 ,1,2, 王佰翠2, 马晓兰2, 贾子苗2, 叶兴国2, 林志珊
,1,2, 王佰翠2, 马晓兰2, 贾子苗2, 叶兴国2, 林志珊 ,2, 胡汉桥
,2, 胡汉桥 ,1
,1Polymorphism Analysis Among Chromosomes of Dasypyrum villosum 6V#2 and 6V#4 and Wheat 6A and 6D Based on Wheat SNP Chip
XU ZhiYing ,1,2, WANG BaiCui2, MA XiaoLan2, JIA ZiMiao2, YE XingGuo2, LIN ZhiShan
,1,2, WANG BaiCui2, MA XiaoLan2, JIA ZiMiao2, YE XingGuo2, LIN ZhiShan ,2, HU HanQiao
,2, HU HanQiao ,1
,1通讯作者:
责任编辑: 李莉
收稿日期:2020-08-24接受日期:2020-10-28网络出版日期:2021-04-16
| 基金资助: |
Received:2020-08-24Accepted:2020-10-28Online:2021-04-16
作者简介 About authors
许志英,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1234KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
许志英, 王佰翠, 马晓兰, 贾子苗, 叶兴国, 林志珊, 胡汉桥. 基于小麦SNP芯片对簇毛麦6V#2和6V#4染色体及其与小麦6A、6D染色体的多态性分析[J]. 中国农业科学, 2021, 54(8): 1579-1589 doi:10.3864/j.issn.0578-1752.2021.08.001
XU ZhiYing, WANG BaiCui, MA XiaoLan, JIA ZiMiao, YE XingGuo, LIN ZhiShan, HU HanQiao.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】簇毛麦(Dasypyrum villosum,2n= 14,VV)是小麦的二倍体近缘种属,耐寒、耐旱,抗全蚀病、眼斑病、黄花叶病、白粉病等多种小麦病害,且籽粒蛋白质含量高,是小麦品种改良的重要种质资源[1,2,3]。由于簇毛麦与小麦染色体之间不能自由交换,给育种带来了极大的困难。基于大数据,从DNA水平上分析不同簇毛麦的同源染色体及其与小麦部分同源染色体间的多态性,为靶向易位设计育种提供理论依据。【前人研究进展】据报道,已收集超过300份的簇毛麦种质资源,一些不同来源的簇毛麦染色体导入小麦基因组中已育成各种异附加系、代换系和易位系[4,5,6],其中,簇毛麦6V#2和6V#4染色体对白粉菌表现明显的抗性,具有良好的应用潜力,因此得到了较为深入的研究。在过去的研究中,基于表型和部分基因序列的比较均表明6V#2和6V#4染色体存在差异。如携带不同来源6V染色体的代换系和易位系对小麦瘿螨及其传播的病毒的抗性表现不同[7];基于表达序列的引物设计与PCR扩增片段的比较,表明6V#2和6V#4上某些基因序列存在差异,并开发了可用于区分6V#2和6V#4的特异PCR标记,其中,刘畅等[8]开发的P461-5a在6V#2和6V#4间存在扩增片段长度的多态性,而P259-1只能检测6V#4;LI等[9]基于转录组测序序列开发了25个6V特异性标记,3个在6V#2和6V#4间存在多态性;BIE等[10]开发了可同时鉴定6AS/6DS/6V#2S/6V#4S的多态性标记MBH1;陈竟男等[11]利用238对EST-SSR引物对小麦中国春与簇毛麦Dv#4和簇毛麦Dv#2的基因组DNA进行扩增,结果显示,47对在簇毛麦#2和簇毛麦#4之间存在多态性。最近,MA等[12]观察到在非易位的完整染色体状态下,小麦背景中的簇毛麦6V#2和6V#4染色体中期I不配对的频率达20.24%,且在配对的二价体中,形成环状二价体的频率低于棒状二价体,说明染色体6V#2和6V#4在染色体水平上也存在较大差异。但在整条染色体的DNA水平上的差异目前尚缺乏相关的信息,难以确定簇毛麦6V#2和6V#4染色体之间及其与小麦部分同源染色体之间的多态性。【本研究切入点】随着单核苷酸多态性(single nucleotide polymorphism,SNP)分析检测技术的迅猛发展,基于Illumina技术平台的9K和90K高通量SNP芯片以及最新开发的660K、55K和820K芯片已被应用于目标基因定位、高密度遗传图谱构建和群体结构分类等小麦诸多研究领域[13,14,15]。RAZ等[16]利用90K iSelect SNP对硬粒小麦和野生二粒小麦杂交构建的140个重组自交系(recombinant inbred lines,RILs)进行全基因组扫描,构建了全长为2 110 cM,包含14 088个SNP位点的遗传连锁图谱,其中,相邻标记之间的平均距离为0.92 cM。STUART等[17]对384HT系列硬粒小麦群体进行研究,构建了包含1 345个SNP标记的复合遗传图谱。ZHOU等[18]利用660K芯片对2个异花授粉的二倍体冰草Z1842和Z2098种间杂交产生的F1代以及35个小麦-冰草附加系/代换系进行基因分型,利用分布在跨839.7 cM 7个连锁群中的913个SNP标记构建遗传连锁图谱,确定了P基因组和小麦基因组之间的同源关系。曹延杰等[19]利用Illumina 90K iSelect SNP标记技术对96个小麦品种进行全基因组扫描,表明不同品种间亲缘关系较近,在育种中迫切需要引入新的种质资源以拓宽遗传背景。目前,利用高通量SNP芯片在DNA水平上对不同来源簇毛麦6V染色体间及其与小麦6A、6D染色体间的多态性分析鲜见报道。【拟解决的关键问题】不同来源的簇毛麦同源染色体需要在相同代换类型的条件下,才可能利用共同的小麦探针对其进行相互比较,因此,本研究通过分子标记和基因组原位杂交技术,对2个不同的异代换系南87-88和RW15杂交的F2群体筛选出的6V#4(6A)异代换系19EL124和6V#2(6D)异代换系19EL134在F3代进行确认,进一步利用小麦55K芯片中的6A、6D探针对异代换系与相同代换类型的双亲分别进行多态性分析,同时整合660K芯片上第6同源群探针对2份簇毛麦的分析结果,筛选6V染色体特异的标记,拟为靶向易位的精准设计育种提供理论依据。1 材料与方法
1.1 试验材料
源于英国剑桥的簇毛麦Dv#2、6V#2(6A)异代换系南87-88(2n=42=21II)由南京农业大学陈佩度教授惠赠。引自俄罗斯的簇毛麦No.1026、6V#4(6D)异代换系RW15(2n=42=21II)、6V#4S·6DL易位系Pm97033和6V#2S·6AL易位系10SR3109以及普通小麦品种宛7107均由中国农业科学院作物科学研究所陈孝研究员提供。19EL124和19EL134来自杂交组合南87-88×RW15 F3的2个株系,系谱号分别为18GL101-2-5和18GL101-2-29。1.2 样本DNA的提取
用植物材料新鲜叶片提取gDNA,方法参照DNA提取试剂盒(北京康为世纪生物科技有限公司)提供的步骤进行,并最终稀释成100 ng·μL-1,-20℃保存备用。1.3 PCR扩增反应
PCR反应体系见表1,反应条件为95℃ 5 min;95℃ 30 s,60℃ 30 s,72℃ 1 min,35个循环;72℃ 8 min,4℃保存。扩增产物经1.5%琼脂糖凝胶电泳,在紫外成像系统下扫描拍照。所用分子标记相关信息列于表2。Table 1
表1
表1PCR 扩增反应体系
Table 1
| 组分 Component | 用量 Dosage (μL) |
|---|---|
| 2×Taq master mix | 7.5 |
| dd H2O | 5.9 |
| 正向引物 Forward primer (10 μmol·L-1) | 0.3 |
| 反向引物 Reverse primer (10 μmol·L-1) | 0.3 |
| DNA (100 ng·μL-1) | 1.0 |
| 总计Total | 15.0 |
新窗口打开|下载CSV
Table 2
表2
表2PCR分子标记引物序列信息
Table 2
| 引物名称 Primer name | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') | 检测类型 Types detected | 参考文献 References |
|---|---|---|---|---|
| 6VS-06 | GACAGGCAGCTATGAGGC | AATCGTCGTTTGGAGTGG | 6V#4S | [20] |
| 6VS-09 | GTAAGAACAAGAGGCTAAACAG | CCAGATGACGGTTATTACATAG | 6V#4S | [20] |
| 6VS-10 | GCCATAAGTGACGCTGAT | GCATCCTGTGAAGTTGTTG | 6V#4S | [20] |
| 6VS-12 | TGTTGCCTCTCCTCATCA | ATTGCTGTCCGCTCATAC | 6V#2S 6V#4S | [20] |
| 6VS-15 | AGGACCATACATTCACAGAG | TTCCATGAGCAGATTAGCA | 6V#2S 6V#4S | [20] |
| 6VS-18 | AGCCAGTAAGATTCCGTATG | TCTAACCTTCCTCACAACAC | 6V#4S | [20] |
| MBH1 | GCCATTATAGTCAAGAGTGCACTAGCTGT | AGCTCCTCTCGTTCTCCAATGCT | 6V#2S 6V#4S 6AS;6DS | [10] |
| P259-1 | CGTGATTCAGGAAATGCGATAC | TTGCGCCGCCATGTTAG | 6V#4S | [8] |
| P461-5a | GCGTCATCCGCGCCCGTCAGGT | GAGTGCTAATGATAGATGTG | 6V#2S 6V#4S | [8] |
| N-P5 | GCGACCTGTTAGAATGCTATTACGATTAC | ATGCTACTCTACCGATGCTTTGAACC | 6AS 6DS | [12] |
| 6V-1 | CCGTGCGACAGAACAGAAGTGA | GCAATCAGCCACATACAGGTCATC | 6VL | [20] |
新窗口打开|下载CSV
1.4 原位杂交
GISH参照WEI等[21]描述的方法,以簇毛麦gDNA作为探针,高温煮沸打断的中国春gDNA作为封阻,用地高辛试剂盒标记探针(Roche,德国),荧光素异硫氰酸酯(fluorescein isothiocyanate,FITC)地高辛试剂盒(Roche,德国)检测杂交信号,在荧光显微镜40×下(BX51,OLYMPUS,日本)观察拍照。1.5 芯片分析
取4份异代换系(RW15、南87-88、19EL124、19EL134)和2份簇毛麦(CMM和No.1026)的叶片提取DNA,委托中玉金标记(北京)生物技术股份有限公司分别进行55K和660K芯片分析:设阈值DQC≥0.82,异代换系CR≥94,簇毛麦CR≥60,用Affymetrix(Thermo Fisher)AxiomAnalysisSuite软件的apt- genotype-axiom、ps-metrics和ps-classification模块进行基因分型分析,对标记类型为otv的标记进行otv- caller分析后,得到样品原始的数字型基因型;利用软件的apt-format-result模块得到原始的基因型数据。本次分析所用的参考基因组为:IWGSC_RefSeq_v1.0(2 结果
2.1 19EL124和19EL134的细胞学鉴定
以簇毛麦gDNA为探针,对2个亲本及衍生的19EL124、19EL134的根尖体细胞进行GISH鉴定(图1),发现探针的杂交信号显示清晰,亲本RW15、南87-88及19EL124、19EL134均为带有2条外源染色体,体细胞的染色体数目2n=42的正常整倍体。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1异代换系的根尖体细胞染色体的GISH图谱
红色表示用碘化丙啶(PI)复染的小麦染色体,箭头所示黄绿色表示外源染色体。a—d:RW15、南87-88、19EL124和19EL134,2n=42
Fig. 1GISH patterns of somatic chromosomes in root tips of alien substitution lines
Wheat chromosomes were stained with propidium iodide (PI) in red, and alien chromosomes in yellow-green, which were indicated by the arrows. a-d: RW15, Nan 87-88, 19EL124, 19EL134, 2n=42
2.2 19EL124和19EL134的分子标记鉴定
以2个亲本和宛7107、Pm97033、10SR3109为对照,用11个特异分子标记,包括5个外源染色体短臂6V#4S特异标记6VS-06、6VS-09、6VS-10、6VS-18和P259-1,3个6V#2S/6#4S共显性标记6VS-12、6VS-15和P461-5a,1个6V#2S/6#4S/6AS/6DS共显性标记MBH1,1个外源染色体长臂标记6V-1以及1个小麦6AS/6DS共显性标记N-P5对新筛选的材料19EL124和19EL134进行PCR检测(图2),19EL124与RW15有6V#4染色体特异带而缺失6V#2染色体特异带,19EL134和南87-88有6V#2染色体特异带而缺失6V#4染色体特异带;19EL124和南87-88有6D染色体特异带而缺失6A染色体特异带;19EL134和RW15有6A染色体特异带而缺失6D染色体特异带。综上所述,RW15为6V#4(6D)异代换系;南87-88为6V#2(6A)异代换系;19EL124为6V#4(6A)代换系;19EL134为6V#2(6D)代换系。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2异代换系的分子标记扩增图谱
M:DL2000 marker;1—7:19EL124、19EL134、RW15、南87-88、宛7107、Pm97033和10SR3109
Fig. 2Amplification patterns of alien substitution lines by molecular markers
M: DL2000 marker; 1-7: 19EL124, 19EL134, RW15, Nan87-88, Wan7107, Pm97033, and 10SR3109
2.3 6A、6D探针对双亲南87-88和RW15基因分型的差异
1 177个高质量的6A探针用于检测分析。如图3A-a所示,探针的物理位置为1.74—617.99 Mb,涵盖小麦6A染色体约616.25 Mb的物理区域,探针平均相距约523.58 kb;1 010个探针在南87-88和RW15间显示不同的分型结果,占6A总探针85.81%。其中包括碱基差异(图3A-b)及位点缺失差异(图3A-c和图3A-d),位点缺失差异是指特定探针对一方不能进行基因分型(not available,NA),而对另一方可以分型(如6V#4NA6V#2+表示探针不能对6V#4分型而可对6V#2分型)。从图3A-c和图3A-d可知,较多的6A探针可在含有6A染色体的6V#4(6D)异代换系RW15中正常分型,而相应的探针大多对不含6A染色体的6V#2(6A)异代换系南87-88不能分型。图3
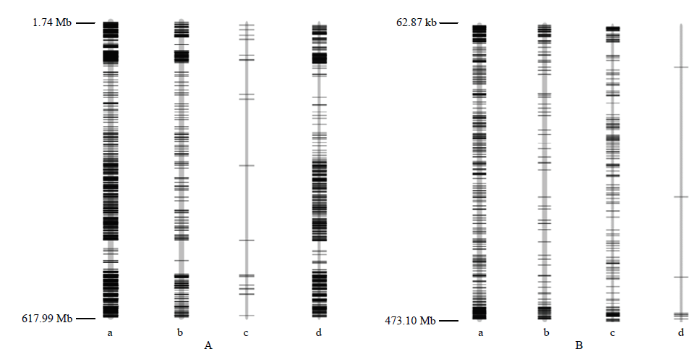 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图36A、6D探针在RW15和南87-88间检测到的SNP
A:6A探针;B:6D探针。a:总探针数及其在染色体上的分布;b:碱基替换型SNP;c:缺失型SNP(RW15-);d:缺失型SNP(南87-88-)
Fig. 3The SNPs between Nan87-88 and RW15 detected by 6A and 6D-specific probes
A: 6A probe; B: 6D probes. a: Total probes and their distribution on the chromosomes; b: Base substitution-type SNP; c: Missing-type SNP(RW15-); d: Missing-type SNP (Nan87-88-)
479个高质量的6D探针用于检测分析。如图3B-a所示,探针物理位置从62.87 kb—473.10 Mb,涵盖小麦6D染色体约473.03 Mb的物理区域,探针平均相距约每987.55 kb;其中372个探针(77.66%)在南87-88和RW15的分型不同。二者的分型差异主要分为碱基替换的差异(图3B-b)以及NA的差异(图3B-c和图3B-d),从图3B-b和图3B-c可知,在南87-88中较少出现分型失败,而6V#4(6D)异代换系RW15,缺失6D染色体,大量6D探针不能在RW15分型。
上述结果表明,NA可能是替换小麦6A或6D的外源染色体分型的特征,为了验证这一点,调查了55K芯片所有探针中NA分型的探针在基因组各染色体的占比(表3和图4)。结果表明,在异代换系南87-88和19EL124中,6A的NA分型探针的占比最高,分别为63.21%和68.90%;在异代换系RW15和19EL134中,6D的NA分型探针的占比最高,分别为49.48%和53.44%。与6A、6D探针相比较,其余染色体探针在南87-88、RW15、19EL124和19EL134检测到的NA数量较少,占比极低,平均为0.94%。这些结果与验证的南87-88和19EL124为6A代换系、RW15和19EL134为6D代换系的结果一致。值得注意的是,尽管19EL124为6A代换系,含有6D染色体,但6D探针检测到其上的NA占比达6.89%,高于其他小麦染色体的平均值。
Table 3
表3
表3探针在异代换系间检测到的NA数量
Table 3
| 探针数量 No. of probes | RW15 | 南87-88 Nan87-88 | 19EL124 | 19EL134 |
|---|---|---|---|---|
| 1A(1797) | 20 | 10 | 51 | 51 |
| 1B(1808) | 20 | 10 | 22 | 26 |
| 1D(1124) | 10 | 6 | 8 | 6 |
| 2A(1597) | 11 | 8 | 9 | 13 |
| 2B(1686) | 27 | 14 | 27 | 35 |
| 2D(1034) | 4 | 3 | 10 | 11 |
| 3A(1108) | 4 | 7 | 11 | 12 |
| 3B(1804) | 13 | 12 | 16 | 17 |
| 3D(945) | 11 | 6 | 8 | 7 |
| 4A(1368) | 8 | 11 | 8 | 5 |
| 4B(1334) | 11 | 6 | 10 | 26 |
| 4D(477) | 6 | 1 | 6 | 6 |
| 5A(1791) | 25 | 9 | 28 | 6 |
| 5B(1637) | 18 | 9 | 21 | 14 |
| 5D(870) | 8 | 6 | 19 | 9 |
| 6A(1177) | 29 | 744 | 811 | 30 |
| 6B(1628) | 22 | 26 | 21 | 32 |
| 6D(479) | 237 | 13 | 33 | 256 |
| 7A(1628) | 9 | 13 | 30 | 15 |
| 7B(1601) | 9 | 13 | 12 | 20 |
| 7D(1090) | 7 | 4 | 16 | 10 |
新窗口打开|下载CSV
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图428 020个物理定位的小麦55K探针检测到的NA在4个异代换系染色体的占比
Fig. 4The NA proportion of each chromosome of four alien substitution lines detected by 28020 probes of 55K chip with physically located on wheat chromosomes
2.4 6A、6D探针对同类型异代换系分型的比较
将6A、6D探针对19EL124和19EL134与相同代换类型的双亲分型进行比较。19EL124与母本南87-88同属于6A异代换类型,1 177个6A探针中,有265个显示2个系间分型不同,占6A总探针22.51%。同时,6A异代换系都含有6D染色体,479个6D探针中,有72个在2个系间表现分型差异,占6D总探针15.03%,表明二者间的6D染色体具有一定的差异。19EL134与父本RW15同属于6D异代换类型,479个6D探针中,共80个探针显示2个系间分型不同,占6D总探针16.70%。同时,6D异代换系都含有6A染色体,1 177个6A探针检测到二者间SNP 54个,占6A总探针的4.56%,说明二者间的6A染色体高度相似。2.5 6A和6D探针对6V#2和6V#4的SNP分析
在1 177个6A探针中,有22.51%显示6V#2(南87-88)和6V#4(19EL124)间分型的差异(图5A-a)。进一步分析发现,其中,仅有22个(8.3%)碱基替换型差异(图5A-b),其余是NA的差异,如19EL124与南87-88比较,显示6V#4NA6V#2+的分型差异有155个(图5A-c),而6V#4+6V#2NA的分型差异有88个(图5A-d)。这表明了2条外源染色体相比,6A探针可以更多地检测6V#2。在479个6D探针中,有80个(15.03%)探针显示19EL134和RW15间分型的不同(图5B-a),进一步分析发现,15个(18.75%)为碱基替换型的差异(图5B-b),其余为缺失型差异(图5B-c和图5B-d),其中6V#2NA6V#4+和6V#2+6V#4NA的多态性探针各有42和23个。与6A探针相比,6D探针可以更多地检测6V#4。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图56A、6D染色体探针检测同类代换系中2条外源染色体6V#2及6V#4间分型的差异
A:6A探针检测6V(6A)异代换系19EL124与南87-88的分型差异,a:总差异探针的分布;b:碱基替换型差异探针的分布;c:缺失型(19EL124-)差异探针的分布;d:缺失型(南87-88-)差异探针的分布。B:6D探针检测6D异代换系19EL134与RW15,a:总差异探针的分布;b:碱基替换型差异探针的分布;c:缺失型(19EL134-)差异探针的分布;d:缺失型(RW15-)差异探针的分布
Fig. 5Differential genotyping between the two alien chromosomes 6V#2 and 6V#4 in the same type of substitution lines detected by 6A and 6D-specific probes, respectively
A: Differential genotyping between two 6V(6A) substitution lines of 19EL124 and Nan 87-88 detected by 6A probes. a: Distribution of total differential probes; b: Distribution of differential probes with base substitution; c: Distribution of differential probes with missing-type (19EL124 -); d: Distribution of differential probes with missing-type (Nan 87-88 -). B: Differential genotyping between two 6V(6A) substitution lines of 19EL134 and RW15 detected by 6D probes. a: Distribution of total differential probes; b: Distribution of differential probes with base substitution; c: Distribution of differential probes with missing-type (19EL134 -); d: Distribution of differential probes with missing-type (RW15 -)
2.6 660K芯片小麦第6同源群染色体探针对2份簇毛麦的SNP分析及6V特异标记的筛选
用小麦660K芯片第6群染色体的探针分析2份簇毛麦CMM(含有6V#2染色体)和No.1026(含有6V#4染色体)间的SNP(表4),结果表明,4 796个6A探针中,1 710个显示CMM和No.1026间的分型差异,占6A总探针的35.65%,其中NA分型多态性探针在6A差异分型中占48.07%,CMM和No.1026各占24.21%和23.86%。14 352个6B探针中,5 626个显示二者间的分型差异,占6B总探针的39.20%,其中NA分型多态性探针占6B差异分型探针的46.02%,CMM和No.1026各占24.46%和21.56%。3 051个6D探针中,1 206个显示二者间的分型差异,占6D总探针的39.53%,其中NA多态性探针占6D差异分型探针的39.88%,CMM和No.1026各占20.48%和19.40%。可见,6A、6D的NA分型差异探针的比例在2份簇毛麦间是比较接近的。Table 4
表4
表4660K芯片6A、6D探针对簇毛麦CMM和No.1026的检测
Table 4
| 染色体 Chr. | 探针数量 No. of probes | 总SNP(占比) Total SNP (percentage) | 碱基替换型SNP(占比) SNP of base replacement (percentage) | CMM-NA SNP(占比) CMM-NA SNP (percentage) | No.1026-NASNP(占比) No.1026-NA SNP (percentage) |
|---|---|---|---|---|---|
| 6A | 4710 | 1710(36.31%) | 888(18.85%) | 414(8.79%) | 408(8.66%) |
| 6B | 14352 | 5626(39.20%) | 3037(21.16%) | 1376(9.59%) | 1213(8.45%) |
| 6D | 3051 | 1206(39.53%) | 725(23.76%) | 247(8.10%) | 234(7.67%) |
新窗口打开|下载CSV
将用于2份簇毛麦和4份异代换系分析的6A、6D探针进行整合,2份芯片中共享的6A探针有395个,其中65个探针在CMM和南87-88中显示一致的分型,61个在No.1026和19EL124中显示一致分型,二者共享的探针25个,占6A总共享探针的6.33%。这些探针在2条外源染色体间有3个探针显示SNP,均为NA多态性,其中CMM有1个,No.1026有2个。与2份6D异代换系的6A染色体分型相比,这25个探针有22个显示SNP,占25个总共享探针的88%。2份芯片中共享的6D探针有231个,其中CMM和19EL134分型相同的探针有40个,No.1026和RW15间相同分型的探针数41个,二者共享的探针15个,占6D 231个总共享探针的6.49%。这些探针在2条外源染色体的分型完全一致,没有多态性。但与2份6A异代换系相比,所有分型都不相同,显示出100%的差异。
2.7 6A和6D探针对双亲与衍生系中6A与6D染色体的SNP分析
RW15和19EL134都含有6A染色体,6A的探针检测到二者碱基替换型SNP有17个,显示NA多态性的探针各有18和19个,各占总探针的1.44%、1.53%和1.61%。南87-88和19EL124中都含有6D染色体,6D探针检测到二者碱基替换型SNP有38个,显示NA多态性的探针各有7和27个(图5A-d),各占总探针的7.93%、1.46%和5.64%。可见,6D染色体在子代与亲代间的差异比较明显。3 讨论
3.1 GISH及染色体特异标记鉴定的必要性
GISH是以外源物种的基因组DNA为探针的DNA分子杂交技术,是鉴定外源染色体最为直观有效的方法,可鉴定各种附加系、代换系及易位系等材料中的外源染色体状况。GISH已成功应用于鉴定小麦遗传背景中的簇毛麦、大麦、新麦草、大赖草、黑麦、拟斯卑尔山羊草等外源染色体[22,23,24,25]。在2份异代换系的杂种后代分离群体中,笔者曾筛选到缺失6VS标记、仅含6VL的材料,说明在小麦-外源异染色体系的杂交过程中,一些外源染色体可能通过着丝粒的错分裂与小麦染色体产生臂间的易位或端体。由于GISH只能检测到外源染色体,但不能确定具体的外源染色体的身份,在筛选异代换系早期,主要利用了6VS特异的分子标记跟踪6V染色体,本研究利用6V长短臂的分子标记及原位杂交相结合的方法追踪外源染色体,同时通过细胞学观察体细胞染色体数目,并利用相应的6AS、6DS特异分子标记确认缺失的小麦染色体以鉴定筛选目标材料,确保用于芯片分析的材料是准确的染色体异代换系。3.2 大量6A和6D探针对外源染色体分型的失败,反映了小麦与近缘种部分同源染色体在DNA水平上存在较大差异
本研究利用小麦55K芯片6A和6D上的探针进行分析,发现小麦与外源部分同源染色体间极高的差异。6A探针检测出双亲间的差异实际上是南87-88中6V#2染色体与RW15中的6A染色体间的多态性。6D探针检测出的差异实际上是南87-88中的6D染色体与RW15中的6V#4染色体间的多态性,而无论6A或6D探针,绝大多数的差异属于小麦染色体可明确分型而外源染色体无法分型的缺失型多态性。此外,从与660K芯片整合的数据中,筛选到40个簇毛麦与代换系中6V染色体分型一致的6A和6D探针,其中有37个(92.5%)显示6V与小麦6A或6D间的SNP,这些结果表明了小麦染色体和外源部分同源染色体间在DNA水平上存在较大差异,反映了二者具有较远的亲缘关系。NA多态性探针在整条染色体上较为均匀地分布(图5)也与整条染色体替换的事实相符合。这些结果与BAI等[26]利用660K芯片对小麦华山新麦草2Ns/2D异代换系H139检测表明其2D上染色体丢失标记率显著高于对照7182的结果极为相似。最近LAUREN等[27]基于35K SNP芯片对17个中国春-长穗偃麦草附加系进行了分析,发现1 594个标记在小麦和长穗偃麦草之间显示出多态性。通过Flapjack?分析删除了497个错误标记,并移除834个单分离或分离不一致的标记后仅筛选到263个用于构建遗传连锁图谱的SNP标记。3.3 同类异代换系间外源同源染色体的NA多态性,反映了2条不同来源的6V染色体在DNA水平上存在差异
QI等[28]和LIU等[29]曾报道,利用6V#3异附加系与小麦6D单体杂交,希望通过双单体诱导6V#3与6D染色体间发生罗伯逊易位,但在其后代中选择到的易位全部发生在6V#3与6A之间,尚未发现发生在预期的靶标染色体之间的例子,作者推测可能与所涉及易位的染色体着丝粒间的亲缘关系或与遗传背景相关。在迄今报道所获得的易位系中,也只存在6V#2S·6AL和6V#4S·6DL 2种类型,虽然最近通过2对不同易位系杂交已在其后代中获得2个外源染色体臂的重组子[30]。本研究中,2对不同的外源同源染色体分别取代了相同的小麦染色体,因此,应用小麦芯片靶标染色体上的探针尝试对不同来源的外源染色体进行多态性分析。我们注意到,缺失小麦6A或6D染色体的异代换系,不能被相应的小麦SNP芯片上的6A或6D染色体的大多数探针所分型,NA比例显著增加是4份异代换系的共同特征(表3和图3),2份6A异代换系用6A探针检测时有22.51%的探针显示分型的多态性,其中91.70%的多态性分型由NA的差异构成;2份6D异代换系用6D探针检测时有16.70%的探针显示分型的多态性,其中81.25%的多态性分型由NA的差异构成,NA的多态性表明不同外源染色体DNA水平上存在较大的差异。对于6A异代换系,与6V#4染色体相比,6A染色体探针可以更多地检测6V#2染色体;反之,对于6D异代换系,与6V#2染色体相比,6D染色体探针则可以更多地检测6V#4染色体。相比之下,660K芯片6A和6D探针对2份不同簇毛麦的基因分型结果,NA多态性的比例较低,占6A和6D染色体总多态性探针的48.07%和39.88%,说明不同遗传背景对基因分型存在影响。异源六倍体小麦基因组含有3个基因序列和排序十分近似的亚基因组,存在大量高度相似的同源基因或序列,虽然SNP芯片的探针在设计上是染色体特异的和唯一的,但染色体的缺失和异源染色体的替换可能增加了基因分型的复杂性。本研究由于2组芯片共有且在异代换系和相应簇毛麦间分型一致的6A、6D探针数只有40个,仅获得37个6V染色体特异标记,有3个标记可区分2条外源同源染色体。
3.4 19EL124/19EL134与双亲6A/6D染色体的SNP
6D异代换系19EL134与RW15中,6A染色体间SNP占总探针的4.59%,而6D探针检测2个6A异代换系19EL124和南87-88的SNP所占比例达15.03%。因为父本RW15缺失6D染色体,19EL124的6D染色体理论上来源于母本南87-88,但检测结果显示,南87-88和19EL124中的6D间存在较大的差异,以此联想到,在南87-88和RW15的杂种F1减数分裂中期I,没有观察到6A和6D 2个预期的单价体,而是观察到一个单价体和1个小麦染色体构成的三价体,鉴于其F2代6D的出现频率高于6A,因而推测6D染色体参与了三价体的形成[12]。本研究芯片分析的结果似乎进一步支持了这一推断,由于杂种中6D参与了三价体的形成,意味着6D染色体发生了交换和重组,因此,后代的6D染色体与亲本的6D染色体间存在明显的差异。4 结论
小麦芯片上的探针对近缘种簇毛麦部分同源染色体的检测效率比小麦显著降低,揭示出2个属间具有较远的亲缘关系;相同染色体探针对不同的簇毛麦同源染色体的检测效率不同,6A探针可以更好地检测6V#2,而6D探针可更好地检测6V#4;通过簇毛麦与异代换系6V染色体分型的关联分析,筛选获得6V染色体特异的SNP标记37个。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-018-3176-5URLPMID:30167758 [本文引用: 1]

KEY MESSAGE: Pm62, a novel adult-plant resistance (APR) gene against powdery mildew, was transferred from D. villosum into common wheat in the form of Robertsonian translocation T2BS.2VL#5. Powdery mildew, which is caused by the fungus Blumeria graminis f. sp. tritici, is a major disease of wheat resulting in substantial yield and quality losses in many wheat production regions of the world. Introgression of resistance from wild species into common wheat has application for controlling this disease. A Triticum durum-Dasypyrum villosum chromosome 2V#5 disomic addition line, N59B-1 (2n = 30), improved resistance to powdery mildew at the adult-plant stage, which was attributable to chromosome 2V#5. To transfer this resistance into bread wheat, a total of 298 BC1F1 plants derived from the crossing between N59B-1 and Chinese Spring were screened by combined genomic in situ hybridization and fluorescent in situ hybridization, 2V-specific marker analysis, and reaction to powdery mildew to confirm that a dominant adult-plant resistance gene, designated as Pm62, was located on chromosome 2VL#5. Subsequently, the 2VL#5 (2D) disomic substitution line (NAU1825) and the homozygous T2BS.2VL#5 Robertsonian translocation line (NAU1823), with normal plant vigor and full fertility, were identified by molecular and cytogenetic analyses of the BC1F2 generation. The effects of the T2BS.2VL#5 recombinant chromosome on agronomic traits were also evaluated in the F2 segregation population. The results suggest that the translocated chromosome may have no distinct effect on plant height, 1000-kernel weight or flowering period, but a slight effect on spike length and seeds per spike. The translocation line NAU1823 has being utilized as a novel germplasm in breeding for powdery mildew resistance, and the effects of the T2BS.2VL#5 recombinant chromosome on yield-related and flour quality characters will be further assessed.
[本文引用: 1]
[本文引用: 1]
DOI:10.1094/PDIS.2002.86.4.423URLPMID:30818719 [本文引用: 1]

Wheat curl mite (WCM), Aceria tosichella, is the vector of Wheat streak mosaic virus (WSMV), a destructive viral pathogen in wheat (Triticum aestivum). Genetic resistance to WCM colonization can reduce the incidence of wheat streak mosaic. Chromosome 6V in Hay-naldia villosa is a new source of WCM resistance. We compared variation in resistance among different sources of H. villosa chromosome 6V and 6VS lines to WCM and WSMV and their effectiveness in controlling the incidence of WSMV following exposure to viruliferous WCM. WCM resistance varied among the 6V and 6VS lines depending on the H. villosa parent. The 6V substitution lines Yi80928, GN21, and GN22 derived from an accession of H. villosa from China, and the 6VS translocation lines 92R137, 92R178, and Sub6V from an H. villosa accession collected from the United Kingdom were uniformly resistant to WCM colonization. In contrast, the 6V substitution line RW15 and a 6VS translocation line Pm33 developed from an H. villosa collection from the former Union of Soviet Socialist Republics were susceptible to WCM. All 6V and 6VS lines were susceptible to WSMV when manually inoculated. However, symptom expression was delayed in the WCM-resistant 6V and 6VS lines after exposure to viruliferous WCM. The 6V and 6VS lines differed in their ability to control WSMV infection. WCM-susceptible lines RW15 and Pm33 had no effect on controlling the infection by WSMV. Lines GN21 and GN22 were the most effective of the three H. villosa sources in limiting the spread of WSMV. Their high yield potential and protein content, in combination with resistance to stripe rust (Puccinia striiformis f. sp. tritici) and powdery mildew (Erysiphe graminis f. sp. tritici), make GN21 and GN22 promising sources of WCM resistance.
[本文引用: 3]
[本文引用: 3]
URLPMID:30987602 [本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:28550605 [本文引用: 1]
DOI:10.1111/pbi.12831URLPMID:28921769 [本文引用: 1]

Agropyron Gaertn. (P genome) is a wild relative of wheat that harbours many genetic variations that could be used to increase the genetic diversity of wheat. To agronomically transfer important genes from the P genome to a wheat chromosome by induced homoeologous pairing and recombination, it is necessary to determine the chromosomal relationships between Agropyron and wheat. Here, we report using the wheat 660K single nucleotide polymorphism (SNP) array to genotype a segregating Agropyron F1 population derived from an interspecific cross between two cross-pollinated diploid collections 'Z1842' [A. cristatum (L.) Beauv.] (male parent) and 'Z2098' [A. mongolicum Keng] (female parent) and 35 wheat-A. cristatum addition/substitution lines. Genetic linkage maps were constructed using 913 SNP markers distributed among seven linkage groups spanning 839.7 cM. The average distance between adjacent markers was 1.8 cM. The maps identified the homoeologous relationship between the P genome and wheat and revealed that the P and wheat genomes are collinear and relatively conserved. In addition, obvious rearrangements and introgression spread were observed throughout the P genome compared with the wheat genome. Combined with genotyping data, the complete set of wheat-A. cristatum addition/substitution lines was characterized according to their homoeologous relationships. In this study, the homoeologous relationship between the P genome and wheat was identified using genetic linkage maps, and the detection mean for wheat-A. cristatum introgressions might significantly accelerate the introgression of genetic variation from Agropyron into wheat for exploitation in wheat improvement programmes.
[本文引用: 1]
[本文引用: 1]
URLPMID:28653149 [本文引用: 7]
DOI:10.1034/j.1601-5223.2003.01544.xURLPMID:12830981 [本文引用: 1]

In this study, the tested four alloplasmic inbred lines, a2-4, a2-5, b1-1 and b2-1 were propagated from the same disease resistant individual in the parthenogenetic progenies of Zea mays L. cv. Lu 9 x Zea diploperennis (DP). All the lines except a2-5 were resistant to Helminthosporium turcium Pass and H. maydis Nisik. Introgressed DP segments in these lines were detected by both Southern hybridization and genomic in situ hybridization (GISH). The results of Southern hybridization showed that DP species-specific DNA sequences had been introgressed into the genomes of alloplasmic lines. The Southern hybridization band patterns in all of the tested lines were consistent with those of DP. Genomic in situ hybridization (GISH) signals were detected on 7 different chromosome pairs in lines a2-4 and a2-5, on 5 chromosome pairs in b1-1 and on 4 chromosome pairs in b2-1. The features of introgression, and disease resistant genes in the introgressed segments, as well as the gene silence or elimination in some alloplasmic lines are discussed.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
URLPMID:32210998 [本文引用: 1]
DOI:10.1007/s00122-020-03591-3URLPMID:32313991 [本文引用: 1]

KEY MESSAGE: One hundred and thirty four introgressions from Thinopyrum elongatum have been transferred into a wheat background and were characterised using 263 SNP markers. Species within the genus Thinopyrum have been shown to carry genetic variation for a very wide range of traits including biotic and abiotic stresses and quality. Research has shown that one of the species within this genus, Th. elongatum, has a close relationship with the genomes of wheat making it a highly suitable candidate to expand the gene pool of wheat. Homoeologous recombination, in the absence of the Ph1 gene, has been exploited to transfer an estimated 134 introgressions from Th. elongatum into a hexaploid wheat background. The introgressions were detected and characterised using 263 single nucleotide polymorphism markers from a 35 K Axiom((R)) Wheat-Relative Genotyping Array, spread across seven linkage groups and validated using genomic in situ hybridisation. The genetic map had a total length of 187.8 cM and the average chromosome length was 26.8 cM. Comparative analyses of the genetic map of Th. elongatum and the physical map of hexaploid wheat confirmed previous work that indicated good synteny at the macro-level, although Th. elongatum does not contain the 4A/5A/7B translocation found in wheat.
URLPMID:21437597 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
