 ,, 王南, 高界, 刘婷婷, 文静, 易斌, 涂金星, 傅廷栋, 沈金雄*华中农业大学作物遗传改良国家重点实验室 / 国家油菜工程技术研究中心, 湖北武汉 430070
,, 王南, 高界, 刘婷婷, 文静, 易斌, 涂金星, 傅廷栋, 沈金雄*华中农业大学作物遗传改良国家重点实验室 / 国家油菜工程技术研究中心, 湖北武汉 430070Bioinformatics analysis of SnRK gene family and its relation with seed oil content of Brassica napus L.
TANG Jing-Quan ,, WANG Nan, GAO Jie, LIU Ting-Ting, WEN Jing, YI Bin, TU Jin-Xing, FU Ting-Dong, SHEN Jin-Xiong*National Key Laboratory of Crop Genetic Improvement / National Engineering Research Center of Rapeseed, Huazhong Agricultural University, Wuhan 430070, Hubei, China
,, WANG Nan, GAO Jie, LIU Ting-Ting, WEN Jing, YI Bin, TU Jin-Xing, FU Ting-Dong, SHEN Jin-Xiong*National Key Laboratory of Crop Genetic Improvement / National Engineering Research Center of Rapeseed, Huazhong Agricultural University, Wuhan 430070, Hubei, China通讯作者:
收稿日期:2020-05-15接受日期:2020-08-19网络出版日期:2021-03-12
| 基金资助: |
Received:2020-05-15Accepted:2020-08-19Online:2021-03-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (4328KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
唐婧泉, 王南, 高界, 刘婷婷, 文静, 易斌, 涂金星, 傅廷栋, 沈金雄. 甘蓝型油菜SnRK基因家族生物信息学分析及其与种子含油量的关系[J]. 作物学报, 2021, 47(3): 416-426. doi:10.3724/SP.J.1006.2021.04108
TANG Jing-Quan, WANG Nan, GAO Jie, LIU Ting-Ting, WEN Jing, YI Bin, TU Jin-Xing, FU Ting-Dong, SHEN Jin-Xiong.
蔗糖非发酵相关蛋白激酶(sucrose non-ferment ing-1-related protein kinase, SnRK)是一类丝氨酸/苏氨酸蛋白激酶, 这类蛋白激酶都包含1个典型的真核Ser-Thr激酶的催化结构域, 广泛参与调控植物的生长发育。1998年, Halford和Hardie[1]提出了SNF1 (sucrose non-fermenting protein kinase 1)相关激酶(SnRK)的名称。它与酵母(Saccharomyces cerevisiae)中调控碳代谢的SNF1具有同源性和功能相似性[2]。Harmon等[3]基于序列相似性和结构域将其分成SnRK1、SnRK2和SnRK3 三个亚组。研究表明, SnRK1与糖、ABA信号传导途径有关, 并且在代谢调节中发挥核心作用[4,5]。SnRK2和SnRK3是植物特有的亚家族[6], SnRK2在植物对非生物胁迫和养分限制的反应中以ABA依赖和独立的方式发挥关键作用[7,8,9], 其分子机制已有报道[10]。SnRK3也称为钙调磷酸酶B (calcineurin B-like protein, CBL)相互作用蛋白激酶(calcineurin B-like interacting protein kinase, CIPK), 也参与ABA信号的转导和非生物胁迫响应[11,12,13]。SnRK3蛋白家族还参与多种生物胁迫反应, 调节植物细胞内离子平衡, 在植物生长发育中起到至关重要的作用[14,15]。非生物胁迫反应功能可能影响脂质代谢, 群体在适应环境时既要面临环境的威胁又要进行世代的传递, 因此能量分配在两者之间发生调节, 使群体更好地适应环境[16]。在甘蓝型油菜和拟南芥中, 不同拷贝的干旱反应基因LEA3和VOC过表达增强了耐旱性和油含量[17]。
SnRK在油脂调控中的功能研究仍处于起步阶段, 酵母中SNF1家族是真核生物中参与脂质代谢和碳水化合物存储的调节剂之一, 而且从酵母(SNF1)到植物(SnRK1)的进化上是保守的[18]。哺乳动物AMP依赖性蛋白激酶(adenosine 5’-monophosphate(AMP)-activated protein kinase, AMPK)通过控制酶的磷酸化来抑制包括脂质的一些合成代谢过程, 并激活脂肪酸分解氧化和糖酵解的代谢过程[19]。酿酒酵母的SNF1激酶和哺乳动物AMPK与植物SnRKs同源。拟南芥中, SnRK1能磷酸化WR1, 促进其降解, 负调控油脂代谢[20]。棉花GhCIPK6过表达植株与野生型相比, 种子总油含量减少, C18:1等含量增加[21]。
在甘蓝型油菜中, 过表达CIPK9其含油量显著低于野生型对照[22]。因此SnRK家族可能在调控油脂代谢和能量平衡中发挥关键作用。本研究对甘蓝型油菜中SnRK基因家族进行了全面的比较分析, 包括基因家族成员鉴定、理化性质分析、含油量相关成员鉴定和与白菜甘蓝之间的共线性等分析。这项研究是关于甘蓝型油菜中BnSnRK基因家族的首次报道, 扩展了研究者对SnRK基因家族在结构、功能和进化中的理解。
1 材料与方法
1.1 甘蓝型油菜中SnRK基因家族成员的鉴定和进化树构建
本研究以拟南芥(Arabidopsis thaliana) SNRK蛋白序列作为查询序列, 对甘蓝型油菜(Brassica napus)基因组进行BlastP搜索与筛选, 通过tblastn补充, 手动去除冗余, 并将候选蛋白序列逐一提交NCBI网站在线工具Conserved Domain Search (http:// www.ncbi.nlm.nih.gov/cdd/)进行结构域确认。提取92个SNRK基因家族成员的氨基酸序列, 利用软件MEGA 7.0, 采用邻接法(Neighbor Joining)构建系统进化树, Bootstrap值为500, 其他参数使用默认值。1.2 甘蓝型油菜SnRK基因家族相关信息学分析
将基因GFF3注释文件与基因组序列导入软件TBtools[23]绘制外显子—内含子结构图。利用MEME在线工具(http://meme-suite.org/tools/meme)和NCBI- CDD数据库进行氨基酸保守基序分析。利用Expasy (https://web.expasy.org/)在线工具ProtParam分析蛋白理化特征。通过SPSS软件进行多重比较, 利用R绘制箱线图。使用WoLFPSORT (http://www.genscript. com/wolf-psort.html)在线工具预测亚细胞定位。利用eggNOG (http://eggnogdb.embl.de/#/app/emapper)对92个甘蓝型油菜SnRK基因进行富集分析, 选择HMMER算法和十字花科数据库进行注释, 其他选项默认。使用R软件包clusterProfiler和ggplot2进行二级分类并绘制GO富集柱形图。采用PRABI (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html)分析蛋白二级结构; 采用plantCARE (http://bioinfor-matics.psb.ugent.be/webtools/ plantcare/html/)分析启动子上游 3 kb区域的顺式作用元件。1.3 甘蓝型油菜SnRK基因家族与含油量性状的关联分析
530份油菜材料的重测序基因型数据和江苏、湖北武汉、重庆3个地区的表型数据由华中农业大学油菜研究室郭亮教授提供, 通过SPAGedi软件计算各材料间的亲缘关系, 最后用R语言mrMLM软件包中mrMLM和FASTmrMLM方法进行关联分析。通过软件SPSS进行单倍型分析中的方差分析和多重比较。1.4 基因家族成员在染色体上的分布和共线性分析
利用甘蓝型油菜‘damor-bzh’、拟南芥、白菜和甘蓝的基因组序列和GFF3注释文件, 通过MCscanX [24]工具进行比对分析并利用TBtools软件作图。1.5 qRT-PCR验证分析
高油材料P1是华中农业大学油菜研究室选育的半冬性甘蓝型油菜新品系, P2为一个来源于加拿大的春性甘蓝型油菜DH系, 两者种子含油量平均值分别为52.53%和46.59%。亲本材料P1、P2及其构建的F2分离群体中高、低油单株各8个衍生的F2:3家系材料在2017年10月种植于华中农业大学油菜转基因基地试验田。高、低油材料开花35 d的种子含油量分别为49.49%和45.39%。表达量分析中, 取开花后15 d和35 d的高低油材料角果, 取其幼嫩种子进行检测, 使用NanoDrop 2000超微量核酸检测仪检测RNA样品的质量与浓度。RNA样品原液进行反转录(Prime ScriptTMRT reagent Kit, TaKaRa), 按照试剂盒的使用标准步骤合成cDNA第1链。对关联分析筛选到的12个甘蓝型油菜含油量关联基因设计引物, 依据2-ΔΔCt方法分析基因的相对表达量。甘蓝型油菜内参基因为Actin。样本间表达量用双尾t测验进行差异显著性分析。2 结果与分析
2.1 甘蓝型油菜SnRKs成员鉴定及性质分析
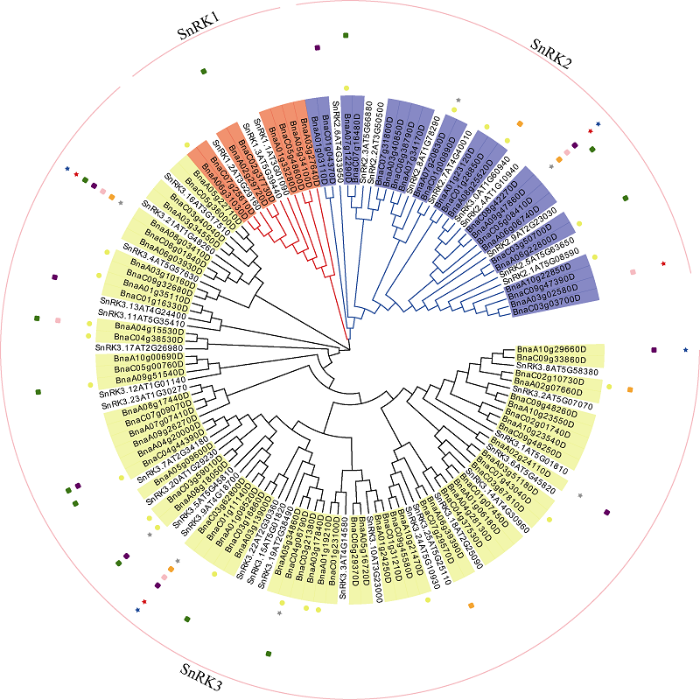
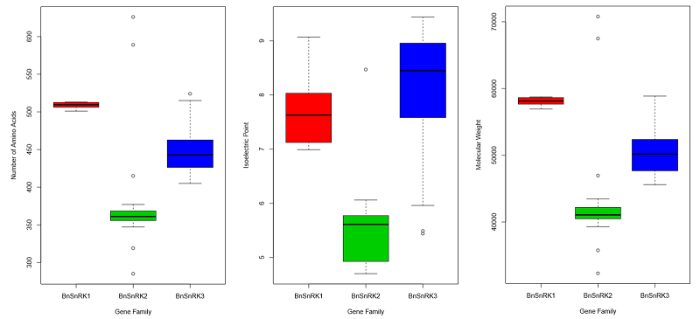
在甘蓝型油菜中共鉴定出92个SnRK基因, 其中SnRK1类成员8个, SnRK2类成员23个, SnRK3类成员61个(图1), 成员比例和SnRK基因家族在白菜中的比例相似[25]。预测的氨基酸数在285 (BnaC06g38790D)到626 (BnaA07g20630D)范围内不等, 基因全长介于858 bp (BnaA06g22800D)到8540 bp (BnaC01g16330D)之间。BnSnRK的理论等电点(pI)为4.70~9.44, 分子量为32.31275~70.7989 kD。亚族间在分子量、等电点和氨基酸数上存在显著差异(附图2)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于BnSnRK的油菜种子含油量和脂肪酸组分的关联分析
图示为较宽松筛选条件下的关联分析结果汇总。点状标注共8层, 从内圈到外圈的表型为含油量、棕榈酸、硬脂酸、油酸、亚油酸、亚麻酸、芥酸、花生烯酸。正方形标注表示18碳类脂肪酸组分。
Fig. 1Association analysis of seed oil content and fatty acid components based on BnSnRK in rapeseed
The figure shows the summary of the association analysis results under the looser screening conditions. There were eight layers of dotted labeling, and the phenotypes from inner to outer circles in order were oil content, palmitic acid, stearic acid, oleic acid, linoleic acid, linoleic acid, linolenic acid, erucic acid, and arachidonic acid. The square label shows the composition of 18 carbon fatty acids.
附图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图2italic>SnRK亚家族的理化性质比较
Fig. s2Comparison of physical and chemical properties of SnRK subfamilies
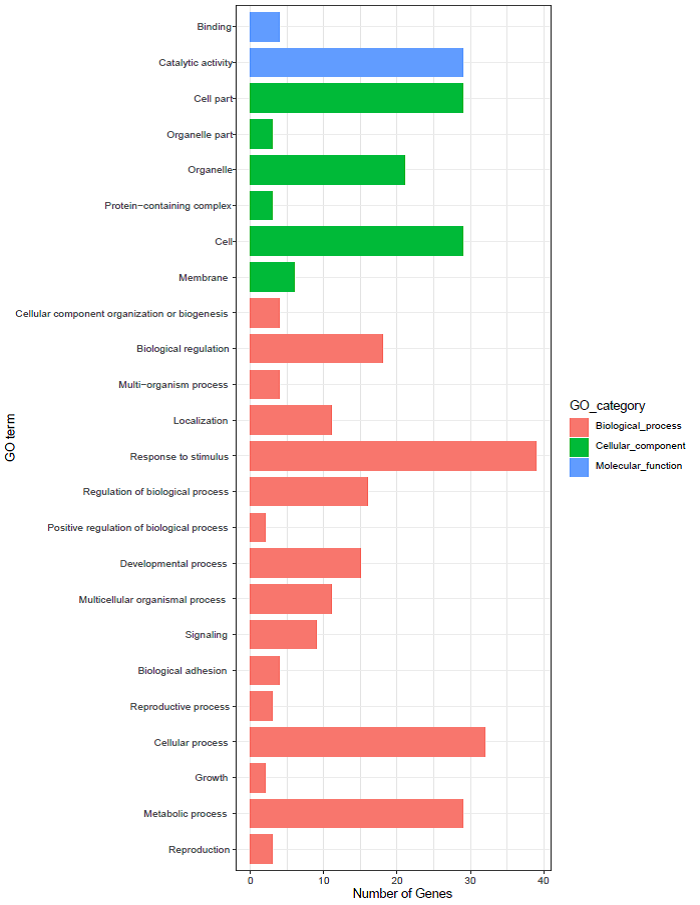
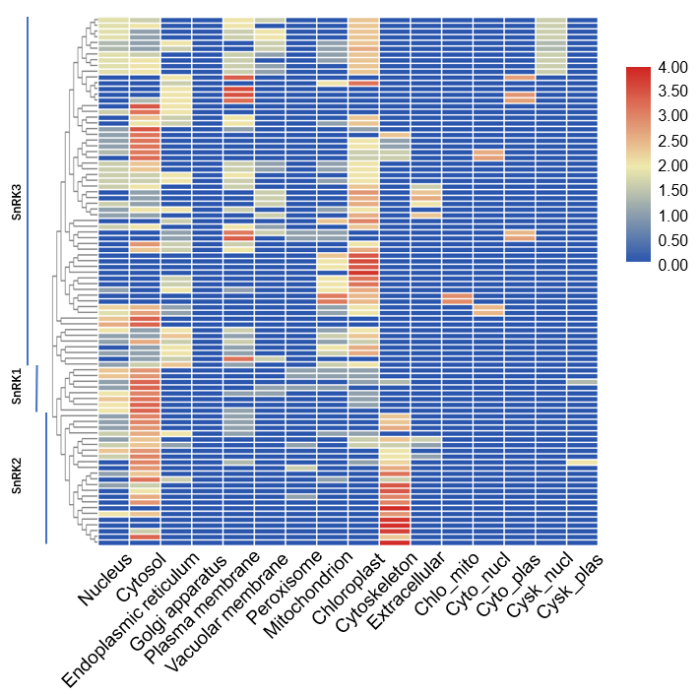
BnSnRK所编码蛋白质的二级结构主要有α-螺旋、β-转角与不规则卷曲(附表1), 与前人研究结果相似[26]。在92个编码蛋白中, 主要是α-螺旋(31.58%~45.50%)和不规则卷曲(29.95%~44.92%), β-转角较少(4.14%~12.56%), 亚族间的蛋白质二级结构有显著差异。甘蓝型油菜中SnRK基因家族的分子功能主要体现在催化活性, 涉及到的生物学过程主要包括应激响应、细胞过程和代谢过程(附图3)。亚细胞定位预测(附图1)表明, 大部分BnSnRK成员在细胞核和细胞质中均会表达, 所有BnSnRK1成员都预测到细胞核和细胞质中的定位信号。BnSnRK3亚族定位预测明显区别于其他2个亚族, 在叶绿体中有很高的表达参数。BnSnRK2亚族在细胞骨架中有很高的表达参数。此外, 与前人发现的SnRK3成员在膜系统上参与离子平衡的研究结果一致[15,27], 有部分BnSnRK基因也在内质网、质膜、液泡膜和线粒体中表达, 以BnSnRK3亚族成员为主。
Table s1
附表1
附表1BnSnRK蛋白二级结构分析
Table s1
| 结构类型 Structure type | BnSnRK1 | BnSnRK2 | BnSnRK3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 最小值 Minimum | 最大值 Maximum | 标准差 Standard deviation | 最小值 Minimum | 最大值 Maximum | 标准差 Standard deviation | 最小值 Minimum | 最大值 Maximum | 标准差 Standard deviation | |
| α-螺旋 Alpha helix | 31.62 | 34.83 | 0.98 | 31.58 | 45.50 | 3.52 | 31.62 | 41.12 | 2.14 |
| β-转角 Beta angle | 5.85 | 7.44 | 0.58 | 4.14 | 8.65 | 1.29 | 8.14 | 12.56 | 0.96 |
| 不规则卷曲 Irregular crimp | 40.70 | 44.92 | 1.30 | 34.05 | 43.09 | 2.50 | 29.95 | 39.92 | 2.50 |
| 延伸链 Extending chain | 15.82 | 18.96 | 0.91 | 13.01 | 20.35 | 1.83 | 15.73 | 21.10 | 1.22 |
新窗口打开|下载CSV
附图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图3BnSnRK基因家族成员富集分析
Fig. s3Enrichment analysis of BnSnRK gene family members
附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1SnRK基因家族亚细胞定位预测
Fig. s1Prediction of subcellular localization of SnRK gene family
SnRK基因家族在植物非生物胁迫应答反应和油脂调控中具有重要作用[28,29,30]。对BnSnRK基因家族成员启动子上游3 kb区域的顺式作用元件进行分析, 得到9种可能影响植物种子含油量的顺式作用元件(附图4)。BnSnRK所有成员中均有MYB类转录因子应答元件; 除BnaA01g23120D和BnaC05 g29370D外, 其余基因中都有MYC类转录因子应答元件; 除基因BnaA06g39390D、BnaA05g16720D和BnaC05g29370D外, ABA响应元件存在于其他所有BnSnRK成员中。此外, 53个成员存在低温响应元件, 44个成员存在干旱响应元件。
附图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图4BnSnRK家族启动子顺式作用元件分析
MYB: MYB转录因子响应元件;MYC: MYC转录因子响应元件;MBS: MYB结合位点。
Fig. s4Copert-acting element analysis of BnSnRK family promoter
MYB: MYB tran-scription factor response element; MYC: MYC transcription factor response element; MBS: MYB binding site.
基于BnSnRK氨基酸序列构建系统发育进化树并分析基因结构和保守基序发现, SnRK3亚族部分成员只有1个或2个外显子, 且极少有内含子, SnRK1所有成员都具有10个外显子, 说明SnRK1亚族的基因结构相对比较稳定。SnRK2亚族成员的基因结构会随着其在进化树中表现出的聚类关系而相似, 但整个亚族的外显子数量跨越了7~14的范围(图2)。这些结果与前人研究结果相似[31], 即BnSnRK基因家族激酶结构域位于N端附近, 中间是连接结构域, C端是调节结构域(图2)。进一步分析发现, BnSnRK1的结构域十分保守, BnSnRK2亚族的上游可能表现有3种, 而BnSnRK3成员最多, 但它的下游非常保守, 只有CIPK_C一种结构域, 上游存在2种结构域, 分别为STKc_SnRK3和PKc_like superfamily (图2)。92条BnSnRK蛋白共预测到61个保守基序(附图5)。其中motif4、motif7、motif10、motif3、motif19、motif2、motif1和motif5存在于3个亚族中, 具有非常高的保守性。3个亚族保守基序的区别主要集中在蛋白质下游区域, 结合系统进化树可发现, 亲缘关系较近的BnSnRK蛋白具有更相似的保守基序构成。
图2
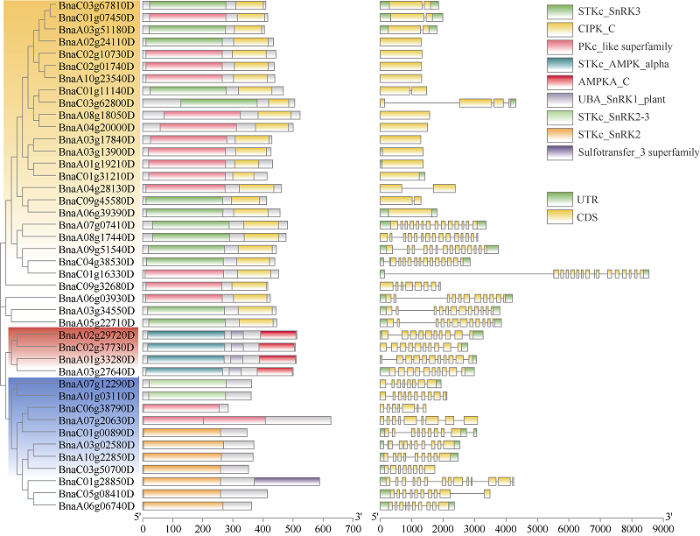 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2BnSnRK基因家族代表性成员的系统进化树、CDD保守结构和基因结构构成
BnSnRK基因家族分为3个亚族, 红色区域为SnRK1亚族, 蓝色区域为SnRK2亚族, 黄色区域为SnRK3亚族。
Fig. 2Phylogenetic tree, CDD conservative structures, and gene structures of representative members of BnSnRK gene family
The BnSnRK gene family are divided into three subfamilies: the red region represents SnRK1 subfamily, the blue region represents SnRK2 subfamily, and the yellow region represents SnRK3 subfamily.
附图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图5BnSnRK部分成员的保守基序分析
Fig. s5Conservative motif analysis of some members of BnSnRK
2.2 BnSnRK与含油量性状的关联分析
2.2.1 显著关联的SNP及其对应基因 对5个环境的表型数据和基因型数据进行分析, 设置阈值-lg (P-value) = 4.11, 共检测到14个与种子含油量显著关联的SNP位点。这些标记对应8个甘蓝型油菜SnRK基因家族成员, 其中2个基因存在环境重复, 分别为BnaA07g12290D和BnaA10g22850D (附图6)。标记为s7970的位点表型贡献率最大, 为12.95%, 对应基因为BnaA10g22850D (表1)。图10
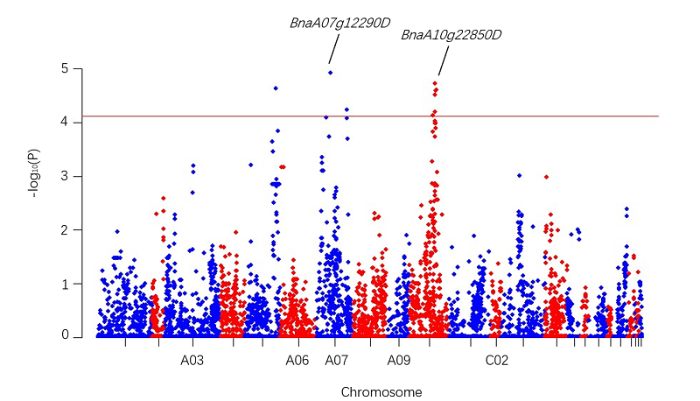 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10附
Fig. 10Fig. S6 Manhattan diagram of association analysis of seed oil content in E4 environment
Table 1
表1
表1重复检测到的油菜种子含油量关联位点
Table 1
| 基因名称 Gene name | 环境 Environment | 标记 Marker | 染色体 Chromosome | LOD值 LOD value | 阈值 -log10 (P) | 表型贡献率 R2 (%) | 单倍型频率 Haplotype frequency |
|---|---|---|---|---|---|---|---|
| BnaA07g12290D | E2 | s5667 | A7 | 3.79 | 4.53 | 7.79 | 0.42 |
| BnaA10g22850D | E2 | s7970 | A10 | 4.50 | 5.33 | 12.95 | 0.30 |
| BnaA07g12290D | E4 | s5664 | A7 | 3.42 | 4.14 | 1.99 | 0.48 |
| BnaA07g12290D | E4 | s5675 | A7 | 4.46 | 5.23 | 3.07 | 0.28 |
| BnaA10g22850D | E4 | s8059 | A10 | 5.44 | 6.26 | 6.70 | 0.11 |
| BnaA10g22850D | E4 | s8018 | A10 | 8.16 | 9.06 | 6.34 | 0.09 |
| BnaA07g12290D | E5 | s5674 | A7 | 5.07 | 5.87 | 4.16 | 0.28 |
新窗口打开|下载CSV
2.2.2 单倍型分析 随后对环境重复基因进行单倍型分析, 将每个基因上的标记按位置顺序进行组合(表1), 得到BnaA07g12290D的4个单倍型和BnaA10g22850D的3个单倍型。每个环境中含油量最高和最低的单倍型相同(附图7), 基因BnaA07g12290D的单倍型TAAC拥有最高含油量, 平均值为42.00%, 表型贡献率为50.01%。基因BnaA10g22850的单倍型CTC中含油量最高, 为41.82% (附表2), 表型贡献率为50.00%。另外BnaA10g21470D、BnaA07g20630D、BnaA06g31030D、BnaA01g35110D、BnaA03g17840D和BnaA03g10160D共6个基因具有环境特异性。采用LSD法对含油量表型进行方差分析发现, 5个环境的含油量有显著差异。这些基因可能在特异环境下才对种子含油量产生影响, 表明这些基因对种子含油量的贡献受环境影响。
附图7
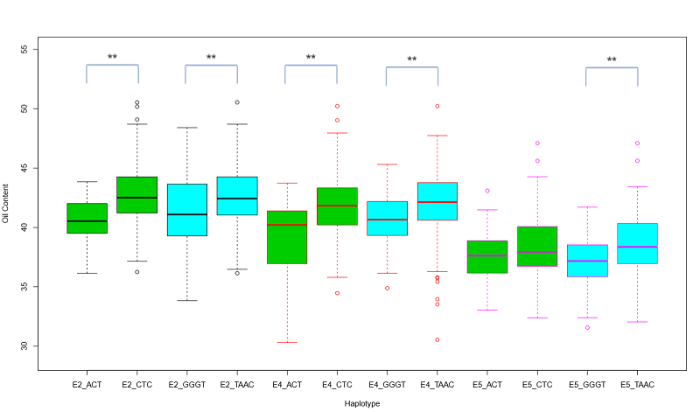 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图7单倍型分析箱线图
单倍型ACT和单倍型CTC分别为基因BnaA10g28850D含油量最低和最高的单倍型,在E2、E4、E5环境中用绿箱体表示;单倍型GGGT和TAAC分别为基因BnaA07g12290D的含油量最低和最高单倍型,在3个环境中用蓝色箱体表示;每个环境下的边框和线用不同颜色区分。
Fig. s7Haplotype analysis boxplot
Haplotype ACT and haplotype CTC were the haplotypes with the lowest and highest oil content of gene BnaA10g28850D, respectively, and were represented by green boxes in E2, E4, and E5 environments. Haplotype GGGT and TAAC were the haplotypes with the lowest and highest oil content of gene BnaA07g12290D, respectively. The borders of the boxes in each environment are colored differently.
Table s2
附表2
附表2单倍型分析的多重比较
Table s2
| 因变量: 含油量 Dependent variable: oil content | (I) 单倍型 Haploid type | (J) 单倍型 Haploid type | 平均差异 Average difference | 标准误 Standard error | 显著性 Significant |
|---|---|---|---|---|---|
| BnaA07g12290D | GAGT | GGAC | -0.48587 | 0.59894 | 0.418 |
| GGGT | 0.31362 | 0.59408 | 0.598 | ||
| TAAC | -1.04697 | 0.56187 | 0.063 | ||
| GGAC | GAGT | 0.48587 | 0.59894 | 0.418 | |
| GGGT | 0.79949* | 0.37903 | 0.035 | ||
| TAAC | -0.5611 | 0.32624 | 0.086 | ||
| GGGT | GAGT | -0.31362 | 0.59408 | 0.598 | |
| GGAC | -0.79949* | 0.37903 | 0.035 | ||
| TAAC | -1.36059* | 0.31722 | 0.000 | ||
| TAAC | GAGT | 1.04697 | 0.56187 | 0.063 | |
| GGAC | 0.5611 | 0.32624 | 0.086 | ||
| GGGT | 1.36059* | 0.31722 | 0.000 | ||
| BnaA10g22850D | ACT | CTC | -2.57628* | 0.44087 | 0.000 |
| CTT | -1.01843 | 0.85095 | 0.232 | ||
| CTC | ACT | 2.57628* | 0.44087 | 0.000 | |
| CTT | 1.55785* | 0.76008 | 0.041 | ||
| CTT | ACT | 1.01843 | 0.85095 | 0.232 | |
| CTC | -1.55785* | 0.76008 | 0.041 |
新窗口打开|下载CSV
2.2.3 脂肪酸组分关联分析 为筛选到更多与含油量相关的候选基因, 调低P-value的阈值至1e-3, 表型设为总含油量和重庆环境的脂肪酸组分, 筛选同时关联总含油量与组分的基因, 得到更多显著关联位点(图1)。它们分别对应11个基因: BnaA04g15530D、BnaA06g22800D、BnaA07g12290D、BnaA07g20630D、BnaA08g18050D、BnaA08g25520D、BnaA09g51540D、BnaC02g10730D、BnaC03g16860D、BnaC04g06790D、BnaC04g44390D。
2.3 高低油材料中候选基因表达量的验证分析
为了解这些基因在甘蓝型油菜种子发育过程中的活跃程度, 本研究针对BnaA04g15530D、BnaA06g22800D、BnaA07g12290D、BnaA07g20630D、BnaA08g18050D、BnaA08g25520D、BnaA09g51540D、BnaC02g10730D、BnaC03g16860D、BnaC04g06790D、BnaC04g44390D和BnaA10g22850D 共12个基因进行了表达量分析。12个基因中, BnaA07g12290D、BnaA08g18050D、BnaC02g10730D、BnaC04g44390D和BnaA10g22850D的t检验差异显著。相较于低油材料, 基因BnaC02g10730D在高油材料中的相对表达量为0.235, 表达水平较低。基因BnaA07g12290D、BnaA10g22850D、BnaA08g18050D、BnaC04g44390D在高油材料中的表达量分别是低油材料的2.854、1.769、4.429和116.705倍(图3-A)。值得注意的是, 除基因BnaA07g12290D和基因BnaA04g15530D在2个时期表现为不同的调控方向外, 其他基因在2个时期中对高低油材料的调控正负方向一致(图3-B)。图3
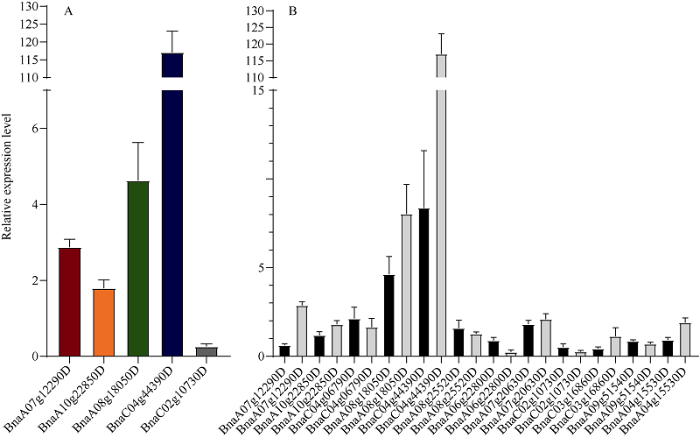 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3含油量相关候选基因在甘蓝型油菜种子中的表达分析
A: qRT-PCR分析BnSnRK基因家族中含油量相关候选基因在油菜幼嫩种子中的表达; B: 全部候选基因的相对表达。
Fig. 3Expression levels of oil-related candidate genes in the seed of Brassica napus
A: expression of oil-related candidate genes of BnSnRK gene family in young seeds of rapeseed detected by qRT-PCR; B: relative expression of all candidate genes.
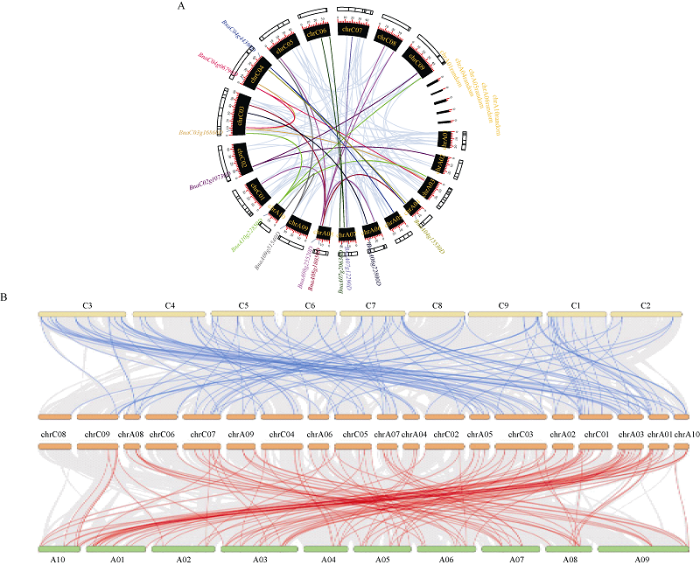
2.4 BnSnRK基因家族的同源共线性分析
BnSnRK基因在A染色体组1~10, C染色体组1~9上都有分布, 另有BnaA01g35110D、BnaA04g28130D、BnaA05g34860D、BnaA06g39390D和BnaA10g29660D 5个基因注释到Random染色体上, 成员在染色体组上的分布没有按亚族分类明显聚集(附图8)。在甘蓝型油菜基因组内, 共得到119个旁系同源基因对, 然而尚未发现BnSnRK中距离小于100 kb且覆盖度和比对相似性均>75%的串联重复基因。在12个候选基因中, 位于A染色体组上的数量是C染色体组的2倍(图4-A)。附图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图8SnRK基因家族染色体位置分布图
图中BnSnRK1、BnSnRK2和BnSnRK3亚族的成员名称分别用绿、红、黑表示,没有成员分布的Random染色体未出现在图中。
Fig. s8Chromosome location distribution of SnRK gene family
In the figure, the member names of BnSnRK1, BnSnRK2, and BnSnRK3 subgroup are represented by green, red, and black respectively. Random chromosome without member distribution does not appear in the figure.
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4甘蓝型油菜SnRK基因家族同源比对图(A)及甘蓝、甘蓝型油菜和白菜3个物种的共线联合比对关系(B)
A图中浅蓝色背景线为甘蓝型油菜SnRK基因家族成员之间的共线性基因对, 彩色线条将可能与甘蓝型油菜种子含油量关联的基因成员高亮表示。B图中灰色背景线代表全基因组中的共线性区块, 高亮线条代表BnSnRK基因家族成员在3个物种间的共线性关系。
Fig. 4Homologous comparison of SnRK gene family in Brassica napus (A) and collinear association of cabbage, Brassica napus, and Chinese cabbage (B)
The light blue background line in figure A is a collinearity gene pair between SnRK gene family members of Brassica napus. The color line will highlight gene member that may be related to the seed oil content of Brassica napus. In figure B, the gray background line represents the collinear block in the whole genome, and the highlighted line represents the collinear relationship between the BnSnRK gene family members in three species.
在甘蓝型油菜和甘蓝的基因组间共得到了177个BnSnRK共线基因对, 其中112个基因对位于2个物种间至少50个基因的大片段重复上。物种间包含较多基因对的同源片段主要存在于甘蓝的C03、C01和甘蓝型油菜的ChrC03、ChrA01中, 分别包含了35、31、19和18个BnSnRK家族的直系同源基因对(图4-B)。在甘蓝型油菜和白菜的基因组间共得到了187个BnSnRK基因对, 其中115个基因对位于2个物种间至少50个基因的大片段重复上, 包含较多基因对的同源片段主要存在于白菜的A01和甘蓝型油菜ChrA01、ChrA03、chrC03染色体中, 包含了32、18、18、18个直系同源基因对。
3 讨论
3.1 甘蓝型油菜SnRK不同亚族间的比较
拟南芥SnRK 3个亚族的不同结构早在2003年就有报道[31]。大多数SnRK2在细胞核和细胞质中均能检测到定位信号, 而部分BnSnRK3亚族成员的亚细胞定位也证明其存在核或者质膜等其他位置[12] (附图1)。已有研究表明, SnRK家族成员的磷酸化作用是其他蛋白质进入细胞核的关键[32]。保守结构域分析表明, BnSnRK存在一些亚族特异的保守基序, 如motif23、motif25和motif14等仅出现在BnSNRK2亚族。可以推测, 这些亚族特异的保守基序可能与亚族共有的核导入识别等特征无关, 它们更可能参与到每个亚族独特的互作蛋白等的结合中。3个亚族的结构域在蛋白质N端相对保守, 而在C端明显因亚族分类而变化, 同一亚族的保守结构域和基因结构更相似(图2)。基因家族之间的基因结构与理化性质存在不同, 是由于基因家族在进化过程中为了能更好地适应环境而作出的结构和性质的变化[33,34]。3.2 BnSnRK的进化
甘蓝型油菜在大约7500年前形成, 经白菜和甘蓝杂交后染色体加倍[35]。油菜中BnSnRK成员的同源基因数量都小于6, 和Chalhoub等[35]报道的拟南芥基因在甘蓝型油菜中普遍存在2~6个直系同源基因的研究结果一致。该基因家族在甘蓝型油菜中不存在串联重复, 分别有63.28%和61.50%的物种间共线基因对来自于白菜和甘蓝基因组上>50个基因的片段重复。在92个BnSnRK成员中, 77个来自于上述大片段重复, 占家族成员的83.70%。在关联分析结果中, 属于BnSnRK3亚族的基因BnaA08g18050D、BnaC02g10730D、BnaC03g16860D、BnaC04g06790D、BnaC04g44390D仅有1个外显子。研究者在白菜中也发现了22个BrSnRK3基因没有内含子[25]。拟南芥中众多SnRK是基因组复制事件后产生的旁系同源物[31]。而甘蓝型油菜中, BnSnRK基本上来自白菜和甘蓝的大片段重复, 这些大片段重复的基因又可能溯源到拟南芥中[35]。表达量分析中t检验显著的5个含油量相关候选基因都在其中。3.3 BnSnRK与油菜种子含油量
3.3.1 重复出现在不同环境条件下的总含油量关联基因 环境重复的含油量关联基因分别为BnSnRK2亚族的BnaA07g12290D和BnaA10g22850D。有研究者在双低油菜的差异表达基因中找到BnaA07g12290D在拟南芥中的同源基因SnRK2.3, 它产生的激素信号可能调节胚胎发育[36]。AtSnRK2.3产生的蛋白激酶的破坏, 导致ABA抑制性基因表达的上调以及ABA诱导型基因表达的下调[37], 而植物激素ABA的含量被证明可以对十字花科油籽油脂积累进行不同方向的调节, 且这种调节可能会改变油脂组分的比例[38]。Zheng等[39]的研究表明, SnRK2.6参与调节拟南芥种子中的油合成、蔗糖合成和叶片中的脂肪酸去饱和, 而SnRK2.6与基因BnaA07g12290D在进化树上相邻。基因BnaA07g12290D中存在和亚麻酸含量显著关联的位点(图1), 推测其通过对ABA的影响间接改变脂肪酸组分中亚麻酸的含量, 进而影响种子总含油量。基因BnaA10g22850D不存在和脂肪酸组分关联的位点, 但其与总油量的显著关联位点位于启动子区域, 该区域存在参与胚乳表达的顺式作用元件(附图4)。推测基因BnaA10g22850D对种子含油量的调控不受ABA的影响, 且作用于种子脂肪酸延伸和去饱和之前。3.3.2 环境特异的含油量关联基因 表达量差异显著的3个环境特异基因都是只具有1个外显子的BnSnRK3亚族基因。基因BnaA04g20000D同时作为候选基因BnaA08g18050D和BnaC04g44390D的共线基因。前者和ATSnRK2.0同源, 与BnaA04g20000D分支相近。后者和BnaA04g20000D同为AtSnRK3.7的直系同源基因。候选基因BnaA08g18050D和BnaC04g44390D都存在种子亚麻酸含量的关联标记, 它们都表现出正调控种子含油量(图3-A)。上述结果表明, 这2个基因可能在种子发育过程中发挥相似的功能。基因BnaA09g51540D的表达量差异不显著, 但在2个时期的相对表达量分别为0.853和0.688 (图3-B), 可能负调控种子含油量。其在拟南芥中的同源基因AT1g01140也被证明负调控种子总含油量, 且CIPK9突变体种子C18:2的比例显著低于野生型[22]。基因BnaA09g51540D也与总含油量和亚麻酸关联(图1), 这进一步验证了本研究分析方法是可靠的。CIPK9可以响应非生物胁迫[40], 这可能导致BnaA09g51540D在高低油材料中表达量差异不显著。
当植物受到非生物逆境胁迫时, 会通过一系列信号转导激活与逆境相关的转录因子, 并结合相应基因的顺式作用调控元件来启动相应基因的表达, 进而对逆境胁迫做出应答反应[41,42,43]。BnSnRK2和BnSnRK3亚族在功能上都参与植物逆境胁迫的应激反应[12,44]。关联分析得到的11个同时影响脂肪酸组分和总含油量的候选基因在2个亚族都有分布。其中, 8个基因(BnaA06g22800D、BnaA07g20630D、BnaA08g18050D、BnaA08g25520D、BnaA09g51540D、BnaC03g16860D、BnaC04g06790D、BnaC04g44390D)的关联标记位于启动子区域, 它们包括在高油材料中表达量较高的BnaA08g18050D和BnaC04g44390D。这些基因都含有MYB、MYC和ABA响应元件(附图4), 推测它们参与植物抗寒、抗旱等胁迫应答机制。高油材料中表达量较低的基因BnaC02g10730D的关联标记位于基因上非启动子区域, 其在葡萄上的同源基因VvCIPK10能够响应干旱、低温和盐胁迫[45]。2011年, Malekzadeh等[46]的研究证实, 水飞蓟在干旱胁迫下会提高籽粒中多不饱和脂肪酸的含量。水分胁迫和温度胁迫也被证明对甘蓝型油菜种子的脂肪酸组成和含油量有影响[47,48]。本研究以油酸、亚油酸和亚麻酸为关联分析表型时发现, SnRK基因家族中影响亚麻酸含量的基因不是随机分布, 而与总含油量相关。因此甘蓝型油菜SnRK基因家族对种子含油量的影响可能与ABA等参与抗逆胁迫的激素信号密切相关。
4 结论
本研究在甘蓝型油菜中鉴定出92个BnSnRK基因, 其中, SnRK1类成员8个, SnRK2类成员23个, SnRK3类成员61个。不同亚族在理化性质上有显著差异。BnSnRK家族主要在细胞质中表达, 蛋白质二级结构主要以α-螺旋和不规则卷曲为主。候选基因关联分析筛选出与油菜种子含油量相关的12个家族成员, 基因BnaA07g12290D的单倍型TAAC拥有最大含油量, 平均值为42.0034%。12个含油量关联基因均含有MYB、MYC和ABA响应元件, 可能参与油菜胁迫应答机制。BnSnRK成员可以对种子油脂积累进行不同方向上的调节。附图和附表 请见网络版: 1) 本刊网站
致谢
华中农业大学油菜研究室郭亮教授提供了530份油菜材料重测序基因型数据和含油量表型鉴定结果, 加拿大国家科学研究委员会萨斯卡通研究中心Zou Jitao博士提供油菜90K基因芯片等鉴定出了油菜参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1023/a:1006024231305URLPMID:9678569 [本文引用: 1]
DOI:10.1093/jxb/erg038URLPMID:12508057 [本文引用: 1]

A protein kinase that plays a key role in the global control of plant carbon metabolism is SnRK1 (sucrose non-fermenting-1-related protein kinase 1), so-called because of its homology and functional similarity with sucrose non-fermenting 1 (SNF1) of yeast. This article reviews studies on the characterization of SnRK1 gene families, SnRK1 regulation and function, interacting proteins, and the effects of manipulating SnRK1 activity on carbon metabolism and development.
[本文引用: 1]
DOI:10.1111/j.1365-313X.2009.03871.xURLPMID:19302419 [本文引用: 1]

The proteins kinases SNF1/AMPK/SnRK1 are a subfamily of serine/threonine kinases that act as metabolite sensors to constantly adapt metabolism to the supply of, and demand for, energy. In the yeast Saccharomyces cerevisiae, the SNF1 complex is a central component of the regulatory response to glucose starvation. AMP activated protein kinase (AMPK) the mammalian homologue of SNF1, plays a central role in the regulation of energy homeostasis at the cellular as well as the whole-body levels. In Arabidopsis thaliana, SnRK1.1 and SnRK1.2 have recently been described as central integrators of a transcription network for stress and energy signalling. In this study, biochemical analysis established SnRK1.1 as the major SnRK1 isoform both in isolated cells and leaves. In order to elucidate the function of SnRK1.1 in Arabidopsis thaliana, transgenic plants over-expressing SnRK1.1 were produced. Genetic, biochemical, physiological and molecular analyses of these plants revealed that SnRK1.1 is implicated in sugar and ABA signalling pathways. Modifications of the starch and soluble sugar content were observed in the 35S:SnRK1.1 transgenic lines. Our studies also revealed modifications of the activity of essential enzymes such as nitrate reductase or ADP-glucose pyrophosphorylase, and of the expression of several sugar-regulated genes, confirming the central role of the protein kinase SnRK1 in the regulation of metabolism.
DOI:10.1042/BJ20082408URLPMID:19309312 [本文引用: 1]

The phosphorylation and dephosphorylation of proteins, catalysed by protein kinases and phosphatases, is the major mechanism for the transduction of intracellular signals in eukaryotic organisms. Signalling pathways often comprise multiple phosphorylation/dephosphorylation steps and a long-standing hypothesis to explain this phenomenon is that of the protein kinase cascade, in which a signal is amplified as it is passed from one step in a pathway to the next. This review represents a re-evaluation of this hypothesis, using the signalling network in which the SnRKs [Snf1 (sucrose non-fermenting-1)-related protein kinases] function as an example, but drawing also on the related signalling systems involving Snf1 itself in fungi and AMPK (AMP-activated protein kinase) in animals. In plants, the SnRK family comprises not only SnRK1, but also two other subfamilies, SnRK2 and SnRK3, with a total of 38 members in the model plant Arabidopsis. This may have occurred to enable linking of metabolic and stress signalling. It is concluded that signalling pathways comprise multiple levels not to allow for signal amplification, but to enable linking between pathways to form networks in which key protein kinases, phosphatases and target transcription factors represent hubs on/from which multiple pathways converge and emerge.
URLPMID:25736509 [本文引用: 1]
URLPMID:22136638 [本文引用: 1]
[本文引用: 1]
DOI:10.1111/pce.12421URLPMID:25074439 [本文引用: 1]

Phosphatidic acid (PA) is an important signalling lipid involved in various stress-induced signalling cascades. Two SnRK2 protein kinases (SnRK2.4 and SnRK2.10), previously identified as PA-binding proteins, are shown here to prefer binding to PA over other anionic phospholipids and to associate with cellular membranes in response to salt stress in Arabidopsis roots. A 42 amino acid sequence was identified as the primary PA-binding domain (PABD) of SnRK2.4. Unlike the full-length SnRK2.4, neither the PABD-YFP fusion protein nor the SnRK2.10 re-localized into punctate structures upon salt stress treatment, showing that additional domains of the SnRK2.4 protein are required for its re-localization during salt stress. Within the PABD, five basic amino acids, conserved in class 1 SnRK2s, were found to be necessary for PA binding. Remarkably, plants overexpressing the PABD, but not a non-PA-binding mutant version, showed a severe reduction in root growth. Together, this study biochemically characterizes the PA-SnRK2.4 interaction and shows that functionality of the SnRK2.4 PABD affects root development.
URLPMID:29898403 [本文引用: 1]
URLPMID:11115898 [本文引用: 1]
URLPMID:24397480 [本文引用: 3]
DOI:10.1371/journal.pone.0225091URLPMID:31714948 [本文引用: 1]

CBL-interacting protein kinases (CIPKs) have been shown to regulate a variety of environmental stress-related signalling pathways in plants. Foxtail millet (Setaria italica (L.) P. Beauv) is known worldwide as a relatively stress-tolerant C4 crop species. Although the foxtail millet genome sequence has been released, little is known about the functions of CIPKs in foxtail millet. Therefore, a systematic genome-wide analysis of CIPK genes in foxtail millet was performed. In total, 35 CIPK members were identified in foxtail millet and divided into four subgroups (I to IV) on the basis of their phylogenetic relationships. Phylogenetic and gene structure analyses clearly divided all SiCIPKs into intron-poor and intron-rich clades. Cis-element analysis subsequently indicated that these SiCIPKs may be involved in responses to abiotic stimuli, hormones, and light signalling during plant growth and development, and stress-induced expression profile analysis revealed that all the SiCIPKs are involved in various stress signalling pathways. These results suggest that the CIPK genes in foxtail millet exhibit the basic characteristics of CIPK family members and play important roles in response to abiotic stresses. The results of this study will contribute to future functional characterization of abiotic stress responses mediated by CIPKs in foxtail millet.
URLPMID:23903322 [本文引用: 1]
URLPMID:25646412 [本文引用: 2]
[本文引用: 1]
URLPMID:30972883 [本文引用: 1]
URLPMID:21883929 [本文引用: 1]
DOI:10.1038/nrm2249URLPMID:17712357 [本文引用: 1]

The SNF1/AMP-activated protein kinase (AMPK) family maintains the balance between ATP production and consumption in all eukaryotic cells. The kinases are heterotrimers that comprise a catalytic subunit and regulatory subunits that sense cellular energy levels. When energy status is compromised, the system activates catabolic pathways and switches off protein, carbohydrate and lipid biosynthesis, as well as cell growth and proliferation. Surprisingly, recent results indicate that the AMPK system is also important in functions that go beyond the regulation of energy homeostasis, such as the maintenance of cell polarity in epithelial cells.
URLPMID:28314829 [本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOI:10.1101/2021.01.05.425441URLPMID:33442695 [本文引用: 1]

The majority of crystal structures are determined by the method of molecular replacement (MR). The range of application of MR is limited mainly by the need for an accurate search model. In most cases, pre-existing experimentally determined structures are used as search models. In favorable cases, ab initio predicted structures have yielded search models adequate for molecular replacement. The ORF8 protein of SARS-CoV-2 represents a challenging case for MR using an ab initio prediction because ORF8 has an all beta-sheet fold and few orthologs. We previously determined experimentally the structure of ORF8 using the single anomalous dispersion (SAD) phasing method, having been unable to find an MR solution to the crystallographic phase problem. Following a report of an accurate prediction of the ORF8 structure, we assessed whether the predicted model would have succeeded as an MR search model. A phase problem solution was found, and the resulting structure was refined, yielding structural parameters equivalent to the original experimental solution.
[本文引用: 1]
DOI:10.3389/fpls.2017.00162URLPMID:28239387 [本文引用: 2]

The CDPK-SnRK (calcium-dependent protein kinase/Snf1-related protein kinase) gene superfamily plays important roles in signaling pathways for disease resistance and various stress responses, as indicated by emerging evidence. In this study, we constructed comparative analyses of gene structure, retention, expansion, whole-genome duplication (WGD) and expression patterns of CDPK-SnRK genes in Brassica rapa and their evolution in plants. A total of 49 BrCPKs, 14 BrCRKs, 3 BrPPCKs, 5 BrPEPRKs, and 56 BrSnRKs were identified in B. rapa. All BrCDPK-SnRK proteins had highly conserved kinase domains. By statistical analysis of the number of CDPK-SnRK genes in each species, we found that the expansion of the CDPK-SnRK gene family started from angiosperms. Segmental duplication played a predominant role in CDPK-SnRK gene expansion. The analysis showed that PEPRK was more preferentially retained than other subfamilies and that CPK was retained similarly to SnRK. Among the CPKs and SnRKs, CPKIII and SnRK1 genes were more preferentially retained than other groups. CRK was closest to CPK, which may share a common evolutionary origin. In addition, we identified 196 CPK genes and 252 SnRK genes in 6 species, and their different expansion and evolution types were discovered. Furthermore, the expression of BrCDPK-SnRK genes is dynamic in different tissues as well as in response to abiotic stresses, demonstrating their important roles in development in B. rapa. In summary, this study provides genome-wide insight into the evolutionary history and mechanisms of CDPK-SnRK genes following whole-genome triplication in B. rapa.
[本文引用: 1]
[本文引用: 1]
URLPMID:21445098 [本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.006858URLPMID:12566581 [本文引用: 1]

Plants respond to environmental stress by activating
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.102.011999URLPMID:12805596 [本文引用: 3]

The CDPK-SnRK superfamily consists of seven types of serine-threonine protein kinases: calcium-dependent protein kinase (CDPKs), CDPK-related kinases (CRKs), phosphoenolpyruvate carboxylase kinases (PPCKs), PEP carboxylase kinase-related kinases (PEPRKs), calmodulin-dependent protein kinases (CaMKs), calcium and calmodulin-dependent protein kinases (CCaMKs), and SnRKs. Within this superfamily, individual isoforms and subfamilies contain distinct regulatory domains, subcellular targeting information, and substrate specificities. Our analysis of the Arabidopsis genome identified 34 CDPKs, eight CRKs, two PPCKs, two PEPRKs, and 38 SnRKs. No definitive examples were found for a CCaMK similar to those previously identified in lily (Lilium longiflorum) and tobacco (Nicotiana tabacum) or for a CaMK similar to those in animals or yeast. CDPKs are present in plants and a specific subgroup of protists, but CRKs, PPCKs, PEPRKs, and two of the SnRK subgroups have been found only in plants. CDPKs and at least one SnRK have been implicated in decoding calcium signals in Arabidopsis. Analysis of intron placements supports the hypothesis that CDPKs, CRKs, PPCKs and PEPRKs have a common evolutionary origin; however there are no conserved intron positions between these kinases and the SnRK subgroup. CDPKs and SnRKs are found on all five Arabidopsis chromosomes. The presence of closely related kinases in regions of the genome known to have arisen by genome duplication indicates that these kinases probably arose by divergence from common ancestors. The PlantsP database provides a resource of continuously updated information on protein kinases from Arabidopsis and other plants.
DOI:10.1105/tpc.15.00371URL [本文引用: 1]
[本文引用: 1]
DOI:10.1086/509078URL [本文引用: 1]
DOI:10.1126/science.1253435URLPMID:25146293 [本文引用: 3]

Oilseed rape (Brassica napus L.) was formed ~7500 years ago by hybridization between B. rapa and B. oleracea, followed by chromosome doubling, a process known as allopolyploidy. Together with more ancient polyploidizations, this conferred an aggregate 72x genome multiplication since the origin of angiosperms and high gene content. We examined the B. napus genome and the consequences of its recent duplication. The constituent An and Cn subgenomes are engaged in subtle structural, functional, and epigenetic cross-talk, with abundant homeologous exchanges. Incipient gene loss and expression divergence have begun. Selection in B. napus oilseed types has accelerated the loss of glucosinolate genes, while preserving expansion of oil biosynthesis genes. These processes provide insights into allopolyploid evolution and its relationship with crop domestication and improvement.
DOI:10.1093/pcp/pcv074URLPMID:26092973 [本文引用: 1]

Canola is an important vegetable oil crop globally, and the understanding of the molecular mechanism underlying fatty acids biosynthesis during seed embryo development is an important research goal. Here we report the transcriptional profiling analysis of developing canola embryos using RNA-sequencing (RNA-Seq) method. RNA-Seq analysis generated 58,579,451 sequence reads aligned with 32,243 genes. It was found that a total of 55 differential expression genes (DEGs) encoding 28 enzymes function in carbon flow to fatty acids of storage TAG. Most of the DEGs encoding above enzymes showed similar expression pattern, indicating the DEGs are cooperatively involved in carbon flow into fatty acids. In addition, 41 DEGs associated with signal transductions, transport and metabolic processing of auxin, gibberellin, abscisic acid, cytokinin and salicylic acids were found in the RNA-Seq database, which indicates the important roles of the phytohormones in controlling embryo development and fatty acids synthesis. 122 DEGs encoding transcriptional factor family members were found in developing canola embryos. Furthermore, BnDOF5.6, a zinc finger transcriptional factor gene, found in RNA-Seq database was down-regulated in developing canola embryos. The transgenic plants displayed reduced embryo sizes, decreased fatty acids contents and altered seed fatty acids composition in canola. Down-regulated of BnDof5.6 also changed the expression levels of genes involved in fatty acids synthesis and desaturation. Our results indicate that BnDof5.6 is required for embryo development and fatty acids synthesis in canola. Overall this study presents new information on the global expression patterns of genes during embryo development and will expand our understanding of the complex molecular mechanism of carbon flow into fatty acids and embryo development in canola.
URLPMID:19541597 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:20200070 [本文引用: 1]
DOI:10.1038/cr.2007.39URLPMID:17486125 [本文引用: 1]

Potassium is one of the major macro-nutrients essential for a number of cellular processes in plants. Low potassium level in the soil represents a limiting factor for crop production. Recent studies have identified potassium transporters that are involved in potassium acquisition, and some of them are critical for potassium nutrition under low potassium conditions. However, little is understood on the molecular components involved in low potassium signaling and responses. We report here the identification of a calcineurin B-like protein-interacting protein kinase (CIPK9) as a critical regulator of low potassium response in Arabidopsis. The CIPK9 gene was responsive to abiotic stress conditions, and its transcript was inducible in both roots and shoots by potassium deprivation. Disruption of CIPK9 function rendered the mutant plants hypersensitive to low potassium media. Further analysis indicated that K(+) uptake and content were not affected in the mutant plants, implying CIPK9 in the regulation of potassium utilization or sensing processes.
DOI:10.1021/pr050291gURLPMID:16457606 [本文引用: 1]

Drought is one of the most severe limitations on the productivity of rainfed lowland and upland rice. To investigate the initial response of rice to drought stress, changes in protein expression were analyzed using a proteomic approach. Two-week-old rice seedlings were exposed to drought conditions from 2 to 6 days, and proteins were extracted from leaf sheaths, separated by two-dimensional polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. After drought stress for 2 to 6 days, 10 proteins increased in abundance and the level of 2 proteins decreased. The functional categories of these proteins were identified as defense, energy, metabolism, cell structure, and signal transduction. In addition to drought stress, accumulations of protein were analyzed under several different stress conditions. The levels of an actin depolymerizing factor, a light harvesting complex chain II, a superoxidase dismutase and a salt-induced protein were changed by drought and osmotic stresses, but not cold or salt stresses, or abscisic acid treatment. The effect of drought stress on protein in the leaf sheaths of drought-tolerant rice cultivar was also analyzed. The light harvesting complex chain II and the actin depolymerizing factor were present at high levels in a drought-tolerant rice cultivar before stress application. With drought stress, actin depolymerizing factor was expressed in leaf blades, leaf sheaths, and roots. These results suggest that actin depolymerizing factor is one of the target proteins induced by drought stress.
DOI:10.1007/s00299-010-0956-zURLPMID:21132431 [本文引用: 1]

Rice (Oryza sativa L.), one of the most agronomically important crops, supplies staple food for more than half of the world's population, especially those living in developing countries. The intensively increasing world population has put a great burden on rice production. Drought as one of the major limiting factors for rice productivity has challenged researchers to improve both the water management system and rice characteristics. Biotechnology has assisted researchers to identify genes that are responsive toward drought. This review consolidates the recent studies that expose a number of drought-responsive genes in rice, which are potential candidates for development of improved drought-tolerant transgenic rice cultivars. In addition, examples are provided of how various drought-responsive genes, such as transcription factor and protein kinase encoding genes, were explored to engineer rice plants for enhanced drought tolerance using transgenic approach. Furthermore, the involvement of various phytohormones in regulation of drought response as well as the complexity of drought-responsive networks, which is indicated by the crosstalks with other stress-responsive networks such as cold and salt stresses, will be discussed. It is hoped that by understanding how rice responds to drought, crop performance can be stabilized and protected under water deficit conditions.
URLPMID:12223810 [本文引用: 1]
DOI:10.1126/science.287.5451.300URLPMID:10634783 [本文引用: 1]

Abscisic acid (ABA) stimulates stomatal closure and thus supports water conservation by plants during drought. Mass spectrometry-generated peptide sequence information was used to clone a Vicia faba complementary DNA, AAPK, encoding a guard cell-specific ABA-activated serine-threonine protein kinase (AAPK). Expression in transformed guard cells of AAPK altered by one amino acid (lysine 43 to alanine 43) renders stomata insensitive to ABA-induced closure by eliminating ABA activation of plasma membrane anion channels. This information should allow cell-specific, targeted biotechnological manipulation of crop water status.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
