 ,1,2,*
,1,2,*Identification of thaumatin-like protein family in Saccharum spontaneum and functional analysis of its homologous gene in sugarcane cultivar
SU Ya-Chun1,2, LI Cong-Na1, SU Wei-Hua1, YOU Chui-Huai3, CEN Guang-Li1, ZHANG Chang1, REN Yong-Juan1, QUE You-Xiong ,1,2,*
,1,2,*通讯作者:
收稿日期:2020-08-21接受日期:2020-12-1网络出版日期:2021-01-04
| 基金资助: |
Received:2020-08-21Accepted:2020-12-1Online:2021-01-04
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (4110KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
苏亚春, 李聪娜, 苏炜华, 尤垂淮, 岑光莉, 张畅, 任永娟, 阙友雄. 甘蔗割手密种类甜蛋白家族鉴定及栽培种同源基因功能分析[J]. 作物学报, 2021, 47(7): 1275-1296. doi:10.3724/SP.J.1006.2021.04192
SU Ya-Chun, LI Cong-Na, SU Wei-Hua, YOU Chui-Huai, CEN Guang-Li, ZHANG Chang, REN Yong-Juan, QUE You-Xiong.
病程相关蛋白(pathogenesis-related proteins, PRs)属于一种典型的生物防御型蛋白, 在植物抵抗外界胁迫因子过程中发挥重要作用[1]。Van根据PRs蛋白的理化性质及其基因序列特征, 将PRs分为17类, 即PR1~PR17, 每类PRs蛋白都存在功能分化[2]。类甜蛋白(thaumatin-like protein, TLP)属于PRs的第5类, 又名PR-5蛋白, 其氨基酸序列与西非竹芋(Thaumatococcus daniellii)的甜蛋白高度相似, 故得此名[3]。TLP广泛分布在动植物和微生物中, 当植物遇到外界因子, 比如病原物、化学元素和物理因子的刺激或者在植物生长发育的关键时期, 该蛋白在植物体内快速积累[4]。植物TLP蛋白的相对分子量一般为16~40 kD, 多数TLP氨基酸序列中含有1个索马甜标签G-X-[GF]-X-C-X-T-[GA]-D-C-X- (1,2)-G-X-(2,3)-C, 同时包含16个半胱氨酸, 并且会形成8个二硫键, 逐个分布在TLP蛋白的功能结构域中, 从而极大地提高了PR-5蛋白的稳定性, 最终强化了PR-5蛋白对蛋白酶、热激以及极端环境的抵抗作用[5,6,7]。某些TLPs的上游起始区段还含有一段信号肽序列, 可以帮助物质进行跨膜运输活动[5]。
TLP可以抵抗病害及其他因素对作物生长造成的不良影响, 进而提高作物抗逆性[4]。目前, 对于TLP家族基因的研究已经在多种作物中广泛开展, 如在水稻(Oryza sativa)[8]、杨树(Populus)[3]、山松(Pinus monticola D. Don)[9]和沙梨(Pyrus serotina)[10]等已经挖掘与鉴定到了多个TLP家族基因。王树军等[11]发现, 被胶孢炭疽菌(Colletotrichum gloeosporioides)侵染后, 荔枝(Litchi chinensis Sonn) TcTLP基因表达量显著上升。转TLP基因的小麦(Triticum aestivum)在受到禾谷镰刀菌(Fusarium graminearum)侵染时, TLP蛋白表达量上升, 说明通过转TLP基因创制抗病作物新品种是可行和有效的[12]。研究还发现, 在极度寒冷的自然环境中, 挪威云杉(Picea abies)中的TLP表达量增加, 是抵御冷害胁迫的潜在基因资源[13]。对拟南芥(Arabidopsis thaliana)进行高浓度盐处理, 与对照组相比, 转TLP基因的植株生长情况良好, 说明TLP蛋白有助于植物抵御盐胁迫[14]。上述研究表明, TLP蛋白积极参与植物对生物和非生物胁迫的防御效应。但是, 关于TLP的抗性机制、作用机理, 以及TLP是如何诱导植物抗性信号传递等方面的研究仍亟待开展, 这也在一定程度上限制了TLP家族基因在作物遗传育种上的利用。
甘蔗(Saccharum spp.)是世界上最重要的糖料和能源作物。在生长过程中, 甘蔗受黑穗病、低温、干旱、高盐等逆境胁迫, 极大地制约了蔗糖产业的健康发展。甘蔗抗逆基因的挖掘和功能鉴定将为基于基因工程技术提升甘蔗抗逆育种水平提供有效候选基因。目前, 关于甘蔗TLP家族基因成员的鉴定、序列分析及其蛋白结构与功能的研究尚未系统开展。因此, 本研究一方面基于甘蔗野生种割手密(Saccharum spontaneum)基因组[15], 挖掘SsTLP家族基因, 对其进行相关生物信息学分析, 以了解该基因家族各成员的序列和结构特征。另一方面, 从我国大陆主栽甘蔗品种ROC22中克隆获得1个SsTLP同源基因ScTLP1的cDNA全长, 并对其进行基因表达模式、亚细胞定位、瞬时表达及转录自激活活性等功能探究, 研究有望为深入解析甘蔗TLP基因功能及其作用模式提供科学依据。
1 材料与方法
1.1 甘蔗野生种割手密SsTLP家族基因的挖掘与生物信息学分析
在Pfam数据库中下载拟南芥和水稻的Thaumatin.hmm文件, 利用hmmsearch对割手密基因组数据库[15]进行检索, 获得待确认的TLP蛋白序列。采用NCBI (National Center of Biotechnology Information)的Conserved Domain (1.2 甘蔗ScTLP1基因的克隆与序列分析
从低氮胁迫下的甘蔗转录组注释库[18]挖掘出1条基因表达量无显著变化的割手密SsTLP87的同源基因unigene序列。为进一步探究该基因在其他逆境条件下的分子生物学功能, 我们利用Primer premier 5.0设计该基因的克隆引物(上游引物序列: 5°-AACTTGCTTGTGAGCGACCTA-3°, 下游引物序列: 5°-TACGCGTGTGTAGTTGTACTCC-3°)。采用PCR技术, 以我国大陆主栽甘蔗品种ROC22的组培苗叶片cDNA为模板进行扩增, 将含有目的片段的PCR产物进行胶回收、连接、转化和测序验证, 克隆获得割手密SsTLP87的同源基因, 命名为ScTLP1。通过Conserved Domain、SignalP-5.0 Server和DNAMAN软件, 分别对ScTLP1序列进行保守结构域、信号肽和氨基酸序列相似性等分析。1.3 ScTLP1基因的组织特异性表达及在不同逆境胁迫下的表达
1.3.1 甘蔗组织取样 于福建农林大学农业农村部福建甘蔗生物学与遗传育种重点实验室基地采集12月份长势一致的ROC22成熟蔗株, 分别切取白色蔗根、+1叶、第7至第8节处的蔗芽、蔗皮和蔗肉, 液氮速冻后置-80℃保存[19]。每3株为1次生物学重复, 每个样品设3次生物学重复。1.3.2 外源激素处理与取样 参照Liu等[19]的方法, 分别用0.1 mmol L-1脱落酸(abscisic acid, ABA)、5 mmol L-1水杨酸(salicylic acid, SA)和0.1 mmol L-1茉莉酸甲酯(methyl jasmonate, MeJA)喷施ROC22组培苗叶片, 采集处理0、3、6和12 h的叶片样品, 液氮速冻后置-80℃保存。每3株为1次生物学重复, 每个样品设3次生物学重复。
1.3.3 甘蔗黑穗病菌(Sporisorium scitaminea)接种与取样 参照Wang等[20]的方法, 以5×106个 mL-1甘蔗黑穗病菌混合孢子针刺接种ROC22蔗芽, 对照组接种无菌水。分别于接种0、1、2和3 d切取蔗芽, 液氮速冻后置-80℃保存。每5个单芽为1次生物学重复, 每个样品设3次生物学重复。
1.3.4 非生物胁迫与取样 将ROC22成熟茎段分段培养于盛有高压灭菌的潮湿沙土中, 置30℃光照培养箱(光照13 h/黑暗11 h; 光照22,000 μmol m-2 s-1/黑暗0 μmol m-2 s-1; 湿度80%)催芽培养, 待蔗苗生长至1片完全展开叶时, 将蔗苗去土和复水后, 移至Hongland营养液[21]中继续培养至3~4片叶。然后将蔗苗分别置含有20%聚乙二醇(polyethylene glycol)、PEG-8000和250 mmol L-1氯化钠(sodium chloride, NaCl)的Hongland营养液, 另一部分材料置4℃培养箱(光照13 h/黑暗11 h; 光照22,000 μmol m-2 s-1/黑暗0 μmol m-2 s-1; 湿度80%)中进行胁迫处理。分别收集经PEG处理0、6和24 h, NaCl处理0、12和24 h, 以及4℃处理0、6、12、24和48 h的+1叶材料, 液氮速冻后置-80℃保存。每3株为1次生物学重复, 每个样品设3次生物学重复。
1.3.5 基因表达检测 采用TRIzol试剂(Invitrogen, 美国, 卡尔斯巴德)提取甘蔗不同组织以及受不同外源胁迫的样品RNA, 根据RNase-Free DNase (Promega, 中国上海)和PrimeScript RT Reagent Kit (TaKaRa, 中国大连)的操作说明将RNA反转录合成cDNA第一链。借助Primer Premier 5.0设计ScTLP1基因定量检测引物(上游引物序列: 5°-AGTACACGCTCAACAACAAC-3°, 下游引物序列: 5°-AGGACATGGGCACGTTGA-3°), 采用qRT- PCR技术分析ScTLP1基因的组织特异性表达及在不同逆境胁迫下的表达, 以磷酸甘油醛脱氢酶(glyceraldehyde-phosphate dehydrogenase, GAPDH)基因为内参。qRT-PCR反应体系包括10 μL Hieff qPCR SYBR Green Master Mix、1.0 μL模板(10×cDNA稀释液)、0.4 μL上游/下游引物(10 μmol L-1)和8.2 μL无菌水, 试验设3次技术重复。qRT-PCR扩增程序为95℃ 5 min; 95℃ 15 s, 60℃ 20 s, 72℃ 20 s, 共40个循环。熔解曲线收集程序为95℃ 15 s; 60℃ 1 min; 95℃ 15 s。采用2-ΔΔCt法[22]计算ScTLP1基因的相对表达量, 采用DPS 9.50软件进行数据显著性差异分析, 采用Origin 8.0软件作图。
1.4 ScTLP1蛋白的亚细胞定位分析
根据亚细胞定位载体PCAMBIA 2300-GFP的载体图谱设计ScTLP1基因的亚细胞定位引物(上游引物序列: 5°-TGCTCTAGAAACTTGCTTGTGAGCGA-3°, 下游引物序列: 5°-CGGGATCCTGGGCAGAG CACGATAT-3°), 以pMD19-T-ScTLP1克隆质粒为模板进行PCR、胶回收、Xba I和Bam H I双酶切、连接、转化及重组质粒双酶切和单酶切验证。将上述获得的阳性质粒35S::ScTLP1::GFP转化根癌农杆菌GV3101, 根据Liu等[23]的方法, 分别将携带ScTLP1基因和pCAMBIA 2300-GFP空载的农杆菌GV3101菌液针刺注射3~4片叶龄的本氏烟(Nicotiana benthamiana)叶片, 置28℃下, 光照16 h/黑暗8 h培养2 d, 使用Leica TCS SP8激光共聚焦显微镜(莱卡, 德国, 曼海姆)观察ScTLP1蛋白的亚细胞定位结果。1.5 ScTLP1基因的瞬时表达分析
根据ScTLP1基因序列设计gateway BP反应引物(上游引物序列: 5°-GGGGACAAGTTTGTACA AAAAAGCAGGCTTCATGGCGTCGTCTCTAGCCAAG-3°, 下游引物序列: 5°-GGGGACCACTTTGTACA AGAAAGCTGGGTCCTACTCATGGGCAGAGCACGA-3°), 参照Liu等[19]的方法, 构建植物过表达载体pEarleyGate 203-ScTLP1。将空载质粒pEarleyGate 203 (N-Myc)和重组阳性质粒pEarleyGate 203-ScTLP1分别转化农杆菌GV3101菌株, 参考Liu等[23]的本氏烟叶片瞬时表达方法, 通过qRT-PCR检测经瞬时过表达ScTLP1基因1 d后的目标基因和烟草免疫相关标记基因的表达情况, 其中烟草免疫相关标记基因包括NtHSR201、NtHSR203、NtPR3、NtPR2、NtEFE26和NtHSR515, 这些基因分别参与植物的过敏反应、茉莉酸代谢途径和乙烯合成反应, 内参基因为NtEF-1α[23]。对瞬时过表达ScTLP1基因2 d的本氏烟叶片进行DAB染色, 观察过氧化氢(hydrogen peroxide, H2O2)的累积情况。另取一份瞬时过表达ScTLP1基因1 d的本氏烟材料, 参考Liu等[23]的方法分别接种烟草青枯菌(Pseudomonas solanacearum)和烟草茄病镰刀菌蓝色变种(Fusarium solani var. coeruleum), 置28℃下, 光照16 h/黑暗8 h培养7 d, 观察叶片症状变化, 拍照保存, 试验设3次生物学重复。1.6 ScTLP1的转录自激活活性验证
利用Primer Premier 5.0软件设计ScTLP1基因的酵母载体构建引物(上游引物序列: 5°-CCATGGAG GCCAGTGAATTCATGGCGTCGTCTCTAGCCAA-3°, 下游引物序列: 5°-CGCTGCAGGTCGACGGATCCT GGGCAGAGCACGATATCGT-3°), 进行PCR扩增和胶回收纯化。通过In-Fusion HD酶将经EcoR I酶切后的PGBKT7载体与胶回收的目的片段进行连接, 转化大肠杆菌感受态细胞。分别将上述构建好的重组阳性质粒PGBKT7-ScTLP1、阳性对照质粒PGBKT7-53-T和阴性对照质粒pGBKT7转化酵母Y2H Gold感受态细胞, 然后参照王玲等[24]的方法分析ScTLP1的转录自激活活性情况。2 结果与分析
2.1 甘蔗野生种割手密SsTLP家族基因的鉴定
基于甘蔗野生种割手密基因组, 鉴定到122个含有典型THN结构域的SsTLP家族基因(SsTLP1~SsTLP122) (表1)。生物信息学分析显示, 122个SsTLP家族成员的氨基酸数目介于95~1136之间, 氨基酸序列一致性仅为7.70%, 蛋白相对分子量介于10.02~124.24 kD之间(表1)。SsTLPs蛋白的理论等电点介于4.09~11.44之间, 大多数家族成员被预测为定位于细胞质的酸性疏水性不稳定蛋白, 其中, 89个SsTLP为酸性蛋白, 86个SsTLP的不稳定系数大于40, 69个SsTLP的平均疏水性数值大于0, 83个SsTLP蛋白含有信号肽, 104个SsTLP蛋白预测定位于细胞质, 而其余18个SsTLP蛋白预测可能定位于细胞膜、细胞核、细胞壁和液泡(表1)。SsTLP家族蛋白的二级结构以不规则卷曲为主(45.74%~ 87.37%), α-螺旋(0~40.50%)和延伸链(0.92%~ 25.54%)次之(附表1)。Table 1
表1
表1SsTLP家族蛋白的理化性质分析及亚细胞定位和信号肽预测
Table 1
| 基因名称 | 基因ID | 氨基酸数目 | 分子量 | 等电点 | 不稳定系数 | 平均疏水性 | 亚细胞定位 | 信号肽 |
|---|---|---|---|---|---|---|---|---|
| Gene name | Gene ID | Amino acids number | Molecular weight (kD) | Isoelectric point | Instability index | Hydrophobicity | Subcellular localization | Signal peptide |
| SsTLP1 | Sspon.01G0006490-1A | 294 | 29.00 | 4.81 | 27.25 | 0.13 | 细胞质Cytoplasm | 无No |
| SsTLP2 | Sspon.01G0006490-2C | 324 | 32.03 | 5.59 | 29.88 | 0.24 | 细胞质Cytoplasm | 有Yes |
| SsTLP3 | Sspon.01G0006490-3D | 325 | 32.05 | 5.23 | 30.42 | 0.26 | 细胞质Cytoplasm | 有Yes |
| SsTLP4 | Sspon.01G0016190-1A | 625 | 64.44 | 5.20 | 56.53 | -0.10 | 细胞质Cytoplasm | 无No |
| SsTLP5 | Sspon.01G0016190-2B | 464 | 46.16 | 5.02 | 59.63 | 0.10 | 细胞膜, 细胞质Cell membrane, Cytoplasm | 无No |
| SsTLP6 | Sspon.01G0016190-3C | 479 | 47.69 | 4.96 | 59.17 | 0.10 | 细胞膜, 细胞质Cell membrane, Cytoplasm | 无No |
| SsTLP7 | Sspon.01G0016190-4D | 275 | 27.67 | 4.70 | 54.56 | 0.16 | 细胞质Cytoplasm | 有Yes |
| SsTLP8 | Sspon.01G0016220-1A | 331 | 32.82 | 5.40 | 41.00 | 0.24 | 细胞质Cytoplasm | 有Yes |
| SsTLP9 | Sspon.01G0016220-2C | 305 | 31.16 | 8.33 | 59.42 | 0.08 | 细胞质Cytoplasm | 有Yes |
| SsTLP10 | Sspon.01G0016220-3D | 331 | 32.82 | 5.37 | 40.32 | 0.25 | 细胞质Cytoplasm | 有Yes |
| SsTLP11 | Sspon.01G0018320-1A | 236 | 23.36 | 4.30 | 32.97 | 0.18 | 细胞质Cytoplasm | 有Yes |
| SsTLP12 | Sspon.01G0018320-2B | 236 | 23.40 | 4.30 | 31.18 | 0.20 | 细胞质Cytoplasm | 有Yes |
| SsTLP13 | Sspon.01G0018320-3C | 236 | 23.41 | 4.30 | 34.15 | 0.15 | 细胞质Cytoplasm | 有Yes |
| SsTLP14 | Sspon.01G0020410-2C | 1053 | 113.01 | 6.79 | 45.32 | -0.14 | 细胞核Nucleus | 无No |
| SsTLP15 | Sspon.01G0020420-1A | 297 | 30.21 | 5.60 | 47.32 | 0.08 | 细胞质Cytoplasm | 无No |
| SsTLP16 | Sspon.01G0020420-2B | 298 | 30.25 | 5.40 | 45.49 | 0.10 | 细胞质Cytoplasm | 无No |
| SsTLP17 | Sspon.01G0037110-1B | 334 | 33.68 | 5.03 | 46.39 | 0.06 | 细胞质Cytoplasm | 有Yes |
| SsTLP18 | Sspon.01G0037110-1T | 334 | 33.68 | 5.03 | 46.39 | 0.06 | 细胞质Cytoplasm | 有Yes |
| SsTLP19 | Sspon.01G0037110-2C | 334 | 33.68 | 5.03 | 46.39 | 0.06 | 细胞质Cytoplasm | 有Yes |
| SsTLP20 | Sspon.01G0043210-1B | 289 | 29.55 | 7.44 | 40.68 | 0.12 | 细胞质Cytoplasm | 有Yes |
| SsTLP21 | Sspon.01G0043210-2C | 424 | 44.00 | 9.26 | 51.46 | -0.17 | 细胞质Cytoplasm | 有Yes |
| SsTLP22 | Sspon.01G0043210-3D | 292 | 29.81 | 7.44 | 41.03 | 0.12 | 细胞质Cytoplasm | 有Yes |
| SsTLP23 | Sspon.01G0044270-1B | 109 | 11.37 | 8.28 | 71.74 | -0.05 | 细胞质Cytoplasm | 无No |
| SsTLP24 | Sspon.01G0056200-1C | 95 | 10.02 | 8.89 | 73.11 | -0.62 | 细胞质Cytoplasm | 无No |
| SsTLP25 | Sspon.01G0056200-2D | 144 | 14.95 | 6.51 | 44.39 | -0.24 | 细胞质Cytoplasm | 无No |
| SsTLP26 | Sspon.02G0009140-1A | 246 | 24.96 | 4.87 | 43.80 | -0.16 | 细胞质Cytoplasm | 无No |
| SsTLP27 | Sspon.02G0009140-1P | 326 | 34.51 | 6.21 | 46.96 | -0.22 | 细胞质Cytoplasm | 无No |
| SsTLP28 | Sspon.02G0009140-2B | 236 | 23.98 | 4.87 | 43.62 | -0.12 | 细胞质Cytoplasm | 无No |
| SsTLP29 | Sspon.02G0009140-2P | 308 | 32.49 | 4.99 | 43.67 | -0.10 | 细胞质Cytoplasm | 无No |
| SsTLP30 | Sspon.02G0009140-3C | 236 | 23.97 | 4.87 | 44.87 | -0.13 | 细胞质Cytoplasm | 无No |
| SsTLP31 | Sspon.02G0009140-3P | 316 | 33.34 | 5.36 | 45.83 | -0.15 | 细胞质Cytoplasm | 有Yes |
| SsTLP32 | Sspon.02G0009140-4D | 319 | 33.65 | 5.36 | 45.49 | -0.13 | 细胞质Cytoplasm | 有Yes |
| SsTLP33 | Sspon.02G0009150-1A | 328 | 32.84 | 4.91 | 47.87 | 0.16 | 细胞质Cytoplasm | 有Yes |
| SsTLP34 | Sspon.02G0009150-2D | 318 | 31.83 | 4.79 | 46.30 | 0.10 | 细胞质Cytoplasm | 无No |
| SsTLP35 | Sspon.02G0011230-1A | 323 | 34.14 | 6.06 | 43.42 | 0.00 | 细胞质Cytoplasm | 有Yes |
| SsTLP36 | Sspon.02G0011230-1P | 220 | 22.52 | 4.44 | 61.16 | -0.17 | 细胞质Cytoplasm | 无No |
| SsTLP37 | Sspon.02G0011230-2B | 289 | 30.43 | 7.36 | 44.32 | -0.11 | 细胞质Cytoplasm | 无No |
| SsTLP38 | Sspon.02G0011230-2P | 231 | 23.89 | 4.87 | 43.93 | 0.03 | 细胞质Cytoplasm | 有Yes |
| SsTLP39 | Sspon.02G0011230-3C | 382 | 38.73 | 4.45 | 51.65 | -0.12 | 细胞质Cytoplasm | 有Yes |
| SsTLP40 | Sspon.02G0011230-4D | 323 | 34.23 | 6.18 | 42.60 | 0.00 | 细胞质Cytoplasm | 有Yes |
| SsTLP41 | Sspon.02G0020000-1A | 137 | 14.35 | 8.32 | 61.73 | -0.36 | 细胞质Cytoplasm | 无No |
| SsTLP42 | Sspon.02G0020000-1P | 288 | 29.63 | 7.39 | 48.63 | -0.12 | 细胞质Cytoplasm | 有Yes |
| SsTLP43 | Sspon.02G0029050-1A | 135 | 13.93 | 6.52 | 46.66 | 0.35 | 细胞质Cytoplasm | 有Yes |
| SsTLP44 | Sspon.02G0029050-2B | 227 | 23.17 | 4.74 | 39.91 | 0.25 | 细胞质Cytoplasm | 有Yes |
| SsTLP45 | Sspon.02G0029050-3C | 227 | 23.23 | 4.78 | 39.64 | 0.26 | 细胞质Cytoplasm | 有Yes |
| SsTLP46 | Sspon.02G0029050-4D | 215 | 21.88 | 4.96 | 40.74 | 0.39 | 细胞质Cytoplasm | 有Yes |
| SsTLP47 | Sspon.02G0029070-1A | 190 | 19.81 | 11.44 | 56.10 | -0.22 | 细胞壁, 细胞质Cell wall, Cytoplasm | 无No |
| SsTLP48 | Sspon.02G0029080-1A | 459 | 47.73 | 8.61 | 60.12 | -0.25 | 细胞质Cytoplasm | 无No |
| SsTLP49 | Sspon.02G0032190-1A | 163 | 17.00 | 5.76 | 46.52 | -0.29 | 细胞壁, 细胞质Cell wall, Cytoplasm | 无No |
| SsTLP50 | Sspon.02G0032190-1P | 170 | 17.33 | 4.94 | 40.68 | 0.02 | 细胞质Cytoplasm | 有Yes |
| SsTLP51 | Sspon.02G0032190-2B | 168 | 17.20 | 5.54 | 42.99 | 0.04 | 细胞质Cytoplasm | 有Yes |
| SsTLP52 | Sspon.02G0032190-3C | 163 | 17.02 | 5.52 | 45.34 | -0.34 | 细胞壁, 细胞质Cell wall, Cytoplasm | 无No |
| SsTLP53 | Sspon.02G0032190-4D | 168 | 17.20 | 5.54 | 42.99 | 0.04 | 细胞质Cytoplasm | 有Yes |
| SsTLP54 | Sspon.02G0032200-1A | 179 | 18.78 | 9.85 | 50.54 | 0.04 | 细胞质Cytoplasm | 有Yes |
| SsTLP55 | Sspon.02G0032220-1A | 171 | 17.61 | 4.68 | 29.57 | -0.06 | 细胞质Cytoplasm | 有Yes |
| SsTLP56 | Sspon.02G0032220-2B | 171 | 17.38 | 4.35 | 44.70 | 0.22 | 细胞质Cytoplasm | 有Yes |
| SsTLP57 | Sspon.02G0032220-3C | 132 | 13.56 | 4.12 | 39.94 | 0.09 | 细胞质Cytoplasm | 有Yes |
| SsTLP58 | Sspon.02G0032230-1A | 168 | 17.20 | 5.54 | 42.99 | 0.04 | 细胞质Cytoplasm | 有Yes |
| SsTLP59 | Sspon.02G0032230-2C | 409 | 43.36 | 8.98 | 48.87 | -0.31 | 细胞质, 细胞核Cytoplasm, Nucleus | 有Yes |
| SsTLP60 | Sspon.02G0032240-1A | 201 | 20.71 | 6.38 | 26.29 | -0.09 | 细胞质Cytoplasm | 有Yes |
| SsTLP61 | Sspon.02G0032240-2C | 175 | 17.72 | 4.60 | 43.06 | 0.09 | 细胞质Cytoplasm | 有Yes |
| SsTLP62 | Sspon.02G0034650-1B | 230 | 24.19 | 7.31 | 24.08 | -0.01 | 细胞质Cytoplasm | 有Yes |
| SsTLP63 | Sspon.02G0034650-2D | 224 | 23.68 | 7.30 | 22.53 | 0.02 | 细胞质Cytoplasm | 有Yes |
| SsTLP64 | Sspon.02G0036340-1B | 317 | 32.21 | 4.51 | 39.86 | 0.12 | 细胞质Cytoplasm | 无No |
| SsTLP65 | Sspon.02G0036340-2C | 322 | 32.67 | 4.51 | 38.40 | 0.13 | 细胞质Cytoplasm | 有Yes |
| SsTLP66 | Sspon.02G0036340-3D | 316 | 32.12 | 4.51 | 39.95 | 0.12 | 细胞质Cytoplasm | 无No |
| SsTLP67 | Sspon.02G0041150-1B | 291 | 30.58 | 8.03 | 51.00 | 0.04 | 细胞质Cytoplasm | 有Yes |
| SsTLP68 | Sspon.02G0041150-2C | 383 | 40.61 | 8.39 | 48.90 | -0.01 | 细胞质Cytoplasm | 有Yes |
| SsTLP69 | Sspon.02G0045690-2D | 171 | 17.51 | 4.47 | 29.08 | -0.03 | 细胞质Cytoplasm | 有Yes |
| SsTLP70 | Sspon.02G0045700-1B | 173 | 17.67 | 4.26 | 31.34 | 0.10 | 细胞质Cytoplasm | 有Yes |
| SsTLP71 | Sspon.02G0045700-1P | 416 | 42.99 | 9.33 | 35.28 | -0.38 | 细胞质Cytoplasm | 有Yes |
| SsTLP72 | Sspon.02G0045700-2C | 173 | 17.68 | 4.43 | 31.87 | 0.14 | 细胞质Cytoplasm | 有Yes |
| SsTLP73 | Sspon.02G0045700-3D | 173 | 17.73 | 4.43 | 33.23 | 0.09 | 细胞质Cytoplasm | 有Yes |
| SsTLP74 | Sspon.02G0053220-1C | 245 | 25.68 | 6.27 | 27.29 | -0.32 | 细胞质Cytoplasm | 有Yes |
| SsTLP75 | Sspon.02G0053220-2D | 271 | 28.16 | 5.87 | 38.49 | -0.29 | 细胞质Cytoplasm | 有Yes |
| SsTLP76 | Sspon.02G0053240-1C | 166 | 17.35 | 6.66 | 30.90 | -0.17 | 细胞质Cytoplasm | 有Yes |
| SsTLP77 | Sspon.02G0053250-2D | 233 | 24.46 | 8.92 | 60.54 | -0.45 | 细胞质, 细胞核Cytoplasm, Nucleus | 无No |
| SsTLP78 | Sspon.02G0059040-1D | 124 | 12.61 | 6.50 | 44.52 | 0.23 | 细胞膜, 细胞质Cell membrane, Cytoplasm | 有Yes |
| SsTLP79 | Sspon.02G0059050-1D | 127 | 12.95 | 4.27 | 27.47 | 0.47 | 细胞质Cytoplasm | 有Yes |
| SsTLP80 | Sspon.03G0003060-1A | 252 | 26.62 | 7.86 | 47.70 | -0.05 | 细胞质Cytoplasm | 有Yes |
| SsTLP81 | Sspon.03G0003060-1P | 253 | 26.73 | 7.86 | 47.55 | -0.04 | 细胞质Cytoplasm | 有Yes |
| SsTLP82 | Sspon.03G0003060-2C | 251 | 26.48 | 7.86 | 46.89 | -0.06 | 细胞质Cytoplasm | 有Yes |
| SsTLP83 | Sspon.03G0014170-1A | 633 | 69.61 | 6.63 | 48.05 | -0.21 | 细胞核Nucleus | 有Yes |
| SsTLP84 | Sspon.03G0018890-1A | 1028 | 111.78 | 9.42 | 52.82 | -0.72 | 细胞核Nucleus | 无No |
| SsTLP85 | Sspon.03G0018890-2C | 516 | 57.24 | 6.15 | 38.71 | -0.30 | 细胞核Nucleus | 无No |
| SsTLP86 | Sspon.03G0018890-3D | 727 | 80.10 | 6.23 | 49.20 | -0.33 | 细胞核Nucleus | 有Yes |
| SsTLP87 | Sspon.03G0040280-1C | 227 | 23.49 | 7.33 | 31.85 | -0.08 | 细胞质Cytoplasm | 有Yes |
| SsTLP88 | Sspon.03G0040280-1P | 227 | 23.51 | 7.33 | 31.48 | -0.07 | 细胞质Cytoplasm | 有Yes |
| SsTLP89 | Sspon.04G0003480-1A | 186 | 18.49 | 4.09 | 48.17 | 0.20 | 细胞质Cytoplasm | 无No |
| SsTLP90 | Sspon.04G0003480-2C | 186 | 18.49 | 4.09 | 48.17 | 0.20 | 细胞质Cytoplasm | 无No |
| SsTLP91 | Sspon.04G0003480-3D | 191 | 18.97 | 4.41 | 47.57 | 0.21 | 细胞质Cytoplasm | 无No |
| SsTLP92 | Sspon.05G0007890-1A | 1136 | 124.24 | 5.26 | 52.74 | -0.44 | 细胞核Nucleus | 无No |
| SsTLP93 | Sspon.05G0014480-1A | 280 | 27.85 | 5.42 | 47.70 | 0.28 | 细胞质Cytoplasm | 有Yes |
| SsTLP94 | Sspon.05G0014480-1P | 267 | 26.55 | 5.68 | 46.13 | 0.23 | 细胞质Cytoplasm | 有Yes |
| SsTLP95 | Sspon.05G0014480-2B | 280 | 27.78 | 5.35 | 45.12 | 0.34 | 细胞质Cytoplasm | 有Yes |
| SsTLP96 | Sspon.05G0014480-3D | 277 | 27.62 | 5.35 | 44.31 | 0.32 | 细胞质Cytoplasm | 有Yes |
| SsTLP97 | Sspon.05G0029070-1B | 550 | 60.87 | 6.45 | 43.06 | -0.32 | 细胞核Nucleus | 有Yes |
| SsTLP98 | Sspon.05G0029070-2C | 514 | 56.87 | 6.33 | 45.88 | -0.35 | 细胞核Nucleus | 有Yes |
| SsTLP99 | Sspon.05G0032300-1C | 244 | 24.70 | 4.62 | 36.67 | 0.09 | 细胞质Cytoplasm | 有Yes |
| SsTLP100 | Sspon.05G0032300-2D | 392 | 40.73 | 7.03 | 46.75 | -0.08 | 细胞质Cytoplasm | 无No |
| SsTLP101 | Sspon.05G0032310-1C | 242 | 25.22 | 4.53 | 47.73 | -0.18 | 细胞质Cytoplasm | 无No |
| SsTLP102 | Sspon.05G0032310-2D | 242 | 25.19 | 4.53 | 47.73 | -0.17 | 细胞质Cytoplasm | 无No |
| SsTLP103 | Sspon.06G0027890-1B | 241 | 25.13 | 6.50 | 41.80 | -0.14 | 细胞质Cytoplasm | 无No |
| SsTLP104 | Sspon.06G0027890-1T | 217 | 22.57 | 8.36 | 44.97 | -0.11 | 细胞质Cytoplasm | 无No |
| SsTLP105 | Sspon.06G0027890-2C | 251 | 26.21 | 8.10 | 39.51 | -0.11 | 细胞质Cytoplasm | 无No |
| SsTLP106 | Sspon.07G0015550-1A | 318 | 32.05 | 4.60 | 49.86 | 0.08 | 细胞质Cytoplasm | 有Yes |
| SsTLP107 | Sspon.07G0015550-2B | 326 | 33.01 | 7.35 | 38.08 | 0.30 | 细胞质Cytoplasm | 有Yes |
| SsTLP108 | Sspon.07G0015560-1A | 517 | 54.47 | 4.77 | 46.95 | -0.11 | 细胞质, 细胞核, 液泡Cytoplasm, Nucleus, Vacuole | 有Yes |
| SsTLP109 | Sspon.07G0017710-1A | 267 | 26.60 | 4.10 | 43.87 | 0.22 | 细胞质Cytoplasm | 有Yes |
| SsTLP110 | Sspon.07G0017710-2B | 329 | 32.60 | 4.21 | 47.78 | 0.28 | 细胞膜, 细胞质, 液泡Cell membrane, Cytoplasm, Vacuole | 有Yes |
| SsTLP111 | Sspon.08G0001280-1A | 260 | 25.63 | 4.82 | 46.65 | 0.35 | 细胞质Cytoplasm | 有Yes |
| SsTLP112 | Sspon.08G0001280-2C | 260 | 25.58 | 4.65 | 46.15 | 0.36 | 细胞质Cytoplasm | 有Yes |
| SsTLP113 | Sspon.08G0001280-3D | 279 | 27.61 | 4.92 | 47.72 | 0.10 | 细胞质Cytoplasm | 无No |
| SsTLP114 | Sspon.08G0003010-1A | 248 | 25.25 | 8.59 | 38.06 | 0.13 | 细胞质Cytoplasm | 有Yes |
| SsTLP115 | Sspon.08G0003010-2B | 254 | 25.87 | 8.59 | 36.97 | 0.14 | 细胞质Cytoplasm | 有Yes |
| SsTLP116 | Sspon.08G0003010-3C | 248 | 25.26 | 8.59 | 37.63 | 0.15 | 细胞质Cytoplasm | 有Yes |
| SsTLP117 | Sspon.08G0007410-1A | 313 | 31.28 | 4.74 | 33.53 | 0.25 | 细胞质Cytoplasm | 有Yes |
| SsTLP118 | Sspon.08G0007410-2C | 313 | 31.25 | 4.74 | 34.28 | 0.24 | 细胞质Cytoplasm | 有Yes |
| SsTLP119 | Sspon.08G0015670-1A | 309 | 31.95 | 6.13 | 49.19 | 0.07 | 细胞质Cytoplasm | 有Yes |
| SsTLP120 | Sspon.08G0015670-2B | 309 | 31.92 | 6.20 | 46.42 | 0.07 | 细胞质Cytoplasm | 有Yes |
| SsTLP121 | Sspon.08G0015670-3D | 251 | 26.04 | 9.12 | 55.78 | -0.23 | 细胞质Cytoplasm | 有Yes |
| SsTLP122 | Sspon.08G0030460-1D | 275 | 28.40 | 6.12 | 54.61 | 0.10 | 细胞质Cytoplasm | 有Yes |
新窗口打开|下载CSV
2.2 SsTLP家族基因的染色体分布
染色体定位分析显示, 除了染色体Chr3B、Chr4B、Chr7C和Chr7D, 其余28条割手密染色体分布1~13个数目不等的SsTLP基因, 其中, Chr3D、Chr4A、Chr4C、Chr4D和Chr6A只分布1个SsTLP基因, 分别为SsTLP86、SsTLP89、SsTLP90、SsTLP91和SsTLP27, Chr2A和Chr2C分布的SsTLP基因数目多达13个(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1SsTLP家族基因的染色体定位
Fig. 1Chromosome location of SsTLP family genes
2.3 SsTLP家族基因结构及系统进化分析
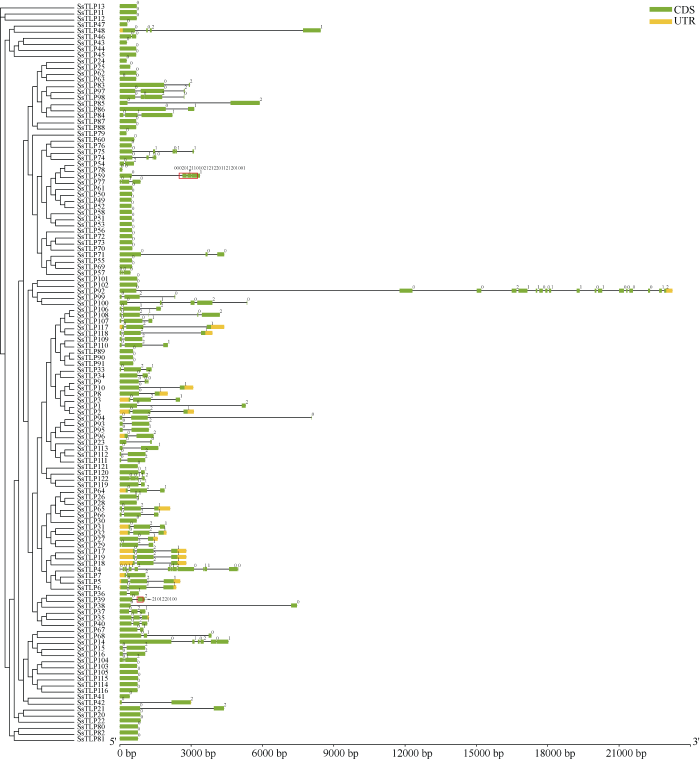
基因结构分析显示, SsTLP家族基因结构类型丰富, 其中不含内含子和含有1、2和3 (及以上)个内含子的基因数目分别为48、30、32和12, 含有1个内含子的基因均为0型相位, 含有2个内含子的基因主要为0型和2型相位(图2)。此外, SsTLP48、SsTLP74和SsTLP98基因含有3个内含子, SsTLP75、SsTLP100、SsTLP108和SsTLP122基因含有4个内含子, SsTLP14基因含有5个内含子, SsTLP39基因含有11个内含子, SsTLP4基因含有12个内含子, SsTLP92基因含有17个内含子, SsTLP59基因含有30个内含子, 上述基因的内含子相位存在多种类型(图2)。Motif分析显示, 69个SsTLP蛋白均含有5种Motif, 其余53个SsTLP蛋白含有1~4种Motif (图3)。系统进化树分析发现, 122个SsTLP与拟南芥AtTLPs和水稻OsTLPs聚类在TLP家族的I~X分支上, SsTLP家族蛋白在10个分类中均有分布, 其中, V类的TLP最多, 包含36个SsTLP, 其次是VI类, 包含25个SsTLP, 而IV和X类的TLP最少, 分别仅包含2个SsTLP (图4)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2SsTLP家族基因结构
绿色方框代表编码序列(CDS); 黄色方框代表非编码序列(UTR); 方框中间的横线代表内含子; 0、1和2代表内含子相位。
Fig. 2Gene structure of SsTLP family
The green box representes the coding sequence (CDS); the yellow box representes the untranslated region (UTR); the horizontal line in the middle of the box representes the intron; 0, 1, and 2 representes the intron phase.
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3SsTLP家族保守基序分析 不同颜色的方框代表基序种类与位置。
Fig. 3Conservation motif analysis of SsTLP family The boxes of different colors represent the motif types and positions.
2.4 ScTLP1基因的克隆与序列分析
通过RT-PCR技术, 从我国大陆主栽甘蔗栽培种ROC22中克隆获得割手密SsTLP87的同源基因ScTLP1。ScTLP1基因的开放读码框长度为681 bp, 编码227个氨基酸(图5), 且与SsTLP87蛋白的氨基酸序列相似性高达99.56%。ScTLP1基因推导的编码蛋白包含16个半胱氨酸残基(蓝色字体标注), 在N端存在一段信号肽(黄色阴影标注), 从第35位至第227位氨基酸(方框标注序列)为典型THN结构域。以ScTLP1基因推导的氨基酸序列为探针, 从NCBI数据库分别查找获得粗山羊草TLP (Aegilops tauschii, XP_020192437.1)、高粱TLP (Sorghum bicolor, XP_002456520.1)、谷子TLP (Setaria italica, XP_004970321.2)、黍TLP (Panicum miliaceum, RLN21715.1)和玉米TLP (Zea mays, PWZ30741.1)等甘蔗近缘物种TLPs的氨基酸序列。多序列比对结果显示, ScTLP1蛋白与高粱TLP、玉米TLP、黍TLP、谷子TLP和粗山羊草TLP的氨基酸序列相似性分别为94.76%、90.31%、83.26%、77.27%和64.94% (图6)。鉴于ScTLP1蛋白与高粱TLP的相似性最高, 推测甘蔗ScTLP1蛋白在物种进化上具有保守性。2.5 ScTLP1基因的组织特异性表达及在不同逆境胁迫下的表达
qRT-PCR分析结果显示, ScTLP1基因在ROC22蔗芽中的表达量最高, 约为蔗根中的13.72倍, 其在蔗根、蔗肉、蔗叶和蔗皮中的基因表达量次之, 且四者之间的差异不显著(图7-A)。受甘蔗黑穗病菌侵染后, 在1 d时ScTLP1基因表达量在ROC22中基本不变, 但在2 d时该基因显著上调表达, 其表达量约为对照的6.00倍, 3 d时ScTLP1基因表达量恢复至对照水平(图7-B)。在ABA和SA处理下, ScTLP1基因的表达量无显著变化(图7-C, D)。在MeJA处理下, ScTLP1基因的表达量在12 h时达到峰值, 为对照的2.61倍, 而该基因在其余时间点的表达量与对照组持平(图7-E)。在NaCl和PEG胁迫下, ScTLP1基因的表达量在6 h和12 h时均与对照差异不显著, 而在24 h时显著上调, 分别为对照的137.14倍和32.20倍(图7-F, G)。在4℃低温胁迫下, ScTLP1基因的表达量在6 h时显著上调, 为对照的44.08倍, 在12 h, ScTLP1基因的表达量有所下降, 但仍显著高于对照水平, 24~48 h时, ScTLP1基因的表达量达到峰值且维持在一定的水平, 分别为对照的51.52倍和44.36倍(图7-H)。表明, ScTLP1基因在甘蔗ROC22中为组成型表达, 在蔗芽中的表达量最高, 该基因在ABA和SA处理下的表达量无显著变化, 但受NaCl、4℃低温、PEG、甘蔗黑穗病菌以及MeJA诱导上调表达。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4SsTLP家族系统进化树
红色圆点代表割手密SsTLP蛋白; 绿色圆点代表拟南芥AtTLP蛋白; 黑色圆点代表水稻OsTLP蛋白。
Fig. 4Phylogenetic tree of SsTLP family
The red dot representes the Saccharum spontaneum SsTLP protein; the green dot representes the Arabidopsis thaliana AtTLP protein; the black dot representes the Oryza sativa OsTLP protein.
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5ScTLP1基因的核酸序列及其编码的氨基酸序列
蓝色字体为半胱氨酸位点; 方框序列为THN保守结构域; 黄色序列为信号肽; *为终止密码子。
Fig. 5Nucleic acid sequence and the coding amino acid sequence of ScTLP1 gene
The blue font is the cysteine site; the box sequence is the THN conservative domain; the yellow sequence is the signal peptide; * representes stop codon.
图6
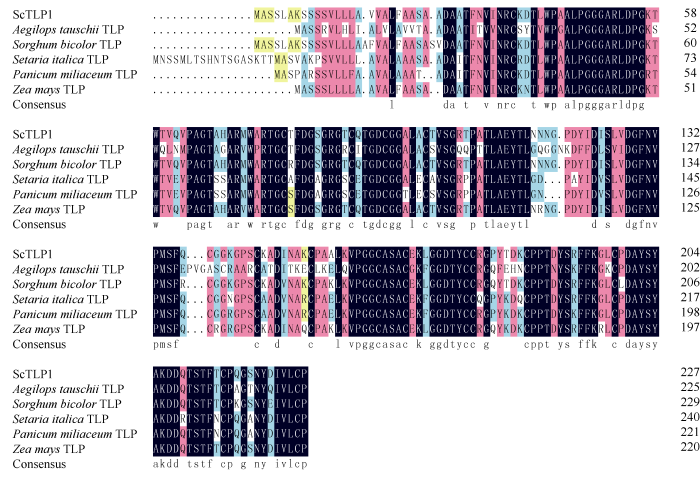 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6甘蔗ScTLP1蛋白与其他近缘物种TLP蛋白的氨基酸序列多重比对
Fig. 6Multiple alignment of amino acid sequences of sugarcane ScTLP1 protein and TLP proteins of other homologous species
2.6 ScTLP1基因的瞬时表达分析
将构建好的含有pEarleyGate 203-ScTLP1重组载体的GV3101农杆菌注射本氏烟叶片, 以携带pEarleyGate 203空载的GV3101为对照组。从图8-A和C可以看出, 与对照组相比, 处理组叶片中ScTLP1基因的表达量在注射24 h后出现上调, 且DAB染色加深, 表现出过敏反应症状。烟草免疫相关标记基因表达分析显示, 在瞬时过表达ScTLP1基因的本氏烟叶片中, 烟草过敏反应相关基因NtHSR201、NtHSR203和NtHSR515的表达量没有明显变化, 而乙烯合成相关基因NtEFE26上调表达, 茉莉酸代谢途径相关基因NtPR3的表达量也显著高于对照组(图8-B)。观察接种烟草青枯菌和茄病镰刀菌蓝色变种的本氏烟发病状况, 发现接种青枯菌7 d后的对照组发病情况明显, 叶片出现明显病斑和枯萎现象,而过表达ScTLP1基因的本氏烟叶片状况相对较好(图8-D); 接种烟草茄病镰刀菌蓝色变种的对照组叶片发病状况也较为严重, 整片叶片枯萎皱缩, 而过表达了ScTLP1基因的本氏烟只有个别叶片边缘发生轻微枯萎, 出现局部病斑(图8-E)。表明, ScTLP1基因可以诱发本氏烟的过敏反应, 诱导乙烯合成相关基因NtEFE26和茉莉酸代谢途径相关基因NtPR3上调表达, 增强本氏烟对烟草青枯菌和茄病镰刀菌蓝色变种的防御效应。图7
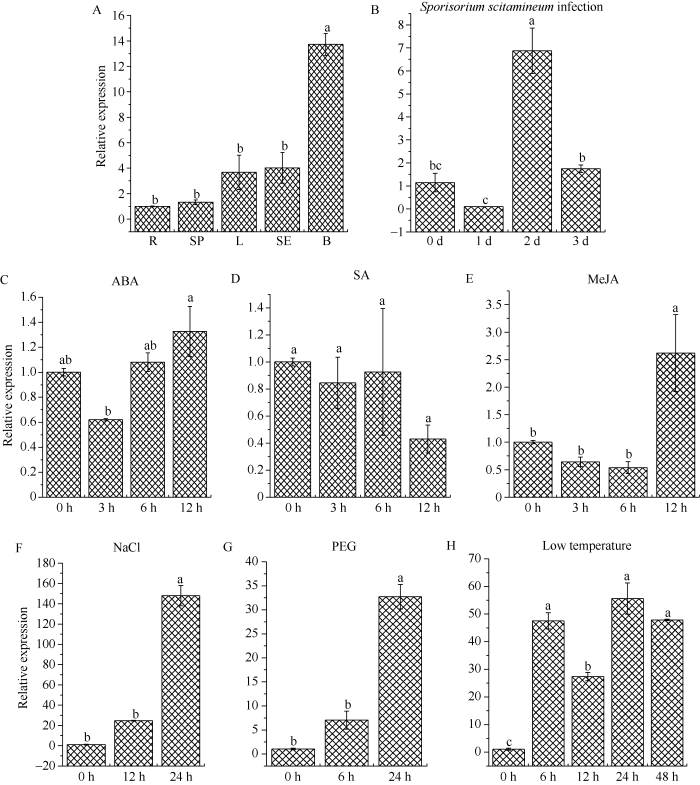 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7ScTLP1基因的组织特异性表达及在不同逆境胁迫下的表达
A: ScTLP1基因在甘蔗ROC22中的组织特异性表达。R: 蔗根; SP: 蔗肉; L: 蔗叶; SE: 蔗皮; B: 蔗芽。B: ScTLP1基因在甘蔗黑穗病菌侵染下的表达。C~H: ScTLP1基因在外源激素和非生物胁迫下的表达。ABA: 脱落酸; SA: 水杨酸; MeJA: 茉莉酸甲酯; NaCl: 氯化钠; PEG: 聚乙二醇; Low temperature: 4℃低温处理。误差线代表每组处理的标准误差(n = 3)。柱上标以不同小写字母表示在0.05水平显著差异。
Fig. 7Tissue specific expression of ScTLP1 gene and its expression under different environmental stresses
A: tissue-specific expression of ScTLP1 gene in sugarcane ROC22. R: root; SP: stem pith; L: leaf; SE: stem epidermis; B: bud. B: expression of ScTLP1 gene after inoculation with Sporisorium scitamineum. C-H: expression of ScTLP1 gene under exogenous hormones and abiotic stresses. ABA: abscisic acid; SA: salicylic acid; MeJA: methyl jasmonate; NaCl: sodium chloride; PEG: polyethylene glycol; Low temperature: 4°C low temperature treatment. The error bars represent the standard error of each group treatment (n = 3). Different lowercase letters above the bars mean significant differences at the 0.05 probability level.
图8
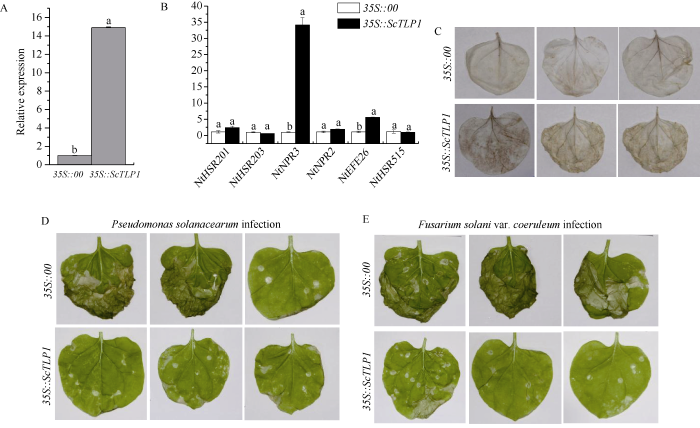 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8ScTLP1基因在本氏烟叶片中的瞬时表达效应
A: ScTLP1在瞬时表达1 d后的本氏烟中的基因表达量。B: 烟草免疫相关基因在瞬时表达1 d后的对照组和实验组的基因表达量。C: 瞬时表达2 d后的本氏烟叶片的DAB染色结果。D和E: 本氏烟接种烟草青枯菌和烟草茄病镰刀菌蓝色变种7 d的叶片表型。实验数据以NtEF-1α基因的表达水平进行归一化处理。误差线代表每组处理的标准误差(n = 3)。柱上标以不同小写字母表示在0.05水平显著差异。
Fig. 8Transient expression effect of ScTLP1 gene in Nicotiana benthamiana leaves
A: gene expression level of ScTLP1 in N. benthamiana after transient expression for 1 d. B: gene expression level of tobacco immune related genes in control group and experimental group after transient expression for 1 d. C: the DAB staining result of N. benthamiana leaves after transient expression for 2 d. D-E: the leaf phenotypes of N. benthamiana inoculated with Pseudomonas solanacearum and Fusarium solani var. coeruleum for seven days, respectively. The experimental data were normalized by the expression level of NtEF-1α gene. The error bars represent the standard errors of each group treatment (n = 3). Different lowercase letters above the bars mean significant differences at the 0.05 probability level.
2.7 ScTLP1蛋白的亚细胞定位分析
利用激光共聚焦显微镜观察ScTLP1蛋白的亚细胞定位情况发现, 对照组中的绿色荧光信号在细胞的各个部位均有分布, 而ScTLP1-GFP融合蛋白主要定位在细胞膜上(图9)。图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9ScTLP1蛋白在本氏烟叶片表皮的亚细胞定位
该结果包含明场、绿色荧光、叠加场3个视野下拍摄的照片。箭头a、b和c分别表示细胞核、细胞膜和细胞质。35S::GFP为含有亚细胞定位空载的GV3101菌液注射本氏烟的结果; 35S::ScTLP1::GFP为含有目的基因ScTLP1的重组亚细胞定位载体的GV3101菌液注射本氏烟的结果。标尺为50 μm。
Fig. 9Subcellular localization of ScTLP1 protein in the epidermis of Nicotiana benthamiana
The results include photographs taken from three perspectives, including visual field, green fluorescence, and merged field. The arrows a, b, and c represent the nucleus, cell membrane, and cytoplasm, respectively. 35S::GFP is the result of N. benthamiana injection with GV3101 bacterial solution containing subcellular localization empty vector; 35S::ScTLP1::GFP is the result of N. benthamiana injection with GV3101 bacterial solution containing and the recombinant subcellular localization vector with the target gene ScTLP1. Bar: 50 μm.
2.8 ScTLP1蛋白的转录自激活活性验证
图10-A显示, 涂布在SD-Trp培养基上的PGBKT7-ScTLP1的Y2H Gold酵母菌液长出白色菌落, 与阳性对照pGADT7-T+pGBKT7-p53和阴性对照pGBKT7的菌落情况一致, 证明ScTLP1基因成功转化酵母Y2H Gold且成功表达色氨酸。利用X-α-gal对SD-Trp培养基上的菌落染色, 阳性菌落成蓝色, 阴性对照和PGBKT7-ScTLP1的菌落均为白色(图10-B), 说明编码α-半乳糖苷酶的MEL1基因未被激活, 表示ScTLP1蛋白不具有转录自激活活性。在SD-Trp-AbA培养基上, 只有阳性对照pGADT7-T+pGBKT7-p53的Y2H Gold菌落生长, 阴性对照和PGBKT7-ScTLP1的菌落均不生长, 且用X-α-gal染色后, 阳性菌落呈蓝色(图10-C), 进一步证明ScTLP1蛋白无转录自激活活性, 可以用于后续互作蛋白的筛选工作。图10
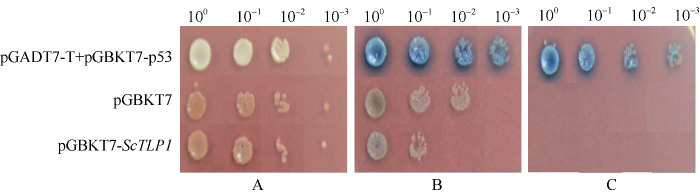 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10甘蔗ScTLP1蛋白的转录自激活活性验证
A: 不同浓度的阳性对照、阴性对照和pGBKT7-ScTLP1的Y2H Gold酵母菌液涂布在SD-Trp固体培养基上的生长情况。B: 利用X-α-gal染色液对SD-Trp固体培养基上的菌落的染色结果。C: X-α-gal染色液对添加了金担子素A抑制剂SD-Trp固体培养基上生长的菌落的染色结果。pGADT7-T+pGBKT7-p53: 阳性对照; pGBKT7: 阴性对照。
Fig. 10Validation of transcriptional self-activation activity of ScTLP1 protein
A: the growth of Y2H Gold yeast solution sprayed on SD-Trp solid medium with different concentrations of positive control, negative control and pGBKT7-ScTLP1. B: colonial staining result of SD-Trp solid culture medium with X-α-gal staining solution. C: colonial staining result after adding SD-Trp solid culture with X-α-gal staining solution. pGADT7-T+pGBKT7-p53: positive control; pGBKT7: negative control.
3 讨论
TLP基因在植物生长发育和应答生物和非生物胁迫方面起作用。有关TLP家族基因的研究已经在多种植物中被开展, 基于植物基因组和EST数据库, TLP超家族至少包含31个水稻TLP基因、28个拟南芥TLP基因、13个白云杉(Picea glauca Moench) TLP基因、10个白松(Pinus monticola Dougl. ex D. Don) TLP基因等[25]。Zhao等[17]预测与分析了50个毛果杨(Populus trichocarpa) TLP家族基因的分子进化模式与功能。刘潮等[26]对28个辣椒(Capsicum annuum) TLP家族基因的序列特征及时空表达特性进行了分析。本研究从甘蔗野生种割手密基因组中共发现122个SsTLP家族基因, 多数为定位于细胞质的酸性疏水性不稳定蛋白。28条割手密染色体分布1~13个数目不等的SsTLP基因, 其中有多条染色体分布多个SsTLP基因成员, 表明割手密SsTLP家族基因与其它大多数基因类似, 存在基因聚类现象, 推测与其内部结构和功能有关。植物TLPs蛋白的分子量大小不一, 如30个胡萝卜(Daucus carota) TLP家族蛋白的氨基酸数介于220~330之间, 相对分子量为18.64~68.39 kD[27]; 28个辣椒TLP家族成员的氨基酸数为191~400, 相对分子量为20.72~40.54 kD[26]; 88个陆地棉(Gossypium hirsutum) TLP家族成员的氨基酸数在200~817之间, 相对分子量为20.58~90.27 kD[28]。本研究中, 122个割手密SsTLP家族成员的氨基酸数目为95~1136, 蛋白相对分子量介于10.02~124.24 kD之间, 推测不论TLPs分子量如何变化, 同源性高的TLPs可能具有相似的生理学特征[5]。常见的TLP蛋白的三维结构包含3个保守结构域, 即Domain I、Domain II和Domain III, 其中Domain I是由2个反向平行的β片层构成的N端核心功能域, Domain II由半胱氨酸二硫键形成的3个α螺旋组成, Domain III由1个大环和2个β折叠构成, 且Domain I和Domain II之间存在1个“V”字形裂缝, 是TLP蛋白行使抗真菌性活性功能的重要位点[29,30]。本研究中, SsTLP蛋白的Motif 3 (TFTITNNCGYTVWPGILPNAG)位于Domain I上, Motif 2 (TCRPTAYSQFFKSACPRAYSYAYDDATSTF)位于Domain I和Domain II的结合部位, Motif 4 (APAGWSGRVWARTGCSFDGSG)位于Domain II和Domain III的结合部位, Motif 1 (GQDFYDVSL VDGYNLPMLVSP)位于TLP蛋白“V”字形裂缝内部, Motif 5 (GGGVVACRSACEAFGTPZYCCSGAY)属于Domain II, 位于“V”字形裂缝外缘。尤其需要强调的是, 本研究结果显示, SsTLP蛋白的Motif 1、2、4和5共同构成了SsTLP蛋白的“V”字形裂缝, 可能有助于确保蛋白催化功能的实现, 该研究结果与刘潮等[27]的研究结果一致。
据文献报道, TLPs共分为10个聚类组, 每个分类组功能各不相同[16]。在本研究中, 分布于TLP家族第V类的SsTLP蛋白数目最多。前人对毛果杨和拟南芥TLP蛋白家族的系统进化树分析表明, 位于TLP家族第V类的成员居多, 该分类组成员对于病原菌胁迫有明显应答效应, 而对于其它分类组的基因功能尚待研究[14,17]。本研究从甘蔗栽培种ROC22中克隆获得割手密SsTLP87的同源基因ScTLP1, 两者编码蛋白的氨基酸序列相似性高达99.56%, 均属于TLP家族的第V类成员, 推测ScTLP1具有抑菌活性。此外, GH64-TLP-SF超级家族含有β-1,3-葡聚糖酶活性, 可以降解真菌细胞壁, THN结构域主要含有带负电的16个半胱氨酸的基团, 保证TLP蛋白具有抗真菌活性, TLP-PA保守结构域是PR-5蛋白家族的一类特定保守结构域, 对植物的抗逆反应有应答作用[31]。ScTLP1基因同样属于GH64-TLP-SF超级家族成员, 其编码蛋白含有THN和TLP-PA保守结构域, 包含16个半胱氨酸位点, 暗示该基因在抵抗生物胁迫和非生物胁迫尤其是防御真菌性病害方面可能起作用。亚细胞定位结果显示, 柠檬(Citrus limon) RlemTLP蛋白定位于细胞膜和细胞质上[32], 荔枝TLP蛋白定位在细胞内[33]。本研究通过烟草亚细胞定位试验发现, 甘蔗ScTLP1蛋白定位在细胞膜上, 由于ScTLP1存在信号肽和跨膜结构域区段, 所以推测ScTLP1的蛋白定位和物质的跨膜运输存在关联。
前人研究显示, TLP基因在植物组织中组成型表达[5]。在湖北海棠(Malus hupehensis)和黄冠梨(Pyrus bretschneideri)中, TLP基因在树根中的表达量最高, 而在叶中最低[34,35]。对草莓(Fragaria ananassa) TLP基因的研究显示, 该基因在草莓叶部的表达量高于其他部位[36]。对于杨树TLP的研究表明, PeTLP基因在茎组织部位的表达量明显高于叶和根部位[37]。TLP作为一种病程防御相关蛋白, 能够应答病原菌的侵染。利用胶孢炭疽菌侵染荔枝果皮发现, TcTLP基因产生积极响应, 其表达量显著上升[11]。Ta-Tlp基因在高抗白粉病的小麦6VS/6AL易位系中高水平表达, 且其在小麦中的超量表达增强了转基因植株对白粉病的抗性[38]。冠腐病菌(Fusarium pseudograminearum)侵染4 d后, 小麦TLPs基因在抗病品种中的表达量显著高于感病品种[39]。本研究发现, ScTLP1基因在不同甘蔗组织部位均有表达, 但在蔗芽中的积累量最高, 且该基因在黑穗病菌侵染甘蔗2 d后的表达量显著升高。据报道, 蔗芽是黑穗病侵染甘蔗的唯一通道[40], ScTLP1基因在该组织部位的高表达量及其受黑穗病菌诱导上调表达, 表明其参与应答甘蔗黑穗病菌的胁迫反应。
植物激素SA和MeJA可以作为植物的潜在抗性促进剂, 诱导植物产生系统获得抗性(systemic acquired resistance, SAR), 甚至可以起到直接抑制病原菌生长和增殖的作用等[41]。前人对薇甘菊(Mikania micrantha)的研究发现, 在SA、MeJA和ABA处理后, 薇甘菊MmTLP1基因随着激素处理时间的增长, 诱导表达量逐渐上升[42]。对枣(Ziziphus jujuba)和谷子TLP家族基因的研究发现, 部分TLP基因的启动子区包含激素响应元件, 证明该TLP基因具有参与激素信号传导的作用[43]。在植物与病原菌的互作过程中会伴随着过敏反应的产生。烟草TLP蛋白通过RAS2/cAMP通路介导的胞内活性氧的累积, 诱导酵母细胞凋亡, 起到抗菌作用[44]。在本研究中, ScTLP1基因受MeJA诱导上调表达, 此外, 本氏烟瞬时表达结果显示, ScTLP1基因可以有效抑制烟草青枯菌和茄病镰刀菌蓝色变种的发病效应, 并且DAB染色加深, 乙烯合成相关基因NtEFE26和茉莉酸途径相关基因NtPR3都上调表达, 表明ScTLP1基因可能通过参与植物乙烯和茉莉酸信号传导途径, 诱发活性氧积累以促使过敏反应发生, 积极参与植物病害的防御作用。
TLP基因在响应非生物胁迫方面也发挥重要作用。在冷胁迫处理下, 挪威云杉TLP表达量累积[13]。白菜型冬油菜(Brassica rapa)陇油7号和天油2号的TLP基因的表达水平在4℃低温处理时均达到峰值, 分别较对照组增加了347.60%和92.70%[45]。转TLP基因的拟南芥植株在高盐浓度胁迫下的长势优于对照组, 表明TLP蛋白有助于植物抵御盐胁迫[14]。经200 mmol L-1 NaCl和4℃处理后, 湖北海棠(Malus hupehensis) TLP基因MhPR5的表达量显著增加[33]。茶树(Camellia sinensis) TLP基因受干旱胁迫诱导表达[46]。本研究中, 20% PEG、250 mmol L-1 NaCl和4℃低温处理可以诱导ScTLP1基因上调表达, 说明ScTLP1基因参与调控甘蔗对干旱、盐和低温胁迫的适应性响应。
酵母杂交技术是目前探究蛋白互作机理的主要技术, 随着酵母文库技术的日益完善, 越来越多的酵母互作研究相继开展。Nakahara等[47]在盐胁迫下构建的盐角草(Salicornia europaea)酵母文库中发现了一个TLP基因。麻楠等[48]通过酵母双杂实验技术验证了TaTLP基因与调控HR-PCD信号转导网络有关的TaTCTP基因之间存在互作机制。本研究验证了ScTLP1蛋白无转录自激活活性, 可以为后续利用酵母文库筛选ScTLP1蛋白的互作蛋白奠定基础, 有利于通过蛋白互作网络的研究探讨ScTLP1基因参与植物防御的作用机制。
4 结论
本研究鉴定获得了122个甘蔗野生种割手密SsTLP家族基因, 分布在28条割手密染色体上, 且存在多基因聚集现象, 其基因结构类型丰富, 内含子数目、内含子相位和Motif存在多种类型和分布。SsTLP基因编码蛋白分别聚类在TLP家族的I~X分支上, 其中具有抑菌潜力的第V类SsTLP最多。从甘蔗栽培种ROC22中克隆到属于V类的割手密SsTLP87的同源基因ScTLP1, 该基因定位在细胞膜上, 在甘蔗中为组成型表达, 且其在蔗芽中的积累量最多。ScTLP1基因的表达量在ABA和SA处理下无显著变化, 但在甘蔗黑穗病菌、MeJA、NaCl、PEG和4℃低温处理下升高, 表明该基因可以被生物和非生物胁迫诱导表达。此外, ScTLP1基因可以诱发本氏烟的过敏反应, 参与乙烯和茉莉酸信号传导途径, 增强本氏烟对烟草青枯菌和茄病镰刀菌蓝色变种的防御效应。酵母自激活验证试验表明, ScTLP1基因编码的蛋白无转录自激活活性, 可用于下一步的互作蛋白筛选。以上结果为深入探究甘蔗TLP家族基因的结构特征和解析其功能特性奠定了良好的基础。Table 1
表1
表1附SsTLP家族蛋白的二级结构预测
Table 1
| 蛋白名称 Protein name | α-螺旋 Alpha helix | 延伸链 Extended strand | 不规则卷曲 Random coil | 蛋白名称 Protein name | α-螺旋 Alpha helix | 延伸链 Extended strand | 不规则卷曲 Random coil |
|---|---|---|---|---|---|---|---|
| SsTLP1 | 19.05 | 15.99 | 64.97 | SsTLP62 | 18.70 | 15.65 | 65.65 |
| SsTLP2 | 22.84 | 16.05 | 61.11 | SsTLP63 | 19.20 | 16.52 | 64.29 |
| SsTLP3 | 23.08 | 16.00 | 60.92 | SsTLP64 | 24.92 | 14.20 | 60.88 |
| SsTLP4 | 20.80 | 15.36 | 63.84 | SsTLP65 | 23.91 | 14.91 | 61.18 |
| SsTLP5 | 20.47 | 10.13 | 69.40 | SsTLP66 | 24.68 | 14.24 | 61.08 |
| SsTLP6 | 22.55 | 9.39 | 68.06 | SsTLP67 | 14.78 | 20.27 | 64.95 |
| SsTLP7 | 22.91 | 11.64 | 65.45 | SsTLP68 | 18.28 | 15.93 | 65.80 |
| SsTLP8 | 27.79 | 14.20 | 58.01 | SsTLP69 | 9.36 | 22.81 | 67.84 |
| SsTLP9 | 26.56 | 13.11 | 60.33 | SsTLP70 | 9.25 | 18.50 | 72.25 |
| SsTLP10 | 28.70 | 13.90 | 57.40 | SsTLP71 | 14.90 | 18.51 | 66.59 |
| SsTLP11 | 12.71 | 16.95 | 70.34 | SsTLP72 | 9.25 | 20.23 | 70.52 |
| SsTLP12 | 12.71 | 19.07 | 68.22 | SsTLP73 | 9.25 | 18.50 | 72.25 |
| SsTLP13 | 9.75 | 16.95 | 73.31 | SsTLP74 | 7.76 | 19.59 | 72.65 |
| SsTLP14 | 31.15 | 15.10 | 53.75 | SsTLP75 | 10.33 | 13.65 | 76.01 |
| SsTLP15 | 12.79 | 19.19 | 68.01 | SsTLP76 | 7.83 | 21.08 | 71.08 |
| SsTLP16 | 11.07 | 19.13 | 69.80 | SsTLP77 | 19.74 | 12.45 | 67.81 |
| SsTLP17 | 11.98 | 15.27 | 72.75 | SsTLP78 | 26.61 | 20.97 | 52.42 |
| SsTLP18 | 11.98 | 15.27 | 72.75 | SsTLP79 | 18.11 | 22.83 | 59.06 |
| SsTLP19 | 11.98 | 15.27 | 72.75 | SsTLP80 | 17.86 | 15.87 | 66.27 |
| SsTLP20 | 15.22 | 17.99 | 66.78 | SsTLP81 | 18.18 | 15.81 | 66.01 |
| SsTLP21 | 17.92 | 16.75 | 65.33 | SsTLP82 | 17.93 | 16.33 | 65.74 |
| SsTLP22 | 15.75 | 17.81 | 66.44 | SsTLP83 | 29.07 | 17.85 | 53.08 |
| SsTLP23 | 36.70 | 0.92 | 62.39 | SsTLP84 | 27.04 | 9.92 | 63.04 |
| SsTLP24 | 3.16 | 9.47 | 87.37 | SsTLP85 | 40.50 | 13.76 | 45.74 |
| SsTLP25 | 9.72 | 14.58 | 75.69 | SsTLP86 | 30.54 | 17.06 | 52.41 |
| SsTLP26 | 8.13 | 18.70 | 73.17 | SsTLP87 | 24.23 | 11.01 | 64.76 |
| SsTLP27 | 19.02 | 13.50 | 67.48 | SsTLP88 | 24.23 | 11.45 | 64.32 |
| SsTLP28 | 11.02 | 19.07 | 69.92 | SsTLP89 | 9.68 | 21.51 | 68.82 |
| SsTLP29 | 19.81 | 15.91 | 64.29 | SsTLP90 | 9.68 | 21.51 | 68.82 |
| SsTLP30 | 8.90 | 19.07 | 72.03 | SsTLP91 | 9.95 | 23.56 | 66.49 |
| SsTLP31 | 22.78 | 10.76 | 66.46 | SsTLP92 | 40.49 | 10.56 | 48.94 |
| SsTLP32 | 22.57 | 11.29 | 66.14 | SsTLP93 | 13.21 | 22.86 | 63.93 |
| SsTLP33 | 13.41 | 21.34 | 65.24 | SsTLP94 | 11.99 | 20.97 | 67.04 |
| SsTLP34 | 11.32 | 22.96 | 65.72 | SsTLP95 | 13.93 | 22.14 | 63.93 |
| SsTLP35 | 17.65 | 16.41 | 65.94 | SsTLP96 | 15.88 | 21.30 | 62.82 |
| SsTLP36 | 11.82 | 15.45 | 72.73 | SsTLP97 | 29.09 | 18.18 | 52.73 |
| SsTLP37 | 12.46 | 15.57 | 71.97 | SsTLP98 | 29.77 | 16.54 | 53.70 |
| SsTLP38 | 9.52 | 25.54 | 64.94 | SsTLP99 | 12.30 | 21.72 | 65.98 |
| SsTLP39 | 12.57 | 14.40 | 73.04 | SsTLP100 | 15.31 | 20.66 | 64.03 |
| SsTLP40 | 17.65 | 16.10 | 66.25 | SsTLP101 | 12.40 | 24.79 | 62.81 |
| SsTLP41 | 13.14 | 12.41 | 74.45 | SsTLP102 | 12.40 | 24.79 | 62.81 |
| SsTLP42 | 13.19 | 15.62 | 71.18 | SsTLP103 | 13.69 | 12.03 | 74.27 |
| SsTLP43 | 17.78 | 20.00 | 62.22 | SsTLP104 | 16.59 | 14.75 | 68.66 |
| SsTLP44 | 12.78 | 20.26 | 66.96 | SsTLP105 | 16.33 | 11.16 | 72.51 |
| SsTLP45 | 12.33 | 19.82 | 67.84 | SsTLP106 | 17.30 | 16.98 | 65.72 |
| SsTLP46 | 16.28 | 20.93 | 62.79 | SsTLP107 | 26.99 | 21.17 | 51.84 |
| SsTLP47 | 13.68 | 13.68 | 72.63 | SsTLP108 | 31.53 | 12.77 | 55.71 |
| SsTLP48 | 15.25 | 19.17 | 65.58 | SsTLP109 | 16.48 | 19.48 | 64.04 |
| SsTLP49 | 0.00 | 23.93 | 76.07 | SsTLP110 | 19.45 | 17.33 | 63.22 |
| SsTLP50 | 9.41 | 21.18 | 69.41 | SsTLP111 | 29.23 | 12.69 | 58.08 |
| SsTLP51 | 11.31 | 19.64 | 69.05 | SsTLP112 | 28.46 | 13.46 | 58.08 |
| SsTLP52 | 0.00 | 22.09 | 77.91 | SsTLP113 | 22.94 | 13.98 | 63.08 |
| SsTLP53 | 11.31 | 19.64 | 69.05 | SsTLP114 | 17.74 | 20.97 | 61.29 |
| SsTLP54 | 29.61 | 20.11 | 50.28 | SsTLP115 | 21.26 | 20.47 | 58.27 |
| SsTLP55 | 9.36 | 22.81 | 67.84 | SsTLP116 | 18.95 | 21.37 | 59.68 |
| SsTLP56 | 11.70 | 22.22 | 66.08 | SsTLP117 | 22.68 | 18.85 | 58.47 |
| SsTLP57 | 21.97 | 18.18 | 59.85 | SsTLP118 | 22.36 | 18.85 | 58.79 |
| SsTLP58 | 11.31 | 19.64 | 69.05 | SsTLP119 | 20.39 | 16.50 | 63.11 |
| SsTLP59 | 20.29 | 11.49 | 68.22 | SsTLP120 | 19.09 | 16.50 | 64.40 |
| SsTLP60 | 15.42 | 14.93 | 69.65 | SsTLP121 | 17.13 | 10.36 | 72.51 |
| SsTLP61 | 10.29 | 21.14 | 68.57 | SsTLP122 | 21.45 | 14.55 | 64.00 |
新窗口打开|下载CSV
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOI:10.1016/j.plaphy.2008.06.011URLPMID:18674922 [本文引用: 1]

The novel classes of plant pathogenesis-related (PR) proteins identified during the last decade also include novel peptide families. This review specifically focuses on these pathogenesis-related peptides, including proteinase inhibitors (PR-6 family), plant defensins (PR-12 family), thionins (PR-13 family) and lipid transfer proteins (PR-14 family). For each family of PR peptides, the general features concerning occurrence, expression and possible functions of their members are described. Next, more specifically the occurrence of each PR peptide family in the model plant Arabidopsis thaliana is discussed. Single-gene studies performed on particular gene members of a PR peptide family are reported. In addition, expression data of yet undescribed gene members of that particular PR peptide family are presented by consultation of publicly available micro-array databases. Finally an update is provided on the potential role of these PR peptides in A. thaliana, with a focus on their possible involvement in plant defense.
DOI:10.1186/1471-2229-11-33URLPMID:21324123 [本文引用: 2]

BACKGROUND: Plant inducible immunity includes the accumulation of a set of defense proteins during infection called pathogenesis-related (PR) proteins, which are grouped into families termed PR-1 to PR-17. The PR-5 family is composed of thaumatin-like proteins (TLPs), which are responsive to biotic and abiotic stress and are widely studied in plants. TLPs were also recently discovered in fungi and animals. In the poplar genome, TLPs are over-represented compared with annual species and their transcripts strongly accumulate during stress conditions. RESULTS: Our analysis of the poplar TLP family suggests that the expansion of this gene family was followed by diversification, as differences in expression patterns and predicted properties correlate with phylogeny. In particular, we identified a clade of poplar TLPs that cluster to a single 350 kb locus of chromosome I and that are up-regulated by poplar leaf rust infection. A wider phylogenetic analysis of eukaryote TLPs - including plant, animal and fungi sequences - shows that TLP gene content and diversity increased markedly during land plant evolution. Mapping the reported functions of characterized TLPs to the eukaryote phylogenetic tree showed that antifungal or glycan-lytic properties are widespread across eukaryote phylogeny, suggesting that these properties are shared by most TLPs and are likely associated with the presence of a conserved acidic cleft in their 3D structure. Also, we established an exhaustive catalog of TLPs with atypical architectures such as small-TLPs, TLP-kinases and small-TLP-kinases, which have potentially developed alternative functions (such as putative receptor kinases for pathogen sensing and signaling). CONCLUSION: Our study, based on the most recent plant genome sequences, provides evidence for TLP gene family diversification during land plant evolution. We have shown that the diverse functions described for TLPs are not restricted to specific clades but seem to be universal among eukaryotes, with some exceptions likely attributable to atypical protein structures. In the perennial plant model Populus, we unravelled the TLPs likely involved in leaf rust resistance, which will provide the foundation for further functional investigations.
DOI:10.1155/2017/5046451URLPMID:28875151 [本文引用: 2]

A thaumatin-like protein gene from Basrai banana was cloned and expressed in Escherichia coli. Amplified gene product was cloned into pTZ57R/T vector and subcloned into expression vector pET22b(+) and resulting pET22b-basrai TLP construct was introduced into E. coli BL21. Maximum protein expression was obtained at 0.7 mM IPTG concentration after 6 hours at 37 degrees C. Western blot analysis showed the presence of approximately 20 kDa protein in induced cells. Basrai antifungal TLP was tried as pharmacological agent against fungal disease. Independently Basrai antifungal protein and amphotericin B exhibited their antifungal activity against A. fumigatus; however combined effect of both agents maximized activity against the pathogen. Docking studies were performed to evaluate the antimicrobial potential of TLP against A. fumigatus by probing binding pattern of antifungal protein with plasma membrane ergosterol of targeted fungal strain. Ice crystallization primarily damages frozen food items; however addition of antifreeze proteins limits the growth of ice crystal in frozen foods. The potential of Basrai TLP protein, as an antifreezing agent, in controlling the ice crystal formation in frozen yogurt was also studied. The scope of this study ranges from cost effective production of pharmaceutics to antifreezing and food preserving agent as well as other real life applications.
[本文引用: 4]
[本文引用: 4]
DOI:10.1016/j.jplph.2006.01.006URLPMID:16542753 [本文引用: 1]

A full-length 910bp cDNA encoding osmotin-like protein with an open reading frame of 744bp encoding a protein of 247 amino acids with a calculated molecular mass of 26.8kDa was cloned from Solanum nigrum (SniOLP). Phylogenetic analysis revealed the evolutionary conservation of this protein among diverse taxa. The genomic DNA gel blot showed that SniOLP belongs to a small multigene family and it showed organ-specific expression. Time-course studies revealed that the expression of SniOLP was upregulated by treatment with various signaling molecules, osmotic and oxidative stress inducers. Recombinant protein purified from overexpressed Escherichia coli cells showed hyphal growth inhibition in Rhizoctonia batiticola and Sclerotinia sclerotiorum but without any endo-beta-1,3-glucanase activity. Model built by homology modeling showed that the protein consists of an acidic cleft region that is capable of interacting with the carbohydrate components of the fungal cell walls. Analysis of the structure and functional relationship was carried out by docking of the beta-(1,3)-glucan onto the acidic cleft region on the surface of the protein (SniOLP).
URLPMID:18951281 [本文引用: 1]
DOI:10.1104/pp.101.3.1113URLPMID:8310049 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 2]
DOI:10.1038/srep25340URLPMID:27150014 [本文引用: 3]

Plant often responds to fungal pathogens by expressing a group of proteins known as pathogenesis-related proteins (PRs). The expression of PR is mediated through pathogen-induced signal-transduction pathways that are fine-tuned by phytohormones such as methyl jasmonate (MeJA). Here, we report functional characterization of an Ocimum basilicum PR5 family member (ObTLP1) that was identified from a MeJA-responsive expression sequence tag collection. ObTLP1 encodes a 226 amino acid polypeptide that showed sequence and structural similarities with a sweet-tasting protein thaumatin of Thaumatococcus danielli and also with a stress-responsive protein osmotin of Nicotiana tabacum. The expression of ObTLP1 in O. basilicum was found to be organ-preferential under unstressed condition, and responsive to biotic and abiotic stresses, and multiple phytohormone elicitations. Bacterially-expressed recombinant ObTLP1 inhibited mycelial growth of the phytopathogenic fungi, Scleretonia sclerotiorum and Botrytis cinerea; thereby, suggesting its antifungal activity. Ectopic expression of ObTLP1 in Arabidopsis led to enhanced tolerance to S. sclerotiorum and B. cinerea infections, and also to dehydration and salt stress. Moreover, induced expression of the defense marker genes suggested up-regulation of the defense-response pathways in ObTLP1-expressing Arabidopsis upon fungal challenge. Thus, ObTLP1 might be useful for providing tolerance to the fungal pathogens and abiotic stresses in crops.
DOI:10.1038/s41588-018-0237-2URLPMID:30297971 [本文引用: 2]

Modern sugarcanes are polyploid interspecific hybrids, combining high sugar content from Saccharum officinarum with hardiness, disease resistance and ratooning of Saccharum spontaneum. Sequencing of a haploid S. spontaneum, AP85-441, facilitated the assembly of 32 pseudo-chromosomes comprising 8 homologous groups of 4 members each, bearing 35,525 genes with alleles defined. The reduction of basic chromosome number from 10 to 8 in S. spontaneum was caused by fissions of 2 ancestral chromosomes followed by translocations to 4 chromosomes. Surprisingly, 80% of nucleotide binding site-encoding genes associated with disease resistance are located in 4 rearranged chromosomes and 51% of those in rearranged regions. Resequencing of 64 S. spontaneum genomes identified balancing selection in rearranged regions, maintaining their diversity. Introgressed S. spontaneum chromosomes in modern sugarcanes are randomly distributed in AP85-441 genome, indicating random recombination among homologs in different S. spontaneum accessions. The allele-defined Saccharum genome offers new knowledge and resources to accelerate sugarcane improvement.
DOI:10.1007/s00239-005-0053-zURLPMID:16736102 [本文引用: 2]

Pathogenesis-related group 5 (PR5) plant proteins include thaumatin, osmotin, and related proteins, many of which have antimicrobial activity. The recent discovery of PR5-like (PR5-L) sequences in nematodes and insects raises questions about their evolutionary relationships. Using complete plant genome data and discovery of multiple insect PR5-L sequences, phylogenetic comparisons among plants and animals were performed. All PR5/PR5-L protein sequences were mined from genome data of a member of each of two main angiosperm groups-the eudicots (Arabidoposis thaliana) and the monocots (Oryza sativa)-and from the Caenorhabditis nematode (C. elegans and C. briggsase). Insect PR5-L sequences were mined from EST databases and GenBank submissions from four insect orders: Coleoptera (Diaprepes abbreviatus and Biphyllus lunatus), Orthoptera (Schistocerca gregaria), Hymenoptera (Lysiphlebus testaceipes), and Hemiptera (Toxoptera citricida). Parsimony and Bayesian phylogenetic analyses showed that the PR5 family is paraphyletic in plants, likely arising from 10 genes in a common ancestor to monocots and eudicots. After evolutionary divergence of monocots and eudicots, PR5 genes increased asymmetrically among the 10 clades. Insects and nematodes contain multiple sequences (seven PR5-Ls in nematodes and at least three in some insects) all related to the same plant clade, with nematode and insect sequences separating as two clades. Protein structural homology modeling showed strong similarity among animal and plant PR5/PR5-Ls, with divergence only in surface-exposed loops. Sequence and structural conservation among PR5/PR5-Ls suggests an important and conserved role throughout the evolutionary divergence of the diverse organisms from which they reside.
DOI:10.1007/s00425-010-1218-6URLPMID:20645107 [本文引用: 3]

Some pathogenesis-related proteins (PR proteins) are subject to positive selection, while others are under negative selection. Here, we report the patterns of molecular evolution in thaumatin-like protein (TLP, PR5 protein) genes of Populus trichocarpa. Signs of positive selection were found in 20 out of 55 Populus TLPs using the likelihood ratio test and ML-based Bayesian methods. Due to the connection between the acidic cleft and the antifungal activity, the secondary structure and three-dimensional structure analyses predicted antifungal activity beta-1,3-glucanase activities in these TLPs. Moreover, the coincidence with variable basic sites in the acidic cleft and positively selected sites suggested that fungal diseases may have been the main environmental stress that drove rapid adaptive evolution in Populus.
[本文引用: 1]
DOI:10.3389/fpls.2017.02262URLPMID:29387074 [本文引用: 3]

The L-ascorbate peroxidase 6 gene (APX6) is one of the most important genes for scavenging H2O2 and plays a vital role in plant resistance to environmental stresses. In this study, a novel ScAPX6 gene (GenBank Accession No. KT907352) was obtained from a sugarcane variety (ROC22). Bioinformatics analysis showed that ScAPX6 has a cDNA length of 1,086 bp and encoded 333 amino acid residues. Subcellular localization confirmed that ScAPX6 was located in the chloroplast. Enhanced growth of Escherichia coli BL21 cells that expressed ScAPX6 showed high tolerance under copper (Cu) stress. Real-time quantitative PCR analysis revealed that ScAPX6 was constitutively expressed wherein with the highest expression levels in sugarcane pith and leaf and the lowest in the root. ScAPX6 was down-regulated by salicylic acid (SA), hydrogen peroxide (H2O2), polyethylene glycol (PEG) and sodium chloride (NaCl) stimuli. Interestingly, it was significantly up-regulated under the stresses of abscisic acid (ABA) and methyl jasmonate (MeJA) wherein with the highest inducible expression levels at 6 h at 6.0- and 70.0-times higher, respectively than that of control. Overexpression of ScAPX6 in Nicotiana benthamiana leaves enhanced the resistance to the infection of tobacco pathogens Pseudomonas solanacearum and Fusarium solani var. coeruleum. These results implied that ScAPX6 might positively respond to ABA, MeJA, and Cu, but might negatively respond to the stresses of SA, H2O2, PEG, and NaCl. Keeping in view the current investigation, ScAPX6 could be associated with the hypersensitive response (HR) or immunity of sugarcane, which will provide a baseline for the function identification of sugarcane ScAPX6.
[本文引用: 1]
[本文引用: 1]
DOI:10.1006/meth.2001.1262URLPMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.
DOI:10.1186/s12864-017-4142-3URLPMID:29020924 [本文引用: 4]

BACKGROUND: Sugarcane smut caused by Sporisorium scitamineum is one of the most severe fungal diseases in the sugarcane industry. Using a molecular biological technique to mine sugarcane resistance genes can provide gene resources for further genetic engineering of sugarcane disease-resistant breeding. Jasmonate ZIM (zinc-finger inflorescence meristem) domain (JAZ) proteins, which involved in the responses to plant pathogens and abiotic stresses, are important signaling molecules of the jasmonic acid (JA) pathway. RESULTS: Seven differentially expressed sugarcane JAZ genes, ScJAZ1-ScJAZ7, were mined from the transcriptome of sugarcane after inoculation with S. scitamineum. Bioinformatic analyses revealed that these seven ScJAZ genes encoded basic proteins that contain the TIFY and CCT_2 domains. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis demonstrated that the ScJAZ1-ScJAZ7 genes were tissue specific and differentially expressed under adverse stress. During S. scitamineum infection, the transcripts of ScJAZ4 and ScJAZ5 were both upregulated in the susceptible genotype ROC22 and the resistant genotype Yacheng05-179; ScJAZ1, ScJAZ2, ScJAZ3, and ScJAZ7 were downregulated in Yacheng05-179 and upregulated in ROC22; and the expression of ScJAZ6 did not change in ROC22, but was upregulated in Yacheng05-179. The transcripts of the seven ScJAZ genes were increased by the stimuli of salicylic acid and abscisic acid, particularly methyl jasmonate. The expression of the genes ScJAZ1-ScJAZ7 was immediately upregulated by the stressors hydrogen peroxide, sodium chloride, and copper chloride, whereas slightly induced after treatment with calcium chloride and polyethylene glycol. In addition, the expression of ScJAZ6, as well as seven tobacco immunity-associated marker genes were upregulated, and antimicrobial activity against Pseudomonas solanacearum and Fusarium solani var. coeruleum was observed during the transient overexpression of ScJAZ6 in Nicotiana benthamiana, suggesting that the ScJAZ6 gene is associated with plant immunity. CONCLUSIONS: The different expression profiles of the ScJAZ1-ScJAZ7 genes during S. scitamineum infection, the positive response of ScJAZ1-ScJAZ7 to hormones and abiotic treatments, and the function analysis of the ScJAZ6 gene revealed their involvement in the defense against biotic and abiotic stresses. The findings of the present study facilitate further research on the ScJAZ gene family especially their regulatory mechanism in sugarcane.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00299-010-0826-8URLPMID:20204373 [本文引用: 1]

Thaumatin-like proteins (TLPs) are the products of a large, highly complex gene family involved in host defence and a wide range of developmental processes in fungi, plants, and animals. Despite their dramatic diversification in organisms, TLPs appear to have originated in early eukaryotes and share a well-defined TLP domain. Nonetheless, determination of the roles of individual members of the TLP superfamily remains largely undone. This review summarizes recent advances made in elucidating the varied TLP activities related to host resistance to pathogens and other physiological processes. Also discussed is the current state of knowledge on the origins and types of TLPs, regulation of gene expression, and potential biotechnological applications for TLPs.
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.biochi.2005.07.001URLPMID:16085352 [本文引用: 1]

The structure of a thaumatin-like protein from banana (Musa acuminata) fruit, an allergen with antifungal properties, was solved at 1.7-A-resolution, by X-ray crystallography. Though the banana protein exhibits a very similar overall fold as thaumatin it markedly differs from the sweet-tasting protein by the presence of a surface exposed electronegative cleft. Due to the presence of this electronegative cleft, the banana thaumatin-like protein (Ban-TLP) acquires a strong (local) electronegative character that eventually explains the observed antifungal activity. Our structural analysis also revealed the presence of conserved residues of exposed epitopic determinants that are presumably responsible for the allergenic properties of banana fruit towards susceptible individuals, and provided evidence that the Ban-TLP shares some structurally highly conserved IgE-binding epitopes with thaumatin-like proteins from fruits or pollen from other plants. In addition, some overlap was detected between the predicted IgE-binding epitopes of the Ban-TLP and IgE-binding epitopes previously identified in the mountain cedar Jun a 3 TLP aeroallergen. The presence of these common epitopes offers a molecular basis for the cross-reactivity between aeroallergens and fruit allergens.
DOI:10.1006/jmbi.1998.2540URLPMID:10047487 [本文引用: 1]

The crystal structure of tobacco PR-5d, an antifungal thaumatin-like protein isolated from cultured tobacco cells, was determined at the resolution of 1.8 A. The structure consists of 208 amino acid residues and 89 water molecules with a crystallographic R-factor of 0.169. The model has good stereochemistry, with respective root-mean-square deviations from the ideal values for bond and angle distances of 0.007 A and 1.542 degrees. Of the homologous PR-5 proteins, only those with antifungal activity had a common motif, a negatively charged surface cleft. This cleft is at the boundary between domains I and II, with a bottom part consisting of a three-stranded antiparallel beta-sheet in domain I. The acidic residues located in the hollow of the cleft form the beta-sheet region. Sequence and secondary structure analyses showed that the amino acid residues comprising the acidic cleft of PR-5d are conserved among other antifungal PR-5 proteins. This is the first report on the high-resolution crystal structure of an antifungal PR-5 protein. This structure provides insight into the function of pathogenesis-related proteins.
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.jplph.2006.02.002URLPMID:16603274 [本文引用: 1]

A strawberry genomic clone containing an osmotin-like protein (OLP) gene, designated FaOLP2, was isolated and sequenced. FaOLP2 is predicted to encode a precursor protein of 229 amino acid residues, and its sequence shares high degrees of homology with a number of other OLPs. Genomic DNA hybridization analysis indicated that FaOLP2 represents a multi-gene family. The expression of FaOP2 in different strawberry organs was analyzed using real-time PCR. The results showed that FaOLP2 expressed at different levels in leaves, crowns, roots, green fruits and ripe red fruits. In addition, the expression of FaOLP2 under different abiotic stresses was analyzed at different time points. All of the three tested abiotic stimuli, abscisic acid, salicylic acid and mechanical wounding, triggered a significant induction of FaOLP2 within 2-6h post-treatment. Moreover, FaOLP2 was more prominently induced by salicylic acid than by abscisic acid or mechanical wounding. The positive responses of FaOLP2 to the three abiotic stimuli suggested that strawberry FaOLP2 may help to protect against osmotic-related environmental stresses and that it may also be involved in plant defense system against pathogens.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11033-009-9823-9URLPMID:19757158 [本文引用: 1]

b-1,3-Glucanases are a group of pathogenesis related proteins that have been reported to be involved in plant defense against pathogens in many other plant pathogen systems. However, it was not clear if these genes play similar role in wheat (Triticum aestivum L.) against Puccinia striiformis f. sp. tritici (Pst), the stripe rust pathogen. To investigate the role of b-1,3-glucanase (EC3.2.1.39) in the resistance response of wheat (cv. Suwon11) to stripe rust, a wheat b-1,3-glucanase gene induced by Pst, designated as TaGlu, was cloned and characterized.TaGlu was predicted to encode a basic protein of 334 amino acids. Quantitative real-time PCR analyses revealed that the transcription of TaGlu was induced during both compatible and incompatible interactions with Pst, but the transcription level was much higher in the incompatible interaction than that in the compatible interaction. TaGlu also showed noticeable induction of gene expression in young green leaf tissues treated with salicylic acid, methyl jasmonate or ethylene. Immunogold labeling assays showed that the enzyme were localized mainly in the host cell wall and over the extra haustorial matrix, and the labeling densities were found significantly higher in the incompatible interaction than those in the compatible interaction.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/s1097-2765(01)00365-3URLPMID:11684026 [本文引用: 1]

Osmotin is a tobacco PR-5 protein that has antifungal activity and is implicated in host-plant defense. We show here that osmotin induces apoptosis in Saccharomyces cerevisiae. Induction of apoptosis was correlated with intracellular accumulation of reactive oxygen species and was mediated by RAS2, but not RAS1. Osmotin treatment resulted in suppression of transcription of stress-responsive genes via the RAS2/cAMP pathway. It was therefore concluded that osmotin induced proapoptotic signaling in yeast. The results indicate that the ability of antimicrobial proteins to induce microbial apoptosis could be an important factor in determining a pathogen's virulence and could therefore be targeted for the design of new antifungal drugs.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/BF02703703URLPMID:15886459 [本文引用: 1]

There is no information on drought-modulated gene(s) in tea [Camellia sinensis (L.) O. Kuntze], a woody and perennial plant of commercial importance. Using differential display of mRNA, three drought-modulated expressed sequence tags (ESTs) were identified. Northern and BLAST analysis revealed that clone dr1 (drought-responsive), induced only by drought but not by ABA, showed significant scores with PR-5 (pathogenesis related) family of PR-protein gene. Another clone dr2, repressed by drought but not by ABA, had nucleotide repeats for polyasparate that are also present in chicken calsequestrin-like mRNA. Clone dr3, responded similarly to clone dr2 but did not show significant homology with the reported genes, hence appears to be novel. Identification of these ESTs is an initial step to clone the full length genes and their promoters.
DOI:10.3389/fpls.2015.00920URLPMID:26579166 [本文引用: 1]

Salinity is a critical environmental factor that adversely affects crop productivity. Halophytes have evolved various mechanisms to adapt to saline environments. Salicornia europaea L. is one of the most salt-tolerant plant species. It does not have special salt-secreting structures like a salt gland or salt bladder, and is therefore a good model for studying the common mechanisms underlying plant salt tolerance. To identify candidate genes encoding key proteins in the mediation of salt tolerance in S. europaea, we performed a functional screen of a cDNA library in yeast. The library was screened for genes that allowed the yeast to grow in the presence of 1.3 M NaCl. We obtained three full-length S. europaea genes that confer salt tolerance. The genes are predicted to encode (1) a novel protein highly homologous to thaumatin-like proteins, (2) a novel coiled-coil protein of unknown function, and (3) a novel short peptide of 32 residues. Exogenous application of a synthetic peptide corresponding to the 32 residues improved salt tolerance of Arabidopsis. The approach described in this report provides a rapid assay system for large-scale screening of S. europaea genes involved in salt stress tolerance and supports the identification of genes responsible for such mechanisms. These genes may be useful candidates for improving crop salt tolerance by genetic transformation.
[本文引用: 1]
[本文引用: 1]
