罗瑞1,2, 潘力2, 孙元2

 , 黄淑坚1
, 黄淑坚1
 , 仇华吉1,2
, 仇华吉1,2

1. 佛山科学技术学院生命科学与工程学院, 广东 佛山 528231;
2. 中国农业科学院哈尔滨兽医研究所, 兽医生物技术国家重点实验室, 黑龙江 哈尔滨 150069
收稿日期:2021-03-15;修回日期:2021-06-07;网络出版日期:2021-09-28
基金项目:国家自然科学基金(U20A2060,32072854,32072855,32072866);广东省重点领域研发计划(2019B020211003)
*通信作者:孙元, E-mail: sunyuan@caas.cn;
黄淑坚, E-mail: sjhuang.foshan@163.com;
仇华吉, Tel/Fax: +86-451-51051708, E-mail: qiuhuaji@caas.cn.
摘要:非洲猪瘟(African swine fever,ASF)是由非洲猪瘟病毒(African swine fever virus,ASFV)引起的一种出血性、致死性的猪烈性传染病。ASF在全球广泛传播,给养猪业造成重大的经济损失。ASFV基因组庞大,可编码150多种蛋白,一些非必需基因编码的蛋白与调控病毒毒力、复制和免疫逃逸等相关。通过删除ASFV毒力相关的非必需基因所构建的减毒株是当前比较有前景的疫苗,然而其安全性有待提高。系统地鉴定ASFV非必需基因及其功能,不仅有助于ASF基因缺失疫苗的研发,也有益于ASFV致病机制研究。本文对目前已鉴定的ASFV非必需基因及其功能研究进行了总结分析,着重讨论了影响ASFV毒力、调控病毒复制、参与免疫逃逸的非必需基因及其编码蛋白的功能,旨在加深对ASFV病原学的认识,为新的ASFV非必需基因的鉴定和功能研究提供参考。
关键词:非洲猪瘟非洲猪瘟病毒非必需基因毒力免疫逃逸
Nonessential genes of African swine fever virus: nothing or something?
Rui Luo1,2, Li Pan2, Yuan Sun2

 , Shujian Huang1
, Shujian Huang1
 , Hua-Ji Qiu1,2
, Hua-Ji Qiu1,2

1. College of Life Science and Engineering, Foshan University, Foshan 528231, Guangdong Province, China;
2. State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin 150069, Heilongjiang Province, China
Received: 15 March 2021; Revised: 7 June 2021; Published online: 28 September 2021
*Corresponding author: Yuan Sun, E-mail: sunyuan@caas.cn;
Shujian Huang, E-mail: sjhuang.foshan@163.com;
Hua-Ji Qiu, Tel/Fax: +86-451-51051708, E-mail: qiuhuaji@caas.cn.
Foundation item: Supported by the National Natural Science Foundation of China (U20A2060, 32072854, 32072855, 32072866) and by the Key Realm R & D Program of Guangdong Province (2019B020211003)
Abstract: African swine fever (ASF) is a hemorrhagic and fatal infectious disease caused by African swine fever virus (ASFV). ASF is endemic or epidemic in Africa, Asia, and Europe and causes huge economic losses to the pig industry. ASFV has a large DNA genome encoding more than 150 proteins, including many nonessential genes-encoded proteins associating with ASFV virulence, viral replication, immunoescape and unknown functions. Currently, a number of ASF live attenuated vaccines have been developed by deleting virulence-related nonessential genes. Generally, these vaccines have safety concerns, although they are able to provide partial to full protection. Systematic identification of more nonessential genes, especially virulence-related genes, will not only contribute to the development of safer gene-deleted ASF vaccines, but also benefit the understanding of the ASF pathogenesis. This review systematically summarizes the functions of known nonessential genes of ASFV, with focus on those involved in virulence, regulation of viral replication and escape of host antiviral immunity, and puts forward suggestions for the identification and functional study of unknown nonessential genes of ASFV.
Keywords: African swine feverAfrican swine fever virusnonessential genesvirulenceimmunoevasion
非洲猪瘟(African swine fever,ASF)是由非洲猪瘟病毒(African swine fever virus,ASFV)引起的一种出血性、致死性猪烈性传染病[1]。ASF于1921年在肯尼亚首次暴发,主要在撒哈拉以南的非洲地区流行,上世纪中叶传入欧洲,随后传至南美洲地区,2007年格鲁吉亚暴发ASF疫情并迅速波及俄罗斯、立陶宛等多个欧洲国家[2–4]。2018年8月我国辽宁省沈阳市首次报告该病,随后的几个月内,ASF几乎席卷了我国所有省份,给我国养猪行业带来了沉重打击[5]。ASFV的天然宿主为家猪、野猪和钝缘蜱,ASF在家猪-野猪、野猪-软蜱-家猪间循环传播,疣猪、钝缘蜱等自然宿主感染后无明显临床表现,是本病的传播媒介之一[6–7]。ASFV可通过直接、间接接触感染ASFV的病猪、排泄物、污染物及节肢动物媒介(蝇、蚊子、软蜱)等进行传播[8]。
通过生物信息学预测发现,ASFV基因组编码的151–174个基因中,有86个基因为必需基因,其余为非必需基因[9]。虽然众多****开展了相关研究,但仍有超过半数的ASFV基因功能不清楚。ASFV的非必需基因对病毒毒力、免疫逃逸等方面具有重要作用[10],因此,对非必需基因的研究十分重要。本文着重介绍了ASFV的非必需基因研究概况,并将其分类汇总,以期为ASFV的致病机制与疫苗研发提供思路。
1 非洲猪瘟病毒 ASFV是非洲猪瘟相关病毒科(Asfarviridae)非洲猪瘟病毒属(Asfivirus)的成员,是目前已知的唯一虫媒DNA病毒,成熟病毒粒子直径260–300 nm,自内向外分别是病毒基因组、内核心壳、内膜、衣壳和外膜(图 1)[11–12]。根据ASFV p72 (B646L)基因末端约500 bp核苷酸的差异,现已鉴定出24种基因型,格鲁吉亚、俄罗斯、中国、东南亚和东欧地区流行的主要是基因Ⅱ型,其他基因型主要流行于非洲和南美洲等地区[13–15]。
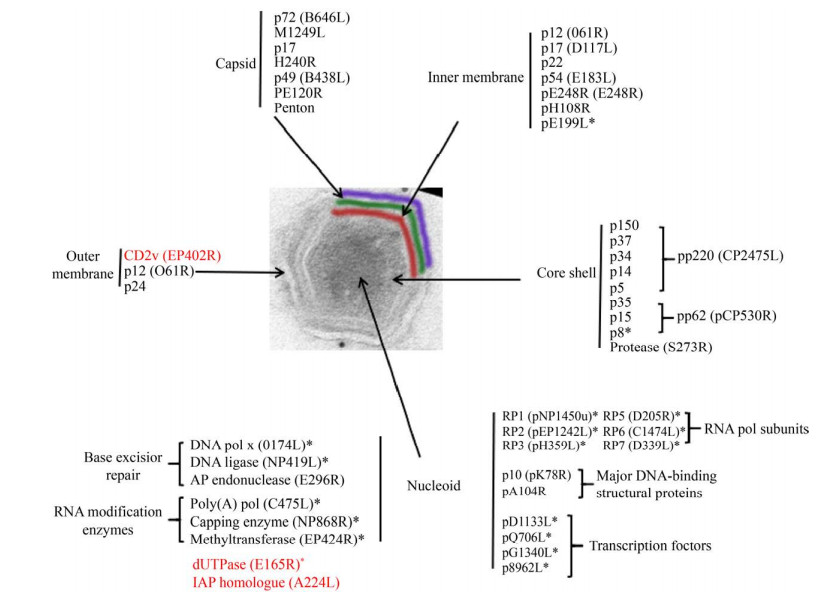 |
| 图 1 ASFV蛋白分布(改自Alejo等[16]) Figure 1 ASFV protein atlas (adapted from Alejo et al.[16]). The distribution of proteins marked with an asterisk (*) was inferred from the predicted or known roles; the genes marked in red are nonessential genes. |
| 图选项 |
ASFV是一种有囊膜的双链DNA病毒,基因组全长170–194 kb,基因组两端具有串联重复序列和多基因家族构成的可变区,不同毒株的两端可变区不同[11, 17–19]。ASFV的基因组分析对病毒基因功能、发病机制等研究具有重要意义。
根据基因缺失是否导致生物活性丧失,基因可分为必需基因和非必需基因,必需基因是生物生存和增殖所必需的,而非必需基因对生物生存和增殖是非必需的[20]。2020年Wang等对46株ASFV基因组进行了综合分析发现,ASFV具有开放性泛基因组,同时在ASFV中发现的151–174个基因中,只有86个基因被鉴定为必需基因,其余为非必需基因[9]。
ASFV基因组包含151–174个开放阅读框,可编码68种结构蛋白和100多种非结构蛋白[11]。据报道,24%的蛋白与ASFV形态相关,19%的蛋白与ASFV基因转录相关,6%的蛋白与维持ASFV基因组完整性有关,4%的蛋白介导ASFV侵入细胞,3%的蛋白与免疫逃逸有关,其他功能已知的蛋白占10%,功能未知的蛋白约占34%[16]。大部分蛋白功能未知,限制了ASFV复制相关基因和毒力相关基因的鉴定以及疫苗的研制。
在ASFV结构蛋白中,囊膜蛋白包括CD2v、p12和p24等,其中CD2v参与吸附红细胞、干扰宿主免疫防御[21],p12属于黏附蛋白,参与病毒吸附入侵[22];二十面体的蛋白质衣壳由p72、p49、M1249L、p17、H240R和PE120R等组成,其中p72为主要衣壳蛋白[23];内膜至少由7种已知的蛋白组成,分别为p17(pD117L)、pE183L、p12(pO61R)、p22(pKP177L)、pH108R、pE199L和pE248R;核壳主要由多蛋白pp220(CP2475L)[24]和pp62(CP530R)[25]的蛋白水解产物和病毒蛋白酶组成[26] (图 1),这些蛋白约占病毒粒子总质量的1/3[25]。
除上述结构蛋白外,ASFV还存在多种不同功能的非结构蛋白:维持基因组完整性的碱基切除修复(BER)系统的AP内切酶(PE296R)[27]、DNA连接酶(PNP419L)[28]、DNA聚合酶(Q174L)[29];干扰宿主防御机制的pA224L和CD2v等蛋白[30];细胞凋亡相关蛋白A179L、EP153R、DP71L和E183L;自噬相关蛋白A179L;调控蛋白合成的DP71L、A224L和D250R;调控MHC表达的EP153R[31]。
2 ASFV非必需基因的鉴定方法 ASFV非必需基因主要通过分析ASFV基因序列,预测出非必需基因后,通过构建基因缺失病毒进行验证,并研究其编码蛋白的功能。
2.1 非必需基因的预测 可用Sanger和Roary软件对ASFV基因组的必需基因和非必需基因进行分析。使用InterProScan等软件在NCBI、TIGRFAM、Panther、Gene3D、PRINTS、Pfam和ProDom等数据库中进行同源性搜索,对ASFV基因组序列进行比较。利用微阵列技术寻找目的基因和评估ASFV基因的潜在功能,利用SCOARY v1.6.16软件进行全基因组关联研究(PANGWAS)分析基因型与表型之间的关系[32],利用上述生物信息学方法进行预测后,需要试验验证,能够缺失的基因为非必需基因。
2.2 非必需基因的验证 分析出待验证的非必需基因并对其功能预测以后,可用TALEN、ZFN和CRISPR/Cas9等基因编辑技术构建基因缺失病毒。CRISPR/Cas9技术可对不同的真核和原核生物进行遗传操作,有针对性地对病毒基因进行编辑,目前已经利用CRISPR/Cas9系统研究了ASFV的EP402R、9GL基因[33]。其主要步骤分为:构建CRISPR/Cas9载体、构建筛选表达盒、构建同源重组转移载体、细胞转染和重组病毒筛选与纯化[34]。
除了上述基因编辑方法外,还可以采用其他基因编辑手段使非必需基因编码的蛋白功能丧失,例如移码突变、点突变、缺失和插入等。基因缺失病毒构建成功后,可利用测序技术把基因缺失病毒和亲本病毒的全基因组序列进行比较,评价基因修饰的准确性、基因组的完整性和重组病毒的纯度。
2.3 非必需基因功能研究 基因缺失(突变)病毒构建后,可进行体内或体外的验证,以明确非必需基因对病毒毒力、病毒复制的影响以及免疫逃逸等方面的作用机制。在体外培养,可观察病毒在细胞中的复制水平和致细胞病变(CPE)情况;也可进行体内实验,观察动物接种基因缺失毒后的临床症状、检测抗体水平并评价免疫攻毒保护效果等。
3 ASFV非必需基因编码蛋白的功能 3.1 影响病毒毒力 与其他类型的疫苗相比,通过删除与毒力相关的非必需基因而构建的基因缺失活疫苗,是当前最具有前景的疫苗研发方案。
3.1.1 非必需基因单个缺失对病毒复制与毒力的影响:: 一些非必需基因的单个缺失可在不同毒株中产生减毒作用,为基因缺失苗的开发提供研究思路,例如I177L、TK(A240L)、UK(DP96R)、9GL、NL(DP71L)、CD2v和DP148R等基因[35–39],这些基因缺失后对病毒毒力、病毒复制的影响以及同源保护效果见表 1。值得注意的是,ASFV-BA71V-ΔCD2可诱导交叉保护,这与特异性T细胞识别BA71V株和E75株病毒有关[40];同时CD2v蛋白可增强ASFV在蜱中的复制[41],因CD2v和C型凝集素蛋白与HAI的血清学特异性有关,可用CD2v/C型凝集素基因进行ASFV血清型的分型[42];在不同ASFV毒株中,缺失9GL对降低病毒毒力效果不同[43],Malawi Lil-20/1株缺失9GL后毒力显著降低[44],而ASFV Georgia/2007株缺失9GL后并未充分致弱[45]。此外,研究证实9GL编码的晚期病毒蛋白p14在核苷酸和氨基酸水平上均高度保守,且9GL与酵母ERV1和ALR基因相似,同时9GL影响正常的病毒粒子成熟[44]。
表 1. ASFV的非必需基因 Table 1. The nonessential genes of ASFV
| Genes | Function | Isolate | Virulence | Virus replication in cells | Homologous protection effect | References |
| I177L | – | Georgia | Completely attenuated | Reduced | Good protection | [35] |
| DP148R | – | Benin 97/1 | Attenuated | No effect | – | [39] |
| 9GL (B119L) | Morphogenesis | Georgia | Attenuated | Reduced replication | Good protection | |
| Malawi Lil-20/1 | Attenuated | Reduced replication | Good protection | [44–45] | ||
| Pretoriuskop/96/4 | Attenuated | Reduced replication | Good protection | |||
| CD2v (EP402R) | Binding to red blood cells | BA71V | Attenuated | Reduced replication | Resist BA71v E75 attacks | [40] |
| NL-S | – | E70 | Attenuated | No effect | Good protection | [38] |
| UK (DP96R) | IFN inhibitor | E70 | Attenuated | No effect | – | [37] |
| A238L | IFN inhibitor | NH/P68 | Attenuated | Reduced replication | Good protection | [66] |
| A240L (TK) | Thymidine kinase | Georgia | Completely attenuated | Reduced replication | No protection | [36] |
| C962R | Encode late expression protein | Georgia | No effect | No effect | – | [49] |
| X69R | Encode early expression protein | Georgia | No effect | No effect | – | [46] |
| MGF360-1L | – | Georgia | No effect | No effect | – | [50] |
| MGF360-16R | Interaction with host proteins SERTAD3 and SDCBP | Georgia | No effect | No effect | – | [47] |
| L83L | IL-1beta binding protein | Georgia | No effect | No effect | – | [51] |
| 8DR | Binding to red blood cells | Georgia | No effect | – | – | [48] |
| 4CL (A224L) | IAP apoptosis inhibitor | MalawiLIL-20/1 | No effect | No effect | – | [58] |
| I329L | IFN inhibitor | OURT88/3 | No effect | No effect | – | [59] |
| MGF360-12L | IFN inhibitor | – | – | – | – | [67] |
| NL (DP71L) | IFN inhibitor | MalawiLil-20/, Pretoriuskop/96/4 | No effect | No effect | – | [53] |
| EP153R | C-type lectin | BA71V | No effect | – | – | [68] |
| 8CR | – | Malawi Lil-20/1 | No effect | No effect | – | [54] |
| 11L | Transmembrane | BA71V | No effect | No effect | – | [56] |
| 5EL | – | MalawiLIL-20/1 | No effect | No effect | – | [52] |
| E165R | dUTPase | BA71V | No effect | No effect | – | [57] |
| O174L | Polymerase X | BA71V | No effect | Reduced replication | – | [29] |
| E296R | AP endonuclease | BA71V | No effect | Reduced replication | – | [27] |
| Nonessential gene combination deletion | ||||||
| MGF505-1R, 2R, 3R, MGF360-12L, 13L, 14L, CD2v | IFN inhibitor | HLJ/18 | Completely attenuated | Reduced replication | Good protection | [61] |
| MGF505-1R, 2R, 3R MGF360-12L, 13L, 14L | IFN inhibitor | Georgia | Completely attenuated | No effect | Good protection | [63] |
| MGF360-9L, 10L, 11L, 12L, 13L, 14L, MGF530/505-1R, 2R, 3R, 4R, | IFN inhibitor | Benin 97/1 | Attenuated | No effect | Good protection | [62] |
| 9GL, CD2v, EP153R | Binding to red blood | Georgia | Attenuated | Reduced replication | No protection | |
| 9GL, UK | Morphogenesis | Georgia | Attenuated | Reduced replication | Good protection | [60] |
| 9GL, NL, UK | IFN inhibitor | Georgia | Attenuated | Cause replication defects | No protection | [64] |
| 9GL, MGF360/505 | IFN inhibitor | Georgia | Attenuated | Reduced replication | No protection | [65] |
| L7L-L11L | – | SY18 | Attenuated | No effect | Good protection | [55] |
| CD2v, UK | IFN inhibitor binding to red blood | SY18 | Attenuated | No effect | Good protection | [69] |
| –: no reports. | ||||||
表选项
但是一些非必需基因,如X69R、MGF360-16R、8DR、MGF360-1L、C962R、11L、8CR、5EL、4CL、I329L和E165R等,与病毒毒力无关[46–59]。
3.1.2 非必需基因组合缺失对病毒复制与毒力的影响:: 虽然单基因缺失毒在诱导保护方面是有效的,但其安全性令人担忧。因此,同时删除多个非必需基因构建的多基因缺失毒,不仅可以起到保护作用,也降低了毒力返强的风险[60]。截至目前,最有商业化前景的多基因缺失疫苗候选株有ASFV-HLJ/18-Δ7GD、ASFV-G- Δ9GL/ΔUK、ASFV-Benin97/1-ΔMGF360/MGF530、ASFV-Benin97/1-MGFΔ505/ MGF360-9L/MGF530等[61–63]。其中,我国科学家以ASFV HLJ/18株为骨架,构建了7个基因(MGF505-1R、MGF505-2R、MGF505-3R、MGF360-12L、MGF360-13L、MGF360-14L和CD2v)缺失的突变株,临床试验初步证实ASFV-HLJ/18-Δ7GD在猪体内可完全致弱。可以预测,ASFV-HLJ/-18-Δ7GD是一种安全有效的ASF候选疫苗株,有望在控制ASF中发挥重要作用,但还需进一步评估[61]。O’Donnell等构建了ASFV-Georgia-Δ9GL/ΔUK缺失毒,临床试验表明,ASFV-Georgia-Δ9GL/ΔUK对猪无致病性并且提供了良好的同源保护,为设计新型ASF候选疫苗株提供了新思路[60]。
除此之外,多基因家族的基因缺失也能产生良好的减毒效果并提供完全的同源保护。例如,ASFV-Benin 97/1分离株缺失MGF360 (MGF360-10L、11L、12L、13L和14L)和MGF530/505 (MGF530/505-1R、2R和3R)以及(MGF360-9L和MGF530/505-4R)基因,能降低病毒的毒力,并诱导同源保护[62]。ASFV Georgia株缺失MGF360 (MGF360-12L、MGF360-13L和MGF360-14L)与MGF505 (MGF505-1R、MGF505-2R和MGF505-3R)后也可降低毒力并可提供同源保护,但对病毒的复制无影响[63]。
但一些非必需基因组合缺失时,存在减毒效果不佳、同源保护降低的情况。目前发现的这些组合缺失有ASFV-Georgia-Δ9GL/ΔCD2v/ΔEP153R、ASFV-Georgia-Δ9GL/ΔNL/ΔUK、ASFV-Georgia- Δ9GL/ΔMGF360/505等。ASFV Georgia株的9GL、CD2v和EP153R同时缺失后不能提供同源保护,而单独缺失9GL基因则可提供同源保护,据此推测,CD2v的缺失会拮抗9GL的致弱效果[43]。
ASFV-Georgia-Δ9GL/ΔNL/ΔUK无同源保护作用,ASFV-Georgia的UK基因缺失并不会降低毒力,NL基因的缺失反而使病毒毒力增强[64]。在ASFV-Georgia株中同时缺失9GL和MGF360/505基因可显著降低病毒毒力和复制水平,但不能提供同源保护[65]。以上结果表明,研发ASF疫苗株时,要结合前人的研究以及非必需基因的特性谨慎组合缺失,切勿盲目操作,总结前人的经验可以发现,利用I177L、9GL、CD2v和MGF相关基因构建基因缺失苗的成功率是比较大的,但需要进一步证实。
3.2 调控病毒免疫逃逸
3.2.1 抑制细胞凋亡:: ASFV能编码类似凋亡抑制剂的蛋白来抑制细胞凋亡,以促进细胞存活,保证病毒在细胞中的复制。编码抗凋亡蛋白的基因包括EP153R、A224L(4CL)、DP71L、A179L和E183L等,除E183L外,其他均为非必需基因[31, 68, 70–76]。
EP153R基因可抑制ASFV感染所引起的细胞凋亡。在病毒感染或星孢菌素诱导的Vero细胞中,EP153R蛋白抑制细胞蛋白p53的反式激活,从而抑制细胞凋亡。EP153R是第一个被发现的具有抗凋亡特性的病毒C型凝集素,并且参与ASFV感染细胞的红细胞吸附过程。除此之外,EP153R能够抑制MHC-I分子的表达,这一抑制作用可能是通过破坏胞吐过程实现的,不影响MHC与抗原的合成或糖基化[68]。
A224L基因是凋亡抑制蛋白(IAP)家族的成员。Dixon等发现,A224L IAP样蛋白可通过抑制caspase-3以及激活NF-κB转录因子调控的抗凋亡基因来抑制细胞凋亡[70]。Nogal等利用缺失A224L基因的缺失毒,发现A224L基因编码的蛋白与caspase-3蛋白水解酶片段相互作用,抑制酶的活性从而诱导细胞凋亡。此外,A224L能显著抑制肿瘤坏死因子α (TNF-α)、放线菌酮或星形孢菌素在Vero细胞中过表达时的caspase活性和细胞凋亡[71]。
DP71L编码的蛋白与宿主的生长抑制DNA损伤基因34 (GADD34)具有类似的功能,能够利用蛋白激酶1将真核转录起始因子2α (eIF2α)去磷酸化,促进宿主细胞蛋白合成,同时还能够抑制促细胞凋亡因子(CHOP)的活化,和细胞凋亡[72–73]。
A179L编码的蛋白能与几种促凋亡的Bcl-2蛋白结合[74],可抑制各类细胞的细胞凋亡,例如,它能抑制双链RNA激活蛋白激酶(P68)诱导的HeLa和BSC-40细胞的凋亡[75],以及大分子合成抑制剂诱导K562的凋亡[76]。除了具有抑制细胞凋亡作用外,还能通过与Beclin-1的相互作用调节自噬,抑制自噬小体的形成[77]。
3.2.2 参与干扰素的调节:: ASFV基因组编码许多不是病毒复制所必需的基因,如A238L、I329L、MGF360-12L、DP96R、MGF360、MGF530/505和A276R基因等,但可以调控干扰素(IFN)的表达,影响宿主防御病毒感染从而实现免疫逃逸[78–86]。
A238L蛋白是免疫逃逸相关蛋白[78]。A238L蛋白能够抑制炎症反应以及核转录因子NF-κB和活化T细胞核因子(NAFT)依赖性基因的表达,从而实现免疫逃逸。A238L能下调TNF-α的表达或者抑制环氧化(cox-2)的表达从而下调前列腺素E2的表达量,而前列腺素E2是一种炎症脂质介质和免疫反应调节剂,参与炎症反应与免疫应答。除此之外,A238L抑制p65/relA乙酰化以及抑制p300反式激活对诱导型一氧化氮合酶(INOS)表达的调节,致使INOS产生一氧化氮(NO),对宿主细胞造成损害,利于病毒的传播[79]。
ASFV OURT88/3株的I329L基因能抑制Ⅰ型IFN的产生,通过靶向不同的细胞内信号中间产物来降低Ⅰ型IFN的表达[80]。I329L基因编码一种高度糖基化蛋白,I329L蛋白抑制IFN-β和CCL5的激活,也能抑制双链RNA诱导的NF-κB和IRF3激活[81]。A276R与I329L基因都可通过TLR3抑制IFN-β的产生[66]。
MGF360-12L基因通过阻断Importin α介导的p65入核以及NF-κB信号通路来抑制Ⅰ型IFN的产生。研究显示,ASFV-MGF360-12L可以降低IRF3、STING、TBK1、ISG54、ISG56和AP-1的mRNA转录,MGF360-12L还可抑制经典核定位信号(NLS)介导的p50和p65的核定位,此外,MGF360-12L可以竞争性地抑制NF-κB与核转运蛋白的相互作用,从而干扰NF-κB的核转移,这也为ASFV实现其免疫逃逸提供了一种新策略[67]。
ASFV-China 2018/1株的DP96R通过cGAS- STING-TBK1信号通路干扰Ⅰ型IFN的产生。DP96R可抑制cGAS/STING和TBK1,选择性地阻断cGAS/STING和TBK1诱导的NF-κB启动子的激活。此外,还可抑制cGAS/STING激活的TBK1磷酸化以及TBK1诱导的抗病毒应答[82]。
MGF360和MGF505基因可直接或间接抑制Ⅰ型IFN的表达[83],MGF360和MGF505与ASFV宿主范围特异性、抑制宿主先天免疫和病毒毒力有关[63]。MGF360家族成员A276R可以抑制Poly(1:C)刺激的Ⅰ型IFN上调表达,而对JAK-STAT途径和NF-κB信号通路没有抑制作用;MGF505家族成员A528R可以抑制Poly(I: C)刺激的IFN诱导表达,同时对JAK-STAT途径具有抑制作用[84]。
3.3 其他功能 维持基因组完整性的碱基切除修复(BER)系统对于消除许多类型的碱基损伤、修复碱基位点至关重要,可在宿主细胞高度氧化的环境中保护病毒基因组,BER主要包括AP内切酶(E296R)[27]、DNA连接酶(NP419L)[28]、DNA聚合酶(O174L)[29],E296R与O174L是ASFV在Vero细胞中增殖非必需的基因[27–28]。
ASFV E296R基因编码一种Ⅱ类无嘌呤和无嘧啶(AP)核酸内切酶,具有AP位点特异性的核酸内切活性。ASFV AP内切酶具有AP内切酶、3′–5′外切酶和核苷酸切割修复(NIR)活性[85–86]。3′–5′核酸外切酶可以参与基因组复制过程中校对,也可以消除出现在单链断裂中的错配,这些特性使该酶适合参与BER[87]。当AP核酸内切酶基因缺失时,病毒在猪巨噬细胞中的复制受损,并导致Vero细胞对氧化和烷基化DNA损伤化合物的敏感性增高[27]。
ASFV O174L基因编码DNA聚合酶Pol X,为一种晚期结构蛋白,缺失该基因的病毒在Vero细胞中复制时对氧化损伤很敏感,该基因的缺失会导致基因突变频率增加[29]。NP419L基因编码一种Ⅰ型DNA连接酶,是病毒BER系统的一部分,所以它并不是非必需基因[28]。由于这些基因具有某些独特的功能,可以据此设计小分子抑制剂,以干扰ASFV基因组的修复过程,达到抗病毒的目的。
ASFV-Georgia编码的MGF360-16R与宿主蛋白SERTAD3和SDCBP相互作用。利用酵母双杂交技术分析发现,MGF360-16R与宿主蛋白SERTAD3的Serta结构域和Syndecan结合蛋白(SDCBP)相结合,SERTAD3和SDCBP都参与核转录,且SDCBP参与宿主细胞内的病毒运输[47]。因而,推测MGF360-16R可能与核转录和宿主细胞内的病毒运输有关,但需要进行进一步验证。
ASFV的E165R基因是dUTP核苷酸水解酶(dUTPase)家族的成员,E165R的活性中心与dUTPase非常相似。dUTPase在该蛋白家族的基序中是保守的,该酶在感染的早期和晚期都有表达,并定位于感染细胞的细胞质中。dUTPase在细胞质中降解dUTP,从而减少尿嘧啶与病毒DNA的错误结合,这在维持基因的保真度方面起着至关重要的作用。抑制dUTPase的活性可能不利于ASFV的复制。因此,可将dUTPase作为抗病毒药物设计的靶点[88–89]。
4 总结和展望 ASF是一种致死率可高达100%的猪烈性传染病。ASF的传播方式复杂多样,目前无商品化的疫苗可用,仅能依靠生物安全防护、快速诊断及扑杀等措施进行防控,严重威胁各国养猪及相关行业的健康发展。对此,笔者作出以下总结与展望。
首先,ASFV非必需基因鉴定与研究十分必要。ASFV具有复杂的基因组结构、编码的蛋白众多,而非必需基因在ASFV基因组中所占比重大;ASFV的非必需基因对病毒毒力、病毒复制、免疫逃逸、参与病毒-宿主相互作用等功能具有重要的影响;ASFV某些基因表现出遗传多样性或在某些区域具有明显的复杂性,这表明ASFV可能利用多种机制,从而获得新的表型特征,如抗原和毒力的变化。
其次,ASFV非必需基因的筛选鉴定与功能研究具有很大的潜在价值,尤其在疫苗研究方面。有关****尝试了各种类型疫苗和各种构建疫苗的策略,但这些疫苗均未商品化。大部分基因缺失疫苗的保护率高,免疫后可以抵抗亲本毒株的攻击,有的可以抵抗异源毒株的攻击。笔者认为,基因缺失疫苗是目前比较有希望成功的疫苗,通过敲除与毒力、免疫逃逸相关的非必需基因构建基因缺失病毒,如ASFV-HLJ/-18-7GD、ASFV-Georgia-Δ9GL/ΔUK和ASFV-SY18-ΔCD2v/ UK[60–61, 69]等候选株毒力完全被致弱,给ASF防控带来曙光。遗憾的是,减毒活疫苗可能存在重组、突变和毒力返强的风险,这也是各国对现有ASF基因缺失活疫苗商品化慎之又慎的主要因素。既然缺失ASFV非必需基因可将病毒致弱且能提供免疫保护,因而今后要深入解析未知的非必需基因,最终达到ASF疫苗株即使小范围内重组或突变也不影响该疫苗株的安全性和免疫原性的目的。因此,积极探究ASFV非必需基因的功能,尤其是未知非必需基因的功能,将为揭开ASFV的神秘“面纱”打下基础。
再次,ASFV非必需基因的研究策略需要改进。未来应充分利用基因组学、生物信息学、合成生物学和空间转录组学等预测出具有研究意义的非必需基因并对其功能进行验证。利用这些技术构建ASFV突变体,敲除与毒力相关的非必需基因以实现预期目的;代谢组学可通过对ASFV感染宿主代谢产物的识别,有利于揭示ASFV的致病机制;空间转录组学将有助于我们研究ASFV非必需基因的功能,通过结合影像组学和测序技术,可以对特定条件下的非必需基因转录产物进行定位,从而明确ASFV非必需基因编码蛋白与免疫系统之间的相互作用。完成基因预测后,利用CRISPR/Cas9等基因编辑技术构建基因缺失突变株,从而确定是否为非必需基因。
功能已被初步解析的非必需基因仅占少数,是否还有其他非必需基因与病毒毒力和病毒复制相关?还有哪些非必需基因在免疫逃逸过程中发挥作用?非必需基因之间存在怎样的协同作用?以上几个问题值得继续探究。
致谢
感谢本团队王涛博士和孙茂文硕士所提出的专业性修改建议。
References
| [1] | Lokhandwala S, Petrovan V, Popescu L, Sangewar N, Elijah C, Stoian A, Olcha M, Ennen L, Bray J, Bishop RP, Waghela SD, Sheahan M, Rowland RRR, Mwangi W. Adenovirus-vectored African Swine Fever Virus antigen cocktails are immunogenic but not protective against intranasal challenge with Georgia 2007/1 isolate. Veterinary Microbiology, 2019, 235: 10-20. DOI:10.1016/j.vetmic.2019.06.006 |
| [2] | Simulundu E, Lubaba CH, van Heerden J, Kajihara M, Mataa L, Chambaro HM, Sinkala Y, Munjita SM, Munang'andu HM, Nalubamba KS, Samui K, Pandey GS, Takada A, Mweene AS. The epidemiology of African swine fever in "nonendemic" regions of Zambia (1989-2015): implications for disease prevention and control. Viruses, 2017, 9(9): E236. DOI:10.3390/v9090236 |
| [3] | Rowlands RJ, Michaud V, Heath L, Hutchings G, Oura C, Vosloo W, Dwarka R, Onashvili T, Albina E, Dixon LK. African swine fever virus isolate, Georgia, 2007. Emerging Infectious Diseases, 2008, 14(12): 1870-1874. DOI:10.3201/eid1412.080591 |
| [4] | Gogin A, Gerasimov V, Malogolovkin A, Kolbasov D. African swine fever in the North Caucasus region and the Russian Federation in years 2007-2012. Virus Research, 2013, 173(1): 198-203. DOI:10.1016/j.virusres.2012.12.007 |
| [5] | Zhou XT, Li N, Luo YZ, Liu Y, Miao FM, Chen T, Zhang SF, Cao PL, Li XD, Tian KG, Qiu HJ, Hu RL. Emergence of African swine fever in China, 2018. Transboundary and Emerging Diseases, 2018, 65(6): 1482-1484. DOI:10.1111/tbed.12989 |
| [6] | Olesen AS, Lohse L, Boklund A, Halasa T, Gallardo C, Pejsak Z, Belsham GJ, Rasmussen TB, B?tner A. Transmission of African swine fever virus from infected pigs by direct contact and aerosol routes. Veterinary Microbiology, 2017, 211: 92-102. DOI:10.1016/j.vetmic.2017.10.004 |
| [7] | Luo YZ, Sun Y, Wang T, Qiu HJ. African swine fever: A major threat to the Chinese swine industry. Scientia Agricultura Sinica, 2018, 51(21): 4177-4187. (in Chinese) 罗玉子, 孙元, 王涛, 仇华吉. 非洲猪瘟——我国养猪业的重大威胁. 中国农业科学, 2018, 51(21): 4177-4187. DOI:10.3864/j.issn.0578-1752.2018.21.016 |
| [8] | Saegerman C, Bonnet S, Bouhsira E, De Regge N, Fite J, Etoré F, Garigliany MM, Jori F, Lempereur L, Le Potier MF, Quillery E, Vergne T, Vial L. An expert opinion assessment of blood-feeding arthropods based on their capacity to transmit African swine fever virus in Metropolitan France. Transboundary and Emerging Diseases, 2021, 68(3): 1190-1204. DOI:10.1111/tbed.13769 |
| [9] | Wang L, Luo YZ, Zhao YH, Gao GF, Bi YH, Qiu HJ. Comparative genomic analysis reveals an 'open' Pan-genome of African swine fever virus. Transboundary and Emerging Diseases, 2020, 67(4): 1553-1562. DOI:10.1111/tbed.13489 |
| [10] | Wu KK, Liu JM, Wang LX, Fan SQ, Li ZY, Li YW, Yi L, Ding HX, Zhao MQ, Chen JD. Current state of global African swine fever vaccine development under the prevalence and transmission of ASF in China. Vaccines, 2020, 8(3): 531. DOI:10.3390/vaccines8030531 |
| [11] | Dixon LK, Chapman DAG, Netherton CL, Upton C. African swine fever virus replication and genomics. Virus Research, 2013, 173(1): 3-14. DOI:10.1016/j.virusres.2012.10.020 |
| [12] | Alonso C, Borca M, Dixon L, Revilla Y, Rodriguez F, Escribano JM, Consortium IR. ICTV virus taxonomy profile: Asfarviridae. Journal of General Virology, 2018, 99(5): 613-614. DOI:10.1099/jgv.0.001049 |
| [13] | Quembo CJ, Jori F, Vosloo W, Heath L. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transboundary and Emerging Diseases, 2018, 65(2): 420-431. DOI:10.1111/tbed.12700 |
| [14] | Bisimwa PN, Ongus JR, Tiambo CK, Machuka EM, Bisimwa EB, Steinaa L, Pelle R. First detection of African swine fever (ASF) virus genotype X and serogroup 7 in symptomatic pigs in the Democratic Republic of Congo. Virology Journal, 2020, 17(1): 135. DOI:10.1186/s12985-020-01398-8 |
| [15] | Hakizimana JN, Nyabongo L, Ntirandekura JB, Yona C, Ntakirutimana D, Kamana O, Nauwynck H, Misinzo G. Genetic analysis of African swine fever virus from the 2018 outbreak in south-eastern Burundi. Frontiers in Veterinary Science, 2020, 7: 578474. DOI:10.3389/fvets.2020.578474 |
| [16] | Alejo A, Matamoros T, Guerra M, Andrés G. A proteomic atlas of the African swine fever virus particle. Journal of Virology, 2018, 92(23): e01293-18. |
| [17] | Gallardo MC, Reoyo ADLT, Fernández-Pinero J, Iglesias I, Mu?oz MJ, Arias ML. African swine fever: a global view of the current challenge. Porcine Health Management, 2015, 1(1): 1-14. DOI:10.1186/2055-5660-1-1 |
| [18] | Wang N, Zhao DM, Wang JL, Zhang YL, Wang M, Gao Y, Li F, Wang JF, Bu ZG, Rao ZH, Wang XX. Architecture of African swine fever virus and implications for viral assembly. Science, 2019, 366(6465): 640-644. DOI:10.1126/science.aaz1439 |
| [19] | Salas ML, Andrés G. African swine fever virus morphogenesis. Virus Research, 2013, 173(1): 29-41. DOI:10.1016/j.virusres.2012.09.016 |
| [20] | Li J, Wang HT, Wang WT, Zhang XR, Suo F, Ren JY, Bi Y, Xue YX, Hu W, Dong MQ, Du LL. Systematic analysis reveals the prevalence and principles of by-passable gene essentiality. Nature Communications, 2019, 10: 1002. DOI:10.1038/s41467-019-08928-1 |
| [21] | Goatley LC, Dixon LK. Processing and localization of the African swine fever virus CD2v transmembrane protein. Journal of Virology, 2011, 85(7): 3294-3305. DOI:10.1128/JVI.01994-10 |
| [22] | Suárez C, Gutiérrez-Berzal J, Andrés G, Salas ML, Rodríguez JM. African swine fever virus protein p17 is essential for the progression of viral membrane precursors toward icosahedral intermediates. Journal of Virology, 2010, 84(15): 7484-7499. DOI:10.1128/JVI.00600-10 |
| [23] | Liu S, Luo YZ, Wang YJ, Li SH, Zhao ZN, Bi YH, Sun JQ, Peng RC, Song H, Zhu DJ, Sun Y, Li S, Zhang L, Wang W, Sun YP, Qi JX, Yan JH, Shi Y, Gao GF. Cryo-EM structure of the African swine fever virus. Cell Host & Microbe, 2019, 26(6): 836-843. |
| [24] | Andrés G, Simón-Mateo C, Vi?uela E. Assembly of African swine fever virus: role of polyprotein pp220. Journal of Virology, 1997, 71(3): 2331-2341. DOI:10.1128/jvi.71.3.2331-2341.1997 |
| [25] | Andrés G, Alejo A, Salas J, Salas ML. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. Journal of Virology, 2002, 76(24): 12473-12482. DOI:10.1128/JVI.76.24.12473-12482.2002 |
| [26] | Andrés G, Alejo A, Simón-Mateo C, Salas ML. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. Journal of Biological Chemistry, 2001, 276(1): 780-787. DOI:10.1074/jbc.M006844200 |
| [27] | Redrejo-Rodríguez M, García-Escudero R, Yá?ez-Mu?oz RJ, Salas ML, Salas J. African swine fever virus protein pE296R is a DNA repair apurinic/apyrimidinic endonuclease required for virus growth in swine macrophages. Journal of Virology, 2006, 80(10): 4847-4857. DOI:10.1128/JVI.80.10.4847-4857.2006 |
| [28] | Lamarche BJ, Showalter AK, Tsai MD. An error-prone viral DNA ligase. Biochemistry, 2005, 44(23): 8408-8417. DOI:10.1021/bi047706g |
| [29] | Redrejo-Rodríguez M, Rodríguez JM, Suárez C, Salas J, Salas ML. Involvement of the reparative DNA polymerase Pol X of African swine fever virus in the maintenance of viral genome stability in vivo. Journal of Virology, 2013, 87(17): 9780-9787. DOI:10.1128/JVI.01173-13 |
| [30] | Ge SQ, Li JM, Fan XX, Liu FX, Li L, Wang QH, Ren WJ, Bao JY, Liu CJ, Wang H, Liu YT, Zhang YQ, Xu TG, Wu XD, Wang ZL. Molecular characterization of African swine fever virus, China, 2018. Emerging Infectious Diseases, 2018, 24(11): 2131-2133. DOI:10.3201/eid2411.181274 |
| [31] | Sun MW, Wang T, Sun Y, Yang YY, Qiu HJ. Immunoevasion strategies of African Swine fever virus. Acta Microbiologica Sinica, 2021, 61(2): 249-262. (in Chinese) 孙茂文, 王涛, 孙元, 杨玉莹, 仇华吉. 非洲猪瘟病毒的免疫逃逸策略. 微生物学报, 2021, 61(2): 249-262. |
| [32] | Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. Roary: rapid large-scale prokaryote Pan genome analysis. Bioinformatics, 2015, 31(22): 3691-3693. DOI:10.1093/bioinformatics/btv421 |
| [33] | Wo?niakowski G, Mazur-Panasiuk N, Walczak M, Juszkiewicz M, Frant M, Niemczuk K. Attempts at the development of a recombinant African swine fever virus strain with abrogated EP402R, 9GL, and A238L gene structure using the CRISPR/Cas9 system. Journal of Veterinary Research, 2020, 64(2): 197-205. DOI:10.2478/jvetres-2020-0039 |
| [34] | 郑海学, 李攀, 冯涛, 齐晓兰, 刘华南, 张克山, 李丹, 吴盼雪, 马昭, 党文, 刘迎琦, 石正旺, 杨波, 田宏, 郭建宏, 刘湘涛. 非洲猪瘟基因缺失弱毒株的构建及其作为疫苗的应用. 中国专利: CN202010661680. 7. 2020-07-08. |
| [35] | Borca MV, Medina ER, Silva E, Vuono E, Rai A, Pruitt S, Holinka LG, Salinas LV, Zhu J, Gladue DP. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. bioRxiv, 2019. DOI:10.1101/861666 |
| [36] | Sanford B, Holinka LG, O'Donnell V, Krug PW, Carlson J, Alfano M, Carrillo C, Wu P, Lowe A, Risatti GR, Gladue DP, Borca MV. Deletion of the thymidine kinase gene induces complete attenuation of the Georgia isolate of African swine fever virus. Virus Research, 2016, 213: 165-171. DOI:10.1016/j.virusres.2015.12.002 |
| [37] | Zsak L, Caler E, Lu Z, Kutish GF, Neilan JG, Rock DL. A nonessential African swine fever virus gene UK is a significant virulence determinant in domestic swine. Journal of Virology, 1998, 72(2): 1028-1035. DOI:10.1128/JVI.72.2.1028-1035.1998 |
| [38] | Zsak L, Lu Z, Kutish GF, Neilan JG, Rock DL. An African swine fever virus virulence-associated gene NL-S with similarity to the Herpes simplex virus ICP34.5 gene. Journal of Virology, 1996, 70(12): 8865-8871. DOI:10.1128/jvi.70.12.8865-8871.1996 |
| [39] | Reis AL, Goatley LC, Jabbar T, Sanchez-Cordon PJ, Netherton CL, Chapman DAG, Dixon LK. Deletion of the African swine fever virus gene DP148R does not reduce virus replication in culture but reduces virus virulence in pigs and induces high levels of protection against challenge. Journal of Virology, 2017, 91(24): e01428-17. |
| [40] | Monteagudo PL, Lacasta A, López E, Bosch L, Collado J, Pina-Pedrero S, Correa-Fiz F, Accensi F, Navas MJ, Vidal E, Bustos MJ, Rodríguez JM, Gallei A, Nikolin V, Salas ML, Rodríguez F. BA71ΔCD2:a new recombinant live attenuated African swine fever virus with cross-protective capabilities. Journal of Virology, 2017, 91(21): e01058-17. |
| [41] | Rowlands RJ, Duarte MM, Boinas F, Hutchings G, Dixon LK. The CD2v protein enhances African swine fever virus replication in the tick vector, Ornithodoros erraticus. Virology, 2009, 393(2): 319-328. DOI:10.1016/j.virol.2009.07.040 |
| [42] | Malogolovkin A, Burmakina G, Tulman ER, Delhon G, Diel DG, Salnikov N, Kutish GF, Kolbasov D, Rock DL. African swine fever virus CD2v and C-type lectin gene loci mediate serological specificity. Journal of General Virology, 2015, 96(4): 866-873. DOI:10.1099/jgv.0.000024 |
| [43] | Gladue DP, O'Donnell V, Ramirez-Medina E, Rai A, Pruitt S, Vuono EA, Silva E, Velazquez-Salinas L, Borca MV. Deletion of CD2-like (CD2v) and C-type lectin-like (EP153R) genes from African swine fever virus Georgia-?9GL abrogates its effectiveness as an experimental vaccine. Viruses, 2020, 12(10): 1185. DOI:10.3390/v12101185 |
| [44] | Lewis T, Zsak L, Burrage TG, Lu Z, Kutish GF, Neilan JG, Rock DL. An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine. Journal of Virology, 2000, 74(3): 1275-1285. DOI:10.1128/JVI.74.3.1275-1285.2000 |
| [45] | O'Donnell V, Holinka LG, Krug PW, Gladue DP, Carlson J, Sanford B, Alfano M, Kramer E, Lu ZQ, Arzt J, Reese B, Carrillo C, Risatti GR, Borca MV. African swine fever virus Georgia 2007 with a deletion of virulence-associated Gene9GL(B119L), when administered at low doses, leads to virus attenuation in swine and induces an effective protection against homologous challenge. Journal of Virology, 2015, 89(16): 8556-8566. DOI:10.1128/JVI.00969-15 |
| [46] | Ramirez-Medina E, Vuono E, Pruitt S, Rai A, Silva E, Zhu J, Velazquez-Salinas L, Gladue DP, Borca MV. X69R is a non-essential gene that, when deleted from African swine fever, does not affect virulence in swine. Viruses, 2020, 12(9): 918. DOI:10.3390/v12090918 |
| [47] | Ramírez-Medina E, Vuono EA, Velazquez-Salinas L, Silva E, Rai A, Pruitt S, Berggren KA, Zhu J, Borca MV, Gladue DP. The MGF360-16R ORF of African swine fever virus strain Georgia encodes for a nonessential gene that interacts with host proteins SERTAD3 and SDCBP. Viruses, 2020, 12(1): 60. DOI:10.3390/v12010060 |
| [48] | Borca MV, O'Donnell V, Holinka LG, Risatti GR, Ramirez-Medina E, Vuono EA, Shi JS, Pruitt S, Rai A, Silva E, Velazquez-Salinas L, Gladue DP. Deletion of CD2-like gene from the genome of African swine fever virus strain Georgia does not attenuate virulence in swine. Scientific Reports, 2020, 10: 494. DOI:10.1038/s41598-020-57455-3 |
| [49] | Ramirez-Medina E, Vuono EA, Rai A, Pruitt S, Silva E, Velazquez-Salinas L, Zhu J, Borca MV, Gladue DP. The C962R ORF of African swine fever strain Georgia is non-essential and not required for virulence in swine. Viruses, 2020, 12(6): 676. DOI:10.3390/v12060676 |
| [50] | Ramirez-Medina E, Vuono EA, Rai A, Pruitt S, Silva E, Velazquez-Salinas L, Zhu J, Gladue DP, Borca MV. Evaluation in swine of a recombinant African swine fever virus lacking the MGF-360-1L gene. Viruses, 2020, 12(10): 1193. DOI:10.3390/v12101193 |
| [51] | Borca MV, O'Donnell V, Holinka LG, Ramírez-Medina E, Clark BA, Vuono EA, Berggren K, Alfano M, Carey LB, Richt JA, Risatti GR, Gladue DP. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Research, 2018, 249: 116-123. DOI:10.1016/j.virusres.2018.03.017 |
| [52] | Neilan JG, Lu Z, Kutish GF, Zsak L, Lewis TL, Rock DL. A conserved African swine fever virus IkappaB homolog, 5EL, is nonessential for growth in vitro and virulence in domestic swine. Virology, 1997, 235(2): 377-385. DOI:10.1006/viro.1997.8693 |
| [53] | Afonso CL, Zsak L, Carrillo C, Borca MV, Rock DL. African swine fever virus NL gene is not required for virus virulence. The Journal of General Virology, 1998, 79(Pt 10): 2543-2547. |
| [54] | Neilan JG, Borca MV, Lu Z, Kutish GF, Kleiboeker SB, Carrillo C, Zsak L, Rock DL. An African swine fever virus ORF with similarity to C-type lectins is non-essential for growth in swine macrophages in vitro and for virus virulence in domestic swine. The Journal of General Virology, 1999, 80(Pt 10): 2693-2697. |
| [55] | Zhang JY, Zhang YY, Chen T, Yang JJ, Yue HX, Wang LD, Zhou XT, Qi Y, Han X, Ke JN, Wang SC, Yang JM, Miao FM, Zhang SF, Zhang F, Wang Y, Li M, Hu RL. Deletion of the L7L-L11L genes attenuates ASFV and induces protection against homologous challenge. Viruses, 2021, 13(2): 255. DOI:10.3390/v13020255 |
| [56] | Kleiboeker SB, Kutish GF, Neilan JG, Lu Z, Zsak L, Rock DL. A conserved African swine fever virus right variable region gene, l11L, is non-essential for growth in vitro and virulence in domestic swine. The Journal of General Virology, 1998, 79(Pt 5): 1189-1195. |
| [57] | Oliveros M, García-Escudero R, Alejo A, Vi?uela E, Salas ML, Salas J. African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. Journal of Virology, 1999, 73(11): 8934-8943. DOI:10.1128/JVI.73.11.8934-8943.1999 |
| [58] | Neilan JG, Lu Z, Kutish GF, Zsak L, Burrage TG, Borca MV, Carrillo C, Rock DL. A BIR motif containing gene of African swine fever virus, 4CL, is nonessential for growthin vitroand viral virulence. Virology, 1997, 230(2): 252-264. DOI:10.1006/viro.1997.8481 |
| [59] | Reis AL, Goatley LC, Jabbar T, Lopez E, Rathakrishnan A, Dixon LK. Deletion of the gene for the type I interferon inhibitor I329L from the attenuated African swine fever virus OURT88/3 strain reduces protection induced in pigs. Vaccines, 2020, 8(2): 262. DOI:10.3390/vaccines8020262 |
| [60] | O'Donnell V, Risatti GR, Holinka LG, Krug PW, Carlson J, Velazquez-Salinas L, Azzinaro PA, Gladue DP, Borca MV. Simultaneous deletion of the 9GL and UK genes from the African swine fever virus Georgia 2007 isolate offers increased safety and protection against homologous challenge. Journal of Virology, 2017, 91(1): e01760-16. |
| [61] | Chen WY, Zhao DM, He XJ, Liu RQ, Wang ZL, Zhang XF, Li F, Shan D, Chen HF, Zhang JW, Wang LL, Wen ZY, Wang XJ, Guan YT, Liu JX, Bu ZG. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Science China Life Sciences, 2020, 63(5): 623-634. DOI:10.1007/s11427-020-1657-9 |
| [62] | Reis AL, Abrams CC, Goatley LC, Netherton C, Chapman DG, Sanchez-Cordon P, Dixon LK. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine, 2016, 34(39): 4698-4705. DOI:10.1016/j.vaccine.2016.08.011 |
| [63] | O'Donnell V, Holinka LG, Gladue DP, Sanford B, Krug PW, Lu XQ, Arzt J, Reese B, Carrillo C, Risatti GR, Borca MV. African swine fever virus Georgia isolate harboring deletions of MGF360 and MGF505 genes is attenuated in swine and confers protection against challenge with virulent parental virus. Journal of Virology, 2015, 89(11): 6048-6056. DOI:10.1128/JVI.00554-15 |
| [64] | Ramirez-Medina E, Vuono E, O'Donnell V, Holinka LG, Silva E, Rai A, Pruitt S, Carrillo C, Gladue DP, Borca MV. Differential effect of the deletion of African swine fever virus virulence-associated genes in the induction of attenuation of the highly virulent Georgia strain. Viruses, 2019, 11(7): 599. DOI:10.3390/v11070599 |
| [65] | O'Donnell V, Holinka LG, Sanford B, Krug PW, Carlson J, Pacheco JM, Reese B, Risatti GR, Gladue DP, Borca MV. African swine fever virus Georgia isolate harboring deletions of 9GL and MGF360/505 genes is highly attenuated in swine but does not confer protection against parental virus challenge. Virus Research, 2016, 221: 8-14. DOI:10.1016/j.virusres.2016.05.014 |
| [66] | Gallardo C, Sánchez EG, Pérez-Nú?ez D, Nogal M, de León P, Carrascosa áL, Nieto R, Soler A, Arias ML, Revilla Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine, 2018, 36(19): 2694-2704. DOI:10.1016/j.vaccine.2018.03.040 |
| [67] | Zhuo YS, Guo ZH, Ba TT, Zhang C, He LH, Zeng CP, Dai HC. African swine fever virus MGF360-12L inhibits type I interferon production by blocking the interaction of importin α and NF-κB signaling pathway. Virologica Sinica, 2021, 36(2): 176-186. DOI:10.1007/s12250-020-00304-4 |
| [68] | Hurtado C, Granja AG, Bustos MJ, Nogal ML, González de Buitrago G, de Yébenes VG, Salas ML, Revilla Y, Carrascosa AL. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology, 2004, 326(1): 160-170. DOI:10.1016/j.virol.2004.05.019 |
| [69] | Teklue T, Wang T, Luo YZ, Hu RL, Sun Y, Qiu HJ. Generation and evaluation of an African swine fever virus mutant with deletion of the CD2v and UK genes. Vaccines, 2020, 8(4): 763. DOI:10.3390/vaccines8040763 |
| [70] | Dixon L, Sánchez-Cordón P, Galindo I, Alonso C. Investigations of pro- and anti-apoptotic factors affecting African swine fever virus replication and pathogenesis. Viruses, 2017, 9(9): 241. DOI:10.3390/v9090241 |
| [71] | Nogal ML, González de Buitrago G, Rodríguez C, Cubelos B, Carrascosa AL, Salas ML, Revilla Y. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. Journal of Virology, 2001, 75(6): 2535-2543. DOI:10.1128/JVI.75.6.2535-2543.2001 |
| [72] | Zhang FQ, Moon A, Childs K, Goodbourn S, Dixon LK. The African swine fever virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2alpha and inhibits CHOP induction but is dispensable for these activities during virus infection. Journal of Virology, 2010, 84(20): 10681-10689. DOI:10.1128/JVI.01027-10 |
| [73] | Barber C, Netherton C, Goatley L, Moon A, Goodbourn S, Dixon L. Identification of residues within the African swine fever virus DP71L protein required for dephosphorylation of translation initiation factor eIF2α and inhibiting activation of pro-apoptotic CHOP. Virology, 2017, 504: 107-113. DOI:10.1016/j.virol.2017.02.002 |
| [74] | Banjara S, Shimmon GL, Dixon LK, Netherton CL, Hinds MG, Kvansakul M. Crystal structure of African swine fever virus A179L with the autophagy regulator beclin. Viruses, 2019, 11(9): 789. DOI:10.3390/v11090789 |
| [75] | Brun A, Rivas C, Esteban M, Escribano JM, Alonso C. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology, 1996, 225(1): 227-230. DOI:10.1006/viro.1996.0592 |
| [76] | Revilla Y, Cebrián A, Baixerás E, Mart??nez-A C, Vi?uela E, Salas ML. Inhibition of apoptosis by the African swine fever virus bcl-2 homologue: role of the BH1 domain. Virology, 1997, 228(2): 400-404. DOI:10.1006/viro.1996.8395 |
| [77] | Hernaez B, Cabezas M, Mu?oz-Moreno R, Galindo I, Cuesta-Geijo MA, Alonso C. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Current Molecular Medicine, 2013, 13(2): 305-316. DOI:10.2174/156652413804810736 |
| [78] | Abrams CC, Chapman DAG, Silk R, Liverani E, Dixon LK. Domains involved in calcineurin phosphatase inhibition and nuclear localisation in the African swine fever virus A238L protein. Virology, 2008, 374(2): 477-486. DOI:10.1016/j.virol.2008.01.005 |
| [79] | Granja AG, Sabina P, Salas ML, Fresno M, Revilla Y. Regulation of inducible nitric oxide synthase expression by viral A238L-mediated inhibition of p65/RelA acetylation and p300 transactivation. Journal of Virology, 2006, 80(21): 10487-10496. DOI:10.1128/JVI.00862-06 |
| [80] | Correia S, Ventura S, Parkhouse RM. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Research, 2013, 173(1): 87-100. DOI:10.1016/j.virusres.2012.10.013 |
| [81] | Oliveira VL, Almeida SCP, Soares HR, Crespo A, Marshall-Clarke S, Parkhouse RME. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Archives of Virology, 2011, 156(4): 597-609. DOI:10.1007/s00705-010-0894-7 |
| [82] | Wang XX, Wu J, Wu YT, Chen HJ, Zhang SF, Li JX, Xin T, Jia H, Hou SH, Jiang YT, Zhu HF, Guo XY. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochemical and Biophysical Research Communications, 2018, 506(3): 437-443. DOI:10.1016/j.bbrc.2018.10.103 |
| [83] | Afonso CL, Piccone ME, Zaffuto KM, Neilan J, Kutish GF, Lu Z, Balinsky CA, Gibb TR, Bean TJ, Zsak L, Rock DL. African swine fever virus multigene family 360 and 530 genes affect host interferon response. Journal of Virology, 2004, 78(4): 1858-1864. DOI:10.1128/JVI.78.4.1858-1864.2004 |
| [84] | Golding JP, Goatley L, Goodbourn S, Dixon LK, Taylor G, Netherton CL. Sensitivity of African swine fever virus to type I interferon is linked to genes within multigene families 360 and 505. Virology, 2016, 493: 154-161. DOI:10.1016/j.virol.2016.03.019 |
| [85] | Redrejo-Rodríguez M, Ishchenko AA, Saparbaev MK, Salas ML, Salas J. African swine fever virus AP endonuclease is a redox-sensitive enzyme that repairs alkylating and oxidative damage to DNA. Virology, 2009, 390(1): 102-109. DOI:10.1016/j.virol.2009.04.021 |
| [86] | Oliveros M, Yá?ez RJ, Salas ML, Salas J, Vi?uela E, Blanco L. Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. Journal of Biological Chemistry, 1997, 272(49): 30899-30910. DOI:10.1074/jbc.272.49.30899 |
| [87] | Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature, 2002, 415(6868): 183-187. DOI:10.1038/415183a |
| [88] | Li CY, Chai Y, Song H, Weng CJ, Qi JX, Sun YP, Gao GF. Crystal structure of African swine fever virus dUTPase reveals a potential drug target. mBio, 2019, 10(5): e02483-19. |
| [89] | Liang R, Wang G, Zhang D, Ye G, Li MX, Shi YJ, Shi JL, Chen HC, Peng GQ. Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development. Journal of Biological Chemistry, 2021, 296: 100015. DOI:10.1074/jbc.RA120.014005 |
