 ,1,*
,1,*Comparative proteomic analysis of two wheat genotypes with contrasting grain softness index
LIU Pei-Xun1, MA Xiao-Fei2, WAN Hong-Shen1, ZHENG Jian-Min1, LUO Jiang-Tao1, PU Zong-Jun ,1,*
,1,*通讯作者:
收稿日期:2019-11-23接受日期:2020-03-30网络出版日期:2020-08-12
| 基金资助: |
Received:2019-11-23Accepted:2020-03-30Online:2020-08-12
| Fund supported: |
作者简介 About authors
E-mail:littlefarmer@163.com;Tel:028-84504231。

摘要
关键词:
Abstract
Keywords:
PDF (636KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘培勋, 马小飞, 万洪深, 郑建敏, 罗江陶, 蒲宗君. 两个不同籽粒硬度小麦的比较蛋白组学分析[J]. 作物学报, 2020, 46(8): 1275-1282. doi:10.3724/SP.J.1006.2020.91068
LIU Pei-Xun, MA Xiao-Fei, WAN Hong-Shen, ZHENG Jian-Min, LUO Jiang-Tao, PU Zong-Jun.
小麦硬度是评价小麦品质和影响小麦最终用途的重要性状指标。硬质小麦适用于制作面包和面条等, 软质小麦适用于制作饼干、糕点和酿酒等, 西南地区酿酒企业较多, 酒曲以及酿酒过程对软质小麦的需求较大。弄清小麦硬度遗传基础, 对于改良小麦品质, 培育满足不同需要的小麦品种具有重大意义。
小麦籽粒硬度与蛋白质含量高度相关, 硬质小麦蛋白质含量比软质小麦高, 且在小麦籽粒中蛋白质和淀粉以水溶性糖蛋白方式紧密结合。因此, 籽粒硬度与蛋白质含量、湿面筋含量呈显著正相关[1]。硬质、高蛋白常被看作是强筋小麦的标志, 而软质、低蛋白则被作为评价弱筋小麦的依据[2]。小麦种子蛋白主要由麦谷蛋白、醇溶蛋白、球蛋白和清蛋白组成, 其中谷蛋白和醇溶蛋白共同构成种子贮藏蛋白。李硕碧等利用扫描电镜观察了小麦籽粒的断面, 发现软质品种的淀粉近似球形, 清晰可见; 硬麦品种籽粒的断面则很少见到完整淀粉粒, 破碎淀粉粒较多, 胚乳结构相对致密[3]。
普通小麦籽粒硬度由主效基因puroindoline控制[4], puroindoline分为Pina和Pinb两种类型, 位于5D染色体短臂上, 二者高度同源且紧密连锁。软质对硬质为显性, 当2个基因同时存在时, 小麦籽粒表现为软质, 当Pina缺失或Pinb发生变异时, 小麦籽粒表现为硬质, 能够解释60%左右的变异[5]。Pina和Pinb均编码148个氨基酸序列, 其氨基酸同源性约60%, 分子量约为13 kD。研究发现, 这种蛋白只存在于种子的胚乳细胞中, 而在其他组织中没有[6]。随着对puroindoline认识的逐步加深, 发现其除了调节籽粒硬度之外, 还具有一定的抗病性[7]。
与硬度相关的gsp-1 (grain softness protein-1)基因与puroindoline基因共同存在于5DS染色体上, 且高度同源, 但研究表明其对籽粒硬度影响不显著[8]。Wilkinson等[9]在小麦cDNA中发现一种与puroindoline b核苷酸序列相似性高达70%的类PIN基因, 称为puroindoline b-2, 对籽粒硬度存在一定影响, Chen等[10]进一步发现该基因有多种变异类型。随着基因组学的发展, 研究发现了一种新型的贮藏蛋白基因avenin like b, 其所表达的蛋白与燕麦蛋白相似, 为小麦品质的改良提供了参考[11,12]。Furtadoet等[13]在小麦种子中发现一类新的基因wheat bread making (wbm), 对面筋质量、强度和延伸性有显著影响[14]。对puroindoline同源蛋白家族分析发现, gsp-l、puroindolineb- 2、avenin like b蛋白与puroindoline同属于AAI_LTSS超级家族, 推测除puroindoline外, 存在其他基因, 尤其是其同源基因, 也对籽粒硬度起作用[15]。
目前, 对于小麦籽粒硬度的研究主要集中于puroindoline基因(Pina和Pinb), 但该基因尚不能完全解释小麦籽粒硬度变异的机制[16,17]。本研究利用TMT (tandem mass tags)定量蛋白质组学技术, 结合小麦最新基因组(iwgsc_refseqv1.0 2018)注释信息[18]和UniProt蛋白数据库[19], 快速高通量地鉴定2个不同籽粒硬度小麦种子中蛋白差异表达情况, 并通过生物信息学分析, 对差异表达蛋白进行功能和通路等富集分析, 以期挖掘影响籽粒硬度的潜在的新基因, 为小麦品质改良提供理论参考。
1 材料与方法
1.1 试验设计
四川主推小麦品种川麦66 (系谱: 川麦42/98-226// 01-3570)是西南地区筋力最弱、硬度最低的品种之一[20], 蜀麦969 (SHW-L1/SW8188//川育18/3/川麦42)是四川主推品种中筋力最强、硬度最高的品种之一[21], 两者都是川麦42的衍生品种, 其亲缘关系相对较近。2018年10月至2019年5月将其种植于四川省农业科学院德阳试验基地(31°14′N, 104°24′E), 3个重复, 随机区组设计, 每个小区行长1.5 m, 行距0.3 m, 共10行。播种时各施75 kg hm?2的磷肥(过磷酸钙)和钾肥(硫酸钾)作为基肥, 氮肥施尿素150 kg hm?2, 分2次施用, 一半基施, 另一半作拔节肥。收取成熟小麦籽粒(含水量12%), 进行品质检测和蛋白表达分析。使用单粒谷物测定仪KSCS4100检测小麦籽粒硬度指数, 使用近红外谷物品质分析仪Infratec 1241检测粗蛋白含量、湿面筋含量和淀粉含量。1.2 种子蛋白提取
将冷冻的种子样品低温研磨成粉, 加入蛋白裂解液(100 mmol L?1碳酸氢铵、6 mol L?1尿素、0.2%SDS, pH 8.0)充分裂解, 然后在4℃下12,000 × g离心15 min, 取上清液, 加入终浓度10 mmol L?1 DTTred于56℃反应1 h, 再加入足量IAM, 于室温避光反应1 h。加入4倍体积的?20℃预冷丙酮, 于?20℃条件下沉淀2 h, 最后在4℃下12,000×g离心15 min, 收集沉淀。经重悬清洗后, 加入适量蛋白溶解液(6 mol L?1尿素、100 mmol L?1 TEAB, pH 8.5)溶解蛋白沉淀。使用Bradford蛋白质定量试剂盒, 绘制标准曲线, 计算待测样品的蛋白浓度。1.3 TMT标记和液质检测
2个品种, 3次重复, 共6个蛋白样品经酶切与除盐[22]后, 分别加入0.1 mol L?1 TEAB缓冲液和乙腈溶解的含不同同位素标签的TMT (Tandem Mass Tags)标记[23]试剂, 室温下颠倒混匀反应2 h。加入终浓度8%氨水终止反应, 取等体积标记后的样品混合, 除盐后冻干。使用L-3000HPLC系统进行馏分分离, 每分钟收集1管, 合并为10个馏分, 冻干后各加入0.1%甲酸溶解。使用EASY-nLCTM1200纳升级UHPLC系统, 进行液相色谱检测。使用QExactiveTMHF-X质谱仪, NanosprayFlex ESI)离子源, 数据依赖型采集模式进行质谱检测, 设一级质谱分辨率为60,000 (200 m z?1)。选取全扫描中离子强度TOP40的母离子使用高能碰撞裂解(HCD)方法碎裂, 进行二级质谱检测, 设二级质谱分辨率为15,000 (200 m z?1), 最终生成质谱检测原始数据(.raw)。1.4 蛋白鉴定
合并UniProt数据库[19]和最新小麦基因组[18]注释文件IWGSC RefSeq v1.0 annotation (154,254 sequences)作为最终的搜索数据库。利用Proteome Discoverer 2.2软件对检索结果进一步过滤: 可信度在99%以上的谱肽为可信PSMs (peptide spectrum matches), 至少包含一个unique肽段(特有肽段)的蛋白为可信蛋白, 只保留可信的谱肽和蛋白, 通过FDR验证, 去除掉FDR大于1%的肽段和蛋白。对鉴定到的蛋白进行蛋白注释, 包括GO注释[24]、KEGG注释[25]、COG注释[26]和结构域注释(IPR)[27]。1.5 蛋白定量分析和差异表达蛋白功能分析
根据原始下机的谱图峰面积可以得到各个样品中每个PSM的相对定量值, 后再根据鉴定出的Unique肽段中所包含所有PSM的定量信息校正得到Unique肽段的相对定量值, 最后根据每个蛋白质包含的所有Unique肽段的定量信息校正得到每个蛋白的相对定量值。将每个蛋白在比较样品对中的所有生物重复定量值的均值的比值作为差异倍数(Fold Change, FC)。将每个蛋白在样品中的相对定量值进行T-test检验, P值作为显著性指标。当FC≥1.5, 且P≤0.05时, 蛋白表现为表达量上调, 当FC≤0.67, 同时P≤0.05时, 蛋白表现为表达量下调。针对筛选出来的差异蛋白进行GO、KEGG功能富集分析, 与所有鉴定到的蛋白背景相比, 应用超几何检验计算P值, 以P≤0.05为阈值, 满足此条件的条目(或路径)定义为在差异蛋白质中显著富集的条目(或路径)。1.6 差异表达蛋白与puroindolines蛋白系统发育分析
前人对于小麦籽粒硬度的研究主要集中于puroindoline基因(Pina和Pinb), 该蛋白主要在软质麦中存在和表达。从UniProt数据库下载小麦Pina和Pinb蛋白。将本研究中鉴定出的在软质麦川麦66中较硬质麦蜀麦969中显著上调表达的蛋白与puroindolines蛋白一起, 利用Clustal X 2.0进行序列比对, 利用MEGA 7.0软件邻接法构建系统发育树, 分析差异表达蛋白与puroindolines蛋白的同源关系[28]。2 结果与分析
2.1 2个小麦籽粒样品质分析
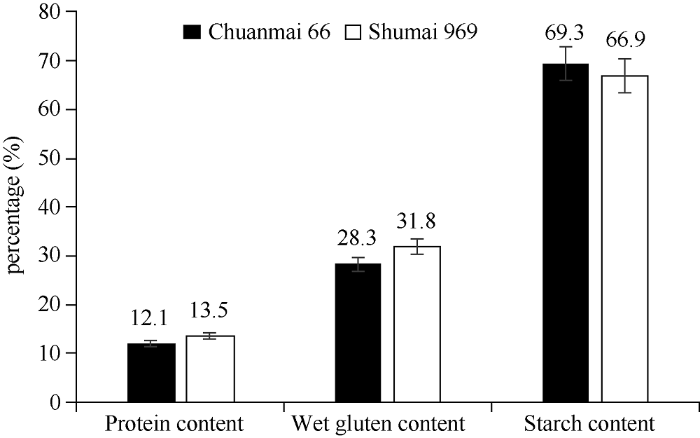
经检测,川麦66的平均籽粒硬度指数为25.2,蜀麦969的平均籽粒硬度指数为40.2。籽粒粗蛋白含量、湿面筋含量和淀粉含量检测主要结果见图1,川麦66的籽粒硬度指数、粗蛋白含量和湿面筋含量均低于蜀麦969, 淀粉含量则高于蜀麦969, 经t检验, 差异均达到显著水平。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1川麦66和蜀麦969籽粒主要品质指标比较
Fig. 1Comparison of the grain quality between Chuanmai 66 and Shumai 969
2.2 蛋白鉴定
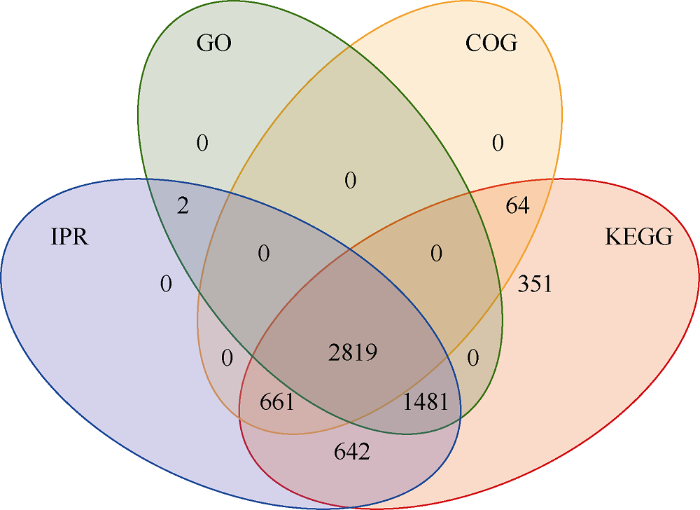
通过数据库搜索, 鉴定到二级谱图(total spectra)数量为85,497个, 肽段(Peptide)数为32,552个, 蛋白数(protein)为6061个。利用GO、KEGG和COG数据库对鉴定到的蛋白质进行功能注释, 利用Pfam、ProDom、SMART等结构域的数据库进行结构域注释(IPR), 利用模式结构或特征进行功能未知蛋白的结构域注释, 共注释到6020个蛋白, 注释结果统计见图2, 有2819个蛋白可同时被4种数据库注释。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2蛋白质功能注释结果
Fig. 2Result of protein functional annotations
2.3 差异表达蛋白
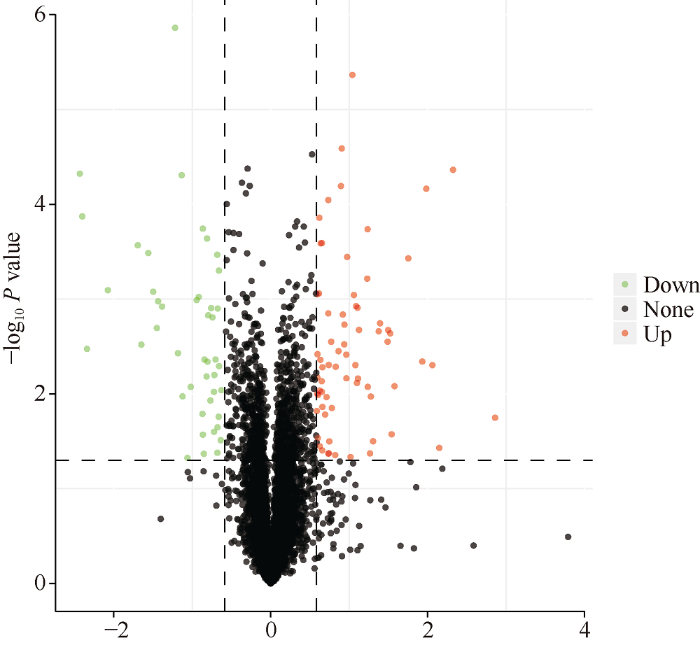
根据限制条件筛选的差异表达蛋白(DEP), 在注释到的6020个蛋白中, 对每个蛋白质差异倍数以2为底取对数, 将P值以10为底取对数的绝对值, 做出火山图, 观察不同蛋白在不同样品间比较时的上调、下调情况, 结果见图3。2个样品差异表达蛋白(DEP)总数为113, 其中软质麦川麦66中较硬质麦蜀麦969中显著上调的蛋白为69个, 显著下调的蛋白为44个。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3差异表达蛋白火山图
Fig. 3Differential protein volcano map
2.4 差异蛋白GO富集分析
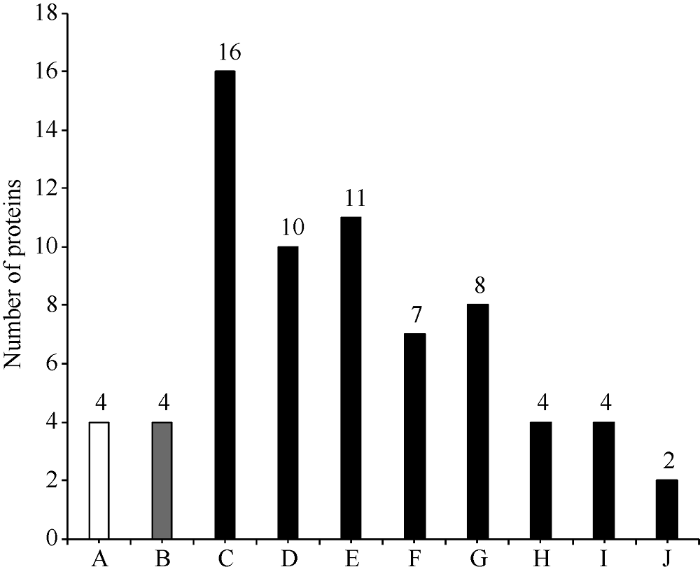
GO显著性富集分析能分析差异蛋白行使的主要生物学功能。2个小麦样品中, 表达差异显著的113个蛋白经GO富集分析, 富集到65个GO条目中, 去除仅包含1个差异蛋白的条目, 剩余33个可靠GO条目。其中, 达到显著富集(P<0.05)水平的GO条目为10个, 属于生物过程1个, 细胞组分1个, 分子功能的8个(图4)。达到极显著富集(P<0.01)水平的条目为8个, 生物过程条目有1个, 为防御反应过程(defense response); 细胞组分条目有1个, 为胞外区(extracellular region); 分子功能条目有6个, 包括营养库活性(nutrient reservoir activity)、酶抑制剂活性(enzyme inhibitor activity)、酶调节活性(enzyme regulator activity)、丝氨酸型内肽酶抑制剂活性(serine-type endopeptidase inhibitor activity)、肽链内切酶抑制剂活性(endopeptidase inhibitor activity)和天冬氨酸型内肽酶活性类(aspartic-type endopeptidase activity)。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4GO富集柱状图
A属于生物过程(BP), B属于细胞组分(CC), C、D、E、F、G、H、I、J属于分子功能(MF)。A: 防御反应; B: 胞外区; C: 营养库活性; D: 酶抑制剂活性; E: 酶调节活性; F: 丝氨酸型内肽酶抑制剂活性; G: 肽链内切酶抑制剂活性; H: 天冬氨酸型内肽酶活性; I: 电子载体活性; J: 几丁质结合。
Fig. 4Differential protein GO enrichment results
A belongs to BP (biological process), B belongs to CC (cellular component), C, D, E, F, G, H, I, J belong to MF (molecular function). A: defense response; B: extracellular region; C: nutrient reservoir activity; D: enzyme inhibitor activity; E: enzyme regulator activity; F: serine-type endopeptidase inhibitor activity; G: endopeptidase inhibitor activity; H: aspartic-type endopeptidase activity; I: electron carrier activity; J: chitin binding.
达到极显著富集水平的分子功能类的6个GO条目, 有1个为营养库活性, 其余5个条目均涉及酶抑制剂活性。营养库活性条目中包含16个差异表达蛋白(表1), 其中在川麦66中显著上调表达的有7个, 下调表达的9个。该类别中包含的蛋白主要有低分子谷蛋白亚基、燕麦相似蛋白、α和γ-醇溶蛋白及萌发素类蛋白。5个涉及酶抑制剂活性的条目中, 多个蛋白均同时属于不同的GO条目。经整理, 属于酶抑制剂活性类的差异表达蛋白共15个, 包括α-淀粉酶、胰蛋白酶、转化酶、糜蛋白酶等(表2)。其中, 10个蛋白在川麦66中显著上调表达, 5个下调表达, puroindoline b蛋白富集于该类别中, 在川麦66中较蜀麦969差异表达倍数达到2.8倍。
Table 1
表1
表1营养库活性类相关差异表达蛋白(川麦66 vs. 蜀麦969)
Table 1
| 蛋白名称 Protein ID | 描述 Description | 差异倍数 Fold change | 上调/下调 Up/down |
|---|---|---|---|
| B2Y2Q6 | B2Y2Q6_WHEAT LMW-B2 | 1.5425 | Up |
| B2BZC7 | B2BZC7_WHEAT LMW-m glutenin subunit 0154A5-M | 1.8737 | Up |
| A0A173DQZ1 | A0A173DQZ1_WHEAT type-b avenin-like protein | 3.8163 | Up |
| Q8H0J5 | Q8H0J5_WHEAT low molecular weight glutenin subunit (fragment) | 5.0056 | Up |
| TraesCS7A01G035300.1 | Gliadin-like avenin | 2.9853 | Up |
| Q6WZC3 | Q6WZC3_WHEAT low molecular weight glutenin subunit | 2.1622 | Up |
| TraesCS4A01G453400.1 | Gamma-gliadin | 7.2606 | Up |
| A0A286QTK1 | A0A286QTK1_WHEAT avenin-like protein A2 | 0.5984 | Down |
| F8SGL3 | F8SGL3_WHEAT low-molecular-weight glutenin subunit | 0.5493 | Down |
| Q0QBR3 | Q0QBR3_WHEAT LMW-glutenin P3-5 | 0.3190 | Down |
| B2Y2R3 | B2Y2R3_WHEAT low molecular weight glutenin subunit | 0.5725 | Down |
| B6UKN9 | B6UKN9_WHEAT gamma-gliadin | 0.6340 | Down |
| TraesCS2A01G211800.1 | Germin-like protein 1-1 | 0.5875 | Down |
| A0A2P1H6A2 | A0A2P1H6A2_WHEAT alpha-gliadin | 0.5908 | Down |
| A0A0K2QJY6 | A0A0K2QJY6_WHEAT alpha/beta-gliadin | 0.6081 | Down |
| I0IT65 | I0IT65_WHEAT alpha-gliadin | 0.6241 | Down |
新窗口打开|下载CSV
Table 2
表2
表2酶抑制剂类相关差异表达蛋白(川麦66 vs. 蜀麦969)
Table 2
| 蛋白名称 Protein ID | 描述 Description | 差异倍数 Fold change | 上调/下调 Up/down |
|---|---|---|---|
| Q5UHH6 | Q5UHH6_WHEAT 0.19 dimeric alpha-amylase inhibitor (fragment) | 1.6788 | Up |
| TraesCS5D01G004300.1 | Puroindoline-b, protease inhibitor/seed storage/LTP family | 2.8227 | Up |
| P81713 | IBB3_WHEAT Bowman-Birk type trypsin inhibitor | 1.5092 | Up |
| TraesCS4A01G460900.1 | Invertase inhibitor, plant invertase/pectin methylesterase inhibitor | 1.5169 | Up |
| TraesCS1D01G028500.1 | Chymotrypsin inhibitor | 2.3465 | Up |
| TraesCS5D01G561800.1 | Invertase inhibitor, plant invertase/pectin methylesterase inhibitor | 2.0582 | Up |
| A0A080YTU1 | A0A080YTU1_WHEAT uncharacterized protein | 1.5779 | Up |
| A0A2X0S1F0 | A0A2X0S1F0_WHEAT peptidase A1 domain-containing protein | 1.9149 | Up |
| TraesCS4D01G205800.1 | ADP-ribosylation factor GTPase-activating protein | 1.6650 | Up |
| TraesCS7A01G502500.1 | Eukaryotic aspartyl protease family protein | 2.3542 | Up |
| TraesCS3D01G467500.1 | Eukaryotic aspartyl protease family protein | 0.3700 | Down |
| TraesCS4D01G250000.1 | Dimeric alpha-amylase inhibitor | 0.3659 | Down |
| TraesCS1D01G265900.1 | Wound-induced protease inhibitor | 0.5474 | Down |
| TraesCS3D01G025700.1 | Trypsin inhibitor | 0.1974 | Down |
| TraesCS6B01G407700.1 | Aspartic proteinase nepenthesin-1 | 0.6473 | Down |
新窗口打开|下载CSV
2.5 差异蛋白KEGG富集分析
通过KEGG Pathway显著性富集分析, 可了解差异蛋白参与的最主要生化代谢途径和信号转导途径。通过KEGG富集, 113个差异蛋白中有48个差异蛋白富集到26个生化代谢途径, 达到显著富集的有谷胱甘肽代谢(Glutathione metabolism)、光合作用-天线蛋白(phtosynthesis- antenna proteins)和类胡萝卜素代谢(carotenoid biosynthesis)。但光合作用-天线蛋白代谢和类胡萝卜素代谢分别仅有1个差异蛋白, 可靠性较差。谷胱甘肽代谢条目达到极显著富集水平, 包含6个差异表达蛋白, 其中4个上调表达, 2个下调表达(表3)。Table 3
表3
表3谷胱甘肽代谢途径相关差异表达蛋白(川麦66 vs. 蜀麦969)
Table 3
| 蛋白名称 Protein ID | 描述 Description | 差异倍数 Fold change | 上调/下调 Up/down |
|---|---|---|---|
| TraesCS3D01G491400.1 | 6-phosphogluconate dehydrogenase | 2.5968 | Up |
| TraesCS2B01G096200.1 | Ascorbate peroxidase | 1.5695 | Up |
| TraesCS3A01G488200.1 | Glutathione S-transferase | 1.5215 | Up |
| TraesCS3B01G536100.1 | Glutathione S-transferase | 1.9499 | Up |
| TraesCS3D01G445400.1 | Glutathione S-transferase | 0.6058 | Down |
| W5D4D9 | W5D4D9_WHEAT uncharacterized protein | 0.5675 | Down |
新窗口打开|下载CSV
2.6 上调表达蛋白与puroindolines蛋白同源关系分析
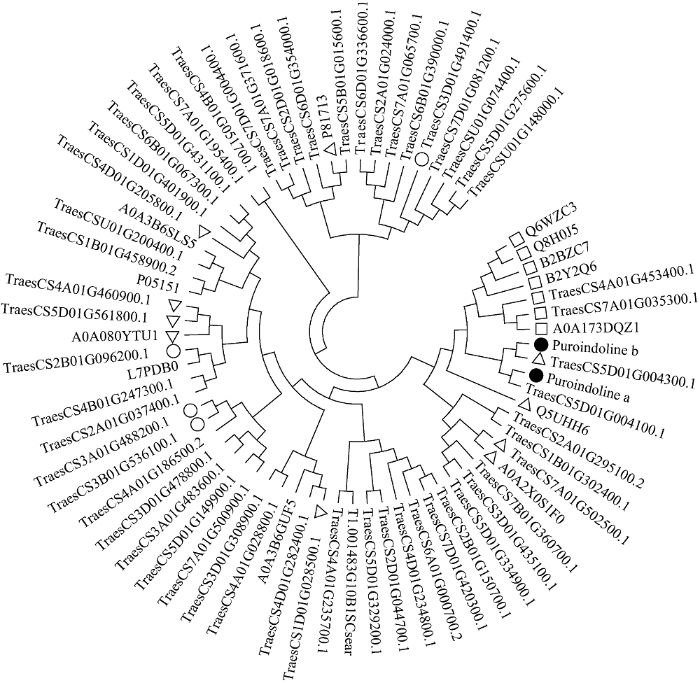
将提取出的69个在软质麦川麦66中显著上调表达的蛋白与从UniProt数据库下载的小麦puroindolines蛋白(puroindoline a和puroindoline b)共同构建系统发育树(图5)。对属于营养库活性类别、酶抑制剂活性类别和谷胱甘肽代谢途径的蛋白分别用空心正方形、三角形和圆形标记。可见, 7个营养库活性类蛋白在进化树中, 与puroindolines蛋白聚到一起; 10个酶抑制剂活性类蛋白中有4个与puroindolines蛋白聚到一起; 谷胱甘肽代谢类蛋白则没有与puroindolines蛋白聚到一起。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5软质麦中上调表达蛋白与puroindolines蛋白共同构建的系统发育树
带实心圆形的为puroindolines蛋白; 带空心正方形的为营养库活性类别; 带空心三角形的为酶抑制剂活性类别; 带空心圆形的为谷胱甘肽代谢途径蛋白。
Fig. 5Phylogenetic tree constructed by differentially expressed proteins and the puroindolines
The proteins with solid circles are puroindolines, the proteins with squares belong to nutrient reservoir activity category, the proteins with triangles belong to enzyme inhibitor activity category, and the proteins with circles belong to glutathione metabolism category.
3 讨论
本研究中所用2个硬度差异较大的品种, 都是来自西南麦区, 并且是同一个亲本的衍生品种, 其亲缘关系较近, 一定程度上可以排除不同麦区品种之间对习性、抗性的差异以及部分遗传背景的差异。通过定量蛋白质组学TMT标记结合液相色谱质谱联用技术和生物信息学分析, 共鉴定出6020个蛋白, 在2个不同籽粒硬度小麦种子中表达差异显著的蛋白113个, 69个蛋白在川麦66中高表达, 44个蛋白在蜀麦969中高表达。鉴定的蛋白数量和差异表达蛋白数量均较高, 证明TMT技术是一种高通量高分辨率的蛋白定量检测技术[29]。通过GO富集方法, 2个样品中表达差异显著的蛋白, 极显著富集到生物过程、细胞组成和分子功能类别的数量分别为1、1和6个。1个生物过程条目为防御反应类别, 1个细胞组分条目为胞外区类别。推测小麦籽粒硬度相关蛋白与小麦抗逆反应有关, 可能主要分布于细胞胞外区。Kim等研究表明, 影响小麦籽粒硬度最重要的蛋白puroindolines具有抗菌作用[30,31,32], 与本研究推测硬度相关蛋白参与抗逆作用可互为支撑。6个分子功能类GO条目中, P值最小的条目为营养库活性类别, 另外5个均与酶抑制剂活性相关。
营养库活性类别, 主要是低分子谷蛋白和醇溶蛋白等贮藏蛋白, 是小麦种子的重要结构组成成分。贮藏蛋白的种类和表达量, 决定着小麦种子中蛋白的含量和组成, 从而影响小麦硬度。酶抑制剂类蛋白对硬粒的影响目前未见专门报道。以puroindolines蛋白为例, 该蛋白具有α-淀粉酶抑制剂功能, 假设puroindolines高表达可有效抑制α-淀粉酶, 从而防止种子中淀粉水解。由此, 可解释软质小麦中淀粉粒较完整、近似球形, 而硬质小麦淀粉粒破坏严重、胚乳结构致密[3]。硬质麦中, 蛋白质和淀粉以水溶性糖蛋白方式紧密结合[1], 可能与缺少淀粉酶抑制剂, 造成淀粉表面水解成糖有关。不管是营养库类别还是酶抑制剂类别, 均有部分蛋白相对表达量与籽粒柔软度呈正相关, 另一部分则为负相关, 其作用机制有待进一步研究。
通过KEGG Pathway显著性富集分析, 富集到26个生化代谢途径, 其中只有3个途径达到显著富集水平, 其中谷胱甘肽代谢途径达到极显著富集水平, 推测相关蛋白对籽粒硬度有一定的作用。TraesCS3A01G 488200.1、TraesCS3B01G536100.1和TraesCS3D01G 445400.1蛋白描述均为谷胱甘肽转化酶, 且分别位于3A、3B和3D染色体, 应该是分布于3号染色体群的同源基因。但之前未见相关报道, 其影响机制有待进一步研究。
Puroindolines基因(Pina和Pinb)目前被认为是影响小麦籽粒硬度的最主要基因, 但尚不能完全解释小麦籽粒硬度变异机制[16,33]。经序列比对和查阅小麦基因组浏览器(JBrowse)[34], TraesCS5D01G004100.1和TraesCS5D01G 004300.1分别就是Pina和Pinb蛋白, 该实验中puroindolines蛋白在软质麦中显著高表达, 与前人研究结果一致[35]。将puroindolines蛋白与川麦66中显著上调表达的蛋白共同构建进化树, 7个营养库活性类蛋白和4个酶抑制剂蛋白与puroindolines蛋白聚为一类。可见, 从蛋白序列同源性角度, 营养库活性类蛋白和部分酶抑制剂类蛋白与puroindolines蛋白有着高度同源性, 可视为同一蛋白家族成员, 因此与puroindolines蛋白具有相似的分子功能。某些小麦籽粒贮藏蛋白, 可能兼具酶抑制剂功能。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1073/pnas.95.11.6262URL [本文引用: 1]
DOI:10.1023/A:1014837431178URL [本文引用: 1]

The variation in grain hardness is the single most important trait that determines end-use quality of wheat. Grain texture classification is based primarily on either the resistance of kernels to crushing or the particle size distribution of ground grain or flour. Recently, the molecular genetic basis of grain hardness has become known, and it is the focus of this review. The puroindoline proteins a and b form the molecular basis of wheat grain hardness or texture. When both puroindolines are in their `functional' wild state, grain texture is soft. When either one of the puroindolines is absent or altered by mutation, then the result is hard texture. In the case of durum wheat which lacks puroindolines, the texture is very hard. Puroindolines represent the molecular-genetic basis of the Hardness locus on chromosome 5DS and the soft (Ha) and hard (ha) alleles present in hexaploid bread wheat varieties. To date, seven discrete hardness alleles have been described for wheat. All involve puroindoline a or b and have been designated Pina-D1b and Pinb-D1b through Pinb-D1g. A direct role of a related protein, grain softness protein (as currently defined), in wheat grain texture has yet to be demonstrated.
DOI:10.1007/s11103-007-9139-xURLPMID:17294254 [本文引用: 1]

The purolindolines are small cysteine-rich proteins which are present in the grain of wheat. They have a major impact on the utilisation of the grain as they are the major determinants of grain texture, which affects both milling and baking properties. Bread and durum wheats were transformed with constructs comprising the promoter regions of the Puroindoline a (Pina) and Puroindoline b (Pinb) genes fused to the uidA (GUS) reporter gene. Nine lines showing 3:1 segregation for the transgene and comprising all transgene/species combinations were selected for detailed analysis of transgene expression during grain development. This showed that transgene expression occurred only in the starchy endosperm cells and was not observed in any other seed or vegetative tissues. The location of the puroindoline proteins in these cells was confirmed by tissue printing of developing grain, using a highly specific monoclonal antibody for detection and an antibody to the aleurone-localised 8S globulin as a control. This provides clear evidence that puroindolines are only synthesised and accumulated in the starchy endosperm cells of the wheat grain.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.jcs.2008.03.007URL [本文引用: 1]
DOI:10.1007/s00122-009-1195-yURL [本文引用: 1]

Puroindoline a and b proteins soften the endosperm of wheat kernels. When the underlying puroindoline genes are altered by mutation or are deleted, kernels become harder. Thus, puroindoline a and b (Pina and Pinb) play an important role in wheat quality and utilization. Recently, additional Pinb genes have been reported. In the present report, we provide corroborating coding and additional 5′ and 3′ flanking sequence for three Pinb variants: Pinb-2v1, Pinb-2v2, and Pinb-2v3. Additionally, a new Pinb variant, Pinb-2v4, is reported. All four variants were physically mapped using Chinese Spring (CS) diteolosomics, nullisomic–tetrasomics, and CS-Cheyenne disomic substitution lines. Results place Pinb-2v1 on 7DL, Pinb-2v2 on 7BL, Pinb-2v3 on 7B, and Pinb-2v4 on 7AL. Pinb-2v1 and Pinb-2v4 were present in all cvs. examined: CS, Cheyenne, Recital, Wichita and Winsome. Pinb-2v2 was present in CS and Recital; Pinb-2v3 was present in Cheyenne, Wichita, and Winsome. These results are not wholly consistent with prior research and additional studies will be required to reconcile discrepancies. The discovery of Pinb-2v4 and the mapping of all four variants will contribute to a better understanding of gene duplication events in wheat and their bearing on wheat kernel texture and grain utilization.
DOI:10.1007/s001220100576URL [本文引用: 1]

During the initial phases of a wheat endosperm Expressed-Sequence-Tag (EST) project, several clones were determined to be related to wheat gliadin sequences, but not similar enough to be classified into any of the traditional gliadin families [α-, γ-, and ω-gliadins, low-molecular-weight (LMW) glutenins]. Complete sequences of these cDNA clones revealed four new classes of gliadin-related endosperm proteins, but lacking a prominent repeat domain which until now has been characteristic of the gliadins. Two of these classes are related to different minimally described groups of Triticeae endosperm proteins. One class of proteins, which has N-terminal amino-acid sequences matching members of a reported 25-kDa globulin family from wheat, is shown by amino-acid sequencing to match to a family of 25-kDa endosperm proteins, is encoded by a multigene family, and is most similar to the LMW-glutenins. A second new class shows N-terminal homologies to LMW secalins from rye, and has an amino-acid composition similar to wheat and barley LMW proteins with extraction properties similar to prolamins. The third class is most similar to α-gliadins, and the fourth class has no close association to previously described wheat endosperm proteins.
DOI:10.1073/pnas.1812855115URLPMID:30530679 [本文引用: 1]

Fifteen full-length wheat grain avenin-like protein coding genes (TaALP) were identified on chromosome arms 7AS, 4AL, and 7DS of bread wheat with each containing five genes. Besides the a- and b-type ALPs, a c type was identified in the current paper. Both a and b types have two subunits, named x and y types. The five genes on each of the three chromosome arms consisted of two x-type genes, two y-type genes, and one c-type gene. The a-type genes were typically of 520 bp in length, whereas the b types were of 850 bp in length, and the c type was of 470 bp in length. The ALP gene transcript levels were significantly up-regulated in Blumeria graminis f. sp. tritici (Bgt)-infected wheat grain caryopsis at early grain filling. Wild emmer wheat [(WEW), Triticum dicoccoides] populations were focused on in our paper to identify allelic variations of ALP genes and to study the influence of natural selection on certain alleles. Consequently, 25 alleles were identified for TdALP-bx-7AS, 13 alleles were identified for TdALP-ax-7AS, 7 alleles were identified for TdALP-ay-7AS, and 4 alleles were identified for TdALP-ax-4AL Correlation studies on TdALP gene diversity and ecological stresses suggested that environmental factors contribute to the ALP polymorphism formation in WEW. Many allelic variants of ALPs in the endosperm of WEW are not present in bread wheat and therefore could be utilized in breeding bread wheat varieties for better quality and elite plant defense characteristics.
URLPMID:26011437 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOI:10.1007/s00122-005-0047-7URLPMID:16133313 [本文引用: 1]

DNA from six hexaploid, tetraploid and diploid species of Aegilops with the C, D, S, M and U genomes was amplified with specific PCR primers to identify sequences encoding puroindolines (Pins) a and b and grain softness protein (GSP), all of which are encoded by genes at the Ha (hardness) locus, with Ae. tauschii (DD) and bread wheat (T. aestivum) (AABBDD) cv Hiline being studied as controls. Seven new allelic forms of Pin a and Pin b were identified, including forms with mutations within or close to the tryptophan motif. In addition, five new forms of GSP were detected. In all species both genomic DNA from leaves and cDNA from developing grain were analysed. This revealed the presence of both silent genes (with premature stop codons) and multiple genes, with the latter being confirmed by Southern blot analysis. Freeze fracture analysis demonstrated that all except one accession (Ae. sharonensis) were soft textured. However, this difference cannot be accounted for by the sequences of the Pin alleles present in this line.
DOI:10.1186/s13059-018-1491-4URLPMID:30115101 [本文引用: 2]

The Wheat@URGI portal has been developed to provide the international community of researchers and breeders with access to the bread wheat reference genome sequence produced by the International Wheat Genome Sequencing Consortium. Genome browsers, BLAST, and InterMine tools have been established for in-depth exploration of the genome sequence together with additional linked datasets including physical maps, sequence variations, gene expression, and genetic and phenomic data from other international collaborative projects already stored in the GnpIS information system. The portal provides enhanced search and browser features that will facilitate the deployment of the latest genomics resources in wheat improvement.
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.ab.2009.10.047URLPMID:19891953 [本文引用: 1]

Serum low-molecular-weight proteins (LMWPs, molecular weight<30kDa) are closely related to the body physiological and pathological situations, whereas many difficulties are encountered when enriching and fractionating them. Using C(18) absorbent (100 A) enrichment and fractionation under urea/dithiothreitol (DTT) denatured environment followed by 60% acetonitrile (ACN) elution, serum LMWPs could be enriched more than 100-fold and were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), two-dimensional gel electrophoresis (2-DE), and isotope-coded affinity tag (ICAT) labeling quantification. Proteins existing in human serum at low nanograms/milliliter (ng/ml) levels, such as myeloid-related proteins (MRPs), could be identified directly from 2-DE coupled with matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI-TOF/TOF MS) and LTQ-Orbitrap MS. Sixteen proteins were confidentially identified and quantified using ICAT labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS). By virtue of its easy operation and high reproducibility to process large quantity complex serum samples, this method has potential uses in enriching LMWPs either in serum or in cell and tissue samples.
DOI:10.1021/ac0262560URLPMID:12713048 [本文引用: 1]

A novel MS/MS-based analysis strategy using isotopomer labels, referred to as
DOI:10.1038/75556URLPMID:10802651 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:32652019 [本文引用: 1]
DOI:10.1093/molbev/msw054URLPMID:27004904 [本文引用: 1]

We present the latest version of the Molecular Evolutionary Genetics Analysis (Mega) software, which contains many sophisticated methods and tools for phylogenomics and phylomedicine. In this major upgrade, Mega has been optimized for use on 64-bit computing systems for analyzing larger datasets. Researchers can now explore and analyze tens of thousands of sequences in Mega The new version also provides an advanced wizard for building timetrees and includes a new functionality to automatically predict gene duplication events in gene family trees. The 64-bit Mega is made available in two interfaces: graphical and command line. The graphical user interface (GUI) is a native Microsoft Windows application that can also be used on Mac OS X. The command line Mega is available as native applications for Windows, Linux, and Mac OS X. They are intended for use in high-throughput and scripted analysis. Both versions are available from www.megasoftware.net free of charge.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1371/journal.pone.0164746URLPMID:27741295 [本文引用: 1]

Puroindoline (Pina and Pinb) genes control grain texture or hardness in wheat. Wild-type/soft alleles lead to softer grain while a mutation in one or both of these genes results in a hard grain. Variation in hardness in genotypes with identical Pin alleles (wild-type or mutant) is known but the molecular basis of this is not known. We now report the identification of wheat genotypes with hard grain texture and wild-type/soft Pin alleles indicating that hardness in wheat may be controlled by factors other than mutations in the coding region of the Pin genes. RNA-Seq analysis was used to determine the variation in the transcriptome of developing grains of thirty three diverse wheat genotypes including hard (mutant Pin) and soft (wild type) and those that were hard without having Pin mutations. This defined the role of pin gene expression and identified other candidate genes associated with hardness. Pina was not expressed in hard wheat with a mutation in the Pina gene. The ratio of Pina to Pinb expression was generally lower in the hard non mutant genotypes. Hardness may be associated with differences in Pin expression and other factors and is not simply associated with mutations in the PIN protein coding sequences.
URLPMID:19570905 [本文引用: 1]
DOI:10.1007/s11032-013-9971-4URL [本文引用: 1]
