潘晖1, 刘欣欣1, 孔令慧1, 夏永军1, 印伯星2, 艾连中1, 熊智强1


1. 上海理工大学医疗器械与食品学院, 上海食品微生物工程技术研究中心, 上海 200093;
2. 扬州市扬大康源乳业有限公司, 江苏 扬州 225004
收稿日期:2019-12-31;修回日期:2020-02-06;网络出版日期:2020-02-29
基金项目:国家自然科学基金(31871776, 31771956);上海市自然科学基金(18ZR1426800)
*通信作者:熊智强, Tel:+86-21-55803272;E-mail:xiongzq@hotmail.com.
摘要:[目的] 研究精氨酸代谢调控蛋白ArgR对嗜热链球菌胞外多糖(EPS)合成的调控作用。[方法] 利用大肠杆菌异源表达嗜热链球菌ArgR蛋白,通过尿素变性-复性和Ni2+亲和层析纯化。采用凝胶电泳迁移(EMSA)和生物膜层干涉(BLI)分析ArgR和eps基因簇中PepsA启动子的相互作用和动力学信息。构建过表达和弱化argR基因菌株,利用苯酚-硫酸法测定其合成EPS差异。[结果] 大肠杆菌异源表达的ArgR为包涵体,使用尿素变性-复性纯化可获得2.95 mg/mL可溶性蛋白;EMSA和BLI结果显示ArgR和启动子PepsA有特异性结合,且结合因解离水平低而稳定;过表达argR基因可显著降低嗜热链球菌EPS合成,而弱化argR基因则提高EPS合成。[结论] 本研究表明ArgR能特异性结合嗜热链球菌eps基因簇启动子,并负调控EPS生物合成。
关键词:嗜热链球菌ArgR胞外多糖生物合成转录调控
Arginine regulator ArgR regulates exopolysaccharides biosynthesis of Streptococcus thermophilus
Pan Hui1, Liu Xinxin1, Kong Linghui1, Xia Yongjun1, Yin Boxing2, Ai Lianzhong1, Xiong Zhiqiang1


1. Shanghai Engineering Research Center of Food Microbiology, School of Medical Instrument and Food Engineering, University of Shanghai for Science and Technology, Shanghai 200093, China;
2. Kangyuan Dairy Co. Ltd., Yangzhou University, Yangzhou 225004, Jiangsu Province, China
Received: 31 December 2019; Revised: 6 February 2020; Published online: 29 February 2020
*Corresponding author: Zhiqiang Xiong. Tel:+86-21-55803272; E-mail: xiongzq@hotmail.com.
Foundation item: Supported by the National Natural Science Foundation of China (31871776, 31771956) and by the Natural Science Foundation of Shanghai (18ZR1426800)
Abstract: [Objective] The regulatory effect of arginine regulator ArgR on the biosynthesis of exopolysaccharides (EPS) was studied in Streptococcus thermophilus. [Methods] ArgR from S. thermophilus was heterologously expressed by Escherichia coli, and purified by urea denaturation refolding and Ni2+ affinity chromatography. The interaction and kinetic information between ArgR and eps promoter PepsA were detected by electrophoretic mobility shift assays (EMSA) and biolayer interferometry (BLI). The yield alteration of EPS was determined by phenol-sulfuric acid assay when the gene argR overexpressed or repressed. [Results] Heterologous expression of ArgR was formed inclusion body, and 2.95 mg/mL of soluble protein was achieved by urea denaturation refolding. EMSA and BLI analysis showed that ArgR can specifically bind with the promoter PepsA and their affinity was high because of the low dissociation. Increased expression of argR gene reduced EPS synthesis and the suppression raised. [Conclusion] It is the first time to report that ArgR can specifically bind the promoter of eps gene cluster and negatively regulate EPS synthesis in S. thermophilus.
Keywords: Streptococcus thermophilusArgRexopolysaccharide biosynthesistranscriptional regulation
乳酸菌胞外多糖(exopolysaccharides, EPS)分为同聚多糖和杂聚多糖[1],作为食品稳定剂和增稠剂能显著增加粘度防止乳清沉淀。EPS不但有利于益生菌肠内定植,还具有增强机体免疫、抗肿瘤、抗氧化和降血脂等活性功能[2]。嗜热链球菌(Streptococcus thermophilus)是一种产酸性能优良的乳酸菌,其代谢合成EPS能显著改善食品质地和风味[3-4],广泛应用于乳制品发酵。嗜热链球菌EPS主要由不同比例的半乳糖和葡萄糖组成,伴有少量乙酰半乳糖、乙酰半乳糖胺、鼠李糖和岩藻糖等单糖[5]。嗜热链球菌EPS生物合成由基因组中大小为15–36 kb的eps基因簇控制,其中epsA和epsB编码EPS合成调控蛋白,epsC和epsD决定EPS链长,epsE、epsF、epsG、epsH和epsI是功能性重复糖单元,epsJ、epsK、epsL和epsM是聚合和输出单元。功能性重复糖单元中,epsE编码半乳糖基转移酶,受epsD调控;epsF、epsG和epsI编码不同的糖基转移酶,epsH编码乙酰转移酶[6]。
革兰氏阳性菌胞内精氨酸生物合成受argCJBDFRGH基因簇控制,其中argR编码精氨酸合成代谢的主要调控蛋白ArgR;结合L-精氨酸的ArgR能和启动子区域18 bp回文结构序列(ARG boxes)结合[7],强烈抑制操纵子上游启动子活性,负调控整个精氨酸合成通路[8]。ArgR蛋白N端含有Ser-57和Arg-58构成的“SR”序列,用于DNA结合[8];C端保守区域是与L-精氨酸和部分DNA结合的结构域[9-10],6个精氨酸分子结合在2个ArgR三聚体的C端结构域界面,起到协同调控因子的作用[11]。ArgR与ARG boxes序列结合引起基因结构的拓扑形变,诱发调控作用[12-13]。除调控精氨酸合成,ArgR在天蓝色链霉菌中直接或间接控制452个基因,涉及氮代谢、嘌呤和嘧啶生物合成、细胞形态和抗生素基因等众多代谢通路[14];在谷氨酸棒状杆菌中ArgR已证明调控谷氨酸合成代谢[15],而在嗜热栖热菌中还调控赖氨酸合成代谢[16]。这些研究表明ArgR可能是一个全局性调控因子,对非精氨酸代谢也存在广泛调控,但目前尚未见对EPS合成调控的报道。
嗜热链球菌S-3是本实验室筛选的一株具有良好产粘性和口感辅助效果的益生乳酸菌。对S-3中EPS结构解析,发现其单糖组成为N-乙酰半乳糖胺、半乳糖和葡萄糖,摩尔比为1:2:1[17],并通过全基因组测序和生物信息学分析完成eps基因簇功能注释[6]。目前国内外对EPS生物合成的转录调控研究比较欠缺,而本研究前期使用S-3的eps基因簇启动子结合磁珠后作为诱饵,通过Pull-down方法富集得到ArgR,因此本研究首次探究ArgR对嗜热链球菌EPS合成的调控作用。
1 材料和方法 1.1 材料
1.1.1 菌株、质粒以及细菌培养条件: 所用菌株与质粒如表 1所示。大肠杆菌(Escherichia coli)在LB培养基中37 ℃、200 r/min培养;嗜热链球菌在LM17培养基中37 ℃厌氧静置培养。本研究根据菌株所含质粒的抗性添加抗生素,其工作浓度为卡那霉素100 μg/mL、红霉素300 μg/Ml (大肠杆菌)和20 μg/mL (嗜热链球菌)。
表 1. 本研究使用的菌株和质粒 Table 1. Strains and plasmids in this study
| Strains and plasmids | Characterizations | Sources |
| Strains | ||
| Top10 | E. coli K-12, F–, mcrAΔ (mrr-hsd RMS-mcrBC), φ80, lacZΔM15, △lacX74, recA1, araΔ139Δ (ara-leu)7697, galU, galK, rps, (Strr) endA1, nupG. | Invitrogen |
| BL21 (DE3) | E. coli B, F–, dcm, ompT hsdS (rB-mB-), gal, λ(DE3) | Invitrogen |
| S-3 | S. thermophilus | Our lab |
| Plasmids | ||
| pET30α | Expressional vector, KanR, 5.4 kb | Novagen |
| pIB184 | Shuttle vector, EmR, containing constitutive promoter P23 | BioVector |
| pPH12 | pET30α containing N-6His-argR gene, KanR | This study |
| pPH13 | pET30α containing N, C-6His-argR gene, KanR | This study |
| pPH14 | pET30α containing C-6His-argR gene, KanR | This study |
| pPH15 | pIB184 containing argR gene, EmR | This study |
| pPH16 | pIB184 containing antisense argR gene, EmR | This study |
表选项
1.1.2 试剂与仪器: 质粒提取、DNA和PCR纯化回收试剂盒购自Axygen公司;高保真酶、限制性内切酶、T4连接酶和蛋白酶抑制剂购自宝生物工程(大连)有限公司和南京诺唯赞生物科技有限公司;Ni-NTA琼脂糖树脂购自QIAGEN公司;EMSA化学发光试剂盒购自上海碧云天生物技术有限公司;电泳仪设备及超灵敏凝胶成像仪购自Bio-Rad公司;分子互作仪及链霉亲和素传感器购自Fortebio公司。
1.1.3 引物合成及序列测定: 引物由生工生物工程(上海)股份有限公司合成(表 2),质粒由华大基因股份有限公司测序。
表 2. 本研究使用的引物序列 Table 2. Primers used in this study
| Primer name | Sequence (5′→3′) |
| ArgR-1-F | GGAATTCCATATGTTGGAGTTAATCCGAAAGATTGTCC |
| ArgR-1-R | CCGCTCGAGTTCTTCAACCCACTTGACGATTTG |
| ArgR-2-F | GGAATTCCATATG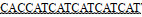 TTGGAGTTAATCCGAAAGATTGTCC TTGGAGTTAATCCGAAAGATTGTCC |
| ArgR-2-R | CCGCTCGAGTTCTTCAACCCACTTGACGATTTG |
| ArgR-3-F | GGAATTCCATATG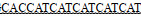 TTGGAGTTAATCCGAAAGATTGTCC TTGGAGTTAATCCGAAAGATTGTCC |
| ArgR-3-R | CCGCTCGAGCTATTCTTCAACCCACTTGACGATTTG |
| box-F1 | AGCCAGTGGCGATAAGTTTGTAAAAGGACGCCATTT |
| box-R1 | AGCCAGTGGCGATAAGATAAATTGCTCCTAAAAATTAAAATTAGGTATTCCCCATAATACAACCTCATTTCAAT |
| box-R2 | ATATCAATCATTTAAATATTGTGAACTATCTTTTA |
| box-F2 | TTAAATGATTGATATCATAATG |
| box-R3 | AGCCAGTGGCGATAAGATAAATTGCTCCTAAAAATT |
| Biotin-F-R | AGCCAGTGGCGATAAG |
| ArgR-pIB-F | TATGAATGACAATGATGTTGGATCCATGTTGGAGTTAATCCGAAAGATTGT |
| ArgR-pIB-R | CGATAGATCTCGAGCTCTAGAATTCCTATTCTTCAACCCACTTGACGATTTG |
| antiArgR-pIB-R | TATGAATGACAATGATGTTGGATCCCTATTCTTCAACCCACTTGACGATTTG |
| antiArgR-pIB-R | CGATAGATCTCGAGCTCTAGAATTCATGTTGGAGTTAATCCGAAAGATTGT |
| Underlined parts are restriction sites, underlined parts are His-tag sequences, and underlined parts are homologous arms. | |
表选项
1.2 质粒构建 以S-3基因组为模板,使用引物ArgR-1-F/R、ArgR-2-F/R和ArgR-3-F/R扩增3种argR基因片段,经Nde I和Xho I酶切后通过T4连接酶插入质粒pET30α,转化至E. coli Top 10,在抗性平板筛选阳性转化子,测序正确的重组质粒分别命名为pPH12、pPH13和pPH14。使用引物ArgR-pIB-F/R、antiArgR-pIB-F/R扩增得到目标基因,质粒pIB184经EcoR I和BamH I酶切,酶切产物和目标基因连接后转化,测序正确的质粒分别命名为pPH15和pPH16。
1.3 诱导表达ArgR 将质粒pET30α、pPH12、pPH13和pPH14分别转化至E. coli BL21 (DE3),将种子液接种至新鲜培养基,37 ℃培养至OD550=0.6,加入终浓度0.4 mmol/L IPTG后20 ℃诱导8 h。使用磷酸盐缓冲液(PBS)离心洗涤菌体,加入1% (V/V)蛋白酶抑制剂,低温超声破碎(800 W功率,5 min),经离心获得上清液和沉淀。使用SDS-PAGE检测菌液、上清液和沉淀悬浮液。
1.4 低温初始诱导ArgR E. coli BL21(DE3)/pPH13种子液接种至新鲜培养基,分别加入终浓度为0.2、0.4、0.6 mmol/L的IPTG,20 ℃培养;分别在2、4、6、8、10、12和24 h收集菌液,SDS-PAGE检测蛋白表达,二喹啉甲酸法测定总蛋白。
1.5 ArgR包涵体纯化 按照前述方法培养和诱导菌体,破碎离心后获得包涵体,使用包涵体溶解液(8 mol/L Urea,50 mmol/L NaH2PO4,300 mmol/L NaCl;pH 8.0)将其低温溶解,经0.45 μm滤膜过滤;用2个柱体积洗涤液(5 mmol/L Imidazole,50 mmol/L NaH2PO4,300 mmol/L NaCl;pH 8.0)平衡Ni-NTA琼脂糖树脂后,包涵体溶解液以0.5 mL/min的流速流过柱体;用5个柱体积平衡液(10 mmol/L Imidazole,8 mol/L Urea,50 mmol/L NaH2PO4,300 mmol/L NaCl;pH 8.0)以1 mL/min的流速洗去杂蛋白;用2个柱体积洗脱液(250 mmol/L Imidazole,8 mol/L Urea,50 mmol/L NaH2PO4,300 mmol/L NaCl;pH 8.0)洗脱出目的蛋白。使用3 kDa超滤管低温脱盐浓缩,测定蛋白质浓度,–80 ℃保存备用。
1.6 凝胶迁移检测ArgR与PepsA启动子结合 使用引物box-F1/R3以及Biotin-F-R扩增得到两端含有生物素标记的PepsA片段,同时获得未标记片段。凝胶阻滞实验(EMSA,electrophoresis mobility shift assays)如下:10 μL反应体系包含25 ng标记探针,不同浓度ArgR (0–10 μmol/L)和2 μL结合缓冲液,对照组需额外加入1.25 μg未标记探针;25 ℃孵育20 min后上样至4% (W/V)非变性聚丙烯酰胺胶,使用Tris-硼酸缓冲液(TBE)电泳; 经电泳迁移的凝胶条带随后转移至尼龙膜, 在紫外交联、封闭后结合Streptavidin-HRP,使用化学发光成像。
1.7 ArgR与PepsA启动子结合解离系数 生物素标记的PepsA片段使用DNA缓冲液(10 mmol/L HEPES,2 mmol/L MgCl2,0.1 mmol/L EDTA,200 mmol/L KCl;pH 8.0)溶解,ArgR溶液中加入终浓度1‰ (W/V)BSA和2‰ (V/V) Tween-20作为母液,使用蛋白质缓冲液(1‰ BSA,2‰ Tween-20,PBS)稀释至62.5、125、250、500和1000 mg/L。溶液处理后加样至检测孔板,使用生物膜层干涉仪(BIL)测定:传感器在DNA缓冲液中平衡10 min,在生物素标记DNA溶液中饱和10 min,蛋白质缓冲液中平衡8 min,蛋白样品中饱和10 min,蛋白质缓冲液解离15 min。
1.8 ArgR结合位点突变验证 PepsA启动子上预测含有2个ArgR结合位点(ARG-box),其中ARG-box1位于PepsA启动子上游38 bp,ARG-box1和ARG-box2间距79 bp。为得到PepsA上结合位点的突变DNA,以PepsA启动子为模板,使用引物box-F1/R1扩增得到ARG-box1缺失突变DNA;以引物box-F1/R2及box-F2/R3的扩增产物为模板,用引物box-F1/R3扩增得到ARG-box2缺失突变DNA;以引物box-F1/R2及box-F2/R1扩增产物为模板,用引物box-F1/R1扩增得到ARG-box1及ARG-box2缺失突变DNA。3种缺失突变DNA通过引物Biotin-F-R扩增标记生物素,采用EMSA检测ArgR与3种缺失突变DNA的结合情况。10 μL EMSA反应体系含25 ng标记探针和10 μmol/L ArgR,对照组不引入蛋白,竞争探针为1.25 μg。
1.9 测定EPS合成量变化 重组菌株S-3/pPH15、S-3/pPH16和S-3/pIB184过夜培养的种子液分别以3%接种量接种于LM17培养基,培养至6、12、24 h离心收集上清液。根据已报道方法[18]纯化EPS,上清液经终浓度6% (W/V)三氯乙酸处理10–12 h后离心去除蛋白质,上清液用纤维素透析袋(8000–1000 Da)透析3 d,每4 h更换去离子水。定容后采用苯酚硫酸法[19]测定EPS含量,以半乳糖为标准,每组6个平行,数据应用SPSS软件进行独立样本t检验分析。
2 结果和分析 2.1 ArgR异源表达和蛋白纯化 本研究前期将S-3中eps基因簇启动子PepsA生物素标记,结合链霉亲和素磁珠,通过Pull-down方法捕获到结合蛋白ArgR。在此基础上,为探究ArgR对嗜热链球菌EPS合成的转录调控作用,我们首先在大肠杆菌中异源表达ArgR。argR基因为432 bp,其蛋白分子量为16123 Da。采用TMHMN在线分析(http://www.cbs.dtu.dk)显示其无跨膜结构。pPH12、pPH13和pPH14分别为C端、NC两端和N端含有His标签的argR表达质粒(图 1-A)。但将含不同位置His标签的ArgR表达质粒pPH12、pPH13和pPH14分别转入大肠杆菌BL21 (DE3)后,37 ℃培养,0.4 mmol/L IPTG诱导后发现,3种工程菌表达的可溶性ArgR蛋白含量较低,主要形成包涵体(图 1-B)。
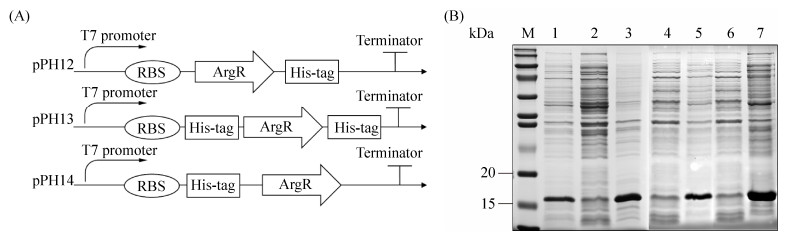 |
| 图 1 质粒示意图以及ArgR表达和纯化 Figure 1 Plasmid structure and protein expression of ArgR. A: Structures of plasmid pPH12, pPH13 and pPH14. B: ArgR expression in E. coli by SDS-PAGE. M: protein marker; lane 1, 2, 3: bacterial culture, supernatant and precipitation from the plasmid pPH12; lane 4, 5: supernatant and precipitation from pPH13; lane 6, 7: supernatant and precipitation from pPH14. |
| 图选项 |
为增加可溶性蛋白表达,我们采用低温诱导降低ArgR蛋白合成速度,减少包涵体形成。在20 ℃使用不同IPTG浓度和诱导时间表达ArgR,分析条带灰度值的结果表明与其他两种IPTG浓度相比,在0.4 mmol/L IPTG浓度下ArgR可溶性表达量最高,在12 h时ArgR占比总蛋白达到82% (图 2-A);在此条件下,可溶性蛋白浓度达到0.15 mg/mL (图 2-B)。尽管低温诱导获得更多的可溶性ArgR,但对于蛋白纯化而言,其浓度仍然过低,而包涵体含量较高。因此,我们选择Ni2+柱结合效率最高的E. coli BL21(DE3)/pPH13包涵体,在尿素溶解后纯化获得可溶性ArgR (图 2-C),通过超滤管脱盐浓缩后浓度达到2.95 mg/mL,满足后续实验要求。
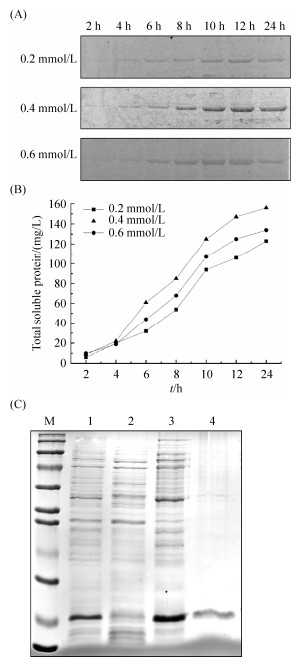 |
| 图 2 不同诱导条件的ArgR蛋白表达及尿素变性纯化包涵体 Figure 2 ArgR expression and purification using urea. A: SDS-PAGE of ArgR expression at different IPTG concentrations and induction time. B: total soluble protein content under different induction conditions. C: purification of inclusion body protein of ArgR by urea. M: protein marker; lane 1: bacterial culture; lane 2, 3: supernatant and precipitation after fragmentation and centrifugation; lane 4: purified protein by urea. |
| 图选项 |
2.2 ArgR与PepsA启动子的结合 为研究ArgR对eps基因簇的调控,采用EMSA和BIL实验分别检测ArgR对eps基因簇PepsA启动子的结合特异性和结合亲和力(图 3),PepsA启动子DNA结合ArgR蛋白后发生阻滞迁移,在8 μmol/L浓度时达到饱和;加入竞争探针后标记探针的阻滞迁移基本消失,说明结合具有特异性(图 3-A)。ArgR与PepsA启动子的BIL分析表明,结合系数(Ka)为1.45×103±1.10×101 M–1S–1,解离系数(Kd)为2.57×10–5±4.83×10–6 S–1,亲和力系数(KD)为1.78×10–8±3.60×10–10 M,数据拟合度0.978 (图 3-B),说明ArgR与PepsA的解离水平低,分子间结合稳定。上述两种检测结果都显示ArgR可稳定结合PepsA,说明ArgR可以调控eps基因簇。
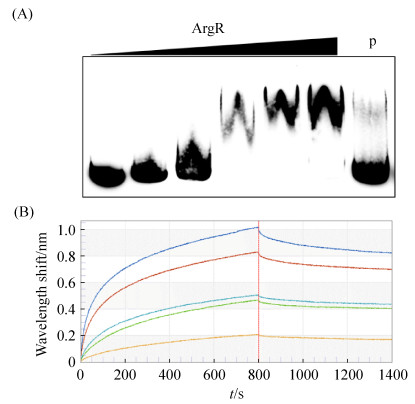 |
| 图 3 ArgR和PepsA启动子的相互作用 Figure 3 Interaction between ArgR and Promoter PepsA. A: The interaction by EMSA; Increasing amount of ArgR concentrations (0, 2, 4, 6, 8, 10 μmol/L) were used with the biotin-labeled probe at 25 ng; lane P: 1.25 μg unlabeled probe was added as the control. B: Kinetic curves measured by BLI with ArgR concentrations from 1 mg/mL to 0.0625 mg/mL, and all baselines leveled when balance and dissociation. |
| 图选项 |
为进一步证明DNA结合位点对ArgR结合的影响,我们将PepsA中2个预测的ArgR结合位点ARG-box1和ARG-box2进行缺失突变后,与ArgR进行EMSA结合验证(图 4)。泳道1, 2, 3显示PepsA可特异性结合ArgR;ARG-box1缺失突变(泳道5)和ARG-box2缺失突变(泳道7)DNA仍能结合ArgR,但在相同蛋白浓度条件下,与泳道2相比出现略微拖尾现象,ARG-box单缺失突变可能表现为结合作用的减弱;ARG-box1和ARG-box2双缺失突变DNA (泳道9)无法与同浓度ArgR产生阻滞迁移。突变结果直接证明ArgR结合在PepsA的ARG-box上。
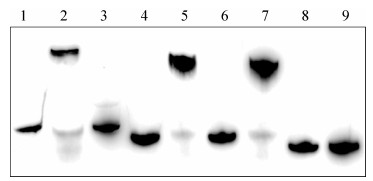 |
| 图 4 PepsA中ARG-box缺失突变对结合ArgR的影响 Figure 4 Effects of ARG-box mutation of PepsA on ArgR binding. Lane 1: original PepsA without ArgR; lane 2: original PepsA with ArgR; lane 3: original PepsA with ArgR and competitive DNA; lane 4: ARG-box1 deleted PepsA without ArgR; lane 5: ARG-box1 deleted PepsA with ArgR and competitive DNA; lane 6: ARG-box2 deleted PepsA without ArgR; lane 7: ARG-box2 deleted PepsA with ArgR and competitive DNA; lane 8: ARG-box1 and ARG-box2 deleted PepsA without ArgR; lane 9: ARG-box1 and ARG-box2 deleted PepsA with ArgR and competitive DNA. |
| 图选项 |
2.3 过表达和弱化argR基因对EPS合成的影响 为进一步证明eps基因簇受AgrR调控,我们测定argR基因过表达或弱化后对EPS合成的影响(图 4-B)。与含空质粒pIB184菌株(对照)相比,argR基因过表达或弱化菌株生长延迟期和倍增时间并无显著性差异(图 5-A),说明细菌初级代谢可能未受较大影响。培养到6 h时这3种菌株的EPS产量均无显著差别,但培养12 h时过表达argR菌株EPS产量仅为0.089 g/L,24 h时0.116 g/L,与对照相比均显著降低;而采用反义RNA弱化argR菌株,EPS产量在12 h与对照组无显著性差异,而在24 h时与对照相比显著上升,达到0.152 g/L (图 5-B)。上述结果表明EPS合成受AgrR负调控。
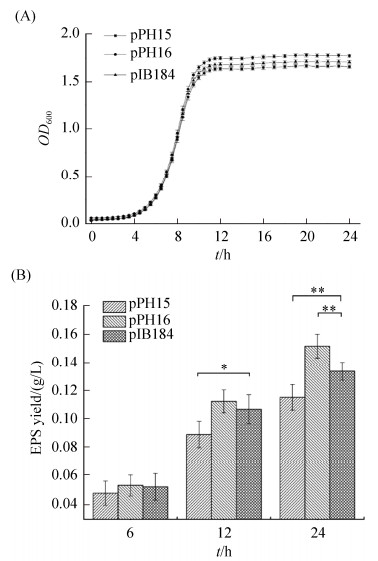 |
| 图 5 过表达和弱化argR对生长和EPS合成的影响 Figure 5 Effects on cell growth and EPS yield by overexpressed or weakened argR. A: Growth of engineered strains containing plasmids pPH15 (overexpressed), pPH16 (weakened), and pIB184 (the control), respectively. B: EPS yield of engineered strains containing plasmids pPH15, pPH16, and pIB184 (the control), respectively. *: 0.01 < P≤0.05. |
| 图选项 |
3 讨论 本实验采用E. coli BL21(DE3)/pET系统异源表达ArgR,但主要形成包涵体,这可能是由于多肽合成时速率较高,缺乏有效折叠[20],导致异源表达ArgR蛋白溶解度低,形成有利于避免蛋白酶降解的包涵体。我们通过尿素变性复性后纯化获得高浓度可溶性蛋白,可能是由于尿素可破坏蛋白质氢键,打开肽链,使亲水基团暴露,提高可溶性;尿素变性蛋白一般不破坏一级结构,当其除去或浓度降低后可复性[21]。此外,也可使用对二级结构更温和的溶解剂取代尿素和盐酸胍[22]。本实验中ArgR经变复性以及超滤脱盐浓缩处理,在SDS-PAGE检测中尽管出现微弱的杂条带,但并不干扰后续的特异性结合实验。文献报道利用本研究相同的系统表达嗜热栖热菌ArgR时,并未形成包涵体[16, 23];使用E.coli M15/pQE30系统同样表达出猪链球菌可溶性ArgR[24],与本研究结果不同。通过NCBI数据库对嗜热链球菌与其他链球菌属和嗜热菌属的ArgR进行序列比对,发现它们的相似度仅为40%–60%,这可能是嗜热链球菌ArgR主要以包涵体形成表达的重要因素。
乳酸菌存在CodY、ArgR、GlnR、AhrC、FhuR等众多氮代谢调控蛋白,其中大多数调控因子受生长介质影响,例如CodY在嗜热链球菌中调控作用受支链氨基酸浓度影响[25]。精氨酸代谢调控蛋白ArgR通过与ARG-box靶序列结合,参与精氨酸合成代谢,在结构和DNA结合位点与AhrC高度相似[26];ArgR与启动子区域存在2–3个ARG-box的结构结合更紧密[27]。本研究中,EMSA结果显示加入50倍浓度竞争探针后,标记探针的阻滞迁移消失,表明PepsA与ArgR存在特异性结合;通过BIL检测PepsA与ArgR结合情况显示,两者解离系数低,结合稳定,两种检测结果都与先前Pull-down方法得到的结果一致。为寻找ArgR结合启动子的DNA序列,基于公布在Regprecise网站(http://regprecise.lbl.gov)上嗜热链球菌CNRZ1066的ArgR结合序列,使用WebLogo在线服务器(http://weblogo.berkeley.edu)生成motif信息:5′-WwTdWATAAWwATAvAdW-3′(图 6);同时综合嗜热栖热菌已报道的ARG-box[23]:5′-nkT GyATAnTtTTnCnnG-3′,大肠杆菌的ARG-box[28]:5′-WnTGnATWWWWATnCAnW-3′,以及乳酸乳球菌的ARG-box[29]:5′-AwwGwATAAWWATrCWnw-3′,通过MEME网站(http://meme-suite.org/)对eps基因簇中PepsA启动子中ARG-box位点进行预测(表 3),分析结果表明PepsA上可能存在2个ArgR结合位点,分别为ARG-box1:TTATTATTATTAT AACAT和ARG-box2:TTTATATCATTTTCCATT。ARG-box缺失突变实验(图 4)表明PepsA仅缺失1个ARG-box仍与ArgR有结合作用,但缺失上述2个ARG-box时结合阻滞现象消失,证明了预测的准确性。存在2个结合位点可能使ArgR与PepsA有更强的结合效果,具有显著影响嗜热链球菌eps基因簇转录调控的潜力。
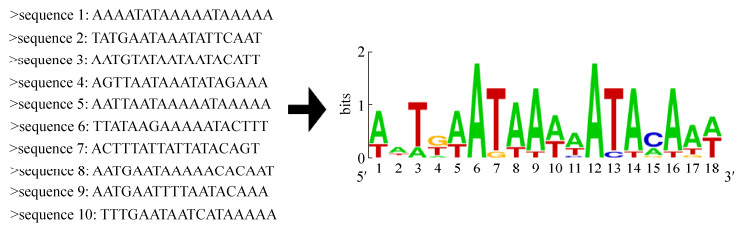 |
| 图 6 嗜热链球菌CNRZ1066的ARG-box motif Figure 6 The motif of ARG-box generated based on S. thermophilus CNRZ1066. |
| 图选项 |
表 3. eps基因簇ArgR结合位点分析 Table 3. Analysis of ArgR binding sites on eps biosynthetic gene cluster
| Strains | Site (in S-3) | P-value | Sequence (5′→3′) | Motif (5′→3′) |
| Streptococcus thermophilus CNRZ1066 | 16471–16488 | 2.2×10–5 | TTATTATTATTATAACAT | WwTdWATAAWwATAvAdW |
| 16568–16585 | 3.9×10–5 | TTTATATCATTTTCCATT | ||
| Escherichia coli | 16568–16585 | 9.8×10–5 | TTTATATCATTTTCCATT | WnTGnATWWWWATnCAnW |
| Thermus thermophilus | 16568–16585 | 4.8×10–5 | TTTATATCATTTTCCATT | nkTGyATAnTtTTnCnnG |
| 16600–16617 | 4.8×10–5 | CTTTTACATTTTTAGTAG | ||
| Lactococcus lactis | 16471–16488 | 1.4×10–4 | TTATTATTATTATAACAT | AwwGwATAAWWATrCWnw |
| 16568–16585 | 5.0×10–4 | TTTATATCATTTTCCATT |
表选项
ArgR蛋白作为转录调控因子在不同代谢途径中调控作用不同。例如,在猪链球菌中ArgR蛋白作为转录激活因子,激活精氨酸合成代谢中arc操纵子的结构基因表达[24];而在嗜热栖热菌、谷氨酸棒杆菌和乳酸乳球菌等细菌中作为转录抑制子,结合操纵子上不同启动子,负调控精氨酸合成与分解代谢。在嗜热栖热菌中ArgR通过结合argG启动子,负调控精氨酸合成代谢[23];在谷氨酸棒杆菌中ArgR并非结合argG启动子,而是通过结合argC启动子,负调控精氨酸合成代谢[8];在乳酸乳球菌中ArgR结合argC、argG和arcA启动子,负调控精氨酸代谢[26]。本研究为进一步证实ArgR功能,我们采用基因弱化策略来替代argR基因敲除,通过反义RNA和正常RNA碱基互补来弱化基因表达[30];反义RNA对基因表达抑制效果受多重因素影响,同一区域不同组合的反义序列在嗜热链球菌中效果也差异明显[31]。通过导入argR反义序列弱化argR基因表达,弱化后的EPS产量在12 h未变化,在24 h显著提高;而过表达argR使EPS合成显著减少,证实ArgR作为一个转录抑制子,负调控嗜热链球菌EPS合成。本研究表明ArgR通过结合eps基因簇中PepsA启动子的ARG-box位点,负调控嗜热链球菌EPS合成,为阐明嗜热链球菌EPS生物合成的转录调控奠定基础。
References
| [1] | Cui YH, Xu TT, Qu XJ, Hu T, Jiang X, Zhao CY. New insights into various production characteristics of Streptococcus thermophilus strains. International Journal of Molecular Sciences, 2016, 17(10): 1701. DOI:10.3390/ijms17101701 |
| [2] | Ruas-Madiedo P, Hugenholtz J, Zoon P. An overview of the functionality of exopolysaccharides produced by lactic acid bacteria. International Dairy Journal, 2002, 12(2/3): 163-171. |
| [3] | Dan T, Jin RL, Ren WY, Li T, Chen HY, Sun TS. Characteristics of milk fermented by Streptococcus thermophilus MGA45-4 and the profiles of associated volatile compounds during fermentation and storage. Molecules, 2018, 23(4): 878. DOI:10.3390/molecules23040878 |
| [4] | Li YH, Zhang LW, Wang WJ, Zhang LL, Han X, Jiao YH. The flavor property of soft cheese fermented by two stains of Streptococcus thermophilus and made of reconstituted milk. Advanced Materials Research, 2012, 396-398: 1536-1540. |
| [5] | Stingele F, Neeser JR, Mollet B. Identification and characterization of the eps (Exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. Journal of Bacteriology, 1996, 178(6): 1680-1690. DOI:10.1128/JB.178.6.1680-1690.1996 |
| [6] | Xiong ZQ, Kong LH, Lai PFH, Xia YJ, Liu JC, Li QY, Ai LZ. Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3. Journal of Dairy Science, 2019, 102(6): 4925-4934. DOI:10.3168/jds.2018-15572 |
| [7] | Cho S, Cho YB, Kang TJ, Kim SC, Palsson B, Cho BK. The architecture of ArgR-DNA complexes at the genome-scale in Escherichia coli. Nucleic Acids Research, 2015, 43(6): 3079-3088. DOI:10.1093/nar/gkv150 |
| [8] | Yim SH, Jung S, Lee SK, Cheon CI, Song E, Lee SS, Shin J, Lee MS. Purification and characterization of an arginine regulatory protein, ArgR, in Corynebacterium glutamicum. Journal of Industrial Microbiology & Biotechnology, 2011, 38(12): 1911-1920. |
| [9] | Tian G, Maas WK. Mutational analysis of the arginine repressor of Escherichia coli. Molecular Microbiology, 1994, 13(4): 599-608. DOI:10.1111/j.1365-2958.1994.tb00454.x |
| [10] | Burke M, Merican AF, Sherratt DJ. Mutant Escherichia coli arginine repressor proteins that fail to bind L-arginine, yet retain the ability to bind their normal DNA-binding sites. Molecular Microbiology, 1994, 13(4): 609-618. DOI:10.1111/j.1365-2958.1994.tb00455.x |
| [11] | Van Duyne GD, Ghosh G, Maas WK, Sigler PB. Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli. Journal of Molecular Biology, 1996, 256(2): 377-391. DOI:10.1006/jmbi.1996.0093 |
| [12] | Lu CD, Houghton JE, Abdelal AT. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. Journal of Molecular Biology, 1992, 225(1): 11-24. DOI:10.1016/0022-2836(92)91022-H |
| [13] | Thompson JF, Landy A. Empirical estimation of protein-induced DNA bending angles:applications to λ site-specific recombination complexes. Nucleic Acids Research, 1988, 16(20): 9687-9705. DOI:10.1093/nar/16.20.9687 |
| [14] | Pérez-Redondo R, Rodríguez-García A, Botas A, Santamarta I, Martín JF, Liras P. ArgR of Streptomyces coelicolor is a versatile regulator. PLoS One, 2012, 7(3): e32697. DOI:10.1371/journal.pone.0032697 |
| [15] | Lee SY, Kim YH, Min J. Conversion of phenol to glutamate and proline in Corynebacterium glutamicum is regulated by transcriptional regulator ArgR. Applied Microbiology and Biotechnology, 2010, 85(3): 713-720. |
| [16] | Fujiwara K, Tsubouchi T, Kuzuyama T, Nishiyama M. Involvement of the arginine repressor in lysine biosynthesis of Thermus thermophilus. Microbiology, 2006, 152(12): 3585-3594. DOI:10.1099/mic.0.29222-0 |
| [17] | Xu ZY, Guo QB, Zhang H, Wu Y, Hang XM, Ai LZ. Exopolysaccharide produced by Streptococcus thermophiles S-3:Molecular, partial structural and rheological properties. Carbohydrate Polymers, 2018, 194: 132-138. DOI:10.1016/j.carbpol.2018.04.014 |
| [18] | Ai LZ, Zhang H, Guo BH, Chen W, Wu ZJ, Wu Y. Preparation, partial characterization and bioactivity of exopolysaccharides from Lactobacillus casei LC2W. Carbohydrate Polymers, 2008, 74(3): 353-357. DOI:10.1016/j.carbpol.2008.03.004 |
| [19] | Cuesta G, Suarez N, Bessio MI, Ferreira F, Massaldi H. Quantitative determination of pneumococcal capsular polysaccharide serotype 14 using a modification of phenol-sulfuric acid method. Journal of Microbiological Methods, 2003, 52(1): 69-73. DOI:10.1016/S0167-7012(02)00151-3 |
| [20] | Sunitha K, Chung BH, Jang KH, Song KB, Kim CH, Rhee SK. Refolding and purification of Zymomonas mobilis levansucrase produced as inclusion bodies in fed-batch culture of recombinant Escherichia coli. Protein Expression & Purification, 2000, 18(3): 388-393. |
| [21] | Smith VR, Walker JE. Purification and folding of recombinant bovine oxoglutarate/malate carrier by immobilized metal-ion affinity chromatography. Protein Expression & Purification, 2003, 29(2): 209-216. |
| [22] | Puri NK, Crivelli E, Cardamone M, Fiddes R, Bertolini J, Ninham B, Brandon MR. Solubilization of growth hormone and other recombinant proteins from Escherichia coli inclusion bodies by using a cationic surfactant. The Biochemical Journal, 1992, 285(3): 871-879. |
| [23] | Iwanaga N, Ide K, Nagashima T, Tomita T, Agari Y, Shinkai A, Kuramitsu S, Okada-Hatakeyema M, Kuzuyama T, Nishiyama M. Genome-wide comprehensive analysis of transcriptional regulation by ArgR in Thermus thermophilus. Extremophiles, 2014, 18(6): 995-1008. DOI:10.1007/s00792-014-0669-2 |
| [24] | Fulde M, Willenborg J, de Greeff A, Benga L, Smith HE, Valentin-Weigand P, Goethe R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology, 2011, 157(2): 572-582. |
| [25] | Qi MX, Mei F, Wang H, Sun M, Wang GJ, Yu ZN, Je Y, Li MS. Function of global regulator CodY in Bacillus thuringiensis BMB171 by comparative proteomic analysis. Journal of Microbiology and Biotechnology, 2015, 25(2): 152-161. |
| [26] | Larsen R, Buist G, Kuipers OP, Kok J. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. Journal of Bacteriology, 2004, 186(4): 1147-1157. DOI:10.1128/JB.186.4.1147-1157.2004 |
| [27] | Cunin R, Eckhardt T, Piette J, Boyen A, Pièrard A, Glansdorff N. Molecular basis for modulated regulation of gene expression in the arginine regulon of Escherichia coli K-12. Nucleic Acids Research, 1983, 11(15): 5007-5019. DOI:10.1093/nar/11.15.5007 |
| [28] | Maas WK. The arginine repressor of Escherichia coli. Microbiological Reviews, 1994, 58(4): 631-640. DOI:10.1128/MMBR.58.4.631-640.1994 |
| [29] | Larsen R, Van Hijum SAFT, Martinussen J, Kuipers OP, Kok J. Transcriptome analysis of the Lactococcus lactis ArgR and AhrC regulons. Applied and Environmental Microbiology, 2008, 74(15): 4768-4771. DOI:10.1128/AEM.00117-08 |
| [30] | Schmiedel JM, Klemm SL, Zheng YN, Sahay A, Blüthgen N, Marks DS, Van Oudenaarden A. Gene expression. MicroRNA control of protein expression noise. Science, 2015, 348(6230): 128-132. DOI:10.1126/science.aaa1738 |
| [31] | Sturino JM, Klaenhammer TR. Antisense RNA targeting of primase interferes with bacteriophage replication in Streptococcus thermophilus. Applied and Environmental Microbiology, 2004, 70(3): 1735-1743. |
