高旭1,2, 白晓晔1,2, 郑慧娟1,2, 孙志宏1,2, 张和平1,2, 钟智1,2


1. 内蒙古农业大学乳品生物技术与工程教育部重点实验室, 内蒙古 呼和浩特 010018;
2. 内蒙古农业大学农业农村部奶制品加工重点实验室, 内蒙古 呼和浩特 010018
收稿日期:2019-05-22;修回日期:2019-07-21;网络出版日期:2019-12-05
基金项目:国家自然科学基金(31720103911,31601451);内蒙古自治区科技重大专项(ZDZX2018018);内蒙古自治区自然科学基金(2019MS03020)
*通信作者:钟智, Tel/Fax:+86-471-4301591;E-mail:imu150zhongzhi@163.com.
摘要:[目的] 旨在设计一对双歧杆菌属特异性引物以检测不同样品中低丰度双歧杆菌的含量。[方法] 在NCBI中下载57株双歧杆菌全基因组序列,以其共有单拷贝核心基因为目的片段设计双歧杆菌属特异性引物;并对引物进行PCR初筛和特异性复筛;之后借助ddPCR(Droplet Digital PCR,微滴式数字PCR)依次对筛选出的引物进行特异性、灵敏度和实用性验证。[结果] 引物Bif-D-9特异性最好,可扩增出4株双歧杆菌而不能扩增20株非双歧杆菌中的任何一株菌;同时通过ddPCR仪定量稀释后的DNA,其扩增结果呈线性下降趋势,证明其灵敏度较好;另外,Bif-D-9结合ddPCR定量出婴儿粪便中双歧杆菌的拷贝数为71 copies/μL,母亲粪便中双歧杆菌的拷贝数为2.7 copies/μL,证明了该方法的实用性。[结论] 引物Bif-D-9具有双歧杆菌属特异性,且灵敏度较高、实用性较好,适用于复杂样品中双歧杆菌属定量。
关键词:双歧杆菌属特异性引物定量验证微滴式数字PCR
Design and evaluation of Bifidobacterium genus-specific primer for quantification
Xu Gao1,2, Xiaoye Bai1,2, Huijuan Zheng1,2, Zhihong Sun1,2, Heping Zhang1,2, Zhi Zhong1,2


1. Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University, Hohhot 010018, Inner Mongolia Autonomous Region, China;
2. Key Laboratory of Dairy Products Processing, Ministry of Agriculture, Inner Mongolia Agricultural University, Hohhot 010018, Inner Mongolia Autonomous Region, China
Received: 22 May 2019; Revised: 21 July 2019; Published online: 5 December 2019
*Corresponding author: Zhi Zhong, Tel/Fax: +86-471-4301591; E-mail:imu150zhongzhi@163.com.
Foundation item: Supported by the National Natural Science Foundation of China (31720103911, 31601451), by the Major Science and Technology Projects in Inner Mongolia Autonomous Region (ZDZX2018018) and by the Natural Science Foundation of Inner Mongolia (2019MS03020)
Abstract: [Objective] The aim of this study was to design a pair of absolute quantitative specific primers for detecting the number of the low content Bifidobacterium in different samples. [Methods] The complete genome sequence of 57 strains of Bifidobacterium was downloaded from NCBI, and the specific primers of Bifidobacterium were designed based on their common single-copy core genes. Primers were screened by PCR and rescreened by specificity. Then ddPCR (Droplet Digital PCR) was used to verify the specificity, sensitivity and practicability of the selected primers. [Results] The specificity of Bif-D-9 primer was best, and 4 strains Bifidobacterium were amplified but 20 strains of non-Bifidobacterium could not be amplified. The diluted DNA was quantitatively by ddPCR, and the amplification results show a linear decreasing trend, which proves that the sensitivity is better. At the same time, Bif-D-9 combined with ddPCR can quantitatively determine the copy number of Bifidobacterium in infant feces is 71 copies/μL, and the maternal feces contained 2.7 copies/μL, which proves the practicality is good. [Conclusion] The primer Bif-D-9 was high specificity, high sensitivity and accuracy of Bifidobacterium, and is suitable for the quantitative study of Bifidobacterium in complex environmental samples.
Keywords: Bifidobacteriumgenus-specific primerquantitativeevaluationdroplet digital PCR
双歧杆菌作为一种革兰氏阳性厌氧菌,呈杆状,最早于1899年由法国巴斯德研究院的Tissier从母乳喂养的健康婴儿粪便中发现并分离出来,后根据其形态被命名为双歧杆菌[1]。肠道内的双歧杆菌作为判断人体是否健康的标志性菌株之一,研究发现,母乳喂养的婴儿体内双歧杆菌含量占肠道微生物总数的91%,而健康中青年肠道内双歧杆菌相对含量约3%-7%[2-3]。Nagpal等[4]通过研究不同生产方式对婴儿肠道菌群的影响,发现剖腹产婴儿3岁前粪便中双歧杆菌定殖率远低于顺产婴儿,得出剖腹产影响婴儿肠道内双歧杆菌的定殖这一结论。Kerckhoffs等[5]使用FISH法和q-PCR法(实时荧光定量PCR,Quantitative Real-time PCR)对41例IBS (irritable bowel syndrome)患者和26例正常人的粪便及十二指肠粘膜样品进行分析,发现IBD患者粪便及十二指肠粘膜中双歧杆菌含量较正常人下降了2倍。Fukuda等[6]通过喂食小鼠出血性大肠杆菌,发现肠道内定殖有双歧杆菌的小鼠存活率远高于无菌小鼠,且含有双歧杆菌的小鼠粪便中乙酸含量明显高于无菌小鼠,得出肠道内双歧杆菌可通过产乙酸来促进宿主上皮细胞的防御功能,从而保护宿主免受致命感染。鉴于双歧杆菌在人体肠道内具有重要作用,因此检测其含量对评估宿主健康具有重要意义。
肠道中双歧杆菌定量常用的方法是q-PCR法[7-9]。q-PCR虽然融合了第一代PCR技术的灵敏度与准确度,同时克服了易产生假阳性的缺点,但其定量准确度需要参考标准曲线,使得定量准确度降低。近年来,随着技术的发展,数字PCR (digital PCR,dPCR)凭借其高灵敏度、高精确度、高耐受性和不依靠标准曲线等优点而得到广泛应用[10-13],其中又以微滴式数字PCR应用最广。但是,无论基于qPCR还是dPCR进行定量,引物特异性依然是定量准确度的关键。目前已发表文章中的引物质量参差不齐,Youn等[14]通过对37份双歧杆菌引物进行PCR扩增,发现仅有10对引物对双歧杆菌表现出较好的特异性,而其余引物的重复性或特异性较差。另外,由于双歧杆菌引物设计大多基于16S rRNA基因序列[15-16],它虽然具有高度的保守性和特异性,但16S rRNA基因序列为多拷贝基因[17],且不同物种各拷贝之间存在差异,甚至同一菌种中的不同拷贝序列也可能存在差异[18];另外,由于生存环境不同,不同环境中的同一种细菌,其多重拷贝基因之间会根据环境因素产生异化[19-21],使得定量拷贝数的变化导致准确度降低。
本试验下载57株双歧杆菌全基因组序列,通过比对筛选出其单拷贝共有核心基因,并以该单拷贝核心基因为目的片段设计特异性引物,同时对设计出的引物进行特异性筛选,旨在筛选出一对双歧杆菌属特异性引物,应用于复杂样品中低丰度双歧杆菌属的定量。
1 材料和方法 1.1 试验菌株 本试验共使用24株菌,其中包括4株双歧杆菌和20株非双歧杆菌,这20株非双歧杆菌分别隶属于乳酸杆菌属、链球菌属、乳球菌属、片球菌属、肠球菌属、肠膜明串珠菌属、魏斯氏菌属,属于双歧杆菌亲缘菌。以上菌株由内蒙古农业大学乳酸菌菌种资源库(LABCC)提供,具体信息见表 1。
表 1. 试验菌株信息表 Table 1. Information of experimental type strains
| Type strain | Numbers |
| Bifidobacterium gallicum | DSM 20093T |
| Bifidobacterium longum subsp. infantis | DSM 20088 T |
| Bifidobacterium animalis subsp. animalis | ATCC 25527T |
| Enterococcus asini | DSM 11492 T |
| Enterococcus faecium | ATCC 19434T |
| Enterococcus italicus | DSM 15952T |
| Lactobacillus casei | ATCC 334T |
| Lactobacillus salivarius | DSM 20555T |
| Lactobacillus helveticus | DSM 20075T |
| Streptococcus thermophilus | NM-81-2 |
| Lactococcus plantarum | DSM 20686T |
| Leuconostoc psedomesenteroides | DSM 20193T |
| Weissella beninensis | DSM 22752T |
| Weissella confusa | IMAU 10245 |
| Bifidobacterium bifidum | DSM20456T |
| Weissella kandleri | DSM 20593T |
| Pediococcus acidilactici | JCM 8791T |
| Pediococcus pentosaceus | DSM 20336T |
| Pseudomonas aeruginosa | ATCC 10145T |
| Salmonella typhimurium | ATCC 19430T |
| Escherichia coli | ATCC 11775T |
| Staphylococcus aureus | ATCC 25923T |
| Shigella Castellani | ATCC 29903T |
| Listeria monocytogenes | ATCC 19115T |
表选项
1.2 仪器及试剂 高速离心机(Centrifuge,5810 R),德国Eppendorf公司;PCR扩增仪(Veriti),美国AppliedBiosystems公司;电泳仪(DYY-12),北京六一仪器厂;微量紫外分光光度计(ND-1000),美国NanoDrop公司;微滴式数字PCR(QX200 Droplet Digital PCR)系统,美国Bio-Rad公司。
QX200 ddPCR EvaGreen Supermix、QX200 DropletGeneration Oil、Droplet Reader Oil,均购自美国Bio-Rad公司。
1.3 引物设计及理论筛选 通过NCBI(www.ncbi.nih.gov)下载57株双歧杆菌的全基因组序列,采用Prokka(v1.11)软件对菌株基因组进行基因预测,之后采用Roary(v3.6.1)软件以编码蛋白氨基酸相似性95%识别全部双歧杆菌核心基因。将核心基因代表性序列采用BLAST(v2.2.24+)将全部核心基因序列与核酸序列数据库(nucleotide sequence database,nt)进行比对,删除非双歧杆菌相似度大于97%的共有核心序列,筛选得到双歧杆菌属特异性共有核酸序列。
以双歧杆菌共有序列为目的片段,使用Primer5.0软件设计双歧杆菌属引物。将设计好的引物序列重新在NCBI中进行比对,确定引物序列均来自于双歧杆菌;借助MEGA 6.0软件将得到的双歧杆菌特异性共有核酸序列与双歧杆菌亲缘菌(乳酸杆菌属、链球菌属、乳球菌属、片球菌属、肠球菌属、肠膜明串珠菌属、魏斯氏菌属)进行比对,查看引物与双歧杆菌亲缘菌的匹配率,已确定引物特异性。引物由上海桑尼生物科技有限公司合成。
1.4 DNA提取 取1 mL活化第3代的菌株培养液振荡混匀后移入2 mL无菌离心管中,12000×g离心10 min,按照天根细菌基因组DNA提取试剂盒(天根生化科技有限公司,Cat#DP302-02)的操作说明进行DNA提取,称取2 g粪便置于2 mL无菌离心管,加入1 mL DNA保护液,充分振荡30 min,4 ℃、10000×g离心10 min,弃上清,之后按照QIAamp DNA stool mini kit试剂盒(德国QIAGEN公司)操作说明进行DNA提取。将提取质量合格的DNA置于-20°冰箱备用。
1.5 PCR扩增 PCR扩增条件:扩增体系均为50 μL;基因组模板DNA2 μL (20-50 ng),10×buffer 5 μL,dNTPsmix (2.5 mmol/L) 4 μL,Taq酶(2.5 U/μL) 0.5 μL,上下游引物(20 pmol/L)各1.5 μL,ddH2O补足50 μL,PCR扩增试剂购自美国TaKaRa公司。
扩增反应程序:预变性94 ℃ 5 min;变性94 ℃ 1 min,退火(温度待测) 45 s,延伸72 ℃ 2 min,30个循环;再72 ℃延伸10 min。
1.6 引物初筛 以双歧杆菌模式菌株Bifidobacterium animalis subsp. animalis (ATCC 25527T)和Bifidobacterium longum subsp. infantis (DSM 20088T)为模板,以Primer 5.0软件提供的理论退火温度为依据,结合上下游引物的不同理论退火温度进行常规PCR扩增,并以1%琼脂糖凝胶电泳进行初筛。
1.7 特异性试验 以另外2株双歧杆菌Bifidobacterium bifidum (DSM 20456T)、Bifidobacterium gallicum (DSM 20093T)为阳性检测菌株,以20株双歧杆菌亲缘菌株(表 2)为阴性检测菌株进行PCR扩增,通过1%琼脂糖凝胶电泳进行验证。
表 2. 试验引物对两株双歧杆菌的扩增结果 Table 2. The amplification results of two strains of Bifidobacterium were tested
| Primer | Product result |
| BIF-D-1 | - |
| BIF-D-2 | ++ |
| BIF-D-3 | ++ |
| BIF-D-4 | ++ |
| BIF-D-5 | - |
| BIF-D-6 | ++ |
| BIF-D-7 | - |
| BIF-D-8 | ++ |
| BIF-D-9 | ++ |
| BIF-D-10 | - |
| BIF-D-11 | - |
| BIF-D-12 | - |
| “++” represents bright positive amplification for Bifidobacterium animalis subsp. animalis and Bifidobacterium longum subsp. infantis DNA; “-” represents no amplification for one or both strains. | |
表选项
1.8 ddPCR定量验证
1.8.1 引物特异性验证: 为进一步验证引物特异性,我们采用更灵敏的ddPCR,以双歧杆菌Bifidobacterium longum subsp. infantis DSM 20088T为阳性对照,以Lactobacillus salivarius DSM 20555T、Enterococcus faecium ATCC 19434T、Pediococcus pentosaceus DSM 20336T、Weissella confusa IMAU 10245、Streptococcus thermophilus NM-81-2、Leuconostoc psedomesenteroides DSM 20193T和Lactococcus plantarum DSM 20686T 7株双歧杆菌亲缘菌为阴性对照进行特异性验证。所有试验菌株DNA浓度均稀释至相同浓度。
1.8.2 引物灵敏度验证: 将双歧杆菌Bifidobacterium longum subsp. infantis DNA用灭菌过滤后的ddH2O进行连续10倍稀释,稀释最低浓度为10-2 ng/μL数量级,取浓度最低的4次DNA稀释液样品进行试验。以ddH2O为阴性对照,将稀释后的双歧杆菌DNA进行ddPCR扩增,以检测ddPCR的灵敏度。扩增结果将以拷贝数的形式给出。
1.8.3 引物实用性验证: 使用ddPCR对一对母婴粪便样品中的双歧杆菌进行定量,以验证引物在定量复杂样品中双歧杆菌的实用性,其中母亲生育年龄28岁,婴儿约3个月。为进一步检测引物的实用性,我们将粪便样品DNA浓度稀释为10-2 ng/μL,用以检测引物在低浓度环境样品中定量双歧杆菌的能力。
2 结果和分析 2.1 引物设计及筛选结果 本试验通过下载57株双歧杆菌全基因组序列,通过比对筛选出3个双歧杆菌属共有单拷贝核心基因,分别为RpsK、RpsG、rpsL。将上述3个基因序列在NCBI中与NT库进行比对,发现其均为双歧杆菌属特异性片段;在此基础上根据双歧杆菌模式菌株B. longum subsp. infantis NCC2705序列中的上述3个基因进行特异性引物设计,在NCBI中,RpsK、RpsG、rpsL基因在B. longum subsp. infantis NCC2705中的编号分别为:ID: 2767483、ID: 1022641、ID: 1022640;我们根据3个特异性基因共设计出12对双歧杆菌属特异性引物;将此12对引物再次在NCBI中进行BLAST比对后进行后续试验。12对引物中,其中RpsK基因4对(BIF-D-1至BIF-D-4),RpsG基因3对(BIF-D-5至BIF-D-7),rpsL基因5对(BIF-D-8至BIF-D-12)。
使用Bifidobacterium animalis subsp. animalis、Bifidobacterium longum subsp. infantis对上述12对引物进行PCR初筛发现,BIF-D-2、BIF-D-3、BIF-D-4、BIF-D-6、BIF-D-8和BIF-D-9这6对引物在退火温度为60 ℃时,可有效扩增2株双歧杆菌,且条带清晰明亮,无拖尾,扩增片段长度符合预期片段大小。而剩余6对引物对上述2株双歧杆菌中的1株或2株扩增结果呈阴性,因此排除进一步验证。对双歧杆菌呈阳性扩增的6对引物信息见表 3。
表 3. 双歧杆菌引物信息 Table 3. Primer information table of Bifidobacterium
| Primer | Gene | Primer f/r | Sequence (5′→3′) | Size/bp |
| BIF-D-2 | RpsK | BIF-D2f | GGTAAGAGTCGGACGCTGTGCRATAA | 119 |
| BIF-D2r | TCGCGTCTGAGCACTACTAGACAATAT | |||
| BIF-D-3 | RpsK | BIF-D3f | GTGCAATAAGCACTGTGGTATCC | 102 |
| BIF-D3r | TCGCGTCTGAGCACTACTAGACAATAT | |||
| BIF-D-4 | RpsK | BIF-D4f | CGGTGATCGAGATGATCGTGTTRT | 144 |
| BIF-D4r | AATTCGTTACTAGGAAACGAGGG | |||
| BIF-D-6 | RpsG | BIF-D6f | CTCATAAGATGGCGGAGGCYAACA | 196 |
| BIF-D6r | AGCTCTTGCCGGTGTAGTACAGRA | |||
| BIF-D-8 | RpsL | BIF-D8f | TATCAAGAGCGGAATACACGATG | 170 |
| BIF-D8r | GAGCAAAGAAGGCGAAGTAAGAG | |||
| BIF-D-9 | RpsL | BIF-D9f | CTTACTTCGCCTTCTTTGCTCCRTAC | 324 |
| BIF-D9r | AGAAGTCCAAGACTTTGGCCCTGA |
表选项
2.2 引物的特异性验证结果
2.2.1 引物特异性验证: 通过上述筛选,以双歧杆菌Bifidobacterium bifidum、Bifidobacterium gallicum为阳性模板,20株双歧杆菌亲缘菌为阴性模板,退火温度为60 ℃,对筛选出的6对引物进行扩增,结果如表 4所示。BIF-D-8对Bifidobacterium bifidum、Bifidobacterium gallicum均无扩增;BIF-D-2、BIF-D-3对双歧杆菌Bifidobacterium bifidum无扩增;BIF-D-6对双歧杆菌Bifidobacterium gallicum扩增条带微弱。BIF-D-4对双歧杆菌Bifidobacterium bifidum、Bifidobacterium gallicum扩增微弱;引物BIF-D-9对双歧杆菌Bifidobacterium bifidum、Bifidobacterium gallicum呈阳性扩增,且条带清晰明亮,无拖尾,对20株非双歧杆菌无特异性。引物BIF-D-9对双歧杆菌Bifidobacterium bifidum、Bifidobacterium gallicum及20株非双歧杆菌扩增结果如图 1。
表 4. 双歧杆菌属引物特异性扩增结果 Table 4. Results of specific amplification of Bifidobacterium primers
| Type strain | Product result | |||||
| BIF-D-2 | BIF-D-3 | BIF-D-4 | BIF-D-6 | BIF-D-8 | BIF-D-9 | |
| Bifidobacterium gallicum | ++ | ++ | + | + | - | ++ |
| Bifidobacterium bifidum | - | - | + | - | - | ++ |
| Enterococcus asini | - | - | - | - | - | - |
| Enterococcus faecium | - | - | - | - | - | - |
| Enterococcus italicus | - | - | - | - | - | - |
| Lactobacillus casei | - | - | - | - | - | - |
| Lactobacillus salivarius | - | - | - | - | - | - |
| Lactobacillus helveticus | - | - | - | - | - | - |
| Streptococcus thermophilus | - | - | - | - | - | - |
| Lactococcus plantarum | - | - | - | - | - | - |
| Leuconostoc psedomesenteroides | - | - | - | - | - | - |
| Streptococcus thermophilus | - | - | - | - | - | - |
| Lactococcus plantarum | - | - | - | - | - | - |
| Leuconostoc psedomesenteroides | - | - | - | - | - | - |
| Streptococcus thermophilus | - | - | - | - | - | - |
| Lactococcus plantarum | - | - | - | - | - | - |
| Leuconostoc psedomesenteroides | - | - | - | - | - | - |
| Streptococcus thermophilus | - | - | - | - | - | - |
| Lactococcus plantarum | - | - | - | - | - | - |
| Staphylococcus aureus | - | - | - | - | - | - |
| Shigella Castellani | - | - | - | - | - | - |
| Listeria monocytogenes | - | - | - | - | - | - |
| “++” expression is bright positive amplification; “+” expression is low positive amplification; “-” expression is negative amplification. | ||||||
表选项
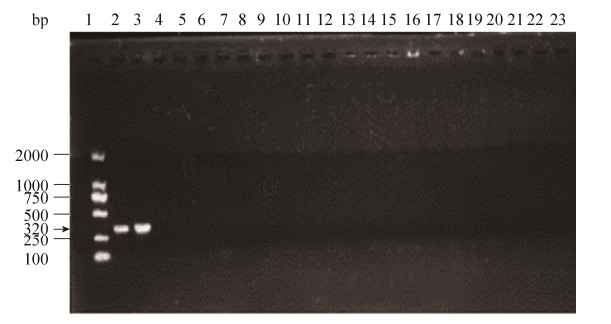 |
| 图 1 BIF-D-9引物的特异性验证胶图 Figure 1 Specificity verification map of BIF-D-9 primer. 1: marker; 2: Bifidobacterium bifidum; 3: Bifidobacterium gallicum; 4: Weissella beninensis; 5: Weissella confuse; 6: Weissella kandleri; 7: Lactobacillus casei; 8: Lactobacillus helveticus; 9: Lactobacillus salivarius; 10: Enterococcus asini; 11: Enterococcus faecium; 12: Enterococcus italicus; 13: Pediococcus acidilactici; 14: Pediococcus pentosaceus; 15: Leuconostoc psedomesenteroides; 16: Streptococcus thermophilus; 17: Lactococcus plantarum; 18: Salmonella typhimurium; 19: Escherichia coli; 20: Staphylococcus aureus; 21: Shigella Castellani; 22: Listeria monocytogenes; 23: Pseudomonas aeruginosa. |
| 图选项 |
2.2.2 ddPCR验证引物特异性: BIF-D-9引物ddPCR验证引物特异性结果如图 2所示:7份样品所产生的总微滴数均大于10000微滴,满足泊松分布;试验样品中,双歧杆菌(图 2,A06)产生的阳性微滴明显区分于阴性微滴,且阳性微滴较多;与其他样品相比,BIF-D-9对其他6株非双歧杆菌扩增则几乎无阳性微滴产生,证明引物BIF-D-9对双歧杆菌属具有较好特异性。
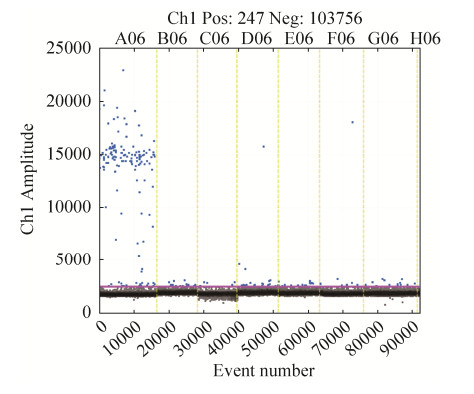 |
| 图 2 ddPCR验证引物BIF-D-9特异性 Figure 2 Validation of primer BIF-D-9 specificity using ddPCR. A06: Bifidobacterium longum subsp. infantis DSM 20088T; B06: Lactobacillus salivarius DSM20555T; C06: Enterococcus faecium ATCC 19434T; D06: Pediococcus pentosaceus DSM 20336T; E06: Weissella confusa IMAU 10245; F06: Streptococcus thermophilus NM-81-2; G06: Leuconostoc psedomesenteroides DSM 20193T; H06: Lactococcus plantarum DSM 20686T. |
| 图选项 |
2.3 ddPCR引物灵敏度验证 将双歧杆菌Bifidobacterium longum subsp. infantis DNA连续稀释,取其浓度最低的4次稀释液2、0.2、0.2×10-1、0.2×10-2 ng/μL进行ddPCR扩增,结果由图 3可知:ddH2O (图 3,A11)几乎无阳性微滴产生;而Bifidobacterium longum subsp. infantis阳性扩增逐渐降低,QuantaSoft系统显示其拷贝数分别为4810、488、51.3、17.7 copys/μL,拷贝数随着DNA浓度呈逐级下降趋势。
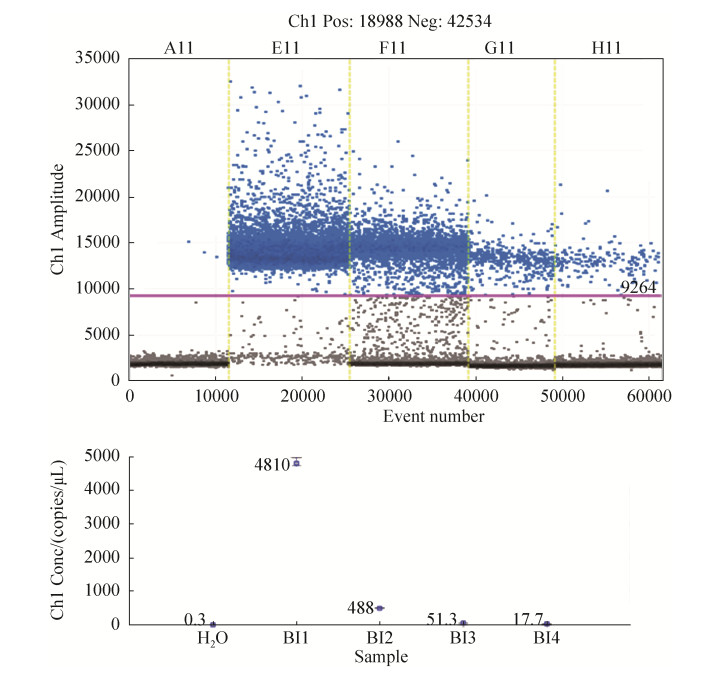 |
| 图 3 使用ddPCR验证引物BIF-D-9灵敏度 Figure 3 Verification of primer BIF-D-9 sensitivity using ddPCR. A11: H2O; E11: Bifidobacterium longum subsp. infantis (0.2×101 ng/μL); F11: Bifidobacterium longum subsp. infantis (0.2 ng/μL); G11: Bifidobacterium longum subsp. infantis (0.2×10-1 ng/μL); H11: Bifidobacterium longum subsp. infantis (0.2×10-2 ng/μL). |
| 图选项 |
2.4 ddPCR引物实用性验证 实用性验证结果如图 4所示,在母亲粪便样品(A07)中,阳性微滴与阴性微滴具有较明显区分,且无非特异性扩增,其拷贝数为2.7 copies/μL;在婴儿粪便样品(A08)中,阳性微滴与阴性微滴区分明显,拷贝数为71 copies/μL。BIF-D-9可从2份低浓度DNA粪便样品中扩增出双歧杆菌,且阳性微滴较为集中,几乎无非特异性扩增,证明该引物可扩增出复杂环境样品中较低浓度的双歧杆菌。
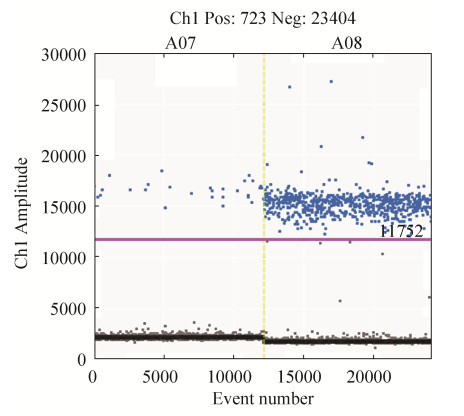 |
| 图 4 使用ddPCR验证引物BIF-D-9实用性 Figure 4 The practicality of bif-d-9 primer was verified by ddPCR. A07 is the DNA of the mother's fecal, A08 is the DNA of the child's feces. |
| 图选项 |
综上所述,本试验基于双歧杆菌单拷贝基因rpsL设计的引物BIF-D-9对双歧杆菌具有较好特异性及灵敏度,且能够应用于微量复杂样品中双歧杆菌的检测。
3 讨论 本试验筛选出的BIF-D-9引物具有较好的双歧杆菌特异性,对乳酸杆菌属、链球菌属、乳球菌属、片球菌属、肠球菌属、肠膜明串珠菌属、魏斯氏菌属DNA扩增均呈阴性。引物BIF-D-9是基于双歧杆菌共有基因rpsL基因序列设计,这个基因为单拷贝核心基因,可编码糖体蛋白S12[22]。单拷贝核心基因主要编码蛋白质,其序列含有系统发育信息,且多以点突变的方式存在,具有中等程度的进化速率和适合系统发育分析的基因结构[23],使得其基因较为稳定,不会轻易随着环境及复制次数而发生改变;并且这些基因无核酸偏向性,也不存在使序列比对变得复杂的内部重复序列[24],这些特点使得rpsL基因适用于双歧杆菌属特异性定量。且在2008年,Shenai等[26]就曾基于rpsL基因设计特异性引物并快速检测出120株突变结核分枝杆菌,证明了该基因可用于菌株的定性检测。此外,定量过程中常以单位样品中的拷贝数计算定量结果,且拷贝数由系统直接读出,默认一个拷贝基因即为一株菌,在定量多拷贝基因时(例如16S rRNA)无形中扩大了目标菌的数量,而本试验引物设计使用共有核心基因rpsL为单拷贝基因,可使结果更接近真实值。
人体肠道中双歧杆菌相对含量是判断人体是否健康的指标之一,健康人肠道约含有1014细菌,其中双歧杆菌约109-1010个[25]。纯培养法的检测限为108个/g细胞数,且只能检测出肠道内双歧杆菌的优势种,对于数量较少的非优势种则较难检测到[26];qPCR法的检测限较低为106个/g[26]细胞数,而本试验设计引物与ddPCR联合使用可检测到粪便中相对含量 < 0.1%的双歧杆菌。此外,粪便提取试剂盒中存在的多种PCR反应抑制剂使得PCR的效率减弱甚至不发生PCR反应[27],而ddPCR可将PCR反应体系通过微滴生成仪形成上万个微滴,每个微滴作为一个PCR反应体系,这使得PCR抑制剂被无限稀释,从而减弱了抑制剂对PCR反应的影响。
引物特异性是决定定量准确度的关键因素之一。然而事实上,无论基于16SrRNA基因、管家基因还是其他基因设计引物,都很难设计出绝对特异的引物[28-29]。本文设计的双歧杆菌属特异性引物BIF-D-9,在对粪便样品中的双歧杆菌进行定量时,也会产生不同程度非特异性扩增。但是由于双歧杆菌的G+C含量较高,可通过提高退火温度来减少非特异性扩增。
本试验基于双歧杆菌属核心基因设计的特异性引物BIF-D-9可灵敏地检测出复杂样品中丰度较低的双歧杆菌属,这为人体肠道、益生菌酸奶、益生菌活性补充剂等复杂样品中双歧杆菌的定量提供了有效工具;同时本试验还为其他引物设计提供了新的思路。
References
| [1] | Gomes AMP, Malcata FX. Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends in Science & Technology, 1999, 10(4/5): 139-157. |
| [2] | Ballongue J. Bifidobacteria and probiotic action//Salinen S, Von Wright A. Lactic Acid Bacteria. New York: Marcel Dekker, Inc., 2004. |
| [3] | Cheikhyoussef A, Pogori N, Chen HQ, Tian FW, Chen W, Tang J, Zhang H. Antimicrobial activity and partial characterization of bacteriocin-like inhibitory substances (BLIS) produced by Bifidobacterium infantis BCRC 14602. Food Control, 2009, 20(6): 553-559. DOI:10.1016/j.foodcont.2008.08.003 |
| [4] | Nagpal R, Kurakawa T, Tsuji H, Takahashi T, Kawashima K, Nagata S, Nomoto K, Yamashiro Y. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: a quantitative assessment. Scientific Reports, 2017, 7(1): 10097. DOI:10.1038/s41598-017-10711-5 |
| [5] | Kerckhoffs APM, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, Akkermans LMA. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World Journal of Gastroenterology, 2009, 15(23): 2887-2892. DOI:10.3748/wjg.15.2887 |
| [6] | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature, 2012, 469(7331): 543-547. |
| [7] | Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science, 2006, 314(5804): 1464-1467. DOI:10.1126/science.1131370 |
| [8] | Pohl G, Shih IM. Principle and applications of digital PCR. Expert Review of Molecular Diagnostics, 2004, 4(1): 41-47. DOI:10.1586/14737159.4.1.41 |
| [9] | Zimmermann BG, Grill S, Holzgreve W, Zhong XY, Jackson LG, Hahn S. Digital PCR: a powerful new tool for noninvasive prenatal diagnosis?. Prenatal Diagnosis, 2010, 28(12): 1087-1093. |
| [10] | Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang SL, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry, 2011, 83(22): 8604-8610. DOI:10.1021/ac202028g |
| [11] | Zhang YN, Tang ET, Du ZQ. Detection of MET gene copy number in cancer samples using the droplet digital PCR method. PLoS One, 2016, 11(1): e0146784. DOI:10.1371/journal.pone.0146784 |
| [12] | Miotto E, Saccenti E, Lupini L, Callegari E, Negrini M, Ferracin M. Quantification of circulating miRNAs by droplet digital PCR: comparison of EvaGreen- and TaqMan-based chemistries. Cancer Epidemiology, Biomarkers & Prevention, 2014, 23(12): 2638-2642. |
| [13] | Dobnik D, Spilsberg B, Ko?ir AB, Holst-Jensen A, ?el J. Multiplex quantification of 12 European Union authorized genetically modified maize lines with droplet digital polymerase chain reaction. Analytical Chemistry, 2015, 87(16): 8218-8226. DOI:10.1021/acs.analchem.5b01208 |
| [14] | Youn SY, Seo JM, Ji GE. Evaluation of the PCR method for identification of Bifidobacterium species. Letters in Applied Microbiology, 2008, 46(1): 7-13. |
| [15] | Deng SP, Chen ZD, Chen L, Ding GH, Li YX. An integrated strategy for functional analysis in 16S rRNA gene-based research of microbial communities by gene ontology. International Journal of Data Mining and Bioinformatics, 2015, 11(1): 3276-3282. |
| [16] | Xia LP, Bian LY, Xu M, Liu Y, Tang AL, Ye WQ. 16S rRNA gene sequencing is a non-culture method of defining the specific bacterial etiology of ventilator-associated pneumonia. International Journal of Clinical and Experimental Medicine, 2015, 8(10): 18560-18570. |
| [17] | Yan YW, Zhang DM. Multiplicity and heterogeneity of 16S rRNA genes in prokaryotes-a mini review. Journal of Biology, 2013, 30(4): 63-66. (in Chinese) 阎永伟, 张德民. 原核生物16S rRNA基因多重拷贝及其序列异化. 生物学杂志, 2013, 30(4): 63-66. |
| [18] | Wang Y, Zhang Z, Ramanan N. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. Journal of Bacteriology, 1997, 179(10): 3270-3276. DOI:10.1128/JB.179.10.3270-3276.1997 |
| [19] | Nilsson WB, Paranjype RN, DePaola A, Strom MS. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. Journal of Clinical Microbiology, 2003, 41(1): 442-446. DOI:10.1128/JCM.41.1.442-446.2003 |
| [20] | Lin ML, Schwarz JR. Seasonal shifts in population structure of Vibrio vulnificus in an estuarine environment as revealed by partial 16S ribosomal DNA sequencing. FEMS Microbiology Ecology, 2010, 45(1): 23-27. |
| [21] | Guinebretière MH, Thompson FL, Sorokin A, Normand P, Dawyndt P, Ehling-Schulz M, Svensson B, Sanchis V, Nguyen-The C, Heyndrickx M, De Vos P. Ecological diversification in the Bacillus cereus Group. Environmental Microbiology, 2008, 10(4): 851-865. DOI:10.1111/j.1462-2920.2007.01495.x |
| [22] | Nair J, Rouse DA, Bai GH, Morris SL. The rpsL gene and streptomycin resistance in single and multiple drug‐resistant strains of Mycobacterium tuberculosis. Molecular Microbiology, 1993, 10(3): 521-527. DOI:10.1111/j.1365-2958.1993.tb00924.x |
| [23] | Ma L, Huang Y. The application of single-copy nuclear gene in molecular phylogenetics of insects. Chinese Bulletin of Entomology, 2006, 43(1): 6-9. (in Chinese) 马兰, 黄原. 单拷贝核基因在昆虫分子系统学中的应用. 应用昆虫学报, 2006, 43(1): 6-9. |
| [24] | Friedlander TP, Regier JC, Mitter R. Phylogenetic information content of five nuclear gene sequences in animals: initial assessment of character sets from concordance and divergence studies. Systematic Biology, 1994, 43(4): 511-525. DOI:10.1093/sysbio/43.4.511 |
| [25] | Shenai S, Rodrigues C, Mehta A. Rapid speciation of 15 clinically relevant mycobacteria with simultaneous detection of resistance to rifampin, isoniazid, and streptomycin in Mycobacterium tuberculosis complex. International Journal of Infectious Diseases, 2009, 13(1): 46-58. DOI:10.1016/j.ijid.2008.03.025 |
| [26] | Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. Quantitative PCR with 16S rRNA-Gene-targeted species-specific primers for analysis of human intestinal Bifidobacteria. Applied and Environmental Microbiology, 2004, 70(1): 167-173. DOI:10.1128/AEM.70.1.167-173.2004 |
| [27] | Satake S, Clark N, Rimland D, Nolte FS, Tenover FC. Detection of vancomycin-resistant enterococci in fecal samples by PCR. Journal of Clinical Microbiology, 1997, 35(9): 2325-2330. DOI:10.1128/JCM.35.9.2325-2330.1997 |
| [28] | Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 2001, 67(6): 2578-2585. DOI:10.1128/AEM.67.6.2578-2585.2001 |
| [29] | Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, VanderGheynst J, Mills DA. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 2003, 69(11): 6801-6807. DOI:10.1128/AEM.69.11.6801-6807.2003 |
