 ,1,2, 任毅1,2, 张新忠1,3, 王继庆1,2, 张志辉1,2, 石书兵1,2, 耿洪伟
,1,2, 任毅1,2, 张新忠1,3, 王继庆1,2, 张志辉1,2, 石书兵1,2, 耿洪伟 ,1,2,*
,1,2,*Genome-wide association study of pre-harvest sprouting traits in wheat
XIE Lei ,1,2, REN Yi1,2, ZHANG Xin-Zhong1,3, WANG Ji-Qing1,2, ZHANG Zhi-Hui1,2, SHI Shu-Bing1,2, GENG Hong-Wei
,1,2, REN Yi1,2, ZHANG Xin-Zhong1,3, WANG Ji-Qing1,2, ZHANG Zhi-Hui1,2, SHI Shu-Bing1,2, GENG Hong-Wei ,1,2,*
,1,2,*通讯作者: *耿洪伟, E-mail:hw-geng@163.com
收稿日期:2020-10-4接受日期:2021-01-13网络出版日期:2021-02-24
| 基金资助: |
Corresponding authors: *E-mail:hw-geng@163.com
Received:2020-10-4Accepted:2021-01-13Published online:2021-02-24
| Fund supported: |
作者简介 About authors
E-mail:1784462634@qq.com

摘要
为了解小麦穗发芽的遗传机制, 发掘与小麦穗发芽相关的候选基因, 采用整穗发芽法对来自全国的207份小麦品种(系)进行表型鉴定, 并结合小麦90K SNP基因芯片, 通过TASSLE软件的MLM (Q+K)模型对小麦的整穗发芽率进行全基因组关联分析(genome-wide association study, GWAS)。研究结果表明, 不同年份间小麦品种(系)的穗发芽表现出丰富的表型变异, 变异系数为0.34和0.25, 多态性信息含量(polymorphic information content, PIC)为0.01~0.38, 全基因组LD衰减距离为3 Mb。群体结构分析和主成分分析表明, 207份小麦品种(系)所构成的自然群体结构简单, 可分为3个亚群。GWAS检测结果显示, 在不同环境间共检测到34个与小麦穗发芽显著关联的SNP标记位点(P≤0.001), 分布在小麦3A、3B、4A、4B、5D、6A、6B、6D、7B和7D染色体上, 单个位点可解释5.55%~11.63%的表型变异, 16个标记位点在两个及以上环境下均被检测到, 其中6B染色体上的标记wsnp_Ex_c14101_22012676在E1、E2及平均环境下被共同检测到, 属稳定遗传的位点。通过对表型效应值大且稳定遗传的关联位点进行挖掘, 共筛选到13个与小麦穗发芽相关的候选基因。其中TraesCS3A01G589400LC、TraesCS6B01G138600/ TraeCS6B01G516 700LC/TraesCS6B01G548900LC、TraesCS6D01G103600及TraesCS7B01G200100等基因通过调控植物内源激素-脱落酸(abscisic acid, ABA)的灵敏性进而影响种子的休眠; TraesCS3B01G415900LC、TraesCS6A01G144700LC及TraesCS6B01G294800等基因编码的F-box蛋白在植物激素的信号转导、光信号转导以及花器官发育等生理过程中起重要作用; TraesCS6A01G108800、TraesCS6B01G138200/TraesCS6B01G293700基因编码Myb转录因子家族蛋白能调控种子中类黄酮的生物合成, 对籽粒颜色有重要影响, 这些候选基因是与小麦穗发芽相关的重要基因。
关键词:
Abstract
To understand the genetic mechanism of wheat pre-harvest sprouting (PHS) in wheat breeding, it is significant to explore marker loci and candidate genes associated with PHS resistance using intact spikes. In this study, a total of 207 wheat varieties (lines) from China and 16,686 SNP markers were analyzed in wheat whole genome. The mixed liner model (Q + K) was used to analyze PHS phenotypic data in three environments. Genome-wide association study showed that there were abundant phenotypic variations in different environments and wheat varieties (lines). The coefficient of variation was 0.34 and 0.25, the polymorphic information content of value (PIC) was from 0.01 to 0.38, and the attenuation distance of whole genome LD was 3 Mb. The population structure and principal component analysis revealed that 207 wheat varieties (lines) could be divided into three subgroups. GWAS results indicated that 34 SNP markers were detected, which were significantly associated with pre-harvest sprouting at P < 0.001. They were located on chromosomes 3A, 3B, 4A, 4B, 5D, 6A, 6B, 6D, 7B, and 7D, and each explained 5.55%-11.63% of phenotypic variation. There were 16 markers loci detected in more than two environments, and the marker Np_Ex_c14101_22,012,676 on 6B chromosome detected in E1, E2, and average environment. Meanwhile, 13 candidate genes were screened out by mining association loci with large phenotypic effect value and stable inheritance. TraesCS3A01G589400LC, TraesCS6B01G138600/TraesCS6B01G516700LC/TraesCS6B01G548900LC, TraesCS6D01G103600, and TraesCS7B01G200100 could affect seed dormancy by regulating the sensitivity of endogenous ABA in plants. The F-box proteins were encoded by TraesCS3B01G415900LC, TraesCS6A01G144700LC, and TraesCS6B01G294800, which played major roles in plant hormone signal transduction, light signal transduction, and flower organ development. TraesCS6A01G108800, TraesCS6B01G138200/ TraesCS6B01G293700 encoded Myb transcription factor family. These candidate genes are important genes related to wheat sprouting.
Keywords:
PDF (5692KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
谢磊, 任毅, 张新忠, 王继庆, 张志辉, 石书兵, 耿洪伟. 小麦穗发芽性状的全基因组关联分析[J]. 作物学报, 2021, 47(10): 1891-1902 DOI:10.3724/SP.J.1006.2021.01078
XIE Lei, REN Yi, ZHANG Xin-Zhong, WANG Ji-Qing, ZHANG Zhi-Hui, SHI Shu-Bing, GENG Hong-Wei.
小麦是我国乃至世界最重要的粮食作物之一, 其产量与品质备受关注[1,2]。近年来, 随着全球气候变暖以及极端天气频发, 小麦成熟至收获期间穗上发芽现象越发频繁, 严重降低小麦产量, 劣化小麦品质, 穗发芽已成为全球性危害[3]。穗发芽促使籽粒内部水解酶活性增加, 籽粒内部储藏物质被分解消耗, 从而造成籽粒容重和千粒重下降, 导致小麦减产[4]。同时, 由于蛋白质被降减, 小麦制粉后的SDS沉降值和面筋含量逐渐降低, 用发芽小麦粉加工的面制品如馒头、面包等, 外形和口感较差, 严重影响其营养品质和加工品质[5,6]。全球每年因穗发芽而造成的直接经济损失约10亿美元[7]。培育抗穗发芽小麦新品种是解决这一问题最有效的途径之一。
小麦穗发芽是受种子休眠水平、种子结构、种皮颜色等内部因素以及温度、湿度、光照等外界环境因素共同作用的结果[8,9]。种子的休眠特性是影响小麦穗发芽抗性的主要遗传因素[10]。目前对于小麦穗发芽鉴定的方法主要有籽粒发芽法、整穗发芽法和α-淀粉酶活性测定法[10]。籽粒发芽主要反映小麦籽粒的休眠情况, 并不能表现出颖壳、麦芒等抑制物的作用[10]。α-淀粉酶活性测定可以鉴定胚乳抑制物对穗发芽的影响, 但操作复杂, 不适合进行批量化测定[11]。整穗发芽不仅能综合性反映小麦穗发芽的抗性, 而且操作简单, 适合批量化测定, 因此常用来鉴定小麦的穗发芽抗性。测定小麦整穗发芽常用的方法有沙培法、发芽纸发芽法、塑料袋保湿法、模拟降雨法、纱布保湿法等[11,12]。
小麦穗发芽是受多基因调控的数量性状, 存在微效和主效基因[13]。连锁分析与全基因组关联分析目前已成为数量性状基因定位和挖掘的重要途径[14]。许多研究通过双单倍体(doubled haploid, DH)或重组自交系(recombinant inbred lines, RIL)等人工作图群体, 将穗发芽相关基因定位于2AL、2BS、3AS、3AL、4AL、5D、6BS及7DL染色体上[14,15,16,17,18,19,20]。但连锁作图受限于遗传群体亲本间的差异度及标记密度等的不同, QTL定位的区间较大, 仅能分析部分等位基因。全基因组关联分析(genome-wide association study, GWAS)与连锁分析互补, 可以高效定位和挖掘多个小麦穗发芽相关的优异等位基因[10]。Kulwal等[21]利用1166个简单序列重复(simple sequence repeat, SSR)和多样性芯片(diversity arrays technology, DArT)分子标记, 对208份白皮冬小麦品种4年的穗发芽率进行测定, 定位了8个与穗发芽相关的数量性状基因座(quantitative trait locus, QTL), 分布在1BS、2DS、4AL、6DL、7BS和7DS染色体上, 并证实了7BS是新的QTL位点。Jaiswal等[22]通过对242份普通小麦的穗发芽率进行鉴定, 并结合250个SSR标记对穗发芽性状进行QTL分析, 发现了30个小麦穗发芽抗性相关的主效位点, 其中仅有8个标记位点与前人研究结果相同, 其余22个标记位点均处在前人未报道的染色体区域。Lin等[23]首次使用9K和90K单核苷酸多态性(single nucleotide polymorphism, SNP)基因芯片对155份美国冬小麦品种(系)的穗发芽率进行全基因组关联分析, 共检测到12个与穗发芽相关的QTL, 其中3AS、3AL、3B、4AL和7A是QTL分布频率较大的区域。Zhu等[24]利用90K SNP基因芯片对192份小麦品种(系)的穗发芽率进行全基因组关联分析, 检测到23个显著关联位点, 解释了6.0%~18.9%的表型变异, 其中1A、3B和6B是抗性位点的热点区域。Zuo等[25]对周8425B×中国春重组自交系群体家系和166份普通小麦的籽粒发芽率进行了QTL定位及GWAS分析, 研究结果除发现位于3AS、6AL、7BL和3DL上已报道的QTL外, 还挖掘了位于5A、7A、4B、3D、6D染色体上的8个新位点。
虽然前人在小麦穗发芽的鉴定、基因的筛选及其定位等方面取得了一定进展, 但由于小麦穗发芽的遗传机制较为复杂, 遗传背景来源不同的材料携带着不同的抗性因子[10]。因此, 为了进一步发掘穗发芽相关基因并开发相关功能标记, 本研究对207份小麦品种(系)的整穗发芽率进行鉴定, 结合SNP标记进行全基因组关联分析, 以期为小麦穗发芽遗传机制研究和分子育种提供参考。
1 材料与方法
1.1 供试材料
供试小麦品种(系)共计207份, 是我国各麦区主推品种(系)、部分骨干亲本和历史品种。其中国内品种(系) 165份, 包含黄淮麦区93份, 西南麦区12份, 长江麦区29份, 北部麦区31份以及国外品种(系) 42份, 供试材料在不同区域间的分布频率依次为: 45%、6%、14%、15%和20%, 上述供试材料均由中国农业科学院作物科学研究所小麦品质课题组夏先春研究员惠赠。供试材料分别于2017—2018 (简称E1)和2018—2019年度(简称E2)种植于新疆农业科学院玛纳斯试验站, 单行种植, 行长2 m, 行距25 cm, 施肥、滴灌、防虫及除草同当地田间管理。以连续2年的均值为平均环境(简称E3)。1.2 整穗发芽的表型鉴定及分析
2017—2018和2018—2019年度的供试材料均在室内进行表型鉴定。在小麦生长到蜡熟后期时, 剪取每份材料的10个主茎穗(带穗下节), 每5穗结扎为一个重复, 室内风干1 d, 立即存放于-20℃的冰箱内以维持其休眠性, 待收获全部材料后统一进行穗发芽实验。先将整穗在蒸馏水中浸泡10~12 h, 再用0.1%次氯酸钠溶液浸泡消毒15 min后, 用无菌水冲洗干净, 用发芽纸卷裹, 保鲜袋保湿, 置于人工气候培养箱(温度20℃, 光周期为16 h昼/8 h夜, 相对湿度为80%)培养7 d后取出, 在电热恒温烘箱(150℃)内快速烘干, 阻止其继续发芽, 手工脱粒, 以种胚破裂为鉴定标准[26], 记录每份材料的穗发芽率, 2次重复的平均值为此材料在该年度的穗发芽率。整穗发芽率(sprouting rate, SR)=5穗的发芽粒数/总粒数×100%。采用Microsoft Excel 2016进行基本数据统计与分析, 采用SPSS 21.0、QTL IciMapping V4.1软件进行描述性统计分析和方差分析[27]。
1.3 群体结构分析和主成分分析
应用Tassel 5.0软件计算多态性信息量(PIC, polymorphic information content), PIC = 1-ΣP2ij (P2ij表示第i个位点的第j个等位变异出现的频率)[28]。从筛选过的标记中, 选取2000个最小等位基因频率大于10%且在染色体上均匀分布的SNP标记, 利用Structure 2.3.4软件进行群体结构分析。参数设置: Length of Burn-Period=10,000, MCMC Reps after Burn-in=100,000, 选择Admixture Ancestry模型和Dependent Allele Frequencies模式, 令K=2~12, 每个K值重复运行5次。将不同K值下重复运行5次的结果上传至Structure Harvester(利用TASSEL5.0软件进行主成分分析(principle component analysis, PCA), 利用ggplot2工具绘制主成分分析图。
1.4 全基因组关联分析
实验室前期使用90K SNP基因芯片对207份自然群体材料进行了基因分型, 共获得16,686个高质量SNP标记。基于这些高质量SNP标记, 使用Tassel 5.0软件中的MLM (Q+K)模型对207份小麦品种(系)在不同环境下的整穗发芽率进行全基因组关联分析, 以P=1.0×10-3为阈值, 判定SNP标记与目标性状关联的显著性[30]。1.5 连锁不平衡分析
以位点间的相关系数平方(r2)作为衡量多态性位点两两之间的连锁不平衡(linkage disequilibrium, LD)参数。采用Tassel 5.0软件计算r2, 以第95百分位的r2值作为阈值估测LD衰减距离。考虑连锁群(linkage group)间连锁不平衡背景的影响, 对连锁群间的r2值进行平方根转换, 以大于此分布95%的参数值为阈值, 用来截取同一连锁群内LD的衰减距离[31]。关联分析中, 超过LD衰减距离的2个位点则认为是2个不同的位点。2 结果与分析
2.1 小麦穗发芽的表型分析
本研究采用整穗发芽法对207份供试材料进行表型鉴定(图1和图2), 研究结果表明, 在2个环境下, 整穗发芽率的变异幅度分别为12.98%~100.00%和13.84%~100.00%, 变异系数为0.34和0.25, 平均值均较高, 分别为74.31%和84.80%, 表明遗传背景来源不同的材料在2个环境下均表现出丰富的表型变异, 但自然群体内多数材料的穗发芽抗性较弱, 具有较大的遗传改良潜力。不同年份间相关系数为0.51。方差分析(表1)显示, 2个环境下测得供试材料间整穗发芽率差异极显著(P = 0.00001), 基因型、环境及基因型与环境互作皆影响小麦穗发芽的抗性。其遗传力为0.60, 表明小麦穗发芽抗性主要由基因型决定, 但也受到环境因素的影响, 遗传因素是导致其表型变异的主要原因。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1部分供试材料的整穗发芽图
A: 京冬8号; B: 淮麦20号; C: 郑麦2号。
Fig. 1Pre-harvest sprouting diagram of some tested materials
A: Jingdong 8; B: Huaimai 20; C: Zhengmai 2.
图2
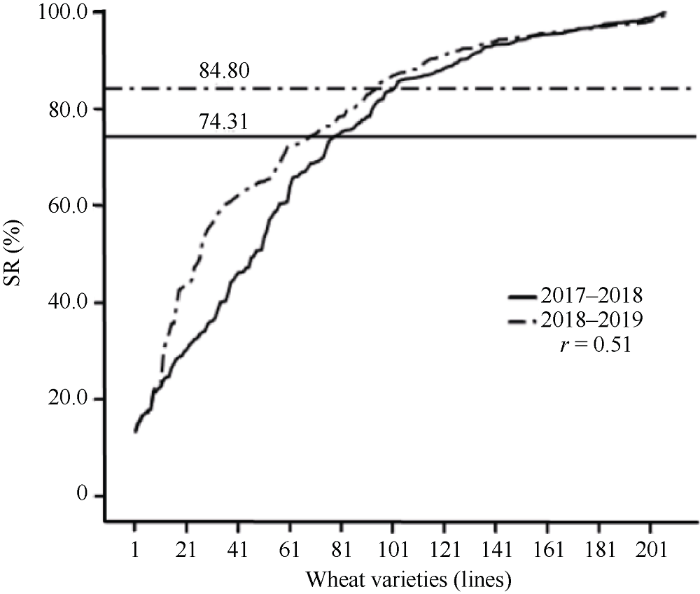 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2207份材料在2个环境下的整穗发芽率变异
SR: 整穗发芽率。
Fig. 2Variation of germination rate of intact spike in 207 wheat genotypes detected in two environments
SR: sprouting rate.
Table 1
表1
表1207份自然群体小麦穗发芽方差分析
Table 1
| 变异来源 Source of variance | 自由度 DF | 平方和 SS | 均方 MS | F值 F-value | P值 P-value | 遗传力 h2 |
|---|---|---|---|---|---|---|
| 基因型Genotype (G) | 206 | 95,102,272.00 | 312,836.40 | 540.38 | 0.00001 | 0.60 |
| 环境Environment (E) | 1 | 756,525,632.00 | 378,262,816.00 | 653,396.18 | 0.00001 | |
| 基因型×环境G×E | 443 | 94,041,216.00 | 212,282.65 | 366.68 | 0.00001 | |
| 误差Error | 747 | 432,451.71 | 578.91 | |||
| 总变异Total | 1397 | 946,109,312.00 |
新窗口打开|下载CSV
2.2 SNP多态性及分布
使用90K SNP基因芯片对E1、E2及平均环境(E3)下的207份小麦品种(系)的整穗发芽率进行检测, 经过优质筛选, 最终采用16,686个具有多态性的SNP标记进行GWAS分析, 平均单条染色体包含795个SNP标记, 其中分布在A、B和D基因组染色体上的SNP标记数分别为7145、7343和2198。A、B基因组的染色体组所用标记数目、标记密度和多态性信息含量均高于D染色体组。物理图谱总长度为14,043.30 Mb, 每个标记间的平均物理距离为1.27 Mb, SNP标记的PIC值介于0.01~0.38之间(表2)。Table 2
表2
表2标记的分布及多态性
Table 2
| 染色体 Chromosome | 标记数目 No. of markers | 染色体长度 Chromosome length (Mb) | 标记密度 Density of marker | 多态信息量 PIC | |
|---|---|---|---|---|---|
| 平均值Mean | 变异范围Range | ||||
| 1A | 1263 | 592.38 | 0.47 | 0.27 | 0.01-0.38 |
| 2A | 1145 | 780.46 | 0.68 | 0.24 | 0.01-0.38 |
| 3A | 891 | 749.46 | 0.84 | 0.25 | 0.01-0.38 |
| 4A | 708 | 741.73 | 1.05 | 0.25 | 0.01-0.38 |
| 5A | 960 | 709.43 | 0.74 | 0.28 | 0.01-0.38 |
| 6A | 1006 | 617.40 | 0.61 | 0.27 | 0.03-0.38 |
| 7A | 1172 | 736.44 | 0.63 | 0.25 | 0.01-0.38 |
| 1B | 1292 | 688.60 | 0.53 | 0.27 | 0.01-0.38 |
| 2B | 1291 | 799.62 | 0.62 | 0.26 | 0.01-0.38 |
| 3B | 1087 | 829.32 | 0.76 | 0.27 | 0.01-0.38 |
| 4B | 571 | 672.56 | 1.18 | 0.26 | 0.01-0.38 |
| 5B | 1156 | 712.82 | 0.62 | 0.28 | 0.01-0.38 |
| 6B | 1085 | 720.82 | 0.66 | 0.25 | 0.01-0.38 |
| 7B | 861 | 750.49 | 0.87 | 0.26 | 0.02-0.38 |
| 1D | 568 | 495.14 | 0.87 | 0.13 | 0.01-0.38 |
| 2D | 482 | 650.94 | 1.35 | 0.25 | 0.02-0.38 |
| 3D | 232 | 613.92 | 2.65 | 0.24 | 0.01-0.38 |
| 4D | 99 | 508.58 | 5.14 | 0.25 | 0.08-0.38 |
| 5D | 240 | 563.41 | 2.35 | 0.26 | 0.02-0.38 |
| 6D | 319 | 472.61 | 1.48 | 0.25 | 0.03-0.38 |
| 7D | 260 | 637.17 | 2.45 | 0.23 | 0.01-0.38 |
| A染色体组 Total genome A | 7145 | 4927.30 | 0.72 | 0.26 | 0.01-0.38 |
| B染色体组 Total genome B | 7343 | 5174.23 | 0.75 | 0.27 | 0.01-0.38 |
| D染色体组 Total genome D | 2198 | 3941.77 | 2.33 | 0.23 | 0.01-0.38 |
| 总计Total | 16686 | 14043.30 | 1.27 | 0.25 | 0.01-0.38 |
新窗口打开|下载CSV
2.3 群体结构及连锁不平衡分析
利用Structure 2.3.4软件对207份供试材料进行群体结构分析(图3-A, B)。结果显示, 当K=3时, ΔK达到峰值, 曲线变化程度最大。PCA分析(图3-C)与Structure分析结果一致。供试材料可分为3个亚群, 其中, 亚群1有60份品种(系), 主要来自法国(14份)和中国北京(13份); 亚群2有62份品种(系), 主要来自中国河南(27份); 亚群3有85份品种(系), 主要来自中国河南(17份)、陕西(16份)和江苏(13份)。3个亚群所含材料的分布频率依次为: 28.99%、29.95%和41.06%。经计算得到207份小麦品种(系)在基因组A、B、D和全基因组的LD衰减距离分别为5、3、1和3 Mb, 依据全基因组的LD衰减距离, 将在物理图谱上前后3 Mb区间内的位点认定为一个候选位点。图3
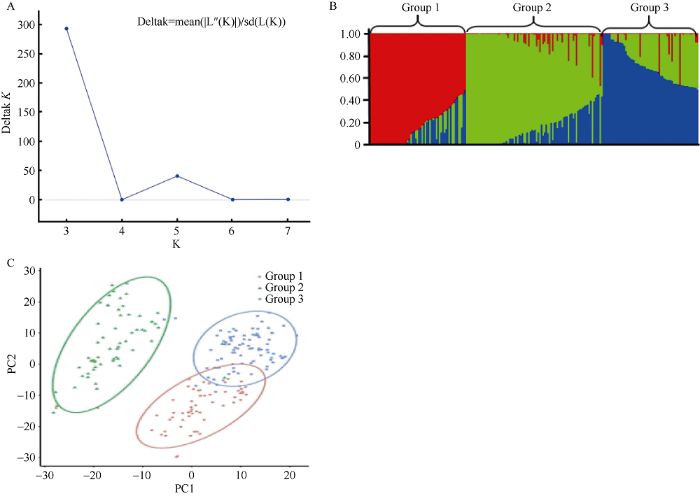 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3207份小麦品种(系)群体结构分析
Fig. 3Population structure analysis of 207 wheat varieties (lines)
2.4 全基因组关联分析
利用TASSEL5.0软件将207份小麦品种(系)的整穗发芽率结合90K SNP芯片筛选分型出的16,686个高质量SNP标记进行全基因组关联分析。基于MLM (Q+K)模型, 当P≤0.001时, 认为该标记与性状显著关联, 在多个环境下检测到的位点视为可稳定遗传的位点(图4)。GWAS分析结果表明, 共检测到34个显著性位点, 分布于小麦3A、3B、4A、4B、5D、6A、6B、6D、7B和7D染色体上, 单个位点可解释5.55%~11.63%的表型变异。其中, 定位于6B染色体上的标记wsnp_Ex_c14101_22012676在E1、E2及平均环境下被共同检测到, 对表型的贡献率为5.86%~11.61%; 另有15个标记位点均在1个环境及平均环境下被检测到, 分布于3A、6A、6B及7B染色体上, 其贡献率为5.55%~11.63%, 这16个可重复检测到的位点属稳定遗传位点, 其中6B染色体是携带小麦穗发芽相关基因的热点区域; 其余的18个标记位点均仅在1个环境或平均环境下被检测到。表明在影响小麦穗发芽的众多基因中, 存在效应稳定的QTL, 同时也受到基因型与环境互作的影响, 方差分析的结果也证实了这一点。将GWAS检测到的34个显著性位点与前人的定位结果进行比较, 发现10个位点被定位到前人报道的基因/标记/QTL区间附近, 其余24个标记位点均处在前人未报道的染色体区域(表3)。图4
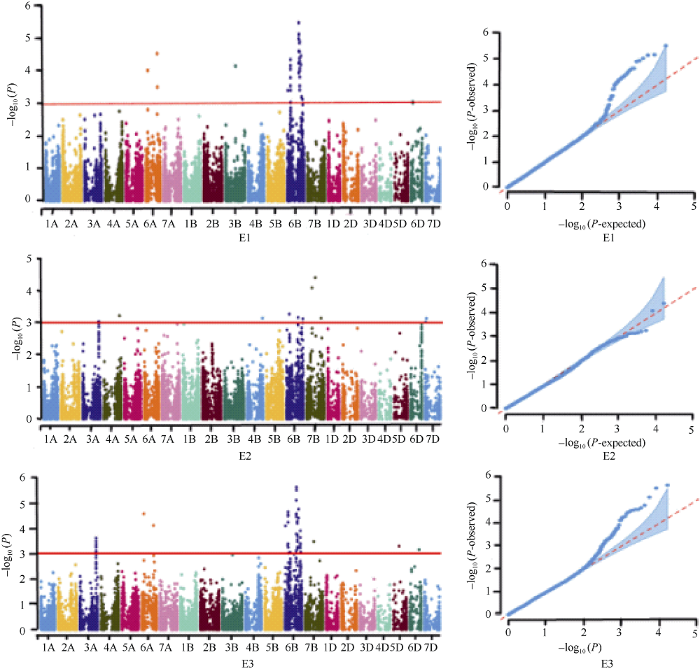 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4不同环境中穗发芽的曼哈顿图和Q-Q图
E1: 2017-2018年, 玛纳斯; E2: 2018-2019年, 玛纳斯; E3: 平均环境。
Fig. 4Manhattan and Q-Q diagrams of pre-harvest sprouting traits in different environments
E1: 2017 and 2018 in Manasi; E2: 2018 and 2019 in Manasi; E3: average environment.
Table 3
表3
表3小麦穗发芽显著关联位点信息
Table 3
| 标记 Marker | 染色体 Chr. | 位置 Position (Mb) | MLM | 环境 Environment | 已报道的QTL/标记/基因Known QTL/marker/gene | |
|---|---|---|---|---|---|---|
| P值 P-value | 贡献率 R2 (%) | |||||
| Kukri_c8465_54 | 3A | 684.04-685.36 | 2.35E-04-9.14E-04 | 5.55-8.75 | E2/E3 | TaVp-1[32] |
| Kukri_c9259_421 | 4A | 673.07 | 5.95E-04 | 6.34 | E2 | IWB23723[23] |
| RAC875_rep_c107892_142 | 6A | 77.78 | 2.56E-05-9.28E-05 | 8.04-9.25 | E1/E3 | |
| Kukri_c5744_92 | 6A | 483.90 | 2.78E-05-7.41E-04 | 8.34-9.25 | E1/E3 | |
| RAC875_c43536_193 | 6A | 490.50 | 3.03E-04 | 6.94 | E1 | |
| IAAV8536 | 3B | 422.52 | 6.78E-05 | 9.81 | E1 | TaDFR[33] |
| Tdurum_contig60051_838 | 4B | 644.68 | 7.10E-04 | 9.42 | E2 | Xwmc349[34] |
| CAP11_c2542_147 | 6B | 24.92 | 7.71E-05-3.83E-04 | 6.71-8.29 | E1/E3 | TaCYP707A1[35] |
| Kukri_c33668_877 | 6B | 119.53 | 4.55E-04-5.32E-04 | 6.05-6.16 | E2/E3 | AX-108844376[24] |
| GENE-3171_203 | 6B | 135.11-138.65 | 2.15E-05-5.50E-04 | 6.09-9.71 | E1/E3 | |
| BS00069412_51 | 6B | 143.20 | 9.03E-04 | 5.64 | E1 | |
| Excalibur_c18382_760 | 6B | 151.13 | 9.24E-04 | 5.60 | E3 | |
| wsnp_Ku_c2614_4970880 | 6B | 226.87 | 8.44E-04 | 5.59 | E3 | |
| Tdurum_contig13698_141 | 6B | 475.73 | 7.54E-04 | 5.70 | E3 | |
| RFL_Contig6050_941 | 6B | 481.47-481.86 | 3.10E-06-1.63E-04 | 8.83-11.63 | E1/E3 | |
| wsnp_Ex_c3640_6644345 | 6B | 485.54 | 2.36E-05-1.08E-04 | 8.21-11.04 | E1/E3 | IWB2831[24] |
| wsnp_Ex_c14101_22012676 | 6B | 492.02 | 2.26E-06-6.77E-04 | 5.86-11.61 | E1/E2/E3 | |
| wsnp_Ex_c6143_10747643 | 6B | 516.08-519.15 | 1.30E-05-1.65E-04 | 8.30-10.65 | E1/E3 | |
| Kukri_c16568_287 | 6B | 522.49 | 4.81E-05-1.69E-04 | 7.21-8.45 | E1/E3 | |
| Excalibur_c7785_123 | 6B | 526.48-528.92 | 1.19E-056.21E-04 | 5.94-10.02 | E1/E3 | |
| Excalibur_c11245_880 | 6B | 530.57 | 2.79E-04-5.34E-04 | 6.06-6.83 | E1/E3 | |
| wsnp_Ex_c3990_7223090 | 6B | 577.48 | 1.67E-05-6.19E-05 | 8.45-9.95 | E1/E3 | Qphs.ahau-6B[36] |
| RAC875_c57261_265 | 6B | 609.38 | 6.10E-04-6.34E-04 | 5.99-6.02 | E1 | |
| 标记 Marker | 染色体 Chr. | 位置 Position (Mb) | MLM | 环境 Environment | 已报道的QTL/标记/基因Known QTL/marker/gene | |
| P值 P-value | 贡献率 R2 (%) | |||||
| RAC875_rep_c117796_352 | 6B | 632.97 | 5.23E-04-5.35E-04 | 6.19-6.21 | E3 | |
| wsnp_CV776265A_Ta_2_1 | 6B | 651.01-652.00 | 1.21E-04-7.21E-04 | 5.78-7.51 | E1/E3 | |
| BS00023032_51 | 6B | 664.38 | 8.28E-04 | 5.90 | E3 | |
| IAE36519 | 6B | 701.98-704.04 | 7.44E-04-9.92E-04 | 5.87-6.09 | E2 | |
| wsnp_Ku_c18780_28136150 | 7B | 223.61 | 8.12E-05 | 10.12 | E2 | |
| Excalibur_c25719_238 | 7B | 363.24 | 3.92E-05-3.24E-04 | 6.64-8.89 | E2/E3 | |
| RAC875_c4834_694 | 7B | 613.38 | 7.14E-04 | 5.87 | E2 | Dorm-1[37] |
| BobWhite_c8092_726 | 5D | 246.30 | 4.94E-04 | 6.22 | E3 | |
| wsnp_Ex_c1249_2399894 | 6D | 67.39-68.10 | 8.81E-04-9.23E-04 | 5.72-5.78 | E1 | |
| D_wsnpbe403818_Contig1_1 | 6D | 389.70 | 6.71E-04 | 5.79 | E3 | |
| D_contig10382_335 | 7D | 23.50-55.42 | 7.24E-04-9.41E-04 | 5.70-6.15 | E2 | MST101[37] |
新窗口打开|下载CSV
2.5 小麦穗发芽候选基因功能预测
将表型效应值大且能稳定遗传的SNP标记在普通小麦中国春基因组数据库中进行检索, 并在NCBI数据库中进行BLASTx序列比对, 共挖掘了13个最有可能与穗发芽相关的候选基因(表4)。穗发芽候选基因主要与种子的休眠、植物激素的生物合成及信号转导相关。其中, 定位于3A、6B、6D和7B染色体上的基因TraesCS3A01G589400LC、TraesCS6B01G138600/TraesCS6B01G516700LC/ TraesCS6B01G548900LC、TraesCS6D01G103600及TraesCS7B01G200100均编码锌指结构蛋白; 定位于3B、6A和6B染色体上的基因TraesCS3B01G415 900LC、TraesCS6A01G144700LC及TraesCS6B01G2 94800均编码F-box家族蛋白; 定位于6B染色体上的基因TraesCS6B01G511400LC功能注释为厌氧型一氧化氮还原酶转录调控因子。Table 4
表4
表4候选基因信息
Table 4
| 位点 Marker | 染色体 Chr. | 物理位置 Position (Mb) | 基因 Gene | 基因注释或编码蛋白 Gene annotation or coding protein |
|---|---|---|---|---|
| Kukri_c8465_54 | 3A | 684.76 | TraesCS3A01G589400LC | Zinc finger MYM-type-like protein |
| IAAV8536 | 3B | 421.53 | TraesCS3B01G415900LC | F-box protein |
| RAC875_rep_c107892_142 | 6A | 77.61 | TraesCS6A01G144700LC | F-box family protein |
| 6A | 77.54 | TraesCS6A01G108800 | Myb-like transcription factor family protein | |
| GENE-3171_203 | 6B | 135.71 | TraesCS6B01G138200 | Myb family transcription factor-like |
| 6B | 135.94 | TraesCS6B01G138600 | RING/U-box superfamily protein | |
| RFL_Contig6050_941 | 6B | 481.79 | TraesCS6B01G511400LC | Anaerobic nitric oxide reductase transcription regulator NorR |
| wsnp_Ex_c14101_22012676 | 6B | 491.55 | TraesCS6B01G516700LC | Zinc finger (C3HC4-type RING finger) family protein |
| Excalibur_c7785_123 | 6B | 528.92 | TraesCS6B01G294800 | F-box protein |
| 6B | 527.93 | TraesCS6B01G293700 | Myb family transcription factor-like | |
| 6B | 526.49 | TraesCS6B01G548900LC | Zinc finger MYM-type-like protein | |
| Excalibur_c25719_238 | 7B | 363.00 | TraesCS7B01G200100 | Zinc finger C-x8-C-x5-C-x3-H type family protein |
| wsnp_Ex_c1249_2399894 | 6D | 67.39 | TraesCS6D01G103600 | Zinc finger BED domain-containing protein DAYSLEEPER |
新窗口打开|下载CSV
3 讨论
3.1 小麦穗发芽性状的分析
小麦穗发芽的抗性机制较为复杂, 主要受籽粒休眠性、穗部形态、种皮颜色及环境等因素的影响[38,39]。朱冬梅等[40]对33份长江中下游麦区主推品种的穗发芽抗性进行评价, 结果表明, 品种的休眠特性与其穗发芽抗性呈正相关。苗西磊等[41]利用相关的分子标记并结合整穗发芽实验, 证实穗部形态如麦芒、颖壳及其抑制物对小麦穗发芽具有抑制作用。Warner等[42]的研究指出, 种皮颜色与小麦穗发芽抗性呈正相关。在众多的环境因素中, 温度和水分是影响穗发芽的主要因素。温度在26℃以上时, 籽粒的休眠程度会降低或丧失; 在15℃以下时, 籽粒的休眠程度加深[43,44]; 20℃时, 小麦的发芽率最高, 温度继续升高, 发芽率降低[45]。从穗发芽的定义可以看出, 水分是导致小麦穗发芽的直接外界因素。本研究利用整穗发芽法在人工气候培养箱(温度20℃, 相对湿度为80%)进行发芽实验, 不仅能反映出各品种(系)间穗发芽受种子本身的休眠特性及穗部形态等的影响, 还能体现出环境等因素对其抗性的作用, 能较好的反映出各小麦品种(系)间的综合抗性。研究结果显示, 在同一年份及不同年份间, 整穗发芽率的变异幅度较大(12.98%~100.00%和13.84%~100.00%), 且平均值均高于标准差, 表明不同小麦品种(系)可能有着不同的抗性因子, 抗性材料间的抗性因子也不同, 因而表现出了丰富的表型变异。与2018年相比, 207份小麦品种(系)在2019年整穗发芽率的变异范围较小, 平均值较大, 究其原因可能是受小麦成熟期间的温度及收获前降雨的影响, 导致供试品种(系)整体的穗发芽抗性水平下降。此外, 本研究在2个环境下的方差分析与朱玉磊等[13]利用264份供试材料在3个环境下分析整穗发芽率的结果一致, 表明小麦穗发芽抗性既受基因型调控, 也易受环境因素的影响, 同时基因与环境互作效应对其影响显著, 这也是导致不同年份间的相关系数较低的原因。3.2 小麦穗发芽关联分析
随着分子生物学及生物信息学的迅速发展, GWAS分析与QTL定位已成为研究植物数量性状的重要途径, 小麦穗发芽相关基因的挖掘也得到了较大程度的推动。Munkvold等[14]、Osa等[15]、Lin等[17]利用不同的RIL群体结合基因芯片对小麦穗发芽进行连锁分析, 定位了2AL、2BS、3AS、3AL、4AL、5D、6B及7D上的主效QTL位点, 其中3A、5D、6B及7D位点与本研究相同, 验证了这些位点的重要性, 同时也进一步证明了关联分析与连锁分析均能检测到与目标性状显著相关的QTL位点。将GWAS检测结果与前人定位的结果相比较, 发现一些已报道的基因/标记位点/QTL在本研究中被重复定位到, 如本研究定位于3A染色体上的显著性位点Kukri_c8465_54, 根据染色体上位置的比对结果发现, 该位点与Bailey等[46]报道的调控种胚休眠性的TaVp-1基因距离较近。定位于4B染色体上的标记位点Tdurum_contig60051_838 (644.68 Mb)与Rasul等[34]定位在4B染色体上的标记位点Xwmc349 (640.9 Mb)相距约4 Mb, 可能为同一位点。Zhu等[24]定位了3个与穗发芽抗性相关的QTL, 且6B位点是携带穗发芽抗性基因的热点区域, 这一结果与本研究一致。同时, 本研究发现定位在6B染色体上的标记wsnp_Ex_c3990_7223090 (577.48 Mb)与Zhu等[24]定位的Qphs.ahau-6B (574.6~576.8 Mb)物理位置相近, 可以推测这一区段内可能存在与穗发芽抗性相关的基因。但本研究鉴定结果中有24个显著性位点与相同染色体上已报道的位点在物理位置上相距较远, 如7B染色体上的wsnp_Ku_c18780_28136150 (223.61 Mb)与王焕雪等[47]定位在7B染色体上的AX-109349257 (718.5~720.0 Mb)相距约500 Mb。这些小麦穗发芽相关位点可能均为新位点。此外, 本研究检测到在E1、E2及平均环境下被共同检测到, 物理位置为492.02 Mb, 对表型解释效应为5.86%~11.61%。该位点可能是与小麦穗发芽抗性相关的重要位点。对其进行候选基因的挖掘, 筛选到的候选基因TraesCS6B01G516700LC, 其基因功能注释为锌指(C3HC4型指环)家族蛋白, 该蛋白的功能是通过调控植物内源激素-脱落酸(abscisic acid, ABA)的灵敏性进而影响种子的休眠。今后将着手开发多种标记(EST、EST-SSR、CAPS等)用于加密标记图谱, 同时构建适当的作图群体, 以准确定位这些关联位点, 并对效应值大且稳定遗传的候选基因进行图位克隆、功能验证等, 以期进一步剖析小麦穗发芽的遗传机制。
3.3 候选基因的功能分析
本研究利用GWAS检测到34个与小麦穗发芽显著关联的SNP标记位点, 并在普通小麦中国春基因组数据库筛选获得13个可能与穗发芽相关的候选基因。位于3A、6B、6D和7B上的基因TraesCS3蛋白, 该蛋白的功能是通过调控植物内源激素-脱落酸(abscisic acid, ABA)的灵敏性对种子的休眠进行调控[48]; 位于3B、6A和6B上的基因TraesCS3B01 G415900LC、TraesCS6A01G144700LC及TraesCS6B 01G294800编码F-box家族蛋白, 该家族蛋白在植物激素的信号转导, 光信号转导以及花器官发育等生理过程有重要作用[49]; 6B染色体上的基因TraesCS6B01G511400LC注释为厌氧型一氧化氮还原酶转录调控因子, 一氧化氮对植物种子的休眠解除和萌发有促进作用[50]; 位于6A和6B染色体上的基因TraesCS6A01G108800、TraesCS6B01G13 8200/TraesCS6B01G293700编码Myb转录因子家族蛋白, 该蛋白调控种子中类黄酮的生物合成, 对籽粒颜色有重要影响[48]。Himi等[51]证明R基因能够促进类黄酮基因的转录, 但在小麦白皮品种中并不表达, 红皮小麦因受控制种皮颜色的三重R基因的调控导致其穗发芽抗性较强, R基因与休眠位点紧密连锁, 在对应区间检测到Myb家族转录因子, 并成功开发相关标记。这对本研究在6A和6B染色上检测到Myb转录因子区段具有借鉴作用, 但能否据此开发有效的功能标记, 是我们下一步工作的重点。4 结论
本研究采用Q+K混合线性模型, 使用90K SNP芯片对207份国内外小麦品种(系)的整穗发芽率进行全基因组关联分析, 获得34个显著关联的SNP标记位点, 其中6B是携带穗发芽抗性基因的热点区域, 定位于6B染色体上的wsnp_Ex_c14101_22012676在E1、E2及平均环境下被共同检测到, 对表型的贡献率为5.86%~11.61%, 该位点可能是与小麦穗发芽抗性相关的重要位点。将表型效应值大且能稳定遗传的SNP标记在普通小麦中国春基因组数据库中进行检索, 共挖掘了13个最有可能与穗发芽抗性相关的候选基因。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 5]
[本文引用: 5]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]
URL [本文引用: 2]
DOIPMID [本文引用: 3]

The premature germination of seeds before harvest, known as preharvest sprouting (PHS), is a serious problem in all wheat growing regions of the world. In order to determine genetic control of PHS resistance in white wheat from the relatively uncharacterized North American germplasm, a doubled haploid population consisting of 209 lines from a cross between the PHS resistant variety Cayuga and the PHS susceptible variety Caledonia was used for QTL mapping. A total of 16 environments were used to detect 15 different PHS QTL including a major QTL, QPhs.cnl-2B.1, that was significant in all environments tested and explained from 5 to 31% of the trait variation in a given environment. Three other QTL QPhs.cnl-2D.1, QPhs.cnl-3D.1, and QPhs.cnl-6D.1 were detected in six, four, and ten environments, respectively. The potentially related traits of heading date (HD), plant height (HT), seed dormancy (DOR), and rate of germination (ROG) were also recorded in a limited number of environments. HD was found to be significantly negatively correlated with PHS score in most environments, likely due to a major HD QTL, QHd.cnl-2B.1, found to be tightly linked to the PHS QTL QPhs.cnl-2B.1. Using greenhouse grown material no overlap was found between seed dormancy and the four most consistent PHS QTL, suggesting that greenhouse environments are not representative of field environments. This study provides valuable information for marker-assisted breeding for PHS resistance, future haplotyping studies, and research into seed dormancy.
[本文引用: 2]
DOIURL [本文引用: 1]
[本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

Association mapping identified quantitative trait loci (QTLs) and the markers linked to pre-harvest sprouting (PHS) resistance in an elite association mapping panel of white winter wheat comprising 198 genotypes. A total of 1,166 marker loci including DArT and SSR markers representing all 21 chromosomes of wheat were used in the analysis. General and mixed linear models were used to analyze PHS data collected over 4 years. Association analysis identified eight QTLs linked with 13 markers mapped on seven chromosomes. A QTL was detected on each arm of chromosome 2B and one each on chromosome arms 1BS, 2DS, 4AL, 6DL, 7BS and 7DS. All except the QTL on 7BS are located in a location similar to previous reports and, if verified, the QTL on 7BS is likely to be novel. Principal components and the kinship matrix were used to account for the presence of population structure but had only a minor effect on the results. Although, none of the QTLs was highly significant across all environments, a QTL on the long arm of chromosome 4A was detected in three different environments and also using the best linear unbiased predictions over years. Although previous reports have identified this as a major QTL, its effects were minor in our biparental mapping populations. The results of this study highlight the benefits of association mapping and the value of using elite material in association mapping for plant breeding programs.
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 5]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

In many wheat ( Triticum aestivumL.) growing areas, pre-harvest sprouting (PHS) may cause important damage, and in particular, it has deleterious effects on bread-making quality. The relationship between PHS and grain color is well known and could be due either to the pleiotropic effect of genes controlling red-testa pigmentation ( R) or to linkage between these genes and other genes affecting PHS. In the present work, we have studied a population of 194 recombinant inbred lines from the cross between two cultivars, 'Renan' and 'Récital', in order to detect QTLs for both PHS resistance and grain color. The variety 'Renan' has red kernels and is resistant to PHS, while 'Récital' has white grain and is highly susceptible to PHS. A molecular-marker linkage map of this cross was constructed using SSRs, RFLPs and AFLPs. The population was evaluated over 2 years at Clermont-Ferrand (France). PHS was evaluated on mature spikes under controlled conditions and red-grain color was measured using a chromameter. Over the 2 years, we detected four QTLs for PHS, all of them being co-localized with QTLs for grain color. Three of them were located on the long arm of chromosomes 3 A, 3B and 3D, close to the loci where the genes R and taVp1 were previously mapped. For these three QTLs, the resistance to PHS is due to the allele of the variety 'Renan'. Another co-located QTL for PHS and grain color was detected on the short arm of chromosome 5 A. The resistance for PHS for this QTL is due to the allele of 'Récital'.
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]
URL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
