龚泽龙, 高雪锋, 李煜彬, 黄媛媛, 伦静娴, 周承星, 陈振辉, 林琼希, 曹虹


南方医科大学公共卫生学院, 广东省热带病研究重点实验室, 微生物学系, 广东 广州 510515
收稿日期:2020-12-14;修回日期:2021-03-18;网络出版日期:2021-04-02
基金项目:国家自然科学基金(81871198)
*通信作者:曹虹, Tel: +86-20-61648723;E-mail: gzhcao@smu.edu.cn.
摘要:[目的] 采用生物信息学方法分析公共数据库来源的细菌性败血症患者全血转录组学表达谱,探讨细菌败血症相关的宿主关键差异基因及意义。[方法] 基于GEO数据库中GSE80496和GSE72829全血转录组基因数据集,采用GEO2R、基因集富集分析(GSEA)联用加权基因共表达网络分析(WGCNA)筛选细菌性败血症患者相比健康人群显著改变的差异基因,通过R软件对交集基因进行GO功能分析和KEGG富集分析。同时,通过String 11.0和Cytoscape分析枢纽基因,验证枢纽基因在数据集GSE72809(Health组52例,Defined sepsis组52例)全血标本中的表达情况,并探讨婴儿性别、月(胎)龄、出生体重、是否接触抗生素等因素与靶基因表达谱间的关系。[结果] 分析GSE80496和GSE72829数据集分别筛选得到932个基因和319个基因,联合WGCNA枢纽模块交集得到与细菌性败血症发病相关的10个枢纽基因(MMP9、ITGAM、CSTD、GAPDH、PGLYRP1、FOLR3、OSCAR、TLR5、IL1RN和TIMP1);GSEA分析获得关键通路(氨基酸糖类-核糖代谢、PPAR信号通路、聚糖生物合成通路、自噬调控通路、补体、凝血因子级联反应、尼古丁和烟酰胺代谢、不饱和脂肪酸生物合成和阿尔兹海默症通路)及生物学过程(类固醇激素分泌、腺苷酸环化酶的激活、细胞外基质降解和金属离子运输)。[结论] 本项研究通过GEO2R、GSEA联用WGCNA分析,筛选出与细菌性败血症发病相关的2个枢纽模块、10个枢纽基因以及一些关键信号通路和生物学过程,可为后续深入研究细菌性败血症致病机制奠定理论依据。
关键词:细菌性败血症基因芯片基因富集分析(GSEA)加权基因共表达网络分析(WGCNA)
Bioinformatics analysis of hub genes and correlative pathways in the host pathogenesis of bacterial sepsis by gene set enrichment analysis and weighted gene co-expression network analysis
Zelong Gong, Xuefeng Gao, Yubin Li, Yuanyuan Huang, Jingxian Lun, Chengxing Zhou, Zhenhui Chen, Qiongxi Lin, Hong Cao


Guangdong Provincial Key Laboratory of Tropical Diseases, Department of Microbiology, School of Public Health, Southern Medical University, Guangzhou 510515, Guangdong Province, China
Received: 14 December 2020; Revised: 18 March 2021; Published online: 2 April 2021
*Corresponding author: Hong Cao, E-mail: gzhcao@smu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (81871198)
Abstract: [Objective] Bioinformatics methods were used to analyze the whole blood transcriptome data of patients diagnosed with bacterial sepsis from public databases, and we explored the hub genes related to bacterial sepsis and their clinical significance. [Methods] The datasets GSE80496 and GSE72829 were obtained from Gene Expression Omnibus (GEO). GEO2R, gene set enrichment analysis (GSEA) and weighted gene co-expression network analysis (WGCNA) were applied to screen out the differentially expressed genes (DEGs) among the patients with bacterial sepsis, when compared to the healthy. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genome (KEGG) enrichment analysis were also performed on the key genes by R software, hubs genes were obtained by using String 11.0 and Cytoscape subsequently. In addition, the dataset GSE72809 was involved to verify the expression profiles of hub genes in whole blood samples among infants. And the relationships between gene expression and samples' baseline data (such as: gender, gestational age, birth weight, and antibiotic exposure) were also discussed in the study. [Results] 932 DEGs were screened out in GSE80496 dataset, while 319 DEGs were screened out in GSE72829 dataset. We identified 10 hub genes (MMP9, ITGAM, CSTD, GAPDH, PGLYRP1, FOLR3, OSCAR, TLR5, IL1RN and TIMP1). GSEA analysis shows key pathways (amino acid carbohydrate ribose metabolism, PPAR signaling pathway, glycan biosynthesis, autophagy regulatory pathway, complement/coagulation factor cascade reaction, nicotine and nicotinamide metabolism, unsaturated fatty acids biosynthesis and Alzheimer's disease pathway) and biological processes (steroid hormone secretion, adenylate cyclase activation, extracellular matrix degradation and metal ion transport) associated with bacterial sepsis. [Conclusion] Through the analysis of GEO2R, GSEA combined with WGCNA, our study screened out 2 pivotal modules, 10 hub genes, several signaling pathways and biological processes closely related to bacterial sepsis, which may lay a theoretical basis for further research on the pathogenesis of bacterial sepsis.
Keywords: bacterial sepsisgene chip techniquegene set enrichment analysis (GSEA)weighted gene co-expression network analysis (WGCNA)
随着高通量检测技术在微生物领域的广泛应用,微生物感染相关的组学数据呈现指数级别增长。合理地运用公共数据库来源的微生物学大数据,将有助于我们更加深入地探讨病原菌感染与宿主免疫之间的互作和机制,以及致病过程中发挥作用的关键基因和信号通路,这也将帮助我们更好地识别病原菌感染对宿主带来的潜在危害。
新生儿死亡率是衡量国家和地区卫生文化水平的重要指标,而感染性疾病目前仍是导致新生儿死亡最常见的原因,尤其在发展中国家以细菌感染引起的败血症(bacterial sepsis)发病最为凶险[1]。目前,广泛应用的抗生素疗法治疗细菌性败血症收效显著,但却造成抗生素耐药问题日趋严峻[2]。越来越多的细菌耐药监测显示,无论是社区感染或院内感染,所分离的抗生素耐药菌株逐年增加,亦渐成困扰临床上治疗的难题。根据最近公布的2019年全国细菌耐药监测报告显示,常见耐药菌金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯、铜绿假单胞菌等分离率仍处高位,且脑脊液标本和血液标本均作为耐药菌检出率出现在较高位次的来源[3]。因此,寻找新的抗生素替代疗法治疗细菌性败血症刻不容缓。
病原菌与宿主细胞间的相互作用,是感染发生的关键步骤[4]。近年来,以宿主与病原菌间相互作用为靶点是研发治疗感染性损伤策略的新思路。本研究从宿主-病原菌相互作用的角度,基于Gene Expression Omnibus公共数据库数据,采用GEO2R、基因富集分析(gene set enrichment analysis,GSEA)联合加权基因共表达网络分析(weighted gene co-expression network analysis,WGCNA)的多种生物信息学分析手段,筛选、挖掘与细菌性败血症发病相关的枢纽基因和关键通路,这有助于更加深入探讨其致病的分子机制,可为后续寻找新型的抗生素替代疗法、规避病原菌感染对宿主带来的潜在危害提供理论依据。
1 材料和方法 1.1 数据获取及候选差异表达基因 本研究通过Gene Expression Omnibus数据库(https://www.ncbi.nlm.nih.gov/geo/)检索并获取GSE80496 (健康21例,败血症21例) (健康样本:GSM2128369–GSM2128389;败血症样本:GSM2128393–GSM2128413;排除芯片中包含的3例确诊Meningitis Sepsis样本)和GSE72829 (健康16例,败血症23例) (健康样本:GSM2126114– GSM2126129;败血症样本:GSM1872657– GSM1872679),样本准入标准为:患者出现发热现象(≥38 ℃)、出现明显临床症状需抽血化验、年龄小于17岁、排除骨髓移植、免疫缺陷或免疫抑制治疗样本。GSE25504 (健康35例,败血症28例) (健康样本:GSM627008–GSM627042;败血症样本:GSM627043–GSM627070)。所有分析样本均为人群全血转录水平表达谱数据集。通过R语言Bioconductor Affy package处理探针数据,基于RMA (Robust Multi Array Average)算法对数据进行标准化处理,转化得到基因表达谱的矩阵数据,以|Log2FoldChange| > 1且P < 0.05为标准对差异表达基因进行筛选。
1.2 GSEA分析 准备表达数据集、表型数据、功能基因集(包括KEGG基因集c2.cp.kegg.v7.2.symbols.gmt和GO基因集c5.all.v7.2.symbols.gmt)文件。设置分析参数“No collapse”、Number of permutations (模拟次数)为“1000”、Permutation type (模拟类型)为“Phenotype”,利用GSEA_4.0.3软件对上述文件进行分析。根据NES (校正富集分数)值和NOM P-value对富集通路进行筛选分析。
1.3 基因共表达网络分析 准备转录组表达谱的矩阵数据,通过R软件WGCNA包和Limma包对样本进行聚类、计算软阈值β和构建无尺度网络、拓扑重叠矩阵TOM构建,通过基因显著性以及模块显著性评价基因/模块和临床症状信息之间的关联性,筛选与临床相关模块的靶基因。
1.4 枢纽基因筛选 枢纽基因(hub gene)是在生物学或信号通路激活过程中发挥至关重要作用的基因,其他相关基因一般会受到枢纽基因的调控。因此,筛选枢纽基因是分析信号通路过程中重要的环节。我们通过GEO-2R在线分析软件对GSE80496和GSE72829分别分析,以|Log2FoldChange| > 1且P < 0.05为标准,筛选得到在Sepsis患者上调显著的基因,分别标记为“GSE80496-diff”和“GSE72829-diff”文件。根据WGCNA分析结果,计算不同模块对应的MM值(module membership)和GS值(gene significance),设置参数|MM| > 0.8和|GS| > 0.5筛选与枢纽模块相关度高的差异基因,分别标记“GSE72829-Brown”和“GSE80496-Blue”文件。通过在线软件Venny2.1绘制上述4个标记文件的交集即交集差异基因。将交集差异基因上传至STRING数据库构建蛋白互作网络,设置“minimum required interaction score”为“medium confidence”参数为0.4进行分析并将PPI数据信息导入Cytoscape (3.7.1版本),加载cytoHubba插件基于MCC模式(Maximal clique centrality)筛选排位靠前的枢纽基因用以后续分析。
1.5 枢纽基因在新生儿败血症患者表达情况分析 通过String Cytoscape蛋白互作分析筛选枢纽基因。基于公共数据库下载GSE25504芯片数据集,该数据集共包含63例婴幼儿(其中健康样本35例,诊断败血症样本28例)外周血标本并进行转录组水平的表达谱测序。我们分析了与败血症发病相关的枢纽基因在健康婴儿和诊断败血症婴儿外周血中的表达情况。同时,整理婴儿性别、胎龄、出生体重和是否使用抗生素等信息,分析上述因素与患儿外周血中枢纽基因水平之间的关联性。
2 结果和分析 2.1 筛选差异表达基因 分析流程如图 1所示。应用R软件对GSE80496和GSE72829数据集进行标准化处理,依据|Log2FoldChange| > 1且P < 0.05为条件对差异表达基因进行筛选,结果如图 2。GSE72829芯片中筛选出319个显著差异表达基因,其中203个上调基因、116个下调基因(图 2-A)。在GSE80496芯片中共筛选出932个显著差异表达基因,其中489个上调基因、443个下调基因(图 2-B)。选择两个不同芯片中在细菌性败血症患者中显著上调的差异表达基因留后续分析用。
 |
| 图 1 应用GEO2R、GSEA联合WGCNA分析细菌性败血症芯片流程 Figure 1 Bioinformatics analysis summary of bacterial sepsis datasets using GEO2R, GSEA and WGCNA. |
| 图选项 |
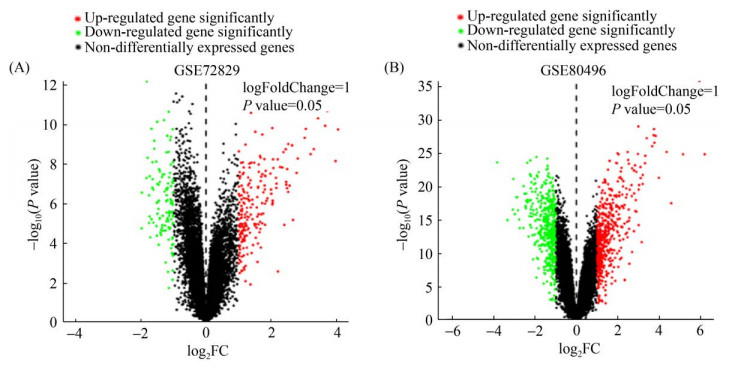 |
| 图 2 全血转录组学芯片中显著差异表达基因 Figure 2 DEGs in datasets of whole blood samples. A: volcano plot of GSE72829, B: volcano plot of GSE80496. |
| 图选项 |
2.2 WGCNA分析及枢纽模块内基因筛选 通过R软件分别对GSE80496和GSE72829数据集进行样本聚类,两芯片样本均展现出较好的聚类趋势(附图 1,http://journals.im.ac.cn/html/actamicrocn/2021/10/20211017.htm)。使用R软件计算芯片软阈值为β(GSE80496、GSE72829)=17,基于软阈值构建基因模块(图 3-A),构建共表达矩阵网络,并通过动态混合剪切得到6个以热图方式呈现基因模块与临床数据的相关性的结果(图 3-B),选择与细菌性败血症呈显著正相关关系的Brown模块标记为“GSE72829-Brown”进行后续分析。该模块包括962个基因与Sepsis感染的相关性为(r=0.67,P < 0.01) (图 3-D)。
 |
| 附图1 GSE80496和GSE72829样本聚类分析结果 S1 Sample cluster diagram of GSE72829 and GSE80496 datasets. |
| 图选项 |
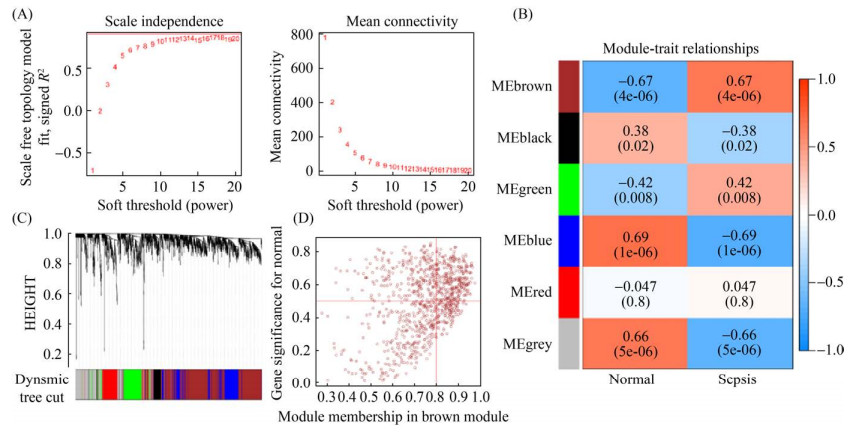 |
| 图 3 基于GSE72829芯片数据的WGCNA共表达模块分析 Figure 3 Identification of key modules correlated with bacterial sepsis in GSE72829 through WGCNA. A: analysis of the scale-free fit index and the mean connectivity for various soft-thresholding powers (β=17); B: heatmap for the correlation between module eigengenes and clinical traits; C: dendrogram of genes clustered based on the dissimilarity measure; D: scatter plot of module eigengenes in the brown module. |
| 图选项 |
同上分析GSE80496数据集获得枢纽模块(Blue模块)与细菌性败血症相关性散点图。选择与细菌性败血症呈显著正相关关系的Blue模块标记为“GSE80496-Blue”进行后续分析,该模块包括563个基因与Sepsis感染的相关性为(r=0.89,P < 0.01) (图 4)。
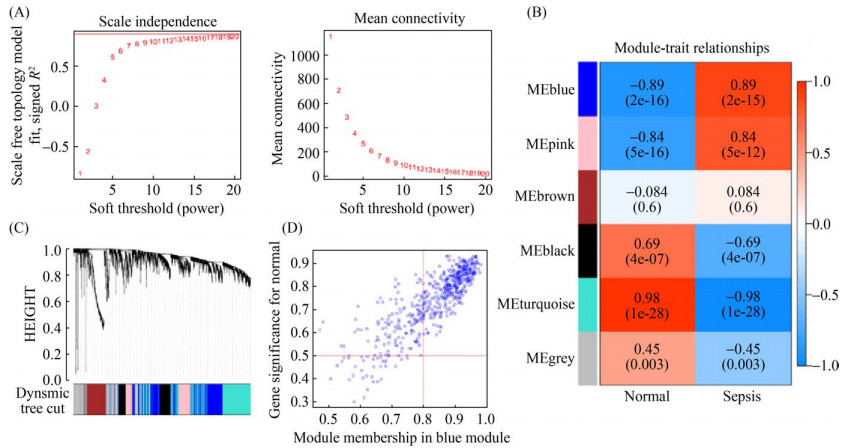 |
| 图 4 基于GSE80496芯片数据的WGCNA共表达模块分析 Figure 4 Identification of key modules correlated with bacterial sepsis in GSE80496 through WGCNA. A: analysis of the scale-free fit index and the mean connectivity for various soft-thresholding powers (β=17); B: heatmap for the correlation between module eigengenes and clinical traits; C: dendrogram of genes clustered based on the dissimilarity measure; D: scatter plot of module eigengenes in the blue module. |
| 图选项 |
2.3 筛选与细菌性败血症相关的枢纽基因 通过韦恩图绘制显著差异表达基因GSE80496-diff、GSE72829-diff和模块差异基因GSE72829-Brown、GSE80496-Blue的共同差异基因交集,筛选到包括ZNF438、ZDHHC19、WIPI1、TSPO和TLR5在内的共60个交集差异基因(图 5)。通过String-Cytoscape-cytoHubba分析60个交集差异基因网络,获得MCC模式下评分TOP10的枢纽基因:MMP9、ITGAM、CSTD、GAPDH、PGLYRP1、FOLR3、OSCAR、TLR5、IL1RN和TIMP1 (图 6)。
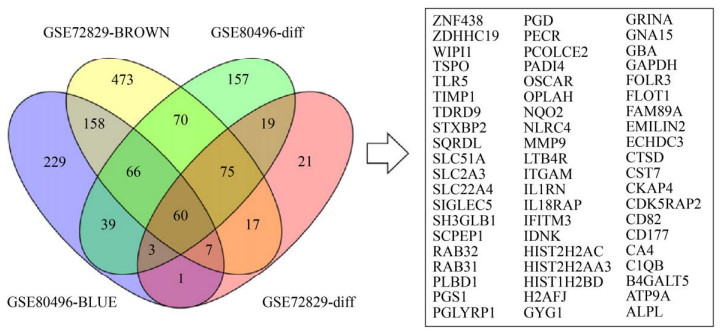 |
| 图 5 交集差异表达基因筛选 Figure 5 Venn diagram of intersection differential expression genes. |
| 图选项 |
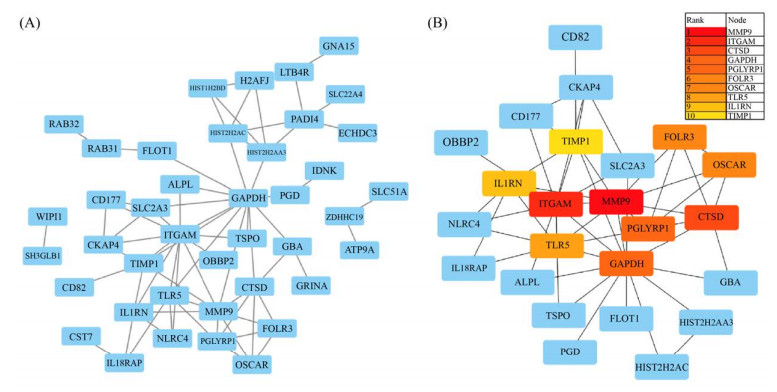 |
| 图 6 PPI网络构建及核心基因筛选 Figure 6 PPI network construction and hub genes screening. A: PPI network construction; B: Hub genes screening based on MCC. |
| 图选项 |
选择细菌性败血症患者中显著升高的60个交集差异基因进行GO基因功能注释,结果提示交集基因主要富集在中性粒细胞激活、中性粒细胞脱颗粒、中性粒细胞活化相关免疫反应、中性粒细胞介导的免疫、正向调控杀伤细胞、细胞死亡、细胞杀伤调控、肽酶活性的调节、活性氧代谢过程的正调控和调节白细胞脱颗粒等生物学功能上(图 7,表 1)。
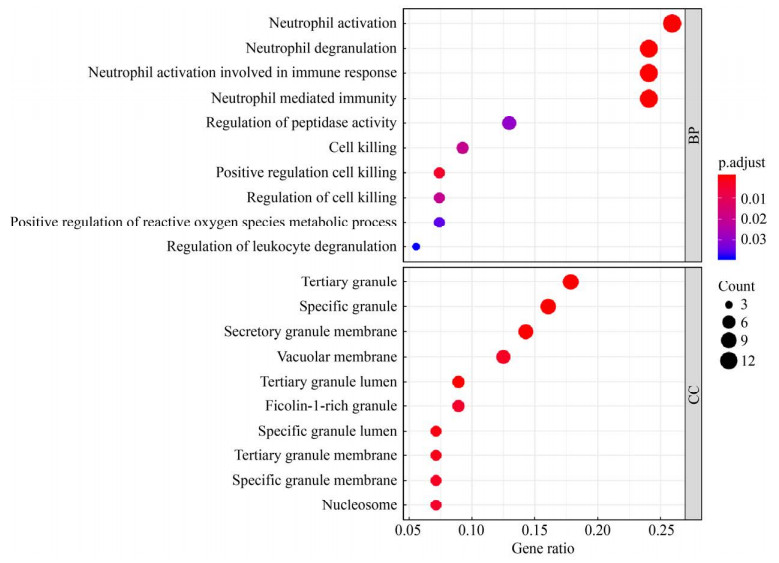 |
| 图 7 GO基因功能注释气泡图 Figure 7 GO analysis of genes in intersection differential expression genes. |
| 图选项 |
表 1. 针对芯片的GO富集分析及相关DEGs Table 1. GO analysis of intersection differential expression genes
| ID | Description | Gene Ratio | BgRatio | P value | p.adjust | q value | Gene ID |
| GO: 0042119 | Neutrophil activation | 10/37 | 499/17913 | 4.56E-08 | 3.84E-05 | 3.24E-05 | STXBP2/SLC2A3/SIGLEC5/PGLYRP1/OSCAR/IL18RAP/GYG1/FOLR3/CKAP4/CD177 |
| GO: 0043312 | Neutrophil degranulation | 9/37 | 485/17913 | 4.60E-07 | 0.000123 | 0.000104 | STXBP2/SLC2A3/SIGLEC5/PGLYRP1/OSCAR/GYG1/FOLR3/CKAP4/CD177 |
| GO: 0002283 | Neutrophil activation involved in immune response | 9/37 | 488/17913 | 4.84E-07 | 0.000123 | 0.000104 | STXBP2/SLC2A3/SIGLEC5/PGLYRP1/OSCAR/GYG1/FOLR3/CKAP4/CD177 |
| GO: 0002446 | Neutrophil mediated immunity | 9/37 | 499/17913 | 5.84E-07 | 0.000123 | 0.000104 | STXBP2/SLC2A3/SIGLEC5/PGLYRP1/OSCAR/GYG1/FOLR3/CKAP4/CD177 |
| GO: 0016045 | Detection of bacterium | 2/37 | 12/17913 | 0.00027 | 0.044794 | 0.037856 | PGLYRP1/NLRC4 |
| GO: 0098543 | Detection of other organism | 2/37 | 13/17913 | 0.000319 | 0.044794 | 0.037856 | PGLYRP1/NLRC4 |
表选项
2.4 枢纽基因在诊断婴儿败血症患者中的表达分析 公共数据库获取婴儿败血症芯片GSE25504 (健康婴儿35例,败血症婴儿28例),且每个样本记录了婴儿基线资料如性别、胎龄、出生体重、是否接触抗生素等信息。基于该芯片,我们分析上述筛选获得的60个交集差异基因和评分TOP10的枢纽基因与不同基线资料之间的关联性(图 8)。结果显示,ATP9A、CD177、GYG1、H2AFJ、ITGAM、NLRC4、PADI4、PGLYRP1、RAB31在不同性别败血症婴儿间存在统计学差异(P < 0.05);相比采样前未接受抗生素治疗的败血症患儿,CDK5RAP2在接受抗生素治疗的患儿中显著升高(P < 0.05),而CDK5RAP2、SIGLEC5在败血症患儿中的表达量与胎龄呈负相关趋势(附图 2,http://journals.im.ac.cn/html/actamicrocn/2021/10/20211017.htm)。
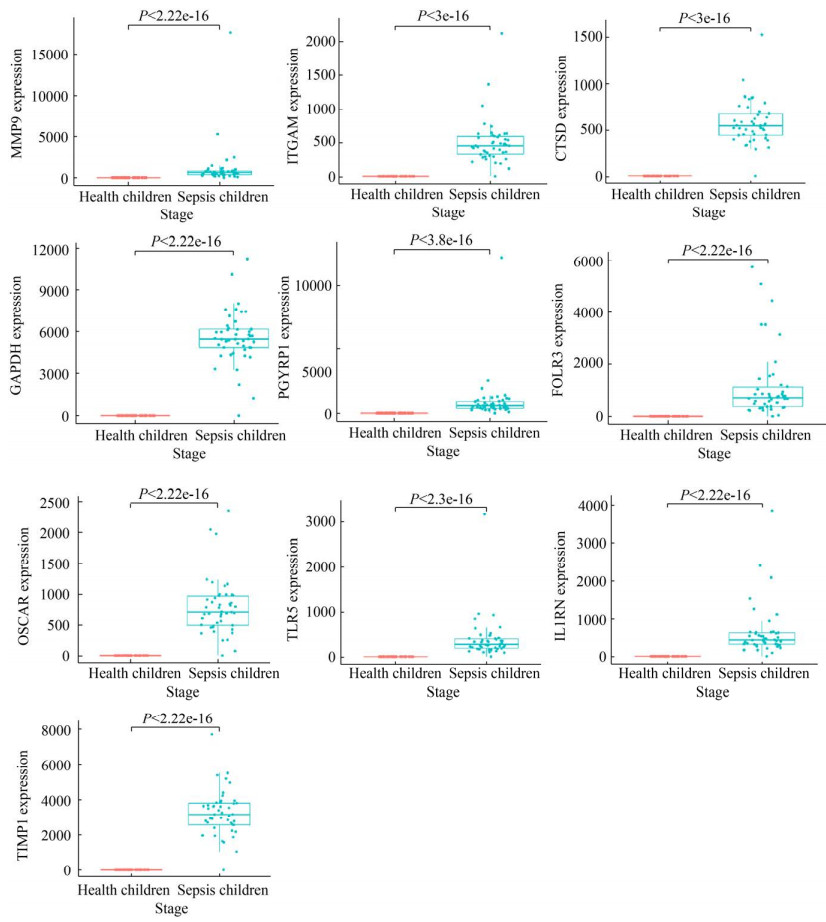 |
| 图 8 前10核心基因在GSE25504婴儿败血症数据集中的表达量 Figure 8 The expression profiles of hub genes in GSE25504 dataset associated with infant sepsis. |
| 图选项 |
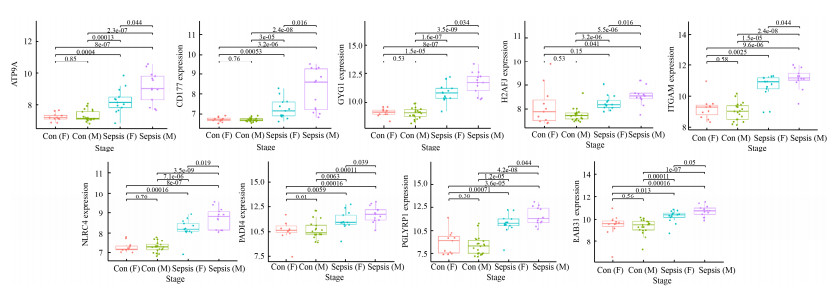 |
| 附图2 不同性别败血症患儿的外周血ATP9A、CD177、GYG1、H2AFJ、ITGAM、NLRC4、PADI4、PGLYRP1、RAB31基因表达谱差异 S2 ATP9A, CD177, GYG1, H2AFJ, ITGAM, NLRC4, PADI4, PGLYRP1, RAB31 expression profiles in peripheral blood among the sepsis infants with different gender. Con (F) means health group female infant; Con (M) means health group male infant; Sepsis (F) means sepsis group female infant; Sepsis (M) means sepsis group male infant. |
| 图选项 |
2.5 GSEA分析 标准化处理GSE80496和GSE72829矩阵数据,分别42个样本包含17516个基因和39个样本包含19573个基因的表达谱,以KEGG pathway基因集为分类标准得到两个芯片的交集KEGG通路有:氨基酸糖类-核糖代谢通路(NES=1.57,P=0.027)、PPAR信号通路(NES=1.48,P=0.037)、聚糖生物合成通路(NES=1.48,P=0.026)、自噬调控通路(NES=1.51,P=0.023)、补体和凝血因子级联反应通路(NES=1.57,P=0.027)、尼古丁和烟酰胺代谢通路(NES=1.35,P=0.035)、不饱和脂肪酸生物合成通路(NES=1.39,P=0.05)和阿尔兹海默症通路(NES=1.44,P=0.02)等。综上,我们可以发现以GO Ontology基因集为标准富集到类固醇激素分泌功能(NES=1.76,P=0.000)、腺苷酸环化酶的激活功能(NES=1.67,P=0.000)、细胞外基质降解功能(NES=1.66,P=0.002)、金属离子运输功能(NES=1.65,P=0.000)等。
3 讨论 随着高通量技术在微生物领域的应用,微生物感染相关的组学数据与日俱增,合理地运用生物信息学方法去分析公共数据库来源的微生物大数据,将有利于我们更为深入地探讨病原菌致病过程和机制。这也可为研发新型治疗感染性疾病的抗生素替代疗法提供思路和依据。一直以来,细菌性败血症与脑膜炎都是导致新生儿死亡率持续高位的主要原因之一[1, 5]。而目前,抗生素耐药菌检出率仍处于高位,尤其是导致新生儿细菌性败血症和脑膜炎(bacterial sepsis and meningitis,BSM)的常见细菌如大肠埃希菌、葡萄球菌和肺炎链球菌等,在最近发布的耐药菌监测数据中都处于排位靠前位置[2-3, 6]。因此,寻找新的抗生素替代疗法具有广阔应用前景和研究意义。
为了探究感染过程中宿主和病原菌之间的相互作用,本研究从分子水平阐明患者体内靶器官或组织中基因表达谱的变化,应用生物信息学的方法分析公共数据库来源的微生物大数据(细菌性败血症)的关键差异基因和信号通路。基于对健康和患病人群间具有差异表达的基因进行分析,并富集到与之相关的信号通路和生物学功能,此举有助于我们从整体的角度较全面地探讨细菌性败血症的潜在机制,并为后续设计实验验证提供依据。
本研究收集包含正常人群和诊断败血症患者的全血转录基因芯片数据集GSE80496和GSE72829进行生物信息学分析。通过GEO2R联用WGCNA构建的方法筛选了60个交集差异基因(其中包括MCC TOP10的枢纽基因:MMP9、ITGAM、CSTD、GAPDH、PGLYRP1、FOLR3、OSCAR、TLR5、IL1RN和TIMP1),GO注释显示这些差异基因主要富集在中性粒细胞激活、脱颗粒和细胞杀伤调控等基因功能上。具体来看,MMP9参与生理或病理过程中细胞外基质的分解,而TIMP1抑制基质分解[7-8];ITGAM编码的整合素可反映白细胞黏附、穿越内皮细胞过程[9];PGLYRP1、FOLR3基因则参与宿主固有免疫[10-11]等。另外,在筛选得到的60个交集基因中也有不少基因在败血症的致病过程中具有一定的研究意义,如ZDHHC19作为一种调节STAT3棕榈酰化的酰基转移酶,细胞因子刺激可通过促进ZDHHC19和STAT3之间的关联来激活JAK2/STAT3相关的宿主炎症反应[12];SLC51A、SLC2A3和SLC22A4作为溶质转运体家族蛋白,协助胆汁酸、葡萄糖和脂质穿越血脑屏障过程[13-14];SIGLEC5结合与GBS直接结合可损伤白细胞吞噬能力、氧化爆发和NETs产生,促进细菌存活[15]。此外通过对GSE25504婴儿败血症数据集分析,我们发现败血症患儿出生体重与血液中SIGLEC5表达呈一定负相关趋势(附图 3,http://journals.im.ac.cn/html/actamicrocn/2021/10/20211017.htm)。同时,我们分析发现相比采样前未接受抗生素治疗的败血症患儿,CDK5RAP2在接受抗生素治疗的患儿中显著升高(P < 0.05) (附图 4,http://journals.im.ac.cn/html/actamicrocn/2021/10/20211017.htm),而CDK5RAP2被证明与DNA损伤和小头畸形有密切联系[16]。此外,ATP9A、CD177、GYG1、H2AFJ、ITGAM、NLRC4、PADI4、PGLYRP1、RAB31在不同性别败血症婴儿间亦存在统计学差异(P < 0.05),上述可为后续深入研究细菌性败血症致病机制提供理论基础。
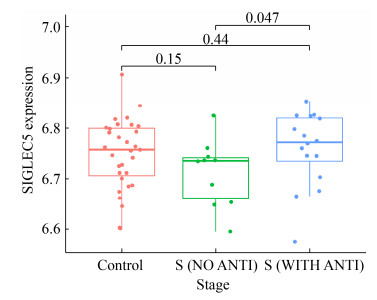 |
| 附图3 不同出生体重的败血症患儿外周血SIGLEC5表达谱差异情况 S3 SIGLEC5 expression profiles in peripheral blood among the sepsis infants with different birth weight.(Control means health group infant; S (NO ANTI) means sepsis group infant without antibiotics exposure; S (ANTI) means sepsis group infant with antibiotics exposure). |
| 图选项 |
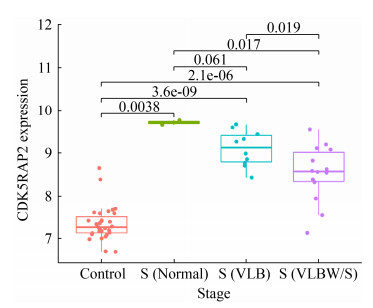 |
| 附图4 不同出生体重的败血症患儿外周血CDK5RAP2表达谱差异情况 S4 CDK5RAP2 expression profiles in peripheral blood among the sepsis infants with/without antibiotic exposure. |
| 图选项 |
针对两个不同基因数据集的GSEA分析结果显示,氨基酸糖类-核糖代谢、PPAR信号通路、聚糖生物合成通路、自噬调控通路、补体、凝血因子级联反应、尼古丁和烟酰胺代谢、不饱和脂肪酸生物合成和阿尔兹海默症通路等为主要富集的KEGG通路。类固醇激素分泌、腺苷酸环化酶的激活、细胞外基质降解和金属离子运输为GO富集的主要生物学功能。针对氨基酸糖类-核糖通路和聚糖生物合成通路的改变,有研究表明中性粒细胞胞外诱捕网(neutrophil extracellular traps,NETs)是败血症发病过程中由中性粒细胞转化形成的特殊的网状结构,该网状结构是以DNA为骨架镶嵌组蛋白、中性粒细胞弹性蛋白酶等效应蛋白为特征,而这个过程已被证明伴随多种糖类、氨基酸代谢变化[17]。过氧化物酶体增殖物激活受体(PPAR)可抑制JAK/STAT和NF-κB信号通路从而抑制炎症损伤[18]。自噬通路激活促进自噬溶酶体形成,进而诱发宿主细胞清除入侵病原菌[18]。上述信号通路均较好地关联了细菌性败血症引发的宿主防御反应。另外,值得关注的是,患者全血样本中尼古丁和烟酰胺代谢通路与血管内皮细胞(包括构成血脑屏障的脑微血管内皮细胞)膜上受体烟碱型乙酰胆碱7受体(α7 nAChR)有密切关系[19-20],越来越多研究证实α7 nAChR可作为免疫调控的关键靶点参与NF-κB等炎症通路激活,并在细菌性败血症发展至脑膜炎的致病过程起到调节作用[21]。最近Chang等研究证明α7 nAChR介导的MAPKs通路导致Aβ相关认知损伤并诱发阿尔兹海默症的发病[22],这提示α7nAChR存在同时介导细菌性败血症与阿尔兹海默症发病的可能性。
综上所述,本研究采用GEO2R、GSEA联合WGCNA的生物信息学分析手段,筛选、挖掘与细菌性败血症发病相关的10个枢纽基因(MMP9、ITGAM、CSTD、GAPDH、PGLYRP1、FOLR3、OSCAR、TLR5、IL1RN和TIMP1)、关键通路(氨基酸糖类-核糖代谢、PPAR信号通路、聚糖生物合成通路、自噬调控通路、补体、凝血因子级联反应、尼古丁和烟酰胺代谢、不饱和脂肪酸生物合成和阿尔兹海默症通路)和生物学过程(类固醇激素分泌、腺苷酸环化酶的激活、细胞外基质降解和金属离子运输为GO富集的主要生物学功能),这些信息将有助于后续更加深入探讨致病机制,为后续临床上诊断、治疗细菌性败血症提供新的思路。
References
| [1] | Schmatz M, Srinivasan L, Grundmeier RW, Elci OU, Weiss SL, Masino AJ, Tremoglie M, Ostapenko S, Harris MC. Surviving Sepsis in a referral neonatal intensive care unit: association between time to antibiotic administration and in-hospital outcomes. The Journal of Pediatrics, 2020, 217: 59-65.e1. DOI:10.1016/j.jpeds.2019.08.023 |
| [2] | Wattal C, Kler N, Oberoi JK, Fursule A, Kumar A, Thakur A. Neonatal Sepsis: mortality and morbidity in neonatal Sepsis due to multidrug-resistant (MDR) organisms: part 1. The Indian Journal of Pediatrics, 2020, 87(2): 117-121. DOI:10.1007/s12098-019-03106-z |
| [3] | Zheng YG, Hu FP, Zhu DM, Guo Y, Yang Y, Dong D, Wu S, Wang RZ, Fang H, Yu BX, Zhao Y, Gong P, Wen KZ, Zhang YR, Liu JS, Liao LF, Gu HQ, Jiang L, He W, Xue SH, Feng J, Dou R, Yue CL. CHINET surveillance of bacterial resistance in secondary care hospitals across China: report of results in 2019. Chinese Journal of Infection and Chemotherapy, 2020, 20(6): 585-593. (in Chinese) 郑永贵, 胡付品, 朱德妹, 郭燕, 杨洋, 董栋, 吴湜, 汪瑞忠, 房华, 俞碧霞, 赵勇, 龚萍, 温开镇, 张贻荣, 刘江山, 廖龙凤, 顾洪芹, 姜琳, 贺雯, 薛顺虹, 冯佼, 窦睿, 岳春雷. 2019年CHINET细菌耐药监测网二级医院监测结果. 中国感染与化疗杂志, 2020, 20(6): 585-593. |
| [4] | Baddley JW, Cantini F, Goletti D, Gómez-Reino JJ, Mylonakis E, San-Juan R, Fernández-Ruiz M, Torre-Cisneros J. ESCMID Study Group for Infections in Compromised Hosts (ESGICH) Consensus Document on the safety of targeted and biological therapies: an infectious diseases perspective (Soluble immune effector molecules. Clinical Microbiology and Infection, 2018, 24: S10-S20. DOI:10.1016/j.cmi.2017.12.025 |
| [5] | Harbeson D, Francis F, Bao W, Amenyogbe NA, Kollmann TR. Energy demands of early life drive a disease tolerant phenotype and dictate outcome in neonatal bacterial Sepsis. Frontiers in Immunology, 2018, 9: 1918. DOI:10.3389/fimmu.2018.01918 |
| [6] | Wang J, Zhang HF, Yan J, Zhang T. Literature review on the distribution characteristics and antimicrobial resistance of bacterial pathogens in neonatal Sepsis. The Journal of Maternal-Fetal & Neonatal Medicine, 2020: 1-10. |
| [7] | Shogan BD, Belogortseva N, Luong PM, Zaborin A, Lax S, Bethel C, Ward M, Muldoon JP, Singer M, An G, Umanskiy K, Konda V, Shakhsheer B, Luo J, Klabbers R, Hancock LE, Gilbert J, Zaborina O, Alverdy JC. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Science Translational Medicine, 2015, 7(286): 286ra68. DOI:10.1126/scitranslmed.3010658 |
| [8] | Ni?o ME, Serrano SE, Ni?o DC, McCosham DM, Cardenas ME, Villareal VP, Lopez M, Pazin-Filho A, Jaimes FA, Cunha F, Schulz R, Torres-Due?as D. TIMP1 and MMP9 are predictors of mortality in septic patients in the emergency department and intensive care unit unlike MMP9/TIMP1 ratio: Multivariate model. PLoS ONE, 2017, 12(2): e0171191. DOI:10.1371/journal.pone.0171191 |
| [9] | Fonseca ICCFE, da Luz FAC, Uehara IA, Silva MJB. Cell-adhesion molecules and their soluble forms: Promising predictors of "tumor progression" and relapse in leukemia. Tumour Biology, 2018, 40(11): 1010428318811525. |
| [10] | Yashin DV, Ivanova OK, Soshnikova NV, Sheludchenkov AA, Romanova EA, Dukhanina EA, Tonevitsky AG, Gnuchev NV, Gabibov AG, Georgiev GP, Sashchenko LP. Tag7 (PGLYRP1) in complex with Hsp70 induces alternative cytotoxic processes in tumor cells via TNFR1 receptor. Journal of Biological Chemistry, 2015, 290(35): 21724-21731. DOI:10.1074/jbc.M115.639732 |
| [11] | Kulohoma BW, Marriage F, Vasieva O, Mankhambo L, Nguyen K, Molyneux ME, Molyneux EM, Day PJR, Carrol ED. Peripheral blood RNA gene expression in children with pneumococcal meningitis: a prospective case-control study. BMJ Paediatrics Open, 2017, 1(1): e000092. DOI:10.1136/bmjpo-2017-000092 |
| [12] | Niu JX, Sun Y, Chen BE, Zheng BH, Jarugumilli GK, Walker SR, Hata AN, Mino-Kenudson M, Frank DA, Wu X. Fatty acids and cancer-amplified ZDHHC19 promote STAT3 activation through S -palmitoylation. Nature, 2019, 573(7772): 139-143. DOI:10.1038/s41586-019-1511-x |
| [13] | Dong C, Wong ML, Licinio J. Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans. Molecular Psychiatry, 2009, 14(12): 1105-1118. DOI:10.1038/mp.2009.92 |
| [14] | D?ring B, Lütteke T, Geyer J, Petzinger E. The SLC10 carrier family: transport functions and molecular structure. Current Topics in Membranes, 2012, 70: 105-168. |
| [15] | B?renwaldt A, L?ubli H. The sialoglycan-Siglec glyco-immune checkpoint - a target for improving innate and adaptive anti-cancer immunity. Expert Opinion on Therapeutic Targets, 2019, 23(10): 839-853. DOI:10.1080/14728222.2019.1667977 |
| [16] | Nasser H, Vera L, Elmaleh-Bergès M, Steindl K, Letard P, Teissier N, Ernault A, Guimiot F, Afenjar A, Moutard ML, Héron D, Alembik Y, Momtchilova M, Milani P, Kubis N, Pouvreau N, Zollino M, Guilmin Crepon S, Kaguelidou F, Gressens P, Verloes A, Rauch A, El Ghouzzi V, Drunat S, Passemard S. CDK5RAP2 primary microcephaly is associated with hypothalamic, retinal and cochlear developmental defects. Journal of Medical Genetics, 2020, 57(6): 389-399. DOI:10.1136/jmedgenet-2019-106474 |
| [17] | Kumar S, Gupta E, Kaushik S, Jyoti A. Neutrophil extracellular traps: formation and involvement in disease progression. Iranian Journal of Allergy, Asthma, and Immunology, 2018, 17(3): 208-220. |
| [18] | Xiang SH, Chen K, Xu L, Wang T, Guo CY. Bergenin exerts hepatoprotective effects by inhibiting the release of inflammatory factors, apoptosis and autophagy via the PPAR-γ pathway. Drug Design, Development and Therapy, 2020, 14: 129-143. DOI:10.2147/DDDT.S229063 |
| [19] | Chi F, Wang L, Zheng XY, Jong A, Huang SH. Recruitment of α7 nicotinic acetylcholine receptor to caveolin-1-enriched lipid rafts is required for nicotine-enhanced Escherichia coli K1 entry into brain endothelial cells. Future Microbiology, 2011, 6(8): 953-966. DOI:10.2217/fmb.11.65 |
| [20] | Yu JY, Zhang B, Peng L, Wu CH, Cao H, Zhong JF, Hoffman J, Huang SH. Repositioning of memantine as a potential novel therapeutic agent against meningitic E. coli-induced pathogenicities through disease-associated Alpha7 cholinergic pathway and RNA sequencing-based transcriptome analysis of host inflammatory responses. PLoS One, 2015, 10(5): e0121911. DOI:10.1371/journal.pone.0121911 |
| [21] | Coughlin JM, Rubin LH, Du Y, Rowe SP, Crawford JL, Rosenthal HB, Frey SM, Marshall ES, Shinehouse LK, Chen A, Speck CL, Wang Y, Lesniak WG, Minn I, Bakker A, Kamath V, Smith GS, Albert MS, Azad BB, Dannals RF, Horti A, Wong DF, Pomper MG. High availability of the α7-nicotinic acetylcholine receptor in brains of individuals with mild cognitive impairment: a pilot study using (18)F-ASEM PET. Journal of Nuclear Medicine, 2020, 61(3): 423-426. DOI:10.2967/jnumed.119.230979 |
| [22] | Chang KW, Zong HF, Rizvi MY, Ma KG, Zhai WY, Wang M, Yang WN, Ji SF, Qian YH. Modulation of the MAPKs pathways affects Aβ-induced cognitive deficits in Alzheimer's disease via activation of α7nAChR. Neurobiology of Learning and Memory, 2020, 168: 107154. DOI:10.1016/j.nlm.2019.107154 |
