 ,*南京农业大学作物遗传与种质创新国家重点实验室 / 江苏省植物基因工程技术研究中心, 江苏南京210095
,*南京农业大学作物遗传与种质创新国家重点实验室 / 江苏省植物基因工程技术研究中心, 江苏南京210095Identification and mapping of round seed related gene in rice (Oryza sativa L.)
CHEN Ya-Ping, MIAO Rong, LIU Xi, CHEN Ben-Jia, LAN Jie, MA Teng-Fei, WANG Yi-Hua, LIU Shi-Jia, JIANG Ling ,*State Key Laboratory of Crop Genetics and Germplasm Enhancement / Research Center of Jiangsu Plant Gene Engineering, Nanjing Agricultural University, Nanjing 210095, Jiangsu, China
,*State Key Laboratory of Crop Genetics and Germplasm Enhancement / Research Center of Jiangsu Plant Gene Engineering, Nanjing Agricultural University, Nanjing 210095, Jiangsu, China通讯作者:
第一联系人:
收稿日期:2018-06-7接受日期:2018-10-8网络出版日期:2018-11-01
| 基金资助: |
Received:2018-06-7Accepted:2018-10-8Online:2018-11-01
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (4722KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
陈雅萍, 缪荣, 刘喜, 陈本佳, 兰杰, 马腾飞, 王益华, 刘世家, 江玲. 水稻圆粒基因RS的鉴定和定位[J]. 作物学报, 2019, 45(1): 1-9. doi:10.3724/SP.J.1006.2019.82032
CHEN Ya-Ping, MIAO Rong, LIU Xi, CHEN Ben-Jia, LAN Jie, MA Teng-Fei, WANG Yi-Hua, LIU Shi-Jia, JIANG Ling.
水稻产量是一个复杂的性状, 与单株穗数、每穗粒数及粒重等有关, 而粒重与粒长、粒宽、粒厚等相关。所以, 优化籽粒形状对提高水稻产量至关重要[1]。
参与水稻粒型大小的调控途径众多, 如植物激素、泛素化蛋白酶体途径、G-蛋白信号、表观遗传学途径等。粒长粒重主效基因GS3对粒重起负调控作用[2]; GW2编码一个环型E3泛素连接酶, 位于细胞质中将其底物锚定到蛋白酶体进行降解, 可负调节细胞的分裂[3]; GS2通过促进细胞分裂和细胞膨大调控籽粒大小[4]; 非洲稻GL4通过调控外颖和内颖纵向细胞的伸长控制非洲栽培稻的粒长, 同时也能调控种子落粒性[5]; GL7位点上17.1-kb的串联重复引起GL7表达水平上调, 并下调GL7邻近负调因子表达, 从而增加水稻粒长并改善稻米外观品质[6]; GS5编码一个丝氨酸羧肽酶, 正向调控水稻籽粒大小, 高表达形成更大的籽粒[7]; HGW作为一个重要的上游调控蛋白促进抽穗和粒重相关基因的表达, 很可能直接通过GIF1控制水稻籽粒的重量[8]; GIF1编码早期籽粒灌浆过程中碳分配所需的细胞壁转化酶, TGW6编码吲哚-3-乙酸(IAA)-葡萄糖水解酶, 影响胚乳合胞体向细胞期的转变, 二者都调节灌浆过程中的源-库关系, 从而影响最终的粒重[9,10]; GW8/OsSPL16基因座上的等位基因变异是其启动子中的10 bp缺失, 显著降低了基因的表达水平并因此降低了谷粒宽度[11]; GW5编码一个钙调素结合蛋白, 与糖原合酶激酶GSK2互作并抑制GSK2激酶活性。GW5抑制GSK2的自磷酸化及GSK2对OsBZR1和DLT的磷酸化, 影响细胞核中未磷酸化的OsBZR1和DLT蛋白的积累, 从而调节油菜素内酯响应基因的表达[12]。水稻粒型基因中除与粒型相关的主效QTL外, 还有许多与作物的生长发育及激素传导相关的基因也影响粒型, 如D2、BG1、OsCDPK13、D61、DSG1等[13,14,15,16,17]。
本研究利用EMS诱变“宁粳3号”筛选到一个籽粒变圆且稳定遗传的突变体round seed (rs)。从表型、生理特性、遗传方式等方面对rs鉴定, 明确了其遗传特性, 并对突变基因进行了精细定位。对rs的研究有利于进一步加深人们对水稻粒型调控机制的认识, 为完善水稻粒型的调控网络奠定基础。
1 材料与方法
1.1 材料
利用rs与“宁粳3号”杂交构建遗传分析群体, 对F1和F2植株进行表型观察与统计。以rs与“滇粳优”杂交构建F2遗传分离群体, 用于基因定位。1.2 突变体表型考察
将野生型和rs在同一时期种植于南京农业大学土桥实验基地。在成熟期分别测定野生型和rs的株高、穗长、分蘖数等主要农艺性状。利用万深SC-G自动考种分析仪测定粒长、粒宽、千粒重, 重复3次, 随机取20个单株测定株高等农艺性状。1.3 颖壳扫描电镜观察
取野生型和rs抽穗后的颖壳, 将其固定在2.5%戊二醛溶液中, 抽真空1 h, 室温避光放置48 h, 而后利用扫描电镜(S-3000N, Hitachi, Tokyo, Japan)观察颖壳外表皮细胞。1.4 基因定位
成熟期从rs与“滇粳优”构建的F2群体中选取与突变体粒型一致的极端个体220株, 采用CTAB法提取叶片DNA, 用于基因型分析。从实验室已有的分子标记中, 筛选突变体rs与“滇粳优”之间有多态性的标记, 最终筛选到均匀分布于12条染色体的有多态性标记256对。利用F2群体中的10个极端个体进行基因型和表型连锁分析, 确定与目的基因连锁的分子标记, 再进一步扩大极端个体数量进行精细定位。精细定位引物序列见表1。Table 1
表1
表1用于精细定位的分子标记
Table 1
| 标记 Marker | 正向引物序列 Forward primer sequence (5°→3°) | 反向引物序列 Reverse primer sequence (5°→3°) |
|---|---|---|
| N3-5 | GTCTGTGCGCTCCTTGTTCTAGC | CCTGGACCAATTTGTATGGTTGG |
| N3-9 | GATGCAGTAGGAACACCAAACAGC | ATCGAGTACCAAGTGCCTGTGC |
| N3-10 | GGTTTGGGAGCCCATAATCT | CTGGGCTTCTTTCACTCGTC |
| RM3413 | TTAGAGGAGATGATGGTGCAACG | AGCAGCCATTGAATGTGTTTGG |
| RM14282 | CCCAAACACAAACACAAAGAGAGC | AACACGCAGGTCCTCTTGAACC |
| I3-2 | TACTTTAATTTTGCAGCTC | TTTTACCCCACTCCATCT |
新窗口打开|下载CSV
1.5 油菜素内酯(BR)处理
选取野生型和rs的饱满种子各100粒, 经浸种与催芽, 选发芽一致的播种, 在30℃培养箱中培养10 d, 培养条件为14 h光照/10 h黑暗, 待小苗长至三叶期后, 剪取包含第二叶的叶片、叶枕和叶鞘的2 cm区段于不同浓度梯度(0、0.1、1.0 μmol L-1)的BR溶液中浸泡, 在光照条件下30℃处理2 d后, 用量角器测量叶夹角, 并照相。1.6 qRT-PCR分析
用总RNA提取试剂盒(康为世纪生物科技有限公司)提取rs和野生型幼苗叶片的总RNA, 用DU800分光光度计检测RNA的浓度及质量, 用琼脂糖凝胶电泳检测RNA的完整性。取2 μg RNA作模板, 用SuperScript II Kit试剂盒(TaKaRa公司)反转录合成cDNA。利用qRT-PCR分析rs及野生型中粒型相关基因、细胞周期相关基因、油菜素内酯合成和信号途径相关基因的表达水平。以UBQ (LOC_Os01g22490)为内参基因, 扩增反应在 7900HT Real-time PCR仪(Applied Biosystems)进行。10 μL实时荧光定量PCR体系含cDNA模板2 μL、2× SYBR Select Master Mix (TOYOBO) 5 μL、正反引物(10 μmol L-1)各0.3 μL, ddH2O补足10 μL。Real-time PCR程序为90℃反应30 s, 95℃反应5 s, 60℃反应34 s, 40个循环。待扩增反应结束后, 利用7900HT Real-time PCR仪(Applied Biosystems)自带的软件分析Ct值, 计算出目的基因的表达量。采用2-ΔΔCt方法分析基因表达, 设置3个生物重复。相关基因所用引物序列参照文献[18,19]。2 结果与分析
2.1 突变体rs的表型鉴定
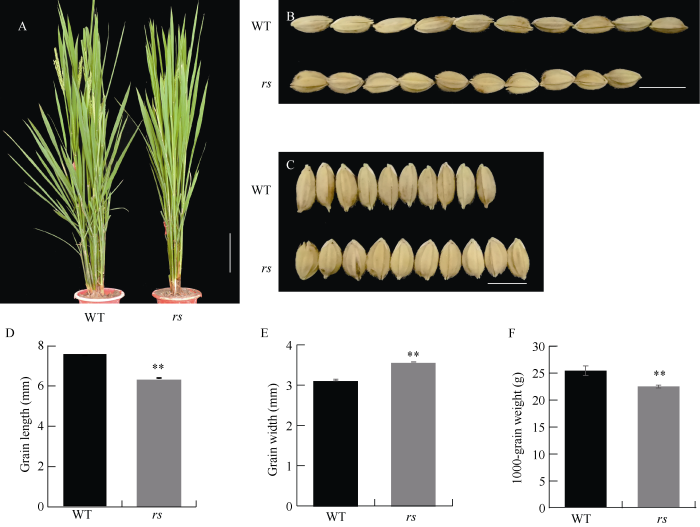
与野生型相比, 突变体rs的粒长变短、粒宽变大, 粒厚增加, 籽粒显著变圆(图1-B, C; 表2), 粒长、粒宽分别为6.39 mm和2.58 mm (图1-D, E), 为野生型的85.1%和114.4%。此外, 突变体rs的千粒重相对于野生型降低了约11.6% (图1-F), 结实率比野生型下降, 但在株高、穗长等方面差异不显著(图1-A和表2)。Table 2
表2
表2野生型和rs的农艺性状比较
Table 2
| 性状 Trait | 野生型 Wild type | 突变体 rs |
|---|---|---|
| 粒长 Grain length (mm) | 7.50±0.009 | 6.39±0.023** |
| 粒宽 Grain width (mm) | 3.13±0.021 | 3.58±0.003** |
| 长/宽比 Length/width ratio | 2.42±0.013 | 1.80±0.009** |
| 粒厚 Grain thickness (mm) | 1.14±0.025 | 1.28±0.062** |
| 千粒重 1000-grain weight (g) | 25.45±0.890 | 22.51±0.252** |
| 株高 Plant height (cm) | 91.40±2.633 | 90.50±2.121 |
| 穗长 Panicle length (cm) | 17.35±0.803 | 17.19±0.709 |
| 分蘖数 Number of tillers | 11.00±2.160 | 10.1±2.025 |
| 结实率 Seed setting rate (%) | 72.69±2.430 | 56.90±4.270** |
新窗口打开|下载CSV
2.2 突变性状的遗传分析
为明确水稻圆粒突变体rs的遗传特性, 对rs与野生型进行正、反交试验。正交与反交F1单株均表现与野生型类似的表型, 说明圆粒是受隐性核基因控制的。通过对各组合F2代分离群体调查发现, F2群体的单株可分为两类, 一类表型正常, 另一类与突变体rs表型相似, 且野生型和突变表型的单株在F2群体中的分离比符合3∶1 (表3), 表明rs的突变表型是由单隐性核基因控制。Table 3
表3
表3突变体rs的遗传分析
Table 3
| 杂交组合 Cross-combination | 总株数 Total number of plants | 正常表现 Wild-type phenotype | 突变表现 Mutant phenotype | χ2(3:1) |
|---|---|---|---|---|
| rs/WT | 174 | 135 | 39 | 0.62 |
| WT/rs | 207 | 161 | 46 | 0.85 |
新窗口打开|下载CSV
图 1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图 1野生型和突变体rs的表型
A: 野生型和rs在抽穗期植株表现, 比例尺为10 cm。B, C: 野生型和rs糙米籽粒表现。比例尺为6 mm。D, F: 野生型和rs粒长、粒宽与千粒重比较。**代表突变体与野生型之间在P = 0.01水平上差异显著。
Fig. 1Phenotypes of the wild type and the rs mutant
A: plants of the wild type and the rs mutant at heading stage. Bar = 10 cm. B, C: grain phenotypes of the wild type and rs. Bar = 6 mm. D, F: comparisons of grain length, width, and the 1000-grain weight. ** Significant difference at P = 0.01 level between the wild type and the mutant rs.
2.3 颖壳外表皮细胞扫描电镜观察
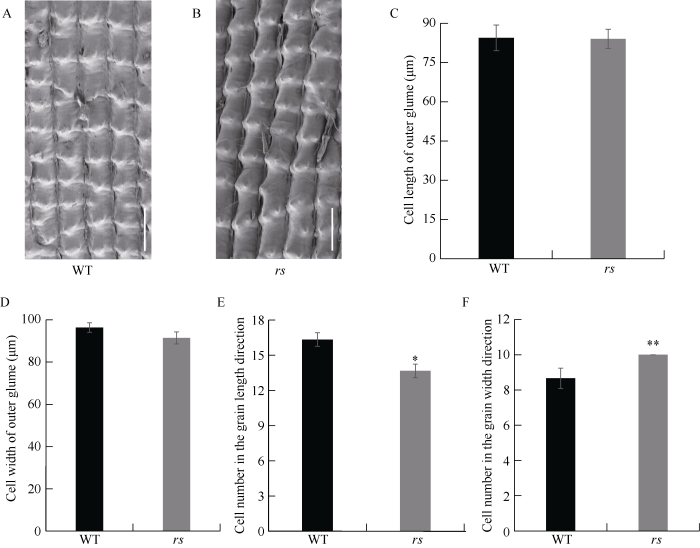
水稻籽粒大小通常由颖壳细胞大小和细胞数目的多少决定。为了明确导致rs圆粒表型的原因, 对rs和野生型成熟籽粒颖壳的外表皮细胞进行扫描电镜观察(图2-A, B)。相比野生型, rs的细胞长度与宽度无显著差异(图2-C, D), 然而, 突变体rs和野生型细胞数目差异显著, 即纵向rs细胞数目减少约16.33% (图2-E), 横向rs细胞数目增加约15.38% (图2-F)。综上, rs籽粒变圆的原因主要是细胞数目改变。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2颖壳组织学分析
A, B: 扫描电镜观察颖壳外表皮。比例尺为200 μm。C, D: 颖壳外表皮细胞长度与宽度的比较。E, F: 颖壳外表皮细胞数目的比较。 *和**分别表示在P= 0.05与P=0.01水平上差异显著。
Fig. 2Histological analysis of spikelet hulls
A, B; scanning electron microscope analysis of the outer surfaces of glumes. Bar=200 μm. C, D: comparison of cell length and width in outer glumes. E, F: comparison of cell number in outer glumes. Data are given as means ± SD. *P = 0.05; **P = 0.01.
2.4 细胞周期相关基因表达分析
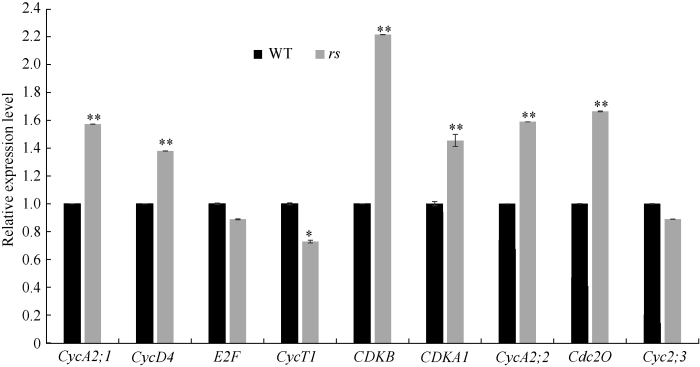
细胞分裂是指活细胞为增殖其数量由一个细胞分裂为两个细胞的过程, 细胞数目的变化通常与细胞分裂的改变有关。为确定突变体rs细胞数目的变化是否与细胞分裂相关, 利用qRT-PCR分析细胞周期相关基因在野生型和rs中的表达水平。结果发现, 相比野生型, 多数与细胞周期相关的基因在突变体中显著上调, 如CycA2;1、CycD4、CDKB、CDKA1、CycA2;2、cdc20 (图3)。综上, 突变体rs圆粒可能是与细胞周期的变化有关。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3细胞周期相关基因(CycA2;1、CycD4、E2F2、CycT1、CDKB、CDKA1、CycA2;2、Cdc20和CycA2;3)在野生型与rs中的表达分析
数据结果为3次生物重复平均值, *和**分别表示在P = 0.05与0.01水平上差异显著。
Fig. 3Expression levels of cell cycle-related genesCycA2;1, CycD4, E2F2, CycT1, CDKB, CDKA1, CycA2;2, Cdc20, and CycA2;3
Values are mean±SD (n = 3). * and ** indicate significant differences between WT and rs at P = 0.05 and P = 0.01 by Student’s t-test.
2.5 基因定位
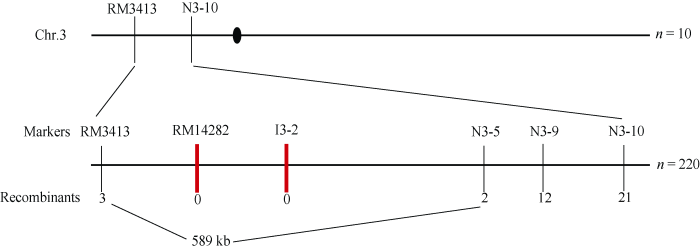
为分离rs基因, 构建rs和“滇粳优”籼粳杂交F2群体, 利用10个具有典型突变性状的F2单株和覆盖水稻12条染色体的SSR标记进行初步连锁分析, 将突变基因初步定位于第3染色体标记RM3413和N3-10之间, 两者物理距离约为1116 kb。而后利用220个F2突变单株并开发新的InDel标记对目的基因进行精细定位。最终将目标基因定位在标记RM3413和N3-5间, 物理距离约589 kb (图4)。根据Gramane和RiceData等网站预测, 该区间内共有85个开放阅读框(ORF)分别为β-膨胀素前体、编码假定的蛋白、编码类似于EL2蛋白、编码含有同源框结构域的蛋白质、编码类似于转录因子同系物BTF3样蛋白质、编码肽酶、编码胰蛋白酶样丝氨酸和半胱氨酸结构域蛋白以及编码未知功能的DUF791家族蛋白等。在该区间内未见已克隆的与籽粒大小相关基因的报道。因此, 推测RS可能是与粒型相关的新基因。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4RS基因的精细定位
黑色圆圈表示着丝粒; 粗竖线表示共分离标记; 标记下方的数字为极端个体的数目。
Fig. 4Fine mapping of the RS gene
The black circle means the centromere; the thick vertical line means a co-segregate marker; the number under the markers means the number of extreme individuals.
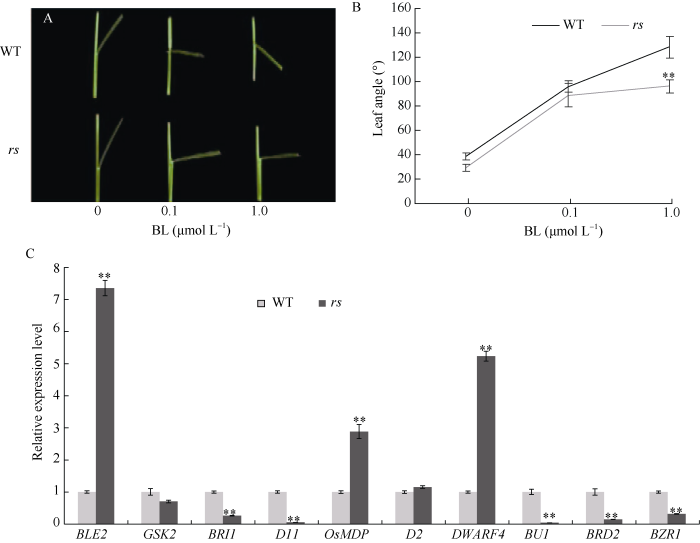
2.6 油菜素内酯(BR)处理
为检测rs是否与油菜素内酯合成或信号转导途径相关, 利用油菜素内酯2,4-epiBL处理野生型和rs幼苗来检测其对BR敏感性。由图5-A可以看出, 野生型的叶夹角随着BR浓度升高呈增大趋势且增大幅度明显, 当2,4-epiBL的浓度达到1 μmol L-1时, 叶夹角为128.5°, 表现为下垂状态。然而, 随2,4-epiBL的浓度升高, 突变体rs叶夹角的增大趋势减缓, 当2,4-epiBL的浓度达到1 μmol L-1时, 叶夹角为96.25°, 表现为敏感性降低(图5-B)。另外, 利用qRT-PCR分析与油菜素内酯合成和信号转导相关基因在野生型和rs中的表达水平表明, 多数与BR合成和信号相关基因(D11、BRD2、BR11、BU1、BZR1)的表达呈下调趋势(图5-C), 而油菜素内酯合成限速酶基因OsDWARF4、油菜素内酯信号相关基因OsBLE2、OsMDP1受到负反馈调节, 其表达呈显著上调。由此推断, RS的突变影响BR合成途径和信号途径。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5野生型和突变体rs 对2,4-epiBL处理的响应
A: 不同2,4-epiBL浓度处理, 上面为野生型, 下面为rs。B: 不同2,4-epiBL浓度处理叶夹角的变化。C: 野生型和rs中BR合成和信号转导途径基因的表达水平。**分别代表突变体与野生型之间差异达0.01显著水平。
Fig. 5Response test to 2,4-epiBL in the wild type and rs
A: performance of lamina joint to 2,4-epiBL in wild-type (up) and rs (down). B: changes of lamina inclination to 2,4-epiBL. C: expression levels of BR synthesis and signal transduction pathway genes in the wild type and rs. ** Significant difference at the 0.01 level between wild type and rs.
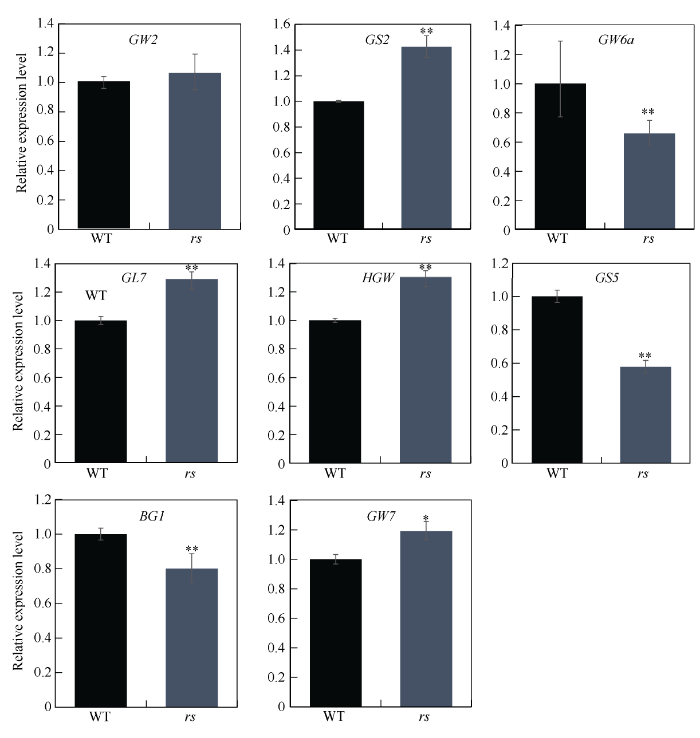
2.7 粒型基因的表达分析
水稻粒型受复杂分子网络的调控, 目前在水稻中已经报道了一些参与调控籽粒大小的基因, 如GW2、GS2、GW6a、GL7、HGW、GS5、BG1、GW7。为探究RS与这些粒型调控因子的相互关系, 我们对这些基因在野生型与突变体rs中的表达进行了qRT-PCR分析。结果发现, 相比野生型, GS2、GL7、GW7等基因在rs中表达上调, GW6a、GS5、BG1等基因表达下调(图6), 这暗示RS的突变影响了粒型相关基因的表达。 RS调控水稻粒型的具体分子机制将是后续研究的重点内容。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6粒型相关基因在野生型和突变体rs中的表达
*与**分别表示在P= 0.05与0.01水平上差异显著。*P= 0.05; **P =0.01.
Fig. 6Expression levels of grain shape-related genes in wild type (WT) and rs
3 讨论
本研究通过EMS诱变的方法从“宁粳3号”为背景的突变体库中筛选到一个籽粒变圆的突变体rs。相关研究指出器官大小由细胞数目和细胞大小共同决定, 与之对应的是两个基本过程, 即细胞增殖和细胞扩张[20,21]。而细胞增殖和细胞扩张与植物体内激素含量相关。调控水稻粒重的TGW6编码IAA-葡萄糖水解酶, 通过控制IAA供应影响合胞体到细胞化阶段的转变, 从而限制了细胞数目和籽粒长度[22]。BG1是生长素的原初响应基因, 参与调节生长素的转运, 是生长素响应和转运的正调控因子, 是植物特异的控制器官大小的调节因子[14]。OsBRI1是一个BR信号受体, 通过控制细胞分裂和伸长, 调控器官发育[23]。油菜素内酯是一种新型植物内源激素, 是第一个被分离出的具有活性的油菜素甾族化合物(brassinosteroids, BRs), 在自然界中已发现超过70种BR[24], 目前育种研究工作者已筛选出一些参与BR调控途径相关的突变体[25,26]。相比野生型, rs对外源BR敏感性降低, BR信号相关基因BRI1、BZR1与BU1在突变体中显著下调, 而BR合成基因DWARF4受负反馈调节而表达上调, 说明RS的突变影响了植株体内正常的BR信号转导过程。细胞学观察显示细胞数目的改变是导致rs籽粒变圆的原因, 遗传分析表明该突变受隐性单基因控制, 并将rs精细定位于第3染色体短臂末端, 位于标记RM3413和N3-5之间大约589 kb的区间内, 而第3染色体上已报道的控制粒型的相关基因GS3、VLN2、sd37等均不在该区间内, 提示RS可能是一个新的控制粒型的基因。植物激素对种子大小起着直接或间接的作用, 尤其是油菜素内酯和生长素。如OsBRI1是一个BR信号受体, 在植物生长和发育中起着重要作用, 通过控制细胞分裂和伸长, 调控器官发育[27]。D11参与油菜素内酯生物合成, 过表达D11会导致粒长、粒宽增加, 同时会引起发育的花药和种子中糖的积累[28]。OsBZR1是油菜素内酯信号传导的下游信号分子, 干扰抑制OsBZR1表达, 导致转基因水稻产生矮秆、直立叶的表型, BR敏感性降低且BR应答基因表达发生改变[29]。因此, 对RS基因进行克隆和功能研究, 可能为阐明BR调控水稻籽粒大小的分子网络提供新的线索。
RS可能影响GS5与BG1的表达, 来控制水稻籽粒大小。GS5是籽粒大小的正向调控因子, 其突变会产生小粒[7]。BG1参与生长素的转运, 正向调控籽粒大小, 其功能性获得突变体粒长和粒宽分别增加15.2%和17.0%[14]。相比野生型, 粒型基因GS5与BG1的表达水平在突变体中显著下调。突变体rs的表型可能与GS5和BG1表达下调有关。因此, 需要进一步克隆RS基因, 并进行功能分析, 阐述RS的分子调控网络, 为调控水稻粒型的分子育种提供基础。
4 结论
鉴定了一个圆粒突变体rs, 相比野生型, rs籽粒变圆, 而其他农艺性状无显著差异。rs对外源BR处理敏感性降低。细胞数目的改变是导致rs籽粒变圆的原因。rs突变是由1对隐性基因控制,该基因被定位在第3染色体标记RM3413和N3-5之间, 共包含80个ORFs。rs突变影响BR合成途径和信号途径, 改变了粒型相关基因的表达。致谢:农业农村部长江中下游粳稻生物学与遗传育种重点实验室、长江流域杂交水稻协同创新中心、江苏省现代作物生产中心。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1146/annurev-arplant-042809-112209URLPMID:20192739 [本文引用: 1]

Abstract Grain yield in rice is a complex trait multiplicatively determined by its three component traits: number of panicles, number of grains per panicle, and grain weight; all of which are typical quantitative traits. The developments in genome mapping, sequencing, and functional genomic research have provided powerful tools for investigating the genetic and molecular bases of these quantitative traits. Dissection of the genetic bases of the yield traits based on molecular marker linkage maps resolved hundreds of quantitative trait loci (QTLs) for these traits. Mutant analyses and map-based cloning of QTLs have identified a large number of genes required for the basic processes underlying the initiation and development of tillers and panicles, as well as genes controlling numbers and sizes of grains and panicles. Molecular characterization of these genes has greatly advanced the mechanistic understanding of the regulation of these rice yield traits. These findings have significant implications in crop genetic improvement.
DOI:10.1073/pnas.1014419107URL [本文引用: 1]
DOI:10.1038/ng2014URLPMID:17417637 [本文引用: 1]

Abstract Grain weight is one of the most important components of grain yield and is controlled by quantitative trait loci (QTLs) derived from natural variations in crops. However, the molecular roles of QTLs in the regulation of grain weight have not been fully elucidated. Here, we report the cloning and characterization of GW2, a new QTL that controls rice grain width and weight. Our data show that GW2 encodes a previously unknown RING-type protein with E3 ubiquitin ligase activity, which is known to function in the degradation by the ubiquitin-proteasome pathway. Loss of GW2 function increased cell numbers, resulting in a larger (wider) spikelet hull, and it accelerated the grain milk filling rate, resulting in enhanced grain width, weight and yield. Our results suggest that GW2 negatively regulates cell division by targeting its substrate(s) to proteasomes for regulated proteolysis. The functional characterization of GW2 provides insight into the mechanism of seed development and is a potential tool for improving grain yield in crops.
DOI:10.1016/j.molp.2015.07.002URLPMID:26187814 [本文引用: 1]

Grain size determines grain weight and affects grain quality.GS2, a dominant QTL, encodes Growth-Regulating Factor 4 (OsGRF4), which acts as a transcriptional regulator. A mutation in this gene at the OsmiR396c target site results in elevated transcript levels ofGS2and accumulation ofGS2leads to enlarged cell size and increased cell number, which in turn results in enhanced grain weight and yield.
DOI:10.1038/nplants.2017.64URLPMID:28481332 [本文引用: 1]

The reason why African cultivated rice has smaller grains than its wild progenitor has remained puzzling.
DOI:10.1038/ng.3346URLPMID:26147619 [本文引用: 1]

Copy number variants (CNVs) are associated with changes in gene expression levels and contribute to various adaptive traits. Here we show that a CNV at the Grain Length on Chromosome 7 (GL7) locus contributes to grain size diversity in rice (Oryza sativa L.). GL7 encodes a protein homologous to Arabidopsis thaliana LONGIFOLIA proteins, which regulate longitudinal cell elongation. Tandem duplication of a 17.1-kb segment at the GL7 locus leads to upregulation of GL7 and downregulation of its nearby negative regulator, resulting in an increase in grain length and improvement of grain appearance quality. Sequence analysis indicates that allelic variants of GL7 and its negative regulator are associated with grain size diversity and that the CNV at the GL7 locus was selected for and used in breeding. Our work suggests that pyramiding beneficial alleles of GL7 and other yield- and quality-related genes may improve the breeding of elite rice varieties.
DOI:10.1093/jxb/erv058URLPMID:25711711 [本文引用: 2]

Two SNPs in the promoter ofGS5are responsible for expression variation controlling grain size. Enhanced expression of GS5 competitively inhibits the interaction between OsBAK1 and OsMSBP1, promoting grain size. Grain weight is a major determinant of grain yield.GS5is a positive regulator of grain size such that grain width, filling, and weight are correlated with its expression level. Previous work suggested that polymorphisms ofGS5in the promoter region might be responsible for the variation in grain size. In this study, two single nucleotide polymorphisms (SNPs) between the wide-grain alleleGS5-1and the narrow-grain alleleGS5-2in the upstream region of the gene that were responsible for the differential expression in developing young panicles were identified. These two polymorphs altered the responses of theGS5alleles to abscisic acid (ABA) treatments, resulting in higher expression ofGS5-1than ofGS5-2in developing young panicles. It was also shown that SNPs in light-responsive elements of the promoter altered the response to light induction, leading to higher expression ofGS5-2thanGS5-1in leaves. Enhanced expression of GS5 competitively inhibits the interaction between OsBAK1-7 and OsMSBP1 by occupying the extracellular leucine-rich repeat (LRR) domain of OsBAK1-7, thus preventing OsBAK1-7 from endocytosis caused by interacting with OsMSBP1, providing an explanation for the positive association between grain size andGS5expression. These results advanced our understanding of the molecular mechanism by whichGS5controls grain size.
DOI:10.1371/journal.pone.0034231URLPMID:22457828 [本文引用: 1]

Heading date and grain weight are two determining agronomic traits of crop yield. To date, molecular factors controlling both heading date and grain weight have not been identified. Here we report the isolation of a hemizygous mutation, heading and grain weight (hgw), which delays heading and reduces grain weight in rice. Analysis of hgw mutant phenotypes indicate that the hemizygous hgw mutation decreases latitudinal cell number in the lemma and palea, both composing the spikelet hull that is known to determine the size and shape of brown grain. Molecular cloning and characterization of the HGW gene showed that it encodes a novel plant-specific ubiquitin-associated (UBA) domain protein localized in the cytoplasm and nucleus, and functions as a key upstream regulator to promote expressions of heading date- and grain weight-related genes. Moreover, co-expression analysis in rice and Arabidopsis indicated that HGW and its Arabidopsis homolog are co-expressed with genes encoding various components of ubiquitination machinery, implying a fundamental role for the ubiquitination pathway in heading date and grain weight control.
DOI:10.1038/ng.220URLPMID:18820698 [本文引用: 1]

Abstract Grain-filling, an important trait that contributes greatly to grain weight, is regulated by quantitative trait loci and is associated with crop domestication syndrome. However, the genes and underlying molecular mechanisms controlling crop grain-filling remain elusive. Here we report the isolation and functional analysis of the rice GIF1 (GRAIN INCOMPLETE FILLING 1) gene that encodes a cell-wall invertase required for carbon partitioning during early grain-filling. The cultivated GIF1 gene shows a restricted expression pattern during grain-filling compared to the wild rice allele, probably a result of accumulated mutations in the gene's regulatory sequence through domestication. Fine mapping with introgression lines revealed that the wild rice GIF1 is responsible for grain weight reduction. Ectopic expression of the cultivated GIF1 gene with the 35S or rice Waxy promoter resulted in smaller grains, whereas overexpression of GIF1 driven by its native promoter increased grain production. These findings, together with the domestication signature that we identified by comparing nucleotide diversity of the GIF1 loci between cultivated and wild rice, strongly suggest that GIF1 is a potential domestication gene and that such a domestication-selected gene can be used for further crop improvement.
DOI:10.1038/ng.2612URLPMID:23583977 [本文引用: 1]

Increases in the yield of rice, a staple crop for more than half of the global population, are imperative to support rapid population growth. Grain weight is a major determining factor of yield. Here, we report the cloning and functional analysis of THOUSAND-GRAIN WEIGHT 6 (TGW6), a gene from the Indian landrace rice Kasalath. TGW6 encodes a novel protein with indole-3-acetic acid (IAA)-glucose hydrolase activity. In sink organs, the Nipponbare tgw6 allele affects the timing of the transition from the syncytial to the cellular phase by controlling IAA supply and limiting cell number and grain length. Most notably, loss of function of the Kasalath allele enhances grain weight through pleiotropic effects on source organs and leads to significant yield increases. Our findings suggest that TGW6 may be useful for further improvements in yield characteristics in most cultivars.
DOI:10.1038/ng.2327URLPMID:22729225 [本文引用: 1]

Abstract Grain size and shape are important components of grain yield and quality and have been under selection since cereals were first domesticated. Here, we show that a quantitative trait locus GW8 is synonymous with OsSPL16, which encodes a protein that is a positive regulator of cell proliferation. Higher expression of this gene promotes cell division and grain filling, with positive consequences for grain width and yield in rice. Conversely, a loss-of-function mutation in Basmati rice is associated with the formation of a more slender grain and better quality of appearance. The correlation between grain size and allelic variation at the GW8 locus suggests that mutations within the promoter region were likely selected in rice breeding programs. We also show that a marker-assisted strategy targeted at elite alleles of GS3 and OsSPL16 underlying grain size and shape can be effectively used to simultaneously improve grain quality and yield.
DOI:10.1038/nplants.2017.43URLPMID:28394310 [本文引用: 1]

Abstract Grain size is a major determinant of grain yield in cereal crops. qSW5/GW5, which exerts the greatest effect on rice grain width and weight, was fine-mapped to a 2,263-bp/21-kb genomic region containing a 1,212-bp deletion, respectively. Here, we show that a gene encoding a calmodulin binding protein, located 5 b downstream of the 1,212-bp deletion, corresponds to qSW5/GW5. GW5 is expressed in various rice organs, with highest expression level detected in young panicles. We provide evidence that the 1,212-bp deletion affects grain width most likely through influencing the expression levels of GW5. GW5 protein is localized to the plasma membrane and can physically interact with and repress the kinase activity of rice GSK2 (glycogen synthase kinase 2), a homologue of Arabidopsis BIN2 (BRASSINOSTEROID INSENSITIVE2) kinase, resulting in accumulation of unphosphorylated OsBZR1 (Oryza sativa BRASSINAZOLE RESISTANT1) and DLT (DWARF AND LOW-TILLERING) proteins in the nucleus to mediate brassinosteroid (BR)-responsive gene expression and growth responses (including grain width and weight). Our results suggest that GW5 is a novel positive regulator of BR signalling and a viable target for genetic manipulation to improve grain yield in rice and perhaps in other cereal crops as well.
DOI:10.1105/tpc.014712URLPMID:14615594 [本文引用: 1]

We characterized a rice dwarf mutant, ebisu dwarf (d2). It showed the pleiotropic abnormal phenotype similar to that of the rice brassinosteroid (BR)-insensitive mutant, d61. The dwarf phenotype of d2 was rescued by exogenous brassinolide treatment. The accumulation profile of BR intermediates in the d2 mutants confirmed that these plants are deficient in late BR biosynthesis. We cloned the D2 gene by map-based cloning. The D2 gene encoded a novel cytochrome P450 classified in CYP90D that is highly similar to the reported BR synthesis enzymes. Introduction of the wild D2 gene into d2-1 rescued the abnormal phenotype of the mutants. In feeding experiments, 3-dehydro-6-deoxoteasterone, 3-dehydroteasterone, and brassinolide effectively caused the lamina joints of the d2 plants to bend, whereas more upstream compounds did not cause bending. Based on these results, we conclude that D2/CYP90D2 catalyzes the steps from 6-deoxoteasterone to 3-dehydro-6-deoxoteasterone and from teasterone to 3-dehydroteasterone in the late BR biosynthesis pathway.
DOI:10.1073/pnas.1512748112URLPMID:26283354 [本文引用: 3]

Grain size is one of the key factors determining grain yield. However, it remains largely unknown how grain size is regulated by developmental signals. Here, we report the identification and characterization of a dominant mutant big grain1 (Bg1-D) that shows an extra-large grain phenotype from our rice T-DNA insertion population. Overexpression of BG1 leads to significantly increased grain size, and the severe lines exhibit obviously perturbed gravitropism. In addition, the mutant has increased sensitivities to both auxin and N-1-naphthylphthalamic acid, an auxin transport inhibitor, whereas knockdown of BG1 results in decreased sensitivities and smaller grains. Moreover, BG1 is specifically induced by auxin treatment, preferentially expresses in the vascular tissue of culms and young panicles, and encodes a novel membrane-localized protein, strongly suggesting its role in regulating auxin transport. Consistent with this finding, the mutant has increased auxin basipetal transport and altered auxin distribution, whereas the knockdown plants have decreased auxin transport. Manipulation of BG1 in both rice and Arabidopsis can enhance plant biomass, seed weight, and yield. Taking these data together, we identify a novel positive regulator of auxin response and transport in a crop plant and demonstrate its role in regulating grain size, thus illuminating a new strategy to improve plant productivity.
DOI:10.1007/s11103-004-1178-yURLPMID:15604699 [本文引用: 1]

Calcium-dependent protein kinases (CDPKs) play an important role in rice signal transduction, but the precise role of each individual CDPK is still largely unknown. Recently, a full-length cDNA encoding OsCDPK13 from rice seedling was isolated. To characterize the function of OsCDPK13, its responses to various stresses and hormones were analyzed in this study. OsCDPK13 accumulated in 2-week-old leaf sheath and callus, and became phosphorylated in response to cold and gibberellin (GA). OsCDPK13 gene expression and protein accumulation were up-regulated in response to GA 3 treatment, but suppressed in response to abscisic acid and brassinolide. Antisense OsCDPK13 transgenic rice lines were shorter than the vector control lines, and the expression of OsCDPK13 was lower in dwarf mutants of rice than in wild type. Furthermore, OsCDPK13 gene expression and protein accumulation were enhanced in response to cold, but suppressed under salt and drought stresses. Sense OsCDPK13 transgenic rice lines had higher recovery rates after cold stress than vector control rice. The expression of OsCDPK13 was stronger in cold-tolerant rice varieties than in cold-sensitive ones. The results suggest that OsCDPK13 might be an important signaling component in the response of rice to GA and cold stress.
[本文引用: 1]
DOI:10.1111/tpj.13025URLPMID:26366992 [本文引用: 1]

Summary Grain size is an important agronomic trait in determining grain yield. However, the molecular mechanisms that determine the final grain size are not well understood. Here, we report the functional analysis of a rice ( Oryza sativa L.) mutant, dwarf and small grain1 ( dsg1 ), which displays pleiotropic phenotypes, including small grains, dwarfism and erect leaves. Cytological observations revealed that the small grain and dwarfism of dsg1 were mainly caused by the inhibition of cell proliferation. Map-based cloning revealed that DSG1 encoded a mitogen-activated protein kinase (MAPK), OsMAPK6. OsMAPK6 was mainly located in the nucleus and cytoplasm, and was ubiquitously distributed in various organs, predominately in spikelets and spikelet hulls, consistent with its role in grain size and biomass production. As a functional kinase, OsMAPK6 interacts strongly with OsMKK4, indicating that OsMKK4 is likely to be the upstream MAPK kinase of OsMAPK6 in rice. In addition, hormone sensitivity tests indicated that the dsg1 mutant was less sensitive to brassinosteroids (BRs). The endogenous BR levels were reduced in dsg1 , and the expression of several BR signaling pathway genes and feedback-inhibited genes was altered in the dsg1 mutant, with or without exogenous BRs, indicating that OsMAPK6 may contribute to influence BR homeostasis and signaling. Thus, OsMAPK6, a MAPK, plays a pivotal role in grain size in rice, via cell proliferation, and BR signaling and homeostasis.
[本文引用: 1]
DOI:10.1007/s11105-015-0962-yURL [本文引用: 1]

Floral organ development is fundamentally important to plant reproduction and seed quality, yet its underlying regulatory mechanisms are still largely unknown, especially in crop plants. In this study
DOI:10.1007/s10265-005-0232-4URLPMID:16284709 [本文引用: 1]

Size is an important parameter in the characterization of organ morphology and function. To understand the mechanisms that control leaf size, we previously isolated a number of Arabidopsis thaliana mutants with altered leaf size. Because leaf morphogenesis depends on determinate cell proliferation, the size of a mature leaf is controlled by variation in cell size and number. Therefore, leaf-size mutants should be classified according to the effects of the mutations on the cell number and/or size. A group of mutants represented by angustifolia3 / grf-interacting factor1 and aintegumenta exhibits an intriguing cellular phenotype termed compensation: when the leaf cell number is decreased due to the mutation, the leaf cell size increases, leading to compensation in leaf area. Several lines of genetic evidence suggest that compensation is probably not a result of the uncoupling of cell division from cell growth. Rather, the evidence suggests an organ-wide mechanism that coordinates cell proliferation with cell expansion during leaf development. Our results provide a key, novel concept that explains how leaf size is controlled at the organ level.
[本文引用: 1]
DOI:10.1038/ng.2612URLPMID:23583977 [本文引用: 1]

Increases in the yield of rice, a staple crop for more than half of the global population, are imperative to support rapid population growth. Grain weight is a major determining factor of yield. Here, we report the cloning and functional analysis of THOUSAND-GRAIN WEIGHT 6 (TGW6), a gene from the Indian landrace rice Kasalath. TGW6 encodes a novel protein with indole-3-acetic acid (IAA)-glucose hydrolase activity. In sink organs, the Nipponbare tgw6 allele affects the timing of the transition from the syncytial to the cellular phase by controlling IAA supply and limiting cell number and grain length. Most notably, loss of function of the Kasalath allele enhances grain weight through pleiotropic effects on source organs and leads to significant yield increases. Our findings suggest that TGW6 may be useful for further improvements in yield characteristics in most cultivars.
URL [本文引用: 1]
DOI:10.1016/j.plaphy.2007.01.002URLPMID:17346983 [本文引用: 1]

Brassinosteroids represent a class of plant hormones. More than 70 compounds have been isolated from plants. Currently 42 brassinosteroid metabolites and their conjugates are known. This review describes the miscellaneous metabolic pathways of brassinosteroids in plants. There are some types of metabolic processes involving brassinosteroids in plants: dehydrogenation, demethylation, epimerization, esterification, glycosylation, hydroxylation, side-chain cleavage and sulfonation. Metabolism of brassinosteroids can be divided into two categories: i) structural changes to the steroidal skeleton; and ii) structural changes to the side-chain.
DOI:10.1046/j.1365-313X.2002.01438.xURL [本文引用: 1]
DOI:10.1111/tpj.12071URLPMID:23146214 [本文引用: 1]

The phytohormones auxins and brassinosteroids are both essential regulators of physiological and developmental processes, and it has been suggested that they act inter-dependently and synergistically. In rice (Oryza sativa), auxin co-application improves the brassinosteroid response in the rice lamina inclination bioassay. Here, we showed that auxins stimulate brassinosteroid perception by regulating the level of brassinosteroid receptor. Auxin treatment increased expression of the rice brassinosteroid receptor gene OsBRI1. The promoter of OsBRI1 contains an auxin-response element (AuxRE) that is targeted by auxin-response factor (ARF) transcription factors. An AuxRE mutation abolished the induction of OsBRI1 expression by auxins, and OsBRI1 expression was down-regulated in an arf mutant. The AuxRE motif in the OsBRI1 promoter, and thus the transient up-regulation of OsBRI1 expression caused by treatment with indole-3-acetic acid, is essential for the indole-3-acetic acid-induced increase in sensitivity to brassinosteroids. These findings demonstrate that some ARFs control the degree of brassinosteroid perception required for normal growth and development in rice. Although multi-level interactions between auxins and brassinosteroids have previously been reported, our findings suggest a mechanism by which auxins control cellular sensitivity to brassinosteroids, and further support the notion that interactions between auxins and brassinosteroids are extensive and complex.
DOI:10.1016/j.devcel.2016.06.011URLPMID:27424498 [本文引用: 1]

The hormones abscisic acid (ABA) and brassinosteroid (BR) exhibit antagonistic interactions during plant development. Gui et al. now identify rice OsREM4.1 as a link between the ABA and BR signaling pathways. OsREM4.1, which is transcriptionally upregulated by ABA, inhibits the formation and activation of a BR receptor kinase (OsBRI1-OsSERK1) complex.
DOI:10.1111/tpj.12820URLPMID:25754973 [本文引用: 1]

Summary Transport of photoassimilates from leaf tissues (source regions) to the sink organs is essential for plant development. Here, we show that a phytohormone, the brassinosteroids (BRs) promotes pollen and seed development in rice by directly promoting expression of Carbon Starved Anther ( CSA ) which encodes a MYB domain protein. Over-expression of the BR-synthesis gene D11 or a BR-signaling factor OsBZR1 results in higher sugar accumulation in developing anthers and seeds, as well as higher grain yield compared with control non-transgenic plants. Conversely, knockdown of D11 or OsBZR1 expression causes defective pollen maturation and reduced seed size and weight, with less accumulation of starch in comparison with the control. Mechanically, OsBZR1 directly promotes CSA expression and CSA directly triggers expression of sugar partitioning and metabolic genes during pollen and seed development. These findings provide insight into how BRs enhance plant reproduction and grain yield in an important agricultural crop.
DOI:10.1073/pnas.0706386104URL [本文引用: 1]
