 ,**, 王蕊
,**, 王蕊 ,**, 张利霞, 宋亚萌, 田晓楠, 葛荣朝
,**, 张利霞, 宋亚萌, 田晓楠, 葛荣朝 ,*河北师范大学生命科学学院, 河北石家庄 050024
,*河北师范大学生命科学学院, 河北石家庄 050024Cloning and functional identification of gene OsATS in rice
LI Xiao-Xu ,**, WANG Rui
,**, WANG Rui ,**, ZHANG Li-Xia, SONG Ya-Meng, TIAN Xiao-Nan, GE Rong-Chao
,**, ZHANG Li-Xia, SONG Ya-Meng, TIAN Xiao-Nan, GE Rong-Chao ,*College of Life Science, Hebei Normal University, Shijiazhuang 050024, Hebei, China
,*College of Life Science, Hebei Normal University, Shijiazhuang 050024, Hebei, China通讯作者: *葛荣朝, E-mail:grcgp@sina.com
第一联系人:
收稿日期:2020-11-21接受日期:2021-03-22网络出版日期:2021-04-07
| 基金资助: |
Corresponding authors: *E-mail:grcgp@sina.com
Received:2020-11-21Accepted:2021-03-22Published online:2021-04-07
| Fund supported: |
作者简介 About authors
李晓旭,E-mail:jiandanxiaoxiao@163.com;
王蕊,E-mail:1915435558@qq.com.

摘要
植物胚胎特异性蛋白ATS3和植物的渗透胁迫响应有密切关系, 本文对水稻OsATS基因的抗逆相关功能进行了初步研究。qRT-PCR检测发现, 水稻在盐胁迫后OsATS基因表达量显著增加。构建OsATS基因过表达载体, 转化拟南芥植株, 抗逆性检测表明, OsATS基因的过表达可以显著提高拟南芥在萌发阶段和成株阶段的耐盐性。随后将过表达载体p1300-35S:OsATS和RNA干涉载体pTCK303-OsATS-RNAi转入水稻, 抗逆性分析表明, OsATS过表达水稻株系在萌发阶段和苗期的耐盐性显著提高, 而OsATS基因RNAi水稻株系耐盐性则明显下降。qRT-PCR和生理指标检测表明, OsATS基因的表达可能通过调节OsP5CS1、OsLEA3-1、OsPDH基因的表达, 调控了水稻细胞中的脯氨酸、LEA蛋白质含量, 进而影响了水稻植株整体的耐盐性。本研究初步揭示了OsATS基因的抗逆功能, 后续可通过调整该基因的表达量, 改良水稻的抗逆性。
关键词:
Abstract
The plant embryo specific protein ATS3 is closely related to osmotic stress response in plants. Here, the stress resistance related gene OsATS was preliminarily studied in rice. Fluorescence quantitative PCR showed that the relative expression level of OsATS increased significantly after salt stress in rice. The overexpression vector of OsATS was constructed and transformed into Arabidopsis thaliana. The stress resistance test revealed that the overexpression of OsATS gene could significantly improve the salt tolerance of Arabidopsis thaliana at germination and adult stages. After that, the overexpression vector p1300-35s:OSATS and RNA interference vector pTCK303-OsATS-RNAi were transferred into rice. The stress tolerance analysis indicated that the salt tolerance of OsATS overexpression rice lines significantly increased at germination stage and seedling stage, while the salt tolerance of OsATS RNAi rice lines significantly decreased. Results of qRT-PCR and physiological index detection demonstrated that the relative expression levels of OSATS gene might regulate the protein content of proline and LEA cells by regulating the expression of OsP5CS1, OsLEA3-1 and OsPDH, thus affecting the salt tolerance in rice. This study preliminarily revealed the stress resistance function of OSATS gene, which laid a foundation for improving rice stress resistance by adjusting the relative expression level of OSATS gene.
Keywords:
PDF (4705KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李晓旭, 王蕊, 张利霞, 宋亚萌, 田晓楠, 葛荣朝. 水稻基因OsATS的克隆及功能鉴定[J]. 作物学报, 2021, 47(10): 2045-2052 DOI:10.3724/SP.J.1006.2021.02079
LI Xiao-Xu, WANG Rui, ZHANG Li-Xia, SONG Ya-Meng, TIAN Xiao-Nan, GE Rong-Chao.
水稻是我国乃至全世界重要的粮食作物之一[1], 水稻生产在国民经济发展中发挥着巨大作用, 直接关系着国民生计[2]。水稻在谷类作物中对盐分胁迫响应最为敏感, 盐胁迫是导致水稻减产的重要原因之一, 是制约其生长发育和产量品质的一个重要环境因素[3]。目前土壤盐渍化在全世界范围严重影响着农业生产[4]。我国人口不断增加, 而可耕地面积不断减少, 这促使人们对盐碱地的开发与利用产生了极大关注[5,6]。因此, 在我国农作物育种工作中选育耐盐碱的优良品种显得尤为重要。其中探究耐盐相关基因及其耐盐内在生理机制, 成为了耐盐碱农作物选育的重要研究方向之一[4]。
生物胁迫和非生物胁迫是影响农作物生长、造成农业减产的重要原因, 植物在自身的生活周期中往往面临着各种胁迫。在胁迫环境下, 植物通过调整特定的抗逆相关基因表达, 来提高自身对环境胁迫的适应能力。近年来, 水稻中已经鉴定了一系列抗逆相关基因, 这些基因的表达产物可以明显影响植物对生物和非生物胁迫的耐受能力。高继平等定位克隆到位于水稻第1染色体的耐盐相关基因SKC1, 其编码的Na+载体蛋白, 在Na+循环、离子长距离运输中起重要作用。当水稻处于高盐环境中时, SKC1蛋白能将过量的Na+集中转运至水稻根部, 从而提高水稻耐盐性[7,8]。彭静静等[9]研究发现水稻OsAQP基因的过表达不仅能够促进拟南芥种子萌发和根系生长, 而且在盐胁迫下通过提高拟南芥内源抗氧化酶活性、降低膜脂过氧化程度, 增强了转基因植株对一定程度盐胁迫的耐受性。Liao等[10]研究表明水稻OsPP1a基因的过表达能够使水稻OsNAC5和OsNAC6表达量上调, 提高水稻对高盐胁迫的耐受性。另外研究发现, 水稻OsNHX1、SIDP361、OsSUV3、PDH45等基因均可通过不同的机制影响水稻的抗逆特性[11,12,13,14,15], OsNAP、DST、SNAC1和SNAC2等转录因子基因则通过调控多种抗逆相关基因的表达从而提高水稻的抗逆性[16,17,18,19,20]。
胚胎发育晚期富集蛋白(late embryogenesis abundant proteins, LEA)是在植物种子成熟后大量积累的一类亲水蛋白[21]。研究表明, LEA家族基因广泛参与植物生长发育、形态建成和衰老等生物学过程。在高盐、干旱等非生物胁迫下, LEA蛋白还能够对植株起到关键的保护作用[22,23,24]。水稻基因Os01g0172800 (OsATS)与辣椒抗逆基因CAPIP2序列有一定同源性, 具有胚胎特异性蛋白ATS3保守结构域和脂氧合酶2基因家族保守域[25]。研究表明, 植物胚胎特异性蛋白ATS3也是在成熟种子大量表达的一种蛋白, 和植物的渗透胁迫响应有密切关系[26]。本文拟通过对基因OsATS在水稻中进行过量表达及RNA干涉, 从而对其抗逆功能和相关机制进行初步研究。
1 材料与方法
1.1 试验材料
供试品种为日本晴水稻(Oryza sativa L. japonica. cv. Nipponbare)、Columbia型拟南芥, 真核表达载体采用pCAMBIA1300 (简称p1300), 植物转化采用GV3101、EHA105型根癌农杆菌, 总RNA提取试剂TRNzol、逆转录试剂盒、Taq酶、限制性内切酶和T4连接酶等, 购自Takara Bio生物技术有限公司, 常用试剂均为分析纯。1.2 试验方法
1.2.1 盐胁迫后OsATS基因的表达模式 将培养14 d的水稻幼苗在140 mmol L-1 NaCl溶液中处理0、1、6和12 h后, 剪取叶片, 提取总RNA, 反转录获得cDNA, 利用P1: 5'-CTGTCAACGACGGTTTCCAAG-3'、P2: 5'- GCATCGTGACCTTGGTGTAC-3', 以Actin基因作为内参, 对不同处理的cDNA进行qRT-PCR检测, 重复检测3次, 确定OsATS基因在盐胁迫后的表达模式。1.2.2 目的基因OsATS的克隆及载体构建 提取日本晴水稻幼苗叶片总RNA, 反转录获得cDNA, 利用高保真DNA聚合酶Prime Star对目的基因OsATS进行PCR扩增, 引物采用RP: 5'-CTCTAGACTCGCCATCTCCTCAGCCAT-3' (Xba I, 下画线为酶切位点, 下同)和LP: 5'-CGAGCTC CGACGTCGCAAACCATCACTG-3' (Sac I)。扩增产物回收后连接到pMD18-T载体。基因测序正确后, 酶切p1300和pMD18-T-OsATS质粒, 回收、连接获得过表达载体p1300-35S:OsATS, 转入根癌农杆菌EHA105、GV3101备用。
以pMD18-T-OsATS质粒为模板, 用引物RNAi RP: 5'-CGGATCCGAGCTCAGTGGGTCAGGGTCTAC-3' (Spe I、Kpn I)和RNAi LP: 5'-CGGTACCACTAGTCAGATCAG AATCAGAATCTGATCTG-3' (Sac I、BamH I), PCR扩增后连接到pMD18-T, 获得阳性克隆pMD18-T-OsATS- RNAi。基因测序后酶切pTCK303和pMD18-T-OsATS- RNAi质粒, 首先采用内侧酶Sac I/Spe I进行双酶切, 电泳、回收、连接、转化到大肠杆菌, 获得正连载体pTCK303-OsATS-RNAi-sense。之后提取pTCK303-OsATS- RNAi-sense和pMD18-T-OsATS-RNAi质粒, 采用外侧酶Kpn I/BamH I进行双酶切, 构建RNAi载体pTCK303- OsATS-RNAi, 转入根癌农杆菌EHA105备用。
以pMD18-T-OsATS质粒为模板, PCR扩增构建pMD18-T-OsATS-GFP, 测序准确后进行酶切、回收、连接获得亚细胞定位载体p2300-OsATS-GFP4, 转入根癌农杆菌GV3101备用。
1.2.3 OsATS蛋白质的亚细胞定位 培养含有p2300-OsATS-GFP4质粒的农杆菌GV3101, 制备浸染液, 通过注射器将农杆菌液注入到烟草叶片内, 做好标记, 在40~48 h之间, 通过激光共聚焦显微镜观察OsATS的亚细胞定位情况。
1.2.4 OsATS基因过表达拟南芥的获得和抗逆性分析 培养含有p1300-35S:OsATS的农杆菌GV3101, 利用浸花法转化拟南芥。取OsATS基因过表达纯合体拟南芥和野生型拟南芥种子4℃春化3 d后, 分别消毒、播种于含150 mmol L-1 NaCl、200 mmol L-1 NaCl的MS培养基上, 正常光照培养3~4 d, 统计萌发率。为检测转基因拟南芥成株的耐盐性, 将培养14 d刚刚抽薹的OsATS基因过表达和野生型拟南芥每隔4 d浇灌200 mmol L-1 NaCl进行胁迫处理, 连续处理15 d。
1.2.5 水稻愈伤组织的遗传转化及抗逆性检测 利用水稻种子诱导愈伤组织, 利用含有p1300-35S:OsATS、pTCK303-OsATS-RNAi的EHA105农杆菌共培养转化获得OsATS基因过表达水稻和RNAi转基因水稻。用含50 mg L-1潮霉素的水浸泡转基因水稻种子至萌发。9 d后选取生根发芽的抗性株系, 分别移栽入微孔板中, Hoagland营养液培养7 d, 然后分别转入含140 mmol L-1 NaCl的Hoagland营养液中处理4~10 d, 恢复培养5 d, 观察表型变化。
1.2.6 转基因水稻的生理指标检测 分别用含有清水和140 mmol L-1 NaCl溶液培养野生型和转基因水稻5 d, 测定其脯氨酸含量、MDA含量。同时选取盐胁迫后、叶龄相似的野生型水稻与转基因水稻的叶片, 剪成小块, 称取0.2 g, 将叶片压入10 mL蒸馏水中, 真空抽气1 h后重新充入空气。将样品置于室温搅动浸提1 h后, 用电导仪测定样品的处理电导率。再将同样的叶片样品放入100℃沸水浴15 min, 转入清水中冷却10 min, 测定其煮沸电导率。电导率公式为: 相对电导率(%) = (处理电导率-空白电导率) / (煮沸电导率-空白电导率)×100。
1.2.7 OsATS基因过量表达水稻中相关基因表达量的检测 利用总RNA提取试剂盒提取水稻总RNA, 反转录获得cDNA, 以水稻Actin-1基因(AK071586)为内参, 对水稻脯氨酸脱氢酶基因OsPDH (proline dehydrogenase, AK121010)、吡咯啉-5-羧酸合酶基因OsP5CS1 (δ-1-pyrroline-5-carboxylate synthase 1, AK101985)、晚期胚胎发生丰富蛋白基因OsLEA3-1 (late embryogenesis abundant protein, AK063984)进行荧光定量PCR检测, 每个样本3次重复, PCR在ABI 7300仪器上进行?
2 结果与分析
2.1 盐胁迫后OsATS基因在水稻中的表达模式
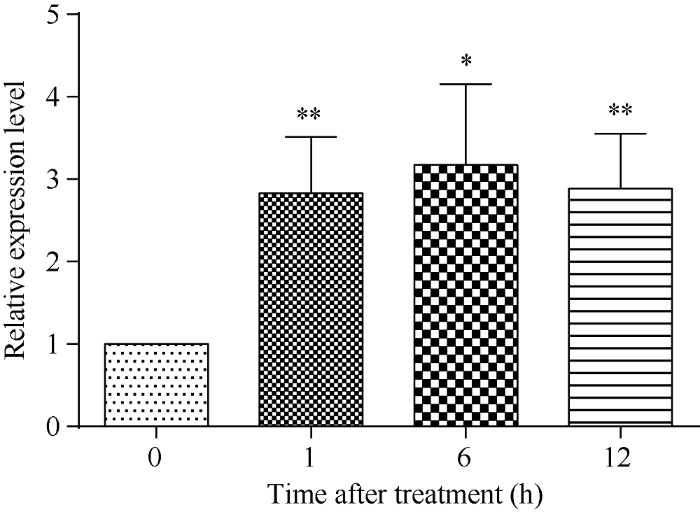
对盐胁迫处理的日本晴水稻幼苗进行qRT-PCR检测结果表明, 在水稻受到140 mmol L-1 NaCl胁迫后, OsATS基因表达量会出现显著增加, 1 h后即上升到胁迫处理前的3倍左右。此后, 在盐胁迫处理6 h和12 h时, 其表达量均维持在较高水平(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1盐胁迫后水稻OsATS基因表达量的变化
差异显著性分析采用t-test 方法, *P < 0.05、**P < 0.01。
Fig. 1Relative expression levels of OsATS genes under NaCl stress in rice
The significant difference is evaluated by the Student’s t-test. *P< 0.05, **P < 0.01.
2.2 OsATS蛋白的亚细胞定位
利用烟草叶片对OsATS蛋白进行亚细胞定位结果显示, 对照试验中注射含有p2300-GFP质粒菌株的烟草叶片在细胞核、细胞质和细胞膜均有绿色荧光, 而注射含有p2300-OsATS-GFP质粒菌株的烟草叶片只在细胞核有绿色荧光(图2), 说明OsATS蛋白在细胞中主要定位于细胞核。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2OsATS在烟草表皮的亚细胞定位
Fig. 2Subcellular localization of OsATS in tobacco epidermis
2.3 OsATS基因的克隆与转基因拟南芥的获得
以日本晴水稻cDNA为模板扩增获得长度为624 bp的OsATS基因序列, 构建过表达载体p1300-35S:OsATS。利用含有p1300-35S:OsATS质粒的农杆菌GV3101通过浸花法转化拟南芥, 在含有潮霉素的MS培养基上逐代筛选获得阳性纯合体转基因拟南芥。对获得的35S:OsATS转基因拟南芥提取叶片DNA进行PCR扩增检测, 过表达植株OX-1、OX-4和OX-5转基因拟南芥均扩增获得了624 bp的特异条带, 确定外源基因OsATS成功插入到3个转基因拟南芥株系的基因组之中(图3)。对3个OsATS过表达拟南芥株系cDNA进行qRT-PCR检测实验中, 由于野生型拟南芥没有, 因此只能对qRT-PCR检测获得的ΔCt进行比较(ΔCt值为样本Ct值根据内参基因Ct值调整后的数值), 最终确认过表达拟南芥株系OX-1、OX-4、OX-5中OsATS基因均获得了高水平的表达, OX-5株系中OsATS基因的表达量最高(图4)。图3
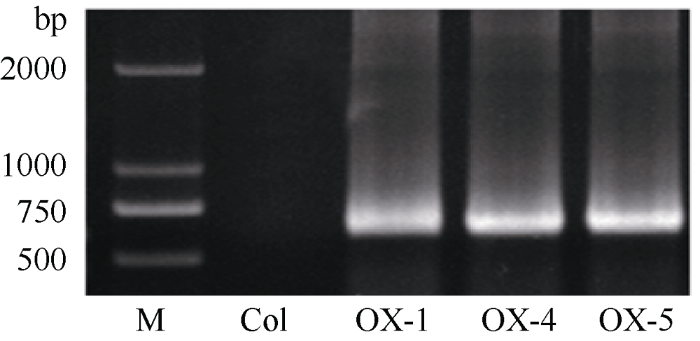 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3OsATS过表达拟南芥的PCR鉴定
Fig. 3PCR detection of OsATS overexpression in Arabidopsis
图4
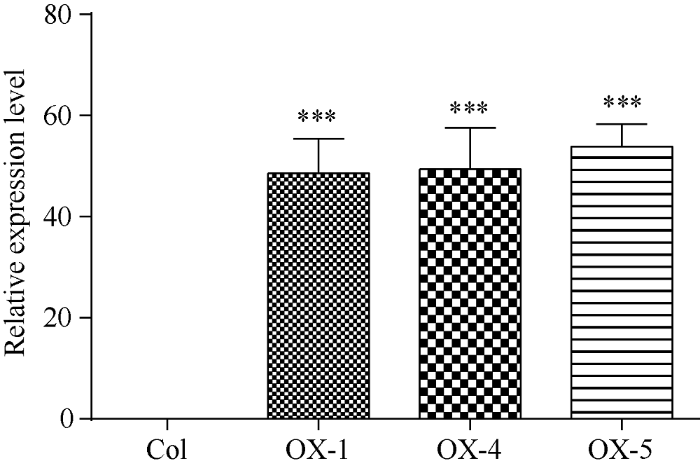 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4OsATS基因表达量的qRT-PCR检测
差异显著性分析采用t-test方法, *** P <0.001。
Fig. 4Relative expression levels of OsATS genes in Arabidopsis
The significant difference is evaluated by the Student’s t-test. *** P < 0.001.
2.4 OsATS转基因拟南芥的抗逆性分析
将OsATS过表达拟南芥的3个纯合体株系种子消毒后接种到含有150 mmol L-1和200 mmol L-1 NaCl的MS培养基, 对其种子萌发阶段的耐盐性检测结果表明, 在2种浓度的盐胁迫下, 过表达OsATS拟南芥萌发率都显著高于野生型, 其中OX-5株系在萌发阶段的耐盐性提高更加明显, 尤其是对200 mmol L-1 NaCl的耐受性提高较另外2个株系更加明显(图5)。图5
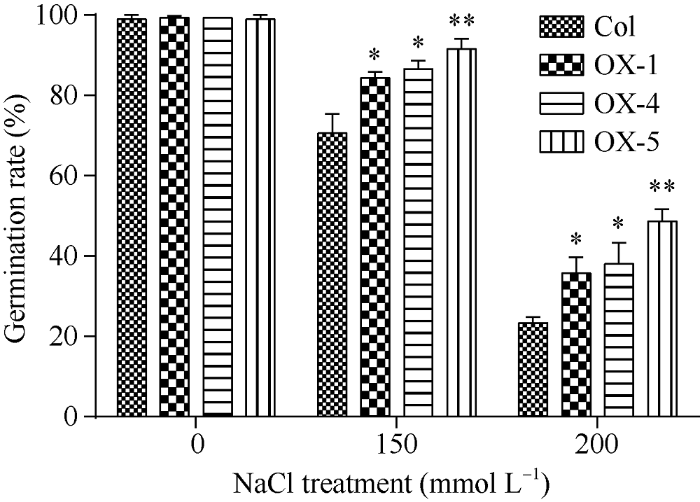 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5盐胁迫下OsATS过表达拟南芥种子萌发率统计
差异显著性分析采用t-test方法, * P < 0.05、** P < 0.01。
Fig. 5Seed germination rate of OsATS overexpression under the salt stress in Arabidopsis
The significant difference is evaluated by the Student's t-test. * P < 0.05, ** P < 0.01.
将培养14 d的Columbia野生型拟南芥、OsATS过表达拟南芥成株进行200 mmol L-1 NaCl盐胁迫处理, 结果表明, 高盐溶液胁迫15 d后, 野生型拟南芥叶片明显枯黄, 植株瘦弱矮小, 而3个OsATS过表达拟南芥株系叶片, 均呈墨绿色, 可以正常抽薹、开花结实, 果荚数目明显比野生型较多(图6)。因此, OsATS基因的过表达明显提高了拟南芥成株对盐胁迫的耐受性。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6OsATS过表达拟南芥成株的耐盐性检测
Fig. 6Salt tolerance detection of OsATS overexpression of adult plants in Arabidopsis
2.5 转基因水稻的获得及其抗逆性鉴定
利用含有pTCK303-OsATS-RNAi或p1300-35S:OsATS载体的农杆菌, 分别对水稻愈伤组织进行浸染, 分化培养后获得OsATS基因过表达水稻和RNAi水稻。通过对OsATS基因过表达株系、RNAi株系和野生型水稻幼苗进行荧光定量PCR检测表明, 过表达转基因水稻OX-3、OX-4和OX-9株系中OsATS基因的表达量显著增高, 而RNAi转基因水稻株系Ri1、Ri2和Ri5的OsATS基因表达量则显著下降(图7)。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7OsATS过表达水稻和RNAi转基因水稻的qRT-PCR检测
WT: 野生型水稻; OX: OsATS过表达转基因水稻株系; Ri: RNAi转基因水稻株系; 差异显著性分析采用t-test方法, *** P < 0.001。
Fig. 7Relative expression levels of OsATS overexpressed and OsATS-RNAi transgenic plants in rice
WT: wild type rice; OX: OsATS overexpression transgenic rice lines; Ri: RNAi transgenic rice lines; The significant difference is evaluated by the Student’s t-test. *** P < 0.001.
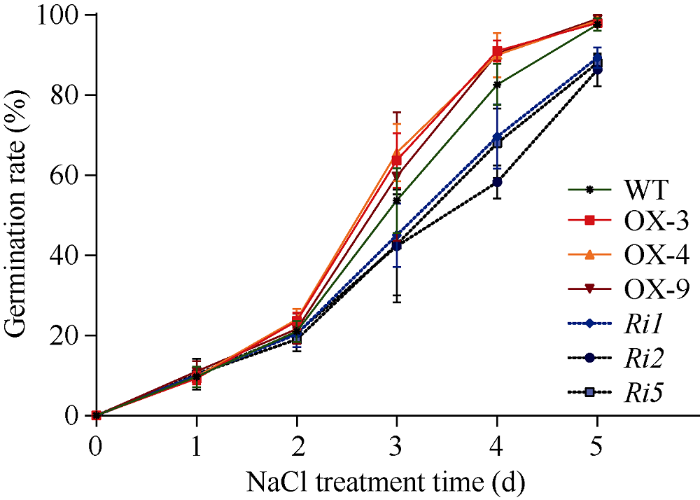
对OsATS过表达水稻的3个纯合体株系OX3、OX4、OX9和RNAi转基因水稻株系Ri1、Ri2、Ri5在种子萌发阶段的耐盐性进行检测, 结果表明, 在140 mmol L-1 NaCl胁迫下, OsATS过表达水稻萌发速度比野生型水稻要快, OsATS RNAi水稻种子的萌发则相对缓慢, 且第5天其萌发率平均仅达到87.9%, 而此时OsATS过表达水稻与野生型水稻种子均全部萌发(图8)。
图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8OsATS过表达水稻和RNAi转基因水稻种子在NaCl胁迫下的萌发率
Fig. 8Seed germination rate of OsATS overexpressed and OsATS-RNAi transgenic plants under NaCl stress in rice
对生长状态一致的日本晴野生水稻植株、p1300- 35S:OsATS转基因株系OX3、OX4、OX9和RNAi转基因水稻株系Ri1、Ri2、Ri5用140 mmol L-1 NaCl溶液进行胁迫培养, OsATS过表达转基因株系和野生型水稻胁迫6 d、恢复5 d后叶片均出现枯萎变黄的现象, 但过表达转基因植株的长势明显强于日本晴植株, OX3、OX4、OX9株系的绿叶率分别为21.3%、17.3%、23.3%, 而野生型水稻绿叶率仅为6%, 说明OsATS基因的过量表达可以显著提高水稻植株的耐盐性(图9)。RNAi转基因株系和野生型水稻盐胁迫处理4 d、恢复5 d后同样均有叶片枯萎的现象, 但是RNAi转基因株系相对萎蔫更为严重, Ri1、Ri2、Ri5株系的绿叶率分别为4.0%、6.7%、3.3%, 相对于野生型水稻20%的绿叶率, 水稻基因OsATS表达被干涉后其耐盐性显著降低(图9)。
图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9OsATS转基因水稻的耐盐性检测
Fig. 9Salt tolerance detection of the OsATS transgenic rice
2.6 OsATS转基因水稻的生理检测
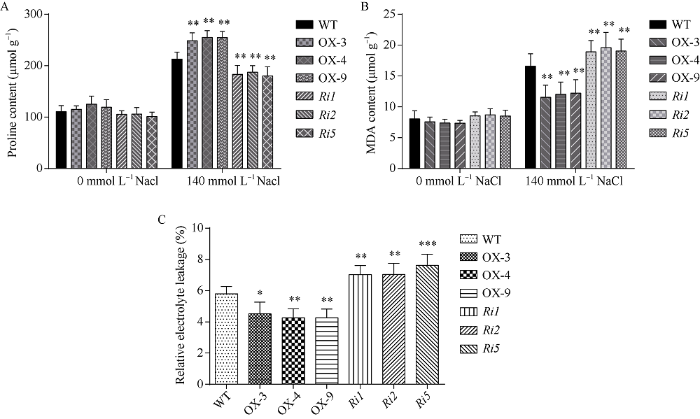
将野生型水稻、OsATS过表达水稻和RNAi转基因水稻幼苗用140 mmol L-1 NaCl处理5 d后, 分别对其叶片的脯氨酸含量、丙二醛含量和细胞质膜透性进行检测, 结果表明, OsATS过表达水稻的脯氨酸含量的增加量显著高于野生型, RNAi株系的脯氨酸在盐胁迫前后较野生型水稻的含量均明显较低。丙二醛含量检测结果表明, OsATS过表达水稻盐胁迫后的丙二醛含量明显低于野生型水稻, RNAi株系在盐胁迫后丙二醛含量则明显较高。盐胁迫后, OsATS过表达水稻的细胞质膜透性明显低于野生型水稻, 而RNAi转基因水稻的细胞质膜透性则显著高于野生型水稻(图10)。图10
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10OsATS过表达和RNAi水稻的生理指标检测
A: 脯氨酸含量; B: 丙二醛含量; C: 细胞质膜透性。差异显著性分析采用t-test方法, * P < 0.05、** P < 0.01、*** P <0.001。
Fig. 10Physiological indexes detection of the OsATS overexpression and OsATS-RNAi transgenic plants in rice
A: proline content; B: MDA content; C: relative electrolyte leakage; The significant difference is evaluated by the Student’s t-test. * P< 0.05, ** P < 0.01, *** P < 0.001.
2.7 OsATS基因表达对水稻抗逆相关基因表达的影响
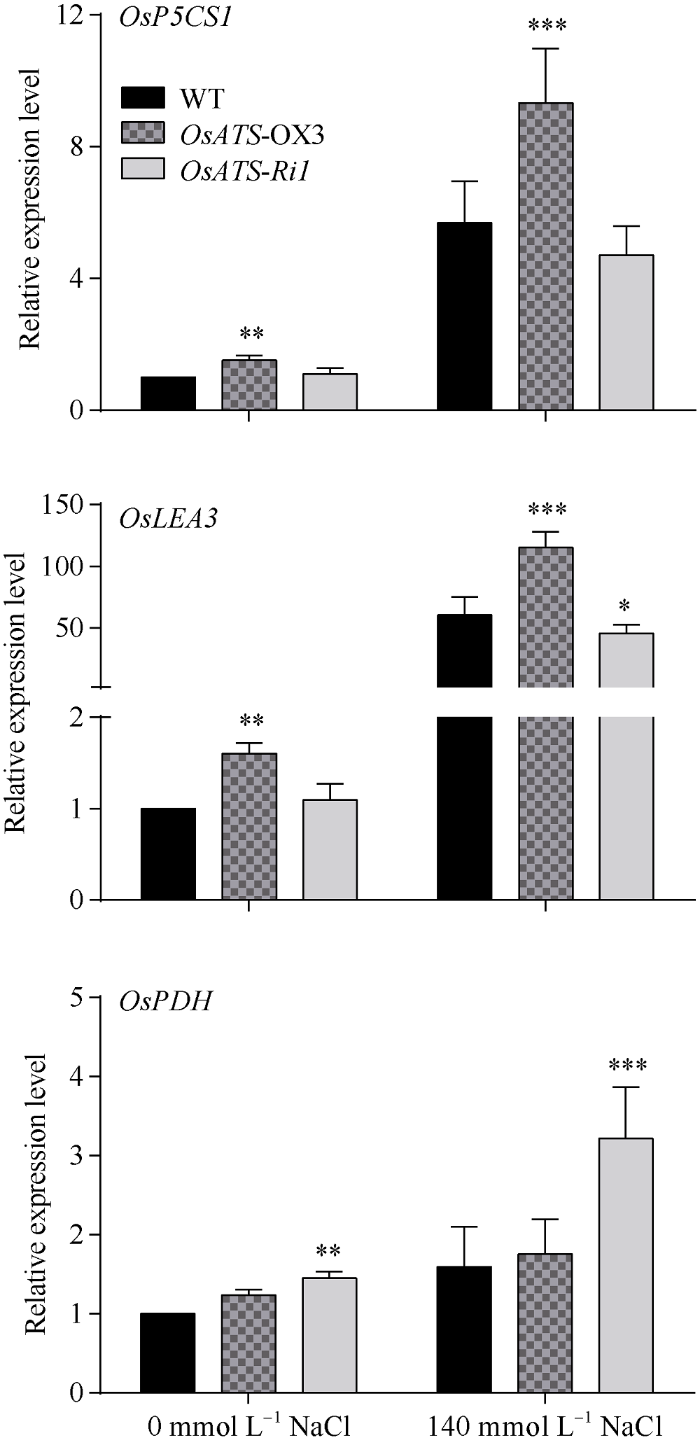
对OsATS基因过表达水稻株系OX3和RNAi水稻株系Ri1进行140 mmol L-1 NaCl处理前后的OsPDH、OsP5CS1、OsLEA3-1等抗逆相关基因表达量进行qRT-PCR检测结果表明, OsATS基因过表达水稻在盐胁迫前后OsP5CS1、OsLEA3-1基因的表达量显著较高。OsATS基因RNAi水稻中的OsPDH表达量在盐胁迫前明显较高, 在盐胁迫后, RNAi水稻中的OsPDH表达量更是极显著高于对照水稻和OsATS基因过表达水稻(图11)。图11
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图11OsATS转基因水稻中耐盐相关基因的表达变化
差异显著性分析采用t-test方法, * P < 0.05、** P < 0.01、*** P < 0.001。
Fig. 11Relative expression levels of the salt tolerance related genes in OsATS transgenic rice
The significant difference is evaluated by the Student’s t-test. * P < 0.05, ** P < 0.01, *** P < 0.001.
3 讨论
OsATS蛋白具有高度同源的植物胚胎特异性蛋白ATS3保守结构域, 可能与水稻抵御生物和非生物胁迫的功能密切相关, 我们对OsATS基因通过烟草花叶病毒CAMV 35S启动子实现过量表达或通过RNAi进行干涉表达, 以期确定该基因对水稻耐盐性的影响。首先, OsATS基因在拟南芥中的过表达明显提高了转基因拟南芥在盐胁迫下的萌发率, 使OsATS过表达拟南芥表现出更好的耐盐性。随后, 通过对OsATS基因过表达水稻和RNAi水稻进行抗逆性检测, 表明无论是在萌发阶段还是在幼苗生长阶段, OsATS基因过表达水稻表现出更好的耐盐性, 而RNAi水稻则表现出明显的敏盐特点。生理指标的检测表明, 在盐胁迫前后OsATS基因过表达水稻细胞中都具有更高的脯氨酸含量, OsATS基因RNAi水稻细胞中的水稻脯氨酸含量则明显较低。脯氨酸作为细胞中一种重要的氨基酸小分子, 对细胞众多代谢过程有着至关重要的影响[27]。在植物遭受盐、旱等非生物胁迫时, 脯氨酸在细胞溶质中能维持稳定的渗透压, 同时可以高效清除胞内由于胁迫产生的大量活性氧, 维持细胞稳态[28]。另外2项的生理指标检测表明, OsATS基因过表达水稻在盐胁迫后胞内丙二醛含量明显较低, 细胞质膜透性也比野生型水稻要低, 表明过量表达的OsATS蛋白质使得水稻植株细胞在盐胁迫后受损明显减轻。而OsATS基因RNAi水稻植株中丙二醛、细胞质膜透性两项指标相对于野生型水稻明显要高, OsATS基因表达量的下降造成水稻细胞受到了更明显的盐胁迫损伤。亚细胞定位检测发现OsATS蛋白明显位于细胞核, 但分析发现其并不具备转录因子的相关特征序列, 因此推测OsATS有可能通过间接调控的方式影响到了其他抗逆相关基因的表达。通过对相关基因的定量PCR检测, OsATS基因表达量的变化明显影响了3个水稻基因OsPDH、OsP5CS1、OsLEA3-1的表达。OsP5CS是一种位于水稻细胞溶质之中、具有谷氨酸激酶(GK)和C-谷氨酰磷酸还原酶(GPR)活性的双功能酶, 是脯氨酸合成途径中重要的限速酶, P5CS表达量的增加可明显提高胞内脯氨酸的大量积累[29]?胚胎发育晚期富集蛋白LEA3则可作为渗透调节物质, 维持细胞膨压, 减轻水分胁迫对植物造成的损伤, 与植物的抗逆性密切相关[30]。脯氨酸脱氢酶PDH属于脯氨酸代谢途径的重要蛋白酶, PDH与细胞中活性氧ROS的形成密切相关。胞内大量ROS的生成则对细胞造成损伤, 促进细胞凋亡[31]。定量PCR检测表明OsATS基因过量表达会造成OsP5CS1表达量上升, 这应该是OsATS过表达水稻胞内脯氨酸积累的主要原因之一。另外, OsATS基因过表达水稻还通过OsLEA3-1基因的表达增强, 更有效的解除了盐胁迫造成的活性氧增高损伤。相对而言, OsATS基因RNAi植株OsP5CS1、OsLEA3-1基因表达的受抑, 减弱了渗透胁迫保护物质脯氨酸、LEA蛋白质的积累。同时, OsATS基因RNAi水稻中OsPDH基因的更强表达使得细胞产生了更多的活性氧物质, 从而降低了转基因水稻的耐盐性。
当然, 本研究只是初步揭示了OsATS基因对水稻耐盐性的影响及其部分的内在机理, 至于该基因的表达对水稻在干旱等其他逆境胁迫下的抗逆性影响, 以及该基因涉及的具体信号传导通路还有待进行进一步深入研究。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 2]
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

Soil salinity is one of the most challenging problems that restricts the normal growth and production of rice worldwide. It has therefore become very important to produce more saline tolerant rice varieties. This study shows constitutive over-expression of the vacuolar Na+/H+ antiporter gene (OsNHX1) from the rice landrace (Pokkali) and attainment of enhanced level of salinity tolerance in transgenic rice plants. It also shows that inclusion of the complete un-translated regions (UTRs) of the alternatively spliced OsNHX1 gene provides a higher level of tolerance to the transgenic rice. Two separate transformation events of the OsNHX1 gene, one with 1.9 kb region containing the 5 ' UTR with CDS and the other of 2.3 kb, including 5 ' UTR, CDS, and the 3 ' UTR regions were performed. The transgenic plants with these two different constructs were advanced to the T-3 generation and physiological and molecular screening of homozygous plants was conducted at seedling and reproductive stages under salinity (NaCl) stress. Both transgenic lines were observed to be tolerant compared to WT plants at both physiological stages. However, the transgenic lines containing the CDS with both the 5 ' and 3 ' UTR were significantly more tolerant compared to the transgenic lines containing OsNHX1 gene without the 3 ' UTR. At the seedling stage at 12 dS/m stress, the chlorophyll content was significantly higher (P < 0.05) and the electrolyte leakage significantly lower (P < 0.05) in the order 2.3 kb > 1.9 kb > and WT lines. Yield in g/plant in the best line from the 2.3 kb plants was significantly more (P < 0.01) compared, respectively, to the best 1.9 kb line and WT plants at stress of 6 dS/m. Transformation with the complete transcripts rather than the CDS may therefore provide more durable level of tolerance.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

Plants respond to environmental stresses by altering gene expression, and several genes have been found to mediate stress-induced expression, but many additional factors are yet to be identified. OsNAP is a member of the NAC transcription factor family; it is localized in the nucleus, and shows transcriptional activator activity in yeast. Analysis of the OsNAP transcript levels in rice showed that this gene was significantly induced by ABA and abiotic stresses, including high salinity, drought and low temperature. Rice plants overexpressing OsNAP did not show growth retardation, but showed a significantly reduced rate of water loss, enhanced tolerance to high salinity, drought and low temperature at the vegetative stage, and improved yield under drought stress at the flowering stage. Microarray analysis of transgenic plants overexpressing OsNAP revealed that many stress-related genes were up-regulated, including OsPP2C06/OsABI2, OsPP2C09, OsPP2C68 and OsSalT, and some genes coding for stress-related transcription factors (OsDREB1A, OsMYB2, OsAP37 and OsAP59). Our data suggest that OsNAP functions as a transcriptional activator that plays a role in mediating abiotic stress responses in rice.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
DOIPMID [本文引用: 1]

Background: Late embryogenesis abundant (LEA) proteins are large groups of hydrophilic proteins with major role in drought and other abiotic stresses tolerance in plants. In-depth study and characterization of LEA protein families have been carried out in other plants, but not in upland cotton. The main aim of this research work was to characterize the late embryogenesis abundant (LEA) protein families and to carry out gene expression analysis to determine their potential role in drought stress tolerance in upland cotton. Increased cotton production in the face of declining precipitation and availability of fresh water for agriculture use is the focus for breeders, cotton being the backbone of textile industries and a cash crop for many countries globally.Results: In this work, a total of 242, 136 and 142 LEA genes were identified in G. hirsutum, G. arboreum and G. raimondii respectively. The identified genes were classified into eight groups based on their conserved domain and phylogenetic tree analysis. LEA 2 were the most abundant, this could be attributed to their hydrophobic character. Upland cotton LEA genes have fewer introns and are distributed in all chromosomes. Majority of the duplicated LEA genes were segmental. Syntenic analysis showed that greater percentages of LEA genes are conserved. Segmental gene duplication played a key role in the expansion of LEA genes. Sixty three miRNAs were found to target 89 genes, such as miR164, ghr-miR394 among others. Gene ontology analysis revealed that LEA genes are involved in desiccation and defense responses. Almost all the LEA genes in their promoters contained ABRE, MBS, W-Box and TAC-elements, functionally known to be involved in drought stress and other stress responses. Majority of the LEA genes were involved in secretory pathways. Expression profile analysis indicated that most of the LEA genes were highly expressed in drought tolerant cultivars Gossypium tomentosum as opposed to drought susceptible, G. hirsutum. The tolerant genotypes have a greater ability to modulate genes under drought stress than the more susceptible upland cotton cultivars.Conclusion: The finding provides comprehensive information on LEA genes in upland cotton, G. hirsutum and possible function in plants under drought stress.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

A modified protocol for differential display of mRNA was used to identify and clone genes expressed in developing Arabidopsis thaliana seeds. Two novel embryo-specific genes designated ATS1 and ATS3 (Arabidopsis thaliana seed gene) were identified. In situ hybridization showed that, spatially, ATS1 is expressed in a pattern similar to the Arabidopsis GEA1 gene and that ATS3 is expressed in a pattern similar to the Arabidopsis seed storage protein genes. Southern analysis of Arabidopsis genomic DNA indicated that ATS1 is a member of a small gene family and that ATS3 is present as a single copy in the diploid genome. Sequence analysis of both genes showed that ATS1 is similar to the rice EFA27 gene and that ATS3 is unique. Western analysis and light level immunocytochemistry using antisera raised against the putative ATS1 and ATS3 translation products verified that ATS1 and ATS3 proteins are seed-specific and accumulate in a spatial pattern similar to their respective transcripts. Taken together, these data show that ATS1 and ATS3 are novel embryo-specific genes in Arabidopsis.
[本文引用: 1]
PMID [本文引用: 1]

Proline (Pro) accumulation is one of the most prominent changes in plant metabolism during drought and low water potential; however, the regulation and function of Pro metabolism remain unclear. We used a combination of forward genetic screening based on a Proline Dehydrogenase1 (PDH1) promoter-luciferase reporter (PDH1:LUC2) and RNA sequencing of the Pro synthesis mutant p5cs1-4 to identify multiple loci affecting Pro accumulation in Arabidopsis (Arabidopsis thaliana). Two mutants having high PDH1:LUC2 expression and increased Pro accumulation at low water potential were found to be alleles of Cytochrome P450, Family 86, Subfamily A, Polypeptide2 (CYP86A2) and Long Chain Acyl Synthetase2 (LACS2), which catalyze two successive steps in very-long-chain fatty acid (VLCFA) synthesis. Reverse genetic experiments found additional VLCFA and lipid metabolism-related mutants with increased Pro accumulation. Altered cellular redox status is a key factor in the coordination of Pro and VLCFA metabolism. The NADPH oxidase inhibitor diphenyleneiodonium (DPI) induced high levels of Pro accumulation and strongly repressed PDH1:LUC2 expression. cyp86a2 and lacs2 mutants were hypersensitive to diphenyleneiodonium but could be reverted to wild-type Pro and PDH1:LUC2 expression by reactive oxygen species scavengers. The coordination of Pro and redox metabolism also was indicated by the altered expression of chloroplast and mitochondria electron transport genes in p5cs1-4 These results show that Pro metabolism is both influenced by and influences cellular redox status via previously unknown coordination with several metabolic pathways. In particular, Pro and VLCFA synthesis share dual roles to help buffer cellular redox status while producing products useful for stress resistance, namely the compatible solute Pro and cuticle lipids.© 2016 American Society of Plant Biologists. All Rights Reserved.
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

Proline fulfils diverse functions in plants. As amino acid it is a structural component of proteins, but it also plays a role as compatible solute under environmental stress conditions. Proline metabolism involves several subcellular compartments and contributes to the redox balance of the cell. Proline synthesis has been associated with tissues undergoing rapid cell divisions, such as shoot apical meristems, and appears to be involved in floral transition and embryo development. High levels of proline can be found in pollen and seeds, where it serves as compatible solute, protecting cellular structures during dehydration. The proline concentrations of cells, tissues and plant organs are regulated by the interplay of biosynthesis, degradation and intra- as well as intercellular transport processes. Among the proline transport proteins characterized so far, both general amino acid permeases and selective compatible solute transporters were identified, reflecting the versatile role of proline under stress and non-stress situations. The review summarizes our current knowledge on proline metabolism and transport in view of plant development, discussing regulatory aspects such as the influence of metabolites and hormones. Additional information from animals, fungi and bacteria is included, showing similarities and differences to proline metabolism and transport in plants.
DOIPMID [本文引用: 1]

Salinity, drought and low temperature are the common forms of abiotic stress encountered by land plants. To cope with these adverse environmental factors, plants execute several physiological and metabolic responses. Both osmotic stress (elicited by water deficit or high salt) and cold stress increase the endogenous level of the phytohormone abscisic acid (ABA). ABA-dependent stomatal closure to reduce water loss is associated with small signaling molecules like nitric oxide, reactive oxygen species and cytosolic free calcium, and mediated by rapidly altering ion fluxes in guard cells. ABA also triggers the expression of osmotic stress-responsive (OR) genes, which usually contain single/multiple copies of cis-acting sequence called abscisic acid-responsive element (ABRE) in their upstream regions, mostly recognized by the basic leucine zipper-transcription factors (TFs), namely, ABA-responsive element-binding protein/ABA-binding factor. Another conserved sequence called the dehydration-responsive element (DRE)/C-repeat, responding to cold or osmotic stress, but not to ABA, occurs in some OR promoters, to which the DRE-binding protein/C-repeat-binding factor binds. In contrast, there are genes or TFs containing both DRE/CRT and ABRE, which can integrate input stimuli from salinity, drought, cold and ABA signaling pathways, thereby enabling cross-tolerance to multiple stresses. A strong candidate that mediates such cross-talk is calcium, which serves as a common second messenger for abiotic stress conditions and ABA. The present review highlights the involvement of both ABA-dependent and ABA-independent signaling components and their interaction or convergence in activating the stress genes. We restrict our discussion to salinity, drought and cold stress.
[本文引用: 1]
