 ,*湖南农业大学农学院 / 国家油料改良中心湖南分中心, 湖南长沙 410128
,*湖南农业大学农学院 / 国家油料改良中心湖南分中心, 湖南长沙 410128Cloning and characterization of phytochrome interacting factor 4 (BnaPIF4) gene from Brassica napus L.
FENG Tao, GUAN Chun-Yun ,*College of Agronomy, Hunan Agricultural University / National Oilseed Crops Improvement Center in Hunan, Changsha 410128, Hunan, China
,*College of Agronomy, Hunan Agricultural University / National Oilseed Crops Improvement Center in Hunan, Changsha 410128, Hunan, China通讯作者:
第一联系人:
收稿日期:2018-06-21接受日期:2018-08-20网络出版日期:2018-09-25
| 基金资助: |
Received:2018-06-21Accepted:2018-08-20Online:2018-09-25
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (4080KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
冯韬, 官春云. 甘蓝型油菜光敏色素互作因子4 (BnaPIF4)基因克隆和功能分析[J]. 作物学报, 2019, 45(2): 204-213. doi:10.3724/SP.J.1006.2019.84085
FENG Tao, GUAN Chun-Yun.
低光寡照影响甘蓝型油菜结实, 是造成油菜减产的重要原因之一[1], 增强油菜对低光胁迫的抗性是生产上面临的重要议题。低光如何影响油菜产量尚无明确的机制研究, 光信号通过何种转化最终影响油菜产量形成尚不明确。有研究显示施用外源油菜素内酯可以增强番茄[2]、水稻[3]等多种作物光合效率、增强弱光抗性、增加产量, 过表达油菜素内酯合成基因DWF4增强甘蓝型油菜非生物胁迫抗性和产量[4], 表明光信号途径与油菜素内酯信号途径的互作可能是光最终影响油菜产量的原因之一。在拟南芥和水稻中均证实光敏色素互作因子4 (phytochrome interacting factor 4, PIF4)与芸薹素唑抗性因子(brassinazole-resistant, BZR/BES)互作介导光信号途径与油菜素内酯信号途径的信号转导[5], 对甘蓝型油菜PIF4基因及其与BZR/BES互作的相关研究是阐明弱光胁迫对油菜产量影响的关键问题之一, 作者前期工作中已克隆了甘蓝型油菜BZR/BES基因, 本文主要关注甘蓝型油菜PIF4基因相关研究。
PIFs属于bHLH转录因子家族第15亚家族, 具有典型的bHLH结构域可自结合形成同源二聚体或与含有HLH结构域的其他蛋白形成异源二聚体参与光、温、激素等响应的下游基因表达调控[6]。PIF4是PIFs家族中重要成员, 是植物光信号途径中的核心转录因子之一, 最早发现其负调控光敏色素B信号参与拟南芥光质响应过程[7]。后续研究发现PIF4不仅参与光质响应, 同样对光强和温度响应[8], PIF4与光敏色素B互作调控光子总量影响的拟南芥气孔发育[9], 调控光敏色素合成介导高温胁迫下蓝光响应[10], 对生长素、赤霉素、油菜素内酯等多种植物激素信号调控或互作。PIF4在高温胁迫下, 通过调控上游基因转录影响生长素合成[11], 通过与DELLA互作影响赤霉素的合成与功能发挥[12], 油菜素内酯与PIF4则存在双向关系, 一方面PIF4通过与BZR/BES互作影响油菜素内酯信号的转导, 另一方面油菜素内酯依赖的磷酸化影响PIF4基因转录, 调控PIF4水平[13]。弱光胁迫下, 菊花PIF4基因表达出现先升高后趋稳缓降的规律, 乙烯、赤霉素和生长素等激素水平同时发生变动, 表明PIF4可能参与植物对弱光胁迫的响应[14]。
甘蓝型油菜为异源多倍体, 基因组结构复杂, 测序完成较晚[15], 因此相关基因功能研究滞后, 目前对甘蓝型油菜中的光信号转导, 以及与其他激素途径之间的互作尚不明确, 信号途径中关键基因的解析不足, 导致相关基因在育种中的应用受限。
本文通过甘蓝型油菜基因组数据库中BnaPIF4基因序列设计引物, 从甘蓝型油菜品种湘油15号中克隆获得2个BnaPIF4基因的编码序列(coding sequence, CDS)、全长mRNA和全长基因序列, 染色体步移证实一个位于A03染色体, 一个位于C03染色体。
1 材料与方法
1.1 试验材料
甘蓝型油菜(Brassica napus L.)双低品系湘油15号由国家油料改良中心湖南分中心提供。1.2 RNA提取与cDNA合成
湘油15号种子在22℃、光周期为16 h/8 h条件下培养至2片真叶, 取幼苗50~100 mg。加液氮充分研磨后, 使用TRIzol RNA提取试剂盒(TransGen生物技术有限公司), 按说明书所示方法提取总RNA, 以此RNA为模板, 使用Easy Script First-Strand cDNA Synthesis SuperMix试剂盒(TransGen生物技术有限公司)按说明书所示方法合成第1链cDNA。1.3 DNA提取
取1.2所述湘油15号叶片50~100 mg, 液氮研磨后加入1 mL CTAB-PVP buffer, 提取总DNA备用于目标基因全长序列和启动子序列的克隆。1.4 BnaPIF4基因的克隆及序列分析
从BRAD数据库(http://brassicadb.org/brad/)甘蓝型油菜全转录组中提取BnaPIF4的转录本序列。分析比对, 设计含起始密码子的5'端引物和含有终止密码子的3'端引物(湖南擎科生物技术有限公司合成), 用于CDS克隆; 根据CDS测序比对结果, 检索全长mRNA和全长基因并设计克隆引物。CDS克隆引物为: BnaPIF4-F: 5'-ATGGAACACCAAGGTTGGAG-3', BnaPIF4-R: 5'-CTAACGGGGACCGTCGGT-3'; 边界序列一致, mRNA克隆与全长基因克隆共用引物对为: dPIF4-F: 5'-TCTGTCTGTACCAAAAGAAG-3', dPIF4-R: 5'-TCTGGACGATTATGAAAAGC-3'; 分别以1.2所述cDNA和1.3所述DNA为模板进行PCR扩增。PCR体系含50 ng μL-1模板1 μL、10 mmol L-1 dNTPs 0.5 μL、2 μmol L-1 BnaPIF4-F/mPIF4-F 0.75 μL、2 μmol L-1 BnaPIF4-R/mPIF4-R 0.75 μL、10×反应缓冲液1 μL、GXL高保真DNA聚合酶0.5 μL和ddH2O 15.5 μL。扩增程序为预变性95℃ 5 min; 95℃ 50 s, 58℃ 45 s, 72℃ 3 min, 35个循环; 72℃后延伸10 min。PCR产物经1.5%琼脂糖凝胶电泳, 用凝胶成像系统(Bio-Rad)检测和记录结果。将PCR产物回收后与pMD18-T载体(TransGen生物技术有限公司)连接, 转化大肠杆菌感受态细胞Trans1-T1 (北京天根生化有限公司), 筛选阳性克隆进行基因测序, 通过BRAD数据库(http://brassicadb.org/brad/)对获得的克隆序列进行染色体步移分析, 随后使用DNAman对克隆获得的BnaPIF4基因拷贝进行序列比对。1.5 BnaPIF4蛋白生物信息学分析
1.5.1 BnaPIF4_A03和BnaPIF4_C03氨基酸序列特征与二级结构预测 通过ANTHEPROT 6.2软件分析来源于甘蓝型油菜的BnaPIF4_A03和BnaPIF4_C03基因所编码的蛋白质。统计编码蛋白的氨基酸残基组分、分析等电点、特异性识别序列和亲疏水特征并使用Gariner模型进行二级结构预测。使用wolf-pSORT (https://www.genscript.com/tools/ wolf-psort)和Plant-mPLoc (http://www.csbio.sjtu.edu. cn/cgi-bin/PlantmPLoc.cgi)进行亚细胞定位预测。1.5.2 BnaPIF4蛋白同源结构域分析 使用DNAman对BnaPIF4蛋白进行序列比对分析; 同时使用NCBI (http://blast.ncbi.nlm.nih.gov/)分别对不同的BnaPIF4蛋白序列进行保守结构域分析; 使用SMART (http://smart.embl-heidelberg.de/)、MOTIF (http://www.genome.jp/tools/motif/)以及InterPro (http://www.ebi.ac.uk/interpro/)分析BnaPIF4包含的特殊结构域。
1.5.3 PIF4蛋白聚类分析 通过NCBI蛋白数据库(http://www.ncbi.nlm.nih.gov/protein), String数据库(http://string.embl.de/), PDB数据库(http://www. pdb.org/pdb/home/home.do)检索来源于不同物种的PIF4蛋白序列, 并使用MEGA6.0对来源不同的PIF4相关蛋白进行聚类分析。
1.6 BnaPIF4互作因子酵母杂交分析
使用pGAD-T7、pGBK-T7和pHIS2载体(本实验室保存), HF7c氨基酸合成缺陷酵母进行酵母单双杂交实验, 分析BnaPIF4与BnaBZR1/BnaBES1互作模式。BnaPIF4、BnaBZR1和BnaBES1/2基因的CDS分别亚克隆到pGAD-T7载体上; BnaPIF4各拷贝CDS亚克隆到pGBK-T7载体; 从湘油15号gDNA中克隆BnaPIF4、BnaBZR1和BnaBES1/2基因启动子(起始密码子上游1 kb序列), 随后将各启动子序列亚克隆到pHIS2载体上; 随后分别将不同组合的AD与BD质粒共转化HF7c酵母并接种到添加浓度10 mmol L-1 3-AT的三缺培养基进行筛选, 用于验证BnaPIF4蛋白与BnaBZR1BnaBES1/2蛋白以及启动子之间的互作关系。1.7 甘蓝型油菜中BnaPIF4_A03和BnaPIF4_ C03基因时空表达分析
以1.2方法提取甘蓝型油菜湘油15号不同时期根、茎、叶、花和角果皮总RNA, 使用宝生物工程(大连)有限公司PrimeScript RTreagent Kit with gDNA Eraser (Perfect Real-time)试剂盒进行反转录, 获得cDNA第1链作为qRT-PCR模板。分析1.4中克隆获得的BnaPIF4片段, 根据5'-UTR序列特征设计两基因分型定量引物如下: PIF4A03-F: 5'-AAGTAAA GAACCTTACCCATCT-3', PIF4A03-R: 5'-TAAAGTT CCACGCCCAAA-3'; PIF4C03-F: 5'-AATCATTATCT CCCTCGCTGTA-3', PIF4C03-R: 5'-TGAAGTTCCA CGCCCA AC-3'。以BnaUBC21作为内参基因, 内参引物为BnUBC21-F: 5'-CCTCTGCAGCCTCCTCAA GT-3', BnUBC21-R: 5'-TATCTCCCCTGTCTTGAAA TGC-3', 用于BnaPIF4_A03和BnaPIF4_C03时空表达分析, 每样品进行3次重复。在ABI 7500荧光定量PCR系统上运行qRT-PCR。PCR体系包含1 μL cDNA、10 μL 2×FastStart Universal SYBR GreenMaster with ROX、10 μmol L-1正向引物和反向引物各0.5 μL、8 μL ddH2O。PCR程序为95℃ 10 min; 95℃ 15 s, 60℃ 15 s, 72℃ 32 s, 35个循环。反应完成后进行95℃ 20 s, 60℃ 20 s, 95℃ 20 s, 59℃ 20 s绘制熔解曲线检测扩增产物的正确性和引物二聚体。2 结果与分析
2.1 BnaPIF4基因克隆
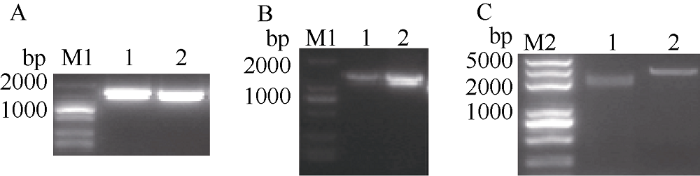
以湘油15号cDNA为模板, 克隆到2个CDS序列(图1-A), 长度分别为1242 bp和1245 bp, 对应的克隆到2个基因全长mRNA (图1-B)分别为1701 bp和1731 bp, 以湘油15号gDNA为模板克隆得到对应基因的全长序列(图1-C)为2527 bp和2665 bp, 分别定位于A03和C03染色体, 命名为BnaPIF4_ A03和BnaPIF4_C03。通过BRAD数据库比对, 确认2个CDS、mRNA和基因全长DNA序列的完整性, 对BRAD数据库检索未发现更多的BnaPIF4基因拷贝, 表明BnaPIF4_A03和BnaPIF4_ C03克隆完整。序列比对显示BnaPIF4_A03和BnaPIF4_C03基因的CDS相似度98.15%, mRNA相似度92.82%, 基因全长相似度94.64%, 表明BnaPIF4_A03和BnaPIF4_C03基因高度同源, 且基因序列变异主要发生在UTR区域和内含子区域。仔细分析BnaPIF4_A03和BnaPIF4_C03基因各序列, 并绘制基因结构示意图(图2)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1BnaPIF4_A03和BnaPIF4_C03基因克隆
A: 编码序列; B: mRNA; C: 基因全长: M1: 2K DNA 标记; M2: 2K plus DNA标记; 1: BnaPIF4_A03: 2: BnaPIF4_C03。
Fig. 1Cloning of BnaPIF4_A03 and BnaPIF4_C03
A: CDS; B: mRNA; C: Full length gene; M1: 2K DNA marker; M2: 2K plus DNA marker; 1: BnaPIF4_A03; 2: BnaPIF4_C03.
图2
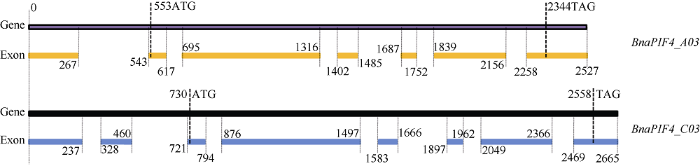 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2BnaPIF4_A03和BnaPIF4_C03基因结构
Fig. 2Gene structure of BnaPIF4_A03 and BnaPIF4_C03
序列分析显示, BnaPIF4_C03基因序列与测序品种中双11号中一致, 存在8个外显子和7个内含子, CDS序列位于第3~8外显子; BnaPIF4_A03基因具有7个外显子和6个内含子, 但序列与测序品种中双11号存在差异, 主要表现为第1内含子上352 bp位置存在单T碱基插入突变, 第4内含子1511 bp位置缺失AT碱基, 1582 bp位置缺失14碱基的富A/T序列, 第六内含子2228 bp处缺失ATTT, 且第7外显子3'端相对中双11号长91 bp。BnaPIF4_A03和BnaPIF4_C03基因间差异主要在5'端的外显子与内含子上, BnaPIF4_A03的5'端相对BnaPIF4_C03缺少C03拷贝具有的第2外显子, 造成其5'-UTR比C03拷贝更短, 二者CDS序列相似度极高, 且与测序品种中双11号中对应拷贝无序列差别。
2.2 BnaPIF4生物信息学分析
BnaPIF4_A03和BnaPIF4_C03基因各自编码长度413和414氨基酸残基的蛋白, 分别命名为BnaPIF4_A03和BnaPIF4_C03, 2个BnaPIF4蛋白的氨基酸组成、摩尔质量和等电点如表1。Table 1
表1
表1BnaPIF4_A03和BnaPIF4_C03蛋白信息汇总表
Table 1
| 蛋白编号 Serial number | 氨基酸残基数 Number of amino acid residues | 摩尔质量 Molar mass (Da) | 等电点 Isoelectric point |
|---|---|---|---|
| BnaPIF4-A03 | 413 | 46,374.937 | 5.535 |
| BnaPIF4-C03 | 414 | 46,260.633 | 5.595 |
新窗口打开|下载CSV
亲疏水性分析显示, BnaPIF4蛋白主要表现疏水性, 在C端约280残基位置存在一个跨膜结构。Wolf-pSORT与Plant-mPLoc亚细胞定位预测显示, BnaPIF4_A03和BnaPIF4_C03均定位于细胞核, 这与PIF4蛋白作为转录因子的功能一致。
BnaPIF4_A03和BnaPIF4_C03的二级结构预测结果如图3, BnaPIF4_A03有23%α螺旋, 21%β折叠, 26%β转角, 30%无规卷曲; BnaPIF4_C03有25%α螺旋, 20%β折叠, 25%β转角和30%无规卷曲。
图3
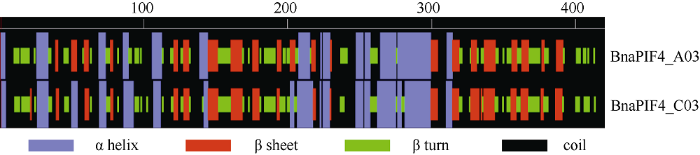 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3BnaPIF4_A03和BnaPIF4_C03蛋白二级结构
Fig. 3Secondary structure of BnaPIF4_A03 and BnaPIF4_C03
通过ANTHEPROT分析了BnaPIF4_A03和BnaPIF4_C03蛋白具有的序列特异性识别位点(表2)。
Table 2
表2
表2BnaPIF4_A03和BnaPIF4_C03蛋白特殊识别位点序列总表
Table 2
| 蛋白编号 Serial number | 位点类型 Domains type | 数量 Number | 位置 Location | 序列 Sequence | 序列模式 Sequence motif |
|---|---|---|---|---|---|
| BnaPIF4_A03 | PS00001 | 4 | 158-161, 185-188, 234-237, 253-256 | NQSQ, NSSS, NKSN, NLSE | N-{P}-[ST]-{P} |
| PS00005 | 7 | 43-45, 61-63, 242-244, 243-245, 255-257, 280-282, 400-402 | THR, TLR, STR, TRR, SER, TDK, SQR | [ST]-x-[RK] | |
| PS00006 | 9 | 7-10, 43-46, 66-69, 97-100, 160-163, 162-165, 175-178, 226-229, 284-287 | SFEE, THRD, TFLE, STID, SQTD, TDLD, TIDE, SQSD, SILE | [ST]-x(2)-[DE] | |
| PS00007 | 1 | 174-181 | KTIDERLY | [RK]-x(2,3)-[DE]-x(2,3)-Y | |
| PS00008 | 8 | 156-161, 189-194, 190-195, 193-198, 232-237, 306-311, 308-313, 393-398 | GSNQSQ, GGSSGC, GSSGCS, GCSLGK, GNNKSN, GSGMAG, GMAGAA, GSPAGQ | G-{EDRKHPFYW}-x(2)-[STAGCN]-{P} | |
| PS00845 | 2 | 371-377, 368-377 | GRYVGLF, DRFGRYVGLF | [GD]-x(1)-[FYWA]-x(1)-G-[LIVM]-x(0)-[LIVMFYD] | |
| BnaPIF4_C03 | PS00001 | 4 | 13-16, 158-161, 185-188, 253-256 | NLSN, NQSQ, NSSS, NLSE | N-{P}-[ST]-{P} |
| PS00005 | 8 | 43-45, 61-63, 109-111, 242-244, 243-245, 255-257, 280-282, 400-402 | THR, TLR, STR, TRR, SER, TDK, SQR, THR, TLR, TVK, STR, TRR, SER, TDK, SQR | [ST]-x-[RK] | |
| PS00006 | 10 | 7-10, 43-46, 66-69, 97-100, 160-163, 162-165, 175-178, 226-229, 278-281, 284-287 | SFEE, THRD, TFLE, STID, SQTD, TDLD, TIDE, SQSD, TKTD, SILE | [ST]-x(2)-[DE] | |
| PS00007 | 1 | 174-181 | KTIDERLY | [RK]-x(2,3)-[DE]-x(2,3)-Y | |
| PS00008 | 9 | 156-161, 189-194, 190-195, 193-198, 232-237, 239-244, 306-311, 308-313, 393-398 | GSNQSQ, GGSSGC, GSSGCS, GCSLGK, GNNKSN, GSGSTR, GSGMAG, GMAGAA, GSPAGQ | G-{EDRKHPFYW}-x(2)-[STAGCN]-{P} | |
| PS00845 | 2 | 371-377, 368-377 | GRYVGLF, DRFGRYVGLF | [GD]-x(1)-[FYWA]-x(1)-G-[LIVM]-x(0)-[LIVMFYD] |
新窗口打开|下载CSV
BnaPIF4_A03和BnaPIF4_C03具有仅含微弱差别的特异性识别序列, BnaPIF4_A03相较BnaPIF4_ C03缺少109~111的蛋白激酶C磷酸化位点、278~ 281的酪蛋白磷酸化位点和239~244的酰化位点。整体来看BnaPIF4_A03和BnaPIF4_C03的特异性识别位点基本一致, 主要是多种激酶的磷酸化修饰位点、小分子结合位点和酰化等蛋白修饰位点, 多数与转录因子功能相关, 这与PIF4蛋白是bHLH家族成员的结论一致。
使用SMART和MOTIF分析结构域(图4)发现, 2个BnaPIF4蛋白具有基本相同的结构域且主要集中于C端, 且多为具有核酸结合功能的结构域, 表现转录因子特性。在第250残基到第310残基之间具有多个重叠的bHLH结构域, 亲疏水性分析发现的跨膜结构域在第280残基左右, 正位于bHLH结构域中段, 这与PIF4作为典型的bHLH家族成员的结论一致。
图4
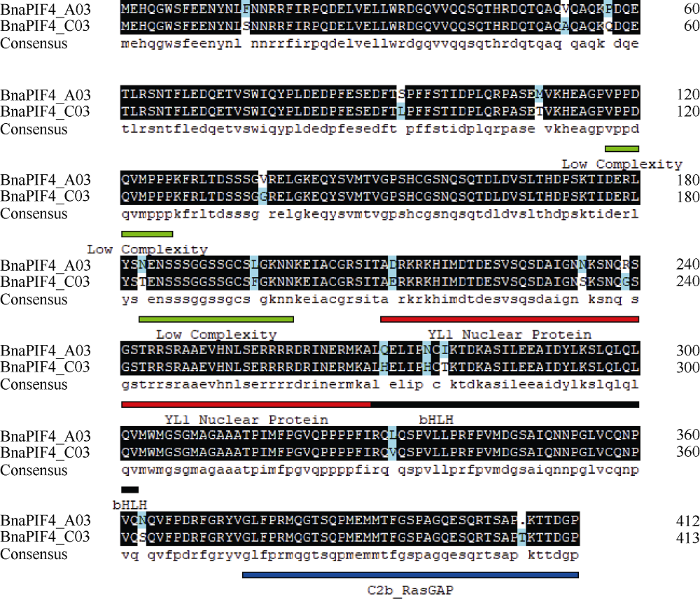 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4BnaPIF4氨基酸序列比对
Fig. 4Comparison of the amino acid sequences of BnaPIF4
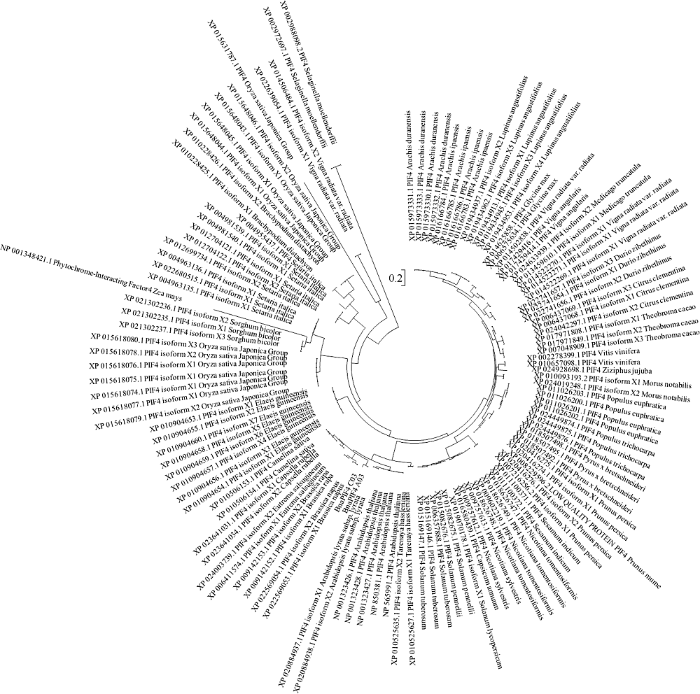
通过不同数据库, 共获得来源于41种不同植物的127条PIF4蛋白序列, 使用MEGA6.0进行聚类分析, 绘制进化树(图5)。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5不同植物PIF4蛋白的进化树分析
Arabidopsis thaliana: 拟南芥; Arabidopsis lyrata subsp. Lyrata: 深山南芥; Arachis ipaensis: 花生; Arachis duranensis: 曼花生; Brachypodium distachyon: 二穗短柄草; Brassica napus L.: 甘蓝型油菜; Brassica rapa L.: 白菜; Camelina sativa: 亚麻荠; Capsicum annuum: 辣椒; Capsella rubella: 碎米荠; Citrus clementina: 克莱门柚; Durio zibethinus: 榴莲; Elaeis guineensis: 油棕; Eutrema salsugineum: 盐芥; Glycine max: 大豆; Medicago truncatula: 苜蓿; Morus notabilis: 桑; Nicotiana sylvestris: 林烟草; Nicotiana tomentosiformis: 绒毛状烟草; Oryza sativa japonica Group: 水稻(粳); Prunus mume: 中国梅; Lupinus angustifolius: 狭叶羽扇豆; Sesamum indicum: 芝麻; Solanum lycopersicum: 番茄; Solanum pennellii: 小番茄; Sorghum bicolor: 高粱; Populus euphratica: 胡杨; Populus trichocarpa : 欧洲大叶杨; Prunus persica: 桃; Pyrus × bretschneideri: 鸭梨; Selaginella moellendorffii: 卷柏; Setaria italica: 狗 草; Tarenaya hassleriana: 醉蝶花; Theobroma cacao: 可可; Vigna angularis: 红豆; Vigna radiata: 绿豆; Vigna radiata var. radiata: 赤小豆; Solanum tuberosum: 马铃薯; Vitis vinifera: 酿酒葡萄; Zea mays: 玉米; Ziziphus jujuba: 枣。
Fig. 5Phylogenetic analysis of PIF4 in different plant species
PIF4蛋白聚类表现出与植物亲缘关系的高度一致性, 大部分植物中均存在PIF4蛋白的高拷贝现象, 且各拷贝之间的相似性高, 基本上处于同一进化分支。PIF4蛋白大体上按植物科属分布聚类, 如豆科的花生、绿豆、大豆等多拷贝PIF4蛋白高度聚类(图5右上), 十字花科的甘蓝型油菜、白菜、拟南芥、亚麻芥等多拷贝PIF4蛋白高度聚类, 但禾本科植物中PIF4蛋白分化比较极端, 水稻、狗 草、玉米等禾本科植物中PIF4蛋白具有较远的分支距离, 且同一物种中不同的PIF4拷贝也表现出较高的分化水平, 如水稻不同亚种、不同拷贝的PIF4蛋白被聚类到完全隔离的分支上(图5左上)。另外, PIF4蛋白表现出木本植物分化程度低于草本植物的规律, 如梨、李、榴莲、可可等木本植物处于不同的科属但PIF4蛋白分支距离相对较小, 且与卷柏中PIF4具有较近的分支距离, 而禾本科的水稻、玉米、狗 草等, 茄科的番茄、马铃薯、烟草等草本植物PIF4蛋白均表现相对较高的分化程度。表明PIF4蛋白在进化过程中相对保守, 其分化发生在进化较晚的阶段, 且分化过程剧烈, 表现出低等植物中PIF4高度一致, 高等植物中PIF4分化程度显著提高, 在进化过程中PIF4蛋白大量发生重复, 表现为大量植物中存在多个PIF4蛋白拷贝。
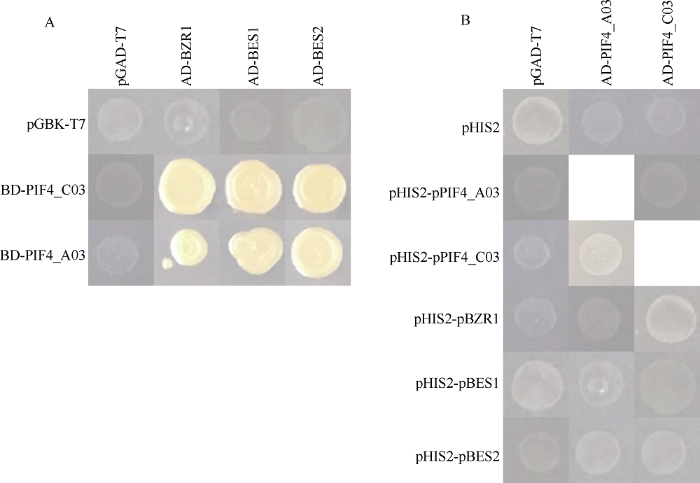
2.3 BnaPIF4和BnaBZR/BES互作
使用酵母双杂检测了BnaPIF4与BnaBZR1, BnaBES1/2蛋白互作, 酵母单杂交检测了BnaPIF4与BnaBZR1, BnaBES1/2基因启动子之间的互作(图6)。从酵母杂交实验结果可见, BnaPIF4_A03与BnaPIF4_C03互作模式一致, 均可以与BnaBZR1, BnaBES1/2蛋白互作, 但不能结合BnaBZR1、BnaBES1/2基因的启动子, BnaPIF4_A03和BnaPIF4_C03也不相互与启动子结合, 显示BnaPIF4主要通过蛋白层面而非转录层面的互作参与甘蓝型油菜的生物学调控。BnaPIF4可能通过HLH结构域结合BnaBZR/BES蛋白, 从而影响后者磷酸化, 调控磷酸化修饰水平介导后者的核质穿梭, 调控与下游靶基因启动子的结合, 共同调控下游基因表达。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6BnaPIF4和BnaBZR/BES互作
Fig. 6Interaction of BnaPIF4 and BnaBZR/BES
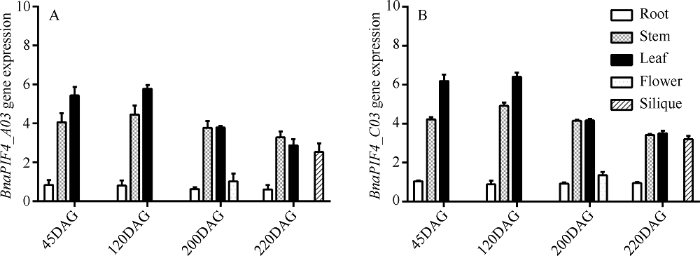
2.4 甘蓝型油菜中BnaPIF4时空表达特征
为进一步了解BnaPIF4在甘蓝型油菜中的功能, 对甘蓝型油菜不同时期, 基于5'-UTR差异对不同组织中BnaPIF4_A03和BnaPIF4_C03表达进行分析。由图7-A与图7-B比较可见, 湘油15号中BnaPIF4_A03和BnaPIF4_C03具有相同的表达模式, 基因表达均表现明显的组织差异和一定的生长发育时间差异。BnaPIF4基因主要表达于油菜的绿色组织, 茎表皮、未成熟角果和叶中表达量较高, 在花和根中表达较低, 叶中BnaPIF4表达水平最高, 其次为茎表皮和未成熟的角果, 这些位置是植物感受光信号刺激、发生光合作用的主要场所, 会聚集大量光敏色素, PIF4作为光信号的负调控因子, 可能受到光敏色素浓度的影响, 改变表达水平以对光信号做出响应。在生育进程中, BnaPIF4基因表达量差异不明显, 整体表现为苗期和薹期表达量稍高, 随生育进程有缓慢降低的趋势, 这可能是由于随油菜生长发育, 光敏色素积累降低, 造成BnaPIF4表达做出调整, 也可能与油菜激素水平有关系, 油菜快速生长的苗期和薹期生长素、油菜素内酯等激素均保持较高水平, 可能通过激素与光信号互作, 影响BnaPIF4表达。PIF4表达与光信号之间存在着反馈调节, 一方面光信号受到PIF4的负调控, 另一方面外源光变化也能诱导PIF4表达变化, 这表明BnaPIF4在油菜光信号途径中具有重要作用。
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7不同发育时期的湘油15号根、茎、叶、花和角果中BnaPIF4_A03和BnaPIF4_C03的表达量
DAG表示种子萌发后天数。
Fig. 7Expression levels of BnaPIF4_A03 and BnaPIF4_C03 in root, stem, leaf, flower, and silique of Xiangyou 15 at different developmental stages
DAG means days after seed germination.
3 讨论
从甘蓝型油菜湘油15号中克隆获得2个BnaPIF4基因拷贝, 其中BnaPIF4_A03拷贝与测序品种中双11号对应拷贝存在内含子和3'-UTR序列差异, CDS序列一致。BnaPIF4_A03和BnaPIF4_C03基因结构基本一致, 基因、全长mRNA和CDS序列相似度均超过90%, 表明BnaPIF4基因高度保守。BnaPIF4_A03和BnaPIF4_C03蛋白具有典型的植物bHLH结构域, 预测亚细胞定位于细胞核, 具有多重转录因子功能的序列结合元件, 存在大量磷酸化和酰化功能位点, 表明克隆获得的BnaPIF4基因属于植物PIF4基因家族, 这与拟南芥、水稻等植物[16]中PIF4结构和功能一致。对植物PIF4蛋白的聚类分析显示其在进化上具有较高的保守性, 且蛋白进化关系与物种进化关系密切, 近缘物种中的PIF4蛋白高度保守, 但高等植物相对低等植物中PIF4蛋白明显高分化, 表明PIF4蛋白分化可能是一个较晚但相对剧烈的进化事件, 这与植物bHLH家族进化保守性基本一致[17]。NCBI数据库中共收录了6条甘蓝型油菜PIF4及其类似序列, 2个位于A染色体组, 4个位于C染色体组, 这表明PIF4家族序列在甘蓝型油菜亚基因组间分布可能不平衡, 但6个BnaPIF4及类似序列保守性较高, 这显示了甘蓝型油菜A、C亚基因组在进化关系上具有较高的同源性但存在各自独立的进化过程, 也间接证实“禹氏三角”中白菜与甘蓝的近缘关系[18]。
植物PIF4蛋白参与植物开花、病原免疫、非生物胁迫抗性等生物学过程, 是光信号途径中重要的转录因子之一。在高温诱导下拟南芥中PIF4可直接激活FT的表达, 正调控拟南芥的开花, pif4-101突变体在持续高温条件下表现小株型和晚花表型, 随着温度的升高, PIF4负调控植物免疫反应、促进拟南芥的生长发育[19]。PIFs转录因子家族是整合光信息与植物激素信号的桥梁[20], 其中PIF4介导光信号和多种植物激素信号的互作[21], PIF4与BZR/BES复合体结合介导光信号与油菜素内酯信号之间的转导。本文通过酵母杂交证实甘蓝型油菜中BnaPIF4和BnaBZR/BES基于蛋白结合而非转录层面的互作介导光信号和油菜素内酯信号转导, 但有研究显示PIF4[22]和BZR1[23]均能对油菜素内酯生物合成产生调控, 表明PIF4对油菜素内酯生物合成的反馈通路不同于PIF4通过与BZR/BES结合介导光信号与油菜素内酯信号互作的通路, 对油菜素内酯合成的反馈可能有更多生理功能。同时本文在PIF4蛋白进化分析中发现植物中大量存在PIF4蛋白重复, 也表明PIF4功能冗余或者具有除介导光信号之外的功能, 尚待进一步研究。
湘油15号中BnaPIF4_A03和BnaPIF4_C03的表达规律一致, 这与BnaPIF4基因的保守性相统一, 2个BnaPIF4基因拷贝CDS、mRNA、全长基因和启动子序列均具有高度相似性, 可能具有共同的上游调控因子, 对植物内、外环境因素可能具有相同的响应模式。BnaPIF4_A03和BnaPIF4_C03序列差异主要发生在5'端的内含子和5'-UTR上, 二者可能存在转录和翻译稳定性上的差异。甘蓝型油菜中BnaPIF4基因的表达调控规律、蛋白翻译稳定性、翻译后修饰过程、基因功能等有待进一步研究。
4 结论
从甘蓝型油菜湘油15号中克隆到2个PIF4基因, 分别定位于A03和C03染色体, 分别命名为BnaPIF4_A03和BnaPIF4_C03, 全长编码序列(coding sequence, CDS)、全长mRNA和全长基因分别为1242 bp和1245 bp、1701 bp和1731 bp、2527 bp和2665 bp, 各自编码413和414个氨基酸。2拷贝BnaPIF4差异序列集中于5'端UTR和内含子, 与测序品种中双11号中对应拷贝存在内含子序列差异。BnaPIF4_A03和BnaPIF4_C03具有典型bHLH结构域, 与白菜、拟南芥等十字花科植物PIF4蛋白高度相似, 植物中PIF4高度保守。甘蓝型油菜中PIF4与BZR/BES存在蛋白层面的互作关系。分析了BnaPIF4_A03和BnaPIF4_C03基因的时空表达。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3724/SP.J.1006.2017.00082URL [本文引用: 1]

收获指数偏低是制约油菜籽粒产量和产油量进一步提高的瓶颈。为解析适合不同产量的环境下油菜高收获指数的构成因素及形成机制,本研究选择在高产环境下的云南临沧和一般产量的长江流域上游主产区重庆北碚均能正常生长和成熟的321份甘蓝型油菜品种(系)为材料,分析其产量相关性状的变异以及两地间的差异,利用相关分析和通径分析研究籽粒产量收获指数(YHI)、产油量收获指数分别与17个产量组成性状的关系。结果表明,云南高产环境下,油菜高产的主要原因是光照充足、昼夜温差大,导致生物产量高、角果多、每角粒数多,特别是二次分枝角果对产量贡献较大。主序和一、二次分枝的角果数与产量收获指数在重庆均呈显著正相关,而云南环境下主序角果数与产量收获指数呈显著负相关。主序和一、二次分枝的每角粒数均以云南环境下极显著高于重庆环境下,且在两种环境下,主序和一次分枝每角粒数都与产量收获指数和产油量收获指数呈显著正相关,表明每角粒数需要充足的光合产物积累及高效的籽粒填充效率来保证。主序、一次和二次分枝千粒重云南均低于重庆,表明寡日照区域的油菜会减少籽粒数,以保证部分种子的干物质填充。云南环境下,各部位千粒重与产量收获指数和产油量收获指数均呈显著正相关,而重庆环境下的相关性不明显,说明在光照充足条件下,光合产物转运能力是导致千粒重差异的主要原因。综合分析表明,在光照充足的环境下,主序角果数和单株经济产量是提高YHI的关键;而寡日照环境下YHI构成复杂,必须将主序和一次分枝的产量组分有机结合,且严格控制生物产量才能实现YHI的提高。
DOI:10.3724/SP.J.1006.2017.00082URL [本文引用: 1]

收获指数偏低是制约油菜籽粒产量和产油量进一步提高的瓶颈。为解析适合不同产量的环境下油菜高收获指数的构成因素及形成机制,本研究选择在高产环境下的云南临沧和一般产量的长江流域上游主产区重庆北碚均能正常生长和成熟的321份甘蓝型油菜品种(系)为材料,分析其产量相关性状的变异以及两地间的差异,利用相关分析和通径分析研究籽粒产量收获指数(YHI)、产油量收获指数分别与17个产量组成性状的关系。结果表明,云南高产环境下,油菜高产的主要原因是光照充足、昼夜温差大,导致生物产量高、角果多、每角粒数多,特别是二次分枝角果对产量贡献较大。主序和一、二次分枝的角果数与产量收获指数在重庆均呈显著正相关,而云南环境下主序角果数与产量收获指数呈显著负相关。主序和一、二次分枝的每角粒数均以云南环境下极显著高于重庆环境下,且在两种环境下,主序和一次分枝每角粒数都与产量收获指数和产油量收获指数呈显著正相关,表明每角粒数需要充足的光合产物积累及高效的籽粒填充效率来保证。主序、一次和二次分枝千粒重云南均低于重庆,表明寡日照区域的油菜会减少籽粒数,以保证部分种子的干物质填充。云南环境下,各部位千粒重与产量收获指数和产油量收获指数均呈显著正相关,而重庆环境下的相关性不明显,说明在光照充足条件下,光合产物转运能力是导致千粒重差异的主要原因。综合分析表明,在光照充足的环境下,主序角果数和单株经济产量是提高YHI的关键;而寡日照环境下YHI构成复杂,必须将主序和一次分枝的产量组分有机结合,且严格控制生物产量才能实现YHI的提高。
[本文引用: 1]
DOI:10.1007/s00344-014-9467-4URL [本文引用: 1]

The application of brassinosteroids (BRs) has been reported to alleviate heat stress. This study investigated the effect of 7,8-dihydro-8α-20-hydroxyecdysone (DHECD)—a BR mimic—by comparison with...
DOI:10.1038/srep28298URLPMID:4915011 [本文引用: 1]

As a resource allocation strategy, plant growth and defense responses are generally mutually antagonistic. Brassinosteroid (BR) regulates many aspects of plant development and stress responses, however, genetic evidence of its integrated effects on plant growth and stress tolerance is lacking. We overexpressed theArabidopsisBR biosynthetic geneAtDWF4in the oilseed plantBrassica napusand scored growth and stress response phenotypes. The transgenicB. napusplants, in comparison to wild type, displayed increased seed yield leading to increased overall oil content per plant, higher root biomass and root length, significantly better tolerance to dehydration and heat stress, and enhanced resistance to necrotrophic fungal pathogensLeptosphaeria maculansandSclerotinia sclerotiorum.Transcriptome analysis supported the integrated effects of BR on growth and stress responses; in addition to BR responses associated with growth, a predominant plant defense signature, likely mediated by BES1/BZR1, was evident in the transgenic plants. These results establish that BR can interactively and simultaneously enhance abiotic and biotic stress tolerance and plant productivity. The ability to confer pleiotropic beneficial effects that are associated with different agronomic traits suggests that BR elated genes may be important targets for simultaneously increasing plant productivity and performance under stress conditions.
DOI:10.1038/ncb2545URLPMID:22820378 [本文引用: 1]

Abstract Plant growth is coordinately regulated by environmental and hormonal signals. Brassinosteroid (BR) plays essential roles in growth regulation by light and temperature, but the interactions between BR and these environmental signals remain poorly understood at the molecular level. Here, we show that direct interaction between the dark- and heat-activated transcription factor phytochrome-interacting factor 4 (PIF4) and the BR-activated transcription factor BZR1 integrates the hormonal and environmental signals. BZR1 and PIF4 interact with each other in vitro and in vivo, bind to nearly 2,000 common target genes, and synergistically regulate many of these target genes, including the PRE family helix-loop-helix factors required for promoting cell elongation. Genetic analysis indicates that BZR1 and PIFs are interdependent in promoting cell elongation in response to BR, darkness or heat. These results show that the BZR1-PIF4 interaction controls a core transcription network, enabling plant growth co-regulation by the steroid and environmental signals.
URL [本文引用: 1]

光敏色素互作因子(PIFs)蛋白家族隶属于bHLH转录因子家族,参与多种信号转导途径,调控植物的生长发育进程,如抑制种子萌发、促进幼苗的暗形态建成和调控开花时间等。作为一个胞内信号调控的重要组分,PIFs广泛参与到由多种植物内源激素如赤霉素、乙烯、生长素、油菜素内酯、脱落酸和外部环境因素如高温、光等所介导的信号网络中。本文从PIFs的结构、参与途径及调控植物发育进程3个方面,简要介绍近年来国内外对PIFs在信号网络调控方面的最新研究进展。
URL [本文引用: 1]

光敏色素互作因子(PIFs)蛋白家族隶属于bHLH转录因子家族,参与多种信号转导途径,调控植物的生长发育进程,如抑制种子萌发、促进幼苗的暗形态建成和调控开花时间等。作为一个胞内信号调控的重要组分,PIFs广泛参与到由多种植物内源激素如赤霉素、乙烯、生长素、油菜素内酯、脱落酸和外部环境因素如高温、光等所介导的信号网络中。本文从PIFs的结构、参与途径及调控植物发育进程3个方面,简要介绍近年来国内外对PIFs在信号网络调控方面的最新研究进展。
DOI:10.1093/emboj/21.10.2441URLPMID:12006496 [本文引用: 1]

Plants sense and respond to red and far-red light using the phytochrome (phy) family of photoreceptors. However, the mechanism of light signal transduction is not well defined. Here, we report the identification of a new mutant Arabidopsis locus, srl2 (short under red-light 2), which confers selective hypersensitivity to continuous red, but not far-red, light. This hypersensitivity is eliminated in srl2phyB, but not srl2phyA, double mutants, indicating that this locus functions selectively and negatively in phyB signaling. The SRL2 gene encodes a bHLH factor, designated PIF4 (phytochrome-interacting factor 4), which binds selectively to the biologically active Pfr form of phyB, but has little affinity for phyA. Despite its hypersensitive morphological phenotype, the srl2 mutant displays no perturbation of light-induced expression of marker genes for chloroplast development. These data suggest that PIF4 may function specifically in a branch of the phyB signaling network that regulates a subset of genes involved in cell expansion. Consistent with this proposal, PIF4 localizes to the nucleus and can bind to a G-box DNA sequence motif found in various light-regulated promoters.
DOI:10.1016/j.tplants.2007.10.001URL [本文引用: 1]
DOI:10.1016/j.cub.2008.12.046URLPMID:19185498 [本文引用: 1]

Stomata are pores on the surfaces of leaves that regulate gas exchange between the plant interior and the atmosphere [1]. Plants adapt to changing environmental conditions in the short term by adjusting the aperture of the stomatal pores, whereas longer-term changes are accomplished by altering the proportion of stomata that develop on the leaf surface []. Although recent work has identified genes involved in the control of stomatal development [4], we know very little about how stomatal development is modulated by environmental signals, such as light. Here, we show that mature leaves of 5] compared to those grown at lower photon irradiances. Light quantity-mediated changes in S.I. occur in red light, suggesting that phytochrome photoreceptors [6] are involved. By using a genetic approach, we demonstrate that this response is dominated by phytochrome B and also identify a role for the transcription factor, PIF4]. In sum, we identify a photoreceptor and downstream signaling protein involved in light-mediated control of stomatal development, thereby establishing a tractable system for investigating how an environmental signal modulates stomatal development.
DOI:10.1016/j.cub.2009.01.046URLPMID:19249207 [本文引用: 1]

Exposure of Arabidopsis plants to high temperature (28 C) results in a dramatic change in plant development. Responses to high temperature include rapid extension of plant axes, leaf hyponasty, and early flowering [1, 2]. These phenotypes parallel plant responses to the threat of vegetational shade and have been shown to involve the hormone auxin [1, 3]. In this work, we demonstrate that high temperature-induced architectural adaptations are mediated through the bHLH transcriptional regulator PHYTOCHROME INTERACTING FACTOR 4 (PIF4). Roles for PIF4 have previously been established in both light and gibberellin (GA) signaling, through interactions with phytochromes and DELLA proteins, respectively [4 6]. Mutants deficient in PIF4 do not display elongation responses or leaf hyponasty upon transfer to high temperature. High temperature-mediated induction of the auxin-responsive gene IAA29 is also abolished in these plants. An early flowering response to high temperature is maintained in pif4 mutants, suggesting that architectural and flowering responses operate via separate signaling pathways. The role of PIF4 in temperature signaling does not, however, appear to operate through interaction with either phytochrome or DELLA proteins, suggesting the existence of a novel regulatory mechanism. We conclude that PIF4 is an important component of plant high temperature signaling and integrates multiple environmental cues during plant development.
DOI:10.1073/pnas.1110682108URL [本文引用: 1]
DOI:10.1016/j.pbi.2014.06.010URLPMID:25061896 [本文引用: 1]

Gibberellic acid (GA) regulates a diversity of processes associated with plant growth and development. The DELLA proteins act as repressors of GA signaling, and are destabilized by GA. Although it is known that GA signaling integrates various endogenous and environmental signals, the molecular basis of their modulation of plant growth and development is only now beginning to be understood. The current suggestion is that the DELLA proteins act as one possible quantitative modulator of plant growth, achieved by integrating multiple environmental and hormonal signals via protein rotein interactions. This review discusses recent elaborations of the de-repression model proposed to describe the GA response, and focuses on integrative networks thought to regulate plant growth, development and the adaptation to a fluctuating environment.
DOI:10.1101/gad.243675.114URLPMID:25085420 [本文引用: 1]

Signaling by the hormones brassinosteroid (BR) and gibberellin (GA) is critical to normal plant growth and development and is required for hypocotyl elongation in response to dark and elevated temperatures. Active BR signaling is essential for GA promotion of hypocotyl growth and suppresses the dwarf phenotype of GA mutants. Cross-talk between these hormones occurs downstream from the DELLAs, as GA-induced destabilization of these GA signaling repressors is not affected by BRs. Here we show that the light-regulated PIF4 (phytochrome-interacting factor 4) factor is a phosphorylation target of the BR signaling kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2), which marks this transcriptional regulator for proteasome degradation. Expression of a mutated PIF41A protein lacking a conserved BIN2 phosphorylation consensus causes a severe elongated phenotype and strongly up-regulated expression of the gene targets. However, PIF41A is not able to suppress the dwarf phenotype of the bin2-1 mutant with constitutive activation of this kinase. PIFs were shown to be required for the constitutive BR response of bes1-D and bzr1-1D mutants, these factors acting in an interdependent manner to promote cell elongation. Here, we show that bes1-D seedlings are still repressed by the inhibitor BRZ in the light and that expression of the nonphosphorylatable PIF41A protein makes this mutant fully insensitive to brassinazole (BRZ). PIF41A is preferentially stabilized at dawn, coinciding with the diurnal time of maximal growth. These results uncover a main role of BRs in antagonizing light signaling by inhibiting BIN2-mediated destabilization of the PIF4 factor. This regulation plays a prevalent role in timing hypocotyl elongation to late night, before light activation of phytochrome B (PHYB) and accumulation of DELLAs restricts PIF4 transcriptional activity.
DOI:10.3864/j.issn.0578-1752.2015.02.12URL [本文引用: 1]

【目的】测量菊花在弱光条件 下,出现庇荫症状过程中乙烯生成速率、赤霉素和生长素含量的变化及相关基因表达变化,从激素和基因水平探究庇荫机理。【方法】以菊花‘清露’为试验材料, 设置55%光照(对照)和15%光照(弱光处理)。利用气相色谱仪Agilent 6890N GC在0、2、4、6和8 d测量乙烯生成速率,利用高效液相色谱仪Agilent HPLC-110在0、2、4、6和8 d测量赤霉素(GA3)和生长素(IAA)含量,以GAPDH为内参基因用荧光定量PCR方法测量0、2、4、6和8 d时PHYA、COP1、PIF4、GAI和IAA1的相对表达量。设置0.5、1.0、2.0和4.0 mmol·L-14个GA3浓度,用适量乙醇溶解,使乙醇终浓度为1%,0 d、4 d时各处理1次,采用地上部喷施;设10、20、50和100μmol·L-14个多效唑(paclobutrazol,PAC)浓度,用适量乙醇溶解, 使乙醇终浓度是1%,0 d和4 d时各处理1次,根据结果以2.0 mmol·L-1GA3,50μmol·L-1PAC对植物生长影响显著,分别作为处理浓度。0 d、7 d各喷1次,14 d时取样进行基因表达分析。【结果】15%光照条件下,第2天和第4天时乙烯生成速率显著增加,第4天最显著,比对照增加了2.04倍,第6天以后对照和 处理的植株乙烯生成速率差异不显著,15%弱光处理条件下植株地上部乙烯生成速率比55%光照条件增加显著;15%光照条件下,第2、4和6天时叶片赤霉 素含量与对照相比显著增加,第6天时出现峰值,达到4 700.56 ng·g-1,比对照高出2.57倍,随后又迅速下降,第8天降到与对照相同的水平;15%光照条件下生长素水平与对照组相比显著下降,随着处理时间的延 长,对照组生长素含量呈增加趋势,弱光处理组生长素含量呈下降趋势,第8天时处理组生长素含量为223.95 ng·g-1FW,对照为489.77 ng·g-1FW,对照比处理的生长素含量高出1.12倍。弱光处理第2天时PHYA表达量迅速增加,2—6 d以略小的速度增加,6—8 d保持不变,对照组从第2天开始下降,4—8 d时处理组PHYA变化量都高于对照组;对照和处理的PIF4表达量变化在0—4天趋势相同,不同的是处理组在4—8 d高于对照,尤其是第6天比对照高出1.85倍。对照组COP1呈缓慢降低趋势,处理组先增加后下降,峰值出现在第2天,第8天降到对照组含量以下。整个 处理过程中,处理组的GAI一直增加,对照组0—4 d小幅增加,后以较快的速度下降。处理组和对照组IAA1在0—2 d均小幅上升,对照组在2—4 d又以较快速度下降,2 d后处理组IAA1表达量变化不大。外源赤霉素使COP1、PIF4的表达量上升,使GAI和IAA1表达量下降;PAC处理抑制COP1、PIF4、 GAI和IAA1的表达。【结论】PHYA、COP1、IAA1和PIF4都受光的诱导,弱光引起GA3含量升高和GAI转录水平上调,赤霉素和乙烯参与 菊花庇荫调节过程。
DOI:10.3864/j.issn.0578-1752.2015.02.12URL [本文引用: 1]

【目的】测量菊花在弱光条件 下,出现庇荫症状过程中乙烯生成速率、赤霉素和生长素含量的变化及相关基因表达变化,从激素和基因水平探究庇荫机理。【方法】以菊花‘清露’为试验材料, 设置55%光照(对照)和15%光照(弱光处理)。利用气相色谱仪Agilent 6890N GC在0、2、4、6和8 d测量乙烯生成速率,利用高效液相色谱仪Agilent HPLC-110在0、2、4、6和8 d测量赤霉素(GA3)和生长素(IAA)含量,以GAPDH为内参基因用荧光定量PCR方法测量0、2、4、6和8 d时PHYA、COP1、PIF4、GAI和IAA1的相对表达量。设置0.5、1.0、2.0和4.0 mmol·L-14个GA3浓度,用适量乙醇溶解,使乙醇终浓度为1%,0 d、4 d时各处理1次,采用地上部喷施;设10、20、50和100μmol·L-14个多效唑(paclobutrazol,PAC)浓度,用适量乙醇溶解, 使乙醇终浓度是1%,0 d和4 d时各处理1次,根据结果以2.0 mmol·L-1GA3,50μmol·L-1PAC对植物生长影响显著,分别作为处理浓度。0 d、7 d各喷1次,14 d时取样进行基因表达分析。【结果】15%光照条件下,第2天和第4天时乙烯生成速率显著增加,第4天最显著,比对照增加了2.04倍,第6天以后对照和 处理的植株乙烯生成速率差异不显著,15%弱光处理条件下植株地上部乙烯生成速率比55%光照条件增加显著;15%光照条件下,第2、4和6天时叶片赤霉 素含量与对照相比显著增加,第6天时出现峰值,达到4 700.56 ng·g-1,比对照高出2.57倍,随后又迅速下降,第8天降到与对照相同的水平;15%光照条件下生长素水平与对照组相比显著下降,随着处理时间的延 长,对照组生长素含量呈增加趋势,弱光处理组生长素含量呈下降趋势,第8天时处理组生长素含量为223.95 ng·g-1FW,对照为489.77 ng·g-1FW,对照比处理的生长素含量高出1.12倍。弱光处理第2天时PHYA表达量迅速增加,2—6 d以略小的速度增加,6—8 d保持不变,对照组从第2天开始下降,4—8 d时处理组PHYA变化量都高于对照组;对照和处理的PIF4表达量变化在0—4天趋势相同,不同的是处理组在4—8 d高于对照,尤其是第6天比对照高出1.85倍。对照组COP1呈缓慢降低趋势,处理组先增加后下降,峰值出现在第2天,第8天降到对照组含量以下。整个 处理过程中,处理组的GAI一直增加,对照组0—4 d小幅增加,后以较快的速度下降。处理组和对照组IAA1在0—2 d均小幅上升,对照组在2—4 d又以较快速度下降,2 d后处理组IAA1表达量变化不大。外源赤霉素使COP1、PIF4的表达量上升,使GAI和IAA1表达量下降;PAC处理抑制COP1、PIF4、 GAI和IAA1的表达。【结论】PHYA、COP1、IAA1和PIF4都受光的诱导,弱光引起GA3含量升高和GAI转录水平上调,赤霉素和乙烯参与 菊花庇荫调节过程。
[本文引用: 1]
DOI:10.1104/pp.110.153593URLPMID:20472752 [本文引用: 1]

Basic helix-loop-helix proteins (bHLHs) are found throughout the three eukaryotic kingdoms and constitute one of the largest families of transcription factors. A growing number of bHLH proteins have been functionally characterized in plants. However, some of these have not been previously classified. We present here an updated and comprehensive classification of the bHLHs encoded by the whole sequenced genomes of Arabidopsis (Arabidopsis thaliana), Populus trichocarpa, Oryza sativa, Physcomitrella patens, and five algae species. We define a plant bHLH consensus motif, which allowed the identification of novel highly diverged atypical bHLHs. Using yeast two-hybrid assays, we confirm that (1) a highly diverged bHLH has retained protein interaction activity and (2) the two most conserved positions in the consensus play an essential role in dimerization. Phylogenetic analysis permitted classification of the 638 bHLH genes identified into 32 subfamilies. Evolutionary and functional relationships within subfamilies are supported by intron patterns, predicted DNA-binding motifs, and the architecture of conserved protein motifs. Our analyses reveal the origin and evolutionary diversification of plant bHLHs through differential expansions, domain shuffling, and extensive sequence divergence. At the functional level, this would translate into different subfamilies evolving specific DNA-binding and protein interaction activities as well as differential transcriptional regulatory roles. Our results suggest a role for bHLH proteins in generating plant phenotypic diversity and provide a solid framework for further investigations into the role carried out in the transcriptional regulation of key growth and developmental processes.
DOI:10.1111/j.1365-313X.2010.04459.xURLPMID:21443626 [本文引用: 1]

The expansion of gene families encoding regulatory proteins is typically associated with the increase in complexity characteristic of multi-cellular organisms. The MYB and basic helix-loop-helix (bHLH) families provide excellent examples of how gene duplication and divergence within particular groups of transcription factors are associated with, if not driven by, the morphological and metabolic diversity that characterize the higher plants. These gene families expanded dramatically in higher plants; for example, there are approximately 339 and 162 MYB and bHLH genes, respectively, in Arabidopsis, and approximately 230 and 111, respectively, in rice. In contrast, the Chlamydomonas genome has only 38 MYB genes and eight bHLH genes. In this review, we compare the MYB and bHLH gene families from structural, evolutionary and functional perspectives. The knowledge acquired on the role of many of these factors in Arabidopsis provides an excellent reference to explore sequence unction relationships in crops and other plants. The physical interaction and regulatory synergy between particular sub-classes of MYB and bHLH factors is perhaps one of the best examples of combinatorial plant gene regulation. However, members of the MYB and bHLH families also interact with a number of other regulatory proteins, forming complexes that either activate or repress the expression of sets of target genes that are increasingly being identified through a diversity of high-throughput genomic approaches. The next few years are likely to witness an increasing understanding of the extent to which conserved transcription factors participate at similar positions in gene regulatory networks across plant species.
[本文引用: 1]
DOI:10.1038/nature10928URLPMID:22437497 [本文引用: 1]

Plant growth and development are strongly affected by small differences in temperature. Current climate change has already altered global plant phenology and distribution, and projected increases in temperature pose a significant challenge to agriculture. Despite the important role of temperature on plant development, the underlying pathways are unknown. It has previously been shown that thermal acceleration of flowering is dependent on the florigen, FLOWERING LOCUS T (FT). How this occurs is, however, not understood, because the major pathway known to upregulate FT, the photoperiod pathway, is not required for thermal acceleration of flowering. Here we demonstrate a direct mechanism by which increasing temperature causes the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) to activate FT. Our findings provide a new understanding of how plants control their timing of reproduction in response to temperature. Flowering time is an important trait in crops as well as affecting the life cycles of pollinator species. A molecular understanding of how temperature affects flowering will be important for mitigating the effects of climate change. 2012 Macmillan Publishers Limited. All rights reserved.
DOI:10.1111/nph.12725URLPMID:24571056 [本文引用: 1]

'Summary'1126I.1126II.1127III.1128IV.1128V.1129VI.1132VII.1135VIII.1136IX.1136 1137 References1137SummaryLight and temperature, in coordination with the endogenous clock and the hormones gibberellin (GA) and brassinosteroids (BRs), modulate plant growth and development by affecting the expression of multiple cell wall- and auxin-related genes. PHYTOCHROME INTERACTING FACTORS (PIFs) play a central role in the activation of these genes, the activity of these factors being regulated by the circadian clock and phytochrome-mediated protein destabilization. GA signaling is also integrated at the level of PIFs; the DELLA repressors are found to bind these factors and impair their DNA-binding ability. The recent finding that PIFs are co-activated by BES1 and BZR1 highlights a further role of these regulators in BR signal integration, and reveals that PIFs act in a concerted manner with the BR-related BES1/BZR1 factors to activate auxin synthesis and transport at the gene expression level, and synergistically activate several genes with a role in cell expansion. Auxins feed back into this growth regulatory module by inducing GA biosynthesis and BES1/BZR1 gene expression, in addition to directly regulating several of these growth pathway gene targets. An exciting challenge in the future will be to understand how this growth program is dynamically regulated in time and space to orchestrate differential organ expansion and to provide plants with adaptation flexibility.
DOI:10.14348/molcells.2016.0126URLPMID:4990750 [本文引用: 1]

As sessile organisms, plants must be able to adapt to the environment. Plants respond to the environment by adjusting their growth and development, which is mediated by sophisticated signaling networks that integrate multiple environmental and endogenous signals. Recently, increasing evidence has shown that a bHLH transcription factor PIF4 plays a major role in the multiple signal integration for plant growth regulation. PIF4 is a positive regulator in cell elongation and its activity is regulated by various environmental signals, including light and temperature, and hormonal signals, including auxin, gibberellic acid and brassinosteroid, both transcriptionally and post-translationally. Moreover, recent studies have shown that the circadian clock and metabolic status regulate endogenous PIF4 level. The PIF4 transcription factor cooperatively regulates the target genes involved in cell elongation with hormone-regulated transcription factors. Therefore, PIF4 is a key integrator of multiple signaling pathways, which optimizes growth in the environment. This review will discuss our current understanding of the PIF4-mediated signaling networks that control plant growth.
DOI:10.1104/pp.16.01778URLPMID:28438793 [本文引用: 1]

Brassinosteroids (BRs) are essential phytohormones regulating various developmental and physiological processes during normal growth and development. cog1-3D (cogwheel1-3D) was identified as an activation-tagged genetic modifier of bri1-5, an intermediate BR receptor mutant in Arabidopsis (Arabidopsis thaliana). COG1 encodes a Dof-type transcription factor found previously to act as a negative regulator of the phytochrome signaling pathway. cog1-3D single mutants show an elongated hypocotyl phenotype under light conditions. A loss-of-function mutant or inducible expression of a dominant negative form of COG1 in the wild type results in an opposite phenotype. A BR profile assay indicated that BR levels are elevated in cog1-3D seedlings. Quantitative reverse transcription-polymerase chain reaction analyses showed that several key BR biosynthetic genes are significantly up-regulated in cog1-3D compared with those of the wild type. Two basic helix-loop-helix transcription factors, PIF4 and PIF5, were found to be transcriptionally up-regulated in cog1-3D. Genetic analysis indicated that PIF4 and PIF5 were required for COG1 to promote BR biosynthesis and hypocotyl elongation. Chromatin immunoprecipitation and electrophoretic mobility shift assays indicated that COG1 binds to the promoter regions of PIF4 and PIF5, and PIF4 and PIF5 bind to the promoter regions of key BR biosynthetic genes, such as DWF4 and BR6ox2, to directly promote their expression. These results demonstrated that COG1 regulates BR biosynthesis via up-regulating the transcription of PIF4 and PIF5.
DOI:10.1016/S1534-5807(02)00153-3URLPMID:11970900 [本文引用: 1]

Plant steroid hormones, brassinosteroids (BRs), are perceived by a cell surface receptor kinase, BRI1, but how BR binding leads to regulation of gene expression in the nucleus is unknown. Here we describe the identification of BZR1 as a nuclear component of the BR signal transduction pathway. A dominant mutation bzr1- 1D suppresses BR-deficient and BR-insensitive ( bri1) phenotypes and enhances feedback inhibition of BR biosynthesis. BZR1 protein accumulates in the nucleus of elongating cells of dark-grown hypocotyls and is stabilized by BR signaling and the bzr1-1D mutation. Our results demonstrate that BZR1 is a positive regulator of the BR signaling pathway that mediates both downstream BR responses and feedback regulation of BR biosynthesis.
