裂素增多、细胞数目增加,叶茎器官增大、侧根形成增加[10,34],在此基础上,由于SiANT1表达的蛋白是转录因子,过表达SiANT1可能也通过增强相关下游耐盐基因表达来提高水稻耐盐性。本文中 SiANT1可能的下游基因OsSOS1、OsZFP182的相对表达量在过表达SiANT1水稻中较野生型增加。SOS1蛋白是一种质膜Na+/H+逆向转运蛋白,是一个重要的耐逆决定簇,参与Na+从细胞中排出[35,36]。MARTíNEZ-ATIENZA等[26]和SCHMIDT等[27]证明了OsSOS1水稻转运蛋白在酵母细胞的质膜囊泡中具有Na+/H+交换能力,降低了细胞中Na+净含量,减轻了Na+对细胞造成的离子毒害。Na+/H+逆向转运蛋白SOS1是迄今为止植物质膜上唯一的Na+流出蛋白[26]。缺少SOS1的拟南芥突变体对盐是极其敏感的,其Na+排出和将该离子从根部长距离运输到茎都有缺陷[36,37]。SOS1蛋白主要表达于植物根尖表皮和木质部共质体边界的木质部薄壁组织[36]。在根-土壤界面处,SOS1将从根表皮细胞排出过量的Na+离子。此外,SOS1突变体中Na+根冠分配的分析表明,在不同盐度条件下SOS1也以复杂的方式参与Na+在根和茎之间的重新分配[36]。在适度的盐(25 mmol·L-1NaCl)胁迫下,SOS1突变体植株在其地上部中积累的Na+比野生型低,这表明SOS1的功能是将Na+装载到木质部用于控制传递至茎。相反,在高盐(100 mmol·L-1NaCl)胁迫下,SOS1突变体植株的根和地上部分比野生型植物积累更多的Na+,这可能是由于根表皮Na+排出以及Na+穿过木质部-共质体边界的电化学梯度受到破坏[36,38]。锌指蛋白(zinc finger Protein)是一类具有“手指状”结构域的转录因子,负责调控基因的表达[32]。锌指蛋白的共同特征是通过结合Zn2+来稳定一种很短的可自我折叠成“手指”的蛋白结构[32]。ZFP182蛋白大小为18.2 kD,含有2个C2H2型锌指基序,一个核定位信号和一个富含Leu的结构域。大多数植物的C2H2型锌指蛋白中存在DLN-box/EAR-motif结构域,但是ZFP182中不存在[28]。ZFP182在成年稻的叶,茎,根和穗中组成型表达,并且水稻幼苗分别在冷(4℃)、150 mmol·L-1NaCl和0.1 mmol·L-1ABA处理下,ZFP182明显被诱导表达[28]。将水稻的ZFP182的启动子区约1.4 kb融合到GUS报告基因中并转化烟草, 组织化学分析结果显示,转基因烟草幼苗在正常条件下不能检测到GUS的表达,但在用NaCl或KCl处理后的烟草叶片和根的维管组织中强烈观察到GUS的表达[28]。HUANG等[28]研究表明水稻中过表达ZFP182提高了盐胁迫的耐受性。目前关于ZFP182的作用机理仍不清楚。本研究证明过表达SiANT1提高了水稻的耐盐性,可能是通过增强水稻中下游内源基因OsSOS1和OsZFP182的表达实现的,为证明谷子SiANT1具有耐盐性进一步提供了依据。
4 结论
谷子SiANT1受100 mmol·L-1 NaCl诱导上调表达,在24 h时相对表达量最高。过表达SiANT1提高水稻耐盐性,其通过增强可能的下游基因OsSOS1和OsZFP182的表达提高耐盐性。The authors have declared that no competing interests exist.
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
| [1] | . The ethylene responsive factors (ERF) belong to a subfamily of the AP2/ERF superfamily in plants. The ERF family was defined by the presence of a conserved ERF domain consisting of 58 or 59 amino acids and has been demonstrated to be widely involved in regulation of various aspects of plant growth and development as well as in responses to different abiotic and biotic stresses. In this minireview, we summarize the functions and mechanisms of the ERF transcription factors in regulation of responses to abiotic and biotic stresses and discuss the future directions of studies on the ERF transcription factors in plants. |
| [2] | . . |
| [3] | . We demonstrated that the GCC box, which is an 11-bp sequence (TAAGAGCCGCC) conserved in the 5鈥 upstream region of ethylene-inducible pathogenesis-related protein genes in Nicotiana spp and in some other plants, is the sequence that is essential for ethylene responsiveness when incorporated into a heterologous promoter. Competitive gel retardation assays showed DNA binding activities to be specific to the GCC box sequence in tobacco nuclear extracts. Four different cDNAs encoding DNA binding proteins specific for the GCC box sequence were isolated, and their products were designated ethylene-responsive element binding proteins (EREBPs). The deduced amino acid sequences of EREBPs exhibited no homology with those of known DNA binding proteins or transcription factors; neither did the deduced proteins contain a basic leucine zipper or zinc finger motif. The DNA binding domain was identified within a region of 59 amino acid residues that was common to all four deduced EREBPs. Regions highly homologous to the DNA binding domain of EREBPs were found in proteins deduced from the cDNAs of various plants, suggesting that this domain is evolutionarily conserved in plants. RNA gel blot analysis revealed that accumulation of mRNAs for EREBPs was induced by ethylene, but individual EREBPs exhibited different patterns of expression. |
| [4] | DRE/CRT is a cis-acting element that is involved in gene expression responsive to drought and low-temperature stress in higher plants. DREB1A/CBF3 and DREB2A are transcription factors that specifically bind to DRE/CRT in Arabidopsis. We precisely analyzed the DNA-binding specificity of DREBs. Both DREBs specifically bound to six nucleotides (A/GCCGAC) of DRE. However, these proteins had different binding specificities to the second or third nucleotides of DRE. Gel mobility shift assay using mutant DREB proteins showed that the two amino acids, valine and glutamic acid conserved in the ERF/AP2 domains, especially valine, have important roles in DNA-binding specificity. In the Arabidopsis genome, 145 DREB/ERF-related proteins are encoded. These proteins were classified into five groups鈥擜P-2 subfamily, RAV subfamily, DREB subfamily, ERF subfamily, and others. The DREB subfamily included three novel DREB1A- and six DREB2A-related proteins. We analyzed expression of novel genes for these proteins and discuss their roles in stress-responsive gene expression. |
| [5] | . 植物AP2/ERF是一个庞大的转录因子基因家族,含有由60~70个氨基酸组成的AP2/ERF结构域而得名,存在于所有的植物中。AP2/ERF转录因子参与多种生物学过程,包括植物生长、花发育、果实发育、种子发育、损伤、病菌防御、高盐、干旱等环境胁迫响应等。AP2/ERF类转录因子参与水杨酸、茉莉酸、乙烯、脱落酸等多种信号转导途径,而且是逆境信号交叉途径中的连接因子。文章对国内外近年来有关植物AP2/ERF类转录因子的分类、生物学功能、基因调控等方面的研究进行了综述。 . 植物AP2/ERF是一个庞大的转录因子基因家族,含有由60~70个氨基酸组成的AP2/ERF结构域而得名,存在于所有的植物中。AP2/ERF转录因子参与多种生物学过程,包括植物生长、花发育、果实发育、种子发育、损伤、病菌防御、高盐、干旱等环境胁迫响应等。AP2/ERF类转录因子参与水杨酸、茉莉酸、乙烯、脱落酸等多种信号转导途径,而且是逆境信号交叉途径中的连接因子。文章对国内外近年来有关植物AP2/ERF类转录因子的分类、生物学功能、基因调控等方面的研究进行了综述。 |
| [6] | |
| [7] | |
| [8] | . |
| [9] | Abstract AINTEGUMENTA ( ANT ) was previously shown to be involved in floral organ initiation and growth in Arabidopsis . ant flowers have fewer and smaller floral organs and possess ovules that lack integuments and a functional embryo sac. The present work shows that young floral meristems of ant plants are smaller than those in wild type. Failure to initiate the full number of organ primordia in ant flowers may result from insufficient numbers of meristematic cells. The decreased size of ant floral organs appears to be a consequence of decreased cell division within organ primordia. Ectopic expression of ANT under the control of the constitutive 35S promoter results in the development of larger floral organs. The number and shape of these organs is not altered and the size of vegetative organs is normal. Microscopic and molecular analyses indicate that the increased size of 35S::ANT sepals is the result of increased cell division, whereas the increased sizes of 35S::ANT petals, stamens, and carpels are primarily attributable to increased cell expansion. In addition, 35S::ANT ovules often exhibit increased growth of the nucellus and the funiculus. These results suggest that ANT stimulates cell growth in floral organs. Dev. Genet. 25:224鈥236, 1999. 漏 1999 Wiley-Liss, Inc. |
| [10] | The control of cell proliferation during organogenesis plays an important role in initiation, growth, and acquisition of the intrinsic size of organs in higher plants. To understand the developmental mechanism that controls intrinsic organ size by regulating the number and extent of cell division during organogenesis, we examined the function of the Arabidopsis regulatory gene AINTEGUMENATA (ANT). Previous observations revealed that ANT regulates cell division in integuments during ovule development and is necessary for floral organ growth. Here we show that ANT controls plant organ cell number and organ size throughout shoot development. Loss of ANT function reduces the size of all lateral shoot organs by decreasing cell number. Conversely, gain of ANT function, via ectopic expression of a 35S::ANT transgene, enlarges embryonic and all shoot organs without altering superficial morphology by increasing cell number in both Arabidopsis and tobacco plants. This hyperplasia results from an extended period of cell proliferation and organ growth. Furthermore, cells ectopically expressing ANT in fully differentiated organs exhibit neoplastic activity by producing calli and adventitious roots and shoots. Based on these results, we propose that ANT regulates cell proliferation and organ growth by maintaining the meristematic competence of cells during organogenesis. |
| [11] | . Lateral organs in flowering plants display polarity along their adaxial-abaxial axis with distinct cell types forming at different positions along this axis. Members of three classes of transcription factors in Arabidopsis (Arabidopsis thaliana; the Class III homeodomain/leucine zipper [HD-ZIP] proteins, KANADI proteins, and YABBY proteins) are expressed in either the adaxial or abaxial domain of organ primordia where they confer these respective identities. Little is known about the factors that act upstream of these polarity-determining genes to regulate their expression. We have investigated the relationship between AINTEGUMENTA (ANT), a gene that promotes initiation and growth of lateral organ primordia, and polarity genes. Although ant single mutants do not display any obvious defects in organ polarity, loss of ANT activity in combination with mutations in one or more YABBY genes results in polarity defects greater than those observed in the yabby mutants alone. Our results suggest that ANT acts in combination with the YABBY gene FILAMENTOUS FLOWER (FIL) to promote organ polarity by upregulating the expression of the adaxial-specifying HD-ZIP gene PHABULOSA. Furthermore, we show that ANT acts with FIL to up-regulate expression of the floral homeotic gene APETALA3. Our work defines new roles for ANT in the development of lateral organs. |
| [12] | |
| [13] | . Abstract We have cloned and characterized two novel DNA binding proteins designated RAV1 and RAV2 from Arabidopsis thaliana. RAV1 and RAV2 contain two distinct amino acid sequence domains found only in higher plant species. The N-terminal regions of RAV1 and RAV2 are homologous to the AP2 DNA-binding domain present in a family of transcription factors represented by the Arabidopsis APETALA2 and tobacco EREBP proteins, while the C-terminal region exhibits homology to the highly conserved C-terminal domain, designated B3, of VP1/ABI3 transcription factors. Binding site selection assays using a recombinant glutathione S-transferase fusion protein have revealed that RAV1 binds specifically to bipartite recognition sequences composed of two unrelated motifs, 5'-CAACA-3' and 5'-CACCTG-3', separated by various spacings in two different relative orientations. Analyses using various deletion derivatives of the RAV1 fusion protein show that the AP2 and B3-like domains of RAV1 bind autonomously to the CAACA and CACCTG motifs, respectively, and together achieve a high affinity and specificity of binding. From these results, we suggest that the AP2 and B3-like domains of RAV1 are connected by a highly flexible structure enabling the two domains to bind to the CAACA and CACCTG motifs in various spacings and orientations. |
| [14] | . Abstract A novel pathogen-induced gene encoding the RAV (Related to ABI3/VP1) transcription factor, CARAV1, was isolated from pepper leaves infected with Xanthomonas campestris pv. vesicatoria. CARAV1 contains two distinct DNA-binding domains AP2 and B3 uniquely found in higher plants. Transient expression analysis of the smGFP:CARAV1 fusion construct in Arabidopsis protoplasts and pepper epidermal cells revealed the CARAV1 protein to be localized in the nucleus. The N-terminal region of CARAV1 fused to the GAL4 DNA-binding domain was required to activate transcription of reporter genes in yeast. In yeast one-hybrid, the recognition of CAACA and CACCTG motifs also were essential for the CARAV1 protein to bind to a specific target gene and activate the reporter gene. The expression of the CARAV1 gene was strongly induced early in pepper leaves during the pathogen infection, abiotic elicitors and environmental stresses. CARAV1 transcripts were localized in the phloem cells of leaf tissues during pathogen infection and ethylene treatment. Ectopic expression of the CARAV1 gene in transgenic Arabidopsis plants induced some PR genes and enhanced resistance against infection by Pseudomonas syringae pv. tomato DC3000 and osmotic stresses by high salinity and dehydration. The CARAV1 promoter activation was induced by P. syringae pv. tabaci, salicylic acid and abscisic acid. These data suggest that pathogen- and abiotic stress-inducible CARAV1 functions as a transcriptional activator triggering resistance to bacterial infection and tolerance to osmotic stresses. |
| [15] | Abstract Two genes, rd29A and rd29B, which are closely located on the Arabidopsis genome, are differentially induced under conditions of dehydration, low temperature, high salt, or treatment with exogenous abscisic acid (ABA). It appears that rd29A has at least two cis-acting elements, one involved in the ABA-associated response to dehydration and the other induced by changes in osmotic potential, and that rd29B contains at least one cis-acting element that is involved in ABA-responsive, slow induction. We analyzed the rd29A promoter in both transgenic Arabidopsis and tobacco and identified a novel cis-acting, dehydration-responsive element (DRE) containing 9 bp, TACCGACAT, that is involved in the first rapid response of rd29A to conditions of dehydration or high salt. DRE is also involved in the induction by low temperature but does not function in the ABA-responsive, slow expression of rd29A. Nuclear proteins that specifically bind to DRE were detected in Arabidopsis plants under either high-salt or normal conditions. Different cis-acting elements seem to function in the two-step induction of rd29A and in the slow induction of rd29B under conditions of dehydration, high salt, or low temperature. |
| [16] | . 74 Abstract68Many plants increase in freezing tolerance upon exposure to low nonfreezing temperatures, a phenomenon known as cold acclimation. In this review, recent advances in determining the nature and function of genes with roles in freezing tolerance and the mechanisms involved in low temperature gene regulation and signal transduction are described. One of the important conclusions to emerge from these studies is that cold acclimation includes the expression of certain cold-induced genes that function to stabilize membranes against freeze-induced injury. In addition, a family of Arabidopsis transcription factors, the CBF/DREB1 proteins, have been identified that control the expression of a regulon of cold-induced genes that increase plant freezing tolerance. These results along with many of the others summarized here further our understanding of the basic mechanisms that plants have evolved to survive freezing temperatures. In addition, the findings have potential practical applications as freezing te... |
| [17] | . The dehydration responsive element binding (DREB) transcription factors play an important role in regulating stress-related genes. OsDREB2A, a member of the DREBP subfamily of AP2/ERF transcription factors in rice (Oryza sativa), is involved in the abiotic stress response. OsDREB2A expression is induced by drought, low-temperature and salt stresses. Here, we report the ability of OsDREB2A to regulate high-salt response in transgenic soybean. Overexpressing OsDREB2A in soybeans enhanced salt tolerance by accumulating osmolytes, such as soluble sugars and free proline, and improving the expression levels of some stress-responsive transcription factors and key genes. The phenotypic characterization of transgenic soybean were significantly better than those of wild-type (WT). Electrophoresis mobility shift assay (EMSA) revealed that the OsDREB2A can bind to the DRE core element in vitro. These results indicate that OsDREB2A may participate in abiotic stress by directly binding with DRE element to regulate the expression of downstream genes. Overexpression of OsDREB2A in soybean might be used to improve tolerance to salt stress. |
| [18] | . Ethylene-responsive element-binding proteins (EREBPs)have novel DNA-binding domains (ERF domains), which are widely conserved in plants, and interact specifically with sequences containing AGCCGCC motifs (GCC box). Deletion experiments show that some flanking region at the N terminus of the conserved 59-amino acid ERF domain is required for stable binding to the GCC box. Three ERF domain-containing fragments of EREBP2, EREBP4, and AtERF1 from tobacco and Arabidopsis, bind to the sequence containing the GCC box with a high binding affinity in the pM range. The high affinity binding is conferred by a monomeric ERF domain fragment, and DNA truncation experiments show that only 11-base pair DNA containing the GCC box is sufficient for stable ERF domain interaction. Systematic DNA mutation analyses demonstrate that the specific amino acid contacts are confined within the 6-base pair GCCGCC region of the GCC box, and the first G, the fourth G, and the sixth C exhibit highest binding specificity common in all three ERF domain-containing fragments studied. Other bases within the GCC box exhibit modulated binding specificity varying from protein to protein, implying that these positions are important for differential binding by different EREBPs. The conserved N-terminal half is likely responsible for formation of a stable complex with the GCC box and the divergent C-terminal half for modulating the specificity. |
| [19] | The AP2/ERF transcription factors play crucial roles in plant growth, development and responses to biotic and abiotic stresses. A total of 119 AP2/ERF genes (JcAP2/ERFs) have been identified in the physic nut genome; they include 16 AP2, 4 RAV, 1 Soloist, and 98 ERF genes. Phylogenetic analysis indicated that physic nut AP2 genes could be divided into 3 subgroups, while ERF genes could be classed into 11 groups or 43 subgroups. The AP2/ERF genes are non-randomly distributed across the 11 linkage groups of the physic nut genome and retain many duplicates which arose from ancient duplication events. The expression patterns of severalJcAP2/ERFduplicates in the physic nut showed differences among four tissues (root, stem, leaf, and seed), and 38JcAP2/ERFgenes responded to at least one abiotic stressor (drought, salinity, phosphate starvation, and nitrogen starvation) in leaves and/or roots according to analysis of digital gene expression tag data. The expression ofJcERF011was downregulated by salinity stress in physic nut roots. Overexpression of theJcERF011gene in rice plants increased its sensitivity to salinity stress. The increased expression levels of several salt tolerance-related genes were impaired in theJcERF011-overexpressing plants under salinity stress. |
| [20] | |
| [21] | SiARDP is a DREB-type transcription factor from foxtail millet. SiARDP is involved in ABA-dependent signal pathways under the control of SiARDP and plays a positive role in plant responses to drought stress. The DREB (dehydration-responsive element binding)-type transcription factors regulate the expression of stress-inducible genes by binding the DRE/CRT cis-elements in promoter regions. The upstream transcription factors that regulate the transcription of DREB transcription factors have not been clearly defined, although the function of DREB transcription factors in abiotic stress is known. In this study, an abscisic acid (ABA)-responsive DREB-binding protein gene (SiARDP) was cloned from foxtail millet (Setaria italica). The transcript level of SiARDP increased not only after drought, high salt, and low temperature stresses, but also after an ABA treatment in foxtail millet seedlings. Two ABA-responsive elements (ABRE1: ACGTGTC; ABRE2: ACGTGGC) exist in the promoter of SiARDP. Further analyses showed that two ABA-responsive element binding (AREB)-type transcription factors, SiAREB1 and SiAREB2, could physically bind to the ABRE core element in vitro and in vivo. The constitutive expression of SiARDP in Arabidopsis thaliana enhanced drought and salt tolerance during seed germination and seedling development, and overexpression of SiARDP in foxtail millet improved drought tolerance. The expression levels of target genes of SiARDP were upregulated in transgenic Arabidopsis and foxtail millet. These results reveal that SiARDP, one of the target genes of SiAREB, is involved in ABA-dependent signal pathways and plays a critical role in the abiotic stress response in plants. |
| [22] | . 【目的】谷子(Setaria italica L.)具有显著耐旱性。研究旨在通过反向遗传学方法分析并鉴定在干旱条件下影响植物萌发过程的重要调控因子,为研究作物干旱条件下种子萌发的调控机制创造条件。【方法】使用Clustal X 2.0和MEGA 5.05软件对谷子SiNAC18蛋白序列及其同源序列进行多序列比对,并构建系统进化树;利用real-time PCR方法检测SiNAC18在不同胁迫条件下的表达模式;通过瞬时转化的方法分析SiNAC18蛋白亚细胞定位;在拟南芥中过表达SiNAC18,分析SiNAC18的生物学功能;分析SiNAC18在转基因拟南芥中可能控制的下游基因。【结果】SiNAC18全长1 074 bp,编码由357个氨基酸组成的亲水性蛋白,分子量约为38.8 k D;系统进化树分析表明SiNAC18属于NAC转录因子家族第Ⅰ组的NAP亚组,与拟南芥基因At NAC29同源性最高;氨基酸序列比对结果显示,SiNAC18与其他物种包括水稻、拟南芥、大豆和玉米中同源性最高的NAC类转录因子蛋白的N端都具有A、B、C、D和E这5个保守结构域,蛋白C端具有高度多态性,证明SiNAC18的N端序列与其结合下游基因启动子元件相关;real-time PCR结果显示,SiNAC18在干旱(PEG)、ABA、高盐(Na Cl)及过氧化氢(H2O2)处理条件下的表达量明显上升;亚细胞定位结果表明SiNAC18蛋白定位于细胞核中;基因功能分析结果显示,在ABA和PEG胁迫处理下,SiNAC18转基因拟南芥与野生型种子的萌发率存在明显差异:在正常生长条件下,野生型拟南芥WT和SiNAC18转基因拟南芥的萌发率基本一致,在PEG浓度为10%和15%的MS培养基上,SiNAC18转基因拟南芥的萌发率显著高于WT。在2和5μmol·L-1 ABA处理条件下,转基因拟南芥的萌发率显著低于WT;下游基因表达分析结果显示,ABA信号途径相关基因At RD29A,脯氨酸合成相关基因At P5CR和At PRODH以及过氧化物酶基因At PRX34在SiNAC18转基因株系中的表达量高于WT中的表达量,表明SiNAC18通过调控这些下游基因影响转基因植物在干旱条件下的萌发率。【结论】谷子NAC类转录因子基因SiNAC18可能通过ABA信号途径、氧化胁迫调控等途径正向调控植物在干旱条件下的萌发过程。 【目的】谷子(Setaria italica L.)具有显著耐旱性。研究旨在通过反向遗传学方法分析并鉴定在干旱条件下影响植物萌发过程的重要调控因子,为研究作物干旱条件下种子萌发的调控机制创造条件。【方法】使用Clustal X 2.0和MEGA 5.05软件对谷子SiNAC18蛋白序列及其同源序列进行多序列比对,并构建系统进化树;利用real-time PCR方法检测SiNAC18在不同胁迫条件下的表达模式;通过瞬时转化的方法分析SiNAC18蛋白亚细胞定位;在拟南芥中过表达SiNAC18,分析SiNAC18的生物学功能;分析SiNAC18在转基因拟南芥中可能控制的下游基因。【结果】SiNAC18全长1 074 bp,编码由357个氨基酸组成的亲水性蛋白,分子量约为38.8 k D;系统进化树分析表明SiNAC18属于NAC转录因子家族第Ⅰ组的NAP亚组,与拟南芥基因At NAC29同源性最高;氨基酸序列比对结果显示,SiNAC18与其他物种包括水稻、拟南芥、大豆和玉米中同源性最高的NAC类转录因子蛋白的N端都具有A、B、C、D和E这5个保守结构域,蛋白C端具有高度多态性,证明SiNAC18的N端序列与其结合下游基因启动子元件相关;real-time PCR结果显示,SiNAC18在干旱(PEG)、ABA、高盐(Na Cl)及过氧化氢(H2O2)处理条件下的表达量明显上升;亚细胞定位结果表明SiNAC18蛋白定位于细胞核中;基因功能分析结果显示,在ABA和PEG胁迫处理下,SiNAC18转基因拟南芥与野生型种子的萌发率存在明显差异:在正常生长条件下,野生型拟南芥WT和SiNAC18转基因拟南芥的萌发率基本一致,在PEG浓度为10%和15%的MS培养基上,SiNAC18转基因拟南芥的萌发率显著高于WT。在2和5μmol·L-1 ABA处理条件下,转基因拟南芥的萌发率显著低于WT;下游基因表达分析结果显示,ABA信号途径相关基因At RD29A,脯氨酸合成相关基因At P5CR和At PRODH以及过氧化物酶基因At PRX34在SiNAC18转基因株系中的表达量高于WT中的表达量,表明SiNAC18通过调控这些下游基因影响转基因植物在干旱条件下的萌发率。【结论】谷子NAC类转录因子基因SiNAC18可能通过ABA信号途径、氧化胁迫调控等途径正向调控植物在干旱条件下的萌发过程。 |
| [23] | . |
| [24] | 【Objective】The homologous genes of Arabidopsis ARGOS(auxin-regulated gene involved in organ size) family were isolated from foxtail millet, which were induced expression by hormone and participate in the regulation of plant organ size. Bioinformatics, including alignment of amino acid sequences and promoter cis acting elements, and the expression pattern in different tissues and plant hormones were analyzed. Functional markers were developed from sequences analysis of gene encoding region and promoter, which will play an important role in genetic improvement of yield traits, accelerate breeding process and increase yield of foxtail millet.【Method】The number of ARGOS family members and the sequence of the ARGOS family in foxtail millet were analyzed through BLAST of conserved domain with the existing ARGOS in other plants. The encoding region and promoter sequences of SiARGOS1 gene, one of the family members of ARGOS in foxtail millet, were obtained with homologous cloning method. The promoter cis acting elements of SiARGOS1 was analyzed by the bioinformatics analysis. Expression patterns in different tissues and plant hormone of SiARGOS1 were analyzed using real-time PCR. Functional markers were developed from sequence analysis of SNP and insertion/deletion in gene encoding region and promoter. Functional markers of elite alleles of yield related traits were tested using data analysis of significant differences between genotypes associated with the yield traits such as panicle weight, grain weight and thousand-grain weight in 85 cultivars. 【Result】A total of six ARGOS family members were found to have a conserved OSR(Organ Size Related) domain consisting of two transmembrane helical structures and a highly conserved proline-rich motif in foxtail millet. The open reading frame(ORF) and the promoter of SiARGOS1, one of the family members homologous to Arabidopsis AtARGOS were cloned. Sequence analysis showed that the ORF of SiARGOS1 on chromosome 8 encoding a putative protein composed of 113 amino acids was 342 bp in length, without intron. The promoter contains a variety of regulation related components including auxin, ethylene and other plant hormone with 2 109 bp in length. SiARGOS1 is expressed in all organs of foxtail millet, at high levels in root, whereas at lowest level in panicle. The gene was insensitive to indole-3-acetic acid(IAA), but was up-regulated by ethephon(ETH). On the basis of the alignment of gene SiARGOS1 genomic DNA sequence, CAPS-AccⅡmarker was developed on the missense polymorphism site(C/G→Ala/Gly) at the 150 bp of the encoding region, two SSR markers, AP-1 and AP-2, were developed at-1652--1651(TA)_(2/3) and-1165--1163(TCA)_(1/2) of promoter, respectively. No significant association was found among the yield associated traits such as panicle weight, grain weight and thousand-grain weight with CAPS-AccⅡ and AP-1 in 85 cultivars. However, AP-2 proved to be highly correlated with panicle weight and grain weight in 85 cultivars, in addition to thousand-grain weight. The average values of panicle weight for two genotypes were 15.52 g and 20.61 g, respectively, while the ones of grain weight for two genotypes were 12.24 g and 16.74 g, respectively. 【Conclusion】Six ARGOS family members were found in foxtail millet, all of which had conserved OSR domain. The open reading frame(ORF) of SiARGOS1, homologous to Arabidopsis thaliana AtARGOS, is 342 bp in length without intron. The SSR marker AP-2, which was developed in the promoter region of the gene, could be used as functional marker for the selection of superior allelic variation in yield traits such as panicle weight and panicle weight of foxtail millet varieties. |
| [24] | . . |
| [25] | . |
| [26] | . |
| [27] | . |
| [28] | . A cDNA for the gene ZFP182, encoding a C2H2-type zinc finger protein, was cloned from rice by RT-PCR. ZFP182 codes an 18.202kDa protein with two C2H2-type zinc finger motifs, one nuclear localization signal and one Leu-rich domain. The DLN-box/EAR-motif, which exists in most of plant C2H2-type zinc finger proteins, does not exist in ZFP182. The expression analysis showed that ZFP182 gene was constitutively expressed in leaves, culms, roots and spikes at the adult rice plants, and markedly induced in the seedlings by cold (402°C), 15002mM NaCl and 0.102mM ABA treatments. The approximate 1.402kb promoter region of ZFP182 gene was fused into GUS reporter gene and transformed into tobacco. The histochemical analysis revealed that GUS expression could not be detected in transformed tobacco seedlings under normal conditions, but strongly observed in tobacco leaf discs and the vascular tissue of roots treated with NaCl or KCl. Expression of ZFP182 in transgenic tobacco and overexpression in rice increased plant tolerance to salt stress. These results demonstrated that ZFP182 might be involved in plant responses to salt stress. |
| [29] | . 植物对盐胁迫的耐受反应是个复杂的过程,在分子水平上它包括对外界盐信号的感应和传递,特异转录因子的激活和下游控制生理生化应答的效应基因的表达.在生化应答中,本文着重讨论负责维持和重建离子平衡的膜转运蛋白、渗调剂的生物合成和功能及水分控制.这些生理生化应答最终使得液泡中离子浓度升高和渗调剂在胞质中积累.近年来,通过对各种盐生植物或盐敏感突变株的研究,阐明了许多盐应答的离子转运途径、水通道和物种特异的渗调剂代谢途径,克隆了其相关基因并能在转基因淡水植物中产生耐盐表型;另一方面,在拟南芥突变体及利用酵母盐敏感突变株功能互补筛选得到一些编码信号传递蛋白的基因,这些都有助于阐明植物盐胁迫应答的分子机制。<br>Abstract:Plant responses to salt stress via a complex mechanism,including sensing and transducing the stress signal,activating the transcription factors and the corresponding metabolizing genes.Since the whole mechanism is still unclear,this review emphasize the biochemical events during the plant adaptation to salt stress referring to an index of importance:the homeostasis in cytoplasm,the biosynthesis of osmolytes and the transport of water.Most of these biochemical events were elucidated by study of halophyte and salt-sensitive mutations,also many important genes involved were cloned and used to generate stress-tolerance phenotypes in transgenic plants.On the other hand,about the molecular mechanism in signal transduction,the research of Arabidopsis mutations and yeast functional complementation provided helpful traces but not full pathway.<br> . 植物对盐胁迫的耐受反应是个复杂的过程,在分子水平上它包括对外界盐信号的感应和传递,特异转录因子的激活和下游控制生理生化应答的效应基因的表达.在生化应答中,本文着重讨论负责维持和重建离子平衡的膜转运蛋白、渗调剂的生物合成和功能及水分控制.这些生理生化应答最终使得液泡中离子浓度升高和渗调剂在胞质中积累.近年来,通过对各种盐生植物或盐敏感突变株的研究,阐明了许多盐应答的离子转运途径、水通道和物种特异的渗调剂代谢途径,克隆了其相关基因并能在转基因淡水植物中产生耐盐表型;另一方面,在拟南芥突变体及利用酵母盐敏感突变株功能互补筛选得到一些编码信号传递蛋白的基因,这些都有助于阐明植物盐胁迫应答的分子机制。<br>Abstract:Plant responses to salt stress via a complex mechanism,including sensing and transducing the stress signal,activating the transcription factors and the corresponding metabolizing genes.Since the whole mechanism is still unclear,this review emphasize the biochemical events during the plant adaptation to salt stress referring to an index of importance:the homeostasis in cytoplasm,the biosynthesis of osmolytes and the transport of water.Most of these biochemical events were elucidated by study of halophyte and salt-sensitive mutations,also many important genes involved were cloned and used to generate stress-tolerance phenotypes in transgenic plants.On the other hand,about the molecular mechanism in signal transduction,the research of Arabidopsis mutations and yeast functional complementation provided helpful traces but not full pathway.<br> |
| [30] | . |
| [31] | . |
| [32] | . . |
| [33] | . This review focuses on the regulation of transcription factors, many of which are DNA-binding proteins that recognize cis-regulatory elements of target genes and are the most direct regulators of gene transcription. Transcription factors serve as integration centres of the different signal-transduction pathways affecting a given gene. It is obvious that the regulation of these regulators themselves is of crucial importance for differential gene expression during development and in terminally differentiated cells. Transcription factors can be regulated at two, principally different, levels, namely concentration and activity, each of which can be modulated in a variety of ways. The concentrations of transcription factors, as of intracellular proteins in general, may be regulated at any of the steps leading from DNA to protein, including transcription, RNA processing, mRNA degradation and translation. The activity of a transcription factor is often regulated by (de)phosphorylation, which may affect different functions, e.g. nuclear localization, DNA binding and trans-activation. Ligand binding is another mode of transcription-factor activation. It is typical for the large superfamily of nuclear hormone receptors. Heterodimerization between transcription factors adds another dimension to the regulatory diversity and signal integration. Finally, non-DNA binding (accessory) factors may mediate a diverse range of functions, e.g. serving as a bridge between the transcription factor and the basal transcription machinery, stabilizing the DNA-binding complex or changing the specificity of the target sequence recognition. The present review presents an overview of different modes of transcription-factor regulation, each illustrated by typical examples. |
| [34] | . |
| [35] | . A mutation of AtSOS1 (Salt Overly Sensitive 1), a plasma membrane Na+/H+-antiporter inArabidopsis thaliana,leads to a salt-sensitive phenotype accompanied by the death of root cells under salt stress. Intracellular events and changes in gene expression were compared during a non-lethal salt stress between the wild type and a representative SOS1 mutant,atsos1-1,by confocal microscopy using ion-specific fluorophores and by quantitative RT-PCR. In addition to the higher accumulation of sodium ions,atsos1-1showed inhibition of endocytosis, abnormalities in vacuolar shape and function, and changes in intracellular pH compared to the wild type in root tip cells under stress. Quantitative RT-PCR revealed a dramatically faster and higher induction of root-specific Ca2+transporters, including several CAXs and CNGCs, and the drastic down-regulation of genes involved in pH-homeostasis and membrane potential maintenance. Differential regulation of genes for functions in intracellular protein trafficking inatsos1-1was also observed. The results suggested roles of the SOS1 protein, in addition to its function as a Na+/H+antiporter, whose disruption affected membrane traffic and vacuolar functions possibly by controlling pH homeostasis in root cells. |
| [36] | . |
| [37] | |
| [38] | . Uptake and translocation of cations play essential roles in plant nutrition, signal transduction, growth, and development. Among them, potassium (K+) and sodium (Na+) have been the focus of numerous physiological studies because K+ is an essential macronutrient and the most abundant inorganic cation in plant cells, whereas Na+ toxicity is a principal component of the deleterious effects associated with salinity stress. Although the homeostasis of these two ions was long surmised to be fine tuned and under complex regulation, the myriad of candidate membrane transporters mediating their uptake, intracellular distribution, and long-distance transport is nevertheless perplexing. Recent advances have shown that, in addition to their function in vacuolar accumulation of Na+, proteins of the NHX family are endosomal transporters that also play critical roles in K+ homeostasis, luminal pH control, and vesicle trafficking. The plasma membrane SOS1 protein from Arabidopsis thaliana, a highly specific Na+/H+ exchanger that catalyses Na+ efflux and that regulates its root/shoot distribution, has also revealed surprising interactions with K+ uptake mechanisms by roots. Finally, the function of individual members of the large CHX family remains largely unknown but two CHX isoforms, AtCHX17 and AtCH23, have been shown to affect K+ homeostasis and the control of chloroplast pH, respectively. Recent advances on the understanding of the physiological processes that are governed by these three families of cation exchangers are reviewed and discussed. |
