 ,1,*
,1,*Genetic Mapping of Bruchid Resistance Gene in Mungbean V1128
LIU Chang-You1, SU Qiu-Zhu1, FAN Bao-Jie1, CAO Zhi-Min1, ZHANG Zhi-Xiao1, WU Jing2, CHENG Xu-Zhen2, TIAN Jing ,1,*
,1,*通讯作者:
第一联系人:
收稿日期:2018-05-8接受日期:2018-08-20网络出版日期:2018-09-18
| 基金资助: |
Received:2018-05-8Accepted:2018-08-20Online:2018-09-18
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (544KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘长友, 苏秋竹, 范保杰, 曹志敏, 张志肖, 武晶, 程须珍, 田静. 栽培绿豆V1128抗豆象基因定位[J]. 作物学报, 2018, 44(12): 1875-1881. doi:10.3724/SP.J.1006.2018.01875
LIU Chang-You, SU Qiu-Zhu, FAN Bao-Jie, CAO Zhi-Min, ZHANG Zhi-Xiao, WU Jing, CHENG Xu-Zhen, TIAN Jing.
绿豆(Vigna radiata)作为一种医食同源的作物, 在健康饮食和膳食结构调整中发挥着越来越重要的作用[1]。近年来, 我国绿豆品种的产量和品质都有大幅提高[2]。然而, 绿豆储藏过程中的豆象危害却一直是困扰农民和商家的主要难题。
豆象是危害绿豆等食用豆类作物的严重仓储害虫。危害绿豆的豆象种类主要有绿豆象(Callosobruchus chinensis)、四纹豆象(Callosobruchus maculates)、鹰嘴豆象(Callosobruchus analis)、巴西豆象(Zabrotes subfasciatus)和灰豆象(Callosobruchus phaseoli)等, 其中在我国存在的主要是绿豆象和四纹豆象, 且绿豆象对生产的危害最严重[3,4], 若处置不及时, 可在3~6个月内造成整仓绿豆全部受损[5,6]。生产上防治豆象危害一般采用磷化铝熏蒸法, 这种处理方法存在农药残留超标和造成环境污染的隐患, 不符合安全生产的目标。培育抗豆象绿豆品种, 利用作物本身的抗性是一种最经济有效的防治豆象危害的手段。然而, 以杂交选育为主要手段的常规育种途径存在选择周期长、抗豆象鉴定工作繁琐等诸多问题。对抗豆象基因进行定位, 开拓抗豆象育种方法, 利用分子手段辅助抗豆象育种, 成为当前十分紧迫的研究课题。
到目前为止, 筛选鉴定出的抗豆象绿豆种质资源主要有TC1966、ACC41、V2802、V2709、V1128和V2817 6份[7,8,9,10]。其中TC1966和ACC41是2份野生绿豆资源。研究表明, TC1966的抗豆象基因Br和ACC41的抗豆象基因Br1均为单显性主基因遗传[8,11-12], 但它们的豆象抗性还可能存在其他微效基因的修饰作用[13]。Talekar等[9]和Somta等[14]先后发现栽培绿豆V2802和V2709高抗绿豆象和四纹豆象。Somta等[10]发现另外2份栽培绿豆V1128和V2817对绿豆象和四纹豆象具有完全抗性。利用不同类型的分子标记和作图群体, TC1966的Br基因和ACC41的Br1基因已经被定位在不同的连锁标记之间[12,15-18]。孙蕾等[19]以V2709为研究材料, 将其抗豆象基因Br2定位在1个RAPD和1个STS标记之间。Chotechung等[20]和Kaewwongwal等[21]分别将V2802和V2709的抗豆象基因定位在绿豆第5染色体上, 并预测VrPGIP2可能是V2802的抗豆象候选基因, 而VrPGIP1和VrPGIP2可能是V2709的抗豆象候选基因。
在抗豆象育种中, 应用抗豆象野生种容易引入一些不利性状, 如蔓生、炸荚等。加强对栽培抗豆象绿豆资源的研究利用可以有效避免这些问题。V1128是对绿豆象和四纹豆象均具有完全抗性的栽培绿豆资源, 在人工种子试验中表现出比V2817更强的抗性[10]。因此, V1128在绿豆抗豆象育种中具有更大的潜力。然而, 到目前为止, V1128抗豆象基因的遗传定位研究尚无人开展。对其进行抗豆象基因定位有利于补充对目前尚不明晰的绿豆抗豆象基因的认识, 并加强其在抗豆象育种中的应用。因此, 本研究利用感豆象亲本冀绿7号和抗豆象亲本V1128杂交组配获得的F2群体对V1128抗豆象基因进行遗传定位分析, 为V1128及其他绿豆抗豆象资源抗性基因来源及基因克隆提供参考。
1 材料与方法
1.1 试验材料
抗豆象材料V1128由泰国农业大学Peerasak Srinnives教授惠赠, 该材料籽粒无光泽; 感豆象材料为河北省农林科学院粮油作物研究所育成品种冀绿7号, 该材料籽粒有光泽。以冀绿7号作母本, V1128作父本, 杂交获得F1。籽粒无光泽相对于籽粒有光泽为显性, 因此可以从F1植株所收获籽粒判断是否为真杂交种。选择来源于同一F1植株的种子繁殖F2分离群体, 共获得158个单株, 用于抗豆象遗传分析和构建遗传连锁图谱。1.2 DNA提取
从每个F2单株取幼嫩的三出复叶, 立即投入液氮, -80°C保存, 以备提取DNA。利用植物基因组提取试剂盒(TIANGEN, 北京)提取DNA。使用1%琼脂糖凝胶检测DNA质量。利用超微量核酸蛋白检测仪(Nanodrop ND-2000, 美国)测定DNA浓度, 并稀释到10 ng μL-1。1.3 抗豆象鉴定及遗传分析
从每个鉴定单株随机选取90粒健康种子, 分成3个重复, 每个重复30粒, 分别放入直径5 cm、高1.8 cm的圆形小塑料盒中。同时, 分别放入抗豆象亲本V1128和感豆象亲本冀绿7号作为对照。将小塑料盒并排放入大塑料箱内(66 cm×40 cm×15 cm), 每个大塑料箱中放入刚羽化1~3 d的成虫作为虫源(平均每份被鉴定材料3~8对), 至每粒种子着卵量3~5粒时, 除去成虫; 养虫室温度(29±2)℃, 湿度60%~70%。待感虫亲本完全被蛀食后(约40~60 d), 调查每份材料的受害粒数。3个重复取平均值用于后续数据分析。参照Young等[15]和Kaewwongwal等[21]的豆象抗性分级方法, 种子被害率0~80%视为抗, 其中0~20%视为纯合抗性, 21%~80%视为杂合抗性; 种子被害率81%~100%视为纯合感豆象。利用Microsoft Excel 2010进行卡方测验, 检测F2群体的抗感分离比是否符合3 (抗)∶1 (感)的遗传分离规律, 利用SPSS 16.0统计分析软件计算F2群体种子被害率的频率分布。
1.4 DNA标记分析
分别选择10个完全抗豆象和10个完全感豆象F2单株的DNA混合组成抗感池。利用两亲本和抗感池进行多态性分子标记筛选, 选择在两亲本间表现多态且抗池带型与V1128相同, 感池带型与冀绿7号相同的标记用于后续F2群体的基因型分析。PCR扩增及电泳分析参照Liu等[22]的方法。在亲本和抗感池间共计筛选了3767个不同类型的分子标记, 包括SSR、EST-SSR、STS和Indel标记。最终获得在两亲本间表现多态且抗池带型与V1128相同, 感池带型与冀绿7号相同的分子标记31个, 其中VRID1和VRID5为InDel标记, 其余均为SSR标记, 多态性标记信息见表1。
Table 1
表1
表1多态性标记信息
Table 1
| 标记名称 Marker | 标记类型 Type | 引物序列 Primer sequence (5′-3′) | 退火温度 Tm | 文献来源 Reference |
|---|---|---|---|---|
| Mchr5-17 | SSR | F: GCTTGCTTATGCTCAAAACT; R: TACAGATAAACCCAAGCCAT | 55 | [23] |
| Mchr5-20 | SSR | F: TCAAAACTTTCACTGGACCT; R: GCTGTTTGTCACATGCATAA | 55 | [23] |
| Mchr5-23 | SSR | F: ATCTTAATCCCATCCTTGGT; R: AACTGGCTTGTAAGGTGAGA | 55 | [23] |
| Mchr5-25 | SSR | F: CCGTTGTGAATCAACTTTTC; R: GAGTCGTCGTGTAATCCTTC | 55 | [23] |
| Mchr5-32 | SSR | F: ACTTGTAGGTGGAAGAGATGA; R: GATTAAGGGCGTGTTTTGT | 55 | [23] |
| Mchr5-33 | SSR | F: CTAATGAAACAGGACAAGGG; R: CTCACTCTTCTCATTCCACC | 55 | [23] |
| Mchr5-36 | SSR | F: ACCTACTGATTGGTGTTTGG; R: CAGTGAATGCTGACAGTGAC | 55 | [23] |
| Mchr5-37 | SSR | F: AACGGTTGGAGTTAGGAAAT; R: TGAACATCACCAACTATCAC | 55 | [23] |
| Mchr3-85 | SSR | F: AACAGCGTTGATTTATGGAC; R: AATCATGTTGGTGTGTTGTG | 55 | [23] |
| Mchr3-86 | SSR | F: TGCCTAAGGGTCAATTTCTA; R: ATGCACCAGAAGACAAAAAC | 55 | [23] |
| Mchr3-87 | SSR | F: AATGAGGTAATGCAGAGGTG; R: GACAAGGGTTGTTGTTCACT | 55 | [23] |
| Mchr3-89 | SSR | F: GAATTAAAGCCCTTGTTCTG; R: GGTGAATTTTCTGTTTCCAC | 55 | [23] |
| Mchr3-96 | SSR | F: CTGATGCTATTTCCATCCAT; R: TAACCTTTTGCATTTGGTGC | 55 | [23] |
| MUS150 | SSR | F: GCTGTTTGTCACATGCATAA; R: TCAAAACTTTCACTGGACCT | 55 | [23] |
| MUS365 | SSR | F: TGAGCCCGATTTTTATCTC;R: GCTCACAGATAACTGACACAAC | 55 | [23] |
| MUS569 | SSR | F: GGAGGGGATTTTTAAGATTG; R: GGATACGATTTTGTCGTGTT | 55 | [23] |
| HAAS_VR_379 | SSR | F: CCTATCCGAATCGACACCAC; R: GTAGCAATAGCAGCCCAAGG | 55 | [22] |
| HAAS_VR_106 | SSR | F: ACGGCTATTCATCGTTTTGC; R: CAACCCGAAGCCAAAAACTA | 55 | [22] |
| HAAS_VR_89 | SSR | F: GCTGGAAGGATCCAATTTCA; R: TCGCCATTCCCAAGATAAAG | 55 | [22] |
| HAAS_VR_1701 | SSR | F: CCGGGGTGAAATTGATACAC; R: CAAAGGGGCTATGAACAGGA | 55 | [22] |
| VRID1 | Indel | F: TCGGTTTCAGCTCGATAGATTC; R: GATGTTGTCTGAAGTAGTGGTA | 55 | [21] |
| VRID5 | Indel | F: AGAATAAAATGAATCTAGAAGACCA; R: TGAATTAATTTTCTTACCCTTGT | 50 | [21] |
| VRBR-SSR016 | SSR | F: GACGGCTAGGTACAACACTGC; R: TTTAGAGCAATTGGGTGGATTT | 55 | [20] |
| VRBR-SSR024 | SSR | F: TTTTGTGGACACTCCTTCCA; R: AAGCGTCACACCCTCAATTC | 55 | [20] |
| VRBR-SSR030 | SSR | F: CAGCTAGGAAACTCACCAAACC; R: CAGCTGGCAGGTGAAATATG | 55 | [20] |
| VRBR-SSR032 | SSR | F: GGTTATTTTGATCTAAAGGGCCA; R: GTGTGAGAAGATTTGGGAATGTAA | 55 | [20] |
| VRBR-SSR033 | SSR | F: CTCAAGTCTTATGTTTCCCCCTAT; R: GCACTAAAGGACTTTCCTTGAAC | 55 | [20] |
| VRBR-SSR039 | SSR | F: AAGTTGGTGTAGCACTTGCAGA; R: AATGAATTAAAAGAAAACACTACTG | 55 | [20] |
| GBssr-MB87 | SSR | F: TCCCTTGTGGGAGATCCT; R: CTTTGCCACACTCCTTGC | 55 | [24] |
| DMB158 | SSR | F: TGGAAAATTTGCAGCAGTTG; R: ATTGATGGAGGGCGGAAGTA | 55 | [25] |
| PVBR201 | SSR | F: GTGATGGTGCTGCTTTTCAA; R: ATGCGTGGGGAGAAGTAAGA | 50 | [26] |
新窗口打开|下载CSV
1.5 抗豆象基因定位
利用QTL IciMapping 4.0构建连锁图谱[27]。在LOD=5.0时进行标记分组, 利用Kosambi作图方法计算相邻标记间的遗传距离[28]。采用两种方法定位抗豆象基因, 第一种, 将抗豆象性状作为质量性状, 根据上述分级方法分类, 纯合抗豆象记作“A”, 纯合感豆象记作“B”, 杂合抗豆象记作“H”, 由此, 将抗豆象基因“Br3”作为一个表型标记构建到连锁图中, 对其进行连锁定位; 第二种, 将抗豆象性状作为数量性状, 以F2群体的种子被害率定位QTL。QTL作图方法为完备区间作图法(ICIM)[29], QTL的显著性LOD阈值在P=0.01时作10 000次置换试验(Permutation test)确定, 其他参数设置采用QTL IciMapping 4.0软件的默认参数。2 结果与分析
2.1 亲本及F2群体的豆象抗性鉴定与遗传分析
经抗豆象鉴定, 亲本V1128表现为完全抗豆象(种子被害率=0), 而亲本冀绿7号表现为完全感豆象(种子被害率=100%)。F2群体的抗性频率分布趋向于二项分布(图1), 抗感分离比为117 (抗)∶41 (感), 卡方测验符合3 (抗)∶1 (感)的分离规律, χ2=0.034 (χ20.05, v=1 = 3.84)。将37份种子被害率0~20%的材料视为纯合抗性个体, 80份种子被害率21%~80%的材料视为杂合抗性个体, 41份种子被害率81%~100%视为纯合感豆象个体, 卡方测验符合1∶2∶1的分离规律, χ2 = 0.228 (χ20.05, v=2 = 5.99)。说明抗豆象亲本V1128对绿豆象的抗性由具有主效作用的显性单基因控制。延续前人的命名规则, 将该基因暂命名为“Br3”。图1
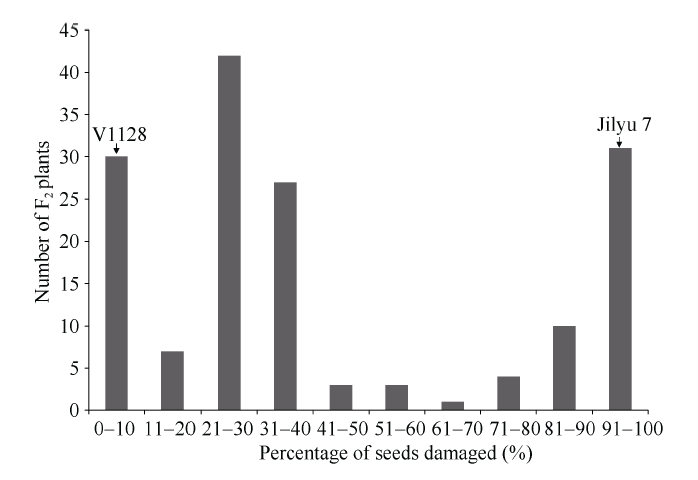 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1F2群体种子被害率分布图
Fig. 1Distribution of seed damage rate in F2 population
2.2 基因定位
利用31个多态性分子标记对158个F2单株进行基因型分析, 获得每个个体的基因型数据。在将抗豆象性状作为质量性状的条件下, 按照显性单基因的定位方法, 抗豆象基因Br3作为一个表型标记与31个多态性分子标记共同进行标记间连锁分析。在LOD=5.0时, 所有31个分子标记和表型标记Br3均位于同一分组内, 形成长度为73.9 cM的连锁群。在该连锁群中, 抗豆象基因Br3被定位于标记DMB158和VRBR-SSR033 (标记VRID5、VRBR-SSR032与VRBR-SSR033的连锁群位置相同)之间, 两侧遗传距离分别为4.4 cM和5.8 cM (图2)。图2
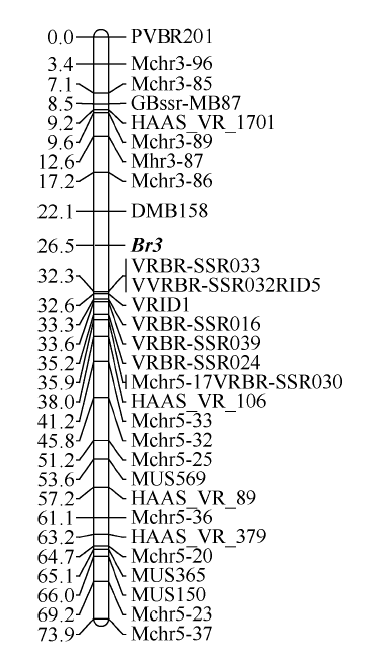 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2抗豆象基因Br3连锁定位分析
利用QTL IciMapping 4.0软件进行标记连锁分析, LOD=5; 连锁群左右两侧分别标示图距(单位为cM)和标记名称。
Fig. 2Positioning analysis of the bruchids resistance gene Br3
Linkage map constructed by using QTL IciMapping 4.0 software, LOD=5; map distances (cM) and marker names are shown on the left and right sides of the linkage group, respectively.
利用F2群体的分子标记基因分型数据单独进行连锁图谱构建, 在LOD=5.0时, 形成1个长度为65.7 cM的连锁群。将抗豆象性状作为数量性状, 采用完备区间作图法(ICIM)对种子被害率进行QTL定位, 在该连锁群上检测到1个主效QTL, 位于标记DMB158和VRID5 (标记VRBR-SSR032、VRBR-SSR033与VRID5的连锁群位置相同)之间, LOD值为38.04, 可以解释表型变异(PVE)的71.64%, 其加性效应和显性效应值分别为57.78和22.92 (图3)。来自父本V1128的等位基因具有明显减少种子被害率的效应。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3抗豆象QTL qBr3的定位图谱
利用QTL IciMapping 4.0软件的完备区间作图法(ICIM)进行QTL 定位分析; 连锁群左右两侧分别标示图距(单位为cM)和标记名称; 竖线标示LOD阈值(3.18)。
Fig. 3Location of QTL qBr3 controlling resistance to bruchids
QTL mapping based on inclusive composite interval mapping method (ICIM) using QTL IciMapping 4.0 software; map distances (cM) and marker names are shown on the left and right sides of the linkage group, respectively; the vertical line shows the LOD threshold (3.18).
为了将qBr3与其他研究的抗豆象QTL定位结果进行比较, 利用7个共有标记将V1128抗豆象基因的QTL分析结果与V2709的QTL进行共线性分析, 所有7个共有标记均表现出极好的共线性。本研究中定位的V1128的抗豆象QTL qBr3与V2709的抗豆象位点QTL qBr5.1位于临近的标记区间内(图4), 2个QTL间存在部分重叠。
图4
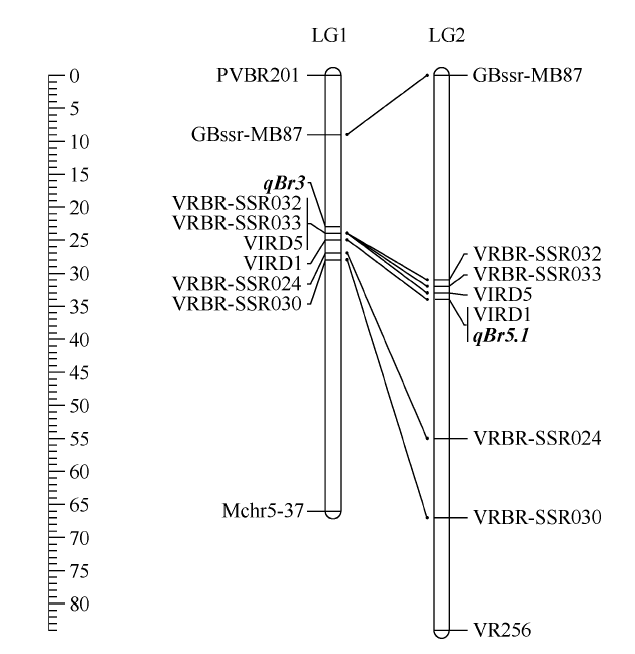 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4QTL定位结果共线性比较分析
基于7个共有标记, 利用MapChart 2.2软件[30]作本研究与Kaewwongwal等[21]构建的连锁图谱的共线性图谱; 左侧标尺标示连锁群长度, 单位为cM; “LG1”为本研究构建的连锁图谱, “LG2”为Kaewwongwal等构建的连锁图谱; QTL名字用斜体字标示。
Fig. 4Collinearity comparison analysis of QTL mapping results
A synteny map between the genetic linkage map from this study and the linkage map constructed by Kaewwongwal et al. [21], this map was drown using MapChart 2.2 software [30] based on seven common markers; the scale bars on the left of the figure indicate the length of each linkage group in cM; “LG1” is the linkage group constructed in this study, “LG2” was constructed by Kaewwongwal et al.; the names of QTLs are shown in italics.
为了确定Br3基因的染色体位置, 利用其两侧标记的引物序列与已经公布的绿豆基因组数据库[31]进行BLAST分析。DMB158位于绿豆第5染色体(Chr5: 5 597 658… 5 597 891), VRID5位于绿豆第5染色体(Chr5: 5 410 272… 5 410 493), VRBR-SSR032位于绿豆第5染色体(Chr5: 5 310 107…5 310 281), VRBR-SSR033位于绿豆第5染色体(Chr5: 5 380 081…5 380 180)。若取4个标记间的最远距离(VRBR-SSR032和DMB158之间), 可以将Br3定位在绿豆第5染色体约288 kb的物理区间内(5 310 107… 5 597 891)。在此区间内共有11个注释基因(表2), 其中有5个基因编码RD22类蛋白, 2个基因编码多聚半乳糖醛酸酶抑制剂类蛋白, 3个基因编码未知蛋白; LOC106760238则为假基因。
Table 2
表2
表2V1128抗豆象基因位点范围内基因注释
Table 2
| 基因序号 Gene ID | 基因位置 Gene location | 基因注释 Gene annotation |
|---|---|---|
| LOC106760111 | 5 338 753…5 340 435 | RD22类蛋白 RD22-like protein |
| LOC106761406 | 5 346 646…5 349 613 | 未知蛋白 Uncharacterized protein |
| LOC106760112 | 5 361 741…5 362 985 | 未知蛋白 Uncharacterized protein |
| LOC106761824 | 5 367 108…5 367 592 | RD22类蛋白 RD22-like protein |
| LOC106761559 | 5 389 064…5 390 972 | RD22类蛋白 RD22-like protein |
| LOC111241568 | 5 403 161…5 404 109 | 未知蛋白 Uncharacterized protein |
| LOC106760880 | 5 409 223…5 413 311 | RD22类蛋白 RD22-like protein |
| LOC106761219 | 5 511 814…5 514 459 | RD22类蛋白 RD22-like protein |
| LOC106760236 | 5 561 973…5 563 546 | 多聚半乳糖醛酸酶抑制剂类蛋白(VrPGIP1) Polygalacturonase inhibitor-like (VrPGIP1) |
| LOC106760237 | 5 590 847…5 591 984 | 多聚半乳糖醛酸酶抑制剂类蛋白(VrPGIP2) Polygalacturonase inhibitor-like (VrPGIP2) |
| LOC106760238 | 5 590 527…5 598 757 | 假基因 Pseudogene |
新窗口打开|下载CSV
3 讨论
从第一份抗豆象野生绿豆资源TC1966被发现以来, 研究者就没有停止过对不同抗豆象资源的遗传规律、基因定位、抗豆象机制等的研究。大量的研究表明, 不同抗豆象绿豆资源中均存在主效的显性抗豆象基因[8,11-12]。同TC1966、ACC41、V2802、V2709的抗豆象遗传规律相似, V1128的抗豆象特性也由主效的显性单基因控制。但从不同抗豆象单株的种子被害率存在差异、抗豆象性状表现出一定程度的数量性状遗传特征来看, 很可能也存在其他微效基因起修饰作用。这与Chen等[13]对TC1966抗豆象遗传规律的分析结论相同。基于抗豆象绿豆资源中存在主效抗性单显性基因, 可以将抗豆象性状作为质量性状定位, 如Young等[15]、Kaga等[12]、Wang等[18]、孙蕾等[19]在对TC1966、ACC41、V2709的抗豆象基因定位研究中均采用了此策略。同时, 抗豆象性状表现出一定程度的数量性状遗传特征, 因此可以将其按照数量性状QTL定位的方法定位。如Chotechung等[20]、Kaewwongwal等[21]、Mei等[17]、Hong等[32]、Chen等[13]均采用数量性状QTL定位的方法, 根据种子被害率对V2802、V2709、ACC41、TC1966的抗豆象性状进行QTL定位。本研究同时采用上述2种方法均将Br3定位在相同的标记区间内。2种方法的结果可以相互验证, 从而增加定位的准确度。
V1128抗豆象基因Br3所在的物理区间(5 310 107… 5 597 891)包含在Kaga等[12]对TC1966抗豆象基因Br的定位标记区间内(Bng143~Bng110, 物理区间为Chr.5: 5 052 031…6 779 027)。同时, 该区间包含Chotechung等[20]对V2802抗豆象基因定位的标记区间(VrSSR013~ DMB158, 物理区间为Chr.5: 5 561 387…5 597 891), 而与Kaewwongwal等[21]对V2709抗豆象基因定位的标记区间(VRID5~VRBR-SSR037, 物理区间为Chr.5: 5 410 272… 5 647 621)存在大部分重叠。表明这些不同的抗豆象种质可能具有相同或紧密连锁的抗豆象基因。.
Chotechung等[20]在V2802的抗豆象分离后代群体中发现标记DMB158与抗豆象性状共分离, 该标记位于LOC106760237 (VrPGIP2)基因内部, 认为VrPGIP2基因很可能是V2802的抗豆象候选基因。另外一个与VrPGIP2紧密连锁的基因LOC106760236 (VrPGIP1)与抗豆象性状不存在共分离, 认为VrPGIP1不是抗豆象基因。然而Liu等[33]在NM92×TC1966的重组自交系群体中发现DMB158与抗豆象性状不完全连锁, 表明VrPGIP2不是TC1966的抗豆象基因。根据绿豆基因组注释结果, Kaewwongwal等[21]确定V2709的定位区间内共有8个注释基因, 其中包括VrPGIP1和VrPGIP2。VrPGIP1和VrPGIP2蛋白同属于多聚半乳糖醛酸酶抑制剂类蛋白, 研究表明该类蛋白在植物抵御微生物病原体侵染及昆虫危害中起作用。如菜豆的PvPGIP基因在抗菌核病中发挥作用[34]; 菜豆PvPGIP3/PvPGIP4蛋白可以抑制盲蝽象的多聚半乳糖醛酸酶[35]。因此, 预测VrPGIP1和VrPGIP2可能是V2709的抗豆象候选基因。本研究构建的连锁图谱与Kaewwongwal等[21]构建的连锁图谱具有很好的共线性。但在本研究中DMB158与抗豆象性状之间并不存在共分离。因此, VrPGIP2可能不是V1128的抗豆象基因。在Br3定位区间内的11个注释基因中, 5个为RD22类蛋白编码基因。研究表明RD22类蛋白可能与植物抵御生物或非生物胁迫有关。如拟南芥的RD22蛋白参与耐脱水胁迫过程[36]。然而, 不能排除这类基因在抗豆象过程中发挥作用的可能性。同样, 在11个注释基因中还有3个未知蛋白的编码基因, 它们是否在抗豆象过程中起作用还无法判断。因此, 目前还不能仅仅依靠定位区间内基因功能分析的结果来判断V1128抗豆象基因的归属。有待进一步利用较大的近等基因系群体, 从中寻找交换单株, 对抗豆象基因Br3精细定位。
致谢:
对泰国农业大学Peerasak Srinnives教授提供抗豆象栽培绿豆种质V1128表示感谢。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.fct.2014.11.024URLPMID:25533792 [本文引用: 1]

Mung bean has been traditionally and widely used as an edible and medicinal plant in the South and Southeast Asia. Bruchid resistance mung bean has more potential in commercial use, but scarcely been evaluated for safety through standard in vivo toxicological studies. In the present study, subchronic oral toxicity studies of bruchid-resistant mung bean were designed and conducted in Sprague-Dawley (SD) rats for 90 days. During the subchronic oral toxicity study, no mortality and toxicologically significant changes in clinical signs, food consumption, opthalmoscopic examination, hematology, clinical biochemistry, macroscopic findings, organ weights and histopathological examination were noted in animal administered diet containing bruchid-resistant mung bean. These results demonstrated that bruchid resistant mung bean is as safe as conventional mung bean.
DOI:10.3864/j.issn.0578-1752.2009.05.003URL [本文引用: 1]

As a traditional crop, mungbean (Vigna radiata L.) is widely cultivated in China. It is also important in modern planting systems because it can improve the environments of soils by nitrogen fixation. However, comparing to the major crops, the genetic study of mungbean is lagged, which has limited the intensive development of this crop. In the paper, studies on breeding and genetics of mungbean both in China and the world were reviwed, the main aim is to supply useful information so as to improve the genetic study and accelerate the development of mungbean in China.
DOI:10.3864/j.issn.0578-1752.2009.05.003URL [本文引用: 1]

As a traditional crop, mungbean (Vigna radiata L.) is widely cultivated in China. It is also important in modern planting systems because it can improve the environments of soils by nitrogen fixation. However, comparing to the major crops, the genetic study of mungbean is lagged, which has limited the intensive development of this crop. In the paper, studies on breeding and genetics of mungbean both in China and the world were reviwed, the main aim is to supply useful information so as to improve the genetic study and accelerate the development of mungbean in China.
DOI:10.3969/j.issn.1672-1810.2001.04.003URL [本文引用: 1]

绿豆象(Callos0bruchun chinensis L.)是豇豆属豆类作物重要的仓库害虫.本研究通过抗豆象杂交育种后代VC1973A/TC1966 F1、F2和VC1973A/(VC1973A/TC1966 F2)BC1F1及TC1966/(VC1973A/TC1966 F2)BC1F1分离群体的遗传分析,发现绿豆抗豆象性状符合3:1的遗传分离规律,证明绿豆对豆象的抗性由1对显性单基因(Aa)控制,抗虫性为显性(A),感虫为隐性(a).
DOI:10.3969/j.issn.1672-1810.2001.04.003URL [本文引用: 1]

绿豆象(Callos0bruchun chinensis L.)是豇豆属豆类作物重要的仓库害虫.本研究通过抗豆象杂交育种后代VC1973A/TC1966 F1、F2和VC1973A/(VC1973A/TC1966 F2)BC1F1及TC1966/(VC1973A/TC1966 F2)BC1F1分离群体的遗传分析,发现绿豆抗豆象性状符合3:1的遗传分离规律,证明绿豆对豆象的抗性由1对显性单基因(Aa)控制,抗虫性为显性(A),感虫为隐性(a).
URL [本文引用: 1]

Mungbean (Vigna radiata), adzuki bean (Vigna angularis) and cowpea (Vigna unguiculata ) are main cultivated Vigna food legumes in China. Bruchids are serious devastating stored pests of those grain legumes. Breeding for bruchid-resistant cultivars is the best way to avoid the harm of bruchids. However, comparing to the major crops, genetic study of these grain legumes are lagged behind in China, which has constrained the intensive development of breeding for bruchid-resistant cultivars. In this paper, studies on bruchid-resistant germplasm collection, genetics and breeding of mungbean, adzuki bean and cowpea were reviewed both in China and the world, in order to supply useful information to improve the genetic study and accelerate the development of breeding for bruchid-resistant cultivars in China.
URL [本文引用: 1]

Mungbean (Vigna radiata), adzuki bean (Vigna angularis) and cowpea (Vigna unguiculata ) are main cultivated Vigna food legumes in China. Bruchids are serious devastating stored pests of those grain legumes. Breeding for bruchid-resistant cultivars is the best way to avoid the harm of bruchids. However, comparing to the major crops, genetic study of these grain legumes are lagged behind in China, which has constrained the intensive development of breeding for bruchid-resistant cultivars. In this paper, studies on bruchid-resistant germplasm collection, genetics and breeding of mungbean, adzuki bean and cowpea were reviewed both in China and the world, in order to supply useful information to improve the genetic study and accelerate the development of breeding for bruchid-resistant cultivars in China.
DOI:10.1023/A:1003906715119URL [本文引用: 1]

A species level germplasm collection representing 76% of known taxa in the genus Vigna subgenus Ceratotropis was evaluated for resistance to two species of bruchid beetles, Callosobruchus chinensis and C. maculatus . Seven taxa consisting of 29 accessions were found to be resistant to C. chinensis and 4 taxa consisting of 24 accessions were found to be resistant to C. maculatus . This compared with no resistant accessions being found in several hundred landrace accessions of mungbean, V. radiata var. radiata , in the same subgenus. Sometimes resistance was found in all accessions of a particular taxon, such as complete resistance to both C. chinensis and C. macualtus in V. umbellata . Other taxa showed intra taxon variation for resistance such as V. reflexo-pilosa and V. minima . The levels and patterns of resistance among taxa were diverse. The results suggest that various factors cause resistance to bruchid in the subgenus Ceratotropis . While the number of eggs laid on seeds generally reflected seed size, one small seeded cultivar of V. mungo var. mungo , black gram, had an unusually high number of eggs laid per seed. No correlation was found between seed size and levels of resistance. The species level germplasm collection, which reflects the core collection concept in trying to maximize genetic diversity in a limited number of accessions, has enabled a large number of potentially useful sources of resistance to bruchid beetles to be found efficiently.
DOI:10.3969/j.issn.0529-1542.2004.06.008URL [本文引用: 1]

Callosobruchus chinensis is the major store pest of adzuki bean. The author investigated the insect in adzuki bean fields through sampling. The results showed that the insect laid eggs on pods at the initial filling stage of grains. The peak ovipositional period covers the first twenty days of September. The eggs then hatched into larvae and intruded into adzuki bean grains. The spatial distribution of the egg-laying pattern was aggregation distribution caused by environmental factors. The best time for controlling egg laying and egg development is Sept 10-15.
DOI:10.3969/j.issn.0529-1542.2004.06.008URL [本文引用: 1]

Callosobruchus chinensis is the major store pest of adzuki bean. The author investigated the insect in adzuki bean fields through sampling. The results showed that the insect laid eggs on pods at the initial filling stage of grains. The peak ovipositional period covers the first twenty days of September. The eggs then hatched into larvae and intruded into adzuki bean grains. The spatial distribution of the egg-laying pattern was aggregation distribution caused by environmental factors. The best time for controlling egg laying and egg development is Sept 10-15.
[本文引用: 1]
[本文引用: 3]
DOI:10.1093/jee/85.4.1150URL [本文引用: 2]

ABSTRACT The characteristics of Callosobruchus chinensis (L.) resistance in two mungbean, Vigna radiata (L.) Wilczek, and one black gram accession, Vigna mungo (L.) Hepper, were investigated in a series of laboratory tests. Oviposition and emergence of first-generation adults of C. chinensis were significantly reduced with the pods of two mungbean accessions, V2709 and V2802, compared with the susceptible check VC 1973A. Smaller seed size of two resistant mungbean accessions and one resistant black gram accession, VM 2164, was not responsible for the resistance. Artificial seeds wereprepared from mixtures of seeds of resistant and susceptible accessions. As the concentration of resistant accessions increased, the number of C. chinensis adults emerging after feeding as larvae in such artificial seeds decreased. Adults that emerged from the artificial seeds made from mixtures of resistant and susceptible accessions were significantly lighter than from artificial seeds made from the susceptible accession alone. Results indicate the possible presence of antibiotic factors in resistant accessions.
[本文引用: 3]
[本文引用: 2]
DOI:10.1007/s004380050744URLPMID:9648742 [本文引用: 5]

Abstract Bruchid resistance, controlled by a single dominant gene (Br) in a wild mungbean accession (TC1966), has been incorporated into cultivated mungbean (Vigna radiata). The resistance gene simultaneously confers inhibitory activity against the bean bug, Riptortus clavatus Thunberg (Hemiptera: Alydidae). The resultant isogenic line (BC20 generation) was characterized by the presence of a group of novel cyclopeptide alkaloids, called vignatic acids. A linkage map was constructed for Br and the vignatic acid gene (Va) using restriction fragment length polymorphism (RFLP) markers and a segregating BC20F2 population. By screening resistant and susceptible parental lines with 479 primers, eight randomly amplified polymorphic DNA (RAPD) markers linked to Br were identified and cloned for use as RFLP probes. All eight RAPD-based markers, one mungbean, and four common bean genomic clones were effectively integrated around Br within a 3.7-cM interval. Br was mapped to a 0.7-cM segment between a cluster consisting of six markers and a common bean RFLP marker, Bng110. The six markers are closest to the bruchid resistance gene, approximately 0.2 cM away. The vignatic acid gene, Va, cosegregated with bruchid resistance. However, one individual was identified in the BC20F2 population that retained vignatic acids in spite of its bruchid susceptibility. Consequently, Va was mapped to a single locus at the same position as the cluster of markers and 0.2 cM away from Br. These results suggest that the vignatic acids are not the principal factors responsible for bruchid resistance in V. radiata but will facilitate the use of map-based cloning strategies to isolate the Br gene.
DOI:10.1007/s10681-012-0831-9URL [本文引用: 3]

Mungbean yellow mosaic Indian virus (MYMIV) and bruchid infestation are severe production constraints of mungbean in South Asia, a major global mungbean production area. Marker-assisted selection for resistance against these disorders while maintaining or even improving agronomic traits is an important step toward breeding elite mungbean varieties. This study employed recombinant inbred lines (F12) derived from a cross between MYMIV-tolerant Vigna radiata NM92 and bruchid-resistant V. radiata ssp. sublobata TC1966 to identify chromosomal locations associated with disease and insect pest resistance and seed traits. A linkage map comprising 11 linkage groups was constructed with random amplified polymorphic DNA (RAPD), sequence characterized amplified regions (SCAR), cleaved amplified polymorphic DNA (CAP), amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR) markers. Quantitative trait loci (QTLs) for MYMIV and bruchid resistance, 100 seed weight and seed germination rate were identified. Three major QTLs for MYMIV and one major bruchid resistance locus were mapped on LG 9. The resistance alleles were contributed by the MYMIV tolerant parent NM92 and the bruchid resistant parent TC1966 respectively. One of the MYMIV QTLs was tightly linked in repulsion phase to the bruchid resistance locus. In addition, three minor QTLs for MYMIV resistance were found, where the resistance alleles were contributed by TC1966. Lines combining MYMV resistance alleles from both parents have greater resistance to MYMIV than the tolerant parent. Two minor bruchid resistance QTLs were identified in TC1966. Furthermore, three QTLs each for 100 seed weight and germination rate were detected. The markers defining the QTLs identified in this study will be useful in marker-assisted breeding of improved mungbean varieties in the future.
DOI:10.1007/s10681-006-9299-9URL [本文引用: 1]

Bruchid beetles or seed weevils are the most devastating stored pests of grain legumes causing considerable loss to mungbean ( Vigna radiata (L.) Wilczek). Breeding for bruchid resistance is a major goal in mungbean improvement. Few sources of resistance in cultivated genepool were identified and characterized, however, there has been no study on the genetic control of the resistance. In this study, we investigated the inheritance of seed resistance to Callosobruchus chinensis (L.) and C. maculatus (F.) in two landrace mungbean accessions, V2709BG and V2802BG. The F 1 , F 2 and BC generations were developed from crosses between the resistant and susceptible accessions and evaluated for resistance to the insects. It was found that resistance to bruchids in seeds is controlled by maternal plant genotype. All F 1 plants derived from both direct and reciprocal crosses exhibited resistance to the bruchids. Segregation pattern of reaction to the beetles in the F 2 and backcross populations showed that the resistance is controlled by a major gene, with resistance is dominant at varying degrees of expressivity. Although the presence of modifiers was also observed. The gene is likely the same locus in both V2709BG and V2802BG. The resistant gene is considered very useful in breeding for seed resistance to bruchids in mungbean.
DOI:10.1007/BF00227394URLPMID:24201484 [本文引用: 3]

Abstract Bruchids (genus Callosobruchus) are among the most destructive insect pests of mungbeans and other members of the genus, Vigna. Genetic resistance to bruchids was previously identified in a wild mungbean relative, TC1966. To analyze the underlying genetics, accelerate breeding, and provide a basis for map-based cloning of this gene, we have mapped the TC1966 bruchid resistance gene using restriction fragment length polymorphism (RFLP) markers. Fifty-eight F2 progeny from a cross between TC1966 and a susceptible mungbean cultivar were analyzed with 153 RFLP markers. Resistance mapped to a single locus on linkage group VIII, approximately 3.6 centimorgans from the nearest RFLP marker. Because the genome of mungbean is relatively small (estimated to be between 470 and 560 million base pairs), this RFLP marker may be suitable as a starting point for chromosome walking. Based on RFLP analysis, an individual was also identified in the F2 population that retained the bruchid resistance gene within a tightly linked double crossover. This individual will be valuable in developing resistant mungbean lines free of linkage drag.
DOI:10.1007/s00122-004-1821-7URLPMID:15490104

Bacterial artificial chromosome (BAC) libraries have been widely used in different aspects of genome research. In this paper we report the construction of the first mungbean ( Vigna radiata L. Wilczek) BAC libraries. These BAC clones were obtained from two ligations and represent an estimated 3.5 genome equivalents. This correlated well with the screening of nine random single-copy restriction fragment length polymorphism probes, which detected on average three BACs each. These mungbean clones were successfully used in the development of two PCR-based markers linked closely with a major locus conditioning bruchid ( Callosobruchus chinesis ) resistance. These markers will be invaluable in facilitating the introgression of bruchid resistance into breeding programmes as well as the further characterisation of the resistance locus.
DOI:10.1139/G09-031URLPMID:19767890 [本文引用: 1]

Abstract Bruchids (Coleoptera: Bruchidae) can cause serious damage to mungbean and several other leguminous crops and there is a strong association between small seed size and bruchid resistance. In investigating the feasibility of breeding large-seeded cultivars with high levels of bruchid resistance, we studied the relationship between these two traits by QTL analysis. A major locus conferring resistance to Callosobruchus chinensis was identified from a wild mungbean genotype, 'ACC41' (belonging to Vigna radiata var. sublobata), collected in Australia. The proportion of the C. chinensis resistance response that could be attributed to this single QTL varied among four different resistance assays. The highest value reached was 98.5%, suggesting that bruchid resistance in this genotype is likely conditioned by this single locus. The QTL was robust and its detection was not affected by the use of different sources of the insect, different lengths and conditions of seed storage, or different bruchid resistance assay methods. This bruchid resistance QTL was coincident with one of the loci conferring seed mass detected from the three seed sources produced in Australia. However, such a co-location was not detected for the seed source produced in China. Covariance analysis revealed a complex relationship between seed mass and bruchid resistance. Nevertheless, the effect of the bruchid resistance QTL remained highly significant for all four assays after the effect of seed mass was accounted for. These results, together with the relationship between the bruchid resistance QTL identified in this study and a second one detected previously in a wild mungbean genotype from Madagascar, are discussed.
DOI:10.1016/j.cj.2016.06.010URL [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s00122-016-2731-1URLPMID:27220975 [本文引用: 4]

Abstract KEY MESSAGE: The Br locus confers bruchid resistance in mungbean; VrPGIP2 (encoding a polygalacturonase inhibitor) is a strong candidate gene for this resistance. The VrPGIP2 sequence differs between resistant and susceptible lines. Azuki bean weevil (Callosobruchus chinensis) and cowpea weevil (Callosobruchus maculatus) are serious insect pests of mungbean during storage. Bruchid resistance in mungbean is controlled by a single dominant locus, Br. Although the Br locus has been located on a genetic map, molecular basis and function of the gene remain unknown. In this study, high-resolution mapping using a BC11F2 population of 418 plants derived from a cross between 'Kamphaeng Saen 1' (KPS1; susceptible) and 'V2802' (resistant) using simple sequence repeat (SSR) markers delimited the Br locus to a genomic region of 38 Kb of chromosome 5 containing two annotated genes. EST-SSR marker DMB-SSR158 co-segregated perfectly with the Br locus. Bioinformatics analyses revealed that DMB-SSR158 corresponds to a gene encoding a polygalacturonase inhibitor (polygalacturonase-inhibiting protein PGIP) and was designated as VrPGIP2. Comparison of VrPGIP2 coding sequences between four bruchid-resistant (V2802, V1128, V2817 and TC1966) and four bruchid-susceptible (KPS1, Sulu-1, CM and an unknown accession) mungbean lines revealed six single nucleotide polymorphisms (SNPs) between the resistant and susceptible groups. Three of the six SNPs resulted in amino acid changes; namely, alanine (A) to serine (S) at position 320, leucine (L) to proline (P) at position 332, and threonine (T) to P at position 335 of the VrPGIP2 sequence in resistant lines, compared with that in susceptible lines. The A to S change at position 320 may affect the interaction between PGIP and polygalacuronase. These results indicate that VrPGIP2 is very likely the gene at the Br locus responsible for bruchid resistance in mungbean.
DOI:10.3389/fpls.2017.01692URLPMID:29033965 [本文引用: 8]

Nearly all mungbean cultivars are completely susceptible to seed bruchids (Callosobruchus chinensisandCallosobruchus maculatus). Breeding bruchid-resistant mungbean is a major goal in mungbean breeding programs. Recently, we demonstrated in mungbean (Vigna radiata) accessionV2802thatVrPGIP2, which encodes a polygalacturonase inhibiting protein (PGIP), is theBrlocus responsible for resistance toC. chinensisandC. maculatus. In this study, mapping in mungbean accessionV2709using a BC11F2population of 355 individuals revealed that a single major quantitative trait locus, which controlled resistance to bothC. chinensisandC. maculatus, was located in a 237.35 Kb region of mungbean chromosome 5 that contained eight annotated genes, includingVrPGIP1(LOC106760236) andVrPGIP2(LOC106760237).VrPGIP1andVrPGIP2are located next to each other and are only 27.56 Kb apart. SequencingVrPGIP1andVrPGIP2in “V2709” revealed new alleles for both VrPGIP1 and VrPGIP2, namedVrPGIP1-1andVrPGIP2-2, respectively.VrPGIP2-2has one single nucleotide polymorphism (SNP) at position 554 of wild typeVrPGIP2. This SNP is a guanine to cystine substitution and causes a proline to arginine change at residue 185 in the VrPGIP2 of “V2709”.VrPGIP1-1has 43 SNPs compared with wild type and “V2802”, and 20 cause amino acid changes in VrPGIP1. One change is threonine to proline at residue 185 in VrPGIP1, which is the same as in VrPGIP2. Sequence alignments of VrPGIP2 and VrPGIP1 from “V2709” with common bean (Phaseolus vulgaris) PGIP2 revealed that residue 185 in VrPGIP2 and VrPGIP1 contributes to the secondary structures of proteins that affect interactions between PGIP and polygalacturonase, and that some amino acid changes in VrPGIP1 also affect interactions between PGIP and polygalacturonase. Thus, tightly linkedVrPGIP1andVrPGIP2are the likely genes at theBrlocus that confer bruchid resistance in mungbean “V2709”.
DOI:10.1007/s12041-016-0663-9URLPMID:27659323 [本文引用: 1]

Mungbean ( Vigna radiata L. Wilczek) is one of the most important leguminous food crops in Asia. We employed Illumina paired-end sequencing to analyse transcriptomes of three different mungbean genotypes. A total of 38.3 39.8 million paired-end reads with 73 bp lengths were generated. The pooled reads from the three libraries were assembled into 56,471 transcripts. Following a cluster analysis, 43,293 unigenes were obtained with an average length of 739 bp and N50 length of 1176 bp. Of the unigenes, 34,903 (80.6%) had significant similarity to known proteins in the NCBI nonredundant protein database (Nr), while 21,450 (58.4%) had BLAST hits in the Swiss-Prot database (E-value<10 5 ). Further, 1245 differential expression genes were detected among three mungbean genotypes. In addition, we identified 3788 expressed sequence tag-simple sequence repeat (EST-SSR) motifs that could be used as potential molecular markers. Among 320 tested loci, 310 (96.5%) yielded amplification products, and 151 (47.0%) exhibited polymorphisms among six mungbean accessions. These transcriptome data and mungbean EST-SSRs could serve as a valuable resource for novel gene discovery and the marker-assisted selective breeding of this species.
DOI:10.1007/s00122-017-2965-6URLPMID:28831522

Abstract KEY MESSAGE: A novel genetic linkage map was constructed using SSR markers and stable QTLs were identified for six drought tolerance related-traits using single-environment analysis under irrigation and drought treatments. Mungbean (Vigna radiata L.) is one of the most important leguminous food crops. However, mungbean production is seriously constrained by drought. Isolation of drought-responsive genetic elements and marker-assisted selection breeding will benefit from the detection of quantitative trait locus (QTLs) for traits related to drought tolerance. In this study, we developed a full-coverage genetic linkage map based on simple sequence repeat (SSR) markers using a recombinant inbred line (RIL) population derived from an intra-specific cross between two drought-resistant varieties. This novel map was anchored with 313 markers. The total map length was 1010.18 cM across 11 linkage groups, covering the entire genome of mungbean with a saturation of one marker every 3.23 cM. We subsequently detected 58 QTLs for plant height (PH), maximum leaf area (MLA), biomass (BM), relative water content, days to first flowering, and seed yield (Yield) and 5 for the drought tolerance index of 3 traits in irrigated and drought environments at 2 locations. Thirty-eight of these QTLs were consistently detected two or more times at similar linkage positions. Notably, qPH5A and qMLA2A were consistently identified in marker intervals from GMES5773 to MUS128 in LG05 and from Mchr11-34 to the HAAS_VR_1812 region in LG02 in four environments, contributing 6.40-20.06% and 6.97-7.94% of the observed phenotypic variation, respectively. None of these QTLs shared loci with previously identified drought-related loci from mungbean. The results of these analyses might facilitate the isolation of drought-related genes and help to clarify the mechanism of drought tolerance in mungbean.
DOI:10.1111/j.1471-8286.2006.01461.xURL

The present work reports the isolation and characterization of new polymorphic microsatellites in mung bean ( Vigna radiata L.). Of 93 designed primer pairs, seven were found to amplify polymorphic microsatellite loci, which were then characterized using 34 mung bean accessions. The number of alleles ranged from two to five alleles per locus with an average of three alleles. Observed and expected heterozygosity values ranged from 0 to 0.088 and from 0.275 to 0.683, respectively. All seven loci showed significant deviations from Hardy einberg equilibrium, whereas only one pairwise combination (GBssr-MB77 and GBssr-MB91) exhibited significant departure from linkage disequilibrium. These newly developed markers are currently being utilized for diversity assessment within the mung bean germplasm collection of the Korean Gene Bank.
DOI:10.1007/s10592-009-9860-xURL

In this paper, we report the development and characterization of genic microsatellite markers for mungbean by mining sequence database, and transferability of the markers to Asian Vigna species. A total of 157 markers were designed upon searching for SSR in 830 transcript sequences. Thirty-three loci revealed single copy polymorphism in a panel of 17 mungbean accessions with number of alleles ranging from 2 to 8, and observed heterozygosity from 0 to 0.3125. Seven loci deviated from Hardy einberg Equilibrium. Two pairwise combinations of allele frequencies implied linkage disequilibrium. Cross-species amplification in 19 taxa of Asian Vigna using 85 primers showed that amplification rates varied between 80% ( V. aconitifolia ) to 95.3% ( V. reflexo-pilosa ). These mungbean genic microsatellite markers will be useful to study genetic resource and conservation of Asian Vigna species.
DOI:10.1590/S1415-47572007000600029URLPMID:18050090

The present study describes a new set of 61 polymorphic microsatellite markers for beans and the construction of a genetic map using the BAT93 x Jalo EEP558 (BJ) population for the purpose of developing a reference linkage map for common bean (Phaseolus vulgaris). The main objectives were to integrate new microsatellites on the existing framework map of the BJ population, and to develop the first linkage map for the BJ population based exclusively on microsatellites. Of the total of 264 microsatellites evaluated for polymorphism, 42.8% showed polymorphism between the genitors. An integrated map was created totaling 199 mapped markers in 13 linkage groups, with an observed length of 1358 cM and a mean distance between markers of 7.23 cM. For the map constructed exclusively with microsatellites, 106 markers were placed in 12 groups with a total length of 606.8 cM and average distance of 6.8 cM. Linkage group designation and marker order for BM microsatellites generally agreed with previous mapping, while the new microsatellites were well distributed across the genome, corroborating the utility of the BJ population for a reference map. The extensive use of the microsatellites and the availability of a reference map can help in the development of other genetic maps for common bean through the transfer of information of marker order and linkage, which will allow comparative analysis and map integration, especially for future quantitative trait loci and association mapping studies.
DOI:10.1016/j.cj.2015.01.001URL [本文引用: 1]
DOI:10.1111/j.1469-1809.1943.tb02321.xURL [本文引用: 1]

The articles published by the Annals of Eugenics (1925–1954) have been made available online as an historical archive intended for scholarly use. The work of eugenicists was often pervaded by prejudice against racial, ethnic and disabled groups. The online publication of this material for scholarly research purposes is not an endorsement of those views nor a promotion of eugenics in any way.
DOI:10.1007/s00122-007-0663-5URLPMID:17985112 [本文引用: 1]

Abstract It has long been recognized that epistasis or interactions between non-allelic genes plays an important role in the genetic control and evolution of quantitative traits. However, the detection of epistasis and estimation of epistatic effects are difficult due to the complexity of epistatic patterns, insufficient sample size of mapping populations and lack of efficient statistical methods. Under the assumption of additivity of QTL effects on the phenotype of a trait in interest, the additive effect of a QTL can be completely absorbed by the flanking marker variables, and the epistatic effect between two QTL can be completely absorbed by the four marker-pair multiplication variables between the two pairs of flanking markers. Based on this property, we proposed an inclusive composite interval mapping (ICIM) by simultaneously considering marker variables and marker-pair multiplications in a linear model. Stepwise regression was applied to identify the most significant markers and marker-pair multiplications. Then a two-dimensional scanning (or interval mapping) was conducted to identify QTL with significant digenic epistasis using adjusted phenotypic values based on the best multiple regression model. The adjusted values retain the information of QTL on the two current mapping intervals but exclude the influence of QTL on other intervals and chromosomes. Epistatic QTL can be identified by ICIM, no matter whether the two interacting QTL have any additive effects. Simulated populations and one barley doubled haploids (DH) population were used to demonstrate the efficiency of ICIM in mapping both additive QTL and digenic interactions.
DOI:10.1093/jhered/93.1.77URLPMID:12011185 [本文引用: 2]

Provides information on MapChart software designed for the graphical representation of linkage maps and quantitative trait loci. Features of the software; Details of how the software works; Discussion on data and formatting of the software.
URLPMID:25384727 [本文引用: 1]

Abstract Mungbean (Vigna radiata) is a fast-growing, warm-season legume crop that is primarily cultivated in developing countries of Asia. Here we construct a draft genome sequence of mungbean to facilitate genome research into the subgenus Ceratotropis, which includes several important dietary legumes in Asia, and to enable a better understanding of the evolution of leguminous species. Based on the de novo assembly of additional wild mungbean species, the divergence of what was eventually domesticated and the sampled wild mungbean species appears to have predated domestication. Moreover, the de novo assembly of a tetraploid Vigna species (V. reflexo-pilosa var. glabra) provides genomic evidence of a recent allopolyploid event. The species tree is constructed using de novo RNA-seq assemblies of 22 accessions of 18 Vigna species and protein sets of Glycine max. The present assembly of V. radiata var. radiata will facilitate genome research and accelerate molecular breeding of the subgenus Ceratotropis.
DOI:10.9787/PBB.2015.3.1.039URL [本文引用: 1]

Bruchid (L.) and pod sucking bug (Thunberg) are serious insect pests during the reproduction stage and seed storage period of legume crops worldwide. However, few sources of resistance to each of these insects have been identified and characterized, and no genetic studies have been carried out with simultaneous tests of these two insects. In this study, the inheritance of seed resistance to L. and Thunberg was examined in a mungbean cultivar, Jangan mungbean, which was developed by backcrossing with the V2709 resistant donor. The F, F, and Fseed generations were developed from the cross between susceptible and resistant parents, and evaluated for resistance to the two insects. It was found that resistance to bruchid and bean bug was controlled by a single dominant gene in the Fand Fseeds. However, the segregation pattern of reciprocal reaction to each insect in Fseeds showed seeds were susceptible to each insect. These results suggest that the resistance genes in Jangan mungbean to bug and weevil are either different or closely linked with each other. A genetic linkage map 13.7 cM in length with 6 markers was successfully constructed. Two QTLs were identified for bruchid resistance, and a QTL for bean bug resistance was detected. One of the QTLs for resistance to bruchid was shared with the QTL for bean bug. These newly developed closely linked markers will be used for cloning of the resistance genes to bruchid and bean bug in the future.
DOI:10.1186/s12870-016-0736-1URLPMID:26887961 [本文引用: 1]

Mungbean (Vigna radiata [L.] R. Wilczek) is an important legume crop with high nutritional value in South and Southeast Asia. The crop plant is susceptible to a storage pest caused by bruchids (Callosobruchus spp.). Some wild and cultivated mungbean accessions show resistance to bruchids. Genomic and transcriptomic comparison of bruchid-resistant and -susceptible mungbean could reveal bruchid-resistant genes (Br) for this pest and give insights into the bruchid resistance of mungbean. Flow cytometry showed that the genome size varied by 61 Mb (mega base pairs) among the tested mungbean accessions. Next generation sequencing followed by de novo assembly of the genome of the bruchid-resistant recombinant inbred line 59 (RIL59) revealed more than 42,000 genes. Transcriptomic comparison of bruchid-resistant and -susceptible parental lines and their offspring identified 91 differentially expressed genes (DEGs) classified into 17 major and 74 minor bruchid-resistance ssociated genes. We found 408 nucleotide variations (NVs) between bruchid-resistant and -susceptible lines in regions spanning 2 kb (kilo base pairs) of the promoters of 68 DEGs. Furthermore, 282 NVs were identified on exons of 148 sequence-changed-protein genes (SCPs). DEGs and SCPs comprised genes involved in resistant-related, transposable elements (TEs) and conserved metabolic pathways. A large number of these genes were mapped to a region on chromosome 5. Molecular markers designed for variants of putative bruchid-resistance ssociated genes were highly diagnostic for the bruchid-resistant genotype. In addition to identifying bruchid-resistance-associated genes, we found that conserved metabolism and TEs may be modifier factors for bruchid resistance of mungbean. The genome sequence of a bruchid-resistant inbred line, candidate genes and sequence variations in promoter regions and exons putatively conditioning resistance as well as markers detecting these variants could be used for development of bruchid-resistant mungbean varieties.
[本文引用: 1]
DOI:10.1104/pp.104.044644URLPMID:15299124 [本文引用: 1]

Polygalacturonase-inhibiting proteins (PGIPs) are extracellular plant inhibitors of fungal endopolygalacturonases (PGs) that belong to the superfamily of Leu-rich repeat proteins. We have characterized the full complement of pgip genes in the bean (Phaseolus vulgaris) genotype BAT93. This comprises four clustered members that span a 50-kb region and, based on their similarity, form two pairs (Pvpgip1/Pvpgip2 and Pvpgip3/Pvpgip4). Characterization of the encoded products revealed both partial redundancy and subfunctionalization against fungal-derived PGs. Notably, the pair PvPGIP3/PvPGIP4 also inhibited PGs of two mirid bugs (Lygus rugulipennis and Adelphocoris lineolatus). Characterization of Pvpgip genes of Pinto bean showed variations limited to single synonymous substitutions or small deletions. A three-amino acid deletion encompassing a residue previously identified as crucial for recognition of PG of Fusarium moniliforme was responsible for the inability of BAT93 PvPGIP2 to inhibit this enzyme. Consistent with the large variations observed in the promoter sequences, reverse transcription-PCR expression analysis revealed that the different family members differentially respond to elicitors, wounding, and salicylic acid. We conclude that both biochemical and regulatory redundancy and subfunctionalization of pgip genes are important for the adaptation of plants to pathogenic fungi and phytophagous insects.
[本文引用: 1]
