 ,, 刘朝显, 陈秋栏, 王文琴, 姚顺, 赵子堃, 朱思颖, 洪祥德, 熊雨涵, 蔡一林
,, 刘朝显, 陈秋栏, 王文琴, 姚顺, 赵子堃, 朱思颖, 洪祥德, 熊雨涵, 蔡一林 ,*西南大学玉米研究所 / 农业科学研究院 / 南方山地作物逆境生物学国家级培育基地, 重庆400715
,*西南大学玉米研究所 / 农业科学研究院 / 南方山地作物逆境生物学国家级培育基地, 重庆400715Fine mapping and candidate gene analysis of maize defective kernel mutant dek54
ZHOU Lian ,, LIU Chao-Xian, CHEN Qiu-Lan, WANG Wen-Qin, YAO Shun, ZHAO Zi-Kun, ZHU Si-Ying, HONG Xiang-De, XIONG Yu-Han, CAI Yi-Lin
,, LIU Chao-Xian, CHEN Qiu-Lan, WANG Wen-Qin, YAO Shun, ZHAO Zi-Kun, ZHU Si-Ying, HONG Xiang-De, XIONG Yu-Han, CAI Yi-Lin ,*Maize Research Institute / Academy of Agricultural Sciences / State Cultivation Base of Crop Stress Biology for Southern Mountainous Land, Southwest University, Chongqing 400715, China
,*Maize Research Institute / Academy of Agricultural Sciences / State Cultivation Base of Crop Stress Biology for Southern Mountainous Land, Southwest University, Chongqing 400715, China通讯作者: *蔡一林, E-mail:caiyilin1789@163.com
收稿日期:2020-10-15接受日期:2021-01-13网络出版日期:2021-03-02
| 基金资助: |
Corresponding authors: *E-mail:caiyilin1789@163.com
Received:2020-10-15Accepted:2021-01-13Published online:2021-03-02
| Fund supported: |
作者简介 About authors
E-mail:zhoulianjojo@swu.edu.cn

摘要
玉米籽粒与产量和营养品质密切相关, 控制籽粒发育基因的功能研究对解析籽粒发育分子机制, 提高玉米产量, 改善籽粒营养品质提供重要依据。利用甲基磺酸乙酯(ethyl methanesulfonate, EMS)处理B73花粉, 筛选到一个玉米籽粒缺陷突变体defective kernel 54 (dek54)。dek54表现出成熟籽粒变小、皱缩、颜色发白等特征; 遗传分析表明dek54是一个单基因控制的隐性突变体。石蜡切片显示dek54淀粉胚乳细胞形状不规则且排列致密, 扫描电镜观察成熟籽粒胚乳中心区域发现dek54淀粉粒周围蛋白体比野生型少且排列疏松。dek54成熟籽粒的总蛋白、醇溶蛋白、各氨基酸组分和全氮含量相比野生型都显著降低。利用F2分离群体中的1566个dek54单株, 把dek54定位在7号染色体标记SSR6和SSR7之间, 物理位置约为290 kb。该区间有3个基因, 基因测序发现Zm00001d019294基因第2个外显子上第351个碱基由G突变为A, 从而导致蛋白翻译的提前终止。该基因在玉米籽粒中特异性表达, 且在12 DAP (days after pollination)籽粒中表达量最高。通过CRISPR/Cas9系统进行靶向突变确定候选基因Zm00001d019294导致该突变表型。Dek54编码一个与ZmNRT1.5 (nitrate transporter)具有较高同源性的MFS (major facilitator superfamily)家族蛋白并定位在玉米原生质体的细胞质膜。该研究为揭示dek54在玉米籽粒发育的分子机制奠定了重要基础。
关键词:
Abstract
Maize kernel is closely related to yield and nutritive quality. Study on the function of maize kernel development relative genes provides important basis for the molecular mechanism analysis, yield increasing and nutritive quality improving. B73 pollen was treated with ethyl methylmethanesulfonate (EMS) and a defective maize kernel defective kernel 54 (dek54) was screened. dek54 had small mature kernel, wrinkled and whitened seed coat phenotype. Genetic analysis indicated that dek54 is a recessive mutant controlled by a single gene. Paraffin sections showed starchy endosperm cells of dek54 had irregular shape and dense arrangement at developmental stage. Scanning electron microscopy observation indicated that protein bodies around starch granules in the central region of the dek54 mature kernel endosperm were fewer and arranged more loosely compare to wild type. Total protein, zein, amino acids components contents and total nitrogen content of dek54 mature kernel were significantly lowered compared with the wild type. dek54 was located on chromosome 7 within the interval of the physical distance of about 290 kb between markers SSR6 and SSR7. Sequencing revealed that the 351th base G on the 2nd exon of Zm00001d019294 gene changed into A, which led to the premature termination of the protein translation. Zm00001d019294 gene was specifically expressed in immature maize kernel, and has the highest expression in 12 DAP (days after pollination) immature kernel. Targeted mutation was performed using CRISPR/Cas9 system to identify that mutant phenotype was caused by candidate gene Zm00001d019294. Dek54 encoded an MFS (major facilitator superfamily) protein and had high homology with ZmNRT1.5 (nitrate transporter). Besides, Dek54 protein was localized in the plasma membrane of maize protoplasts. The study of dek54 laid the foundation for the molecular mechanism analysis of maize kernel development.
Keywords:
PDF (3857KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
周练, 刘朝显, 陈秋栏, 王文琴, 姚顺, 赵子堃, 朱思颖, 洪祥德, 熊雨涵, 蔡一林. 玉米籽粒缺陷突变基因dek54的精细定位及候选基因分析[J]. 作物学报, 2021, 47(10): 1903-1912 DOI:10.3724/SP.J.1006.2021.03060
ZHOU Lian, LIU Chao-Xian, CHEN Qiu-Lan, WANG Wen-Qin, YAO Shun, ZHAO Zi-Kun, ZHU Si-Ying, HONG Xiang-De, XIONG Yu-Han, CAI Yi-Lin.
玉米(Zea mays L.)作为一年生禾本科植物, 是我国重要的粮食作物。玉米生产对保障我国粮食安全有着重要的战略意义。随着世界人口和经济水平的不断增长, 对玉米产量和营养品质的要求日益增长。玉米的胚和胚乳作为籽粒的主要组成部分, 来自植物双受精作用, 与玉米产量和营养品质密切相关。玉米籽粒主要是由淀粉、蛋白质和油脂组成。其中淀粉和蛋白质主要储存在胚乳中, 油脂主要存在胚中。玉米胚乳由4种不同类型的细胞组成, 包括淀粉胚乳细胞、基部胚乳转移层细胞、糊粉层细胞和胚包围层细胞[1,2]。8~14 DAP胚乳细胞具有很强的有丝分裂活性, 到20~25 DAP时有丝分裂细胞只在包含糊粉层和亚糊粉层的外周细胞层增殖。淀粉胚乳细胞在胚乳的中央区域, 10 DAP淀粉胚乳细胞从有丝分裂转变为核内复制[1]。淀粉胚乳细胞储存籽粒中大部分的淀粉和蛋白质, 有效地决定籽粒质地和营养品质。玉米籽粒胚乳的外侧是蛋白体含量较多的透明区域, 而中心则是由较多淀粉粒和少量蛋白体组成的不透明区域。淀粉粒和蛋白体之间的比例决定着玉米籽粒的硬度, 也是玉米品质的重要影响因素。玉米胚乳蛋白的质量也会影响玉米营养品质和病虫害抗性。胚乳蛋白中醇溶蛋白(zein)的含量占籽粒总蛋白量的70%左右, 但醇溶蛋白中人体必需的赖氨酸含量低, 极大的降低了玉米的营养价值。因此, 醇溶蛋白和非醇溶蛋白的比例也是决定玉米籽粒品质的重要因素之一。
随着玉米全基因组测序的完成, 玉米籽粒发育及产量相关的基因功能研究逐渐成为热点。玉米具有大量的籽粒突变体, 这些突变体籽粒表型表现异常。根据遗传学分析, 可以分为3种不同的类型, 包括: (1) 单基因隐性突变体: defective kernel突变体dek1[3,4,5,6]、dek2[7]、dek15[8]、dek36[9]、dek37[10]、dek39[11]和dek44[12]等, dek突变体影响籽粒胚和胚乳的发育, 使籽粒产生严重缺陷。例如dek44突变体产生小粒的胚致死表型, Dek44基因编码一种线粒体核糖体蛋白L9, 调节细胞生长和籽粒发育[12]。opaque突变体o1[13]、o2[14,15]、o5[16]、o7[17]、o9-11[18]和o13-17[19]等, opaque突变体籽粒粉质化, 通过改变醇溶蛋白含量从而影响胚乳质地和蛋白品质。例如o7突变体基因编码蛋白影响19-kD和22-kD α-zein以及16-kD γ-zein的合成, 从而引起蛋白体数量减少和变小, 出现粉质胚乳表型[17]。(2) 显性突变体Mucuronate (Mc)[20]和Defective endosperm (De-B30)[21], DeB30突变体籽粒的非醇溶蛋白比正常籽粒增加了约70%。(3) 半显性floury突变体f1[22]和f2[23], floury突变体也会引起胚乳粉质化。例如fl2由于单个氨基酸的突变使得22 kD α-zein的信号肽不能正常剪切从而导致蛋白体异常[23]。由于玉米基因组大, 且转座子元件较多, 相比水稻等主要粮食作物, 仍有大量籽粒突变基因的功能及分子机制仍然不清楚。
本研究利用EMS处理玉米自交系B73花粉得到一个籽粒缺陷突变体dek54 (根据已发表的dek突变体顺序命名)。本研究观察了dek54籽粒形态学和组织学结构, 分析了dek54籽粒相关生化成分; 通过与Mo17构建的F2分离群体对dek54进行了基因定位, 并通过CRISPR/Cas9系统靶向突变确定了候选基因Dek54; 分析了Dek54基因在不同组织的时空表达情况及其编码蛋白结构和同源性比对, 明确了Dek54蛋白的细胞质膜定位结果。该研究解析Dek54在玉米籽粒发育中的分子机制奠定了重要基础。
1 材料与方法
1.1 dek54突变体获得和分离群体构建
2015年春, 于重庆北碚田间种植玉米自交系B73和Mo17, 收集B73花粉进行EMS (终浓度0.67 mL L-1)诱变处理; 将诱变处理后的花粉与Mo17植株进行杂交。同年下半年, 把获得的M0代籽粒种植于云南元江, 自交后收获果穗, 观察表型, 筛选出了籽粒皱缩突变体dek54。继续用dek54为母本与Mo17杂交, 构建F2分离群体。提取1566个dek54植株DNA, 鉴定其基因型, 筛选重组单株, 对dek54进行精细定位。1.2 石蜡切片和扫描电镜观察
分别取16 DAP果穗上的野生型和dek54未成熟籽粒, 在福尔马林-乙酸-乙醇固定液(FAA, 50%乙醇, 0.9 mmol L-1冰醋酸, 3.7%甲醛)中4℃固定过夜。将样品放置在一系列乙醇和二甲苯梯度溶液中脱水, 在石蜡中进行包埋。将石蜡样本切成10 μm的厚度,经烤片并脱水后, 用1%番红和固绿(Amresco)染色。荧光显微镜下(Leica, D3000)进行观察。分别取F2果穗上野生型和dek54的成熟籽粒, 经临界干燥、断面喷金后, 在扫描电镜下(Hitachi, SU3500)观察dek54与野生型的籽粒相同部位胚乳形态结构差异。1.3 籽粒蛋白、淀粉、氨基酸和全氮含量测定
分别取F2群体中同一个果穗上野生型和dek54的成熟籽粒, 用水浸泡5 min, 并去除种皮和胚。将剩余胚乳部分在液氮中研磨成细粉。将真空抽干粉末各称取50 mg两份, 加入1 mL石油醚, 置于37℃摇床1 h; 去除石油醚, 加入硼酸钠蛋白提取液和巯基乙醇, 37℃摇床过夜。分别提取每个样品的总蛋白、醇溶蛋白和非醇溶蛋白, 并进行蛋白含量测定[17]。总淀粉含量的测量按照总淀粉测定试剂盒(Megazyme, K-TSTA-100A)使用说明进行。将样品进行蛋白质水解, 去除脂质、色素等, 使用全自动氨基酸分析仪(Hitachi, L-8900)测定氨基酸含量, 每个样品3次重复。将样品用浓硫酸/双氧水进行消煮, 使用凯氏定氮仪(Alva, KN580)测定全氮含量。1.4 dek54连锁标记筛选及精细定位
在F2群体中分别选择10株野生型和10株突变体, 提取基因组DNA, 并等量混合, 构建突变体DNA池(mutant DNA pool, MP)和野生型DNA池(wild-type DNA pool, WP)。利用实验室已有的覆盖玉米全基因组, 在B73和Mo17之间有多态性的432个SSR标记对这2个DNA进行扩增。筛选在2个DNA池之间具有多态性的分子标记。通过Gramene网站(1.5 qRT-PCR
取B73根、茎、叶、未成熟雌穗(1~2 cm)和雄穗(1~2 cm)及12、14、16、18、20 DAP的未成熟籽粒和成熟籽粒, 迅速冻于液氮。提取总RNA, 进行反转录生成cDNA[25]。qRT-PCR (quantitative reverse transcription- PCR)中利用玉米ZmACTIN1基因作为内参, 样品间同一基因的相对表达量通过下例公式计算: 2-ΔΔCt。1.6 载体构建及玉米遗传转化
通过对候选基因Zm00001d019294 gDNA序列进行分析, 选择第2个外显子的19 bp (5'-GCCGAG CTACCTCTGCGAC-3')的靶向序列。合成包括靶向序列和sgRNA在内的寡核苷酸引物, 采用单编辑策略将该序列克隆到pCAMBIA3301-Cas9载体, 并将构建好的载体转入农杆菌EHA105菌株。通过农杆菌介导的玉米未成熟胚遗传转化, 获得玉米转基因植株。通过测序来确定CRISPR/Cas9编辑玉米植株目标位置附近的编辑位点, 获得5个独立的CRISPR/ Cas9基因敲除转基因株系, 选择2个转基因株系进行后续实验。1.7 基因注释及系统进化分析
通过NCBI(1.8 亚细胞定位
以玉米B73的12 DAP未成熟籽粒cDNA为模板, 通过PCR扩增Dek54的全长CDS序列(除去终止密码子)连接到pAN580载体, 构建Dek54-GFP融合表达载体。将含有正确插入片段的载体和质膜定位标记载体PM107 (AtPIP1; 2-mCherry)分别通过PEG介导共转化玉米原生质体。在激光共聚焦显微镜(LSM800, Carl Zeiss)下观察荧光。2 结果与分析
2.1 dek54表型鉴定与遗传分析
利用EMS化学诱变技术获得了玉米籽粒缺陷突变体dek54。该突变体与Mo17杂交, F1群体表型表现正常, 自交后F2果穗上产生分离(图1-A)。统计5个F2果穗, 野生型(+/+和dek54/+)与突变型(dek54/ dek54)籽粒分别有1549粒和499粒, 适合度测验结果显示, 野生型和突变体籽粒的分离比接近3∶1 (χ2 = 0.16 < χ20.05 = 3.84), 说明dek54是一个单基因控制的隐性突变体。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1dek54突变体表型鉴定
A: 成熟果穗F2分离群体; B: 籽粒表型; C: 籽粒纵切; D: 籽粒横切; E: F2分离群体上野生型和dek54突变体籽粒的百粒重、十粒长和十粒宽。标尺为0.5 cm。Em: 胚; En: 胚乳; SE: 粉质胚乳; VE: 玻璃质胚乳。
Fig. 1Phenotypic characterization of dek54 mutant
A: F2 segregating population of mature ear; B: phenotypes of kernels; C: cross section of kernels; D: longitudinally section of kernels; E: 100-kernel weight, ten-kernel length, and ten-kernel width of WT and dek54 mutant kernels from F2 segregating population. Bar: 0.5 cm. Em: embryo; En: endosperm; SE: starchy endosperm; VE: vitreous endosperm.
dek54与野生型相比表现为, 成熟籽粒变小, 皱缩干瘪, 且颜色发白(图1-A, B)。对野生型和dek54成熟籽粒进行徒手切片并在体式显微镜下观察籽粒胚和胚乳的形态。籽粒纵切面显示, 突变体胚的形状正常, 但显著小于野生型(图1-C), 突变体籽粒较野生型萌发延迟但后续生长正常(附图1)。籽粒横切面显示, 突变体籽粒胚乳中间不透明的淀粉胚乳面积缩小, 分布在粉质胚乳周围的透明玻璃质胚乳面积显著增大(图1-D)。野生型和dek54的百粒重分别为31.2 g和10.0 g, dek54仅为野生型的32.0%; dek54十粒长和十粒宽分别为野生型的79.0%和66.7%, 均显著低于野生型(图1-E)。由此可见, dek54突变显著影响了籽粒胚和胚乳的发育。
附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1野生型和dek54 成熟籽粒的发芽测试(发芽后5 d)
标尺为1 cm。
Fig. S1Germination test of wildtype and dek54 mature kernels (5 days after germination)
Bar: 1 cm.
2.2 dek54籽粒组织学观察
取16 DAP的野生型和突变体籽粒进行石蜡切片观察。结果显示, dek54的种皮与胚乳脱离, 胚乳细胞相比野生型发生了明显变化, 表现在糊粉层细胞相比野生型层数增加, 并且淀粉胚乳细胞组织形态不规则并且相比野生型排列较密且较小(图2-A)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2dek54突变体的组织学观察
A: 16 DAP野生型和dek54突变体籽粒的石蜡切片纵切观察。标尺为100 µm。AL: 糊粉层; SE: 淀粉胚乳。B: 野生型和dek54突变体成熟籽粒胚乳中心区域扫描电镜观察。标尺为50 µm。SG: 淀粉粒; PB: 蛋白体。
Fig. 2Histological observatin of dek54 mutant
A: longitudinal paraffin sections observation of developing WT and dek54 mutant kernels at 16 DAP. Bar: 100 µm. AL: aleurone layer; SG: starch endosperm. B: scanning electron microscopy observation of the central regions of WT and dek54 mutant mature kernel endosperm. Bar: 50 µm. SG: starch granule; PB: protein body.
说明dek54突变显著地影响胚乳细胞的发育。推测由于胚乳细胞发育的不完全使得dek54突变体籽粒表现出皱缩的表型。对成熟籽粒横截面的扫描电镜观察显示, 相同位置的野生型胚乳中淀粉粒周围有密集的蛋白体包围, 而dek54突变体的淀粉周围蛋白体数量较少, 且排列较疏松(图2-B)。
2.3 dek54相关生化成分检测
分别提取野生型和dek54成熟籽粒胚乳中的总蛋白、醇溶蛋白和非醇溶蛋白。结果显示, 突变体的总蛋白和醇溶蛋白含量相比野生型分别降低了29.2%和27.6%, 而非醇溶蛋白没有显著差异(图3-A)。成熟籽粒胚乳中的淀粉含量测定发现, 突变体的胚乳淀粉含量与野生型没有显著差异(图3-B)。总氨基酸含量测定显示, 突变体籽粒胚乳中各类氨基酸含量相比野生型均显著降低, 其中赖氨酸含量仅为野生型的63.0% (图3-C)。进一步全氮含量测定结果表明, dek54突变体的成熟籽粒中全氮含量显著低于野生型, 降低了16.5% (图3-D)。图3
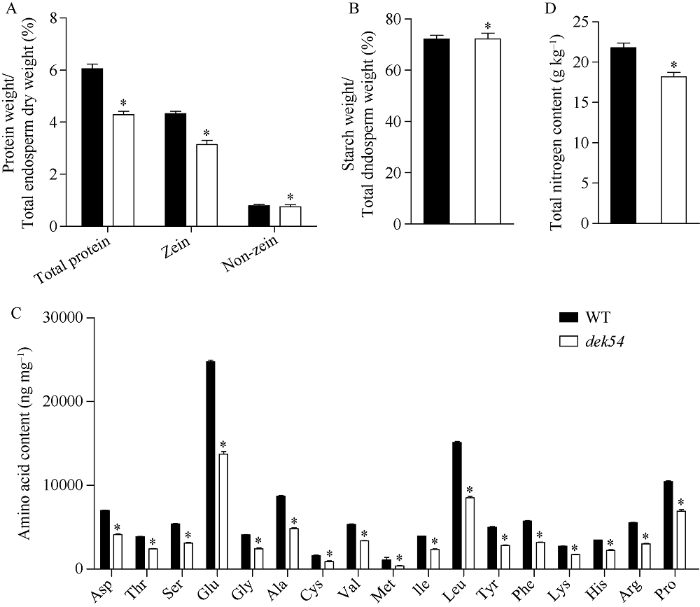 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3dek54突变体的生化成分分析
野生型和dek54突变体成熟籽粒的蛋白(A)、淀粉(B)、各氨基酸组分(C)和全氮含量(D)。
Fig. 3Biochemical component analysis of dek54 mutant
Protein (A), starch (B), amino acid components (C), and total nitrogen contents (D) of WT and dek54 mutant kernels.
2.4 dek54突变体的精细定位
利用432个在B73和Mo17基因组间有多态性的SSR标记对突变体DNA池和野生型DNA池进行筛选。发现SSR标记umc2160在2个DNA池中检Table 1
表1
表1dek54定位SSR标记引物序列
Table 1
| 标记类型 Type | 标记名称 Name | 正向引物序列 Forward sequence (5′-3′) | 反向引物序列 Reverse sequence (5′-3′) |
|---|---|---|---|
| SSR | SSR23403 | ACTAGTATGTGGATTGCTCGTCG | GCTGCTGAGCTGTATGTACCA |
| SSR | SSR1 | CCCTGACGAAAGCATGAATGAG | ACAGATGACTCTGCACCTCAAG |
| SSR | SSR2 | TGCCCATAAGAGCTTCGAGGATA | AGCCTTAGTTGTCAGTCCATCG |
| SSR | SSR3 | TGCCCATAAGAGCTTCGAGGATA | AGCCTTAGTTGTCAGTCCATCG |
| SSR | SSR4 | CCATCTGTACTAATGGCACCTGA | GCCAGCAAAGCTTTTCAAGAGT |
| SSR | SSR5 | GGCTACATAAGATGCAAAGCGG | TACCCTTTGACAGAGCCTACCT |
| SSR | SSR6 | GGTGCCAAAAACATCTCCCAAC | TAGCGTGGGGTCATAGCAACA |
| SSR | SSR7 | CATGGCCAAAATATCGCACGAG | TGACGTACATGAACACCTCGG |
| SSR | SSR8 | GCTAGGTGCAGTGTCTCTGCTT | CCTTGAACGTGGGGTAGGCT |
| SSR | SSR10308 | GCAAATGTTCTGTGCAAGGCTA | GCCCCACAAGAACTCCATCTAT |
新窗口打开|下载CSV
Table 2
表2
表2 dek54定位区间基因注释
Table 2
| 编号 No. | 基因ID Gene ID | 功能注释 Function annotation |
|---|---|---|
| 1 | Zm00001d019292 | 无注释 No annotation |
| 2 | Zm00001d019293 | 异柠檬酸/异丙基苹果酸脱氢酶 Isocitric acid/isopropyl malate dehydrogenase |
| 3 | Zm00001d019294 | MFS家族 Major facilitator superfamily |
新窗口打开|下载CSV
测出多态性(图4-A), 该标记位于第7号染色体的7.01 bin。使用umc2160检测33株dek54的基因型, 结果显示30株dek54的基因型与表型相一致, 3株(17、25和27) dek54因为染色体重组表现为杂合基因型(图4-B), 由此证实了该标记与dek54基因连锁。在umc2160标记邻近的区间开发新的SSR标记(表1)用于dek54定位。利用F2群体中1566个单株将dek54初步定位在与SSR标记SSR23403和SSR0308相邻的8.52 Mb的物理区间内(图4-C)。进一步通过重组植株的筛选, 开发并筛选到8个具有多态性的SSR标记。在SSR1~8标记位置各筛选到交换单株20、11、11、3、2、2、2和11株。最终, dek54定位在玉米7号染色体标记SSR6和SSR7之间物理距离约为290 kb的区间内(图4-C)。
图4
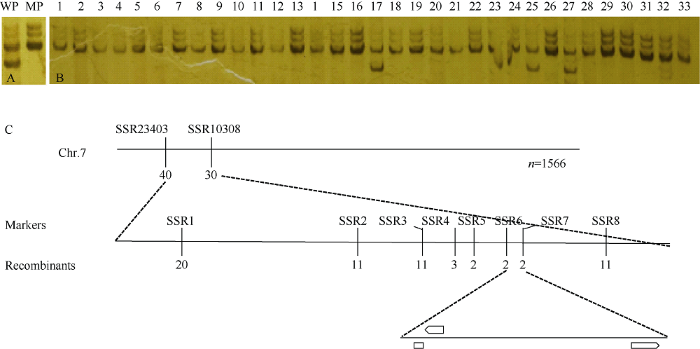 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4dek54的连锁SSR标记筛选和精细定位
A: umc2160的PCR扩增产物在WP和MP之间具有多态性; B: 通过33个dek54单株的基因分型确定SSR连锁标记umc2160; C: 通过F2群体1566个单株对dek54进行精细定位。dek54定位标记SSR6和SSR7之间物理距离约为290 kb的区间内。黑色垂直实线上方为分子标记名称, 下方数字为该标记鉴定的交换单株数量。
Fig. 4Linkage SSR marker screening and fine mapping of dek54
A: the polymorphic PCR products amplified by umc2160 between WP and MP; B: confirmation of linkage SSR marker umc2160 by genotyping 33 dek54 mutant plants; C: fine mapping of dek54 using F2 population including 1566 individuals. dek54 was mapped to an interval about 290 kb flanked by SSR6 and SSR7. The symbols above and below the black vertical solid lines represent the molecular marker and the number of recombinants, respectively.
2.5 候选基因序列及表达分析
Gramene玉米数据库显示SSR6和SSR7之间物理距离约为290 kb, 定位区间内仅包括3个基因(表2), 通过扩增并测序, 比较这3个基因在dek54与对照B73的DNA序列, 发现Zm00001d019294基因第2个外显子上第351个碱基位点由G突变转换为A, 导致密码子由TGG变为TAG, 从而造成蛋白翻译的提前终止(图5-A); 而其他2个基因的DNA序列没有差异。因此推测Zm00001d019294基因的突变是导致dek54表型产生的原因。图5
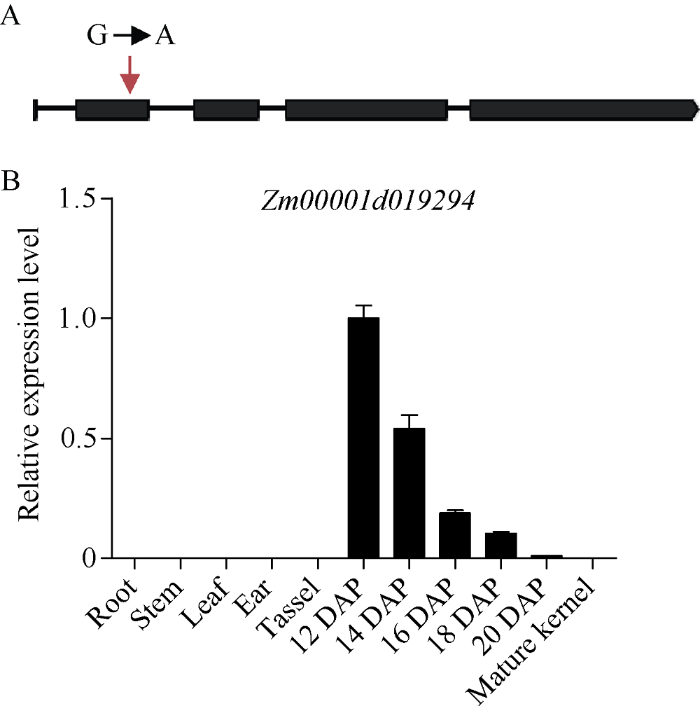 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5Zm00001d019294基因突变位点及时空表达分析
A: Zm00001d019294基因结构图, 红色箭头所示为突变位点; B: Zm00001d019294基因在不同组织及发育中的未成熟籽粒中的表达分析。
Fig. 5Mutation site and spatio-temporal expression analysis of Zm00001d019294
A: gene structure diagram of Zm00001d019294. The red arrow indicates mutation sites. B: the relative expression level of Zm00001d019294 in different tissues and developing immature kernel.
通过qRT-PCR的方法, 检测Zm00001d019294基因在玉米自交系B73的不同组织的表达情况, 包括根、茎、叶、雄穗、花丝、12、14、16、18、20 DAP的未成熟籽粒和成熟籽粒。检测表明, Zm00001d019294基因在未成熟籽粒(12~20 DAP)中特异性表达; 且在12 DAP未成熟籽粒中表达量最高, 并随着籽粒发育表达量逐渐下降, 在其他组织和成熟籽粒中均不能检测到其表达(图5-B)。Zm00001d019294基因在未成熟籽粒中的特异性表达说明其在籽粒发育过程中发挥作用。
2.6 CRISPR/Cas9靶向突变Zm00001d019294基因表型鉴定
利用CRISPR/Cas9系统对Zm00001d019294基因进行靶向突变。以Zm00001d019294的第2个外显子为靶标设计sgRNA间隔序列。获得了5个独立的CRISPR/Cas9转基因株系, 通过测序确定其中2个(dek-cas9-1和dek-cas9-2)含有由目标序列插入或缺失引起的密码子移位突变(图6-A)。CRISPR/Cas9转基因株系的成熟籽粒与dek54表现出相似的缺陷表型(图6-B~D)。该结果表明Zm00001d019294基因是引起该籽粒突变表型的目标基因。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6Zm00001d019294 CRISPR/Cas9靶向突变表型鉴定
A: Zm00001d019294的CRISPR/Cas9靶位点序列。sgRNA靶序列为绿色, PAM (protospacer-adjacent motif)序列为橙色, 红色字母和短线分别代表插入和缺失; B: dek54-cas9-1xMo17的F2成熟果穗; C: 成熟的WT和dek54-cas9-1籽粒; D: 成熟的WT和dek54-cas9-1籽粒的纵切。Em: 胚; En: 胚乳。标尺为0.5 cm。
Fig. 6Phenotypic characterization of CRISPR/Cas9 targeted mutation of Zm00001d019294
A: the sequence in the Zm00001d019294 locus targeted using CRISPR/Cas9. The sgRNA target sequence is green. Protospacer-adjacent motif (PAM) is blue. Red letters and dashes represent insertions and deletions, respectively. B: mature F2 ear of dek54-cas9-1xMo17. C: mature WT and dek54-cas9-1 kernels. D: longitudinal paraffin sections of mature WT and dek54-cas9-1 kernels. Em: embryo; En: endosperm. Bars: 0.5 cm.
2.7 Dek54蛋白预测及亚细胞定位
Dek54 (Zm00001d019294)由5个外显子和4个内含子组成。Dek54的成熟转录本编码序列长1923 bp, 编码一个由640个氨基酸组成的蛋白。通过BLAST CD-Search比对分析该蛋白的保守序列, 结果表明, Dek54蛋白序列中包含MFS保守结构域(图7-A)。蛋白序列比对及系统进化分析发现, Dek54蛋白与玉米和拟南芥硝酸盐转运体NRT1类蛋白具有较高的同源性, 且与ZmNRT1.5B和ZmNRT1.5A蛋白序列同源性最高(图7-B)。通过PEG介导的玉米原生质体共转化及荧光显微镜观察显示, Dek54蛋白的绿色荧光与质膜标记蛋白红色荧光完全重合(图8), 表明Dek54定位在细胞质膜上。推测Dek54可能是一个位于细胞质膜的硝酸盐转运体。图7
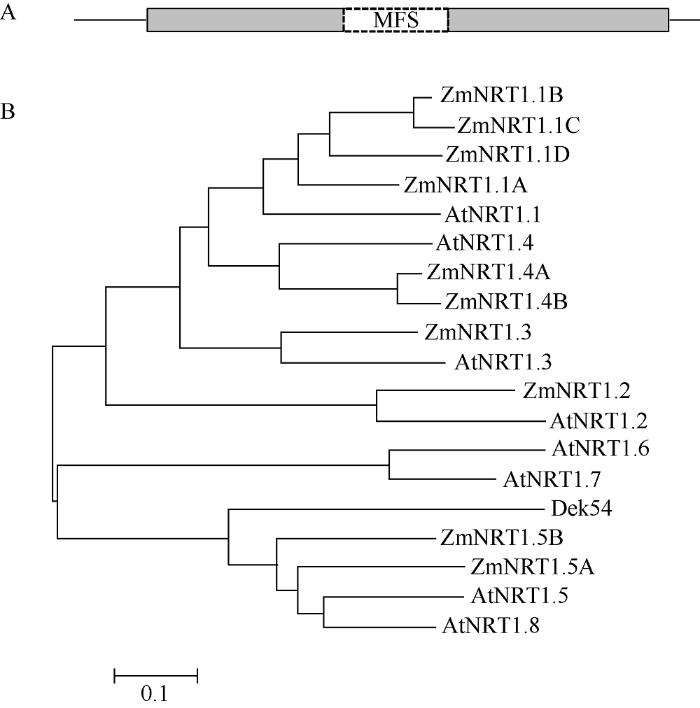 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7Dek54蛋白保守结构域(A)及进化(B)分析
Fig. 7Conserved domain (A) and phylogenetic (B) analysis of Dek54 protein
图8
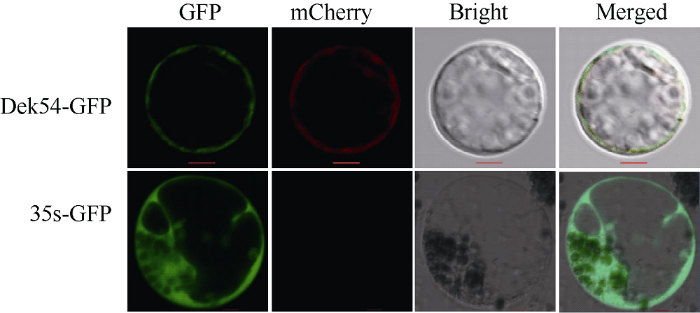 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8玉米原生质体中Dek54的亚细胞定位观察
Dek54-GFP载体与mCherry标记的质膜标记(mCherry-PM; CD3-1007)共定位。标尺为20 μm。
Fig. 8Observation of subcellular localization of Dek54 in maize protoplasts
Dek54-GFP was co-localized with a mCherry-labeled plasma membrane marker (mCherry-PM; CD3-1007). Bar: 20 μm.
3 讨论
籽粒缺陷(defective kernel, dek)突变体是一类主要的玉米籽粒突变体。dek突变体表型变化极为显著, 主要表现为, 胚乳发育缺陷、籽粒较小、种皮与胚乳分离、种皮颜色较浅, 且表面皱缩等特征[26]。本研究中, dek54突变体具有dek突变体的典型表型特征, 其胚发育延迟, 胚乳发育不良; 相比野生型, dek54籽粒的长、宽和粒重都显著降低。dek54是一个单基因控制的隐性突变体, 显示为功能缺失突变。对玉米籽粒缺陷突变体dek54进行精细定位, 并通过CRISPR/Cas9系统靶向突变实验确定导致突变表型的基因Zm00001d019294。测序结果显示, dek54突变体在Zm00001d019294基因的第2个外显子上有一个碱基突变导致该密码子突变为终止子, 提前终止了蛋白序列。qRT-PCR结果表明, Zm00001d019294基因在未成熟籽粒中特异性表达, 并随着籽粒的发育表达降低, 且在成熟籽粒中不表达。说明Dek54基因在玉米籽粒发育过程中起着重要作用。玉米中dek突变体种类多, 涉及的机制非常复杂。至今, 超过20个dek突变体已被鉴定。大部分dek突变体都是由PPR (pentatricopeptide repeat)蛋白突变所引起[7,9-11,27-29]。PPR蛋白在线粒体和质体中特异性作用于RNA编辑、剪辑、稳定性和转录后调控等重要进程[30], 但也有部分dek突变体不是PPR蛋白突变引起。例如Dek1编码一个钙蛋白酶家族蛋白, 影响籽粒胚和胚乳的糊粉层发育[3], 可能在陆地植物表面细胞位置传感和信号传递过程中发挥重要作用[31]; Dek15编码一个黏连蛋白装载复合体亚基, 决定染色体分离和籽粒发育[8]。本研究中, Dek54基因编码一个含有MFS保守结构域的蛋白。MFS是一类广泛存在细胞膜上的转运蛋白家族, 由协同转运体、共转运体和反向转运体共同组成, 能够转运多种底物包括无机和有机离子、核酸、氨基酸、短肽和脂类物质[32], 在多种生理生化途径中起着重要作用。所有的MFS转运体都具有一个典型的MFS折叠。这个折叠包含了2个结构域, 每个结构域都由6个连续的跨膜基序组成, 分别称为N端和C端结构域[33]。Dek1蛋白由一个多跨膜结构域(mutispanning tranmembrane domain, MEM)组成[3]。序列分析发现, MFS和DEK1-MEM的第16到22个跨膜基序具有相似性, 推测MFS蛋白在响应化学渗透梯度促进各种溶质跨膜运输的功能, 与MEM感知相邻细胞表面膜和外部环境差异方面功能的作用是相容的[4]。硝酸盐转运体NRT1是在高等植物中唯一具有典型的MFS结构的蛋白[34]。由于对氨基酸残基T101的磷酸化[35], 使得NRT1蛋白对硝酸根离子NO3-具有双亲和力, 即高亲和力和低亲和力[36]。其中, AtNRT1.5介导NO3-的外排, 并在将NO3-装载进入木质部, 并转运到地上部分起着重要作用[37]。AtNRT1.8负责从根和木质部中提取NO3-, 并与AtNRT1.5协同调控NO3-的远距离运输[38]。将拟南芥和玉米的NRT1家族的蛋白以及Dek54蛋白进行比对及系统分析, Dek54与ZmNRT1.5B、ZmNRT1.5A、AtNRT1.5和AtNRT1.8分在一个组, 且具有最大的序列相似性。进一步研究表明, Dek54蛋白定位在玉米原生质体的细胞质膜上。组织学观察显示, dek54突变体籽粒淀粉胚乳细胞组织形态不规则, 并且相比野生型排列较密且较小, 胚乳中心区域的淀粉粒周围的蛋白体相比野生型较小, 且数量较少。dek54突变成熟体籽粒的总蛋白、醇溶蛋白、各类氨基酸及全氮含量均显著低于野生型。推测dek54突变体可能由于硝酸盐转运体的缺失, 导致在玉米籽粒发育过程中不能有效的转运氮, 从而造成胚乳和胚的发育缺陷。但Dek54如何在玉米籽粒发育中发挥功能的具体作用机制仍然不明确。我们发现新的籽粒突变体丰富了玉米籽粒发育的研究材料, 为进一步解析该基因在玉米籽粒发育过程的分子机制奠定了基础。
4 结论
通过EMS化学诱变, 鉴定了一个新的玉米籽粒缺陷突变体dek54。dek54被定位在玉米7号染色体标记SSR6和SSR7之间物理距离约为290 kb的区间内。测序发现Dek54基因的第2个外显子上第351个碱基由G突变转换为A, 从而引起蛋白翻译的提前终止, 并在玉米未成熟籽粒中特异性表达。CRISPR/Cas9系统靶向突变实验确定与突变表型相关的候选基因Dek54, 该基因编码一个MFS家族蛋白并与ZmNRT1.5具有较高的同源性。此外, Dek54蛋白定位在玉米原生质体的细胞质膜上。dek54突变体为解析玉米籽粒发育的分子机制提供了新的研究材料和理论依据。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOIPMID [本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 3]
DOI [本文引用: 2]

DEFECTIVE KERNEL1 (DEK1) of higher plants plays an essential role in position-dependent signaling and consists of a large transmembrane domain (MEM) linked to a protease catalytic domain and a regulatory domain. Here, we show that the postulated sensory Loop of the MEM domain plays an important role in the developmental regulation of DEK1 activity in the moss Physcomitrella patens. Compared with P. patens lacking DEK1 (Delta dek1), the dek1 Delta loop mutant correctly positions the division plane in the bud apical cell. In contrast with an early developmental arrest of Delta dek1 buds, dek1 Delta loop develops aberrant gametophores lacking expanded phyllids resulting from misregulation of mitotic activity. In contrast with the highly conserved sequence of the protease catalytic domain, the Loop is highly variable in land plants. Functionally, the sequence from Marchantia polymorpha fully complements the dek1 Delta loop phenotype, whereas sequences from maize (Zea mays) and Arabidopsis (Arabidopsis thaliana) give phenotypes with retarded growth and affected phyllid development. Bioinformatic analysis identifies MEM as a member of the Major Facilitator Superfamily, membrane transporters reacting to stimuli from the external environment. Transcriptome analysis comparing wild-type and Delta dek1 tissues identifies an effect on two groups of transcripts connected to dek1 mutant phenotypes: transcripts related to cell wall remodeling and regulation of the AINTEGUMENTA, PLETHORA, and BABY BOOM2 (APB2) and APB3 transcription factors known to regulate bud initiation. Finally, sequence data support the hypothesis that the advanced charophyte algae that evolved into ancestral land plants lost cytosolic calpains, retaining DEK1 as the sole calpain in the evolving land plant lineage.
PMID [本文引用: 1]

Mutants in the maize defective kernel1 (dek1) gene are blocked in embryogenesis and the endosperm is chalky and lacks an aleurone layer. Here we show that intermediate alleles result in embryos that lack a shoot axis while weak alleles result in endosperms with mosaic aleurone and deformed plants with epidermal cells that resemble bulliform cells, a specialized epidermal cell type. This indicates that dek1 functions in embryonic pattern formation, cell fate specification and pattern formation in the leaf epidermis, and cell fate specification in the endosperm. Thus, the dek1 gene product appears to control different cellular-developmental processes depending on cellular context. The phenotype of the weak dek1-Dooner allele resembles the crinkly4 (cr4) mutant phenotype. Double mutants between dek1 and cr4 showed elements of epistasis, additivity and synergy, suggesting that the gene products may function in overlapping developmental processes. cr4 transcript was detectable in dek1 mutant kernels indicating that DEK1 was not required for Cr4 transcript accumulation. To test whether DEK1 regulated the ligand for the CR4 receptor kinase, a genetic mosaic analysis was performed. The dek1 phenotype appeared to be generally cell-autonomous, leading to the conclusion that it was not likely to produce a diffusible signal molecule, and therefore was not likely to regulate the CR4 ligand.
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 2]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

Genetic analyses suggested that the opaque-2 (o2) locus in maize acts as a positive, transacting, transcriptional activator of the zein seed storage-protein genes. Because isolation of the gene is requisite to understanding the molecular details of this regulation, transposon mutagenesis with the transposable element suppressor-mutator (Spm) was carried out, and three mutable o2 alleles were obtained. One of these alleles contained an 8.3-kilobase autonomous Spm, another a 6.8-kilobase nonautonomous Spm, and the third an unidentified transposon that is unrelated to Spm. A DNA sequence flanking the autonomous Spm insertion was verified to be o2-specific and provided a probe to clone a wild-type allele. Northern blots indicated that the gene is expressed in wild-type endosperm but not in leaf tissues or in endosperms homozygous for a mutant allele of the O2 gene. A transcript was detected in endosperms homozygous for mutations at opaque-7 and floury-2, an indication that O2 expression is independent of these two other putative regulators of zein synthesis.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 3]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

The maize (Zea mays) floury1 (fl1) mutant was first reported almost 100 years ago, but its molecular identity has remained unknown. We report the cloning of Fl1, which encodes a novel zein protein body membrane protein with three predicted transmembrane domains and a C-terminal plant-specific domain of unknown function (DUF593). In wild-type endosperm, the FL1 protein accumulates at a high level during the period of zein synthesis and protein body development and declines to a low level at kernel maturity. Immunogold labeling showed that FL1 resides in the endoplasmic reticulum surrounding the protein body. Zein protein bodies in fl1 mutants are of normal size, shape, and abundance. However, mutant protein bodies ectopically accumulate 22-kD alpha-zeins in the gamma-zein-rich periphery and center of the core, rather than their normal discrete location in a ring at outer edge of the core. The 19-kD alpha-zein is uniformly distributed throughout the core in wild-type protein bodies, and this distribution is unaffected in fl1 mutants. Pairwise yeast two-hybrid experiments showed that FL1 DUF593 interacts with the 22-kD alpha-zein. Results of these studies suggest that FL1 participates in protein body formation by facilitating the localization of 22-kD alpha-zein and that this is essential for the formation of vitreous endosperm.
DOIURL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

This report presents the initial results of our study of the immature kernel stage of 150 defective kernel maize mutants. They are single gene, recessive mutants that map throughout the genome, defective in both endosperm and embryo development and, for the most part, lethal (Neuffer and Sheridan 1980). All can be distinguished on immature ears, and 85% of them reveal a mutant phenotype within 11 to 17 days post-pollination. Most have immature kernels that are smaller and lighter in color than their normal counterparts. Forty of the mutants suffer from their defects early in kernel development and are blocked in embryogenesis before their primordia differentiate, or, if primordia are formed, they are unable to germinate when cultured as immature embryos or tested at maturity; a few begin embryo degeneration prior to the time that mutant kernels became visually distinguishable. The others express the associated lesion later in kernel development and form at least one leaf primordium by the time kernels are distinguishable and will germinate when cultured or tested at maturity. In most cases, on a fresh weight basis, the mutants have embryos that are more severely defective than the endosperm; their embryos usually are no more than one-half to two-thirds the size, and lag behind by one or two developmental stages. in comparison with embryos in normal kernels from the same ear. One hundred and two mutants were examined by culturing embryos on basal and enriched media; 21 simply enlarged or completely failed to grow on any of the media tested; and 81 produced shoots and roots on at least one medium. Many grew equally well on basal and enriched media; 16 grew at a faster rate on basal medium and 23 displayed a superior growth on enriched medium. Among the latter group, 10 may be auxotrophs. One of these mutants and another mutant isolated by E. H. Coe are proline-requiring mutants, allelic to pro-1. Considering their diversity of expression as evidenced by their differences in morphological appearance, degree of defectiveness and response to embryo culturing, we believe that they represent many different gene loci.
DOIURL [本文引用: 1]
DOIURL
[本文引用: 1]
DOIPMID [本文引用: 1]

The pentatricopeptide repeat (PPR) is a degenerate 35-amino-acid structural motif identified from analysis of the sequenced genome of the model plant Arabidopsis thaliana. From the wealth of sequence information now available from plant genomes, the PPR protein family is now known to be one of the largest families in angiosperm species, as most genomes encode 400-600 members. As the number of PPR genes is generally only c. 10-20 in other eukaryotic organisms, including green algae, the family has obviously greatly expanded during land plant evolution. This provides a rare opportunity to study selection pressures driving a 50-fold expansion of a single gene family. PPR proteins are sequence-specific RNA-binding proteins involved in many aspects of RNA processing in organelles. In this review, we will summarize our current knowledge about the evolution of PPR genes, and will discuss the relevance of the dramatic expansion in the family to the functional diversification of plant organelles, focusing primarily on RNA editing.© 2011 The Authors. New Phytologist © 2011 New Phytologist Trust.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

Higher plants have both high- and low-affinity nitrate uptake systems. These systems are generally thought to be genetically distinct. Here, we demonstrate that a well-known low-affinity nitrate uptake mutant of Arabidopsis, chl1, is also defective in high-affinity nitrate uptake. Two to 3 hr after nitrate induction, uptake activities of various chl1 mutants at 250 microM nitrate (a high-affinity concentration) were only 18 to 30% of those of wild-type plants. In these mutants, both the inducible phase and the constitutive phase of high-affinity nitrate uptake activities were reduced, with the inducible phase being severely reduced. Expressing a CHL1 cDNA driven by the cauliflower mosaic virus 35S promoter in a transgenic chl1 plant effectively recovered the defect in high-affinity uptake for the constitutive phase but not for the induced phase, which is consistent with the constitutive level of CHL1 expression in the transgenic plant. Kinetic analysis of nitrate uptake by CHL1-injected Xenopus oocytes displayed a biphasic pattern with a Michaelis-Menten Km value of approximately 50 microM for the high-affinity phase and approximately 4 mM for the low-affinity phase. These results indicate that in addition to being a low-affinity nitrate transporter, as previously recognized, CHL1 is also involved in both the inducible and constitutive phases of high-affinity nitrate uptake in Arabidopsis.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
