蒋玲艳1,2,3

 , 周启星1, 王培胜2,3, 江小涵2,3, 冯露2,3
, 周启星1, 王培胜2,3, 江小涵2,3, 冯露2,3 1.南开大学环境科学与工程学院, 天津 300350;
2.南开大学泰达生物技术研究院, 天津 300457;
3.天津市微生物功能基因组学重点实验室, 天津 300457
收稿日期:2016-07-21;修回日期:2016-11-04;网络出版日期:2016-11-29
基金项目:中国博士后科学基金(2016M591381);国家国际科技合作专项(2012DFG31680)
*通信作者:蒋玲艳, Tel/Fax: +86-22-65333286; E-mail: jianglingyan@nankai.edu.cn
摘要: [目的]研究鼠伤寒沙门菌致病岛1(SPI-1)内部的假定调控蛋白STM14_3514的功能及其作用机制。[方法]以鼠伤寒沙门菌模式菌株ATCC 14028为亲本株,构建了STM14_3514基因的缺失突变体及互补菌株,通过小鼠实验、细胞侵袭实验、western blot及实时荧光定量PCR(qRT-PCR)等实验技术,深入研究了STM14_3514基因对鼠伤寒沙门菌致病过程的影响。[结果]STM14_3514突变提高了细菌对小鼠的致病能力,突变体在小鼠肠道、肝和脾中的定殖能力均增强;细胞实验揭示,突变体致病力提升主要由于STM14_3514突变能显著增强细菌对上皮细胞的侵袭力( > 2倍,P < 0.05)。qRT-PCR及western blot分析表明,STM14_3514显著抑制SPI-1内部主要调控因子hilA及侵袭相关基因的表达。此外,STM14_3514对hilA的抑制由HilC介导。[结论]STM14_3514是鼠伤寒沙门菌SPI-1内部的负调控因子,能通过HilC抑制hilA及SPI-1其他入侵基因的表达,该基因的生物学意义可能与细菌进入细胞后对SPI-1的负调控相关。
关键词: 鼠伤寒沙门菌 STM14_3514 侵袭力 HilA HilC
Putative regulatory protein STM14_3514 decreases Salmonella Typhimurium invasion of epithelial cells
Lingyan Jiang1,2,3

 , Qixing Zhou1, Peisheng Wang2,3, Xiaohan Jiang2,3, Lu Feng2,3
, Qixing Zhou1, Peisheng Wang2,3, Xiaohan Jiang2,3, Lu Feng2,3 1.College of Environmental Science and Engineering, Nankai University, Tianjin 300350, China;
2.TEDA Institute of Biological Sciences and Biotechnology, Nankai University, Tianjin 300457, China;
3.Tianjin Key Laboratory of Microbial Functional Genomics, Tianjin 300457, China
Received 21 July 2016; Revised 04 November 2016; Published online 29 November 2016
*Corresponding author: Lingyan Jiang, Tel/Fax: +86-22-65333286; E-mail: jianglingyan@nankai.edu.cn
Supported by China Postdoctoral Science Foundation Funded Project (2016M591381) and by the International Science & Technology Cooperation Program of China (2012DFG31680)
Abstract: [Objective]To study the function and mechanism of STM14_3514 gene that encoded in Salmonella pathogenicity island (SPI)-1 of Salmonella enterica serovar Typhimurium strain ATCC 14028.[Methods]We constructed STM14_3514 mutant strain and a complemented strain of the mutant. Through mice experiment, attachment assays, invasion assays, macrophage replication assays, western blot, and Quantitative real-time PCR analysis (qRT-PCR), we compared the virulence of the mutant strain to that of the wild-type 14028.[Results]STM14_3514 mutant shows increased virulence to mice, and the bacterial number of STM14_3514 mutant in liver, spleen, and ileum was more abundant than that of the wild-type strain. The increased virulence of STM14_3514 mutant is caused by its elevated invasion ability to epithelial cells ( > 2-fold and P < 0.05). qRT-PCR and western blot results show that STM14_3514 reduced the expression of HilA and another SPI-1invasion locus. Moreover, the repression of HilA by STM14_3514 is mediated by HilC.[Conclusion]STM14_3514 is a negative regulator in SPI-1, which can repress HilA and SPI-1invasion locus through HilC, and possibly contribute to the repression on SPI-1 after bacterial invasion.
Key words: Salmonella Typhimurium STM14_3514 invasion ability HilA HilC
沙门菌 (Salmonella) 是一种重要的人畜共患肠道致病菌,能引起肠胃炎、菌血症和伤寒等多种疾病[1]。全球每年沙门菌感染的病例超过9000万,造成约15万人死亡[2],给社会经济和人类健康带来巨大威胁。深入研究沙门菌的致病机制具有重要的公共卫生意义。鼠伤寒沙门菌 (S. enterica serovar Typhimurium,S. Typhimurium) 是引起人类肠胃炎的主要血清型,它对小鼠致死、小鼠感染后症状类似于人伤寒症[3-4]。因此,鼠伤寒沙门菌感染小鼠及小鼠细胞常被用作典型模型,用于研究沙门菌的致病因子及调控机制。
侵袭宿主的肠道上皮细胞是鼠伤寒沙门菌成功致病的第1个关键步骤,该过程所依赖的毒力因子由沙门致病岛1(Salmonella pathogenicity island-1,SPI-1) 编码[5]。SPI-1毒力蛋白会引起宿主细胞发生一系列变化,例如:诱导细胞骨架重排促使细菌进入细胞、诱导细胞凋亡、刺激细胞因子分泌和引起炎症等[6-7]。SPI-1的表达及其相关的侵袭过程受到严格的调控,涉及到许多调控蛋白复杂的相互作用。其中,SPI-1内部的HilA是最主要的调控因子,几乎控制SPI-1所有效应基因的表达,其他的调节蛋白多数通过HilA来控制SPI-1[8]。目前,已经发现了HilA的多个上游调控因子,其中最主要的几个分别为激活因子HilD、HilC和RtsA,以及抑制因子HilE;HilD和HilC由SPI-1内部基因编码,RtsA和HilE由SPI-1外部基因编码[9-10]。
本实验室前期研究鼠伤寒沙门菌Fis蛋白对SPI-1调控的工作中发现,Fis蛋白对SPI-1大多数基因都是正调控,只有少数基因是负调控,其中包括假定的调控蛋白编码基因STM14_3514[11]。由于Fis蛋白对侵袭的正调控作用,推测STM14_3514可能是与侵袭相关的负调控因子。本文通过基因敲除、突变体毒力研究、western blot以及实时荧光定量PCR (qRT-PCR) 等手段,对STM14_3514基因的调节功能进行了深入研究。结果证实基因STM14_3514是鼠伤寒沙门菌SPI-1内部的负调控因子,能通过HilC抑制hilA基因的表达。这个负调控因子在细菌侵入细胞后,在抑制SPI-1的表达过程中发挥作用。
1 材料和方法 1.1 材料
1.1.1 菌株、质粒、细胞系和培养条件: 本实验所用到的菌株及质粒见表 1。鼠伤寒沙门菌野生型菌株ATCC 14028以及质粒pKD3、pKD4、pKD46和pCP20均为本实验室收集保存。本实验所用其他菌株皆通过ATCC 14028构建。LB培养基或琼脂平板用于菌株的培养及保存,实验中所用抗生素浓度为:氨苄青霉素 (Ap) 100 μg/mL,卡那霉素 (Km) 50 μg/mL,氯霉素 (Cm) 10 μg/mL,庆大霉素 (Gm) 10或100 μg/mL。HeLa细胞系和小鼠巨噬细胞系RAW264.7购自上海中国科学院细胞库。细胞培养采用RPMI-1640细胞培养基,补充10%的胎牛血清,于75 cm2细胞培养瓶、5% CO2恒温培养箱中传代培养。细胞用胰酶消化后接种于24孔细胞培养板中,继续孵育约24-48 h,待细胞铺满后直接用于实验。: 表 1. 本文所用菌株和质粒 Table 1. Strains and plasmids used in this study
| Plasmids or strains | Description | Sources |
| Plasmids | ||
| pKD46 | Red recombinase system under an arabinose-inducible promoter; ApR | Lab collection |
| pKD3 | Template plasmid containing the Cm cassette for λ Red recombination; CmR | Lab collection |
| pKD4 | Template plasmid containing the Km cassette for λ Red recombination; KmR | Lab collection |
| pCP20 | a temperature-sensitive replicon expressing the FLP gene to remove antibiotic resistance of mutant strains; ApR | Lab collection |
| pWSK129 | low-copy-number expression vector; KmR | Lab collection |
| pWSK 3×FLAG | pWSK129 carrying 3×FLAG sequence and Cm cassette; CmR, KmR | Lab construction |
| p3514 | pWSK129 carrying the 14028 STM14_3514 gene; KmR | This study |
| pHilC | pWSK129 carrying the 14028 hilC gene; KmR | This study |
| Strains | ||
| 14028 | Wild-type S. Typhimurium strain | Lab collection |
| 14028 pKD46 | 14028 contain plasmid pKD46; ApR | Lab construction |
| ΔSTM14_3514 | 14028 STM14_3514 gene deleted | This study |
| ΔHilC | 14028 hilC gene deleted | This study |
| ΔSTM14_3514 ΔHilC | 14028 STM14_3514 and hilC gene deleted | This study |
| ΔSTM14_3514+ p3514 | ΔSTM14_3514 complemented by plasmid p3514; KmR | This study |
| ΔSTM14_3514ΔHilC+p3514 | ΔSTM14_3514 ΔHilC complemented by plasmid p3514; KmR | This study |
| ΔSTM14_3514ΔHilC+pHilC | ΔSTM14_3514 ΔHilC complemented by plasmid pHilC; KmR | This study |
| 14028 hilA-3×FLAG | 14028 hilA gene added with 3×FLAG tag; CmR | This study |
| ΔSTM14_3514 hilA-3×FLAG | ΔSTM14_3514 hilA gene added with 3×FLAG tag; CmR | This study |
| ΔSTM14_3514+p3514 hilA-3×FLAG | ΔSTM14_3514+p3514 hilA gene added with 3×FLAG tag; CmR, KmR | This study |
| ApR: ampicillin resistance; KmR: kanamycin resistance; CmR: chloramphenicol resistance. | ||
表选项
1.1.2 主要试剂: 限制性内切酶、Taq酶、dNTPs、T4连接酶、cDNA反转录试剂盒及SYBR Green Mix购自TaKaRa公司;RPMI-1640培养基、胰酶和胎牛血清购自Gibco公司;24孔细胞培养板购自Corning公司;Trizol购自Invitrogen公司;RNA纯化试剂盒购自QIAGEN公司;DL2000 DNA Marker、阿拉伯糖及DNA胶回收试剂盒购自上海生工;鼠源单克隆FLAG标签抗体购自Sigma公司;辣根过氧化物酶标记的羊抗鼠二抗购自康为世纪。: 1.2 菌株构建 缺失及标签菌株的构建:通过λ Red重组酶系统构建缺失菌株[12]。设计含有被缺失基因同源序列 (38-40 bp) 的正反向引物,引物由北京英骏生物技术有限公司合成,序列见表 2。STM14_3514和HilC的缺失分别以pKD3和pKD4为模板,扩增出含有同源序列的Cm及Km抗性基因片段。将扩增片段分别电转至14028 pKD46感受态细胞,使抗性片段同源重组至基因组上替换相应基因,通过Cm和Km平板筛选阳性克隆,PCR以及测序验证缺失菌株的正确性。抗性片段的消除通过pCP20质粒完成,pCP20可在42 ℃诱导表达FLP重组酶,重组删除FRT位点之间的抗性片段。构建hilA 3×FLAG标签菌株的原理与缺失菌相同,引物设计稍有差异。上游同源臂设计在hilA基因的终止密码子TAA之前,下游同源臂设计在TAA之后,以实验室前期构建好的质粒pWSK 3×FLAG为模板扩增3×FLAG标签和Cm片段的组合序列,产物经纯化后电转至相应菌株的感受态细胞,从而通过同源重组方式在hilA基因的终止密码子前加入3×FLAG标签序列,Cm平板筛选阳性克隆,PCR以及测序方法验证标签序列的正确性。
表 2. 本文所用引物序列 Table 2. Primers used in this study
| Target genes | Primer sequences (5′→3′) | |
| Construction of mutants and hilA 3×FLAG-tagged strain* | ||
| STM14_3514 | F | TTGACAGTAAAGGAGCTCGCCATGCTCAATTTGCAGCG GTGTAGGCTGGAGCTGCTTCG |
| R | CTACGATGATGGGGCGTGCTCCCCGCAATCCCATTGCG CATATGAATATCCTCCTTAG | |
| hilC | F | ATGGTATTGCCTTCAATGAATAAATCAGTTGAGGCCAT GTGTAGGCTGGAGCTGCTTCG |
| R | TCAATGGTTCATTGTACGCATAAAGCTAAGCGGTGTAA CATATGAATATCCTCCTTAG | |
| hilA | F | AAAGATGGAAACAGGATCCCCGCTTGATTAAATTACGG GACTACAAAGACCATGACGGTG |
| R | CGATGATAAAAAAATAATGCATATCTCCTCTCTCAGATT TTACGCCCCGCCCTGCCACTCA | |
| Identification of the mutants and hilA 3×FLAG-tagged strain | ||
| STM14_3514 | F | AAACCACTGCCACTTTACTGC |
| R | CGTTACTGCCGATTGTTGTT | |
| hilC | F | TGGTGTAGCGATACTGAAAT |
| R | GTGCATAAAGTTTGTAAGGGTA | |
| hilA | F | TTCTGGAAAGTGAACAGCGT |
| R | TGGGCGATAGCGTAAAGTAG | |
| construction of complementation strain# | ||
| STM14_3514 | F | CG GGATCC GTATGCAGATCCACGGAC |
| R | G GAATTC CTACGATGATGGGGCGTG | |
| hilC | F | CG GGATCC ATTAATATTGTAAGCTTTCAT |
| R | G GAATTC TCAATGGTTCATTGTACGC | |
| q RT-PCR primers | ||
| 16S rRNA | F | GAAAGCGTGGGGAGCAAAC |
| R | ACATGCTCCACCGCTTGTG | |
| STM14_3514 | F | ATTACCACTACGCCCGAGTTT |
| R | AACGACTCCAGCGACGACA | |
| hilA | F | CGCTGGCAGAATGCTACCTC |
| R | TGTTTGAATAGCAAACTCCCGA | |
| hilC | F | TTTTCATGCGGACTTGTTGC |
| R | CTCAGCCTGTGACCATTTGC | |
| hilD | F | GCTTTCGGAGCGGTAAACTG |
| R | CCAAGTCGTTGCGTCGGTAT | |
| hilE | F | GCTTACAACCACAACCCGAC |
| R | CAGCACGCCTTCTTTCACC | |
| rtsA | F | TATTACGGCATCAGGGCCA |
| R | ACTCTTGCTACGCCTGTTTCTA | |
| sipA | F | CTGCCAGAAACAAAGAAAGCG |
| R | CTTTGTCAACAAGGTGCGTAAG | |
| spaQ | F | GGTAGGGTTATTCCAGACGGT |
| R | ACTTCGCCATACCAGCCAGA | |
| invI | F | GAGGCGATCCTTGAACAAATAG |
| R | GCGAACAATAGACTGCTTACGT | |
| * Primers were designed carry extensions homologous to 38-40 bp (underlined) of the target gene; # Restriction enzyme cutting site were marked in italics, EcoRⅠ: GAATTC, BamHⅠ: GGATCC. F: forward; R: reverse. | ||
表选项
回补菌株的构建:设计回补引物,以14028基因组为模板,PCR扩增hilC及STM14-3514基因;扩增产物经EcoRⅠ和BamHⅠ双酶切后连接至低拷贝表达载体pWSK129;电转化相应缺失菌株的感受态细胞;Km抗性平板筛选阳性克隆;通过测序验证基因序列的准确性。
1.3 小鼠感染模型 实验用鼠为6-8 w的BALB/c小鼠,小鼠实验依照国家研究委员会 (美国) 规定的法则进行。存活实验中,被感染的小鼠20只分为4个组,其中2组感染14028野生型菌株,另2个组感染STM14_3514突变菌株。每只小鼠经口感染 (灌胃) 约5×107的野生型14028或者STM14_3514突变菌株,监测被感染的小鼠的存活率,监测时长为8 d。
肝、脾及肠道中的细菌计数,被感染的小鼠分为4组,每只小鼠经口感染约5×106的野生型14028或者STM14_3514突变菌株,感染后5 h,处死2组小鼠,分离回肠打碎匀浆,稀释涂板统计细菌数目。5 d后,处死另外2组小鼠,分离肝脏和脾脏打碎匀浆,稀释涂板统计细菌数目。
1.4 粘附及侵袭力检测 细菌对上皮细胞的粘附及侵袭力检测参考文献[13]中的方法。对数中期 (OD600=0.6-0.8) 的细菌用细胞培养基重悬,平板计数初始菌量。细菌按照感染复数 (multiplicity of infection,MOI)=10加入至细胞培养板,1000×g离心5 min,然后在37 ℃、5% CO2恒温培养箱中共培养1 h。吸去菌液,用磷酸 (PBS) 缓冲液洗3次,加入1 mL 0.1% SDS裂解细胞,裂解液梯度稀释后涂板计数。细菌粘附率 (%)=粘附细菌数/每孔中加入的细菌数×100。
侵袭力检测方法基本同上。细菌与细胞共培养1 h,吸去菌液并用PBS洗3遍,然后在含细菌和细胞的24孔板中加入含有浓度为100 μg/mL Gm的培养基,继续培养1 h,以杀死未进入细胞的细菌。裂解细胞,稀释后涂平板计数。细菌侵袭率 (%)=细胞内细菌数/粘附细菌总数×100。
1.5 复制能力检测 在巨噬细胞内的复制力检测参考文献[14]中的方法。对数中期的细菌用细胞培养基重悬,按照MOI=10加入至细胞培养板,1000×g离心5 min,37 ℃、5% CO2恒温培养箱培养30 min。PBS洗3遍,此时的时间点定为T0。向孔板中加入终浓度为100 μg/mL Gm的细胞培养基,继续孵育2 h,裂解部分细胞,涂板计数,此时即T2的细菌数目为初始胞内菌量。将培养基换成新的含10 μg/mL庆大霉素的1640,继续孵育14 h,裂解剩余细胞涂板计数细胞内T16时的胞内菌量。细菌复制力=T16胞内菌量/T2胞内菌量。
1.6 qRT-PCR 提取各测试样品的RNA并用RNA纯化试剂盒纯化,进行反转录,程序为:37 ℃ 15 min,85 ℃ 5 s,4 ℃保存。反转录得到的cDNA进行qRT-PCR检测,反应在ABI 7500荧光定量PCR仪上进行,反应程序为:95 ℃ 10 min,95 ℃ 15 s,60 ℃ 1 min,40个循环。以16S rRNA作为内参基因。以2-ΔΔCt方法计算基因的差异表达倍数。
1.7 Western blot 参考文献[15]和[16]中的方法进行western blot实验。应用构建好的FLAG标签菌株,收集细胞侵袭阶段的细菌,通过测定OD600及平板计数检测各样品中细菌含量,PBS稀释最终使各样品的菌浓度保持一致。对浓度调节后的样品进行超声破碎,与4×载样缓冲液混合后95 ℃煮10 min,12000×g离心10 min,收集上清中蛋白样品。电泳前测定总蛋白浓度,确保各样品蛋白浓度一致,取18 μL样品进行SDS-PAGE电泳 (5%浓缩胶,电压80 V;10%分离胶,电压120 V,电泳时间2-3 h),电转移至醋酸纤维素膜上,丽春红染色观察蛋白质转移情况。经水冲洗后用含5%脱脂奶粉的TBST溶液封闭1 h,后加入500倍稀释的鼠源FLAG抗体,4 ℃孵育过夜;TBST洗涤3次,每5 min换液1次;加入1000倍稀释的辣根过氧化物酶标记的羊抗鼠二抗,室温孵育1.5 h后显影分析。
2 结果和分析 2.1 STM14_3514突变增强鼠伤寒沙门菌对小鼠的致病性 比较野生型14028和ΔSTM14_3514对小鼠的毒性,发现与野生型14028相比,STM14_3514突变体对小鼠的毒性明显增强。如图 1-A所示,感染14028野生型菌株的小鼠在感染后第5天开始死亡,8 d内全部死亡;感染ΔSTM14_3514菌株的小鼠在感染后4 d开始死亡,6 d内全部死亡;说明STM14_3514突变导致鼠伤寒沙门菌对小鼠的致死能力提高。由于小肠末端、肝、脾是鼠伤寒沙门菌感染后的主要定殖位点,定殖能力越强,相应的菌株毒力越强,因此我们比较了野生型和突变体在这些位点的定殖细菌数目。结果如图 1-B-D,突变株ΔSTM14_3514在回肠、肝和脾的定殖细菌量均高于野生型。以上结果综合表明,STM14_3514突变能增强鼠伤寒沙门菌对小鼠的致病性,因此STM14_3514基因对鼠伤寒沙门菌的毒力起抑制作用。
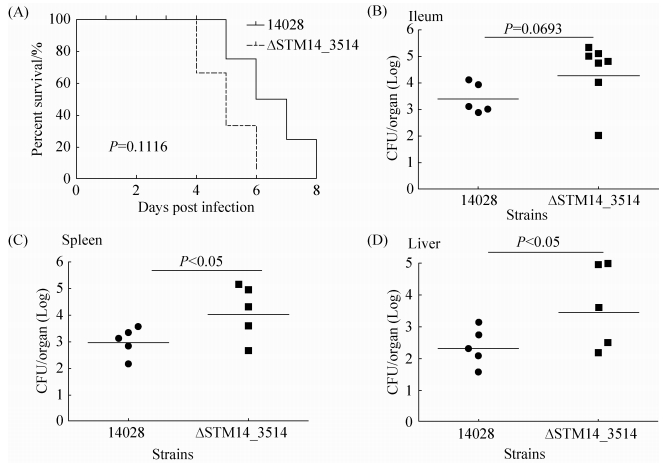 |
| 图 1. STM14_3514突变增强鼠伤寒沙门菌对小鼠的致病性 Figure 1. ΔSTM14_3514 are more virulent to BABL/C mice than wild-type S. Typhimurium 14028. A: survival plots of mice intragastriclly inoculated with 5×107 CFUs S. typhimurium 14028 or ΔSTM14_3514 strains. Data are representative of at least two independent experiments. B-D: bacterial counts recovered from ileum and systemic organs. Mice were intragastrically inoculated with 5×106 CFUs S. typhimurium 14028 or ΔSTM14_3514 strain. Five hours post-infection ileum were harvested and homogenized for colony enumeration; five days post-infection liver and spleen were harvested and homogenized for colony enumeration. Data are representative of three independent experiments. Bars represent mean CFUs of all mice, with data significance determined by student's t test. |
| 图选项 |
2.2 STM14_3514突变增强鼠伤寒沙门菌对上皮细胞的侵袭力 粘附并侵入宿主的肠道上皮细胞以及在宿主巨噬细胞内进行生存复制是鼠伤寒沙门菌致病过程中的关键因素[4-6],我们检测了STM14_3514突变对这些过程的影响。为避免由缺失菌生长能力差异造成的实验误差,我们首先检测了基因缺失及回补对细菌生长的影响。如图 2-A和2-B所示,STM14_3514突变株及回补菌株ΔSTM14_3514+p3514在普通LB培养基和细胞培养基1640中的生长能力都与野生型14028类似。沙门菌对HeLa细胞的粘附侵袭力能反映该细菌在体内对肠道上皮细胞的粘附侵袭能力[17],已被广泛用于沙门菌的侵袭实验[18-20],因此本研究以HeLa细胞系为模型检测缺失菌的粘附及侵袭力变化。对HeLa细胞的粘附及侵袭结果表明:在生长情况基本一致的前提下,STM14_3514突变不影响细菌的粘附率 (图 2-C),但显著增强细菌侵入细胞的能力 (P < 0.05),回补STM14_3514后细菌的侵袭力又回复到野生型水平 (图 2-D),说明确实是由于STM14_3514突变导致的侵袭力提高。此外,巨噬细胞内生存复制实验结果显示,突变株在鼠巨噬细胞RAW264.7内的复制能力略低于野生株,但差异不显著 (图 2-E)。以上结果表明,STM14_3514基因突变主要影响致病过程中对上皮细胞的侵袭阶段,因此ΔSTM14_3514对小鼠毒力的提高源于其增强的上皮细胞侵袭能力。由于STM14_3514基因是位于SPI-1内部的假定调控因子,并能抑制侵袭,推测STM14_3514基因可能对与侵袭密切相关的SPI-1基因具有调控作用。
 |
| 图 2. STM14_3514突变增强鼠伤寒沙门菌对上皮细胞的侵袭力,对粘附及巨噬细胞内复制力影响不显著 Figure 2. STM14_3514 mutant increased bacterial invasion ability to epithelial cells, while did not significantly affect attachment ability and replication ability in macrophages. The growth assay for 24 h time frame in LB (A) and RPMI-1640 (B) mediums were determined for the strains used. For cell attachment (C) and invasion assays (D), HeLa cells were infected with bacteria strains at the multiplicity of infection (MOI) of 10. Attachment ability=attached bacteria/input bacteria ×100; invasion ability=intracellular bacteria at 1 h post-infection/attached bacteria ×100. For the intracellular replication assays (E), bacteria were added to RAW264.7 macrophages at the MOI of 10. Bacterial replication was determined by the ratio of the number of intracellular bacteria at 16 h post-infection to the initial number of bacteria at time 2 h. Data from each graph represent the average of three independent experiments. Error bars indicate standard deviations of the mean, with data significance determined by student's t test. *P < 0.05. |
| 图选项 |
2.3 STM14_3514基因抑制HilA表达 为检验STM14_3514基因是否对SPI-1有调控作用,我们选择了SPI-1的中心调节因子hilA及其下游3个侵袭相关蛋白编码基因 (sipA,spaQ和invI),通过qRT-PCR检测了STM14_3514基因缺失后这些基因的表达变化。收取HeLa细胞侵袭阶段的细菌进行RNA提取及qRT-PCR分析,以野生型菌株侵袭阶段的基因表达水平为参照。如图 3-A所示,STM14_3514基因缺失显著增强4个检测基因的转录 (P < 0.05),STM14_3514基因回补后被检测基因的转录回复到野生型水平,说明STM14_3514基因能抑制hilA及其他SPI-1基因的转录表达。此外,为进一步证明hilA与STM14-3514基因的表达相关性,我们检测了在HeLa细胞整个感染阶段 (包括侵袭及胞内生存),14028野生型菌株中hilA与STM14-3514基因的表达变化。收取14028侵袭阶段 (0 h) 及侵染细胞后2 h和6 h的细菌进行RNA提取和qRT-PCR分析,以细胞悬浮液中的细菌RNA样品为对照。如图 3-B所示,hilA与STM14_3514的转录呈负相关性,STM14_3514在侵袭时表达最低,hilA表达最高,这一表达方式也最利于细菌入侵。细菌进入细胞后,STM14_3514基因表达提高,hilA表达相应降低,这种表达趋势在整个胞内阶段基本趋于一致。另外,我们通过Western blot检测了缺失菌在侵袭阶段的HilA蛋白表达水平,通过λ Red重组酶系统构建了带有3×FLAG标签的14028野生型、STM14-3514缺失菌及互补菌菌株,收取在侵袭阶段的不同类型3×FLAG标签菌株进行总蛋白提取及Western blot检测。如图 3-C所示,STM14_3514缺失菌的HilA蛋白表达水平显著高于野生型14028,STM14_3514基因回补菌株HilA表达水平与野生型相似。以上结果从RNA及蛋白水平综合表明,STM14_3514基因能抑制HilA表达,并且通过抑制HilA抑制SPI-1其他基因的表达。
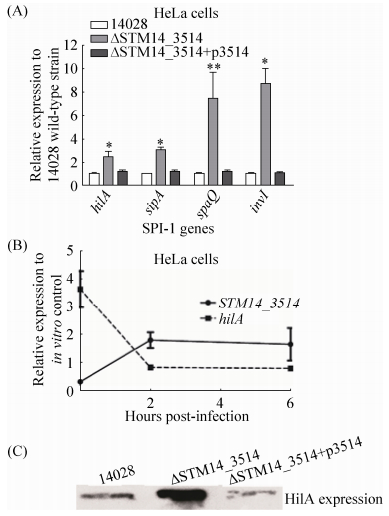 |
| 图 3. STM14_3514抑制HilA表达 Figure 3. Expression levels of hilA gene and other SPI-1 genes are significantly repressed by the mutation of STM14_3514. A: hilA and other SPI-1 genes transcript in 14028, ΔSTM14_3514 andΔSTM14_3514 +p3514. RNA was harvested from bacteria at the invasion stage. B:hilA and STM14_3514 genes transcript in 14028 wild-type strain during the infection course. RNA was extracted from the intracellular bacteria at 0 h, 2 h and 6 h post-infection of HeLa cells. RNA extracted from bacteria in the cell suspension was used as in vitro control. For A and B, the expression was normalized using the 16S rRNA gene as the internal control. The data represents the average of three independent experiments. Error bars indicate standard deviations of the mean. Student's t-test was used to calculate P value. *P < 0.05, **P < 0.01. C: western blot result of the HilA protein in different strains. Bacteria proteins were extracted from strain 14028 hilA-3×FLAG, ΔSTM14_3514 hilA-3×FLAG and ΔSTM14_3514+p3514 hilA-3×FLAG at the invasion stage, respectively. |
| 图选项 |
2.4 STM14_3514基因通过HilC调控HilA表达 为了进一步了解STM14_3514基因对HilA的调控方式,是直接调控还是间接调控,我们通过qRT-PCR检测了STM14_3514基因缺失后对几个熟知的HilA上游调节因子 (hilC,hilD,hilE和rtsA) 的表达影响。收取HeLa细胞侵袭阶段的细菌进行RNA提取及qRT-PCR分析,以野生型菌株侵袭阶段的基因表达水平为对照。如图 4-A所示,STM14_3514基因缺失显著增强了hilC基因的转录 (P < 0.01),对其他3个调节基因的转录没有影响,说明STM14_3514能抑制hilC的转录表达,并可能通过hilC抑制HilA。为进一步证实上述结论,我们构建了hilC基因的缺失菌株ΔHilC,hilC和STM14_3514基因的双缺失菌株ΔSTM14_3514 ΔHilC,以及相应的回补菌株,qRT-PCR检测了这些菌株中hilA基因的转录表达变化。结果如图 4-B所示,STM14_3514基因缺失能显著增强hilA转录,hilC和STM14_3514双缺失后hilA转录水平与hilC单缺失菌类似,说明HilC不存在的情况下,STM14_3514基因不影响hilA的转录。另外,双缺菌回补STM14_3514(ΔSTM14_3514ΔHilC+p3514) 不能调控hilA,而回补HilC (ΔSTM14_3514ΔHilC+ pHilC) 才能影响hilA的转录。以上结果表明,STM14_3514在HilC存在的条件下才能调控HilA表达,因此STM14_3514基因是通过HilC来调控HilA。
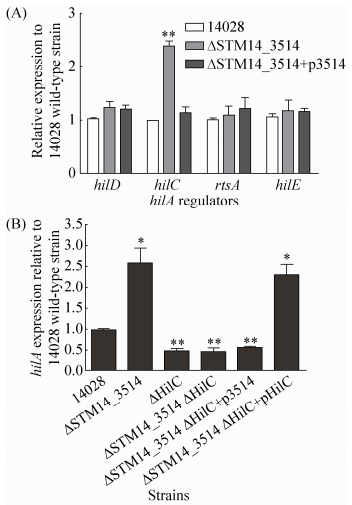 |
| 图 4. STM14_3514基因通过HilC调控HilA表达 Figure 4. The repression of hilA by STM14_3514 gene is mediated by HilC. A: the transcript levels of hilA regulators (hilC, hilD, hilE, and rtsA) in 14028, ΔSTM14_3514 and ΔSTM14_3514+p3514. RNA was harvested from bacteria at the invasion stage. B: hilA transcript in different strain at the invasion stage. The expression was normalized using the 16S rRNA gene as the internal control. The data represents the average of three independent experiments. Error bars indicate standard deviations of the mean. Student's t-test was used to calculate P value. *P < 0.05, **P < 0.01. |
| 图选项 |
3 讨论 作为致病过程中的主要毒力因子之一,鼠伤寒沙门菌的SPI-1表达受到多种环境因素和调节因子的调节,从而保证入侵系统能适时适地地表达[9-10]。目前已经发现的SPI-1调控因子主要包括HilA、HilC、HilD、HilE和RtsA。最近一些研究发现,某些调节其他代谢的调节因子也加入至SPI-1的调控网络,它们对SPI-1的调控大多数是通过调节上述几个调控因子控制SPI-1侵袭位点的表达。如:调控蛋白Fur和鞭毛调控基因FliZ都能通过控制HilD来调控SPI-1[21-22];糖代谢的调节基因Mlc通过抑制HilE调节SPI-1表达[23]。这些调节因子新调控功能的发现无疑证明了SPI-1及其入侵系统调控机制的复杂性,可能仍有许多未知的SPI-1调节机制等待研究发掘。
本文为了研究SPI-1内部假定调控因子STM14_3514的功能,构建了STM14_3514的突变株,检测了STM14_3514突变对鼠伤寒沙门菌毒力的影响及其作用机制。研究结果证明STM14_3514是负调控因子,STM14_3514突变能增强细菌对肠道上皮细胞的侵袭力,提高细菌对小鼠的致病性。进一步研究STM14_3514调控侵袭的机制,我们发现STM14_3514能通过HilC抑制hilA及SPI-1其他入侵基因的表达 (图 5)。该研究首次发现并证实了SPI-1内部负调节因子的存在。
 |
| 图 5. STM14_3514通过HilC调节SPI-1模式图 Figure 5. Model for STM14_3514 regulation of SPI-1, STM14_3514 acts through HilC to control HilA function. |
| 图选项 |
基于沙门菌致病过程的精确调控,STM14_3514负调控因子的存在也必然有其特定的生物学意义。值得注意的是,STM14_3514基因的表达在致病过程中能被很好地调控 (图 3-B),在侵袭时被抑制,此时促进hilC、hilA及其他入侵相关基因的表达,有利于细菌侵入细胞。细菌进入细胞后,开启细胞内的生存模式,这时SPI-1的大部分基因已经不再需要,STM14_3514基因在侵入细胞后上调,能适时降低SPI-1基因的表达。细菌进入细胞后,对SPI-1侵袭基因的降低可能具有重要的意义,推测有以下两方面原因:(ⅰ) 减少合成不必要的SPI-1效应蛋白而造成的能源浪费。在不需要的情况下如果合成大量的“无用”蛋白,会造成能源浪费,可能降低细菌的生存力。Sturm等发现,SPI-1的表达本身会降低沙门菌的生长速率。在体外,表达SPI-1的沙门菌个体呈现出缓慢生长的状态,他们推测这可能与表达SPI-1过程中大量的能量消耗相关,这些消耗对生长造成负担[24]。对乳糖操纵子的转录表达研究也证明上述结论。一般来讲,乳糖操纵子的转录表达受到严格的调控。Stoebel等研究发现,当环境中没有乳糖时,组成型表达乳糖操纵子会显著降低大肠杆菌的生存力,这是由于不必要的转录和翻译过程造成了大量的能源浪费[25];Jiang等发现沙门菌表达乳糖操纵子会降低其对上皮细胞的侵袭力,一部分原因也是因为能源浪费[26]。在体外,H-NS能有效抑制SPI-1等毒力基因的表达,从而消除了毒力基因的无意义表达对细菌生长的不利效应[27-28]。相应的,宿主细胞尤其是巨噬细胞内部存在巨大的选择压力,鼠伤寒沙门菌进入细胞后有效抑制“无用的”SPI-1可能有利于致病菌在胞内的生存复制行为。本研究结果中STM_3514突变株在巨噬细胞内的复制力稍微低于野生型 (图 2-E),证明由于STM_3514突变造成的SPI-1过度表达确实不利于细菌的胞内生存复制。(ⅱ) SPI-1效应蛋白包括SopB、SopE、SopE2、SipA、SipC和SopA是刺激被感染的细胞分泌白细胞介素等炎症因子的主要蛋白[29-30],因此STM14_3514基因对SPI-1的抑制作用可能与细菌介导的降低细胞炎症因子的释放,从而降低免疫细胞的募集及杀伤相关,该假设需要进一步研究证实。
综上所述,本实验研究了鼠伤寒沙门菌STM14_3514突变对鼠伤寒沙门菌致病过程的影响,证实了STM14_3514是SPI-1内部的负调控因子,通过HilC抑制HilA和SPI-1入侵基因表达,推测该基因在细菌侵袭后抑制SPI-1,此过程具有重要的生物学意义。但是STM14_3514对HilC的调控方式以及STM14_3514的上游调控因子目前并不清楚,仍需进一步研究。
参考文献
| [1] | Fàbrega AF, Vila J. Salmonella entericaserovar Typhimurium skills to succeed in the host: virulence and regulation.Clinical Microbiology Reviews, 2013, 26(2): 308–341DOI:10.1128/CMR.00066-12. |
| [2] | Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. The global burden of nontyphoidal Salmonella gastroenteritis.Clinical Infectious Diseases, 2010, 50(6): 882–889DOI:10.1086/649513. |
| [3] | Kr?ger C, Colgan A, Srikumar S, H?ndler K, Sivasankaran SK, Hammarl?f DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JCD. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium.Cell Host & Microbe, 2013, 14(6): 683–695. |
| [4] | Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review.Immunology and Cell Biology, 2007, 85(2): 112–118DOI:10.1038/sj.icb.7100007. |
| [5] | LaRock DL, Chaudhary A, Miller SI. Salmonellaeinteractions with host processes.Nature Reviews Microbiology, 2015, 13(4): 191–205DOI:10.1038/nrmicro3420. |
| [6] | Valdez Y, Ferreira RBR, Finlay BB. Molecular mechanisms ofSalmonella virulence and host resistance//Sasakawa C. Molecular mechanisms of bacterial infection via the gut. Berlin Heidelberg: Springer, 2009, 337: 93-127. |
| [7] | Schlumberger MC, Hardt WD. Salmonella type Ⅲ secretion effectors: pulling the host cell's strings.Current Opinion in Microbiology, 2006, 9(1): 46–54DOI:10.1016/j.mib.2005.12.006. |
| [8] | Lostroh CP, Lee CA. The Salmonellapathogenicity island-1 type Ⅲ secretion system.Microbes and Infection, 2001, 3(14/15): 1281–1291. |
| [9] | Altie C. Genetic and environmental control of salmonella invasion.The Journal of Microbiology, 2005, 43(1): 85–92. |
| [10] | Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA inSalmonella entericaserovar Typhimurium.Molecular Microbiology, 2005, 57(3): 691–705DOI:10.1111/mmi.2005.57.issue-3. |
| [11] | Wang H, Liu B, Wang Q, Wang L. Genome-wide analysis of the Salmonella Fis regulon and its regulatory mechanism on pathogenicity islands.PLoS One, 2013, 8(5): e64688DOI:10.1371/journal.pone.0064688. |
| [12] | Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products.Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(12): 6640–6645DOI:10.1073/pnas.120163297. |
| [13] | Isberg RR, Falkow S. A single genetic locus encoded byYersinia pseudotuberculosispermits invasion of cultured animal cells by Escherichia coliK-12.Nature, 1985, 317(6034): 262–264DOI:10.1038/317262a0. |
| [14] | Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium.Molecular Microbiology, 2007, 63(1): 193–217DOI:10.1111/mmi.2007.63.issue-1. |
| [15] | Yang B, Feng L, Wang F, Wang L. EnterohemorrhagicEscherichia coli senses low biotin status in the large intestine for colonization and infection.Nature Communications, 2015, 6: 6592DOI:10.1038/ncomms7592. |
| [16] | Marshall JM, Gunn JS. The O-antigen capsule of Salmonella enterica serovar Typhimurium facilitates serum resistance and surface expression of FliC.Infection and Immunity, 2015, 83(10): 3946–3959DOI:10.1128/IAI.00634-15. |
| [17] | Giannella RA, Washington O, Gemski P, Formal SB. Invasion of HeLa cells by Salmonellatyphimurium: a model for study of invasiveness of Salmonella.The Journal of Infectious Diseases, 1973, 128(1): 69–75DOI:10.1093/infdis/128.1.69. |
| [18] | Elhadad D, Desai P, Grassl GA, McClelland M, Rahav G, Gal-Mor O. Differences in host cell invasion and Salmonella pathogenicity island 1 expression between Salmonella enterica serovar paratyphi A and nontyphoidal S.Typhimurium.Infection and Immunity, 2016, 84(4): 1150–1165DOI:10.1128/IAI.01461-15. |
| [19] | Gong H, Vu GP, Bai Y, Chan E, Wu RB, Yang E, Liu FY, Lu SW. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors.PLoS Pathogens, 2011, 7(9): e1002120DOI:10.1371/journal.ppat.1002120. |
| [20] | Winter SE, Winter MG, Poon V, Keestra AM, Sterzenbach T, Faber F, Costa LF, Cassou F, Costa EA, Alves GES, Paix?o TA, Santos RL, B?umler AJ. Salmonella entericaserovar typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation.PLoS Pathogens, 2014, 10(7): e1004207DOI:10.1371/journal.ppat.1004207. |
| [21] | Ellermeier JR, Slauch JM. Fur regulates expression of the Salmonella pathogenicity island 1 type Ⅲ secretion system through HilD.Journal of Bacteriology, 2008, 190(2): 476–486DOI:10.1128/JB.00926-07. |
| [22] | Chubiz JEC, Golubeva YA, Lin DX, Miller LD, Slauch JM. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella entericaserovar typhimurium.Journal of Bacteriology, 2010, 192(23): 6261–6270DOI:10.1128/JB.00635-10. |
| [23] | Lim S, Yun J, Yoon H, Park C, Kim B, Jeon B, Kim D, Ryu S. Mlc regulation of Salmonellapathogenicity island Ⅰ gene expression via hilErepression.Nucleic Acids Research, 2007, 35(6): 1822–1832DOI:10.1093/nar/gkm060. |
| [24] | Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type Ⅲ secretion system 1.PLoS Pathogens, 2011, 7(7): e1002143DOI:10.1371/journal.ppat.1002143. |
| [25] | Stoebel DM, Dean AM, Dykhuizen DE. The cost of expression of Escherichia coli lacoperon proteins is in the process, not in the products.Genetics, 2008, 178(3): 1653–1660DOI:10.1534/genetics.107.085399. |
| [26] | Jiang LY, Ni ZW, Wang L, Feng L, Liu B. Loss of the lac operon contributes to Salmonella invasion of epithelial cells through derepression of flagellar synthesis.Current Microbiology, 2015, 70(3): 315–323DOI:10.1007/s00284-014-0720-7. |
| [27] | Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria.PLoS Pathogens, 2006, 2(8): e81DOI:10.1371/journal.ppat.0020081. |
| [28] | Navarre WW, Porwollik S, Wang YP, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella.Science, 2006, 313(5784): 236–238DOI:10.1126/science.1128794. |
| [29] | Zhang SP, Santos RL, Tsolis RM, Stender S, Hardt WD, B?umler AJ, Adams LG. The Salmonella entericaserotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves.Infection and Immunity, 2002, 70(7): 3843–3855DOI:10.1128/IAI.70.7.3843-3855.2002. |
| [30] | Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice.Infection and Immunity, 2004, 72(2): 795–809DOI:10.1128/IAI.72.2.795-809.2004. |
