黄在兴1,2, 陈华3, 翁伯琦4, 刘斌2, 王义祥4

 , 刘朋虎1,4
, 刘朋虎1,4

1. 福建农林大学国家菌草工程技术研究中心, 福建 福州 350002;
2. 广西大学农学院, 广西大学食用菌研究所, 广西 南宁 530004;
3. 福建省农业科学院科研处, 福建 福州 350003;
4. 福建省红壤山地农业生态过程重点实验室, 福建 福州 350003
收稿日期:2021-01-21;修回日期:2021-04-13;网络出版日期:2021-05-10
基金项目:中央引导地方科技发展专项(2020L3030);福建省科技厅农业引导性(重点)项目(2020N0007);福建省红壤山地农业生态过程重点实验室开放课题(Aephrs-202001);福建农林大学学科交叉融合项目(XKJC-71202103C)
*通信作者:王义祥, E-mail: sd-wolong@163.com;
刘朋虎, Tel/Fax: +86-591-83789223, E-mail: phliu1982@163.com.
摘要:[目的] 筛选不同实验条件下,尤其是镉胁迫下的姬松茸内参基因,为研究姬松茸镉富集相关基因功能研究奠定基础。[方法] 根据不同浓度Cd(0、2、5 mg/L)胁迫下菌丝中转录组表达量数据,筛选出18个候选基因,设计了30对引物;根据引物扩增的特异性、扩增效率初步筛选出来自不同基因的5对引物;运用qRT-PCR检测了5个基因在不同浓度Cd胁迫下的菌丝样品、原基、菌柄和菌盖中的Ct值,利用geNorm、NormFinder和BestKeeper对这些基因的表达稳定性进行评价。[结果] SGT2和STK是菌丝阶段Cd胁迫下最稳定的2个候选内参基因,GAPDH是不同组织中最稳定的内参基因,SGT2、STK和GAPDH是所有实验条件下最稳定的3个内参基因。[结论] 在姬松茸镉胁迫条件下,本研究筛选出的SGT2和STK比常用的内参基因表现出更强的稳定性。
关键词:巴西蘑菇镉胁迫不同组织荧光定量PCR
Screening of reference genes under cadmium stress in Agaricus brasiliensis based on transcriptome sequencing
Zaixing Huang1,2, Hua Chen3, Boqi Weng4, Bin Liu2, Yixiang Wang4

 , Penghu Liu1,4
, Penghu Liu1,4

1. China National Engineering Research Center of JUNCAO Technology, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian Province, China;
2. College of Agriculture, Guangxi University, Institute of Mushroom, Nanning 530004, Guangxi Province, China;
3. Scientific Research Office of Fujian Academy of Agricultural Sciences, Fuzhou 350003, Fujian Province, China;
4. Fujian Province Key Laboratory of Agro-Ecological Processes in Hilly Red Soil, Fuzhou 350003, Fujian Province, China
Received: 21 January 2021; Revised: 13 April 2021; Published online: 10 May 2021
*Corresponding author: Yixiang Wang, E-mail: sd-wolong@163.com;
Penghu Liu, Tel/Fax: +86-591-83789223, E-mail: phliu1982@163.com.
Foundation item: Supported by the Special Program for Central Government to Guide Local Science and Technology Development (2020L3030), by the Agricultural Guidance (Key) Project of Fujian Science and Technology Department (2020N0007), by the Fujian Province Key Laboratory of Agro-Ecological Processes in Hilly Red Soil Open Project (Aephrs-202001) and by the Fujian Agriculture and Forestry University Interdisciplinary Integration (XKJC-71202103C)
Abstract: [Objective] The aim of this study was to screen the reference genes of Agaricus brasiliensis under different experimental conditions, especially under cadmium (Cd) stress, and to lay the foundation for studying the functional studies of cadmium enrichment-related genes in A. brasiliensis. [Methods] We screened 18 candidate genes and designed 30 primer pairs based on the transcriptomes of mycelia under the stress of different concentrations of Cd (0, 2, 5 mg/L). According to the specificity and amplification efficiency of primers, 5 pairs of primers from different genes were selected. The Ct values of mycelia, primordium, stipe, and pileus of 5 genes under different concentrations of cadmium stress were detected by QRT-PCR, and the expression stability of these genes was evaluated by geNorm, NormFinder, and BestKeeper. [Results] The results showed that Small glutamine-rich tetratricopeptide repeat-containing protein 2 (SGT2) and Serine/threonine-protein kinase (STK) were the two most stable candidate reference genes under Cd stress in the mycelial stage, GAPDH was the most stable reference gene in different tissues and SGT2, STK and GAPDH were the three most stable reference genes under all experimental conditions. [Conclusion] Under Cd stress conditions in A. brasiliensis, SGT2 and STK screened in this study were more stable compared to the commonly used reference genes.
Keywords: Agaricus brasiliensiscadmium stressdifferent tissuesqRT-PCR
姬松茸(Agaricus brasiliensis),又名巴西蘑菇,是一种原产于巴西的重要食药用菌,具有抗肿瘤作用、抗炎作用、调节免疫作用、抗突变作用,还可用于治疗糖尿病、高脂血症等[1],但姬松茸镉(Cd)富集能力强,对消费者的健康构成潜在威胁。Huang等[2]对姬松茸的Cd积累模式及Cd积累与磷之间的关系进行了研究;Wang等[3]则通过抑制消减杂交技术发现了39个与姬松茸Cd响应有关的基因,已知26个基因参与代谢、细胞运输、运输促进、运输途径和转录等,还有13个基因是编码未知功能的假定蛋白,通过qRT-PCR验证了已知基因中的6个基因,可见,姬松茸对Cd的富集是多方面调控的,其过程极其复杂,以上证据还不足以揭示姬松茸富集Cd的机制。
基因的表达分析对于理解生物某些特定功能至关重要[4-5],更深入和更广泛地分析Cd胁迫下姬松茸相关基因表达量的变化,有助于揭示姬松茸Cd富集分子机制。实时荧光定量PCR (qRT-PCR),具有灵敏、准确、可重复性等优点,被广泛应用于基因表达分析,qRT-PCR通过实时检测扩增过程中达到规定荧光强度所需的周期数,即循环阈值(Ct),基因起始拷贝数越多Ct值越小[6-9],可通过Ct值比较样本之间的转录水平,并可计算出样本中不同基因的表达水平[10]。
选择合适的内参基因来标准化是保证qRT-PCR结果可靠性的关键[3, 11],一些管家基因,如actin (ACT)、TUB、18S rRNA、3-磷酸甘油醛脱氢酶(GAPDH)等被用作内参基因[3, 12-14],但许多研究表明,这些管家基因在特殊的实验条件下,特别是在Cd胁迫[15-16]、高温胁迫[17-18]、干旱胁迫[19]和盐胁迫[20]等实验条件下表达不稳定,可见,在特定的实验条件下找出表达稳定的内参基因对目标基因的表达量进行校准至关重要[21]。但目前未见有关于姬松茸在Cd胁迫下内参基因研究的报道,为了更准确分析姬松茸功能基因的表达,有必要系统地筛选和评价适合姬松茸在Cd胁迫下的内参基因。
本课题组前期已经完成了两个姬松茸菌株在不同Cd浓度胁迫下的转录组数据分析[22],在此基础上本研究从转录组数据中初步筛选出在不同Cd浓度胁迫下18个表达量较高且变化不大的基因,为了评价这18个基因在不同Cd浓度胁迫下作为内参基因的可能,设计了30对特异性引物(表 1),通过qRT-PCR检测了候选内参基因在不同Cd浓度胁迫(0、2、5 mg/L)下和子实体不同发育阶段的表达,并运用geNorm、NormFinder和BestKeeper对以上候选内参基因进行了表达稳定性评价,旨在为姬松茸在Cd胁迫下提供可靠的内参基因,以便为更好理解姬松茸Cd富集相关功能基因和揭示相关分子机制奠定基础。
表 1. 引物信息 Table 1. Primer information
| Primers number | The name of the genes | Gene annotation | Primer sequences (5′—3′) | Tm/℃ | Length of product/bp |
| 1 | c32039.graph_c0 | Endopolyphosphatase | 60-S: GCACTGTATCTCGCCTCGGTAG 1-A: GAACCAGTGCGAAATACCTTGAC | 61.6±0.5 | 245 |
| 2 | c32039.graph_c0 | Endopolyphosphatase | 2-S: TGATTGCGGGCTTTACTTGC 2-A: CCGTTCCGAAACCCAGATTG | 61.95±0.65 | 302 |
| 3 | c32039.graph_c0 | Endopolyphosphatase | 3-S: CCTCATAACATCATGGCTCCTG 3-A: GGGTCATCCCTATCTCCGTATTG | 60.6±1.0 | 224 |
| 4 | c32039.graph_c0 | Endopolyphosphatase | 4-S: TCCGACCCACAATCGTGTCC 4-A: TGCATGGTCTCGGGGCTAGG | 64.4±1.0 | 174 |
| 5 | c32039.graph_c0 | Endopolyphosphatase | 5-S: TTCTTTTTTCGGACCACTACACC 5-A: GTCACTGAGGACCGTGCTCG | 60.65±0.35 | 299 |
| 6 | c28585.graph_c0 | Small glutamine-rich tetratricopeptide repeat-containing protein 2 (SGT2) | 6-S: ATGTTTCTTCGCCTGTCGATC 6-A: TACGGCTCGTCGGAGTGTTC | 60.25±0.85 | 325 |
| 7 | c28585.graph_c0 | Small glutamine-rich tetratricopeptide repeat-containing protein 2 (SGT2) | 7-S: CCGTTCAAGTCCACCATTAGC 7-A: CCTAATAACAGCGGTCTCAAATCC | 60.65±0.95 | 258 |
| 8 | c28618.graph_c0 | Serine/threonine-protein kinase (STK) | 8-S: CAAGCCCTACTGCCTCTATCTCG 8-A: GGGGACCCATACCAAGTTCATC | 62.25±0.25 | 335 |
| 9 | c28618.graph_c0 | Serine/threonine-protein kinase (STK) | 9-S: AAAATCCAAACGAGCGACAATG 9-A: CCGTTCGTCGCAATCAAATAC | 61.3±0.6 | 189 |
| 10 | c28651.graph_c0 | FAD linked oxidases, C-terminal domain; FAD binding domain | 10-S: CGCACAACTCGGACTGGATG 10-A: GCCGCCAATCTGACAACTCC | 62.3±0.3 | 347 |
| 11 | c30371.graph_c0 | hypothetical protein | 11-S: TCTAATTCCTCACTACGGGTTCG 11-A: TGCCTCGCCTACATCTCCATC | 61.45±0.95 | 288 |
| 12 | c30543.graph_c0 | Calcium permeable stress-gated cation channel | 12-S: AGGTGAACGGGCTCTCGCAG 12-A: CGGACGAGGTTTATGAGGAACG | 64.2±0.7 | 268 |
| 13 | c30806.graph_c0 | DNA replication ATP-dependent helicase | 13-S: GGTGAGAGAAAGACTCCGTGG 13-A: GAATCAACAACCTCCGCAATC | 59.05±0.45 | 203 |
| 14 | c30901.graph_c1 | Heavy metal tolerance protein | 14-S: CCTCTGCTCTCTTCCCTTGC 14-A: AGGACAAGGGGGACAAGAGTC | 59.25±0.25 | 232 |
| 15 | c30933.graph_c0 | Vacuolar protein sorting-associated protein | 15-S: GACGAGGCTCAAAGTGGACAAG 15-A: CATTCATCTCTAAGGGGGCATC | 60.85±0.55 | 196 |
| 16 | c31128.graph_c0 | Rho guanine nucleotide exchange factor scd1 | 16-S: GGCGGTAATGAATTGGAAGGTC 16-A: GGACAGTCTTCTCAACGGTTCTTC | 61.9±0.6 | 192 |
| 17 | c31395.graph_c0 | Cyclin | 17-S: TCAAAAACTCCTCCGGTCTAAAG 17-A: TCGCCACTCAAGTCGTCTGC | 60.85±0.95 | 193 |
| 18 | c31397.graph_c0 | Nuclease 1, mitochondrial (Precursor) (pnu1) | 18-S: AGTGGGCACAGAAACATTTGG 18-A: GCAGGAAGCGATGGATGAGAC | 60.9±1.0 | 235 |
| 19 | c31695.graph_c0 | DNA polymerase family B | 19-S: GCCAGGTCACAAAGCATACG 19-A: TGTCACCAACGAGACTATGCTTC | 59.2±0.3 | 243 |
| 20 | c32021.graph_c0 | Rho-type GTPase-activating protein 1 | 20-S: GGCGTTGAGGCGGTTGTATC 20-A: GAGCTGTCGCCGATGATTTAC | 61.3±1.4 | 167 |
| 21 | c4736.graph_c0 | 18S rRNA | S: AAGCAGTATTCAGTATGGCAACC; A: AATGGCTTGTTTCTTTGGGAG | 58.5±0.2 | 211 |
| 22 | c4736.graph_c0 | 18S rRNA | S: TCTCTCCCAAAGAAACAAGCC A: AAGGTTTAGCAAATTGGTTGTCC | 59.2±0.5 | 293 |
| 23 | c14447.graph_c0 | α-tubulin | S: GAAAGTGAGACCAGTCTGTTGTTG A: TCTTTGTGTGTTCAGTGTTCGTTG | 59.35±1.05 | 253 |
| 24 | c31752.graph_c1 | GAPDH | S: ATTGCCACAAAATGGGAAAGAC A: GTTGTGTATTTGTTGGTGCTCG | 59.55±1.25 | 209 |
| 25 | c31752.graph_c1 | GAPDH | S: AGAAACGACCTGATCCTCTGTG A: AGCAGCCCAGAACATCATCC | 59±0.7 | 250 |
| 26 | c31752.graph_c1 | GAPDH | S: AGAAACGACCTGATCCTCTGTG A: AGCAGCCCAGAACATCATCC | 59.25±0.45 | 216 |
| 27 | c31752.graph_c1 | GAPDH | S: TAACGGTTTCGGTCGTATCG A: CGTTCGGCAAAGACTTTCATAG | 59.8±1.1 | 269 |
| 28 | c31752.graph_c1 | GAPDH | S: TTCGGCTCATTTGAAGGGTG A: CACGCCAGTCCTTGTGAGAG | 61.3±0 | 244 |
| 29 | c55882.graph_c0 | α-actinin | S: ATCCCGTTAGCCTGGTCTTG A: TCCCAGGAGTGGATAGGTAAAG | 58.45±0.65 | 167 |
| 30 | c25638.graph_c0 | Actin-related protein | S: TTTGAACATCTGCTCTGTGTGG A: TGAAAGACACAACAAACCAGAAG | 57.95±0.65 | 295 |
表选项
1 材料和方法 1.1 实验分组及培养条件 A组——姬松茸的不同组织(原基、菌柄及菌盖):采用熟料袋栽方式出菇。将配方中各种培养料混合均匀,按1:1.25的比例计算加水量,混合均匀的培养料分装到聚丙烯袋中,每袋装料0.8 kg,放入高压灭菌锅中,121 ℃灭菌2 h,接种后在26 ℃恒温培养室中培养,待菌丝走满袋后,移到栽培室,开袋覆土。依次收集原基、菌柄和菌盖备用。
培养料配方:棉籽壳20%,玉米芯12.5%,牛粪35%,麸皮10%,稻草20%,CaCO3 1%,石灰1.5%。
B组——不同Cd浓度胁迫下的姬松茸菌丝:姬松茸菌株福姬77 (J77)与福姬1 (J1)分别在Cd浓度为0、2、5 mg/L的液体培养基中培养20 d,收集菌丝备用。
液体培养基组成:土豆230 g/L、蔗糖20 g /L、磷酸二氢钾2 g/L、硫酸镁0.5 g/L、VB1 10 mg/L。
C组:A组样品+B组样品。
1.2 总RNA和cDNA第一链合成 使用植物总RNA提取试剂盒(天根生化科技(北京)有限公司)对各样品的总RNA进行提取,使用OMEGA公司DNase I (RNaseFree)处理消除基因组DNA,通过1%琼脂糖凝胶在6 V/cm条件下电泳检测RNA完整性和质量。使用cDNA合成试剂盒EasyScript? First-Strand cDNA Synthesis SuperMix (北京全式金生物技术有限公司)合成各样品cDNA的第一链。
1.3 引物设计及初步筛选 根据本实验课题组前期转录组数据,采用Primer Premier 5.0设计引物(表 1),以姬松茸菌丝cDNA为模板,进行PCR扩增,PCR扩增总体系为20 μL:2×Taq PCR Mix (北京全式金生物技术有限公司) 10 μL,Forward Primer 0.5 μL,Reverse Primer 0.5 μL,ddH2O 8 μL,DNA 1 μL;扩增条件为:94 ℃预变性2 min,94 ℃变性30 s,56 ℃退火30 s,72 ℃延伸2 min,35个循环,72 ℃最终延伸10 min。PCR扩增产物通过1%琼脂糖凝胶在5 V/cm条件下电泳40 min,在凝胶成像系统中观察PCR产物条带。
1.4 实时荧光定量PCR 以5倍稀释cDNA样品,共稀释6个梯度,使用含有1、1/5、1/25、1/125、1/625、1/3125倍的cDNA样品作为反应模板,在Bio-Rad CFX96TM Real-Time System上进行荧光定量PCR,仪器自动获得内参基因的扩增效率E、相关系数R2及熔解温度等系列参数。
qRT-PCR反应体系为25 μL,包括:2×SYBR Green qRT-PCR Master Mix 12.5 μL,Forward Primer 0.5 μL,Reverse Primer 0.5 μL,ddH2O 10.5 μL,各样品cDNA 1 μL;反应条件为:94 ℃预变性2 min,94 ℃变性20 s,56 ℃退火20 s,72 ℃延伸1 min,40个循环,95 ℃ 5 s,65 ℃ 1 min,97 ℃ 10 s。形成溶解曲线,每个基因设3个重复,反应后根据溶解曲线确保PCR产物具有特异性,导出内参基因的Ct值。
1.5 数据分析 用Excel计算候选内参基因的Cp值(所有样品Ct值的平均值)及其标准差(SD),采用GraphPad Prism 5.0对候选内参基因的Cp值进作图,以显示候选内参基因的稳定性,运用geNorm软件通过在内参基因的矩阵内进行成对比较确定候选内参基因的稳定值(M值)[23];利用NormFinder软件将内参基因的组合和组间方差相结合来确定稳定值[24];通过BestKeeper来计算各内参基因的Ct值的标准差(SD)和变异系数(CV)[25],以上3种方法对应的结果值越小就表明内参基因的稳定性越好。
2 结果和分析 2.1 候选内参基因引物扩增效果和扩增特异性 表 1对应的各编号引物的普通PCR扩增结果如图 1所示,结合qRT-PCR扩增结果分析,最终6号(SGT2)、8号(STK)、12号(Calcium permeable stress-gated cation channel)、18号(pnu1)和27号(GAPDH)的引物普通PCR扩增产物条带单一、清晰,与预期大小一致,qRT-PCR扩增产物的溶解峰单一。除以上5对引物外,其他引物的普通PCR扩增条带不单一或不清晰,qRT-PCR扩增产物的溶解峰不单一。常用的内参基因18S rRNA、α-tubulin和α-actinin在本实验中表现出引物特异性不强,以上内参基因在姬松茸qRT-PCR研究中也很少见,多以GAPDH基因作为内参基因[3, 26]。因此以27号引物片段作为GAPDH基因进行后续的具体研究和评价。
 |
| 图 1 各引物普通PCR扩增产物条带 Figure 1 PCR product bands of primers. The numbers in the figure correspond to the numbers in Table 1. Lane 1–20: No. 1–20; B: No. 25–27, 22, No. 30; C: No. 21, No. 23–24, No. 28–29. |
| 图选项 |
6号、8号、12号、18号、27号引物的扩增效率(E)和相关性(R2)分别为99.9%和0.999、90.6%和1.000、100.9%和0.988、107.7%和0.998、101.3%和0.993,以上结果说明5对候选引物符合qRT-PCR实验要求。
2.2 候选内参基因表达丰度分析 如图 2所示,5个候选内参基因的Cp值在18.65–23.36之间,具有较高的表达量,GAPDH (18.65±0.96)、SGT2 (21.16±0.97)、STK (22.36±0.922)、Calcium permeable stress-gated cation channel (22.94±1.76)和pnu1 (23.36±1.36)的表达量依次降低,其中GAPDH中值线在框中间且标准差较小,因此GAPDH可能是较好的内参基因,但在不同的实验条件下,以上5个候选内参基因的稳定性可能会发生变化,因此为了在特定的实验条件下得到最优的内参基因,我们需要结合具体实验条件来评价候选内参基因的稳定性。
 |
| 图 2 候选内参基因表达量比较 Figure 2 Comparison of expression levels of candidate reference genes. The middle line represents the median value, the box represents the 10th/90th percentile, and the error line represents the maximum and minimum Cp values. |
| 图选项 |
2.3 geNorm评价候选内参基因的稳定性及最佳内参基因数量的确定 geNorm是Vandesompele等[23]编写的内参基因筛选程序,可确定2个或2个以上的内参基因组合来校正qRT-PCR数据。在数据输入geNorm之前,需要先算出候选内参基因的2–△Ct,△Ct=候选内参基因在样品中的Ct值–该基因在样品中最小的Ct值(表达量最高)。如图 3 (A1、B1、C1)所示,通过geNorm软件分析,在3组实验中,稳定性最好的2个内参基因均为GAPDH和SGT2,且STK、Calcium permeable stress-gated cation channel、pnu1稳定性依次降低。通过geNorm软件进一步分析最适数据归一化内参基因数目发现(图 3-A2、B2、C2),A组和B组的V2/3值分别为0.035和0.081,均小于0.15,因此,在A组和B组试验条件下的最适内参基因数目均为2,C组的V2/3=0.166 > 0.15,所以在C组实验条件下最适内参基因数目为n+1=3。
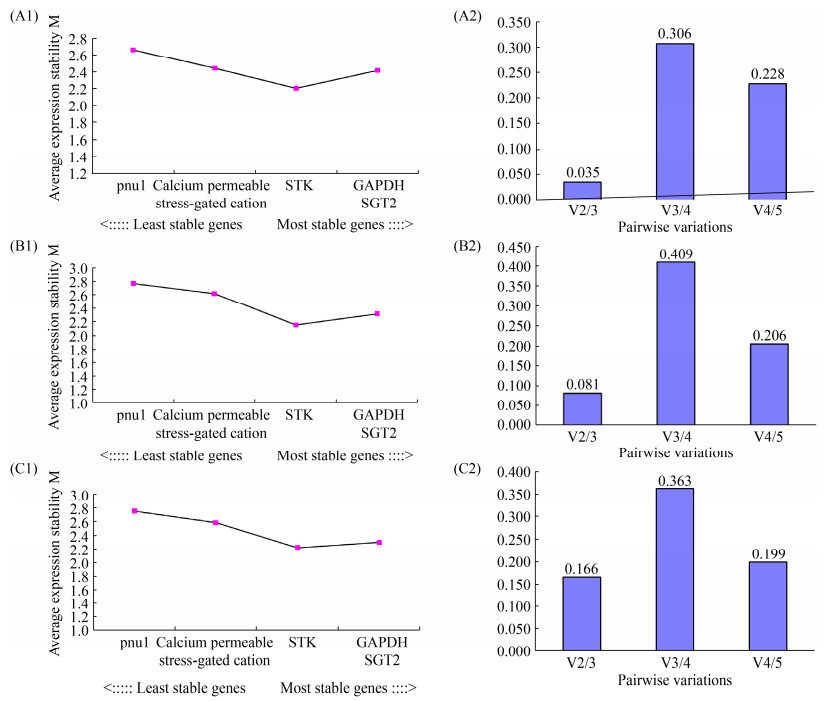 |
| 图 3 候选内参基因geNorm分析 Figure 3 Analysis of candidate reference genes by geNorm. A1, B1, C1 represent the expression stability of candidate reference genes in groups A, B and C, respectively. A2, B2, C2 represent the determination of the number of reference genes normalized to the most suitable data in groups A, B and C, respectively. |
| 图选项 |
2.4 NormFinder评价候选内参基因的稳定性 NormFinder是由Andersen等[24]编写的内参基因稳定性评价程序,数据预处理与geNorm一样,先计算出各候选内参基因的2–△Ct,但该程序只能筛选出一个最合适的内参基因。经NormFinder对候选内参基因的稳定性分析发现(表 2),在3组实验条件下STK的值最小,即最优的内参基因为STK,在B组和C组实验条件下候选内参基因稳定性排序依次为STK、SGT2、GAPDH、pnu1和Calcium permeable stress-gated cation channel,在A组实验条件下候选内参基因稳定性排序依次为STK、GAPDH、SGT2、pnu1和Ca-permeable stress-gated cation channel。
表 2. 候选内参基因NormFinder分析 Table 2. NormFinder analysis of the candidate reference genes
| Candidate reference genes | Group A | Group B | Group C |
| SGT2 | 0.507 | 0.359 | 0.371 |
| STK | 0.205 | 0.193 | 0.239 |
| Ca-permeable stress-gated cation channel | 0.963 | 1.183 | 1.056 |
| pnu1 | 0.740 | 0.635 | 0.620 |
| GAPDH | 0.334 | 0.449 | 0.416 |
表选项
2.5 BestKeeper评价候选内参基因的稳定性 BestKeeper通过在特定实验条件下Cp值的标准差(SD)和变异系数(CV)来评价候选内参基因的稳定性,SD和CV越小内参基因的稳定性越好。如表 3所示,5个候选内参基因在A组实验条件下GAPDH的稳定性最高,其次是Calcium permeable stress-gated cation channel、STK、SGT2和pnu1;在B组实验条件下SGT2表达稳定性最好,其次是STK、GAPDH、pnu1和Calcium permeable stress-gated cation channel;而在C组实验条件下候选内参基因STK表达稳定性最高,其次是GAPDH、SGT2、pnu1和Calcium permeable stress-gated cation channel,可见在不同实验条件下基因表达稳定性会发生较大的变化。
表 3. 候选内参基因BestKeeper分析 Table 3. BestKeeper analysis of the candidate reference genes
| Candidate reference genes | Group A | Group B | Group C | |||||
| SD | CV | SD | CV | SD | CV | |||
| SGT2 | 1.11 | 5.2 | 0.63 | 3.01 | 0.79 | 3.75 | ||
| STK | 0.66 | 2.85 | 0.73 | 3.31 | 0.71 | 3.17 | ||
| Calcium permeable stress-gated cation channel | 0.4 | 1.73 | 1.76 | 7.76 | 1.31 | 5.69 | ||
| pnu1 | 1.3 | 5.57 | 1.12 | 4.82 | 1.18 | 5.07 | ||
| GAPDH | 0.26 | 1.4 | 0.97 | 5.21 | 0.73 | 3.89 | ||
表选项
通过geNorm、NormFinder和BestKeeper对候选内参基因的稳定性进行了分析,虽然每个软件得出的结果稍有不同,但通过以上结果不难看出,在B组和C组实验条件下Calcium permeable stress-gated cation channel和pnu1的候选内参基因稳定性差,不推荐作为B组和C组实验条件下的内参基因;在A组实验条件下,GAPDH是最佳的内参基因,pnu1是最不稳定的基因;结合NormFinder和BestKeeper分析结果以及geNorm对最佳内参基因数目的确定,在B组实验条件下,SGT2和STK是2个最佳的内参基因;在C组实验条件下,V2/3=0.166 > 1.5,因此需要3个内参基因,则SGT2、STK和GAPDH是最佳的内参基因组合。
3 讨论 实时荧光定量PCR (qRT-PCR)被认为是用于基因表达分析的最新技术之一[27],选择合适的内参基因越来越受到关注[20],也是影响qRT-PCR结果可靠性的主要因素之一[18],任何一个基因不可能作为所有实验条件下的内参基因[28-29]。因此,很有必要系统地筛选和评价生物在不同生长时期和生物或非生物胁迫下的内参基因[30-31]。
GAPDH经常被用作食药用菌基因表达分析的内参基因,如分析灵芝中三萜合成基因的表达[14]、姬松茸中Cd响应的相关基因[3]及转录组数据验证[26]等。然而,Xu等[32]的研究表明GAPDH因序列高相似性和引物低特异性在灵芝中不适合作为内参基因。Zhao等[17]系统评价了在高温胁迫下香菇中10种传统内参基因(TUB,TUA,GADPH,EF1,18S,GTP,ACT,UBI,UBC和H2A)的稳定性,发现GAPDH的标准偏差(SD)值大于1,不适合作为该实验条件下的内参基因;此外,GAPDH也不适合作为冷杉(松杉)干旱胁迫下的内参基因[19]。Xiang等[33]的研究分析表明18S是香菇不同发育阶段的最佳内参基因,18S也属于常用的内参基因之一,但该研究未评论GAPDH的稳定性。本研究运用geNorm、NormFinder和BestKeeper对姬松茸中常用的内参基因(GAPDH)以及其他4个候选内参基因(SGT2、STK、Calcium permeable stress-gated cation channel、pnu1)表达的稳定性进行分析,以上3种软件可以足够预测候选内参基因的稳定性[34],结果表明姬松茸菌丝在Cd胁迫下GAPDH的SD值接近1 (0.97),其稳定性不如SGT2和STK,但它是不同组织中表达最稳定的基因。
经常被用作内参基因的管家基因表达量在Cd胁迫下会发生明显变化[16],Gao等[15]的研究表明,在Cd胁迫下大豆的根和叶中60S和UKN2是表达最稳定的基因,而Gu等[35]发现马蔺(Iris lacteal var. chinensis)中UBC和EF1b是Cd胁迫下表达最稳定的基因。可见,在Cd胁迫下不同生物体内表达稳定的基因也不同。在本研究中,3种软件的评价结果虽然稍有不同,在Tian等[36]的研究中也存在同样现象,但综合3个软件的分析结果不难发现姬松茸菌丝在不同浓度Cd胁迫下,GAPDH的表达不如SGT2和STK稳定;在所有实验条件下(不同浓度Cd胁迫和不同组织中) GAPDH、SGT2和STK是最佳的内参基因组合。因此,在使用qRT-PCR对姬松茸进行基因表达分析时,需要根据不同的实验条件选择合适的内参基因,这与上述文献结果相似。
SGT2是一种伴侣蛋白,其TPR结构域高度保守[37];STK在细胞生长过程中有着关键作用[38],Azad等[39]将其归为稳定类蛋白;GAPDH是参与糖酵解的一种关键酶,具有保守序列[40],这说明SGT2、STK和GAPDH在一定程度上是稳定表达的,并且在本研究中它们都具有很好的扩增效率和相关性。此外,一个良好的内参基因需要具备合适的表达水平,通常情况下内参基因的Cp值在15–30具有准确的标准化功能[41],本研究分析中5个候选内参基因的Cp值为18.65–23.36,其中GAPDH、SGT2和STK分别为18.65、21.16和22.36,这表明GAPDH、SGT2和STK符合优良内参基因的要求。
本研究首次报道SGT2和STK可作为姬松茸在Cd胁迫下的候选内参基因,研究结果可为更好地理解姬松茸相关分子生物学机制奠定基础,也为其他食药用菌的内参基因选择提供参考。
References
| [1] | Firenzuoli F, Gori L, Lombardo G. The medicinal mushroom Agaricus blazei Murrill: review of literature and pharmaco-toxicological problems. Evidence-Based Complementary and Alternative Medicine: ECAM, 2008, 5(1): 3-15. DOI:10.1093/ecam/nem007 |
| [2] | Huang JC, Li KB, Yu YR, Wu HW, Liu DL. Cadmium accumulation in Agaricus blazei Murrill. Journal of the Science of Food and Agriculture, 2008, 88(8): 1369-1375. DOI:10.1002/jsfa.3225 |
| [3] | Wang LL, Li HB, Wei HL, Wu XQ, Ke LQ. Identification of cadmium-induced Agaricus blazei genes through suppression subtractive hybridization. Food and Chemical Toxicology, 2014, 63: 84-90. DOI:10.1016/j.fct.2013.10.036 |
| [4] | Boavida LC, Borges F, Becker JD, Feijó JA. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiology, 2011, 155(4): 2066-2080. DOI:10.1104/pp.110.169813 |
| [5] | Xu YY, Zhu XW, Gong YQ, Xu L, Wang Y, Liu LW. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochemical and Biophysical Research Communications, 2012, 424(3): 398-403. DOI:10.1016/j.bbrc.2012.06.119 |
| [6] | Xu J, Xu ZC, Zhu YJ, Luo HM, Qian J, Ji AJ, Hu YL, Sun W, Wang B, Song JY, Sun C, Chen SL. Identification and evaluation of reference genes for qRT-PCR normalization in Ganoderma lucidum. Current Microbiology, 2014, 68(1): 120-126. DOI:10.1007/s00284-013-0442-2 |
| [7] | Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. Journal of Experimental Botany, 2009, 60(2): 487-493. DOI:10.1093/jxb/ern305 |
| [8] | Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology, 1993, 11(9): 1026-1030. |
| [9] | Xiao Z, Sun XB, Liu XQ, Li C, He LS, Chen SP, Su JL. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. don. Frontiers in Plant Science, 2016, 7: 1547. |
| [10] | Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics, 2006, 7: 85. DOI:10.1186/1471-2105-7-85 |
| [11] | Liu X, Guan HR, Song M, Fu YP, Han XM, Lei M, Ren JY, Guo B, He W, Wei YH. Reference gene selection for qRT-PCR assays in Stellera chamaejasme subjected to abiotic stresses and hormone treatments based on transcriptome datasets. PeerJ, 2018, 6: e4535. DOI:10.7717/peerj.4535 |
| [12] | Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnology Letters, 2003, 25(21): 1869-1872. DOI:10.1023/A:1026298032009 |
| [13] | Liang CX, Li YB, Xu JW, Wang JL, Miao XL, Tang YJ, Gu TY, Zhong JJ. Enhanced biosynthetic gene expressions and production of ganoderic acids in static liquid culture of Ganoderma lucidum under phenobarbital induction. Applied Microbiology and Biotechnology, 2010, 86(5): 1367-1374. DOI:10.1007/s00253-009-2415-8 |
| [14] | Shang CH, Shi L, Ren A, Qin L, Zhao MW. Molecular cloning, characterization, and differential expression of a lanosterol synthase gene from Ganoderma lucidum. Bioscience, Biotechnology, and Biochemistry, 2010, 74(5): 974-978. DOI:10.1271/bbb.90833 |
| [15] | Gao MM, Liu YP, Ma X, Shuai Q, Gai JY, Li Y. Evaluation of reference genes for normalization of gene expression using quantitative RT-PCR under aluminum, cadmium, and heat stresses in soybean. PLoS ONE, 2017, 12(1): e0168965. DOI:10.1371/journal.pone.0168965 |
| [16] | Basa B, Solti á, Sárvári é, Tamás L. Housekeeping gene selection in poplar plants under Cd-stress: comparative study for real-time PCR normalization. Functional Plant Biology: FPB, 2010, 36(12): 1079-1087. |
| [17] | Zhao X, Yang HL, Chen MJ, Song XX, Yu CX, Zhao Y, Wu YJ. Reference gene selection for quantitative real-time PCR of mycelia from Lentinula edodes under high-temperature stress. BioMed Research International, 2018, 2018: 1670328. |
| [18] | Xu H, Bao JD, Dai JS, Li YQ, Zhu Y. Genome-wide identification of new reference genes for qRT-PCR normalization under high temperature stress in rice endosperm. PLoS ONE, 2015, 10(11): e0142015. |
| [19] | Behringer D, Zimmermann H, Ziegenhagen B, Liepelt S. Differential gene expression reveals candidate genes for drought stress response in Abies alba (Pinaceae). PLoS ONE, 2015, 10(4): e0124564. |
| [20] | Xiao XL, Wu XM, Ma JB, Li PB, Li TT, Yao YN. Systematic assessment of reference genes for RT-qPCR across plant species under salt stress and drought stress. Acta Physiologiae Plantarum, 2015, 37(9): 1-9. |
| [21] | Zhang Y, Zhang XD, Liu X, Li YS, Ding JP, Zhang XR, Zhang YH. Reference gene screening for analyzing gene expression across goat tissue. Asian-Australasian Journal of Animal Sciences, 2013, 26(12): 1665-1671. DOI:10.5713/ajas.2013.13199 |
| [22] | Liu PH, Huang ZX, Luo XH, Chen H, Weng BQ, Wang YX, Chen LS. Comparative transcriptome analysis reveals candidate genes related to cadmium accumulation and tolerance in two almond mushroom (Agaricus brasiliensis) strains with contrasting cadmium tolerance. PLoS ONE, 2020, 15(9): e0239617. DOI:10.1371/journal.pone.0239617 |
| [23] | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 2002, 3(7): RESEARCH0034. DOI:10.1186/gb-2002-3-7-reports0034 |
| [24] | Andersen CL, Jensen JL, ?rntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 2004, 64(15): 5245-5250. |
| [25] | Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnology Letters, 2004, 26(6): 509-515. |
| [26] | Lu YP, Liao JH, Guo ZJ, Cai ZX, Chen MY. Genome survey and transcriptome analysis on mycelia and primordia of Agaricus blazei. BioMed Research International, 2020: 1824183. |
| [27] | Silveira TLR, Domingues WB, Remi?o MH, Santos L, Barreto B, Lessa IM, Varela Junior AS, Martins Pires D, Corcini C, Collares T, Seixas FK, Robaldo RB, Campos VF. Evaluation of reference genes to analyze gene expression in silverside Odontesthes humensis under different environmental conditions. Frontiers in Genetics, 2018, 9: 75. |
| [28] | Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal, 2008, 6(6): 609-618. |
| [29] | Tao YX, van Peer AF, Huang QH, Shao YP, Zhang L, Xie B, Jiang YJ, Zhu J, Xie BG. Identification of novel and robust internal control genes from Volvariella volvacea that are suitable for RT-qPCR in filamentous fungi. Scientific Reports, 2016, 6: 29236. |
| [30] | Wu ZJ, Tian C, Jiang Q, Li XH, Zhuang J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Scientific Reports, 2016, 6: 19748. |
| [31] | Bansal R, Mittapelly P, Cassone BJ, Mamidala P, Redinbaugh MG, Michel A. Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS ONE, 2015, 10(8): e0134890. |
| [32] | Xu ZC, Xu J, Ji AJ, Zhu YJ, Zhang X, Hu YL, Song JY, Chen SL. Genome-wide selection of superior reference genes for expression studies in Ganoderma lucidum. Gene, 2015, 574(2): 352-358. |
| [33] | Xiang QJ, Li J, Qin P, He ML, Yu XM, Zhao K, Zhang XP, Ma MG, Chen Q, Chen XQ, Zeng XF, Gu YF. Identification and evaluation of reference genes for qRT-PCR studies in Lentinula edodes. PLoS ONE, 2018, 13(1): e0190226. |
| [34] | Qian J, Gao YN, Wang Y, Wu YY, Wang Y, Zhao YC, Chen HY, Bao DP, Xu JY, Bian XH. Selection and evaluation of appropriate reference genes for RT-qPCR normalization of Volvariella volvacea gene expression under different conditions. BioMed Research International, 2018, 2018: 6125706. |
| [35] | Gu CS, Liu LQ, Xu C, Zhao YH, Zhu XD, Huang SZ. Reference gene selection for quantitative real-time RT-PCR normalization in Iris. lacteal var. chinensis roots under cadmium, lead, and salt stress conditions. The Scientific World Journal, 2014, 2014: 1-7. |
| [36] | Tian C, Jiang Q, Wang F, Wang GL, Xu ZS, Xiong AS. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE, 2015, 10(2): e0117569. |
| [37] | Krysztofinska EM, Evans NJ, Thapaliya A, Murray JW, Morgan RML, Martinez-Lumbreras S, Isaacson RL. Structure and interactions of the TPR domain of Sgt2 with yeast chaperones and Ybr137wp. Frontiers in Molecular Biosciences, 2017, 4: 68. |
| [38] | Wu FL, Liu Y, Jiang HW, Luan YZ, Zhang HN, He X, Xu ZW, Hou JL, Ji LY, Xie Z, Czajkowsky DM, Yan W, Deng JY, Bi LJ, Zhang XN, Tao SC. The Ser/Thr protein kinase protein-protein interaction map of M. tuberculosis. Molecular & Cellular Proteomics, 2017, 16(8): 1491-1506. |
| [39] | Azad I, Alemzadeh A. Bioinformatic and empirical analysis of a gene encoding serine/threonine protein kinase regulated in response to chemical and biological fertilizers in two maize (Zea mays L.) cultivars. Molecular Biology Research Communications, 2017, 6(2): 65-75. |
| [40] | Bao WL, Qu YL, Shan XY, Wan YL. Screening and validation of housekeeping genes of the root and cotyledon of Cunninghamia lanceolata under abiotic stresses by using quantitative real-time PCR. International Journal of Molecular Sciences, 2016, 17(8): 1198. |
| [41] | Wan HJ, Zhao ZG, Qian CT, Sui YH, Malik AA, Chen JF. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Analytical Biochemistry, 2010, 399(2): 257-261. |
