 ,**, 农保选
,**, 农保选 ,**, 夏秀忠, 张宗琼, 曾宇, 冯锐, 郭辉, 邓国富, 李丹婷
,**, 夏秀忠, 张宗琼, 曾宇, 冯锐, 郭辉, 邓国富, 李丹婷 ,*, 杨行海
,*, 杨行海 ,*广西农业科学院水稻研究所/广西水稻遗传育种重点实验室, 广西南宁 530007
,*广西农业科学院水稻研究所/广西水稻遗传育种重点实验室, 广西南宁 530007Genome-wide association study of blast resistance loci in the core germplasm of rice landraces from Guangxi
CHEN Can ,**, NONG Bao-Xuan
,**, NONG Bao-Xuan ,**, XIA Xiu-Zhong, ZHANG Zong-Qiong, ZENG Yu, FENG Rui, GUO Hui, DENG Guo-Fu, LI Dan-Ting
,**, XIA Xiu-Zhong, ZHANG Zong-Qiong, ZENG Yu, FENG Rui, GUO Hui, DENG Guo-Fu, LI Dan-Ting ,*, YANG Xing-Hai
,*, YANG Xing-Hai ,*Rice Research Institute, Guangxi Academy of Agricultural Sciences/Guangxi Key Laboratory of Rice Genetics and Breeding, Nanning 530007, Guangxi, China
,*Rice Research Institute, Guangxi Academy of Agricultural Sciences/Guangxi Key Laboratory of Rice Genetics and Breeding, Nanning 530007, Guangxi, China通讯作者:
收稿日期:2020-07-12接受日期:2020-12-1网络出版日期:2021-06-12
| 基金资助: |
First author contact:
Received:2020-07-12Accepted:2020-12-1Online:2021-06-12
| Fund supported: |
作者简介 About authors
陈灿, E-mail:
农保选, E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2361KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
陈灿, 农保选, 夏秀忠, 张宗琼, 曾宇, 冯锐, 郭辉, 邓国富, 李丹婷, 杨行海. 广西水稻地方品种核心种质稻瘟病抗性位点全基因组关联分析[J]. 作物学报, 2021, 47(6): 1114-1123. doi:10.3724/SP.J.1006.2021.02047
CHEN Can, NONG Bao-Xuan, XIA Xiu-Zhong, ZHANG Zong-Qiong, ZENG Yu, FENG Rui, GUO Hui, DENG Guo-Fu, LI Dan-Ting, YANG Xing-Hai.
稻瘟病是由稻瘟菌(Magnaporthe grisea)引起的一种水稻毁灭性真菌病害, 严重影响世界水稻种植区的产量与品质, 从而威胁全球粮食安全[1,2]。全球每年由稻瘟病引起的水稻产量损失约为总量的10%~30%, 它可以养活至少6000万人, 经济价值高达660亿美元[3]。化学防治作为一种主要、传统的病虫害防治手段, 长期使用既污染环境, 也增加经济成本。种质资源对抗性具有广泛遗传变异, 因此利用寄主植物自身抗性是防治该病最有效、最经济和最环保的方法[4]。但病原真菌对宿主的适应能力的频繁突变, 会使得品种抗性在3~5年内丧失[5]。因此, 需要不断挖掘鉴定新的稻瘟病基因, 这对于开展抗病选育有重要的理论与实践意义。
水稻抗性基因包括2种类型, 一种是提供种系特异性抗性的抗性基因(resistance gene, R), 另一种是控制部分非种系特异性抗性的微效基因, 又称数量性状位点(quantitative trait locus, QTL)。鉴定抗性基因(R)/数量性状位点(QTL)及开展抗病机制的研究对抗病品种的选育尤为重要。水稻稻瘟病抗性基因挖掘一直是水稻抗病育种的热点问题。迄今为止, 已经鉴定了110多个抗稻瘟病基因, 其中克隆了36个[6,7,8]。根据克隆基因的编码蛋白类型, 可以分为5类。(1) 核苷酸结合位点(nucleotide binding site, NBS)-富含亮氨酸重复序列(leucine rich repeat, LRR)-蛋白(NBS-LRR或NLR), 根据N端结构域可分为2个子类TIR (Toll/Interleukin-1receptor)-NBS- LRR和CC (coiled-coil)-NBS-LRR。Pi2、Pi9、Pi35、Pi37、Pi50、Pi56、Piz-t、Pita、Pib、Pid3、Pi-CO39、Pigm、Pi54、Pid3-A4、Pi54rh、Pik-m等属于前者, Pi1、Pi25、Pi36、Pi5、Pit、Pi-k、Pik-p、Pik-e、Pia、Pi63、Pi64、Pb1、Pish等属于后者; (2) 凝集素受体(lectin receptor), 如Pid2; (3) 富含脯氨酸结构域蛋白(proline-rich domain proteins), 如pi21; (4) 富含Armadillo重复序列蛋白, 如Ptr; (5) 富含四肽重复序列(tetratricopeptide repeats, TPRs)蛋白, 如bsr-k1。这些编码蛋白类型为稻瘟病候选基因的筛选提供了重要理论依据。
大多数植物基因/QTL是通过连锁作图鉴定的, 因此基因/QTL的检测受到使用的双亲材料的限制, 不能够解释自然界广泛存在的遗传变异。随着高密度遗传作图技术的发展, 基于连锁不平衡的全基因组关联分析(genome-wide association analysis, GWAS)已成为自然群体基因/QTL鉴定的重要工具。GWAS克服了利用来自双亲材料的群体进行连锁映射的缺点。通过与高通量测序、分群分析法(bulked segregant analysis, BSA)等技术手段相结合, GWAS在检测水稻稻瘟病性状关联位点及筛选候选基因方面得到了大量快速应用[9,10,11,12]。如最近Li等[13]利用湖南省3个分离菌株和234份水稻多样性小组1进行了水稻抗稻瘟病的全基因组关联研究(GWAS), 共鉴定出56个QTL。其中1个QTL定位于抗性基因Pik位点, 该位点对3个分离菌株均具有抗性。抗性品种基因组序列分析结果表明, 该位点是一个新的Pik等位基因, 将其命名为Pikx。Lu等[14]利用全基因组关联研究与RNA测序分析, 鉴定出127个水稻抗稻瘟病相关位点。此外, 在一个200 kb的基因组区域中预测了2341个非冗余候选基因, 其中45个基因与抗病相关。
本研究供试材料为419份广西地方品种核心种质, 利用简化基因组测序(specific-locus amplified fragment sequencing, SLAF-seq), 已经获得了高质量的单核苷酸多态性位点(single nucleotide polymorphism, SNP) 208,993个[15]。利用该阵列SNP已对蒸煮食味、糯性、种皮颜色和南方黑条矮缩病等性状进行了GWAS研究[15,16,17,18]。但利用GWAS检测该群体的稻瘟病抗性位点尚未报道。
为此, 我们拟使用基于419份广西地方品种核心种质的SNP数据, 并结合7个生理小种的苗期叶瘟抗性表型数据, 利用GWAS挖掘出该群体材料的稻瘟病关联位点, 并预测显著关联位点附近区域的候选基因, 为下一步候选基因验证及基因克隆提供理论依据。
1 材料与方法
1.1 供试材料及菌种
实验材料来源于广西农业科学院水稻研究所库存的地方稻种资源核心种质, 共计419份, 包括330份籼稻、78份粳稻及11份其他类型品种。丽江新团黑谷和Tetep分别为感病及抗病对照。ZA 9、ZA 13、ZB 1、ZB 9、ZB 13、ZC 3和ZC 13等7个小种均由广西农业科学院植物保护研究所提供。水稻材料及对照品种浸种催芽后播到塑料盘中, 每份材料播20粒, 待长至三至五叶龄时(大约2~3周)用配制好的孢子液人工喷雾接种。接种菌液浓度为1×105~2×105个 mL-1的孢子悬浮液[19], 然后在26℃及相对湿度95%左右的培养室培养24 h, 接种大约7 d后, 根据描述的病斑大小和面积比(diseased leaf area, DLA), 对其进行0到9分的评分[20]。每份材料鉴定3次。0~3级为抗病(resistance, 用R表示), 4~9级为感病(susceptibility, 用S表示), 取3次平均值为鉴定结果。利用SPSS 19统计分析软件进行描述性统计及相关作图分析。1.2 SLAF测序和SNP基因分型
在Illumina Hiseq 2500系统上进行SLAF测序。利用BLAT软件对clean reads进行聚类, 得到多态SLAF标签。再使用BWA软件, 将多态SLAF标签序列比对至日本晴参考基因组上(http://plants. ensembl.org/Oryza_sativa/Info/Index)。然后利用GATK and SAM工具包分析SNP calling。根据最小等位基因频率(minor allele frequency, MAF) > 0.05和完整性>0.5, 共获得208,993个SNP[15]。1.3 全基因组关联分析的方法
使用软件TASSEL V3对208,993个SNP基因型和苗期叶瘟表型数据进行GWAS关联分析[21], 混合线性模型MLM为(Q+K)模型, Q为群体结构, K为亲缘系数。采用MEGA5软件构建系统发育树。采用SPAGeDi软件进行两两亲属关系分析。采用ADMIXTURE软件分析种群结构[15]。P < 4.79×10-6 (1/208,993 = 4.79E-6)被认为具有显著相关性。根据R环境生成Manhattan和Q-Q plot。1.4 候选基因的选择
以日本晴为参考基因组, 基于水稻的连锁不平衡(linkage disequilibrium, LD)衰减情况, 参考杨行海等[16]研究结果, 选择峰值SNP上下游各150 kb区间为候选基因区域。所有植物中的抗性基因, 包括NLR、凝集素受体等被认为是候选基因[22,23]。2 结果与分析
2.1 苗期稻瘟病抗性评价
利用7个不同类型的稻瘟病生理小种对419份水稻材料进行苗期喷雾接种, 根据DLA评价其抗性, 并对抗性级别进行统计分析。DLA评估的抗性等级从“ZC13”的2.93到“ZA13”的5.72, 平均为4.49, 变异系数从“ZA13”的0.24到“ZC13”的0.48, 平均为0.34 (表1)。根据抗性级别均值, 可以发现ZA种群毒性最强, 其次是ZB种群, ZC种群较弱, 这与实际情况相一致, 这说明这3类小种对实验材料有很好的鉴别性。从图1可以看出, 在接种7个不同类型的稻瘟病生理小种下, 叶瘟抗性级别分布具有较好的拟合正态分布, 利于开展GWAS关联分析。Table 1
表1
表1不同稻瘟病小种接种下水稻苗期叶瘟抗性级别统计分析
Table 1
| 小种 Strain | 抗级 Range | 平均值±标准差 Mean±SE | 变异系数 CV (%) |
|---|---|---|---|
| ZA9 | 1-9 | 5.03±1.49 | 29.62 |
| ZA13 | 1-8 | 5.72±1.36 | 23.78 |
| ZB1 | 1-7 | 4.29±1.44 | 33.57 |
| ZB9 | 1-8 | 4.83±1.54 | 31.88 |
| ZB13 | 1-7 | 3.80±1.53 | 40.26 |
| ZC3 | 1-8 | 4.84±1.62 | 33.47 |
| ZC13 | 1-7 | 2.93±1.40 | 47.78 |
新窗口打开|下载CSV
图1
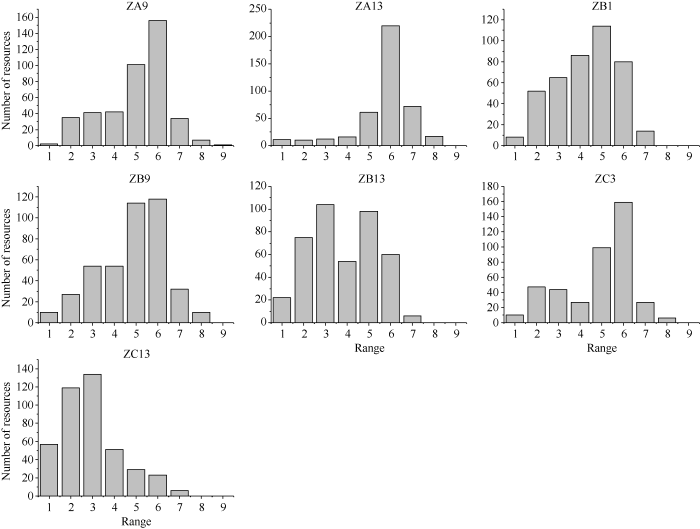 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1关联群体在7个不同稻瘟病生理小种接种下的叶瘟抗性级别分布
Fig. 1Distribution of resistance levels of leaf blast among the associated population inoculated with seven strains
2.2 全基因组关联分析
亲缘关系与群体结构分析表明, 系统发育树聚集为2个主要类群, 其与籼稻、粳稻亚群相一致。419份地方种质分为6个类型群体结构, 包括籼稻、粳稻亚类[15]。在P<4.79×10-6 (4.79E-6)水平下, 使用一般线性模型(GLM)共检测到20个稻瘟病相关SNP (图2-A~C和表2), 其中位点10,803,913在MLM下也被检测到, 3个位点为新位点, 分别为3号染色体上的18,302,718、18,302,744位点(Chr3_18302718, Chr3_18302744)及5号染色体上10,379,127位点(Chr5_10379127), 其他17个位点附近均有已知定位基因/QTL。在接种的7个小种中, 只有3个小种关联到显著性位点, 平均每个小种关联到6.7个位点。ZC3关联到最多的位点, 共16个, ZC13、ZB9各关联到2个位点。从关联位点在染色体上的分布来看, 12号染色体关联的位点最多, 为13个, 其次是1号和3号染色体, 分别为3个和2个, 5号和8号染色体最少, 均为1个。
图2
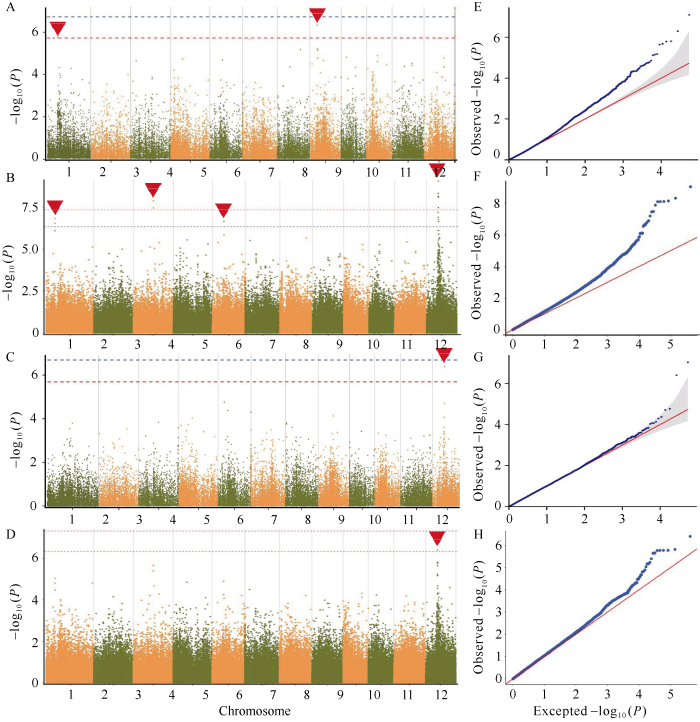 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图23个菌种下水稻稻瘟病抗性的全基因组关联研究
A、B、C分别表示GLM模型ZB9、ZC3、ZC13的曼哈顿图。D表示MLM模型ZC3的曼哈顿图。E、F、G分别表示GLM模型ZB9、ZC3、ZC13的QQ图。H表示MLM模型ZC3的QQ图。图中实心倒三角形为本文中显著性关联位点。
Fig. 2Genome-wide association study of rice blast resistance to three strains
A, B, and C represent Manhattan plot of GLM model ZB9, ZC3, and ZC13, respectively. D represents Manhattan plot ZC3 in MLM model. D, E, and F represent Quantitle-Quantitle plot of GLM model ZB9, ZC3, and ZC13, respectively. H represents Quantitle-Quantitle plot ZC3 in MLM model. The solid inverse triangle in the figure is the significant correlation point in this study.
Table 2
表2
表2水稻稻瘟病显著关联的SNP位点及已定位的基因/QTL
Table 2
| 小种 Strain | 染色体 Chr. | 位置 Position | P值 P-value | 上游位点 Upstream loci | 下游位点 Downstream loci | 模型 Model | 已知基因/QTL Known genes/QTLs |
|---|---|---|---|---|---|---|---|
| ZB9 | 1 | 10,443,043 | 1.63E-06 | 10,293,043 | 10,593,043 | GLM | Pi-h2(t) |
| ZC3 | 1 | 8,116,727 | 1.54E-07 | 7,966,727 | 8,266,727 | GLM | Pi-sj9 |
| ZC3 | 1 | 8,116,887 | 2.91E-07 | 7,966,887 | 8,266,887 | GLM | Pi-sj9 |
| ZC3 | 3 | 18,302,718 | 3.60E-08 | 18,152,718 | 18,452,718 | GLM | |
| ZC3 | 3 | 18,302,744 | 1.35E-08 | 18,152,744 | 18,452,744 | GLM | |
| ZC3 | 5 | 10,379,127 | 2.32E-07 | 10,229,127 | 10,529,127 | GLM | |
| ZB9 | 8 | 6,776,078 | 4.99E-07 | 6,626,078 | 6,926,078 | GLM | Pizh, Pi42 |
| 小种 Strain | 染色体 Chr. | 位置 Position | P值 P-value | 上游位点 Upstream loci | 下游位点 Downstream loci | 模型 Model | 已知基因/QTL Known genes/QTLs |
| ZC13 | 12 | 10,917,077 | 9.44E-08 | 10,767,077 | 11,067,077 | GLM | Pi12, Pi157, Pi19(t), Pi20, Pi31(t), Pi42(t), Pi6(t), Pita, Pita2, Pi67, Pi39(t), Pi58(t), Pi57, Ptr |
| ZC13 | 12 | 10,919,541 | 3.98E-07 | 10,769,541 | 11,069,541 | GLM | |
| ZC3 | 12 | 10,629,609 | 6.56E-08 | 10,479,609 | 107,79,609 | GLM | |
| ZC3 | 12 | 10,796,961 | 1.84E-07 | 10,646,961 | 10,946,961 | GLM | |
| ZC3 | 12 | 10,801,871 | 1.30E-07 | 10,651,871 | 10,951,871 | GLM | |
| ZC3 | 12 | 10,803,744 | 7.39E-09 | 10,653,744 | 10,953,744 | GLM | |
| ZC3 | 12 | 10,803,791 | 3.45E-08 | 10,653,791 | 10,953,791 | GLM | |
| ZC3 | 12 | 10,803,913 | 9.20E-10 | 10,653,913 | 10,953,913 | GLM | |
| ZC3 | 12 | 10,816,142 | 4.90E-09 | 10,666,142 | 10,966,142 | GLM | |
| ZC3 | 12 | 10,816,145 | 8.24E-09 | 10,666,145 | 10,966,145 | GLM | |
| ZC3 | 12 | 10,816,166 | 8.24E-09 | 10,666,166 | 10,966,166 | GLM | |
| ZC3 | 12 | 10,816,338 | 8.01E-09 | 10,666,338 | 10,966,338 | GLM | |
| ZC3 | 12 | 10,926,790 | 2.35E-07 | 10,776,790 | 11,076,790 | GLM | |
| ZC3 | 12 | 10,803,913 | 3.93E-07 | 10,653,913 | 10,953,913 | MLM |
新窗口打开|下载CSV
在P<4.79×10-6 (4.79E-6)水平下, 使用混合线性模型(MLM)仅检测到1个稻瘟病相关SNP (图2-D和表2), 该位点10,803,913位于12号染色体(Chr12_10803913)。
2.3 候选基因分析
根据水稻基因组注释(http://rice.plantbiology. msu.edu/)及LD衰退水平, 在20个关联位点300 kb的基因组区域中共鉴定出候选基因323个。根据已知克隆稻瘟病抗性基因的编码蛋白类型、候选基因功能注释信息及利用植物比较基因组学资源库(https://phytozome.Jgi.doe.gov/pz/portal.html)蛋白序列同源比对分析, 初步筛选到8个候选基因与稻瘟病相关, 其中LOC_Os12g18360 (Pita)、LOC_ Os12g18729 (Ptr)为已知克隆基因(表3)。本研究鉴定到的一些SNP位点与这2个克隆基因距离非常接近。如12号染色体上的位点10,629,609 (P = 6.56E-08)与Pita[24,25] (10,606,359~10,612,068)相距仅约17.5 kb; 该染色体上的另一个位点10,816,338 (P = 8.01E-09)与Ptr[26] (10,822,534~10,833,768)相距仅约6.2 kb。LOC_Os01g14550和LOC_Os01g14590功能注释为病理相关蛋白, LOC_Os12g18374编码含NB-ARC结构蛋白。说明这3个基因可能是抗病候选基因。Table 3
表3
表3候选基因信息
Table 3
| 基因名称 Gene name | 物理位置 Physical position | 基因注释 Gene annotation |
|---|---|---|
| LOC_Os01g14550 | 8,159,849-8,161,554 | Pathogen-related protein, putative, expressed |
| LOC_Os01g14590 | 8,175,375-8,177,889 | Pathogen-related protein, putative, expressed |
| LOC_Os03g32100 | 18,367,156-18,368,378 | Spotted leaf 11, putative, expressed |
| LOC_Os03g32180 | 18,410,063-18,411,482 | Polygalacturonase inhibitor 1 precursor, putative, expressed |
| LOC_Os05g18090 | 10,404,235-10,407,571 | SHR5-receptor-like kinase, putative, expressed |
| LOC_Os12g18360 | 10,606,359-10,612,068 | NB-ARC domain containing protein, expressed |
| LOC_Os12g18374 | 10,624,037-10,633,368 | NB-ARC domain containing protein, expressed |
| LOC_Os12g18729 | 10,822,534-10,833,768 | Expressed protein |
新窗口打开|下载CSV
通过蛋白序列同源比对, 在3个新的关联位点附近筛选到3个可能与稻瘟病相关的候选基因, 分别为LOC_Os03g32100、LOC_Os03g32180和LOC_Os05g18090。LOC_Os03g32100编码蛋白与玉米的2个基因GRMZM2G09049、GRMZM2G151649编码产物相似度为66.8%、69.0%, 这2个基因都与ARM重复超家族蛋白相关, 推测此基因可能属于含ARM重复序列蛋白抗病类型。LOC_Os03g32180编码蛋白与菠萝的1个基因Aco021559.1编码产物相似度为60.8%。该基因编码富亮氨酸重复受体蛋白激酶家族蛋白, 推测此基因可能属于受体蛋白激酶抗病类型。LOC_Os05g18090编码蛋白与玉米的1个基因GRMZM2G151567编码产物相似度为57.9%。该基因编码富亮氨酸重复跨膜蛋白激酶, 推测此基因可能属于受体蛋白激酶抗病类型。
3 讨论
GWAS已广泛应用于植物各种性状的基因位点挖掘。在稻瘟病抗性基因方面, 前人已经开展了大量的研究工作, 并关联到一些与已知基因/QTL相重叠的显著性关联位点[9,10,11,12,13,14]。这些基因/QTL主要是Pita、Ptr、Pi-6和Pi42等。本研究也在这些基因/QTL附近关联到一些显著性位点, 这说明这些基因/QTL可能广泛存在于自然群体材料中。在1号染色体上, 本研究共鉴定出3个显著关联位点, 这些位点与该染色体上的2个基因相重叠(图3-A), 分别是Pi-sj9[27] (7.86 Mb~4.97 Mb)和Pi-h2(t)[28] (8.41 Mb~18.83 Mb)。在8号染色体上, 本研究共鉴定出1个显著关联位点, 该位点与该染色体上的2个基因相重叠, 分别是Pi-zh[29] (4.37 Mb~21.01 Mb)和Pi42[30] (5.11 Mb~6.76 Mb)。在12号染色体上, 本研究共鉴定出13个显著关联位点, 这些位点与该染色体上的14个基因相重叠或位于其附近(图3-B)其分别为: Pita[24-25] (10.60 Mb~10.60 Mb)、Ptr[26] (10.82 Mb~10.83 Mb)、Pi31(t)[31] (7.73 Mb~11.91 Mb)、Pi12[32] (6.98 Mb~15.12 Mb)、Pi157[33] (8.82 Mb~18.05 Mb)、Pi19(t)[34] (10.73 Mb~12.04 Mb)、Pi20[35] (6.98 Mb~10.60 Mb)、Pi42(t)[36] (9.36 Mb~12.24 Mb)、Pi6(t)[37] (4.05 Mb~18.86 Mb)、Pita2[38] (10.07 Mb~13.21 Mb)、Pi67[39] (10.60 Mb~ 12.63 Mb)、Pi39(t)[40] (10.61 Mb~10.65 Mb)、Pi58(t)[41] (7.46 Mb~11.26 Mb)和Pi57[42] (10.79 Mb~10.85 Mb)。以上说明本研究群体可能含有上述基因。
图3
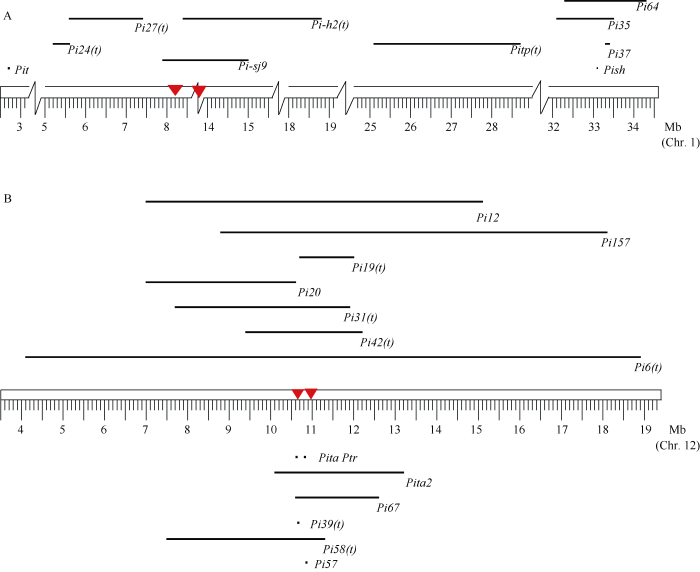 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3已定位的稻瘟病抗性基因与关联位点
倒三角形所指关联位点或关联位点所在区域。A: 1号染色体上已定位的稻瘟病基因; B: 12号染色体上与显著关联位点区域重叠的稻瘟病基因。
Fig. 3Identified blast resistance genes and their associated loci
An inverted triangle refers to the region of an association point or associated bit. A: the rice blast genes located on chromosome 1; B: the rice blast genes on chromosome 12 overlapped with the significantly associated locus region.
在3号染色体及5号染色体上, 本研究分别鉴定出2个(Chr3_18302718、Chr3_18302744)和1个(Chr5_ 10379127)显著关联位点, 这些位点都与染色体上已经定位的基因(pi66(t)[43] (22.92 Mb~27.89 Mb)和Pi68[44] (9.33 Mb~9.78 Mb)在3号染色体上; Pi10[45] (14.52 Mb~18.85 Mb)、Pi23[46] (10.75 Mb~19.17 Mb)和Pi26(t)[31] (2.06 Mb~2.76 Mb)在5号染色体上)不重叠。据此推测, 这些位点附近可能含有新的抗稻瘟病基因。
进一步候选基因分析表明, 初步筛选到了8个与稻瘟病相关的候选基因, 其中LOC_Os12g18360(Pita)、LOC_Os12g18729 (Ptr)为已知克隆基因。LOC_Os12g18374也被其他研究者认为是稻瘟病相关的候选基因[47], 这与本研究结果相一致。LOC_Os03g32100、LOC_Os03g32180和LOC_ Os05g18090为新关联位点附近筛选到的候选基因, 其中LOC_Os03g32100在接种稻瘟菌后基因表达量显著上调[48]。其他候选基因如LOC_Os03g32230和LOC_Os03g32220在接种稻瘟菌后基因表达量也显著上调[48,49], 其中LOC_Os03g32230 (bsr-d1/ZOS3- 12)是一个非小种特异性的C2H2转录因子, 协调减弱过氧化氢的降解, 表现出对稻瘟病的广谱抗性[50]。本研究发现, 虽然LOC_Os03g32230在关联位点Chr3_18302718和Chr3_18302744的区间内, 但是该位点仅在接种ZC3小种后鉴定到, 即该位点上的候选基因不具有广谱抗性, 故不含有bsr-d1/ZOS3-12。以上候选基因是否在本研究中参与稻瘟病抗性过程, 还需要作进一步的实时定量PCR验证。
4 结论
基于GLM和MLM模型的GWAS结果, 共检测到20个与稻瘟病抗性显著相关的SNP位点, 其中17个位点与前人定位的基因/QTLs重叠, 其余3个是新位点。在关联位点区域初步确定8个候选基因与抗病相关, 包括2个克隆的基因LOC_ Os12g18360(Pita)、LOC_Os12g18729 (Ptr)及3个新位点附近筛选到的基因LOC_Os03g32100、LOC_ Os03g32180和LOC_Os05g18090。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s13205-019-1738-0URLPMID:31093479 [本文引用: 1]

Blast disease caused by fungal pathogen Pyricularia oryzae is a major threat to rice productivity worldwide. The rice-blast pathogen can infect both leaves and panicle neck nodes. Nearly, 118 genes for resistance to leaf blast have been identified and 25 of these have been molecularly characterized. A great majority of these genes encode nucleotide-binding site-leucine-rich repeat (NBS-LRR) proteins and are organized into clusters as allelic or tightly linked genes. Compared to ever expanding list of leaf-blast-resistance genes, a few major genes mediating protection to neck blast have been identified. A great majority of the genetic studies conducted with the genotypes differing in the degree of susceptibility/resistance to neck blast have suggested quantitative inheritance for the trait. Several reports on co-localization of gene/QTLs for leaf- and neck-blast resistance in rice genome have suggested the existence of common genes for resistance to both phases of the disease albeit inconsistencies in the genomic positions leaf- and neck-blast-resistance genes in some instances have presented the contrasting scenario. There is a strong evidence to suggest that developmentally regulated expression of many blast-resistance genes is a key determinant deciding their effectiveness against leaf or neck blast. Testing of currently characterized leaf-blast-resistance genes for their reaction to neck blast is required to expand the existing repertoire resistance genes against neck blast. Current developments in the understanding of molecular basis of host-pathogen interactions in rice-blast pathosystem offer novel possibilities for achieving durable resistance to blast through exploitation of natural or genetically engineered loss-of-function alleles of host susceptibility genes.
[本文引用: 2]
DOI:10.1186/s12870-014-0311-6URLPMID:25403621 [本文引用: 2]

BACKGROUND: Rice blast disease is one of the most serious and recurrent problems in rice-growing regions worldwide. Most resistance genes were identified by linkage mapping using genetic populations. We extensively examined 16 rice blast strains and a further genome-wide association study based on genotyping 0.8 million single nucleotide polymorphism variants across 366 diverse indica accessions. RESULTS: Totally, thirty associated loci were identified. The strongest signal (Chr11_6526998, P =1.17 x 10-17) was located within the gene Os11g0225100, one of the rice Pia-blast resistance gene. Another association signal (Chr11_30606558) was detected around the QTL Pif. Our study identified the gene Os11g0704100, a disease resistance protein containing nucleotide binding site-leucine rich repeat domain, as the main candidate gene of Pif. In order to explore the potential mechanism underlying the blast resistance, we further examined a locus in chromosome 12, which was associated with CH149 (P =7.53 x 10-15). The genes, Os12g0424700 and Os12g0427000, both described as kinase-like domain containing protein, were presumed to be required for the full function of this locus. Furthermore, we found some association on chromosome 3, in which it has not been reported any loci associated with rice blast resistance. In addition, we identified novel functional candidate genes, which might participate in the resistance regulation. CONCLUSIONS: This work provides the basis of further study of the potential function of these candidate genes. A subset of true associations would be weakly associated with outcome in any given GWAS; therefore, large-scale replication is necessary to confirm our results. Future research will focus on validating the effects of these candidate genes and their functional variants using genetic transformation and transferred DNA insertion mutant screens, to verify that these genes engender resistance to blast disease in rice.
[本文引用: 2]
[本文引用: 2]
URLPMID:31309315 [本文引用: 2]
[本文引用: 2]
URLPMID:29746484 [本文引用: 5]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:24906128 [本文引用: 1]
[本文引用: 1]
URLPMID:11090206 [本文引用: 2]
URLPMID:11090207 [本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:16791691 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:29016662 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
URLPMID:28666113 [本文引用: 1]
