 ,1,2, 李德芳
,1,2, 李德芳 ,2,*, 邓勇2, 潘根2, 陈安国2, 赵立宁2, 唐慧娟2
,2,*, 邓勇2, 潘根2, 陈安国2, 赵立宁2, 唐慧娟2Expression analysis of abiotic stress response gene HcWRKY71 in kenaf and transformation of Arabidopsis
LI Hui ,1,2, LI De-Fang
,1,2, LI De-Fang ,2,*, DENG Yong2, PAN Gen2, CHEN An-Guo2, ZHAO Li-Ning2, TANG Hui-Juan2
,2,*, DENG Yong2, PAN Gen2, CHEN An-Guo2, ZHAO Li-Ning2, TANG Hui-Juan2通讯作者:
收稿日期:2019-12-19接受日期:2020-10-14网络出版日期:2021-06-12
| 基金资助: |
Received:2019-12-19Accepted:2020-10-14Online:2021-06-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (8740KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李辉, 李德芳, 邓勇, 潘根, 陈安国, 赵立宁, 唐慧娟. 红麻非生物逆境胁迫响应基因HCWRKY71表达分析及转化拟南芥 [J]. 作物学报, 2021, 47(6): 1090-1099. doi:10.3724/SP.J.1006.2021.04201
LI Hui, LI De-Fang, DENG Yong, PAN Gen, CHEN An-Guo, ZHAO Li-Ning, TANG Hui-Juan.
红麻(Hibiscus cannabinus L.)是一年速生草本纤维作物。随着人们环保意识的增强, 全球对于天然纤维的需求日益增加, 大力发展红麻产业是满足天然纤维需求的重要途径之一[1]。然而随着全球人口的增加, 人口、粮食、耕地之间的矛盾日益加剧, 红麻的种植必须转向盐碱地、干旱坡地、不能生产粮食的重金属污染土地。因此, 研究红麻的耐逆机制, 选育红麻耐逆品种对维持红麻产业的可持续发展具有重要意义。
高等植物在进化过程中形成了精细的调控网络来应对不同的逆境胁迫。转录因子如MYB[2]、WRKY[3]、NAC[4]在调控网络中发挥极其重要的作用, 它们通过激活/抑制下游基因表达来提高植物本身对逆境的适应能力。其中WRKY转录因子是植物体内特有的转录因子家族, 其特点是DNA结合域的N-端至少包含一段保守的WRKYGQK序列, C-端是一个类锌指结构域。DNA结合域通过与启动子中的W-box序列结合来调节靶标基因的表达[5]。
WRKY转录因子家族在植物响应干旱、盐、重金属、温度、病害等多种非生物/生物胁迫过程中具有重要的调控功能, 且不断有新的家族成员被鉴定[6]。Wu等[7]在菜豆基因组中发现了19个WRKY响应基因, 在干旱胁迫条件下, 其中11个表现为下调表达, 8个表现为上调表达。王岩岩等[8]研究表明, 野生大豆GsWRKY57基因表达受干旱胁迫的诱导, 且随干旱胁迫时间的增加, GsWRKY57基因表达呈现先上升后下降的趋势。柯丹霞等[9]研究表明, 大豆GmWRKY6基因表达受盐胁迫诱导, 并且能够显著提高转基因百脉根的耐盐性。甘蔗ScWRKY4基因在NaCl胁迫下显著上调表达, 推测该基因在甘蔗响应NaCl胁迫过程中具有一定的调控作用[10]。在低温胁迫条件下, 转CsWRKY46基因的拟南芥与野生型相比, 具有较高的存活率。在低温胁迫和外源ABA诱导下, CsWRKY46基因出现上调表达[11]。王影等[12]研究表明, 东南景天SaWRKY7基因表达受镉胁迫的诱导, 且随镉胁迫时间的增加, 其表达量逐渐降低, 推测其在镉胁迫响应早期具有重要作用。以上研究证实, WRKY转录因子在植物抵御各种非生物胁迫过程中具有正向或负向的调控功能。但目前关于WRKY基因家族成员WRKY71在红麻非生物逆境胁迫响应过程中的作用鲜见报道。
本课题组前期通过转录组测序获得unigene序列(CL3883.Contig4)[13], 通过NCBI的在线工具Blast分析发现, 其编码产物为转录因子WRKY71, 且该unigene受盐胁迫的诱导。因此, 本研究克隆了WRKY71基因序列, 利用实时荧光定量PCR的方法分析了其在盐、干旱和重金属不同逆境胁迫下的时空表达特征, 并转化拟南芥进行初步功能验证, 旨在为进一步研究该基因耐盐的调控机制奠定基础。
1 材料与方法
1.1 试验材料
红麻16号由中国农业科学院麻类研究所提供。挑选籽粒饱满、大小均匀的红麻种子, 用0.1%的氯化汞消毒10 min, 蒸馏水冲洗3次, 然后播种于发芽盒中, 置光照培养箱中培养。培养10 d后, 选取植株健壮、长势均匀的红麻幼苗, 移植到1/2 Hoagland营养液中培养。红麻幼苗植株长到四叶一心时, 选取20株幼苗进行NaCl胁迫处理, 每个处理5株, 1/2 Hoagland营养液中NaCl的最终浓度分别为0、50、100、200 mmol L-1; 同样选取20株幼苗进行干旱处理, 将选取的幼苗根部暴露在空气中, 不进行供水处理, 处理时间为0、0.5、1.0、2.0、4.0 h, 每个处理5株; 同样选取20株幼苗进行重金属镉胁迫处理, 每个处理5株, 1/2 Hoagland营养液中氯化镉(CdCl2)的最终浓度分别为0、50、100、150 μmol L-1。分别取每个处理组红麻植株的根、茎、叶, 液氮速冻, 放置于-80℃冰箱备用。1.2 红麻HcWRKY71基因的克隆及测序
利用植物总RNA提取试剂盒(DP432) (天根生化科技北京有限公司)提取200 mmol L-1 NaCl处理的红麻植株叶片总RNA, 然后逆转录合成cDNA, 进行PCR扩增。根据课题组转录组unigene序列(编号: CL3883.Contig4), 利用PrimerPremier 5.0设计HcWRKY71基因全长扩增引物HcWRKY71-F/R (表1)。PCR反应的体系包括2.5 μL 10× PCR buffer (Mg2+ plus)、0.25 μL Taq酶 (TaKaRa, 中国大连)、1 μL 2.5 mmol L-1 dNTP、0.25 μL 10 μmol L-1 HcWRKY71-F、0.25 μL 10 μmol L-1 HcWRKY71-R、20 ng cDNA, ddH2O补足25 μL。PCR扩增的反应程序为94℃预变性2 min; 94℃变性30 s, 55℃退火30 s, 72℃延伸60 s, 30个循环; 72℃延伸3 min, 4℃终止反应。反应产物经1.2%琼脂糖凝胶电泳检测。参照琼脂糖凝胶DNA回收试剂盒(天根生化科技北京有限公司)的说明书切胶回收, 然后将回收片段连接到pMD19-T载体上, 转化大肠杆菌DH5α感受态细胞, 挑取阳性菌落送生工生物工程(上海)股份有限公司测序。Table 1
表1
表1本研究所用引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequences (5°-3°) | 引物用途 Primer usage |
|---|---|---|
| HcWRKY71-F | ATGTCGGATCATGGATTTA | 基因克隆 |
| HcWRKY71-R | TCATGGTTCGTGTTTAAGGA | Gene cloning |
| HcWRKY71-QF | GGTGCGGAGGAATTAGTA | 实时荧光定量PCR |
| HcWRKY71-QR | ATGAAAGCAAATCGTGGT | qRT-PCR |
| Actin-QF | CAGGCAGTTCTTTCTTTGT | 内参基因 |
| Actin- QR | ATCCTCCAATCCAGACACT | Reference gene |
| PC1301-35S-HcWRKY71-GFP-F | GAGAACACGGGGGACTGGTACCCGGGGATCCATGTCGGATCATGGATT | 目的基因与表达载体的连接 Ligation of target gene with expression vector |
| PC1301-35S-HcWRKY71-GFP-R | ACAGCTCCTCGCCCTTGCTCACCATGTCGACTGGTTCGTGTTTAAGGAA | |
| HcWRKY71-PF | AGATCAGAAGATGGTGGCG | 转基因拟南PCR鉴定及实时荧光定量分析 |
| HcWRKY71-PR | TCGGATATGGGCTGTTCTT | PCR identification and real-time fluorescence quantitative analysis of transgenic Arabidopsis |
新窗口打开|下载CSV
1.3 红麻HcWRKY71基因的生物信息学分析
利用NCBI在线程序ORF finder (http://www. ncbi.nlm.nih.gov/gorf/gorf.html)和NCBI Conservered Domain (https//www.ncbi.nlm.nih.gov/cdd)预测基因序列的最大开放读码框及其保守功能结构域; 运用在线工具(http://www.expasy.ch/tools/pi_tool.html)和NetPhos2.0 (http://www.cbs.dtu.dk/services/NetPhos/)分别预测HcWRKY71基因编码蛋白质的分子量和等电点及其蛋白磷酸化位点; 利用在线工具BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi)对HcWRKY71的氨基酸序列进行在线序列比对; 利用DNAMAN软件进行HcWRKY71基因氨基酸序列一致性比对; 利用MEGA6.0软件构建HcWRKY71基因系统进化树。1.4 红麻HcWRKY71基因在盐、干旱、重金属镉胁迫下的表达分析
红麻幼苗植株分别在NaCl和CdCl2胁迫下生长2周后, 剪取每个处理的红麻植株叶片, 进行浓度梯度的表达分析, 同时剪取红麻植株的根、茎、叶进行组织特异性表达分析。干旱胁迫条件下, 分别剪取不同处理时间的红麻植株叶片, 进行不同胁迫时间的表达分析; 红麻干旱处理8 h时, 剪取红麻植株的根、茎、叶, 进行组织特异性表达分析。每个处理取样后, 分别提取总RNA, 反转录合成cDNA, 利用荧光定量PCR检测基因表达量。荧光定量PCR引物为HcWRKY71-QF/QR (表1), 内参基因为β-actin, 内参引物为Actin-QF/QR (表1)。荧光定量PCR反应体系10 μL, 包括Power 2×SYBR PCR Premixture 5 μL、上下游引物各0.3 μL、cDNA 2 μL, ddH2O补足10 μL。荧光定量PCR反应程序为95℃预变性2 min, 95℃变性15 s, 61℃退火15 s, 72℃ 延伸20 s, 40个循环。每个反应设置3个生物学重复, 每个生物学重复设置3个技术重复。1.5 红麻HcWRKY71基因通过花序浸染法转化拟南芥
设计含有Bam H I和Sal I 2个限制性内切酶位点的表达引物PC1301-35S-HcWRKY71-GFP-F/R (表1), 以阳性克隆载体pMD19-T为模板进行PCR扩增, 并回收PCR扩增产物。利用限制性内切酶Bam H I, Sal I分别对表达载体pC1301-35S-GFP和PCR产物进行双酶切, 利用T4 DNA聚合酶将目的基因片段和载体连接。将连接好的重组产物转化到DH5α感受态细胞中, LB培养基培养1 h, 涂含有卡那霉素的平板, 37℃倒置培养18 h, 经菌落PCR测序验证获得重组表达质粒pC1301-35S-HcWRKY71- GFP。将重组质粒通过电击法转化到农杆菌EHA105中, 用含有卡那霉素(50 mg L-1)和利福平(50 mg L-1)双抗的LB固体培养基划线培养、筛选, 获得含有功能质粒的农杆菌。挑取单菌落接种到含有卡那霉素(50 mg L-1)和利福平(50 mg L-1)的LB液体培养基, 培养至OD600为1.2。离心收集菌液, 然后加入50 mL悬浮液(1/2 MS培养基, 50 g L-1蔗糖, 0.5 g L-1 2-吗啉乙磺酸, 用NaOH (1 mol L-1)溶液调pH 5.7, 加表面活性剂Silwet至终浓度为0.02%)悬浮菌液。将培养4周左右刚开花的拟南芥植株上的果荚去掉, 将整个花序浸入悬浮好的菌液10 s, 暗培养24 h, 正常培养1周后再浸染1次。果荚变黄后收获T0代转基因种子。1.6 转基因拟南芥纯合株系的筛选及幼苗耐盐性鉴定
用次氯酸钠消毒T0代转基因种子6~7 min, 然后用蒸馏水洗涤3~5次。用移液枪将消毒后的种子点种在含有40 mg L-1潮霉素的MS培养基上, 对转基因种子进行抗性筛选。将健壮绿色的转基因幼苗移栽到灭菌的营养土培养, 然后单株收获T1代种子。根据孟德尔遗传定律T1代转基因种子含有杂合种子需要进一步进行筛选, 筛选方法同上, 然后单株收获T2代种子。T2代种子继续筛选, 如果T2代单株收获的种子没有发生分离, 则该单株为纯合植株, 此时筛选单株收获的种子为T3代, 则该单株收获的T3代种子为纯合株系。将筛选获得纯合的转基因拟南芥种子和野生型拟南芥种子消毒后分别播种于MS培养基上, 待拟南芥幼苗长到四叶一心时移栽到装有灭菌营养土(营养土与蛭石按3︰1的比例混合)的塑料花盆。光照培养室培养5 d后用100 mmol L-1 NaCl溶液胁迫处理, 对转基因幼苗耐盐性进行鉴定。
1.7 转基因拟南芥目的基因PCR鉴定及荧光定量分析
将筛选获得纯合的转基因拟南芥种子和野生型拟南芥种子消毒后分别播种于MS培养基上, 待拟南芥幼苗长到四叶一心时移栽到装有灭菌营养土(营养土与蛭石按3︰1的比例混合)的塑料花盆。光照培养室培养7 d后用100 mmol L-1 NaCl溶液胁迫处理, 24 h后分别提取对照条件下野生型拟南芥植株和转基因拟南芥植株的总RNA以及盐胁迫下野生型拟南芥植株和转基因拟南芥植株的总RNA, 进行目的基因检测和荧光定量分析。用于目的基因PCR检测和荧光定量分析的引物HcWRKY71-PF/R (表1)。2 结果与分析
2.1 红麻HcWRKY71基因的克隆和测序
根据红麻转录组unigene序列(编号: CL3883.Contig4), 设计特异性引物, 进行PCR扩增, 获得长度为957 bp的cDNA序列, 扩增产物经1.2%琼脂糖凝胶电泳检测, 结果如图1所示。对PCR产物进行切胶回收, 然后连接到pMD19-T载体上, 送生工生物工程(上海)股份有限公司测序。图1
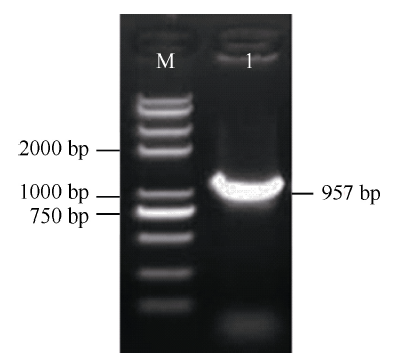 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1HcWRKY71 cDNA全长琼脂糖凝胶电泳
M: 2K Plus II; 1: PCR产物。
Fig. 1Agarose gel electrophoresis of cDNA full length of HcWRKY71
M: DNA marker 2K Plus II; 1: PCR product.
2.2 红麻HcWRKY71基因的生物信息学分析
PCR产物经sanger测序, 获得长度为957 bp cDNA序列, 经NCBI ORF Finder在线预测, HcWRKY71基因的开放读码框为957 bp, 编码1个含有318个氨基酸的蛋白(图2)。NCBI在线预测该蛋白的保守功能域发现, 该蛋白含有1个WRKY功能保守结构域(172~229位) (附图1), 属于第二类WRKY转录因子。利用在线工具预测该蛋白的理论分子量为35.5 kD, 等电点为7.24。蛋白质磷酸化位点预测结果(附图2)显示, 该蛋白质含有32个丝氨酸、7个酪氨酸、18个苏氨酸, 其中位于第189位、194位、212位的丝氨酸和位于第199位、200位、204位、218位和221位的苏氨酸位的磷酸化位点, 位于WRKY的保守功能域内, 推测这8个磷酸化位点可能对HcWRKY71转录因子活性的调控具有重要作用。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2HcWRKY71基因的核酸序列及其编码的氨基酸序列
红色框为WRKYGQK基序; 黑色框为C2H2基序(CX4CX23HXH)。
Fig. 2Nucleotide sequence and encoded amino acids sequence of HcWRKY71 gene
The sequence of WRKYGQK motif is highlighted in red box, that of C2H2 motif (CX4CX23HXH) in black box.
附图1
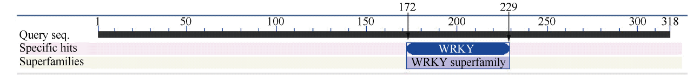 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1红麻HcWRKY71蛋白的保守功能结构域预测
Fig. S1Conserved domain prediction of HcWRKY71 protein in kenaf
附图2
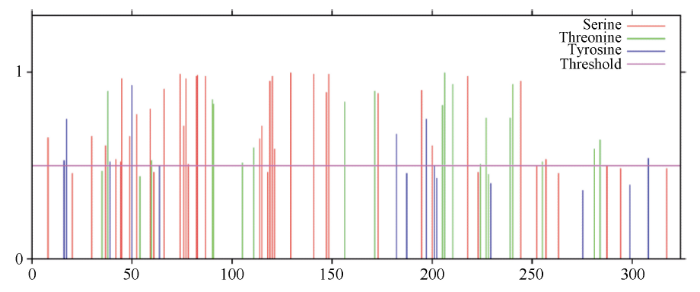 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图2HcWRKY71转录因子磷酸化位点的预测
Fig. S2Prediction of phosphorylation sites of transcription factor HcWRKY71
HcWRKY71转录因子氨基酸序列相似性通过NCBI BlastP比对发现, 红麻与雷蒙德氏棉(XP_012445471.1)、可可(XP_017978107.1)、榴莲(XP_022720946.1)、哥伦比亚锦葵(XP_021293950.1)、葡萄(XP_002272089.1)、枣(XP_015876832.1)、梨(XP_009353824.1)、苹果(XP_008383508.1)、橄榄(XP_022879638.1)的氨基酸序列相似性分别为84%、80%、79%、79%、60%、60%、57%、57%、53%。DNAMAN的分析结果(图3)表明, 上述物种氨基酸序列的一致性为67.92%。WRKY的功能结构域高度保守, 其他位置氨基酸序列, 由于物种的不同表现出不同的变化。WRKY71转录因子可能在不同的物种中具有相似的生物学功能。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3红麻HcWRKY71与其他植物WRKY71蛋白氨基酸序列一致性比对
不同颜色代表不同氨基酸残基的保守性。蓝色表示氨基酸完全保守; 粉红色、青色、黄色分别表示氨基酸的保守性为75%以上、50%以上及33%以上; 白色表示氨基酸的保守性不足33%。
Fig. 3Amino acid sequence alignment of HcWRKY71 and WRKY71 proteins from other plants
The different color indicates that the different levels of the conservation amino acid residues. The blue means amino acids are complete conservation, the pink, cyan, yellow means that the conservation of amino acids is no less than 75%, no less than 50%, no less than 33% and white means no more than 33%.
利用MEGA6.0软件分析HcWRKY71转录因子氨基酸序列与具有代表性的雷蒙德氏棉(XP_012445471.1)、可可(XP_017978107.1)、榴莲(XP_022720946.1)、哥伦比亚锦葵(XP_ 021293950.1)、葡萄(XP_002272089.1)、枣(XP_ 015876832.1)、梨(XP_009353824.1)、苹果(XP_ 008383508.1)、橄榄(XP_022879638.1)氨基酸序列的进化关系(图4)发现, 红麻与雷蒙德氏棉的关系最近, 这可能是两者同属于锦葵科的缘故。而红麻与橄榄的关系最远。
图4
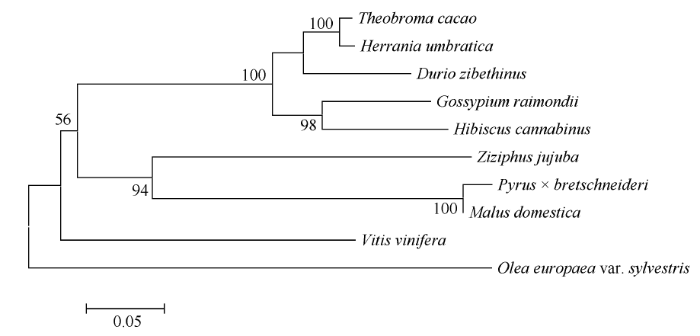 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4红麻HcWRKY71蛋白与其他植物WRKY71蛋白的系统进化树
Fig. 4Phylogenetic tree of HcWRKY71 and WRKY71 from other plants
2.3 盐胁迫下红麻HcWRKY71基因的表达分析
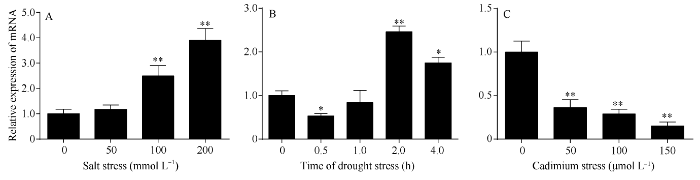
分别提取0、50、100、200 mmol L-1 NaCl胁迫下红麻叶片总RNA, 并反转录合成cDNA作为荧光定量PCR反应的模板, 进行定量分析。HcWRKY71基因表达量随NaCl浓度的增加而增加。当NaCl浓度为100 mmol L-1和200 mmol L-1时HcWRKY71基因表达量与对照表达量相比, 达到极显著差异水平(P < 0.01) (图5-A)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5HcWRKY71基因在盐、干旱和镉胁迫下的表达
A: 盐胁迫; B: 干旱胁迫; C: 镉胁迫。*和*分别表示在0.05和0.01水平上差异显著。误差线为每组处理的标准误差(n = 3)。
Fig. 5Expression of HcWRKY71 under salt, drought and cadmium stress
A: salt stress; B: drought stress; C: cadmium stress. *, ** mean significant difference at the 0.05 and 0.01 probability levels, respectively. Error bars represent the standard error of each treating group (n = 3).
分别提取对照和200 mmol L-1 NaCl胁迫下红麻根、茎、叶的总RNA, 反转录合成cDNA进行荧光定量PCR分析。HcWRKY71基因在根、茎、叶中均有表达, 其表达量没有显著差异。在NaCl胁迫下, 根、茎、叶中HcWRKY71基因表达量均有增加, 其中根部的表达量上升最多, 与对照表达量相比, 具有极显著差异(P < 0.01) (图6-A)。可见HcWRKY71基因的表达是受NaCl胁迫诱导。
图6
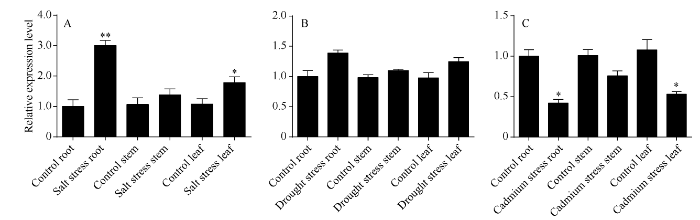 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6HcWRKY71基因在盐、干旱和镉胁迫下不同器官的表达
A: 盐胁迫; B: 干旱胁迫; C: 镉胁迫。*和*分别表示在0.05和0.01水平上差异显著。误差线为每组处理的标准误差(n = 3)。
Fig. 6HcWRKY71 expression in different organs under salt, drought and cadmium stress
A: salt stress; B: drought stress; C: cadmium stress. *, ** mean significant difference at the 0.05 and 0.01 probability levels, respectively. Error bars represent the standard error of each treating group (n = 3).
2.4 干旱胁迫下红麻HcWRKY71基因的表达分析
分别提取0、0.5、1.0、2.0、4.0 h干旱胁迫下红麻叶片总RNA, 并反转录合成cDNA作为荧光定量PCR反应的模板, 进行定量分析。HcWRKY71基因表达量随干旱胁迫时间的增加先下降再上升然后再下降。干旱胁迫0.5 h时, HcWRKY71基因的表达量下调, 其表达量与对照的表达量相比, 达到显著差异水平(P < 0.05); 干旱胁迫2 h时, HcWRKY71基因的表达量达到最高, 其表达量与对照的表达量相比, 达到极显著差异水平(P < 0.01) (图5-B)。分别提取对照和干旱胁迫8 h时红麻植株根、茎、叶的总RNA, 反转录合成cDNA进行荧光定量PCR分析。HcWRKY71基因在根、茎、叶中均有表达, 其表达量没有显著差异。在干旱胁迫条件下, 根、茎、叶中HcWRKY71基因表达量均有增加, 且根中表达量增加最多, 与对照相比差异不显著(图6-B)。这可能是干旱胁迫后期HcWRKY71基因表达量出现下降的原因。综上所述, HcWRKY71基因的表达受干旱胁迫诱导。
2.5 CdCl2胁迫下红麻HcWRKY71基因的表达分析
分别提取0、50、100、150 μmol L-1 CdCl2胁迫下红麻叶片总RNA, 并反转录合成cDNA作为荧光定量PCR反应的模板, 进行定量分析。HcWRKY71基因的表达量随着CdCl2浓度的增加而减少, 其表达量与对照的表达量相比, 达到极显著差异水平(P < 0.01) (图5-C)。分别提取对照和CdCl2 (100 μmol L-1)胁迫下红麻根、茎、叶的总RNA, 反转录合成cDNA进行荧光定量PCR分析。HcWRKY71基因在根、茎、叶中均有表达, 其表达量没有显著差异; 在CdCl2胁迫条件下, 根、茎、叶中HcWRKY71基因表达量均有下降, 与对照相比根和叶中的表达量下降达显著水平, 其中根部的表达量下降最多(图6-C)。由此可见, HcWRKY71基因的表达受CdCl2胁迫诱导。
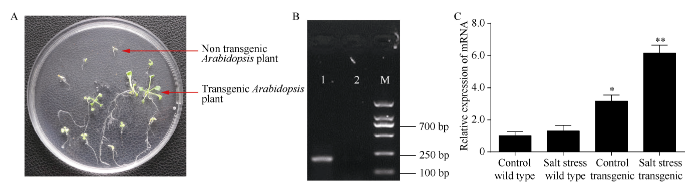
2.6 转基因拟南芥纯合株系的获得及幼苗耐盐性鉴定
通过农杆菌浸染花序法将红麻HcWRKY71基因转化拟南芥, 收获转基因种子。利用含有40 mg L-1潮霉素的MS培养基对收获的转基因拟南芥种子进行筛选, 结果如图7-A所示。绿色健壮的幼苗为转基因苗, 黄色弱小的幼苗为非转基因苗。转基因种子T0代、T1代、T2代经过潮霉素筛选最终获得纯合的T3代转基因株系。纯合转基因植株的获得为进一步研究HcWRKY71基因的功能奠定坚实的基础。野生型和转HcWRKY71基因拟南芥幼苗在100 mmol L-1 NaCl溶液胁迫下生长10 d后发现, 转HcWRKY71基因拟南芥幼苗的耐盐性显著高于野生型拟南芥幼苗的耐盐性。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7转基因拟南芥鉴定
A: 潮霉素鉴定; B: PCR鉴定; C: 实时定量PCR鉴定。*和*分别表示在0.05和0.01水平上差异显著。误差线为每组处理的标准误差(n = 3)。
Fig. 7Identification of transgenic Arabidopsis
A: identification of transgenic Arabidopsis by hygromycin; B: PCR identification of transgenic Arabidopsis plant; C: real time quantitative PCR identification. *, ** mean significant difference at the 0.05 and 0.01 probability levels, respectively. Error bars represent the standard error of each treating group (n = 3).
2.7 转基因拟南芥HcWRKY71基因的PCR鉴定和实时定量PCR分析
在转基因拟南芥植株的泳道能够观察到明显的条带, 而在野生型拟南芥植株的泳道没有看到明显的电泳条带(图7-B)。这表明HcWRKY71基因已经整合到转基因拟南芥植株的基因组内。HcWRKY71基因在对照野生型拟南芥植株和盐胁迫下拟南芥植株中的表达量没有显著差异。HcWRKY71基因在转基因拟南芥植株中的表达量显著高于其在野生型拟南芥植株中的表达量(P < 0.05), 在盐胁迫下转基因拟南芥植株中的表达量极显著高于其在野生型拟南芥植株中的表达量(P < 0.01) (图7-C)。由此可见, HcWRKY71基因在转基因拟南芥中同样受到盐胁迫的诱导, 在转基因拟南芥响应盐胁迫的过程中同样具有重要的作用。3 讨论
WRKY转录因子家族是植物体内最大的转录因子家族之一, 在多种植物中都发现了WRKY转录因子家族成员。本课题组通过对转录组序列CL3883.Contig4进行序列分析发现, CL3883.Contig4中间含有1个完整的阅读框, 即包含具有完整的起始密码子和终止密码子序列。根据完整的阅读框序列设计引物、扩增、测序获得HcWRKY71基因完整的cDNA序列。根据生物信息学分析发现, HcWRKY71基因的编码产物为红麻HcWRKY71转录因子。转录因子WRKY71在植物生长发育[14]、逆境胁迫[15]、防御[16]等方面具有重要的调控功能。目前有关转录因子WRKY71的调控功能主要集中在植物的生长发育和生物逆境胁迫响应方面。在拟南芥中, AtWRKY71通过调控RAX基因提高腋芽分生组织的活性, 从而增加拟南芥的分枝数; 通过直接激活开花基因FT和LFY加速拟南芥开花[17,18,19]。Qin等[20]研究表明, 在转基因拟南芥中, 通过小麦基因TaWRKY71的过表达调控拟南芥植株内IAA-MeIAA的激素水平来调控拟南芥下部叶片的发育。邹克琴等[21]研究表明, 水稻OsWRKY71基因的表达受水稻稻曲菌的侵染诱导, 表现出先升高后下降的趋势, 推测其可能在稻曲菌防御过程中发挥重要作用。
在植物响应非生物胁迫过程中WRKY71转录因子同样具有重要的调控作用。在拟南芥中, AtWRKY71基因表达受盐胁迫的诱导, 其过程快速而短暂。在转AtWRKY71基因的拟南芥超表达株系中, AtWRKY71通过抑制基因SUC8和SUC9的表达促进拟南芥开花[22]。Guo和Qin[18]的研究表明, AtWRKY71基因可能参与多种非生物胁迫的生理响应过程。在水稻中, OsWRKY71通过调控下游基因OsTGFR和WSI76的表达来提高转基因水稻植株的耐寒性[23]。本研究通过荧光定量PCR分析发现, 在NaCl溶液胁迫下, HcWRKY71基因的表达量随着NaCl溶液浓度的提高而增加, 这与耐盐转录组的测序结果[13]相一致; 在干旱胁迫下, HcWRKY71基因的表达量随着时间的延长呈现出先下降在上升然后在下降的规律; 在CdCl2溶液胁迫下, HcWRKY71基因的表达量随着CdCl2溶液浓度的增加而降低。在转基因拟南芥植株中, HcWRKY71基因的表达同样受到盐胁迫的诱导, 其表达量与野生型拟南芥的表达量相比显著上调。表明, HcWRKY71基因同时参与红麻对盐胁迫、干旱胁迫和重金属镉胁迫的响应过程, 且在不同的逆境胁迫下具有不同的表达方式, 推测该基因在红麻非生物逆境胁迫调控网络中具有关键枢纽作用。
在NaCl溶液、干旱和CdCl2溶液不同逆境胁迫下, HcWRKY71基因在根中的表达量变化最大(图6), 推测该基因在逆境胁迫响应过程中主要通过调控根部的基因表达来提高红麻自身的抗逆性。
4 结论
从红麻16号中成功克隆出1个HcWRKY71基因。该基因编码1个含有318个氨基酸的II类WRKY转录因子。HcWRKY71基因是一个组成型且受盐、干旱和重金属镉胁迫诱导表达的基因。通过农杆菌介导的花序浸染法和潮霉素筛选最终获得了转HcWRKY71基因拟南芥的纯合植株。苗期耐盐性鉴定发现, HcWRKY71基因提高了转基因拟南芥幼苗的耐盐性。这为进一步研究该基因耐盐的调控机制奠定了基础。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1111/pbi.13341URLPMID:31975524 [本文引用: 1]

Kenaf is an annual crop that is widely cultivated as a source of bast (phloem) fibres, the phytoremediation of heavy metal-contaminated farmlands and textile-relevant compounds. Leaf shape played a unique role in kenaf improvement, due to the inheritance as a single locus and the association with fibre development in typical lobed-leaf varieties. Here we report a high-quality genome assembly and annotation for var. 'Fuhong 952' with 1078 Mbp genome and 66 004 protein-coding genes integrating single-molecule real-time sequencing, a high-density genetic map and high-throughput chromosome conformation capture techniques. Gene mapping assists the identification of a homeobox transcription factor LATE MERISTEM IDENTITY 1 (HcLMI1) gene controlling lobed-leaf. Virus-induced gene silencing (VIGS) of HcLMI1 in a lobed-leaf variety was critical to induce round (entire)-like leaf formation. Candidate genes involved in cell wall formation were found in quantitative trait loci (QTL) for fibre yield and quality-related traits. Comparative genomic and transcriptome analyses revealed key genes involved in bast fibre formation, among which there are twice as many cellulose synthase A (CesA) genes due to a recent whole-genome duplication after divergence from Gossypium. Population genomic analysis showed two recent population bottlenecks in kenaf, suggesting domestication and improvement process have led to an increase in fibre biogenesis and yield. This chromosome-scale genome provides an important framework and toolkit for sequence-directed genetic improvement of fibre crops.
DOI:10.1093/jxb/err431URLPMID:22301384 [本文引用: 1]

MYB-type transcription factors play a diverse role in plant development and response to abiotic stress. This study isolated a rice R2R3-type MYB gene, OsMYB2, and functionally characterized its role in tolerance to abiotic stress by generating transgenic rice plants with overexpressing and RNA interference OsMYB2. Expression of OsMYB2 was up-regulated by salt, cold, and dehydration stress. OsMYB2 was localized in the nucleus with transactivation activity. No difference in growth and development between the OsMYB2-overexpressing and wild-type plants was observed under normal growth conditions, but the OsMYB2-overexpressing plants were more tolerant to salt, cold, and dehydration stresses and more sensitive to abscisic acid than wild-type plants. The OsMYB2-overexpressing plants accumulated greater amounts of soluble sugars and proline than wild-type plants under salt stress. Overexpression of OsMYB2 enhanced up-regulation of genes encoding proline synthase and transporters. The OsMYB2-overexpressing plants accumulated less amounts of H(2)O(2) and malondialdehyde. The enhanced activities of antioxidant enzymes, including peroxidase, superoxide dismutase, and catalase, may underlie the lower H(2)O(2) contents in OsMYB2-overexpressing plants. There was greater up-regulation of stress-related genes, including OsLEA3, OsRab16A, and OsDREB2A, in the OsMYB2-overexpressing plants. Microarray analysis showed that expression of numerous genes involving diverse functions in stress response was altered in the OsMYB2-overexpressing plants. These findings suggest that OsMYB2 encodes a stress-responsive MYB transcription factor that plays a regulatory role in tolerance of rice to salt, cold, and dehydration stress.
DOI:10.1007/s00299-015-1745-5URLPMID:25627252 [本文引用: 1]

KEY MESSAGE: A salicylic acid-inducible WRKY gene, PtrWRKY73, from Populus trichocarpa , was isolated and characterized. Overexpression of PtrWRKY73 in Arabidopsis thaliana increased resistance to biotrophic pathogens but reduced resistance against necrotrophic pathogens. WRKY transcription factors are commonly involved in plant defense responses. However, limited information is available about the roles of the WRKY genes in poplar defense. In this study, we isolated a salicylic acid (SA)-inducible WRKY gene, PtrWRKY73, from Populus trichocarpa, belonging to group I family and containing two WRKY domains, a D domain and an SP cluster. PtrWRKY73 was expressed predominantly in roots, old leaves, sprouts and stems, especially in phloem and its expression was induced in response to treatment with exogenous SA. PtrWRKY73 was localized to the nucleus of plant cells and exhibited transcriptional activation. Overexpression of PtrWRKY73 in Arabidopsis thaliana resulted in increased resistance to a virulent strain of the bacterial pathogen Pseudomonas syringae (PstDC3000), but more sensitivity to the necrotrophic fungal pathogen Botrytis cinerea. The SA-mediated defense-associated genes, such as PR1, PR2 and PAD4, were markedly up-regulated in transgenic plants overexpressing PtrWRKY73. Arabidopsis non-expressor of PR1 (NPR1) was not affected, whereas a defense-related gene PAL4 had reduced in PtrWRKY73 overexpressor plants. Together, these results indicated that PtrWRKY73 plays a positive role in plant resistance to biotrophic pathogens but a negative effect on resistance against necrotrophic pathogens.
URLPMID:26991441 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.3389/fpls.2017.00380URLPMID:28386267 [本文引用: 1]

WRKY transcription factor plays a key role in drought stress. However, the characteristics of the WRKY gene family in the common bean (Phaseolus vulgaris L.) are unknown. In this study, we identified 88 complete WRKY proteins from the draft genome sequence of the
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.plaphy.2016.08.013URLPMID:27592172 [本文引用: 1]

Plant WRKY transcription factors are trans-regulatory proteins that are involved in plant immune responses, development and senescence; however, their roles in abiotic stress are still not well understood, especially in the horticultural crop cucumber. In this study, a novel cucumber WRKY gene, CsWRKY46 was cloned and identified, which was up-regulated in response to cold stress and exogenous abscisic acid (ABA) treatment. CsWRKY46 is belonging to group II of the WRKY family, CsWRKY46 was found exclusively in the nucleus, as indicated by a transient expression assay. Yeast one-hybrid assay shown that CsWRKY46 interact with the W-box in the promoter of ABI5. Transgenic Arabidopsis lines over-expressing CsWRKY46, WRK46-OE1 and WRK46-OE5 had higher seedling survival rates upon freezing treatment compared with that of the wild-type. The above over-expression lines also showed much a higher proline accumulation, less electrolyte leakage and lower malondialdehyde (MDA) levels. Furthermore, the CsWRKY46 overexpression lines were hypersensitive to ABA during seed germination, but the seedlings were not. Quantitative RT-PCR analyses revealed that the expression levels of the ABA-responsive transcription factor ABI5 were higher in the WRKY46-OE lines than in wild-type and that the overexpression of CsWRKY46 increased the expression of stress-inducible genes, including RD29A and COR47. Taken together, our results demonstrated that CsWRKY46 from cucumber conferred cold tolerance to transgenic plants and positively regulated the cold signaling pathway in an ABA-dependent manner.
[本文引用: 1]
[本文引用: 1]
URLPMID:27999968 [本文引用: 2]
[本文引用: 1]
DOI:10.1371/journal.pone.0065120URLPMID:23762295 [本文引用: 1]

WRKY transcription factors are reported to be involved in defense regulation, stress response and plant growth and development. However, the precise role of WRKY transcription factors in abiotic stress tolerance is not completely understood, especially in crops. In this study, we identified and cloned 10 WRKY genes from genome of wheat (Triticum aestivum L.). TaWRKY10, a gene induced by multiple stresses, was selected for further investigation. TaWRKY10 was upregulated by treatment with polyethylene glycol, NaCl, cold and H2O2. Result of Southern blot indicates that the wheat genome contains three copies of TaWRKY10. The TaWRKY10 protein is localized in the nucleus and functions as a transcriptional activator. Overexpression of TaWRKY10 in tobacco (Nicotiana tabacum L.) resulted in enhanced drought and salt stress tolerance, mainly demonstrated by the transgenic plants exhibiting of increased germination rate, root length, survival rate, and relative water content under these stress conditions. Further investigation showed that transgenic plants also retained higher proline and soluble sugar contents, and lower reactive oxygen species and malonaldehyde contents. Moreover, overexpression of the TaWRKY10 regulated the expression of a series of stress related genes. Taken together, our results indicate that TaWRKY10 functions as a positive factor under drought and salt stresses by regulating the osmotic balance, ROS scavenging and transcription of stress related genes.
[本文引用: 1]
[本文引用: 1]
URLPMID:26578700 [本文引用: 1]
[本文引用: 2]
DOI:10.1111/tpj.13092URLPMID:26643131 [本文引用: 1]

Flowering is crucial for achieving reproductive success. A large number of well-delineated factors affecting flowering are involved in complex genetic networks in Arabidopsis thaliana. However, the underlying part played by the WRKY transcription factors in this process is not yet clear. Here, we report that WRKY71 is able to accelerate flowering in Arabidopsis. An activation-tagged mutant WRKY71-1D and a constitutive over-expresser of WRKY71 both flowered earlier than the wild type (WT). In contrast, both the RNA interference-based multiple WRKY knock-out mutant (w71w8 + 28RNAi) and the dominant repression line (W71-SRDX) flowered later. Gene expression analysis showed that the transcript abundance of the flowering time integrator gene FLOWERING LOCUS T (FT) and the floral meristem identity genes LEAFY (LFY), APETALA1 (AP1) and FRUITFULL (FUL) were greater in WRKY71-1D than in the WT, but lower in w71w8 + 28RNAi and W71-SRDX. Further, WRKY71 was shown to bind to the W-boxes in the FT and LFY promoters in vitro and in vivo. The suggestion is that WRKY71 activity hastens flowering via the direct activation of FT and LFY.
DOI:10.1371/journal.pone.0063033URLPMID:23671653 [本文引用: 1]

Leaf type is an important trait that closely associates with crop yield. WRKY transcription factors exert diverse regulatory effects in plants, but their roles in the determination of leaf type have not been reported so far. In this work, we isolated a WRKY transcription factor gene TaWRKY71-1 from a wheat introgression line SR3, which has larger leaves, superior growth capacity and higher yield than its parent common wheat JN177. TaWRKY71-1 specifically expressed in leaves, and produced more mRNA in SR3 than in JN177. TaWRKY71-1 localized in the nucleus and had no transcriptional activation activity. TaWRKY71-1 overexpression in Arabidopsis resulted in hyponastic rosette leaves, and the hyponastic strength was closely correlative with the transcription level of the transgene. The spongy mesophyll cells at abaxial side of leaves were drastically compacted by TaWRKY71-1 overexpression. In TaWRKY71-1 overexpression Arabidopsis, the expression of IAMT1 that encodes a methyltransferase converting free indole-3-acetic acid (IAA) to methyl-IAA ester (MeIAA) to alter auxin homeostatic level was induced, and the induction level was dependent on the abundance of TaWRKY71-1 transcripts. Besides, several TCP genes that had found to be restricted by IAMT1 had lower expression levels as well. Our results suggest that TaWRKY71-1 causes hyponastic leaves through altering auxin homeostatic level by promoting the conversion of IAA to MeIAA.
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/pcp/pcx201URLPMID:29272465 [本文引用: 1]

Soil salinity affects various aspects of plant growth and development including flowering. Usually, plants show a delayed flowering phenotype under high salinity conditions, whereas some plants will risk their life to continue to grow, thereby escaping serious salt stress to achieve reproductive success. However, the molecular mechanisms of the escape strategies are not clear yet. In this work, we report that the transcription factor WRKY71 helps escape salt stress in Arabidopsis. The expression of the WRKY71 wild-type (WT) allele was salinity inducible. Compared with Col-0, high salt stress caused only a marginal delay in the flowering time of the activation-tagged mutant WRKY71-1D. However, flowering in the RNA interference (RNAi)-based multiple WRKY knock-out mutant (w71w8 + 28RNAi) was dramatically later than in the WT under high salinity conditions. Meanwhile, expression of FLOWERING LOCUS T (FT) and LEAFY (LFY) was greater in WRKY71-1D than in the WT, and lower in w71w8 + 28RNAi under salinity-stressed conditions. The suggestion is that WRKY71 activity hastens flowering, thereby providing a means for the plant to complete its life cycle in the presence of salt stress.
[本文引用: 1]
