 ,1, 程仲杰1, 陈晓杰1, 王嘉欢1, 陈锋2, 范家霖1, 张建伟
,1, 程仲杰1, 陈晓杰1, 王嘉欢1, 陈锋2, 范家霖1, 张建伟 ,1,*, 杨保安
,1,*, 杨保安 ,1,*
,1,*Molecular identification and breeding application of allelic variation of grain weight gene in wheat from the Yellow-Huai-River Valley
ZHANG Fu-Yan ,1, CHENG Zhong-Jie1, CHEN Xiao-Jie1, WANG Jia-Huan1, CHEN Feng2, FAN Jia-Lin1, ZHANG Jian-Wei
,1, CHENG Zhong-Jie1, CHEN Xiao-Jie1, WANG Jia-Huan1, CHEN Feng2, FAN Jia-Lin1, ZHANG Jian-Wei ,1,*, YANG Bao-An
,1,*, YANG Bao-An ,1,*
,1,*通讯作者: * 张建伟, E-mail:zjw10308@163.com;杨保安, E-mail:yangcorn@163.com
收稿日期:2020-11-3接受日期:2021-03-19网络出版日期:2021-04-14
| 基金资助: |
Corresponding authors: * E-mail:zjw10308@163.com;E-mail:yangcorn@163.com
Received:2020-11-3Accepted:2021-03-19Published online:2021-04-14
| Fund supported: |
作者简介 About authors
E-mail:zhangfuyan704@163.com

摘要
采用特异性引物PCR扩增方法对183份黄淮海麦区的小麦品种(系)的粒重基因TaCwi-A1、TaGw8-B1和TaGS-D1的等位变异进行分子鉴定, 并结合2016—2017和2017—2018年度的千粒重表型数据, 分析不同等位变异类型对小麦粒重的影响, 从而找出优势基因型组合。结果表明, 不同年份间参试品种(系)的千粒重差异达到极显著水平(P<0.01); TaCwi-A1位点上发现TaCwi-A1a和TaCwi-A1b两种等位变异, 其分布频率分别为66.7%和33.3%; TaGw8-B1位点上TaGw8-B1a等位变异分布频率较高, 为94.5%, 而TaGw8-B1b等位变异分布频率极低, 仅为5.5%; TaGS-D1位点上发现TaGS-D1a和TaGS-D1b两种等位变异, 其分布频率分别为79.8%和20.2%。不同等位变异组合的品种千粒重存在显著差异(P<0.05), 其中具有3个高千粒重等位变异组合TaCwi-A1a/TaGS-D1a/TaGw8-B1a品种的平均千粒重最高, 与具有TaCwi-A1b/TaGS-D1a/TaGw8-B1a品种的平均千粒重差异不显著, 但是显著高于其他组合(P<0.05)。TaCwi-A1a/TaGS-D1a/TaGw8-B1b基因型组合小麦品种(系)的平均千粒重最低。TaCwi-A1、TaGw8-B1和TaGS-D1位点上的不同等位变异均会导致小麦千粒重的显著变化, 其中TaGw8-B1和TaGS-D1位点上的等位变异对小麦粒重的影响更为重要。在参试材料中没有发现具有3个低千粒重等位变异组合TaCwi-A1b/TaGS-D1b/TaGw8-B1b的品种, 在7种不同等位变异组合中, 具有3个高千粒重等位变异组合TaCwi-A1a/TaGS-D1a/TaGw8-B1a品种的平均千粒重最高, 是优势基因型组合。
关键词:
Abstract
Grain weight is one of the most important yield traits in wheat. To accelerate the application process of dominant allelic variation of grain weight gene in wheat breeding, the allelic variation of grain weight gene was identified by using functional markers and the combinations of dominant grain weight genotypes were investigated the allelic variations of grain weight genes TaCwi-A1, TaGw8-B1, and TaGS-D1 in 183 wheat varieties (lines) from the Yellow-Huai-River Valley were identified by PCR amplification with specific primers. To identify the dominant genotype combinations, the effects of different allelic variation genotypes on wheat grain weight were studied by combining the phenotypic data of the 1000-grain weight (TGW) from 2016-2017 and 2017-2018. The results showed that the difference of TGW between different years was highly significant at P < 0.01. Two alleles, TaCwi-A1a and TaCwi-A1b, were detected at TaCwi-A1 locus, with the frequencies of 66.7% and 33.3%, respectively. The frequency of TaGw8-B1a allele on TaGw8-B1 locus was up to 94.5%, while the frequency of TaGw8-B1b allele was only 5.5%. In addition, two alleles, TaGS-D1a and TaGS-D1b, were found in TaGS-D1 locus, and their frequencies were 79.8% and 20.2%, respectively. Further results indicated that there were significant differences in TGW among different allelic variation combinations at P < 0.05. Among them, the average TGW of varieties with TaCwi-A1a/TaGS-D1a/TaGw8-B1a genotype was the highest. There was not significant difference in TGW between TaCwi-A1a/TaGS-D1a/TaGw8-B1a and TaCwi-A1b/ TaGS-D1a/TaGw8-B1a genotype, but it was significantly higher than other genotypes at P < 0.05. The average TGW of TaCwi-A1a/TaGS-D1a/TaGw8-B1b genotype was the lowest. The allelic variations at the loci of TaCwi-A1, TaGw8-B1, and TaGS-D1 all led to significant changes in TGW, and the allelic variations of TaGw8-B1 and TaGS-D1 loci were more important to TGW in wheat. There were no varieties with three low TGW allelic variation combinations TaCwi-A1b/TaGS-D1b/TaGw8-B1b in the tested materials. Among the seven different allele combinations, the average TGW of three high TGW allelic variation combinations TaCwi-A1a/TaGS-D1a/TaGw8-B1a was the highest, which was the dominant genotype combination.
Keywords:
PDF (595KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张福彦, 程仲杰, 陈晓杰, 王嘉欢, 陈锋, 范家霖, 张建伟, 杨保安. 黄淮麦区小麦粒重基因等位变异的分子鉴定及育种应用. 作物学报, 2021, 47(11): 2091-2098 DOI:10.3724/SP.J.1006.2021.01083
ZHANG Fu-Yan, CHENG Zhong-Jie, CHEN Xiao-Jie, WANG Jia-Huan, CHEN Feng, FAN Jia-Lin, ZHANG Jian-Wei, YANG Bao-An.
小麦是全球最为重要的粮食作物之一, 世界上约40%的人口以小麦为主要食粮, 我国小麦的丰歉直接影响到国家乃至世界粮食安全, 因此高产、稳产、抗病、优质、广适是现阶段我国小麦育种中最主要的育种目标之一[1]。小麦产量由单位面积穗数、穗粒数以及粒重三因素构成, 粒重是三要素中遗传相对稳定的性状, 主要受遗传因子影响且主要受加性效应的基因控制[2]。粒重是影响小麦产量最为重要因素之一, 粒重在我国不同的小麦生态区均被不同程度的正向选择和提高, 是现代小麦育种中遗传改良最为显著的产量性状, 对我国小麦单产水平的提高做出了较大贡献[3,4], 但目前关于小麦粒重基因的遗传基础解析和粒重相关的功能标记在育种中的应用仍不够深入。
近年来, 随着小麦全基因组序列信息的公布及现代分子克隆技术的快速发展, 关于小麦粒重基因的克隆和功能标记开发也取得较快发展。Ma等[5]在普通小麦的2AL染色体上克隆得到细胞壁转化酶TaCwi-A1基因, 发现该基因的2种等位变异, 分别命名为TaCwi-A1a和TaCwi-A1b, 并根据二者序列差异开发了一对互补的显性功能标记CWI21和CWI22, 进一步研究发现TaCwi-A1a变异类型与高千粒重的品种相关, 而TaCwi-A1b类型则与低千粒重的品种相关。之后, Jiang等[6]在普通小麦中克隆出TaCWI基因, 其中TaCWI-4A和TaCWI-5D基因也都发现了相应的单倍型, 进一步研究发现具有Hap-4A-T和Hap-5D-C单倍型的品种千粒重显著高于具有Hap-4A-C和Hap-5D-G单倍型的品种。Zhang等[7]利用同源克隆从普通小麦7DS染色体上克隆得到TaGS-D1基因, 发现其含有TaGS-D1a和TaGS-D1b两种等位变异, 根据TaGS-D1基因序列多态性开发的共显性标记GS7D, 能够准确检测TaGS-D1a和TaGS-D1b类型, 发现拥有不同等位变异类型品种的千粒重间存在显著性差异。Wang等[8]利用同源克隆技术从普通小麦中克隆了一个控制籽粒大小的TaGS5-A1基因, 该基因被定位在3AS染色体上, 并发现其至少存在2种等位变异, 分别命名为TaGS5-A1a和TaGS5-A1b。之后, 又在该基因启动子上游顺式作用元件Sp1内发现单碱基G的插入, 这种变化对小麦的粒宽和粒重具有显著影响[9]。Hanif等[10]和Hu等[11]利用水稻中的TGW6基因在普通小麦进行同源克隆, Hanif等[10]克隆得到位于普通小麦3AL染色体上的TaTGW6-A1基因, 并发现该基因在不同品种中存在单核苷酸多态性, 分别命名为TaTGW6-A1a和TaTGW6-A1b, 而Hu等[11]克隆得到TaTGW6基因, 并将其定位在普通小麦4AL染色体上, 且在TaTGW6位点发现3种等位变异类型, 分别命名为: TaTGW6-a、TaTGW6-b和TaTGW6-c, 并开发了共显性标记TG23, 进一步研究发现TaTGW6-b和TaTGW6-c变异类型的品种在粒径和粒重方面显著高于TaTGW6-a类型, 但TaTGW6-b和TaTGW6-c变异类型在当前品种中分布概率较低。最近, Yang等[12]同源克隆发现了控制小麦粒长和粒重的基因TaGL3-5A, 被定位在5AL染色体上, 根据多单核苷酸多态性发现存在2种等位变异, 分别命名为TaGL3-5A-A和TaGL3-5A-G, 后者是小麦粒长和粒重的优势等位变异, 但在我国小麦中分布频率较低, 认为该类型在我国小麦育种利用上具有较大潜力。Ma等[13]利用同源克隆的方法获得小麦TaCYP78A5基因, 并利用瞬时沉默和过表达技术揭示TaCYP78A5表达水平与小麦粒重呈正相关。之后, 司文洁等[14]又根据不同小麦品种TaCYP78A5启动子区序列内SNP位点差异开发TaCYP78A5-2A启动子区功能标记CAPS-5Ap, 并对其进行了验证。Yan等[15]利用同源克隆的方法获得控制小麦粒重的基因, 命名为TaGW8, 位于7BS上。不同籽粒大小的小麦品种基因多态性分析, 发现TaGW8-B1基因存在2种等位变异, 命名为TaGW8-B1a和TaGW8-B1b, 并开发了功能标记TaGW8-7B, 并发现不同等位变异对小麦粒重有显著影响。高产和稳产是我国乃至世界小麦育种家永恒的育种目标, 粒重的增加为推动我国乃至世界小麦单产水平的整体提升做出重要贡献, 而关于小麦粒重遗传基础解析已成为当前研究热点之一。
截至目前, 尽管已有关于小麦粒重基因等位变异检测的相关报道[16,17], 但同时对多个粒重基因进行分子鉴定及粒重功能标记育种应用的研究少之甚少。本研究以183份黄淮海麦区的小麦品种(系)为材料, 利用功能标记CWI21、CWI22、TaGW8-7B以及GS7D分别对2AL、7BS和7DS染色体上的粒重基因等位变异进行检测, 同时结合2016—2017和2017—2018年度的千粒重表型数据, 分析不同等位变异类型对小麦粒重的影响, 并找出优势的粒重基因型组合, 旨在为培育出高产、稳产的小麦新品种提供一些优异亲本资源, 也为粒重基因功能标记辅助育种提供参考依据。
1 材料与方法
1.1 试验材料
豫丰11、郑品麦22、百农207、济麦22、石4185、西农511、西农979、周麦27、周麦32、周麦36、豫同194、豫农202、鲁原502、良星99、豫同843、丰德存麦1号、丰德存麦5号等183份小麦品种(系)来自河南省科学院同位素研究所小麦育种室收集的种质资源圃, 这些材料主要涵盖了黄淮海麦区近20年来生产上种植的小麦品种(系)。1.2 表型测定
2016—2017和2017—2018年度在河南省科学院新郑试验基地种植, 每个品种(系)种植2行, 行长2.5 m, 行距0.25 m, 每行播50粒种子, 待出苗后每行定苗至30株左右。田间管理同大田生产, 正常成熟后人工收获, 及时晾晒, 储藏备用。千粒重测定, 采用德国福弗(pfeuffer)公司生产的康达得型(contador)全自动数粒仪进行种子计数, 每份材料均匀取1000粒, 重复3次, 同时采用常熟市天量仪器有限责任公司生产的LT1002E型电子天平进行称重。
1.3 基因型检测
每个材料选取3粒有代表性的种子, 粉碎后分别放入3个2 mL离心管中, 根据Chen等[18]提取籽粒DNA的方法, 采用SLS (十二酰肌氨酸钠)快速提取小麦籽粒DNA法提取参试材料DNA, 并根据每个品种(系) 3种功能标记检测判断该品种(系)的基因型。利用已开发功能标记CWI21、CWI22、TaGW8- 7B和GS7D对所有参试材料中粒重基因TaCwi-A1、TaGw8-B1和TaGS-D1的等位变异进行分子检测(表1)。PCR反应体系为20 μL, 主要包括buffer缓冲液2.5 μL, Taq DNA 聚合酶(北京天根生化科技有限公司) (2.5 U μL-1) 0.4 μL, dNTP (200 μmol L-1) 1.0 μL, 上下游引物(10 pmol L-1)各0.5 μL, 模板DNA 2.0 μL, ddH2O 13.1 μL。在Tprofessional standard型PCR仪(Biometra)中进行扩增, PCR反应程序为94℃预变性5 min, 94℃变性30 s, 退火温度具体参照(表1)进行, 72℃延伸30 s, 30个循环; 72℃延伸10 min, 反应程序结束后取5 μL PCR扩增产物用1.5%琼脂糖凝胶电泳分离、利用GeneGreen Nucleic Acid DyeGeneGreen核酸染料进行染色, Fire Reader凝胶成像系统下进行扫描成像, 并保存至计算机以备统计分析。
Table 1
表1
表1检测目的基因等位变异的功能标记及其扩增信息
Table 1
| 基因 Genes | 等位变异 Alleles | 标记引物 Mark primer | 引物序列 Primer sequences (5′-3′) | 退火温度 Annealing temperature (℃) | 扩增片段 Targeted fragment (bp) | 参考文献 Reference |
|---|---|---|---|---|---|---|
| TaGS-D1 | TaGS-D1a | GS7D-F | AACTTAGGGAGCGAAAACAA | 58 | 562 | [7] |
| TaGS-D1b | GS7D-R | CACCAAGACTGGAGATGAAA | 522 | [7] | ||
| TaCwi-A1 | TaCwi-A1a | CWI22-F | GGTGATGAGTTCATGGTTAAT | 56 | 402 | [5] |
| CWI22-R | AGAAGCCCAACATTAAATCAAC | |||||
| TaCwi-A1b | CWI21-F | GTGGTGATGAGTTCATGGTTAAG | 56 | 404 | [5] | |
| CWI21-R | AGAAGCCCAACATTAAATCAAC | |||||
| TaGw8-B1 | TaGW8-B1a | TaGW8-7B-F | CGCTCATCCATTCCTTCATCG | 60 | 1097 | [15] |
| TaGW8-B1b | TaGW8-7B-R | GCTATATGGGTTGGTGTCGC | 1373 | [15] |
新窗口打开|下载CSV
1.4 统计分析
采用SPSS 18.0软件进行数据分析,对具有不同粒重基因型的小麦品种(系)间千粒重表型进行差异显著性分析和方差分析(ANOVA),并利用LSD法进行多重比较。2结果与分析
2.1 不同年份间粒重的差异分析
从表2可知,不同年份间供试品种(系)的千粒重差异达到极显著水平。两个年度间供试品种(系)的千粒重平均值分别为44.11 g和46.29 g,变幅分别为34.50~53.35 g和34.20~58.45 g,其变异系数均较高,分别为8.48%和8.75%。可见,黄淮海地区小麦粒重的遗传变异非常丰富,具有较大的遗传改良潜力。Table 2
表2
表2不同年份中供试材料的千粒重方差分析
Table 2
| 年份 Year | 千粒重 Thousand-grain weight (g) | 变幅 Range | 方差 Variance | 标准差 Standard deviation | 变异系数 Coefficient of variation (%) |
|---|---|---|---|---|---|
| 2016-2017 | 44.11 | 34.50-53.35 | 14.00** | 3.74 | 8.48 |
| 2017-2018 | 46.29 | 34.20-58.45 | 16.42** | 4.05 | 8.75 |
新窗口打开|下载CSV
2.2 粒重基因不同等位变异分子检测
利用显性互补功能标记CWI21和CWI22检测TaCwi-A1基因在供试材料中的不同等位变异, 发现CWI21标记在石4185、郑品麦22、郑育麦16、豫丰11、农大2011、轮选166、豫农186和淮麦19等61份材料中可扩增出404 bp的片段, 属于TaCwi-A1b类型(图1-B), 而CWI22标记在商麦167、中育1220、涡麦66、泛育麦17、郑麦103、郑麦369和周麦36等122个材料中扩增出402 bp的片段, 属于TaCwi-A1a类型(图1-A)。图1
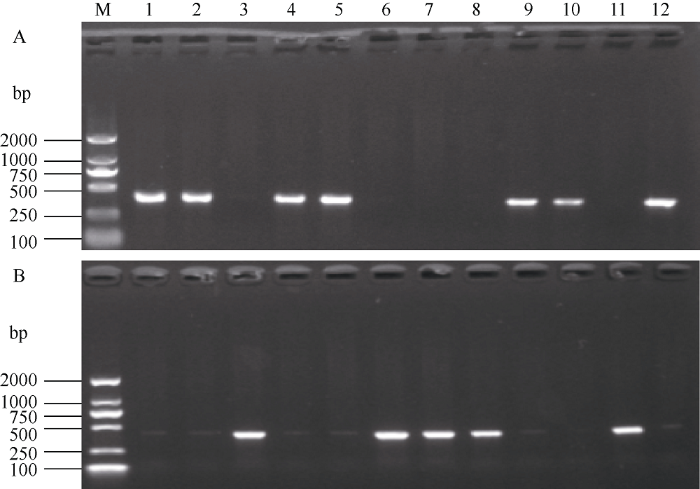 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1标记CWI21 (A)和CWI22 (B)在部分小麦品种中扩增结果
M: DL2000; 1: 石4185; 2: 郑品麦22; 3: 商麦167; 4: 郑育麦16; 5: 豫丰11; 6: 中育1220; 7: 涡麦66; 8: 泛育麦17; 9: 豫农186; 10: 赛德麦1号; 11: 郑麦369; 12: 淮麦19。
Fig. 1Amplification of TaCwi-A1 alleles by functional markers CWI21 (A) and CWI22 (B) in partial wheat varieties
M: DL2000; 1: Shi 4185; 2: Zhengpinmai 22: 3: Shangmai 167; 4: Zhengyumai 16; 5: Yufeng 11; 6: Zhongyu 1220; 7: Guomai 66; 8: Fanyumai 17; 9: Yunong 186; 10: Saidemai 1; 11: Zhengmai 369; 12: Huaimai 19.
利用共显性功能标记TaGW8-7B对所有参试材料进行分子检测(图2), 发现在商麦167、鑫农518、中新16、豫丰11、皖科06725、轮选166、泉麦29、农大2011、郑麦1860等173份材料中扩增出1097 bp的片段, 属于TaGW8-B1a类型, 而在温麦6号、郑麦518、豫农202、石84-7111、淮麦19等10份材料中扩增出1373 bp的片段, 属于TaGW8-B1b类型。
图2
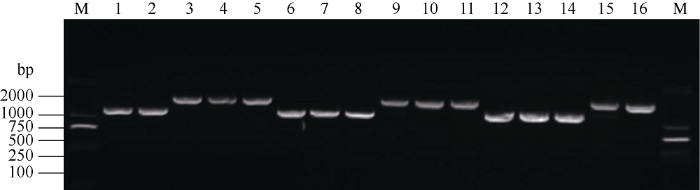 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2TaGW8-7B标记在部分小麦品种中扩增结果
1: 郑品麦22; 2: 商麦167; 3: 温麦6号; 4: 郑麦518; 5: 豫农202; 6: 中新16; 7: 豫丰11; 8: 皖科06725; 9: 豫麦2号; 10: 冀麦38; 11: 石84-7111; 12: 丰德存麦5号; 13: 良星99; 14: 洛麦28; 15: 淮麦19; 16: 百农3217; M: DL2000。
Fig. 2Amplification of TaGW8 genes by functional marker TaGW8-7B in partial wheat varieties
1: Zhengpinmai 22; 2: Shangmai 167; 3: Wenmai 6; 4: Zhengmai 518; 5: Yunong 202; 6: Zhongxin 16; 7: Yufeng 11; 8: Wanke 06725; 9: Yumai 2; 10: Jimai 38; 11: Shi 84-7111; 12: Fengdecunmai 5; 13: Liangxing 99; 14: Luomai 28; 15: Huaimai 19; 16: Bainong 3217; M: DL2000.
利用共显性标记GS7D对183份小麦品种(系)进行检测(图3), 发现在鲁原502、濮兴0369、轮选987、周8425B、兰考906、周麦27、西农511、中育1220、豫丰11等146份材料中扩增出562 bp的片段, 属于TaGS-D1a类型, 而在豫农186、赛德麦1号、龙科1221、中育9302等37份材料中可扩增出522 bp的片段, 属于TaGS-D1b类型。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3GS7D标记检测部分小麦品种
1: 豫农186; 2: 赛德麦1号; 3: 鲁原502; 4: 郑麦366; 5: 中育9302; 6: 龙科1221; 7: 濮兴0369; 8: 豫丰11; 9: 农大2011; 10: 郑品麦8号; 11: 高麦6号; 12: 丰德存麦5号; M: DL2000。
Fig. 3Molecular detection for partial wheat varieties by GS7D
1: Yunong 186; 2: Saidemai 1; 3: Luyuan 502; 4: Zhengmai 366; 5: Zhongyu 9302; 6: Longke 1221; 7: Puxing 0369; 8: Yufeng 11; 9: Nongda 2011; 10: Zhengpinmai 8; 11: Gaomai 6; 12: Fengdecunmai 5; M: DL2000.
2.3 粒重基因不同等位变异的分布及其对小麦粒重的影响
在所有参试材料中, TaCwi-A1、TaGw8-B1和TaGS-D1位点具上均发现2种等位变异。从表3可知, TaCwi-A1位点上鉴定发现183份参试材料中TaCwi- A1a等位变异的分布频率为66.67%, TaCwi-A1b为33.33%, 同时利用SPSS 18.0软件对不同等位变异对应的千粒重进行方差分析, 发现具有TaCwi-A1a等位变异类型品种的千粒重显著高于具有TaCwi- A1b等位变异类型品种的千粒重(P<0.05), TaCwi- A1a为优异等位基因。TaGw8-B1位点上鉴定所有参试材料中TaGw8-B1a等位变异分布频率较高, 为94.54%, 而TaGw8-B1b等位变异分布频率极低, 仅为5.46%, 方差分析发现具有TaGw8-B1a等位变异类型品种的千粒重与具有TaGw8-B1b等位变异类型品种的千粒重, 二者差异达到极显著水平(P<0.01), TaGw8-B1a为优异等位基因。TaGS-D1位点上鉴定所有参试材料中TaGS-D1a等位变异分布频率为79.78%, TaGS-D1b为20.22%, 方差分析发现具有TaGS-D1a等位变异类型品种的千粒重与具有TaGS-D1b等位变异类型品种的千粒重, 二者差异也达到极显著水平(P<0.01), TaGS-D1a为优异等位基因。说明TaCwi-A1、TaGw8-B1和TaGS-D1位点上不同等位变异均会导致小麦千粒重的显著变化, 而TaGw8-B1和TaGS-D1位点上的等位变异对粒重具有更为重要的影响。Table 3
表3
表3粒重基因的不同等位变异对小麦千粒重影响
Table 3
| 基因位点 Gene locus | 等位变异 Allele | 品种数量 Number of varieties | 频率 Frequency (%) | 2016年千粒重 TGW in 2016 (g) | 2017年千粒重 TGW in 2017 (g) | 均值 Mean (g) |
|---|---|---|---|---|---|---|
| TaCwi-A1 | TaCwi-A1a | 122 | 66.7 | 44.46* | 46.61* | 45.54* |
| TaCwi-A1b | 61 | 33.3 | 43.40 | 45.65 | 44.52 | |
| TaGw8-B1 | TaGw8-B1a | 173 | 94.5 | 44.27** | 46.51** | 45.39** |
| TaGw8-B1b | 10 | 5.5 | 41.34 | 42.46 | 41.90 | |
| TaGS-D1 | TaGS-D1a | 146 | 79.8 | 44.44** | 46.71** | 45.58** |
| TaGS-D1b | 37 | 20.2 | 42.79 | 44.63 | 43.71 |
新窗口打开|下载CSV
2.4 不同基因型组合与小麦粒重的关系
利用功能标记CWI21、CWI22、TaGW8-7B和GS7D对所有参试材料进行检测。在TaCwi-A1、TaGw8-B1和TaGS-D1位点上共发现TaCwi-A1a/ TaGS-D1a/TaGw8-B1a、TaCwi-A1b/TaGS-D1a/ TaGw8-B1a、TaCwi-A1b/TaGS-D1b/TaGw8-B1a、TaCwi-A1a/TaGS-D1b/TaGw8-B1a、TaCwi-A1b/TaGS- D1a/TaGw8-B1b、TaCwi-A1a/TaGS-D1b/TaGw8-B1b和TaCwi-A1a/TaGS-D1a/TaGw8-B1b共7种不同的等位变异组合, 分别占参试材料的52.5%、23.5%、7.1%、11.5%、2.7%、1.6%和1.1%。从表4可知, 具有不同等位变异组合品种的千粒重间存在显著差异。具有3个高千粒重等位变异组合TaCwi-A1a/ TaGS-D1a/TaGw8-B1a品种的平均千粒重最高, 为44.69 g, 与具有TaCwi-A1b/TaGS-D1a/TaGw8-B1a品种的平均千粒重差异不显著, 但是显著高于其他组合; 在参试材料中没有发现具有3个低千粒重等位变异组合TaCwi-A1b/TaGS-D1b/TaGw8-B1b的品种;具有TaCwi-A1b/TaGS-D1b/TaGw8-B1a、TaCwi-A1a/ TaGS-D1b/TaGw8-B1a、TaCwi-A1b/TaGS-D1a/TaGw8- B1b和TaCwi-A1a/TaGS-D1b/TaGw8-B1b等位变异组合品种(系)间的平均千粒重差异也不显著; 而TaCwi-A1a/TaGS-D1a/TaGw8-B1b基因型组合小麦品种(系)的平均千粒重最低, 仅为41.39 g。可见, TaCwi-A1、TaGw8-B1和TaGS-D1位点上不同等位变异组合会引起小麦粒重的显著变化。Table 4
表4
表4TaCwi-A1、TaGS-D1和TaGw8-B1位点上不同等位变异组合对小麦千粒重的影响
Table 4
| 等位变异组合 Allelic variation combination | 品种数量 Number of varieties | 频率 Frequency (%) | 表型† Phenotype† | 2016年千粒重 2016 TGW (g) | 2017年千粒重 2017 TGW (g) | 均值‡ Mean (g)‡ |
|---|---|---|---|---|---|---|
| TaCwi-A1a/TaGS-D1a/TaGw8-B1a | 96 | 52.5 | H | 43.67 | 45.71 | 44.69 a |
| TaCwi-A1b/TaGS-D1a/TaGw8-B1a | 43 | 23.5 | H | 43.28 | 45.73 | 44.50 a |
| TaCwi-A1b/TaGS-D1b/TaGw8-B1a | 13 | 7.1 | M | 41.56 | 43.53 | 42.54 b |
| TaCwi-A1a/TaGS-D1b/TaGw8-B1a | 21 | 11.5 | M | 41.61 | 43.39 | 42.50 b |
| TaCwi-A1b/TaGS-D1a/TaGw8-B1b | 5 | 2.7 | ML | 41.69 | 42.90 | 42.30 b |
| TaCwi-A1a/TaGS-D1b/TaGw8-B1b | 3 | 1.6 | ML | 41.13 | 42.92 | 42.03 bc |
| TaCwi-A1a/TaGS-D1a/TaGw8-B1b | 2 | 1.1 | L | 41.18 | 41.60 | 41.39 c |
新窗口打开|下载CSV
3 讨论
小麦产量由单位面积穗数、穗粒数以及千粒重三因素构成, 其中穗粒数的增加是建立在单位面积穗数减少的基础上, 而千粒重的增加则是相对独立的, 且受遗传特性影响最大, 其遗传力高达89% [19]。Wu等[20]研究发现1945年至2010年以来我国不同的小麦主产区小麦产量平均每年增产幅度都在0.33%~1.42%之间, 而粒重和单穗重的显著增加对小麦产量的持续增长起到了举足轻重的作用。Rasheed等[21]研究表明, 在普通小麦中目前已知的有TaCwi-2A、TaSus-2B、TaCKX6-3D、TaGS-D1、TaCKX6a02和TaGw2-6A等控制粒长、粒宽、粒重的基因均具有正向遗传效应, 且对小麦粒重高低具有极其重要的影响。同时, 除了基因型影响外, 脱落酸、赤霉素等内源激素以及光照、温度、水分、养分等外界环境等多种因素对小麦粒重也产生重要影响。本文仅对粒重控制基因对小麦粒重的影响以及利用粒重功能标记进行育种应用进行了研究和讨论。小麦粒重是一个受多基因控制的复杂数量性状, 与其相关的大多数QTL位点的表型贡献率较小, 重复性较差, 且较为分散。Gupta等[22]对小麦粒长、粒宽和千粒重的QTL遗传定位的研究结果进行了系统总结, 发现受亲本遗传背景差异、QTL作图方法、标记类型等因素的影响, 在不同的作图群体中小麦千粒重的QTL定位的结果存在较大的差异, 且远远不能满足分子标记辅助选择育种和粒重相关基因克隆的需要。随着图位克隆、同源克隆等现代生物学技术和标记开发技术的快速发展, 目前已先后克隆出TaGS-D1、TaTGW6、TaGW8-B1、TaGL3-5A、TaSus2-B1、TaCwi-A1、TaCWI-D1、TaCKX6-D1和TaCYP78A5等小麦粒重形成的相关基因, 并开发出GS7D、TG23、TaGW8-7B、TaSus2-2Btgw、CWI22、CWI21和TKX3D等相应的功能标记, 可快速准确鉴定不同小麦品种中控制粒重的相关基因及其变异类型的分布情况。相吉山等[23]和刘永伟等[24]利用CWI22、CWI21标记分别对新疆小麦品种资源和黄淮麦区的小麦品种资源的TaCwi-A1等位变异类型进行了分子检测, 均发现具有TaCwi-A1a基因型材料的千粒重显著高于TaCwi-A1b类型, 进一步验证了TaCwi-A1a是粒重的优异等位变异。Lu等[25]在普通小麦3DS染色体上电子克隆获得了细胞分裂素氧化酶TaCKX6a02-D1基因, 并根据其序列开发共显性功能标记TKX3D, 在不同小麦品种能够分子检测出TaCKX6a02-D1a和TaCKX6a02-D1b变异类型, 发现利用该标记能区分小麦粒重的高低, 可满足小麦粒重高低的分子标记辅助选择需求。之后, 简大为等[26]利用上述功能标记揭示了新疆小麦改良品种与地方品种的遗传变异, 发现高千粒重等位变异在新疆改良品种中分布频率明显高于地方品种, 而且大部分优异等位变异分布频率随着育种时期的推进呈现不连续性上升的趋势。本研究从基因型的角度来分析黄淮地区小麦品种(系)粒重的变化情况, 结果在所有参试材料中共发现7种不同的等位变异组合, 且具有不同等位变异组合的品种千粒重存在显著差异。此外, 没有发现具有3个低千粒重等位变异组合TaCwi-A1b/TaGS-D1b/TaGw8-B1b的品种, 这可能在长期的人工选择过程中低千粒重的劣势等位变异类型逐渐被淘汰, 也可能是由于本研究所选择试验材料的代表性存在一定的局限性。
本研究利用小麦粒重功能标记进行分子检测, 并结合其粒重表型测定进行分析, 结果发现少数品种基因型与千粒重表型不一致的情况, 如具有2个高千粒重和1个低千粒重等位变异组合TaCwi-A1a/ TaGS-D1a/TaGw8-B1b品种的千粒重也较低, 这可能是由于粒重除受上述检测位点控制外, 还存在其他控制小麦粒重基因位点及其相关基因, 也可能是由于本试验材料该类型品种数量较少(仅2份), 不具有代表性。从本研究的粒重表型结果来看, 黄淮麦区品种(系)的千粒重变异范围较大, 且变异系数高, 遗传变异非常丰富, 具有较大的遗传改良潜力, 也为筛选较高粒重的优异种质资源创造了有利条件。
4 结论
利用功能标记分子鉴定的方法对来自于黄淮麦区的183份小麦品种(系)检测, 发现在TaCwi-A1、TaGw8-B1和TaGS-D1位点上不同等位变异均会导致小麦千粒重的显著变化, 而TaGw8-B1和TaGS-D1位点上的等位变异则对小麦粒重的影响更为重要。参试材料中发现共7种不同的等位变异组合, 不同等位变异组合的品种千粒重存在显著性差异, 没有发现具有3个低千粒重等位变异组合TaCwi-A1b/ TaGS-D1b/TaGw8-B1b的品种, 同时发现具有3个高千粒重等位变异组合TaCwi-A1a/TaGS-D1a/TaGw8- B1a品种的平均千粒重最高, 是优势基因型组合, 所占比例为52.5%, 具有这些优势基因型组合的优异材料将为我国小麦高产育种过程中亲本的选配提供有效资源。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 3]
DOIURL [本文引用: 1]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

Kernel dimensions (KD) contribute greatly to thousand-kernel weight (TKW) in wheat. In the present study, quantitative trait loci (QTL) for TKW, kernel length (KL), kernel width (KW) and kernel diameter ratio (KDR) were detected by both conditional and unconditional QTL mapping methods. Two related F(8:9) recombinant inbred line (RIL) populations, comprising 485 and 229 lines, respectively, were used in this study, and the trait phenotypes were evaluated in four environments. Unconditional QTL mapping analysis detected 77 additive QTL for four traits in two populations. Of these, 24 QTL were verified in at least three trials, and five of them were major QTL, thus being of great value for marker assisted selection in breeding programmes. Conditional QTL mapping analysis, compared with unconditional QTL mapping analysis, resulted in reduction in the number of QTL for TKW due to the elimination of TKW variations caused by its conditional traits; based on which we first dissected genetic control system involved in the synthetic process between TKW and KD at an individual QTL level. Results indicated that, at the QTL level, KW had the strongest influence on TKW, followed by KL, and KDR had the lowest level contribution to TKW. In addition, the present study proved that it is not all-inclusive to determine genetic relationships of a pairwise QTL for two related/causal traits based on whether they were co-located. Thus, conditional QTL mapping method should be used to evaluate possible genetic relationships of two related/causal traits.
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

Background: Grain size and shape greatly influence grain weight which ultimately enhances grain yield in wheat. Digital imaging (DI) based phenomic characterization can capture the three dimensional variation in grain size and shape than has hitherto been possible. In this study, we report the results from using digital imaging of grain size and shape to understand the relationship among different components of this trait, their contribution to enhance grain weight, and to identify genomic regions (QTLs) controlling grain morphology using genome wide association mapping with high density diversity array technology (DArT) and allele-specific markers. Results: Significant positive correlations were observed between grain weight and grain size measurements such as grain length (r = 0.43), width, thickness (r = 0.64) and factor from density (FFD) (r = 0.69). A total of 231 synthetic hexaploid wheats (SHWs) were grouped into five different sub-clusters by Bayesian structure analysis using unlinked DArT markers. Linkage disequilibrium (LD) decay was observed among DArT loci > 10 cM distance and approximately 28% marker pairs were in significant LD. In total, 197 loci over 60 chromosomal regions and 79 loci over 31 chromosomal regions were associated with grain morphology by genome wide analysis using general linear model (GLM) and mixed linear model (MLM) approaches, respectively. They were mainly distributed on homoeologous group 2, 3, 6 and 7 chromosomes. Twenty eight marker-trait associations (MTAs) on the D genome chromosomes 2D, 3D and 6D may carry novel alleles with potential to enhance grain weight due to the use of untapped wild accessions of Aegilops tauschii. Statistical simulations showed that favorable alleles for thousand kernel weight (TKW), grain length, width and thickness have additive genetic effects. Allelic variations for known genes controlling grain size and weight, viz. TaCwi-2A, TaSus-2B, TaCKX6-3D and TaGw2-6A, were also associated with TKW, grain width and thickness. In silico functional analysis predicted a range of biological functions for 32 DArT loci and receptor like kinase, known to affect plant development, appeared to be common protein family encoded by several loci responsible for grain size and shape. Conclusion: Conclusively, we demonstrated the application and integration of multiple approaches including high throughput phenotyping using DI, genome wide association studies (GWAS) and in silico functional analysis of candidate loci to analyze target traits, and identify candidate genomic regions underlying these traits. These approaches provided great opportunity to understand the breeding value of SHWs for improving grain weight and enhanced our deep understanding on molecular genetics of grain weight in wheat.
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
