 ,1,*, 李剑峰1, 张博1, 全建章2, 王永芳2, 赵渊1, 张小梅1, 王振山1, 桑璐曼1, 董志平
,1,*, 李剑峰1, 张博1, 全建章2, 王永芳2, 赵渊1, 张小梅1, 王振山1, 桑璐曼1, 董志平 ,2,*
,2,*Responsive features of SiPRR37 to photoperiod and temperature, abiotic stress and identification of its favourable allelic variations in foxtail millet (Setaria italica L.)
JIA Xiao-Ping ,1,*, LI Jian-Feng1, ZHANG Bo1, QUAN Jian-Zhang2, WANG Yong-Fang2, ZHAO Yuan1, ZHANG Xiao-Mei1, WANG Zhen-Shan1, SANG Lu-Man1, DONG Zhi-Ping
,1,*, LI Jian-Feng1, ZHANG Bo1, QUAN Jian-Zhang2, WANG Yong-Fang2, ZHAO Yuan1, ZHANG Xiao-Mei1, WANG Zhen-Shan1, SANG Lu-Man1, DONG Zhi-Ping ,2,*
,2,*通讯作者:
收稿日期:2020-06-25接受日期:2020-10-14网络出版日期:2021-04-12
| 基金资助: |
Received:2020-06-25Accepted:2020-10-14Online:2021-04-12
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (1129KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
贾小平, 李剑峰, 张博, 全建章, 王永芳, 赵渊, 张小梅, 王振山, 桑璐曼, 董志平. 谷子SiPRR37基因对光温、非生物胁迫的响应特点及其有利等位变异鉴定[J]. 作物学报, 2021, 47(4): 638-649. doi:10.3724/SP.J.1006.2021.04139
JIA Xiao-Ping, LI Jian-Feng, ZHANG Bo, QUAN Jian-Zhang, WANG Yong-Fang, ZHAO Yuan, ZHANG Xiao-Mei, WANG Zhen-Shan, SANG Lu-Man, DONG Zhi-Ping.
伪应答调控蛋白(pseudo-response regulators, PRRs)基因属于CCT基因家族中的PRR亚家族, 所编码蛋白的氨基端含有1个PRR结构域, 羧基端含有1个CCT结构域, 这2个结构域被1个不太保守的可变域分割[1,2]。作为生物钟核心组分, PRRs基因广泛参与植物光周期调控开花、抵御非生物胁迫及生物量积累等多个生长发育过程[3,4,5]。
OsPRR37是从水稻中克隆的1个非常关键的生物钟基因, 与控制光周期敏感性的主效QTL Hd2密切相关, 影响株高、开花期和每穗小穗数[6,7]; OsPRR37的天然突变促进了水稻向亚洲高纬度地区的扩张, 使其地理适应性和种植面积都得到提升[8,9,10]。大麦Ppd-H1、小麦Ppd-D1和高粱SbPRR37与OsP RR37同源, 同样在对应作物光周期调控开花过程中发挥重要功能, 影响区域适应性[11,12,13]。PRR37蛋白提前终止、CCT结构域和PRR结构域中的错义突变会影响作物对光周期的敏感性, 进而影响表型性状, 因此可以把PRR37作为改变生态适应性的靶点, 通过选择特定的碱基突变类型(单倍型)从而达到所需的适应性表型, 发掘该基因有利等位变异, 在作物广适应性高产分子辅助选择育种中展现良好应用前景[5,11,14-16]。如水稻开花期与OsPRR37表达水平显著相关, 利用OsPRR37可以增强水稻光温敏感性, 从而实现对开花期的调节[17]。除了与生态适应性密切相关, PRR37的直系同源基因PRR7还参与了拟南芥对干旱胁迫的抵御过程, 其许多靶基因能够负调控植物ABA以响应干旱胁迫[18,19,20]。PRR7基因的作用靶点还包括fer1、fer3和fer4 三种铁蛋白, 在植物铁过量应激反应中具有保护作用[21]。此外低温诱导可使PRR37及其他生物钟基因形成不同的可变剪切体, 导致不同的转录本产生, 通过负调控抗冷基因CBF (C-repeat-binding factor)表达来响应冷胁迫[20,22-23]。
目前对PRR37基因的研究主要集中于光周期调控开花、逆境胁迫响应机制方面, 有关该基因在禾本科作物光温互作调控中的作用机制尚未见报道, 同时PRR37在非生物胁迫中的作用机制研究局限于拟南芥、水稻几种模式植物, 在C4作物中的研究比较缺乏。谷子(Setaria italica L.)是我国的传统杂粮作物, 具有基因组较小(约515 Mb)、籽粒营养价值高、生育期短、抗旱性强、耐贫瘠和高光效等优点, 已经成为北方干旱半干旱地区重要的粮食作物以及C4禾谷类作物的理想模型[24,25,26,27]。谷子对光温反应敏感, 自然界存在丰富的光温敏感型变异材料, 是揭示C4作物光温敏感性形成分子机制的合适对象[28]。有关谷子光温敏感性研究较少, 主要包括光温敏感性指标筛选、光周期敏感性相关性状QTL定位等方面[29,30,31,32,33,34,35,36]。谷子全基因组序列测定及大量转录组数据的公布为利用反向遗传学方法快速克隆光温敏感性关键基因提供了便利, 这对揭示谷子光温敏感性形成的分子机制具有极大的促进作用, 因此本研究利用RT-PCR技术克隆谷子生物钟关键基因SiPRR37, 设置不同光温组合条件、不同非生物胁迫条件分析该基因的表达规律, 并通过对160份谷子材料重测序检测基因突变位点, 进行单倍型效应分析, 以期揭示SiPRR37参与谷子光温互作调控和抵御非生物胁迫的可能机制, 确定其对主要农艺性状的作用。
1 材料与方法
1.1 试验材料
本研究用于SiPRR37基因克隆及表达分析的谷子品种为来自陕西的延谷11号, 该品种对光温敏感, 是研究春谷品种光温敏感性的理想材料。用于重测序及单倍型分析的160份谷子材料见附表1。Table S1
附表1
附表1本研究所用的160份谷子资源
Table S1
| 序号 Serial number | 国家统一编号 National unified number | 品种名 Variety name | 来源地 Origin region |
|---|---|---|---|
| 1 | 00027956 | 郑谷2 Zhenggu 2 | 中国河南 Henan, China |
| 2 | 00027954 | 豫谷6 Yugu 6 | 中国河南 Henan, China |
| 3 | 00024231 | 郑06-6 Zheng 06-6 | 中国河南 Henan, China |
| 4 | 00024221 | 豫谷3 Yugu3 | 中国河南 Henan, China |
| 5 | 00027933 | 郑05-2 Zheng 05-2 | 中国河南 Henan, China |
| 6 | 00024226 | 郑州12 Zhengzhou 12 | 中国河南 Henan, China |
| 7 | 00024231 | 郑315 Zheng 315 | 中国河南 Henan, China |
| 8 | 00024262 | 郑8041 Zheng 8041 | 中国河南 Henan, China |
| 9 | 00024195 | 安5424 An 5424 | 中国河南 Henan, China |
| 10 | 00027946 | 豫谷15 Yugu 15 | 中国河南 Henan, China |
| 11 | 00027948 | 豫谷11 Yugu 11 | 中国河南 Henan, China |
| 12 | 00028386 | 豫谷16 Yugu 16 | 中国河南 Henan, China |
| 13 | 00027937 | 安4117 An 4117 | 中国河南 Henan, China |
| 14 | 00027952 | 豫谷13号Yugu 13 | 中国河南 Henan, China |
| 15 | 00028387 | 豫谷18号Yugu 18 | 中国河南 Henan, China |
| 16 | 00027937 | 安04-5014 An 04-5014 | 中国河南Henan, China |
| 17 | 00024170 | 豫谷2号Yugu2 | 中国河南 Henan, China |
| 18 | 00010182 | 十里香Shilixiang | 中国河南 Henan, China |
| 19 | 00007844 | 毛毛亮Maomaoliang | 中国河南 Henan, China |
| 20 | 00020503 | 大黄糯谷Dahuangnuogu | 中国河南 Henan, China |
| 21 | 00027920 | 冀谷27 Jigu 27 | 中国河北 Hebei, China |
| 22 | 00027921 | 冀谷28 Jigu 28 | 中国河北 Hebei, China |
| 23 | 00027925 | 冀谷17 Jigu 17 | 中国河北 Hebei, China |
| 24 | 00027919 | 金谷1号Jingu 1 | 中国河北 Hebei, China |
| 25 | 冀特5号Jite 5 | 中国河北 Hebei, China | |
| 26 | 00027910 | 冀谷24 Jigu 24 | 中国河北 Hebei, China |
| 27 | 00027909 | 冀谷22 Jigu 22 | 中国河北 Hebei, China |
| 28 | 00027907 | 冀谷18 Jigu 18 | 中国河北 Hebei, China |
| 29 | 00027906 | 冀谷31 Jigu 31 | 中国河北 Hebei, China |
| 30 | 00027905 | 复12 Fu 12 | 中国河北 Hebei, China |
| 31 | 00027924 | 冀谷15 Jigu 15 | 中国河北 Hebei, China |
| 32 | 00027912 | 冀谷29 Jigu 29 | 中国河北 Hebei, China |
| 33 | 00027911 | 冀谷26 Jigu 26 | 中国河北 Hebei, China |
| 34 | 00027922 | 冀谷30 Jigu 30 | 中国河北 Hebei, China |
| 35 | 00027733 | 谷丰1号Gufeng 1 | 中国河北 Hebei, China |
| 36 | 532 | ||
| 37 | 00007619 | 张农8 Zhangnong 8 | 中国河北 Hebei, China |
| 38 | 00022106 | 坝谷6 Bagu 6 | 中国河北 Hebei, China |
| 39 | 00022109 | 坝谷81 Bagu 81 | 中国河北 Hebei, China |
| 40 | 00022123 | 坝谷210 Bagu 210 | 中国河北 Hebei, China |
| 41 | 00022173 | 坝91-0130 Ba 91-0130 | 中国河北 Hebei, China |
| 42 | 00022116 | 坝谷139 Bagu 139 | 中国河北 Hebei, China |
| 43 | 00022162 | 坝91-0053 Ba 91-0053 | 中国河北 Hebei, China |
| 44 | 00022166 | 坝91-0079 Ba 91-0079 | 中国河北 Hebei, China |
| 45 | 00007036 | 毛毛谷Maomaogu | 中国河北 Hebei, China |
| 46 | 00007040 | 沙粒滚Shaligun | 中国河北 Hebei, China |
| 47 | 00019241 | 大九根齐Dajiugenqi | 中国河北 Hebei, China |
| 48 | 00022090 | 矮41 Ai 41 | 中国河北 Hebei, China |
| 49 | 000027926 | 衡谷9号Henggu 9 | 中国河北 Hebei, China |
| 50 | 00022052 | 毛谷2号Maogu 2 | 中国河北 Hebei, China |
| 51 | 00022608 | 南育3号Nanyu 3 | 中国河北 Hebei, China |
| 52 | 00027898 | 承谷11 Chenggu 11 | 中国河北 Hebei, China |
| 53 | 00028046 | 矮88 Ai 88 | 中国河北 Hebei, China |
| 54 | 00012329 | 小青谷Xiaoqinggu | 中国河北 Hebei, China |
| 55 | 00012587 | 黄毛谷Huangmaogu | 中国河北 Hebei, China |
| 56 | 00022881 | 齐头白Qitoubai | 中国河北 Hebei, China |
| 57 | 00022525 | 老绳头Laoshengtou | 中国河北 Hebei, China |
| 58 | 00008960 | 龙爪谷Longzhuagu | 中国河北 Hebei, China |
| 59 | 00008461 | 小白苗Xiaobaimiao | 中国河北 Hebei, China |
| 60 | 00007568 | 黑色腰Heiseyao | 中国河北 Hebei, China |
| 61 | 00024082 | 济叶冲4 Jiyechong 4 | 中国山东Shandong, China |
| 62 | 00019597 | 鲁谷3号Lugu 3 | 中国山东Shandong, China |
| 63 | 00023553 | 早白糯Zaobainuo | 中国山东Shandong, China |
| 64 | 00012887 | 钱串子Qianchuanzi | 中国山东Shandong, China |
| 65 | 00014233 | 拔谷Bagu | 中国山东Shandong, China |
| 66 | 00012877 | 红根子谷Honggenzigu | 中国山东Shandong, China |
| 67 | 00011078 | 早谷Zaogu | 中国山东Shandong, China |
| 68 | 00006337 | 晋汾13 Jinfen 13 | 中国山西Shanxi, China |
| 69 | 00027993 | 晋谷35号Jingu 35 | 中国山西Shanxi, China |
| 70 | 00027995 | 汾选5号Fenxuan 5 | 中国山西Shanxi, China |
| 71 | 00018008 | 皇龙谷Huanglonggu | 中国山西Shanxi, China |
| 72 | 00017042 | 红腿谷Hongtuigu | 中国山西Shanxi, China |
| 73 | 00024502 | 大同黄Datonghuang | 中国山西Shanxi, China |
| 74 | 00020590 | 龙谷26 Longgu 26 | 中国黑龙江Heilongjiang, China |
| 75 | 00027854 | 白谷9号Baigu 9 | 中国吉林Jilin, China |
| 76 | 00027862 | 公谷66号Gonggu 66 | 中国吉林Jilin, China |
| 77 | 00027699 | 公谷69号Gonggu 69 | 中国吉林Jilin, China |
| 78 | 00027704 | 公矮3号Gong’ai 3 | 中国吉林Jilin, China |
| 79 | 00000785 | 白杆白沙Baiganbaisha | 中国吉林Jilin, China |
| 80 | 00000077 | 糟皮一把奇Zaopiyibaqi | 中国黑龙江Heilongjiang, China |
| 81 | 00015441 | 龙爪粘Longzhuanian | 中国黑龙江Heilongjiang, China |
| 82 | 00015445 | 安丰Anfeng | 中国黑龙江Heilongjiang, China |
| 83 | 00000011 | 老来变Laolaibian | 中国黑龙江Heilongjiang, China |
| 84 | 00027903 | 06-766 | 中国北京 Beijing, China |
| 85 | 00012037 | 小早谷Xiaozaogu | 中国北京Beijing, China |
| 86 | 00018398 | 红杆谷Honggangu | 中国陕西Shaanxi, China |
| 87 | 00026203 | 呼和浩特大毛谷Huhehaotedamaogu | 中国陕西Shaanxi, China |
| 88 | 00028011 | 延谷11号Yangu 11 | 中国陕西Shaanxi, China |
| 89 | 11郄1071 11qie1071 | ||
| 90 | 00014610 | 米泉谷Miquangu | 中国新疆Xinjiang, China |
| 91 | 00014612 | 沙湾谷子Shawanguzi | 中国新疆Xinjiang, China |
| 92 | 00018817 | 谷子Guzi | 中国新疆Xinjiang, China |
| 93 | 00018811 | 谷子Guzi | 中国新疆Xinjiang, China |
| 94 | 00021709 | 谷上谷Gushanggu | 中国甘肃 Gansu, China |
| 95 | 00028014 | 陇谷11号Longgu 11 | 中国甘肃Gansu, China |
| 96 | 00027972 | 塞外香谷子Saiwaixiangguzi | 中国宁夏Ningxia, China |
| 97 | 00021671 | 红燃谷Hongrangu | 中国宁夏Ningxia, China |
| 98 | 00018751 | 小苗谷Xiaomiaogu | 中国宁夏Ningxia, China |
| 99 | 00003251 | 大青苗鱼刺Daqingmiaoyuci | 中国甘肃Gansu, China |
| 100 | 00018675 | 尕红谷Gahonggu | 中国甘肃Gansu, China |
| 101 | 00002910 | 黄玉3 Huangyu 3 | 中国内蒙古 Inner Mongolia, China |
| 102 | 00003005 | 籼紫灰谷Xianzihuigu | 中国内蒙古Inner Mongolia, China |
| 103 | 00003008 | 二白谷Erbaigu | 中国内蒙古Inner Mongolia, China |
| 104 | 00022053 | 蒜皮白Suanpibai | 中国内蒙古Inner Mongolia, China |
| 105 | 00015547 | 金香玉Jinxiangyu | 中国内蒙古Inner Mongolia, China |
| 106 | 00014740 | 朝鲜谷子Chaoxianguzi | 朝鲜Korea |
| 107 | 00015037 | SET3/80 | 德国Germany |
| 108 | 00015049 | ISE-430 | 美国 America |
| 109 | 00014718 | 大王国Dawangguo | 日本Japan |
| 110 | 00014938 | ISE-245 | 印度 India |
| 111 | 00015012 | 法谷28-81 Fagu 28-81 | 法国 France |
| 112 | 00014729 | 骨绿早1 Gulyuzao 1 | 朝鲜DPRK |
| 113 | 00022367 | ISE 770 | 国际半干旱研究所ICRISAT |
| 114 | 00022370 | ISE 775 | 国际半干旱研究所ICRISAT |
| 115 | 00014697 | 岛原Daoyuan | 日本Japan |
| 116 | 00014706 | 六十日Liushiri | 日本Japan |
| 117 | 00015402 | 金德Jinde | 美国America |
| 118 | 00022430 | Red manna | 南非South Africa |
| 119 | 00026837 | K-3606 | 俄罗斯Russia |
| 120 | 00014949 | Ise-455 | 美国America |
| 121 | 00015029 | Set 64/82 | 德国Germany |
| 122 | 00027931 | 8322-14 | |
| 123 | 00027915 | 白米1号Baimi 1 | 中国辽宁 Liaoning, China |
| 124 | 00027927 | 2013 | |
| 125 | 0027917 | 谷丰2号Gufeng 2 | 中国河北Hebei, China |
| 126 | 00015128 | 芝麻粟Zhimasu | |
| 127 | 00014623 | 大头糯Datounuo | 中国湖南 Hunan, China |
| 128 | 00022285 | 乐山白糯Leshanbainuo | 中国四川 Sichuan, China |
| 129 | 00022313 | 黄谷Huanggu | 中国西藏 Tibet, China |
| 130 | 00026488 | 黄粟Huangsu | 中国广西 Guangxi, China |
| 131 | 00025646 | 黄谷子Huangguzi | 中国青海Qinghai, China |
| 132 | 00025962 | 喇叭黄Labahuang | 中国青海Qinghai, China |
| 133 | 15HN-206 | 中国河北Hebei, China | |
| 134 | 15HN-138 | 中国河北Hebei, China | |
| 135 | 15HN-79 | 中国河北Hebei, China | |
| 136 | 六十天还家 Liushitianhuanjia | 中国吉林Jilin, China | |
| 137 | 谷子Guzi | 中国青海Qinghai, China | |
| 138 | 红糯谷Hongnuogu | 中国宁夏Ningxia, China | |
| 139 | 小明谷子Xiaomingguzi | 中国甘肃Gansu, China | |
| 140 | 粘子糯Nianzinuo | ||
| 141 | 毛粟Maosu | ||
| 142 | 00018651 | 茄谷Qiegu | 中国甘肃Gansu, China |
| 143 | 然谷Rangu | 中国陕西Shaanxi, China | |
| 144 | 铁谷4号Tiegu 4 | 中国辽宁 Liaoning, China | |
| 145 | 红苗2 Hongmiao 2 | 中国吉林Jilin, China | |
| 146 | 叩根Kougen | 中国吉林Jilin, China | |
| 147 | 00021176 | 铁7924 Tie 7924 | 中国辽宁 Liaoning, China |
| 148 | 00020837 | 小金苗Xiaojinmiao | 中国吉林Jilin, China |
| 149 | 00015437 | 嫩选十号Nenxuanshi | 中国黑龙江Heilongjiang, China |
| 150 | 00028013 | 陇谷10号Longgu 10 | 中国甘肃Gansu, China |
| 151 | 00027871 | 辽谷1号Liaogu 1 | 中国辽宁 Liaoning, China |
| 152 | 00027852 | 嫩选十六Nenxuanshiliu | 中国黑龙江Heilongjiang, China |
| 153 | 00027882 | 蒙早谷9号Mengzaogu 9 | 中国内蒙古Inner Mongolia, China |
| 154 | 红钙谷Honggaigu | 中国天津Tianjin, China | |
| 155 | 00007028 | 黑谷子Heiguzi | 中国河北Hebei, China |
| 156 | 00021303 | 赤谷6号Chigu 6 | 中国内蒙古Inner Mongolia, China |
| 157 | 二不黄Erbuhuang | 中国山西Shanxi, China | |
| 158 | 白罗砂Bailuosha | 中国河北Hebei, China | |
| 159 | 竹叶青Zhuyeqing | 中国河北Hebei, China | |
| 160 | 压塌车Yatache | 中国河北Hebei, China |
新窗口打开|下载CSV
1.2 谷子SiPRR37基因的克隆
首先通过检索NCBI数据库获得1条注释为PRR37基因的谷子mRNA序列(XM_022824614), 将该基因命名为SiPRR37, 以此序列为模板设计3对特异引物用于基因扩增(表1)。将延谷11号饱满种子种植于口径为10 cm×10 cm装有营养土的塑料盆中, 在自然条件下生长至五叶期时采集顶叶, 液氮速冻, 然后用Ultrapure RNA Kit (康为世纪生物科技有限公司)提取总RNA, 用PrimeScript II 1st Strand cDNA Synthesis Kit (宝日医生物技术(北京)有限公司)反转录合成第一链cDNA, 用3对特异性引物分段扩增目的基因, 扩增体系包括稀释10倍的cDNA模板1 μL、2×Es Taq MasterMix (Dye) (康为世纪生物科技有限公司) 10 μL、10 μmol L-1正、反向引物各0.5 μL, 最后用ddH2O补足到20 μL。PCR扩增程序为94℃预变性5 min; 94℃变性30 s、58℃退火30 s、72℃延伸90 s, 35个循环; 72℃延伸5 min。用EasyPure Quick Gel Extraction Kit (北京全式金生物技术有限公司)纯化回收PCR产物, 然后连接到pBM16A克隆载体(北京艾德莱生物科技有限公司), 再转化大肠杆菌DH5α感受态细胞, 挑选阳性克隆菌液送生工生物工程(上海)股份有限公司测序。Table 1
表1
表1特异性引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequences (5°-3°) | 预期片段大小 Expected fragment size (bp) | 用途 Purpose |
|---|---|---|---|
| PRR37-1 | F: TGACAACGACGAGGACG R: CGTTAGCAATCTCCGTGT | 1295 | 克隆SiPRR37 Cloning SiPRR37 |
| PRR37-2 | F: CCATTTGCTGACTCGCCTAC R: TCCTGTCCCGCCTTGAT | 1586 | |
| PRR37-3 | F: AAGGCTCCAATGGCAGTAG R: GAACCAGCGAAGGAAGAATC | 1267 | |
| SiActin | F: GGCAAACAGGGAGAAGATGA R: GAGGTTGTCGGTAAGGTCACG | 229 | 内参荧光定量 Internal reference fluorescence quantification |
| RtPRR37 | F: CACCACTTTCGTCTACCTCTT R: CTGGCATCTCTTCTAACGG | 91 | SiPRR37荧光定量 SiPRR37 fluorescence quantification |
新窗口打开|下载CSV
1.3 谷子SiPRR37基因的生物信息学分析
利用在线分析工具SubLoc软件(1.4 谷子SiPRR37基因的表达分析
将延谷11号种子种于口径10 cm×10 cm装有营养土的塑料方盆中, 总计种植115盆, 每盆种植4株。将其中3盆放在低温短日照(low temperature short day, LTSD, 22℃、9 h光/15 h暗)条件下培养至抽穗, 然后用剪刀剪取穗、穗颈、茎秆、顶叶、次顶叶和根, 放入液氮速冻, 每个样品3次重复, 用于SiPRR37基因的组织特异性表达分析。将64盆放在自然条件下长至三叶期后分别转至高温短日照(high temperature short day, HTSD, 27℃、9 h光/15 h暗)、低温短日照(low temperature short day, LTSD, 22℃、9 h光/15 h暗)、高温长日照(high temperature long day, HTLD, 27℃、15 h光/9 h暗)、低温长日照(low temperature long day, LTLD, 22℃、15 h光/9 h暗) 4个培养箱培养, 每个培养箱16盆, 2个重复, 3周后昼夜24 h内每隔3 h取植株顶端2片叶液氮速冻, 取样时间点为6:00、9:00、12:00、15:00、18:00、21:00、0:00、3:00, 所取样品用于SiPRR37基因昼夜表达分析。
剩余的48盆分为6组, 1个对照(CK)组, 5个胁迫处理组, 每组8盆, 在培养室萌发3周后分别进行200 mmol L-1 NaCl、100 μmol L-1 ABA、20% PEG-6000、600 μmol L-1 EDTA-Fe和15℃冷胁迫处理。NaCl、EDTA-Fe和PEG-6000处理采用浇灌法, ABA处理采用叶面喷洒法, 将整盆植株移入15℃冰箱进行冷胁迫处理。分别取各处理组和对照组(CK)在处理前(0 h)和处理后0.5、1、2、4、8、16、24 h每株顶端2片叶于液氮速冻, 培养室温度为25℃, 光照条件为14 h光/10 h暗, 所取样品用于SiPRR37基因在非生物胁迫条件下的表达分析。
上述所有样品RNA提取、反转录同1.2, 将反转录好的cDNA作为Real-time PCR的模板, 用表1中的SiActin引物和RtPRR37引物分别扩增内参基因和SiPRR37基因特异片段, 每个样品做3个重复, 扩增体系及循环程序参照宝日医生物技术(北京)有限公司的TB Green Premix Ex Taq II (Tli RNaseH Plus)试剂说明书, 首先配制10 μL扩增体系, 包含cDNA模板1 μL、TB Green Premix Ex Taq II (Tli RNaseH Plus) 5 μL、10 μmol L-1的RtPRR37正向和反向引物各1 μL、ddH2O 2 μL。使用Roche Light Cycler 96实时定量PCR仪, 采用两步法PCR反应程序, 扩增程序为95℃预变性30 s; 95℃变性30 s, 58℃退火30 s, 共40个循环。分析得到的扩增曲线、溶解曲线, 并计算??CT值, 采用2-ΔΔCT计算相对表达量。
1.5 SiPRR37基因单倍型效应分析
从160份谷子材料重测序数据中获得SiPRR37基因SNP位点, 利用dnasp 5软件对SiPRR37基因编码区的SNP位点进行单倍型分析, 利用本实验室于2015、2016连续2年在海南乐东、河南洛阳、吉林吉林市、公主岭市调查的160份谷子材料8个农艺性状表型数据进行单倍型效应分析[30]。2 结果与分析
2.1 谷子SiPRR37基因的克隆及生物信息学分析
首先提取延谷11号叶片总RNA, 经1%的琼脂糖凝胶电泳检测所提RNA质量较好, 无明显降解, 可以用于后续的反转录试验(图1)。以反转录合成的第一链cDNA为模板, 用PRR37-1、PRR37-2和PRR37-3 3对特异性引物分段扩增SiPRR37基因, 均获得预期大小片段(图2-A, B)。将3个扩增片段测序后经过拼接得到谷子SiPRR37基因含有完整编码区域的cDNA序列, 该序列长度为2953 bp, 包含2247 bp的CDS区域, 编码748个氨基酸(附图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1延谷11号叶片总RNA电泳结果
1, 2: 提取的两管RNA。
Fig. 1Electrophoresis results of total RNA extracted from leaves in Yangu 11
1, 2: two tubes of RNA extracted.
图2
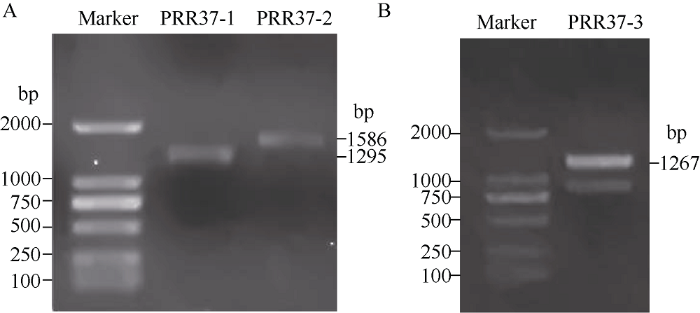 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2SiPRR37基因的RT-PCR产物电泳结果
M: marker DL2000; A: PRR37-1, PRR37-2扩增产物; B: PRR37-3扩增产物。
Fig. 2RT-PCR results of SiPRR37 genes
M: marker DL2000; A: amplified products of PRR37-1 and PRR37-2; B: amplified product of PRR37-3.
附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1谷子 SiPRR37 基因的核苷酸序列及其编码的氨基酸序列
下画线部分为起始密码子和终止密码子。
Fig. S1cDNA sequences of SiPRR37 gene in millet
The underlined parts are initiation codon and termination codon.
生物信息学分析发现, 谷子SiPRR37蛋白定位于细胞核中, 该蛋白含有2个明显的结构域, 即REC和CCT结构域(图3); SiPRR37蛋白的多肽链中只有丝氨酸(Ser)可能发生磷酸化, 85个丝氨酸残基中较易发生磷酸化的有14个, 占16.47% (附图2)。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3谷子SiPRR37蛋白的结构域分析
Fig. 3Domain analysis of SiPRR37 protein in millet
附图2
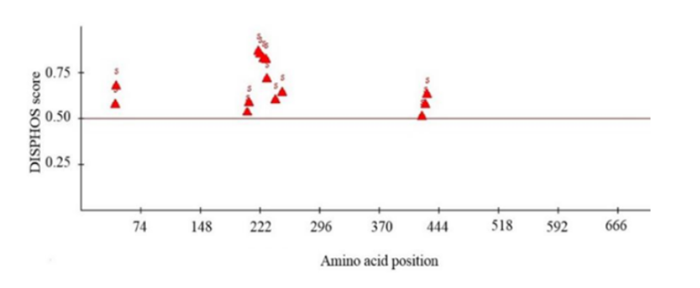 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图2谷子SiPRR37 蛋白磷酸化位点分析
Fig. S2Analysis of phosphorylation site of millet SiPRR37 protein
在NCBI数据库中下载与谷子SiPRR37蛋白具有直系同源关系的大麻(Cannabis sativa XP_ 030488228.1)、枣(Ziziphus jujuba XP_015877113.1)、蓖麻(Ricinus communis XP_015578380.1)、澳洲棉(Gossypium australe KAA3464109.1)、番木瓜(Caricapapaya XP_021910105.1)、葡萄(Vitis vinifera XP_010658157.1)等32种植物的PRR37蛋白序列, 采用NJ法构建分子系统进化树发现, 谷子与糜子亲缘关系最近, 其次是高粱和玉米, 这4种C4作物聚为一个小亚群, 与同为禾本科作物的小麦和水稻聚为一个较大的群, 其余物种聚在另一个大群里(图4)。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4PRR37蛋白的分子系统进化树
Fig. 4Molecular phylogenetic tree of PRR37 protein
图5
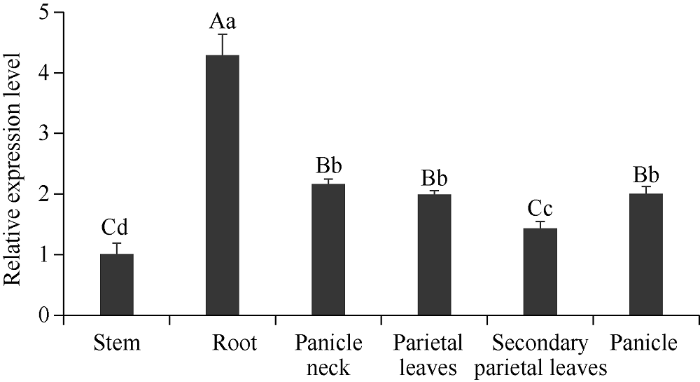 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5SiPRR37基因的组织特异性表达分析
柱上不同大小写字母分别表示在0.01和0.05水平差异显著。
Fig. 5Tissue-specific expression analysis of SiPRR37 gene
Values followed by different uppercase and lowercase letters above the bar represent significant difference at the 0.01 and 0.05 probability levels, respectively.
分析SiPRR37基因上游启动子区发现, 除了TATA-box、CCAAT-box核心启动子元件外, 还有8个光响应元件、1个低温响应元件、2个赤霉素响应元件、1个生长素响应元件、2个ABA响应元件和2个茉莉酸甲酯响应元件, 说明SiPRR37基因受光温和多种激素调控。此外启动子区还发现2个参与干旱、盐等逆境胁迫的作用元件, 说明SiPRR37在谷子应对逆境胁迫中可能发挥作用(表2)。
Table 2
表2
表2SiPRR37启动子区顺式作用元件分析
Table 2
| 调控元件 Regulatory element | 核心序列 Core sequence | 数目 Number | 分布区域 Distribution region (bp) | 功能 Function |
|---|---|---|---|---|
| LTR | CCGAAA | 1 | 658-663 | 参与低温响应顺式激活元件 cis-acting element involved in low-temperature responsiveness |
| G-box | CACGTG, TCCACATGGCA, CACGAC | 4 | 123-128, 1870-1879, 1103-1108, 1517-1522 | 参与光响应顺式作用元件 cis-acting regulatory element involved in light responsiveness |
| AE-box | AGAAACTT | 1 | 409-416 | 部分光响应元件 Part of a light responsive element |
| Box II | CCACGTGGC | 1 | 122-130 | 部分光响应元件 Part of a light responsive element |
| Sp1 | GGGCGG | 1 | 23-28 | 光响应元件 Light responsive element |
| 3-AF1 binding site | TAAGAGAGGAA | 1 | 1045-1054 | 光响应元件 Light responsive element |
| AuxRR-core | GGTCCAT | 1 | 1536-1542 | 参与生长素响应顺式作用元件 cis-acting regulatory element involved in auxin responsiveness |
| TGACG-motif | TGACG, CGTCA | 2 | 29-33, 1563-1567 | 参与茉莉酸甲酯响应的顺式激活元件 cis-acting regulatory element involved in the MeJA- responsiveness |
| TATC-box | TATCCCA | 1 | 925-931 | 参与赤霉素响应顺式激活元件 cis-acting element involved in gibberellin-responsiveness |
| P-box | CCTTTTG | 1 | 1726-1732 | 赤霉素响应元件 Gibberellin-responsive element |
| Myb-binding site | CAACAG | 1 | 1154-1159 | 参与干旱、盐胁迫等逆境响应 Involved in drought, salt dresses responsiveness |
| DRE1 | ACCGAGA | 1 | 342-348 | 干旱胁迫响应元件 |
| ABRE | ACGTG, CACGTG | 2 | 31-35, 123-128 | 参与ABA响应的顺式激活元件 cis-acting element involved in the abscisic acid responsiveness |
新窗口打开|下载CSV
2.2 SiPRR37基因的组织特异性表达分析
通过分析SiPRR37基因在延谷11茎秆、根、穗颈、顶叶、次顶叶和穗6个部位的相对表达量发现, 在抽穗后, SiPRR37基因在根中表达量最高, 茎秆中表达量最低, 相对表达量从高到低依次为根>穗颈>穗>顶叶>次顶叶>茎秆(图5)。2.3 SiPRR37基因在4种光温组合条件下的表达分析
在4种光温组合条件下SiPRR37基因均在光照期出现1个表达峰, 且光照期的表达量明显高于黑暗期, 说明该基因受光诱导(图6-A~D)。此外无论低温(22℃)还是高温(27℃), SiPRR37在短日照条件的表达峰均比长日照提前, 低温条件短日照在光照6 h表达量最高, 长日照在光照9 h表达量最高; 高温条件短日照在光照9 h表达量最高, 长日照在光照12 h表达量最高。同时可以看出, 温度的提高使SiPRR37的表达峰无论长日照条件还是短日照条件均发生推迟。说明SiPRR37在受光周期调控的同时也受温度的影响。图6
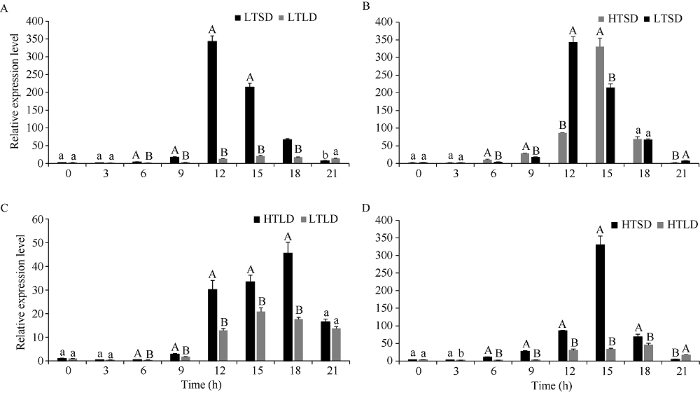 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不同光温组合处理SiPRR37基因表达特性
A: 低温条件不同光周期对SiPRR37基因的影响; B: 短日照条件不同温度对SiPRR37基因的影响; C: 长日照条件不同温度对SiPRR37基因的影响; D: 高温条件不同光周期对SiPRR37基因的影响。长日照: 6:00-21:00光照, 0:00-6:00和21:00-0:00为黑暗; 短日照: 6:00-15:00光照, 0:00-6:00和15:00-0:00为黑暗。LTSD: 低温短日照; LTLD: 低温长日照; HTSD: 高温短日照; HTLD: 高温长日照。柱上不同大小写字母分别表示在0.01和0.05水平差异显著。
Fig. 6Expression characteristics of SiPRR37 gene under different photo-thermal combinational treatments
A: the effect of different photoperiods on SiPRR37 gene under low temperature condition; B: the effect of different temperatures on SiPRR37 gene under short-day condition; C: the effect of different temperatures on SiPRR37 gene under long-day condition; D: the effect of different photoperiods on SiPRR37 gene under high temperature condition. Long day: 6:00-21:00 light, 0:00-6:00 and 21:00-0:00 dark; Short day: 6:00-15:00 light, 0:00-6:00 and 15:00-0:00 dark. LTSD: low temperature short day; LTLD: low temperature long day; HTSD: high temperature short day; HTLD: high temperature long day. Different uppercase and lowercase letters above the bar represent significant difference at the 0.01 and 0.05 probability levels, respectively.
2.4 谷子SiPRR37基因对非生物胁迫的响应
SiPRR37基因在NaCl胁迫处理24 h内表达量均显著或极显著低于对照, 说明盐胁迫能够明显抑制SiPRR37基因的表达(图7-A); 在ABA处理2~12 h SiPRR37表达水平和对照产生极显著差异, 说明SiPRR37参与了ABA信号传导过程(图7-B); 在PEG胁迫处理4 h、24 h与对照表达量无显著差异, 其余时间点表达量均极显著高于对照, 说明SiPRR37可以被干旱胁迫诱导表达(图7-C)。图7
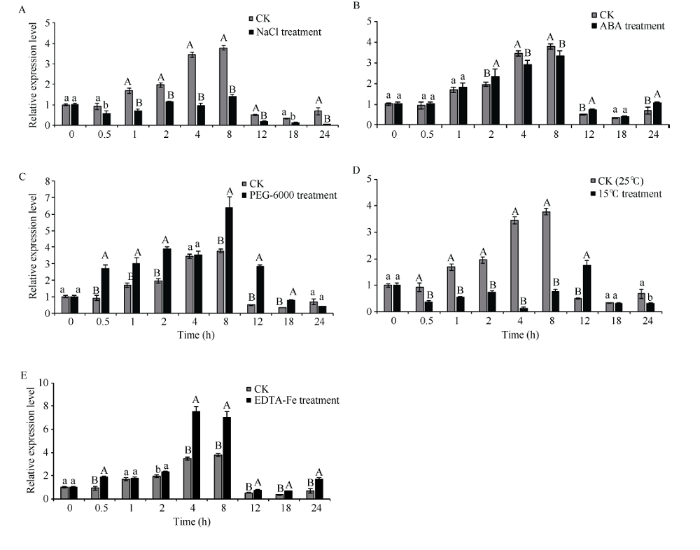 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7谷子SiPRR37基因对非生物胁迫的响应
A: 盐胁迫; B: ABA渗透胁迫; C: PEG模拟干旱胁迫; D: 低温胁迫; E: 铁胁迫。柱上不同大小写字母分别表示在0.01和0.05水平差异显著。
Fig. 7Response of SiPRR37 gene to abiotic stresses in millet
A: salt stress; B: ABA osmotic stress; C: PEG simulated drought stress; D: low-temperature stress; E: iron stress. Different uppercase and lowercase letters above the bars represent significant difference at the 0.01 and 0.05 probability levels, respectively.
15℃低温胁迫处理除了在处理12 h SiPRR37被诱导表达、处理18 h 表达水平接近于对照, 其余时间点表达量均显著或极显著低于对照, 说明总体上15℃低温胁迫处理使SiPRR37表达受到抑制(图7-D)。Fe胁迫处理的24 h内除了处理1 h SiPRR37基因的表达量与对照无显著差异, 其余时间点表达水平均显著或极显著高于对照(图7-E), 说明铁胁迫处理可以诱导SiPRR37基因的表达, SiPRR37蛋白有fer1、fer3和fer4 3种铁蛋白作用靶点, 推测其在铁过量应激反应中具有保护作用。
2.5 SiPRR37基因单倍型效应分析
从160份谷子重测序数据中提取获得14,343 bp的SiPRR37基因核苷酸序列, 包括978 bp的5°非翻译区、2247 bp的编码区、10,440 bp的内含子区和678 bp的3°非翻译区。对SiPRR37基因结构进行分析发现, 该基因编码区有7个外显子、6个内含子。经对160份谷子材料基因序列进行比对共获得47个SNPs, 其中外显子区10个SNPs, 包括8个错义突变, 错义突变频率为80% (表3)。Table 3
表3
表3SiPRR37基因在160份谷子材料中检测到的SNP位点
Table 3
| 参数 Parameter | 5°非翻译区 5° UTR | 外显子区 Exon region | 内含子区 Intron region | 3°非翻译区 3° UTR | 全长 Full length |
|---|---|---|---|---|---|
| 序列长度Sequence length (bp) | 978 | 2247 | 10,440 | 678 | 14,343 |
| SNP数目 Number of SNPs | 6 | 10 | 31 | 0 | 47 |
| SNP频率SNP frequency (bp SNP-1) | 163 | 224.7 | 336.77 | 0 | 305.17 |
新窗口打开|下载CSV
对SiPRR37基因外显子区的10个SNP位点进行单倍型分析, 共发现19个单倍型, 160份谷子材料在19个单倍型中分布不均衡, 出现频率最高的单倍型是Hap_1, 包括84个谷子品种, 占所有品种的52.5%; 8个单倍型(Hap_3、Hap_4、Hap_8、Hap_10、Hap_12、Hap_17、Hap_18和Hap_19)只包含1个品种, 占所有品种的5% (附表2)。对19个单倍型进行表型效应分析发现, Hap_19的抽穗期、叶片数、穗长、穗码数显著高于其他单倍型, 但是千粒重最低(图8-A, C, D, F, H); Hap_10的穗粒重、穗粗、穗重显著高于其他单倍型(图8-B, E, G); Hap_7的千粒重最高(图8-H)。
Table S2
附表2
附表2 SiPRR37基因编码区序列的单倍型在160份谷子品种中的分布
Table S2
| 单倍型 Haplotype | 个数 Number | 品种 Variety |
|---|---|---|
| Hap_1 | 84 | 郑谷2, 豫谷6, 郑06-6, 郑05-2, 郑州12, 郑315, 安5424, 豫谷11, 豫谷16, 安4117, 豫谷13号, 安04-5014,大黄糯谷, 冀谷27, 冀谷28, 冀谷17, 金谷1号, 冀谷24, 冀谷22, 冀谷18, 冀谷31, 复12, 冀谷15, 冀谷29号, 冀谷26号, 冀谷30, 谷丰1号, 532, 坝91-0130, 坝91-0053, 大九根齐, 矮41, 衡谷9号, 南育3号, 承谷11, 矮88, 黄毛谷, 齐头白, 老绳头, 济叶冲4, 早白糯, 钱串子, 拔谷, 红根子谷, 晋谷35号, 皇龙谷, 龙谷26, 白谷9号, 公谷66号, 公谷69号, 公矮3号, 白杆白沙, 安丰, 老来变, 06-766, 延谷11号, 黄玉3, 金香玉, ISE-430, 大王国, ISE-245, 骨绿早1, ISE775, 岛原, 六十日, 金德, Set64/82, 8322-14, 白米1号, 2013, 芝麻粟, 大头糯, 乐山白糯, 黄谷, 喇叭黄, 15HN-138, 然谷, 红苗2, 铁7924, 小金苗, 嫩选十六, 黑谷子, 赤谷6号, 压塌车 Zhenggu 2, Yugu 6, Zheng 06-6, Zheng 05-2, Zhengzhou 12, Zheng 315, An 5424, Yugu 11, Yugu 16, An 4117, Yugu 13, An 04-5014, Dahuangnuogu, Jigu 27, Jigu 28, Jigu 17, Jingu 1, Jigu 24, Jigu 22, Jigu 18, Jigu 31, Fu 12, Jigu 15, Jigu 29, Jigu 26, Jigu 30, Gufeng 1, 532, Ba 91-0130, Ba 91-0053, Dajiugenqi, Ai 41, Henggu 9, Nanyu 3, Chenggu 11, Ai 88, Huangmaogu, Qitoubai, Laoshengtou, Jiyechong 4, Zaobainuo, Qianchuanzi, Bagu, Honggenzigu, Jingu 35, Huanglonggu, Longgu 26, Baigu 9, Gonggu 66, Gonggu 69, Gong’ai 3, Baiganbaisha, Anfeng, Laolaibian, 06-766, Yangu 11, Huangyu 3, Jinxiangyu, ISE-430, Dawangguo, ISE-245, Gulyzao 1, ISE775, Daoyuan, Liushiri, Jinde, Set64/82, 8322-14, Baimi1, 2013, Zhimasu, Datounuo, Leshanbainuo, Huanggu, Labahuang, 15HN-138, Rangu, Hongmiao 2, Tie7924, Xiaojinmiao, Nenxuanshiliu, Heiguzi, Chigu 6, Yatache |
| Hap_2 | 2 | 豫谷3号, 鲁谷3号 Yugu 3, Lugu 3 |
| Hap_3 | 1 | 郑8041 Zheng 8041 |
| Hap_4 | 1 | 豫谷15 Yugu 15 |
| Hap_5 | 2 | 豫谷18号, 沙粒滚 Yugu 18, Shaligun |
| Hap_6 | 48 | 豫谷2号, 十里香, 坝谷6, 坝谷81, 坝谷210, 坝91-0079, 毛毛谷, 毛谷2号, 小青谷, 龙爪谷, 黑色腰, 早谷,晋汾13, 汾选5号, 红腿谷, 龙爪粘, 红杆谷, 呼和浩特大毛谷, 11郄1071, 米泉谷, 谷上谷, 塞外香谷子, 红燃谷, 小苗谷, 大青苗鱼刺, 尕红谷, 籼紫灰谷, 二白谷, Red manna, Ise-455, 黄粟, 黄谷子, 15HN-206, 15HN-79,六十天还家, 谷子, 红糯谷, 毛粟, 茄谷, 铁谷4号, 嫩选十号, 陇谷10号, 辽谷1号, 蒙早谷9号, 红钙谷, 二不黄, 白罗砂, 竹叶青 Yugu 2, Shilixiang, Bagu 6, Bagu 81, Bagu 210, Ba 91-0079, Maomaogu, Maogu 2, Xiaoqinggu, Longzhuagu, Heiseyao, Zaogu, Jinfen 13, Fenxuan 5, Hongtuigu, Longzhuanian, Honggangu, Huhehaotedamaogu, 11qie1071, Miquangu, Gushanggu, Saiwaixiangguzi, Hongrangu, Xiaomiaogu, Daqingmiaoyuci, Gahonggu, Xianzihuigu, Erbaigu, Red manna, Ise-455, Huangsu, Huangguzi, 15HN-206, 15HN-79, Liushitianhuanjia, Guzi, Hongnuogu, Maosu, Qiegu, Tiegu 4, Nenxuanshi, Longgu 10, Liaogu1, Mengzaogu 9, Honggaigu, Erbuhuang, Bailuosha, Zhuyeqing |
| Hap_7 | 2 | 毛毛亮, 陇谷11号 Maomaoliang, Longgu 11 |
| Hap_8 | 1 | 冀特5号 Jite 5 |
| Hap_9 | 3 | 张农8, 谷子, 蒜皮白 Zhangnong8, Guzi, Suanpibai |
| Hap_10 | 1 | 坝谷139 Bagu 139 |
| Hap_11 | 2 | 小白苗, K-3606 Xiaobaimiao, K-3606 |
| Hap_12 | 1 | 大同黄 Datonghuang |
| Hap_13 | 3 | 糟皮一把奇, 朝鲜谷子, SET3/80 Zaopiyibaqi, Chaoxianguzi, SET3/80 |
| Hap_14 | 2 | 小早谷, 谷子 Xiaozaogu, Guzi |
| Hap_15 | 2 | 沙湾谷子, 谷丰2号 Shawanguzi, Gufeng 2 |
| Hap_16 | 2 | 法谷28-81, 叩根 Fagu 28-81, Kougen |
| Hap_17 | 1 | ISE770 ISE770 |
| Hap_18 | 1 | 小明谷子 Xiaomingguzi |
| Hap_19 | 1 | 粘子糯 Nianzinuo |
新窗口打开|下载CSV
图8
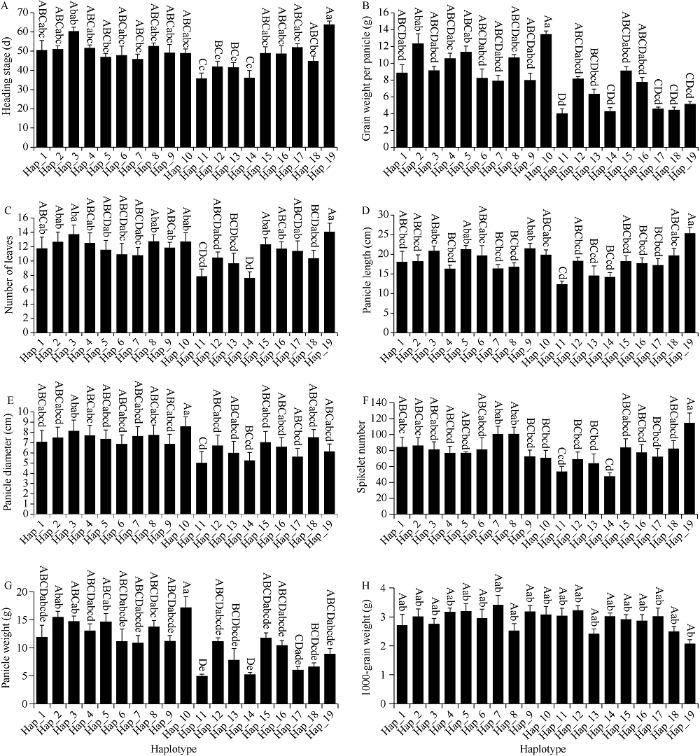 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8基于SiPRR37基因SNP的单倍型效应分析
柱上不同大小写字母分别表示在0.01和0.05水平差异显著。
Fig. 8Haplotypes effects analysis based on SNPs of SiPRR37 gene
Bars marked with different uppercase and lowercase letters represent significant difference at the 0.01 and 0.05 probability levels, respectively.
3 讨论
本研究从延谷11克隆的SiPRR37基因与在水稻和高粱中已报道的CO、OsPRR37、SbPRR37等基因一样, 均属于CCT基因家族中的PRR亚家族[3,13,16,37]。基于PRR37蛋白序列的系统进化分析表明, OsPRR37、SbPRR37与谷子SiPRR37有较近的进化关系, 推测它们可能具有相同或类似的功能, 能对节律性变化的环境条件作出应答反应, 参与植物开花期的调控[8]。OsPRR37基因通过感受外界光周期和温度的变化而调控抽穗开花, 对水稻的环境适应性有重要影响, 是控制水稻抽穗期、株高和每穗小穗数的主效基因[5,6,7,8]; 而高粱SbPRR37与籽粒成熟度和茎秆甜度相关[17]; 小麦TaPRR37对抽穗期、株高和千粒重有显著影响[38]。表明PRR37是一个多效基因, 同时控制多个农艺性状。本研究通过单倍型效应分析发现, SiPRR37基因与谷子抽穗期、穗码数、穗粒重、千粒重等性状相关, 同样具有多效性, 与其他作物报道的PRR37基因研究结果一致[2,5-6,17,38,40]。但是SiPRR37的表达模式与SbPRR37、OsPRR37存在差异, 虽然三者的表达都是光依赖性的, 属于光周期途径中调控作物开花的主要基因[7,13], SbPRR37在长日照下具有早晨和黄昏2个表达峰, 在短日照下仅有早晨表达峰, 而SiPRR37无论高温还是低温、长日照还是短日照, 只在光照期出现1个表达峰, 在暗期表达峰消失, 其昼夜节律性表达与拟南芥直系同源基因APRR7相似[37]。产生这种结果的原因可能和取样间隔时间有关, 本研究昼夜取样间隔时间为3 h, 较疏的取样密度可能会漏掉潜在的表达峰。本研究发现, 温度的提高使SiPRR37的表达峰无论长日照条件还是短日照条件均发生推迟, 但不影响短日照条件SiPRR37的表达峰比长日照提前, 说明光周期对SiPRR37表达起主要调控作用, 但其同时也受温度的影响, SiPRR37可能参与了谷子光温互作调节过程。我们近期发现光周期和温度对谷子生长发育具有明显的互作效应, 长日照条件温度升高使抽穗期延迟, 而短日照条件温度升高使抽穗期缩短, 高温的作用受到光周期的制约[39], SiPRR37基因在此互作过程中起到怎样的作用值得深入研究。PRRs家族基因还广泛参与逆境胁迫调节过程。有研究表明温度主要影响生物钟基因的可变剪切方式, 低温可以使PRRs家族基因形成不同的转录本来影响生物钟功能和增强植物对冷胁迫的耐受性[22]。在冷胁迫中, PRR9、PRR7、PRR5作为负调节因子来调节抗冷基因CBF的表达[20,23]。有研究发现, 水稻受到冷胁迫刺激后, 会促进OsPRR37的表达[41], 但是本研究中SiPRR37基因在15℃低温胁迫下表达被显著抑制, 这表明PRRs家族基因在不同的作物中对低温胁迫的应对方式存在差异。铁也是一种非生物胁迫因子, 土壤中过量的铁会损害作物根系细胞, 降低生长能力, 比缺铁时更影响产量。有研究表明PRR7具有fer1、fer3和fer4三种铁蛋白作用靶点, 而铁蛋白在作物中能够储存过量的铁, 因此推测PRR7参与了铁胁迫调节过程[18,21]。本研究发现, 铁胁迫处理可诱导SiPRR37基因的表达, 推测该基因也具有铁蛋白作用靶点, 在作物铁过量应激反应中具有保护作用。PRRs家族基因也参与植物对干旱胁迫的响应[42]。在水稻中, 干旱可以抑制OsPRR73的表达, 诱导OsTOC1的表达, 对OsPRR59、OsPRR95和OsPRR37基因的影响较小[43]; 在大豆中, OsPRR37的直系同源基因GmPRR7在干旱胁迫下表达减弱, 仅GmPRR3的表达增高, 在响应干旱胁迫中发挥作用。本研究发现, 干旱胁迫可以诱导SiPRR37的表达, 说明SiPRR37基因参与了谷子干旱胁迫的应答反应, 与水稻OsPRR37不同。ABA在植物对逆境的适应性反应中起着重要作用, 已有的研究表明, PRRs家族的TOC1基因与ABA调控基因有部分重叠, TOC1能够结合到ABA调控基因启动子上抑制 ABAR、CBF、ABI3等基因的表达, 这些基因的表达又能促进TOC1表达, 从而能够形成调节ABA信号传导的调节环, 以增加植物抗逆性[44]。本研究发现, SiPRR37在ABA胁迫1 h后开始响应表达, 随后多数时间点表达受到抑制, 推测SiPRR37通过某种作用机制参与了ABA介导的信号传导过程。
4 结论
从延谷11克隆得到SiPRR37基因2953 bp的cDNA序列, 包含2247 bp的完整CDS区域, 编码748个氨基酸; SiPRR37基因含有REC和CCT 2个结构域, 属于CCT基因家族的PRR亚家族成员, 与玉米PRR37、高粱SbPRR37、糜子PRR37蛋白亲缘关系较近; SiPRR37基因表达具有昼夜节律性, 且同时受光周期和温度调控, 可能参与了谷子光温互作调节过程; SiPRR37基因受干旱胁迫和铁胁迫诱导, 受盐胁迫和低温胁迫抑制, 参与了对非生物胁迫的应答; SiPRR37基因和抽穗期及多个穗部性状相关, 具有多效性, 鉴定出Hap_19、Hap_10、Hap_7三个用于谷子生育期和穗部性状改良的有利单倍型。附表和附图 请见网络版: 1) 本刊网站
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1093/pcp/41.6.791URLPMID:10945350 [本文引用: 1]

In the higher plant, Arabidopsis thaliana, results from recent intensive studies suggested that His-to-Asp phosphorelay mechanisms are involved presumably in propagation of environmental stimuli, such as phytohormones (e.g. ethylene and cytokinin). Here we identified and characterized a set of novel Arabidopsis genes whose products considerably resemble the authentic response regulators (ARR-series) of Arabidopsis in the sense that they have a phospho-accepting receiver-like domain. However, they should be discriminated from the classical ones in the strict sense that they lack the invariant phospho-accepting aspartate site. They were thus named APRRs (Arabidopsis pseudo-response regulators). Two such representatives, APRR1 and APRR2, were characterized extensively through cloning of the corresponding cDNAs, in terms of their structural designs, biochemical properties, subcellular localization in plant cells, and expression profiles at the transcriptional level. The result of in vitro phosphorylation experiment with the Arabidopsis AHP phosphotransmitter suggested that the pseudo-receivers have no ability to undergo phosphorylation. The result of transient expression assay with onion epidermal cells showed that the GFP-APRR1 fusion protein has an ability to enter into the nuclei. The C-terminal domain of APRR1, termed CONSTANS-motif, appears to be responsible for the nuclear-localization. The most intriguing result was that the accumulation of APRR1 transcript is subjected to a circadian rhythm. The APRR1 protein is identical to the one that was recently suggested to interact with the ABI3 (ABISCISIC ACID INSENSITIVE3) protein. These are discussed with special reference to the His-to-Asp phosphorelay signal transduction and circadian rhythm in Arabidopsis thaliana.
[本文引用: 2]
[本文引用: 2]
DOI:10.1111/j.1365-313X.2007.03258.xURLPMID:17877705 [本文引用: 2]

Interlocking transcriptional loops and regulated protein degradation are the principal mechanisms involved in the generation of self-sustaining circadian rhythms in many organisms. In Arabidopsis the first proposed regulatory transcriptional loop involved the transcription factors CIRCADIAN CLOCK ASSOCIATED (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) and the pseudo-response regulator TIMING OF CHLOROPHYLL A/B BINDING PROTEIN (TOC1/PRR1). Recent findings indicate that the TOC1 homologues PRR7 and PRR9 might also be involved in transcriptional regulatory loops with CCA1 and LHY. In this study we show that the overexpression of PRR7 in Arabidopsis leads to severely compromised circadian rhythms. These transgenic lines display significantly reduced levels of CCA1 and LHY RNA, providing further evidence for a transcriptional feedback loop between PRR7 and these transcription factors. In addition, we show that the PRR7 protein is phosphorylated in a circadian regulated manner and that its levels are post-translationally regulated by both diurnal and circadian mechanisms. The Arabidopsis circadian oscillator is therefore likely to be entrained to light/dark cycles both through transcriptional and post-transcriptional mechanisms.
DOI:10.1093/pcp/pcd043URLPMID:11100772 [本文引用: 1]

The Arabidopsis pseudo-response regulator, APRR1, has a unique structural design containing a pseudo-receiver domain and a C-terminal CONSTANS motif. This protein was originally characterized as a presumed component of the His-to-Asp phosphorelay systems in Arabidopsis thaliana. Recently, it was reported that APRR1 is identical to the TOC1 gene product, a mutational lesion of which affects the periods of many circadian rhythms in Arabidopsis plants. TOC1 is believed to be a component of the presumed circadian clock (or central oscillator). Based on these facts, in this study four more genes, each encoding a member of the APRR1/TOC1 family of pseudo-response regulators were identified and characterized with special reference to circadian rhythms. It was found that all these members of the APRR1/TOC1 family (APRR1, APRR3, APRR5, APRR7, and APRR9) are subjected to a circadian rhythm at the level of transcription. Furthermore, in a given 24 h period, the APRR-mRNAs started accumulating sequentially after dawn with 2-3 h intervals in the order of APRR9-->APRR7-->APRR5-->APRR3-->APRR1. These sequential events of transcription, termed 'circadian waves of APRR1/TOCI', were not significantly affected by the photoperiod conditions, if any (e.g. both long and short days), and the expression of APRR9 was first boosted always after dawn. Among these APRRs, in fact, only the expression of APRR9 was rapidly and transiently induced also by white light, whereas such light responses of others were very dull, if any. These results collectively support the view that these members of the APRR1/TOC1 family are together all involved in an as yet unknown mechanism underlying the Arabidopsis circadian clock. Here we propose that the circadian waves of the APRR1/TOC1 family members are most likely a molecular basis of such a biological clock in higher plants.
DOI:10.1093/mp/sst088URL [本文引用: 4]

Rice Pseudo-Response Regulator 37 (OsPRR37) is the Hd2 quantitative trait locus (QTL) gene and natural variation in OsPRR37 has contributed to global adaptability of rice by reducing photoperiod sensitivity and allowing early flowering, especially under long-day environments in temperate and cooler regions.Heading date and photoperiod sensitivity are fundamental traits that determine rice adaptation to a wide range of geographic environments. By quantitative trait locus (QTL) mapping and candidate gene analysis using whole-genome re-sequencing, we found that Oryza sativa Pseudo-Response Regulator37 (OsPRR37; hereafter PRR37) is responsible for the Early heading7-2 (EH7-2)/Heading date2 (Hd2) QTL which was identified from a cross of late-heading rice Milyang23 (M23) and early-heading rice H143. H143 contains a missense mutation of an invariantly conserved amino acid in the CCT (CONSTANS, CO-like, and TOC1) domain of PRR37 protein. In the world rice collection, different types of nonfunctional PRR37 alleles were found in many European and Asian rice cultivars. Notably, the japonica varieties harboring nonfunctional alleles of both Ghd7/Hd4 and PRR37/Hd2 flower extremely early under natural long-day conditions, and are adapted to the northernmost regions of rice cultivation, up to 53 N latitude. Genetic analysis revealed that the effects of PRR37 and Ghd7 alleles on heading date are additive, and PRR37 down-regulates Hd3a expression to suppress flowering under long-day conditions. Our results demonstrate that natural variations in PRR37/Hd2 and Ghd7/Hd4 have contributed to the expansion of rice cultivation to temperate and cooler regions.
DOI:10.1007/s00122-004-1905-4URLPMID:15723276 [本文引用: 3]

In this study we have attempted to quantify the thermal and photoperiodical responses of rice (Oryza sativa L.) flowering time QTLs jointly by a 'date-of-planting' field experiment of a mapping population, and a 'phenological model' analysis that separately parameterizes the two responses, based on daily temperature, daily photoperiod and flowering date. For this purpose, the 'three-stage Beta model', which parameterizes the sensitivity to temperature (parameter alpha), the sensitivity to photoperiod (parameter beta), and earliness under optimal conditions (10 h photoperiod at 30 degrees C) (parameter G), was applied to 'Nipponbare' x 'Kasalath' backcross inbred lines that were transplanted on five dates. QTLs for the beta value were detected in the four known flowering time QTL (Hd1, Hd2, Hd6 and Hd8) regions, while QTLs for the G value were detected only in the Hd1 and Hd2 regions. This result was consistent with previous reports on near-isogenic lines (NILs) of Hd1, Hd2 and Hd6, where these loci were involved in photoperiod sensitivity, and where Hd1 and Hd2 conferred altered flowering under both 10 and 14 h photoperiods, while Hd6 action was only affected by the 14 h photoperiod. Hd8 was shown to control photoperiod sensitivity for the first time. Interestingly, Hd1 and Hd2 were associated with a QTL for the alpha value, which might support the previous hypothesis that the process of photoinduction depends on temperature. These results demonstrate that our approach can effectively quantify environmental responses of flowering time QTLs without controlled environments or NILs.
DOI:10.1038/srep12803URLPMID:26238949 [本文引用: 3]

Artificial selection of high yield crops and better livestock is paramount importance in breeding programs. Selection of elite parents with preferred traits from a phalanx of inbred lines is extremely laborious, time-consuming and highly random. General combining ability (GCA) was proposed and has been widely used for the evaluation of parents in hybrid breeding for more than half a century. However, the genetic and molecular basis of GCA has been largely overlooked. Here, we present two pleotropic QTLs are accounting for GCA of days to heading (DTH), plant height (PH) and spikelet per panicle (SPP) using an F2-based NCII design, the BC3F2 population as well as a set of nearly isogenic lines (NILs) with five testers. Both GCA1 and GCA2 were loss-of-function gene in low-GCA parent and gain-of-function gene in high-GCA parent, encoding the putative Pseudo-Response Regulators, OsPRR37 and Ghd7, respectively. Overexpression of GCA1 in low-GCA parent significantly increases GCA effects in three traits. Our results demonstrate that two GCA loci associate with OsPRR37 and Ghd7 and reveal that the genes responsible for important agronomic traits could simultaneously account for GCA effects.
DOI:10.1073/pnas.1418204111URLPMID:25378698 [本文引用: 3]

Success of modern agriculture relies heavily on breeding of crops with maximal regional adaptability and yield potentials. A major limiting factor for crop cultivation is their flowering time, which is strongly regulated by day length (photoperiod) and temperature. Here we report identification and characterization of Days to heading 7 (DTH7), a major genetic locus underlying photoperiod sensitivity and grain yield in rice. Map-based cloning reveals that DTH7 encodes a pseudo-response regulator protein and its expression is regulated by photoperiod. We show that in long days DTH7 acts downstream of the photoreceptor phytochrome B to repress the expression of Ehd1, an up-regulator of the
.
DOI:10.1038/ng.143URLPMID:18454147 [本文引用: 1]

Yield potential, plant height and heading date are three classes of traits that determine the productivity of many crop plants. Here we show that the quantitative trait locus (QTL) Ghd7, isolated from an elite rice hybrid and encoding a CCT domain protein, has major effects on an array of traits in rice, including number of grains per panicle, plant height and heading date. Enhanced expression of Ghd7 under long-day conditions delays heading and increases plant height and panicle size. Natural mutants with reduced function enable rice to be cultivated in temperate and cooler regions. Thus, Ghd7 has played crucial roles for increasing productivity and adaptability of rice globally.
URLPMID:23090144 [本文引用: 1]
DOI:10.1126/science.1117619URLPMID:16284181 [本文引用: 2]

Plants commonly use photoperiod (day length) to control the timing of flowering during the year, and variation in photoperiod response has been selected in many crops to provide adaptation to different environments and farming practices. Positional cloning identified Ppd-H1, the major determinant of barley photoperiod response, as a pseudo-response regulator, a class of genes involved in circadian clock function. Reduced photoperiod responsiveness of the ppd-H1 mutant, which is highly advantageous in spring-sown varieties, is explained by altered circadian expression of the photoperiod pathway gene CONSTANS and reduced expression of its downstream target, FT, a key regulator of flowering.
DOI:10.1007/s00122-007-0603-4URLPMID:17634915 [本文引用: 1]

Ppd-D1 on chromosome 2D is the major photoperiod response locus in hexaploid wheat (Triticum aestivum). A semi-dominant mutation widely used in the
DOI:10.1073/pnas.1106212108URLPMID:21930910 [本文引用: 3]

Optimal flowering time is critical to the success of modern agriculture. Sorghum is a short-day tropical species that exhibits substantial photoperiod sensitivity and delayed flowering in long days. Genotypes with reduced photoperiod sensitivity enabled sorghum's utilization as a grain crop in temperate zones worldwide. In the present study, Ma(1), the major repressor of sorghum flowering in long days, was identified as the pseudoresponse regulator protein 37 (PRR37) through positional cloning and analysis of SbPRR37 alleles that modulate flowering time in grain and energy sorghum. Several allelic variants of SbPRR37 were identified in early flowering grain sorghum germplasm that contain unique loss-of-function mutations. We show that in long days SbPRR37 activates expression of the floral inhibitor CONSTANS and represses expression of the floral activators Early Heading Date 1, FLOWERING LOCUS T, Zea mays CENTRORADIALIS 8, and floral induction. Expression of SbPRR37 is light dependent and regulated by the circadian clock, with peaks of RNA abundance in the morning and evening in long days. In short days, the evening-phase expression of SbPRR37 does not occur due to darkness, allowing sorghum to flower in this photoperiod. This study provides insight into an external coincidence mechanism of photoperiodic regulation of flowering time mediated by PRR37 in the short-day grass sorghum and identifies important alleles of SbPRR37 that are critical for the utilization of this tropical grass in temperate zone grain and bioenergy production.
DOI:10.1093/aob/mcu032URL [本文引用: 1]

This review summarizes recent knowledge on the molecular mechanisms regulating daylength perception and flowering time control in arabidopsis and rice. Similarities and differences are discussed between the regulatory networks of the two species and they are compared with the regulatory networks of temperate cereals, which are evolutionarily more similar to rice but have evolved in regions where exposure to low temperatures is crucial to confer competence to flower. Finally, the role of flowering time genes in expansion of rice cultivation to Northern latitudes is discussed.Understanding the mechanisms involved in photoperiodic flowering and comparing the regulatory networks of dicots and monocots has revealed how plants respond to environmental cues and adapt to seasonal changes. The molecular architecture of such regulation shows striking similarities across diverse species. However, integration of specific pathways on a basal scheme is essential for adaptation to different environments. Artificial manipulation of flowering time by means of natural genetic resources is essential for expanding the cultivation of cereals across different environments.]]>
DOI:10.1016/j.jas.2008.12.012URL

AbstractBetween ca. 6000 BC and ca. 500 BC, barley cultivation spread across the continent of Europe from the extreme south to the extreme north. Carbon-dating would suggest that this spread, and indeed the spread of crop cultivation generally, varied in its pace, with ‘delays’ at certain points along its route. Such delays in the spread of agriculture have been explained as resulting from the slow assimilation of agricultural practices by existing indigenous human populations or as the time taken for the crops to adapt to novel climatic conditions, such as altered temperature regimes and day-lengths. A mutant form of the photoperiod response gene, Ppd-H1, causes barley to be non-responsive to long days, while the wild-type responsive form allows plants to flower in response to long days. We sequenced this gene in 65 ‘historic’ barley accessions, from the late 19th and early 20th centuries, in order to explore the potential role of environmental adaptation in the spread of agriculture. We chose to use ‘historic’ material, to complement the richer patterns in extant genetic lines, by spreading the data range in both time and space. Our ‘historic’ barley data shows a latitudinal divide in the Ppd-H1 gene similar to that found in extant lines, but with clearer geographical resolution, and extending northwards into the Arctic Circle. We discuss the implications of our results in relation to the dynamics of agricultural spread across Europe.]]>
DOI:10.1007/s00122-015-2523-zURLPMID:25982128 [本文引用: 2]

KEY MESSAGE: Allele phylogenetic analysis of the sorghum flowering-time gene PRR37 provided new insight into the human-mediated selection of a key adaptive gene that occurred during sorghum's diversification and worldwide dispersal. The domestication and spread of the tropical cereal sorghum is associated with the historic movement of humans. We show that an allelic series at PRR37 (pseudo-response regulator 37), a circadian clock-associated transcription factor, was selected in long-day ecosystems worldwide to permit floral initiation and grain production. We identified a series of loss-of-function (photoperiod-insensitive) alleles encoding truncated PRR37 proteins, alleles with key amino acid substitutions in the pseudo-receiver domain, and a novel splice variant in which the pseudo-receiver domain is truncated. Each PRR37 allelic variant was traced to a specific geographic location or specialized agronomic type. We present a graphical model that shows evidence of human selection and gene flow of the PRR37 allelic variants during the global dispersal and agronomic diversification of sorghum. With the recent identification of the Ghd7 gene as an important regulator of flowering date in sorghum, we briefly examine whether loss-of-function Ghd7 allelic variants were selected prior to the human-mediated movement of sorghum from its equatorial center of origin to temperate climates worldwide.
URLPMID:29314052 [本文引用: 3]
URLPMID:23808423 [本文引用: 2]
DOI:10.1073/pnas.0900952106URLPMID:19359492 [本文引用: 1]

In higher plants, the circadian clock controls a wide range of cellular processes such as photosynthesis and stress responses. Understanding metabolic changes in arrhythmic plants and determining output-related function of clock genes would help in elucidating circadian-clock mechanisms underlying plant growth and development. In this work, we investigated physiological relevance of PSEUDO-RESPONSE REGULATORS (PRR 9, 7, and 5) in Arabidopsis thaliana by transcriptomic and metabolomic analyses. Metabolite profiling using gas chromatography-time-of-flight mass spectrometry demonstrated well-differentiated metabolite phenotypes of seven mutants, including two arrhythmic plants with similar morphology, a PRR 9, 7, and 5 triple mutant and a CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1)-overexpressor line. Despite different light and time conditions, the triple mutant exhibited a dramatic increase in intermediates in the tricarboxylic acid cycle. This suggests that proteins PRR 9, 7, and 5 are involved in maintaining mitochondrial homeostasis. Integrated analysis of transcriptomics and metabolomics revealed that PRR 9, 7, and 5 negatively regulate the biosynthetic pathways of chlorophyll, carotenoid and abscisic acid, and alpha-tocopherol, highlighting them as additional outputs of pseudo-response regulators. These findings indicated that mitochondrial functions are coupled with the circadian system in plants.
DOI:10.1093/pcp/pcp004URLPMID:19131357 [本文引用: 3]

Arabidopsis PSEUDO RESPONSE REGULATOR (PRR) genes are components of the circadian clock mechanism. In order to understand the scope of genome-wide transcriptional regulation by PRR genes, a comparison survey of gene expression in wild-type Arabidopsis and a prr9-11 prr7-10 prr5-10 triple mutant (d975) using mRNA collected during late daytime was conducted using an Affymetrix ATH-1 GeneChip. The expression of 'night genes' increased and the expression of 'day genes' decreased toward the end of the diurnal light phase, but expression of these genes was essentially constant in d975. The expression levels of 'night genes' were lower, whereas the expression of 'day genes' was higher in d975 than in the wild type. Bioinformatics approaches have indicated that the set of up-regulated genes in d975 and the set of cold-responsive genes have significant overlap. We found that d975 is more tolerant to cold, high salinity and drought stresses than the wild type. In addition, dehydration-responsive element B1/C-repeat-binding factor (DREB1/CBF), which is expressed around mid-day, is more highly expressed in d975. Raffinose and L-proline accumulated at higher levels in d975 even when plants were grown under normal conditions. These results suggest that PRR9, PRR7 and PRR5 are involved in a mechanism that anticipates diurnal cold stress and which initiates a stress response by mediating cyclic expression of stress response genes, including DREB1/CBF.
[本文引用: 2]
URLPMID:25653664 [本文引用: 2]
URLPMID:25631123 [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.3109/07388551.2012.716809URLPMID:22985089 [本文引用: 1]

Foxtail millet is one of the oldest domesticated diploid C4 Panicoid crops having a comparatively small genome size of approximately 515 Mb, short life cycle, and inbreeding nature. Its two species, Setaria italica (domesticated) and Setaria viridis (wild progenitor), have characteristics that classify them as excellent model systems to examine several aspects of architectural, evolutionary, and physiological importance in Panicoid grasses especially the biofuel crops such as switchgrass and napiergrass. Foxtail millet is a staple crop used extensively for food and fodder in parts of Asia and Africa. In its long history of cultivation, it has been adapted to arid and semi-arid areas of Asia, North Africa, South and North America. Foxtail millet has one of the largest collections of cultivated as well as wild-type germplasm rich with phenotypic variations and hence provides prospects for association mapping and allele-mining of elite and novel variants to be incorporated in crop improvement programs. Most of the foxtail millet accessions can be primarily abiotic stress tolerant particularly to drought and salinity, and therefore exploiting these agronomic traits can enhance its efficacy in marker-aided breeding as well as in genetic engineering for abiotic stress tolerance. In addition, the release of draft genome sequence of foxtail millet would be useful to the researchers worldwide in not only discerning the molecular basis of biomass production in biofuel crops and the methods to improve it, but also for the introgression of beneficial agronomically important characteristics in foxtail millet as well as in related Panicoid bioenergy grasses.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/pcp/pci087URLPMID:15767264 [本文引用: 2]

In Arabidopsis thaliana, AUTHENTIC RESPONSE REGULATORS (ARRs) act as downstream components of the His-to-Asp phosphorelay (two-component) signaling pathway that is propagated primarily by the cytokinin receptor kinases, AUTHENTIC HIS-KINASES (AHK2, AHK3 and AHK4/CRE1). Thus, this bacterial type of signaling system is essential for responses to a class of hormones in plants. Interestingly, this higher plant has also evolved its own atypical (or unique) variants of two-component signal transducers, PSEUDO-RESPONSE REGULATORS (PRRs). Several lines of recent results suggest that the functions of PRRs are closely relevant to the plant clock (oscillator) that is central to circadian rhythms, the underlying mechanisms of which have long been the subject of debate. Through an overview of recent results, the main issue addressed here is whether or not the pseudo-response regulators (PRRs) are true oscillator components (TOCs).
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
.
URLPMID:10743655 [本文引用: 1]
