 ,1,3,*, 张国平1
,1,3,*, 张国平1Genome-wide association study of β-glucan content in barley grains
GENG La1, HUANG Ye-Chang2, LI Meng-Di1, XIE Shang-Geng1, YE Ling-Zhen ,1,3,*, ZHANG Guo-Ping1
,1,3,*, ZHANG Guo-Ping1通讯作者:
收稿日期:2020-08-23接受日期:2020-12-1网络出版日期:2020-12-30
| 基金资助: |
Received:2020-08-23Accepted:2020-12-1Online:2020-12-30
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (2090KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
耿腊, 黄业昌, 李梦迪, 谢尚耿, 叶玲珍, 张国平. 大麦籽粒β-葡聚糖含量的全基因组关联分析[J]. 作物学报, 2020, 47(7): 1205-1214. doi:10.3724/SP.J.1006.2021.01074
GENG La, HUANG Ye-Chang, LI Meng-Di, XIE Shang-Geng, YE Ling-Zhen, ZHANG Guo-Ping.
大麦是全球广泛栽培的主要谷类作物, 且用途多样, 集饲用、啤用和食用于一体。与水稻、玉米和小麦等其他禾谷类作物相比, 大麦籽粒的化学成分独特, 其中富含β-葡聚糖是其主要特点之一, 平均含量高于小麦近10倍[1]。β-葡聚糖是大麦籽粒胚乳和糊粉层细胞壁的主要成份, 无论在啤用、饲用还是食用上都是一个重要的品质性状。用于酿造啤酒时, 大麦籽粒β-葡聚糖会增加麦汁黏度从而影响麦汁的过滤速度, 降低麦芽浸出物产量, 同时还因产生凝胶沉淀作用而使啤酒易发生混浊[2]; 用于饲料原料时, β-葡聚糖会增加非反刍畜禽动物肠液的黏度, 影响食料的消化和吸收, 从而降低饲用价值, 故被认为是畜禽的一种抗营养因子[3,4]。相反, 大麦用于人类食粮时, β-葡聚糖则是一种有益健康因子, 它具有降低血液胆固醇及预防结肠癌、直肠癌的作用[5]。因此, 明确大麦籽粒β-葡聚糖合成的遗传及其调控基因, 筛选与鉴定相关分子标记, 根据大麦用途调控β-葡聚糖的合成与积累, 对于提高专用大麦品种的品质具有重要意义。
首个发现与大麦β-葡聚糖合成相关的基因是纤维素合成酶类基因(CesA)[6]。后续研究显示, 大麦β-葡聚糖由纤维素合成酶类F (CslF)和纤维素合成酶类H (CslH)基因家族成员共同调控合成[7,8]。CslF基因家族是纤维素合成酶基因超家族的一部分, 由位于大麦基因组1H、2H、5H和7H染色体上的10个成员组成[8,9]; 其中CslF6基因在决定β-葡聚糖含量中发挥着重要作用。大麦过表达由胚乳特异性启动子控制的CslF6基因可显著增加籽粒β-葡聚糖含量[10], 而小麦中下调该基因表达可显著降低籽粒β-葡聚糖含量[11]。虽然大麦β-葡聚糖合成相关的基因已有报道, 但有关大麦籽粒β-葡聚糖积累的遗传调控机制仍不十分清楚。
近年来, 全基因组关联研究(genome-wide association study, GWAS)分析作为重要的遗传解析方法, 已广泛应用于水稻、玉米和小麦等作物重要农艺性状的基因鉴定研究[12,13,14]。在大麦中, 利用这一方法已对籽粒密度[15]、花期[16]、棱型[17]等重要性状进行过相关研究, 但对籽粒β-葡聚糖等重要品质性状仍有待于开展更深入和广泛的研究。本研究拟以119份大麦种质资源(包括西藏半野生大麦、国内栽培品种及国际常见商业品种)为材料, 分别种植在土壤和气候条件存在一定差异的2个试点, 对籽粒β-葡聚糖含量进行GWAS分析; 旨在获得与β-葡聚糖含量显著连锁的SNP位点, 挖掘其优异等位变异及预测候选基因, 从而为大麦β-葡聚糖的遗传改良提供理论指导与优异基因资源。
1 材料与方法
1.1 供试材料与田间试验
本研究供试大麦材料共119份, 含我国西藏半野生大麦44份、我国栽培品种59份、国外栽培品种16份。所有材料均于2018年11月初播于浙江大学长兴农业科学试验站(CHX)和浙江省慈溪农业科学试验站(CX)。长兴和慈溪的土壤pH分别为6.0和8.29, 全氮含量分别为1.22 g kg?1和1.31 g kg?1, 速效磷含量分别为8.9 mg kg?1和102 mg kg?1, 速效钾含量分别为12.5 mg kg?1和163 mg kg?1。每份材料种5行, 每行播50粒种子, 行长2 m, 行间距0.25 m。所有农艺管理措施, 包括施肥、除草和病虫害防治等与当地大田生产相同。大麦成熟时每份材料收中间3行的种子, 干燥后储存于4℃冰箱备用。1.2 β-葡聚糖含量测定
大麦种子用旋风磨粉机粉碎, 过40目筛, 密闭储存。β-葡聚糖按Houston等[18]介绍的方法, 采用Megazyme公司生产的β-葡聚糖试剂盒(K-BGLU 02/17)测定, 每份材料测定重复3次。1.3 DNA提取和基因型分析
供试大麦材料培养至二叶一心, 取叶片, 采用CTAB法提取全基因组DNA; 用1%琼脂糖凝胶电泳检测DNA质量及浓度。委托澳大利亚Dart公司(1.4 全基因组关联分析、候选基因鉴定与基因表达分析
用Structure 2.3.3软件进行群体结构Q矩阵的计算, 用SPAGeDi计算亲缘关系K矩阵。将基因型、表型、Q矩阵和K矩阵导入Tassel 5.0后, 以Q矩阵和Q+K矩阵作为协变量, 分别进行基于一般线性模型(General Linear Model, GLM)和混合线性模型 (Mixed Linear Model, MLM)的全基因组关联分析。在关联分析过程中计算获得每个SNP位点的P值和R2。将显著性阈值设置为P≤1×10-4和P≤1×10-3, 即-log10(P)≥和-log10(P)≥3, 使用R软件将GWAS分析结果绘制成曼哈顿图和QQ图。利用大麦全基因组数据库, 对GWAS分析所获的显著性标记附近提取±100 kb范围内基因作为候选基因; 进一步在数据库中提取注释信息和基因表达谱信息。
1.5 数据分析
采用SPSS软件进行频次分布图制作、相关和方差分析。使用的模型为混合线性模型: y = μ+基因型+环境+基因型×环境+误差(基因型和环境为固定因子); 遗传力按H2 = Vg/[Vg+VGL/L+Ve/(L×R)]估算, 其中Vg表示品种方差, VGL表示地点和品种的互作, Ve表示残差的方差组分, L为2 (2个地点), R为3 (3次重复)。2 结果与分析
2.1 β-葡聚糖含量的基因型与环境差异
供试的119份大麦材料的β-葡聚糖含量在2个环境中存在显著的差异, 长兴和慈溪点β-葡聚糖含量分别变动于1.66%~5.48%和1.98%~6.49%, 平均含量分别是3.03%和3.39% (图1)。所有材料在2个环境种植, β-葡聚糖含量的相关系数为0.75 (P≤0.001), 达极显著水平, 且呈正态分布。方差分析表明, 基因型、环境、互作和误差可分别解释表型变异的76.46%、8.18%、12.40%和2.96% (表1), 且β-葡聚糖含量在2个环境下的广义遗传力为73.9%, 说明大麦籽粒β-葡聚糖含量主要受遗传控制。图1
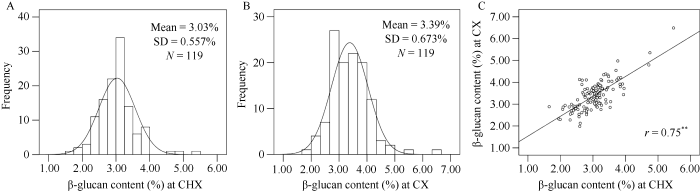 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1119份大麦材料在长兴(A)和慈溪(B)的β-葡聚糖含量分布以及2个地点的β-葡聚糖含量相关性(C)
**表示在0.01水平差异显著。** Significant differences at P = 0.01.
Fig. 1Distribution frequency of β-glucan content of the examined 119 barley accessions at CHX (A) and CX (B) and correlation between two different sites (C)
Table 1
表1
表1119份栽培品种在2个不同环境中的β-葡聚糖含量方差分析
Table 1
| 变异源 Source of variation | III型平方和 SS | 自由度 DF | 均方 MS | F值 F-value | R2 (%) |
|---|---|---|---|---|---|
| 基因型 Genotype | 235.00 | 118 | 1.98 | 104.14*** | 76.46 |
| 环境 Environment | 25.13 | 1 | 25.13 | 1325.35*** | 8.18 |
| 环境×基因型 Genotype × Environment | 38.12 | 118 | 0.32 | 16.90*** | 12.40 |
| 误差 Residual error | 9.10 | 476 | 0.02 | 2.96 | |
| 总计 Total | 307.35 | 713 |
新窗口打开|下载CSV
2.2 β-葡聚糖含量的全基因组关联分析
利用MLM模型对2个环境下的β-葡聚糖含量进行全基因组关联分析, 当阈值为-log10(P)≥3时, 在长兴和慈溪2个环境型中分别检测到7个和2个显著位点(图2和表2), 其中位于5号染色体的标记M-3924095 (标记编号4)在2个环境中均被检测到。合并重叠的位点后得到8个显著性位点, 分布在2号~7号染色体, 可解释13.5%~16.98%的表型变异。图2
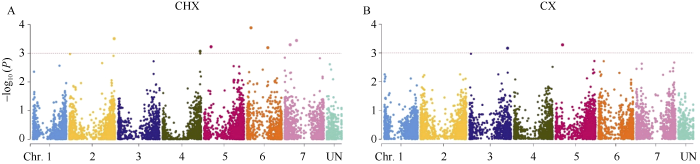 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2MLM模型下β-葡聚糖含量的曼哈顿图
A: 长兴β-葡聚糖含量MLM曼哈顿图; B: 慈溪β-葡聚糖含量MLM曼哈顿图。水平线代表显著相关的1E-03阈值。
Fig. 2Manhattan plot of grain β-glucan content in MLM model
A: Manhattan plot of MLM in CHX; B: Manhattan plot of MLM in CX. The horizontal line depicts the 1E-03 threshold for significant association.
Table 2
表2
表2MLM模型下β-葡聚糖含量显著相关的SNP位点(-log10(P) > 3)
Table 2
| 标记编号 MTA No. | 标记名称 Marker | 染色体 Chr. | 位置 Position | 峰值 -log10(P) | 表型贡献率 R2 (%) | 环境 Environment | 最佳等位基因a SNP a | 最小等位基因频率 MAF | 候选基因 Putative candidated genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M-4172115 | 2 | 753357365 | 3.50 | 16.98 | CHX | A/G | 0.14 | HORVU2Hr1G122510, HORVU2Hr1G122540, HORVU2Hr1G122560, HORVU2Hr1G122570, HORVU2Hr1G122580 |
| 2 | M-3262394 | 3 | 640508648 | 3.16 | 13.50 | CX | A/G | 0.12 | HORVU3Hr1G092890 |
| 3 | M-3432496 | 4 | 629360418 | 3.07 | 13.55 | CHX | C/T | 0.14 | HORVU4Hr1G084590 |
| 4 | M-3924095 | 5 | 116486105 | 3.28 3.22 | 14.20 14.40 | CX CHX | A/G | 0.07 | HORVU5Hr1G022710 |
| 5 | M-3263056 | 6 | 68979147 | 3.88 | 13.65 | CHX | G/T | 0.05 | HORVU6Hr1G022070, HORVU6Hr1G022080 |
| 6 | M-4792001 | 6 | 350443314 | 3.19 | 15.62 | CHX | C/G | 0.11 | HORVU6Hr1G055090 |
| 7 | M-3911599 | 7 | 93157425 | 3.29 | 14.13 | CHX | A/C | 0.05 | HORVU7Hr1G038040, HORVU7Hr1G038060, HORVU7Hr1G038070, HORVU7Hr1G038080 |
| 8 | M-3665085 | 7 | 199549260 | 3.44 | 14.79 | CHX | A/G | 0.17 | HORVU7Hr1G052390, HORVU7Hr1G052400 |
新窗口打开|下载CSV
同时, 利用GLM模型对2个环境下的β-葡聚糖含量进行全基因组关联分析, 当阈值为-log10(P)≥4时, 在长兴和慈溪分别检测到32个和17个显著位点(图3和表3), 其中有9个位点在2个环境中均被检测到。合并重叠的位点后得到40个显著性位点, 分布在各条染色体, 可解释12.24%~22.93%的表型变异。
比较以上2个关联模型的结果, MLM模型和GLM模型有4个位点重叠, 分别是M-3262394、M-3924095、M-3263056和M-3911599; 合并2个模型的结果, 共得到了44个显著位点。
2.3 最佳等位基因对β-葡聚糖含量的影响
为了明确检测到的显著性位点在大麦育种上利用价值, 在此我们引入最佳等位基因。对119份大麦籽粒β-葡聚糖含量和等位基因信息进行方差分析, 可显著提高β-葡聚糖含量的等位基因视为最佳等位基因(表2和表3)。在MLM模型和GLM模型中, 最佳等位基因的数量分别为0~7和3~30 (附表1), 且与β-葡聚糖含量呈显著正相关(图4)。2.4 候选基因鉴定
在显著性位点上下游100 kb范围内寻找候选基因, 基于MLM模型和GLM模型分别鉴定到17个和117个基因, 合并重合基因, 2个模型共鉴定到126个基因(表2和表3)。HORVU5Hr1G022710基因在2个模型、2个地点均被鉴定到, 可认为是和β-葡聚糖含量显著相关的候选基因。该基因的注释信息为MYB21转录因子。此外,基于基因注释, 共鉴定到10个与糖合成、转运及分解相关的酶类基因, 这些基因可能与籽粒中β-葡聚糖的合成、积累和分解紧密相关。结合基因转录表达信息, 发现这些候选基因在大麦不同组织和不同发育时期均有表达(图5和附表2); 在后续研究可通过转基因或者CRISPR/Cas9敲除, 进一步验证这些候选基因的功能。Table 3
表3
表3GLM模型下β-葡聚糖含量显著相关的SNP位点(-lg (P)>4)
Table 3
| 标记编号 MTA No. | 标记名称 Marker | 染色体 Chr. | 位置 Positon | 峰值 -lg (P) | 表型贡献率 R2 (%) | 环境 Environment | 最佳等位基因 SNP a | 最小等位基因频率 MAF | 候选基因 Putative candidate genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M-3256305 | 1 | 20684690 | 6.55 4.18 | 21.39 14.56 | CHX CX | C/T | 0.235 | HORVU1Hr1G009320, HORVU1Hr1G009330 |
| 2 | M-3432526 | 1 | 443440212 | 4.89 4.08 | 14.79 12.24 | CHX CX | A/G | 0.050 | HORVU1Hr1G061160, HORVU1Hr1G061160 |
| 3 | M-3259980 | 2 | 40851287 | 4.89 | 19.40 | CX | A/G | 0.143 | HORVU2Hr1G017350 |
| 4 | M-5249622 | 2 | 41956883 | 4.10 | 15.89 | CX | C/G | 0.286 | HORVU2Hr1G017400 |
| 5 | M-29425925 | 2 | 42674382 | 4.45 | 17.86 | CX | C/T | 0.235 | HORVU2Hr1G017470 |
| 6 | M-3259183 | 2 | 752978172 | 4.79 4.18 | 19.75 16.80 | CHX CX | C/T | 0.370 | HORVU2Hr1G122270, HORVU2Hr1G122280 HORVU2Hr1G122300, HORVU2Hr1G122310 HORVU2Hr1G122320 |
| 7 | M-3918521 | 3 | 27028266 | 4.42 | 19.16 | CX | A/G | 0.168 | HORVU3Hr1G012510, HORVU3Hr1G012520 HORVU3Hr1G012530, HORVU3Hr1G012550 |
| 8 | M-4014347 | 3 | 597391541 | 4.56 | 13.78 | CHX | G/T | 0.076 | HORVU3Hr1G082030 |
| 9 | M-3917069 | 3 | 601025304 | 4.23 | 15.20 | CX | C/T | 0.067 | HORVU3Hr1G082940, HORVU3Hr1G082980 |
| 10 | M-3262394 | 3 | 640508648 | 5.48 | 19.22 | CX | A/G | 0.124 | HORVU3Hr1G092890 |
| 11 | M-3258241 | 4 | 594473630 | 4.56 | 16.27 | CHX | C/G | 0.092 | HORVU4Hr1G074460, HORVU4Hr1G074470 |
| 12 | M-3398576 | 5 | 103430100 | 4.55 | 13.72 | CHX | C/T | 0.092 | HORVU5Hr1G021420, HORVU5Hr1G021430 |
| 13 | M-3924095 | 5 | 116486105 | 5.68 5.03 | 20.83 18.82 | CHX CX | A/G | 0.067 | HORVU5Hr1G022710 |
| 14 | M-4330916 | 5 | 554243179 | 4.58 | 16.84 | CHX | G/T | 0.370 | HORVU5Hr1G077910, HORVU5Hr1G077920 HORVU5Hr1G077930, HORVU5Hr1G077940 |
| 15 | M-3257651 | 5 | 569528889 | 4.94 | 18.77 | CHX | A/C | 0.092 | HORVU5Hr1G083170, HORVU5Hr1G083210 HORVU5Hr1G083240, HORVU5Hr1G083260 |
| 16 | M-3272118 | 5 | 573026756 | 4.53 | 13.80 | CHX | A/G | 0.092 | HORVU5Hr1G084630, HORVU5Hr1G084650 |
| 17 | M-3923379 | 5 | 573026822 | 4.66 | 14.19 | CHX | A/G | 0.092 | HORVU5Hr1G084630, HORVU5Hr1G084650 |
| 18 | M-4000027 | 5 | 573026822 | 4.55 | 13.72 | CHX | A/G | 0.092 | HORVU5Hr1G084630, HORVU5Hr1G084650 |
| 19 | M-3254699 | 5 | 573766812 | 5.23 | 15.85 | CHX | C/T | 0.080 | HORVU5Hr1G085020 |
| 20 | M-5255433 | 5 | 596702361 | 5.60 | 16.99 | CHX | C/G | 0.403 | HORVU5Hr1G094660, HORVU5Hr1G094670 HORVU5Hr1G094680, HORVU5Hr1G094690 HORVU5Hr1G094700, HORVU5Hr1G094710 |
| 21 | M-3255521 | 5 | 597561678 | 4.79 | 16.99 | CHX | C/T | 0.395 | HORVU5Hr1G095040, HORVU5Hr1G095050 HORVU5Hr1G095080, HORVU5Hr1G095090 |
| 22 | M-3254781 | 5 | 597650244 | 4.55 | 14.12 | CHX | G/T | 0.387 | HORVU5Hr1G095060, HORVU5Hr1G095090 HORVU5Hr1G095100, HORVU5Hr1G095110 HORVU5Hr1G095120 |
| 23 | M-3264230 | 5 | 646301855 | 4.48 | 16.04 | CX | G/T | 0.059 | HORVU5Hr1G115310, HORVU5Hr1G115320 HORVU5Hr1G115340 |
| 24 | M-3265903 | 6 | 17194598 | 4.67 | 17.25 | CHX | C/T | 0.303 | HORVU6Hr1G009620, HORVU6Hr1G009670 HORVU6Hr1G009690, HORVU6Hr1G009700 |
| 25 | M-3261129 | 6 | 42557400 | 6.03 | 21.68 | CHX | A/G | 0.361 | HORVU6Hr1G017720, HORVU6Hr1G017740 |
| 26 | M-4185933 | 6 | 48980591 | 4.47 4.82 | 15.96 17.11 | CHX CX | A/G | 0.051 | HORVU6Hr1G018830 |
| 27 | M-3257746 | 6 | 49613603 | 4.31 | 15.55 | CHX | A/C | 0.420 | HORVU6Hr1G018930, HORVU6Hr1G018950 |
| 28 | M-3263056 | 6 | 68979147 | 6.08 | 18.45 | CHX | G/T | 0.050 | HORVU6Hr1G022070, HORVU6Hr1G022080 |
| 29 | M-3920842 | 6 | 264916265 | 4.64 | 16.9 | CHX | A/G | 0.050 | HORVU6Hr1G045550, HORVU6Hr1G045590 HORVU6Hr1G045600 |
| 30 | M-3257940 | 6 | 545380415 | 4.30 | 17.26 | CHX | A/G | 0.353 | HORVU6Hr1G081290, HORVU6Hr1G081310 |
| 31 | M-3662578 | 7 | 3735940 | 6.55 4.68 | 22.93 17.46 | CHX CX | A/G | 0.303 | HORVU7Hr1G001750, HORVU7Hr1G001760 HORVU7Hr1G001790, HORVU7Hr1G001800 |
| 32 | M-3262082 | 7 | 4764013 | 5.25 5.12 | 19.29 18.53 | CHX CX | G/T | 0.387 | HORVU7Hr1G002490, HORVU7Hr1G002500 HORVU7Hr1G002530 |
| 33 | M-4016281 | 7 | 4834039 | 4.74 4.64 | 18.97 18.67 | CHX CX | A/G | 0.336 | HORVU7Hr1G002530, HORVU7Hr1G002540 HORVU7Hr1G002550, HORVU7Hr1G002560 HORVU7Hr1G002570 |
| 34 | M-3256657 | 7 | 7391399 | 4.44 | 20.21 | CX | A/C | 0.328 | HORVU7Hr1G005200 |
| 35 | M-6284081 | 7 | 21620741 | 5.41 | 16.70 | CHX | C/T | 0.050 | HORVU7Hr1G016920 |
| 36 | M-3911599 | 7 | 93157425 | 4.68 | 16.66 | CHX | A/C | 0.050 | HORVU7Hr1G038040, HORVU7Hr1G038060 HORVU7Hr1G038070, HORVU7Hr1G038080 |
| 37 | M-9772928 | 7 | 636164939 | 4.71 4.36 | 16.73 15.64 | CHX CX | C/T | 0.076 | HORVU7Hr1G112900, HORVU7Hr1G112910 HORVU7Hr1G112920, HORVU7Hr1G112930 HORVU7Hr1G112940, HORVU7Hr1G112950 |
| 38 | M-4015782 | 7 | 636588717 | 5.84 | 20.31 | CHX | C/T | 0.479 | HORVU7Hr1G113200, HORVU7Hr1G113210 HORVU7Hr1G113230, HORVU7Hr1G113270 HORVU7Hr1G113290, HORVU7Hr1G113300 HORVU7Hr1G113320 |
| 39 | M-3258181 | 7 | 641059108 | 5.15 | 18.14 | CHX | A/G | 0.437 | HORVU7Hr1G115230, HORVU7Hr1G115240 HORVU7Hr1G115250, HORVU7Hr1G115260 HORVU7Hr1G115270, HORVU7Hr1G115280 |
| 40 | M-5333232 | 7 | 641079051 | 4.65 | 16.74 | CHX | G/T | 0.370 | HORVU7Hr1G115230, HORVU7Hr1G115240 HORVU7Hr1G115250, HORVU7Hr1G115260 HORVU7Hr1G115270, HORVU7Hr1G115280 |
新窗口打开|下载CSV
图3
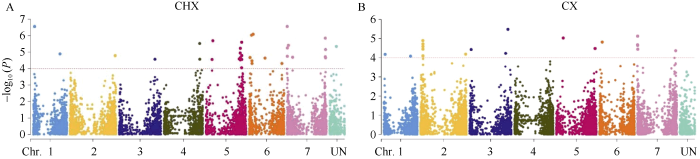 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3GLM模型下β-葡聚糖含量的曼哈顿图
A: 长兴β-葡聚糖含量GLM曼哈顿图; B: 慈溪β-葡聚糖含量GLM曼哈顿图。水平线代表显著相关的1E-04阈值。
Fig. 3Manhattan plot of grain β-glucan content in GLM model
A: Manhattan plot of GLM in CHX; B: Manhattan plot of GLM in CX. The horizontal line depicts the 1E-04 threshold for significant association.
图4
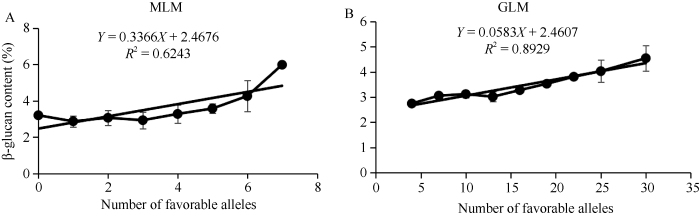 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4最佳等位基因个数和β-葡聚糖含量的线性回归分析
A: MLM模型结果; B: GLM模型结果。
Fig. 4Linear regression analysis of the number of favorable alleles and β-glucan content
A: MLM model results; B: GLM model results.
图5
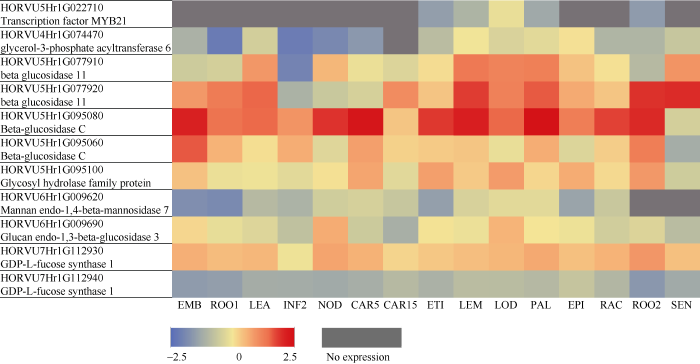 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图511个候选基因转录本表达的热图和注释
基因在各个生长发育期的转录表达值以对数进行标准化后的取值范围是-2.5~2.5, 红色表示高表达, 蓝色表示低表达, 灰色表示无表达。
Fig. 5Heat map and annotations of transcript expression of 11 putative candidate genes
For each period, the transcript expression values are calculated as LOG ranged from -2.5 to 2.5. Red blocks indicate high expression, blue indicate low expression, and gray mean no expression.
3 讨论
本研究以119份大麦基因型为材料, 种植在土壤和气候条件存在明显差异的2个试点, 测定分析籽粒β-葡聚糖含量的基因型与环境差异。结果显示, 供试的大麦群体中β-葡聚糖含量的基因型差异显著, 其广义遗传力为73.9%, 证明β-葡聚糖含量主要受遗传控制, 环境条件虽有一定影响, 但作用相对较小[18]。因此, 根据大麦的用途, 通过遗传改良手段定向调控籽粒β-葡聚糖含量是现实可行的, 也是改善这一品质性状的有效途径。本研究利用2个关联模型(MLM和GLM)鉴定调控籽粒β-葡聚糖含量的相关基因或位点, 共检测到44个显著位点, 分布于7条染色体上。在各个模型下, 两试点中均有位点重合, 且2个模型之间也有部分位点重合。这些在不同环境或模型下均检测到的位点, 可视为调控大麦籽粒β-葡聚糖含量的遗传位点。另外, 发现了可显著提高β-葡聚糖含量的等位基因, 即最佳等位基因。以上结果可为大麦分子标记辅助育种提供相关信息, 即最佳等位基因数较多的大麦品种, 如藏青320和Ticn, 适用于食用大麦育种; 相反, 那些最佳等位基因数较少的大麦品种, 如崇穗刺毛大麦和岗2, 则适用于饲用或者啤用大麦品种的育种。
国际上已有一些关于大麦籽粒β-葡聚糖含量的关联和连锁分析研究[21,22], 结合前人研究结果, 发现本研究中所发掘的位点大多数为新位点。已有大量文献显示纤维素合成酶类基因对大麦籽粒β-葡聚糖含量具有显著效应, 尤其是CslF6和CslH[7,10,23]。CslF6被认为是调控β-葡聚糖合成的主要基因, 但本研究中未鉴定到该基因。这可能是由于CslF6基因在β-葡聚糖合成中十分重要, 其编码区和启动子区的核苷酸高度保守[24]; 也有可能是其有效等位基因的比率过低(低于0.05, 被过滤)以及本研究的群体数目和SNP标记密度不够等原因引起。不过, 类似的大麦β-葡聚糖含量的遗传定位研究也有未鉴定到该基因的报道[18,25-27]。因此, CslF6在大麦β-葡聚糖合成中的作用是否存在基因型依赖性, 值得深入研究。
虽然没有鉴定到已知与β-葡聚糖合成相关的基因, 但本研究鉴定到一些和糖代谢有关的酶类基因, 主要包括一些与糖转运和糖降解有关的酶, 它们可能与β-葡聚糖含量存在一定的关联。有研究表明, 水解酶和转运酶在决定最终β-葡聚糖含量具有重要作用[28,29,30]。在β-葡聚糖合成过程中, 转运酶负责催化糖基化反应[28], 但是水解酶的具体作用尚不清楚, 仍需进一步研究。同时也有报道显示, 除纤维素合成酶类基因, 一些参与β-葡聚糖组装的酶类基因也可能对β-葡聚糖最终含量具有显著影响[31]。
本研究通过对大麦籽粒β-葡聚糖含量的GWAS分析, 其中大多数为新位点, 进一步挖掘到到一些与糖转运、降解相关的基因, 这与前人关注纤维素合成酶类基因的研究不同, 可为进一步研究β-葡聚糖的遗传调控提供了新的视角。
4 结论
本文采用GWAS方法, 分别用GLM和MLM两种模型开展了大麦籽粒β-葡聚糖含量有关的遗传调控机制解析及优良等位基因鉴定的系统研究, 共鉴定到44个重合位点, 126个候选基因。基于鉴定到的位点稳定性和基因功能注释, 共鉴定到11个可能与大麦籽粒β-葡聚糖合成、积累和分解紧密相关的酶类基因。该结果对于大麦品质性状形成和育种实践具有较好的理论和应用参考价值。附表 请见网络版: 1) 本刊网站
Supporting Information Table S1:
附表1信息:
附表1信息:119份大麦栽培品种的信息,长兴和慈溪的β-葡聚糖含量,该含量是三个重复的平均值,以及在两种模型下的最佳等位基因个数。
Supporting Information Table S1:
| 品种名 | 类型 | β-葡聚糖含量(%) β-glucan content (%) | 最佳等位基因个数 Number of favorable alleles | |||
|---|---|---|---|---|---|---|
| 长兴(CHX) | 慈溪(CX) | 均值 | MLM | GLM | ||
| X1 | 西藏野生、半野生 | 2.86% | 3.23% | 3.04% | 4 | 5 |
| X5 | 西藏野生、半野生 | 2.39% | 2.92% | 2.65% | 4 | 6 |
| X10 | 西藏野生、半野生 | 2.69% | 3.53% | 3.11% | 5 | 20 |
| X15 | 西藏野生、半野生 | 3.46% | 4.33% | 3.90% | 5 | 21 |
| X20 | 西藏野生、半野生 | 3.39% | 4.38% | 3.89% | 5 | 19 |
| X25 | 西藏野生、半野生 | 3.26% | 2.95% | 3.11% | 0 | 10 |
| X30 | 西藏野生、半野生 | 3.94% | 3.44% | 3.69% | 5 | 11 |
| X36 | 西藏野生、半野生 | 2.42% | 2.75% | 2.59% | 4 | 3 |
| X40 | 西藏野生、半野生 | 3.16% | 3.93% | 3.54% | 4 | 23 |
| X45 | 西藏野生、半野生 | 3.08% | 2.98% | 3.03% | 4 | 3 |
| X51 | 西藏野生、半野生 | 3.84% | 3.80% | 3.82% | 4 | 18 |
| X55 | 西藏野生、半野生 | 3.86% | 4.21% | 4.03% | 4 | 30 |
| X60 | 西藏野生、半野生 | 3.62% | 3.68% | 3.65% | 4 | 22 |
| X65 | 西藏野生、半野生 | 3.22% | 3.30% | 3.26% | 4 | 22 |
| X70 | 西藏野生、半野生 | 3.14% | 3.81% | 3.48% | 5 | 17 |
| X76 | 西藏野生、半野生 | 3.91% | 3.92% | 3.92% | 4 | 23 |
| X80 | 西藏野生、半野生 | 3.24% | 2.75% | 3.00% | 4 | 3 |
| X85 | 西藏野生、半野生 | 3.04% | 3.08% | 3.06% | 4 | 17 |
| X90 | 西藏野生、半野生 | 3.21% | 3.64% | 3.43% | 4 | 15 |
| X95 | 西藏野生、半野生 | 3.58% | 4.05% | 3.81% | 4 | 23 |
| X100 | 西藏野生、半野生 | 2.83% | 3.71% | 3.27% | 4 | 27 |
| X104 | 西藏野生、半野生 | 3.18% | 3.48% | 3.33% | 4 | 19 |
| X110 | 西藏野生、半野生 | 3.21% | 3.51% | 3.36% | 6 | 18 |
| X116 | 西藏野生、半野生 | 2.91% | 3.48% | 3.19% | 4 | 15 |
| X120 | 西藏野生、半野生 | 2.30% | 2.28% | 2.29% | 2 | 5 |
| X124 | 西藏野生、半野生 | 3.71% | 4.54% | 4.13% | 4 | 25 |
| X130 | 西藏野生、半野生 | 3.30% | 3.71% | 3.50% | 4 | 21 |
| X135 | 西藏野生、半野生 | 2.59% | 1.98% | 2.29% | 2 | 5 |
| X140 | 西藏野生、半野生 | 2.07% | 2.76% | 2.42% | 4 | 3 |
| X145 | 西藏野生、半野生 | 3.78% | 4.20% | 3.99% | 4 | 23 |
| X150 | 西藏野生、半野生 | 2.87% | 3.51% | 3.19% | 4 | 17 |
| X155 | 西藏野生、半野生 | 3.11% | 3.31% | 3.21% | 5 | 18 |
| X159 | 西藏野生、半野生 | 4.75% | 5.35% | 5.05% | 6 | 30 |
| X165 | 西藏野生、半野生 | 3.16% | 3.76% | 3.46% | 5 | 17 |
| X170 | 西藏野生、半野生 | 2.34% | 2.99% | 2.66% | 4 | 3 |
| X180 | 西藏野生、半野生 | 2.81% | 2.58% | 2.69% | 3 | 7 |
| X185 | 西藏野生、半野生 | 2.66% | 3.90% | 3.28% | 4 | 14 |
| X189 | 西藏野生、半野生 | 2.47% | 2.80% | 2.64% | 4 | 6 |
| X54 | 西藏野生、半野生 | 3.18% | 3.91% | 3.55% | 4 | 20 |
| X82 | 西藏野生、半野生 | 3.48% | 4.05% | 3.76% | 4 | 21 |
| X32 | 西藏野生、半野生 | 3.73% | 4.11% | 3.92% | 5 | 23 |
| X127 | 西藏野生、半野生 | 2.88% | 3.54% | 3.21% | 4 | 15 |
| X173 | 西藏野生、半野生 | 4.72% | 4.80% | 4.76% | 4 | 21 |
| X8 | 西藏野生、半野生 | 3.11% | 3.05% | 3.08% | 4 | 5 |
| 处麦1号 | 国内栽培品种 | 3.54% | 3.11% | 3.33% | 4 | 8 |
| 仙居早78 | 国内栽培品种 | 2.76% | 3.35% | 3.05% | 3 | 4 |
| 浙皮1号 | 国内栽培品种 | 2.27% | 2.87% | 2.57% | 3 | 8 |
| 白茎二棱 | 国内栽培品种 | 2.82% | 3.48% | 3.15% | 4 | 15 |
| 新登蒙古麦 | 国内栽培品种 | 3.01% | 3.60% | 3.31% | 2 | 7 |
| 农大3号 | 国内栽培品种 | 2.51% | 2.91% | 2.71% | 3 | 3 |
| 农大5号 | 国内栽培品种 | 3.10% | 3.61% | 3.36% | 4 | 10 |
| 早熟3号 | 国内栽培品种 | 3.05% | 3.47% | 3.26% | 4 | 9 |
| 舟麦1号 | 国内栽培品种 | 1.95% | 2.33% | 2.14% | 3 | 4 |
| 奉矮2号 | 国内栽培品种 | 2.14% | 2.94% | 2.54% | 3 | 4 |
| 宜山大麦 | 国内栽培品种 | 1.99% | 2.30% | 2.15% | 3 | 3 |
| 海安大麦 | 国内栽培品种 | 3.45% | 3.76% | 3.60% | 4 | 8 |
| 早矮白 | 国内栽培品种 | 3.17% | 3.09% | 3.13% | 3 | 9 |
| 余杭糯大麦 | 国内栽培品种 | 2.57% | 2.43% | 2.50% | 1 | 7 |
| 崇穗刺毛大麦 | 国内栽培品种 | 2.36% | 2.58% | 2.47% | 1 | 4 |
| 义乌二棱 | 国内栽培品种 | 2.54% | 2.76% | 2.65% | 3 | 4 |
| 浙农12 | 国内栽培品种 | 3.32% | 3.43% | 3.37% | 5 | 16 |
| 牛古特 | 国内栽培品种 | 3.19% | 3.41% | 3.30% | 3 | 8 |
| 盐66 | 国内栽培品种 | 2.59% | 2.97% | 2.78% | 3 | 6 |
| 岗2 | 国内栽培品种 | 2.80% | 2.52% | 2.66% | 2 | 4 |
| 光芒二棱 | 国内栽培品种 | 2.99% | 3.75% | 3.37% | 3 | 18 |
| 海紫2号 | 国内栽培品种 | 2.45% | 2.90% | 2.68% | 1 | 6 |
| 衢州裸麦 | 国内栽培品种 | 3.40% | 3.05% | 3.23% | 1 | 6 |
| 御岛裸 | 国内栽培品种 | 3.00% | 3.21% | 3.11% | 1 | 5 |
| 大罗锤 | 国内栽培品种 | 3.11% | 2.90% | 3.01% | 2 | 8 |
| 天台白四棱 | 国内栽培品种 | 2.94% | 2.73% | 2.84% | 2 | 8 |
| 安吉芒毛大麦 | 国内栽培品种 | 2.81% | 2.81% | 2.81% | 2 | 7 |
| 黄青裸 | 国内栽培品种 | 3.96% | 3.72% | 3.84% | 5 | 19 |
| 六棱黑麦稞 | 国内栽培品种 | 3.05% | 3.02% | 3.03% | 3 | 6 |
| 香川裸1号 | 国内栽培品种 | 3.39% | 3.20% | 3.30% | 0 | 7 |
| 嵊县江颈四棱大麦 | 国内栽培品种 | 3.06% | 3.82% | 3.44% | 2 | 4 |
| 如皋早六棱 | 国内栽培品种 | 2.82% | 2.75% | 2.79% | 1 | 8 |
| 农大2号 | 国内栽培品种 | 3.53% | 2.85% | 3.19% | 3 | 5 |
| 甬87-031 | 国内栽培品种 | 2.61% | 2.12% | 2.37% | 3 | 6 |
| 秀87-8 | 国内栽培品种 | 2.30% | 2.55% | 2.43% | 3 | 4 |
| 早熟7号 | 国内栽培品种 | 2.57% | 2.54% | 2.55% | 3 | 4 |
| 无芒六棱 | 国内栽培品种 | 3.13% | 2.86% | 3.00% | 1 | 6 |
| 春风皮大麦 | 国内栽培品种 | 3.27% | 4.08% | 3.68% | 5 | 21 |
| 肚里黄 | 国内栽培品种 | 2.76% | 3.31% | 3.03% | 4 | 24 |
| 丰产二棱 | 国内栽培品种 | 3.04% | 3.37% | 3.20% | 3 | 9 |
| 玉环洋大麦 | 国内栽培品种 | 3.13% | 3.29% | 3.21% | 1 | 7 |
| 永加仰天冲 | 国内栽培品种 | 2.84% | 3.07% | 2.96% | 2 | 7 |
| 秀87-6 | 国内栽培品种 | 2.80% | 2.69% | 2.74% | 2 | 3 |
| 浙农14 | 国内栽培品种 | 3.24% | 3.84% | 3.54% | 3 | 19 |
| 矮脚二棱 | 国内栽培品种 | 3.23% | 3.04% | 3.13% | 2 | 5 |
| 岗33 | 国内栽培品种 | 3.09% | 3.64% | 3.37% | 2 | 15 |
| 戈贝纳 | 国内栽培品种 | 2.93% | 3.09% | 3.01% | 3 | 14 |
| 浙农白壳 | 国内栽培品种 | 2.66% | 2.61% | 2.63% | 4 | 11 |
| 矮早3 | 国内栽培品种 | 2.74% | 2.71% | 2.72% | 3 | 3 |
| 红光二棱 | 国内栽培品种 | 2.95% | 3.65% | 3.30% | 2 | 7 |
| 奉矮二棱 | 国内栽培品种 | 2.95% | 3.28% | 3.12% | 2 | 3 |
| 中熟1号 | 国内栽培品种 | 2.59% | 4.13% | 3.36% | 4 | 8 |
| 浙皮2号 | 国内栽培品种 | 3.02% | 3.73% | 3.37% | 3 | 6 |
| 不知梅雨 | 国内栽培品种 | 2.75% | 3.67% | 3.21% | 3 | 16 |
| 沪麦1号 | 国内栽培品种 | 3.41% | 3.66% | 3.53% | 5 | 16 |
| Gairdner | 国外栽培品种 | 2.71% | 3.46% | 3.08% | 3 | 16 |
| Metcalfe | 国外栽培品种 | 3.19% | 3.04% | 3.11% | 4 | 17 |
| harington | 国外栽培品种 | 3.33% | 4.35% | 3.84% | 2 | 8 |
| triumph | 国外栽培品种 | 1.66% | 2.88% | 2.27% | 4 | 13 |
| boluke | 国外栽培品种 | 2.72% | 2.87% | 2.80% | 4 | 14 |
| Franklin | 国外栽培品种 | 3.25% | 4.13% | 3.69% | 2 | 12 |
| Yerong | 国外栽培品种 | 2.67% | 4.10% | 3.38% | 5 | 25 |
| morex | 国外栽培品种 | 3.83% | 3.91% | 3.87% | 3 | 20 |
| Vlamingh | 国外栽培品种 | 3.22% | 3.86% | 3.54% | 4 | 12 |
| Golden Promise | 国外栽培品种 | 2.33% | 2.70% | 2.51% | 4 | 12 |
| 藏青320 | 国内栽培品种 | 5.48% | 6.49% | 5.98% | 7 | 24 |
| haruna nijo | 国外栽培品种 | 2.40% | 2.72% | 2.56% | 3 | 9 |
| ZU9 | 国内栽培品种 | 3.10% | 3.46% | 3.28% | 2 | 7 |
| 花30 | 国内栽培品种 | 3.28% | 3.71% | 3.50% | 3 | 7 |
| 单二 | 国内栽培品种 | 3.09% | 3.19% | 3.14% | 2 | 8 |
| Tadmor | 国外栽培品种 | 2.80% | 4.11% | 3.45% | 4 | 20 |
| Mikamogold | 国外栽培品种 | 2.53% | 3.20% | 2.86% | 3 | 5 |
| 日本二条2号 | 国外栽培品种 | 2.08% | 2.86% | 2.47% | 3 | 7 |
| 拉一把 | 国内栽培品种 | 3.48% | 4.26% | 3.87% | 3 | 8 |
| Ticn | 国外栽培品种 | 3.76% | 4.98% | 4.37% | 6 | 27 |
新窗口打开|下载CSV
Supporting Information Table S2:
附表2:
附表2: 11个候选基因在不同发育时期和组织转录本表达值的对数值作为标准化值。
Supporting Information Table S2:
| 候选基因 putative candidate genes | EMB | ROO1 | LEA | INF2 | NOD | CAR5 | CAR15 | ETI | LEM | LOD | PAL | EPI | RAC | ROO2 | SEN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HORVU5Hr1G022710 Transcriptionfactor MYB21 | NE | NE | NE | NE | NE | NE | NE | -1.198 | -0.620 | 0.577 | -0.946 | NE | NE | -1.276 | NE |
| HORVU4Hr1G074470 glycerol-3-phosphate acyltransferase 6 | -0.690 | -2.088 | 0.131 | -2.060 | -1.756 | -1.369 | NE | -0.377 | 0.721 | 0.259 | 0.431 | 0.777 | -0.592 | -0.584 | -0.113 |
| HORVU5Hr1G077910 beta glucosidase 11 | 0.018 | 0.164 | 1.380 | -1.873 | 1.112 | 0.606 | 0.333 | 0.806 | 1.673 | 1.571 | 1.610 | 1.011 | 0.785 | -0.463 | 1.427 |
| HORVU5Hr1G077920 beta glucosidase 11 | 1.373 | 1.652 | 1.819 | -0.679 | -0.062 | 0.186 | 1.491 | 0.999 | 2.055 | 1.603 | 1.876 | 1.222 | 0.986 | 2.120 | 2.165 |
| HORVU5Hr1G095080 Beta-glucosidase C | 2.261 | 1.731 | 1.810 | 1.414 | 2.109 | 2.429 | 0.990 | 2.076 | 2.297 | 1.813 | 2.492 | 1.634 | 2.012 | 2.182 | 0.133 |
| HORVU5Hr1G095060 Beta-glucosidase C | 1.879 | 1.156 | 0.797 | 1.198 | 0.361 | 1.308 | 0.900 | 0.953 | 0.989 | 0.575 | 1.200 | 0.335 | 0.933 | 1.632 | -0.812 |
| HORVU5Hr1G095100 Glycosyl hydrolase family protein | 1.028 | 0.609 | 0.663 | 0.441 | 0.786 | 1.262 | 0.413 | 1.321 | 0.962 | 1.338 | 0.862 | 1.207 | 0.842 | 1.372 | 0.045 |
| HORVU6Hr1G009620 Mannan endo-1,4-beta-mannosidase 7 | -1.591 | -1.721 | -0.460 | -0.635 | 0.073 | 0.179 | -0.111 | -1.140 | 0.204 | 0.442 | 0.436 | -1.040 | -0.066 | NE | NE |
| HORVU6Hr1G009690 Glucan endo-1,3-beta-glucosidase 3 | 0.853 | 0.537 | 0.355 | -0.265 | 1.207 | -0.004 | -0.756 | 0.771 | 0.631 | 1.149 | 0.756 | 0.632 | 0.089 | 0.310 | -0.400 |
| HORVU7Hr1G112930 GDP-L-fucose synthase 1 | 1.177 | 1.076 | 1.125 | 0.666 | 1.276 | 1.161 | 0.870 | 0.976 | 1.049 | 1.119 | 1.229 | 1.034 | 1.156 | 1.385 | 1.036 |
| HORVU7Hr1G112940 GDP-L-fucose synthase 1 | -1.267 | -1.196 | -0.819 | -0.847 | -0.896 | -0.769 | -0.460 | -0.685 | -0.376 | -0.395 | -0.486 | -0.131 | -0.529 | -1.393 | -0.911 |
| NE:无表达 NE : No expression |
新窗口打开|下载CSV
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/ajcn/78.2.221URLPMID:12885701 [本文引用: 1]

BACKGROUND: Findings about the effects of beta-glucan on serum lipoproteins are conflicting. OBJECTIVE: The study investigated the effects of beta-glucan from oat bran in bread and cookies (study 1) and in orange juice (study 2) on serum lipoproteins in mildly hypercholesterolemic subjects. DESIGN: In study 1, 48 subjects (21 men, 27 women) received for 3 wk control bread and cookies rich in wheat fiber. For the next 4 wk, by random assignment, 23 subjects continued to consume the control products, and 25 received bread and cookies rich in beta-glucan. Mean daily intake of beta-glucan was 5.9 g. Total dietary fiber intake did not differ significantly between the groups. In study 2, the same sources of control fiber and beta-glucan (5 g/d) as in study 1 were provided. For 2 wk, 25 of the original 48 subjects (10 men, 15 women) were randomly assigned to consume orange juice containing either wheat fiber (n = 13) or beta-glucan from oat bran (n = 12). After a washout period of 1 wk, dietary regimens were crossed over. RESULTS: In study 1, the change in LDL cholesterol did not differ significantly (-0.12 mmol/L; P = 0.173) between the 2 groups. In study 2, the drink rich in beta-glucan decreased LDL cholesterol by 0.26 +/- 0.07 mmol/L (6.7 +/- 1.8%; P = 0.001) and the ratio of total to HDL cholesterol by 0.26 +/- 0.11 (5.4 +/- 2.1%; P = 0.029) compared with the other drink. HDL-cholesterol and triacylglycerol concentrations did not change significantly. CONCLUSIONS: The food matrix or the food processing, or both, could have adverse effects on the hypocholesterolemic properties of oat beta-glucan.
DOI:10.1073/pnas.93.22.12637URLPMID:8901635 [本文引用: 1]

In spite of much effort, no one has succeeded in isolating and characterizing the enzyme(s) responsible for synthesis of cellulose, the major cell wall polymer of plants. We have characterized two cotton (Gossypium hirsutum) cDNA clones and identified one rice (Oryza sativa) cDNA that are homologs of the bacterial celA genes that encode the catalytic subunit of cellulose synthase. Three regions in the deduced amino acid sequences of the plant celA gene products are conserved with respect to the proteins encoded by bacterial celA genes. Within these conserved regions, there are four highly conserved subdomains previously suggested to be critical for catalysis and/or binding of the substrate UDP-glucose (UDP-Glc). An overexpressed DNA segment of the cotton celA1 gene encodes a polypeptide fragment that spans these domains and binds UDP-Glc, while a similar fragment having one of these domains deleted does not. The plant celA genes show little homology at the N- and C-terminal regions and also contain two internal insertions of sequence, one conserved and one hypervariable, that are not found in the bacterial gene sequences. Cotton celA1 and celA2 genes are expressed at high levels during active secondary wall cellulose synthesis in developing cotton fibers. Genomic Southern blot analyses in cotton demonstrate that celA forms a small gene family.
DOI:10.1073/pnas.0902019106URLPMID:19321749 [本文引用: 2]

The walls of grasses and related members of the Poales are characterized by the presence of the polysaccharide (1,3, 1,4)-beta-D-glucan (beta-glucan). To date, only members of the grass-specific cellulose synthase-like F (CSLF) gene family have been implicated in its synthesis. Assuming that other grass-specific CSL genes also might encode synthases for this polysaccharide, we cloned HvCSLH1, a CSLH gene from barley (Hordeum vulgare L.), and expressed an epitope-tagged version of the cDNA in Arabidopsis, a species with no CSLH genes and no beta-glucan in its walls. Transgenic Arabidopsis lines that had detectable amounts of the epitope-tagged HvCSLH1 protein accumulated beta-glucan in their walls. The presence of beta-glucan was confirmed by immunoelectron microscopy (immuno-EM) of sectioned tissues and chemical analysis of wall extracts. In the chemical analysis, characteristic tri- and tetra-saccharides were identified by high-performance anion-exchange chromatography and MALDI-TOF MS following their release from transgenic Arabidopsis walls by a specific beta-glucan hydrolase. Immuno-EM also was used to show that the epitope-tagged HvCSLH1 protein was in the endoplasmic reticulum and Golgi-associated vesicles, but not in the plasma membrane. In barley, HvCSLH1 was expressed at very low levels in leaf, floral tissues, and the developing grain. In leaf, expression was highest in xylem and interfascicular fiber cells that have walls with secondary thickenings containing beta-glucan. Thus both the CSLH and CSLF families contribute to beta-glucan synthesis in grasses and probably do so independently of each other, because there is no significant transcriptional correlation between these genes in the barley tissues surveyed.
DOI:10.1126/science.1122975URLPMID:16574868 [本文引用: 2]

A characteristic feature of grasses and commercially important cereals is the presence of (1,3;1,4)-beta-d-glucans in their cell walls. We have used comparative genomics to link a major quantitative trait locus for (1,3;1,4)-beta-d-glucan content in barley grain to a cluster of cellulose synthase-like CslF genes in rice. After insertion of rice CslF genes into Arabidopsis, we detected (1,3;1,4)-beta-d-glucan in walls of transgenic plants using specific monoclonal antibodies and enzymatic analysis. Because wild-type Arabidopsis does not contain CslF genes or have (1,3;1,4)-beta-d-glucans in its walls, these experiments provide direct, gain-of-function evidence for the participation of rice CslF genes in (1,3;1,4)-beta-d-glucan biosynthesis.
URLPMID:24595438 [本文引用: 1]
DOI:10.1111/j.1467-7652.2010.00532.xURLPMID:20497371 [本文引用: 2]

Cell walls in commercially important cereals and grasses are characterized by the presence of (1,3;1,4)-beta-d-glucans. These polysaccharides are beneficial constituents of human diets, where they can reduce the risk of hypercholesterolemia, type II diabetes, obesity and colorectal cancer. The biosynthesis of cell wall (1,3;1,4)-beta-d-glucans in the Poaceae is mediated, in part at least, by the cellulose synthase-like CslF family of genes. Over-expression of the barley CslF6 gene under the control of an endosperm-specific oat globulin promoter results in increases of more than 80% in (1,3;1,4)-beta-d-glucan content in grain of transgenic barley. Analyses of (1,3;1,4)-beta-d-glucan fine structure indicate that individual CslF enzymes might direct the synthesis of (1,3;1,4)-beta-d-glucans with different structures. When expression of the CslF6 transgene is driven by the Pro35S promoter, the transgenic lines have up to sixfold higher levels of (1,3;1,4)-beta-d-glucan in leaves, but similar levels as controls in the grain. Some transgenic lines of Pro35S:CslF4 also show increased levels of (1,3;1,4)-beta-d-glucans in grain, but not in leaves. Thus, the effects of CslF genes on (1,3;1,4)-beta-d-glucan levels are dependent not only on the promoter used, but also on the specific member of the CslF gene family that is inserted into the transgenic barley lines. Altering (1,3;1,4)-beta-d-glucan levels in grain and vegetative tissues will have potential applications in human health, where (1,3;1,4)-beta-d-glucans contribute to dietary fibre, and in tailoring the composition of biomass cell walls for the production of bioethanol from cereal crop residues and grasses.
DOI:10.1104/pp.109.151712URLPMID:20089768 [本文引用: 1]

(1,3;1,4)-beta-d-Glucan (beta-glucan) accounts for 20% of the total cell walls in the starchy endosperm of wheat (Triticum aestivum) and is an important source of dietary fiber for human nutrition with potential health benefits. Bioinformatic and array analyses of gene expression profiles in developing caryopses identified the CELLULOSE SYNTHASE-LIKE F6 (CSLF6) gene as encoding a putative beta-glucan synthase. RNA interference constructs were therefore designed to down-regulate CSLF6 gene expression and expressed in transgenic wheat under the control of a starchy endosperm-specific HMW subunit gene promoter. Analysis of wholemeal flours using an enzyme-based kit and by high-performance anion-exchange chromatography after digestion with lichenase showed decreases in total beta-glucan of between 30% and 52% and between 36% and 53%, respectively, in five transgenic lines compared to three control lines. The content of water-extractable beta-glucan was also reduced by about 50% in the transgenic lines, and the M(r) distribution of the fraction was decreased from an average of 79 to 85 x 10(4) g/mol in the controls and 36 to 57 x 10(4) g/mol in the transgenics. Immunolocalization of beta-glucan in semithin sections of mature and developing grains confirmed that the impact of the transgene was confined to the starchy endosperm with little or no effect on the aleurone or outer layers of the grain. The results confirm that the CSLF6 gene of wheat encodes a beta-glucan synthase and indicate that transgenic manipulation can be used to enhance the health benefits of wheat products.
DOI:10.1007/s00122-017-2867-7URLPMID:28215025 [本文引用: 1]

4) in an association panel of 240 maize inbred lines. Combining the two mapping populations, a total of 17 pleiotropic QTL/SNPs (pQTL/SNPs) were associated with various traits across multiple environments. PKS2, a stable locus influencing kernel shape identified on chromosome 2 in a genome-wide association study (GWAS), was within the QTL confidence interval defined by the RILs. The candidate region harbored a short 13-Kb LD block encompassing four SNPs (SYN11386, PHM14783.16, SYN11392, and SYN11378). In the association panel, 13 lines derived from the hybrid PI78599 possessed the same allele as Qi319 at the PHM14783.16 (GG) locus, with an average value of 0.21 for KS, significantly lower than that of the 34 lines derived from Ye478 that carried a different allele (0.25, P
[本文引用: 1]
[本文引用: 1]
DOI:10.1073/pnas.1311681110URLPMID:24065816 [本文引用: 1]

Within the cereal grasses, variation in inflorescence architecture results in a conspicuous morphological diversity that in crop species influences the yield of cereal grains. Although significant progress has been made in identifying some of the genes underlying this variation in maize and rice, in the temperate cereals, a group that includes wheat, barley, and rye, only the dosage-dependent and highly pleiotropic Q locus in hexaploid wheat has been molecularly characterized. Here we show that the characteristic variation in the density of grains along the inflorescence, or spike, of modern cultivated barley (Hordeum vulgare) is largely the consequence of a perturbed interaction between microRNA172 and its corresponding binding site in the mRNA of an APELATA2 (AP2)-like transcription factor, HvAP2. We used genome-wide association and biparental mapping to identify HvAP2. By comparing inflorescence development and HvAP2 transcript abundance in an extreme dense-spike mutant and its nearly isogenic WT line, we show that HvAP2 turnover driven by microRNA 172 regulates the length of a critical developmental window that is required for elongation of the inflorescence internodes. Our data indicate that this heterochronic change, an altered timing of developmental events caused by specific temporal variation in the efficiency of HvAP2 turnover, leads to the striking differences in the size and shape of the barley spike.
DOI:10.1038/ng.2447URLPMID:23160098 [本文引用: 1]

As early farming spread from the Fertile Crescent in the Near East around 10,000 years before the present, domesticated crops encountered considerable ecological and environmental change. Spring-sown crops that flowered without the need for an extended period of cold to promote flowering and day length-insensitive crops able to exploit the longer, cooler days of higher latitudes emerged and became established. To investigate the genetic consequences of adaptation to these new environments, we identified signatures of divergent selection in the highly differentiated modern-day spring and winter barleys. In one genetically divergent region, we identify a natural variant of the barley homolog of Antirrhinum CENTRORADIALIS (HvCEN) as a contributor to successful environmental adaptation. The distribution of HvCEN alleles in a large collection of wild and landrace accessions indicates that this involved selection and enrichment of preexisting genetic variants rather than the acquisition of mutations after domestication.
DOI:10.1038/ng.745URLPMID:21217754 [本文引用: 1]

The domestication of cereals has involved common changes in morphological features, such as seed size, seed retention and modification of vegetative and inflorescence architecture that ultimately contributed to an increase in harvested yield. In barley, this process has resulted in two different cultivated types, two-rowed and six-rowed forms, both derived from the wild two-rowed ancestor, with archaeo-botanical evidence indicating the origin of six-rowed barley early in the domestication of the species, some 8,600-8,000 years ago. Variation at SIX-ROWED SPIKE 1 (VRS1) is sufficient to control this phenotype. However, phenotypes imposed by VRS1 alleles are modified by alleles at the INTERMEDIUM-C (INT-C) locus. Here we show that INT-C is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1 (TB1) and identify 17 coding mutations in barley TB1 correlated with lateral spikelet fertility phenotypes.
DOI:10.1186/1471-2164-15-907URLPMID:25326272 [本文引用: 3]

BACKGROUND: (1,3;1,4)-beta-Glucan is an important component of the cell walls of barley grain as it affects processability during the production of alcoholic beverages and has significant human health benefits when consumed above recommended threshold levels. This leads to diametrically opposed quality requirements for different applications as low levels of (1,3;1,4)-beta-glucan are required for brewing and distilling and high levels for positive impacts on human health. RESULTS: We quantified grain (1,3;1,4)-beta-glucan content in a collection of 399 2-row Spring-type, and 204 2-row Winter-type elite barley cultivars originating mainly from north western Europe. We combined these data with genotypic information derived using a 9 K Illumina iSelect SNP platform and subsequently carried out a Genome Wide Association Scan (GWAS). Statistical analysis accounting for residual genetic structure within the germplasm collection allowed us to identify significant associations between molecular markers and the phenotypic data. By anchoring the regions that contain these associations to the barley genome assembly we catalogued genes underlying the associations. Based on gene annotations and transcript abundance data we identified candidate genes. CONCLUSIONS: We show that a region of the genome on chromosome 2 containing a cluster of CELLULOSE SYNTHASE-LIKE (Csl) genes, including CslF3, CslF4, CslF8, CslF10, CslF12 and CslH, as well as a region on chromosome 1H containing CslF9, are associated with the phenotype in this germplasm. We also observed that several regions identified by GWAS contain glycoside hydrolases that are possibly involved in (1,3;1,4)-beta-glucan breakdown, together with other genes that might participate in (1,3;1,4)-beta-glucan synthesis, re-modelling or regulation. This analysis provides new opportunities for understanding the genes related to the regulation of (1,3;1,4)-beta-glucan content in cereal grains.
DOI:10.1186/1471-2164-15-907URLPMID:25326272 [本文引用: 1]

BACKGROUND: (1,3;1,4)-beta-Glucan is an important component of the cell walls of barley grain as it affects processability during the production of alcoholic beverages and has significant human health benefits when consumed above recommended threshold levels. This leads to diametrically opposed quality requirements for different applications as low levels of (1,3;1,4)-beta-glucan are required for brewing and distilling and high levels for positive impacts on human health. RESULTS: We quantified grain (1,3;1,4)-beta-glucan content in a collection of 399 2-row Spring-type, and 204 2-row Winter-type elite barley cultivars originating mainly from north western Europe. We combined these data with genotypic information derived using a 9 K Illumina iSelect SNP platform and subsequently carried out a Genome Wide Association Scan (GWAS). Statistical analysis accounting for residual genetic structure within the germplasm collection allowed us to identify significant associations between molecular markers and the phenotypic data. By anchoring the regions that contain these associations to the barley genome assembly we catalogued genes underlying the associations. Based on gene annotations and transcript abundance data we identified candidate genes. CONCLUSIONS: We show that a region of the genome on chromosome 2 containing a cluster of CELLULOSE SYNTHASE-LIKE (Csl) genes, including CslF3, CslF4, CslF8, CslF10, CslF12 and CslH, as well as a region on chromosome 1H containing CslF9, are associated with the phenotype in this germplasm. We also observed that several regions identified by GWAS contain glycoside hydrolases that are possibly involved in (1,3;1,4)-beta-glucan breakdown, together with other genes that might participate in (1,3;1,4)-beta-glucan synthesis, re-modelling or regulation. This analysis provides new opportunities for understanding the genes related to the regulation of (1,3;1,4)-beta-glucan content in cereal grains.
[本文引用: 1]
DOI:10.1104/pp.17.01718URLPMID:29780036 [本文引用: 1]

Cell walls are crucial for the integrity and function of all land plants and are of central importance in human health, livestock production, and as a source of renewable bioenergy. Many enzymes that mediate the biosynthesis of cell wall polysaccharides are encoded by members of the large cellulose synthase (CesA) gene superfamily. Here, we analyzed 29 sequenced genomes and 17 transcriptomes to revise the phylogeny of the CesA gene superfamily in angiosperms. Our results identify ancestral gene clusters that predate the monocot-eudicot divergence and reveal several novel evolutionary observations, including the expansion of the Poaceae-specific cellulose synthase-like CslF family to the graminids and restiids and the characterization of a previously unreported eudicot lineage, CslM, that forms a reciprocally monophyletic eudicot-monocot grouping with the CslJ clade. The CslM lineage is widely distributed in eudicots, and the CslJ clade, which was thought previously to be restricted to the Poales, is widely distributed in monocots. Our analyses show that some members of the CslJ lineage, but not the newly identified CslM genes, are capable of directing (1,3;1,4)-beta-glucan biosynthesis, which, contrary to current dogma, is not restricted to Poaceae.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOI:10.1016/s0141-8130(97)00043-3URLPMID:9283018 [本文引用: 1]

Cell wall degradation is an important event during endosperm mobilization in the germinated barley grain. A battery of polysaccharide and oligosaccharide hydrolases is required for the complete depolymerization of the arabinoxylans and (1 --> 3,1 --> 4)-beta-glucans which comprise in excess of 90% by weight of these walls. The (1 --> 3,1 --> 4)-beta-glucan endohydrolases release oligosaccharides from their substrate and are probably of central importance for the initial solubilization of the (1 --> 3,1 --> 4)-beta-glucans, but beta-glucan exohydrolases and beta-glucosidases may be important additional enzymes for the conversion of released oligosaccharides to glucose. The latter enzymes have recently been purified from germinated barley and characterized. There is an increasing body of evidence to support the notion that the (1 --> 3,1 --> 4)-beta-glucan endohydrolases from germinated barley evolved from the pathogenesis-related (1 --> 3)-beta-glucanases which are widely distributed in plants and which hydrolyse polysaccharides that are abundant in fungal cell walls. Arabinoxylan depolymerization is also mediated by a family of enzymes, but these are less well characterized. (1 --> 4)-beta-Xylan endohydrolases have been purified and the corresponding cDNAs and genes isolated. While the presence of (1 --> 4)-beta-xylan exohydrolases and alpha-L-arabinofuranosidases has been reported many times, the enzymes have not yet been studied in detail. Here, recent advances in the enzymology and physiology of cell wall degradation in the germinated barley grain are briefly reviewed.
[本文引用: 1]
[本文引用: 1]
