 ,1,2,**, 白建芳2,**, 马有志1, 郭昊宇2, 王永波2, 陈兆波2, 赵昌平
,1,2,**, 白建芳2,**, 马有志1, 郭昊宇2, 王永波2, 陈兆波2, 赵昌平 ,2,*, 张立平
,2,*, 张立平 ,2,*
,2,*Cloning and expression analysis of lncRNA27195 and its target gene TaRTS in wheat (Triticum aestivum L.)
WANG Na ,1,2,**, BAI Jian-Fang2,**, MA You-Zhi1, GUO Hao-Yu2, WANG Yong-Bo2, CHEN Zhao-Bo2, ZHAO Chang-Ping
,1,2,**, BAI Jian-Fang2,**, MA You-Zhi1, GUO Hao-Yu2, WANG Yong-Bo2, CHEN Zhao-Bo2, ZHAO Chang-Ping ,2,*, ZHANG Ling-Ping
,2,*, ZHANG Ling-Ping ,2,*
,2,*通讯作者: * 张立平, E-mail:lpzhang8@126.com;赵昌平, E-mail:cp_zhao@vip.sohu.com
第一联系人:
收稿日期:2020-09-2接受日期:2021-01-13网络出版日期:2021-08-12
| 基金资助: |
Received:2020-09-2Accepted:2021-01-13Online:2021-08-12
| Fund supported: |
作者简介 About authors
王娜,E-mail:wangna_00@sina.com

摘要
长链非编码RNA (long non-coding RNA, lncRNA)是一种大于200 bp的非编码RNA, 大量存在于植物体中, 其可通过调节基因表达或蛋白功能, 在植物生长发育与胁迫应答中发挥重要作用。前期在研究光照和温度对小麦光温敏核雄性不育系BS366育性诱导中, 利用转录组测序筛选获得与育性相关的lncRNA (lncRNA2719)。为研究小麦lncRNA27195的功能, 本研究在BS366中克隆lncRNA27195及其靶基因TaRTS, 并对TaRTS进行生物信息学分析, 结果显示, TaRTS基因全长315 bp, 编码104个氨基酸, 且发现该RTS蛋白仅存在于禾本科植物中。通过实时荧光定量PCR, 对lncRNA27195及TaRTS基因在不同组织不同光温处理及茉莉酸甲酯处理下进行表达模式分析, 发现lncRNA27195和TaRTS均在雄蕊中高表达, 呈显著的正相关, 且两者在不同光温条件下呈现出不同的表达模式。光照和温度均对lncRNA27195和TaRTS有调控作用, 适当浓度的MeJA促进两者的表达, SA抑制两者表达。以上结果表明, 在光周期、温度和植物激素的诱导下, lncRNA27195正向调节TaRTS基因表达, 进而影响花粉育性, 本研究结果有助于促进对小麦光温敏核型雄性不育系的机理研究和生产应用。
关键词:
Abstract
Long non-coding RNA (lncRNA) is a non-coding RNA length over 200 bp, which is abundant in plants. It plays important roles in plant growth, development, and stress response by regulating gene expression or protein function. In the previous study, a fertility-related lncRNA named lncRNA27195 was screened and obtained by transcriptome sequencing from the anther of wheat Photoperiod-thermo Sensitive Genic Male Sterility (PTGMS) line BS366. To investigate the function of lncRNA27195 in wheat, the lncRNA27195 gene and its target gene TaRTS were cloned from BS366. Bioinformatics analysis were performed on TaRTS. The expressions of lncRNA27195 and TaRTS in different tissues and their expression correlation between them were analyzed by qRT-PCR. Meanwhile, the expression patterns of lncRNA27195 and TaRTS under different light and temperature treatments, and methyl jasmonate (MeJA) treatments were investigated. The results showed that the TaRTS gene with 315 bp length, encoded 104 amino acids. Additionally, RTS proteins were only found as anther-specific proteins in gramineae plants. Both lncRNA27195 and TaRTS with a significantly positive correlation were highly expressed in stamens, and revealed different expression patterns in different fertility environments. The results demonstrated that the expression of lncRNA27195 and TaRTS were also regulated by light and temperature. In addition, we found that the appropriate concentration of MeJA could promote the expression of lncRNA27195 and TaRTS while SA could inhibit the expression. The results indicated that under the induction of photoperiod, temperature, and plant hormones, IncRNA27195 positively regulated TaRTS gene expression, resulting in affecting pollen development and male fertility. This study contributed to the mechanism research and production application of PTGMS wheat.
Keywords:
PDF (3770KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王娜, 白建芳, 马有志, 郭昊宇, 王永波, 陈兆波, 赵昌平, 张立平. 小麦lncRNA27195及其靶基因TaRTS克隆及表达分析. 作物学报[J], 2021, 47(8): 1417-1426 DOI:10.3724/SP.J.1006.2021.01071
WANG Na, BAI Jian-Fang, MA You-Zhi, GUO Hao-Yu, WANG Yong-Bo, CHEN Zhao-Bo, ZHAO Chang-Ping, ZHANG Ling-Ping.
长链非编码RNA (long non-cording RNA), 是指长度超过200个核苷酸的非编码RNA, 通过RNA聚合酶Ⅱ转录[1]。研究表明lncRNA可与DNA、RNA或蛋白质分子相互作用, 从而调节基因表达, 并通过多种机制发挥功能。在转录水平上, lncRNA可以招募转录因子, 调控相邻基因的表达[2,3]; 在表观遗传水平上, lncRNA可以通过影响染色质结构域调节目的基因的表达[4,5]; 在转录后水平上, lncRNA可通过与靶mRNA的互补碱基配对调控mRNA各方面的功能, 类似于微小RNA, 如microRNA和snoRNA[6]。目前, 在拟南芥[7]、小麦[8]、水稻[9,10]、玉米[11]、番茄[12]等植物体内均发现了lncRNA, 并发现他们具有重要的作用, 可调控一系列重要生物途径, 包括生长代谢、激素信号、逆境胁迫响应、生殖发育和维护基因组完整性等。在水稻中, 研究人员发现可调控光敏雄性不育水稻的lncRNA (LDMA), 在长日条件下高表达可保持花粉正常发育。而LDMA突变体中, LDMAR的二级结构发生改变, 导致LDMAR启动子区域的甲基化程度提高, 从而降低LDMAR转录水平, 导致长日条件下花药绒毡层细胞提早发生程序性死亡, 进而引起光敏雄性不育[9,10]。二系杂交小麦主要是通过环境条件的改变可以调控植物的雄性育性转换[13]。光温敏核雄性不育小麦既可作为雄性不育系, 也可发挥保持系的作用, 是目前杂种优势利用的主要方式, 因此小麦光温敏核雄性不育基因的挖掘和研究具有重要意义。
RTS蛋白, 是一个花药特异性蛋白, 只在绒毡层细胞中特异性表达。研究发现水稻OsRTS基因的下调表达会导致花粉败育, 从而引起水稻雄性不育[14]。绒毡层位于四层花药细胞的最内层, 对花粉的发育至关重要[15]。在绒毡层发育过程中绒毡细胞的程序性死亡对原染色体的形成、孢粉素的合成至关重要[16]。绒毡层结构破损会导致花粉失活, 引起雄性不育。例如, 拟南芥的SERK1、SERK2双突变体的花药缺乏绒毡层的生成, 导致小孢子母细胞无法在减数分裂后正常发育, 导致小孢子败育和雄性不育[17]。前期通过对光照和温度诱导的可育环境和不育环境的BS366光温敏核雄性不育小麦的花药进行lncRNA测序, 并对育性相关差异表达的lncRNA进行筛选。转录组结果发现lncRNA27195及其靶基因TaRTS (TraesCS3D02G436700.1)在可育和不育环境中表达差异较明显。因此, 本研究克隆lncRNA27195及其靶基因TaRTS, 通过对其进行生信分析、组织特异性和时空表达分析, 并分析其在不同育性条件下和非生物胁迫下的表达模式, 为进一步阐明长链非编码RNA在光温敏雄性不育小麦育性转换过程中的调控模式提供一定的理论基础。
1 材料与方法
1.1 试验材料
试验材料为光温敏核雄性不育系冬小麦BS366, 由北京市农林科学院北京杂交小麦工程技术研究中心提供。BS366表现出光温敏感性育性, 在不育环境(花粉发育阶段日平均温度12℃, 12 h光照/12 h黑暗)下, 花粉失活, 结实率<5%; 在可育环境(花粉发育阶段日平均温度20℃, 14 h光照/10 h黑暗)下, 花粉发育正常, 育性恢复至40%~55%。所有植物均种植于花盆, 放置在北京的实验田, 并进行常规管理, 至四叶一心期放入人工气候箱。1.2 试验方法
1.2.1 lncRNA27195基因及TaRTS基因的克隆 各组织RNA提取按照TRIzol Reagent (Invitrogen)说明书进行。RNA经浓度和纯度鉴定后合格放置-80℃备用。利用TaKaRa PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time)试剂盒将总RNA反转录为cDNA。根据前期转录组学的分析结果, 根据lncRNA27195基因序列和TaRTS基因(TraesCS3D02G436700.1)序列利用Primer premier 5软件设计引物(表1), 以BS366花药组织样品cDNA为模板克隆lncRNA27195及TaRTS基因全长。扩增体系为: 2× Phanta Max Buffera 25 μL, dNTP Mix (10 mmol L-1 each) 1 μL, 上游引物(10 μmol L-1) 1 μL, 下游引物(10 μmol L-1) 1 μL, Phanta Max Super-Fidelity DNA Polymerase 1 μL, cDNA 2 μL, ddH2O 19 μL。PCR扩增程序为: 95℃预变性3 min; 95℃变性15 s, 55℃退火15 s, 72℃延伸15 s; 72℃延伸10 min, 4℃保存。按照pBackZero-T Vector Cloning Kit说明书, 将lncRNA27195全长序列与TaRTS基因的CDS序列克隆至pBackZero-T载体上。转化大肠杆菌DH-5α, 挑取阳性单克隆菌, 对目标片段进行测序, 并将甘油菌保存于-80℃冰箱。Table 1
表1
表1引物设计
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 产物大小 Product length (bp) |
|---|---|---|
| lncRNA27195-F | TTTAATCTTCCGAGCTCACCCA | 396 |
| lncRNA27195-R | TTTAAATGTTAAAACATGTTATATTTTTGA | |
| TaRTS-F | TTTAAATGGCACGCACCGGC | 315 |
| TaRTS-R | TTTAATCAGAGCCCCTGGGTGGT | |
| lncRNA27195-qRT-F | GCATACGCACCGCATGTATAAT | 109 |
| lncRNA27195-qRT-R | CTAGTCGCACAGGTCGACTGAA | |
| TaRTS-qRT-F | AGGTCGCATCGTGCCTCCT | 114 |
| TaRTS-qRT-R | CCCCTGGGTGGTGCAGT | |
| Actin1-F | TTGGCATCTCTCAGCACCTTC | 138 |
| Actin1-R | TCAGCCTTCGCAATCCACA |
新窗口打开|下载CSV
1.2.2 TaRTS基因生物信息学分析 利用ExPASy ProtParam (
1.2.3 TaRTS基因亚细胞定位 将TaRTS的ORF序列克隆到16318-GFP载体上, 构建TaRTS-GFP融合表达载体。将融合载体转化到新制备的小麦原生质体中, 以16318-GFP转化的小麦原生质体作为对照, 将融合载体TaRTS-GFP转化的小麦原生质体细胞在激光共聚焦显微镜(Leica, 德国)下观察绿色荧光信号在原生质体中的分布。
1.2.4 表达模式分析 组织特异性分析: 在抽穗期采集BS366小麦的根、茎、叶、颖壳、雄蕊和雌蕊样品, 置于-80℃冰箱保存备用, 进行组织特异性表达分析。不同育性环境对基因表达模式影响分析: 在四叶期结束后(育性敏感期), 将长势良好且一致BS366植株分为两组, 分别在可育环境(20℃, 14 h L/10 h D; L/D为光照/黑暗)和不育环境(12℃, 12 h L/12 h D)条件下, 相对湿度为60%~80%的人工气候培养箱(CLC-BIV-M/CLC404-TV, MMM, 德国)中培养。根据叶龄和花药长度, 采集BS366在两个不同生育条件下的孢子母细胞时期(S1)、二分体时期(S2)、四分体时期(S3)和单核期(S4)的花药组织, 置于-80℃冰箱保存备用。不同光温对基因表达模式影响分析: 将育性敏感期长势良好且一致BS366植株分为4组, 分别在低温长日环境(12℃, 14 h L/10 h D)、低温短日环境环境(12℃, 12 h L/12 h D)、高温长日环境(20℃, 14 h L/10 h D)、和高温短日环境(20℃, 12 h L/12 h D)条件下中培养约2周, 至抽穗期, 采集花药组织, 置于-80℃冰箱保存备用。茉莉酸甲酯对基因表达模式调控分析: BS366正常生长20℃ (12 h L/12 h D)至抽穗期, 每天喷施茉莉酸甲酯(MeJA), 浓度分别为0、0.5、2和4 mmol L-1, 连续5 d后, 同时用10 mmol L-1水杨酸(SA)喷施MeJA处理后的小穗, 采集花药组织, 置于-80℃冰箱保存备用。根据lncRNA27195基因和TaRTS基因的序列设计qPCR引物(表1), 以上述样品RNA反转录的cDNA为模板, 以TaActin1为内参基因, 在(CFX96; Bio-Rad, Hercules, CA, USA)平台使用TB Green Premix Ex Taq (Tli RNaseH Plus)试剂盒进行qPCR分析。反应体系为: TB GreenPremix Ex Taq (Tli RNaseH Plus) (2×) 5 μL, cDNA 1 μL, 上游引物(10 μmol L-1) 0.5 μL, 下游引物(10 μmol L-1) 0.5 μL, ddH2O 3 μL。反应程序为: 预变性95℃5 s; 95℃ 5 s, 60℃ 30 s, 45个循环; 溶解曲线分析95℃ 15 s, 55℃ 1 min, 95℃ 15 s。数据按照2-ΔΔCt法计算相对表达量, 生物学重复和技术重复均为3次。
2 结果与分析
2.1 lncRNA27195及TaRTS基因的克隆
以BS366的花药cDNA为模板克隆lncRNA27195与TaRTS基因的CDS序列, 电泳结果与网上公布的同序列大小基本一致,分别为396 bp和315 bp (图1)。将克隆至pBackZero-T载体上, 转化大肠杆菌DH5α, 挑取阳性单克隆菌, 并进行测序,结果同前期转录组测序结果(图2)。图1
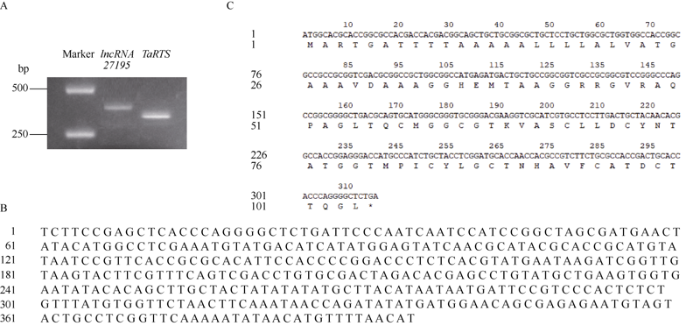 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1lncRNA27195基因和TaRTS基因扩增片段(A)和序列(B, C)
Fig. 1Amplified fragment of lncRNA27195 and TaRTS (A) and sequence of lncRNA27195 (B), and TaRTS (C)
图2
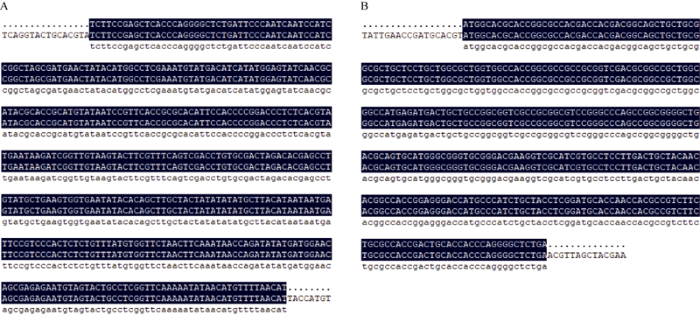 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2lncRNA27195基因(A)和TaRTS基因(B)序列比对
Fig. 2Comparison of the prokaryotic expression sequence of lncRNA27195 (A) and TaRTS (B)
2.2 TaRTS基因的蛋白质结构分析
小麦的TaRTS基因位于3D染色体上, CDS序列长315 bp, 编码104个氨基酸。通过Ensambl搜索比对, 发现其在3A和3B上存在等位基因TraesCS3A02G444400.1和TraesCS3B02G477900.1。TaRTS蛋白分子量为10.08 kD; 等电点为7.54, 为碱性蛋白质; 共1389个原子; 半衰期为30 h; 不稳定指数为30.16, 为稳定蛋白; 脂肪族指数为77.31; 总平均疏水指数(grand average of hydropathicity)为0.497; 有4个带负电荷的氨基酸残基, 包括3个天冬氨酸(Asp)和1个谷氨酸(Glu), 有5个带正电荷的氨基酸残基, 包括4个精氨酸(Arg)和1个赖氨酸(Lys)。利用Prot Scale在线软件测得TaRTS蛋白的氨基酸残基整体为疏水性, 同理化性质分析一致, 推测为疏水性蛋白(图3-A)。蛋白二级结构分析显示, 该蛋白由41个α-螺旋(39.42%)、12个延伸链(11.25%)、9个β-转角(8.65%)和42个无规则卷曲(40.38%)组成(图3-B)。蛋白三级结构结果显示主要结构为α-螺旋(图3-C)。亚细胞定位预测结果显示TaRTS蛋白为分泌蛋白。此外, 小麦原生质体亚细胞定位显示, 绿色荧光蛋白GFP在小麦原生质体中瞬时高效表达, 表明TaRTS蛋白定位于原生质体的细胞质部分, 符合预测结果(图4)。图3
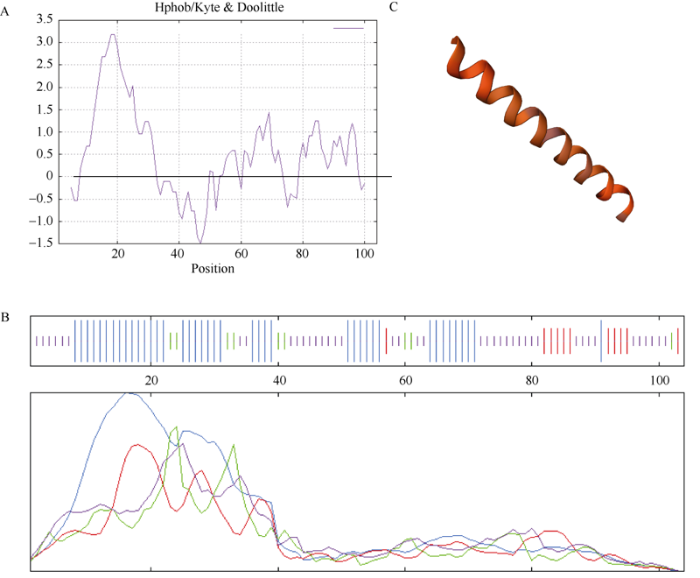 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3TaRTS蛋白的亲水性(A)、二级结构(B)和三级结构(C)
蓝色为α-螺旋, 绿色为β-转角, 红色为延伸链, 紫色为无规则卷曲。
Fig. 3The hydrophilicity (A), secondary structure (B), and tertiary structure (C) of TaRTS
Blue line means alpha helix, green line means beta turn, red line means extended strand, and purple line means random coil.
图4
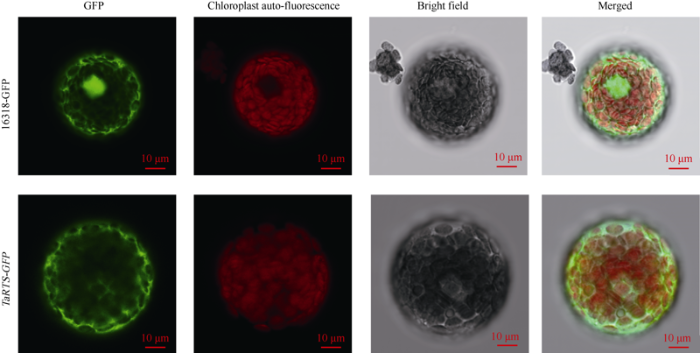 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4TaRTS蛋白的小麦原生质体亚细胞定位
Fig. 4Subcellular localization of TaRTS in wheat protoplasts
2.3 TaRTS基因编码蛋白的系统发育分析
研究结果显示仅在水稻(Oryza sativa japonica Group)、粟(Setaria italica)、山羊草(Aegilops tauschii)、二穗短柄草(Brachypodium distachyon)、哈氏黍(Panicum hallii)和狗尾草(Setaria viridis)这些禾本科植物中发现同源基因, 表明RTS基因家族是一种禾本科特有基因。多序列比对结果显示, RTS基因家族的保守性较差, 但具有保守的半胱氨酸位点(图5-A)。利用MEGA 6.0软件用邻接法构建系统进化树, 显示TaRTS基因与二穗短柄草的同源性较高, 同粟、狗尾草同源性较低(图5-B)。图5
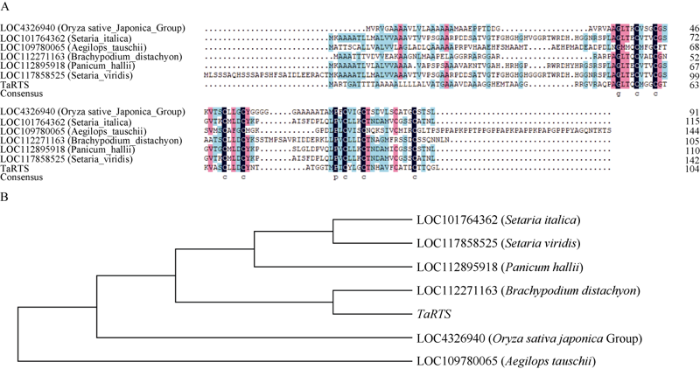 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5TaRTS的多序列比对(A)和进化树分析(B)
Fig. 5Multiple alignments of protein sequences (A) and phylogenetic tree analysis (B) of TaRTS
2.4 TaRTS基因启动子元件分析
对TaRTS基因启动子区前2000 bp序列的转录因子的预测分析发现, 基因的启动子序列中包含: 光响应元件(ACE, G-box, GT1-motif, T-box, Sp1)、 分生组织表达有关(CAT-box)、MYBHv1结合位点(CCAAT-box)、 茉莉酸甲酯响应元件(CGTCA-motif, TGACG-motif)、低温响应元件(LTR)、细胞周期调控元件(MSA-like)等元件。推测TaRTS基因与光、温胁迫, 调节细胞周期相关(表2)。Table 2
表2
表2启动子元件分析
Table 2
| 上游启动子元件 Upstream promoter elements | 功能 Function | |
|---|---|---|
| CAAT-box, TATA-box | 核心启动子元件 Core promoter element | |
| ACE, G-box, GT1-motif, T-box, Sp1 | 光响应元件 Light responsiveness element | |
| ABRE | 脱落酸响应元件 Abscisic acid responsiveness element | |
| CAT-box | 分生组织表达相关元件 Related to meristem expression element | |
| CCAAT-box | MYBHv1结合位点 MYBHv1 binding site | |
| CGTCA-motif, TGACG-motif | 茉莉酸甲酯响应元件 MeJA-responsiveness responsiveness element | |
| LTR | 低温响应元件 Low-temperature responsiveness element | |
| MSA-like | 细胞周期调控元件 Cell cycle regulation element | |
新窗口打开|下载CSV
2.5 lncRNA27195及TaRTS组织特异性分析
本实验研究了lncRNA27195与TaRTS基因在小麦孕穗期(挑旗)根、茎、旗叶、颖壳、雄蕊和雌蕊不同组织的表达谱。结果显示, lncRNA27195与TaRTS基因在小麦雄蕊中表达量最高, 显著高于其他组织(P<0.01), 而其他组织中的表达量极低, 表明lncRNA27195与TaRTS在雄蕊上特异性表达, 主要参与花药发育的调控(图6)。图6
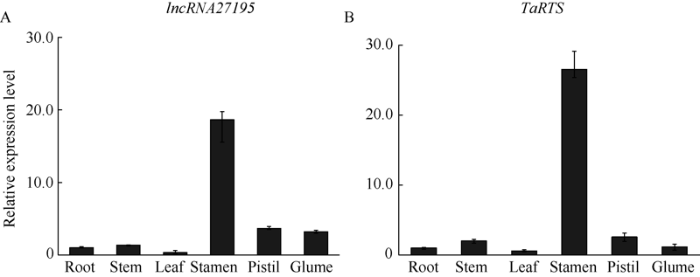 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6lncRNA27195基因(A)及TaRTS基因(B)组织特异性表达分析
Fig. 6Relative expression levels of lnc27195 (A) and TaRTS (B) in different tissues
2.6 lncRNA27195及TaRTS不同花药发育时期表达分析
本研究对lncRNA27195与TaRTS基因在不同育性条件下的4个发育时期的花药中的表达模式进行了分析。结果表明, TaRTS基因在二分体时期、四分体时期和单核期, 可育环境下的表达量均极显著高于不育环境(P<0.01)。lncRNA27195在二分体时期可育环境下的表达量极显著高于不育环境(P<0.01); 在四分体时期和单核期, 可育环境下的表达量均显著高于不育环境(P<0.05); 在小孢子母细胞时期, 不育环境的表达量显著高于可育环境(P<0.05) (图7-A)。皮尔逊相关性分析结果显示, lnc27195同TaRTS基因的表达量在小麦花药中呈显著正相关(R2=0.8724) (图7-B)。图7
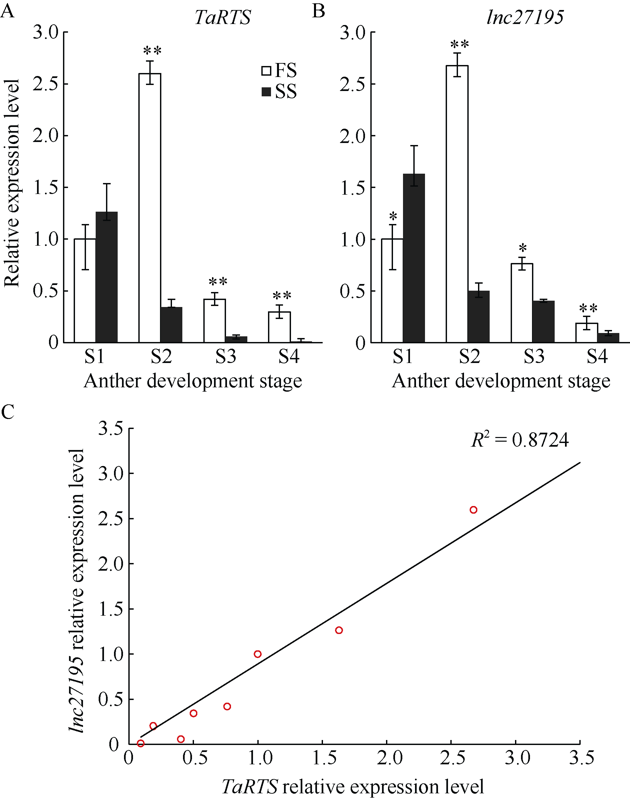 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7TaRTS(A)和lncRNA27195(B)表达模式分析及两者表达量相关性分析(C)
*代表显著差异(P<0.05), **代表极显著差异(P<0.01)。
Fig. 7Relative expression levels of TaRTS (A), lncRNA27195 (B), and the correlation analysis (C) of TaRTS and lncRNA27 195
* means significantly different at P<0.05, and ** means highly significant difference at P<0.01.
2.7 lncRNA27195及TaRTS在不同光温处理下的表达模式分析
前期发现BS366的育性受光照和温度的影响, 且生信分析发现TaRTS基因启动子区包含光和低温响应元件(表2), 因此对不同光照和温度条件下, lncRNA27195与TaRTS基因在花药中的表达水平进行分析。结果显示, 在高温长日条件(20℃, 14 h L/10 h D)、高温短日条件(20℃, 12 h L/12 h D)、低温长日条件(12℃, 14 h L/10 h D)和低温短日(12℃, 12 h L/12 h D)条件下, 小麦lncRNA27195的相对表达量依次下降, 四个条件下存在显著性差异; 小麦TaRTS基因的表达量依次下降, 高温长日条件、高温短日和低温长日条件、低温短日条件三组间存在显著性差异(图8)。此外, 小麦lncRNA27195与TaRTS基因在不同温度(相同光照)和不同光照(相同温度)的组别间的数据分析中显示, 两者皆存在显著性差异, 且受温度影响较大。结果表明, 光照和温度均对小麦lncRNA27195与TaRTS基因的表达存在显著影响, 高温和长日照可以促进lncRNA27195与TaRTS基因的表达, 进而促进育性恢复。图8
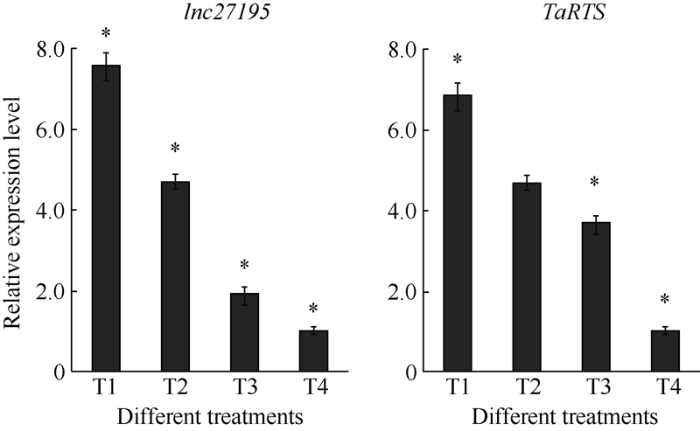 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8TaRTS及lncRNA27195在不同光照温度条件下表达模式分析
T1: 温长日环境(20℃, 14 h L/10 h D); T2: 高温短日环境(20℃, 12 h L/12 h D); T3: 低温长日环境(12℃, 14 h L/10 h D); T4为低温短日(12℃, 12 h L/12 h D)环境。*代表各组间存在显著差异(P < 0.05)。
Fig. 8Relative expression levels of TaRTS and lncRNA27195 in different environments
T1: long-light environment with high temperature (20℃, 14 h L/10 h D); T2: short-light environment with high temperature (20℃, 12 h L/12 h D); T3: long-light environment with low temperature (12℃, 14 h L/10 h D); T4: short-light environment with low temperature (12℃, 12 h L/12 h D). * means significantly different at P < 0.05.
2.8 lncRNA27195及TaRTS在MeJA及SA处理下的表达模式分析
前期研究表明, MeJA参与小麦雄性不育, 可促进花药开裂, 同时SA对该功能有一定的拮抗作用; 本文前期的生物信息学分析中发现TaRTS基因启动子区存在MeJA响应元件。为研究MeJA与TaRTS之间的调控方式, 以及对TaRTS的调控作用, 本研究利用不同浓度的MeJA对抽穗后开花前的花药进行处理, 同时研究SA喷施MeJA处理后的花药中, lncRNA27195及TaRTS的表达模式。在MeJA处理后, 随着浓度的上升, lncRNA27195及TaRTS的表达量先上升后下降, 在MeJA浓度为0.5 mmol L-1时达到峰值; 通过SA喷施处理后, 发现lncRNA 27195及TaRTS的表达量较正常植株显著下降, 且随着MeJA浓度的上升, lncRNA27195及TaRTS的表达量不断下降(图9)。实验表明, 适量的MeJA可以促进lncRNA27195及TaRTS的表达, 但过高浓度的MeJA反而会抑制其表达; SA可以抑制MeJA的功能, 从而抑制lncRNA27195及TaRTS的表达。图9
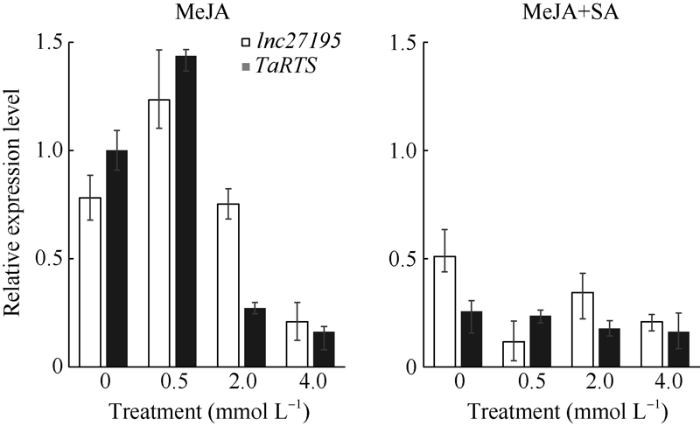 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9TaRTS及lncRNA27195在MeJA和SA处理下的表达模式分析
Fig. 9Relative expression levels of TaRTS and lncRNA27195 under MeJA and SA treatments
3 讨论
在水稻中, 研究表明OsRTS基因仅在绒毡层中表达, 对花粉发育至关重要, 且抑制其表达会导致雄性不育[14]。TaRTS蛋白结构分析发现, 其丙氨酸含量较高(图1-A), 其他植物中发现的一些功能未知的花药特异性表达蛋白也具有这一特征, 如水稻中的RA8蛋白[18], 拟南芥[19]和油菜花[20]中的Bcp1蛋白。拟南芥Bcp1基因, 在绒毡层和小孢子中都有表达, 也被证明是花粉发育所必需的, 其在二倍体绒毡层或单倍体小孢子中的低表达或不表达会导致花粉发育停滞[19]。本文研究发现TaRTS蛋白中富含半胱氨酸, 同家族蛋白也同样如此(图5)。半胱氨酸残基可以通过二硫键提供分子内和分子间的交联, 从而形成蛋白质的三级和四级结构, 表明该特征对蛋白质的功能的发挥具有重要作用。此外, RTS蛋白的N端存在较强的疏水结构域(图3), 这种结构域主要与蛋白靶向到膜部分或细胞外空间过程相关。这些特征也在其他一些花药特异性蛋白中被观察到, 如水稻的PS1[21]、玉米的Zmc13[22]和番茄的LAT52[23]。绒毡层主要参与孢粉素的合成和沉积进而释放到花粉中[24], 本研究中TaRTS蛋白被预测为分泌蛋白, 亚细胞定位到细胞质中(图4), 因而TaRTS蛋白可能被绒毡层细胞分泌到花粉中, 为花粉壁的形成提供必要的组分, 或者为花粉的生长发育提供营养。本研究克隆的基因TaRTS, 其开放阅读框(open reading frame, ORF) GC含量较高(72.7%)。Tsuchiya等[25]分离得到的花药特异性的水稻基因的ORF中显示出较高的GC含量(70.3%和69.6%)。ORF在其他花药特异性基因中是否存在GC含量高的现象尚不清楚。然而, 可以推测高GC含量提高植物对环境胁迫的耐受性, 因为GC之间的氢键比AT更稳定, 而花药是环境胁迫下最脆弱的器官之一。BS366属于光温敏型核雄性不育, 在低温短日照条件下不育, 高温长日下可育。本研究表明TaRTS的启动子序列同时存在光响应元件和低温响应元件(表1), 而lncRNA27195及TaRTS在不同光温处理下的表达模式显示, lncRNA27195及TaRTS的表达量存在显著性差异, 高温和长日照可以促进lncRNA27195与TaRTS基因的表达(图8), 可见其同BS366的育性转换存在着密切的联系。
茉莉酸甲酯(MeJA)广泛分布在植物中, 影响果实成熟、失活花粉的产生、根的生长、卷须卷曲、植物对伤害和非生物胁迫的反应, 以及对昆虫和病原体的防御等过程[26]。Mandaokor等[27]的研究显示, 控制拟南芥的雄蕊发育相关转录因子MYB21和MYB24受茉莉酸甲酯调控。在拟南芥dad1突变体中发现茉莉酸甲酯含量低, 花药开裂异常, 花丝伸长未能正常进行、花粉发育延迟, 导致雄性不育[28], 表明茉莉酸与花药育性直接相关。同时, 水杨酸(SA)和JA途径相互影响, SA的积累可以抑制JA的表达[29], WIPK蛋白的转录和表达可以激活JA的生成, 且抑制SA通路的信号传导[30]。本研究中利用MeJA和SA对抽穗期的小麦进行喷施处理的实验结果发现, 0.5 mmol L-1的MeJA可以促进lncRNA27195及TaRTS的表达, SA的喷施可以抑制MeJA的功能, 从而抑制lncRNA27195及TaRTS的表达, 表明了lncRNA27195及TaRTS受到MeJA介导的JA信号通路调控(图9)。
本研究通过系统进化树分析发现, RTS蛋白目前仅在禾本科植物中有报道, 其他的植物中无相关序列报道。其原因可能是由于, 禾本科的进化路线是由虫媒传粉向风媒传粉演化并且高度特化,花被和雄蕊简化, 因而推测RTS蛋白的出现是为了保证雄蕊的正常发育, 但其具体功能机制尚不可知[31]。此外, lncRNA27195与TaRTS基因均在雄蕊中特异性表达(图6), 且两者间存在显著的正相关关系(图7-B), 说明两者参与花粉的生长发育。另外lncRNA27195与TaRTS基因在可育环境的表达量二分体时期, 达到峰值后表达量下调; 在不育环境则是在小孢子母细胞时期表达量达到峰值, 之后表达量下调(图7-A), 表达峰值较正常植株提前一个时期。另外前人研究表明, 绒毡层程序性死亡的提前和推迟会导致花粉失活, 植株发生雄性不育[32,33]。光周期敏感基因雄性不育水稻(农研58)的花粉退化过程与绒毡层的异常行为密切相关, 光敏雄性不育(PGMS)植株较正常植株提前进行绒毡层程序性死亡, 但绒毡层退化进展缓慢, 直到花粉分解才完全退化[34]。本研究表明, TaRTS蛋白作为分泌蛋白, 其基因表达峰值较正常植株提前, 从而可能导致绒毡层提前降解, 导致花粉败育。
4 结论
lncRNA27195和其靶基因TaRTS是小麦花药特异性基因, 通过生物信息学分析及表达水平分析显示, 两者表达量呈正相关关系, 参与花粉育性调节, 为分泌蛋白, 且受MeJA调控, 可伴随绒毡层凋亡分泌到花粉中, 为花粉壁的形成提供必要的组分, 为花粉的生长发育提供营养。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
PMID [本文引用: 1]

We previously found that the SINE-encoded mouse B2 RNA binds RNA polymerase II and represses mRNA transcription during the cellular heat-shock response. In vitro B2 RNA assembles into preinitiation complexes on promoter DNA via its interaction with the polymerase, thus rendering the complexes inactive. With the goal of understanding which regions of B2 RNA are important for high-affinity binding to RNA polymerase II and repression of transcription, we performed a structural and deletion analysis of a 178 nucleotide (nt) B2 RNA. We describe an experimentally derived secondary structure model for B2 RNA, and show that RNA polymerase II protects a specific region from RNase digestion. Deletion studies revealed that a 51-nt region of B2 RNA is sufficient for high-affinity binding to RNA polymerase II, association with preinitiation complexes, and repression of transcription in vitro, the latter of which involves a large predominately single-stranded region within the RNA. In addition, this piece of B2 RNA blocked the polymerase from properly associating with template DNA during the assembly of elongation complexes. Further deletion revealed that a 33-nt piece of B2 RNA binds RNA polymerase II, associates with preinitiation complexes, but cannot repress transcription. These results support a model in which RNA polymerase II contains a high-affinity ncRNA docking site to which a distinct region of B2 RNA binds, thereby allowing another region of the RNA to repress transcription. Moreover, the mechanism of transcriptional repression by B2 RNA likely involves disrupting critical contacts between RNA polymerase II and the promoter DNA.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

Transcription in eukaryotic genomes generates an extensive array of non-protein-coding RNA, the functional significance of which is mostly unknown. We are investigating the link between non-coding RNA and chromatin regulation through analysis of FLC - a regulator of flowering time in Arabidopsis and a target of several chromatin pathways. Here we use an unbiased strategy to characterize non-coding transcripts of FLC and show that sense/antisense transcript levels correlate in a range of mutants and treatments, but change independently in cold-treated plants. Prolonged cold epigenetically silences FLC in a Polycomb-mediated process called vernalization. Our data indicate that upregulation of long non-coding antisense transcripts covering the entire FLC locus may be part of the cold-sensing mechanism. Induction of these antisense transcripts occurs earlier than, and is independent of, other vernalization markers and coincides with a reduction in sense transcription. We show that addition of the FLC antisense promoter sequences to a reporter gene is sufficient to confer cold-induced silencing of the reporter. Our data indicate that cold-induced FLC antisense transcripts have an early role in the epigenetic silencing of FLC, acting to silence FLC transcription transiently. Recruitment of the Polycomb machinery then confers the epigenetic memory. Antisense transcription events originating from 3' ends of genes might be a general mechanism to regulate the corresponding sense transcription in a condition/stage-dependent manner.
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 2]
[本文引用: 1]
PMID [本文引用: 1]

Plants respond to phosphate (Pi) deficiency through adaptive mechanisms involving several morphological, biochemical and molecular changes. In this study, we have characterized the structure and expression of a tomato (Lycopersicon esculentum L.) phosphate starvation-induced gene (TPSI1). A 3.5 kb genomic fragment containing the 474 bp TPSI1 transcript was isolated. The TPSI1 transcript contains an open reading frame of 174 nucleotides encoding a 58 amino acid polypeptide. TPSI1 is an intron-less gene and only one copy could be detected in the tomato genome. The promoter region of TPSI1 contains several conserved sequences found in phosphate starvation induced genes of yeast. The TPSI1 transcripts are rapidly induced in roots and leaves during Pi starvation. A significant increase in the TPSI1 mRNA was observed in cell cultures and roots after 3 and 12 h of Pi starvation, respectively. Induction of the TPSI1 gene appears to be a response specific to Pi starvation since it is not affected by starvation of other nutrients (nitrogen, potassium and iron). The amount of TPSI1 transcript decreased rapidly when Pi-starved tomato plants were resupplied with Pi. These results suggest that TPSI1 gene expression may be a part of the early Pi starvation response mechanism in plants.
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

An anther-specific cDNA clone of rice, RA8, was isolated from an anther cDNA library by differential screening. RNA blot analysis indicated that the RA8 transcript is present specifically in anthers and the transcript level increased as flowers matured, reaching the highest level in mature flowers. The RA8 clone contains an open reading frame of 264 amino acid residues with a hydrophobic N-terminal region. The deduced amino acid sequences did not show significant homology to any known sequences. Genomic DNA blot analysis showed that RA8 is a single-copy gene. A genomic clone corresponding to the RA8 cDNA was isolated and its promoter region was fused to the beta-glucuronidase (GUS) gene. Transgenic rice plants exhibited anther-specific expression of the GUS reporter gene. Histochemical GUS analysis showed that the RA8 promoter was active in the tapetum, endothecium, and connective tissues of anthers. Experiments showed that expression of the gene starts when microspores are released from tetrads, and it reaches to the maximum level at the late vacuolated-pollen stage. The RA8 promoter may be useful for controlling gene expression in anthers of cereal plants and for generating male-sterile plants.
DOIURL [本文引用: 2]
PMID [本文引用: 1]

Differential screening of a mature Brassica campestris pollen cDNA library has identified five cDNA clones that represent transcripts expressed exclusively, or at elevated levels, in pollen. We show here that the expression of one of these, clone Bcp1, is tissue specific and temporally regulated. The gene is activated during microspore development, as detected by in situ hybridization. Expression is enhanced at the time of pollen maturation and during pollen germination. In situ hybridization has also shown that Bcp 1 is activated in the tapetal cells in early anther development and continues to be expressed until tapetal dissolution. Homologous transcripts are present in pollen of other taxa of Brassicaceae including Arabidopsis, but not in pollen of any other families tested.
DOIURL [本文引用: 1]
PMID [本文引用: 1]

A pollen-specific cDNA clone, Zmc13, has been isolated from a cDNA library constructed to poly(A) RNA from mature maize pollen. The cDNA as shown by primer extension analysis is a full-length copy of the mRNA. The cDNA has been sequenced and is 929 nucleotides in length plus a 47-nucleotide poly(A) tail. Putative polyadenylation signals are identifiable in the 3'-nontranslated region. The mRNA codes for a predicted polypeptide containing 170 amino acid residues and with a molecular mass of 18.3 kilodaltons. The hydropathy profile suggests a possible signal sequence on the amino terminus. A comparison of the nucleotide and deduced amino acid sequence with sequences in data banks has not shown homology to known molecules. In situ hybridizations using RNA probes show that the mRNA is located in the cytoplasm of the vegetative cell of the pollen grain and after germination is distributed throughout the pollen tube cytoplasm.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

A cDNA library of rice (Oryza sativa L.) has been constructed from anthers at an early stage of pollen development. By differential screening of the library, we have isolated cDNAs of two genes, designated as Osc4 and Osc6, that are abundantly expressed in anthers containing tetrads and uninucleate microspores, but are not expressed in leaves or roots. Expression of Osc4 is absent in mature anthers, while Osc6 is present although the expression decays during pollen maturation. A comparison of the nucleotide and deduced amino acid sequences with those in data banks has not shown significant homology to known molecules.
[本文引用: 1]
PMID [本文引用: 1]

In Arabidopsis, jasmonate is required for stamen and pollen maturation. Mutants deficient in jasmonate synthesis, such as opr3, are male-sterile but become fertile when jasmonate is applied to developing flower buds. We have used ATH1 oligonucleotide arrays to follow gene expression in opr3 stamens for 22 h following jasmonate treatment. In these experiments, a total of 821 genes were specifically induced by jasmonate and 480 genes were repressed. Comparisons with data from previous studies indicate that these genes constitute a stamen-specific jasmonate transcriptome, with a large proportion (70%) of the genes expressed in the sporophytic tissue but not in the pollen. Bioinformatics tools allowed us to associate many of the induced genes with metabolic pathways that are probably upregulated during jasmonate-induced maturation. Our pathway analysis led to the identification of specific genes within larger families of homologues that apparently encode stamen-specific isozymes. Extensive additional analysis of our dataset identified 13 transcription factors that may be key regulators of the stamen maturation processes triggered by jasmonate. Two of these transcription factors, MYB21 and MYB24, are the only members of subgroup 19 of the R2R3 family of MYB proteins. A myb21 mutant obtained by reverse genetics exhibited shorter anther filaments, delayed anther dehiscence and greatly reduced male fertility. A myb24 mutant was phenotypically wild-type, but production of a myb21myb24 double mutant indicated that introduction of the myb24 mutation exacerbated all three aspects of the myb21 phenotype. Exogenous jasmonate could not restore fertility to myb21 or myb21myb24 mutant plants. Together with the data from transcriptional profiling, these results indicate that MYB21 and MYB24 are induced by jasmonate and mediate important aspects of the jasmonate response during stamen development.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
PMID [本文引用: 1]

A gene encoding a tobacco mitogen-activated protein kinase (WIPK) is transcriptionally activated in response to wounding. Transgenic tobacco plants, in which expression of endogenous wipk was suppressed, did not accumulate jasmonic acid or its methyl ester when wounded, suggesting that WIPK is involved in jasmonate-mediated wound signal transduction. Here, we demonstrate that activation of WIPK is required for triggering the jasmonate-mediated signal transduction cascade that occurs when wild-type tobacco plants are wounded. We also show that when plants are wounded, WIPK is rapidly and transiently activated, whereas the quantity of WIPK protein is maintained at a constant level. A transgenic tobacco plant in which the wipk gene was constitutively expressed at a high level showed constitutive enzymatic activation of WIPK and exhibited three- to fourfold higher levels of jasmonate than did its wild-type counterpart. This plant also showed constitutive accumulation of jasmonate-inducible proteinase inhibitor II transcripts. These results show that WIPK is activated in response to wounding, which subsequently causes an increase in jasmonate synthesis.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
