 ,*广州大学生命科学学院, 广东广州 510006
,*广州大学生命科学学院, 广东广州 510006Identification of CRISPR/Cas9 knockout targets and tissue expression analysis of circadian clock genes GmLNK1/2, GmRVE4/8, and GmTOC1 in soybean
GAN Zhuo-Ran, SHI Wen-Qian, LI Yong-Li, HOU Zhi-Hong, LI Hai-Yang, CHENG Qun, DONG Li-Dong, LIU Bao-Hui, LU Si-Jia ,*School of Life Sciences, Guangzhou University, Guangzhou 510006, Guangdong, China
,*School of Life Sciences, Guangzhou University, Guangzhou 510006, Guangdong, China通讯作者:
收稿日期:2019-11-7接受日期:2020-03-24网络出版日期:2020-08-12
| 基金资助: |
Received:2019-11-7Accepted:2020-03-24Online:2020-08-12
| Fund supported: |
作者简介 About authors
E-mail:gggzzr@126.com。

摘要
关键词:
Abstract
Keywords:
PDF (2939KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
甘卓然, 石文茜, 黎永力, 侯智红, 李海洋, 程群, 董利东, 刘宝辉, 芦思佳. 大豆生物钟基因GmLNK1/2、GmRVE4/8和GmTOC1 CRISPR/Cas9组织表达分析及敲除靶点的鉴定[J]. 作物学报, 2020, 46(8): 1291-1300. doi:10.3724/SP.J.1006.2020.94169
GAN Zhuo-Ran, SHI Wen-Qian, LI Yong-Li, HOU Zhi-Hong, LI Hai-Yang, CHENG Qun, DONG Li-Dong, LIU Bao-Hui, LU Si-Jia.
单产低是我国大豆面临的最大问题, 因此提高大豆单产是首要任务[1]。影响大豆产量的因素可归为两大类, 一是外部因素, 如耕作技术、气候、土壤和自然灾害等; 二是内部因素, 即基因, 两方面的因素相互影响、协调, 共同调控大豆的产量[2,3]。大豆作为固着生物, 必须准确地感知环境信号, 如光和温度, 以适应其生长发育的周围环境[4]。实际上, 大多数生物并不是简单地对外部环境做出反应, 而是对外部环境的变化做出预测, 并根据自己的生物习性进行调整[5]。这种内源性生物节律就是生物钟。生物钟使生物体能够预测和准备应对周围环境中的变化[4]。生物种基因在控制植物开花、产量、生物胁迫和非生物胁迫中都扮演重要的角色。我们前期的研究发现, 大豆长童期性状关键位点J是拟南芥生物钟关键基因ELF3 (EARLY FLOWERING 3)的一个同源基因, 并证明在短日照下J的等位变异能够延迟大豆开花, 使大豆产量提高30%以上[6]。植物生物钟由许多基因共同调控, 如LHY (LATE ELONGATED HYPOCOTYL)、CCA1 (CIRCADIAN CLOCK -ASSOCIATED)、TOC1 (TIMING OF CAB EXPRESSION1)、PRR (PSEUDO-RESPONSE REGULATOR)、LUX (LUX ARRHYTHMO), ELF3 (EARLY FLOWERING 3)、ELF4 (EARLY FLOWERING 4)、RVE (REVEILLE)、LNK (NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED)等[7]。在拟南芥中, LHY /CCA1 与TOC1是植物生物钟的核心元件, 并且形成反馈调节回路[8,9]。RVE4、RVE6和RVE8是与LHY/CCA1同源的生物钟基因, 作为生物钟激活因子调控下游基因的表达[9,10]。LNK1与LNK2作为转录共激活因子, 能够与RVE4及RVE8相互作用, 共同调控TOC1基因的表达[11]。番茄LNK2基因的突变可导致番茄生物钟周期延迟[12]。这些生物钟基因在不同植物中引起其他性状的变化也开始被解析。RVE功能研究中发现, 拟南芥RVE4/RVE6/RVE8三突变体中, 光敏色素互作因子PIF4 (PHYTOCHROME INTERACTING FACTOR 4)和PIF5 (PHYTOCHROME INTERACTING FACTOR 5)表达量提高, 从而导致其叶片面积显著大于野生型拟南芥植株[10]。LNK2功能研究中发现, 番茄LNK2基因突变可导致番茄生物钟周期延后, 叶绿素含量明显增加, 有效防止番茄晒伤, 该研究指出昼夜节律中的这种条件变化可能对各物种(包括植物和动物)的纬度适应性至关重要[12,13]。尽管这些重要的生物钟基因在拟南芥、番茄等植物中的功能相继被解析, 但在大豆中仍不清楚。
CRISPR/Cas9技术能够精准快速地进行基因编辑, 因此被广泛应用在动物和植物中[14,15,16,17]。近些年在大豆中利用CRISPR/Cas9技术已经成功对多个大豆基因进行定点编辑。例如GmFT2a、GmFT5a、GmSPL9、GmFAD2-1a、GmFAD2-1b等, 为更加深入研究这些基因的功能, 提供了良好的遗传材料[18,19,20]。大豆的基因组复杂, 生长周期长, 转化效率低, 所以我们在利用CRISPR/Cas9技术进行大豆稳定转化前, 确定目的基因靶点(gRNA)的有效性是十分重要的。本研究首先利用蛋白同源比对和进化树分析的方法成功鉴定出与拟南芥同源的大豆生物钟基因GmLNK1、GmLNK2、GmTOC1、GmRVE4和GmRVE8。并利用qRT-PCR技术, 检测这些基因在大豆植株的各个组织中的表达情况。随后, 利用CRISPR/Cas9技术, 构建了大豆GmLNK1a/b/c/d、GmLNK2a/b/c/d、GmTOC1 a/b/c/d、GmRVE4/8a/b/c/d基因的敲除载体, 并利用大豆发根体系成功检测出大部分基因的有效编辑靶点。为利用CRISPR/Cas9技术创造这些生物钟基因的稳定大豆突变体和进一步研究这些生物钟基因的功能提供了理论基础。
1 材料与方法
1.1 材料
大豆发根转化材料为大豆品种Williams 82 (W82), 大肠杆菌菌株DH5α、发根农杆菌菌株K599均由本实验室保存, 载体构建质粒AtU3d、AtU3b、AtU6-1、AtU6-29、Cas9载体由华南农业大学刘耀光院士提供。1.2 植物材料处理及RNA提取
将大豆品种W82种子播于钵中(草炭土:蛭石=2:1), 每钵6粒, 在温度25℃、光照16 h/8 h (光照/黑暗)的温室内培养。大约14 d后, 大豆幼苗长到V1期, 取大豆的根、茎、子叶、下胚轴、初生叶、三出复叶和生长点于液氮中速冻。大豆生长到R1期(约48 d), 取大豆的花于液氮中速冻。参照RNA提取试剂盒说明书(RNA pure Plant Kit, China)提取RNA。qRT-PCR实验分别进行3次生物重复。1.3 大豆生物钟基因的鉴定和进化树分析
分别利用拟南芥生物钟基因AtLNK1、AtLNK2、AtTOC1、AtRVE4、AtRVE8的蛋白序列, 在Phytozome 12数据库进行BLAST分析, 分别鉴定与拟南芥同源的大豆生物种基因。根据同源性由高到低, 分别将每个拟南芥生物钟基因在大豆中4个同源基因命名为a、b、c和d。利用DANMAN软件进行同源比对分析。1.4 靶点设计
利用CRISPR direct在线网站(Table 1
表1
表1基因靶点引物列表
Table 1
| 靶点引物 Target primer | 引物序列 Primer sequence (5°-3°) | 靶点对应基因 Target of gene |
|---|---|---|
| LNK1T1F | gtcaGGAGACAAGTGTGTGGTGG | GmLNK1a, GmLNK1b |
| LNK1T1R | aaacCCACCACACACTTGTCTCC | GmLNK1a, GmLNK1b |
| LNK1T2F | gtcaAGAGGAGTTCTGCTGGCTC | GmLNK1a, GmLNK1b |
| LNK1T2R | aaacGAGCCAGCAGAACTCCTCT | GmLNK1a, GmLNK1b |
| LNK1T3F | attgCTTGGGAGACAAGTGTGTGG | GmLNK1c, GmLNK1d |
| LNK1T3R | aaacCCACACACTTGTCTCCCAAG | GmLNK1c, GmLNK1d |
| LNK1T4F | attgAGACTTTGAAGATGTTGAC | GmLNK1c, GmLNK1d |
| LNK1T4R | aaacGTCAACATCTTCAAAGTCT | GmLNK1c, GmLNK1d |
| LNK2T1F | gtcaACATAATATGGGGTGAAGG | GmLNK2a, GmLNK2b |
| LNK2T1R | aaacCCTTCACCCCATATTATGT | GmLNK2a, GmLNK2b |
| LNK2T2F | gtcaAAACTGATCAGGGTTCCCT | GmLNK2c, GmLNK2d |
| LNK2T2R | aaacAGGGAACCCTGATCAGTTT | GmLNK2c, GmLNK2d |
| LNK2T3F | attgTTTGATTGGAACGACGAAG | GmLNK2a, 2b, 2c, 2d |
| LNK2T3R | aaacCTTCGTCGTTCCAATCAAA | GmLNK2a, 2b, 2c, 2d |
| LNK2T4F | attgTCATATTGTGCCTTATCCGG | GmLNK2c, GmLNK2d |
| LNK2T4R | aaacCCGGATAAGGCACAATATGA | GmLNK2c, GmLNK2d |
| RVE48T1F | gtcaCTTCCCTGCTGATGAATGC | GmRVE4/8b, GmRVE4/8c |
| RVE48T1R | aaacGCATTCATCAGCAGGGAAG | GmRVE4/8b, GmRVE4/8c |
| RVE48T2F | gtcaTCATCCCATGTGACATACCC | GmRVE4/8a, GmRVE4/8d |
| RVE48T2R | aaacGGGTATGTCACATGGGATGA | GmRVE4/8a, GmRVE4/8d |
| RVE48T3F | attgCAGCTTTGCGCTTTGGACG | GmRVE4/8b, GmRVE4/8c |
| RVE48T3R | aaacCGTCCAAAGCGCAAAGCTG | GmRVE4/8b, GmRVE4/8c |
| RVE48T4F | attgAAGCTTTGCGCTTAGGCCG | GmRVE4/8a, GmRVE4/8d |
| RVE48T4R | aaacCGGCCTAAGCGCAAAGCTT | GmRVE4/8a, GmRVE4/8d |
| TOC1T1F | gtcaCGATTCCAAGAGTTCTCAAG | GmTOC1a, GmTOC1b |
| TOC1T1R | aaacCTTGAGAACTCTTGGAATCG | GmTOC1a, GmTOC1b |
| TOC1T2F | gtcaGTGATGTCCGCACAAGATG | GmTOC1a, GmTOC1b |
| TOC1T2R | aaacCATCTTGTGCGGACATCAC | GmTOC1a, GmTOC1b |
| TOC1T3F | attgGTGGGAATAATAGTAAGAG | GmTOC1c, GmTOC1d |
| TOC1T3R | aaacCTCTTACTATTATTCCCAC | GmTOC1c, GmTOC1d |
| TOC1T4F | attgTTGTAAAGTGCTTGAGGCT | GmTOC1c, GmTOC1d |
| TOC1T4R | aaacAGCCTCAAGCACTTTACAA | GmTOC1c, GmTOC1d |
新窗口打开|下载CSV
1.5 CRISPR/Cas9载体构建
参照曾栋昌等[21]、侯智红等[22]研究方法进行载体构建。实验步骤如下: (1)靶点引物退火。(2)已退火的靶点与小载体AtU3d、AtU3b、AtU6-1和AtU6-29一一对应连接, 并进行第一次边切边连。(3)使用第一次边切边连产物进行第一次PCR扩增。(4)以第一次PCR产物为模板, 进行第二次PCR扩增。(5)回收第二次PCR的产物, 进行第二次边切边连, 连接Cas9载体。(6)第二次边切边连产物转化大肠杆菌DH5α。(7)挑取阳性单菌落, 摇菌, 提取质粒, 转化农杆菌K599。1.6 大豆根毛转化
参照表2配制萌发及根诱导培养基, 并参照Cheng等[23]方法进行根毛转化。步骤如下: (1)将大豆种子灭菌, 取饱满没有病斑的籽粒置干燥器中, 在干燥器中放一个200 mL烧杯, 盛96 mL次氯酸钠和4 mL浓盐酸, 灭菌12~16 h。(2)根据表2配制大豆萌发培养基, 在无菌操作台中将已灭菌的大豆种子均匀摆放到萌发培养基。(3)大豆种子萌发5 d左右, 根据表2配制发根培养基, 在无菌操作台中利用手术刀在大豆的子叶上面切一个小洞。然后将处理的大豆子叶均匀放置到发根培养基上, 取20 μL含有目的载体的农杆菌K599放到大豆子叶的小洞中。(4)将培养基放在大豆无菌培养室中, 培养15 d左右, 从大豆子叶的侵染部位会有根毛长出, 分别收集大豆的根毛于液氮中速冻。Table 2
表2
表2萌发培养基与根诱导培养基
Table 2
| 培养基类型 Medium type | 药品名称 Name of the medicine | 药品用量 Medicine dosage |
|---|---|---|
| 萌发培养基 Germination medium | B5盐 B5 salt mixture | 1× |
| 蔗糖 Sucrose (g L-1) | 20 | |
| 琼脂 Agar (g L-1) | 8 | |
| 发根培养基 Rooting medium | MS合成盐 MS salt mixture | 1× |
| 蔗糖 Sucrose (g L-1) | 30 | |
| 2-(4-吗啉)乙磺酸 MES | 0.6 | |
| 琼脂 Agar (g L-1) | 8 | |
| 头孢霉素 Cef (mg L-1) | 250 | |
| 羧苄青霉素 Car (mg L-1) | 250 |
新窗口打开|下载CSV
1.7 基因靶点编辑有效性检测
大豆生物钟基因靶点编辑有效性检测步骤如下: (1)用1.6中收集的大豆根毛提取DNA。(2)利用CRISPR/Cas9载体通用引物SP1和SP3 (表3)检测大豆根毛是否为转基因根毛(目的片段的长度约为2000 bp左右), 检测每个载体20个转基因事件以检测靶点编辑效率。(3)根据每个基因的DNA序列, 分别设计扩增目的基因的靶点检测特异引物。利用每个基因的靶点特异引物(表3)进行PCR扩增目的片段, 将PCR产物送广州天一辉远测序服务公司测序。(4)分析测序结果的峰图, 如果在靶点附近发生套峰, 证明目的基因的靶点是有效的。Table 3
表3
表3靶点检测引物
Table 3
| 靶点检测引物 Target primer | 引物序列 Primer sequence (5°-3°) | 靶点检测引物 Target primer | 引物序列 Primer sequence (5°-3°) |
|---|---|---|---|
| SP3 | GTCGTGCTCCACATGTTGACCGG | LNK2cT4&2R | TCCTGAGGTTAGTAGTTCTCCACT |
| SP1 | GAAGTTATTGCATCTATGTCGGG | LNK2dT3F | CTCTCCGTCGCCGTTATAGCA |
| LNK1aF | TGACGCCAGGGTATCTTAAA | LNK2dT3R | ACCAAACGAAGCACGAACA |
| LNK1aR | TCTCCATGTGTGTGTTTTGGTA | LNK2dT4&2F | GGTGGTGGAGGGAGAAGATGAG |
| LNK1bF | CTCAGGGTAGGGAGGACTTG | LNK2dT4&2R | AGACACTTATTGCCGCTACAACTG |
| LNK1bR | CGGTAAAGTTGAGCCTTGGT | RVE4/8aF | ACAGCTCTTCAGCTAGGTGTT |
| LNK1cF | TCATATAGTGCCCCATGCCA | RVE4/8aR | GAGGAGAGGGGGTATGGGTT |
| LNK1cR | AGTTCTATAGCAGCTCATGACA | RVE4/8bF | AGCGAAGAACTCTGCAATCCA |
| LNK1dF | GCCCGATCATTGCTTCAAGAG | RVE4/8bR | CTACCACCTTGGGCCGAAAT |
| LNK1dR | AGTTCTATAGCAGCTCGTGGC | RVE4/8cF | TTCGAAGCCATGCTCAGAAG |
| LNK2aT3F | GAATTCGGCGATGTGTGAGC | RVE4/8cR | CCAGCAACAAGGTTCGTAGT |
| LNK2aT3R | ACAGCTACACAAAGACACACA | RVE4/8dF | TTCGTTGGTCATCTTGCTGGT |
| LNK2aT1F | CCGTCCAAGGAGATTGTCACTGA | RVE4/8dR | CTGGACATGGCCTTCTGTGT |
| LNK2aT1R | TCCTGAGGTAGGTAGTTCTCCACT | TOC1aF | TCCCTCAACGATGCTG |
| LNK2bT3F | TGATGGAGTGCGTTTCTCTG | TOC1aR | GCCTCCGTCTCCACAT |
| LNK2bT3R | CCCTGATTTTCCTGGCGTAA | TOC1bF | TTGAGCAAGTCCAGGGTT |
| LNK2bT1F | CCGTCCAAGGAGATTGTCACTGA | TOC1bR | ATGGCTGTGATGGTAACTCG |
| LNK2bT1R | TCCTGAGGTAGGTAGTTCTCCACT | TOC1cF | CTCTAACTAACTATCCAGACCCTA |
| LNK2cT3F | TTCTCCGTCGATCAGTGAAGTG | TOC1cR | ATGGCTGGTGGGTTGA |
| LNK2cT3R | GCTAAGAGTCACGCCTCCTTG | TOC1dF | TACGCCCTCCCTCTTT |
| LNK2cT4&2F | CGTGATGCCAAATTAGTTGGGTAT | TOC1dR | GGGACTTGGGAAATACA |
新窗口打开|下载CSV
2 结果与分析
2.1 进化树与同源性比对分析
在Phytozome 12数据库中, 搜索并下载与拟南芥生物钟基因AtLNK1 (AT5G64170.2)、AtLNK2 (AT3G54500.3)、AtTOC1 (AT5G61380.1)、AtRVE4 (AT5G02840.1)、AtRVE8 (AT3G09600.1)同源的大豆基因序列, 发现大豆中分别含有4个AtLNK1同源拷贝, 命名为GmLNK1a (Glyma. 05G118300.1)、GmLNK1b (Glyma.08G073300.1)、GmLNK1c (Glyma.08G179400.1)、GmLNK1d (Glyma.15G053000.1); 大豆中分别含有4个AtLNK2同源拷贝, 命名为GmLNK2a (Glyma.04G141400.1)、GmLNK2b (Glyma.11G154700.1)、GmLNK2c (Glyma.13G199300.1)、GmLNK2d (Glyma. 15G237600.1); 大豆中分别含有4个AtTOC1同源拷贝, 命名为 GmTOC1a (Glyma.04G166300.1)、GmTOC1b (Glyma.06G196200.1)、GmTOC1c (Glyma.05G025000.1)、GmTOC1d (Glyma.17G102200.1); 大豆中分别含有4个AtRVE4与AtRVE8同源拷贝, 命名为GmRVE4/8a (Glyma.03G177300.1)、GmRVE4/8b (Glyma.10G048500.1)、GmRVE4/8c (Glyma.13G136300.1)、GmRVE4/8d (Glyma. 19G178000.1)。利用 DNAMAN软件, 分别与AtLNK1、AtLNK2、AtTOC1、AtRVE4、AtRVE8进行同源性比对。表明, GmLNK1a/b/c/d在第503~618位氨基酸比较保守, 保守率约为63.38% (图1-A)。GmLNK2a/b/c/d在第176~224、438~471和511~620位氨基酸比较保守, 保守率约为68.81% (图1-B)。GmTOC1a/b/c/d在第27~205、553~622位氨基酸比较保守, 保守率约为72.07% (图1-C)。GmRVE4/8a/b/c/d在第35~124、226~282位氨基酸比较保守, 与AtRVE4比对, 保守率约为73.37%, 与AtRVE8比对, 则保守率约为76.66% (图1-D)。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1同源性比对
A: AtLNK1与GmLNK1a、GmLNK1b、GmLNK1c、GmLNK1d氨基酸序列对比; B: AtLNK2与GmLNK2a、GmLNK2b、GmLNK2c、GmLNK2d氨基酸序列对比; C: AtTOC1与GmTOC1a、GmTOC1b、GmTOC1c、GmTOC1d氨基酸序列对比; D: AtRVE4、AtRVE8与GmRVE4/8a、GmRVE4/8b、GmRVE4/8c、GmRVE4/8d氨基酸序列对比。
Fig. 1Homology comparison
A: amino acid sequences comparison of AtLNK1, GmLNK1a, GmLNK1b, GmLNK1c, and GmLNK1d; B: amino acid sequences comparison of AtLNK2, GmLNK2a, GmLNK2b, GmLNK2c, and GmLNK2d; C: amino acid sequences comparison of AtTOC1, GmTOC1a, GmTOC1b, GmTOC1c, and GmTOC1d; D: amino acid sequences comparison of AtRVE4, AtRVE8, GmRVE4/8a, GmRVE4/8b, GmRVE4/8c, and GmRVE4/8d.
2.2 组织特异性表达分析
生物钟基因在整个植物生命进程及应答环境胁迫过程中扮演至关重要的角色, 因此检测生物钟基因在植物中的表达部位非常重要。通过qRT-PCR技术检测显示, GmLNK1a、GmLNK1b和GmLNK1c、GmLNK1d在大豆各个组织中均有表达, 但GmLNK1a/b/c表达量较低, 而GmLNK1d在下胚轴、子叶、茎、生长点中表达较高(图2-A); GmLNK2a/b/c/d在大豆各个组织中均有表达, 且在初生叶中表达最高(图2-B); GmTOC1家族中4个GmTOC1的表达情况与GmLNK1家族相似, GmTOC1a、GmTOC1b和GmTOC1c的表达量都很低, GmTOC1d在下胚轴的表达量最高(图2-C); GmRVE4/8家族则没有明显的规律, GmRVE4/8a在三出复叶处的表达量最高, GmRVE4/8b在花处的表达量最高, GmRVE4/8c在生长点处的表达量最高, GmRVE4/8d在花处的表达量最高(图2-D)。图2
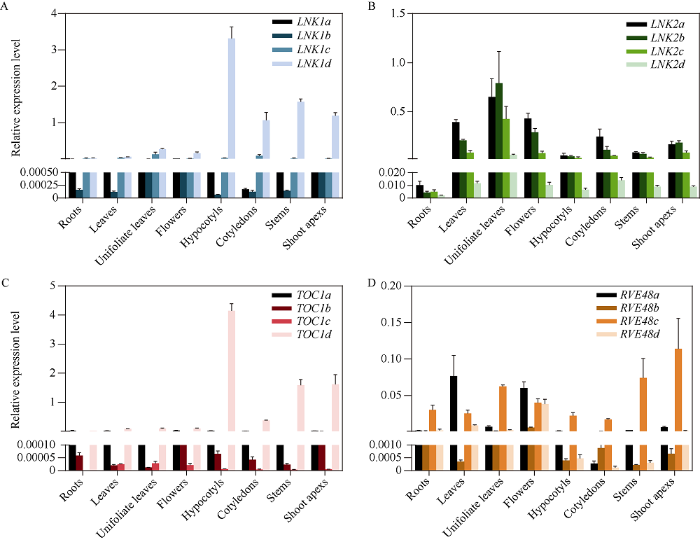 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图24个生物钟基因组织特异表达
A: GmLNK1a/b/c/d在W82中的组织特异表达; B: GmLNK2a/b/c/d在W82中的组织特异表达; C: GmTOC1a/b/c/d在W82中的组织特异表达; D: GmRVE4/8a/b/c/d在W82中的组织特异表达。
Fig. 2Expression of four clock genes in soybean tissues
A: the expression of GmLNK1a/b/c/d in soybean W82 tissues; B: the expression of GmLNK2a/b/c/d in soybean W82 tissues; C: the expression of GmTOC1a/b/c/d in soybean W82 tissues; D: the expression of GmRVE4/8a/b/c/d in soybean W82 tissues.
2.3 CRISPR/Cas9载体构建
为了鉴定这些基因有效的基因编辑靶点, 分别将GmLNK1a/b/c/d、GmLNK2a/b/c/d、GmTOC1a/b/c/d、GmRVE4/8 a/b/c/d 16个基因的16个靶点构建到4个CRISPR/Cas9载体上, 每个基因包含2~3个靶点, 各基因所对应靶点如表1所示。载体结构如图3所示, GmLNK1、GmLNK2、GmTOC1和GmRVE4/8基因的T1靶点连接拟南芥的AtU3d启动子, T2靶点连接拟南芥的AtU3b启动子, T3靶点连接拟南芥的AtU6-1启动子, T4靶点连接拟南芥的AtU6-29启动子。图3
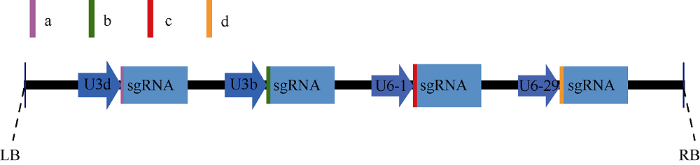 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3Cas9载体结构图
a: 靶点LNK1T1、LNK2T1、TOC1T1和RVE4/8T1; b: 靶点LNK1T2、LNK2T2、TOC1T2和RVE4/8T2; c: 靶点LNK1T3、LNK2T3、TOC1T3和RVE4/8T3; d: 靶点LNK1T4、LNK2T4、TOC1T4和RVE4/8T4。
Fig. 3Schematic diagram of the Cas9 vector construction
a: target of LNK1T1, LNK2T1, TOC1T1, and RVE4/8T1; b: target of LNK1T2, LNK2T2, TOC1T2, and RVE4/8T2; c: target of LNK1T3, LNK2T3, TOC1T3, and RVE4/8T3; d: target LNK1T4, LNK2T4, TOC1T4, and RVE4/8T4.
2.4 GmLNK1、GmLNK2、GmTOC1和GmRVE4/8基因的靶点有效性检测
从图4-A可以看出, GmLNK1基因的T1靶点测序峰图在靶点附近发生套峰, 说明这个靶点是有效的。根据测序发现的16个基因中, 有13个被成功编辑, GmLNK1a的LNK1T1靶点、GmLNK1b的LNK1T2靶点、GmLNK1c和GmLNK1d的LNK1T4靶点可被有效编辑(图4-A); GmLNK2a和GmLNK2b的靶点LNK2T1靶点, GmLNK2c和GmLNK2d的LNK2T4可被有效编辑(图4-B), GmRVE4/8b的RVE48T1靶点和GmRVE4/8d的RVE48T4靶点可被有效编辑(图4-C), GmTOC1b的TOC1T2靶点, 以及GmTOC1c和GmTOC1d的TOC1T4可被有效编辑(图4-D)。图4
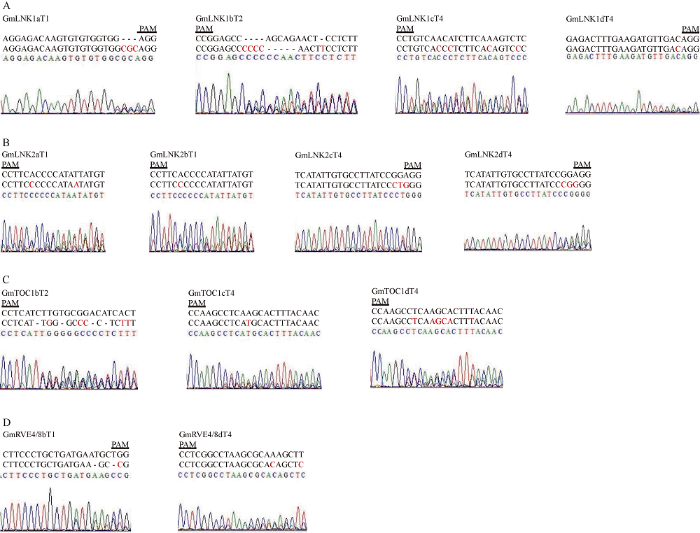 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4发生编辑靶点的测序峰图
A: GmLNK1a的靶点LNK1T1、GmLNK1b的靶点LNK1T2、GmLNK1c的靶点LNK1T4和GmLNK1d的靶点LNK1T4突变情况; B: GmLNK2a的靶点LNK2T1、GmLNK2b的靶点LNK2T1、GmLNK2c的靶点LNK2T4和GmLNK2d的靶点LNK2T4突变情况; C: GmTOC1b的靶点TOC1T2、GmTOC1c的靶点TOC1T4和GmTOC1d的靶点TOC1T4突变情况; D: GmRVE4/8b的靶点RVE4/8T1和GmRVE4/8d的靶点RVE4/8T4突变情况。
Fig. 4Detailed sequence of the targets site in the transgenic soybean hairy roots
A: mutations were found in target LNK1T1 of GmLNK1a, target LNK1T2 of GmLNK1b, target LNK1T4 of GmLNK1c and target LNK1T4 of GmLNK1d; B: mutations were found in target LNK2T1 of GmLNK2a, target LNK2T1 of GmLNK2b, target LNK2T4 of GmLNK2c and target LNK2T4 of GmLNK2d; C: mutations were found in target TOC1T2 of GmTOC1b, target TOC1T4 of GmTOC1c and target TOC1T4 of GmTOC1d; D: mutations were found in target RVE4/8T1 of GmRVE4/8b and target RVE4/8T4 of GmRVE4/8d.
3 讨论
基因组定点编辑是将外源的DNA导入受体细胞染色体的特定位点上, 特异地改造基因组, 是目前研究基因功能的重要方法之一[24]。2013年, 科学家发现了一个突破性的基因编辑技术CRISPR/Cas系统, 这个系统作为防御系统存在于部分古生菌和细菌的基因组中[24,25,26,27]。在植物中, CRISPR/Cas9 系统已成功应用于包括拟南芥、烟草、水稻、烟草、高粱、小麦、玉米、柑橘和苔类植物在内的多种植物中[28,29,30,31,32,33,34,35]。CRISPR/Cas9系统较传统基因编辑技术更加便捷高效, 应用广泛[36]。大豆是古四倍体植物, 约75%的基因重复[37]。在大豆中一个基因往往存在2~4个拷贝, 而且这些基因大多是功能冗余的, 随机突变一个基因往往不会产生表型[38]。随着对CRISPR/Cas9系统的不断完善, 现已成功应用到大豆中, 实现对目标基因的定点编辑[18-19,39]。但由于大豆的转化效率低, 基因组复杂, 生长周期长等特点, CRISPR/ Cas9技术在大豆中的应用仍然会出现靶点无效的问题。生物钟基因在植物的生长发育、生物胁迫和非生物胁迫中都扮演着重要的角色。但大部分生物钟基因在大豆中的功能都没有被解析。本试验通过蛋白序列比对和进化树分析鉴定了大豆中的GmLNK1、GmLNK2、GmTOC1、GmRVE4/8同源基因, 初步分析了它们在大豆植株不同组织中的表达情况, 为进一步预测这些基因在大豆的不同组织中是否具有功能提供了理论基础。例如, GmRVE4/8a在三出复叶中的表达量最高, 推测GmRVE4/8a在大豆的叶片中具有重要功能; GmRVE4/8b和GmRVE4/8d在大豆的花中表达量最高, 这2个基因可能对大豆花的发育具有重要作用; 而GmRVE4/8c在生长点处的表达量最高, GmRVE4/8c可能与大豆的开花和生长习性有关。
CRISPR/Cas9系统在应用于大豆的过程中靶点的编辑效率存在相对低的问题。本研究利用CRISPR/Cas9系统成功构建多个用于诱导大豆GmLNK1、GmLNK2、GmTOC1、GmRVE4/8突变的CRISPR/Cas9载体。共设计了16个靶点, 其中9个能有效编辑目的基因, 同时也发现9个有效的靶点能够有效敲除13个基因。本研究设计大豆多个拷贝的同源基因CRISPR/Cas9基因敲除靶点的策略尽可能设计一个能够同时敲除2个或2个以上基因的靶点, 在构建一个多基因敲除载体时保证每个基因含有2个或2个以上的靶点。本研究中, 构建的CRISPR/Cas9载体能够同时敲除GmLNK1的4个同源基因和GmLNK2的4个同源基因证明CRISPR/Cas9靶点设计策略是有效的。
本研究提供了一个简单高效的大豆多拷贝同源基因CRISPR/Cas9靶点设计策略, 保证每个基因设计2个或2个以上靶点, 同时利用大豆发根转化体系快速检测靶点的编辑效率, 为更加快速创造大豆关键基因的稳定突变体提供了理论基础。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/erw348URLPMID:27660480 [本文引用: 1]

Climate change has brought severe challenges to agriculture. It is anticipated that there will be a drop in crop yield - including that of soybean - due to climatic stress factors that include drastic fluctuations in temperature, drought, flooding and high salinity. Genomic information on soybean has been accumulating rapidly since initial publication of its reference genome, providing a valuable tool for the improvement of cultivated soybean. Not only are many molecular markers that are associated with important quantitative trait loci now identified, but we also have a more detailed picture of the genomic variations among soybean germplasms, enabling us to utilize these as tools to assist crop breeding. In this review, we will summarize and discuss the currently available soybean genomic approaches, including whole-genome sequencing, sequencing-based genotyping, functional genomics, proteomics, and epigenomics. The information uncovered through these techniques will help further pinpoint important gene candidates and genomic loci associated with adaptive traits, as well as achieving a better understanding of how soybeans cope with the changing climate.
[本文引用: 1]
[本文引用: 2]
DOI:10.1105/tpc.106.040980URLPMID:16595397 [本文引用: 1]
DOI:10.1038/ng.3819URLPMID:28319089 [本文引用: 1]

Soybean is a major legume crop originating in temperate regions, and photoperiod responsiveness is a key factor in its latitudinal adaptation. Varieties from temperate regions introduced to lower latitudes mature early and have extremely low grain yields. Introduction of the long-juvenile (LJ) trait extends the vegetative phase and improves yield under short-day conditions, thereby enabling expansion of cultivation in tropical regions. Here we report the cloning and characterization of J, the major classical locus conferring the LJ trait, and identify J as the ortholog of Arabidopsis thaliana EARLY FLOWERING 3 (ELF3). J depends genetically on the legume-specific flowering repressor E1, and J protein physically associates with the E1 promoter to downregulate its transcription, relieving repression of two important FLOWERING LOCUS T (FT) genes and promoting flowering under short days. Our findings identify an important new component in flowering-time control in soybean and provide new insight into soybean adaptation to tropical regions.
URLPMID:29134443 [本文引用: 1]
URLPMID:10097183 [本文引用: 1]
URLPMID:25772379 [本文引用: 2]
DOI:10.1104/pp.17.00109URLPMID:28254761 [本文引用: 2]

The circadian clock is a complex regulatory network that enhances plant growth and fitness in a constantly changing environment. In Arabidopsis (Arabidopsis thaliana), the clock is composed of numerous regulatory feedback loops in which REVEILLE8 (RVE8) and its homologs RVE4 and RVE6 act in a partially redundant manner to promote clock pace. Here, we report that the remaining members of the RVE8 clade, RVE3 and RVE5, play only minor roles in the regulation of clock function. However, we find that RVE8 clade proteins have unexpected functions in the modulation of light input to the clock and the control of plant growth at multiple stages of development. In seedlings, these proteins repress hypocotyl elongation in a daylength- and sucrose-dependent manner. Strikingly, adult rve4 6 8 and rve3 4 5 6 8 mutants are much larger than wild-type plants, with both increased leaf area and biomass. This size phenotype is associated with a faster growth rate and larger cell size and is not simply due to a delay in the transition to flowering. Gene expression and epistasis analysis reveal that the growth phenotypes of rve mutants are due to the misregulation of PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5 expression. Our results show that even small changes in PIF gene expression caused by the perturbation of clock gene function can have large effects on the growth of adult plants.
DOI:10.1105/tpc.114.126573URL [本文引用: 1]

Transcriptional feedback loops are central to the architecture of eukaryotic circadian clocks. Models of the Arabidopsis thaliana circadian clock have emphasized transcriptional repressors, but recently, Myb-like REVEILLE (RVE) transcription factors have been established as transcriptional activators of central clock components, including PSEUDO-RESPONSE REGULATOR5 (PRR5) and TIMING OF CAB EXPRESSION1 (TOC1). We show here that NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED1 (LNK1) and LNK2, members of a small family of four LNK proteins, dynamically interact with morning-expressed oscillator components, including RVE4 and RVE8. Mutational disruption of LNK1 and LNK2 function prevents transcriptional activation of PRR5 by RVE8. The LNKs lack known DNA binding domains, yet LNK1 acts as a transcriptional activator in yeast and in planta. Chromatin immunoprecipitation shows that LNK1 is recruited to the PRR5 and TOC1 promoters in planta. We conclude that LNK1 is a transcriptional coactivator necessary for expression of the clock genes PRR5 and TOC1 through recruitment to their promoters via interaction with bona fide DNA binding proteins such as RVE4 and RVE8.
DOI:10.1073/pnas.1801862115URLPMID:29789384 [本文引用: 2]

Circadian period and phase of cultivated tomato (Solanum lycopersicum) were changed during domestication, likely adapting the species to its new agricultural environments. Whereas the delayed circadian phase is mainly caused by allelic variation of EID1, the genetic basis of the long circadian period has remained elusive. Here we show that a partial deletion of the clock gene LNK2 is responsible for the period lengthening in cultivated tomatoes. We use resequencing data to phylogenetically classify hundreds of tomato accessions and investigate the evolution of the eid1 and lnk2 mutations along successive domestication steps. We reveal signatures of selection across the genomic region of LNK2 and different patterns of fixation of the mutant alleles. Strikingly, LNK2 and EID1 are both involved in light input to the circadian clock, indicating that domestication specifically targeted this input pathway. In line with this, we show that the clock deceleration in the cultivated tomato is light-dependent and requires the phytochrome B1 photoreceptor. Such conditional variation in circadian rhythms may be key for latitudinal adaptation in a variety of species, including crop plants and livestock.
DOI:10.1038/ng.3447URLPMID:26569124 [本文引用: 1]

The circadian clock is a critical regulator of plant physiology and development, controlling key agricultural traits in crop plants. In addition, natural variation in circadian rhythms is important for local adaptation. However, quantitative modulation of circadian rhythms due to artificial selection has not yet been reported. Here we show that the circadian clock of cultivated tomato (Solanum lycopersicum) has slowed during domestication. Allelic variation of the tomato homolog of the Arabidopsis gene EID1 is responsible for a phase delay. Notably, the genomic region harboring EID1 shows signatures of a selective sweep. We find that the EID1 allele in cultivated tomatoes enhances plant performance specifically under long day photoperiods, suggesting that humans selected slower circadian rhythms to adapt the cultivated species to the long summer days it encountered as it was moved away from the equator.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s12010-019-02984-5URLPMID:30859452 [本文引用: 1]

Since the birth of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, the new genome engineering technology has become a hot topic in the scientific community. However, for swine, the system of pig cells' homology directed repair (HDR) is generally unstable and costly. Here, we aim to make knock-in of porcine cells more realizable. The Rosa26 locus was chosen for gene editing. Through the optimization of strategy, an efficient sgRNA was selected by TIDE analysis. Correspondingly, a vector system was constructed for gene insertion in pRosa26 locus by homologous recombination. A large percentage of cells whose gene is edited easily result in apoptosis. To improve the positive rate, culturing systems have been optimized. Sequence alignment and nuclear transfer confirmed that we got two knock-in cell lines and transgene primary porcine fetal fibroblasts (PFFs) successfully. Results showed that the gene editing platform we used can obtain genetically modified pig cells stably and efficiently. This system can contribute to pig gene research and production of transgenic pigs.
DOI:10.1016/j.ydbio.2015.11.018URLPMID:26632489 [本文引用: 1]

Sea urchin embryos are a useful model system for investigating early developmental processes and the underlying gene regulatory networks. Most functional studies using sea urchin embryos rely on antisense morpholino oligonucleotides to knockdown gene functions. However, major concerns related to this technique include off-target effects, variations in morpholino efficiency, and potential morpholino toxicity; furthermore, such problems are difficult to discern. Recent advances in genome editing technologies have introduced the prospect of not only generating sequence-specific knockouts, but also providing genome-engineering applications. Two genome editing tools, zinc-finger nuclease (ZFN) and transcription activator-like effector nucleases (TALENs), have been utilized in sea urchin embryos, but the resulting efficiencies are far from satisfactory. The CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9 (CRISPR-associated nuclease 9) system serves as an easy and efficient method with which to edit the genomes of several established and emerging model organisms in the field of developmental biology. Here, we apply the CRISPR/Cas9 system to the sea urchin embryo. We designed six guide RNAs (gRNAs) against the well-studied nodal gene and discovered that five of the gRNAs induced the expected phenotype in 60-80% of the injected embryos. In addition, we developed a simple method for isolating genomic DNA from individual embryos, enabling phenotype to be precisely linked to genotype, and revealed that the mutation rates were 67-100% among the sequenced clones. Of the two potential off-target sites we examined, no off-target effects were observed. The detailed procedures described herein promise to accelerate the usage of CRISPR/Cas9 system for genome editing in sea urchin embryos.
DOI:10.1111/pbi.13199URLPMID:31240772 [本文引用: 2]

Flowering time is a key agronomic trait that directly influences the successful adaptation of soybean (Glycine max) to diverse latitudes and farming systems. GmFT2a and GmFT5a have been extensively identified as flowering activators and integrators in soybean. Here, we identified two quantitative trait loci (QTLs) regions harbouring GmFT2a and GmFT5a, respectively, associated with different genetic effects on flowering under different photoperiods. We analysed the flowering time of transgenic plants overexpressing GmFT2a or GmFT5a, ft2a mutants, ft5a mutants and ft2aft5a double mutants under long-day (LD) and short-day (SD) conditions. We confirmed that GmFT2a and GmFT5a are not redundant, they collectively regulate flowering time, and the effect of GmFT2a is more prominent than that of GmFT5a under SD conditions whereas GmFT5a has more significant effects than GmFT2a under LD conditions. GmFT5a, not GmFT2a, was essential for soybean to adapt to high latitude regions. The ft2aft5a double mutants showed late flowering by about 31.3 days under SD conditions and produced significantly increased numbers of pods and seeds per plant compared to the wild type. We speculate that these mutants may have enormous yield potential for the tropics. In addition, we examined the sequences of these two loci in 202 soybean accessions and investigated the flowering phenotypes, geographical distributions and maturity groups within major haplotypes. These results will contribute to soybean breeding and regional adaptability.
DOI:10.1186/s12870-019-1746-6URLPMID:30961525 [本文引用: 2]

BACKGROUND: The plant architecture has significant effects on grain yield of various crops, including soybean (Glycine max), but the knowledge on optimization of plant architecture in order to increase yield potential is still limited. Recently, CRISPR/Cas9 system has revolutionized genome editing, and has been widely utilized to edit the genomes of a diverse range of crop plants. RESULTS: In the present study, we employed the CRISPR/Cas9 system to mutate four genes encoding SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors of the SPL9 family in soybean. These four GmSPL9 genes are negatively regulated by GmmiR156b, a target for the improvement of soybean plant architecture and yields. The soybean Williams 82 was transformed with the binary CRISPR/Cas9 plasmid, assembled with four sgRNA expression cassettes driven by the Arabidopsis thaliana U3 or U6 promoter, targeting different sites of these four SPL9 genes via Agrobacterium tumefaciens-mediated transformation. A 1-bp deletion was detected in one target site of the GmSPL9a and one target site of the GmSPL9b, respectively, by DNA sequencing analysis of two T0-generation plants. T2-generation spl9a and spl9b homozygous single mutants exhibited no obvious phenotype changes; but the T2 double homozygous mutant spl9a/spl9b possessed shorter plastochron length. In T4 generation, higher-order mutant plants carrying various combinations of mutations showed increased node number on the main stem and branch number, consequently increased total node number per plants at different levels. In addition, the expression levels of the examined GmSPL9 genes were higher in the spl9b-1 single mutant than wild-type plants, which might suggest a feedback regulation on the expression of the investigated GmSPL9 genes in soybean. CONCLUSIONS: Our results showed that CRISPR/Cas9-mediated targeted mutagenesis of four GmSPL9 genes in different combinations altered plant architecture in soybean. The findings demonstrated that GmSPL9a, GmSPL9b, GmSPL9c and GmSPL9 function as redundant transcription factors in regulating plant architecture in soybean.
DOI:10.1186/s12870-019-1906-8URLPMID:31307375 [本文引用: 1]

BACKGROUND: CRISPR/Cas9 gene editing is now revolutionizing the ability to effectively modify plant genomes in the absence of efficient homologous recombination mechanisms that exist in other organisms. However, soybean is allotetraploid and is commonly viewed as difficult and inefficient to transform. In this study, we demonstrate the utility of CRISPR/Cas9 gene editing in soybean at relatively high efficiency. This was shown by specifically targeting the Fatty Acid Desaturase 2 (GmFAD2) that converts the monounsaturated oleic acid (C18:1) to the polyunsaturated linoleic acid (C18:2), therefore, regulating the content of monounsaturated fats in soybean seeds. RESULTS: We designed two gRNAs to guide Cas9 to simultaneously cleave two sites, spaced 1Kb apart, within the second exons of GmFAD2-1A and GmFAD2-1B. In order to test whether the Cas9 and gRNAs would perform properly in transgenic soybean plants, we first tested the CRISPR construct we developed by transient hairy root transformation using Agrobacterium rhizogenesis strain K599. Once confirmed, we performed stable soybean transformation and characterized ten, randomly selected T0 events. Genotyping of CRISPR/Cas9 T0 transgenic lines detected a variety of mutations including large and small DNA deletions, insertions and inversions in the GmFAD2 genes. We detected CRISPR- edited DNA in all the tested T0 plants and 77.8% of the events transmitted the GmFAD2 mutant alleles to T1 progenies. More importantly, null mutants for both GmFAD2 genes were obtained in 40% of the T0 plants we genotyped. The fatty acid profile analysis of T1 seeds derived from CRISPR-edited plants homozygous for both GmFAD2 genes showed dramatic increases in oleic acid content to over 80%, whereas linoleic acid decreased to 1.3-1.7%. In addition, transgene-free high oleic soybean homozygous genotypes were created as early as the T1 generation. CONCLUSIONS: Overall, our data showed that dual gRNA CRISPR/Cas9 system offers a rapid and highly efficient method to simultaneously edit homeologous soybean genes, which can greatly facilitate breeding and gene discovery in this important crop plant.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/ery103URLPMID:29579245 [本文引用: 1]

Phytophthora sojae Kaufmann and Gerdemann causes Phytophthora root rot, a destructive soybean disease worldwide. A basic helix-loop-helix (bHLH) transcription factor is thought to be involved in the response to P. sojae infection in soybean, as revealed by RNA sequencing (RNA-seq). However, the molecular mechanism underlying this response is currently unclear. Here, we explored the function and underlying mechanisms of a bHLH transcription factor in soybean, designated GmPIB1 (P. sojae-inducible bHLH transcription factor), during host responses to P. sojae. GmPIB1 was significantly induced by P. sojae in the resistant soybean cultivar 'L77-1863'. Analysis of transgenic soybean hairy roots with elevated or reduced expression of GmPIB1 demonstrated that GmPIB1 enhances resistance to P. sojae and reduces reactive oxygen species (ROS) accumulation. Quantitative reverse transcription PCR and chromatin immunoprecipitation-quantitative PCR assays revealed that GmPIB1 binds directly to the promoter of GmSPOD1 and represses its expression; this gene encodes a key enzyme in ROS production. Moreover, transgenic soybean hairy roots with GmSPOD1 silencing through RNA interference exhibited improved resistance to P. sojae and reduced ROS generation. These findings suggest that GmPIB1 enhances resistance to P. sojae by repressing the expression of GmSPOD1.
[本文引用: 2]
[本文引用: 2]
DOI:10.1126/science.1225829URLPMID:22745249 [本文引用: 1]

Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by using CRISPR RNAs (crRNAs) to guide the silencing of invading nucleic acids. We show here that in a subset of these systems, the mature crRNA that is base-paired to trans-activating crRNA (tracrRNA) forms a two-RNA structure that directs the CRISPR-associated protein Cas9 to introduce double-stranded (ds) breaks in target DNA. At sites complementary to the crRNA-guide sequence, the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like domain cleaves the noncomplementary strand. The dual-tracrRNA:crRNA, when engineered as a single RNA chimera, also directs sequence-specific Cas9 dsDNA cleavage. Our study reveals a family of endonucleases that use dual-RNAs for site-specific DNA cleavage and highlights the potential to exploit the system for RNA-programmable genome editing.
DOI:10.1146/annurev-genet-110410-132430URL [本文引用: 1]

Bacteria and archaea have evolved defense and regulatory mechanisms to cope with various environmental stressors, including virus attack. This arsenal has been expanded by the recent discovery of the versatile CRISPR-Cas system, which has two novel features. First, the host can specifically incorporate short sequences from invading genetic elements (virus or plasmid) into a region of its genome that is distinguished by clustered regularly interspaced short palindromic repeats (CRISPRs). Second, when these sequences are transcribed and precisely processed into small RNAs, they guide a multifunctional protein complex (Gas proteins) to recognize and cleave incoming foreign genetic material. This adaptive immunity system, which uses a library of small noncoding RNAs as a potent weapon against fast-evolving viruses, is also used as a regulatory system by the host. Exciting breakthroughs in understanding the mechanisms of the CRISPR-Cas system and its potential for biotechnological applications and understanding evolutionary dynamics are discussed.
DOI:10.1038/nature10886URL [本文引用: 1]

Clustered regularly interspaced short palindromic repeat (CRISPR) are essential components of nucleic-acid-based adaptive immune systems that are widespread in bacteria and archaea. Similar to RNA interference (RNAi) pathways in eukaryotes, CRISPR-mediated immune systems rely on small RNAs for sequence-specific detection and silencing of foreign nucleic acids, including viruses and plasmids. However, the mechanism of RNA-based bacterial immunity is distinct from RNAi. Understanding how small RNAs are used to find and destroy foreign nucleic acids will provide new insights into the diverse mechanisms of RNA-controlled genetic silencing systems.
DOI:10.1038/nbt.2655URLPMID:23929340 [本文引用: 1]
DOI:10.1038/nbt.2650URLPMID:23929338 [本文引用: 1]
DOI:10.1038/nbt.2654URLPMID:23929339 [本文引用: 1]
DOI:10.1038/cr.2013.114URLPMID:23958582 [本文引用: 1]
DOI:10.1038/cr.2013.123URL [本文引用: 1]
DOI:10.1093/nar/gkt780URLPMID:23999092 [本文引用: 1]

The type II CRISPR/Cas system from Streptococcus pyogenes and its simplified derivative, the Cas9/single guide RNA (sgRNA) system, have emerged as potent new tools for targeted gene knockout in bacteria, yeast, fruit fly, zebrafish and human cells. Here, we describe adaptations of these systems leading to successful expression of the Cas9/sgRNA system in two dicot plant species, Arabidopsis and tobacco, and two monocot crop species, rice and sorghum. Agrobacterium tumefaciens was used for delivery of genes encoding Cas9, sgRNA and a non-fuctional, mutant green fluorescence protein (GFP) to Arabidopsis and tobacco. The mutant GFP gene contained target sites in its 5' coding regions that were successfully cleaved by a CAS9/sgRNA complex that, along with error-prone DNA repair, resulted in creation of functional GFP genes. DNA sequencing confirmed Cas9/sgRNA-mediated mutagenesis at the target site. Rice protoplast cells transformed with Cas9/sgRNA constructs targeting the promoter region of the bacterial blight susceptibility genes, OsSWEET14 and OsSWEET11, were confirmed by DNA sequencing to contain mutated DNA sequences at the target sites. Successful demonstration of the Cas9/sgRNA system in model plant and crop species bodes well for its near-term use as a facile and powerful means of plant genetic engineering for scientific and agricultural applications.
URLPMID:24710347 [本文引用: 1]
DOI:10.1111/tpj.12554URLPMID:24836556 [本文引用: 1]

Engineered nucleases can be used to induce site-specific double-strand breaks (DSBs) in plant genomes. Thus, homologous recombination (HR) can be enhanced and targeted mutagenesis can be achieved by error-prone non-homologous end-joining (NHEJ). Recently, the bacterial CRISPR/Cas9 system was used for DSB induction in plants to promote HR and NHEJ. Cas9 can also be engineered to work as a nickase inducing single-strand breaks (SSBs). Here we show that only the nuclease but not the nickase is an efficient tool for NHEJ-mediated mutagenesis in plants. We demonstrate the stable inheritance of nuclease-induced targeted mutagenesis events in the ADH1 and TT4 genes of Arabidopsis thaliana at frequencies from 2.5 up to 70.0%. Deep sequencing analysis revealed NHEJ-mediated DSB repair in about a third of all reads in T1 plants. In contrast, applying the nickase resulted in the reduction of mutation frequency by at least 740-fold. Nevertheless, the nickase is able to induce HR at similar efficiencies as the nuclease or the homing endonuclease I-SceI. Two different types of somatic HR mechanisms, recombination between tandemly arranged direct repeats as well as gene conversion using the information on an inverted repeat could be enhanced by the nickase to a similar extent as by DSB-inducing enzymes. Thus, the Cas9 nickase has the potential to become an important tool for genome engineering in plants. It should not only be applicable for HR-mediated gene targeting systems but also by the combined action of two nickases as DSB-inducing agents excluding off-target effects in homologous genomic regions.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00299-013-1539-6URLPMID:24277082 [本文引用: 1]

Recent advances in genome engineering indicate that innovative crops developed by targeted genome modification (TGM) using site-specific nucleases (SSNs) have the potential to avoid the regulatory issues raised by genetically modified organisms. These powerful SSNs tools, comprising zinc-finger nucleases, transcription activator-like effector nucleases, and clustered regulatory interspaced short palindromic repeats/CRISPR-associated systems, enable precise genome engineering by introducing DNA double-strand breaks that subsequently trigger DNA repair pathways involving either non-homologous end-joining or homologous recombination. Here, we review developments in genome-editing tools, summarize their applications in crop organisms, and discuss future prospects. We also highlight the ability of these tools to create non-transgenic TGM plants for next-generation crop breeding.
DOI:10.1104/pp.111.172981URLPMID:21464476 [本文引用: 1]

We performed targeted mutagenesis of a transgene and nine endogenous soybean (Glycine max) genes using zinc-finger nucleases (ZFNs). A suite of ZFNs were engineered by the recently described context-dependent assembly platform--a rapid, open-source method for generating zinc-finger arrays. Specific ZFNs targeting dicer-like (DCL) genes and other genes involved in RNA silencing were cloned into a vector under an estrogen-inducible promoter. A hairy-root transformation system was employed to investigate the efficiency of ZFN mutagenesis at each target locus. Transgenic roots exhibited somatic mutations localized at the ZFN target sites for seven out of nine targeted genes. We next introduced a ZFN into soybean via whole-plant transformation and generated independent mutations in the paralogous genes DCL4a and DCL4b. The dcl4b mutation showed efficient heritable transmission of the ZFN-induced mutation in the subsequent generation. These findings indicate that ZFN-based mutagenesis provides an efficient method for making mutations in duplicate genes that are otherwise difficult to study due to redundancy. We also developed a publicly accessible Web-based tool to identify sites suitable for engineering context-dependent assembly ZFNs in the soybean genome.
URLPMID:26284791 [本文引用: 1]
