 ,1, 任毅1, 曹俊梅2, 李法计3, 夏先春4, 耿洪伟
,1, 任毅1, 曹俊梅2, 李法计3, 夏先春4, 耿洪伟 ,1
,1Genome-wide Association Analysis of Wheat Grain Size Related Traits Based on SNP Markers
ZHANG Fang ,1, REN Yi1, CAO JunMei2, LI FaJi3, XIA XianChun4, GENG HongWei
,1, REN Yi1, CAO JunMei2, LI FaJi3, XIA XianChun4, GENG HongWei ,1
,1通讯作者:
责任编辑: 李莉
收稿日期:2020-10-28接受日期:2020-12-2网络出版日期:2021-05-16
| 基金资助: |
Received:2020-10-28Accepted:2020-12-2Online:2021-05-16
作者简介 About authors
张芳,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1525KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张芳, 任毅, 曹俊梅, 李法计, 夏先春, 耿洪伟. 基于SNP标记的小麦籽粒性状全基因组关联分析[J]. 中国农业科学, 2021, 54(10): 2053-2063 doi:10.3864/j.issn.0578-1752.2021.10.002
ZHANG Fang, REN Yi, CAO JunMei, LI FaJi, XIA XianChun, GENG HongWei.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】小麦(Triticum aestivum L.)是全球播种面积最广的粮食作物之一,也是中国重要的粮食作物之一,在粮食安全中具有举足轻重的地位[1]。目前,小麦的需求增长速度逐渐快于产量的提高速度,使得粮食安全面临严峻考验。为此,在耕地面积有限的现状下,提高小麦单产至关重要[2]。小麦籽粒大小决定粒重高低,其中籽粒的长、宽、面积、周长、长宽比等参数是构成小麦产量的重要指标[3]。基于此,研究小麦籽粒性状的遗传特性对提高小麦产量及品质至关重要。【前人研究进展】高产一直是育种家最为关注的育种目标之一,而籽粒性状的遗传改良对产量潜力的提升具有显著影响[4,5,6]。随着分子标记技术的快速发展,关联分析被应用于数量性状的遗传研究,作为基因发掘的重要手段,为分子标记辅助选择提供了重要依据[7]。SNP标记具有遗传稳定性高、位点丰富且分布广等特点,其密度远远高于传统的分子标记,对于数量性状定位更为高效和便捷[8,9,10]。目前,小麦相继开发出9K、90K、660K、15K和50K等满足不同育种需求的SNP芯片[11,12,13]。小麦籽粒性状遗传结构复杂,受微效多基因控制,国内外已有大量****对小麦籽粒相关性状进行了遗传研究,并对调控小麦籽粒大小性状的QTL区段进行了定位和遗传解析。小麦籽粒性状已挖掘到QTL位点主要位于1A、1B、1D、2A、2B、2D、3B、4B、5A、6B、7B和7D等染色体[14,15,16]。LI等[17]利用W7984×Opata85重组自交系进行连锁分析,在2个环境下均发现7D染色上控制粒长的主效QTL,可解释13.5%—13.9%的表型变异。全基因组关联分析(genome-wide association study,GWAS)作为分析复杂性状遗传结构的有力工具,在水稻、玉米和拟南芥中得到了广泛的应用[18,19,20,21]。BHATTA等[22]以143份面包小麦品种为材料,采用GWAS关联分析,在1B、1D、2A、2B、3A、3B、4A、4B、4D、5A、5B、5D、6D、7A、7B染色体上共检测到38个籽粒性状相关位点,其贡献率为0.3%—19.6%。SUN等[23]利用90K SNP芯片对163份小麦主载品种(系)进行关联分析,在1A、3A、3D、5B、5D、6A、6B和7A染色体上检测到多个千粒重相关位点,贡献率为15.7%—31.6%。DABA等[24]利用200份冬小麦群体在2个环境下对粒重和粒长进行关联分析,检测到26个粒重和27个粒长显著关联的标记。LI等[25]采用SNP-GWAS方法对166份黄淮麦区小麦品种进行产量相关性状分析,分别挖掘出20个粒长和31个粒宽显著关联的标记,贡献率分别为7.1%—14.2%和6.8%—15.0%,其中位于2A染色体的IWB32119标记与粒重基因TaFlo2-A1[26]的位点一致。【本研究切入点】新疆作为中国主要的小麦产区之一,对产量的提升至关重要。然而,该地区小麦生长关键阶段常面临干旱少雨和干热风等不利环境的影响,常规育种手段依旧有效,但靠其进行单产的提升,跟不上生产需求,需对分子标记等新技术进行充分使用,但基于高密度芯片技术对新疆小麦籽粒性状的全基因组关联分析鲜有报道。【拟解决的关键问题】本研究以121份新疆种植的小麦品种(系)为材料,结合50K SNP芯片对千粒重、粒长、粒宽、籽粒长宽比、籽粒面积和籽粒周长等6个籽粒性状进行全基因组关联分析,以期获得新的与籽粒性状关联的SNP位点,为小麦聚合育种及复杂数量性状位点的研究提供参考依据。1 材料与方法
1.1 试验材料及田间种植
选用121份新疆种植小麦品种(系),其中引进品种(系)36份,分别来自前苏联、乌克兰、罗马尼亚、北京和河北;自育品种(系)68份;地方品种(系)17份。上述材料由新疆农业大学农学院小麦遗传育种课题组提供。参试材料于2016—2017、2017—2018和2018—2019年度种植于新疆农业科学院玛纳斯农业试验站,随机区组设计,三行区,行长2 m,行距25 cm,3次重复。田间管理遵循当地基本栽培管理方式。待小麦生理成熟期后,按每品种取10株收获脱粒,在自然晒干后均通过杭州万深SC-G拷种仪器检测千粒重(thousand kernel weight,TKW)、粒长(grain Length,GL)、粒宽(grain width,GW)、粒长宽比(length width ratio,LWR)、籽粒面积(grain area,GA)、籽粒周长(grain perimeter,GC)。利用IciMapping的ANOVA功能进行田间数据分析及遗传力的估算。广义遗传力(h2),计算公式:h2=σg2/(σg2+σe2),σg2为遗传方差,σe2为环境方差。
1.2 SNP标记检测
50K标记利用Illumina SNP Genotyping技术测试平台(北京博奥生物有限公司),使用微珠芯片技术(Bead Array)进行检测,利用Genomestudio v1.0软件进行多态性分析。获得的基因型数据进行质量控制,删除缺失率>20%的标记,过滤掉最小等位基因频率(minor allele frequency,MAF)<5%的标记。1.3 群体结构分析
采用PowerMarker V3.25软件对参试材料进行遗传多样性和多态性信息含量分析。其中,PIC=1- ∑(Pij)2,Pij表示i位点的第j个等位变异出现的频率。选取在小麦21条染色体上均匀分布的2 000个SNP标记,利用Structure 2.3.4软件进行群体结构分析。参数设置:Burn-in period为10 000,MCMC为100 000,选择Admixed模型,设K=2—12,每个K重复5次,结果上传Structure Harvester,获得最佳K值作为亚群数目。
1.4 关联分析
应用Tassel V5.0中的混合线性模型(mixed linear model,MLM)对SNP标记与性状进行关联分析,并利用Structure 2.3.4软件计算Q值,结合Tassel V5.0软件计算Kinship值。在P<0.001水平下认为标记与性状存在关联,以第95百分位的r2值作为阈值估测LD衰减距离。2 结果
2.1 表型分析
通过对千粒重、粒长、粒宽、籽粒长宽比、籽粒面积和籽粒周长等6个籽粒相关性状的表型进行统计分析(表1),结果表明,不同环境间各性状均表现较大差异,其中,千粒重变异系数最大。各指标的峰度和偏度均接近1,说明均符合正态分布,是多基因控制的数量性状。方差分析(表2)表明,各性状在基因型、环境及基因型与环境互作间均呈现出极显著差异(P<0.001)。6个籽粒相关性状的广义遗传力(h2)为0.85—0.90,其中,长宽比和粒宽的遗传力较高,分别达到0.90和0.89,表明小麦籽粒性状主要受遗传变异影响。相关性分析(表3)表明,粒长与籽粒周长相关性最大(r=0.94),其次为粒宽与千粒重相关性最好(r=0.93);长宽比与千粒重、粒宽和籽粒面积呈极显著负相关(P<0.001),进一步说明小麦籽粒性状间存在协同变化关系。Table 1
表1
表1小麦自然群体籽粒性状基本统计信息
Table 1
| 性状 Trait | 环境 Environment | 平均值 Mean | 范围 Range | 标准差 SD | 变异系数 CV (%) | 偏度 Ske. | 峰度 Kur. |
|---|---|---|---|---|---|---|---|
| 千粒重 TKW | 2017 | 22.92 | 14.86—32.28 | 4.08 | 17.79 | -0.18 | -0.43 |
| 2018 | 35.55 | 23.13—49.03 | 4.95 | 13.91 | 0.05 | -0.02 | |
| 2019 | 37.84 | 19.15—54.15 | 5.62 | 14.85 | -0.02 | 0.41 | |
| 粒长 GL | 2017 | 6.44 | 5.66—7.59 | 0.34 | 5.24 | 0.54 | 0.88 |
| 2018 | 6.63 | 5.67—7.48 | 0.31 | 4.66 | 0.20 | 0.54 | |
| 2019 | 6.61 | 5.78—7.51 | 0.31 | 4.69 | 0.16 | 0.06 | |
| 粒宽 GW | 2017 | 3.14 | 2.61—3.61 | 0.24 | 7.52 | -0.39 | -0.28 |
| 2018 | 3.15 | 2.61—3.65 | 0.22 | 6.95 | -0.19 | -0.65 | |
| 2019 | 3.27 | 2.69—3.64 | 0.18 | 5.50 | -0.43 | 0.03 | |
| 长宽比 LWR | 2017 | 2.09 | 1.75—2.67 | 0.19 | 9.12 | 0.74 | 0.20 |
| 2018 | 2.13 | 1.81—2.58 | 0.16 | 7.39 | 0.59 | -0.06 | |
| 2019 | 2.03 | 1.75—2.41 | 0.13 | 6.40 | 0.68 | 0.43 | |
| 籽粒面积 GA | 2017 | 15.82 | 11.88—19.59 | 1.54 | 9.74 | -0.11 | 0.16 |
| 2018 | 16.24 | 12.55—21.34 | 1.59 | 9.76 | 0.23 | 0.02 | |
| 2019 | 16.67 | 12.65—20.84 | 1.38 | 8.28 | -0.01 | 0.63 | |
| 籽粒周长 GC | 2017 | 16.61 | 14.52—18.62 | 0.76 | 4.56 | 0.07 | 0.67 |
| 2018 | 16.78 | 14.61—19.11 | 0.72 | 4.30 | 0.22 | 0.58 | |
| 2019 | 17.27 | 15.73—19.80 | 0.72 | 4.17 | 0.25 | 0.44 |
新窗口打开|下载CSV
Table 2
表2
表2小麦籽粒性状方差分析
Table 2
| 性状 Trait | 均方MS | F值F value | 遗传力 h2 | |||||
|---|---|---|---|---|---|---|---|---|
| 基因型 Genotype | 环境 Environment | 基因型×环境 G×E | 误差 Error | 基因型 Genotype | 环境 Environment | 基因型×环境 G×E | ||
| 千粒重TKW | 157.46 | 17050.92 | 27.47 | 4.43 | 35.54*** | 3848.31*** | 6.20*** | 0.85 |
| 粒长GL | 0.60 | 2.74 | 0.10 | 0.02 | 37.54*** | 170.47*** | 6.18*** | 0.86 |
| 粒宽GW | 0.29 | 1.63 | 0.04 | 0.01 | 22.98*** | 131.03*** | 2.84*** | 0.89 |
| 长宽比LWR | 0.16 | 0.96 | 0.02 | 0.00 | 40.15*** | 236.77*** | 4.53*** | 0.90 |
| 籽粒面积GA | 14.19 | 52.10 | 1.96 | 0.53 | 26.59*** | 97.66*** | 3.68*** | 0.87 |
| 籽粒周长GC | 3.28 | 36.25 | 0.52 | 0.14 | 23.40*** | 258.40*** | 3.71*** | 0.86 |
新窗口打开|下载CSV
Table 3
表3
表3小麦籽粒性状间相关性分析
Table 3
| 性状Trait | 千粒重TKW | 粒长GL | 粒宽GW | 长宽比LWR | 籽粒面积GA |
|---|---|---|---|---|---|
| 粒长GL | 0.36*** | ||||
| 粒宽GW | 0.93*** | 0.22* | |||
| 长宽比LWR | -0.64*** | 0.42*** | -0.78*** | ||
| 籽粒面积GA | 0.90*** | 0.64*** | 0.88*** | -0.41*** | |
| 籽粒周长GC | 0.58*** | 0.94*** | 0.49*** | 0.15ns | 0.83*** |
新窗口打开|下载CSV
2.2 标记遗传多样性及群体结构
利用50K SNP芯片对121份新疆种植的小麦品种(系)进行检测,经过质量控制最终筛选出36 873个SNP标记进行GWAS分析,其中,A基因组、B基因组和D基因组染色体平均包含16 514、11 490和8 869个SNP标记,每个标记间的平均距离为0.39 Mb(电子附表1)。A与B基因组的染色体组所用标记密度、标记数目、遗传多样性和多态信息含量均高于D染色体组。SNP标记的PIC值为0.09—0.38,平均值为0.32。Supplemental table 1
附表1
附表1标记分布及多态性
Supplemental table 1
| 染色体 Chromosome | 标记数量 No. of markers | 物理长度 Physical distance (Mb) | 标记密度Density of marker | 遗传多样性Genetic diversity | 最小等位基因频率MAF | 多态信含量 PIC | ||
|---|---|---|---|---|---|---|---|---|
| 平均 Mean | 范围 Range | 平均 Mean | 范围 Range | |||||
| 1A | 2544 | 602.45 | 0.24 | 0.35 | 0.25 | 0.05—0.50 | 0.28 | 0.09—0.38 |
| 1B | 1469 | 746.65 | 0.29 | 0.34 | 0.25 | 0.05—0.50 | 0.27 | 0.09—0.38 |
| 1D | 1486 | 498.09 | 0.20 | 0.36 | 0.26 | 0.05—0.50 | 0.29 | 0.09—0.38 |
| 2A | 2682 | 787.57 | 0.31 | 0.38 | 0.29 | 0.05—0.50 | 0.3 | 0.09—0.38 |
| 2B | 2177 | 802.80 | 0.32 | 0.37 | 0.27 | 0.05—0.50 | 0.29 | 0.09—0.38 |
| 2D | 1308 | 655.89 | 0.26 | 0.37 | 0.27 | 0.05—0.50 | 0.29 | 0.09—0.38 |
| 3A | 2252 | 750.52 | 0.29 | 0.46 | 0.37 | 0.25—0.50 | 0.35 | 0.30—0.38 |
| 3B | 2275 | 868.70 | 0.34 | 0.42 | 0.33 | 0.17—0.50 | 0.33 | 0.24—0.38 |
| 3D | 1201 | 626.21 | 0.25 | 0.36 | 0.27 | 0.05—0.50 | 0.29 | 0.09—0.38 |
| 4A | 2063 | 750.43 | 0.29 | 0.46 | 0.39 | 0.26—0.50 | 0.36 | 0.30—0.38 |
| 4B | 1327 | 672.85 | 0.26 | 0.39 | 0.29 | 0.11—0.50 | 0.31 | 0.18—0.38 |
| 4D | 817 | 521.81 | 0.20 | 0.39 | 0.29 | 0.05—0.50 | 0.31 | 0.09—0.38 |
| 5A | 2251 | 709.53 | 0.28 | 0.49 | 0.43 | 0.35—0.50 | 0.37 | 0.35—0.38 |
| 5B | 1605 | 713.57 | 0.28 | 0.46 | 0.38 | 0.26—0.50 | 0.35 | 0.29—0.38 |
| 5D | 1373 | 573.30 | 0.22 | 0.41 | 0.31 | 0.16—0.50 | 0.32 | 0.23—0.38 |
| 6A | 2315 | 625.50 | 0.24 | 0.49 | 0.43 | 0.34—0.50 | 0.37 | 0.35—0.38 |
| 6B | 1305 | 725.17 | 0.28 | 0.45 | 0.37 | 0.21—0.50 | 0.35 | 0.28—0.38 |
| 6D | 1330 | 498.56 | 0.19 | 0.45 | 0.36 | 0.20—0.50 | 0.34 | 0.27—0.38 |
| 7A | 2407 | 745.91 | 0.29 | 0.48 | 0.41 | 0.31—0.50 | 0.36 | 0.34—0.38 |
| 7B | 1332 | 750.19 | 0.29 | 0.45 | 0.37 | 0.22—0.50 | 0.35 | 0.28—0.38 |
| 7D | 1354 | 643.15 | 0.25 | 0.44 | 0.32 | 0.19—0.50 | 0.33 | 0.26—0.38 |
| A基因组A genome | 16514 | 4971.91 | 0.30 | 0.44 | 0.37 | 0.05—0.50 | 0.34 | 0.09—0.38 |
| B基因组 B genome | 11490 | 5279.93 | 0.46 | 0.41 | 0.32 | 0.05—0.50 | 0.32 | 0.09—0.38 |
| D基因组 D genome | 8869 | 4017.01 | 0.45 | 0.40 | 0.30 | 0.05—0.50 | 0.31 | 0.09—0.38 |
| 总计Total | 36873 | 14268.85 | 0.39 | 0.42 | 0.33 | 0.05—0.50 | 0.32 | 0.09—0.38 |
新窗口打开|下载CSV
利用Structure 2.3.4软件分析,获得121份新疆种植的小麦自然群体结构Q值与极大似然值LnP(D),进一步确定最佳K值。当K=4时(图1-A),ΔK值最大,由图1-B可知,参试材料被划分为4个亚群(sub- population),其中,亚群Ⅰ(SubgroupⅠ)包含46份品种(系),主要来自自育和引进品种(系),分别占比47.83%和41.30%,引进品种多为前苏联品种(系);亚群Ⅱ(SubgroupⅡ)包含29份品种(系),主要来自自育品种(系),占比79.31%;亚群Ⅲ(SubgroupⅢ)包含41份品种(系),包含56.10%的自育品种(系)、26.83%的引进品种(系)和17.07%的地方品种(系);亚群Ⅳ(SubgroupⅣ)包含5份品种(系),均来自地方品种(系);4个亚群所含品种数分别占总材料份数的38.02%、23.97%、33.88%和4.13%。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1121份小麦材料的群体结构
Fig. 1Population structure of 121 wheat accessions
2.3 籽粒相关性状关联分析
基于LD衰减距离计算,将在物理图谱上前后3 Mb区间内的位点认定为一个候选位点。用36 873个分子标记对3个环境及平均环境下的表型数据进行关联分析(图2),6个小麦籽粒相关性状共检测到592个显著SNP位点(P<0.001),单个关联标记位点解释的表型变异(R2)范围为9.3%—28.6%。其中29个SNP至少在2个环境中被检测到,分布在1A(5)、1B(2)、1D、2A(5)、3B、5A、5D、6B(4)、6D、7B和7D(7)染色体上,可解释9.3%—22.7%的表型变异(表4)。检测到6个与千粒重稳定关联的标记,分布在1B、2A和6B染色体上,其中在6B染色体检测到4个千粒重位点(AX-109874065、AX-110446017、AX-110936500和AX-109493716),且效应值较大,介于9.7%—12.9%,说明6B染色体对千粒重影响较大,可作为籽粒性状重要的染色体。检测到6个与粒长稳定关联的标记,分布在1A、2A和7D染色体上,其中AX-111082947和AX-94497666在3个环境和均值下同时被检测到,表型变异解释范围为9.9%—22.7%。检测到2个与粒宽稳定关联的标记,分布在3B和5D染色体上的AX-108948870与AX-179477405,表型变异解释范围为9.6%—12.2%。检测到6个与长宽比稳定关联的标记,分布在2A(2)、5A、7B和7D(2)染色体上,其中位于2A染色体的AX-111722425标记在全部环境下均被重复检测到,可解释10.1%—18.3%表型变异。检测到3个与籽粒面积稳定关联的标记,分布在1A、1B和1D染色体上,表型变异解释率范围为9.9%—18.2%,表明1号同源群对籽粒面积具有重要作用。检测到6个与籽粒周长稳定关联的标记,分布在1A(2)、2A、6D和7D(2)染色体上,其中,位于1A染色体的AX-111082947标记在多环境下同时被检测到,且贡献率最大,可解释10.1%—22.6%表型变异。图2
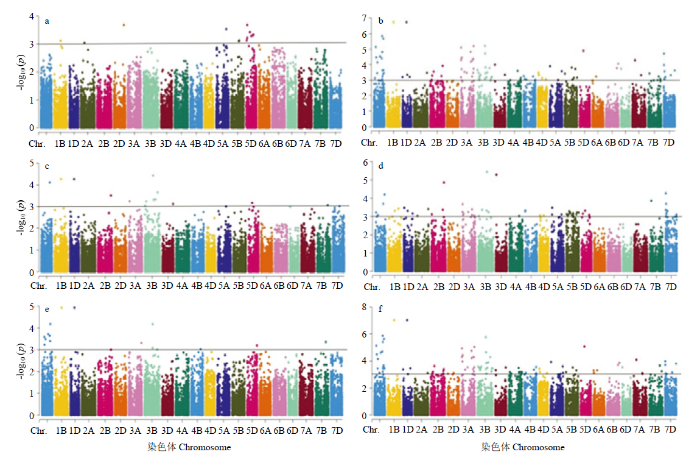 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2平均环境下籽粒相关性状的曼哈顿图
a:粒长;b:粒宽;c:籽粒长宽比;d:籽粒面积;e:籽粒周长;f:千粒重
Fig. 2Manhattan plots for grain size related traits of the diverse panel at average environment
A: Grain length; b: Grain width; c: Length width ratio; d: Grain area; e: Grain circle; f: Thousand kernel weight
Table 4
表4
表4SNP-GWAS检测到的产量性状相关位点
Table 4
| 性状 Trait | 标记 Marker | 染色体 Chr. | 位置 Position (Mb) | 混合线性模型MLM | 环境 Environment | 前人报道 Previously reported | |
|---|---|---|---|---|---|---|---|
| P值P value | 贡献率R2 (%) | ||||||
| 千粒重 TKW | AX-111669426 | 1B | 375.4 | 5.94E-04-7.67E-04 | 10.4—10.5 | E1/E4 | Xwmc269-Xwmc33[27] |
| AX-111531320 | 2A | 204.5 | 5.88E-04-8.98E-04 | 10.3—11.3 | E2/E4 | ||
| AX-109874065 | 6B | 34.0 | 7.73E-04-9.84E-04 | 9.7—10.4 | E1/E2 | TKW-xgwm533[28] | |
| AX-110446017 | 6B | 486.1 | 3.98E-04-7.98E-04 | 10.3—12.9 | E1/E2 | QKWpur-6B[24] AX_110368497 [25] | |
| AX-110936500 | 6B | 569.4 | 4.97E-04-6.86E-04 | 10.5—10.6 | E1/E2 | ||
| AX-109493716 | 6B | 573.6 | 4.92E-04-6.58E-04 | 11.0—11.8 | E1/E2 | ||
| 粒长 GL | AX-94757616 | 1A | 64.6 | 3.56E-04-7.55E-04 | 10.7—11.9 | E2/E4 | |
| AX-111082947 | 1A | 473.7 | 1.41E-06-9.42E-04 | 9.9—22.7 | E1/E2/E3/E4 | ||
| AX-94497666 | 2A | 748.4 | 3.30E-04-7.90E-04 | 10.3—11.8 | E1/E2/E3/E4 | TaFlo2[25-26] | |
| AX-111614568 | 7D | 13.7—13.9 | 1.96E-05-2.03E-04 | 12.9—17.3 | E3/E4 | TaGS-D1[29] | |
| AX-95633409 | 7D | 33.6 | 4.46E-04-8.05E-04 | 10.7—11.9 | E3/E4 | QKLpur-7D.1[24] | |
| AX-111197303 | 7D | 47.4—49.2 | 2.70E-04-8.32E-04 | 10.9—12.6 | E1/E4 | QKL.caas-7DS [30] QGw.ccsu-7D.1 [31] | |
| 粒宽 GW | AX-108948870 | 3B | 24.6 | 5.20E-04-9.21E-04 | 10.6—12.2 | E1/E4 | Kukri_c14642_917[23] |
| AX-179477405 | 5D | 210.6 | 6.78E-04-8.33E-04 | 9.6—10.6 | E2/E4 | ||
| 粒长宽比 LWR | AX-111722425 | 2A | 131.7 | 1.38E-05-9.53E-04 | 10.1—18.3 | E1/E2/E3/E4 | |
| AX-111531320 | 2A | 204.5 | 3.57E-05-3.28E-04 | 13.0—17.1 | E2/E3 | ||
| AX-179560109 | 5A | 141.9 | 1.02E-04-1.08E-04 | 14.7—15.2 | E2/E3 | ||
| AX-112286258 | 7B | 709.1 | 1.48E-05-4.09E-04 | 11.1—19.4 | E1/E3 | QGlwr.ccsu-7B.1[32] | |
| AX-111614568 | 7D | 13.7—13.9 | 4.98E-05-5.51E-05 | 10.9—15.9 | E1/E4 | TaGS-D1[29] | |
| AX-158554015 | 7D | 403.7 | 6.09E-05-6.42E-04 | 10.8—15.4 | E1/E2 | ||
| 籽粒面积 GA | AX-95176275 | 1A | 146.6 | 5.92E-04-9.82E-04 | 9.9—11.2 | E1/E4 | |
| AX-179476290 | 1B | 377.4 | 1.21E-05-8.31E-04 | 10.0—18.2 | E3/E4 | qKA1B-1[33] | |
| AX-179477117 | 1D | 220.5 | 1.21E-05-8.31E-04 | 10.0—18.2 | E3/E4 | ||
| 籽粒周长 GC | AX-111082947 | 1A | 473.7 | 1.34E-06-5.56E-04 | 10.1—22.6 | E1/E3/E4 | |
| AX-109382284 | 1A | 546.8 | 2.27E-06-3.55E-04 | 11.8—21.3 | E3/E4 | ||
| AX-94497666 | 2A | 748.4 | 7.62E-04-8.68E-04 | 9.3—10.3 | E1/E4 | TaFlo2[25,26] | |
| AX-111583179 | 6D | 242.7 | 2.11E-04-5.00E-04 | 12.0—13.2 | E2/E3 | ||
| AX-111614568 | 7D | 13.7—13.9 | 1.04E-04-3.72E-04 | 11.7—14.2 | E3/E4 | TaGS-D1[29] | |
| AX-111197303 | 7D | 47.4—49.2 | 7.15E-04-8.05E-04 | 10.0—10.4 | E1/E4 | QKL.caas-7DS[30] QGw.ccsu-7D.1[31] | |
新窗口打开|下载CSV
29个稳定关联的位点中,位于1A染色体上的AX-111082947和位于2A染色体上的AX-94497666标记均与粒长和籽粒周长显著关联,贡献率分别为9.3%—22.7%和9.3%—11.8%;位于2A染色体上的AX- 111531320标记均与千粒重和籽粒长宽比显著关联,贡献率分别为10.3%—17.1%;位于7D染色体上的AX-111614568标记同时与粒长、籽粒周长和籽粒长宽比显著关联,贡献率为10.9%—17.3%;位于7D染色体上的标记AX-111197303同时与粒长和籽粒周长显著关联,贡献率为10.0%—12.6%,说明这5个位点是一因多效位点。
3 讨论
3.1 小麦籽粒性状表型
小麦籽粒性状包括粒长、粒宽、长宽比、粒厚、籽粒面积、籽粒周长和千粒重等指标,其高低是衡量产量的重要依据之一,且性状便于观察,作为小麦育种选择最直观的性状,往往受到研究者的高度关注[34]。DHOLAKIA等[35]认为小麦籽粒性状不但对产量具有影响,也决定小麦的商品价值。粒重受粒长、粒宽、粒厚以及籽粒周长等方面的直接影响[36],其中粒宽与粒重呈显著正相关,相关性较大。陈佳慧等[37]、余曼丽等[38]发现千粒重与粒长、粒宽、籽粒面积、籽粒周长均呈极显著正相关,与籽粒长宽比呈负相关,这与本研究结果基本一致。此外发现粒长与籽粒周长、粒宽与籽粒面积存在协同促进关系,说明各性状在生长发育过程中由单个基因多效性引起或多个基因紧密连锁共同控制某些性状来作为性状相关的遗传基础。不同类型新疆种植的小麦材料中,自育品种的大粒表现较高,地方品种次之,引进品种较低,这一结果与马艳明等[39]基本一致,充分体现了新疆自育品种间籽粒性状差异较丰富。究其原因一方面是自育品种经过育种家多年以大粒选择为主有关,其重视度和选择度较强;另一方面是结合当地环境的育种需求,大粒小麦品种对小麦粒重遗传改良具有较大的提升空间,将更有利于中国小麦高产优质新品种培育。另外,本研究还发现籽粒长宽比地方品种显著高于自育品种和引进品种,这可能由于地方品种一般为长而尖粒型,其粒长较粒宽发育较早,而育成品种在选育过程中,着重于粒长粒宽进行改良,呈现出粒宽的增加同时粒长未发生明显变化[39]。这一结果充分体现出千粒重提高主要由粒宽的增加而影响,而小麦产量的提升主要由千粒重来贡献。3.2 小麦籽粒性状GWAS分析
在多个环境下均能被检测到的关联位点可视为相对稳定的位点,本研究共检测到29个籽粒性状稳定关联的SNP位点。粒长稳定位点分布于1A、2A和7D染色体上,LI等[25]通过166份黄淮麦区材料结合90K SNP芯片进行关联分析,在2A染色体上发现一个稳定位点,并确定为粒重基因TaFlo2-A1的位点,这与本研究在2A染色体检测到的标记AX-94497666所在位点相距仅约3.4 Mb,可能为相同位点,该位点控制的粒长效应可能来自于该基因的作用。同时位于7D染色体上的标记AX-111614568检测到一个粒长相关位点,这与粒重基因TaGS-D1位点一致,也为相同位点[29]。粒长标记AX-95633409与DABA等[24]发现位点QKLpur-7D.1较近,相距0.5 Mb,LI等[30]与MIR等[31]分别也在7D染色体上检测到标记QKL.caas-7DS和QGw.ccsu-7D.1与本研究粒长标记AX-111197303均位于短臂区域。本研究在3B染色体上发现的控制粒宽的SNP标记AX-108948870与王瑞霞等[27]、SUN等[23]报道的粒宽QTL位置相似,其表型贡献率较大,CABRAL等[40]也证实了在3B染色体上相近的位置上发现了一个QTL位点。位于7B染色体上发现籽粒长宽比标记AX-112286258贡献率最大,KUMARI等[32]检测到的QTL位点相距31.6 Mb。XIN等[33]利用Dalibao×BYL8 RIL进行籽粒性状QTL定位,获得qKA1B-1籽粒面积位点与本研究发现的标记AX-179476290位置相近。对于千粒重,在6B染色体上鉴定了4个稳定显著SNP标记(AX-109874065、AX-110446017、AX-110936500和AX-109493716),其贡献率为9.7%—12.9%。其中,AX-109874065标记与张坤普[28]报告的千粒重的位点TKW-xgwm533仅为1.3 Mb,因此,笔者认为上述属于同一位点控制千粒重高低。DABA等[24]和LI等[25]分别检测到QKWpur-6B和AX_110368497与本研究标记AX-110446017相距31.7和31.9 Mb,说明该染色体上存在粒重密切相关位点。MUHAMMAD等[41]也在6B染色体检测到控制千粒重的位点,其贡献率为12.0%—14.0%,后期将对该染色体的重要位点进行SNP标记开发,为分子标记辅助选择提供有效的选择标记。本研究在位于1A、1B、2A、5A、5D、6B和7D染色体上发现可能与籽粒相关性状稳定遗传的新位点。其中,在2A染色体上发现控制千粒重的新位点SNP标记AX-111531320,同时也控制籽粒长宽比,其效应值大于10%且稳定。共位点区域可能存在着控制不同性状的多个基因,或者是存在一个基因控制多个性状[42]。位于1A染色体上标记AX-111082947被发现是新位点且贡献率最大,为9.8%—22.7%,同时影响粒长和籽粒周长,有可能为控制小麦粒重主效位点,该基因可在小麦产量育种中起到重要的作用,可优选1A染色体关联位点进行功能标记开发,为分子标记辅助育种提供有效信息。在7D染色体上接触到的位点AX-111197303标记,也可同时影响粒长和籽粒周长,表明这两个位点控制粒长和籽粒周长的基因可能为同一基因且验证了粒长与籽粒周长具有协同作用。后期研究可聚焦这些多效应位点且在多环境下均被检测到的稳定位点,进一步开展精细定位和分子标记开发等研究。
4 结论
本研究材料遗传多样性丰富,在2个及以上环境中共检测到29个与6个籽粒性状稳定显著关联位点,其中标记AX-94497666和AX-111614568与粒长和籽粒周长显著关联,与前人发现的TaFlo2-A1和TaGS-D1相吻合。检测到5个显著位点为多效性位点,贡献率为9.9%—22.7%。标记AX-111082947(1A)和AX-111197303(7D)与粒长和籽粒周长显著关联,贡献率分别为9.8%—22.7%和10.0%—12.6%。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
DOI:10.1126/science.1185383URL [本文引用: 1]
DOI:10.1038/s41598-019-55410-5URL [本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/erq311URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S0378-1119(99)00219-XURL [本文引用: 1]
DOI:10.1016/S1369-5266(02)00240-6URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/pbi.2014.12.issue-6URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-018-3164-9URL [本文引用: 1]
DOI:10.2135/cropsci1999.0011183X003900040039xURL [本文引用: 1]
DOI:10.1007/s001220051703URL [本文引用: 1]
.
DOI:10.1016/j.fcr.2006.11.008URL [本文引用: 1]
[本文引用: 1]
DOI:10.1007/s10709-016-9932-zURL [本文引用: 1]
DOI:10.1038/ng.3596URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.3390/genes10010001URL [本文引用: 1]
DOI:10.1111/pbi.2017.15.issue-8URL [本文引用: 3]
DOI:10.3389/fpls.2018.01045URL [本文引用: 5]
DOI:10.1186/s12870-019-1781-3URL [本文引用: 6]
URL [本文引用: 3]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s11032-014-0102-7URL [本文引用: 4]
DOI:10.1007/s00122-018-3122-6URL [本文引用: 3]
DOI:10.1007/s11032-011-9693-4URL [本文引用: 3]
DOI:10.1007/s12298-018-0552-1URL [本文引用: 2]
DOI:10.1038/s41598-019-56979-7URL [本文引用: 2]
DOI:10.1534/genetics.104.034769URL [本文引用: 1]
DOI:10.1046/j.1439-0523.2003.00896.xURL [本文引用: 1]
DOI:10.1105/tpc.110.074153URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
DOI:10.1186/s12870-019-1754-6URL [本文引用: 1]
[本文引用: 1]
