郭睿#, 陈大福#

 , 黄枳腱, 梁勤, 熊翠玲, 徐细建, 郑燕珍, 张曌楠, 解彦玲, 童新宇, 侯志贤, 江亮亮, 刀晨
, 黄枳腱, 梁勤, 熊翠玲, 徐细建, 郑燕珍, 张曌楠, 解彦玲, 童新宇, 侯志贤, 江亮亮, 刀晨 福建农林大学蜂学学院, 福建 福州 350002
收稿日期:2016-12-31;修回日期:2017-02-23;网络出版日期:2017-05-11
基金项目:现代农业产业技术体系建设专项资金(CARS-45-KXJ7);福建农林大学科技发展资金(KF2015123);国家自然科学基金(30800806);福建省大学生创新创业训练计划(201610389053)
*通信作者:陈大福, Tel:+86-591-83726835;E-mail:dfchen826@163.com
#并列第一作者
摘要:[目的]本研究利用RNA-seq技术对球囊菌胁迫的中华蜜蜂(中蜂)幼虫肠道进行深度测序,经趋势分析得到差异表达基因(DEGs)的显著表达模式,进而对胁迫过程中的球囊菌进行转录组学分析。[方法]利用Illumina HiSeq 2500平台对球囊菌胁迫的中蜂幼虫肠道进行深度测序,并利用相关软件进行了深入分析。最后,通过RT-qPCR对RNA-seq数据进行了验证。[结果]本研究共得到球囊菌的41133932条高质量clean reads。22865个DEGs共聚类为8个基因表达模式,其中,16769个DEGs聚类为2个显著上调趋势与2个显著下调趋势。GO富集分析结果显示,显著上调与显著下调趋势中的DEGs分别富集于40与37个GO term,基因富集数最多的为细胞进程(2486 unigenes)。KEGG代谢通路(pathway)富集分析结果显示,显著上调与显著下调趋势中的DEGs分别富集于119和112个pathway,基因富集数最多的分别是氨基酸生物合成(127 unigenes)与核糖体(98 unigenes)。进一步分析表明球囊菌在胁迫中蜂幼虫肠道的过程中通过提高物质合成促进其增殖,而宿主通过抑制球囊菌的蛋白合成抵御病原入侵。富集在MAPK信号通路的11个DEGs的表达水平随着胁迫时间的延长而逐渐下降,推测中蜂幼虫通过抑制该通路而阻遏球囊菌增殖。[结论]本研究不仅为揭示白垩病过程中的球囊菌-中蜂幼虫互作提供了重要信息,也为阐明不同抗性蜂种的球囊菌抗性差异奠定了基础。
关键词: 球囊菌 中华蜜蜂 幼虫肠道 趋势分析 转录组
Transcriptome analysis of Ascosphaera apis stressing larval gut of Apis cerana cerana
Rui Guo#, Dafu Chen#

 , Zhijian Huang, Qin Liang, Cuiling Xiong, Xijian Xu, Yanzhen Zheng, Zhaonan Zhang, Yanling Xie, Xinyu Tong, Zhixian Hou, Liangliang Jiang, Chen Dao
, Zhijian Huang, Qin Liang, Cuiling Xiong, Xijian Xu, Yanzhen Zheng, Zhaonan Zhang, Yanling Xie, Xinyu Tong, Zhixian Hou, Liangliang Jiang, Chen Dao College of Bee Science, Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian Province, China
Received 31 December 2016; Revised 23 February 2017; Published online 11 May 2017
*Corresponding author: Dafu Chen, Tel:+86-591-83726835;E-mail:dfchen826@163.com
Supported by the Earmarked Fund of China Agriculture Research System (CARS-45-KXJ7), by the Science and Technology Development Fund of Fujian Agriculture and Forestry University (KF2015123), by the National Natural Science Foundation of China (30800806) and by the Undergraduate Innovation and Entrepreneurship Training Program of Fujian Province (201610389053)
#These authors contributed equally to this work
Abstract: [Objective]RNA-seq technology was used to sequence the larval guts of Apis cerana cerana under stress of Ascosphaera apis. Subsequently, trend was analyzed for differentially expressed genes (DEGs) to obtain significant gene expression patterns, followed by transcriptome analysis of A. apis stressing the larval gut.[Methods]Infected honeybee larval guts were sequenced at Illumina HiSeq2500 platform and in-depth analyses were done using corresponding biological software. Finally, RT-qPCR was conducted to validate RNA-seq data.[Results]A total of 41133932 high-quality clean reads were obtained. Trend analysis result showed that 22865 DEGs were grouped into 8 gene expression patterns, among them 16769 DEGs were assigned to 4 significant expression patterns including 2 up-regulated trends and 2 down-regulated trends. GO enrichment analysis result showed that all DEGs within significant up-and down-regulated patterns were enriched in 40 and 37 GO terms, respectively, and the mostly enriched one is cellular process (2486 unigenes). KEGG enrichment analysis result displayed that the DEGs within significant up-and down-regulated trends were enriched in 119 and 112 pathways, respectively, and biosynthesis of amino acids (127 unigenes) and ribosome (98 unigenes) were mostly enriched. A. apis facilitated its proliferation through enhancing the biosynthesis and the host could fight A. apis by inhibiting the protein synthesis of the fungal pathogen during the stress process. Furthermore, expression levels of 11 DEGs enriched in the pathogen's MAPK signaling pathway decreased when the stressing time of A. apis was prolonged, suggesting that A. c. cerana larvae could constrain the pathogen's replication by disturbing this pathway.[Conclusion]This is the first report of transcriptome investigation of A. apis infecting A. c. cerana larvae. Our data provide gene expression profiles of A. apis stressing the larval gut of A. c. cerana, as well lay the foundation of unraveling molecular mechanisms regulating the pathogenesis of A. apis.
Key words: Ascosphaera apis Apis cerana cerana larval gut trend analysis transcriptome
蜜蜂是一种重要的社会学模式昆虫,因其在发育学、神经生物学、行为学和病原-宿主互作研究中的重要性而备受关注[1-5]。同时,蜜蜂作为最重要的授粉昆虫在农业生产和生态维持中也发挥着巨大作用[6]。据报道,蜜蜂为全球70%的作物和野生植物授粉[7-8]。蜜蜂易遭受细菌、真菌、病毒、寄生虫及农药的危害[9]。其中,白垩病(chalkbrood)是一种典型的蜜蜂真菌病,该病于1913年首次在德国发现(Massen ex Claussen)[10],于1990年在中国大陆发生[11]。近年来,随着蜂产品贸易的全球化,白垩病的发病率呈逐年上升趋势[12]。白垩病的病原是蜜蜂球囊菌(Ascosphaera apis),A. apis特异性侵染蜜蜂幼虫,发病时期一般为春季和初夏,虽不对蜂群造成毁灭性打击,但却能导致成年蜜蜂数量的大幅下降,从而严重影响蜂蜜等产品的产量[13],据报道,该病可造成蜂蜜产量下降5%-37%[14]。意大利蜜蜂(Apis mellifera ligustica) (属于西方蜜蜂,简称意蜂)和中华蜜蜂(Apis cerana cerana)(属于东方蜜蜂,简称中蜂)是国内养蜂生产中使用的主要蜂种,前者极易被球囊菌侵染而罹患白垩病,而后者几乎不受影响。球囊菌对蜜蜂幼虫的侵染主要是通过蜜蜂的取食活动,含球囊菌孢子的食物被蜜蜂摄入体内,孢子在蜜蜂消化道萌发生长,继而突破消化道围食膜和中肠屏障,在体腔大量生长繁殖,导致蜜蜂幼虫死亡,最后突破体表,异宗菌丝接触形成大量子代孢子[15]。
近20年来,国内外****在培养方法[16-17]、形态学[18-19]、流行病学[20-21]、增殖方式[22-23]、病理学[24-25]、蜜蜂防御[26-27]以及疾病防治[28-30]等方面对白垩病开展了较多研究并取得一些重要进展。本课题组也在球囊菌的病理和检测方面开展了较为系统的研究,如梁勤等从碳源、氮源、维生素、矿质元素等方面研究了营养生态条件对球囊菌生长及产孢的影响,结果表明营养生态条件的变化对球囊菌的影响极大[31];郑志阳等对健康和患病蜜蜂幼虫血淋巴进行SDS-PAGE分析和蛋白酶、酯酶的活性染色,发现健康蜜蜂幼虫血淋巴中的蛋白含量丰富,主要由4种高分子质量的蛋白组成,而患病幼虫血淋巴中的蛋白含量很少,主要蛋白组分被降解,多种蛋白酶和酯酶的活性在患病幼虫血淋巴中可以检测到,而在健康幼虫中检测不到[32]。然而,由于基因组信息的缺失,球囊菌的分子研究进展缓慢。2006年Qin等公布了球囊菌ARSEF7406菌株的基因组序列[33],为球囊菌的分子研究奠定了基础,但作者并没有同时公布基因的位置和功能注释信息。Cornman等利用Roche 454焦磷酸测序技术对来自培养基的球囊菌菌丝和来自蜜蜂幼虫感染组织的球囊菌菌丝进行了转录组测序,功能分析表明球囊菌的差异表达基因参与了交配类型、细胞内信号转导和应激反应[34]。
前期研究中,本课题组在实验室条件下同时对意蜂幼虫和中蜂幼虫进行人工饲养并接种球囊菌,发现后者能够被球囊菌侵染而发生白垩病,但后者的发病率(16.67%)远低于前者(70.83%)(未发表数据)。目前,有关球囊菌的组学研究极少,白垩病过程中的病原-宿主互作机制仍不明确,也无球囊菌侵染中蜂幼虫的分子研究报道。此前,本课题组已成功组装中华蜜蜂幼虫肠道及球囊菌的参考转录组并对它们进行功能及代谢通路注释,为本研究利用RNA-seq技术对球囊菌胁迫中蜂幼虫肠道过程中的病原进行转录组学研究,研究结果可为阐明球囊菌-中蜂幼虫的互作机制提供重要信息,也为白垩病的防治奠定基础。
1 材料和方法 1.1 供试中华蜜蜂幼虫与蜜蜂球囊菌 本研究使用的中蜂幼虫取自福建农林大学蜂学学院教学蜂场,球囊菌菌株由福建农林大学蜂学学院蜜蜂保护实验室保存并活化。
1.2 主要实验试剂及仪器 DEPC水购自生工生物工程(上海)股份有限公司,Dynal M280磁珠购自美国Invitrogen公司,高碘酸钠购自美国Sigma公司,DNA ligase购自美国Thermo公司,RNAiso Reagent抽提试剂盒、Ex Taq Polymerase及Superscript II Reverse Transcriptase均购自日本TaKaRa公司,DNase I和Oligotex mRNA Kits Midi试剂盒购自德国Qiagen公司。纯化cDNA的Agencourt AMPure XPbeads为美国Beckman公司产品,cDNA文库构建试剂盒TruSeqTM DNA Sample Prep Kit v2-Set A为美国Illumina公司产品。DNA marker为日本TaKaRa公司产品。其他试剂均为国产分析纯。
恒温恒湿气候箱购自中国宁波江南仪器厂,超纯水仪购自中国四川沃特尔水处理设备有限公司,倒置显微镜购自中国上海光学仪器五厂,超净工作台购自中国苏州安泰空气技术有限公司,凝胶成像仪购自上海培清科技有限公司,超低温冰箱购自合肥中科美菱低温科技股份有限公司,PCR仪购自美国Bio-Rad公司,高速冷冻离心机购自德国Eppendorf公司,实时荧光定量PCR仪(ABI 7500)为美国ABI公司产品。
1.3 中华蜜蜂幼虫饲养与球囊菌孢子纯化 中蜂幼虫的饲料配方参照王倩等的方法[35]并进行改良,将D-果糖和D-葡萄糖换为新鲜蜂蜜。将2日龄幼虫移至24孔板培养板,置于恒温恒湿培养箱,35 ℃、70%相对湿度(RH)条件下饲养,每24 h更换饲料。预实验结果显示7日龄中蜂幼虫的成活率可达70%以上。
将4 ℃保存的球囊菌培养皿置于已紫外灭菌的超净台,用酒精灯上灼烧片刻的镊子夹取少量菌丝在平板中央2 cm左右圆心区域内划线操作,盖上培养皿,置于37 ℃生化箱恒温培养,3 d后观察球囊菌生长情况。接种8-10 d以后,待培养皿上黑色孢子较多时,将孢子刮至干净的EP管中,用玻璃棒充分研磨,按照Jensen等[36]的方法离心纯化球囊菌孢子,梯度稀释后用血球计数板对孢子进行计数。
1.4 深度测序样品准备、cDNA文库构建及Illumina测序 将高浓度的球囊菌纯化孢子混入人工饲料,稀释至1×107孢子/mL,饲喂3日龄中蜂幼虫,4日龄开始饲喂不含球囊菌孢子的正常饲料。分别剖取球囊菌胁迫的4日龄幼虫肠道(AacT1),5日龄幼虫肠道(AacT2)和6日龄幼虫肠道(AacT3),液氮速冻后迅速转移至超低温冰箱保存备用。上述各样品均设3个生物学重复(AacT1:AacT1-1,AacT1-2,AacT1-3;AacT2:AacT2-1,AacT2-2,AacT2-3;AacT3:AacT3-1,AacT3-2,AacT3-3)。
利用RNAiso Reagent试剂盒抽提上述9个幼虫肠道样品的总RNA,然后用RNase-free DNase I去除基因组DNA残留。RNA的质量通过琼脂糖凝胶电泳和NanoDrop ND-2000 (NanoDrop,Wilmington,DE,USA)进行检测。利用Oligotex mRNA Kits Midi试剂盒说明书,纯化各样品总RNA中的mRNA。以10 μg mRNA作为模板,GsuI-oligo dT作为反转录引物,用1000 U Superscript II reverse transcriptase在42 ℃下孵育1 h合成第1链cDNA;随后利用高碘酸钠氧化mRNA的5′端帽子结构,并连接生物素;通过Dynal M280磁珠筛选连接了生物素的mRNA/cDNA,并通过碱裂解释放第1链cDNA;然后通过DNA ligase在第1链cDNA的5′末端加上接头,利用Ex Taq polymerase合成第2链cDNA。最后,通过Gsu I酶切去除polyA和5′端接头。利用Ampure beads对上述cDNA进行纯化,cDNA文库通过TruSeq? DNA Sample Prep Kit-Set A进行构建和TruSeq PE Cluster Kit进行扩增。通过Illumina Hiseq 2500平台对上述9个样品进行双端(PE125)测序。本研究测得的转录组数据已上传美国国家生物技术信息中心(NCBI) SRA数据库,SRA号:SRA454366。
1.5 测序数据的质控与定位 对于下机数据,利用Perl脚本去除含有adaptor、未知核苷酸比例大于5%和低质量reads,获得有效读段(clean reads)。为获得纯净的球囊菌转录组数据,首先使用短reads比对工具bowtie[37]将各测序样品的clean reads比对(mapping)到核糖体数据库(最多允许5个错配),去除mapping上核糖体的reads,再将保留下来的数据进一步比对中蜂的参考转录组(本课题组已组装并注释,数据已上传NCBI SRA数据库,SRA号:SRA456721)。进而,利用SOAP aligner/soap2软件[38]将过滤后(未mapping上中蜂幼虫肠道参考转录组)的reads mapping到本课题组已组装并注释的球囊菌参考转录组(本课题组已组装并注释,数据已上传NCBI SRA数据库,SRA号:SRA464366)。
1.6 趋势分析 利用FPKM (Fragments per kilobase of transcript per million mapped reads)法计算基因表达量。利用R软件(version 2.16.2)计算各样品之间的相关性系数。利用edgeR软件[39]进行DEG分析(AacT2 VS AacT1、AacT3 VS AacT1)。DEG的筛选标准为FDR≤0.05且|log2Fold change|≥1。
为了对各样品4、5、6三个时间点的基因表达模式进行聚类,首先将基因表达量数据归一化为log2 (AacT1/AacT1)、log2 (AacT2/AacT1)和log2 (AacT3/AacT1),利用STEM (Short Time-series Expression Miner,v1.3.8)软件[40]进一步分析基因表达模式。在所有的基因表达模式中,P value≤0.05为显著表达模式。
1.7 Gene Ontology (GO)与KEGG pathway富集分析 利用WEGO软件[41]对上调表达模式和下调表达模式中的DEGs进行GO富集分析。利用BLASTall将上调表达模式和下调表达模式中的DEGs比对KEGG (Kyoto Encyclopaedia of Genes and Genomes)数据库,进行pathway富集分析。
1.8 实时荧光定量PCR (RT-qPCR)验证 为了验证RNA-seq数据,随机选取6个DEGs进行RT-qPCR。RT-qPCR反应按照SYBR Green Dye试剂盒(Vazyme公司,中国)操作说明书进行,每个反应进行3次重复。20 μL的反应体系中含SYBR Green Dye 10 μL,10 μmol/L正、反向引物各1 μL,cDNA模板DNA 1 μL,DEPC水8 μL。RT-qPCR反应在ABI 7500荧光定量PCR仪(ABI公司,美国)上进行,反应条件:95 ℃ 1 min;95 ℃ 15 s,60 ℃ 30 s,共40个循环;72 ℃ 45 s。所选基因的相对表达量采用2-??Ct法[42]计算。
2 结果和分析 2.1 RNA-seq数据质控与评估 本研究的技术流程如图 1所示。将球囊菌胁迫中蜂幼虫肠道样品的Illumina测序数据先mapping核糖体数据库,再mapping中蜂参考转录组,过滤得到的raw reads进一步去除低质量数据得到41133932条clean reads,各样品clean reads数均在2799036条以上,两端Q20 (99%碱基正确率)均在97.08%以上,两段Q30为(99.9%碱基正确率)为93.46%以上(表 1),说明本研究中的RNA-seq数据质量良好。
 |
| 图 1 本研究的技术流程 Figure 1 Flowchart of this research. |
| 图选项 |
表 1. RNA-seq数据统计 Table 1. Summary of RNA-seq datasets
| Sample | Clean reads | Q20/% | Q30/% |
| AacT1-1 | 2799036 | 339882032(97.14%) | 327064715(93.48%) |
| AacT1-2 | 3196972 | 388799225(97.29%) | 374866896(93.81%) |
| AacT1-3 | 3189982 | 387118651(97.08%) | 372254496(93.36%) |
| AacT2-1 | 4146604 | 508034743(98.01%) | 493998421(95.31%) |
| AacT2-2 | 3424686 | 418310975(97.72%) | 405312848(94.68%) |
| AacT2-3 | 3653650 | 447152102(97.91%) | 434141314(95.06%) |
| AacT3-1 | 9455236 | 1158489428(98.02%) | 1122984422(95.01%) |
| AacT3-2 | 4398766 | 537205684(97.70%) | 520643109(94.69%) |
| AacT3-3 | 6869000 | 841027167(97.95%) | 816721210(95.12%) |
表选项
Pearson相关性分析结果显示AacT1、AacT2与AacT3的组内各生物学重复之间的相关性均在0.8134以上(0.8134-0.9948),说明样本的重复性较高(图 2)。进一步对AacT1、AacT2与AacT3进行主成分分析(PCA),结果显示主成分1 (PC1)与主成分2 (PC2)可分别解释样品基因表达总体差异的91.1%和4.9%,各样品组内各生物学重复聚类良好(图 3-A),表明AacT1、AacT2与AacT3的总体基因表达模式差别显著,聚类结果同样显示各样品的生物学重复之间聚类情况较好(图 3-B)。
 |
| 图 2 AacT1、AacT2与AacT3不同生物学重复间的相关性 Figure 2 Pearson correlation between every two biological repeats within each sample. A: pearson correlation between AacT1-1 and AacT1-2; B: pearson correlation between AacT1-2 and AacT1-3; C: pearson correlation between AacT1-1 and AacT1-3; D: pearson correlation between AacT2-1 and AacT2-2; E: pearson correlation between AacT2-2 and AacT2-3; F: pearson correlation between AacT2-1 and AacT2-3; G: pearson correlation between AacT3-1 and AacT3-2; H: pearson correlation between AacT3-2 and AacT3-3; I: pearson correlation between AacT3-1 and AacT3-3. |
| 图选项 |
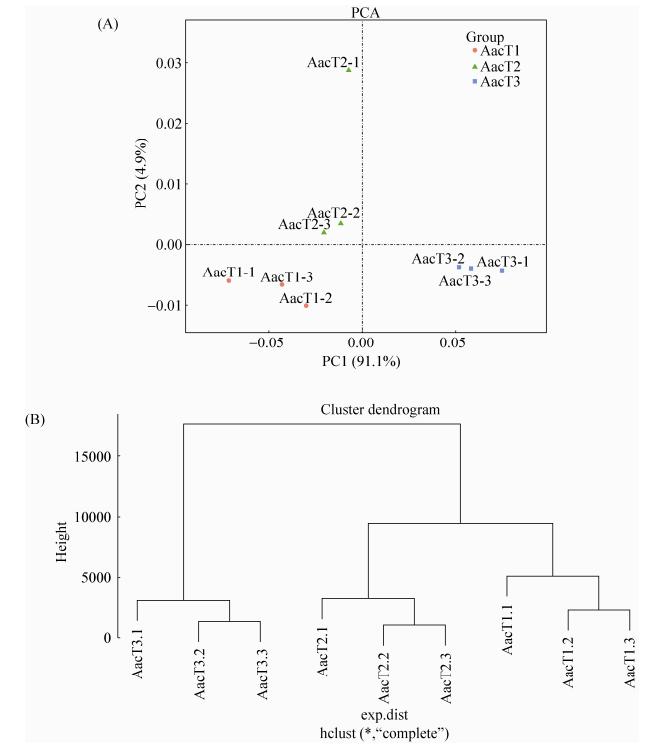 |
| 图 3 AacT1、AacT2与AacT3的主成分分析与聚类分析 Figure 3 PCA and clustering analyses of AacT1, AacT2 and AacT3 samples. A: PCA analysis of each sample; B: clustering analysis of each sample. |
| 图选项 |
RNA-seq数据比对球囊菌参考转录组情况统计显示,各样品clean reads比对上参考转录组的比例均在88.52%以上(表 2),AacT1、AacT2与AacT3的表达基因数目分别为15881、17682和20026个,分别占球囊菌参考转录组的37.28%、41.50%和47.01% (表 3)。
表 2. RNA-seq数据比对参考转录组信息统计 Table 2. Mapping of RNA-seq data to the reference transcriptome for A. apis
| Sample | Total reads | Unmapped reads | Unique mapped reads | Multiple mapped reads | Mapping ratio/% |
| AacT1-1 | 2762984 | 353036 | 2297342 | 112606 | 87.22 |
| AacT1-2 | 3071046 | 402523 | 2550540 | 117983 | 86.89 |
| AacT1-3 | 3014772 | 387972 | 2498276 | 128524 | 87.13 |
| AacT2-1 | 3517220 | 443484 | 2832966 | 240770 | 87.39 |
| AacT2-2 | 3398820 | 414293 | 2726215 | 258312 | 87.81 |
| AacT2-3 | 3632592 | 412844 | 2980136 | 239612 | 88.64 |
| AacT3-1 | 8129694 | 943372 | 6563486 | 622836 | 88.40 |
| AacT3-2 | 4198356 | 482132 | 3398515 | 317709 | 88.52 |
| AacT3-3 | 6540552 | 705934 | 5370084 | 464534 | 89.21 |
表选项
表 3. 各样品的表达基因数量统计 Table 3. Summary of the expressed genes of each sample
| Sample | Expressed gene number | Ratio/% |
| AacT1 | 18945 | 44.47 |
| AacT2 | 22996 | 53.98 |
| AacT3 | 24867 | 58.37 |
表选项
2.2 趋势分析 利用STEM软件对AacT1、AacT2与AacT3的表达基因进行趋势分析,结果显示22865个DEGs共聚类为8个表达模式(profile0、profile1、profile2、profile3、profile4、profile5、profile6和profile7) (图 4),其中有16769个DEGs聚类为4个显著表达模式(P < 0.05),包括2个显著上调表达模式(profile6和profile7)和2个显著上调表达模式(profile0和profile1)(图 4)。profile0、profile1、profile6和profile7分别包含3217、2756、9094和1702个DEGs。上述结果表明在胁迫中蜂幼虫肠道的过程中,球囊菌的部分基因被抑制,但更多的基因被激活。
 |
| 图 4 AacT1、AacT2与AacT3差异表达基因的趋势分析 Figure 4 Trend analysis of DEGs in AacT1, AacT2 and AacT3. A: cluster trajectory profiles across stages of A. apis-treatment; B: expression profiles of profile0; C: expression profiles of profile1; D: expression profiles of profile6; E: expression profiles of profile7. |
| 图选项 |
2.3 GO富集分析 分别对显著上调趋势和显著下调趋势包含的DEGs进行GO富集分析,结果显示显著上调趋势包含的DEGs富集在40个GO term上,基因富集数量前10位的GO term分别为细胞进程(cellular process)(2486 unigenes)、细胞(cell)(2367 unigenes)、细胞组件(cell part)(2367 unigenes)、代谢进程(metabolic process)(2359 unigenes)、氧化磷酸化(catalytic activity)(2270 unigenes)、单组织进程(single-organism process)(1924 unigenes)、结合(binding)(1835 unigenes)、细胞器(organelle)(1686 unigenes)、大分子复合物(macromolecular complex) (822 unigenes)及细胞器组件(organelle part)(780 unigenes)(图 5-A);显著下调趋势包含的DEGs富集在37个GO term上,基因富集数量前10位的GO term分别为细胞(cell)(708 unigenes)、细胞组件(cell part)(708 unigenes)、代谢进程(metabolic process)(668 unigenes)、细胞进程(cellular process) (666 unigenes)、氧化磷酸化(catalytic activity)(620 unigenes)、结合(binding)(586 unigenes)、单组织进程(single-organism process)(467 unigenes)、细胞器(organelle)(380 unigenes)、大分子复合物(macromolecular complex)(281 unigenes)及定位(localization)(199 unigenes)(图 5-B)。
 |
| 图 5 显著上调趋势和显著下调趋势包含基因的GO富集分析 Figure 5 GO enrichment analysis for all DEGs involved in the significant up-and down-regulated trends. A: GO enrichment analysis of all DEGs involved in the significant up-regulated trends; B: GO enrichment analysis of all DEGs involved in the significant down-regulated trends. |
| 图选项 |
2.4 KEGG pathway富集分析 对显著上调趋势中所有DEGs进行KEGG pathway富集分析,结果表明这些DEGs富集在119个pathway上,其中基因富集数量最多的是氨基酸生物合成(biosynthesis of amino acids)(127 unigenes),其次为细胞周期(cell cycle)(111 unigenes)和嘌呤代谢(purine metabolism)(111 unigenes)(图 6-A),说明球囊菌在胁迫中蜂幼虫肠道的过程中通过提高物质合成促进增殖过程;显著下调趋势中所有DEGs的KEGG pathway分析结果显示,这些DEGs富集在112个pathway上,其中基因富集数量最多的是核糖体(ribosome)(98 unigenes),其次是剪接体(spliceosome)(80 unigenes)和内质网蛋白加工(protein processing in endoplasmic reticulum)(72 unigenes) (图 6-B),说明宿主通过抑制球囊菌的蛋白合成抵御其入侵。进一步分析显示,共有11个DEGs富集在MAPK信号通路,这些DEGs的表达量随着球囊菌胁迫时间的延长而呈下降趋势(图 7),说明该通路受到宿主的抑制。
 |
| 图 6 显著上调趋势与显著下调趋势包含基因的KEGG代谢通路富集分析 Figure 6 KEGG pathway enrichment analysis of all DEGs associated with the significant up-/down-regulated profiles. A: KEGG enrichment analysis of all DEGs involved in the significant up-regulated profiles; B: KEGG enrichment analysis of all DEGs involved in the significant down-regulated profiles. |
| 图选项 |
 |
| 图 7 MAPK信号通路富集基因的热图 Figure 7 Heatmap of unigenes enriched in MAPK signaling pathway. |
| 图选项 |
2.5 RT-qPCR验证 利用RT-qPCR检测随机选取的6个DEGs,结果显示这些DEGs的表达水平变化趋势与RNA-seq数据中的基因表达水平变化趋势一致(图 8),证明了本研究获得的转录组数据真实可靠。
 |
| 图 8 RNA-seq数据的RT-qPCR验证 Figure 8 RT-qPCR validation of RNA-seq data. A: unknown gene; B: NADH-ubiquinone oxidoreductase 49 kDa subunit encoded gene; C: multidrug transporter encoded gene; D: eukaryotic translation initiation factor 3 subunit-like protein encoded gene; E: unknown gene; F: unknown gene. |
| 图选项 |
3 讨论 白垩病是困扰养蜂生产的顽疾,尚无有效防治策略。养蜂生产中,意蜂幼虫极易被球囊菌侵染而爆发白垩病,而中蜂幼虫几乎不受该病的影响,偶尔也能发现因白垩病而死亡的中蜂幼虫虫尸。通常认为中蜂具有更强的群体防御[43],清理行为在蜂群抵御白垩病方面发挥重要作用[44]。本课题组在前期研究中也发现中蜂幼虫能被球囊菌侵染,但发病率远低于意蜂幼虫,说明二者在个体水平也存在球囊菌抗性差异。Cornman等[34]曾对来自培养基的球囊菌菌丝和来自蜜蜂幼虫感染组织的球囊菌菌丝进行转录组测序,但后者是感染幼虫肠道内萌发的菌丝突破肠壁后继续生长最后突破体表的菌丝,其转录组变化并不能精确反映侵染过程中球囊菌的基因表达情况,因为此时的菌丝已经不与宿主互作。目前,尚无球囊菌侵染中蜂幼虫的相关分子研究报道。
本研究中,测序材料是球囊菌胁迫的中蜂幼虫肠道,肠道内的球囊菌处于动态增殖过程,故通过比对中蜂幼虫肠道参考转录组过滤得到的球囊菌转录组数据,能更精确地反映病原在侵染过程中的基因表达谱。趋势分析结果显示显著上调趋势(profile6和profile7)与显著下调趋势(profile0和profile1)均为2个;而在另一研究中,对球囊菌胁迫的中蜂幼虫肠道的DEGs进行趋势分析,结果显示基因的显著表达模式共有3个且皆为上调趋势,表明中蜂幼虫通过病原-宿主互作对球囊菌产生一定程度的抑制。其中,显著下调趋势包含5973个DEGs,大量基因(668 unigenes)富集在代谢进程(metabolic process),包括碳代谢(carbon metabolism,ko01200)、脂肪酸降解(fatty acid degradation,ko00071)、甘氨酸、丝氨酸及苏氨酸代谢(glycine, serine and threonine metabolism,ko00260)等74个物质代谢通路,还包括氧化磷酸化(oxidative phosphorylation,ko00190)、氮代谢(nitrogen metabolism,ko00910)等4个能量代谢通路,表明中蜂幼虫在受球囊菌胁迫的过程中,可通过病原-宿主互作对球囊菌的物质和能量代谢产生抑制,从而阻碍其增殖。
真菌具有精细而保守的信号通路,可通过调节黑色素形成、配合、毒力以及形态发生等来感知外界环境并迅速作出应答[33]。在真菌的信号通路中,MAPK信号通路至关重要,在真菌致病性及响应环境胁迫方面发挥关键作用[45]。本研究发现有11个富集在该信号通路上的DEGs表现出下调趋势,如TKL蛋白激酶编码基因(unigene0000726,TKL protein kinase)、p38 MAP激酶编码基因(unigene0012136,p38 MAP kinase)及蛋白激酶编码基因(unigene0021854,protein kinase)等,推测中蜂幼虫可通过抑制球囊菌的MAPK信号通路影响病原增殖,从而赋予其球囊菌抗性。前期研究发现,在球囊菌胁迫意蜂幼虫肠道的过程中,有48个DEGs富集在MAPK信号通路上且都呈现出上调趋势。推测中蜂幼虫与意蜂幼虫的球囊菌抗性差异与球囊菌在胁迫过程中MAPK信号通路的表现密切相关。该信号通路上的基因有望成为白垩病治疗的分子靶点,可利用RNAi技术进行验证。
本研究利用RNA-seq技术对球囊菌胁迫的中蜂幼虫肠道进行深度测序,基于趋势分析进一步对胁迫过程中的球囊菌进行转录组学分析,研究结果不仅为揭示白垩病过程中的球囊菌-中蜂幼虫互作提供了重要信息,也为阐明不同抗性蜂种的球囊菌抗性差异奠定了基础。
References
| [1] | Galizia CG, Eisenhardt D, Giurfa M. Honeybee neurobiology and behavior: a tribute to randolf menzel. Netherlands: Springer, 2012. http://www.mendeley.com/research/honeybee-neurobiology-behavior-tribute-randolf-menzel/ |
| [2] | Begna D, Han B, Feng M, Fang Y, Li JK. Differential expressions of nuclear proteomes between honeybee (Apis mellifera L.) queen and worker larvae: a deep insight into caste pathway decisions. Journal of Proteome Research, 2012, 11(2): 1317-1329. DOI:10.1021/pr200974a |
| [3] | Zayed A, Robinson GE. Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annual Review of Genetics, 2012, 46: 591-615. DOI:10.1146/annurev-genet-110711-155517 |
| [4] | Foret S, Kucharski R, Pellegrini M, Feng SH, Jacobsen SE, Robinson GE, Maleszka R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(13): 4968-4973. DOI:10.1073/pnas.1202392109 |
| [5] | Kurze C, Routtu J, Moritz RFA. Parasite resistance and tolerance in honeybees at the individual and social level. Zoology, 2016, 119(4): 290-297. DOI:10.1016/j.zool.2016.03.007 |
| [6] | National Research Council, Division on Earth and Life Studies, Board on Agriculture and Natural Resources, Board on Life Sciences. Committee on the Status of Pollinators in North America. Washington, D.C.: National Academies Press, 2007. |
| [7] | Klein AM, Vaissière BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C, Tscharntke T. Importance of pollinators in changing landscapes for world crops. Proceedings of the Royal Society B: Biological Sciences, 2007, 274(1608): 303-313. DOI:10.1098/rspb.2006.3721 |
| [8] | Genersch E. Honey bee pathology: current threats to honey bees and beekeeping. Applied Microbiology and Biotechnology, 2010, 87(1): 87-97. DOI:10.1007/s00253-010-2573-8 |
| [9] | Evans JD, Spivak M. Socialized medicine: individual and communal disease barriers in honey bees. Journal of Invertebrate Pathology, 2010, 103(S1): S62-S72. |
| [10] | Aronstein KA, Murray KD. Chalkbrood disease in honey bees. Journal of Invertebrate Pathology, 2010, 103: S20-S29. DOI:10.1016/j.jip.2009.06.018 |
| [11] | 梁勤, 陈大福. 蜜蜂保护学. 第2版. 北京: 中国农业出版社, 2009. |
| [12] | Aizen MA, Garibaldi LA, Cunningham SA, Klein AM. How much does agriculture depend on pollinators? Lessons from long-term trends in crop production. Annals of Botany, 2009, 103(9): 1579-1588. DOI:10.1093/aob/mcp076 |
| [13] | Bailey L. Infectious Diseases of the Honey-Bee. London, UK: Land Books, 1963. |
| [14] | Zaghloul OA, Mourad AK, El Kady MB, Nemat FM, Morsy ME. Assessment of losses in honey yield due to the chalkbrood disease, with reference to the determination of its economic injury levels in Egypt. Communications in Agricultural and Applied Biological Sciences, 2005, 70(4): 703-714. |
| [15] | Li JH, Zheng ZY, Chen DF, Liang Q. Factors influencing Ascosphaera apis infection on honeybee larvae and observation on the infection process. Acta Entomologica Sinica, 2012, 55(7): 790-797. (in Chinese) 李江红, 郑志阳, 陈大福, 梁勤. 影响蜜蜂球囊菌侵染蜜蜂幼虫的因素及侵染过程观察. 昆虫学报, 2012, 55(7): 790-797. |
| [16] | Heath LAF. Chalk brood pathogens: a review. Bee World, 1982, 63(3): 130-135. DOI:10.1080/0005772X.1982.11097877 |
| [17] | Anderson DL, Gibson NL. New species and isolates of spore-cyst fungi (Plectomycetes: Ascosphaerales) from Australia. Australian Systematic Botany, 1998, 11(1): 53-72. DOI:10.1071/SB96026 |
| [18] | Spiltoir CF. Life cycle of Ascosphaera apis (Pericystis apis). American Journal of Botany, 1955, 42(6): 501-508. DOI:10.2307/2438686 |
| [19] | Skou JP. More details in support of the class Ascosphaeromycetes. Mycotaxon, 1988, 31(1): 191-198. |
| [20] | Flores JM, Spivak M, Gutiérrez I. Spores of Ascosphaera apis contained in wax foundation can infect honeybee brood. Veterinary Microbiology, 2005, 108(1/2): 141-144. |
| [21] | Flores JM, Gutiérrez I, Espejo R. The role of pollen in chalkbrood disease in Apis mellifera: transmission and predisposing conditions. Mycologia, 2005, 97(6): 1171-1176. DOI:10.1080/15572536.2006.11832727 |
| [22] | P?ggeler S. Mating-type genes for classical strain improvements of Ascomycetes. Applied Microbiology and Biotechnology, 2001, 56(5/6): 589-601. |
| [23] | Chorbiński P. Enzymatic activity of strains of Ascosphaera apis. Medycyna Weterynaryjna, 2003, 59(11): 1019-1022. |
| [24] | Bailey L, Ball BV. Honey bee Pathology. London: Academic Press, 1991. |
| [25] | Theantana T, Chantawannakul P. Protease and β-N-acetylglucosaminidase of honey bee chalkbrood pathogen Ascosphaera apis. Journal of Apicultural Research, 2008, 47(1): 68-76. DOI:10.1080/00218839.2008.11101426 |
| [26] | Stanley D, Miller J, Tunaz H. Eicosanoid actions in insect immunity. Journal of Innate Immunity, 2009, 1(4): 282-290. DOI:10.1159/000210371 |
| [27] | Evans JD, Spivak M. Socialized medicine: individual and communal disease barriers in honey bees. Journal of Invertebrate Pathology, 2010, 103(S1): S62-S72. |
| [28] | Hornitzky M. Literature Review of Chalkbrood: A Fungal Disease of Honeybees. Kingston ACT, Australia: Rural Industries Research and Development Corporation, 2001. |
| [29] | Tarpy DR. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proceedings of the Royal Society B: Biological Sciences, 2003, 270(1510): 99-103. DOI:10.1098/rspb.2002.2199 |
| [30] | Mourad AK, Zaghloul OA, El Kady MB, Nemat FM, Morsy ME. A novel approach for the management of the chalkbrood disease infesting honeybee Apis mellifera L. (Hymenoptera: Apidae) colonies in Egypt. Communications in Agricultural and Applied Biological Sciences, 2005, 70(4): 601-611. |
| [31] | Liang Q, Chen DF, Wang JD. Effects on the mycelia growth and spore-forming of Ascosphaera apis under ecological condition of nutrients. Chinese Journal of Eco-Agriculture, 2001, 9(4): 31-34. (in Chinese) 梁勤, 陈大福, 王建鼎. 营养生态条件对蜜蜂球囊菌生长及产孢的影响. 中国生态农业学报, 2001, 9(4): 31-34. |
| [32] | Zheng ZY, Li JH, Liang Q, Chen DF. Ascosphaera apis secretes multiple extracellular enzymes to infect honeybee larvae. Journal of Fujian Agriculture and Forestry University (Natural Science Edition), 2011, 40(3): 280-284. (in Chinese) 郑志阳, 李江红, 梁勤, 陈大福. 蜜蜂球囊菌分泌多种胞外酶侵染蜜蜂幼虫. 福建农林大学学报(自然科学版), 2011, 40(3): 280-284. |
| [33] | Qin X, Evans JD, Aronstein KA, Murray KD, Weinstock GM. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Molecular Biology, 2006, 15(5): 715-718. DOI:10.1111/imb.2006.15.issue-5 |
| [34] | Cornman RS, Bennett AK, Murray KD, Evans JD, Elsik CG, Aronstein K. Transcriptome analysis of the honey bee fungal pathogen, Ascosphaera apis: implications for host pathogenesis. BMC Genomics, 2012, 13(1): 285. DOI:10.1186/1471-2164-13-285 |
| [35] | Wang Q, Sun LX, Xiao PX, Liu F, Kang MJ, Xu BH. Study on technology for indoor artificial feeding of Apis cerana cerana larvae. Shandong Agricultural Sciences, 2009, 41(11): 113-116. (in Chinese) 王倩, 孙亮先, 肖培新, 刘锋, 康明江, 胥宝华. 室内人工培育中华蜜蜂幼虫技术研究. 山东农业科学, 2009, 41(11): 113-116. DOI:10.3969/j.issn.1001-4942.2009.11.037 |
| [36] | Jensen AB, Aronstein K, Flores JM, et al. Standard methods for fungal brood disease research. Journal of Apicultural Research, 2013, 52(1): 1-20. |
| [37] | Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 2009, 10(3): R25. DOI:10.1186/gb-2009-10-3-r25 |
| [38] | Hurgobin B. Short read alignment using SOAP2//Edwards D. Plant Bioinformatics: Methods and Protocols. 2nd ed. New York, United States: Springer, 2016: 241-252. |
| [39] | Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010, 26(1): 139-140. DOI:10.1093/bioinformatics/btp616 |
| [40] | Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics, 2006, 7(1): 191. DOI:10.1186/1471-2105-7-191 |
| [41] | Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Research, 2006, 34(S2): W293-W297. |
| [42] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2T-ΔΔC method. Methods, 2001, 25(4): 402-408. DOI:10.1006/meth.2001.1262 |
| [43] | Park D, Jung JW, Choi BS, Jayakodi M, Lee J, Lim J, Yu Y, Choi YS, Lee ML, Park Y, Choi IY, Yang TJ, Edwards OR, Nah G, Kwon HW. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics, 2015, 16(1): 1. DOI:10.1186/1471-2164-16-1 |
| [44] | Invernizzi C, Rivas F, Bettucci L. Resistance to chalkbrood disease in Apis mellifera L. (Hymenoptera: Apidae) colonies with different hygienic behaviour. Neotropical Entomology, 40(1): 28-34. DOI:10.1590/S1519-566X2011000100004 |
| [45] | Perez P, Cansado J. Cell integrity signaling and response to stress in fission yeast. Current Protein & Peptide Science, 2010, 11(8): 680-692. |
