 ,, 杨大为, 唐慧娟, 潘根, 李德芳, 赵立宁, 黄思齐
,, 杨大为, 唐慧娟, 潘根, 李德芳, 赵立宁, 黄思齐 ,*中国农业科学院麻类研究所/农业农村部麻类生物学与加工重点实验室, 湖南长沙 410205
,*中国农业科学院麻类研究所/农业农村部麻类生物学与加工重点实验室, 湖南长沙 410205Genome-wide identification of GRAS transcription factor and expression analysis in response to cadmium stresses in hemp (Cannabis sativa L.)
YIN Ming ,, YANG Da-Wei, TANG Hui-Juan, PAN Gen, LI De-Fang, ZHAO Li-Ning, HUANG Si-Qi
,, YANG Da-Wei, TANG Hui-Juan, PAN Gen, LI De-Fang, ZHAO Li-Ning, HUANG Si-Qi ,*Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences/Key Laboratory of Biological and Processing for Bast Fiber Crops, Ministry of Agriculture and Rural Affairs, Changsha 410205, Hunan, China
,*Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences/Key Laboratory of Biological and Processing for Bast Fiber Crops, Ministry of Agriculture and Rural Affairs, Changsha 410205, Hunan, China通讯作者:
收稿日期:2020-03-27接受日期:2020-11-13网络出版日期:2021-06-12
| 基金资助: |
Received:2020-03-27Accepted:2020-11-13Online:2021-06-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (5747KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
尹明, 杨大为, 唐慧娟, 潘根, 李德芳, 赵立宁, 黄思齐. 大麻GRAS转录因子家族的全基因组鉴定及镉胁迫下表达分析[J]. 作物学报, 2021, 47(6): 1054-1069. doi:10.3724/SP.J.1006.2021.04078
YIN Ming, YANG Da-Wei, TANG Hui-Juan, PAN Gen, LI De-Fang, ZHAO Li-Ning, HUANG Si-Qi.
转录因子是一类能与基因序列上游特定序列结合的蛋白质, 通过改变基因表达来调节生物学过程, 在植物生长发育及对外界胁迫的响应中发挥重要作用[1,2]。GRAS转录因子由GAI、RGA和SCR 3个早期鉴定的功能基因而命名, 被认为在植物的生长与抗逆境胁迫中具有重要作用, 早期被认为只存在于植物中, 近期发现细菌中也含有该基因家族, 并有研究提出GRAS基因家族最早出现在细菌中的折叠甲基转移酶超家族中[3,4]。GRAS蛋白质一般至少由350个氨基酸组成[5], 包括1个高度保守的C端和1个可变的N端, 其C端由SAW、LRI、LRII、PFYRE和VHIID 5个保守基序组成, 其中VHIID最为保守, 是GRAS蛋白的核心结构, 存在于所有亚家族成员中[6,7,8]。有研究发现, N端是GRAS蛋白发挥特异性功能的重要部位, 因为N末端含有IDRs序列, 该序列表达时会与不同蛋白质结合, 而后能够表现出蛋白质的特异性[9,10]。早期的拟南芥GRAS基因分为LS、PAT1、SCL3、DELLA、LISCL、SCR、HAM和SHR 8个亚家族[11], 随后有研究在8个亚家族的基础上新增了DLT和SCL4/7亚家族, 将GRAS基因分为10个亚家族[12], 不同的GRAS亚家族导致了该基因家族功能的多样性。有研究发现, GRAS基因在植物根的形态建成、茎段分生组织的分化、激素信号转导、逆境胁迫响应等过程中发挥重要作用[13,14]。比如SCR和SHR亚家族参与拟南芥侧根的发育, SHR/SCR/SCL3在番茄丛枝菌根化过程中可能参与调控赤霉素[15,16]; PAT1分支中的PAT1和SCL21参与了光敏色素A (phytochrome, phyA)的信号转导, 使下胚轴在脱黄化时逐渐拉长[17]; OsDLT和OsGRAS19在水稻的油菜素类固醇(brassinosteroids, BR)信号转导中作为正向调节因子发挥作用[18]。
目前多种植物的GRAS基因家族都已进行了全基因的鉴定与分析, 在拟南芥[19]、水稻[20]、木薯[21]、棉花[22]和小麦[5]中分别存在34、60、77、150和153个GRAS基因。在各个植物都开始深入研究和挖掘该基因家族时, 众多麻类作物中尤其是大麻都还未见相关报道。大麻(Cannabis sativa L.)又称汉麻, 用于纺织、医药、护肤保健等领域, 其中四氢大麻酚(tetrahydrocannabinol, THC)含量低于0.3%的称为工业大麻, 近年来由于发现大麻对重金属具有较强的耐性, 且具有较高的经济价值, 大麻开始被用于修复重金属污染土壤[23], 有研究发现重金属可以降低大麻中的THC含量, 并提升其大麻二酚(cannabidiol, CBD)含量[24]。随着国际上对工业大麻的解禁及CBD医疗市场的扩大, 培育耐重金属及其他非生物胁迫的工业大麻品种显得十分重要。而GRAS基因与植物生长及耐非生物胁迫有着密切的关系, 已有研究发现, 过表达玉米ZmGRAS31基因, 可以改善玉米的耐盐性[25]; 过表达山葡萄VaPAT1基因可以显著提高了拟南芥的耐旱性、耐寒性和耐盐性[26], 过表达毛竹PeSCL3基因可增强其耐旱性[27], 过表达小麦AtSCL14可增强其耐光氧化胁迫的能力[5], 虽然这些研究已证明GRAS家族与植物的非生物胁迫有关, 但在GRAS家族与重金属镉胁迫之间还未有相关研究。
本研究旨在通过生物信息学方法在全基因组水平上对大麻GRAS基因的组成进行鉴定, 分析其理化性质、进化关系、基因结构、共线性关系, 同时利用2个大麻的镉胁迫下转录组数据及其qPCR验证分析该基因家族表达情况, 以期为后续研究大麻GRAS基因功能提供参考。
1 材料与方法
1.1 大麻GRAS基因的鉴定与理化性质分析
从NCBI数据库(https://www.ncbi.nlm.nih.gov/)中下载大麻基因组[28]文件与基因组注释文件(assembly number: GCA_900626175.2), 从TAIR数据库(https://www.arabidopsis.org/)中下载拟南芥GRAS基因序列。利用软件TBtools将拟南芥GRAS基因与大麻基因组进行blast序列比对(E-vaule<1e-5), 筛选大麻中的GRAS家族候选基因。将候选基因提交至uniprot数据库(https://www.uniprot.org/)进行批量比对, 验证候选基因是否含有GRAS保守结构域, 删除假阳性基因。利用在线分析软件ExPASy Proteomics (http://web.expasy.org/compute_pi)分析理化性质。1.2 大麻GRAS基因系统进化树的构建和基因结构分析
将拟南芥与大麻的GRAS 蛋白序列导入软件MEGA 7.0进行比对, 将比对后的结果采用相邻连接法(Neighbor-Joining, NJ; 执行参数: Bootstrap method 1000)构建系统进化树, 并利用软件FigTree完善美化进化树。利用在线软件NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd/)进行蛋白结构预测, 检测大麻GRAS的蛋白结构域(E-vaule<0.01)。利用在线软件MEME (http://meme-suite.org/)分析大麻GRAS基因保守基序(Motif), 设定预测数为10。利用软件TBtools从大麻基因组注释文件中提取大麻GRAS的相关编码序列(Coding Sequence: CDS)、非翻译区(Untranslated Region: UTR)。利用TBtools将进化树、蛋白结构域、基因保守基序、CDS、UTR结合, 构建GRAS进化关系与结构的整体比对图。1.3 大麻GRAS基因的染色体分布与共线性预测
利用软件TBtools从大麻基因组文件与基因注释文件中提取GRAS基因位置信息, 并构建GRAS基因在染色体上的物理图谱。利用软件TBtools、MCScanX和Circos计算并绘制大麻GRAS在染色体上存在的串联重复, 大麻不同染色体之间存在的共线性基因, 大麻与大豆(Glycine max)、雷蒙德氏棉(Gossypium raimondii)、玉米(Zea mays L.)、水稻(Oryza sativa L.)不同物种间存在的共线性基因。1.4 材料培养及处理与转录组测序
本试验选取2个大麻品种籽粒大小较一致的种子, 置于黑暗条件下25℃萌发3 d, 将幼苗移至1/4 Hoagland营养液中生长5 d, 昼夜温度24℃/16℃, 光周期16 h/8 h (明/暗), 相对湿度60%, 光强度700 μmol m-2 s-1。对生长5 d的幼苗进行处理: 对照(仅营养液) 12 h, 100 μmol L-1的CdCl2处理12 h。在2个处理下取长势一致的植株, 用TRIzol试剂(美国Invitrogen)提取总RNA, 并溶于无RNase的水中(TransGen Biotech, 中国), 用NanDrop 2000 (Thermo Scientific, 美国)检测RNA的质量、纯度和完整性。使用Illumina Truseq RNA样品制备试剂盒构建cDNA文库, 设3个生物学重复, 之后进行转录组测序。通过Illumina测序平台(Illumina HiSeq 4000)进行测序, 通过CASAVA Base Calling分析将原始测序数据转换为Sequenced Reads或Raw Reads, 并再次过滤以获取高质量的纯净读数。利用TopHat2与HISAT2两个软件将质控后的数据与Cannabis sativa (版本号GCF_900626175.2)参考基因组[28]进行对比, 使用String Tie或Cufflinks软件对Mapped Reads进行拼接。将转录组原始数据匹配至全基因组分析得到的工业大麻GRAS基因并统计reads数量, 并进一步换算成FPKM值进行比对分析。1.5 实时荧光定量 PCR 验证
本试验随机选取转录组中12个GRAS家族基因进行实时荧光定量PCR验证, 利用Primer 3 plus设计荧光定量PCR引物, 内参基因选取Hemp actin[29], 其上下游引物序列为Sense: 5°-CCAATAGCCTTG CATTCCAT-3°; Anti-sense: 5°-TCGATTGGAAAGC CGAATAC-3°。试验仪器使用Bio-Rad (伯乐) CFX Connect荧光定量PCR仪, 所取样品与RNA-Seq所选样品一致, 使用TIANGEN试剂盒提取RNA, 用TaKaRa反转录试剂盒合成cDNA, 利用Aidlab试剂盒进行实时荧光定量PCR验证。反应条件为95℃预变性2 min; 95℃变性15 s, 60℃退火30 s, 40个循环。采用2-ΔΔCt法计算目的基因的相对表达量。2 结果与分析
2.1 GRAS基因成员的鉴定及其理化性质分析
利用拟南芥GRAS基因与大麻基因组进行序列比对, 将获得的基因在Swissport数据库中进行结构域分析, 筛选出具有GRAS保守结构域的基因, 最终鉴定出54个大麻GRAS基因, 根据大麻基因CDS片段, 将54个GRAS基因命名为CsGRAS01~CsGRAS54 (表1)。大麻GRAS蛋白长度为415~757 aa, 平均长度586 aa; 分子质量为46,405.05~85,748.52 kD, 平均分子质量66,199.41 kD; 等电点为4.77~8.54, 其中等电点小于7的GRAS蛋白有52个, 偏酸性; 其余的2个GRAS蛋白等电点大于7, 偏碱性。Table 1
表1
表1GRAS基因及相关信息
Table 1
| 基因名称 Gene name | NCBI基因登录号 NCBI gene accession | 蛋白长度 Length (aa) | 等电点 pI | 分子量 Molecular weight (kD) |
|---|---|---|---|---|
| CsGRAS1 | LOC115696991 | 458 | 4.77 | 52,006.96 |
| CsGRAS2 | LOC115698980 | 494 | 5.15 | 54,901.55 |
| CsGRAS3 | LOC115698988 | 724 | 5.99 | 81,047.24 |
| CsGRAS4 | LOC115700662 | 455 | 5.66 | 49,596.06 |
| CsGRAS5 | LOC115701004 | 454 | 6.68 | 50,178.21 |
| CsGRAS6 | LOC115702418 | 528 | 5.24 | 57,541.56 |
| CsGRAS7 | LOC115703632 | 559 | 6.21 | 62,215.78 |
| CsGRAS8 | LOC115705736 | 722 | 5.77 | 78,760.91 |
| CsGRAS9 | LOC115705815 | 722 | 5.72 | 78,817.91 |
| CsGRAS10 | LOC115705915 | 721 | 5.74 | 78,730.89 |
| CsGRAS11 | LOC115706185 | 539 | 5.51 | 59,352.41 |
| CsGRAS12 | LOC115707313 | 544 | 6.33 | 61,258.62 |
| CsGRAS13 | LOC115710202 | 582 | 4.92 | 64,716.73 |
| CsGRAS14 | LOC115710247 | 582 | 4.92 | 64,716.73 |
| CsGRAS15 | LOC115710247 | 582 | 4.92 | 64,716.73 |
| CsGRAS16 | LOC115710247 | 582 | 4.92 | 64,716.73 |
| CsGRAS17 | LOC115710247 | 582 | 4.92 | 64,716.73 |
| CsGRAS18 | LOC115710247 | 582 | 4.92 | 64,716.73 |
| CsGRAS19 | LOC115710247 | 582 | 4.92 | 64,716.73 |
| CsGRAS20 | LOC115710262 | 514 | 4.93 | 57,833.13 |
| CsGRAS21 | LOC115711337 | 467 | 5.82 | 52,554.66 |
| CsGRAS22 | LOC115711473 | 549 | 6.13 | 61,322.10 |
| CsGRAS23 | LOC115712444 | 497 | 5.28 | 56,735.16 |
| CsGRAS24 | LOC115712525 | 504 | 5.68 | 56,355.13 |
| CsGRAS25 | LOC115712712 | 613 | 5.08 | 67,527.83 |
| CsGRAS26 | LOC115712875 | 508 | 6.30 | 56,609.53 |
| CsGRAS27 | LOC115714404 | 622 | 5.33 | 68,325.01 |
| CsGRAS28 | LOC115714408 | 600 | 5.95 | 66,439.36 |
| CsGRAS29 | LOC115715793 | 628 | 5.08 | 71,316.90 |
| CsGRAS30 | LOC115716239 | 617 | 6.03 | 68,009.17 |
| CsGRAS31 | LOC115716331 | 700 | 5.99 | 76,887.41 |
| CsGRAS32 | LOC115716409 | 616 | 6.19 | 67,882.02 |
| CsGRAS33 | LOC115716431 | 752 | 6.29 | 82,128.85 |
| CsGRAS34 | LOC115717012 | 697 | 5.36 | 77,185.61 |
| CsGRAS35 | LOC115718811 | 645 | 6.73 | 73,634.61 |
| CsGRAS36 | LOC115718865 | 746 | 5.80 | 85,106.83 |
| CsGRAS37 | LOC115718865 | 709 | 5.66 | 80,996.82 |
| CsGRAS38 | LOC115718866 | 630 | 7.24 | 72,454.10 |
| CsGRAS39 | LOC115718867 | 621 | 5.69 | 71,491.35 |
| CsGRAS40 | LOC115718867 | 551 | 6.37 | 63,913.20 |
| CsGRAS41 | LOC115718867 | 506 | 8.54 | 58,824.80 |
| CsGRAS42 | LOC115719316 | 749 | 5.03 | 84,965.93 |
| CsGRAS43 | LOC115719656 | 757 | 5.10 | 85,748.52 |
| CsGRAS44 | LOC115719656 | 636 | 5.20 | 73,211.22 |
| 基因名称 Gene name | NCBI基因登录号 NCBI gene accession | 蛋白长度 Length (aa) | 等电点 pI | 分子量 Molecular weight (kD) |
| CsGRAS45 | LOC115719730 | 698 | 5.68 | 78,688.43 |
| CsGRAS46 | LOC115722004 | 577 | 4.94 | 66,178.24 |
| CsGRAS47 | LOC115723037 | 575 | 5.14 | 64,688.92 |
| CsGRAS48 | LOC115723133 | 436 | 5.58 | 49,164.54 |
| CsGRAS49 | LOC115723500 | 454 | 6.00 | 51,420.59 |
| CsGRAS50 | LOC115724710 | 577 | 6.05 | 64,503.06 |
| CsGRAS51 | LOC115724919 | 548 | 5.14 | 61,008.73 |
| CsGRAS52 | LOC115724919 | 415 | 5.20 | 46,405.05 |
| CsGRAS53 | LOC115725148 | 463 | 5.64 | 52,596.41 |
| CsGRAS54 | LOC115725491 | 756 | 6.16 | 85,229.91 |
新窗口打开|下载CSV
2.2 GRAS基因的系统进化树构建和基因结构分析
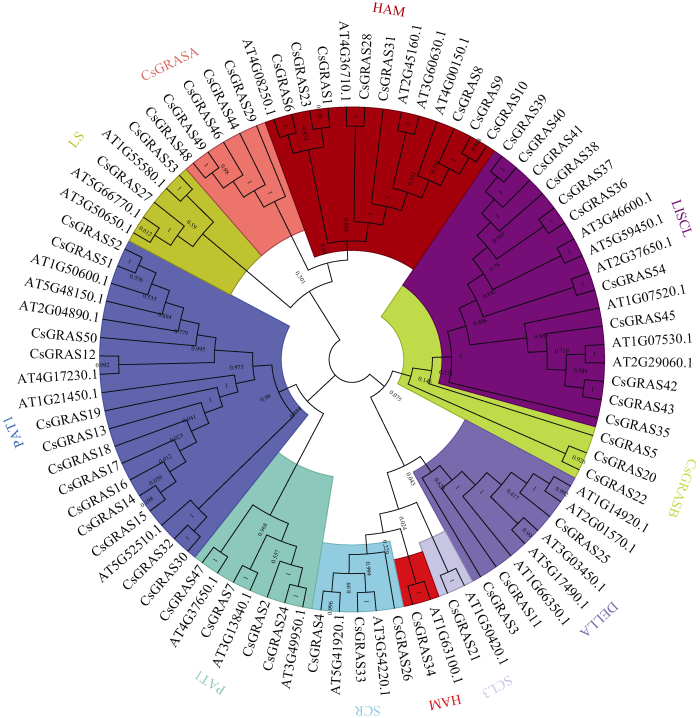
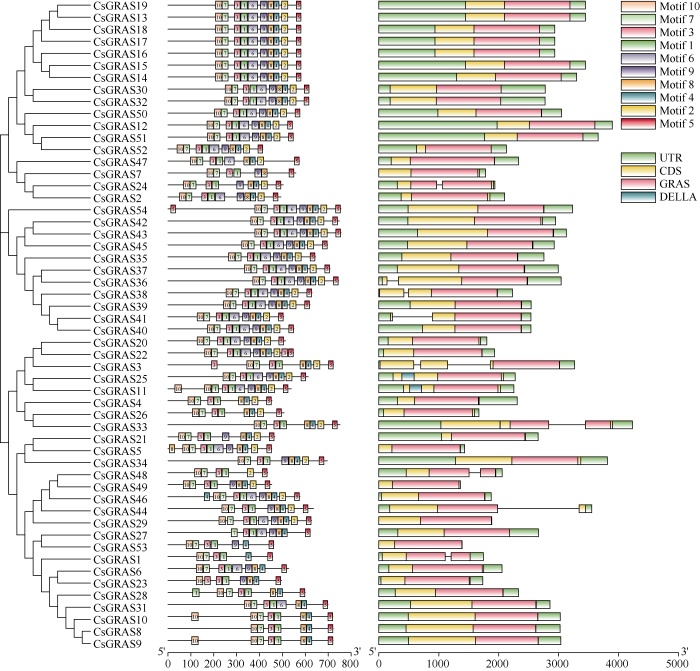
为深入理解大麻GRAS不同基因间的进化关系, 通过构建大麻与拟南芥中GRAS基因进化树, 将大麻中54个GRAS基因分成LS、LISCL、SCR、HAM、SCL3、DELLA、PAT1、SHR、CsGRASA、CsGRASB共10个亚家族(图1)。其中PAT1亚家族成员最多, 含13个基因; SCL3亚家族成员最少, 含1个基因。大麻中新鉴定出2个亚家族, CsGRASA含有5个成员, CsGRASB含有3个成员, 2个亚家族都不含任何拟南芥GRAS成员, 单独形成一个分支, 而且亲缘关系较远。从图2可以看出, 相同亚家族的基因结构基本一致, 相同亚家族的保守基序也具有相似性, 表明同一个亚家族的基因具有相似的功能。在大麻GRAS基因中, 83.33% (45/54)的基因无内含子,DELLA亚家族有3个基因, 其中CsGRAS3的Motif分析中未发现DELLA结构域, 可能是相邻连接法的算法将基因错误划分到了DELLA亚家族, 也有可能是其高度可变的N端序列差异导致其不能与同一亚家族的基因进行聚类, 水稻中GRAS家族成员也有相似的结果[30]。在所有大麻GRAS基因成员中都含有基序1、3、5, 表明这3个基序在GRAS中高度保守(图3), 具有重要作用。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1大麻、拟南芥的GRAS基因系统进化树
不同颜色表示不同的亚家族, 同一颜色的不同基因均属同一亚家族。
Fig. 1Phylogenetic tree of GRAS family in hemp and Arabidopsis
Different colors indicate different subfamily, and different genes under the same color belong to the same subfamily.
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2大麻GRAS基因结构分析
Fig. 2Gene structure analysis of GRAS genes in hemp
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3大麻GRAS基因基序
Fig. 3Gene motifs of GRAS in hemp
2.3 大麻GRAS基因的染色体分布与共线性分析
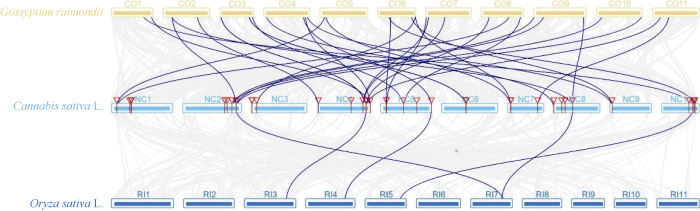
大麻染色体上GRAS基因位置分析表明, 大麻GRAS基因分布在10个染色体上, 与染色体长短无显著相关性(图4)。染色体NC6包含10个GRAS基因, NC1包含9个GRAS基因, 为含GRAS基因最多的2个染色体; NC9包含的GRAS基因最少, 只有1个; 其他染色体包含3~6个GRAS基因。其中5个染色体上出现了基因的串联重复情况, 尤其在NC1、NC6、NC7三个染色体上, 出现了大量GRAS基因的串联重复(相同染色体上基因有红色线段连接, 表示相关基因为串联重复基因), 串联重复基因占全部GRAS基因的44.44%, 表明大麻GRAS家族发生过明显的基因串联复制事件, 也表明基因串联重复事件在大麻GRAS家族的扩展中具有关键作用。通过大麻染色体间的共线性分析(有红色线段连接的为共线性基因)发现4对共线性基因(图5), 除CsGRAS8与CsGRAS9属HAM亚家族, 其他均分属不同亚家族, 这些基因是通过复制而得到相同连续顺序的旁系同源基因[28]。通过旁系同源基因的预测, 我们了解到NC5、NC6、NC8、NC10等4个染色体可能经历过染色体的片段复制, 而这4个染色体都能联系到NC10, 表明NC10可能是4个染色体中GRAS家族的关键染色体。为进一步了解大麻GRAS家族的进化机制, 构建了大麻与雷蒙德氏棉、大豆、水稻、玉米的GRAS家族共线性关系图(图6和图7), 这4种植物分别属于双子叶与单子叶植物。最终鉴定出大麻与雷蒙德氏棉有32对直系同源基因、与大豆有45对直系同源基因、与水稻有5对直系同源基因、与玉米有2对直系同源基因, 这也符合大麻与4种对应植物的进化关系。对5种植物进行共线性分析发现, GRAS6、GRAS8~10、GRAS14~19、GRAS23、GRAS33在5种植物中未被鉴定到, 其中大部分都是串联重复基因, 可能是大麻原有GRAS基因进行串联重复扩增而来, 这些基因可能是双子叶与单子叶植物GRAS基因功能差异的相关基因[31], 而GRAS12在5种植物中都被鉴定到, 且该基因包含GRAS基因中的motif 1~10, 推测该基因可能是双子叶植物与单子叶植物分化前就存在的GRAS基因。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4大麻GRAS基因的染色体定位
Fig. 4Chromosomal locations of GRAS genes in hemp
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5大麻染色体间GRAS基因共线性分析
Fig. 5GRAS synteny analysis among chromosomes in hemp
图 6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图 6雷蒙德氏棉、大麻、水稻间GRAS基因共线性分析
Fig. 6Synteny analysis of GRAS genes between Gossypium raimondii, Cannabis sativa L., and Oryza sativa L.
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7大豆、大麻、玉米间GRAS基因共线性分析
Fig. 7Synteny analysis of GRAS genes between Glycine max, Cannabis sativa L., and Zea mays L.
2.4 镉胁迫下不同大麻耐性筛选
将云麻1号、甘肃大麻、内蒙古小粒大麻、内蒙古油用大麻、内蒙古土右旗大麻、皖大麻6种大麻进行镉胁迫萌发试验发现, 云麻1号发芽率最高, 内蒙古小粒大麻发芽率最低。在镉胁迫水培试验中, 云麻1号镉处理组和对照组平均株高分别为9.4 cm、10.5 cm, 平均鲜重分别为7.4 g、7.9 g; 内蒙古小粒大麻镉处理组和对照组平均株高分别为3.8 cm、11.2 cm, 平均鲜重分别为4.3 g、7.5 g。在重度镉污染耕地中, 云麻1号鲜重产量达21,134.40 kg hm-2, 内蒙古小粒大麻鲜重产量达11,173.95 kg hm-2, 表明云麻1号较内蒙古小粒大麻更耐镉胁迫[32]。2.5 两个大麻品种在镉胁迫下GRAS基因的表达分析及qPCR验证
为了解大麻GRAS基因在正常条件及镉胁迫条件下的表达情况, 对云麻1号及内蒙古小粒大麻在2个环境下的植株进行了转录组测定, 表中各组基因的表达量均为3个重复的平均数。结果显示, 在云麻1号的转录组数据中(表2), 11个基因表达下降(占比20.37%), 下降倍数为0.13~0.91, 42个基因表达上调(占比77.78%), 上调倍数为1.05~18.10; 1个基因表达未发生变化(占比1.85%); 其中28个基因变化值差异显著。在内蒙古小粒大麻的转录组数据中(表3), 27个基因表达下降(占比50.00%), 下降倍数为0.30~0.96, 27个基因表达上调(占比50.00%), 上调倍数为1.01~6.46; 其中18个基因变化值差异显著。将表2与表3中GRAS基因按照增长倍数递增而排列, 云麻1号与内蒙古小粒大麻中GRAS基因家族部分基因对照组(CK组)表达量均为0, 无法计算倍数, 改为增加量(Cd-Ck)。Table 2
表2
表2云麻1号GRAS基因表达
Table 2
| 基因名称 Gene name | 云麻1号对照组 Yunma 1 control check | 云麻镉胁迫组 Yunma 1 cadmium stress | 变化值 Fold change | 基因名称 Gene name | 云麻对照组 Yunma 1 control check | 云麻镉胁迫组 Yunma 1 cadmium stress | 变化值 Fold change |
|---|---|---|---|---|---|---|---|
| CsGRAS24 | 2.44 | 0.33 | 0.13* | CsGRAS28 | 2.31 | 4.07 | 1.76* |
| CsGRAS46 | 4.02 | 0.58 | 0.14* | CsGRAS13 | 4.74 | 9.76 | 2.06* |
| CsGRAS25 | 42.77 | 20.24 | 0.47* | CsGRAS32 | 13.12 | 27.08 | 2.06* |
| CsGRAS33 | 6.56 | 3.12 | 0.48* | CsGRAS52 | 8.33 | 18.67 | 2.24* |
| CsGRAS2 | 1.62 | 0.78 | 0.48 | CsGRAS1 | 0.21 | 0.46 | 2.24 |
| CsGRAS11 | 30.07 | 14.91 | 0.50* | CsGRAS29 | 0.21 | 0.46 | 2.24 |
| CsGRAS22 | 1.63 | 0.91 | 0.56 | CsGRAS12 | 19.52 | 44.15 | 2.26* |
| CsGRAS54 | 6.20 | 4.59 | 0.74 | CsGRAS30 | 7.67 | 18.41 | 2.40* |
| CsGRAS6 | 0.32 | 0.28 | 0.88 | CsGRAS5 | 0.37 | 0.97 | 2.65 |
| CsGRAS42 | 9.74 | 8.87 | 0.91 | CsGRAS7 | 1.00 | 2.68 | 2.69* |
| CsGRAS48 | 0.25 | 0.26 | 1.05 | CsGRAS38 | 2.22 | 6.10 | 2.75* |
| CsGRAS9 | 6.16 | 6.88 | 1.12 | CsGRAS43 | 0.74 | 2.10 | 2.85* |
| CsGRAS4 | 8.31 | 9.43 | 1.14 | CsGRAS47 | 0.04 | 0.11 | 2.91 |
| CsGRAS21 | 2.66 | 3.16 | 1.19 | CsGRAS40 | 0.31 | 0.99 | 3.19 |
| CsGRAS20 | 0.76 | 0.93 | 1.21 | CsGRAS17 | 3.35 | 11.74 | 3.51* |
| CsGRAS10 | 7.49 | 9.54 | 1.27 | CsGRAS37 | 12.42 | 61.52 | 4.95* |
| CsGRAS36 | 8.79 | 11.82 | 1.34 | CsGRAS41 | 0.21 | 1.19 | 5.59* |
| CsGRAS31 | 14.46 | 19.51 | 1.35 | CsGRAS26 | 0.04 | 0.21 | 5.64 |
| CsGRAS23 | 0.20 | 0.27 | 1.37 | CsGRAS50 | 8.33 | 57.94 | 6.96* |
| 基因名称 Gene name | 云麻1号对照组 Yunma 1 control check | 云麻镉胁迫组 Yunma 1 cadmium stress | 变化值 Fold change | 基因名称 Gene name | 云麻对照组 Yunma 1 control check | 云麻镉胁迫组 Yunma 1 cadmium stress | 变化值 Fold change |
| CsGRAS15 | 3.04 | 4.43 | 1.46 | CsGRAS3 | 0.01 | 0.14 | 10.50* |
| CsGRAS49 | 0.97 | 1.44 | 1.49 | CsGRAS39 | 0.03 | 0.60 | 18.10* |
| CsGRAS8 | 8.98 | 13.70 | 1.53* | CsGRAS18 | 0.00 | 0.30 | 0.30* |
| CsGRAS27 | 18.49 | 28.66 | 1.55* | CsGRAS19 | 0.00 | 0.17 | 0.17* |
| CsGRAS44 | 4.33 | 6.75 | 1.56 | CsGRAS34 | 0.00 | 0.03 | 0.03 |
| CsGRAS16 | 0.32 | 0.51 | 1.59 | CsGRAS35 | 0.00 | 0.32 | 0.32* |
| CsGRAS14 | 0.76 | 1.24 | 1.62* | CsGRAS45 | 0.00 | 0.00 | 0.00 |
| CsGRAS51 | 6.82 | 11.11 | 1.63* | CsGRAS53 | 0.27 | 0.00 | -0.27* |
新窗口打开|下载CSV
Table 3
表3
表3内蒙古小粒大麻GRAS基因表达
Table 3
| 基因名称 Gene name | 内蒙大麻对照组 Inner Mongolia Xiaolidama control check | 内蒙大麻镉胁迫组 Inner Mongolia Xiaolidama cadmium stress | 变化值 Fold change | 基因名称 Gene name | 内蒙大麻对照组 Inner Mongolia Xiaolidama control check | 内蒙大麻镉胁迫组 Inner Mongolia Xiaolidama cadmium stress | 变化值 Fold change |
|---|---|---|---|---|---|---|---|
| CsGRAS24 | 5.40 | 1.61 | 0.30* | CsGRAS31 | 20.57 | 20.70 | 1.01 |
| CsGRAS53 | 0.84 | 0.37 | 0.44* | CsGRAS40 | 0.60 | 0.62 | 1.02 |
| CsGRAS22 | 2.70 | 1.35 | 0.50* | CsGRAS39 | 0.31 | 0.33 | 1.05 |
| CsGRAS48 | 0.34 | 0.17 | 0.50 | CsGRAS42 | 9.72 | 10.66 | 1.10 |
| CsGRAS25 | 49.36 | 28.23 | 0.57* | CsGRAS4 | 8.38 | 9.73 | 1.16 |
| CsGRAS21 | 2.54 | 1.61 | 0.63 | CsGRAS44 | 4.87 | 5.91 | 1.21 |
| CsGRAS33 | 12.12 | 7.76 | 0.64* | CsGRAS52 | 12.38 | 15.14 | 1.22 |
| CsGRAS36 | 10.98 | 7.07 | 0.64 | CsGRAS13 | 6.41 | 8.78 | 1.37 |
| CsGRAS41 | 1.27 | 0.97 | 0.76 | CsGRAS47 | 0.08 | 0.11 | 1.38 |
| CsGRAS17 | 8.55 | 6.64 | 0.78 | CsGRAS28 | 1.85 | 2.58 | 1.39 |
| CsGRAS11 | 37.71 | 29.54 | 0.78 | CsGRAS30 | 12.06 | 17.68 | 1.47* |
| CsGRAS18 | 0.22 | 0.18 | 0.78* | CsGRAS9 | 6.75 | 9.90 | 1.47* |
| CsGRAS20 | 0.28 | 0.23 | 0.79 | CsGRAS51 | 14.04 | 21.05 | 1.50* |
| CsGRAS7 | 1.70 | 1.39 | 0.82 | CsGRAS38 | 2.68 | 4.34 | 1.62 |
| CsGRAS6 | 0.28 | 0.23 | 0.82 | CsGRAS12 | 17.93 | 31.35 | 1.75* |
| CsGRAS1 | 0.48 | 0.40 | 0.83 | CsGRAS15 | 1.82 | 3.65 | 2.00 |
| CsGRAS29 | 0.48 | 0.40 | 0.83 | CsGRAS45 | 0.04 | 0.10 | 2.73 |
| CsGRAS14 | 1.08 | 0.94 | 0.87 | CsGRAS16 | 0.10 | 0.30 | 3.00 |
| CsGRAS27 | 20.63 | 18.12 | 0.88 | CsGRAS50 | 13.12 | 41.91 | 3.19* |
| CsGRAS23 | 0.29 | 0.26 | 0.89 | CsGRAS3 | 0.01 | 0.05 | 3.38 |
| CsGRAS49 | 1.10 | 0.99 | 0.89 | CsGRAS5 | 0.31 | 1.20 | 3.87* |
| CsGRAS32 | 13.96 | 12.66 | 0.91 | CsGRAS37 | 8.00 | 34.34 | 4.29* |
| CsGRAS8 | 14.56 | 13.35 | 0.92 | CsGRAS43 | 2.10 | 9.65 | 4.59* |
| CsGRAS10 | 4.19 | 3.87 | 0.92 | CsGRAS26 | 0.08 | 0.50 | 6.46* |
| CsGRAS54 | 7.36 | 6.96 | 0.94 | CsGRAS19 | 0.00 | 0.17 | 0.17* |
| CsGRAS46 | 1.17 | 1.12 | 0.95 | CsGRAS34 | 0.00 | 0.23 | 0.23* |
| CsGRAS2 | 2.82 | 2.71 | 0.96 | CsGRAS35 | 0.00 | 0.89 | 0.89* |
新窗口打开|下载CSV
2.6 两个大麻品种中同源基因对比及qPCR验证
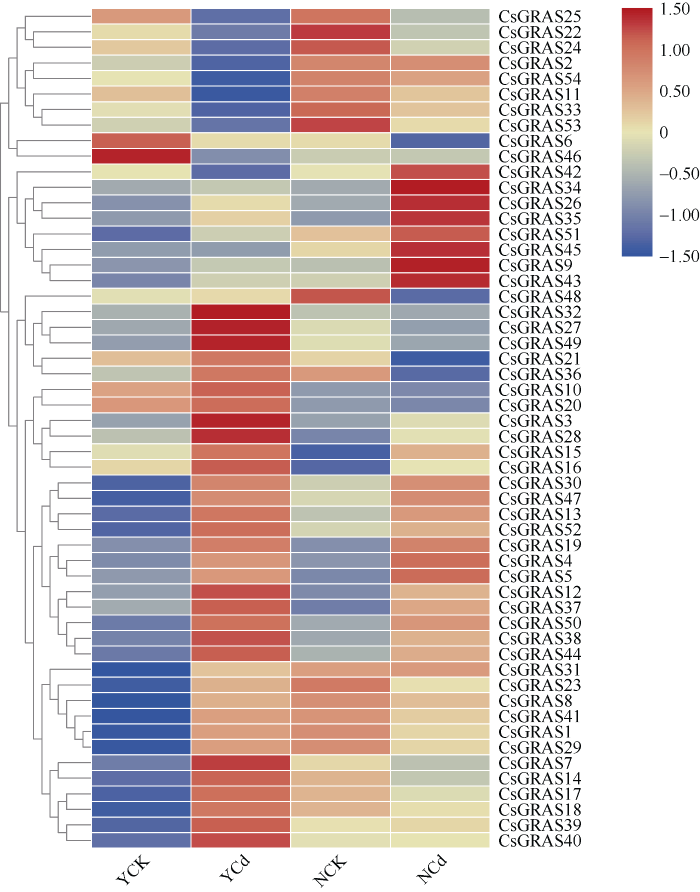
为进一步对比2个品种GRAS基因的表达情况变化, 本研究将2个品种的GRAS家族基因在正常环境下(YCK与NCK)及镉胁迫环境下(YCd与NCd)表达进行比对(图9), 并对大麻GRAS基因与镉胁迫进行关联分析。根据GRAS基因在不同品种大麻不同环境下的表达量, 将图9中的54个GRAS基因从上往下划分, CsGRAS25~CsGRAS46在2种大麻中都表达下调; CsGRAS34~CsGRAS43与CsGRAS3~ CsGRAS44在2种大麻中都表达上调; CsGRAS48~ CsGRAS20与CsGRAS31~CsGRAS40在云麻1号中表达上调, 在内蒙古小粒大麻中表达下调, 其中33个基因在云麻1号中的上调与下调幅度都比内蒙古小粒大麻更加明显。从2个大麻转录组数据中挑选了12个基因进行RT-qPCR验证(图9), 只有YMCsGRAS44表达趋势与转录组结果不一致, 其他基因的qPCR验证均与转录组一致, 表明转录组结果较为可靠。3 讨论
随着高通量测序技术的发展,越来越多的植物基因组完成了测序, 而基因家族分析也成了育种方向挖掘基因功能有效的生物信息学手段。目前, WRKY[33]、核苷酸结合位点基因家族(nucleotide binding site, NBS)[34]、MADS[35]等基因家族在很多物种中都被鉴定出来, 并进行了系统的研究与分析, 而GRAS基因家族在各种作物中, 尤其是各麻类作物中的研究还相对缓慢。本研究从大麻基因组中鉴定出54个GRAS家族基因, 将拟南芥与大麻GRAS基因构建系统进化树后发现, 有2个亚家族未包含拟南芥GRAS基因。通过系统进化树、基因结构与Motif对比发现, CsGRASB亚家族中的CsGRAS5可归纳到SCL3亚家族, CsGRAS20与CsGRAS22可能为CsGRAS25与CsGRAS11演变得到, 在演变过程中丢失了DELLA结构域, 在大麦GRAS家族鉴定中也未发现含DELLA结构域的GRAS基因, 表明DELLA结构域在不同物种间GRAS家族中的高度可变性[36]。而CsGRASA亚家族虽然与HAM亚家族亲缘关系最近, 但两者差异较大, 无法合并为一个亚家族。故大麻GRAS基因一共鉴定出9个亚家族, 其中CsGRASA亚家族是大麻新GRAS基因亚家族。
本研究通过对大麻染色体间GRAS家族基因进行共线性分析发现, PAT1与SHR、SCR与PAT1、HAM与LISCL等不同亚家族间有共线性基因对, 表明这些亚家族有染色体片段复制的相关性。而通过构建大麻、雷蒙德氏棉、大豆、水稻、玉米基因组GRAS共线性分析发现, PAT1、LS、SHR、HAM在大麻、雷蒙德氏棉、水稻中有直系同源基因, 说明这些家族的保守性较强, 而CsGRAS12在5种植物中都存在, 属于SHR亚家族, 表明其保守性最强, 可能在单双子叶植物分化前就存在[33], 且该基因在2个大麻品种镉胁迫下都上调表达, 可作为后续研究的关键基因之一。而演化的过程中, GRAS家族在PAT1、LISCL、CsGRASA上又发生了大量基因的串联重复, 这表明大麻GRAS的染色体片段复制与导基因串联重复是大麻出现54个GRAS基因的关键步骤。
本研究将镉耐受性不同的2种大麻进行GRAS家族基因表达量比对发现, 在镉胁迫下该基因家族表达量发生了明显的变化, 表明镉胁迫会显著影响大麻GRAS基因表达。其中大部分GRAS基因在更耐镉胁迫的云麻1号中上调表达明显, 表明GRAS基因家族与大麻的镉胁迫耐受性明显相关, 上调大麻GRAS家族基因可能会提高大麻的镉胁迫耐受性。通过图8中2种大麻的形态对比发现, 云麻1号在2种环境中形态未有显著差异, 但内蒙古小粒大麻在镉胁迫环境下根系明显变短, 而GRAS基因可以调控植物的根系生长及维管束的发育[37], 推测是镉胁迫抑制了内蒙古小粒大麻部分GRAS基因表达而影响大麻根系的生长。其中CsGRAS48、CsGRAS21、CsGRAS36、CsGRAS7等 4个基因在云麻1号中表达上调, 但在内蒙古小粒大麻中明显下调, 推测其与大麻的根系发育有关。在植物激素方面, 2个DELLA基因: CsGRAS11与CsGRAS25都表达下调, 而DELLA基因负调控赤霉素途径, 推测大麻在镉胁迫环境下增加赤霉素等植物激素以缓解外界的镉胁迫[38]。而属于LISCL亚家族的CsGRAS39与CsGRAS39在云麻1号中明显表达上调, 在内蒙古小粒大麻中未发生变化, 推测其为改变植物的光转导途径来应对外界的环境变化[36]。将转录组中的12个基因进行qPCR验证发现, 其中91.66%的基因变化趋势与转录组结果相同, 表明转录组的结果较可靠。
图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8两个大麻品种在正常环境与镉胁迫环境的生长状况
Fig. 8Growth status of two hemp varieties under normal and cadmium stress environment
图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9GRAS同源基因表达
Fig. 9Relative expression of GRAS homologous genes
图10
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1012个基因的qPCR验证表达
*, **, ***分别表示在0.05、0.01和0.001水平显著差异。
Fig. 10qPCR expression of 12 genes
*, **, *** mean significant differences at the 0.05, 0.01, and 0.001 probability levels, respectively.
4 结论
通过对大麻全基因组的序列比对, 共获得GRAS家族54个成员, 分属9个不同亚家族, 具有典型的GRAS或DELLA保守结构域。通过生物信息学方法分析大麻GRAS家族的理化指标、系统进化关系、基因结构、共线性关系表明, 染色体片段复制与串联重复复制对大麻形成54个GRAS基因起重要作用, 而转录组数据及qPCR验证都表明, GRAS基因与大麻应对镉胁迫有明显相关性, 但其主要影响的代谢通路及不同基因之间的协同作用还需进一步研究。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S1875-5364(16)30013-9URLPMID:27025363 [本文引用: 1]

Isatis indigotica Fort., belonging to Cruciferae, is one of the most commonly used plants in traditional Chinese medicine. The accumulation of the effective components of I. indigotica is related with its growth conditions. The GRAS genes are members of a multigene family of transcriptional regulators that play a crucial role in plant growth. Although the activities of many GRAS genes have long been recognized, only in recent years were some of them identified and functionally characterized in detail. In the present study, 41 GRAS genes were identified from I. indigotica through bioinformatics methods for the first time. They were classified into ten groups according to the classification of Arabidopsis and rice. The characterization, gene structure, conserved motifs, disordered N-terminal domains, and phylogenetic reconstruction of these GRASs were analyzed. Forty-three orthologous gene pairs were shared by I. indigotica and Arabidopsis, and interaction networks of these orthologous genes were constructed. Furthermore, gene expression patterns were investigated by analysis in methyl jasmonate (MeJA)-treated I. indigotica hairy roots based on RNA-seq data. In conclusion, this comprehensive analysis would provide rich resources for further studies of GRAS protein functions in this plant.
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
URLPMID:26759740 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:26042223 [本文引用: 1]
[本文引用: 1]
URLPMID:31387526 [本文引用: 1]
DOI:10.1016/j.gene.2019.04.038URLPMID:30999026 [本文引用: 1]

BACKGROUND: In recent years, the molecular mechanism of plant growth and development has been reported in detail. GRAS genes, a plant-specific family of transcription factor, play critical roles in the process. GRAS transcription factors are associated with axillary shoot meristem formation, radial root patterning, phytohormones (gibberellins) signal transduction, light signaling, and abiotic or biotic stress. OBJECTIVE: Here, we firstly investigated GRAS gene family in Dendrobium catenatum, an important medicinal and flowering orchid in China. METHODS: The GRAS gene family in D. catenatum was cloned based on RNA-Seq data. Selected GRAS genes were introduced into Escherichia coli to express proteins. RESULTS: Based on phylogenetic relationship with the Arabidopsis and Oryza GRAS family members, 47 GRAS genes from D. catenatum are identified and their deduced proteins are classified into 11 subgroups. Most of these GRAS genes contain one exon and closely related members in the phylogenetic tree have similar motif composition. Our result also reveals that GRAS genes in D. catenatum are widely distributed and expressed in different tissue. In addition, 35 GRAS genes are successfully cloned from different subgroups and 7 DoGRAS fusion proteins are induced using E. coli system. Moreover, 8 genes were up-regulated in different tissue following exposure to heat and salt stresses. CONCLUSION: Our findings provide valuable information and candidate genes for future functional analysis for improving the resistance of D. catenatum growth.
[本文引用: 1]
URLPMID:25998905 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12864-018-4722-xURLPMID:29743013 [本文引用: 1]

BACKGROUND: Cotton is a major fiber and oil crop worldwide. Cotton production, however, is often threatened by abiotic environmental stresses. GRAS family proteins are among the most abundant transcription factors in plants and play important roles in regulating root and shoot development, which can improve plant resistance to abiotic stresses. However, few studies on the GRAS family have been conducted in cotton. Recently, the G. hirsutum genome sequences have been released, which provide us an opportunity to analyze the GRAS family in G. hirsutum. RESULTS: In total, 150 GRAS proteins from G. hirsutum were identified. Phylogenetic analysis showed that these GRAS protins could be classified into 14 subfamilies including SCR, DLT, OS19, LAS, SCL4/7, OS4, OS43, DELLA, PAT1, SHR, HAM, SCL3, LISCL and G_GRAS. The gene structure and motif distribution analysis of the GRAS members in G. hirsutum revealed that many genes of the SHR subfamily have more than one intron, which maybe a kind of form in the evolution of plant by obtaining or losing introns. Chromosomal location and duplication analysis revealed that segment and tandem duplication maybe the reasons of the expension of the GRAS family in cotton. Gene expression analysis confirmed the expression level of GRAS members were up-regulated under different abiotic stresses, suggesting that their possible roles in response to stresses. What's more, higher expression level in root, stem, leaf and pistil also indicated these genes may have effect on the development and breeding of cotton. CONCLUSIONS: This study firstly shows the comprehensive analysis of GRAS members in G. hirsutum. Our results provide important information about GRAS family and a framework for stress-resistant breeding in G. hirsutum.
[本文引用: 1]
[本文引用: 1]
URLPMID:31465423 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
URLPMID:28668094 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
URLPMID:29529275 [本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
