 ,, 裴瑞琴, 杨维丰, 朱海涛, 刘桂富, 张桂权, 王少奎
,, 裴瑞琴, 杨维丰, 朱海涛, 刘桂富, 张桂权, 王少奎 ,*广东省植物分子育种重点实验室/亚热带农业生物资源保护与利用国家重点实验室/华南农业大学, 广东广州 510642
,*广东省植物分子育种重点实验室/亚热带农业生物资源保护与利用国家重点实验室/华南农业大学, 广东广州 510642Mapping and identification QTLs controlling grain size in rice (Oryza sativa L.) by using single segment substitution lines derived from IAPAR9
ZHANG Bo ,, PEI Rui-Qing, YANG Wei-Feng, ZHU Hai-Tao, LIU Gui-Fu, ZHANG Gui-Quan, WANG Shao-Kui
,, PEI Rui-Qing, YANG Wei-Feng, ZHU Hai-Tao, LIU Gui-Fu, ZHANG Gui-Quan, WANG Shao-Kui ,*Guangdong Provincial Key Laboratory of Plant Molecular Breeding/State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources/South China Agricultural University, Guangzhou 510642, Guangdong, China
,*Guangdong Provincial Key Laboratory of Plant Molecular Breeding/State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources/South China Agricultural University, Guangzhou 510642, Guangdong, China通讯作者: * 王少奎, E-mail:shaokuiwang@scau.edu.cn
收稿日期:2020-08-17接受日期:2021-01-13网络出版日期:2021-08-12
| 基金资助: |
Received:2020-08-17Accepted:2021-01-13Online:2021-08-12
| Fund supported: |
作者简介 About authors
E-mail:zhangboscau@163.com

摘要
水稻粒型是由多基因控制的复杂数量性状, 是稻米产量和品质的重要影响因子, 水稻粒型基因的定位与遗传研究有助于稻米产量与品质的协同改良。本研究利用巴西陆稻IAPAR9为供体、以华南地区高产籼稻品种华粳籼74为受体, 构建的153份水稻单片段代换系材料, 连续两季通过单因素方差分析和邓肯氏多重比较, 结合代换片段重叠群作图, 定位了13个控制水稻粒型及粒重的QTL。这13个位点分别分布于水稻1、2、4、5、6、7、9和11号染色体上, 包括9个控制谷粒长的QTL、1个控制谷粒宽的QTL和3个控制千粒重的QTL。其中, qGL1-2、qTGW1-2和qGL11为新鉴定的QTL位点。新的粒型QTL定位将为进一步的基因克隆与粒型遗传调控网络解析提供依据和线索, 也可为稻米产量与品质协同改良提供新的种质资源。
关键词:
Abstract
Rice grain size is a complex quantitative trait controlled by multiple genes. Grain size is an important factor affecting rice yield and quality. The mapping and genetic analysis of genes controlling rice grain size are essential for the concurrent improvement of rice yield and quality. Here 13 QTLs for grain size were detected using 153 rice single-segment substitution lines in rice, which were derived from HJX74 as the receptor parent, and IAPAR9 as the donor parent. One-way ANOVA and Duncan’s multiple comparison were employed to detect the genetic bases of rice grain size in two consecutive years. Based on the substitution mapping using overlapped substitution-fragment in the SSSLs, a total of 13 grain size-related QTLs were detected on chromosomes 1, 2, 4, 5, 6, 7, 9, and 11, including nine QTLs controlling grain length, one QTL controlling grain width, and three QTLs controlling 1000-grain weight. Furthermore, qGL1-2, qTGW1-2, and qGL11 were novel identified QTLs. This study provided new basis for cloning and functional analysis of genes regulating grain size.
Keywords:
PDF (3403KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张波, 裴瑞琴, 杨维丰, 朱海涛, 刘桂富, 张桂权, 王少奎. 利用单片段代换系鉴定巴西陆稻IAPAR9中的粒型基因. 作物学报[J], 2021, 47(8): 1472-1480 DOI:10.3724/SP.J.1006.2021.02056
ZHANG Bo, PEI Rui-Qing, YANG Wei-Feng, ZHU Hai-Tao, LIU Gui-Fu, ZHANG Gui-Quan, WANG Shao-Kui.
水稻是世界上最重要的粮食作物之一, 有超过半数的人口以稻米为食。随着现代生活水平逐步提高, 人们对稻米品质的要求也日益增高, 培育高产、优质的水稻品种已成为水稻育种重要目标。水稻粒型直接决定稻米外观品质, 同时也是影响稻谷千粒重的重要因素, 因此, 粒型是水稻高产优质育种的重要靶标性状, 粒型的遗传分析与分子机制研究备受关注。
水稻粒型是受多基因控制的复杂农艺性状。伴随着近代遗传学与基因组学的不断发展, 通过构建F2、重组自交系(recombinant inbreed line, RIL)等初级作图群体以及染色体片段代换系(chromosome segment substitute line, CSSL)等次级作图群体, 大量控制水稻粒型的QTL (quantitative trait locus)陆续被定位[1,2,3,4,5,6,7,8]; 近年来, 全基因组关联分析(genome-wide association study, GWAS)、突变位点图谱(MutMap)等技术更是加快了水稻粒型QTL定位的速度与效率[9,10,11,12]。截至目前, 已经有超过600个粒型相关的QTL被鉴定, 涵盖粒宽、粒长、粒厚、长宽比和粒重等, 被鉴定到的QTL遍布于水稻12条染色体[1,13]。但目前已被克隆的水稻粒型基因数量还较少, 其中, 利用自然变异材料克隆到的粒型基因, 仅有GW2[14]、GS2[15]、GS3[16]、GL3.1[17]、GL3.3/TGW3[18,19,20]、GW5[21,22]、GS5[23]、TGW6[24]、GW6[25]、GW7[26]/ GL7[27]、GLW7[28]、GW8[29]和GS9[30]等十余个。
已克隆的粒型基因数量远远低于已定位的QTL数量, 这与定位群体的复杂性密切相关。传统定位中用到的F2、RIL等群体背景较为复杂, 后代往往持续分离, 对基因的精细定位造成一定的干扰。近年来, GWAS技术被广泛用于QTL定位, 它能够将目标QTL定位在某一区间, 但由于实验材料的复杂多样, 且缺乏必要的遗传材料对基因进行功能解析, 获得的优良基因也难以直接运用到水稻育种实践中。水稻单片段代换系(single segment substitution lines, SSSL)材料经过多代回交和分子标记辅助选择培育而成, 通常只携带一段来自供体亲本的染色体片段, 遗传背景与受体亲本高度相似, 是鉴定复杂农艺性状QTL, 解析目的基因功能的理想材料[31]。经过长期积累, 本团队已构建供体亲本2360份, 包括亚洲栽培稻、非洲栽培稻以及AA基因组野生稻的SSSL材料[31], 但尚未针对单一亲本为供体的SSSL材料系统考察粒型相关QTL。巴西陆稻IAPAR9谷粒较大, 千粒重较高, 本研究利用以其为供体的153份SSSL材料鉴定粒型QTL, 并对鉴定到的QTL进行初步定位和遗传分析。
1 材料与方法
1.1 试验材料
试验材料为华南地区高产籼稻品种华粳籼74 (HJX74)、巴西陆稻IAPAR9和153份以华粳籼74为受体亲本、IAPAR9为供体亲本的水稻单片段代换系材料。水稻材料由本团队水稻种质资源库低温保存。2份亲本材料及153份单片段代换系材料分别于2018年晚季、2019年早季在广州市华南农业大学启林北试验农场种植。每个材料3个重复, 每个重复1个双垄(20株), 按照10 cm × 15 cm单株插秧, 常规管理。1.2 材料的鉴定
插秧7 d后, 按照单株取水稻幼嫩叶片, 利用TPS法[32]提取基因组DNA。以合格的DNA样品作为模板, 使用SSR标记进行PCR扩增, 结合6%聚丙烯酰胺变性凝胶电泳进行检测。带型与HJX74带型一致的标记为“1”, 与IAPAR9带型一致的标记为“3”, 据此判定代换片段的位置与长度, 并选取携带纯合代换片段的材料用于表型分析。1.3 材料表型考察
种子完熟后, 按单株收种, 充分晾干后, 考察粒长、粒宽和千粒重。粒长和粒宽的考察: 按照不同材料编号随机挑选发育健康、大小均一的种子100粒, 利用彩色平台式扫描仪(MICROTER MRS- 9600TFU2L)扫描记录; 采用种子外观分析软件(万深SC-E型)进行图像处理, 读取粒长和粒宽等数据。千粒重考察方法为: 充分晾干谷粒, 随机取出500粒饱满健康的种子进行称重, 重复3次(每次重量之差小于平均值的3%), 折算成千粒重。1.4 数据分析与QTL定位
实验数据采用SPSS 23.0以及Microsoft Excel 2016进行统计与分析; 图表绘制采用MapChart、Origin 2018和Photoshop CS进行。利用单因素方差分析(ANOVA)和邓肯氏(Duncan’s)多重比较分析单片段代换系材料和HJX74目的性状的差异显著性。在0.01水平下进行多重比较, 与HJX74在同一组别的SSSL材料不携带粒型相关QTL, 而与HJX74不在同一组别的SSSL材料则携带粒型QTL; 在2个测试季节均被检测到表型变异的SSSL被认为携带稳定遗传的QTL, 仅在一个季节检测到的表型变化不记为QTL。采用重叠群作图法, 对代换片段存在重叠区间的粒型QTL进行初步定位[33]。加性效应计算方法为HJX74的表型值与携带粒型基因的SSSL的表型值的差值的一半[34]。2 结果与分析
2.1 IAPAR9的粒型与携带的粒型基因
HJX74是本团队育成的高产品种, 籽粒大小适中。IAPAR9由巴西农业科学院稻豆研究中心于1970年育成, 1992年由中国农业科学院检疫、试种并陆续在江西、贵州和北京等地审定。该品种谷粒长为(9.65 ± 0.05) cm, 较HJX74增长约17.60%, 粒宽增加约4.20%, 千粒重也比HJX74重约10.50%, 相关性状差异均达到极显著水平(图1)。以二者为亲本构建定位群体, 鉴定水稻粒型QTL切实可行。图1
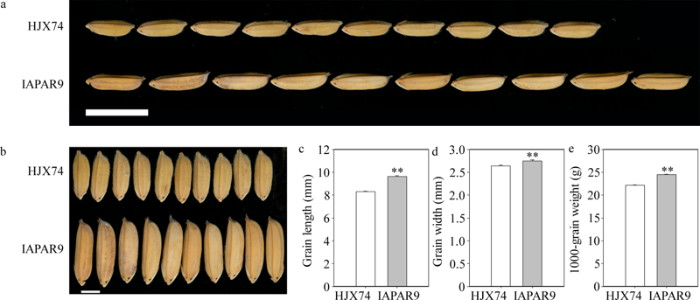 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1IAPAR9与HJX74的粒型
a, b: HJX74和IAPAR9的粒型, 标尺为5 mm; c: HJX74和IAPAR9的粒长; d: HJX74和IAPAR9的粒宽; e: HJX74和IAPAR9的粒重。**表示HJX74的粒长、粒宽和千粒重与IAPAR9之间的差异达到极显著水平(P < 0.01), t检验。
Fig. 1Grain size of IAPAR9 and HJX74
a, b: the grain shape of HJX74 and IAPAR9, Bar: 5 mm; c: the grain length of HJX74 and IAPAR9; d: the grain width of HJX74 and IAPAR9; e: the grain weight of HJX74 and IAPAR9. ** means significant difference in grain length, grain width and grain weight between HJX74 and IAPAR9 by Student’s t-test (P < 0.01).
利用水稻自然变异已克隆了GW2[14]、GS3[16]等十余个粒型基因。本研究利用本团队重测序数据结合Sanger测序, 系统分析了IAPAR9和HJX74中相关粒型基因的功能位点。结果表明, IAPAR9和HJX74二者中的GW2[14]、TGW6[24]、GW6[25]、GLW7[28]和GS9[30]等基因在功能位点上序列一致。而LGY3[35]、gs3[16]、GL3.1[17]、GW5[21,22]、GS5[23]、GW7[26]和gw8[29]等粒型基因在二者中存在功能位点上的序列差异, 上述基因位点在IAPAR9中均为功能变异类型。针对存在功能变异的位点, 开发了功能基因紧密连锁标记(引物序列见附表1), 相关标记可明确区分HJX74和IAPAR9中粒型基因的不同等位变异(图2)。尽管IAPAR9中存在gs3[16]、GW5[21,22]和GW7[26]等多个粒型基因功能变异, 但粒型是极为复杂的数量性状位点, 继续解析造成IAPAR9和HJX74粒型差异的其他基因位点将为系统理解水稻粒型的遗传调控网络提供更多证据。
Table S1
附表1
附表1引物名称及序列
Table S1
| 标记名称 Marker | 基因 Gene | 序列 Sequence (5′-3′) | 标记名称 Marker | 基因 Gene | 序列 Sequence (5′-3′) |
|---|---|---|---|---|---|
| xp14F | LGY3 | TGTCAACAGAAACCAGCAAAA | RMw513F | GW5 | GTATTTGTTTGTCGCATTC |
| xp14R | TGGTCGATCAATTCCTCTGC | RMw513R | TAGGACCATAGATGTGAGTTA | ||
| GS3R1 | gs3 | AGGCTGGCTTACTCGCTG | RM574F | GS5 | GGCGAATTCTTTGCACTTGG |
| GS3R2 | CAGCAGGCTGGCTTACTCTATT | RM574R | ACGGTTTGGTAGGGTGTCAC | ||
| GS3F | CTGTATATATATTTCTTGCAGGGTG | Indel-1F | GW7 | CCATAGTAAGACGACCTT | |
| RM186F | GL3.1 | TCCTCCATCTCCTCCGCTCCCG | Indel-1R | GATATTCTGTCAGCAGTT | |
| RM186R | GGGCGTGGTGGCCTTCTTCGTC | PSM710F | gw8 | GCCAGCCAAGAAAAGCGACA | |
| PSM710R | TCTTGAGATCCCACTCCATG |
新窗口打开|下载CSV
图2
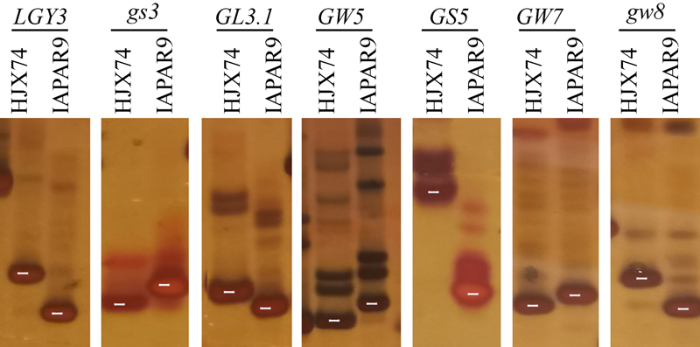 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2HJX74与IAPAR9的粒型基因分子标记多态性
白色线段指示相关引物经PCR扩增获得的目的条带, 其余带纹为引物非特异性扩增条带。
Fig. 2DNA marker polymorphism of grain size between HJX74 and IAPAR9
The white lines indicate the target bands obtained by PCR, and the other bands are non-specific amplifications.
2.2 SSSL材料的代换片段
本团队经过长期积累, 已经建成了供体来源丰富的水稻单片段代换系材料文库, 其中以IAPAR9为供体的材料总计153份。材料中所携带的来自IAPAR9的代换片段分布于水稻12条染色体上, 覆盖了约52.99%的水稻染色体, 每条染色体上的代换片段总长度变幅在3.45~123.65 cM之间, 9号染色体覆盖率最高, 12号染色体覆盖最低(图3和附表2)。每个材料中携带的代换片段长度大小不一, 代换片段平均长度为20.95 cM; 每条染色体上的代换片段数量不同, 位于1号染色体的代换片段最多, 达到30个, 而12号染色体上的代换片段最少, 仅有1个(附表2)。图3
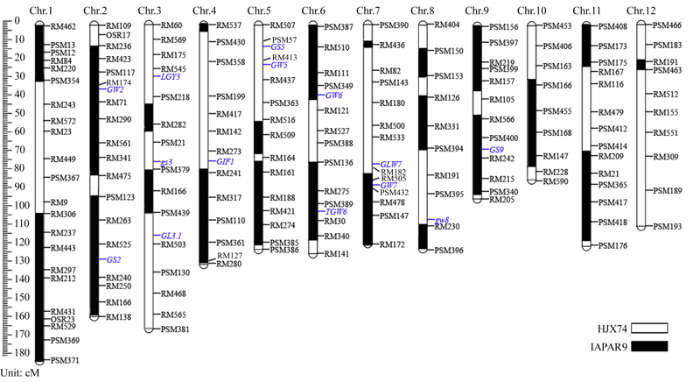 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3代换片段在水稻染色体上的覆盖率
遗传距离(单位: cM)见左侧标尺; 染色体上黑色部分代表该区域可以被来自IAPAR9的代换片段所覆盖; 蓝色字体突出显示相关位点上已克隆的水稻粒型基因。
Fig. 3Coverage of substitution fragments on rice chromosomes
The genetic distance (cM) is shown as rulers on the left margin. Filled and open bars represent chromosomal segments homozygous for IAPAR9 and HJX74 alleles, respectively. The cloned genes are shown in blue font.
Table S2
附表2
附表2单片段代换系代换片段分布
Table S2
| 染色体 Chr. | 单片段材料数量 No. of SSSLs | 代换片段总长度 TL (cM) | 代换片段平均长度 CL (cM) | 代换片段覆盖率 CP (%) |
|---|---|---|---|---|
| Chr. 1 | 30 | 571.00 | 19.03 | 59.16 |
| Chr. 2 | 22 | 526.00 | 23.91 | 78.31 |
| Chr. 3 | 4 | 112.40 | 28.10 | 35.21 |
| Chr. 4 | 14 | 380.70 | 27.19 | 43.90 |
| Chr. 5 | 12 | 179.30 | 14.94 | 53.98 |
| Chr. 6 | 24 | 541.40 | 22.56 | 73.55 |
| Chr. 7 | 7 | 75.20 | 10.74 | 19.27 |
| Chr. 8 | 8 | 181.40 | 22.68 | 66.91 |
| Chr. 9 | 9 | 256.70 | 28.52 | 85.03 |
| Chr. 10 | 6 | 160.60 | 26.77 | 55.93 |
| Chr. 11 | 16 | 320.60 | 20.04 | 60.04 |
| Chr. 12 | 1 | 6.90 | 6.90 | 3.15 |
| 平均Mean | 12.75 | 276.02 | 20.95 | 52.87 |
新窗口打开|下载CSV
2.3 亲本与群体的粒长、粒宽和千粒重
在2018年晚季和2019年早季, 连续两季的粒型分析表明, 153份SSSL材料的粒长、粒宽和千粒重性状变幅很大, 粒宽变幅在2.23~2.87 mm之间, 粒长为7.53~9.04 mm, 千粒重变化区间为14.00~25.44 g。粒长、粒宽和千粒重均变现为连续变异, 接近正态分布; 其中, 粒宽和千粒重在2个种植季节均存在超亲分离; 粒长在2019年早季也观测到超亲分离现象(图4)。图4
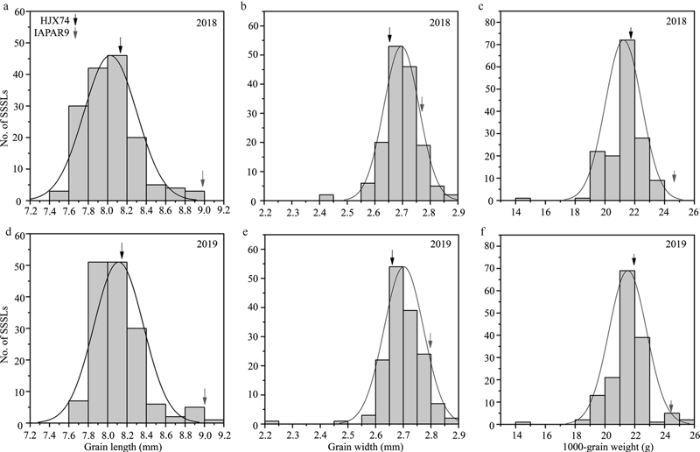 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4亲本和SSSL材料粒长、粒宽和千粒重频次分布
a~c: 2018晚季亲本和SSSL的粒长、粒宽和千粒重的频次分布; d~f: 2019早季亲本和SSSL的粒长、粒宽和千粒重的频次分布。
Fig. 4Frequency distribution of grain length, width and 1000-grain weight in SSSLs and the parents in two seasons
a-c: the frequency distribution of grain length, width, and 1000-grain weight in SSSLs and the parents in later season of 2018; d-f: the frequency distribution of grain length, width, and 1000-grain weight in SSSLs and the parents in early season of 2019.
2.4 QTL定位分析
利用153份SSSL对水稻的粒长、粒宽和千粒重进行QTL检测, 共检测到13个QTL。分别分布在水稻1、2、4、5、6、7、9和11号染色体上(图5和表1), 其中包括9个粒长QTL、1个粒宽QTL和3个控制千粒重的QTL。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图513个粒型及千粒重性状QTL在水稻染色体上的分布
遗传距离见左侧标尺; 染色体右侧的黑色矩形代表QTL及其定位区间。
Fig. 5Chromosome location of 13 QTLs for grain shape and 1000-grain weight in rice
The genetic distance is shown as rulers on the left margin. Black bars on the right of each chromosome are the location intervals of QTLs for grain size with their names on the right.
Table 1
表1
表1利用SSSL定位到的粒型QTL及其加性效应
Table 1
| 性状 Trait | 位点 QTL | 染色体 Chr. | 区间 Marker interval | 加性效应Additive effect | P值P-value | ||
|---|---|---|---|---|---|---|---|
| 2018 | 2019 | 2018 | 2019 | ||||
| 粒长Grain length | qGL1-1 | 1 | PSM13-RM84 | -0.19 | -0.11 | 0.0025 | 0.0188 |
| qGL1-2 | 1 | OSR23-RM104 | 0.30 | 0.31 | 0.0001 | 0.0005 | |
| qGL2 | 2 | RM250-RM166 | -0.19 | -0.15 | <0.0001 | 0.0029 | |
| qGL4 | 4 | RM317-PSM110 | -0.23 | -0.15 | 0.0002 | 0.0081 | |
| qGL5 | 5 | RM164-RM161 | -0.19 | -0.12 | 0.0002 | 0.0289 | |
| qGL6 | 6 | PSM136-RM275 | -0.22 | -0.15 | <0.0001 | 0.0227 | |
| qGL7 | 7 | RM505-PSM432 | 0.37 | 0.43 | 0.0003 | <0.0001 | |
| qGL9 | 9 | PSM399-RM105 | -0.20 | -0.15 | <0.0001 | 0.0084 | |
| qGL11 | 11 | RM209-RM21 | -0.17 | -0.16 | 0.0005 | 0.0023 | |
| 粒宽Grain width | qGW2 | 2 | RM263-RM525 | 0.09 | 0.08 | 0.0003 | 0.0039 |
| 千粒重1000-grain weight | qTGW1-1 | 1 | PSM13-RM84 | -1.46 | -1.32 | <0.0001 | 0.0016 |
| qTGW1-2 | 1 | OSR23-RM104 | 0.65 | 1.25 | 0.0003 | 0.0011 | |
| qTGW4 | 4 | RM127-RM280 | -1.31 | -1.45 | <0.0001 | <0.0001 | |
新窗口打开|下载CSV
控制水稻粒长的9个QTL, 分布于水稻1、2、4、5、6、7、9和11号染色体, 2018晚季对粒长的贡献率在2.12%~4.55%, 2019年早季为1.96%~5.32%。其中, 有6个QTL加性效应为负, 其他3个QTL加性效应为正。本研究在水稻2号染色体上检测到1个控制粒宽的QTL, 其加性效应为正值, 显著增加水稻粒宽, 连续两季对粒型的贡献率均在3.00%以上。检测到的3个千粒重QTL中, 有2个位于水稻1号染色体, 1个位于4号染色体, 其中位于1号染色体长臂端的qTGW1-2加性效应为正, 与控制粒长的qGL1-1共定位; qTGW1-2与qGL1-2共定位, 对表型贡献率均表现为负效应 (表1)。该研究获得的携带优异粒型QTL的SSSL材料有望直接运用于本团队搭建的水稻设计育种平台, 也有助于克隆并解析新的粒型基因, 为阐明水稻粒型的遗传调控网络提供材料基础和理论依据。
3 讨论
本研究利用水稻SSSL材料定位了9个控制水稻粒长、1个控制水稻粒宽和3个影响千粒重的QTL。研究中用到的IAPAR9为耐旱性良好的粳型水稻, 我国已利用该材料育成多个审定品种, 挖掘和研究其中的粒型QTL/基因, 有助于我们深入了解粒型等位基因在水稻籼粳亚种间的遗传特点, 也将直接为水稻外观品质改良提供新的种质和基因资源。本研究定位的多个QTL与前人已有的定位区间重叠, 部分QTL所定位的区间内已有控制水稻粒型的主效基因被克隆。定位于1号染色体上的qGL1-1和qTGW1-1与前人定位到的TGW1.3[36]位于同一区间内, 可能为相同的QTL。定位于2号染色体上的qGW2与已有的研究中定位的GL2[37]、GW2.3[38]和qgl2-2[39]等存在定位区间的重叠, 该区间里包含影响粒型的AFG1[40], AFG1编码转录激活因子, 通过影响调控细胞增殖和伸长基因的表达, 调控水稻籽粒大小。本研究定位到的qGL2与GL2-1[2]、qGW2-2[41]等QTL所在区间重叠, 该区间存在一个已克隆的、负向调控水稻粒宽和粒重的TGW2[42], 该基因的编码蛋白与调控细胞周期的KRP1蛋白存在互作, 影响细胞增殖和细胞生长, 但我们定位到的qGL2主要控制水稻粒长, 二者效应不同, 也有可能是一个控制水稻粒长的新位点。在我们定位的位于4号染色体上的qGL4所在区间, 已有GW4[2]、qGL-4[43]以及qGW4[41]等QTL报道; 位于4号染色体上主要控制千粒重的qTGW4则与已克隆的GL4[44]位于同一区间内, GL4调控水稻籽粒大小, 进而影响水稻的千粒重与单株产量, 二者遗传效应相似, 极有可能为同一基因。5号染色体上的qGL5与邹德堂等[37]定位的GL5有重叠部分, 该区间涵盖一个编码转录因子的SERF1, SERF1是RPBF和GBSSI的直接调控因子, 能够负调控水稻籽粒大小[40], 可能是qGL5的候选基因。qGL6定位于6号染色体, 其所在区间内有已被克隆的、调控水稻粒长和粒重的TGW6[24], 但IAPAR9的TGW6与HJX74并无功能位点上的序列变异, 这暗示了该区间内存在一个尚未被发现的水稻粒型新位点。GW7[26]/GL7[27]是位于水稻7号染色体, 控制粒长和粒宽的主效QTL, 序列分析表明, IAPAR9携带GW7功能变异位点, 本研究定位的qGL7极有可能与GW7[26]/GL7[27]为同一位点。定位于9号染色体上的qGL9与已报道的TGW9.1[2]所在区间重叠。qTGW4位于4号染色体长臂末端, 该位点没有发现粒型、粒宽的变化, 千粒重的变化可能由于谷粒厚度或灌浆饱满程度的影响造成的。qGL1-2、qTGW1-2和qGL11等3个QTL所在区间尚未有利用作图群体定位到粒型QTL的报道, 其中, qGL1-2和qTGW1-2共定位于水稻1号染色体长臂末端, 该位点可能存在一个协同增加水稻粒长和千粒重的正效应因子, 有望为稻米产量与品质协同改良提供新的种质资源。上述发现丰富了我们对水稻粒型遗传调控的认识。
IAPAR9携带gs3[16]、LGY3[35]、GL3.1[17]、GW5[21,22]、GS5[23]和GW7[26]等多个粒型基因功能变异, 但是由于本研究所利用的SSSL材料中的代换片段未能完全覆盖水稻的整个染色体组, 特别是gs3[16]、LGY3[35]、GW5[21,22]、GS5[23]和gw8[29]所在区间均未被代换片段覆盖(图3), 上述基因位点并未在本研究中被检测出来; 而7号染色体上具有覆盖粒型主效基因GW7[26]/GL7[27]所在区间的代换片段, 在该区间鉴定到控制粒长的qGL7, qGL7在2个检测季节中均是具有最高贡献率的粒长QTL。综上, 只要具有足够的代换片段覆盖度, 利用SSSL材料可以全面解析控制相关农艺性状的QTL。另外, SSSL也是研究复杂性状QTL的理想材料, 与受体亲本相比, SSSL只携带一个来自供体的染色体片段, 其遗传背景与受体亲本高度相似, 是受体亲本的天然近等基因系, 能够极大限度地排除遗传背景干扰[31], 这将为后续基因的克隆与功能解析提供扎实的材料基础。而且, 本团队已经建成HJX74背景下, 供体涵盖稻属AA基因组7个种的SSSL文库, 全部代换片段大约覆盖水稻基因组29倍[30], 为功能基因的等位分析提供了充分的材料保障。基于SSSL材料建立的设计育种体系[45,46], 则进一步促进了发掘到的优良基因资源在水稻设计育种中的应用。
4 结论
本研究利用153份以巴西陆稻IAPAR9为供体亲本籼稻品种HJX74为受体亲本构建的SSSL材料, 在2018年晚季和2019年早季鉴定水稻粒长、粒宽和千粒重QTL, 本研究共鉴定到13个在2个不同季节稳定遗传的QTL, 包括9个粒长QTL、1个粒宽QTL和3个千粒重QTL。其中, 仅qGW2、qGL2和qGL7所在的定位区间有已克隆的粒型相关基因被报道。本研究结果为新粒型基因进一步精细定位、克隆和功能解析奠定了良好的基础, 获得的优异种质资源将有助于高产优质水稻设计育种。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
[本文引用: 2]
[本文引用: 4]
[本文引用: 4]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 3]
DOIURL [本文引用: 1]
DOIURL [本文引用: 6]
DOIURL [本文引用: 3]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 5]
DOIURL [本文引用: 5]
DOIURL [本文引用: 4]
DOIURL [本文引用: 3]
DOIURL [本文引用: 2]
DOIURL [本文引用: 7]
DOIURL [本文引用: 4]
DOIURL [本文引用: 2]
DOIURL [本文引用: 3]
DOIURL [本文引用: 3]
[本文引用: 3]
[本文引用: 3]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

Quantitative trait loci (QTLs) have been mapped to small intervals along the chromosomes of tomato (Lycopersicon esculentum), by a method we call substitution mapping. The size of the interval to which a QTL can be mapped is determined primarily by the number and spacing of previously mapped genetic markers in the region surrounding the QTL. We demonstrate the method using tomato genotypes carrying chromosomal segments from Lycopersicon chmielewskii, a wild relative of tomato with high soluble solids concentration but small fruit and low yield. Different L. chmielewskii chromosomal segments carrying a common restriction fragment length polymorphism were identified, and their regions of overlap determined using all available genetic markers. The effect of these chromosomal segments on soluble solids concentration, fruit mass, yield, and pH, was determined in the field. Many overlapping chromosomal segments had very different phenotypic effects, indicating QTLs affecting the phenotype(s) to lie in intervals of as little as 3 cM by which the segments differed. Some associations between different traits were attributed to close linkage between two or more QTLs, rather than pleiotropic effects of a single QTL: in such cases, recombination should separate desirable QTLs from genes with undesirable effects. The prominence of such trait associations in wide crosses appears partly due to infrequent reciprocal recombination between heterozygous chromosomal segments flanked by homozygous regions. Substitution mapping is particularly applicable to gene introgression from wild to domestic species, and generally useful in narrowing the gap between linkage mapping and physical mapping of QTLs.
DOIURL [本文引用: 1]
DOIURL [本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
