 ,1,4, 刘蔚1, 康郁1, 华玮1, 钱论文1,2,3, 官春云1,2,3,*, 何昕1,2,3,*
,1,4, 刘蔚1, 康郁1, 华玮1, 钱论文1,2,3, 官春云1,2,3,*, 何昕1,2,3,*Identification and relative expression analysis of CBF gene family in Brassica napus L.
XIE Pan ,1,4, LIU Wei1, KANG Yu1, HUA Wei1, QIAN Lun-Wen1,2,3, GUAN Chun-Yun1,2,3,*, HE Xin1,2,3,*
,1,4, LIU Wei1, KANG Yu1, HUA Wei1, QIAN Lun-Wen1,2,3, GUAN Chun-Yun1,2,3,*, HE Xin1,2,3,*通讯作者: * 官春云, E-mail:guancy2011@aliyun.com;何昕, E-mail:hexinzhsh@126.com
收稿日期:2020-11-28接受日期:2021-04-14网络出版日期:2021-05-21
| 基金资助: |
Corresponding authors: * E-mail:guancy2011@aliyun.com;E-mail:hexinzhsh@126.com
Received:2020-11-28Accepted:2021-04-14Published online:2021-05-21
| Fund supported: |
作者简介 About authors
E-mail:xiepan@hunau.edu.cn

摘要
低温是影响植物生长发育的重要环境胁迫因子, ICE1 (inducer of CBF expression1)-CBF (C-repeat (CRT)-binding factors)-COR (cold responsive)在植物响应低温胁迫的信号途径中具有重要作用。为探究CBF (C-repeat-binding factors)基因在甘蓝型油菜(Brassica napus L.)中的进化以及在低温胁迫应答反应中的功能, 本研究以4个拟南芥CBF基因为基础序列, 鉴定出11个甘蓝型油菜、6个白菜和5个甘蓝CBF基因, 并对它们的分子特性、蛋白保守结构域和系统进化树、基因结构及基因染色体分布、甘蓝型油菜CBF基因组织表达模式以及不同逆境和激素处理下的表达模式进行系统的比较分析。结果表明, 在11个甘蓝型油菜CBF基因中, 可分为亚组I (CBF1/2/3)和亚组II (CBF4) 2个亚组。转录组测序结果表明, 所有甘蓝型油菜CBF基因受低温诱导表达, 其中亚组Ib的4个基因对冷胁迫响应迅速且持续时间长, 亚组II中2个基因对冷胁迫响应表达较弱, 但它们在根中表达量明显高于叶片, 并且参与盐胁迫和渗透胁迫响应; 甘蓝型油菜中CBF基因家族对冻害响应更强烈, 其中亚组I中的BnaA08g30930D、BnaCnng49280D、BnaAnng34260D、BnaC07g39680D和亚组II中BnaA10g07630D、BnaC09g28190D对冻害响应尤为显著。本研究将为进一步了解甘蓝型油菜CBF家族基因的生物学功能及其对低温尤其是冻害响应奠定基础, 同时为CBF家族基因在其他物种中的生物信息学研究提供参考。
关键词:
Abstract
Low temperature is an important environmental stress factor affecting plant growth and development. The ICE1-CBF-COR [inducer of CBF expression1-c-repeat (CRT)-binding factors-cold responsive] plays an important role in the signaling pathway in response to low temperature stress in plants. To explore the evolution of CBF gene in Brassica napus L. and its function in response to low temperature stress, 11, 6, and 6 CBF genes were detected from Brassica napus, Brassica rapa, and Brassica oleracea, respectively. In addition, the molecular characteristics, protein conserved domains, phylogenetic tree, gene structure, and chromosome distribution were systematically investigated. The results showed that 11 BnaCBF genes could be divided into two subgroups including Subgroup I (CBF1/2/3) and Subgroup II (CBF4). Transcriptome sequencing revealed that all the CBF genes of Brassica napus were induced by low temperature, among which four genes in subgroup Ib responded rapidly and durably to cold stress, and two genes in Subgroup II responded weakly to cold stress, however, the relative expression level of CBF genes in roots was significantly higher than that in leaves, which were involved in salt stress and osmotic stress responses. Further analysis demonstrated that the CBF gene family in Brassica napus responded more strongly to freezing stress, especially BnaA08G30930D, BnaCnng49280D, BnaAnng34260D, BnaC07g39680D in Subsection I, and BnaA10g07630D, BnaC09G28190D in Subsection II. This study lays a foundation for further understanding the biological function of the CBF family gene and its response to low temperature, especially freezing stress in Brassica napus., and provides a reference for bioinformation research of CBF family genes in other species.
Keywords:
PDF (3335KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
解盼, 刘蔚, 康郁, 华玮, 钱论文, 官春云, 何昕. 甘蓝型油菜CBF基因家族的鉴定和表达分析. 作物学报, 2021, 47(12): 2394-2406 DOI:10.3724/SP.J.1006.2021.04259
XIE Pan, LIU Wei, KANG Yu, HUA Wei, QIAN Lun-Wen, GUAN Chun-Yun, HE Xin.
两熟制、三熟制是我国南方地区推广的主要种植模式[1], 为适应南方地区“稻-稻-油”轮作生产模式的变化趋势, 需要种植早熟、特早熟油菜品种。但大量的研究和生产实践表明, 油菜早熟性与抗逆性和丰产性之间始终存在着较大的矛盾, 早熟与低产、抗性差之间存在着连锁关系[2]。早熟品种生育进程快, 现蕾、抽薹、开花都较早, 抗冻性差并且花期耐春寒能力较弱, 一旦遭遇冰冻或持续的低温阴雨天气, 产量损失严重[3]。如2008年1月, 我国遭受了严重的低温冰雪灾害, 对冬油菜生产带来了严重的影响, 受灾面积约占全国冬油菜面积77.8%, 产量损失10.88%。在受灾区随机抽样调查发现, 受冻害影响较严重的品种主要为未经当地审定的早熟品种, 而经过正规审定的油菜品种的受灾程度较轻[4]。此外, 2010年春季“倒春寒”引起的持续低温阴雨天气给我国长江中下游油菜生产带来严重影响, 平均减产10%~20%, 部分地区减产高达30% [5]。因此, 需要我们加强对耐寒、抗冻性强的早熟油菜品种选育研究。
低温作为世界最为严重的自然灾害之一, 影响植物的生长发育以及地理分布, 严重时甚至会造成植物死亡。目前, 低温信号途径研究比较清楚的是ICE1 (inducer of CBF expression 1)-CBF (C-repeat (CRT)-binding factors)-COR (cold responsive)信号途径。研究表明, ICE1通过特异性结合CBF3启动子上的MYC结合位点(CANNTG), 正调控CBF3基因的表达[6]。CBF转录因子中的AP2 domain可直接与冷调节COR基因启动子中的CRT/DRE顺式元件结合, 进而诱导COR基因的表达。在拟南芥中, CBF1/2/3串联排列在第4条染色体上, 受冷诱导表达[7,8,9,10]。而CBF4不受冷诱导, 但在拟南芥中过表达CBF4可增强植物的抗冻性和耐旱性[11]。在拟南芥植株上过表达CBF1/2/3均明显增强其抗冻性[8-9,12]。与野生型相比, 敲除CBF1和CBF3基因拟南芥植株抗冻性降低约60% [13]。CBF1/2/3三突变体在非冷驯化时没有或具有轻微冻敏感表型, 而在冷驯化后表现出强烈的冻敏感表型[14,15,16]。通过对CBF1/2/3三突变体进行RNA-seq分析发现, CBFs突变影响了全转录组10%~20%的COR基因表达[15]。大量研究证明, CBF基因在植物抗寒方面是一个保守的信号路径。抗冷相关基因(CBF4、COR6、COR15和COR25)在抗冻性强的甘蓝型油菜1801中的表达量显著高于在抗冻性弱的C20中的[17]。Maryam等[18]在葡萄中发现4个受冷胁迫诱导表达的CBF基因, 这些CBF在冷处理后几分钟开始表达, 其中CBF2持续表达几个小时, CBF4持续表达到第10天。
研究人员开始利用CBF基因改良作物的抗寒、抗冻性。在冷胁迫下, 超表达拟南芥CBF3基因可以增加木薯、烟草、水稻、油菜和地被石竹中脯氨酸的积累, 减少丙二醛的产生以及电解质外渗, 从而增强它们的抗寒性[19,20,21,22]。此外, OsCBF基因转入水稻后, 脯氨酸及可溶性糖的含量明显提高, 使得水稻的抗寒性、抗旱性和抗盐性都有所提高[23]。但是有研究发现, 超表达甘蓝型油菜BnCBF5和BnCBF17可以提高植株抗冻性, 但会严重抑制植株的生长发育, 并推迟开花时间[24]。
为充分挖掘CBF基因, 利用CBF基因解决甘蓝型油菜早熟品种与抗冻性之间的矛盾, 本研究对拟南芥、甘蓝、白菜以及甘蓝型油菜的CBF家族进行系统地比较分析, 鉴定甘蓝型油菜、白菜和甘蓝CBF基因, 并对这些CBF成员的基因结构、分子特性、基因染色体分布、系统进化树、组织表达、不同逆境、不同激素以及不同低温胁迫处理下的表达模式进行比较分析, 筛选候选抗冻基因, 以期为进一步了解甘蓝型油菜CBF家族基因的生物学功能及其对低温尤其是冻害响应奠定基础, 同时为CBF家族基因在其他物种中的生物信息学研究提供参考。
1 材料与方法
1.1 试验材料
将甘蓝型油菜半冬性品种中双11号(Zhongshuang 11, ZS11)的种子在滤纸上发芽, 移栽到装有土壤或蛭石的花盆中, 在生长室内生长6周。生长室中条件设置如下: 22℃ 14 h光照/20℃10 h黑暗, 相对湿度60%。在8:00使用100 µmol L-1脱落酸(abscisic acid, ABA)、100 μmol L-1茉莉酸甲酯(methyl jasmonate, MeJA)、1 mmol L-1水杨酸(salicylic acid, SA)和10 μg mL-1乙烯利(ethephon, ETH)溶液喷洒叶片进行激素处理, 并在处理后的3、6和12 h时间点采集叶片样品。高温胁迫和低温胁迫, 在8:00—20:00期间, 生长室分别设置为40℃和4℃, 相对湿度60%。在胁迫处理的3、h和12 h时间点采集叶片样品。盐胁迫和PEG胁迫处理, 分别在8:00—20:00期间用200 mmol L-1 NaCl或20%聚乙二醇6000 (PEG-6000)溶液灌溉幼苗, 并在处理期间的3、6和12 h时间点采集根系样本。对照幼苗用去离子水处理, 每个处理重复3次, 每个处理用3~4株幼苗。采集的叶和根样本立即在液氮中冷冻, 然后在-80℃下保存以提取RNA[24]。在光照培养箱中培育2个早熟的半冬性油菜品种(HX17和HX58)的幼苗, 生长室条件设置如下: 20℃14 h光照(6:00—20:00)/16℃10 h黑暗(20:00—6:00), 相对湿度60%, 培养4周; 冷驯化冷害与冻害处理: 在4℃条件下处理14 d后, 分别放入4℃和-4℃处理12 h; 非冷驯化冷害与冻害处理: 20℃ 14 h光照(6:00—20:00)/16℃10 h黑暗(20:00—6:00)处理6周后, 分别放入4℃和-4℃处理12 h; 其中, 4℃冷害/ -4℃冻害处理12 h的时段为20:00—8:00 (黑暗10 h/光照2 h), 在处理后的上午8:00收集从顶部往下数的第3片叶子并立即保存在-80℃, 直到提取RNA[25]。
1.2 白菜、甘蓝及甘蓝型油菜CBF基因的筛选鉴定
从Ensembl Plants (1.3 生物信息学分析
利用DNAMan6.0软件对拟南芥、白菜、甘蓝和甘蓝型油菜CBF蛋白序列比对分析, 并使用MEGA7中的Neighbour-Joining (NJ)系统发育方法构建系统发育树[26]。利用MEME (1.4 表达热图分析
参考He等[24,25]以及本课题组尚未公开的数据进行热图分析, 采用TBtools软件(TBtools.v.6695)作图。1.5 BnaCBF基因表达情况检测
参照植物总RNA提取试剂盒说明书提取植物总RNA, 用反转录试剂盒合成cDNA。根据甘蓝型油菜BnaCBF参考基因序列, 通过Primer premier 6.0软件设计qRT-PCR特异性引物, 引物序列见表1。采用Bio-Rad CFX96-M实时荧光定量PCR仪(Bio-Rad, 美国)检测目标基因的表达。参照荧光定量试剂盒说明书配置20 μL的反应体系, 含2×Trans start, Tip Green qPCR Super Mix 10 μL、cDNA模板2 μL、正反向引物(0.2 μmol L-1)各0.5 μL、去离子水7 μL。qRT-PCR反应程序为95℃ 5 min; 95℃ 10 s, 60℃ 45 s, 40个循环。选用BnaActin作为内参基因, 采用2-ΔΔCt方法分析基因表达变化情况。Table 1
表1
表1本研究所用qRT-PCR引物
Table 1
| 基因名称 Gene name | 正向引物序列 Forward sequence (5°-3°) | 负向引物序列 Reverse sequence (5°-3°) |
|---|---|---|
| BnaCBF4-RT | GAGACCACGGAAGAGAACGG | AGTCGTTATGATTCCATCCAAGTT |
| BnaCBF1-C09-RT | CGACTATGGTTGAGGCAGTGAAA | AAGTCGTTATGATTCCATCCAAGTT |
| BnaCBF3-RT | CCAAGCCGAGTCAGCAAAAT | GAGAAACTCAGGTAAATGGGTGTG |
| BnaCBF1-Cnn-RT | GAGACGACGGTGGAGGGTG | AAGTCATCATTATGTCCCCATTGTA |
| BnaC05g34300D/BnaACTIN | GGTTGGGATGGACCAGAAGG | TCAGGAGCAATACGGAGC |
新窗口打开|下载CSV
2 结果与分析
2.1 甘蓝型油菜、白菜和甘蓝CBF基因鉴定
利用4个AtCBF (At4g25490、At4g25470、At4g25480和AT5G51990)核苷酸和氨基酸序列分别在甘蓝型油菜、白菜和甘蓝的基因组数据库中进行同源序列搜索, 并结合SMART数据库、InterPro和NCBI数据库中的保守域(conservative domain, CD)搜索分析, 最终获得11个甘蓝型油菜CBF基因(BnaCBF)、6个白菜CBF基因(BraCBF)、5个甘蓝CBF基因(BoCBF)。其中11个甘蓝型油菜CBF基因编码区长度在576~663之间, 编码189~220个氨基酸, 预测等电点(Isoelectric point prediction, pI)在4.58~6.14之间, 为酸性蛋白, 分子量在20.57~24.56 kD之间。亚细胞定位预测表明, 这11个甘蓝型油菜CBF基因在细胞核中, 具体信息详见表2。Table 2
表2
表2BnaCBF基因信息汇总
Table 2
| 基因名称 Gene ID | 核苷酸长度Nucleotide length (bp) | 氨基酸长度 Amino Acid length | 等电点预测 Isoelectric point prediction | 分子量Molecular weight (kD) | 内含子数目 Number of introns | 外显子数目Number of exons | 亚细胞定位 Subcellular localization | 染色体定位 Chromosome location | 白菜同源基因Homologous gene in Brassica rape | 甘蓝同源基因Homologous gene in Brassica oleracea |
|---|---|---|---|---|---|---|---|---|---|---|
| BnaA08g30950D | 666 | 221 | 4.97 | 24.56 | 2 | 3 | 核酸Nuclear | chrA08_random:1781546..1782397 | Bra010460 | Bo8g050800 |
| BnaUnng01150D | 651 | 216 | 5.34 | 23.81 | 1 | 3 | 核酸Nuclear | chrUnn_random:1448150..1448983 | ||
| BnaA08g30910D | 654 | 217 | 5.13 | 24.02 | 2 | 3 | 核酸Nuclear | chrA08_random:1768075..1768896 | Bra010461 | |
| BnaA08g30930D | 570 | 189 | 4.73 | 20.57 | 2 | 3 | 核酸Nuclear | chrA08_random:1771443..1772198 | Bra010463 | Bo8g050740 |
| BnaCnng49280D | 576 | 191 | 4.58 | 20.83 | 2 | 3 | 核酸Nuclear | chrCnn_random:48753744..48754496 | ||
| BnaA03g13620D | 645 | 214 | 4.58 | 20.82 | 0 | 1 | 核酸Nuclear | chrA03:6228484..6229128 | Bra022770 | Bo3g024240 |
| BnaC03g71900D | 645 | 214 | 5.03 | 23.79 | 0 | 1 | 核酸Nuclear | chrC03_random:417520..418164 | ||
| BnaAnng34260D | 645 | 214 | 5.21 | 23.83 | 0 | 1 | 核酸Nuclear | chrAnn_random:39020794..39021438 | Bra019162 | Bo7g110320 |
| BnaC07g39680D | 648 | 215 | 5.23 | 23.81 | 0 | 1 | 核酸Nuclear | chrC07:40421468..40422115 | ||
| BnaA10g07630D | 663 | 220 | 5.52 | 24.55 | 0 | 1 | 核酸Nuclear | chrA10:6107845..6108507 | Bra028290 | Bo9g104770 |
| BnaC09g28190D | 663 | 220 | 6.14 | 24.5 | 0 | 1 | 核酸Nuclear | chrC09:30491719..30492381 |
新窗口打开|下载CSV
2.2 拟南芥、白菜、甘蓝及甘蓝型油菜CBF基因的序列比对分析
将鉴定得到的CBF家族蛋白的保守结构域通过DNAMAN软件比对(图1)发现, 蛋白序列保守性较高, 甘蓝型油菜、白菜、甘蓝及拟南芥的26个CBF序列比对一致性为61.08%, 这些基因都具有相同的保守序列AP2 domain (图1红色框出部分)。与其他成员相比, BnaCnng49280D、Bra010463和BnaA08g 30930D 3个基因在AP2 domain的前半部分有27个氨基酸序列缺失。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1甘蓝型油菜、白菜、甘蓝和拟南芥CBF序列比对分析
黑色: 26条序列的共有碱基; 红色: 18~25条序列的共有碱基; 蓝色: 9~17条序列的共有碱基; 白色: 1~8条序列的共有碱基。
Fig. 1Multiple alignment of CBF proteins of Brassica napus, Brassica rapa, Brassica oleracea, and Arabidopsis
Black: the same bases among 26 sequences; Red: the same bases among 18-25 sequences; Blue: the same bases among 9-17 sequences; White: the same bases among 1-8 sequences.
2.3 拟南芥、白菜、甘蓝及甘蓝型油菜的CBF基因进化树分析
为揭示甘蓝型油菜及其二倍体祖先种之间CBF家族基因的进化关系, 本研究利用MEGA7.0软件对拟南芥、白菜、甘蓝和甘蓝型油菜的CBF蛋白序列进行系统发育分析, 构建系统进化树(图2-A)。26个CBF蛋白被分为2个亚组, 与前人研究结果一致。亚组I中含有包括拟南芥AtCBF1/2/3在内的21个基因, 其中, 5个白菜CBF基因、4个甘蓝CBF基因以及9个甘蓝型油菜CBF基因, 它们与拟南芥AtCBF1/2/3基因比对一致性分别为79.77%、68.38%和72.98%; 亚组II中含有包括拟南芥AtCBF4在内的5个基因, 其中2个来自甘蓝型油菜, 白菜和甘蓝各1个, 它们与拟南芥CBF4比对一致性分别为91.07%、80.36%和79.91%。进一步分析发现, 除白菜基因Bra010461外, 其他白菜和甘蓝基因在甘蓝型油菜中均存在直系同源基因; 11个甘蓝型油菜CBF基因中, 除BnaA08g 30910D外, 其他10个基因都能在白菜和甘蓝中找到同源基因。图2
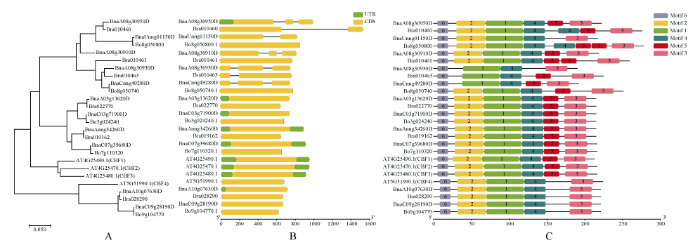 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2拟南芥、白菜、甘蓝、甘蓝型油菜CBF的系统比较分析
A: 系统发育分析; B: 基因结构分析; C: 保守结构MEME分析。
Fig. 2Systematic comparative analysis of CBF of Brassica napus, Brassica rapa, Brassica oleracea, and Arabidopsis chromosomes
A: Neighbor-Joining phylogenetic tree; B: gene structures; C: MEME motifs analysis.
2.4 基因结构分析和MEME分析
为进一步揭示甘蓝型油菜、白菜和甘蓝及拟南芥CBF基因家族的进化关系, 本研究对其基因结构(图2-B)和motif进行了预测分析(图2-C)。CBF基因具有相对保守的基因结构和蛋白功能结构域, CBFs内含子的数目在0~2之间, 亚组I中, 有7个CBF基因有内含子, 其中白菜中2个CBF基因、甘蓝型油菜1个CBF基因有1个内含子和2个外显子, 甘蓝型油菜中4个CBF基因有2个内含子和3个外显子, 其他14个CBF基因只有1个外显子没有内含子, 与拟南芥AtCBF1/2/3基因结构一致, 进化树关系也更近; 亚组II中5个CBF基因没有内含子, 基因结构一致, 具有保守性。进一步分析发现, 亚组I中白菜中Bra010460、Bra010463含有1个内含子, 甘蓝型油菜中与这2个基因进化同源关系BnaA08g30950D和BnaA08g30930D含有2个内含子, 因此推断这4个基因在进化过程中可能发生了变异; 另一方面, 甘蓝型油菜中BnaUnng01150D有1个内含子, BnaCnng49280D有2个内含子, 而它们对应的同源基因Bo8g050800和Bo8g050740没有内含子, 推测这2个甘蓝型油菜CBF基因进化过程可能发生了变异; 同时我们在进化树分析中推断BnaA08g30910D可能是BnaA08g30950D的拷贝基因, 两者基因结构一致。Motif预测分析结果表明, Motif预测分析与进化树比对分析结果吻合, 可明显分为2个亚组, 2组保守基序组成高度相似。图2-C展示了保守性较高的6条基序在26条CBF蛋白序列中的分布情况, 可见各蛋白高度保守, 但不同亚组之间蛋白序列包含的保守基序的种类和数目存在一定的差异, 2个亚组内蛋白保守基序的组成模式更为相似。26条CBF蛋白序列都含有Motif 6、Motif 1、Motif 4和Motif 3, 由此推断这些区域可能是CBF家族蛋白共性的、保守的功能基序。进一步分析发现, 亚组I中的22个成员保守基序组成模式较为一致, 除了亚组I中BnaUnng01150D、BnaA08g30930D不含有Motif5, 其余成员保守基序组成模式与拟南芥CBF1/2/3蛋白保守基序完全一致, 均为Motif6-Motif2-Motif1-Motif4-Motif5- Motif3。亚组II中, 所有成员保守基序组成模式完全一致, 均为Motif6-Motif2-Motif1-Motif4-Motif3; 植物中的CBF家族蛋白含有1个高度保守的AP2 domain, AP2 domain包括Motif2-Motif1-Motif4保守基序。与亚组I相比, 亚组II少了1个保守基序Motif5, 由此推测Motif5为亚组I的特征Motif。
2.5 染色体定位分析
ATCBF1/2/3基因在Chr4染色体上串联, AtCBF4在Chr5染色体上(图3); 6个BrCBF基因分别在A03、A08和A10染色体上分布, 其中Bra010460、Bra010461和Bra010463在A08号染色体上串联(图3); 6个BoCBF基因被定位在C03、C07、C08、C09染色体上, 其中Bo8g050800和Bo8g050740在C08染色体上串联(图3); 根据Region Comparison (http://plants.ensembl.org/)分析, 我们对每个CBF成员之间的共线性进行比较分析发现, BnaA08g 30950D和BnaA03g13620D与AtCBF1同源, BnaUnng01150D、BnaA08g30910D、BnaCnng492 80D、BnaC03g71900D、BnaAnng-34260D和BnaC07g 39680D与AtCBF2同源, BnaA08g30930D与AtCBF3同源, BnaA10g07630D和BnaC09g28190D与AtCBF4同源,这些BnaCBFs基因被定位到19条染色体中的9条上(图3), 其中BnaA08g30910D、BnaA08g30930D和BnaA08g30950D在染色体A08 random上串联重复。图3
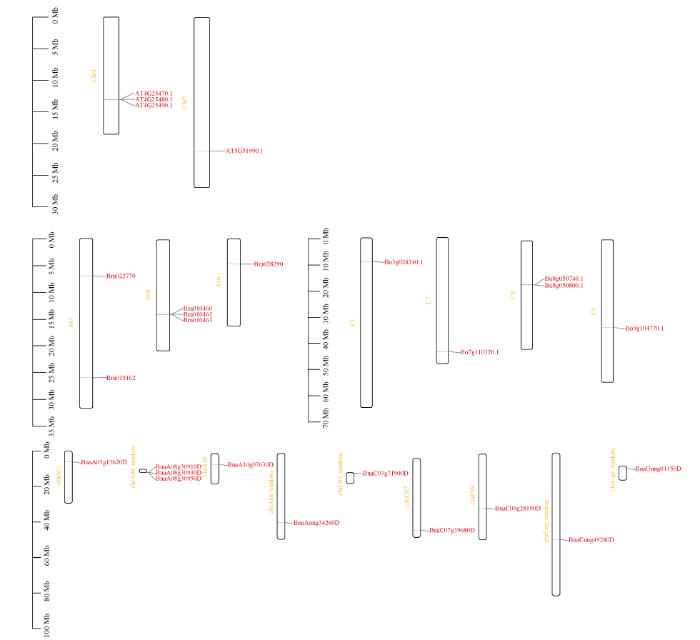 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3拟南芥、白菜、甘蓝、甘蓝型油菜染色体上CBF基因的分布
染色体编号标记在每个染色体的左侧。Chr是拟南芥染色体; A和C分别是白菜和甘蓝的染色体; chrA和chrC分别是甘蓝型油菜的A亚组和C亚组染色体; Random表示该基因不确定在此条染色体的具体位置, chrAnn和chrCnn分别表示属于A亚基因组和C亚基因组但不确定染色体信息, chrUnn表示不确定亚基因组及染色体信息。
Fig. 3Distribution of the CBF genes of Brassica napus, Brassica rapa, Brassica oleracea, and Arabidopsis chromosomes
The numbers of chromosomes are labeled on the left side of each chromosome. Chr is the chromosome of Arabidopsis; A and C are the chromosomes from Brassica rapa, Brassica oleracea; chrA and chrC are the chromosomes of the A-subgenomes and C-subgenomes from Brassica napus, respectively; Random means genes are randomly distributed to the specific chromosome, and chrAnn and chrCnn are unanchored scaffolds that cannot be mapped to a specific chromosome from the A-subgenomes and C-subgenomes, respectively, chrUnn is also an unanchored scaffold.
2.6 甘蓝型油菜BnaCBF基因在不同组织中的表达模式分析
利用本课题组前期完成的中双11材料的转录组数据, 分析了甘蓝型油菜CBF基因家族在各组织器官中表达情况, 以及在不同非生物胁迫和激素处理下的表达情况。在正常情况下, 亚组I中9个BnaCBF成员可分为Ia类和Ib类, 其中Ia类5个CBF成员(BnaA08g30950D、BnaUnng01150D、BnaA08g309 10D、BnaA08g30930D和BnaCnng49280D)在各个组织器官中表达量均较低或者不表达; Ib类4个成员(BnaA03g13620D、BnaC03g71900D、BnaAnng34260D和BnaC07g39680D)分别在茎、叶片和雌蕊中表达, 其中BnaAnng34260D在雌蕊中表达量较高; 亚组II中的2个CBF4成员(BnaA10g07630D和BnaC 09g28190D)在根、叶片、花萼和雌蕊中均有表达, 并且在根部表达量明显高于其他组织器官(图4)。图4
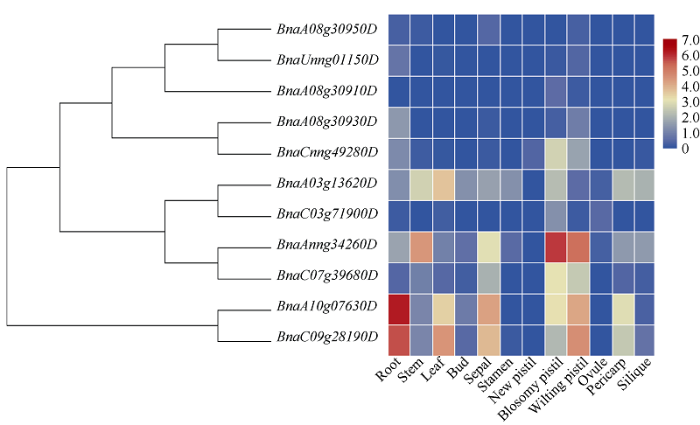 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4BnaCBF基因在不同组织器官中的表达热图
每个样本中每个基因的表达水平(FPKM值)由不同颜色的矩形表示。红色表示高表达; 蓝色表示低表达。
Fig. 4Heat map of relative expression level of BnaCBF genes in the different tissues and organs
The expression levels of each gene (FPKM values) in each sample are indicated by different colored rectangles. Red block indicates relative higher expression level. Blue block indicates relative lower expression level.
2.7 甘蓝型油菜BnaCBF基因在不同非生物逆境和不同激素处理表达模式分析
所有BnaCBF在逆境处理下均存在不同程度的表达。在低温下, Ia类5个基因(BnaA08g30950D、BnaUnng01150D、BnaA08g30910D、BnaA08g30930D和BnaCnng49280D)对冷胁迫迅速响应表达, 并在3 h后表达量显著下降, 意味着这5个基因是早期冷诱导表达基因。Ib类4个基因(BnaA03g13620D、BnaC03g71900D、BnaAnng34260D和BnaC07g39680D)对冷胁迫响应迅速且持续时间长, 冷处理12 h仍有较高表达量; 而亚组II两个CBF4基因(BnaA 10g07630D和BnaC09g28190D)在冷胁迫条件下表现为早期高表达, 而后降低, 6 h后恢复正常。BnaA10g07630D和BnaC09g28190D两个基因在根中表达量明显高于叶片, 并且参与盐胁迫和渗透胁迫响应, 而其他9个基因在根中表达变化不大(图5)。值得注意的是, Ib类4个CBF基因的表达表现出较明显的生物钟节律变化, 它们在11:00 (Leaf_3h)表达量较低, 14:00表达量较之前升高, 20:00表达量几乎为零, 表现出明显的昼夜节律; 亚组II中的2个CBF4基因(BnaA10g07630D和BnaC09g28190D)也呈现出明显的昼夜节律, 不同于Ib类CBF基因, 其在11:00和14:00几乎不表达, 但在20:00有较高的表达量。其他ABA、MeJA和ET等激素处理后表达量与对照无明显变化。图5
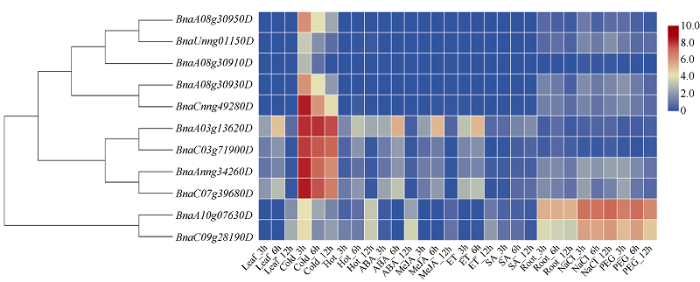 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5不同非生物胁迫和植物激素处理下BnaCBF基因的表达热图
每个样本中每个基因的表达水平(FPKM值)由不同颜色的矩形表示。红色表示高表达; 蓝色表示低表达。Leaf_3h、Leaf_6h、Leaf_12h: 正常生长3 h、6 h、12 h的叶片; Cold_3h、Cold_6h、Cold_12h: 4℃处理3 h、6 h、12 h的叶片; Hot_3h、Hot_6h、Hot_12h: 40℃处理3 h、6 h、12 h的叶片; ABA_3h、ABA_6h、ABA_12h: 脱落酸处理3 h、6 h、12 h的叶片; MeJA_3h、MeJA_6h、MeJA_12h: 茉莉酸甲酯处理3 h、6 h、12 h的叶片; ET_3h、ET_6h、ET_12h: 乙烯处理3 h、6 h、12 h的叶片;SA_3h、SA_6h、SA_12h: 盐胁迫处理3 h、6 h、12 h的叶片; Root_3h、Root_6h、Root_12h: 正常生长3 h、6 h、12 h的根;NaCl_3h、NaCl_6h、NaCl_12h: 氯化钠处理3 h、6 h、12 h的根; PEG_3h、PEG_6h、PEG_12h: 聚乙二醇处理3 h、6 h、12 h的根。
Fig. 5Heat map of the BnaCBF genes under different abiotic stresses and plant hormone treatments
The relative expression levels of each gene (FPKM values) in each sample are indicated by different colored rectangles. Red block indicates relative higher expression level, blue block indicates relative lower expression level. Leaf_3h, Leaf_6h, and Leaf_12h indicates normal growth leaf of 3, 6, and 12 hours, respectively; Hot_3h, Hot_6h, and Hot_12h indicates 40℃ treatment growth leaf of 3, 6, and 12 hours, respectively; ABA_3h, ABA_6h, and ABA_12h indicates abscisic acid treatment growth leaf of 3, 6, and 12 hours, respectively; MeJA_3h, MeJA_6h, and MeJA_12h indicates methy jasmonate treatment growth leaf of 3, 6, and 12 hours, respectively; ET_3h, ET_6h, and ET_12h indicates Ethylene treatment growth leaf of 3, 6, and 12 hours, respectively; SA_3h, SA_6h, and SA_12h indicates salt stress treatment growth leaf of 3, 6, and 12 hours, respectively; Root_3h, Root_6h, and Root_12h indicates normal growth root of 3, 6, and 12 hours, respectively; NaCl_3h, NaCl_6h, and NaCl_12h indicates sodium chloride treatment growth root of 3, 6, and 12 hours, respectively; PEG_3h, PEG_6h, and PEG_12h indicates drought stress treatment growth root of 3, 6, and 12 hours, respectively.
2.8 不同低温处理下甘蓝型油菜BnaCBF基因表达模式分析
为进一步分析这些CBF成员对不同低温的响应, 本研究利用本课题组前期完成的冷驯化和非冷驯化材料中不同低温处理的HX-17和HX-58材料的转录组数据, 对甘蓝型油菜BnaCBF基因家族的表达情况进一步分析(图6)。亚组I中有3个CBF基因在冷胁迫下表达较弱甚至不表达, 冻害胁迫下表达量升高; 其他6个成员在4℃冷害处理和-4℃冻害胁迫下均有不同程度响应, 且-4℃冻害胁迫下表达量更高。亚组II 2个CBF4成员仅在冻害胁迫条件响应表达, 意味着甘蓝型油菜中CBF基因家族对冻害响应更强烈。分析发现, 冷驯化和非冷驯化的材料对低温胁迫和冻害胁迫的响应表达的影响无明显差异。图6
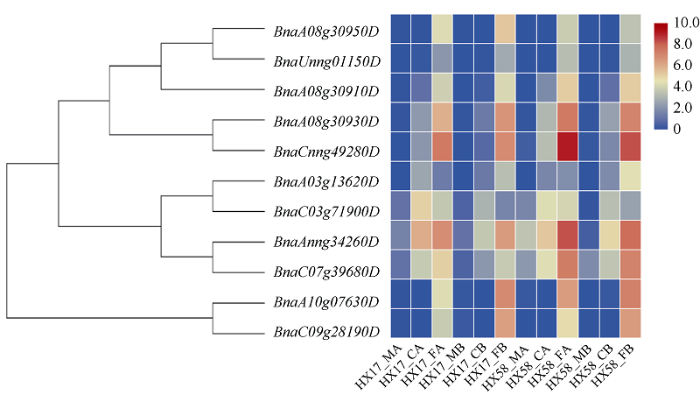 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不同低温处理下BnaCBF基因表达热图的研究
HX17_MA: 正常生长条件下的HX17材料; HX17_CA: 驯化14 d后4℃处理的HX17材料; HX17_FA: 驯化14 d后-4℃处理HX17材料。HX17_MB: 正常生长条件下HX17材料; HX17_CB: 4℃处理HX17材料; HX17_FB: -4℃处理HX17材料。HX58_MA: 正常生长条件下的HX58材料; HX58_CA: 驯化14天后4℃处理的HX58材料; HX58_FA: 驯化14 d后-4℃处理HX58材料。HX58_MB: 正常生长条件下HX58材料; HX58_CB: 4℃处理HX58材料; HX58_FB: -4℃处理HX58材料。
Fig. 6Heat map of the BnaCBF genes under different low-temperature treatments
HX17_MA: mock A, HX17 material under normal growth conditions; HX17_CA: cold A, HX17 material treated at 4℃ after 14 days of domestication; HX17_FA: freezing A, HX17 material treated at -4℃ after 14 days of domestication. HX17_MB: mock B, HX17 material under normal growth condition; HX17_CB: cold B, HX17 material treated at 4℃; HX17_FB: freezing B, HX17 material treated at -4℃. HX58_MA: Mock A, HX58 material under normal growth conditions; HX58_CA: cold A, HX58material treated at 4℃ after 14 days of domestication; HX58_FA: freezing A, HX58 material treated at -4℃ after 14 days of domestication. HX58_MB: mock B, HX58 material under normal growth condition; HX58_CB: cold B, HX58 material treated at 4℃; HX58_FB: freezing B, HX58 material treated at -4℃.
2.9 BnaCBF基因相对表达量结果分析
为验证上图中的RNA-seq数据, 本研究从油菜CBF基因家族亚组Ia、Ib和II中挑选4个具有代表性的基因进行qRT-PCR验证, 分别是亚组Ia类基因BnaCnng49280D, Ib类基因BnaAnng34260D以及亚组II的BnaA10g07630D和BnaC09g28190D。结果显示, 表达情况与图5和图6中完全吻合, 说明表达谱数据可靠(图7和图8)。图7
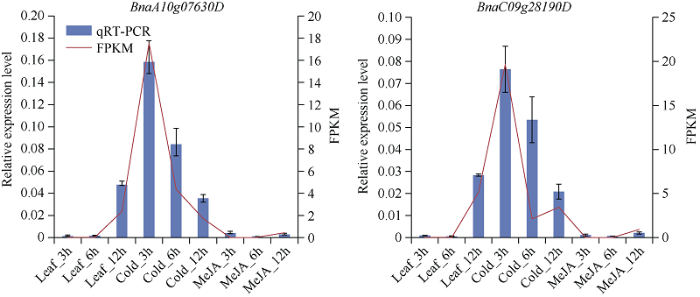 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7低温胁迫与茉莉酸甲酯处理下BnaA10g07630D和BnaC09g28190D基因表达模式的qRT-PCR分析
处理同
Fig. 7Relative expression patterns of BnaCBF genes under cold and MeJA treatments
Treatments are the same as those given in
图8
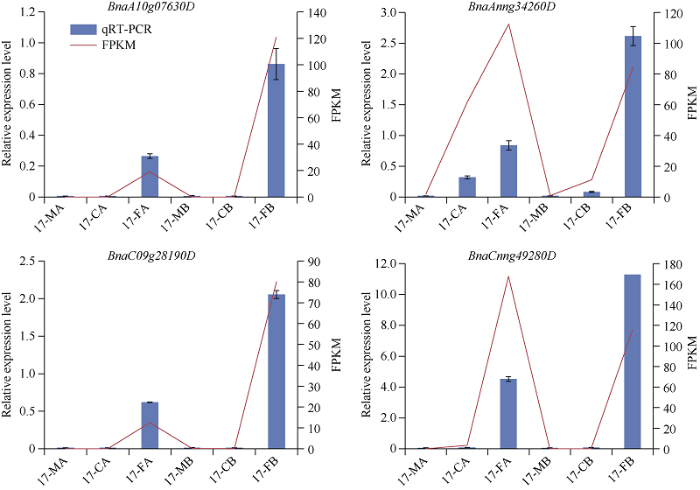 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8低温胁迫处理下BnaCBF基因表达模式的qRT-PCR分析
17_MA: 正常生长条件下的HX17材料; 17_CA: 驯化14 d后4℃处理的HX17材料; 17_FA: 驯化14 d后-4℃处理HX17材料。17_MB: 正常生长条件下HX17材料; 17_CB: 4℃处理HX17材料; 17_FB: -4℃处理HX17材料。
Fig. 8Relative expression patterns of BnaCBF genes under cold stresses treatments
17_MA: mock A, HX17 material under normal growth conditions; 17_CA: cold A, HX17 material treated at 4℃ after 14 days of domestication; 17_FA: freezing A, HX17 material treated at -4℃ after 14 days of domestication. 17_MB: mock B, HX17 material under normal growth condition; 17_CB: cold B, HX17 material treated at 4℃; 17_FB: freezing B, HX17 material treated at -4℃.
3 讨论
甘蓝型油菜是我国最主要的栽培品种, 在湖南、湖北、江西、浙江、云南等多地大面积种植[4]。南方地区推广的稻-稻-油轮作生产模式需要种植早熟、特早熟油菜品种, 但早熟型油菜推广面临抗冻性差、产量受影响等问题[2]。CBF基因家族在植物抗冻性研究中具有重要作用[28], 因此, 系统的比较分析甘蓝型油菜CBF基因以及在油菜二倍体祖先中的进化关系, 对解析CBF基因调控油菜低温及冻害胁迫反应的机制具有重要的意义。CBF/DREB1依赖的冷信号通路在过去的20年中取得了长足的进展。从最早发现的一种对干旱、寒冷和高盐胁迫有响应的顺式作用元件C-反应/脱水响应元件(CRT/DRE)开始[29], 研究者陆续从拟南芥、油菜、小麦(Triticum aestivum)、黑麦(Secale cereale)、番茄(Lycopersicon esculentum)、水稻(Oryza sativa)及玉米(Zea mays)等植物中分离克隆出许多CBF的ESTs序列, 发现其都具有冷诱导特性[30,31,32], 过表达拟南芥及其他CBF基因均可以增强植物的抗冻性[12,19]。大量研究证实, 在植物中这些CBF是参与冷胁迫信号传导, 并在不同物种间具有保守性。
本研究发现, AtCBF在甘蓝型油菜、白菜和甘蓝基因组都存在直系同源基因, 且这些CBF成员序列高度保守, 根据基因结构和Motif等结果分析, 推测BnaA08g30910D可能是BnaA08g30950D的拷贝复制。甘蓝型油菜CBF家族蛋白的结构域保守性较高, 所有成员都有且只有1个AP2 domain, 在此结构域的上下游, 各有一段保守的氨基酸序列PKKP/PKK PAGR (RAGRxxKFx ETRHP)和DSAWR[33], 该保守序列在CBF发挥其转录因子功能中扮演着非常重要的角色。
基因中内含子和外显子的组成对于其进化分析研究具有非常重要的作用[34], 本研究对CBF基因家族各成员的基因结构分析推断含有内含子的CBF基因在进化过程中可能发生了变异。Motif预测分析发现, 所有的CBF基因中Motif1、Motif3、Motif4和Motif6这4个Motif是CBF基因家族的特征性Motif。保守基序分析发现, BnaCnng49280D、BnaA08g 30930D和Bra010463三个基因缺失的27个氨基酸序列位于AP2 domain上游, 结构域分析发现该缺失部分在Motif2区段, 由此推测可能会影响它们结合靶基因启动子中顺式元件CRT/DRE的能力, 从而影响它们激活下游靶基因(如COR)的表达。染色体定位分析表明, 11个甘蓝型油菜CBF基因在染色体上的分布是不均匀的, 这可能是由于在全基因组加倍过程中, 一些同源基因可能发生丢失或扩张。
CBF参与多种逆境响应和植株生长发育等过程[35,36]。本研究发现, 甘蓝型油菜CBF基因的表达强烈受低温诱导, 且具有明显的组织特异性, 由此推断甘蓝型油菜CBF基因可能在功能上产生细化。A亚基因组BnaA03g13620D、BnaAnng34260DCBF的表达量明显高于对应的C亚基因组BnaC03g 71900D、BnaC07g39680D的表达量, 说明A、C亚基因组的基因对表达量存在明显偏移不对称。
Haake等[11]研究表明, CBF1/2/3 (CBF1/ DREB1b、CBF2/DREB1c、CBF3/DREB1a)属于冷诱导CBF基因, 还有一个CBF基因(CBF4)不受冷诱导表达; 但过表达CBF4会增强植物的抗冻性和耐旱性的。本研究发现, 11个BnaCBF中, 与AtCBF1/2/3同源的9个基因对冷胁迫迅速响应, 并且冻害胁迫比冷胁迫响应更为强烈; AtCBF4同源的2个BnaCBF基因BnaA10g07630D和BnaC09g 28190D轻微冷胁迫响应, 但强烈响应冻害胁迫。由于前人主要以非典型的越冬植物如水稻、拟南芥、番茄等作为研究材料, 相关冻害研究开展的较少, 但由于甘蓝型油菜本身是越冬性作物, 生育期通常会经历一段较长的低温冻害时期, 因此BnaCBFs基因在低温时的诱导表达没有冻害胁迫下强烈。另外, 本研究中发现, 冷驯化的冻害和非冷驯化冻害下表达差异不大, 说明BnaCBFs基因驯化和非冷驯化条件下均起作用。
4 结论
本研究鉴定出甘蓝型油菜中11个CBF基因家族成员, 并将BnaCBFs分为亚组I (CBF1/2/3)和亚组II (CBF4) 2个亚组。所有甘蓝型油菜CBF基因受低温诱导表达, 其中亚组Ib的4个基因BnaA03g 13620D、BnaC03g71900D、BnaAnng34260D、BnaC07g 39680D对冷胁迫响应迅速且持续时间长, 亚组II中2个基因BnaA10g07630D和BnaC09g28190D对冷胁迫响应表达较弱, 但它们在根中表达量明显高于叶片, 并且参与盐胁迫和渗透胁迫响应; BnaCBF对冻害响应更强烈。本研究将为进一步了解甘蓝型油菜CBF家族基因的生物学功能、低温胁迫尤其是冻害胁迫研究及耐寒、抗冻品种培育奠定了基础。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
PMID [本文引用: 1]

Transcriptional regulation of gene expression relies on the recognition of promoter elements by transcription factors. In the past several years, a considerable number of (putative) transcription factors have been identified in plants. Some genes coding for these factors were isolated by south-western screening with oligonucleotides as a probe or by homology-based screening, and others were initially isolated by genetic means and subsequently identified as the genes for transcription factors. These transcription factors often form families of structurally related proteins with similar DNA-binding specificities and in addition, they are sometimes involved in related phenomena. Some groups of factors homo- and/or heterodimerize to increase the length and variability of the target sequences. Transcriptional activators, in general, comprise a modular activation domain. The activities of the transcription factors are controlled by post-translational modification, like phosphorylation and glycosylation, as well as at the levels of nuclear transport, oligomerization, etc. In this review, we will summarize the current knowledge of plant transcription factors to help understand the mechanistic aspects of the transcriptional regulation of genes.
DOIURL [本文引用: 1]
PMID [本文引用: 2]

Cold-induced expression of the Arabidopsis COR (cold-regulated) genes is mediated by a DNA regulatory element termed the CRT (C-repeat)/DRE (dehydration-responsive element). Recently, we identified a transcriptional activator, CBF1, that binds to the CRT/DRE and demonstrated that its overexpression in transgenic Arabidopsis plants at non-acclimating temperatures induces COR gene expression and increases plant freezing tolerance. Here we report that CBF1 belongs to a small family of closely related proteins which includes CBF2 and CBF3. DNA sequencing of an 8.7 kb region of the Arabidopsis genome along with genetic mapping experiments indicated that the three CBF genes are organized in direct repeat on chromosome 4 at 72.8 cM, closely linked to molecular markers PG11 and m600. Like CBF1, both CBF2 and CBF3 activated expression of reporter genes in yeast that contained the CRT/DRE as an upstream activator sequence. The transcript levels for all three CBF genes increased within 15 min of transferring plants to low temperature, followed by accumulation of COR gene transcripts at about 2 h. CBF transcripts also accumulated rapidly in response to mechanical agitation. The promoter regions of the CBF genes do not contain the CRT sequence, CCGAC, and overexpression of CBF1 did not have a detectable effect on CBF3 transcript levels, suggesting that the CBF gene family is not subject to autoregulation. We propose that cold-induced expression of CRT/DRE-containing COR genes involves a low temperature-stimulated signalling cascade in which CBF gene induction is an early event.
PMID [本文引用: 2]

Plant growth is greatly affected by drought and low temperature. Expression of a number of genes is induced by both drought and low temperature, although these stresses are quite different. Previous experiments have established that a cis-acting element named DRE (for dehydration-responsive element) plays an important role in both dehydration- and low-temperature-induced gene expression in Arabidopsis. Two cDNA clones that encode DRE binding proteins, DREB1A and DREB2A, were isolated by using the yeast one-hybrid screening technique. The two cDNA libraries were prepared from dehydrated and cold-treated rosette plants, respectively. The deduced amino acid sequences of DREB1A and DREB2A showed no significant sequence similarity, except in the conserved DNA binding domains found in the EREBP and APETALA2 proteins that function in ethylene-responsive expression and floral morphogenesis, respectively. Both the DREB1A and DREB2A proteins specifically bound to the DRE sequence in vitro and activated the transcription of the b-glucuronidase reporter gene driven by the DRE sequence in Arabidopsis leaf protoplasts. Expression of the DREB1A gene and its two homologs was induced by low-temperature stress, whereas expression of the DREB2A gene and its single homolog was induced by dehydration. Overexpression of the DREB1A cDNA in transgenic Arabidopsis plants not only induced strong expression of the target genes under unstressed conditions but also caused dwarfed phenotypes in the transgenic plants. These transgenic plants also revealed freezing and dehydration tolerance. In contrast, overexpression of the DREB2A cDNA induced weak expression of the target genes under unstressed conditions and caused growth retardation of the transgenic plants. These results indicate that two independent families of DREB proteins, DREB1 and DREB2, function as trans-acting factors in two separate signal transduction pathways under low-temperature and dehydration conditions, respectively.
[本文引用: 1]
DOIURL [本文引用: 2]
PMID [本文引用: 2]

Many plants, including Arabidopsis, show increased resistance to freezing after they have been exposed to low nonfreezing temperatures. This response, termed cold acclimation, is associated with the induction of COR (cold-regulated) genes mediated by the C-repeat/drought-responsive element (CRT/DRE) DNA regulatory element. Increased expression of Arabidopsis CBF1, a transcriptional activator that binds to the CRT/DRE sequence, induced COR gene expression and increased the freezing tolerance of nonacclimated Arabidopsis plants. We conclude that CBF1 is a likely regulator of the cold acclimation response, controlling the level of COR gene expression, which in turn promotes tolerance to freezing.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOI [本文引用: 1]

Low temperature is one of the most important abiotic factors inhibiting growth, productivity, and distribution of rapeseed (Brassica napus L.). Therefore, it is important to identify and cultivate cold-tolerant germplasm. The objective of this study was to figure out the mechanism of chilling (4 and 2 degrees C) and freezing (-2 and -4 degrees C) stresses along with a control (22 degrees C) in B. napus cultivars (1801 and C20) under controlled environment (growth chamber). The experiment was arranged in a complete randomized design with three replications. Our results exhibited that under chilling and freezing stresses, the increment of proline accumulation, soluble sugar and protein contents, and antioxidant enzyme activity were enhanced more in 1801 cultivar compared with C20 cultivar. At -2 and -4 degrees C, the seedlings of C20 cultivar died completely compared with 1801 cultivar. Hydrogen peroxide (H2O2) and malondialdehyde contents (MDA) increased in both cultivars, but when the temperature was decreased up to -2 and -4 degrees C, the MDA and H2O2 contents continuously dropped in 1801 cultivar. Moreover, we found that leaf abscisic acid (ABA) was enhanced in 1801 cultivar under chilling and freezing stresses. Additionally, the transcriptional regulations of cold-tolerant genes (COLD1, CBF4, COR6.6, COR15, and COR25) were also determined using real-time quantitative PCR (RT-qPCR). RT-qPCR showed that higher expression of these genes were found in 1801 as compared to C20 under cold stress (chilling and freezing stresses). Therefore, it is concluded from this experiment that 1801 cultivar has a higher ability to respond to cold stress (chilling and freezing stresses) by maintaining hormonal, antioxidative, and osmotic activity along with gene transcription process than C20. The result of this study will provide a solid foundation for understanding physiological and molecular mechanisms of cold stress signaling in rapeseed (B. napus).
DOIURL [本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 3]
[本文引用: 2]
DOIPMID [本文引用: 1]

We present the latest version of the Molecular Evolutionary Genetics Analysis (Mega) software, which contains many sophisticated methods and tools for phylogenomics and phylomedicine. In this major upgrade, Mega has been optimized for use on 64-bit computing systems for analyzing larger datasets. Researchers can now explore and analyze tens of thousands of sequences in Mega The new version also provides an advanced wizard for building timetrees and includes a new functionality to automatically predict gene duplication events in gene family trees. The 64-bit Mega is made available in two interfaces: graphical and command line. The graphical user interface (GUI) is a native Microsoft Windows application that can also be used on Mac OS X. The command line Mega is available as native applications for Windows, Linux, and Mac OS X. They are intended for use in high-throughput and scripted analysis. Both versions are available from www.megasoftware.net free of charge.© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
PMID [本文引用: 1]
PMID [本文引用: 1]

The effects of overexpression of two Brassica CBF/DREB1-like transcription factors (BNCBF5 and 17) in Brassica napus cv. Westar were studied. In addition to developing constitutive freezing tolerance and constitutively accumulating COR gene mRNAs, BNCBF5- and 17-overexpressing plants also accumulate moderate transcript levels of genes involved in photosynthesis and chloroplast development as identified by microarray and Northern analyses. These include GLK1- and GLK2-like transcription factors involved in chloroplast photosynthetic development, chloroplast stroma cyclophilin ROC4 (AtCYP20-3), beta-amylase and triose-P/Pi translocator. In parallel with these changes, increases in photosynthetic efficiency and capacity, pigment pool sizes, increased capacities of the Calvin cycle enzymes, and enzymes of starch and sucrose biosynthesis, as well as glycolysis and oxaloacetate/malate exchange are seen, suggesting that BNCBF overexpression has partially mimicked cold-induced photosynthetic acclimation constitutively. Taken together, these results suggest that BNCBF/DREB1 overexpression in Brassica not only resulted in increased constitutive freezing tolerance but also partially regulated chloroplast development to increase photochemical efficiency and photosynthetic capacity.
DOIURL [本文引用: 1]
PMID [本文引用: 1]

Many plants increase in freezing tolerance in response to low, nonfreezing temperatures, a phenomenon known as cold acclimation. Cold acclimation in Arabidopsis involves rapid cold-induced expression of the C-repeat/dehydration-responsive element binding factor (CBF) transcriptional activators followed by expression of CBF-targeted genes that increase freezing tolerance. Here, we present evidence for a CBF cold-response pathway in Brassica napus. We show that B. napus encodes CBF-like genes and that transcripts for these genes accumulate rapidly in response to low temperature followed closely by expression of the cold-regulated Bn115 gene, an ortholog of the Arabidopsis CBF-targeted COR15a gene. Moreover, we show that constitutive overexpression of the Arabidopsis CBF genes in transgenic B. napus plants induces expression of orthologs of Arabidopsis CBF-targeted genes and increases the freezing tolerance of both nonacclimated and cold-acclimated plants. Transcripts encoding CBF-like proteins were also found to accumulate rapidly in response to low temperature in wheat (Triticum aestivum L. cv Norstar) and rye (Secale cereale L. cv Puma), which cold acclimate, as well as in tomato (Lycopersicon esculentum var. Bonny Best, Castle Mart, Micro-Tom, and D Huang), a freezing-sensitive plant that does not cold acclimate. An alignment of the CBF proteins from Arabidopsis, B. napus, wheat, rye, and tomato revealed the presence of conserved amino acid sequences, PKK/RPAGRxKFxETRHP and DSAWR, that bracket the AP2/EREBP DNA binding domains of the proteins and distinguish them from other members of the AP2/EREBP protein family. We conclude that components of the CBF cold-response pathway are highly conserved in flowering plants and not limited to those that cold acclimate.
PMID [本文引用: 1]

The transcription factor DREB1A/CBF3 specifically interacts with the dehydration responsive element (DRE/CRT) and induces expression of genes involved in environmental stress tolerance in Arabidopsis: Overexpression of DREB1A improved drought- and low-temperature stress tolerance in tobacco. The stress-inducible rd29A promoter minimized the negative effects on the plant growth in tobacco. Furthermore, we detected overexpression of stress-inducible target genes of DREB1A in tobacco. These results indicate that a combination of the rd29A promoter and DREB1A is useful for improvement of various kinds of transgenic plants that are tolerant to environmental stress.
DOIURL [本文引用: 1]
[本文引用: 1]
PMID [本文引用: 1]

We have identified two genes from Arabidopsis that show high similarity with CBF1, a gene encoding an AP2 domain-containing transcriptional activator that binds to the low-temperature-responsive element CCGAC and induces the expression of some cold-regulated genes, increasing plant freezing tolerance. These two genes, which we have named CBF2 and CBF3, also encode proteins containing AP2 DNA-binding motifs. Furthermore, like CBF1, CBF2 and CBF3 proteins also include putative nuclear-localization signals and potential acidic activation domains. The CBF2 and CBF3 genes are linked to CBF1, constituting a cluster on the bottom arm of chromosome IV. The high level of similarity among the three CBF genes, their tandem organization, and the fact that they have the same transcriptional orientation all suggest a common origin. CBF1, CBF2, and CBF3 show identical expression patterns, being induced very rapidly by low-temperature treatment. However, in contrast to most of the cold-induced plant genes characterized, they are not responsive to abscisic acid or dehydration. Taken together, all of these data suggest that CBF2 and CBF3 may function as transcriptional activators, controlling the level of low-temperature gene expression and promoting freezing tolerance through an abscisic acid-independent pathway.
PMID [本文引用: 1]

We further investigated the role of the Arabidopsis CBF regulatory genes in cold acclimation, the process whereby certain plants increase in freezing tolerance upon exposure to low temperature. The CBF genes, which are rapidly induced in response to low temperature, encode transcriptional activators that control the expression of genes containing the C-repeat/dehydration responsive element DNA regulatory element in their promoters. Constitutive expression of either CBF1 or CBF3 (also known as DREB1b and DREB1a, respectively) in transgenic Arabidopsis plants has been shown to induce the expression of target COR (cold-regulated) genes and to enhance freezing tolerance in nonacclimated plants. Here we demonstrate that overexpression of CBF3 in Arabidopsis also increases the freezing tolerance of cold-acclimated plants. Moreover, we show that it results in multiple biochemical changes associated with cold acclimation: CBF3-expressing plants had elevated levels of proline (Pro) and total soluble sugars, including sucrose, raffinose, glucose, and fructose. Plants overexpressing CBF3 also had elevated P5CS transcript levels suggesting that the increase in Pro levels resulted, at least in part, from increased expression of the key Pro biosynthetic enzyme Delta(1)-pyrroline-5-carboxylate synthase. These results lead us to propose that CBF3 integrates the activation of multiple components of the cold acclimation response.
PMID [本文引用: 1]

Many plants increase in freezing tolerance in response to low temperature, a process known as cold acclimation. In Arabidopsis, cold acclimation involves action of the CBF cold response pathway. Key components of the pathway include rapid cold-induced expression of three homologous genes encoding transcriptional activators, CBF1, 2 and 3 (also known as DREB1b, c and a, respectively), followed by expression of CBF-targeted genes, the CBF regulon, that increase freezing tolerance. Unlike Arabidopsis, tomato cannot cold acclimate raising the question of whether it has a functional CBF cold response pathway. Here we show that tomato, like Arabidopsis, encodes three CBF homologs, LeCBF1-3 (Lycopersicon esculentum CBF1-3), that are present in tandem array in the genome. Only the tomato LeCBF1 gene, however, was found to be cold-inducible. As is the case for Arabidopsis CBF1-3, transcripts for LeCBF1-3 did accumulate in response to mechanical agitation, but not in response to drought, ABA or high salinity. Constitutive overexpression of LeCBF1 in transgenic Arabidopsis plants induced expression of CBF-targeted genes and increased freezing tolerance indicating that LeCBF1 encodes a functional homolog of the Arabidopsis CBF1-3 proteins. However, constitutive overexpression of either LeCBF1 or AtCBF3 in transgenic tomato plants did not increase freezing tolerance. Gene expression studies, including the use of a cDNA microarray representing approximately 8000 tomato genes, identified only four genes that were induced 2.5-fold or more in the LeCBF1 or AtCBF3 overexpressing plants, three of which were putative members of the tomato CBF regulon as they were also upregulated in response to low temperature. Additional experiments indicated that of eight tomato genes that were likely orthologs of Arabidopsis CBF regulon genes, none were responsive to CBF overexpression in tomato. From these results, we conclude that tomato has a complete CBF cold response pathway, but that the tomato CBF regulon differs from that of Arabidopsis and appears to be considerably smaller and less diverse in function.
