 ,**, 刘浩
,**, 刘浩 ,**, 洪彦彬, 梁炫强, 陈小平
,**, 洪彦彬, 梁炫强, 陈小平 ,*广东省农作物遗传改良重点实验室 / 国家油料作物改良中心南方分中心 / 广东省农业科学院作物研究所, 广东广州 510640
,*广东省农作物遗传改良重点实验室 / 国家油料作物改良中心南方分中心 / 广东省农业科学院作物研究所, 广东广州 510640Identification and expression analysis of microRNA during peanut (Arachis hypogaea L.) pod development
HU Dong-Xiu ,**, LIU Hao
,**, LIU Hao ,**, HONG Yan-Bin, LIANG Xuan-Qiang, CHEN Xiao-Ping
,**, HONG Yan-Bin, LIANG Xuan-Qiang, CHEN Xiao-Ping ,*Guangdong Provincial Key Laboratory of Crop Genetic Improvement / South China Peanut Sub-Center of National Center of Oilseed Crops Improvement / Crops Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, Guangdong, China
,*Guangdong Provincial Key Laboratory of Crop Genetic Improvement / South China Peanut Sub-Center of National Center of Oilseed Crops Improvement / Crops Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, Guangdong, China通讯作者:
收稿日期:2020-07-2接受日期:2020-10-14网络出版日期:2021-04-12
| 基金资助: |
First author contact:
Received:2020-07-2Accepted:2020-10-14Online:2021-04-12
| Fund supported: |
作者简介 About authors
胡冬秀, E-mail:
刘浩, E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (5437KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
胡冬秀, 刘浩, 洪彦彬, 梁炫强, 陈小平. 花生荚果发育过程中的microRNA鉴定与表达分析[J]. 作物学报, 2021, 47(4): 613-625. doi:10.3724/SP.J.1006.2021.04144
HU Dong-Xiu, LIU Hao, HONG Yan-Bin, LIANG Xuan-Qiang, CHEN Xiao-Ping.
花生作为“地上开花、地下结果”的作物, 其生殖发育过程与其他作物存在差异。花生开花受精后, 子房柄伸长并向地生长形成果针, 位于其顶端的受精胚珠随着果针的伸长被推入土壤中, 果针停止伸长, 胚胎发育在黑暗条件下重新启动, 最终膨大形成荚果。花生荚果发育是一个复杂的生物学过程, 不同水平的调控对花生荚果种子发育均至关重要。研究microRNA如何调控黑暗条件下花生种子发育, 对理解植物果实暗发育调控与作物遗传改良具有重要意义。
microRNA是一类内源性非编码小分子RNA, 在植物中通常由18~24个核苷酸组成, 通过靶向降解或翻译抑制在转录后水平调控基因的表达[1]。植物中非编码的microRNA基因, 在NOT2和CDC5等转录因子的作用下通过RNA聚合酶II转录生成具有茎环结构的初级转录物Pri-microRNA (Primary microRNA)。随后Pri-microRNA经DCL1、HY1和SE组成的切割复合体剪切产生前体pre-microRNA (microRNA前体)[2], pre-microRNA经DCL1与DCL4进一步剪切生成成熟的microRNA[3], 在甲基转移酶HEN1的作用下使其3'端发生2'-O-甲基化来增强其稳定性[4,5]。microRNA在转运蛋白HST作用下从细胞核进入细胞质中, 随后与AGO效应蛋白(argonaute proteins, AGOs)结合形成RNA介导的沉默复合体(RNA-induced silencing complexes, RISCs)中, 结合互补的靶向基因信使RNA后使其被剪切降解[6]。microRNA作为一种重要的转录后调控因子, 参与植物的生长发育、信号转导、胁迫响应、器官分化以及病原菌调控等多种生物学过程[7,8,9,10,11,12,13,14]。拟南芥中, FUL蛋白与ARF (auxin response factor)相互作用并结合到miR172前体序列的启动子区域, 通过抑制miR172下游靶基因AP2与TOE3表达进而促进果实发育[15]; 人为干预miR319的表达活性则导致拟南芥花瓣变短、雄蕊败育、种子缺失等现象[16]。单子叶植物中, 水稻OsIDD2与SLR1相互作用, 通过赤霉素途径调控miR396介导的细胞增殖过程[17]; 而OsmiR408剪切OsUCL8编码序列会影响细胞中的铜离子稳态、质体蓝素丰度与水稻光合效率, 从而增加穗部分枝和粒数[18]。花生受到黄曲霉侵染时, 种子通过降低ayh-miRNA160、ayh-miRNA164对抗病基因TIR-NBS-LRR表达量产生的负调控效应, 从而提高抗病蛋白的表达量以抵御黄曲霉污染[19]。花生子房柄中的ayh-miR160a、ayh-miR171n与ayh-miR156e通过调控其靶基因参与激素信号转导途径, 在花生荚果、种子膨大阶段起关键作用[20]。
花生(Arachis hypogaea L.)是我国重要的油料与经济作物, 荚果暗发育是花生最显著的特征之一, 已有相关研究报道非编码小RNA参与了花生胚胎与荚果早期的发育[21,22], 但对其荚果种子发育的整个过程缺乏全面的了解, 尤其对microRNA方面的相关研究仍然较为欠缺。本研究以‘粤油7号’为试验材料, 对其荚果发育不同时期的材料进行small RNA测序, 并利用qRT-PCR技术验证测序的结果, 鉴定和筛选出花生荚果种子发育过程中相关的microRNA及其在花生荚果种子发育过程中的表达模式。
1 材料与方法
1.1 试验材料
花生品种粤油7号种植于广东省农业科学院白云基地。对粤油7号植株的自交花进行鉴定并用彩色线标记, 在花后第8天用彩色标签系好伸长的果针。花生果针入土前, 被标记为第1个时期, 命名为P0 (Pod 0); 荚果开始膨大后通过测量荚果与种子的直径大小, 将整个荚果发育划分为10个时期, 即P1~P10 (Pod1~Pod10), 其中果壳与种子分开取样。果壳样品以缩写SH (Shell)表示, 种子样本以缩写SD (Seed)表示。本研究共对11个时期, 包括1个地上(果针即将入土)和10个地下发育阶段(图1-A), 以开花后天数为参考, 荚果与种子直径为主要指标来划分生育期, 最后获得20份样品。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1花生荚果发育的表型特征
A: 试验样品信息; B: 花生荚果种子发育的表型特征。P0: 地上果针; P1: 地下果针; P2~P5: 果壳膨大期; P6~P9: 果实充实期; P9~P10: 成熟期。
Fig. 1Phenotype characteristics of pod development in peanut
A: summary of experimental samples; B: phenotype characteristics of peanut pod (shell and seed) development. P0: aerial peg; P1: subterranean peg; P2-P5: pod expansion stage; P6-P9: seed filling stage; P9-P10: mature stage.
1.2 RNA的提取及small RNA测序
利用TRIzol (Invitrogen)法提取20个样品的总RNA, 使用NanoDrop (Thermo Scientific)和安捷伦2100生物分析仪(安捷伦)对每个样品的RNA质量进行评估分析; 利用Illumina small RNA-seq文库构建试剂盒制备small RNA测序文库, 利用Illumina HiSeq 2000平台产生原始测序数据。1.3 small RNA测序结果分析
利用Fastx-Tool kit, 对small RNA序列的原始数据进行过滤筛选, 去除接头序列、低质量序列, 包括冗余碱基或未被识别的碱基冗余序列, 以及小于18 nt的序列。将过滤后的数据利用Rfam 11.0数据库和GenBank非编码RNA数据库去除t/r/sn/snoRNA以及带有ployA尾巴的RNA, 对其他的small RNA序列利用microRNA19.0数据库进行BLASTn搜索, 以检索花生中已知的microRNA。此外, 利用Bowtie将small RNA序列映射到GenBank中的AHGI v2.0和花生GSS中进行比对, 完全匹配的序列将用于进一步分析。如果接近完全匹配的序列小于检索的small RNA或已知small RNA的长度, 则人工挑选检查与比较不匹配部分, 以确定匹配核苷酸的数量。为了有效识别花生中潜在的microRNA前体序列和新的microRNA, 用miREAP预测软件分析microRNA前体序列, 利用MiPred对由此产生的茎环结构进行过滤, 剩余的pre-microRNA的二级发夹结构使用CentroldFold软件进行评估。1.4 花生荚果发育过程中microRNA趋势分析
microRNA测序数据以RPKM (reads per kilobase per million mapped reads)值表示, 将表达量数据归一化分析。差异表达的microRNA以RPKM > 10为筛选标准, 以此数据为基础利用TBtools软件绘制表达热图, 利用STEM软件对花生荚果发育过程中的microRNA和靶基因进行趋势分析, 并获得显著性和非显著性表达模式, 其中P≤0.05的定义为显著表达模式。1.5 茎环RT-PCR荧光定量检测microRNA
取5 μg总RNA加入DNase I去除基因组DNA污染, 利用TaKaRa SMART Scribea Reverse Transcriptase反转录酶对microRNA进行反转录。取2 μg的总RNA样品中加入终浓度为2 μmol L-1的microRNA的茎环引物, 将混合物在72℃预热3 min后置于冰上冷却使RNA变性, 打开二级结构。随后加5×First Strand Buffer、dNTP Mix、20 mmol L-1 DTT、0.15 μL RRI (recombinant RNase inhibitor, 40 U μL-1)。加入1 mL (100 U μL-1) SMART Scribe RT再次混匀, 42℃孵育60 min, 反应结束后70℃加热15 min或加入4 μL 50 mmol L-1 EDTA终止反应。荧光定量PCR方法参照试剂AceQ qPCR SYBR Green Master Mix说明书, 上述反转的cDNA样品浓度稀释为150 ng μL-1。荧光定量PCR的反应体系为20 μL, 包含10 μL SYBR Green Master Mix、2 μL模板、特异性正向引物和通用反向引物各0.4 μL、0.4 μL ROX Reference Dye I、6.8 μL的ddH2O。ABI Step OnePlus仪器检测, 循环反应为95℃ 10 s, 60℃ 30 s, 共40个循环; U6为内参基因, 用2-ΔΔCT法计算microRNA表达水平的相对定量, 每个样品设置3次生物学重复。1.6 Quantative real time-PCR检测靶基因表达量
利用Prime Script RT试剂盒(TaKaRa, 中国大连)将1 μg总RNA反向转录为cDNA, 反应体系为20 μL, 采用ABI Step One Plus系统, 使用SYBR Premix ExTaq (TaKaRa, 中国大连)进行PCR反应, 用2-ΔΔCT法计算目标基因各组织的相对表达量, 以P0作为对照样品。18S rRNA作为内参基因, 每个样品设置3个重复, 数据用平均值±标准差(Mean± SD)表示。2 结果与分析
2.1 花生荚果种子发育的表型特征
花生开花受精后, 子房柄不断伸长形成果针(P0)。随着果针的伸长, 位于其顶端的受精胚珠进入土壤中, 当果针刚进入地下时(P1), 其直径并未发生变化(图1-B)。在黑暗条件下, 果针停止伸长, 顶端迅速膨大形成荚果(P2~P6)。通过测量直径, 果壳在P6时期的直径大小是P2时期的5~6倍, 但是P6阶段过后, 果壳基本不再继续膨大, 因此进入土壤后的第6个时期是果壳发育逐渐停止的时期。种子发育在P2~P6阶段生长缓慢, 在P6期果壳不再发育之后, 种子则以较快的速度开始充实, 其直径从P6到最后的成熟期, 至少增大了3倍以上。表明, 果壳早于种子开始发育, 果壳的提前膨大为后续种子生长提供了空间和能量, 两者间发育的时间差有利于能量物质有效地供应种子生长。2.2 花生荚果发育过程中small RNA分析
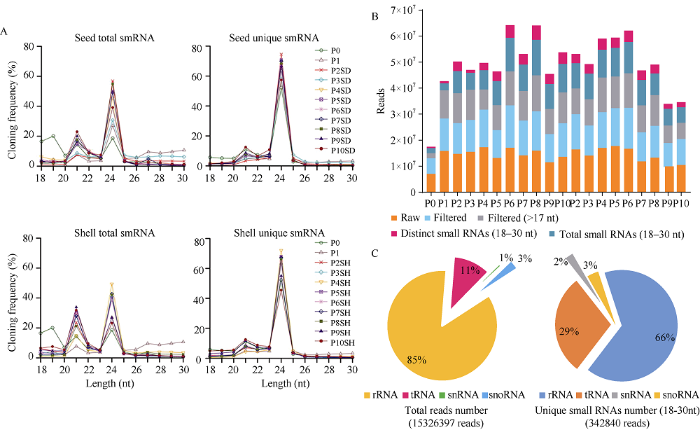
对花生果壳和种子发育的20个样品进行small RNA测序, 共获得282,960,633个reads, 每个文库的reads数量从7,049,655到17,761,513。对接头序列和短序列进行修剪后, 共得到206,940,419个序列(图2-B), 长度大于17 nt。其中长度在18~30 nt的有177,581,859条, 有38,699,203条为花生中的特异small RNA。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2花生荚果small RNA测序结果的分析
A: 全部以及特异性small RNA的长度分布和频率; B: small RNA统计分析; C: small RNA的分类。P0: 地上果针; P1: 地下果针; P2~P5: 果壳膨大期; P6~P9: 果实充实期; P9~P10: 成熟期。
Fig. 2Results of small RNAs sequencing analysis in peanut pod
A: length distribution and frequency of total and unique small RNAs; B: summary statistics of small RNAs; C: the categories of peanut small RNAs. P0: aerial peg; P1: subterranean peg; P2-P5: pod expansion stage; P6-P9: seed filling stage; P9-P10: mature stage.
microRNA属于微小的非编码RNA, 在转录后水平的基因调控中具有重要作用。为全面了解荚果发育过程中microRNA的全基因组信息, 从花生荚果种子发育过程中鉴定了长度为18~30 nt的small RNA序列(图2-A), 在21个和24个核苷酸处观察到了花生地下荚果种子发育(P1~P10)的2个主要高峰。果壳文库(P2SH~P10SH)有2个接近的高峰, 而种子文库(P2SD~P10SD)则以24个核苷酸small RNA为主。值得注意的是, 气生荚果P0中19个核苷酸处出现1个额外的高峰, 进一步分析发现, 这19个核苷酸序列中的大多数来自于几个高度表达的microRNA。此外, 在最初的地下荚果P1中, 总的small RNA在27~30个核苷酸之间高度表达, 但24个核苷酸small RNA在所有样本中均占优势, 在种子和果壳的总small RNA中分别占74%和72%, 表明花生的small RNA是类型复杂且多样的非编码RNA。
2.3 花生荚果发育过程中small RNA的鉴定
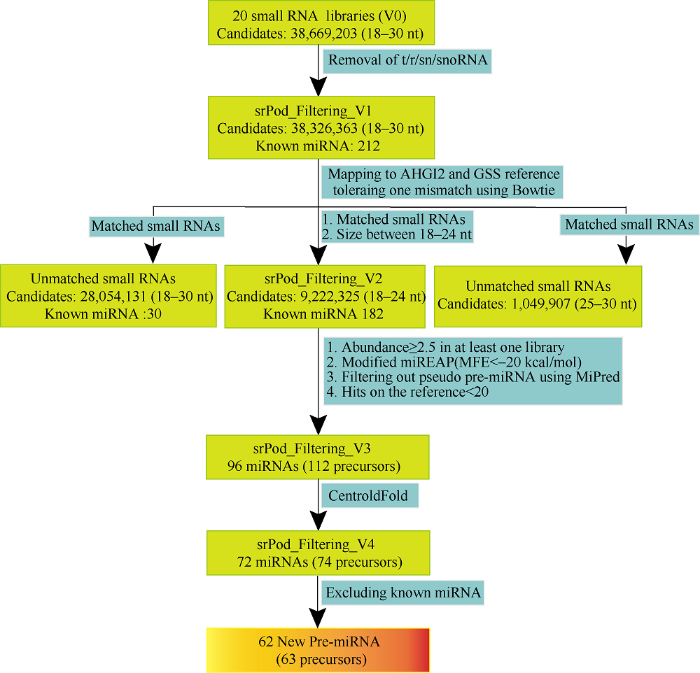
为进一步鉴定花生中保守的和新的特异性microRNA, 我们通过图3所示的策略分析small RNA。对接头和短序列进行过滤后, 共得到38,699,203条花生的特异small RNA, 随后去除t/r/sn/snoRNA共15,326,397条, 其中rRNA的数量最多, 其次是tRNA、snRNA和snoRNA数量相对较少(图2-C)。得到38,326,363条候选的small RNA, 鉴定到212个已知的microRNA, 包括39个microRNA家族的197个保守的microRNA和13个microRNA家族的15个特异microRNA (附表1)。经过逐步筛选, 本研究还获得62个新的microRNA二级前体结构以及其对应生成的112个新的成熟microRNA。将前体命名为ahy-MIR-s (1~62)。利用RNAfold在线软件(图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3microRNA分析流程
Fig. 3Pipeline of microRNA data analysis
图4
 新窗口打开|下载原图ZIP|生成PPT
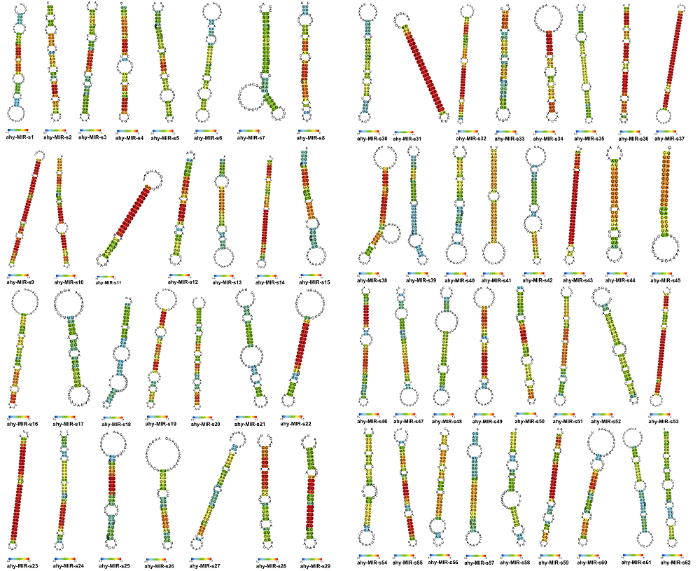
新窗口打开|下载原图ZIP|生成PPT图4新的Pre-microRNA二级结构分析
Fig. 4Secondary structure of new Pre-microRNAs in peanut
Table S1
附表1
附表1花生荚果发育过程中新的和已知的microRNAs鉴定
Table S1
| 编号 ID | 序列 Sequence | 长度 Length(nt) | 编号 ID | 序列 Sequence | 长度 Length(nt) |
|---|---|---|---|---|---|
| New microRNAs | |||||
| ahy-miR-s1-3p | UGUAGAAUUUCUGCUCCUUGG | 21 | ahy-miR-s33-3p | UGUUCAUAGCGGUGCUUGCUG | 21 |
| ahy-miR-s1-5p | UAGGAUGGCAGAUAUUCUUUUCA | 23 | ahy-miR-s33-5p | CAGUGUGGCCGGACUGGAUCAUC | 23 |
| ahy-miR-s2-3p | UCCCUUUGGAUGUCGUCGUGC | 21 | ahy-miR-s34-3p | AUUGCCUACUGAUUGCAUCCCACA | 24 |
| ahy-miR-s2-5p | ACUUCUGACAGACGUAGGGAAG | 22 | ahy-miR-s34-5p | AGGGAUGCUAUCCACAGUCGAUAG | 24 |
| ahy-miR-s3-3p | UUCAUGACAGAGUGCUGGCUC | 21 | ahy-miR-s35-3p | UAACGAGUCGAGCUCGAGCUAGCU | 24 |
| ahy-miR-s3-5p | AAGGUCUCUGUCUCUGUGGU | 20 | ahy-miR-s35-5p | UUUAGGCUCAAGCUUGGCUCACCA | 24 |
| ahy-miR-s4-3p | AACCAAAUGAUGAACCAUUCU | 21 | ahy-miR-s36-3p | UCAGUGUUGCCAGGCGCCUGGG | 22 |
| ahy-miR-s4-5p | AAUGUUGAAGAAUUAGGUUGC | 21 | ahy-miR-s36-5p | CAACGUCUGCCACCACUGAUG | 21 |
| ahy-miR-s5-5p | CAACAGCACCCUCCACCUGAG | 21 | ahy-miR-s37-3p | UUGCGUUGGAGAUGUUGUCCAAUG | 24 |
| ahy-miR-s6-5p | AGCGAUUGUUUCUCUUGAUGCAGU | 24 | ahy-miR-s37-5p | AUUGGACAACAUCUCCAACGUAAC | 24 |
| ahy-miR-s7-3p | GAUGAAUGCAGCUCCCUAUAA | 21 | ahy-miR-s38-3p | UUAAAGUCUGGCCUGACCUAGCCU | 24 |
| ahy-miR-s7-5p | AUGGGGCUGGAGUUUGGCUC | 20 | ahy-miR-s38-5p | GCUACAGGCCAGGCUUAGACC | 21 |
| ahy-miR-s8-3p | AGGGGUGAUCUGCCUAGUC | 19 | ahy-miR-s39-3p | UGAAGGAAGUAGUGGCGACCC | 21 |
| ahy-miR-s8-5p | UGAGGCACGCACUCCAGAUGA | 21 | ahy-miR-s40-3p | UUGAGGACUUGAUAUAUGUCG | 21 |
| ahy-miR-s9-3p | CGUGCGCGGUCUUGCACUGCU | 21 | ahy-miR-s41-3p | GUUAGUAAUUCGAACCACCCUGGU | 24 |
| ahy-miR-s9-5p | CAAGUCGUGCACGGCUGGCCU | 21 | ahy-miR-s41-5p | CACCCUGGUUCGAAUUACGCUUAA | 24 |
| ahy-miR-s10-5p | UUGGAAAUGUAUGGCGAGCCU | 21 | ahy-miR-s42-3p | AGAUAUACUUGUUACGCUUG | 20 |
| ahy-miR-s11-3p | UUUGAGAUUGGAUAAUUGCCU | 21 | ahy-miR-s42-5p | CGGCAGUGAUGAGGCUCUAACAUG | 24 |
| ahy-miR-s12-3p | UUGGAAAAGGUCGGUAGAAAG | 21 | ahy-miR-s43-3p | UGGUAAGAGUUUGAUUAAGUA | 21 |
| ahy-miR-s12-5p | UUCACCGACUCUGUUCAGACU | 21 | ahy-miR-s43-5p | CUUAAUCAAACUCUUAUCAUC | 21 |
| ahy-miR-s13-5p | UUCAGAGGCUUGUUUUAUCUC | 21 | ahy-miR-s44-3p | UACUGAAAUCAGAUCAUCCGAUUA | 24 |
| ahy-miR-s14-3p | GCAAACAGGUUCGGUGAAAAC | 21 | ahy-miR-s44-5p | AUCGGAUGGUCCGAUUUGUGUACC | 24 |
| ahy-miR-s14-5p | UUUACCAGAUCCUGUUUGUUC | 21 | ahy-miR-s45-3p | UCUAACCGAACCGAACCGAAC | 21 |
| ahy-miR-s15-3p | UUGUCUCUAAGUGGUGGUUCCA | 22 | ahy-miR-s45-5p | UCGGUUCGGUUUUCGGUUCUGACC | 24 |
| ahy-miR-s15-5p | GAACUGUUCUUCUUAGUUGCGCCU | 24 | ahy-miR-s46-3p | UGAAGAUUGUUGGGUAGUGUU | 21 |
| ahy-miR-s16-3p | CAGCUGUUUGCGGACAAGUCUGGC | 24 | ahy-miR-s46-5p | AACAUGGUGAAAGGUUUUCAUU | 22 |
| ahy-miR-s16-5p | AGACUGUGCGCCGAUAUGCCCGUG | 24 | ahy-miR-s47-3p | UUGAUAUGGUGUAUUCUGCAGCCU | 24 |
| ahy-miR-s17-5p | UUUGGCAGCUCAAUCACUUGG | 21 | ahy-miR-s47-5p | GUUGUUCAGGUGUAUUAUAUCAGA | 24 |
| ahy-miR-s18-3p | CACUUCUUAUGCUUGUUGGAG | 21 | ahy-miR-s48-3p | AUUUUUAAUCGAUUGUUUUGUGAU | 24 |
| ahy-miR-s18-5p | CGCAUGGAUGGAGAAUCUCGCU | 22 | ahy-miR-s48-5p | GACCAAUCGAUUGAUUUUUG | 20 |
| ahy-miR-s19-5p | UCUUGAAAGCUGAGCAUUGGC | 21 | ahy-miR-s49-3p | CCAACGUUGGAGCAAAAGUUAGA | 23 |
| ahy-miR-s20-3p | ACCGGACUUGCGCCCCAGUCGUA | 23 | ahy-miR-s49-5p | AACUUUUUGGCUAACGUUGGCGCC | 24 |
| ahy-miR-s20-5p | UCACAGGCAGCAUAUCAGGUUG | 22 | ahy-miR-s50-3p | UCCAAGAUGUGGCAUAUCAGUUG | 23 |
| ahy-miR-s21-5p | ACGAAACUGUGUGAAGUAAGGCCU | 24 | ahy-miR-s50-5p | UUUCAUAUGCCGUUCAGAGAGU | 22 |
| ahy-miR-s22-3p | UUAAAUCAGUCUGUGGUACCC | 21 | ahy-miR-s51-3p | UUCUCGGAUUAAUCAGGCUCAGCC | 24 |
| ahy-miR-s22-5p | CCGCGGAUUGAUUUUUACCAGA | 22 | ahy-miR-s51-5p | CUGCCUGUUUAGUUCGCGGACU | 22 |
| ahy-miR-s23-3p | UUGAUUGAGUGACUGUAUAGCU | 22 | ahy-miR-s52-5p | UCAGUGUUUGCUCGCCUCUUC | 21 |
| ahy-miR-s23-5p | CUAUGAGUUACUCAAUCAAAC | 21 | ahy-miR-s53-3p | CUGGAGGCCUUUGAAGGAGAC | 21 |
| ahy-miR-s24-3p | UGGCUUGGUCUGACGUGACUCAGC | 24 | ahy-miR-s53-5p | UCCCUCAAAGGCUUCCAGUA | 20 |
| ahy-miR-s24-5p | AUGAGUCGAGUCGUGAGCCAAGCU | 24 | ahy-miR-s54-3p | AACCGACACGUGAGCUCAUGGCCA | 24 |
| ahy-miR-s25-3p | UUUUCCUCUUAAUGGAUCCUCUGU | 24 | ahy-miR-s54-5p | UUAGCUAUCGGAUGUGUCGAGUCU | 24 |
| ahy-miR-s25-5p | AGAGAGGAUCCUUAUCUGGGAACU | 24 | ahy-miR-s55-3p | UUUGAAACAGAAUCAGUUGGGAUC | 24 |
| ahy-miR-s26-3p | UCCUUGGACCUUGGGUGUGAU | 21 | ahy-miR-s55-5p | UUGGUGACUGAUUUGUGAAUCAAA | 24 |
| ahy-miR-s26-5p | AGGACCCAACUCUCUUGAAGGAGA | 24 | ahy-miR-s56-3p | UUCUGCGCGGUCGCGUCGCUGACG | 24 |
| ahy-miR-s27-3p | UGAGGGGGUUGAGGGUGCUGGGGU | 24 | ahy-miR-s56-5p | UGAGCCAUGAGACCGCGUCACUGC | 24 |
| ahy-miR-s27-5p | UUCAGGAUCUCUAUUACUGGC | 21 | ahy-miR-s57-5p | UUGAGUACCGUCGGAUUUAUCAUC | 24 |
| ahy-miR-s28-3p | GUUACUGAUUUACUGGUUCACG | 22 | ahy-miR-s58-3p | ACUUUAGUGGCUGUUCGCUCACUU | 24 |
| ahy-miR-s28-5p | AGAACCGGACCGGUCAAUAAACCA | 24 | ahy-miR-s58-5p | AUAGAGAGUAGGCCAGUAGAAGCC | 24 |
| ahy-miR-s29-3p | UUCUCCCUUGGUAGUGGCGAAGC | 23 | ahy-miR-s59-3p | UUGUUGUGGCGCCAACGUUUGCCU | 24 |
| ahy-miR-s29-5p | CAUCGCCAACUCCAAGGAAG | 20 | ahy-miR-s59-5p | CAAACGUUGGCGCAAGCUUUUGCU | 24 |
| ahy-miR-s30-3p | GGAAGAUUGUUGGUUAGUGUU | 21 | ahy-miR-s60-3p | AGGAGCUCUGCUGUGUCUUGAUGG | 24 |
| ahy-miR-s30-5p | GACUAAUGACAUUCAACCUC | 20 | ahy-miR-s60-5p | UUCCACGGCAUGACUCUCUAAACC | 24 |
| ahy-miR-s31-3p | AAUUUAGACAAUUCAUCCGAU | 21 | ahy-miR-s61-3p | ACAGUGAGGUUUGUAAGAAAAAGC | 24 |
| ahy-miR-s31-5p | AAAUCGGAUGAAUUGUCUAAAUUC | 24 | ahy-miR-s61-5p | UUUUUCUGUUUAAUUUUGUCU | 21 |
| ahy-miR-s32-3p | AAUAAAACAAGUUUUGACUGU | 21 | ahy-miR-s62-3p | UCAAACGAGGAAAGGCUUAUGG | 22 |
| ahy-miR-s32-5p | AGCUCAAAUUUGCCUUAUUUA | 21 | ahy-miR-s62-5p | AUCUAGCAGCACCUUAGGAUGGCA | 24 |
| Known and Conserved microRNAs | |||||
| ahy-miR156b-5p | UUGACAGAAGAUAGAGAGCAC | 21 | ahy-miR168d | UCGCUUGGUGCAGGUCGGGAC | 21 |
| ahy-miR156b-3p | GCUCUCUAAGCUUCUGUCAUC | 21 | ahy-miR169f | CAGCCAAGGAUGACUUGCCGG | 21 |
| ahy-miR156c | UUGACAGAAGAGAGAGAGCAC | 21 | ahy-miR169h | UGAGCCAAGGAUGGCUUGCCG | 21 |
| ahy-miR156d | UGACAGAAGAAAGUGAGCAC | 20 | ahy-miR169l | CAGCCAAGAAUGACUUGCCGG | 21 |
| ahy-miR156e | UGACAGAAGAGAGUGAGCACA | 21 | ahy-miR169p | UGAGCCAAGGAUGACUUGCCG | 21 |
| ahy-miR156f | UGACAGAAGAGAAUGAGCAC | 20 | ahy-miR169r | UAGCCAAGGAUGACUUGCCU | 20 |
| ahy-miR156g | UGACAGAAGAGAGGGAGCAC | 20 | ahy-miR169s | AAGCCAAGGAUGACUUGCCGG | 21 |
| ahy-miR156h | UGACAGAAGAAAGAGAGCAC | 20 | ahy-miR171a | GGAUAUUGGUACGGUUCAAUC | 21 |
| ahy-miR156i | ACAGAAGAUAGAGAGCACAG | 20 | ahy-miR171b | UUGAGCCGUGCCAAUAUCACG | 21 |
| ahy-miR156j | UGACAGAAGAGGGUGAGCAC | 20 | ahy-miR171c | UAUUGGUGCGGUUCAAUGAGA | 21 |
| ahy-miR156k | UGUGCUCACUCUCUUCUGUCA | 21 | ahy-miR171d | UUGAGCCGUGCCAAUAUCACU | 21 |
| ahy-miR156l | UGUCAGAAGAGAGUGAGCAC | 20 | ahy-miR171e | UGAGCCGUGCCAAUAUCACAU | 21 |
| ahy-miR156m | CGACAGAAGAGAGUGAGCAC | 20 | ahy-miR171f | UGAUUGAGCCGUGCCAAUAUC | 21 |
| ahy-miR156n | UGACAGAGGAGAGUGAGCAC | 20 | ahy-miR171g | CGAUGUUGGUGAGGUUCAAUC | 21 |
| ahy-miR156o | UGACAGAAGAGAGCGAGCAC | 20 | ahy-miR171h | UGAUUGAGCCGCGUCAAUAUC | 21 |
| ahy-miR156q | UGACAGAAGAGAGUGAGCACU | 21 | ahy-miR171i | CGAGCCGAAUCAAUAUCACUC | 21 |
| ahy-miR156s | CUGACAGAAGAUAGAGAGCAC | 21 | ahy-miR171j | UAUUGGCCUGGUUCACUCAGA | 21 |
| ahy-miR157b | GCUCUCUAAGCUUCUGUCAUCA | 22 | ahy-miR171k | UUGAGCCGCGCCAAUAUCACA | 21 |
| ahy-miR157d | UGACAGAAGAUAGAGAGCAC | 20 | ahy-miR171l | UUGAGCCGCGCCAAUAUCACU | 21 |
| ahy-miR159a | UUUGGAUUGAAGGGAGCUCUG | 21 | ahy-miR171n | UUGAGCCGUGCCAAUAUCACA | 21 |
| ahy-miR159b | UUUGGAUUGAAGGGAGCUCUU | 21 | ahy-miR171p | UUGAGCCGCGUCAAUAUCUUA | 21 |
| ahy-miR159c | UUUGGAUUGAAGGGAGCUCCU | 21 | ahy-miR171t | UUGAGCCGCGUCAAUAUCUCA | 21 |
| ahy-miR159d | AGCUGCUUAGCUAUGGAUCCC | 21 | ahy-miR172a | AGAAUCUUGAUGAUGCUGCAU | 21 |
| ahy-miR159e | UUUGGAUUGAAAGGAGCUCUU | 21 | ahy-miR172b | GGAGCAUCAUCAAGAUUCACA | 21 |
| ahy-miR159f | CUUGGAUUGAAGGGAGCUCUA | 21 | ahy-miR172c | GUAGCAUCAUCAAGAUUCACA | 21 |
| ahy-miR159g | UUGGAUUGAAGGGAGCUCCA | 20 | ahy-miR172d | AGAAUCUUGAUGAUGCUGCAG | 21 |
| ahy-miR159h | CUUGGAUUGAAGGGAGCUCU | 20 | ahy-miR172k | UGAAUCUUGAUGAUGCUGCAU | 21 |
| ahy-miR159i | UUUGGACUGAAGGGAGCUCUA | 21 | ahy-miR390a-5p | AAGCUCAGGAGGGAUAGCGCC | 21 |
| ahy-miR159j | AUUGGAGUGAAGGGAGCUCCA | 21 | ahy-miR390b | CGCUAUCCAUCCUGAGUUUCA | 21 |
| ahy-miR159k | UUUGGUUUGAAGGGAGCUCUA | 21 | ahy-miR390c | CGCUAUCCAUCCUGAGUUUC | 20 |
| ahy-miR159l | AUUGGAUUGAAGGGAGCUCCU | 21 | ahy-miR390d | AAGCUCAGGAGGGAUAGCACC | 21 |
| ahy-miR159m | AUUGGAUUGAAGGGAGCUCCA | 21 | ahy-miR390e | AAGCUCAGGAGGGAUAGCGUC | 21 |
| ahy-miR159n | CUUGGAUUGAAGGGAGCUCCC | 21 | ahy-miR390f | GAGCUCAGGAGGGAUAGCGCC | 21 |
| ahy-miR159 | UUUGGAUUGAAGGGAGCUCUA | 21 | ahy-miR390g | CGCUAUCCAUCCUGAGUUCCA | 21 |
| ahy-miR319a | UUGGACUGAAGGGAGCUCCC | 20 | ahy-miR393a | UCCAAAGGGAUCGCAUUGAUCC | 22 |
| ahy-miR319b | UUGGACUGAAGGGAGCUCCCU | 21 | ahy-miR393b | UCCAAAGGGAUCGCAUUGAUCU | 22 |
| ahy-miR319c | UUUGGACUGAAGGGAGCUCCU | 21 | ahy-miR393c | UCCAAAGGGAUCGCAUUGAUC | 21 |
| ahy-miR319d | UUGGACUGAAGGGAGCUCCU | 20 | ahy-miR393d | AUCAUGCUAUCCCUUUGGAUU | 21 |
| ahy-miR319e | CUUGGACUGAAGGGAGCUCCC | 21 | ahy-miR393e | AUCAUGCUAUCUCUUUGGAUU | 21 |
| ahy-miR319f | UUGGACUGAAGGGGCCUCUU | 20 | ahy-miR393k | UUCCAAAGGGAUCGCAUUGAUC | 22 |
| ahy-miR319g | UUGGACUGAAGGGAGCUCCCA | 21 | ahy-miR394 | UUGGCAUUCUGUCCACCUCC | 20 |
| ahy-miR319h | UUGGACUGAAGGGUGCUCCCU | 21 | ahy-miR394c | UUGGCAUUCUGUCCACCUCCAU | 22 |
| ahy-miR319i | UUGGACUGAAGGGGAGCUCCUUC | 23 | ahy-miR396a | UUCCACAGCUUUCUUGAACUA | 21 |
| ahy-miR319j | CUUGGACUGAAGGGAGCUCCU | 21 | ahy-miR396b | UUCCACAGCUUUCUUGAACUG | 21 |
| ahy-miR319k | UUGGGCUGAAGGGAGCUCCC | 20 | ahy-miR396c | UUCCACAGCUUUCUUGAACUU | 21 |
| ahy-miR319l | UUGGACUGAAGGGAGCUCCUUC | 22 | ahy-miR396d | UUCCACGGCUUUCUUGAACUU | 21 |
| ahy-miR160a | GCGUAUGAGGAGCCAAGCAUA | 21 | ahy-miR396e | UUCCACAGCUUUCUUGAACUGU | 22 |
| ahy-miR160b | UGCCUGGCUCCCUGGAUGCCA | 21 | ahy-miR396f | UUCCACGGCUUUCUUGAACUG | 21 |
| ahy-miR160c | UGCCUGGCUCCCUGUAUGCCA | 21 | ahy-miR396g | UCUUCCACAGCUUUCUUGAAC | 21 |
| ahy-miR160d | UGCCUGGCUCCCUGCAUGCCA | 21 | ahy-miR396h | UUCCACAGCUUUCUUGAACAG | 21 |
| ahy-miR160f | UGCCUGGCUCCCUGUAUGCCG | 21 | ahy-miR396i | GUUCAAUAAAGCUGUGGGAAG | 21 |
| ahy-miR160i | CGCCUGGCUCCCUGUAUGCCA | 21 | ahy-miR396j | GUUCAAUAAAGCUGUGGGAAA | 21 |
| ahy-MIR160-5p | UGCCUGGCUCCCUGAAUGCCA | 21 | ahy-miR396k | GCUCAAGAAAGCUGUGGGAGA | 21 |
| ahy-MIR160-3p | GCAUGAAGGGAGUCACGCAGG | 21 | ahy-miR396l | AAGAAAGCUGUGGGAGAAUAUGGC | 24 |
| ahy-miR162a | UCGAUAAACCUCUGCAUCCAG | 21 | ahy-miR397a | UCAUUGAGUGCAGCGUUGAUG | 21 |
| ahy-miR162b | GGAGGCAGCGGUUCAUCGAUC | 21 | ahy-miR397b | UCAUUGAGUGCAGCGUUGAUGU | 22 |
| ahy-miR162c | UCGAUAAACCUCUGCAUCCGG | 21 | ahy-miR397c | CCAUUGAGUGCAGCGUUGAUG | 21 |
| ahy-miR162d | UCGAUAAGCCUCUGCAUCCAG | 21 | ahy-miR398a | UGUGUUCUCAGGUCGCCCCUG | 21 |
| ahy-miR162e | UGGAGGCAGCGGUUCAUCGAUC | 22 | ahy-miR398b | UGUGUUCUCAGGUCACCCCUU | 21 |
| ahy-miR164a | UGGAGAAGCAGGGCACGUGCA | 21 | ahy-miR398c | UGUGUUCUCAGGUCGCCCCCG | 21 |
| ahy-miR164b | UGGAGAAGCAGGGUACGUGCA | 21 | ahy-miR398d | UGUGUUCUCAGGUCACCCCUG | 21 |
| ahy-miR164c | UGGAGAAGCAGGGCACGUGCG | 21 | ahy-miR399a | CGCCAAAGGAGAGUUGCCCUG | 21 |
| ahy-miR164d | UGGAGAAGCAGGGCACGUGCU | 21 | ahy-miR399b | UGCCAAAGGAGAGUUGCCCUA | 21 |
| ahy-miR164e | UGGAGAAGCAGGGCACGUGAA | 21 | ahy-miR399c | UGCCAAAGGAGAUUUGCCCUG | 21 |
| ahy-miR164h | UGGAGAAGCAGGGCACGUGUG | 21 | ahy-miR399d | UGCCAAAGGAGAGCUGCCCUG | 21 |
| ahy-miR166a-5p | GGAAUGUUGUCUGGCUCGAGG | 21 | ahy-miR399e | GGGCUUCUCUUUCUUGGCAGG | 21 |
| ahy-miR166a-3p | UCGGACCAGGCUUCAUUCCCC | 21 | ahy-miR399f | UGCCAAAGGAGAUUUGCCCGG | 21 |
| ahy-miR166b | UCGGACCAGGCUUCAUUCCCCC | 22 | ahy-miR399g | CGGGGCAAAUCUCCUUUGGCA | 21 |
| ahy-miR166c | UCGGACCAGGCUUCAUUCCCGU | 22 | ahy-miR399j | UGCCAAAGGAGAGUUGCCCUG | 21 |
| ahy-miR166d | UCGGACCAGGCUUCAUUCCCU | 21 | ahy-miR399r | UGCCAAAGAAGAUUUGCCCCG | 21 |
| ahy-miR166e | CUCGGACCAGGCUUCAUUCCC | 21 | ahy-miR408-3p | AUGCACUGCCUCUUCCCUGGC | 21 |
| ahy-miR166f | UCGGACCAGGCUUCAUCCCCC | 21 | ahy-miR408a | UGCACUGCCUCUUCCCUGGCU | 21 |
| ahy-miR166g | UCGGACCAGGCUUCAUUCCC | 20 | ahy-miR408b | ACUGGGAACAGGCAGAGCAUGA | 22 |
| ahy-miR166h-3p | UCUCGGACCAGGCUUCAUUCC | 21 | ahy-miR408c | UGCACUGCCUCUUCCCUGGCUG | 22 |
| ahy-miR166i | UCGGACCAGGCUUCAUUCUC | 20 | ahy-miR482c | UUUCCAAUUCCACCCAUUCCUA | 22 |
| ahy-miR166j-3p | UCGGACCAGGCUUCAUUCCCG | 21 | ahy-miR482d-3p | UCUUCCCUACACCUCCCAUACC | 22 |
| ahy-miR166j-5p | GGAAUGUUGUUUGGCUCGAGG | 21 | ahy-miR482d-5p | UAUGGGGGGAUUGGGAAGGAAU | 22 |
| ahy-miR166k | UCGAACCAGGCUUCAUUCCCC | 21 | ahy-miR482e | UAUGGGGGGAUUGGGAAGGAA | 21 |
| ahy-miR166l | UCCGGACCAGGCUUCAUUCCC | 21 | ahy-miR530 | UGCAUUUGCACCUGCACUUUA | 21 |
| ahy-miR166m-5p | GGAAUGUUGGCUGGCUCGAGG | 21 | ahy-miR828 | UCUUGCUCAAAUGAGUAUUCCA | 22 |
| ahy-miR166m-3p | UCGGACCAGGCUUCAUUCCUC | 21 | ahy-miR858 | UCUCGUUGUCUGUUCGACCUU | 21 |
| ahy-miR166n | UCGGACCAGGCUUCAAUCCCU | 21 | ahy-miR894 | CGUUUCACGUCGGGUUCACC | 20 |
| ahy-miR166o | UCGGACCAGGCUUCAUUCCUU | 21 | ahy-miR1310 | AGGCAUCGGGGGCGCAACGCCC | 22 |
| ahy-miR166p | UCGGACCAGGCUUCAUUCCUA | 21 | ahy-miR1507 | CCUCGUUCCAUACAUCAUCUAG | 22 |
| ahy-miR166q | UCGGACCAGGCUUCAUUCCCUU | 22 | ahy-miR1511 | AACCAGGCUCUGAUACCAUG | 20 |
| ahy-miR166r | UCGGAUCAGGCUUCAUUCCUC | 21 | ahy-miR1515 | UCAUUUUUGCGUGCAAUGAUCC | 22 |
| ahy-miR166u | UCUCGGACCAGGCUUCAUUC | 20 | ahy-miR2111 | UAAUCUGCAUCCUGAGGUUUA | 21 |
| ahy-miR167a | AGAUCAUCUGGCAGUUUCACC | 21 | ahy-miR2118a | UUGCCGAUUCCACCCAUUCCUA | 22 |
| ahy-miR167b | UGAAGCUGCCAGCAUGAUCUA | 21 | ahy-miR2118b | UUACCGAUUCCACCCAUUCCUA | 22 |
| ahy-miR167c | UGAAGCUGCCAGCAUGAUCUGG | 22 | ahy-miR3711 | AGGCCCUCCUUCUAGCGCCA | 20 |
| ahy-miR167d | UGAAGCUGCCAGCAUGAUCUGA | 22 | ahy-miR4995 | AGGCAGUGGCUUGGUUAAGGG | 21 |
| ahy-miR167e | UGAAGCUGCCAGCAUGAUCUC | 21 | ahy-miR5139 | AAACCUGGCUCUGAUACCA | 19 |
| ahy-miR167f | UGAAGCUGCCAGCAUGAUCUG | 21 | ahy-miR5141a | UUAUCUGUCAGUCGCGUCGGGUCU | 24 |
| ahy-miR167j | UGAAGCUGCCAGCAUGAUCUUA | 22 | ahy-miR5141b | AGACCCGACGCGACUGACAGAUAA | 24 |
| ahy-miR167-5p | UGAAGCUGCCAGCAUGAUCUU | 21 | ahy-miR5538 | ACUGAACUCAAUCACUUGCUGC | 22 |
| ahy-miR167-3p | AGAUCAUGUGGCAGUUUCACC | 21 | ahy-miR6173a | AGCCGUAAACGAUGGAUACU | 20 |
| ahy-miR168a | UCGCUUGGUGCAGGUCGGGAA | 21 | ahy-miR6173b | AGUAUCCAUCGUUUACGGCU | 20 |
| ahy-miR168b | UCGCUUGGUGCAGGUCGAGAA | 21 | ahy-miR6478 | CCGACCUUAGCUCAGUUGGUG | 21 |
| ahy-miR168c | CCCGCCUUGCAUCAACUGAAU | 21 | |||
| Known and Peanut-Specific | |||||
| ahy-miR3508 | UAGAGGGUCCCCAUGUUCUCA | 21 | ahy-miR3514-3p | UCACCGUUAAUACAGAAUCCUU | 22 |
| ahy-miR3509-5p | AUACUUGAGAGCCGUUAGAUGA | 22 | ahy-miR3515 | AAUGUAGAAAAUGAACGGUAU | 21 |
| ahy-miR3510 | UUAUACCAUCUUGCGAGACUGA | 22 | ahy-miR3516 | GCUGGGUGAUAUUGACAGAAG | 21 |
| ahy-miR3511-3p | UGUUACUAUGGCAUCUGGUAA | 21 | ahy-miR3517 | CUGACCACUGUGAUCCCGGAA | 21 |
| ahy-miR3511-5p | GCCAGGGCCAUGAAUGCAGA | 20 | ahy-miR3518 | UGACCUUUGGGGAUAUUCGUG | 21 |
| ahy-miR3512 | CGCAAAUGAUGACAAAUAGA | 20 | ahy-miR3519 | UCAAUCAAUGACAGCAUUUCA | 21 |
| ahy-miR3513-5p | UUAAUUUCUGAGUUUGUCAUC | 21 | ahy-miR3520-3p | AAGGGAGACGUUUGAAUUAUC | 21 |
| ahy-miR3513-3p | UUGAUAAGAUAGAAAUUGUAU | 21 | |||
新窗口打开|下载CSV
2.4 花生荚果发育过程中microRNA的表达分析
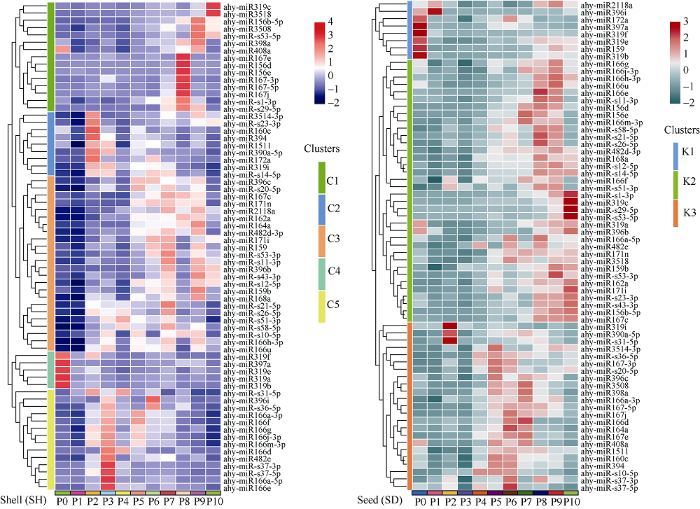
对RPKM大于10的microRNA进行表达分析发现, 在花生荚果11个发育阶段共有67个microRNA差异表达。利用TBtools软件[23]进行聚类分析和绘制热图(图5)发现, microRNA在荚果发育过程动态表达, 且呈时期特异性。在种子中microRNA的表达可分为K1、K2和K3三个簇, K1簇中microRNA在种子发育中表达相对较低。在K2簇中microRNA在种子成熟期(P7~P10)高表达。例如, ahy-miR166e、ahy-miR-s21-5p和ahy-miR-s26-5p在P8具有较高的表达量, 而ahy-miR-s1-3p、ahy-miR319c、ahy- miR-s29-5p和ahy-miR-s53-5p等4个microRNA在P10表达显著高于其他时期; ahy-miR166h-3p、ahy-miR168a和ahy-miR-s12-5p等microRNA在P8~P10都呈现了较高的表达。K3簇中microRNA主要在P5~P7中表达, 其中ahy-miR319i、ahy- miR390a-5p和ahy-miR-s31-5p等3个microRNA在种子初期(P2)有较高表达。ahy-miR167-3p、ahy-miR- s36-5p和ahy-miR-s10-5p等3个microRNA在P4~P6均有较高表达。在种子发育过程中P3时期microRNA的表达都相对较低。microRNA在果壳中的表达, 聚类将其分为了5个簇, C1簇中microRNA在果壳P8~P10高度表达, 例如, ahy-miR319c、ahy-miR156b-5p和ahy-miR167e分别在P10、P9和P8时期高丰度表达。C2簇主要在果壳发育初期(P2)高丰度表达, C3簇中microRNA呈现阶段性表达, 主要在P5~P9期有较高的表达量, 例如ahy- miR167c在P7~P9均表达较高。C4簇中microRNA在果壳中呈现低表达, C5簇主要在P3呈现高表达。表达分析结果进一步表明, microRNA在果壳和种子中都以高度阶段性或时期特异性的方式表达。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5microRNA在花生荚果发育过程中的表达模式分析
P0: 地上果针; P1: 地下果针; P2~P5: 果壳膨大期; P6~P9: 果实充实期; P9~P10: 成熟期。
Fig. 5Expression profile analysis of microRNAs in peanut pod (shell and seed) development
P0: aerial peg; P1: subterranean peg; P2-P5: pod expansion stage; P6-P9: seed filling stage; P9-P10: mature stage.
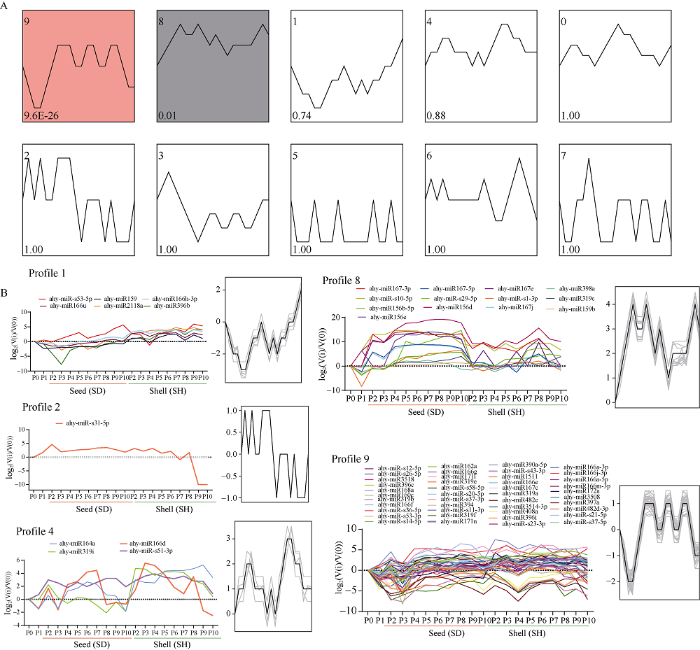
花生荚果发育过程中microRNA的趋势分析结果显示, 花生荚果发育中的microRNA共分为10种表达模式(图6-A), 大约84%的microRNA聚类到两种模式, 其中有43个microRNA聚类为profile.9表达模式(图6-B), 13个microRNA聚类为profile.8表达模式, 说明花生荚果发育过程中多数microRNA归入上述2个显著表达模式。microRNA在花生荚果发育过程中的表达是动态的, 在profile.9表达模式中, 花生荚果发育初期(P2~P5期)中microRNA的表达在种子中呈现下调, 在果壳中表达也较低, 随后microRNA表达上调, 种子和果壳发育中期(P6~P7期) microRNA表达达到最高, 花生荚果发育后期microRNA表达下调。在profile.8表达模式中, 花生荚果发育过程中microRNA的表达均处于上调, 在种子发育P4和P7期表达最高, 而在果壳中P9期表达最高。在其他几个表达模式中, microRNA在花生荚果发育的不同时期的积累水平也存在一定的差异。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6花生荚果发育过程中microRNA的趋势分析
P0: 地上果针; P1: 地下果针; P2~P5: 果壳膨大期; P6~P9: 果实充实期; P9~P10: 成熟期。
Fig. 6Trend analysis of microRNAs during peanut pod (shell and seed) development
P0: aerial peg; P1: subterranean peg; P2-P5: pod expansion stage; P6-P9: seed filling stage; P9-P10: mature stage.
2.5 花生荚果发育过程中靶基因的表达分析
microRNA在转录后水平调控其下游靶基因来发挥生物学功能, 对67个microRNA的靶基因进行预测分析, 按照错配数≤5进行筛选, 共得到552个靶基因, 其中ahy-miR482e的靶基因数目最多(32个), ahy-miR-s51-3p和ahy-miR-s58-5p的靶基因数目最少(仅1个)。大多数保守的microRNA的靶基因是MYB、ARF和BEL1-like等转录因子, 其他microRNA的靶基因包括NB-LRR型抗病蛋白、F-box/FBD/LRR重复蛋白和锌指蛋白等, 此外还有部分未知功能的靶基因。为了进一步了解所预测靶基因的可能功能, 对其进行表达模式分析, 如图7所示, 其靶基因在花生果壳和种子发育过程中也呈现了高度阶段性和特异性的表达方式。例如, AHTC20003756为BEL1-like转录因子, 在种子的P6期高丰度表达, 而AHTC20115371和AHTC20081975分别在果壳的P4和P3期高丰度表达。AHTC20054506编码了NB-LRR抗病蛋白, 在种子中表达量较高, 而在果壳中几乎不表达。AHTC20001034介导丝氨酸羟甲基转移酶(SHMT)途径, 在荚果发育的后期, 即种子和果壳的P7~P10期均有较高的表达。值得注意的是, AHTC20010002在荚果发育的整个过程中均高丰度表达, 但对其功能有待研究。图7
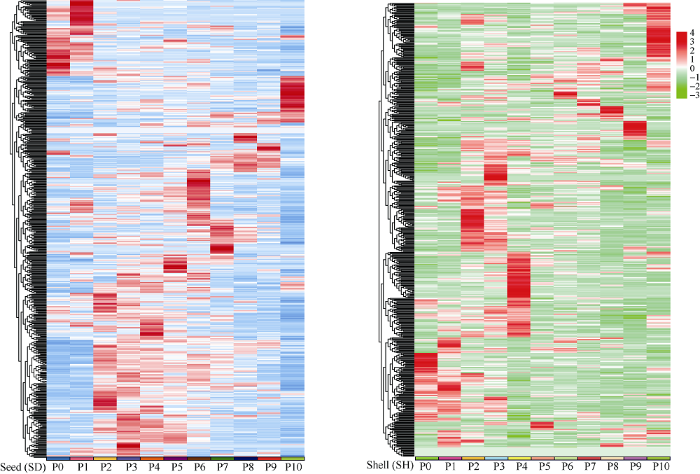 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7靶基因在花生荚果发育过程中的表达模式分析
P0: 地上果针; P1: 地下果针; P2~P5: 果壳膨大期; P6~P9: 果实充实期; P9~P10: 成熟期。
Fig. 7Expression profile analysis of target genes in peanut pod (shell and seed) development
P0: aerial peg; P1: subterranean peg; P2-P5: pod expansion stage; P6-P9: seed filling stage; P9-P10: mature stage.
对花生荚果发育过程中靶基因表达趋势进行分析表明, 花生荚果发育过程中靶基因共有20个表达模式(图8)。其中有profile.7、profile.11、profile.18和profile.19等4个显著表达模式, 在profile.7表达模式中, 基因在种子P6~P9中高丰度表达, 在种子成熟期基因表达逐渐下调, 而在果壳中表达相对较低。在profile.11表达模式中, 基因在种子和果壳的发育早期表达最高。在profile.18中基因在荚果发育的过程中均处于上调表达, 在种子P3和P6以及果壳的P9时期表达量最高。在profile.19中, 种子发育早期基因表达量较低, 在P6时期基因上调表达, 而在果壳发育早期基因表达量较高, P8期开始下调表达。
图8
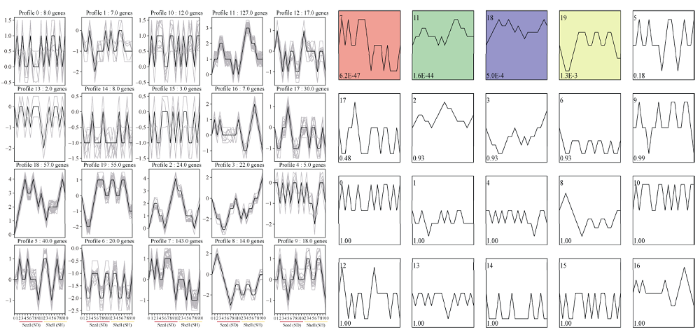 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8花生荚果发育过程中靶基因的表达趋势分析
P0:地上果针;P1:地下果针; P2~P5:果壳膨大期; P6~P9:果实充实期; P9~P10:成熟期。
Fig. 8Expression trend analysis of target genes during peanut pod (shell and seed) development
P0: aerial peg; P1: subterranean peg: P2- P5: pod expansion stage; P6- P9: seed flling stage; P9-P10: mature stage.
2.6 RT-PCR实时定量验证
为了验证测序结果数据的准确性, 选取28个microRNA和30个靶基因分别进行茎环RT-PCR和实时荧光定量PCR检测发现, microRNA和靶基因的表达趋势与测序分析结果基本一致(图9)。例如, 在microRNA中(图9-A), ahy-miR-s29-5p和ahy- miR166d分别在种子P10和果壳P3高丰度表达; ahy-miR2118a在果壳中高丰度表达, 而在种子中表达相对较低; ahy-miR156e在种子发育过程中从P3时期开始有表达, P7时期表达量最高, 随后逐渐降低, 而在果壳中几乎不表达。在靶基因表达中(图9-B), AHTC20084751编码的AUX1蛋白基因在果壳P10时期的表达显著高于其他时期, AHTC20067917为GATA转录因子, 在种子的P8时期高表达, AHTC20022021为乙醇酸氧化酶基因, 该基因在种子P7~P9时期均高丰度表达, 呈现阶段性的表达方式。总体而言, 荧光定量检测结果表明, microRNA及其靶基因的表达量与测序结果基本一致, 该结果为后续深入分析、克隆microRNA及其靶基因调控花生荚果发育奠定了基础。图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9转录组数据的qRT-PCR验证
P0: 地上果针; P1: 地下果针; P2~P5: 果壳膨大期; P6~P9: 果实充实期; P9~P10: 成熟期。
Fig. 9RT-qPCR validation of transcriptome data
P0: aerial peg; P1: subterranean peg; P2-P5: pod expansion stage; P6-P9: seed filling stage; P9-P10: mature stage.
3 讨论
为了解花生荚果发育过程的表型特征, 对花生荚果的1个地上发育时期和10个地下发育时期的研究发现, 果针向地生长过程中(即P0~P1时期)不断伸长但其直径几乎不变, 果针入土后停止生长, 胚胎恢复发育, 荚果迅速膨大, 即P2~P6时期是荚果体积的快速增长阶段, 而种子的发育相对滞后, 即在这一时期生长相对缓慢。在P6时期后荚果大小基本不变, 种子则进入快速生长阶段。因此, P6时期既是荚果停止膨胀的分界点, 也是种子进入快速发育阶段的分界点。本研究获得了62个新的Pre-microRNA和112个新的成熟的microRNA, 对Pre-microRNA进行二级结构预测发现, 所有前体均具有典型的茎环结构, 且多数序列(A+U)含量高于(G+C)含量, 说明鉴定得到的microRNA前体结构较稳定。成熟的microRNA在茎环结构的3°臂或5°臂上, 并能检测到其表达。此外, 对microRNA进行表达模式分析表明, microRNA在荚果发育过程中的表达是动态的, 且呈现阶段性和特异性表达。例如, ahy-miR390a-5p在种子P2时期表达较高, 在甘蓝型油菜中, miR390通过调控靶基因的表达介导早期胚胎发育, 从而影响其种子的含油量, 推测ahy-miR390a-5p可能参与花生荚果发育早期胚胎的发育[24]。ahy-miR156d和ahy-miR156e在花生种子成熟期(P7~P9)的表达显著高于其他时期, 相关研究表明, miR156通过靶向SPL10和SPL11基因, 能够导致不正常的细胞分裂[25], 从而控制种子的发育[26,27]。ahy-miR397a在P0期高度表达, 在水稻中, miR397的过表达通过靶向L-抗坏血酸氧化酶(AO)[28], 抑制抗坏血酸(AA)的氧化, 从而使脱氢抗坏血酸(DHA)维持在较低水平, 进而促进细胞分裂[29]。而种子在早期发育阶段需要具备很高的细胞分裂能力, 推测ahy-miR397a可能通过抗氧化途径而调控花生种子的发育过程。ahy-miR167e在种子P4~P7时期表达显著高于其他时期, ahy-miR167-5p在种子发育过程中均高表达, 在拟南芥和水稻中, miR167通过调控靶基因ARF6和ARF8的表达参与种子的发育[30,31], 因此ahy- miR167可能在花生荚果发育过程中发挥重要作用。microRNA319家族(ahy-miR319a、ahy-miR319b、ahy- miR319e和ahy-miR319f)在P0时期高度表达, miR319在花生荚果发育过程中参与生长素和GA信号传导途径, 能够促进花生胚胎和荚果早期发育[22]。
4 结论
花生荚果种子发育过程中, microRNA以时空特异的方式表达, 表明不同的microRNA在花生荚果、种子发育的不同时期发挥作用, 该结果为阐释花生荚果暗发育的分子机制提供了转录后调控的理论参考依据。附表 请见网络版: 1) 本刊网站
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOI:10.1126/science.1088060URLPMID:12893888 [本文引用: 1]

Plant microRNAs (miRNAs) show a high degree of sequence complementarity to, and are believed to guide the cleavage of, their target messenger RNAs. Here, I show that miRNA172, which can base-pair with the messenger RNA of a floral homeotic gene, APETALA2, regulates APETALA2 expression primarily through translational inhibition. Elevated miRNA172 accumulation results in floral organ identity defects similar to those in loss-of-function apetala2 mutants. Elevated levels of mutant APETALA2 RNA with disrupted miRNA172 base pairing, but not wild-type APETALA2 RNA, result in elevated levels of APETALA2 protein and severe floral patterning defects. Therefore, miRNA172 likely acts in cell-fate specification as a translational repressor of APETALA2 in Arabidopsis flower development.
.
URLPMID:18632569 [本文引用: 1]
DOI:10.1126/science.1107130URLPMID:15705854 [本文引用: 1]

Methylation on the base or the ribose is prevalent in eukaryotic ribosomal RNAs (rRNAs) and is thought to be crucial for ribosome biogenesis and function. Artificially introduced 2'-O-methyl groups in small interfering RNAs (siRNAs) can stabilize siRNAs in serum without affecting their activities in RNA interference in mammalian cells. Here, we show that plant microRNAs (miRNAs) have a naturally occurring methyl group on the ribose of the last nucleotide. Whereas methylation of rRNAs depends on guide RNAs, the methyltransferase protein HEN1 is sufficient to methylate miRNA/miRNA* duplexes. Our studies uncover a new and crucial step in plant miRNA biogenesis and have profound implications in the function of miRNAs.
URLPMID:12225663 [本文引用: 1]
DOI:10.1007/s00018-014-1728-7URLPMID:25209320 [本文引用: 1]

microRNAs (miRNAs) are important regulators of gene expression. After excised from primary miRNA transcript by dicer-like1 (DCL1, an RNAse III enzyme), miRNAs bind and guide their effector protein named argonaute 1 (AGO1) to silence the expression of target RNAs containing their complementary sequences in plants. miRNA levels and activities are tightly controlled to ensure their functions in various biological processes such as development, metabolism and responses to abiotic and biotic stresses. Studies have identified many factors that involve in miRNA accumulation and activities. Characterization of these factors in turn greatly improves our understanding of the processes related to miRNAs. Here, we review recent progress of mechanisms underlying miRNA expression and functions in plants.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12870-019-1936-2URLPMID:31370790 [本文引用: 1]

BACKGROUND: APETALA2-like genes encode plant-specific transcription factors, some of which possess one microRNA172 (miR172) binding site. The miR172 and its target euAP2 genes are involved in the process of phase transformation and flower organ development in many plants. However, the roles of miR172 and its target AP2 genes remain largely unknown in Brassica napus (B. napus). RESULTS: In this study, 19 euAP2 and four miR172 genes were identified in the B. napus genome. A sequence analysis suggested that 17 euAP2 genes were targeted by Bna-miR172 in the 3' coding region. EuAP2s were classified into five major groups in B.napus. This classification was consistent with the exon-intron structure and motif organization. An analysis of the nonsynonymous and synonymous substitution rates revealed that the euAP2 genes had gone through purifying selection. Whole genome duplication (WGD) or segmental duplication events played a major role in the expansion of the euAP2 gene family. A cis-regulatory element (CRE) analysis suggested that the euAP2s were involved in the response to light, hormones, stress, and developmental processes including circadian control, endosperm and meristem expression. Expression analysis of the miR172-targeted euAP2s in nine different tissues showed diverse spatiotemporal expression patterns. Most euAP2 genes were highly expressed in the floral organs, suggesting their specific functions in flower development. BnaAP2-1, BnaAP2-5 and BnaTOE1-2 had higher expression levels in late-flowering material than early-flowering material based on RNA-seq and qRT-PCR, indicating that they may act as floral suppressors. CONCLUSIONS: Overall, analyses of the evolution, structure, tissue specificity and expression of the euAP2 genes were peformed in B.napus. Based on the RNA-seq and experimental data, euAP2 may be involved in flower development. Three euAP2 genes (BnaAP2-1, BnaAP2-5 and BnaTOE1-2) might be regarded as floral suppressors. The results of this study provide insights for further functional characterization of the miR172 /euAP2 module in B.napus.
URLPMID:31871071 [本文引用: 1]
URLPMID:29567662 [本文引用: 1]
DOI:10.1038/nplants.2015.36URLPMID:27247036 [本文引用: 1]

Growth is a major factor in plant organ morphogenesis and is influenced by exogenous and endogenous signals including hormones. Although recent studies have identified regulatory pathways for the control of growth during vegetative development, there is little mechanistic understanding of how growth is controlled during the reproductive phase. Using Arabidopsis fruit morphogenesis as a platform for our studies, we show that the microRNA miR172 is critical for fruit growth, as the growth of fruit is blocked when miR172 activity is compromised. Furthermore, our data are consistent with the FRUITFULL (FUL) MADS-domain protein and Auxin Response Factors (ARFs) directly activating the expression of a miR172-encoding gene to promote fruit valve growth. We have also revealed that MADS-domain (such as FUL) and ARF proteins directly associate in planta. This study defines a novel and conserved microRNA-dependent regulatory module integrating developmental and hormone signalling pathways in the control of plant growth.
DOI:10.1371/journal.pgen.1001031URLPMID:20661442 [本文引用: 1]

Many targets of plant microRNAs (miRNAs) are thought to play important roles in plant physiology and development. However, because plant miRNAs are typically encoded by medium-size gene families, it has often been difficult to assess their precise function. We report the generation of a large-scale collection of knockdowns for Arabidopsis thaliana miRNA families; this has been achieved using artificial miRNA target mimics, a recently developed technique fashioned on an endogenous mechanism of miRNA regulation. Morphological defects in the aerial part were observed for approximately 20% of analyzed families, all of which are deeply conserved in land plants. In addition, we find that non-cleavable mimic sites can confer translational regulation in cis. Phenotypes of plants expressing target mimics directed against miRNAs involved in development were in several cases consistent with previous reports on plants expressing miRNA-resistant forms of individual target genes, indicating that a limited number of targets mediates most effects of these miRNAs. That less conserved miRNAs rarely had obvious effects on plant morphology suggests that most of them do not affect fundamental aspects of development. In addition to insight into modes of miRNA action, this study provides an important resource for the study of miRNA function in plants.
URLPMID:31953375 [本文引用: 1]
DOI:10.1104/pp.17.01169URLPMID:28904074 [本文引用: 1]

Increasing grain yield is the most important object of crop breeding. Here, we report that the elevated expression of a conserved microRNA, OsmiR408, could positively regulate grain yield in rice (Oryza sativa) by increasing panicle branches and grain number. We further showed that OsmiR408 regulates grain yield by down-regulating its downstream target, OsUCL8, which is an uclacyanin (UCL) gene of the phytocyanin family. The knock down or knock out of OsUCL8 also increases grain yield, while the overexpression of OsUCL8 results in an opposite phenotype. Spatial and temporal expression analyses showed that OsUCL8 was highly expressed in pistils, young panicles, developing seeds, and inflorescence meristem and was nearly complementary to that of OsmiR408. Interestingly, the OsUCL8 protein was localized to the cytoplasm, distinct from a majority of phytocyanins, which localize to the plasma membrane. Further studies revealed that the cleavage of OsUCL8 by miR408 affects copper homeostasis in the plant cell, which, in turn, affects the abundance of plastocyanin proteins and photosynthesis in rice. To our knowledge, this is the first report of the effects of miR408-OsUCL8 in regulating rice photosynthesis and grain yield. Our study further broadens the perspective of microRNAs and UCLs and provides important information for breeding high-yielding crops through genetic engineering.
URLPMID:32404101 [本文引用: 1]
DOI:10.3389/fpls.2018.00349URLPMID:29662498 [本文引用: 1]

Seed expansion in peanut is a complex biological process involving many gene regulatory pathways. MicroRNAs (miRNAs) play important regulatory roles in plant growth and development, but little is known about their functions during seed expansion, or how they contribute to seed expansion in different peanut lines. We examined seed miRNA expression patterns at 15 and 35 days after flowering (DAF) in two peanut eighth-generation recombinant inbred lines (RIL8); 8106, a medium-pod variety, and 8107, a super-pod variety. Using high-throughput sequencing, we identified 1,082 miRNAs in developing peanut seeds including 434 novel miRNAs. We identified 316 differentially expressed miRNAs by comparing expression levels between the two peanut lines. Interestingly, 24 miRNAs showed contrasting patterns of expression in the two RILs, and 149 miRNAs were expressed predominantly in only one RIL at 35 DAF. Also, potential target genes for some conserved and novel miRNAs were identified by degradome sequencing; target genes were predicted to be involved in auxin mediated signaling pathways and cell division. We validated the expression patterns of some representative miRNAs and 12 target genes by qPCR, and found negative correlations between the expression level of miRNAs and their targets. miR156e, miR159b, miR160a, miR164a, miR166b, miR168a, miR171n, miR172c-5p, and miR319d and their corresponding target genes may play key roles in seed expansion in peanut. The results of our study also provide novel insights into the dynamic changes in miRNAs that occur during peanut seed development, and increase our understanding of miRNA function in seed expansion.
URLPMID:31113378 [本文引用: 1]
URLPMID:28253861 [本文引用: 2]
[本文引用: 1]
DOI:10.1104/pp.111.187666URLPMID:22138974 [本文引用: 1]

MicroRNAs (miRNAs) and small interfering RNAs are important regulators of plant development and seed formation, yet their population and abundance in the oil crop Brassica napus are still not well understood, especially at different developmental stages and among cultivars with varied seed oil contents. Here, we systematically analyzed the small RNA expression profiles of Brassica napus seeds at early embryonic developmental stages in high-oil-content and low-oil-content B. napus cultivars, both cultured in two environments. A total of 50 conserved miRNAs and 9 new miRNAs were identified, together with some new miRNA targets. Expression analysis revealed some miRNAs with varied expression levels in different seed oil content cultivars or at different embryonic developmental stages. A large number of 23-nucleotide small RNAs with specific nucleotide composition preferences were also identified, which may present new classes of functional small RNAs.
URLPMID:21123653 [本文引用: 1]
URLPMID:12931144 [本文引用: 1]
DOI:10.1038/ng.2327URLPMID:22729225 [本文引用: 1]

Grain size and shape are important components of grain yield and quality and have been under selection since cereals were first domesticated. Here, we show that a quantitative trait locus GW8 is synonymous with OsSPL16, which encodes a protein that is a positive regulator of cell proliferation. Higher expression of this gene promotes cell division and grain filling, with positive consequences for grain width and yield in rice. Conversely, a loss-of-function mutation in Basmati rice is associated with the formation of a more slender grain and better quality of appearance. The correlation between grain size and allelic variation at the GW8 locus suggests that mutations within the promoter region were likely selected in rice breeding programs. We also show that a marker-assisted strategy targeted at elite alleles of GS3 and OsSPL16 underlying grain size and shape can be effectively used to simultaneously improve grain quality and yield.
DOI:10.1104/pp.106.078469URLPMID:16603663 [本文引用: 1]

The role of the redox state of the apoplast in hormone responses, signaling cascades, and gene expression was studied in transgenic tobacco (Nicotiana tabacum) plants with modified cell wall-localized ascorbate oxidase (AO). High AO activity specifically decreased the ascorbic acid (AA) content of the apoplast and altered plant growth responses triggered by hormones. Auxin stimulated shoot growth only when the apoplastic AA pool was reduced in wild-type or AO antisense lines. Oxidation of apoplastic AA in AO sense lines was associated with loss of the auxin response, higher mitogen-activated protein kinase activities, and susceptibility to a virulent strain of the pathogen Pseudomonas syringae. The total leaf glutathione pool, the ratio of reduced glutathione to glutathione disulfide, and glutathione reductase activities were similar in the leaves of all lines. However, AO sense leaves exhibited significantly lower dehydroascorbate reductase and ascorbate peroxidase activities than wild-type and antisense leaves. The abundance of mRNAs encoding antioxidant enzymes was similar in all lines. However, the day/night rhythms in the abundance of transcripts encoding the three catalase isoforms were changed in response to the AA content of the apoplast. Other transcripts influenced by AO included photorespiratory genes and a plasma membrane Ca(2+) channel-associated gene. We conclude that the redox state of the apoplast modulates plant growth and defense responses by regulating signal transduction cascades and gene expression patterns. Hence, AO activity, which modulates the redox state of the apoplastic AA pool, strongly influences the responses of plant cells to external and internal stimuli.
URLPMID:10982417 [本文引用: 1]
DOI:10.1093/nar/gkl118URLPMID:16598073 [本文引用: 1]

MicroRNA167 (miR167) was shown to cleave auxin responsive factor 8 (ARF8) mRNA in cultured rice cells. MiR167 level was found to be controlled by the presence of auxin in the growth medium. When cells grew in auxin-free medium, miR167 level decreased, resulting in an increase in the level of ARF8 mRNA. Cells growing in the normal growth medium containing auxin showed a reversed trend. It was also shown that expression of OsGH3-2, an rice IAA-conjugating enzyme, was positively regulated by ARF8. Delivery of synthesized miR167 into cells led to decrease of both ARF8 mRNA and OsGH3-2 mRNA. This study provides an evidence in which the exogeneous auxin signal is transduced to OsGH3-2 through miR167 and ARF8 in sequence. This proposed auxin signal transduction pathway, auxin-miR167-ARF8-OsGH3-2, could be, in conjunction with the other microRNA-mediated auxin signals, an important one for responding to exogeneous auxin and for determining the cellular free auxin level which guides appropriate auxin responses.
DOI:10.1016/j.molcel.2004.05.027URLPMID:15200956 [本文引用: 1]

MicroRNAs (miRNAs) are approximately 21-nucleotide RNAs, some of which have been shown to play important gene-regulatory roles during plant development. We developed comparative genomic approaches to systematically identify both miRNAs and their targets that are conserved in Arabidopsis thaliana and rice (Oryza sativa). Twenty-three miRNA candidates, representing seven newly identified gene families, were experimentally validated in Arabidopsis, bringing the total number of reported miRNA genes to 92, representing 22 families. Nineteen newly identified target candidates were confirmed by detecting mRNA fragments diagnostic of miRNA-directed cleavage in plants. Overall, plant miRNAs have a strong propensity to target genes controlling development, particularly those of transcription factors and F-box proteins. However, plant miRNAs have conserved regulatory functions extending beyond development, in that they also target superoxide dismutases, laccases, and ATP sulfurylases. The expression of miR395, the sulfurylase-targeting miRNA, increases upon sulfate starvation, showing that miRNAs can be induced by environmental stress.
