王蕊, 吴宪, 李刚

 , 修伟明
, 修伟明
 , 王金鑫, 王欣奕, 王丽丽, 李洁, 张贵龙, 赵建宁, 杨殿林
, 王金鑫, 王欣奕, 王丽丽, 李洁, 张贵龙, 赵建宁, 杨殿林 农业农村部环境保护科研监测所, 天津 300191
收稿日期:2019-11-13;修回日期:2020-04-15;网络出版日期:2020-05-21
基金项目:转基因生物新品种培育重大专项(2015ZX08013002-004,2016ZX08012005-005);公益性行业(农业)科研专项(201503121-04);国家自然科学基金项目(31200424);中国农业科学院科技创新工程项目
*通信作者:李刚,E-mail:ligang06@caas.cn;
修伟明,E-mail:xiuweiming@caas.cn.
摘要:[目的] 通过研究林地转型耕地对土壤编码碱性磷酸酶基因的细菌群落丰度、多样性和结构的影响,为丘陵区耕地长期施肥下农田土壤微生物多样性丧失的影响机制以及未来的退耕还林过程中土壤微生物多样性的提升和土地可持续利用研究提供一些基础数据和技术支撑。[方法] 采用实时荧光定量PCR(real-time quantitative PCR,qPCR)和高通量测序技术解析土壤编码碱性磷酸酶基因的细菌群落的丰度、多样性和结构变化,并耦合土壤化学性质分析,明确土壤编码碱性磷酸酶基因的细菌群落丰度和多样性与土壤化学性质的关系以及关键的驱动因子。[结果] 林地垦殖为农田后,长期施肥导致土壤酸化,pH从5.58降至4.72,而土壤速效磷则从2.49 mg/kg增至49.3 mg/kg。相应地,耕地土壤编码碱性磷酸酶基因的细菌群落的丰度和Shannon指数均显著低于林地。基于编码碱性磷酸酶的phoD基因(alkaline phosphatase-encoding gene)序列的物种分类表明,丘陵区土壤编码碱性磷酸酶基因的细菌群落的优势门为变形菌门(Proteobacteria)、蓝藻门(Cyanobacteria)、浮霉菌门(Planctomycetes)、放线菌门(Actinobacteria)、厚壁菌门(Firmicutes)和疣微菌门(Verrucomicrobia),其中林地土壤的蓝藻门的相对丰度显著高于耕地。耕地土壤的慢生根瘤菌属(Bradyrhizobium)和芽孢杆菌属(Bacillus)的相对丰度显著高于林地,而中慢生根瘤菌属(Mesorhizobium)、假单胞菌属(Pseudomonas)、Chlorogloea属、Gemmata属、Phormidesmis属和Pseudolabrys属的相对丰度显著低于林地。土壤编码碱性磷酸酶基因的细菌群落结构因林地转型耕地而发生显著改变。phoD基因丰度和Shannon指数与pH显著正相关,而与总磷、速效磷、硝态氮和铵态氮均显著负相关,其中土壤速效磷是这些影响因素中影响最强烈的,长期施用无机磷肥导致含碱性磷酸酶的土壤细菌群落对有机磷分解的能力退化。[结论] 林地转型耕地加之长期施肥改变了土壤pH和速效磷,并在其他理化因子的协同驱动下,导致土壤编码碱性磷酸酶基因的细菌群落丰度、多样性和结构的显著变化。
关键词:林地耕地丘陵区phoD基因实时定量PCR高通量测序
Effect of conversion of forest to arable land in the hilly region, Northeast China on soil alkaline phosphatase gene encoding bacterial community
Rui Wang, Xian Wu, Gang Li

 , Weiming Xiu
, Weiming Xiu
 , Jinxin Wang, Xinyi Wang, Lili Wang, Jie Li, Guilong Zhang, Jianning Zhao, Dianlin Yang
, Jinxin Wang, Xinyi Wang, Lili Wang, Jie Li, Guilong Zhang, Jianning Zhao, Dianlin Yang Agro-Environmental Protection Institute, Ministry of Agriculture and Rural Affairs, Tianjin 300191, China
Received: 13 November 2019; Revised: 15 April 2020; Published online: 21 May 2020
*Corresponding author: Gang Li, E-mail:ligang06@caas.cn;
Weiming Xiu, E-mail:xiuweiming@caas.cn.
Foundation item: Supported by the National Transgenic Breeding Project of China(2015ZX08013002-004, 2016ZX08012005-005), by the Agro-scientific Research in the Public Interest(201503121-04), by the National Natural Science Foundation of China(31200424) and by the Agricultural Science and Technology Innovation Program of CAAS
Abstract: [Objective] By studying the effect of conversion of forest to arable land on the abundance, diversity and structure of soil alkaline phosphatase gene encoding bacterial community, to provide basic data of soil microbial diversity, for sustainable land use. [Methods] The abundance, diversity and structure of soil alkaline phosphatase gene encoding bacterial community were investigated using real-time fluorescence quantitative PCR (qPCR) and high-throughput sequencing. Combining the determination and statistical analysis of soil chemical properties, relationships among the soil alkaline phosphatase gene encoding bacterial community abundance, Shannon diversity and soil chemical properties were also evaluated, as well as the key driving factors affecting community structure. [Results] After the forest land was reclaimed as arable land, long-term fertilization led to acidification of the soil, the pH dropped from 5.58 to 4.72, and the soil available phosphorus increased from 2.49 mg/kg to 49.3 mg/kg. Correspondingly, the soil alkaline phosphatase gene encoding bacterial community abundance and diversity significantly decreased with the conversion of forest to arable land. Based on species classification of alkaline phosphatase-encoding gene sequence, Proteobacteria, Cyanobacteria, Planctomycetes, Actinobacteria, Firmicutes and Verrucomicrobia were the dominant phyla, and the relative abundance of Cyanobacteria in forest significantly higher than that in arable land. The relative abundance of Bradyrhizobium and Bacillus in arable land were significantly higher than that in forest, while significantly higher relative abundance of Mesorhizobium, Pseudomonas, Chlorogloea, Gemmata, Phormidesmis and Pseudolabrys was found in forest. The structure of soil alkaline phosphatase gene encoding bacterial community was significantly affected by the land-use change. The abundance and Shannon diversity of soil alkaline phosphatase gene encoding bacterial community were significantly positively correlated with pH, but significantly negatively correlated with the soil available phosphorus, total phosphorus, nitrate nitrogen (NO3-) and ammonium nitrogen (NH4+), the soil available phosphorus is the most affected among these factors. The application of inorganic phosphate fertilizer caused the degradation of organophosphorus decomposition ability of soil bacterial community containing alkaline phosphatase. [Conclusion] The soil available phosphorus and pH changed by land-use change and long-term fertilization cause the alteration of the abundance, diversity and structure of the soil alkaline phosphatase gene encoding bacterial community under the coordinated driving of other physical and chemical factors.
Keywords: forestarable landhilly regionphoD geneqPCRhigh-throughput sequencing
磷是植物必需的常量养分之一,在植物生长和生态系统功能中发挥重要作用[1–2],但只有无机正磷酸盐形态的磷才能够被植物吸收[3]。尽管土壤总磷含量普遍很高[4],但主要以有机形态存在[5]。土壤有机磷不仅含有化合态的肌醇磷酸酯、磷酯、核苷酸、磷蛋白、磷酸糖,而且含有吸附在有机物表面的和与有机物络合的磷酸盐以及微生物生物量磷。磷酸单酯是施肥土壤中有机磷的主要存在形式,约占有机磷总量的60%– 70%[6],磷酸单酯可被磷酸酶水解为正磷酸盐阴离子[7–8],直接被植物以及微生物吸收[9–10]。因此,磷酸酶在提高土壤磷的生物有效性中发挥重要作用。
催化磷酸酯键水解(除肌醇磷酸盐外,如植酸)的磷酸酶包括碱性磷酸酶和酸性磷酸酶[8, 11],在将土壤磷转化成生物可利用的形式中发挥至关重要的作用[12]。此前的研究表明,酸性磷酸酶主要由植物合成和释放[13–14],而碱性磷酸酶主要由土壤微生物分泌,尤其是细菌[8, 15]。碱性磷酸酶是将土壤有机磷水解为可供植物利用的无机磷的关键酶[3, 8]。至今已鉴定了至少3种原核ALP基因家族(例如,phoA、phoD、phoX),构成磷调节子(Pho)的重要组成部分[16]。phoA采用锌和酶作为辅助因子,水解磷酸单酯,而phoX和phoD采用钙作为辅助因子,水解磷酸单酯[17–19]。这三种同源基因的分布随生态系统发生变化。phoA、phoD和phoX基因广泛分布于水生生态系统,如海水和淡水[20–21],与phoA和phoX基因相比,phoD基因在陆地生态系统中出现的频率最高[16],因此被选作最具代表性的碱性磷酸酶基因,已作为关键生物指示物用于研究土壤有机磷的转化。
目前已有多项研究报道了编码碱性磷酸酶基因的细菌群落变化对环境条件的响应。长期施肥试验中贫磷条件改变了编码碱性磷酸酶基因的细菌群落的结构,但编码碱性磷酸酶基因的细菌群落对pH的响应更为强烈[22]。Neal等[23]发现休耕地、耕地和草地间土壤编码碱性磷酸酶基因的细菌群落显著不同。Fraser等[24]研究表明,phoD基因丰度与碱性磷酸单酯酶活性正相关,但却未发现phoD基因丰度与磷有效性间的显著相关性。Tan等[16]发现与贫磷环境相比,长期施用磷肥后牧草土壤编码碱性磷酸酶基因的细菌群落的多样性增加。Jorquera等[25]聚合酶链式反应-变形梯度凝胶电泳(PCR-DGGE)方法发现施氮降低了编码碱性磷酸酶基因的细菌群落的多样性。目前对于编码碱性磷酸酶基因的细菌群落的研究集中于施肥方式[16, 22, 24–31],但对土地利用方式转变下编码碱性磷酸酶基因的细菌群落的存在、多样性和群落结构缺乏研究。
作为人类活动主要方式的土地利用方式转变可以引起陆地生态系统中元素生物地球化学循环过程的变化[32]。森林转型耕地是土地利用方式转变的主要类型,通过改变土壤的物理、化学和生物学性质进而影响土壤质量[33–34]。本课题组前期研究发现,东北丘陵区林地转型耕地显著降低了土壤pH,而显著增加土壤养分(如总磷、速效磷、硝态氮和铵态氮),土壤细菌和真菌的碳源代谢活性和特征也发生了显著变化,土壤细菌16S rRNA基因丰度显著增加,自养细菌群落结构变化显著[35–37]。基于以上前期研究和前人研究的结果,我们推测林地转型耕地将降低土壤细菌phoD基因的丰度和多样性,并且土壤编码碱性磷酸酶基因的细菌群落结构因土地利用的转变而发生改变。本研究采用qPCR和高通量测序技术评价了土壤编码碱性磷酸酶基因的细菌群落对林地转型耕地的响应,并分析了土壤编码碱性磷酸酶基因的细菌群落与土壤化学性质间的关系以探索驱动土壤编码碱性磷酸酶基因的细菌群落变化的关键因子,以期增进对碱性磷酸单酯酶的生物学重要性的认识,并为东北丘陵地区土壤可持续利用及生态环境重建提供科学依据。
1 材料和方法 1.1 试验区概况与试验设计 试验地位于吉林省四平市伊通满族自治县的北京大北农生物技术有限公司试验示范基地和西苇林场(125°20′ N,43°15′ E),地处长白山脉向松辽平原过渡的丘陵地带,为东北典型的低山丘陵区。该地区属中温带湿润季风气候,年均降雨量为627.8 mm,年均气温为4.6 ℃。试验地的土壤类型为白浆土。
试验共设2个处理:林地(F)和耕地(A)。林地的典型林分为红松(Pinus koraiensis),林龄50年以上,在林地中随机选取3个15 m×15 m的小区。耕地为农用坡耕地,与林地毗邻(相距50 m),截至2017年采样时已连续种植26年玉米,在试验田中随机选取3个10 m×15 m的小区。耕地采用单季玉米连作的耕作制度,仅施用化肥,使用N:P:K=15:15:15的复合肥(总氮、有效五氧化二磷、氧化钾),近5年化肥施用量平均为750 kg/hm2,以基肥形式一次性施用在土壤表面,随后翻入土壤表层。玉米于每年5月份播种,深度为5 cm。玉米播种和收获均采取人工方式,收获后将玉米植株的地上部分移出试验田,其他田间管理措施按照当地常规操作进行。
1.2 土壤样品采集与分析 2017年6月使用直径3.5 cm的土钻采集土壤样品(0–15 cm)。从每个处理小区采用W形多点采样法采集15个土芯并混合为1份土壤样品。将土壤样品装入灭菌自封袋中于低温样品储藏箱中运送至实验室。所有样品充分混匀后过筛(2 mm)并分为2份样品。一份样品于–70 ℃保存用于DNA提取,另一份样品风干后用于土壤化学性质分析。本研究中所采用的化学性质分析方法见王蕊等[36]的方法,相关实验数据引用王蕊等[36](表 1)。
表 1. 林地和耕地的土壤化学性质、细菌phoD基因丰度和Shannon指数 Table 1. Soil properties、soil bacterial phoD gene abundance and shannon index of forest and arable land
| Treatment | TN/(g/kg) | TP/(g/kg) | OM/(g/kg) | AP/(mg/kg) | NO3–-N/(mg/kg) | NH4+-N/(mg/kg) | pH | phoD gene abundance/(109 copies/g soil) | Shannon index |
| F | 0.77±0.01a | 0.28±0.02b | 33.13±1.49a | 2.49±0.32b | 5.88±0.60b | 1.16±0.17b | 5.58±0.02a | 5.44±0.53A | 3.72±0.14A |
| A | 0.74±0.02a | 0.52±0.07a | 26.39±5.47a | 79.32±2.61a | 126.46±11.63a | 2.72±0.91a | 4.72±0.06b | 0.86±0.02B | 1.92±0.34B |
| The data is mean±SD (n=3); different lowercase letters indicate that the difference reaches a significant level (P < 0.05); different uppercase letters indicate that the difference reaches a extremely significant level (P < 0.01); F: forest land; A is arable land; TN: total nitrogen; TP: total phosphorus; OM: organic matter; AP: available phosphorus; NO3–-N: nitrate nitrogen; NH4+-N: ammonium nitrogen; pH: pH, the same below. | |||||||||
表选项
1.3 土壤DNA提取与phoD基因丰度的定量 使用DNeasy PowerSoil Kit (QIAGEN,Carlsbad,CA,USA)从0.25 g冻土提取总DNA,按照操作说明书进行。使用NanoDrop ND-2000分光光度计(NanoDrop Technologies, USA)检测总DNA的浓度和质量。
在CXF96 Real-time PCR仪(Bio-Rad, USA)上通过qPCR估算phoD基因的丰度。qPCR采用的试剂为iTaq Universal SYBR Green Supermix (Bio-Rad,USA),引物为phoD-733 (5′-TGGGAYGATCAYGARGT-3′)和phoD-1083 (5′-CTGSGCSAKSACRTTCCA-3′),片段长度约为350 bp左右[19],qPCR反应体系和扩增条件见Chen等[31]的方法。每个样品的DNA均设置3次重复。使用pGEM?-T Easy vector (Promega,USA)构建标准质粒,并对含有目的phoD基因片段的质粒进行测序以证实序列的准确性。使用NanoDrop ND-2000分光光度计测定质粒浓度并转换为单位体积的拷贝数。将质粒进行10倍系列稀释并用于构建标准曲线。每个浓度梯度的标准质粒均设置3次重复。
1.4 phoD基因扩增子的高通量测序与数据处理 phoD基因扩增子的高通量测序的引物、PCR反应体系和程序与qPCR相同,为了区分不同土壤样品产生的扩增子,在引物的5?端添加不同的序列标签。将3次重复反应的PCR产物混合,使用Agencourt AMPure XP(Beckman Coulter, Inc. S. Kraemer Boulevard Brea,CA,USA)清洗,并使用QubitTM ssDNA Assay Kit (Invitrogen,Thermo Fisher Scientific,USA)进行定量。将纯化的扩增子以摩尔体积混合,并使用Illumina MiSeq Reagent Kit v3 (600-cycle) (Illumina Inc.,San Diego,CA)进行成对末端测序。测序由生工生物工程(上海)股份有限公司完成。
采用QIIME(Quantitative Insights Into Microbial Ecology)去除所有长度小于300 bp、含有任何模糊碱基以及平均质量值低于20的序列[38]。采用PEAR根据PE序列之间的重叠关系,将成对的序列拼接成一条序列[39],之后按照barcode标签序列识别并区分样品得到相应样品数据。将拼接的核酸序列进一步去除单体和嵌合体序列[40]。RDP FungGene (http://fungene.cme.msu.edu/)是针对微生物功能基因序列的数据库,其按照功能分为抗生素抗性、植物致病基因、生物地球化学循环、系统进化标志、生物降解、金属循环及其他等7类功能基因,常被用于功能基因高通量测序后的比对[41]。因此,本研究使用RDP function gene pipeline的FrameBot将拼接序列转换为氨基酸序列,并去除所有不能通读、无法匹配到phoD基因的氨基酸序列以及在序列的任何位置存在终止密码子的氨基酸序列。使用USEARCH version 7.1中的USEARCH在75%的相似度阈值下定义操作分类单元(operational taxonomic units,OTUs)[16]。将高通量测序获得的序列原始数据提交至NCBI (National Center for Biotechnology Information)的GenBank数据库,提取号为SRR9878564。
1.5 统计分析 采用SPSS统计软件(version 20.0,IBM SPSS Inc.)进行Pearson相关分析和方差分析。采用典范对应分析(canonical correspondence analysis,CCA)比较样品的β-多样性并评价环境因子对编码碱性磷酸酶基因的细菌群落结构的影响。CCA采用CANOCO for Windows 5.0软件(Microcomputer Power, Inc., Ithaca. NY)进行。使用Monte Carlo检验在CCA中执行手工预选变量程序以确定环境变量的显著性(P < 0.05)。采用Origin 2018绘图。
2 结果和分析 2.1 细菌phoD基因丰度和多样性指数以及与土壤化学性质的相关性 由表 1可见,与林地土壤细菌phoD基因丰度(以土计,下同)(5.44×109 copies/g soil)相比,耕地土壤细菌phoD基因丰度降低了84.1%,为8.64×108 copies/g soil,差异极显著(P < 0.01)。林地转型耕地也显著降低了土壤细菌phoD基因群落的Shannon指数(P < 0.01),耕地的Shannon指数仅为林地的0.52倍。
根据Pearson相关分析,phoD基因丰度和Shannon指数均与总磷(TP*)、速效磷(AP**)和硝态氮(NO3–-N**)显著负相关,而与pH**显著正相关(表 2,*P < 0.05;**P < 0.01)。此外,phoD基因丰度与铵态氮(NH4+-N*)显著负相关(表 2,P < 0.05)。同时,phoD基因丰度与Shannon指数极显著正相关(表 2,P < 0.01)。
表 2. 林地和耕地土壤化学性质、phoD基因丰度和Shannon指数间的相关性 Table 2. Relationship among soil chemical properties, phoD gene abundance and Shannon index of forest and arable land
| Item | phoD gene abundance | Shannon index |
| TN | 0.661 | 0.588 |
| TP | –0.908* | –0.856* |
| OM | 0.674 | 0.784 |
| AP | –0.973** | –0.973** |
| NO3–-N | –0.968** | –0.946** |
| NH4+-N | –0.822* | –0.692 |
| pH | 0.965** | 0.973** |
| phoD gene abundance | 1 | 0.968** |
| Shannon index | 0.968** | 1 |
| *: P < 0.05; **: P < 0.01; F: forest land; A: arable land; TN: total nitrogen; TP: total phosphorus; OM: organic matter; AP: available phosphorus; NO3–-N: nitrate nitrogen; NH4+-N: ammonium nitrogen; pH: pH, the same below. | ||
表选项
2.2 编码碱性磷酸酶基因的细菌群落结构 在去除嵌合体、单体和不能通读以及与phoD基因不匹配的序列后,所有土壤样品共获得191890条高质量序列。每个样品的序列数范围为30837至32629,可聚类为265个OTU。这些OTU可以分为9个已知门、16个已知纲、37个已知目、65个已知科和103个已知属。其中变形菌门(Proteobacteria)、蓝藻门(Cyanobacteria)、浮霉菌门(Planctomycetes)、放线菌门(Actinobacteria)、厚壁菌门(Firmicutes)和疣微菌门(Verrucomicrobia)为优势门(> 1%),占所有序列的98%以上(图 1-A)。在28个优势属(> 1%)中,以慢生根瘤菌属(Bradyrhizobium)和红游动菌属(Rhodoplanes)最为丰富(图 1-B和1-C)。显著性分析发现,优势门中,林地的蓝藻门(Cyanobacteria)(17.67%)的相对丰度显著高于耕地(3.72%)(P < 0.05);优势属中,林地的中慢生根瘤菌属(Mesorhizobium)(1.24%)的相对丰度显著高于耕地(0.11%)(P < 0.05),假单胞菌属(Pseudomonas)(5.76%)的相对丰度显著高于耕地(0.28%)(P < 0.05),Chlorogloea属(0.71%)的相对丰度显著高于耕地(0%)(P < 0.05),Gemmata属(0.68%)的相对丰度显著高于耕地(0.02%)(P < 0.05),Phormidesmis属(5.48%)的相对丰度显著高于耕地(0.02%)(P < 0.05),Pseudolabrys属(4.68%)的相对丰度显著高于耕地(0.25%)(P < 0.05),而慢生根瘤菌属(Bradyrhizobium)(18.15%)的相对丰度显著低于耕地(50.09%)(P < 0.05),芽孢杆菌属(Bacillus) (0.18%)的相对丰度显著低于耕地(2.08%) (P < 0.05),unclassified Isosphaeraceae属(0.20%)的相对丰度显著低于耕地(0.93%)(P < 0.05)。
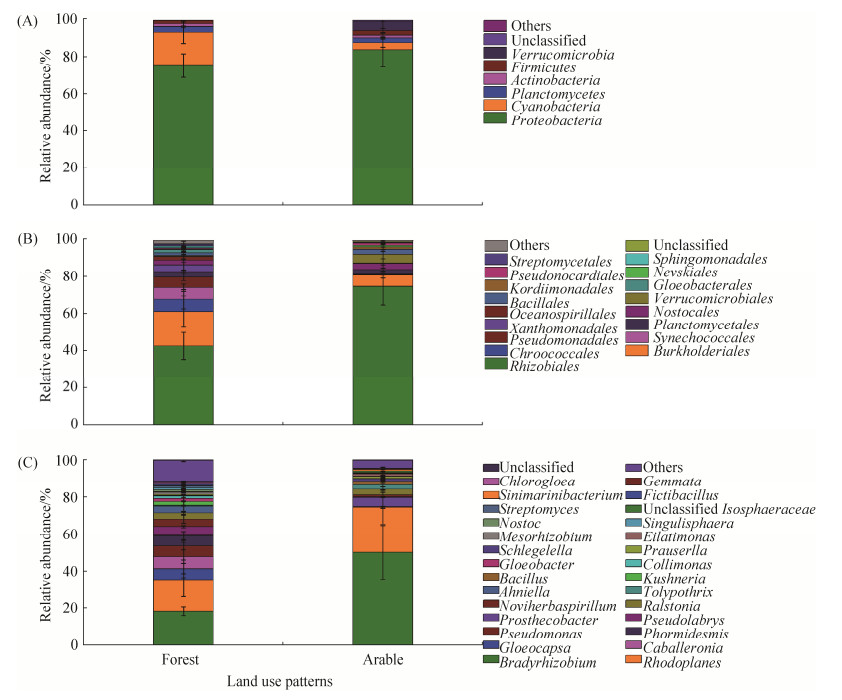 |
| 图 1 林地和耕地土壤编码碱性磷酸酶基因的细菌群落组成 Figure 1 Composition of soil alkaline phosphatase gene encoding bacterial community of forest and arable land. A: phylum; B: order; C: genus. |
| 图选项 |
2.3 土壤理化因子与编码碱性磷酸酶基因的细菌群落的CCA分析 采用CCA分析确定环境因子对土壤编码碱性磷酸酶基因的细菌群落结构的潜在影响。CCA分析的前两轴分别解释了总变异的58.7%和16.9%。林地和耕地土壤编码碱性磷酸酶基因的细菌群落沿第一轴分离明显(图 2),以上结果说明林地和耕地土壤编码碱性磷酸酶基因的细菌群落结构不同,林地转型耕地改变了土壤编码碱性磷酸酶基因的细菌群落结构。根据Monte Carlo排列测试,说明速效磷、pH、硝态氮、总磷和铵态氮对土壤编码碱性磷酸酶基因的细菌群落结构影响显著。其中pH与林地土壤编码碱性磷酸酶基因的细菌群落结构显著正相关;而速效磷、硝态氮、总磷和铵态氮与耕地土壤编码碱性磷酸酶基因的细菌群落结构显著负相关,并且速效磷和硝态氮的影响程度大于总磷和铵态氮,其中速效磷是这些影响因素中影响最强烈的,其他理化因子为协同因子,产生协同效应。
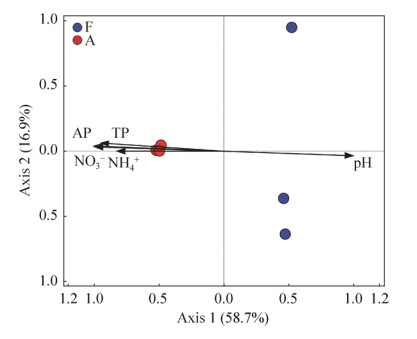 |
| 图 2 土壤编码碱性磷酸酶基因的细菌群落结构的典范对应分析 Figure 2 Soil alkaline phosphatase gene encoding bacterial community structure by CCA analysis. F: forest land; A: arable land; TN: total nitrogen; TP: total phosphorus; OM: organic matter; AP: available phosphorus; NO3–-N: nitrate nitrogen; NH4+-N: ammonium nitrogen; pH: pH. |
| 图选项 |
2.4 土壤编码碱性磷酸酶基因的细菌群落与土壤化学性质间的关系 通过Pearson相关分析评价优势门、目和属的相对丰度与土壤化学性质间的关系。图 3可见,优势门中仅蓝藻门(Cyanobacteria)的相对丰度与pH显著正相关,而与速效磷、硝态氮和总磷显著负相关。目水平下,假单胞菌目(Pseudomonadales)和Synechococcales目与pH极显著正相关,而与速效磷和硝态氮极显著负相关,与总磷显著负相关;鞘脂单胞菌目(Sphingomonadales)同样分别与pH以及速效磷和硝态氮呈显著正相关和负相关(P < 0.05),而根瘤菌目(Rhizobiales)与鞘脂单胞菌目(Sphingomonadales)相反。Nevskiales目和Pseudonocardiales目仅与铵态氮极显著正相关(P < 0.01)。属水平下,慢生根瘤菌属(Bradyrhizobium)、芽孢杆菌属(Bacillus)和unclassified Isosphaeraceae与pH、速效磷和硝态氮的相关关系相似,而且芽孢杆菌属(Bacillus)与总磷和铵态氮显著正相关,而unclassified Isosphaeraceae属仅与总磷显著正相关,却与有机质显著负相关。假单胞菌属(Pseudomonas)、Chlorogloea属、Phormidesmis属和Pseudolabrys属与pH、速效磷、硝态氮和总磷的显著相关关系相似,且Pseudolabrys属与铵态氮显著正相关。中慢生根瘤菌属(Mesorhizobium)和Gemmata与pH、速效磷和硝态氮的相关性与Chlorogloea属和Phormidesmis属相同。念珠藻属(Nostoc)与硝态氮、总磷和铵态氮显著负相关。Sinimarinibacterium属与总磷和铵态氮的相关关系与念珠藻属(Nostoc)相似。Prauserella属与铵态氮显著正相关,而Schlegelella属与有机质显著负相关。这些与不同化学性质表现出显著相关关系的优势目和属的相对丰度总和分别达到49.8%–87.3%和37.7%–66.0%,说明土壤化学性质的变化直接导致土壤编码碱性磷酸酶基因的细菌群落在目和属的变化,最终改变了土壤编码碱性磷酸酶基因的细菌群落结构。
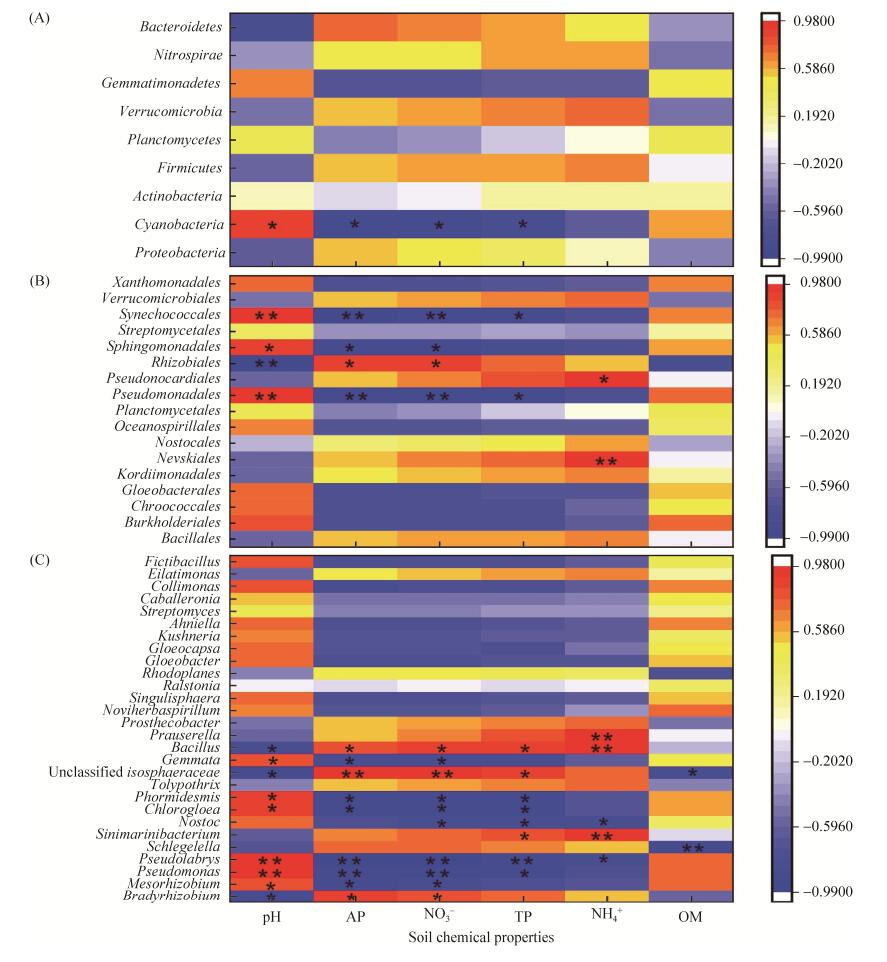 |
| 图 3 优势门、目和属的相对丰度与土壤化学性质的Pearson相关分析 Figure 3 Pearson correlation analysis between the relative abundance of abundant phylum, order and genus and soil chemical properties. A: phylum; B: order; C: genus; *: P < 0.05; **: P < 0.01. TP: total phosphorus; OM: organic matter; AP: available phosphorus; NO3–-N: nitrate nitrogen; NH4+-N: ammonium nitrogen; pH: pH. |
| 图选项 |
3 讨论 3.1 土壤细菌的phoD基因丰度和多样性 耕地土壤细菌的phoD基因丰度显著低于林地,这与笔者基于前人的研究所预期的相符,说明土壤编码碱性磷酸酶基因的细菌的生长受到林地转型耕地的抑制。以连续玉米种植为主的耕作制度在东北地区很常见,并依赖于施用化肥来维持玉米的生产力,而这反过来影响了土壤质量[42]。本研究中,林地转型为耕地后一直用于玉米种植,26年的化肥施用改变了土壤的化学性质(表 1)[36]。Ragot等[22]研究发现pH是驱动编码碱性磷酸酶基因的细菌群落变化的主要因素,土壤编码碱性磷酸酶基因的细菌群落对pH的响应非常强烈。本研究中细菌的phoD基因丰度随pH的减少而显著降低,说明林地转型耕地所造成的土壤酸化是细菌phoD基因丰度变化的重要原因之一。磷肥投入将增加磷有效性,但会降低含phoD的微生物的丰度,本研究发现细菌的phoD基因丰度与速效磷呈极显著负相关关系,这与Tan等[16]、Luo等[28]和Chen等[31]研究发现旱地土壤磷的有效性与phoD基因丰度呈负相关关系一致。而这种负相关关系不仅仅限于旱地土壤,也存在于水稻土,不施肥处理的根际土和非根际土中phoD丰度显著高于施肥处理[30]。而Ikoyi等[43]在仅添加磷的短期温室研究中未发现phoD基因丰度与速效磷间显著相关。产生这种结果的原因与该研究中磷施用的持续时间和数量有关,而长期施肥对土壤编码碱性磷酸酶基因的细菌群落产生的影响更加持久[44]。此外,我们还发现耕地土壤总磷显著增高,说明化肥施用带来了大量的速效磷,但这已远超过植物和微生物生长的需求,因此在这种磷供应充足的条件下,编码碱性磷酸酶基因的细菌能够刺激细胞内磷积累,将磷固定在生物质中[30]。不仅如此,土壤中无机磷被土壤表面活性颗粒快速吸收或沉淀也是造成磷积累的一个主要原因[10]。但本研究中耕地中磷究竟以何种形式留存下来需要进一步研究。phoD基因属于磷饥饿诱导基因,在磷贫化的土壤中可合成速效磷,在土壤总磷含量增高的情况下,抑制phoD基因的表达,使得phoD基因丰度降低。而林地的速效磷含量非常低,处于贫磷状态,但微生物所表现出高度灵活的C︰P和N︰P化学计量可保证其在磷有效性非常低的生境中生存[45–46],高丰度的编码碱性磷酸酶基因的微生物不断地矿化有机磷以减轻磷的限制,从而满足植物和微生物的需要[8, 10]。
林地转型耕地也显著降低了土壤编码碱性磷酸酶基因的细菌群落的多样性。然而,Tan等[16]发现长期施用磷肥增加草地土壤中编码碱性磷酸酶基因的细菌群落的多样性,Chen等[31]也发现长期施用磷肥增加了玉米田土壤编码碱性磷酸酶基因的细菌群落的多样性。这与本研究结果相反,分析发现这些研究均以相同的生态系统(分别为草地生态系统和农田生态系统)为研究对象,而本研究比较的是2种截然不同的生态系统(森林生态系统和农田生态系统)转变下土壤编码碱性磷酸酶基因的细菌群落的变化。作为自然生态系统重要组成部分的森林生态系统比受人类扰动强烈的农田生态系统拥有更高的微生物多样性。本研究发现土壤编码碱性磷酸酶基因的细菌群落的Shannon指数与phoD基因丰度极显著正相关,说明编码碱性磷酸酶基因的细菌群落多样性与丰度呈相同的变化趋势。Shannon指数与pH极显著正相关,而与速效磷极显著负相关,与总磷显著负相关(表 2),这与phoD基因丰度相同,说明编码碱性磷酸酶基因的细菌群落多样性同样受长期施肥的显著影响,并且在响应磷的有效性以及磷积累中发挥重要作用。也有研究表明编码碱性磷酸酶基因的细菌群落的多样性取决于土壤有机碳的数量和质量[19, 23, 28]。Wei等[30]发现编码碱性磷酸酶基因的微生物的多样性与溶解性有机碳含量间呈显著相关关系,磷肥施用通过改变水稻向土壤的有机碳输入间接地影响编码碱性磷酸酶基因的微生物的群落组成。本研究中Shannon指数与有机质也呈正相关关系,但相关性不显著,这可能是试验地位于丘陵地区,玉米的种植和收获均采用人工方式,且在玉米收获时保留全部根茬,根茬的降解向农田输入了有机质,因此耕地土壤有机质含量较林地下降并不显著。但林地与耕地的地表植被群落显著不同,那么二者间土壤有机质组分是否发生变化,且不同组分对土壤编码碱性磷酸酶基因的细菌群落是否有显著影响需要深入研究。施用化肥不仅增加了耕地的速效磷,而且增加了土壤中硝态氮。我们发现Shannon指数与phoD基因丰度均与硝态氮极显著负相关,说明硝态氮对土壤编码碱性磷酸酶基因的细菌群落多样性同样产生了显著的负面影响,而这也与氮肥投入所造成的pH变化密切相关。
3.2 土壤编码碱性磷酸酶基因的细菌群落结构 林地和耕地的土壤编码碱性磷酸酶基因的细菌群落均以变形菌门(Proteobacteria)最为丰富,分别占69.9%–82.3%和74.8%–92.8%,这与多项研究结果一致[16, 24, 29–31, 47]。除林地蓝藻门(Cyanobacteria)的相对丰度(13.2%–24.4%)显著高于耕地(1.7%–6.3%)外,其他优势门的相对丰度在林地和耕地间差异均不显著。Spohn等(2015)研究表明隶属于同一个门的特殊分类群的生态特征也是不同的[48]。笔者在更深层次的分类水平进行分析,结果表明,林地的假单胞菌属(Pseudomonas)、Pseudolabrys属、Phormidesmis属、中慢生根瘤菌属(Mesorhizobium)、Chlorogloea属和Gemmata属的相对丰度显著高于耕地,而慢生根瘤菌属(Bradyrhizobium)和芽孢杆菌属(Bacillus)显著低于耕地。林地中假单胞菌属(Pseudomonas)隶属于变形菌门(Proteobacteria),其相对丰度在4.5%–8.0%,隶属于这个属的细菌具有植物促生长作用[49],其包含了具有磷溶解和磷矿化能力的细菌物种[50–51],有助于有机磷化合物的矿化[52–53],说明该属的细菌在贫磷条件下林地的有机磷矿化中可能发挥重要作用。Pseudolabrys属(3.9%–5.2%)也隶属于变形菌门(Proteobacteria),但其具体的功能未见报道。Phormidesmis属隶属于蓝藻门(Cyanobacteria)(3.2%–8.1%),能进行光合作用将二氧化碳和水转化为糖类,说明这种能够进行光合作用的自养微生物可能将林地的有机磷矿化和碳固定过程相耦合[54]。中慢生根瘤菌属(Mesorhizobium)(0.6%–1.6%)、Chlorogloea属(0.5%–1.1%)和Gemmata属(0.3%–1.0%)分别隶属于变形菌门(Proteobacteria)、蓝藻门(Cyanobacteria)和浮霉菌门(Planctomycetes),虽然相对丰度较其他3个菌属低很多,但其对贫磷条件下的有机磷矿化的作用不应该被忽视,因为经常被忽视的稀有分类群可能对生态功能具有关键作用[55-56],同时这些种细菌在林地土壤的氮、碳和磷的耦合中会发挥一定的作用。而耕地中这6个属的相对丰度相当低。Luo等[28]在不同施肥的农田中检测到高比例的慢生根瘤菌属(Bradyrhizobium),本研究的结果与此相同。慢生根瘤菌属(Bradyrhizobium)是自由生存的共生氮固定微生物,其通过增加碱性磷酸酶活性和磷转运速率响应磷胁迫[57–59],这一属在耦合土壤氮和磷循环中可能发挥重要作用。磷循环与氮循环是紧密联系的[60–61]。植物和微生物竞争土壤中的速效氮[62],长期以来植物通常比土壤微生物更具竞争力,氮的浓度对于微生物的生长通常非常有限[63],那么慢生根瘤菌属(Bradyrhizobium)可能通过利用固定氮气防止氮限制。为何农田生态系统中有如此高比例的慢生根瘤菌属(Bradyrhizobium),以及其在土壤氮循环和磷周转间的耦合中如何发挥作用均是今后研究的重点。而芽孢杆菌属(Bacillus)具有很强的有机质分解力,能够产生丰富的代谢生成物,但其在土壤磷循环中的作用未见报道,其功能同样有待进一步研究。以上这些结果表明林地转型为耕地后,土壤编码碱性磷酸酶基因的细菌群落结构发生了显著变化。
CCA结果表明,速效磷与编码碱性磷酸酶基因的细菌群落结构显著相关,这与Luo等[28]和Chen等[31]报道的结果一致。同时发现编码碱性磷酸酶基因的细菌群落结构与pH、硝态氮、总磷和铵态氮间存在显著的相关性,表明这些土壤化学性质在编码碱性磷酸酶基因的细菌群落变化中发挥重要作用。pH与林地编码碱性磷酸酶基因的细菌群落结构显著正相关,而速效磷、硝态氮、总磷和铵态氮与耕地编码碱性磷酸酶基因的细菌群落结构显著负相关(图 2),说明长期化肥施用造成的土壤酸化以及土壤养分积累塑造了林地和耕地截然不同的编码碱性磷酸酶基因的细菌群落结构,其中pH、速效磷和硝态氮的作用要强于总磷和铵态氮。Wang等[64]和Hu[29]等(2018)分别发现土壤pH是影响种植小麦(Triticum aestivum)以及玉米(Zea maize)和大豆(Glycine max)的卡斯特土壤中编码碱性磷酸酶基因的细菌群落结构的主要因子,这与本研究的结果相似。当编码碱性磷酸酶基因的细菌群落对草地土壤磷状态的应答发生变化时,土壤pH变化所产生的影响更为显著[22],这也说明了土壤pH在塑造编码碱性磷酸酶基因的细菌群落结构中的重要作用。速效磷是影响含土壤编码碱性磷酸酶基因的细菌群落结构的主要因素[22, 27],其与Pho调节子作用,调控磷酸酶的合成与分泌以及无机磷的转运和同化[65–66],因此,本研究中速效磷也产生了非常重要的作用。而硝态氮的增加是导致土壤酸化的主要原因,其间接地影响了编码碱性磷酸酶基因的细菌群落结构。相关分析表明,与土壤化学性质显著相关的目和属中与pH、速效磷和硝态氮显著相关的种类最多,其次为总磷和铵态氮,这也进一步说明了土壤编码碱性磷酸酶基因的细菌群落的变化与这些化学因子密切相关。推荐施用有机肥来改善土壤质量,可以促进微生物多样性和功能,但要仔细考虑有机投入的施用量,以通过改变编码碱性磷酸酶基因的细菌群落来促进磷的利用。
4 结论 与林地相比,耕地土壤编码碱性磷酸酶基因的细菌群落的丰度和多样性均显著降低。pH、速效磷、总磷、硝态氮和铵态氮是决定细菌phoD丰度和多样性的主要环境因子。耕地土壤的慢生根瘤菌属(Bradyrhizobium)和芽孢杆菌属(Bacillus)的相对丰度显著高于林地,而中慢生根瘤菌属(Mesorhizobium)、假单胞菌属(Pseudomonas)、Chlorogloea属、Gemmata属、Phormidesmis属和Pseudolabrys属的相对丰度显著低于林地,林地转型耕地改变了土壤编码碱性磷酸酶基因的细菌群落结构。pH、速效磷、总磷、硝态氮和铵态氮的变化共同引起了土壤编码碱性磷酸酶基因的细菌群落结构的改变。长期化肥投入改变了土壤pH和速效磷,同时也改变了其他的土壤理化因子,造成phoD基因丰度和多样性的变化,进而影响了编码碱性磷酸酶基因的细菌群落结构。
References
| [1] | Condron LM, Turner BL, Cade-Menun BJ. Chemistry and dynamics of soil organic phosphorus//Sims J, Sharpley A. Phosphorus: Agriculture and the Environment. Madison, Wisconsin: Agronomy Monographs, 2005: 87-121. |
| [2] | Turner BL, Cheesman AW, Condron LM, Reitzel K, Richardson AE. Introduction to the special issue:developments in soil organic phosphorus cycling in natural and agricultural ecosystems. Geoderma, 2015, 257-258: 1-3. DOI:10.1016/j.geoderma.2015.06.008 |
| [3] | Schachtman DP, Reid RJ, Ayling SM. Phosphorus uptake by plants:from soil to cell. Plant Physiology, 1998, 116(2): 447-453. |
| [4] | Vitousek PM, Porder S, Houlton BZ, Chadwick OA. Terrestrial phosphorus limitation:mechanisms, implications, and nitrogen-phosphorus interactions. Ecological Applications, 2010, 20(1): 5-15. |
| [5] | Mclaughlin MJ, Baker TG, James TR, Rundle JA. Distribution and forms of phosphorus and aluminum in acidic topsoils under pastures in south-eastern Australia. Australian Journal of Soil Research, 1990, 28(3): 371-385. DOI:10.1071/SR9900371 |
| [6] | Ahlgren J, Djodjic F, B rjesson G, Mattsson L. Identification and quantification of organic phosphorus forms in soils from fertility experiments. Soil Use and Management, 2013, 29(S1): 24-35. |
| [7] | Sharpley AN, Smith SJ, Jones OR, Berg WA, Coleman GA. The transport of bioavailable phosphorus in agricultural runoff. Journal of Environmental Quality, 1992, 21(1): 30-35. |
| [8] | Nannipieri P, Giagnoni L, Landi L, Renella G. Role of phosphatase enzymes in soil//Bünemann E K, Oberson A, Frossard E. Phosphorus in Action. Berlin Heidelberg: Springer International Publishing, 2011: 215-243. |
| [9] | Richardson AE. Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Functional Plant Biology, 2001, 28(9): 897-906. DOI:10.1071/PP01093 |
| [10] | Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant and Soil, 2009, 321(1/2): 305-339. |
| [11] | Juma NG, Tabatabai MA. Distribution of phosphomonoesterases in soils. Soil Science, 1978, 126(2): 101-108. DOI:10.1097/00010694-197808000-00006 |
| [12] | Fraser TD, Lynch DH, Gaiero J, Khosla K, Dunfield KE. Quantification of bacterial non-specific acid (phoC) and alkaline (phoD) phosphatase genes in bulk and rhizosphere soil from organically managed soybean fields. Applied Soil Ecology, 2017, 111: 48-56. DOI:10.1016/j.apsoil.2016.11.013 |
| [13] | Dinkelaker B, Marschner H. In vivo demonstration of acid phosphatase activity in the rhizosphere of soil-grown plants. Plant and Soil, 1992, 144(2): 199-205. DOI:10.1007/BF00012876 |
| [14] | Kr mer S, Green DM. Acid and alkaline phosphatase dynamics and their relationship to soil microclimate in a semiarid woodland. Soil Biology and Biochemistry, 2000, 32(2): 179-188. DOI:10.1016/S0038-0717(99)00140-6 |
| [15] | Romanyà J, Blanco-Moreno JM, Sans FX. Phosphorus mobilization in low-P arable soils may involve soil organic C depletion. Soil Biology and Biochemistry, 2017, 113: 250-259. DOI:10.1016/j.soilbio.2017.06.015 |
| [16] | Tan H, Barret M, Mooij MJ, Rice O, Morrissey JP, Dobson A, Griffiths B, O'Gara F. Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phoD phosphorus mineraliser group in pasture soils. Biology and Fertility of Soils, 2013, 49(6): 661-672. DOI:10.1007/s00374-012-0755-5 |
| [17] | Majumdar A, Ghatak A, Ghosh RK. Identification of the gene for the monomeric alkaline phosphatase of Vibrio cholerae serogroup O1 strain. Gene, 2005, 344: 251-258. DOI:10.1016/j.gene.2004.11.005 |
| [18] | Kageyama H, Tripathi K, Rai AK, Cha-Um S, Waditee-Sirisattha R, Takabe T. An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Applied and Environmental Microbiology, 2011, 77(15): 5178-5183. DOI:10.1128/AEM.00667-11 |
| [19] | Ragot SA, Kertesz MA, Bünemann EK. phoD alkaline phosphatase gene diversity in soil. Applied and Environmental Microbiology, 2015, 81(20): 7281-7289. DOI:10.1128/AEM.01823-15 |
| [20] | Luo HW, Benner R, Long RA, Hu JJ. Subcellular localization of marine bacterial alkaline phosphatases. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(50): 21219-21223. DOI:10.1073/pnas.0907586106 |
| [21] | Dai JY, Chen D, Wu SQ, Wu XF, Zhou J, Tanq XM, Shao KQ, Gao G. Comparative analysis of alkaline phosphatase-encoding genes (phoX) in two contrasting zones of Lake Taihu. Canadian Journal of Microbiology, 2014, 61(3): 227-236. |
| [22] | Ragot SA, Huguenin-Elie O, Kertesz MA, Emmanuel F, Bünemann EK. Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil. Plant and Soil, 2016, 408(1/2): 15-30. |
| [23] | Neal AL, Rossmann M, Brearley C, Akkari E, Guyomar C, Clark IM, Allen E, Hirsch PR. Land-use influences phosphatase gene microdiversity in soils. Environmental Microbiology, 2017, 19(7): 2740-2753. DOI:10.1111/1462-2920.13778 |
| [24] | Fraser TD, Lynch DH, Bent E, Entz MH, Dunfield KE. Soil bacterial phoD gene abundance and expression in response to applied phosphorus and long-term management. Soil Biology and Biochemistry, 2015, 88: 137-147. DOI:10.1016/j.soilbio.2015.04.014 |
| [25] | Jorquera MA, Martínez OA, Marileo LG, Acu a JJ, Saggar S, Mora ML. Effect of nitrogen and phosphorus fertilization on the composition of rhizobacterial communities of two Chilean Andisol pastures. World Journal of Microbiology and Biotechnology, 2014, 30(1): 99-107. |
| [26] | Chhabra S, Brazil D, Morrissey J, Burke J, O'Gara F, Dowling DN. Fertilization management affects the alkaline phosphatase bacterial community in barley rhizosphere soil. Biology and Fertility of Soils, 2013, 49(1): 31-39. DOI:10.1007/s00374-012-0693-2 |
| [27] | Chen XD, Jiang N, Chen ZH, Tian JH, Sun N, Xu MG, Chen LJ. Response of soil phoD phosphatase gene to long-term combined applications of chemical fertilizers and organic materials. Applied Soil Ecology, 2017, 119: 197-204. DOI:10.1016/j.apsoil.2017.06.019 |
| [28] | Luo GW, Ling N, Nannipieri P, Chen H, Raza W, Wang M, Guo SW, Shen QR. Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil fractions. Biology and Fertility of Soils, 2017, 53(4): 375-388. DOI:10.1007/s00374-017-1183-3 |
| [29] | Hu YJ, Xia YH, Sun Q, Liu KP, Chen XB, Ge TD, Zhu BL, Zhu ZK, Zhang ZH, Su YR. Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Science of the Total Environment, 2018, 628-629: 53-63. DOI:10.1016/j.scitotenv.2018.01.314 |
| [30] | Wei XM, Hu YJ, Razavi BS, Zhou J, Shen JL, Nannipieri P, Wu JS, Ge TD. Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biology and Biochemistry, 2019, 131: 62-70. DOI:10.1016/j.soilbio.2018.12.025 |
| [31] | Chen XD, Jiang N, Condron LM, Dunfield KE, Chen ZH, Wang JK, Chen LJ. Impact of long-term phosphorus fertilizer inputs on bacterial phoD gene community in a maize field, Northeast China. Science of the Total Environment, 2019, 669: 1011-1018. DOI:10.1016/j.scitotenv.2019.03.172 |
| [32] | Sun R, Sun BH, Gao MX, Yang XY, Zhang SL. Changes of soil microbial characteristics under long-term different land use patterns on an anthropogenic loess soil. Journal of Plant Nutrition and Fertilizer, 2015, 21(3): 655-663. (in Chinese) 孙瑞, 孙本华, 高明霞, 杨学云, 张树兰.长期不同土地利用方式下锋  土土壤微生物特性的变化.植物营养与肥料学报, 2015, 21(3): 655-663. 土土壤微生物特性的变化.植物营养与肥料学报, 2015, 21(3): 655-663. |
| [33] | Schloter M, Dilly O, Munch JC. Indicators for evaluating soil quality. Agriculture, Ecosystems & Environment, 2003, 98(1/3): 255-262. |
| [34] | Lagomarsino A, Benedetti A, Marinari S, Pompili L, Moscatelli MC, Roggero PP, Lai R, Ledda L, Grego S. Soil organic C variability and microbial functions in a Mediterranean agro-forest ecosystem. Biology and Fertility of Soils, 2011, 47(3): 283-291. DOI:10.1007/s00374-010-0530-4 |
| [35] | Zhu K, Wang R, Li G, Xiu WM, Wang J, Li B, Wang LL, Liu HF, Zhao JN, Yang DL. The response of microbial biomass carbon and metabolic characteristics of albic soil to land use change. Journal of Agro-Environment Science, 2018, 37(10): 2194-2201. (in Chinese) 朱珂, 王蕊, 李刚, 修伟明, 王晶, 李冰, 王丽丽, 刘惠芬, 赵建宁, 杨殿林. 土地利用方式变化对白浆土微生物生物量碳及代谢特征的影响. 农业环境科学学报, 2018, 37(10): 2194-2201. |
| [36] | Wang R, Zhu K, Li G, Wang J, Li B, Xiu WM, Liu HF, Zhao JN, Yang DL. Metabolic characteristics of responses of soil fungi to land-use changes in the hilly regions of northeast China. Journal of Agro-Environment Science, 2018, 37(9): 1925-1932. (in Chinese) 王蕊, 朱珂, 李刚, 王晶, 李冰, 修伟明, 刘惠芬, 赵建宁, 杨殿林. 东北丘陵区林地、耕地和草地土壤真菌群落代谢特征. 农业环境科学学报, 2018, 37(9): 1925-1932. |
| [37] | Wang R, Wu X, Li G, Xiu WM, Wang LL, Zhang GL. Effects of conversion of forest to arable land on the abundance and structure of the cbbL-harboring bacterial community in Albic soil of the hilly region of Northeast China. Environmental Science, 2019, 40(12): 5561-5569. (in Chinese) 王蕊, 吴宪, 李刚, 修伟明, 王丽丽, 张贵龙. 林地转型耕地对东北丘陵区白浆土cbbL细菌群落丰度和结构的影响. 环境科学, 2019, 40(12): 5561-5569. |
| [38] | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(S1): 4516-4522. |
| [39] | Zhang JJ, Kobert K, Flouri T, Stamatakis A. PEAR:a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics, 2013, 30(5): 614-620. |
| [40] | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [41] | Fish JA, Chai BL, Wang Q, Sun YN, Brown CT, Tiedje JM, Cole JR. FunGene:the functional gene pipeline and repository. Frontiers Microbiology, 2013, 4: 291. |
| [42] | Lou YL, Wang JK, Liang WJ. Impacts of 22-year organic and inorganic N managements on soil organic C fractions in a maize field, northeast China. Catena, 2011, 87(3): 386-390. DOI:10.1016/j.catena.2011.07.006 |
| [43] | Ikoyi I, Fowler A, Schmalenberger A. One-time phosphate fertilizer application to grassland columns modifies the soil microbiota and limits its role in ecosystem services. Science of the Total Environment, 2018, 630: 849-858. DOI:10.1016/j.scitotenv.2018.02.263 |
| [44] | Liang YT, Wu LY, Clark IM, Xue K, Yang FY, Van Nostrand JD, Deng Y, He ZL, McGrath S, Storkey J, Hirsch PR, Sun B, Zhou JZ. Over 150 years of long-term fertilization alters spatial scaling of microbial biodiversity. mBio, 2015, 6(2): e00240-15. |
| [45] | Godwin CM, Cotner JB. Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomass stoichiometry. The ISME Journal, 2015, 9(10): 2324-2327. DOI:10.1038/ismej.2015.34 |
| [46] | Wei XM, Hu YJ, Peng PQ, Zhu ZK, Atere CT, O'Donnell AG, Wu J S, Ge TD. Effect of P stoichiometry on the abundance of nitrogen-cycle genes in phosphorus-limited paddy soil. Biology and Fertility of Soils, 2017, 53(7): 767-776. DOI:10.1007/s00374-017-1221-1 |
| [47] | Lagos LM, Acu a JJ, Maruyama F, Ogram A, De La Luz Mora M, Jorquera MA. Effect of phosphorus addition on total and alkaline phosphomonoesterase-harboring bacterial populations in ryegrass rhizosphere microsites. Biology and Fertility of Soils, 2016, 52(7): 1007-1019. DOI:10.1007/s00374-016-1137-1 |
| [48] | Spohn M, Treichel NS, Cormann M, Schloter M, Fischer D. Distribution of phosphatase activity and various bacterial phyla in the rhizosphere of Hordeum vulgare L. depending on P availability. Soil Biology and Biochemistry, 2015, 89: 44-51. |
| [49] | Miller SH, Browne P, Prigent-Combaret C, Combes-Meynet E, Morrissey JP, O'Gara F. Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environmental Microbiology Reports, 2010, 2(3): 403-411. |
| [50] | Rosas SB, Andrés JA, Rovera M, Correa NS. Phosphate-solubilizing Pseudomonas putida can influence the rhizobia-legume symbiosis. Soil Biology and Biochemistry, 2006, 38(12): 3502-3505. DOI:10.1016/j.soilbio.2006.05.008 |
| [51] | Hamdali H, Hafidi M, Virolle MJ, Ouhdouch Y. Rock phosphate-solubilizing Actinomycetes:screening for plant growth-promoting activities. World Journal of Microbiology and Biotechnology, 2008, 24(11): 2565-2575. DOI:10.1007/s11274-008-9817-0 |
| [52] | Giles CD, Hsu PC, Richardson AE, Hurst RH, Hill JE. Plant assimilation of phosphorus from an insoluble organic form is improved by addition of an organic anion producing Pseudomonas sp. Soil Biology and Biochemistry, 2014, 68: 263-269. DOI:10.1016/j.soilbio.2013.09.026 |
| [53] | Alori ET, Glick BR, Babalola OO. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Frontiers in Microbiology, 2017, 8: 971. DOI:10.3389/fmicb.2017.00971 |
| [54] | Chrismas NAM, Barker G, Anesio AM, Sánchez-Baracaldo P. Genomic mechanisms for cold tolerance and production of exopolysaccharides in the Arctic cyanobacterium Phormidesmis priestleyi BC1401. BMC Genomics, 2016, 17: 533. DOI:10.1186/s12864-016-2846-4 |
| [55] | Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N, Gilbert JA. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio, 2014, 5(4). |
| [56] | Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V, Küsel K, Rillig MC, Rivett DW, Salles JF, Van Der Heijden MG, Youssef NH, Zhang XW, Wei Z, Hol WHG. Where less may be more:how the rare biosphere pulls ecosystems strings. The ISME Journal, 2017, 11(4): 853-862. DOI:10.1038/ismej.2016.174 |
| [57] | Smart JB, Dilworth MJ, Robson AD. Effect of phosphorus supply on phosphate uptake and alkaline phosphatase activity in rhizobia. Archives of Microbiology, 1984, 140(2/3): 281-286. |
| [58] | Al-Niemi TS, Summers ML, Elkins JG, Kahn ML, McDermott TR. Regulation of the phosphate stress response in rhizobium meliloti by PhoB. Applied and Environmental Microbiology, 1997, 63(12): 4978-4981. DOI:10.1128/AEM.63.12.4978-4981.1997 |
| [59] | Sakurai M, Wasaki J, Tomizawa Y, Shinano T, Osaki M. Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Science and Plant Nutrition, 2008, 54(1): 62-71. DOI:10.1111/j.1747-0765.2007.00210.x |
| [60] | Zhang T, Li YF, Chang SX, Jiang PK, Zhou GM, Liu J, Lin L. Converting paddy fields to Lei bamboo (Phyllostachys praecox) stands affected soil nutrient concentrations, labile organic carbon pools, and organic carbon chemical compositions. Plant and Soil, 2013, 367(1/2): 249-261. |
| [61] | Xu XY, Liu XR, Li Y, Ran Y, Liu YP, Zhang QC, Li Z, He Y, Xu JM, Di HJ. High temperatures inhibited the growth of soil bacteria and archaea but not that of fungi and altered nitrous oxide production mechanisms from different nitrogen sources in an acidic soil. Soil Biology and Biochemistry, 2017, 107: 168-179. DOI:10.1016/j.soilbio.2017.01.003 |
| [62] | Jackson LE, Burger M, Cavagnaro TR. Roots, nitrogen transformations, and ecosystem services. Annual Review of Plant Biology, 2008, 59: 341-363. DOI:10.1146/annurev.arplant.59.032607.092932 |
| [63] | Kuzyakov Y, Xu XL. Competition between roots and microorganisms for nitrogen:mechanisms and ecological relevance. New Phytologist, 2013, 198(3): 656-669. DOI:10.1111/nph.12235 |
| [64] | Wang Y, Zhang FS, Marschner P. Soil pH is the main factor influencing growth and rhizosphere properties of wheat following different pre-crops. Plant and Soil, 2012, 360(1/2): 271-286. |
| [65] | Vershinina OA, Znamenskaya LV. The Pho regulons of bacteria. Microbiology, 2002, 71(5): 497-511. DOI:10.1023/A:1020547616096 |
| [66] | Apel AK, Sola-Landa A, Rodríguez-García, Martín JF. Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology, 2007, 153(10): 3527-3537. DOI:10.1099/mic.0.2007/007070-0 |
